Abstract
Glycosylation is a major post translational modification of proteins that regulates many biological processes including protein folding, structure stability, receptor activation and immune responses. The glycans attached to proteins represent an important determinant of the protein interaction-specificity and maintain the 3D structure of proteins. Mass spectrometry (MS) is one of the most efficient tools used in the current studies of glycoproteins and structure of their glycoforms. Collision energy (CE) is a crucial instrument parameter that can be exploited to improve structural resolution because different linkages of glycan units show different stability under CID/HCD fragmentation. Here we report the utility of CE modulation for qualitative and quantitative analysis of site- and structure- specific glycoforms of proteins. Using CE modulation, we were able to break selectively specific glycan linkages on intact glycopeptides and get, to some degree, structure-specific mass spectrometric signals. Structure- and CE- specific oxonium ions provide sufficient information for the resolution of outer arm structure motifs with recognized biological functions. The complementary Y-ions, generated under optimized low CE (soft) conditions, provide additional structural information including features specific to the chitobiose core. This methodology of multiple CE fragmentation without merging spectral information can significantly improve confidence of glycopeptide identification and structural resolution by providing additional information to the established gly-copeptide-search algorithms and tools.
Graphical Abstract
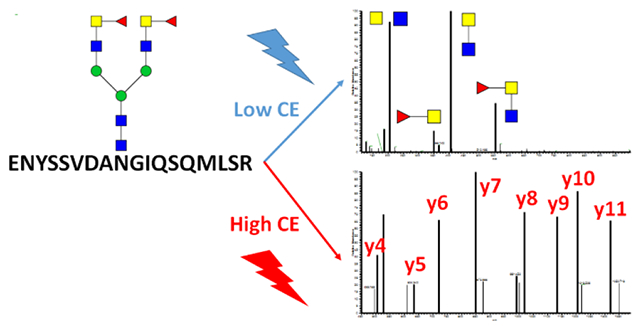
The importance of N-glycosylation in many physiological and pathological processes is well documented in the literature1,2. The utility of mass spectrometry in glycobiological research continues to increase with the development of increasingly efficient analytical workflows 3,4. Scientists were able to improve approaches to the analysis of detached glycans 5, their structure 6,7 and quantity 8. However, site-specific determination of glycan structural motifs on glycopeptides remains a challenge. In glycoproteomics, improved software packages analyze site-specific glycosylation with some degree of specificity but the analysis still requires a good degree of manual curation9-11. Separation techniques remain typically a significant part of the structural elucidation in the current analytical workflows. Recently published research using HILIC or graphitized carbon separation of glycopeptide isomers described selectivity based on the retention time contribution of glycan structure to the separation of glycoptides12-16. However, the lack of glycopeptide and glycan standards complicates unequivocal assignments based on retention time of the analytes. Additional methods need to be developed to assure specificity of the analyses. For example, we and others have reported the use of exoglycosidase digests for structural resolution and recently published work describes efficient use of ion mobility for the separation of glycan isomers 17. Several studies describing glycopeptide fragmentation have been published recently18,19. Here we expand the recently introduced analytical workflows using optimized collision energy for the resolution of specific glycan linkages and we describe a workflow for the determination of specific structural motifs of glycans attached to peptides. To this end, we record glycopeptide spectra at different collision energies which allows us to resolve specific structural motifs of N-glycopeptides under low collision energy settings while high collision energy fragmentation is used for the determination of the site of protein attachment or the assignment of features related to glycan core structures like core-fucosylation. Quantitative structure and site-specific information can be extracted by examining the signals under defined fragmentation condition.
EXPERIMENTAL SECTION
Glycoproteins
Recombinant Human PD-L1/B7-H1 Fc Chimera Protein overexpressed in Mouse myeloma (NS0) cell line (R&D Systems, Minneapolis, MN); Recombinant Human PD-L1/B7-H1 His-tag Protein overexpressed in Human embryonic kidney cell (HEK 293) cell line (R&D Systems), Human Haptoglobin, mixed type isolated from plasma (Athens Research & Technology, Athens, GA), Human Hemopexin isolated from plasma (Athens Research & Technology). His-tagged ProBDNF was expressed in suspension HEK293F cells (Invitrogen, Carlsbad, CA) and purified using Ni-NTA resin (Thermo, Rockford, IL) chromatography on AKTA Start FPLC system (GE Healthcare, Uppsala, Sweden) to > 90% purity as determined by Coomassie Blue stained SDS-PAGE as described 24 Lyophilized proteins were dissolved in 100 mM PBS at a concentration of 0.5 μg/ml and stored at −80°C.
Glycopeptide preparation
Aliquots of dissolved proteins were diluted with 25 mM sodium bicarbonate to a final concentration of 1mg/ml. Protein solution was reduced with 5 mM DTT for 60 min at 60°C and alkylated with 15 mM iodoacetamide for 30 min in the dark. Trypsin Gold (Promega, Madison, WI) digestion (2.5 ng/μl) was carried out at 37°C in Barocycler NEP2320 (Pressure BioSciences, South Easton, MA) for 1 hour. Neuraminidase A and Fucosidase 1-3,4 and Fucosidase 1-2 treated peptides were prepared as previously described by Kozlik et al. 12
Glycopeptide analysis using IDA nano LC-MS/MS on the 6600 TripleTOF
Glycopeptide separation was achieved on a Nanoacquity LC (Waters, Milford, MA) using capillary trap, 180 μm x 0.5 mm, and analytical 75 μm x 150 mm Atlantis DB C18, 3 μm, 300 A columns (Waters) interfaced with the 6600 TripleTOF (Sciex, Framingham, MA). A 3 min trapping step using 2% ACN, 0.1% formic acid at 15 μl/min was followed by chromatographic separation at 0.4 μl/min as follows: starting conditions 5% ACN, 0.1% formic acid; 1-55 min, 5–50% ACN, 0.1% formic acid; 55-60 min, 50–95% ACN, 0.1% formic acid; 60-70 min 95% ACN, 0.1% formic acid followed by equilibration to starting conditions for additional 20 min. For all runs, we have injected 1 μl (0.02 μg of protein) of tryptic digest directly on column. We have used an Information Dependent Acquisition (IDA) workflow with one MS1 full scan (400-1800 m/z) and 50 MS/MS fragmentations (100-1800 m/z), isolation window (0.7 Da), with rolling collision energy using different slope CE calculation for Low CE and High CE fragmentation. MS/MS mass spectra were recorded in the range 100-1800 m/z with resolution 30,000 and mass accuracy less than 15 ppm using the following experimental parameters: declustering potential 80 V, curtain gas 30, ion spray voltage 2,300 V, ion source gas-1 11, interface heater 180°C, entrance potential 10 V, collision exit potential 11 V.
Glycopeptide analysis using DDA nano LC-MS/MS on the Orbitrap Fusion-Lumos
Digested proteins were separated using a 90-minute ACN gradient on a 150 mm x 75 μm C18 pepmap column at a flow rate of 0.3 μl/min. In brief, peptide and glycopeptide separation was achieved by a 5 min trapping/washing step using 99% solvent A (2% acetonitrile, 0.1% formic acid) at 10 μl/min followed by a 90 min acetonitrile gradient at a flow rate of 0.3 μl/min: 0-3 min 2% B (0.1% formic acid in ACN), 3-5 min 2-10% B; 5-60 min 10-45% B; 60-65 min 45-98% B; 65-70 min 98% B, 70-90 min equilibration by 2% B. Glycopeptides were analyzed using Orbitrap Fusion Lumos mass spectrometer with the electrospray ionization voltage at 3 kV and the capillary temperature at 275°C. MSI scans were performed over m/z 400–1800 with the wide quadrupole isolation on a resolution of 120,000 (m/z 200), RF Lens at 40%, intensity threshold for MS2 set to 2.0e4, selected precursors for MS2 with charge state 3-8, and dynamic exclusion 30s. Data-dependent (DDA) HCD tandem mass spectra were collected with a resolution of 15,000 in the Orbitrap with fixed first mass 110 and 4 normalized collision energy 15, 20, 25 and 30%.
Data analysis
Glycopeptide identification
Byonic software (Protein Metrics, Cupertino, CA) was used for the identification of summary formulas of peptide -glycosylation. Independent search was performed on the data with different collision energy settings. All spectra of identified glycopeptides were checked manually for the presence of structure-specific fragments and quantitative information was extracted using Qualbrowser (Thermo) and Peakview (Sciex) software.
RESULTS AND DISCUSSION
Fragmentation of core vs outer arm fucosylated glycopeptides in the analysis of haptoglobin
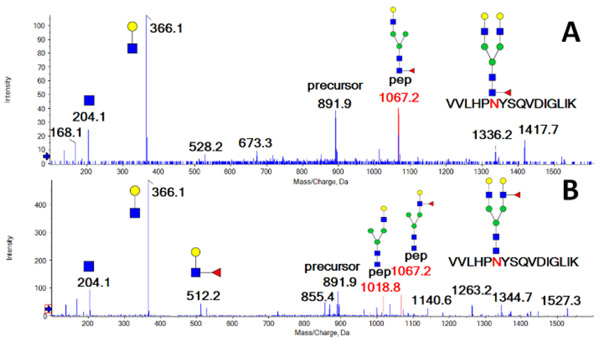
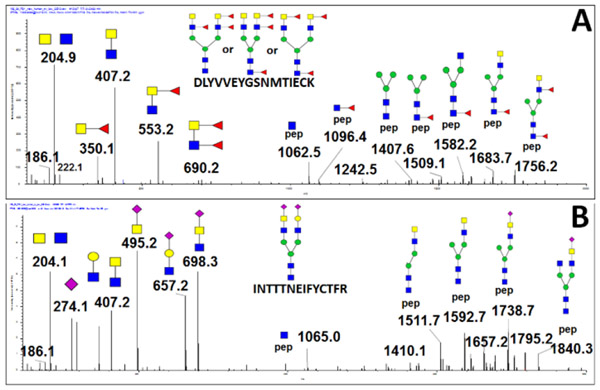
We chose T3 glycopeptide (VVLHPNYSQVDIGLIK) of haptoglobin because it is almost exclusively (99.9%) outer arm fucosylated 22. However, we were able to enrich the core fucosylated glycopeptide using a combination of exoglycosidases (Neuraminidase A, Fucosidase α1-3,4 and Fucosidase α1-2) which verifies by independent methods the structural assignments of the isobaric glycoforms for our fragmentation studies. We have described the use of exoglycosidases or enzyme inhibitors for enrichment of the linkage isoforms in several studies 12,22,24 and apply the methods to several glycopeptides in this study. Fragmentation of the fucosylated glycopeptide of haptoglobin with outer arm fucosylation is shown in0 Figure 1 A. The energy-optimized MS/MS fragmentation spectrum of the bi-antennary fucosylated glycopeptide contains a pair of major fragments (loss of one arm) which shows equivalent fragmentation of the antennae with/out fucosylation under the low collision energy settings (15-20 eV). Fragmentation under higher collision energy (20 eV+) leads to the chitobiose core fragmentation and produces peptide-HexNAc or peptide-HexNAc-Fuc fragments which allowed us to assign core-fucosylation on the peptide backbone. Low collision energy fragmentation of the core-fucosylated glycopeptide (prepared as described above) shows one major Y-ion which is the core fucosylated fragment without one arm and charge state z-1 (Figure 1B), in line with previously published work20,21. Our results show that fragmentation at low CE minimizes false positive results due to fucose rearrangement as described previously 20.
Figure 1.
MS/MS spectrum of isobaric core (A) and outer arm (B) fucosylated VVLHPNYSQVDIGLIK glycopeptides of haptoglobin recorded under low collision energy (20eV) on the 6600 tripleTOF mass spectrometer.
Presence of glycan sulfation based on a characteristic Y-ion neutral loss in low collision energy spectra
Another example of the Y-ion structure-specific information detectable in low collision HCD spectra is outer arm sulfation. Outer arm sulfation is a postranslational modification of the N-glycans of various structures 23. We recently described sulfation on the proBDNF glycoprotein where LacdiNAc structures carry sulfated GalNAc 24. However, the identification of sulfation at conventional (peptide based) collision energies is based on the sulfate loss (80 Da) from the low abundant Y ions. Here we show an example of low collision energy spectra of sulfated LacdiNAc where the loss of sulfate (80 Da) is the major neutral loss (m/z 1132.48 Da, 30% of precursor intensity) followed by the loss of sulfated LacdiNAc (m/z 1455.2 Da) and sulfated GalNAc (m/z 1556.2 Da) with the corresponding B ion (m/z 284.04 Da). These ions are not detectable at higher CE (Supplemental Figure 1).
Outer arm structure-specific oxonium ions
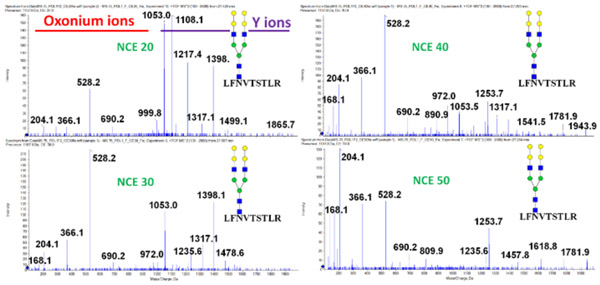
The structure-specific fragments described in this paper are summarized in Table 1. Some of the ions (* labeled) are common N-glycopeptide oxonium ions which are visible in all complex N-glycopeptide tandem mass spectra fragmented at higher collision energy (25+ eV) (Figure 2, Supplemental Figures 2 and 3). However, these ions are structure-specific at low CE as described below. Other ions confirm the localization of fucose or sialic acid on the outer arms of the structures. The last three ions (m/z 1055.6, 1096.4, and 1387.3 Da) in Table 1 are fragments specific for the outer-arm polyLacNAc structural motif. We describe the structure-resolving ability of the ions in the following sections.
Table 1.
Structure specific fragments (oxonium ions) obtained under low collision energy (NCE 20) HCD fragmentation on the Orbitrap Fusion Lumos.
| Fragment m/z |
Structure | Common name | Complementary biantennary Y-ion |
|---|---|---|---|
| *407.2 | LacdiNAc | (M-407.2)z−1 | |
| 350.1 | Fuco-GalNAc | (M-350.1)z−1 | |
| 553.2 | Fuco-LacdiNAc | (M-553.2)z−1 | |
| 699.2 | Difuco-LacdiNAc | (M-699.2)z−1 | |
| 698.3 | Sialo-LacdiNAc | (M-698.3)z−1 | |
| 495.2 | Sialo-GalNAc | (M-495.2)z−1 | |
| 674.3 | Fuco-Galili | (M-674.3)z−1 | |
| *528.2 | Galili | (M-528.2)z−1 | |
| 1055.6 | PolyLacNAc | (M-1055.6)z−1 | |
| 1096.4 | PolyLacNAc | (M-1096.4)z−1 | |
| 1387.3 | Sialo-polyLacNAc | (M-1387.3)z−1 |
Figure 2.
HCD fragmentation spectra of the PD-L1 glycopeptide LFNVTSTLR occupied by a biantennary galili structure obtained at four different collision energies (NCE: 20, 30, 40, 50).
Oxonium ions specific to LacdiNAc and complementary Y ions
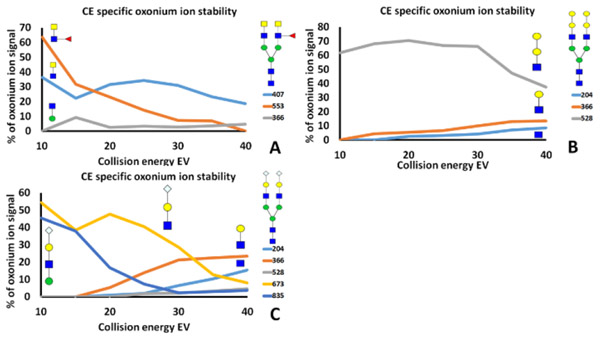
The ion m/z 407.2 Da is specific to LacdiNAc 25 under low collision energy (10-20eV) fragmentation. Fragmentation spectra recorded at higher collision energy produce limited amount of 407.2 by fragmentation of the chitobiose core of N-glycans that produces a GlcNAc-GlcNAc fragment; a specific LacdiNAc ion is produced only under the selective outer arm fragmentation of the N-glycans at low CE. Figure 3B shows the fragmentation spectra of the tryptic glycopeptide (LFNVTSTLR) of human PD-L1 which carries LacdiNAc N-glycan at one antenna. The precursor summary formula is isobaric with the agalactosylated biantennary complex glycan but the structure-specific ion 407.2 confirms the presence of the di-HexNAc outer arm structural motif. The fragmentation spectra of the single arm LacNAc glycopeptide with the peptide backbone (LFNVTSTLR) yields an oxonium ion 366.2 (a charged arm) which distinguishes it from the single arm LacdiNAc glycoform of the same peptide which yields the m/z 407.2 fragment. Figure 4A documents the specificity of the relative intensity of the oxonium ion derived from LacdiNAc (407.2 Da) at 7 different collision energies. Low energy fragmentation is needed for the selectivity of the LacdiNAc ions that are not produced from the tetra antennary agalactosylated glycopeptides isobaric to the symmetric biantennary LacdiNAc structure. Similar trends are documented in the fragmentation of fucosylated LacdiNAc structures (Supplementary Figure 2).
Figure 3.
MS/MS spectra of the following structural motifs of LFNVTSTLR glycopeptide of human PD-L1 recorded under low collision energy NCE 15: A. LacNAc; B. LacdiNAc and their low mass range zoomed spectra showing differences between LacNAc C. and LacdiNAc D. fragment ion spectra.
Figure 4.
Yield of specific oxonium ions at increasing collision energies: A. LacdiNAc containing structure; B. Gal-Gal containing structure; and C. NeuGC-LacNAc containing structure.
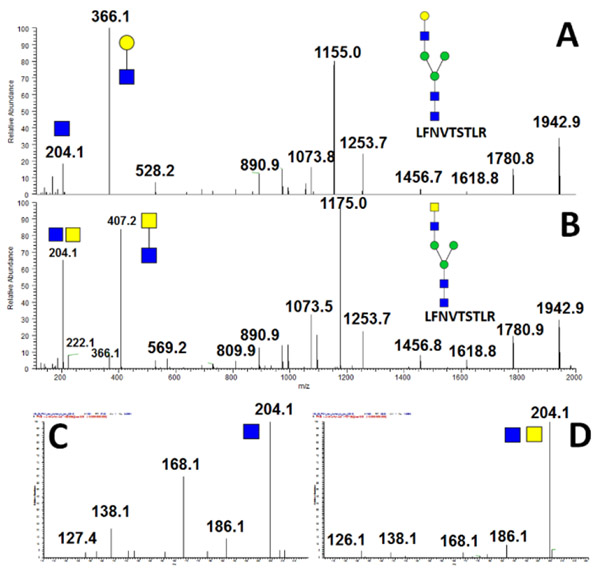
GalNAc containing structure in low mass and CE spectra
The low mass range of the low CE fragmentation spectra confirms the presence of GalNAc (LacdiNAc) in the glycopeptide structure (Figure 3). Peak m/z 186.1 Da, which is dehydrated HexNAc (204.1), dominates the fragmentation spectra of Ga-NAc-containing outer arm structures (Figure 3D, Supplemental Figure 1) as opposed to the peak m/z 138.1 Da which is dominant in the fragmentation spectra of arms containing the GlcNAc (Figure 3C, Supplemental Figure 1) 26. Under high collision energy, GlcNAc contribution in low m/z range is substantially higher due to fragmentation of the second (third etc) arm as well as core structure and the specific GalNAc fragment ion becomes undetectable. Ion abundance is important for structural assignments, especially for the asymmetric and/or multiply branched glycans. We observe resolution of the asymmetric LacdiNAc structures (Supplemental Figure 3) but additional studies will be needed to further resolve the multi-antennary asymmetric structural features. We currently investigate whether ratios of ions or inclusion of ion mobility separations further improves structural resolution.
Presence of glycan sulfation based on the sulfated oxonium ions in low collision energy spectra
As we mentioned above, sulfated glycopeptides can be identified in low collision energy spectra by neutral loss of 80 Da corresponding to the sulfate group (Supplemental Figure 1). Further confirmation of the sulfation of the glycopeptides comes from the oxonium ions confirming the presence of the sulfated glycan building block. Peak m/z 284 corresponds to the sulfo-HexNAc and is well represented in the low collision energy spectra (approximately 8% of relative intensity) (Supplemental Figure 3). This ion can be used for confirmation of sulfation and for quantification of the glycopeptide in a manner analogous to the previously published work 24. This is in contrast to the published study23 where the sulfated oxonium ion is limited to less than 0.1% of relative intensity and becomes undetectable in samples with lower representation of the sulfated glycopeptides (as in complex mixtures).
Outer arm Galabiose structure
Gal1-4Gal-GlcNAc fragment (m/z 528.2 Da) is another example of a structure-specific ion derived from the outer arm at low CE settings. This ion is to some extent visible in all N-glycopeptide tandem mass spectra as a product of fragmentation of the chitobiose core (GlcNAc-Man-Man) or as a fragment of the outer-arm LacNAc structures where one mannose from the chitobiose core remains attached to the fragment. Under limited collision energy, fragmentation of the structure terminated with the Gal1-4Gal provides the signal 528.2 (HexNAc-Hex-Hex) derived from the favored fragmentation of the outer arm (loss of an arm) (Figure 2); the other bonds that could generate an isobaric fragment do not fragment at low CE (Figure 1). This fragment becomes one of the major ions in the spectra comparable to the intensity of the ion m/z 366.2 Da derived from the fragmentation of LacNAc structures.
Sialylated and fucosylated LacNAc vs LacdiNAc
An interesting fragmentation phenomenon is the stability of fucosylated and sialylated GalNAc in comparison to the fucosylated GlcNAc and sialylated Galactose of the outer arm structures. Figure 5A shows the fragmentation of a glycopeptide of human PD-L1 carrying biantennary fucosylated LacdiNAc. Fragment m/z 350.14 is related to the fucosylated HexNAc which is in this case with high probability fucosylated GalNAc derived from the fucosylated LacdiNAc structure. Figure 1B shows fragmentation of a glycopeptide of human haptoglobin carrying fucosylated LacNAc structure which was previously assigned as dominant 1-3,4 linkage to GlcNAc 22. The Fucose-HexNAc fragment m/z 350.14 Da is absent in the fucosylated LacNAc spectra (Figure 1B) but the fragment m/z 512.2 Da confirms fucosylation of the LacNAc structure on the outer arm. The tandem mass spectra of the di-sialo LacNAc/LacdiNAc structure contain non-sialylated (366.2 and 407.2) and sialylated (657.3 and 698.3) outer arm fragments of similar intensities that confirm the asymmetric mixed LacNAc/LacdiNAc biantennary structure. Pair of ions 454.2 (Gal-SA) and 495.2 (GalNAc-SA) 27 show differences in the visibility (1/10) of sialic acid attached to the terminal galactose or terminal GalNAc.
Figure 5.
Spectra of N-glycopeptides of human PD-L1 carrying the following structural motifs: A. multiply fucosylated LacdiNAc structures; B. sialylated LacdiNAc and LacNAc structures.
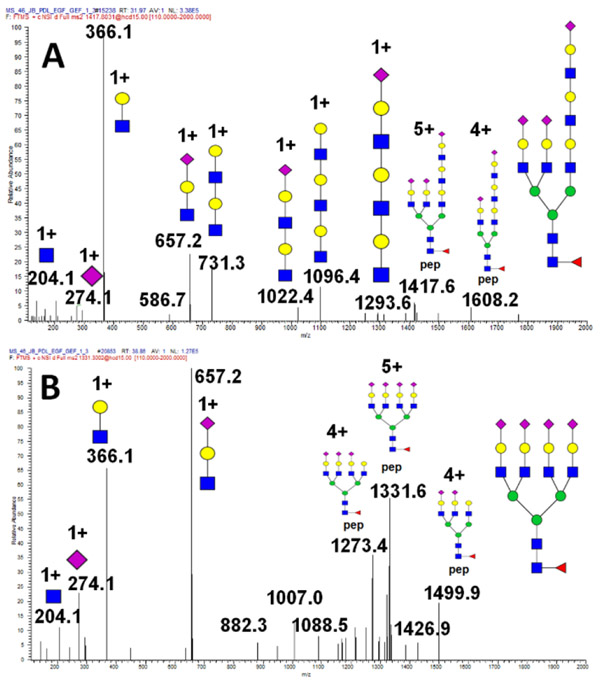
PolyLacNAc structure
Identification of the polyLAcNAc structural motif by mass spectrometry is a challenging task. Here we identify specific oxonium ions which identify the polyLacNAc at low CE (15 NCE) as documented on the glycopeptide of PD-L1 (Figure 6A). The ions such as m/z 1387.3 or 1096.4 are specific for the polyLacNAc motif as documented by comparison to the fragmentation spectra of a tetra-antennary complex glycan under the same fragmentation conditions (Figure 6B). This spectrum does not contains any of the theoretically plausible ions m/z 1022.4, 1096.4, or 731.3 when the low CE fragmentation is used.
Figure 6.
MS/MS spectrum of a poly-LacNAc containing N-glycan observed in glycopeptide RLDPEENHTAELVIPELPLHPPNER of the human PD-L1 containing the following: (A) polyLacNAc carrying tri-antennary and (B) tetra-antennary glycoforms.
Analysis of HEK 293 secretome for glycoprotein carriers of the LacdiNAc structure
HEK 293 cells were originally derived from human embryonic kidney cells and are commonly used for production of proteins in research laboratories and biopharmaceutical industry. We used the HEK 293 cells in our previous studies of glycosylation of ITIH4 28 and proBDNF 24 proteins. In case of proBDNF, our findings show dominant LacdiNAc structures, which are to some degree sulfated, on the N121 sequon. Here we performed a glycoproteomics study of the HEK 293 lysate and the HEK 293 secretome. We have used 3 different collision energies to confirm outer arm LacdiNAc structures from low collision energy spectra. High collision energy spectra were used for reliable identification of the glycosylated peptides. We have found several proteins (Supplemental Table 1) carrying LacdiNAc motifs in the HEK 293 secretome but have not identified any LacdiNAc glycoprotein in the cell lysate even though we identified a large number of glycoproteins/proteins in the cell secretome (n=1791/251). This shows that low CE fragmentation is a reliable method for the identification of the LacdiNAc structural motif ion N-glycoproteins even in complex samples like the HEK 293 secretome.
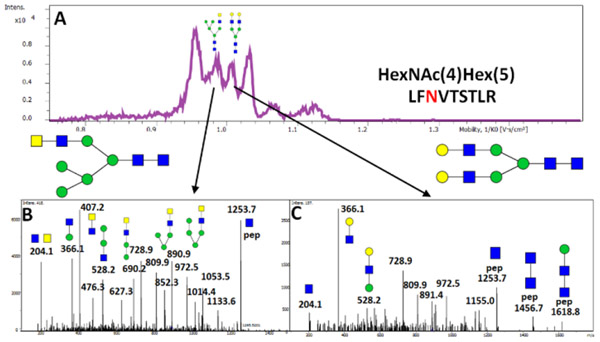
Low energy fragmentation as a technique for the assignment of structural motifs in ion mobility separated isobaric glycopeptides
Ion mobility is a powerful tool for the separation of isobaric species according to their differences in collision cross section (shape differences). Here we used trapping ion mobility spectrometry (TIMS) using timsTOF Pro (Bruker) and method described in the supplemental material for the separation of isobaric glycopeptides. Figure 7 shows differences in the observed fragmentation spectra of the tryptic glycopeptide (HexNAc4Hex5) of PD-L1 containing the LacdiNAc (Figure 7B) and LacNAc (Figure 7C) structural motifs. Structures of this composition are commonly identified as biantennary N-glycans. However, using TIMS separation, we were able to detect 4 peaks of which only peak 3 has fragmentation spectrum consistent with a biantennary complex N-glycan. The remaining 3 peaks are isomeric structures or potentially confomers of a hybrid N-glycan carrying the LacdiNAc structure. We used low collision energy spectra to identify the structural motifs (Figure 7). We expect that, in combination with the TIMS separation, we will be able to complete quantification of the resolved glycopeptide structures in subsequent studies.
Figure 7.
Ion-mobilogram of the LFNVTSTLR glycopeptide carrying isobaric HexNAc(4)Hex(5) N-glycans. This composition, commonly described as biantennary galactosylated complex glycan, is separated (A) into the following peaks with distinct MS/MS spectra: (B) hybrid glycopeptide containing LacdiNAc (second ion mobility peak); and (C) biantennary complex LacNAc glycopeptide (third ion mobility peak).
CONCLUSION
We document that modulation of CID/HCD fragmentation of N-glycans attached to specific peptides provides structure-specific information which is not retrieved at high CE typical of proteomic or glycoproteomic workflows. Under low CE fragmentation conditions, we retrieve fragments diagnostic of common outer arm structural motifs which can resolve isobaric structures attached to the same peptide and we continue to improve methods for their quantification. We expect that glycopeptide analysis, utilizing the structure-specific ions, will allow resolution of isobaric glycopeptides using previously published methods based on B- and Y-ion quantification. Our results suggest that inclusion of the relative intensity of oxonium ions at defined CE settings to the scoring algorithm would improve automated identification of structure-specific ions in current software tools.
Supplementary Material
ACKNOWLEDGMENT
Research reported in this publication was supported by the National Institutes of Health under awards S10OD023557, U01 CA230692, R01CA135069, and R01CA238455 to RG and CCSG Grant P30 CA51008, to Lombardi Comprehensive Cancer Center, supporting the Proteomics and Metabolomics Shared Resource, Georgetown University Medical Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Glycoproteins carrying LacdiNAc structure identified in the secretome of HEK293 cells, Fragmentation spectra of sulfate-containing biantennary LacdiNAc, Oxonium ions in the HCD spectra of the Cochlin glycopeptide ENYSSVDANGIQSQMLSQR carrying the biantennary fucosylated LacdiNAc N-glycan., Oxonium ions in the HCD spectra of Carboxypeptidase E glycopeptide DLQGNPIANATISVEGIDHDVTSAK carrying the mono-sialylated biantennary LacdiNAc/LacNac.
REFERENCES
- 1.Dennis JW; Lau KS; Demetriou M; Nabi IR Adaptive regulation at the cell surface by N-glycosylation. Traffic. 2009, 10 (11), 1569–1578. [DOI] [PubMed] [Google Scholar]
- 2.Varki A; Cummings R; Esko J; Freeze H; Stanley P; Bertozzi CR; Hart GW; Etzler ME Essentials of Glycobiology; 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 2009. [PubMed] [Google Scholar]
- 3.Palaniappan KK; Bertozzi CR Chemical Glycoproteomics. Chem. Rev 2016, 116 (23), 14277–14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldman R; Sanda M Targeted methods for quantitative analysis of protein glycosylation. Proteomics. Clin. Appl 2015, 9 (1-2), 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith DF; Cummings RD; Song X History and future of shotgun glycomics. Biochem. Soc. Trans 2019, 47 (1), 1–11. [DOI] [PubMed] [Google Scholar]
- 6.Mucha E; Stuckmann A; Marianski M; Struwe WB; Meijer G; Pagel K In-depth structural analysis of glycans in the gas phase. Chem. Sci 2019, 10 (5), 1272–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alley WR Jr.; Mann BF; Novotny MV High-sensitivity analytical approaches for the structural characterization of glycoproteins. Chem. Rev 2013, 113 (4), 2668–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbosa EA; Fontes NDC; Santos SCL; Lefeber DJ; Bloch C; Brum JM; Brand GD Relative quantification of plasma N-glycans in type II congenital disorder of glycosylation patients by mass spectrometry. Clin. Chim. Acta 2019, 492, 102–113. [DOI] [PubMed] [Google Scholar]
- 9.Yang Y; Franc V; Heck AJR Glycoproteomics: A Balance between High-Throughput and In-Depth Analysis. Trends Biotechnol. 2017, 35 (7), 598–609. [DOI] [PubMed] [Google Scholar]
- 10.Yu A; Zhao J; Peng W; Banazadeh A; Williamson SD; Goli M; Huang Y; Mechref Y Advances in mass spectrometry-based glycoproteomics. Electrophoresis 2018, 39 (24), 3104–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong X; Huang Y; Cho BG; Zhong J; Gautam S; Peng W; Williamson SD; Banazadeh A; Torres-Ulloa KY; Mechref Y Advances in mass spectrometry-based glycomics. Electrophoresis 2018, 39 (24), 3063–3081. [DOI] [PubMed] [Google Scholar]
- 12.Kozlik P; Goldman R; Sanda M Hydrophilic interaction liquid chromatography in the separation of glycopeptides and their isomers. Anal. Bioanal. Chem 2018, 410 (20), 5001–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozlik P; Sanda M; Goldman R Nano reversed phase versus nano hydrophilic interaction liquid chromatography on a chip in the analysis of hemopexin glycopeptides. J. Chromatogr. A 2017, 1519, 152–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozlik P; Goldman R; Sanda M Study of structure-dependent chromatographic behavior of glycopeptides using reversed phase nanoLC. Electrophoresis 2017, 38 (17), 2193–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinneburg H; Chatterjee S; Schirmeister F; Nguyen-Khuong T; Packer NH; Rapp E; Thaysen-Andersen M Post-Column Make-Up Flow (PCMF) Enhances the Performance of Capillary-Flow PGC-LC-MS/MS-Based Glycomics. Anal. Chem 2019, 91 (7), 4559–4567. [DOI] [PubMed] [Google Scholar]
- 16.Ashwood C; Lin CH; Thaysen-Andersen M; Packer NH Discrimination of Isomers of Released N- and O-Glycans Using Diagnostic Product Ions in Negative Ion PGC-LC-ESI-MS/MS. J. Am. Soc. Mass Spectrom 2018, 29 (6), 1194–1209. [DOI] [PubMed] [Google Scholar]
- 17.Manz C; Grabarics M; Hoberg F; Pugini M; Stuckmann A; Struwe WB; Pagel K Separation of isomeric glycans by ion mobility spectrometry - the impact of fluorescent labelling. Analyst 2019. [DOI] [PubMed] [Google Scholar]
- 18.Kolli V; Dodds ED Energy-resolved collision-induced dissociation pathways of model N-linked glycopeptides: implications for capturing glycan connectivity and peptide sequence in a single experiment. Analyst 2014, 139 (9), 2144–2153. [DOI] [PubMed] [Google Scholar]
- 19.Dodds ED Gas-phase dissociation of glycosylated peptide ions. Mass Spectrom. Rev 2012, 31 (6), 666–682. [DOI] [PubMed] [Google Scholar]
- 20.Acs A; Ozohanics O; Vekey K; Drahos L; Turiak L Distinguishing Core and Antenna Fucosylated Glycopeptides Based on Low-Energy Tandem Mass Spectra. Anal. Chem 2018, 90 (21), 12776–12782. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann M; Pioch M; Pralow A; Hennig R; Kottler R; Reichl U; Rapp E The Fine Art of Destruction: A Guide to In-Depth Glycoproteomic Analyses-Exploiting the Diagnostic Potential of Fragment Ions. Proteomics. 2018, 18 (24), e1800282. [DOI] [PubMed] [Google Scholar]
- 22.Pompach P; Brnakova Z; Sanda M; Wu J; Edwards N; Goldman R Site-specific glycoforms of haptoglobin in liver cirrhosis and hepatocellular carcinoma. Mol. Cell Proteomics 2013, 12 (5), 1281–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo CW; Guu SY; Khoo KH Distinctive and Complementary MS(2) Fragmentation Characteristics for Identification of Sulfated Sialylated N-Glycopeptides by nanoLC- MS/MS Workflow. J. Am. Soc. Mass Spectrom 2018, 29 (6), 1166–1178. [DOI] [PubMed] [Google Scholar]
- 24.Benicky J; Sanda M; Brnakova KZ; Goldman R N-Glycosylation is required for secretion of the precursor to brain-derived neurotrophic factor (proBDNF) carrying sulfated LacdiNAc structures. J. Biol. Chem 2019, 294 (45), 16816–16830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakane T; Angata K; Sato T; Kaji H; Narimatsu H Identification of mammalian glycoproteins with type-I LacdiNAc structures synthesized by the glycosyltransferase B3GALNT2. J. Biol. Chem 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halim A; Westerlind U; Pett C; Schorlemer M; Ruetschi U; Brinkmalm G; Sihlbom C; Lengqvist J; Larson G; Nilsson J Assignment of saccharide identities through analysis of oxonium ion fragmentation profiles in LC-MS/MS of glycopeptides. J. Proteome. Res 2014, 13 (12), 6024–6032. [DOI] [PubMed] [Google Scholar]
- 27.Medzihradszky KF; Kaasik K; Chalkley RJ Characterizing sialic acid variants at the glycopeptide level. Anal. Chem 2015, 87 (5), 3064–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandler KB; Brnakova Z; Sanda M; Wang S; Stalnaker SH; Bridger R; Zhao P; Wells L; Edwards NJ; Goldman R Site-specific glycan microheterogeneity of inter-alpha-trypsin inhibitor heavy chain H4. J. Proteome. Res 2014, 13 (7), 3314–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.