Abstract
A novel series of ciprofloxacin hybrids comprising various heterocycle derivatives has been synthesized and structurally elucidated using 1H NMR, 13C NMR, and elementary analyses. Using ciprofloxacin as a reference, compounds 1–21 were screened in vitro against Gram-positive bacterial strains such as Staphylococcus aureus and Bacillus subtilis and Gram-negative strains such as Escherichia coli and Pseudomonas aeruginosa. As a result, many of the compounds examined had antibacterial activity equivalent to ciprofloxacin against test bacteria. Compounds 2–6, oxadiazole derivatives, were found to have antibacterial activity that was 88 to 120% that of ciprofloxacin against Gram-positive and Gram-negative bacteria. The findings showed that none of the compounds tested had antifungal activity against Aspergillus flavus, but did have poor activity against Candida albicans, ranging from 23% to 33% of fluconazole, with compound 3 being the most active (33% of fluconazole). The most potent compounds, 3, 4, 5, and 6, displayed an IC50 of 86, 42, 92, and 180 nM against E. coli DNA gyrase, respectively (novobiocin, IC50 = 170 nM). Compounds 4, 5, and 6 showed IC50 values (1.47, 6.80, and 8.92 µM, respectively) against E. coli topo IV in comparison to novobiocin (IC50 = 11 µM).
Keywords: ciprofloxacin, heteroaryl, antibacterial, gyrase, topoisomerase IV
1. Introduction
Bacterial infection remains a significant threat to human life due to its increasing resistance to conventional antibiotics, which is a growing public health concern. As a result, there is a critical need to create new antimicrobial agents with potent anti-drug-resistant microorganism activity [1]. That is why antimicrobial agent investigations are so critical and should always be up to date.
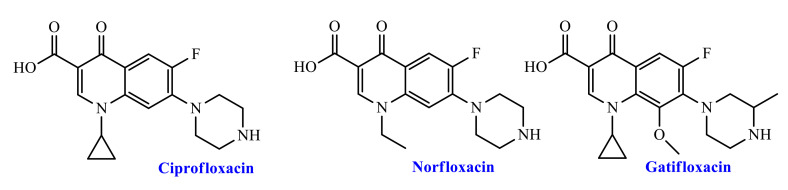
Due to their excellent efficacy, bioavailability, and relatively low toxic and adverse effects, fluoroquinolone (FQ) antibiotics have been one of the most commonly used groups of antibiotics in recent years, (Figure 1). Fluoroquinolones (FQs) are antibiotics that have the ability to treat a range of bacterial infections [2,3]. FQs are inhibitors of S. aureus multidrug efflux pumps [4], lower and upper respiratory infections [5], prostatitis, and urinary tract infections (UTI) [6]. Fluoroquinolone antibiotics co-alter the bacterial DNA gyrase and topoisomerase IV enzymes in a hybrid enzyme–DNA complex [7]. Such a change in bacterial enzyme performance inhibits the desired bacterial growth DNA synthesis. FQ drugs have long been known as a favored structural framework for the development of new commercially available drugs [8]. Changes in the elementary structure of FQs are thought to enhance drug interaction with the target enzyme, which may improve pharmacokinetic properties. This type of structural functioning of the FQs may provide stronger drugs for future generations. Substitution at position 7 of the FQ core has a significant impact on solubility, bioavailability, and antimicrobial activities [9]. With the introduction of various C-7 moieties, researchers have focused on improving their biological range. As a result, different moieties were used to examine its biological profile with a piperazinyl ring, substituted piperazine moiety, heterocyclic ring (especially five or six members). Ciprofloxacin is one of the most potent second-generation fluoroquinolones (FQs). It offers potential anti-infective therapy for a wide range of bacteria, Gram-positive, and Gram-negative (Figure 1) [10,11]. For 12 medical treatments, including veterinary uses, the Food and Drug Administration (FDA) approved ciprofloxacin (CP, see Figure 1) which exhibits antibacterial activity with minimal side effects and good pharmacokinetic properties. In addition, ciprofloxacin has a wide range of biological profiles and has been used to examine its antimalarial, anti-fungal, anti-tumor, and antibacterial properties in different areas of medical research [12,13].
Figure 1.
Chemical structures of some FDA approved marketed fluoroquinolone-based antibiotic drugs.
Ciprofloxacin is currently used to treat a number of Gram-positive and Gram-negative bacterial infections in clinical practice. However, because of the emergency and widespread of drug-resistant bacteria, ciprofloxacin is becoming increasingly ineffective. As a result, novel antibacterial agents are urgently needed [14].
To combat resistance, the production of novel ciprofloxacin derivatives that are effective against both drug-susceptible and drug-resistant pathogens is crucial. Many ciprofloxacin derivatives have been designed and synthesized with excellent in vitro and in vivo potency against both drug-sensitive and drug-resistant species, including fluoroquinolone-resistant, multidrug-resistant pathogens [15,16,17,18,19].
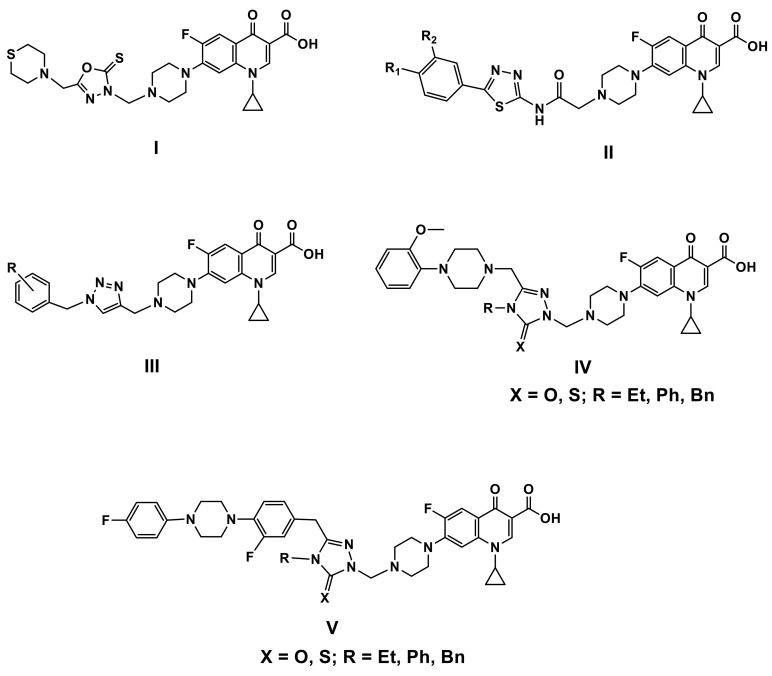
With MICs ranging from 0.28 to 15.8 µM/mL, the 1,3,4-oxadiazole ciprofloxacin hybrid I (Figure 2) was >41 times more potent than ampicillin (MIC: 10–128 mg/mL) against most of the pathogens tested [15].
Figure 2.
Chemical structures of some ciprofloxacin-azole hybrids I–V.
The antibacterial and antifungal activities of the thiadiazole ciprofloxacin hybrids II (Figure 2) were only mild to moderate [16].
Agarwal et al. tested a sequence of bis-1,2,3-triazole-ciprofloxacin hybrids III (Figure 2) in vitro against a panel of clinically important bacteria [17]. A significant portion of the hybrids showed increased activity against both Gram-positive and Gram-negative bacteria relative to ciprofloxacin, and antibacterial activity appears to be related to the nature and position of substituents, as well as their isomeric effects on phenyl rings. Furthermore, the compounds’ low toxicity profile suggests that they may be useful antibiotics in the future [17].
Demirbas et al. found that the 1,2,4-triazole-5(4H)-one/thione ciprofloxacin hybrids IV and V (Figure 2) with substituted piperazine at the C-3 position of the triazole moiety had promising in vitro activity against both Gram-positive and Gram-negative pathogens with MIC 0.24 mg/mL, which was far more potent than Ampicillin (MIC: 3.9–250 mg/mL) [18,19].
Continuing our quest to find a compound with improved antimicrobial properties [20,21,22,23,24,25,26,27], the current study describes the synthesis and structure elucidation of Fluoroquinolone(ciprofloxacin)-based hybrids containing various heterocycle derivatives, as well as antimicrobial activity evaluation using various strains of Gram-positive (S. aureus and B. subtilis), Gram-negative (E. coli and P. aeruginosa), and fungi (A. flavus and C. albicans). In addition, the inhibitory activity of the most active compounds against E. coli DNA gyrase and topoisomerase IV has been identified as a potential molecular target.
2. Results and Discussion
2.1. Chemistry
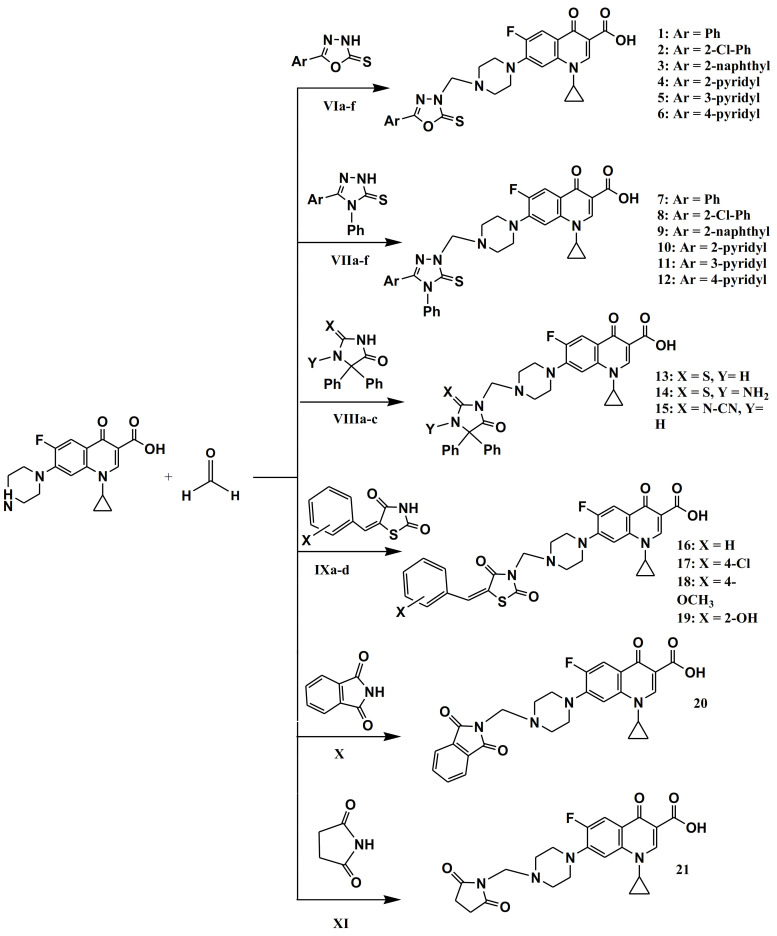
The synthetic strategy towards the synthesis of target compounds 1–21 is outlined in Scheme 1. The intermediates VIa–f [28], VIIa–f [29], VIIIa–c [30], and IXa–d [31] were synthesized, as stated previously, and their structures were confirmed by comparing their physical constants and spectral data to those previously recorded. As shown in Scheme 1, ciprofloxacin undergoes a Mannich reaction with heterocycles I–VI and formaldehyde in refluxing ethanol to yield the target compounds 1–21 in yields ranging from 79% to 97%. The IR spectra of compounds 1–21 showed a stretching band at 3381–3310 cm−1 related to (OH), a medium stretching band at 3062–3004 of CH aromatic and strong stretching band at 1731–1701 cm−1 related to (C=O), which are consistent with the proposed structure. The 1H NMR spectra of 1–21 revealed the appearance of a methylene signal at 5.19–5.16 (s, 2H, N-CH2-N), two sets of triplets at 3.39–3.27 and 3.07–3.03 ppm, which is indicative of piperazinyl protons, three signals at 3.84–3.80 (m, 1H), 1.35–1.30 (q, 2H), and 1.18–1.14 (q, 2H) ppm attributed to cyclopropyl protons, and broad singlet at 15.13–15.05 ppm of the COOH group. Moreover, the 1H NMR spectra of 16–19 revealed the appearance of singlet signal at 8.64 ppm of olefinic CH. The 13C NMR spectra of 1–21 showed the characteristic methyl carbon (N-C-N) at 70 ppm and the (C=O) at 166–160 and 173–178 ppm. At their predicted chemical shifts were the olefin and aromatic carbons (Supplementary File, Figures S1–S21). The purity of 1–21 has been confirmed using elementary analysis, and the results fit the products’ molecular formula.
Scheme 1.
Synthesis of target compounds 1–21. Reagents and conditions: Appropriate heterocyclic compound, EtOH, reflux, 0.5 h, 79–97%.
2.2. Biology
2.2.1. Antimicrobial Sensitivity Test
An updated Kirby-Bauer disc diffusion method was used to assess the antimicrobial activity of the tested samples [32,33,34,35]. Table 1 presents the results of the preliminary antimicrobial testing of final compounds. Using ciprofloxacin as a reference drug, synthetic compounds 1–21 were tested in vitro for antibacterial activity against S. aureus and B. subtilis as Gram-positive strains and E. coli and P. aeruginosa as Gram-negative strains. As a result, the majority of newly synthesized compounds demonstrated promising antibacterial activity comparable to ciprofloxacin against test species (Table 1). The oxadiazole derivatives, compounds 2–6 were found to exhibit pronounced antibacterial activity, which ranged from 88% to 120% that of ciprofloxacin against both Gram-positive and Gram-negative strains. It is worth mentioning that compound 6 showed superior activity (120%) against S. aureus to that of ciprofloxacin. Oxadiazoles 4 and 5 showed equipotent activity to ciprofloxacin against S. aureus and E. coli. The thiazolidine derivative 16 had a ciprofloxacin-like activity of 93% against B. subtilis, E. coli, and P. aeruginosa, but only 85% activity against S. aureus, Table 1. Compounds 1, 7–10, and 17–21 showed moderate antibacterial activity which ranged from 70% to 83% of ciprofloxacin against both Gram-positive and Gram-negative strains.
Table 1.
Inhibitory zone diameter (mm) of compounds 1–21.
| Compound | Inhibition Zone Diameter (mm/mg Sample) | |||||
|---|---|---|---|---|---|---|
| Bacterial Species | Fungi | |||||
| (G+) | (G−) | |||||
| B. subtilis | S. aureus | E. coli | P. aeruginosa | A. flavus | C. albicans | |
| Ciprofloxacin | 40 | 40 | 40 | 40 | -- | -- |
| Fluconazole | -- | -- | -- | -- | 40 | 40 |
| 1 | 31 | 36 | 34 | 31 | 0.0 | 0.0 |
| 2 | 36 | 37 | 36 | 36 | 0.0 | 9 |
| 3 | 36 | 37 | 37 | 35 | 0.0 | 13 |
| 4 | 37 | 41 | 41 | 40 | 0.0 | 10 |
| 5 | 37 | 41 | 41 | 38 | 0.0 | 10 |
| 6 | 38 | 48 | 38 | 37 | 0.0 | 10 |
| 7 | 32 | 35 | 32 | 32 | 0.0 | 0.0 |
| 8 | 32 | 32 | 33 | 31 | 0.0 | 0.0 |
| 9 | 31 | 32 | 30 | 31 | 0.0 | 0.0 |
| 10 | 30 | 34 | 31 | 32 | 0.0 | 0.0 |
| 11 | 35 | 31 | 31 | 35 | 0.0 | 0.0 |
| 12 | 35 | 33 | 35 | 35 | 0.0 | 0.0 |
| 13 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 16 | 37 | 34 | 37 | 37 | 0.0 | 11 |
| 17 | 32 | 28 | 31 | 31 | 0.0 | 0.0 |
| 18 | 32 | 31 | 32 | 33 | 0.0 | 0.0 |
| 19 | 32 | 31 | 31 | 31 | 0.0 | 0.0 |
| 20 | 33 | 41 | 35 | 33 | 0.0 | 0.0 |
| 21 | 31 | 36 | 28 | 31 | 0.0 | 0.0 |
According to the previous findings, the inclusion of oxadiazole and thiadiazole moiety in a compound confers the highest efficacy.
Furthermore, using fluconazole as a reference drug, 1–21 were tested in vitro for antifungal activity against A. flavus and C. albicans (Table 1). The findings showed that the tested compounds had no antifungal activity against A. flavus but had weak antifungal activity against C. albicans, ranging from 23% to 33% of fluconazole, with compound 3 being the most active (33% of fluconazole).
2.2.2. Minimum Inhibitory Concentration Test
A two-fold serial dilution method was used to assess the antimicrobial activity of the most active components, oxadiazole-based hybrids, 2–6 [36]. Table 2, using the reference drug ciprofloxacin, was presented with the MICs (minimal inhibitory concentrations) of these compounds against tested bacteria. Some new compounds have demonstrated good antimicrobial inhibitory activities for Gram-positive and Gram-negative strains. As shown in Table 2, compounds 4, 5, and 6 were the most active and effective against three bacterial strains, in which compound 4 with MIC values of 0.035, 0.062, 0.062 µg/mL against S. aureus, E. coli and P. aureoginosa, compound 5 with MIC values of 0.035, 0.062, 0.125 µg/mL against S. aureus, E. coli and P. aureoginosa, and compound 6 with MIC values of 0.031, 0.125, 0.125 µg/mL against S. aureus, E. coli and P. aureoginosa, respectively. In comparison to ciprofloxacin (MIC of 0.030 g/mL), compound 2 showed the next best activity against S. aureus strains with a MIC value of 0.062 g/mL. Interestingly, all the compounds examined only had a minor inhibitory effect on B. subtilis, a Gram-positive organism. The nature of aromatic substitution in the oxadiazole moiety tends to be correlated with higher antibacterial effects and the activity increased with (Ar) in the order of 2-pyridyl ≥ 3-pyridyl > 4-pyridyl > 2-Cl-Ph > 2-naphthyl.
Table 2.
Antimicrobial activities of compounds 2–6.
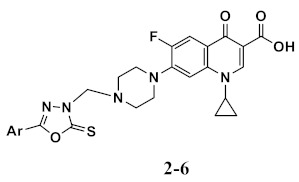
| Minimum Inhibitory Concentration (MIC) in µg/mL | |||||
|---|---|---|---|---|---|
| Compound | Ar | Bacterial Species | |||
| (G+) | (G−) | ||||
| B. subtilis | S. aureus | E. coli | P. aeruginosa | ||
| 2 | 2-Cl-Ph | 0.125 | 0.162 | 0.125 | 0.125 |
| 3 | 2-Naphthyl | 0.125 | 0.125 | 0.125 | 0.05 |
| 4 | 2-Pyridyl | 0.125 | 0.035 | 0.062 | 0.062 |
| 5 | 3-Pyridyl | 0.125 | 0.035 | 0.062 | 0.125 |
| 6 | 4-Pyridyl | 0.125 | 0.031 | 0.125 | 0.125 |
| Ciprofloxacin | -- | 0.010 | 0.030 | 0.060 | 0.060 |
2.2.3. Inhibitory Activity Against E. coli DNA Gyrase and Topoisomerase IV
E. coli DNA gyrase assay [37] was performed to evaluate the inhibitory potency of oxadiazole-based derivatives 2–6 against E. coli DNA gyrase and the results are included in Table 3. Results are presented as residual activities (RAs) of the enzyme at 1 µM of compounds or as IC50 values for compounds with RA <50% (Table 3). The results of the antimicrobial activity study are complemented by the results from this assay. Investigated compounds 3–6 exhibited inhibition of E. coli DNA with IC50 ranging from 42 to 180 nM relative to reference novobiocin (IC50 = 170 nM). Based on the data provided, compounds 3, 4, and 5 were found to be the most active and their inhibitory activities of E. coli DNA gyrase assay (IC50 = 86 ± 9, 42 ± 7, and 92 ± 9, respectively) were superior to positive control novobiocin.
Table 3.
E. coli DNA gyrase and topoisomerase IV inhibitory activities of compounds 2–6 and Novobiocin.
| Compound | IC50 (nM) a or RA (%) b | IC50 (µM) a or RA (%) b |
|---|---|---|
| E. coli DNA Gyrase | E. coli Topo IV | |
| 2 | 57% | 89% |
| 3 | 86 ± 9 nM | 58% |
| 4 | 42 ± 7 nM | 1.47 ± 0.20 µM |
| 5 | 92 ± 9 nM | 6.80 ± 0.20 µM |
| 6 | 180 ± 20 nM | 8.92 ± 0.20 µM |
| Novobiocin | 170 ± 20 nM | 11 ± 2 µM |
a Concentration of compound that inhibits the enzyme activity by 50%; b residual activity of the enzyme at 1 µM of the compound.
Compounds 2–6 were further evaluated against E. coli topoisomerase IV [37], as shown in Table 3. Compounds 4, 5, and 6, which were among the most potent inhibitors of E. coli gyrase also displayed promising results on topoisomerase IV (Table 3). Compounds 4, 5, and 6 had IC50 values = 1.47, 6.80, and 8.92 µM, respectively, which are much lower (more potent) than that for novobiocin (IC50 = 11 µM). From these findings, both 4 and 5, after optimization, appear to be promising dual target inhibitors against DNA gyrase and topoisomerase IV.
2.2.4. Cell Viability Assay
A human mammary gland epithelial cell line (MCF-10A) was used to conduct a cell viability assay [38]. Compounds 2–6 were incubated with MCF-10A cells for four days, and the viability of the cells was determined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay [39]. All compounds had no cytotoxic effects, and the viability of the cells was greater than 85% for most of the compounds examined at 50 M, as shown in Table 4.
Table 4.
Cell viability assay results of compounds 2–6.
| Compound | Cell Viability % |
|---|---|
| 2 | 87 |
| 3 | 85 |
| 4 | 89 |
| 5 | 91 |
| 6 | 87 |
2.3. Drug Likeness Profile
Absorption, delivery, metabolism, and excretion (ADME) testing became popular early in drug development programs, with computer models serving as feasible alternatives to experiments. The Swiss ADME website was used to predict the drug likeness profile of the studied compounds 1–21 [40,41]. Tables S1–S3 (Supplementary File) display the results of the drug likeness profile of these compounds. Many compounds, such as 1, 13, 14, 16, 17, 20, and 21, were expected to have high oral absorption. However, due to the high molecular weight (522) and molar refractivity (143), the others predicted poor oral absorption. Some of the tested compounds as 20 and 21 showed no violation to Lipinski (Pfizer) filters, except for one violation for compounds 1–8 and 11–19. In addition two violation for compounds 9 and 10 (molecular weight >500 [42]) and compound 21 showed no violations to Ghose in addition to two violations for 1–6, three violations for 7, 8, 11, 12, 13, 14 and four violations for 9 and 10 due to high WLOGP and molecular weight [43], no violation to Veber (GSK) for all compounds except one violation for 4, 5, 6, 14, 15, 19 [44], no violation to Egan (Pharmacia) for all compounds except one violation for 4, 5, 6, 14, 15, 18, 19 [45] and no violation to Muegge (Bayer) except one violation for compounds 1, 8, 13, 14, and two violations for 9, 10 [46] filters. The compounds were free from alerts for Pan Assay Interfering substances (PAINS) [47]. Total polar surface area (TPSA) values for most compounds are 103.16–148.69, as shown in Table 1, Table 2 and Table 3. This consists of “good GIT absorption”. There is a correlation between the molecular weight of compounds and their activity. In addition to low rigidity, this pattern highlighted low molecular weight is favorable. Lipophilicity, together with the molecular weight and the number of hydrogen bond donors and the number of hydrogen acceptors shown by these compounds, plays the role of five (see Table S1, Supplementary File).
3. Materials and Methods
3.1. Chemistry
General Details: See Appendix A
3.1.1. General Procedure for the Synthesis of Compounds 1–21
To a mixture of ciprofloxacin HCl (1.28 g, 0.003 mol), formaldehyde (0.2 g, 0.007 mol) and different heterocyclic compounds (0.003 mol) were dissolved in 10 mL of ethanol. The reaction mixture was stirred with reflux for 30 min. The reaction mixture was allowed to cool at room temperature, the separated solid was filtered off, washed with water, and crystallized from methanol.
1-Cyclopropyl-6-fluoro-4-oxo-7-[4-(5-phenyl-2-thioxo[1,3,4]oxadiazol-3-yl-methyl)-piperazin-1-yl]-1,4-dihydro-quinoline-3-carboxylic acid (1)
Yield (94%); m.p. 228 °C; IR: 3350 (OH), 3049 (CH aromatic), 2987, 2946 (CH-aliphatic), 1710 (C=O), 1615 (C=N); 1H NMR: δ 15.13 (brs. OH, COOH), 8.65–7.58 (m, 7H, aromatic), 5.16 (s, 2H, N-CH2-N), 3.83–3.80 (m, J = 2.44 Hz, 1H, CH2-CH-CH2 cyclopropyl-H), 3.38 (t, 4H, piperazinyl-H), 3.04 (t, 4H, piperazinyl-H), 1.34–1.32 (q, J = 3.54 Hz, 2H, CH2-CH-CH2 cyclopropyl-H), 1.18–1.15 (q, J = 4.08 Hz, 2H, CH2-CH-CH2cyclopropyl-H); 13C NMR: δ 178.82., 166.40, 148.28. 145.45, 139.64, 129.88, 111.52, 107.27, 106.96, 70.24, 51.05 49.82, 49.70, 36.27, 8.02. C26H24FN5O5S: C, 59.87; H, 4.64; N, 13.43, Found: C, 59.81; H, 4.62; N, 13.34.
7-{4-[5-(2-Chloro-phenyl)-2-thioxo-[1,3,4]oxadiazol-3-ylmethyl]-piperazin-1-yl}-1-cyclopropyl-6-fluoro-4-oxo-1,4-dihydro-quinoline-3-carboxylic acid (2)
Yield (85%); m.p. 226 °C; IR: 3354 (OH), 3062 (CH aromatic), 2999, 2914 (CH-aliphatic), 1719 (C=O), 1614 (C=N); 1H NMR: δ 15.11 (brs. OH, COOH), 8.65–7.65 (m, 7H, aromatic), 5.19 (s, 2H, N-CH2-N), 3.84–3.80 (m, J = 3.52 Hz, 1H, CH2-CH-CH2 cyclopropyl-H), 3.39 (t, 4H, piperazinyl-H), 3.03 (t, 4H, piperazinyl-H), 1.35–1.30 (q, J = 5.44 Hz, 2H, CH2-CH-CH2 cyclopropyl-H), 1.18–1.15 (q, J = 5.64 Hz, 2H, CH2-CH-CH2cyclopropyl-H); 13C NMR: δ 173.59., 168.75, 166.45, 131.73, 128.38, 111.55, 111.30, 110.09, 109.37, 108.30, 107.17, 70.26, 49.84, 49.62, 36.33, 8.03. C26H23ClFN5O5S: C, 56.16; H, 4.17; N, 12.60, Found: C, 56.08; H, 4.02; N, 12.34.
1-Cyclopropyl-6-fluoro-7-[4-(5-naphthalen-2-yl-2-thioxo-[1,3,4]oxadiazol-3-yl methyl)-piperazin-1-yl]-4-oxo-1,4-dihydro-quinoline-3-carboxylic acid (3)
Yield (96%); m.p. 244 °C; IR: 3367 (OH), 3019 (CH aromatic), 2961, 2957 (CH-aliphatic), 1708 (C=O), 1609 (C=N); 1H NMR: δ 15.11 (brs. OH, COOH), 8.66–7.55 (m, 10H, aromatic), 5.19 (s, 2H, N-CH2-N), 3.83–3.79 (m, J = 5.51 Hz, 1H, CH2-CH-CH2 cyclopropyl-H), 3.39 (t, 4H, piperazinyl-H), 3.07 (t, 4H, piperazinyl-H), 1.35–1.30 (q, 4H, J = 5.69 Hz, CH2-CH-CH2 cyclopropyl-H); 13C NMR: δ 176.74, 166.27, 148.44, 139.39 122.25, 111.72, 107.27, 49.91, 49.79, 36.29, 8.03. C30H26FN5O5S: C, 63.03; H, 4.58; N, 12.25, Found: C, 62.88; H, 4.62; N, 12.01.
1-Cyclopropyl-6-fluoro-4-oxo-7-[4-(5-pyridin-2-yl-2-thioxo-[1,3,4]oxadiazol-3-yl-methyl)-piperazin-1-yl]-1,4-dihydro-quinoline-3-carboxylic acid (4)
Yield (89%); m.p. 218 °C; IR: 3369 (OH), 3049 (CH aromatic), 2967, 2924 (CH-aliphatic), 1721 (C=O), 1619 (C=N); 1H NMR: δ 15.10 (brs. OH, COOH), 9.07–7.55 (m, 7H, aromatic), 5.17 (s, 2H, N-CH2-N), 3.80 (m, J = 2.8 Hz, 1H, CH2-CH-CH2 cyclopropyl-H), 3.27 (t, 4H, piperazinyl-H), 3.04 (t, 4H, piperazinyl-H), 1.30–1.35 (q, J = 5.20 Hz, 2H, CH2-CH-CH2 cyclopropyl-H), 1.14–1.18 (q, J = 6.92 Hz, 2H, CH2-CH-CH2cyclopropyl-H); 13C NMR: δ 176.80., 166.34, 148.40. 142.31, 139.62, 124.25, 119.74, 111.25, 107.25, 107.00, 70.32, 49.84, 49.69, 36.28, 18.95 8.02. C25H24FN6O5S: C, 57.46; H, 4.44; N, 16.08, Found: C, 57.29; H, 4.31; N, 16.01.
1-Cyclopropyl-6-fluoro-4-oxo-7-[4-(5-pyridin-3-yl-2-thioxo-[1,3,4]oxadiazol-3-yl-methyl)-piperazin-1-yl]-1,4-dihydro-quinoline-3-carboxylic acid (5)
Yield (92%); m.p. 230 °C; IR: 3310 (OH), 3019 (CH aromatic), 2984, 2902 (CH-aliphatic), 1707 (C=O), 1609 (C=N); 1H NMR: δ 15.11 (brs. OH, COOH), 8.77–7.55 (m, 1H, aromatic), 5.18 (s, 2H, N-CH2-N), 3.82–3.80 (m, J = 1.80 Hz, 1H, CH2-CH-CH2 cyclopropyl-H), 3.27(t, 4H, piperazinyl-H), 3.04 (t, 4H, piperazinyl-H), 1.33–1.32 (q, J = 1.96 Hz, 2H, CH2-CH-CH2 cyclopropyl-H), 1.17–1.16 (q, J = 1.76 Hz, 2H, CH2-CH-CH2cyclopropyl-H); 13C NMR: δ 176.73, 166.41, 154.62, 152.14, 150.78, 148.37, 145.40, 139.55, 138.32, 127.27, 123.06, 118.96, 111.44, 111.22, 107.13, 106.96, 70.30, 49.79, 49.65, 43.21, 36.28, 18.98, 8.02. C25H24FN6O5S: C, 57.46; H, 4.44; N, 16.08, Found: C, 57.33; H, 4.40; N, 16.41.
1-Cyclopropyl-6-fluoro-4-oxo-7-[4-(5-pyridin-4-yl-2-thioxo-[1,3,4]oxadiazol-3-yl-methyl)-piperazin-1-yl]-1,4-dihydro-quinoline-3-carboxylic acid (6)
Yield (93%); m.p. 208 °C; IR: 3351 (OH), 3004 (CH aromatic), 2995, 2913 (CH-aliphatic), 1731 (C=O), 1607 (C=N); 1H NMR: δ 15.05 (brs. OH, COOH), 8.81–7.57 (m, 7H aromatic), 5.17 (s, 2H, N-CH2-N), 3.82 (m, J = 3 Hz, 1H, CH2-CH-CH2 cyclopropyl-H), 3.22(t, 4H, piperazinyl-H), 3.05 (t, 4H, piperazinyl-H), 1.35–1.33 (q, J = 4.56 Hz, 2H, CH2-CH-CH2 cyclopropyl-H), 1.19–1.17 (q, J = 3.04 Hz, 2H, CH2-CH-CH2cyclopropyl-H); 13C NMR: δ 178.81., 166.31, 157.70, 154.65, 151.34, 140.37, 139.62, 130.27, 120.07, 119.08, 111.51, 111.28, 107.29, 106.99, 70.28, 49.84, 49.68, 36.27, 8.03. C25H24FN6O5S: C, 57.46; H, 4.44; N, 16.08, Found: C, 57.14; H, 4.39; N, 16.15.
1-Cyclopropyl-7-[4-(3,4-diphenyl-5-thioxo-4,5-dihydro-[1,2,4]triazol-1-ylmethyl)-piperazin-1-yl]-6-fluoro-4-oxo-1,4-dihydro-quinoline-3-carboxylic acid (7)
Yield (93%); m.p. 288 °C; IR: 3381 (OH), 3014 (CH aromatic), 2961, 2943, 2906 (CH-aliphatic), 1707 (C=O), 1598 (C=N); 1H NMR: δ 15.12 (brs. OH, COOH), 8.66–7.36 (m, 13H, aromatic), 5.30 (s, 2H, N-CH2-N), 3.80–3.85 (m, J = 7.64 Hz, 1H, CH2-CH-CH2 cyclopropyl-H), 3.3.41 (t, 4H, piperazinyl-H), 3.09 (t, 4H, piperazinyl-H), 1.31–1.36 (q, J = 7.45 Hz, 2H, CH2-CH-CH2 cyclopropyl-H), 1.19–1.15 (q, J = 6.76 Hz, 2H, CH2-CH-CH2cyclopropyl-H); 13C NMR: δ 177.32, 173.40, 172.02, 170.78, 170.66, 150.17, 149.38, 148.54, 143.74, 136.56, 135.68, 135.68, 134.41, 129.07, 128.89, 58.17, 52.68, 50.08, 50.08, 10.64. C32H29FN6O3S: C, 64.41; H, 4.90; N, 14.08, Found: C, 64.22; H, 4.81; N, 14.02.
7-{4-[3-(2-Chloro-phenyl)-4-phenyl-5-thioxo-4,5-dihydro-[1,2,4]triazol-1-ylmethyl]-piperazin-1-yl}-1-cyclopropyl-6-fluoro-4-oxo-1,4-dihydro-quinoline-3-carboxylic acid (8)
Yield (96%); m.p. 214 °C; IR: 3332 (OH), 3056 (CH aromatic), 2984, 2917 (CH-aliphatic), 1701 (C=O), 1614 (C=N); 1H NMR: δ 15.13 (brs. OH, COOH), 8.67–7.33 (m, 12H, aromatic), 5.33 (s, 2H, N-CH2-N), 3.84–3.83 (m, J = 3.00 Hz, 1H, CH2-CH-CH2 cyclopropyl-H), 3.42 (t, 4H, piperazinyl-H), 3.07 (t, 4H, piperazinyl-H), 1.34–1.19 (q, J = 1.40 Hz, 4H, CH2-CH-CH2 cyclopropyl-H); 13C NMR: δ 176.84, 169.33, 166.44, 148.51, 147.50, 145.60, 134.35, 133.45, 129.55, 129.45, 128.57, 127.04, 111.57, 107.19, 107.04, 69.24, 50.06, 49.90, 36.33, 8.08. C32H28ClFN6O3S: C, 60.90; H, 4.47; N, 13.32, Found: C, 60.59; H, 4.28; N, 13.39.
1-Cyclopropyl-6-fluoro-7-[4-(3-naphthalen-2-yl-4-phenyl-5-thioxo-4,5-dihydro-[1,2,4]triazol-1-ylmethyl)-piperazin-1-yl]-4-oxo-1,4-dihydro-quinoline-3-carboxylic acid (9)
Yield (97 %); m.p. 262 °C; IR: 3365 (OH), 3041 (CH aromatic), 2964, 2921 (CH-aliphatic), 1714 (C=O), 1612 (C=N); 1H NMR: δ 15.14 (brs. OH, COOH), 8.65–7.43 (m, 15H, aromatic), 5.34 (s, 2H, N-CH2-N), 3.82 (m, CH2-CH-CH2 cyclopropyl-H), 3.42 (t, 4H, piperazinyl-H), 3.12 (t, 4H, piperazinyl-H), 1.33–1.31 (q, J = 0.74 Hz, 2H, CH2-CH-CH2 cyclopropyl-H), 1.17–1.16 (q, J = 1.52 Hz, 2H, CH2-CH-CH2cyclopropyl-H); 13C NMR: δ 176.81, 170.25, 166.34, 149.70, 148.41, 145.70, 139.62, 135.80, 133.74, 132.42, 130.04, 129.86, 129.28, 129.12, 128.63, 128.25, 127.56, 123.18, 111.53, 111.30, 107.25, 106.97, 69.41, 50.16, 49.96, 36.29, 8.04. C36H31FN6O3S: C, 66.86; H, 4.83; N, 12.99, Found: C, 66.49; H, 4.69; N, 12.58.
1-Cyclopropyl-6-fluoro-4-oxo-7-[4-(4-phenyl-3-pyridin-2-yl-5-thioxo-4,5-dihydro-[1,2,4]triazol-1-ylmethyl)-piperazin-1-yl]-1,4-dihydro-quinoline-3-carboxylic acid (10)
Yield (96%); m.p. 270 °C; IR: 3371 (OH), 3095 (CH aromatic), 2996, 2915 (CH-aliphatic), 1704 (C=O), 1611 (C=N); 1H NMR: δ 15.12 (brs. OH, COOH), 8.63–7.34 (m, 12H, aromatic), 5.33 (s, 2H, N-CH2-N), 3.82–3.79 (m, J = 3.88 Hz, 1H, CH2-CH-CH2 cyclopropyl-H), 3.40(t, 4H, piperazinyl-H), 3.08 (t, 4H, piperazinyl-H), 1.35–1.30 (q, J = 6.49 Hz, 2H, CH2-CH-CH2 cyclopropyl-H), 1.18–1.15 (q, J = 6.44 Hz, 2H, CH2-CH-CH2cyclopropyl-H); 13C NMR: δ 176.80, 170.87, 166.33, 154.88, 152.20, 149.73, 148.67, 148.31, 145.60, 145.28, 139.82, 137.72, 136.05, 129.41, 129.24, 128.87, 125.41, 124.65, 119.08, 111.52, 111.25, 107.33, 106.89, 69.45, 50.16 49.93, 49.53, 36.26, 8.03. C31H28FN7O3S: C, 62.30; H, 4.72; N, 16.41, Found: C, 62.04; H, 4.61; N, 16.21.
1-Cyclopropyl-6-fluoro-4-oxo-7-[4-(4-phenyl-3-pyridin-3-yl-5-thioxo-4,5-dihydro-[1,2,4]triazol-1-ylmethyl)-piperazin-1-yl]-1,4-dihydro-quinoline-3-carboxylic acid (11)
Yield (83%); m.p. 280 °C; IR: 3318 (OH), 3040 (CH aromatic), 2957, 2924 (CH-aliphatic), 1719 (C=O), 1608 (C=N); 1H NMR: δ 15.14 (brs. OH, COOH), 8.66–7.41 (m, 1H, aromatic), 5.32 (s, 2H, N-CH2-N), 3.83 (m, 1H, CH2-CH-CH2 cyclopropyl-H), 3.41–3.40 (t, J = 1.44 Hz, 4H, piperazinyl-H), 3.11–3.10 (t, J = 1.72 Hz, 4H, piperazinyl-H), 1.34–1.32 (q, J = 2.16 Hz, 2H, CH2-CH-CH2 cyclopropyl-H), 1.18 (q, J = 0.84 Hz, 2H, CH2-CH-CH2cyclopropyl-H); 13C NMR: δ 176.81, 170.23, 16.40, 166.40, 149.29, 148.46, 147.66, 145.57, 139.62, 136.53, 135.13, 130.23, 123.92, 122.40, 119.15, 111.52, 107.20, 107.02, 69.39, 50.04, 49.92, 36.32, 19.02, 8.05. C31H28FN7O3S: C, 62.30; H, 4.72; N, 16.41, Found: C, 62.14; H, 4.15; N, 16.20.
1-Cyclopropyl-6-fluoro-4-oxo-7-[4-(4-phenyl-3-pyridin-4-yl-5-thioxo-4,5-dihydro-[1,2,4]triazol-1-ylmethyl)-piperazin-1-yl]-1,4-dihydro-quinoline-3-carboxylic acid (12)
Yield (91%); m.p. 227 °C; IR: 3321 (OH), 3021(CH aromatic), 2968, 2904 (CH-aliphatic), 1717 (C=O), 1607 (C=N);1H NMR: δ 15.06 (brs. OH, COOH), 8.65–7.29 (m, 12H, aromatic), 5.32 (s, 2H, N-CH2-N), 3.83–3.81 (m, J = 3.12 Hz, 1H, CH2-CH-CH2 cyclopropyl-H), 3.41(t, 4H, piperazinyl-H), 3.10 (t, 4H, piperazinyl-H), 1.34–1.32 (q, J = 1.48 Hz, 2H, CH2-CH-CH2 cyclopropyl-H), 1.18–1.16 (q, J = 2.52 Hz, 2H, CH2-CH-CH2cyclopropyl-H); 13C NMR: δ 178.82, 170.70, 166.35, 150.57, 148.43, 147.57, 145.84, 135.84, 135.13, 133.34, 130.37, 130.00, 125.14, 122.57, 111.53, 107.28, 106.99, 67.53, 50.08, 49.94, 36.30, 19.00, 8.04. C31H28FN7O3S: C, 62.30; H, 4.72; N, 16.41, Found: C, 62.16; H, 4.53; N, 16.34.
1-Cyclopropyl-6-fluoro-4-oxo-7-[4-(5-oxo-4,4-diphenyl-2-thioxo-imidazolidin-1-ylmethyl)-piperazin-1-yl]-1,4-dihydro-quinoline-3-carboxylic acid (13)
Yield (84%); m.p. 217 °C; IR: 3412 (NH), 3315 (OH), 3031 (CH aromatic), 2981, 2914 (CH-aliphatic), 1721 (C=O), 1601 (C=N); 1H NMR: δ 15.07 (brs. OH, COOH), 11.65 (brs. NH, NH imidazolyl), 8.61–7.35 (m, 13H, aromatic), 4.85 (s, 2H, N-CH2-N), 3.79–3.77 (m, 1H, CH2-CH-CH2 cyclopropyl-H), 3.49 (t, 4H, piperazinyl-H), 2.83 (t, 4H, piperazinyl-H), 1.31–1.06 (m, J = 4.48 Hz, 4H, CH2-CH-CH2 cyclopropyl-H); 13C NMR: δ 182.95, 175.24, 166.35, 166.36, 139.51, 138.61, 129.28, 128.91, 121.88, 118.91, 111.54, 107.29, 106.12, 71.74, 62.54, 65.53, 49.84, 36.22, 18.93, 8.82. C33H30FN5O4S: C, 64.80; H, 4.94; N, 11.45, Found: C, 64.64; H, 4.77; N, 11.41.
7-[4-(3-Amino-5-oxo-4,4-diphenyl-2-thioxo-imidazolidin-1-ylmethyl)-piperazin-1-yl]-1-cyclopropyl-6-fluoro-4-oxo-1,4-dihydro-quinoline-3-carboxylic acid (14)
Yield (79%); m.p. 234 °C; IR: 3391, 3300 (NH2), 3259 (OH), 3098 (CH aromatic), 2984, 2912, 2847 (CH-aliphatic), 1708 (C=O), 1601 (C=N); 1H NMR: δ 11.81 (brs. OH, COOH), 9.80–9.66 (brs. 2H, NH2), 8.41–7.01 (m, 13H, aromatic), 5.14 (s, 2H, N-CH2-N), 3.14 (m, 1H, CH2-CH-CH2 cyclopropyl-H), 2.95–2.74 (t, 4H, piperazinyl-H), 2.26 (t, 4H, piperazinyl-H), 1.04–0.86 (q, 4H, CH2-CH-CH2 cyclopropyl-H); 13C NMR: δ 176.48, 171.24, 166.32, 154.68, 152.31, 151.54, 141.48, 133.11, 131.43, 131.14, 129.39, 129.11, 122.34, 118.41, 117.42, 116.92, 116.69, 67.24, 61.59, 59.34, 54.43, 51.33, 31.31, 23.39, 22.44, 9.11, 8.91; C33H31FN6O4S: C, 63.24; H, 4.99; N, 13.41, Found: C, 63.12; H, 4.86; N, 13.37.
7-[4-(2-Cyanoimino-5-oxo-4,4-diphenyl-imidazolidin-1-ylmethyl)-piperazin-1-yl]-1-cyclopropyl-6-fluoro-4-oxo-1,4-dihydro-quinoline-3-carboxylic acid (15)
Yield (95%); m.p. 230 °C; IR:3385 (NH), 3330 (OH), 3013 (CH aromatic), 2984, 2901 (CH-aliphatic), 2231 (CN), 1714 (C=O), 1602 (C=N); 1H NMR: δ 15.06 (brs. OH, COOH), 11.20 (brs. NH, NH-CN), 8.63–7.40 (m, 13H, aromatic), 4.61 (s, 2H, N-CH2-N), 3.37 (m, 1H, CH2-CH-CH2 cyclopropyl-H), 3.33 (t, 4H, piperazinyl-H), 2.76 (t, 4H, piperazinyl-H), 1.31–1.11 (q, 2H, CH2-CH-CH2 cyclopropyl-H); 13C NMR: δ 194.38, 176.78, 166.31, 148.31, 145.35, 138.65, 129.32, 121.36, 111.55, 106.16, 62.81, 58.11, 49.16, 36.38, 8.85: C34H30FN7O4: C, 65.90; H, 4.88; N, 15.82; Found: C, 65.71; H, 4.78; N, 15.77.
7-[4-(5-Benzylidene-2,4-dioxo-thiazolidin-3-ylmethyl)-piperazin-1-yl]-1-cyclo-propyl-6-fluoro-4-oxo-1,4-dihydro-quinoline-3-carboxylic acid (16)
Yield (97%); m.p. 237 °C; IR: 3314 (OH), 3017 (CH aromatic), 2984, 2946 (CH-aliphatic), 1711 (C=O), 1603 (C=N); 1H NMR: δ 15.10 (brs. OH, COOH), 8.64 (s, 1H, CH=C, CH olefinic) 8.61–7.49 (m, 8H, aromatic), 4.49 (s, 2H, N-CH2-N), 3.81–3.77 (m, J = 8.00 Hz, 1H, CH2-CH-CH2 cyclopropyl-H), 3.46–3.36 (t, 4H, piperazinyl-H), 3.81–2.73 (t, 4H, piperazinyl-H), 1.35–1.32 (q, J = 1.42 Hz, 2H, CH2-CH-CH2 cyclopropyl-H), 1.19–1.07 (q, J = 2.50 Hz, 2H, CH2-CH-CH2 cyclopropyl-H); 13C NMR: δ 176.34, 168.96, 166.31, 154.63, 152.15, 148.28, 145.58, 139.55, 133.46, 131.81, 130.51, 129.28, 118.91, 111.45, 111.22, 107.21, 106.16, 63.51, 56.53, 50.11, 49.43, 36.25, 18.95, 8.86. C28H25FN4O5S: C, 61.30; H, 4.59; N, 10.21, found: C, 61.25; H, 4.52; N, 10.10.
7-{4-[5-(4-Chloro-benzylidene)-2,4-dioxo-thiazolidin-3-ylmethyl]-piperazin-1-yl}-1-cyclopropyl-6-fluoro-4-oxo-1,4-dihydro-quinoline-3-carboxylic acid (17)
Yield (96%); m.p. 244 °C; IR: 3290 (OH), 3008 (CH aromatic), 2994, 2944 (CH-aliphatic), 1711 (C=O), 1611 (C=N); δ 15.12 (brs. OH, COOH), 8.64 (s, 1H, CH=C, CH olefinic) 7.91–7.54 (m, 7H, aromatic), 4.70 (s, 2H, N-CH2-N), 3.82–3.80 (m, J = 4.00 Hz, 1H, CH2-CH-CH2 cyclopropyl-H), 3.45–3.34 (t, 4H, piperazinyl-H), 2.82–2.75 (t, 4H, piperazinyl-H), 1.34–1.31 (q, J = 4.00 Hz, 2H, CH2-CH-CH2 cyclopropyl-H), 1.20–1.07 (q, J = 8.00 Hz, 2H, CH2-CH-CH2 cyclopropyl-H); 13C NMR: δ 176.19, 167.16,166.30, 148.31, 145.53, 139.61, 132.13, 129.88, 119.15, 111.51, 111.21, 107.25, 106.96, 63.62, 59.15, 49.85, 36.28, 18.99, 8.82; C28H24ClFN4O5S: C, 57.68; H, 4.15; N, 9.61, found: C, 57.52; H, 4.04; N, 9.53.
1-Cyclopropyl-6-fluoro-7-{4-[5-(4-methoxy-benzylidene)-2,4-dioxo-thiazolidin-3-ylmethyl]-piperazin-1-yl}-4-oxo-1,4-dihydro-quinoline-3-carboxylic acid (18)
Yield (92%); m.p. 251 °C; IR: 3349 (OH), 3014 (CH aromatic), 2988, 2908 (CH-aliphatic), 1720 (C=O), 1607 (C=N); 1H NMR: δ 15.10 (brs. OH, COOH), 8.64 (s, 1H, CH=C, CH olefinic) 8.62–7.09 (m, 7H, aromatic), 4.68 (s, 2H, N-CH2-N), 3.83(s, 3H, OCH3), 3.47 (m, 1H, CH2-CH-CH2 cyclopropyl-H), 3.33–3.09 (t,4H, piperazinyl-H), 3.81–2.72 (t, 4H, piperazinyl-H), 1.35–1.32 (q, 2H, CH2-CH-CH2 cyclopropyl-H), 1.33–1.05 (q, 2H, CH2-CH-CH2 cyclopropyl-H); 13C NMR: δ 176.78, 169.81, 167.35, 166.33, 166.01 152.16, 148.39, 145.49, 139.54, 133.52, 132.69, 125.94, 119.19, 118.62, 115.45, 111.49, 111.36, 107.36, 106.91, 63.37, 56.54, 55.91, 54.21, 49.84, 36.26, 19.88, 8.82. C29H27FN4O6S: C, 60.20; H, 4.70; N, 9.68, found: C, 60.13; H, 4.64; N, 9.53.
1-Cyclopropyl-6-fluoro-7-{4-[5-(2-hydroxy-benzylidene)-2,4-dioxo-thiazolidin-3-ylmethyl]-piperazin-1-yl}-4-oxo-1,4-dihydro-quinoline-3-carboxylic acid (19)
Yield (94%); m.p. 244 °C; IR: 3410, 3312 (2OH), 3005 (CH aromatic), 2951, 2910 (CH-aliphatic), 1723 (C=O), 1603 (C=N); 1H NMR: δ 15.10 (brs. OH, COOH), 10.51 (brs. OH, PhOH), 8.63 (s, 1H, CH=C, CH olefinic) 8.61–6.92 (m, 7H, aromatic), 4.67 (s, 2H, N-CH2-N), 3.81–3.79 (m, J = 4.00 Hz, 1H, CH2-CH-CH2 cyclopropyl-H), 3.81–3.77 (t, 4H, piperazinyl-H), 2.81–2.74 (t, 4H, piperazinyl-H), 1.33–1.09 (m, 4H, CH2-CH-CH2cyclopropyl-H); 13C NMR: δ 176.73, 169.24, 167.45, 166.32, 157.88, 154.61, 152.13, 148.26, 145.49, 145.39, 139.54, 132.94, 128.41, 128.11, 119.85, 116.67, 111.45, 111.22, 107.22, 106.84, 63.53, 56.51, 50.21,49.81, 46.15, 36.24, 8.88. C28H25FN4O6S: C, 59.57; H, 4.46; N, 9.92; Found: C, 59.51; H, 4.40; N, 9.78.
1-Cyclopropyl-7-[4-(1,3-dioxo-1,3-dihydro-isoindol-2-ylmethyl)-piperazin-1-yl]-6-fluoro-4-oxo-1,4-dihydro-quinoline-3-carboxylic acid (20)
Yield (89%); m.p. 241 °C; IR: 3341 (OH), 3023 (CH aromatic), 2998, 2812 (CH-aliphatic), 1753, 1721 (2 C=O), 1611 (C=N); 1H NMR: δ 15.11 (brs. OH, COOH), 8.64–7.50 (m, 7H, aromatic), 4.56 (s, 2H, N-CH2-N), 3.81–3.77 (m, J = 3.12 Hz, 1H, CH2-CH-CH2 cyclopropyl-H), 3.42 (t, 4H, piperazinyl-H), 2.91 (t, 4H, piperazinyl-H), 1.34–1.29 (q, J = 4.21 Hz, 2H, CH2-CH-CH2 cyclopropyl-H), 1.19–1.13 (q, J = 4.12 Hz, 2H, CH2-CH-CH2cyclopropyl-H); 13C NMR: δ 176.77, 169.23, 166.38, 154.63, 152.16, 148.25, 145.53, 139.63, 135.83, 134.13,, 131.99, 123.66, 123.33, 118.99, 111.47, 111.29, 107.32, 106.19, 59.39, 54.22, 49.46, 36.23, 7.94. C26H23FN4O5: C, 63.67; H, 4.73; N, 11.42, Found: C, 63.51; H, 4.64; N, 11.33.
1-Cyclopropyl-7-[4-(2,5-dioxo-pyrrolidin-1-ylmethyl)-piperazin-1-yl]-6-fluoro-4-oxo-1,4-dihydro-quinoline-3-carboxylic acid (21)
Yield (88%); m.p. 214 °C; IR: 3412 (OH), 3007 (CH aromatic), 2998, 2987, 2841, 2828, 2810 (CH-aliphatic),1752, 1701 (2 C=O), 1614 (C=N); 1H NMR: δ 15.12 (brs. OH, COOH), 8.63–7.51 (m, 3H, aromatic), 4.34 (s, 2H, N-CH2-N), 3.83–3.80 (m, J = 2.02 Hz, 1H, CH2-CH-CH2 cyclopropyl-H), 3.79–3.77 (t, 2H, CO-CH2-CH2-C=O), 3.49–3.42 (m, 6H, 4H of piperazinyl-H + CO-CH2-CH2-C=O), 2.72–2.58 (t, 4H, piperazinyl-H), 1.34–1.32 (q, J = 1.48 Hz, 2H, CH2-CH-CH2 cyclopropyl-H), 1.17–1.08 (q, J = 2.58 Hz, 2H, CH2-CH-CH2cyclopropyl-H); 13C NMR: δ 179.15, 176.88, 166.32, 154.67, 152.28, 148.28, 145.62, 145.52, 139.64, 119.83, 111.49, 111.36, 107.34, 106.19, 59.52, 56.53, 54.25, 49.87, 36.24, 30.54, 18.93, 8.88.: C22H23FN4O5: C, 59.72; H, 5.24; N, 12.66, Found: C, 59.59; H, 5.08; N, 12.49.
3.2. Antimicrobial Activity
3.2.1. Organisms and Culture Conditions
The cultures used were collected from the Cairo University’s Microanalytical Centre, Faculty of Science. An updated Kirby-Bauer disc diffusion method was applied for antimicrobial activities of the tests 1–21 compounds [32,33,34,35]. See Appendix A.
3.2.2. Minimum Inhibitory Concentration Assay
On 96-well microtiter plates and 50 mL of fresh bacterial culture of a single McFarland unit overnight, a double serial dilution of each compound (100 mL) in sterile standard saline was prepared for every single source well. Ciprofloxacin antibiotic (5 mg/mL−1) and normal saline were included as standard references in each assay [36] (see Appendix A).
3.2.3. Inhibitory Activity Assays on E. coli DNA Gyrase and Topoisomerase IV
IC50 assay determination was carried out in accordance with the procedures previously stated [37] (see Appendix A).
3.2.4. MTT Assay
MTT Assay was performed to investigate the effect of compounds 2–6 on the viability of mammary epithelial cells (MCF-10A) [38,39] (see Appendix A).
4. Conclusions
Several novel heteroaryl-based ciprofloxacin derivatives have been developed. Twenty-one target compounds were synthesized and tested for their in vitro antibacterial activity against bacterial strains of both Gram-positive and Gram-negative using ciprofloxacin as a reference. Most of the compounds examined had evident inhibitory antibacterial activity. Among those compounds, 2–6 were the most potent ones. The findings showed that the compounds tested displayed little or poor antifungal activity against A. flavus and C. albicans. Oxadiazole-based derivatives 4, 5, and 6 were found to be the most active and their inhibitory activity against E. coli DNA gyrase and Topoisomerase IV was superior to novobiocin with no cytotoxic effects. These compounds, after further optimization, form a new class of antibacterial molecules that target DNA gyrase and topoisomerase IV.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ph14050399/s1, Figure S1–S21: 1H, 13C NMR and Dept 135 of compounds 1–20, Table S1–S3: Molecular properties of compounds 1–21.
Appendix A
Appendix A.1. General Details
All the commercially available reagent chemicals were obtained from Sigma-Aldrich Corporation (St. Louis, MO, USA) and were used without further purification. All reactions were monitored by thin layer chromatography (TLC) using precoated plates of silica gel G/UV-254 of 0.25 mm thickness (Merck 60F254-Billerica, MA, USA) using UV light (254 nm/365 nm) for visualization. Melting points were detected with a Koffler melting points apparatus and were uncorrected. Infrared spectra were recorded with a FT-IR-ALPHBROKER-Platinum-ATR spectrometer and are given as cm−1 using the attenuated total reflection (ATR) method. 1H NMR and 13C NMR spectra for all compounds were recorded in DMSO-d6 on a Bruker Bio Spin AG spectrometer at 400 MHz and 100 MHz, respectively. For 1H NMR, chemical shifts (δ) were given in parts per million (ppm) with reference to Tetramethylsilane (TMS) as an internal standard (δ = 0); coupling constants (J) were given in hertz (Hz) and data are reported as follows: chemical shift, integration, multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, dd = doublet of doublets). For 13C NMR, TMS (δ = 0) or DMSO (δ = 39.51) was used as internal standard and spectra were obtained with complete proton decoupling. Elemental analyses were obtained on a Perkin-Elmer CHN-analyzer model. Microwave irradiations were carried out in a Kenstar OM9925E MW oven (2450 MHz, 800 W).
Appendix A.2. Antimicrobial Activity
Appendix A.2.1. Organisms and Culture Conditions
The cultures used were collected from the Cairo University’s Microanalytical Centre, Faculty of Science. An updated Kirby-Bauer disc diffusion method was applied for the antimicrobial activities of the tests for compounds 1–21 [28]. Shortly after, 10 mL of fresh medium was grown to 100 μL bacteria/food until a count of 108 cells/mL or 105 cells/mL was achieved [29]. One hundred μL microbial suspension was spread over agar plates that suited the broth in which it was held. The selected colonies of each organism that may play a pathogenic function should be from the primary agar plates and tested by the disc diffusion method for susceptibility [30,31]. Plates inoculated with filamentous fungi as Aspergillus flavus were incubated at 25 °C for 48 h; Gram positive bacteria as Staphylococcus aureus (ATCC 12600, Microanalytical center, Cairo, Egypt), Bacillus subtilis (ATCC 6051); Gram negative bacteria as Escherichia coli (ATCC 11775), Pseudomonas aeuroginosa (ATCC 10145) were incubated at 35–37 °C for 24–48 h and yeast as Candida albicans (ATCC 7102) was incubated at 30 °C for 24–48 h and, then the diameters of the inhibition zones were measured in millimeters [28]. Standard discs of ciprofloxacin (antibacterial agent), fluconazole (antifungal agent) served as positive controls for antimicrobial activity, but filter discs impregnated with 10 µL of solvent (distilled water, chloroform, DMSO) were used as a negative control. Blank paper disks (Schleicher and Schuell, Cytiva, Spain) with a diameter of 8.0 mm were impregnated with 10 µL of tested concentration of the stock solutions. When a filter paper disc impregnated with a tested chemical is placed on agar, the chemical will diffuse from the disc into the agar. This diffusion will place the chemical in the agar only around the disc. The solubility of the chemical and its molecular size will determine the size of the area of chemical infiltration around the disc. If an organism is placed on the agar, it will not grow in the area around the disc if it is susceptible to the chemical. This area of no growth around the disc is known as a “Zone of inhibition” or" Clear zone". For the disc diffusion, the zone diameters were measured with slipping calipers of the National Committee for Clinical Laboratory Standards [30], and the results are given in Table 1. Agar-based methods such as E-test and disk diffusion can be good alternatives because they are simpler and faster than broth-based methods [30,31].
Appendix A.2.2. Minimum Inhibitory Concentration Assay
In 96-well microtiter plates and 50 mL of fresh bacterial culture of a single McFarland unit overnight, a double serial dilution of each compound (100 mL) in sterile standard saline was prepared for every single source well. Ciprofloxacin antibiotic (5 mg/mL−1) and normal saline were included as standard reference in each assay [32]. The plates were incubated at 37 °C overnight. As an indicator of bacterial growth, 40 mL of p-iodonitrotetrazolium violet (INT) was added to each well and incubated at 37 °C for 30 min. MIC values are recorded as the lowest concentration of the extract that completely inhibited bacterial growth that is clear well. The colorless tetrazolidium salt acts as an electron accepter and is reduced to a red colored formazan product by biological activity organisms. Where bacterial growth was inhibited, the solution in the well remained clear after incubation with INT. The observed MIC values are presented in Table 2.
Appendix A.2.3. Determination of Inhibitory Activities on E. coli and S. aureus DNA Gyrase
Inhibitory activities were determined in an assay from Inspiralis on streptavidin-coated 96-well microtiter plates from Thermo scientific Pierce. First, the plates were rehydrated with a buffer (20 mM TrisHCl with pH 7.6, 0.01% w/v BSA, 0.05% v/v Tween 20, 137 mM NaCl) and the biotinylated oligonucleotide was then immobilized. After washing off the unbound oligonucleotide, the enzyme test was performed. The reaction volume of 30 μL in buffer (35 mM Tris × HCl with pH 7.5, 4 mM MgCl2, 24 mM KCl, 2 mM DTT, 1.8 mM spermidine, 1 mM ATP, 6.5 % w/v glycerol, 0.1 mg/mL albumin) contained 1.5 U of DNA gyrase from E. coli or S. aureus, 0.75 μg of relaxed pNO1 plasmid, and 3 μL solution of the inhibitor in 10% DMSO and 0.008% Tween 20. Reaction solutions were incubated at 37 °C for 30 min. After that, the TF buffer (50 mM NaOAc with pH 5.0, 50 mM NaCl and 50 mM MgCl2) was added to terminate the enzymatic reaction. After additional incubation for 30 min at rt, during which biotin-oligonucleotide-plasmid triplex was 19 formed, the unbound plasmid was washed off using TF buffer and SybrGOLD in T10 buffer (10 mM Tris HCl with pH 8.0 and 1 mM EDTA) was added. The fluorescence was measured with a microplate reader (BioTek Synergy H4, excitation: 485 nm, emission: 535 nm). Initial screening was done at 100 or 10 μM concentration of inhibitors. For the most active inhibitors, IC50 was determined using seven concentrations of tested compounds. GraphPad Prism software was used to calculate the IC50 values. The result is given as the average value of three independent measurements. As the internal standard, novobiocin (IC50 = 0.168 µM for E. coli gyrase and IC50 = 0.041 µM for S. aureus gyrase) was used. Determination of inhibitory activities on E. coli and S. aureus Topoisomerase IV IC50 values were determined in an assay from Inspiralis on streptavidin-coated 96-well microtiter plates from Thermo scientific Pierce. First, the plates were rehydrated with buffer (20 µM Tris-HCl with pH 7.6, 0.01% w/v BSA, 0.05% v/v Tween 20, 137 mM NaCl) and biotinylated oligonucleotide was then immobilized. After washing off the unbound oligonucleotide, the enzyme test was performed. The reaction volume of 30 μL in buffer (40 mM HEPES KOH with pH 7.6, 100 mM potassium glutamate, 10 mM magnesium acetate, 10 mM DTT, 1 mM ATP, 0.05 mg/mL albumin) contained 1.5 U of topoisomerase IV from E. coli or S. aureus, 0.75 μg of pNO1 supercoiled plasmid, and 3 μL solution of the inhibitor in DMSO (10%) and Tween 20 (0.008%). Reaction mixtures were incubated at 37 °C for 30 min and after that, the TF buffer (50 mM NaOAc with pH 5.0, 50 mM NaCl and 50 mM MgCl2) was added to terminate the enzymatic reaction. After additional incubation for 30 min at rt, during which triplex (biotin-oligonucleotide-plasmid) was formed, the unbound plasmid was washed off using TF buffer and SybrGOLD in T10 buffer (10 mM Tris HCl with pH 8.0 and 1 mM EDTA) was added. The fluorescence was measured with a microplate reader (BioTek Synergy H4, excitation: 485 nm, emission: 535 nm). Initial screening was done at 100 or 10 μM concentration of inhibitors. For the most active inhibitors IC50 was determined using seven concentrations of tested compounds. GraphPad Prism software was used to calculate the IC50 values. The result is given as the average value of three independent measurements. As the internal standard, novobiocin (IC50 = 11.1 µM) for E. coli topoisomerase IV and IC50 = 26.7 µM for S. aureus topoisomerase IV) was used.
Author Contributions
Conceptualization, B.G.M.Y., A.A.M. and A.A.A. (Antar A. Abdelhamid); methodology, B.G.M.Y., A.A.M., A.A.A. (Amer A. Amer) and A.A.A. (Antar A. Abdelhamid); software, H.A.M.G.; validation, B.G.M.Y., A.A.M. and A.A.A. (Antar A. Abdelhamid); formal analysis, L.H.A.-W.; investigation, B.G.M.Y.; resources, A.A.M.; data curation, A.A.A. (Amer A. Amer); writing—original draft preparation, B.G.M.Y., A.A.M. and A.A.A. (Antar A. Abdelhamid); writing—review and editing, B.G.M.Y.; visualization, A.A.M. and A.A.A. (Antar A. Abdelhamid); supervision, H.A.M.G.; project administration, H.A.M.G.; funding acquisition, L.H.A.-W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Deanship of Scientific Research at Princess Nourah bint Abdulrahman University through the Fast-track Research Funding Program.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Manav M., Mohit S., Abdul S., Aakash D. New oxadiazole derivatives of isonicotinohydrazide in the search for antimicrobial agents: Synthesis and in vitro evaluation. J. Serb. Chem. Soc. 2012;77:9–16. [Google Scholar]
- 2.Tulkens P.M., van Bambeke F., Zinner S.H. Profile of a Novel Anionic Fluoroquinolone-Delafloxacin. Clin. Infect. Dis. 2019;68:213–222. doi: 10.1093/cid/ciy1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Showalter H.H., Denny W.A. A roadmap for drug discovery and its translation to small molecule agents in clinical development for tuberculosis treatment. Tuberculosis. 2008;88:S3–S17. doi: 10.1016/S1472-9792(08)70032-5. [DOI] [PubMed] [Google Scholar]
- 4.Pieroni M., Dimovska M., Brincat J.P., Sabatini S., Carosati E., Massari S., Kaatz G.W., Fravolini A. From 6-aminoquinolone antibacterials to 6-amino-7-thiopyranopyridinylquinolone ethyl esters as inhibitors of staphylococcus aureus multidrug efflux pumps. J. Med. Chem. 2010;53:4466–4480. doi: 10.1021/jm1003304. [DOI] [PubMed] [Google Scholar]
- 5.Richards G.A., Brink A.J., Feldman C. SAMJ Rational use of the fluoroquinolones. S. Afr. Med. J. 2019;109:378–381. doi: 10.7196/SAMJ.2019.v109i6.14002. [DOI] [PubMed] [Google Scholar]
- 6.Holm A., Cordoba G., Aabenhus R. Scand Prescription of antibiotics for urinary tract infection in general practice in Denmark. J. Prim. Health. 2019;37:83–89. doi: 10.1080/02813432.2019.1569425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohammed H.H.H., Abuo-Rahma G.E.D.A., Abbas S.H., Abdelhafez E.M.N. Current Trends and Future Directions of Fluoroquinolones. Curr. Med. Chem. 2019;26:3132–3149. doi: 10.2174/0929867325666180214122944. [DOI] [PubMed] [Google Scholar]
- 8.Jampilek J. Design and Discovery of New Antibacterial Agents: Advances, Perspectives, Challenges. J. Curr. Med. Chem. 2018;25:4972–5006. doi: 10.2174/0929867324666170918122633. [DOI] [PubMed] [Google Scholar]
- 9.Boothe D.M., Bush K.M., Boothe H.W., Davis H. Pharmacokinetics and pharmacodynamics of oral pradofloxacin administration in dogs. Am. J. Vet. Res. 2018;79:1268–1276. doi: 10.2460/ajvr.79.12.1268. [DOI] [PubMed] [Google Scholar]
- 10.Fedorowicz J., Sączewski J. Modifications of quinolones and fluoroquinolones: Hybrid compounds and dual-action molecules. J. Monatsh. Chem. 2018;149:1199–1245. doi: 10.1007/s00706-018-2215-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berning S.E. The role of fluoroquinolones in tuberculosis today. Drugs. 2001;61:9–18. doi: 10.2165/00003495-200161010-00002. [DOI] [PubMed] [Google Scholar]
- 12.Sharma P.C., Jain A., Jain S., Pahwa R., Yar M.S. Ciprofloxacin: Review on developments in synthetic, analytical, and medicinal aspects. J. Enzyme Inhib. Med. Chem. 2010;25:577–589. doi: 10.3109/14756360903373350. [DOI] [PubMed] [Google Scholar]
- 13.Suresh N., Nagesh H.N., Sekhar K.V.G.C., Kumar A., Shirazi A.N., Parang K. Synthesis of novel ciprofloxacin analogues and evaluation of their anti-proliferative effect on human cancer cell lines. Bioorg. Med. Chem. Lett. 2013;23:6292–6295. doi: 10.1016/j.bmcl.2013.09.077. [DOI] [PubMed] [Google Scholar]
- 14.Zhang G., Liu X., Zhang S., Pan B., Liu M.-L. Ciprofloxacin derivatives and their antibacterial activities. Eur. J. Med. Chem. 2018;146:599–612. doi: 10.1016/j.ejmech.2018.01.078. [DOI] [PubMed] [Google Scholar]
- 15.Demirci S., Mermer A., Ak G., Aksakal F., Colak N., Demirbas A., Ayaz F.A., Demirbas N. Conventional and Microwave-assisted Total Synthesis, Antioxidant Capacity, Biological Activity, and Molecular Docking Studies of New Hybrid Compounds. J. Heterocycl. Chem. 2017;54:1785–1805. doi: 10.1002/jhet.2760. [DOI] [Google Scholar]
- 16.Pandit N., Shah K., Agrawal N., Upmanyu N., Shrivastava S.K., Mishra P. Synthesis, characterization and biological evaluation of some novel fluoroquinolones. P. Med. Chem. Res. 2016;25:843–851. doi: 10.1007/s00044-016-1526-x. [DOI] [Google Scholar]
- 17.Kant R., Singh V., Nath G., Awasthi S.K., Agarwal A. Design, synthesis and biological evaluation of ciprofloxacin tethered bis-1,2,3-triazole conjugates as potent antibacterial agents. Eur. J. Med. Chem. 2016;124:218–228. doi: 10.1016/j.ejmech.2016.08.031. [DOI] [PubMed] [Google Scholar]
- 18.Mermer A., Demirci S., Ozdemir S.B., Demirbas A., Ulker S., Ayaz F.A., Aksakal F., Demirbas N. Conventional and Microwave Irradiated Synthesis, Biological Activity Evaluation and Molecular Docking Studies of Highly Substituted Piperazine-Azole Hybrids. Chin. Chem. Lett. 2017;28:995–1005. doi: 10.1016/j.cclet.2016.12.012. [DOI] [Google Scholar]
- 19.Ozdemir S.B., Cebeci Y.U., Bayrak H., Mermer A., Ceylan S., Demirbas A., Karaoglu S.A., Demirbas N. Synthesis and antimicrobial activity of new piperazine-based heterocyclic compounds. Heterocycl. Commun. 2017;23:43–54. [Google Scholar]
- 20.Marzouk A.A., Mohamed S.K., Aljohani E.T., Abdelhamid A.A. New Method for Synthesis of Multi-substituted Imidazoles. J. Heterocycl. Chem. 2018;55:1775. doi: 10.1002/jhet.3215. [DOI] [Google Scholar]
- 21.Abu-Dief A.M., Abdel-Rahman L.H., Abdelhamid A.A., Marzouk A.A., Shehatad M.R., Bakheet M.A., Nafady A. Synthesis and characterization of new Cr(III), Fe(III) and Cu(II) complexes incorporating multi-substituted aryl imidazole ligand: Structural, DFT, DNA binding, and biological implications. Spectrochim. Acta Part. A. 2020;228:117700. doi: 10.1016/j.saa.2019.117700. [DOI] [PubMed] [Google Scholar]
- 22.Abdelhamid A.A., Salah H.A., Marzouk A.A. Synthesis of imidazole derivatives: Ester and hydrazide compounds with antioxidant activity using ionic liquid as an efficient catalyst. J. Heterocyclic Chem. 2020;57:676–685. [Google Scholar]
- 23.Marzouk A.A., Bass A.K.A., Ahmed M.S., Abdelhamid A.A., Elshaier Y.A.M.M., Salman A.M.M., Aly O.M. Design, synthesis and anticonvulsant activity of new imidazolidindione and imidazole derivatives. Bioorg. Chem. 2020;101:104020. doi: 10.1016/j.bioorg.2020.104020. [DOI] [PubMed] [Google Scholar]
- 24.Shaykoon M.S., Marzouk A.A., Soltan O.M., Wanas A.S., Radwan M.M., Gouda A.M., Youssif B.G.M., Abdel-Aziz M. Design, synthesis and antitrypanosomal activity of heteroaryl-based 1,2,4-triazole and 1,3,4-oxadiazole derivatives. Bioorg. Chem. 2020;100:103933. doi: 10.1016/j.bioorg.2020.103933. [DOI] [PubMed] [Google Scholar]
- 25.Marzouk A.A., Abdel-Aziz S.A., Abdelrahman K.S., Wanas A.S., Gouda A.M., Youssif B.G.M., Abdel-Aziz M. Design and synthesis of new 1,6-dihydropyrimidin-2-thio derivatives targeting VEGFR-2: Molecular docking and antiproliferative evaluation. Bioorg. Chem. 2020;102:104090. doi: 10.1016/j.bioorg.2020.104090. [DOI] [PubMed] [Google Scholar]
- 26.Youssif B.G.M., Mohamed Y.A.M., Salim M.T.A., Inagaki F., Mukai C., Abdu-Allah H.A.M. Synthesis of some benzimidazole derivatives endowed with 1,2,3-triazole as potential inhibitors of hepatitis C virus. Acta Pharm. 2016;66:219–231. doi: 10.1515/acph-2016-0014. [DOI] [PubMed] [Google Scholar]
- 27.Abdu-Allah H.A.M., Youssif B.G.M., Abddelrahman M.H., Abdel-Hamid M.K., Reshma R.S., Yogeeswari P., Aboul-Fadl T., Siriram D. Synthesis and anti-mycobacterial activity of 4-(4-phenyl-1H-1,2,3-triazol-1-yl)salicylhydrazones: Revitalizing an old drug. Arch. Pharm. Res. 2017;40:168–179. doi: 10.1007/s12272-016-0882-x. [DOI] [PubMed] [Google Scholar]
- 28.Jha K.K., Samad A., Kumar Y., Shaharyar M., Khos R.L., Jain J., Kumar V., Singh P. Design, synthesis and biological evaluation of 1,3,4-oxadiazole derivatives. Eur. J. Med. Chem. 2010;45:4963–4967. doi: 10.1016/j.ejmech.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Ghattas A.-B.A.G., Moustafa H.M., Allah O.A.A., Amer A.A. Synthesis and reactions of some 1, 2, 4-triazolo-[4, 3-b]-1, 2, 4-triazoles. Synth. Commun. 2001;31:2447–2456. [Google Scholar]
- 30.Todorov P., Georgieva S., Peneva P., Rusew R., Shivachev B., Georgiev A. Experimental and theoretical study of bidirectional photoswitching behavior of 5,5′-diphenylhydantoin Schiff bases: Synthesis, crystal structure and kinetic approaches. New J. Chem. 2020;44:15081–15099. doi: 10.1039/D0NJ03301D. [DOI] [Google Scholar]
- 31.Tilekar K., Upadhyay N., Schweipert M., Hess J.D., Macias L.H., Mrowka P., Meyer-Almes F., Aguilera R.J., Iancu C.V., Choe J., et al. Permuted 2,4-thiazolidinedione (TZD) analogs as GLUT inhibitors and their in-vitro evaluation in leukemic cells. Eur. J. Pharm. Sci. 2020;154:105512. doi: 10.1016/j.ejps.2020.105512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bauer A.W., Kirby W.M., Sherris C., Turck M. Antibiotic Susceptibility Testing by a Standardized Single Disk Method. Am. J. Clin. Pathol. 1966;45:493–496. doi: 10.1093/ajcp/45.4_ts.493. [DOI] [PubMed] [Google Scholar]
- 33.Pfaller M.A., Burmeister L., Bartlett M.A., Rinaldi M.G. Multicenter evaluation of four methods of yeast inoculum preparation. J. Clin. Microbiol. 1988;26:1437–1441. doi: 10.1128/JCM.26.8.1437-1441.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.National Committee for Clinical Laboratory Standards . Proposed Guideline M 44-P. NCCLS; Wayne, PA, USA: 2003. [Google Scholar]
- 35.National Committee for Clinical Laboratory Standards . Approved Standard M7-A3. National Committee for Clinical Laboratory Standards (NCCLS); Villanova, PA, USA: 1993. [Google Scholar]
- 36.Swamy P.L., Rao T.S., Kumar C.A., Prasad C.S., Rao G.S. IN VITRO—Antimicrobial activity of cashew (Anacardium occidentale, L.) nuts shell liquid against methicillin resistant Staphylococcus Aureus (MRSA) clinical isolates. Int. J. Pharm. Pharm. Sci. 2011;3:436–440. [Google Scholar]
- 37.Durcik M., Tammela P., Barancokova M., Tomasic T., Ilas J., Kikelj D., Zidar N. Synthesis and Evaluation of N-Phenylpyrrolamides as DNA Gyrase B Inhibitors. Chem. Med. Chem. 2017;5:201700549. doi: 10.1002/cmdc.201700549. [DOI] [PubMed] [Google Scholar]
- 38.Youssif B.G., Mohamed A.M., Osman E.E.A., Abou-Ghadir O.F., Elnaggar D.H., Abdelrahman M.H., Treamblu L., Gomaa H.A. 5-Chlorobenzofuran-2-carboxamides: From allosteric CB1 modulators to potential apoptotic antitumor agents. Eur. J. Med. Chem. 2019;177:1–11. doi: 10.1016/j.ejmech.2019.05.040. [DOI] [PubMed] [Google Scholar]
- 39.Abou-Zied H.A., Youssif B.G.M., Mohamed M.F.A., Hayallah A.M., Abdel-Aziz M. EGFR inhibitors and apoptotic inducers: Design, synthesis, anticancer activity and docking studies of novel xanthine derivatives carrying chalcone moiety as hybrid molecules. Bioorg. Chem. 2019;89:102997. doi: 10.1016/j.bioorg.2019.102997. [DOI] [PubMed] [Google Scholar]
- 40.Daina A., Michielin O., Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, druglikeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramadan M., El-Aziz M.A., Elshaier Y.A.M.M., Youssif B.G.M., Brown A.B., Fathy H.M., Aly A.A. Design and synthesis of new pyranoquinolinone heteroannulated to triazolopyrimidine of potential apoptotic antiproliferative activity. Bioorg. Chem. 2020;105:104392. doi: 10.1016/j.bioorg.2020.104392. [DOI] [PubMed] [Google Scholar]
- 42.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 43.Ghose A.K., Viswanadhan V.N., Wendoloski J.J. A Knowledge-Based Approach in Designing Combinatorial or Medicinal Chemistry Libraries for Drug Discovery. 1. A Qualitative and Quantitative Characterization of Known Drug Databases. J. Comb. Chem. 1999;1:55–68. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- 44.Veber D.F., Johnson S.R., Cheng H.-Y., Smith B.R., Ward K.W., Kopple K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 45.Egan W.J., Merz K.M., Jr., Baldwin J.J. Prediction of Drug Absorption Using Multivariate Statistics. J. Med. Chem. 2000;43:3867–3877. doi: 10.1021/jm000292e. [DOI] [PubMed] [Google Scholar]
- 46.Muegge I., Heald S.L., Brittelli D. Simple Selection Criteria for Drug-like Chemical Matter. J. Med. Chem. 2001;44:1841–1846. doi: 10.1021/jm015507e. [DOI] [PubMed] [Google Scholar]
- 47.Baell J.B., Holloway G.A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.