Abstract
Diabetic neuropathic pain (DNP) is one of the most serious complications of diabetes. Patients with DNP always exhibit spontaneous and stimulus-evoked pain. However, the pathogenesis of DNP remains to be fully elucidated. Non-coding RNAs (ncRNAs) serve important roles in several cellular processes and dysregulated expression may result in the development of several diseases, including DNP. Although ncRNAs have been suggested to be involved in the pathogenesis of DNP, their precise roles remain to be determined. In the present study, sequencing analysis was used to investigate the expression patterns of coding genes, microRNAs (miRNAs), long ncRNAs (lncRNAs) and circular RNAs (circRNAs) in the spinal cord of mice with streptozotocin (STZ)-induced DNP. A total of 30 mRNAs, 148 miRNAs, 9 lncRNAs and 135 circRNAs exhibited significantly dysregulated expression 42 days after STZ injection. Functional enrichment analysis indicated that protein digestion and absorption pathways were the most significantly affected pathways of the differentially expressed (DE) mRNAs. The Rap1 signaling pathway, human T-lymphotropic virus-I infection and the MAPK signaling pathway were the three most significant pathways of the DE miRNAs. A total of 2,118 distinct circRNAs were identified and the length of the majority of the circRNAs was <1,000 nucleotides (nt) (1,552 circRNAs were >1,000 nt) with a median length of 620 nt. In the present study, the expression characteristics of coding genes, miRNAs, lncRNAs and circRNAs in DNP mice were determined; it paves the road for further studies on the mechanisms associated with DNP and potentially facilitates the discovery of novel ncRNAs for therapeutic targeting in the management of DNP.
Keywords: diabetes, neuropathic pain, spinal cord, non-coding RNAs, sequence analysis
Introduction
The incidence of diabetes is increasing on a yearly basis worldwide and is frequently attributed to changes in lifestyle (1). Almost 50% of patients with diabetes develop peripheral neuropathy (2). Diabetic neuropathic pain (DNP) is one of the most serious complications of diabetic peripheral neuropathy (3). The majority of patients with DNP experience moderate to severe levels of pain (4). There are various dysregulated mechanisms that may contribute to the pathogenesis of DNP, including hyperglycemia, advanced glycation end-products, oxidative stress, neuroinflammation and endoneural hypoxia (5-7). However, drugs acting on these pathways have limited therapeutic effects in clinical practice. Therefore, identifying alternative mechanisms by which DNP manifests should be explored further.
Non-coding RNAs (ncRNAs) are RNA molecules that do not encode proteins but functionally regulate protein expression (8). An increasing number of studies have indicated that ncRNAs are involved in gene transcription and translation under physiological and pathological conditions (9,10). ncRNAs may be divided into three categories according to their size: Small ncRNAs, <200 nucleotides (nt); long ncRNAs (lncRNAs), >200 nt; and circular RNAs (circRNA) consisting of a closed continuous loop (11). Several studies have indicated that ncRNAs have a crucial role in several types of pain, including neuropathic pain (12,13). Studies have also provided evidence that ncRNAs regulate the occurrence of DNP. For instance, microRNA (miRNA/miR)-190a (14) and miR-193a (15) in the dorsal root ganglion have been indicated to be associated with the induction of DNP. Furthermore, lncRNA NONRATT021972 and lncRNA BC168687 have been indicated to regulate DNP (16). circRNAs are another type of ncRNA that interact with miRNAs (17,18), which regulate gene expression via a circRNA/miRNA/mRNA network (19). A recent study also suggested that altered expression levels of circRNAs accompany the development of neuropathic pain (20). However, to date, the regulatory functions and underlying mechanisms of ncRNAs in DNP have not been systematically reported, to the best of our knowledge. Thus, comprehensive analysis of the ncRNA expression profiles and their association with the pathogenesis of DNP may facilitate the development of effective methods to treat this disease.
In the present study, the expression profiles of ncRNAs in the spinal cord of mice with streptozotocin (STZ)-induced DNP were examined and analyzed using RNA sequencing techniques. The microarray results were subjected to bioinformatics predictions, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses.
Materials and methods
Animals
All experiments were approved by the Animal Use and Care Committee for Research and Education of The First People's Hospital of Foshan (Foshan, China) and were in accordance with the guidelines described in the International Association for the Study of Pain (21). The animal experiments performed in the present study were performed in compliance with the original ARRIVE guidelines.
Periodic changes in gonadal hormone levels may affect pain (22). Therefore, 12 male animals were used in the present study. Experiments were performed on adult male C57BL/6 mice (age, 8 weeks; weight, 25-30 g; obtained from the Center of Laboratory Animal Science of Guangdong), and were housed at a constant ambient temperature of 21±2˚C and relative humidity of 55±5% with a 12-h light/dark cycle and ad libitum access to food and water. The study lasted for 6 weeks. During the entirety of the experimental procedure, staff evaluated the health of the animals every day. Every effort was made to ensure the welfare of the animals, including ensuring sufficient water and food, clean living conditions, suitable temperature and light conditions, and death after anesthesia. When the animals became infected or were unable to eat, the experiment was terminated and the animal was euthanized by administering an overdose of pentobarbital sodium (100 mg/kg, i.p.) followed by cervical dislocation.
STZ-induced DNP model
A total of 12 adult male mice were randomly divided into two groups: N, control mice; and D, DNP mice (n=6 per group), and all mice were fasted for >12 h prior to injection of STZ/citrate buffer. Diabetes mellitus was induced by a single i.p. injection of STZ (150 mg/kg; Sigma-Aldrich; Merck KGaA) freshly dissolved in citrate buffer (pH=4.5). Mice in the control group were injected with an equivalent volume of the vehicle. Diabetes mellitus was defined as hyperglycemia with a plasma glucose concentration of >300 mg/dl (16.7 mmol/l) 3 days after STZ injection. STZ-injected animals were removed from the study if they did not exhibit hyperglycemia at 3 days after injection. Blood glucose levels were measured on a weekly basis to confirm continued hyperglycemia. Blood samples were obtained from the caudal vein and body weight was monitored weekly throughout the experiment. According to a previous study (14), mice should present with mechanical allodynia 6 weeks after STZ injection. In the present study, the mice were left for 6 weeks to allow for the development of neuropathic pain following the STZ injection.
Mechanical sensitivity test
An investigator blinded to the treatments of the mice performed the behavioral tests in a dedicated quiet room under constant conditions. To quantify the mechanical sensitivity of the hind paws, the paw withdrawal threshold (PWT) in response to mechanical stimuli was measured in mice. The behavioral tests were performed 1 day prior to STZ or vehicle injection (baseline) and then on a weekly basis for 6 weeks following STZ injection. The method of assessing mechanical allodynia was performed as described previously (23). Animals were placed in a plexiglas chamber with a 4x3 mm wire mesh grid floor and allowed to acclimatize for 30 min. Calibrated von Frey filaments of different scales (g) were applied perpendicularly to the plantar surface of the right hind paw with sufficient force to bend the filament for 6 sec or until the paw was withdrawn. Rapid withdrawal or paw flinching was interpreted as a positive response. If there was no response, the next higher force filament was applied. Following a response, the next lower force filament was applied.
Tissue collection and RNA isolation
There were 12 mice in both the STZ and vehicle injection groups (6 per group). All animals survived during the study. In order to ensure the stability of the experimental model, preliminary experiments were performed prior to the formal experiments. It has been reported that mice may die after establishing a diabetes model (24). Therefore, the final experiments consisted of 6 mice in each group, and the mice were numbered for further random selection. Finally, 3 mice were randomly selected from each group for statistical analysis. A total of 42 days after STZ/vehicle injection, 3 mice in each group were euthanized using pentobarbital sodium (100 mg/kg, i.p.) followed by cervical dislocation after the final behavioral test. The L4-5 spinal cord tissues were rapidly removed and stored at -80˚C until required.
RNA isolation and RNA quantification
RNA degradation and contamination was monitored on 1% agarose gels. According to the manufacturer's protocol, each tissue sample was washed three times using cold PBS and 1 ml TRIzol® reagent was added (Thermo Fisher Scientific, Inc.) to extract the RNA. The RNA concentration was measured using the Qubit® RNA assay kit in a Qubit® 2.0 Fluorometer (Thermo Fisher Scientific, Inc.). RNA integrity was verified using an RNA Nano 1000 assay kit for the Bioanalyzer 2100 system (Agilent Technologies, Inc.). The method for determining the levels of lncRNAs and miRNAs was the same as that used for mRNAs. For the quantification of circRNAs, exonuclease was used to exclude non-circRNAs. The RNA was divided into two copies. Linear RNA was digested with RNase R (cat. no. RNR07250; Epicentre; Illumina, Inc.) to leave only the circRNAs. The other half of the sample from the same RNA extraction was not treated with RNase R. The two samples of RNA were reverse transcribed according to a previous study (25). The sample treated with RNase R was used to examine the expression of circRNAs and the other sample that was not treated with RNase R was used to measure the expression of β-actin.
Library construction and RNA sequencing
RNA sequencing was performed by Aksomics Inc. A total of 2 µg total RNA from each sample was used for the construction of the sequencing library. According to the manufacturer's protocol, sequencing libraries were built using ribosomal (r)RNA-depleted RNA with an NEB Next® Ultra™ Directional RNA Library Prep Kit for Illumina® (New England BioLabs, Inc.). First, the NEB 3' SR Adaptor was directly ligated to the 3' end of the miRNAs. Subsequently, the SR RT Primer was used to hybridize the excess of 3' SR Adaptor and the single-stranded DNA adaptor was transformed into a double-stranded (ds)DNA molecule. dsDNA cannot ligate to the 5' SR Adaptor in the next ligation step. The 5' end adapter was then ligated to the 5' ends of the miRNAs. Moloney murine leukemiavirus reverse transcriptase was used to synthesize first-strand complementary DNA. PCR amplification was performed using LongAmp Taq 2X Master Mix, SR Primer for Illumina and index (X) primer. PCR products were purified by 8% SDS-PAGE (100 V, 80 min). DNA fragments corresponding to 140-160 bp (the length of small noncoding RNA plus the 3' and 5' adaptors) were recovered and dissolved in 8 µl elution buffer. Finally, library quality was assessed on the Agilent Bioanalyzer 2100 system using DNA High Sensitivity Chips. The method for identifying circRNA in each sample was conducted according to a previous study (26).
GO annotations and KEGG pathway analysis
Fold change (FC) and false discovery rate (FDR) were used to filter DE genes under the following criteria: i) FC >1.5 or <0.5; and ii) FDR<0.05. GO annotations and KEGG pathway analysis were performed to predict the roles of the DE mRNAs and miRNAs. In brief, GO analysis was used to establish genetic regulatory networks of interest of the differentially expressed genes in the GO categories molecular function, cellular component and biological process (geneontology.org). Pathway analysis was performed to select the significant pathways of the differentially expressed genes, according to the KEGG database (genome.jp/kegg/).
Statistical analysis
Values are expressed as the mean ± standard error of the mean. The results of the paw withdrawal thresholds were statistically analyzed using repeated-measures ANOVA in SPSS version 16.0 (SPSS, Inc.). Bonferroni corrections were used for further comparison following ANOVA. A Kolmogorov-Smirnov test and P-P graph were used to test the sample data for normality of distribution and datasets with P>0.05 were considered to be normally distributed. In addition, Levene's test was used to analyze the homogeneity of variance of the data. All measurement data were normally distributed. The blood glucose levels were analyzed using mixed two-way ANOVA with Bonferroni's test. P<0.05 was considered to indicate a statistically significant difference.
Results
Changes in blood glucose levels and PWT of mice with DNP
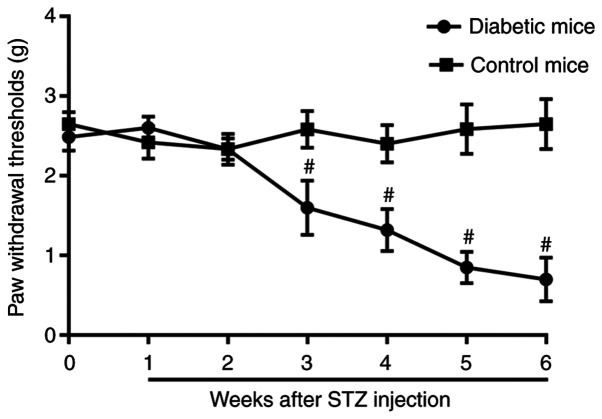
Blood glucose levels were assessed weekly throughout the study. A total of 3 days after STZ injection, diabetic mice presented with significantly increased blood glucose levels (5.0±1.4 mmol/l at baseline vs. 23.5±2.7 mmol/l 3 days after STZ administration) and this was maintained throughout the experiment (23.9±3.1 mmol/l on day 42) (Table I). The mice in the control group did not exhibit hyperglycemia. Mice with STZ-induced diabetes exhibited gradually decreasing PWT values over the 6-week period compared with the baseline (Fig. 1). In the non-diabetic mice, the PWT did not vary during the 6-week period (Fig. 1).
Table I.
Blood glucose levels of the mice during the experiment (mmol/l).
| Days after streptozotocin injection | ||||||||
|---|---|---|---|---|---|---|---|---|
| Group | Baseline | 3 | 7 | 14 | 21 | 28 | 35 | 42 |
| C | 5.6±0.1.1 | 5.00±1.4 | 6.6±1.3 | 6.5±1.5 | 6.9±1.2 | 6.4±1.5 | 5.5±1.6 | 5.9±1.3 |
| D | 5.8±1.4 | 23.5±2.7a | 23.0±2.5a | 21.4±2.6a | 26.4±2.8a | 27.5±2.8a | 27.4±2.9a | 23.9±3.1a |
aP<0.01 vs. group C. The blood glucose levels were analyzed using mixed two-way ANOVA with Bonferroni's test. Groups: C, normal control mice; D, diabetic mice.
Figure 1.
Nociceptive behavior developed in the DNP mice (n=6). #P<0.001 vs.control mice. STZ, streptozotocin.
Expression profile of the coding genes
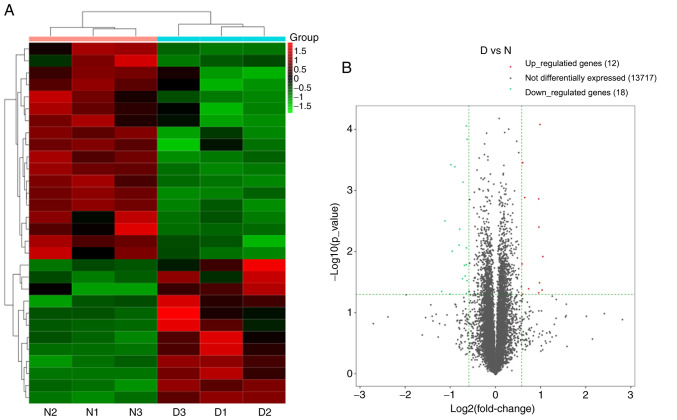
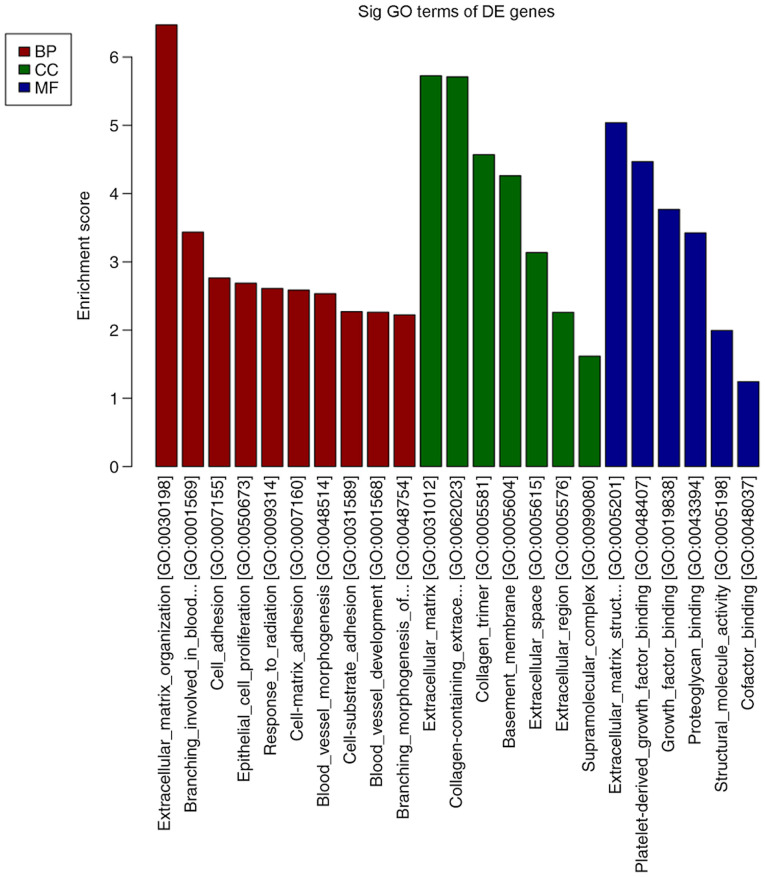
A total of 13,747 mRNAs were detected and 30 were differentially expressed in the spinal cord tissues between the DNP and control groups. At 42 days after STZ injection, there were 12 upregulated mRNAs and 18 downregulated mRNAs in the DNP group compared with those in the control group. The top 10 upregulated and downregulated mRNAs in the DNP group compared with the control group 42 days after STZ injection are listed in Table II. Fig. 2A and B presents the heat map and volcano plot of the DE mRNAs, respectively. Differentially expressed genes were primarily involved in GO molecular function terms of ‘receptor ligand activity’, ‘growth factor binding’ and ‘extracellular matrix structural constituent’ based on the GO analyses (Fig. 3). The products of DE genes were primarily located in the extracellular matrix based on GO cellular component analyses (Fig. 3). GO analysis in the category biological process indicated that ‘hormone metabolic process’, ‘regulation of hormone levels’, ‘regulation of signaling receptor activity’, ‘cell adhesion’, ‘extracellular matrix organization’ and ‘branching involved in blood vessel morphogenesis’ were the most enriched processes amongst the DE mRNAs (Fig. 3). The above-mentioned functional terms were all closely associated with neuropathic pain. KEGG pathway analyses suggested that ‘protein digestion and absorption pathway’, ‘amoebiasis’ and the ‘AGE-RAGE signaling pathway in diabetic complications’ were most enriched amongst the DE genes (Fig. 4).
Table II.
Detailed information of the top 10 upregulated and 10 downregulated mRNAs.
| mRNA | Fold change | P-value | Direction of regulation |
|---|---|---|---|
| Mup3 | 2.072998732 | 0.011983219 | Up |
| Ttr | 2.041955888 | 0.042207033 | Up |
| BC030500 | 1.972604837 | 8.2096E-05 | Up |
| Gm21320 | 1.961106079 | 0.032707565 | Up |
| Gm28036 | 1.944253915 | 0.003957426 | Up |
| Ppp1cb | 1.942959398 | 0.001371697 | Up |
| Srd5a2 | 1.94003704 | 0.046905962 | Up |
| Ccl21b | 1.657051771 | 0.040197116 | Up |
| Gm45713 | 1.565600467 | 0.001308144 | Up |
| Tomm40l | 1.523968534 | 0.000350659 | Up |
| Col3a1 | 0.438021721 | 0.045039029 | Down |
| Sema5a | 0.461710683 | 0.003190532 | Down |
| Pcdhga2 | 0.497370882 | 0.049717412 | Down |
| Mfap4 | 0.50342587 | 0.000375246 | Down |
| Gm27029 | 0.513535783 | 0.009984364 | Down |
| Vkorc1l1 | 0.533680568 | 0.000410166 | Down |
| Slc38a5 | 0.573498342 | 0.007919411 | Down |
| Igfbp4 | 0.577354961 | 0.004239532 | Down |
| Col4a1 | 0.60148317 | 0.027653137 | Down |
| Btbd2 | 0.607906603 | 0.000738708 | Down |
The Balltown function of the R software was used to analyze the differences in gene expression and screen the genes with differential expression between the C and N groups.
Figure 2.
Changes in the mRNA expression profiles in the spinal cord of the DNP mice. (A) Heat map of the mRNAs with hierarchical clustering of the differentially expressed mRNAs between the mice in the D and N groups. In the clustering analysis, up- and downregulated genes are colored in red and blue, respectively. (B) Volcano plot displaying the up- and downregulated mRNAs between the D and N groups. D, diabetic; N, normal control; DNP, diabetic neuropathy.
Figure 3.
GO analysis of the DE mRNAs. BP, biological process; CC, cellular component, MF, molecular function; GO, Gene Ontology; DE, differentially expressed; Sig, significant.
Figure 4.
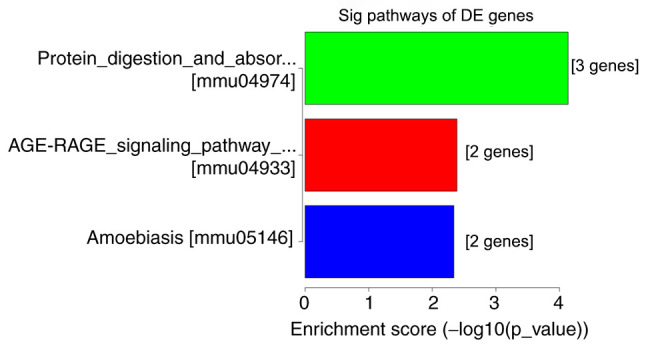
KEGG pathway analyses of the DE mRNAs. KEGG, Kyoto Encyclopedia of Genes and Genomes; Sig, significant; DE, differentially expressed; mmu, Mus musculus.
miRNA expression in DNP
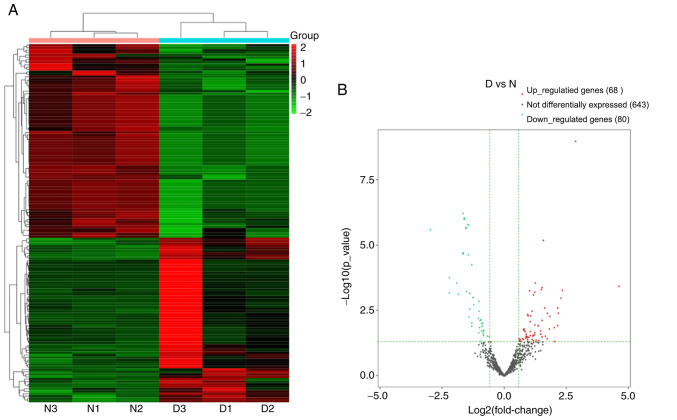
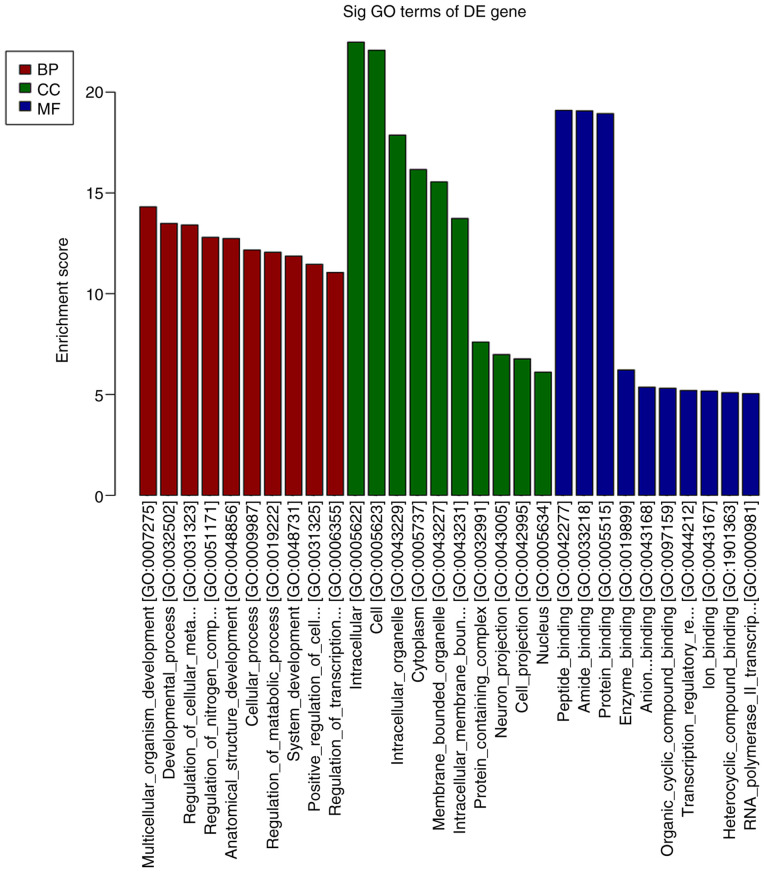
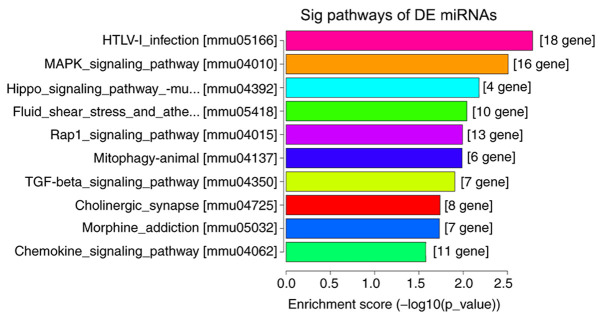
A total of 791 miRNAs detected and 148 miRNAs were differentially expressed in the DNP group compared with those in the control group. At 42 days after STZ injection, 68 upregulated and 80 downregulated miRNAs were detected. Fig. 5A and B present the heat map and volcano plot of the DE miRNAs, respectively. The top 10 upregulated and downregulated miRNAs in the DNP group compared with the control group 42 days after STZ injection are listed in Table III. The products of the target genes of the DE miRNAs were primarily located ‘intracellular and cell’ in the GO cellular component analysis (Fig. 6). GO analysis in the category biological process indicated that ‘multicellular organism development’, ‘developmental process’ and ‘regulation of cellular metabolic process’ were the most enriched processes amongst the DE miRNA target genes (Fig. 6). KEGG pathway analyses suggested that ‘regulation of actin cytoskeleton’, ‘cell adhesion molecules’, ‘Rap1 signaling pathway’, ‘human T-lymphotropic virus-I infection’ and the ‘MAPK signaling pathway’ were the most enriched pathways among the DE genes (Fig. 7).
Figure 5.
Changes in the miRNA expression profiles in the spinal cord of the DNP mice. (A) Heat map of the miRNAs with hierarchical clustering of the differentially expressed mRNAs between the mice in the D and N groups. In the clustering analysis, up- and downregulated genes are colored in red and blue, respectively. (B) Volcano plot displaying the up- and downregulated miRNAs between the D and N groups. D, diabetic; N, normal control; DNP, diabetic neuropathy; miRNA, microRNA.
Table III.
Detailed information of the top 10 upregulated and 10 downregulated miRNAs.
| miRNA | Fold change | P-value | Direction of regulation |
|---|---|---|---|
| mmu-miR-122-5p | 24.70639724 | 0.000371545 | Up |
| mmu-miR-3474 | 7.377313224 | 0.000407508 | Up |
| mmu-miR-342-5p | 5.132108479 | 0.000523721 | Up |
| mmu-miR-376a-3p | 4.896686217 | 0.001069652 | Up |
| mmu-miR-664-5p | 4.514300346 | 0.004094473 | Up |
| mmu-miR-29a-5p | 4.45171166 | 0.002624442 | Up |
| mmu-miR-200a-5p | 4.440473679 | 0.011984776 | Up |
| mmu-miR-378b | 4.102131532 | 0.048569546 | Up |
| mmu-miR-491-5p | 4.064537867 | 0.01467785 | Up |
| mmu-miR-218-1-3p | 3.584196558 | 0.005336551 | Up |
| mmu-miR-669b-3p | 0.128138311 | 0.000262216 | Down |
| mmu-miR-467c-3p | 0.217508283 | 0.000678429 | Down |
| mmu-miR-467e-3p | 0.218846498 | 0.000182836 | Down |
| mmu-miR-215-5p | 0.267746723 | 0.000290494 | Down |
| mmu-miR-3083-5p | 0.278361768 | 0.000762342 | Down |
| mmu-miR-467d-3p | 0.31631178 | 0.000199364 | Down |
| mmu-miR-467a-3p | 0.316889919 | 0.000205482 | Down |
| mmu-miR-466a-3p | 0.317690341 | 0.000612554 | Down |
| mmu-miR-466e-3p | 0.330358608 | 0.000939574 | Down |
| mmu-miR-466b-3p | 0.330809385 | 0.000958902 | Down |
The Balltown function of the R software was used to analyze the differences in gene expression and screen the genes with differential expression between the C and N groups. miRNA/miR, microRNA; mmu, Mus musculus.
Figure 6.
GO analysis of the differentially expressed miRNAs. BP, biological process; CC, cellular component; MF, molecular function; GO, Gene Ontology; miRNA, microRNA; Sig, significant; DE, differentially expressed.
Figure 7.
KEGG pathway analyses of differentially expressed miRNAs. KEGG, Kyoto Encyclopedia of Genes and Genomes; miRNA, microRNA; HTLV-I, human T-lymphotropic virus-I; Sig, significant; DE, differentially expressed; mmu, Mus musculus.
lncRNA expression in DNP
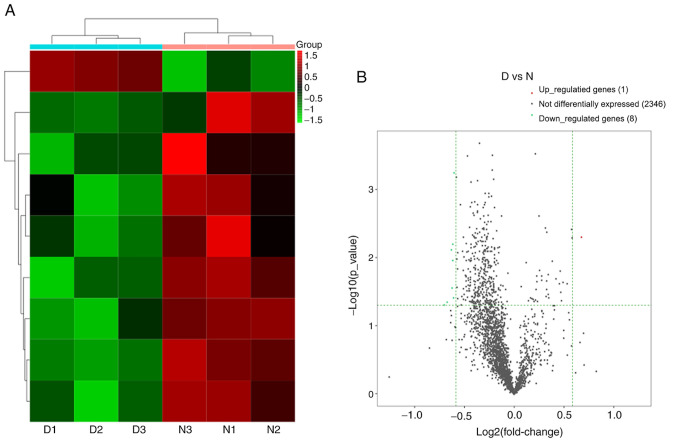
A total of 2,355 lncRNAs detected and 9 lncRNAs were differentially expressed in the DNP group compared with the control group. There were 1 upregulated lncRNA and 8 downregulated lncRNAs at 42 days after STZ injection. Fig. 8A and B provides the heat map and volcano plot of the DE lncRNAs, respectively. Detailed information on the DE lncRNAs is listed in Table IV.
Figure 8.
Changes in the lncRNA expression profiles in the spinal cord of the DNP mice. (A) Heat map of the lncRNAs with hierarchical clustering of the DE mRNAs between the mice in the D and N groups. In the clustering analysis, up- and downregulated genes are colored in red and blue, respectively. (B) Volcano plot displaying the up- and downregulated lncRNAs between the D and N groups. D, diabetic; N, normal control; DNP, diabetic neuropathy; lncRNA, long non-coding RNA.
Table IV.
Detailed information on the upregulated and downregulated long non-coding RNAs.
| Track ID | Gene name | Locus | Log2 (fold change) | Fold change | P-value | Direction of regulation |
|---|---|---|---|---|---|---|
| ENSMUSG00000099759.1 | 1700030C10Rik | chr12:20804381-20815779 | 0.675036717 | 1.596637407 | 0.005038816 | Up |
| ENSMUSG00000084894.1 | Gm13834 | chr6:31087609-31087912 | -0.703372524 | 0.614134892 | 0.049325442 | Down |
| ENSMUSG00000099521.1 | Gm28309 | chr2:74683446-74694194 | -0.670740279 | 0.628184269 | 0.045688601 | Down |
| ENSMUSG00000109359.1 | Gm44797 | chr8:9595109-9596945 | -0.627678166 | 0.647217191 | 0.007719935 | Down |
| ENSMUSG00000108123.1 | Gm43884 | chr6:45329238-45329796 | -0.623406223 | 0.649136497 | 0.028225881 | Down |
| ENSMUSG00000102296.1 | Gm37543 | chr1:25284218-25285248 | -0.614160855 | 0.653309781 | 0.006376142 | Down |
| ENSMUSG00000105791.1 | Gm43341 | chr5:48978278-48979585 | -0.613953524 | 0.653403676 | 0.011026114 | Down |
| ENSMUSG00000103331.1 | Gm37995 | chr6:40026894-40028607 | -0.608815799 | 0.655734725 | 0.039158326 | Down |
| ENSMUSG00000085638.1 | Gm15521 | chr9:29590484-29592510 | -0.605274191 | 0.657346436 | 0.000570587 | Down |
| ENSMUSG00000084894.1 | Gm13834 | chr6:31087609-31087912 | -0.703372524 | 0.614134892 | 0.049325442 | Down |
| ENSMUSG00000099521.1 | Gm28309 | chr2:74683446-74694194 | -0.670740279 | 0.628184269 | 0.045688601 | Down |
| ENSMUSG00000109359.1 | Gm44797 | chr8:9595109-9596945 | -0.627678166 | 0.647217191 | 0.007719935 | Down |
Statistical analysis was performed with the F test. chr, chromosome.
circRNA expression in DNP
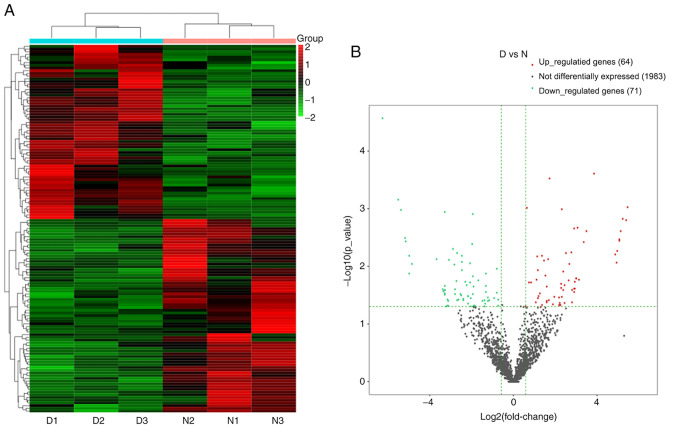
The circRNA prediction algorithm identified 2,118 distinct circRNA candidates (≥2 back-spliced reads). The length of 1,552 circRNAs was <1,000 nt and the median length was 620 nt (Fig. 9), consistent with a previous study (27). According to the filtration criteria (fold change ≥1.5 and P≤0.05), 135 DE circRNAs (64 upregulated and 71 downregulated) between the DNP and control group were identified. A heat map and volcano plot for the DE circRNAs are presented in Fig. 10A and B, respectively. The top 10 upregulated and downregulated circRNAs in the DNP group compared with those in the control group at 42 days after STZ injection are listed in Table V.
Figure 9.
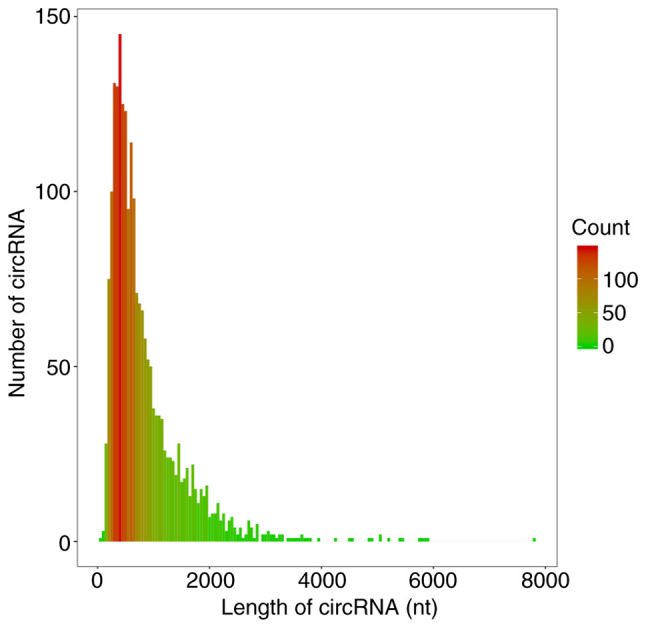
Length of the circRNAs. circRNA, circular RNA.
Figure 10.
Changes in the circRNA expression profiles in the spinal cord of the DNP mice. (A) Heat map of the circRNAs with hierarchical clustering of the differentially expressed mRNAs between the mice in the D and N groups. In the clustering analysis, up- and downregulated genes are colored in red and blue, respectively. (B) Volcano plot displaying the up- and downregulated circRNAs between the D and N groups. D, diabetic; N, normal control; DNP, diabetic neuropathy; circRNA, circular RNA.
Table V.
Detailed information on the top 10 upregulated and 10 downregulated circRNAs.
| circRNA ID | Locus | Gene name | Length | Fold change | P-value | Direction of regulalion |
|---|---|---|---|---|---|---|
| chr4:41226279-41229848:- | Ubap2 | 287 | 43.96713505 | 0.00092825 | Up | |
| mmu_circ_0010794 | chr3:79195934-79215027:- | U6 | 212 | 41.81604666 | 0.001562029 | Up |
| mmu_circ_0006623 | chr17:26142617-26143576:+ | Axin1 | 959 | 37.6311112 | 0.001492146 | Up |
| mmu_circ_0006175 | chr16:29469240-29479902:- | Atp13a4 | 574 | 35.26138418 | 0.002452781 | Up |
| mmu_circ_0007095 | chr17:90362759-90362941:- | Nrxn1 | 182 | 33.46733596 | 0.003380647 | Up |
| chr9:9984062-10172122:- | Cntn5 | 1104 | 33.13425687 | 0.003546726 | Up | |
| mmu_circ_0005297 | chr14:56748834-56764244:- | Pspc1 | 487 | 31.31885528 | 0.005412182 | Up |
| mmu_circ_0012840 | chr5:88934748-88954957:+ | Slc4a4 | 254 | 30.36278728 | 0.00845595 | Up |
| chr19:37044521-37126388:- | Cpeb3 | 755 | 29.27386362 | 0.006105253 | Up | |
| mmu_circ_0001580 | chr7:66125241-66125737:+ | Chsy1 | 496 | 14.39983645 | 0.000247404 | Up |
| chr16:4655009-4655634:- | Coro7 | 170 | 0.012940375 | 0.000268838 | Down | |
| chr13:119381675-119404754:- | Nnt | 1016 | 0.022155209 | 0.000693194 | Down | |
| mmu_circ_0016083 | chrX:113139335-113140786:- | Chm | 198 | 0.02393434 | 0.001058838 | Down |
| chr18:23535161-23545766:+ | Dtna | 149 | 0.027664966 | 0.003177893 | Down | |
| mmu_circ_0006471 | chr16:93799906-93800247:+ | Dopey2 | 257 | 0.028359819 | 0.00367879 | Down |
| mmu_circ_0008757 | chr1:5095614-5124469:+ | Atp6v1h | 564 | 0.031315099 | 0.006494722 | Down |
| chr6:115244145-115263981:+ | Syn2 | 624 | 0.031522436 | 0.013407635 | Down | |
| mmu_circ_0004843 | chr13:8697619-8731971:+ | Adarb2 | 672 | 0.034389942 | 0.009121529 | Down |
| mmu_circ_0013996 | chr7:141588285-141605010:+ | Ap2a2 | 638 | 0.078133536 | 0.007429339 | Down |
| chr1:105640664-105649407:- | Pign | 474 | 0.095120485 | 0.024887204 | Down |
CircRNA was calculated by a negative binomial distribution test, chr, chromosome; circRNA, circular RNA; mmu, Mus musculus.
Discussion
In the present study, the DE mRNAs, miRNAs, lncRNAs and circRNAs in the spinal cord of mice with DNP were comprehensively analyzed using rRNA-depleted RNA sequencing. A total of 30 mRNAs, 148 miRNAs, 9 lncRNAs and 135 circRNAs were determined to be differentially expressed in the DNP mice compared with the control mice. In addition, the potential functions of the DE ncRNAs were determined using GO and KEGG pathway analysis. Based on these results, it was hypothesized that ncRNAs serve a vital role in the development of DNP and that they may serve as potentially novel therapeutic targets for the management of DNP.
The pathogenesis of DNP is complex and remains poorly understood. The spinal cord is the relay station of nociceptive stimuli, which serves an important role in the development of pain (28). However, the underlying mechanisms by which the spinal dorsal horn processes nociceptive stimuli are complex. ncRNAs are genetic, epigenetic and translational regulators. Previous studies have indicated that dysregulation of ncRNAs is associated with a variety of diseases, including neuropathic pain (12,13). However, the role of ncRNAs in the pathogenesis of DNP has remained largely elusive. Thus, in the present study, the DE ncRNAs in DNP were determined and their functions and regulatory interactions were analyzed.
The causal roles of miRNAs in chronic pain have previously been established (29). In the present study, numerous DE miRNAs were detected and miR-122 was the most notably upregulated miRNA. It was previously reported that miR-122 is involved in the regulation of neuropathic pain (30); however, whether miR-122 regulates DNP remains elusive and further studies are required to determine this. Although DE miRNAs in the spinal cord of mice with DNP were screened and the differential expression of certain miRNAs was confirmed in the present study, the underlying mechanisms of miRNAs in DNP are poorly understood. GO analysis may be used to unify the representation of genes and gene product attributes in all species (31). GO terms and GO annotations are good predictors of gene functions and trends (32). The KEGG pathway database is the most widely used enrichment analysis platform and it stores higher-order functional information for systematic analysis of gene functions (33). In the present study, the miRNA-related gene functions and the corresponding pathways in mice with DNP were predicted using GO term and KEGG pathway enrichment analyses. The results indicated that the most significantly involved pathways in the pathogenesis of DNP were the MAPK signaling pathway, Rap1 signaling pathway and TGF-β signaling pathway. Previous studies have demonstrated that these signaling pathways are closely associated with neuropathic pain (34-36). The function of miRNAs in the pathogenesis of DNP should be examined in more detail in future studies.
A growing number of studies have also indicated that noxious stimuli may result in dysregulated expression of lncRNAs and this may be involved in the pain hypersensitivity underlying NP (37,38). To the best of our knowledge, there are no comprehensive studies of the lncRNAs associated with DNP. Thus, second-generation sequencing was used to analyze the DE lncRNAs in the spinal cord of mice with DNP. The results indicated that a total of 9 lncRNAs were significantly dysregulated in mice with DNP compared to control mice.
CircRNAs are a type of highly stable, circularized lncRNA. A previous study indicated that circRNAs are conserved across species and are primarily enriched in the nervous system (39). Various circRNAs have been identified; however, the biological functions of the majority of these circRNAs remain elusive. In the present study, 135 DE circRNAs that may be involved in the pathogenesis of DNP were identified, which may provide further insight into the underlying mechanisms of circRNAs in DNP. The majority of circRNAs detected were derived from exons, similar to the results of a previous study where most circRNAs were derived from coding sequences (39). In the present study, it was also indicated that multiple circRNAs were able to be generated from one host gene. Regulating synaptic membrane exocytosis 2 is able to generate 17 distinct circRNAs and the gene encoding protein tyrosine kinase 2 is able to generate 47 distinct circRNAs. The median length of circRNAs was 620 nt, similar to a previous study in which the median length of circRNAs was around 500 nt (40,41).
The present study had certain limitations. Sequencing analysis was used to investigate the expression patterns of coding genes, miRNAs, lncRNAs and circRNAs in the spinal cord of mice with STZ-induced DNP. In order to verify the effectiveness of the preliminary screening approach, the effects of these DE ncRNAs in DNP will be further assessed in animals and in humans to verify the related functions and pathways of these ncRNAs. In addition, the differential expression of mRNAs, miRNAs, lncRNAs and circRNAs in the spinal cord of DNP mice was assessed in the present study. However, whether these results translate to humans remains unknown. Thus, whether these ncRNAs are of relevance to DNP in humans will next be determined. According to the theory of biological evolution, RNA is conserved to a certain extent. Conservation analysis will be performed on RNAs to identify the human ncRNAs similar to those identified in the mice for further mechanistic research. In addition, the complications and other physiological indicators following STZ injection were not addressed in the present study, and thus, future studies should take this limitation into account. According to a previous study (14), mice exhibit notable chronic DNP 6 weeks after STZ injection; however, whether different time-points affect the results is unknown but worthy of further study.
In conclusion, the differential expression profiles of mRNAs, miRNAs, lncRNAs and circRNAs in the spinal cord of DNP mice were determined using rRNA-depleted RNA sequencing. A total of 30 mRNAs, 148 miRNAs, 9 lncRNAs and 135 circRNAs were differentially expressed between the DNP and control mice. ‘Rap1 signaling pathway’ and ‘MAPK signaling pathway’ were the most enriched pathways among the DE genes. The complete proteomic and relevant signaling pathway of this differential expression ncRNAs is worthy of further study, which may ultimately enable full disclosure of the mechanisms underlying DNP.
Acknowledgements
Not applicable.
Funding Statement
Funding: This study was funded by The National Natural Science Funds of China (grant no. 81771357).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The sequencing data have been submitted to https://bigd.big.ac.cn/databases with the accession ID CRA003943.
Authors' contributions
ZSQ, JH and HBW designed the present study. JH, JJH, LZ and DLL performed the experiments. JH, WYH and QMX analyzed data and wrote the manuscript draft. JH and ZSQ revised the final manuscript. HBW and ZSQ confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All experiments were approved by the Animal Use and Care Committee for Research and Education of The First People's Hospital of Foshan (Foshan, China) and were performed in accordance with the guidelines described in the International Association for the Study of Pain, as well as in compliance with the original ARRIVE guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kesavadev J, Saboo B, Sadikot S, Das AK, Joshi S, Chawla R, Thacker H, Shankar A, Ramachandran L, Kalra S. Unproven therapies for diabetes and their implications. Adv Ther. 2017;34:60–77. doi: 10.1007/s12325-016-0439-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun J, Wang Y, Zhang X, Zhu S, He H. Prevalence of peripheral neuropathy in patients with diabetes: A systematic review and meta-analysis. Prim Care Diabetes. 2020;14:435–444. doi: 10.1016/j.pcd.2019.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Shillo P, Sloan G, Greig M, Hunt L, Selvarajah D, Elliott J, Gandhi R, Wilkinson ID, Tesfaye S. Painful and painless diabetic neuropathies: What is the difference? Curr Diab Rep. 2019;19(32) doi: 10.1007/s11892-019-1150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paisley P, Serpell M. Improving pain control in diabetic neuropathy. Practitioner. 2017;261:23–26. [PubMed] [Google Scholar]
- 5.Dewanjee S, Das S, Das AK, Bhattacharjee N, Dihingia A, Dua TK, Kalita J, Manna P. Molecular mechanism of diabetic neuropathy and its pharmacotherapeutic targets. Eur J Pharmacol. 2018;833:472–523. doi: 10.1016/j.ejphar.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 6.Sloan G, Shillo P, Selvarajah D, Wu J, Wilkinson ID, Tracey I, Anand P, Tesfaye S. A new look at painful diabetic neuropathy. Diabetes Res Clin Pract. 2018;144:177–191. doi: 10.1016/j.diabres.2018.08.020. [DOI] [PubMed] [Google Scholar]
- 7.Yang XD, Fang PF, Xiang DX, Yang YY. Topical treatments for diabetic neuropathic pain. Exp Ther Med. 2019;17:1963–1976. doi: 10.3892/etm.2019.7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 9.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 11.Thum T. Noncoding RNAs and myocardial fibrosis. Nat Rev Cardiol. 2014;11:655–663. doi: 10.1038/nrcardio.2014.125. [DOI] [PubMed] [Google Scholar]
- 12.Li G, Jiang H, Zheng C, Zhu G, Xu Y, Sheng X, Wu B, Guo J, Zhu S, Zhan Y, et al. Long noncoding RNA MRAK009713 is a novel regulator of neuropathic pain in rats. Pain. 2017;158:2042–2052. doi: 10.1097/j.pain.0000000000001013. [DOI] [PubMed] [Google Scholar]
- 13.Liu C, Li C, Deng Z, Du E, Xu C. Long Non-coding RNA BC168687 is involved in TRPV1-mediated diabetic neuropathic pain in rats. Neuroscience. 2018;374:214–222. doi: 10.1016/j.neuroscience.2018.01.049. [DOI] [PubMed] [Google Scholar]
- 14.Yang D, Yang Q, Wei X, Liu Y, Ma D, Li J, Wan Y, Luo Y. The role of miR-190a-5p contributes to diabetic neuropathic pain via targeting SLC17A6. J Pain Res. 2017;10:2395–2403. doi: 10.2147/JPR.S133755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu B, Guo Y, Chen Q, Xiong Q, Min S. MicroRNA-193a Downregulates HMGB1 to alleviate diabetic neuropathic pain in a mouse model. Neuroimmunomodulat. 2019;26:250–257. doi: 10.1159/000503325. [DOI] [PubMed] [Google Scholar]
- 16.Yu W, Zhao GQ, Cao RJ, Zhu ZH, Li K. LncRNA NONRATT021972 was associated with neuropathic pain scoring in patients with Type 2 diabetes. Behav Neurol. 2017;2017(2941297) doi: 10.1155/2017/2941297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440–441. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 19.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Luo T, Bao Z, Li Y, Bu W. Intrathecal circHIPK3 shRNA alleviates neuropathic pain in diabetic rats. Biochem Biophys Res Commun. 2018;505:644–650. doi: 10.1016/j.bbrc.2018.09.158. [DOI] [PubMed] [Google Scholar]
- 21.Zimmermann M. Ethical guidelines for investigations of experimental pain in conscious animals. Pain. 1983;16:109–110. doi: 10.1016/0304-3959(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 22.Stoffel EC, Ulibarri CM, Folk JE, Rice KC, Craft RM. Gonadal hormone modulation of mu, kappa, and delta opioid antinociception in male and female rats. J Pain. 2005;6:261–274. doi: 10.1016/j.jpain.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53:55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 24.Furman BL. Streptozotocin-Induced diabetic models in mice and rats. Curr Protoc Pharmacol. 2015;70:5.47.1–5.47.20. doi: 10.1002/0471141755.ph0547s70. [DOI] [PubMed] [Google Scholar]
- 25.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding X, Zhang S, Li X, Feng C, Huang Q, Wang S, Wang S, Xia W, Yang F, Yin R, et al. Profiling expression of coding genes, long noncoding RNA, and circular RNA in lung adenocarcinoma by ribosomal RNA-depleted RNA sequencing. FEBS Open Bio. 2018;8:544–555. doi: 10.1002/2211-5463.12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prescott SA, Ma Q, De Koninck Y. Normal and abnormal coding of somatosensory stimuli causing pain. Nat Neurosci. 2014;17:183–191. doi: 10.1038/nn.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López-González MJ, Landry M, Favereaux A. MicroRNA and chronic pain: From mechanisms to therapeutic potential. Pharmacol Ther. 2017;180:1–15. doi: 10.1016/j.pharmthera.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 30.Ma F, Wang C, Yoder WE, Westlund KN, Carlson CR, Miller CS, Danaher RJ. Efficacy of herpes simplex virus vector encoding the human preproenkephalin gene for treatment of facial pain in mice. J Oral Facial Pain Headache. 2016;30:42–50. doi: 10.11607/ofph.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Zhang K, Lee J, Cordes S, Davis DP, Tang Z. Discovering cancer genes by integrating network and functional properties. BMC Med Genomics. 2009;2(61) doi: 10.1186/1755-8794-2-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du J, Li M, Yuan Z, Guo M, Song J, Xie X, Chen Y. A decision analysis model for KEGG pathway analysis. BMC Bioinformatics. 2016;17(407) doi: 10.1186/s12859-016-1285-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ni HD, Yao M, Huang B, Xu LS, Zheng Y, Chu YX, Wang HQ, Liu MJ, Xu SJ, Li HB. Glial activation in the periaqueductal gray promotes descending facilitation of neuropathic pain through the p38 MAPK signaling pathway. J Neurosci Res. 2016;94:50–61. doi: 10.1002/jnr.23672. [DOI] [PubMed] [Google Scholar]
- 35.Singhmar P, Huo X, Eijkelkamp N, Berciano SR, Baameur F, Mei FC, Zhu Y, Cheng X, Hawke D, Mayor F Jr, et al. Critical role for Epac1 in inflammatory pain controlled by GRK2-mediated phosphorylation of Epac1. Proc Natl Acad Sci USA. 2016;113:3036–3041. doi: 10.1073/pnas.1516036113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen NF, Huang SY, Chen WF, Chen CH, Lu CH, Chen CL, Yang SN, Wang HM, Wen ZH. TGF-β1 attenuates spinal neuroinflammation and the excitatory amino acid system in rats with neuropathic pain. J Pain. 2013;14:1671–1685. doi: 10.1016/j.jpain.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Wu J, Wang C, Ding H. LncRNA MALAT1 promotes neuropathic pain progression through the miR-154-5p/AQP9 axis in CCI rat models. Mol Med Rep. 2020;21:291–303. doi: 10.3892/mmr.2019.10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren X, Yang R, Li L, Xu X, Liang S. Long non coding RNAs involved in MAPK pathway mechanism mediates diabetic neuropathic pain. Cell Biol Int. 2020;44:2372–2379. doi: 10.1002/cbin.11457. [DOI] [PubMed] [Google Scholar]
- 39.van Rossum D, Verheijen BM, Pasterkamp RJ. Circular RNAs: Novel regulators of neuronal development. Front Mol Neurosci. 2016;9(74) doi: 10.3389/fnmol.2016.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7(11215) doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(409) doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The sequencing data have been submitted to https://bigd.big.ac.cn/databases with the accession ID CRA003943.