Abstract
LncRNA PVT1 (plasmacytoma variant translocation 1) has become a staple of the lncRNA profile in patients with renal cell carcinoma (RCC). Common dysregulation in renal tumors outlines the essential role of PVT1 in the development of RCC. There is already a plethora of publications trying to uncover the cellular mechanisms of PVT1-mediated regulation and its potential exploitation in management of RCC. In this review, we summarize the literature focused on PVT1 in RCC and aim to synthesize the current knowledge on its role in the cells of the kidney. Further, we provide an overview of the lncRNA profiling studies that have identified a more or less significant association of PVT1 with the clinical behavior of RCC. Based on our search, we analyzed the 17 scientific papers discussed in this review that provide robust support for the indispensable role of PVT1 in RCC development and future personalized therapy.
Keywords: biomarker, prognosis, diagnosis, long non-coding RNA, tumorigenesis
1. Introduction
Tumors of the kidney (renal cell carcinoma, RCC) account for 2% of all cancers [1]. Although mortality is slowly decreasing [2], RCC is considered afatal disease, with a relatively high percentage of patients having developed metastasis at the time of the diagnosis and with low chances for 5-year survival [2]. The exact identification of the RCC entity or subtype is necessary for accurate and effective treatment and proper prognosis assessment. The WHO classification system divides RCC into sixteen subtypes, the most common being three of them—clear cell carcinoma (ccRCC) with 75% abundance of all RCC cases, followed by papillary renal cell carcinoma (PRCC; 15–20%) and chromophobe renal cell carcinoma (chRCC; 5%) [3]. CcRCC is also the most aggressive type of renal cancer with the worst prognosis than the less-frequent subtypes [3,4].
Prognosis is assessed based on the TNM classification system (T—the size of the primary tumor, N—the presence of the metastases in regional lymph nodes, and M—distant metastases positivity) describing the tumor’s stage. Other prognostic information is brought in by the histologic type and level of the tumor’s differentiation [5]. From a diagnostic and prognostic point of view, it is possible to discriminate between individual RCC subtypes using immunohistochemistry or using gene expression panels [3]. With the development of targeted therapy, more attention has been turned to predictive biomarkers [6]. However, there is currently no biomarker reaching a sufficient specificity and sensitivity enabling the precise personalization of the therapy [5], although great scientific effort has been focused on searching for biomarkers in many human diseases [7], including RCC [5]. Among the potential molecules feasible as biomarkers, long non-coding RNAs have been studied excessively. Many studies emerged, mainly after expansion of next-generation sequencing, focusing on a lncRNA as diagnostic, prognostic, or predictive biomarkers.
In 2008, one of the first studies described the dysregulation of lncRNA expression levels in renal cell carcinoma. The authors observed deregulated levels of seven intronic lncRNAs in tumor samples compared to nontumor renal tissue [8]. Subsequent microarray profiling showed thousands of lncRNAs with differential expression in RCC tumor tissue [9]. Since then, PVT1 has repeatedly emerged in many profiling studies as a prominently dysregulated lncRNA in renal cell carcinoma, even in comparison with other human tumors [10]. Therefore, we decided to summarize the current knowledge on this lncRNA and its role in the development and progression of RCC, with a particular focus on its potential feasibility as a diagnostic and prognostic biomarker. To review the literature, we searched the PubMed database for relevant studies published from inception to 23 April 2021. Using search terms (“PVT1” and “renal” “and “cell” and “carcinoma”) and excluding studies that were not primarily focused on renal cell carcinoma patients or working with non-RCC cell lines, we identified 17 articles relevant for further discussion. The papers focused on lncRNA profiling, containing biomarker discovery, or the analysis of profiling data as a part of study design are summarized in Table 1. The papers focused on PVT1 in a cellular context or containing an in vitro functional test as a part of the study design are discussed in the subsequent section.
2. General Characterization of Long Non-Coding RNAs
Sequencing of the human genome revealed that the number of protein-coding genes reaches up to 20,000 to 25,000 [11], which makes up around 1.5% of the human genome. The remaining part of the genome serves structural or regulatory functions. Depending on the cell type, up to 85% of the genome is actively transcribed and gives rise to many different types of untranslated transcripts [12,13]. These transcripts were termed non-coding RNAs [14], and new knowledge about their wide range of functions in the human body has emerged in recent decades. The database NONCODE currently states that lncRNAs originate from 96,000 genes producing more than 172,000 transcripts [15]. LncRNAs occur in different tissues, and in contrast to mRNAs, their temporospatial specificity is higher, even though their expression levels usually remain low [16,17,18,19]. They are localized preferably in the cell nucleus, close to chromatin, but can also be found in the cytoplasm [13,19]. According to an extensive analysis of the translational potential of annotated lncRNAs, most of them should not code a functional protein [20], although some rare exceptions have been observed [21,22].
The genomic structure of lncRNAs is similar to mRNA originating from protein-coding genes [19]. LncRNAs are transcribed by RNA polymerase II, usually contain a 5’cap, and are polyadenylated at the 3’ end. However, there are some exceptions [23,24], and several alternative lncRNA-editing pathways have been described [25]. In 98% of all cases, lncRNAs are spliced and usually contain only two exons, much less than in protein-coding mRNA. In more than 25% of all lncRNA transcripts, splicing creates several isoforms [18,19]. Interestingly, lncRNAs can be a source or a precursor of sncRNAs, such as tRNA, miRNA, snRNA, and above all, snoRNA [13,19].
The function can be predicted from the sequence and level of conservation, and because most lncRNAs are less conserved than protein-coding genes, they have long been considered nonfunctional [26,27]. However, the conservation of lncRNAs is higher than those of introns or intergenic regions. Moreover, the promotors of lncRNAs are conserved comparably to the promotors of protein-coding genes [19,23,28,29,30,31]. Taken together with a wide and omnigenous palette of means by which lncRNAs are involved in the life of the cell, their great evolutionary importance is evident [32,33].
LncRNAs regulate cellular gene expression using various mechanisms on the transcriptional, posttranscriptional, and translational level. There are four main archetypes or mechanisms of regulation. LncRNAs function as molecular signals when their presence serves as an indicator of the transcriptional activity, because their expression is time-, stimuli-, or site-specific. Another type is a molecular decoy when lncRNAs bind to other regulatory molecules, such as transcription factors or chromatin remodeling complexes, and lower their availability. Moreover, lncRNAs serve as a guide, interacting with proteins creating ribonucleoprotein complexes guided by lncRNA to specific targets, usually to nearby (cis) or distant (trans) genes. A necessary function is a scaffold, where lncRNA functions as a platform for assembling different molecular components, creating RNP complexes that can activate or inhibit transcription or have a structurally stabilizing effect [34].
The mechanisms of regulation of the gene expression by lncRNAs are inevitably joined with their localization in a cell, which determines what structures or processes will be affected [19]. Nuclear lncRNAs regulate the gene expression on three levels—they affect the chromatin and transcription and posttranscriptional modifications of mRNA. Nuclear lncRNAs can accumulate in cis (close to the side of their own transcription) or in trans (being dislocated from the site of their origin to fulfil their function somewhere else). However, lncRNAs accumulated in cis can also act in trans [35].
An important function of lncRNA is their involvement in epigenetic regulation—specifically, chromatin remodeling. LncRNAs bind to chromatin modification factors such as methyltransferases [36,37] and guide them to the specific loci in chromatin or, on the other hand, prevent them from binding to the chromatin [38,39].
The second level is transcriptional regulation and its activation or repression by three different mechanisms concerning lncRNAs [40]. Firstly, lncRNAs bind activating proteins and protein complexes [41], including transcription factors [42]. Further, they mediate chromatin interactions, including chromosome looping, which enables the interactions of genes and regulatory elements originating from the distant loci on the chromosome [43,44]. LncRNAs also prevent repressive mechanisms from action [45,46] or, conversely, can act as corepressors of transcription [47,48]. On a post-transcriptional level, lncRNAs cofacilitate pre-mRNA splicing [49,50].
In a cytoplasm, lncRNAs regulate the gene expression on a post-transcriptional and post-translational level, influencing the mRNA stability in a positive [51] or negative [52] way. LncRNAs affect even the translation of mRNA itself, both as a suppressor [53] and as an activator [54].
A separate category of lncRNAs is a network of so-called competing endogenous RNAs (ceRNAs), which function as microRNA (miRNA) sponges binding miRNAs and averting them from binding to their target mRNAs [55].
LncRNAs can bind proteins and prevent them from the transcriptional regulation of target genes [56] or maintaining genomic stability [57]. Protein stability and turnover can be affected by the lncRNAs involved in ubiquitin-mediated proteolysis [58]. Conversely, some lncRNAs can prevent proteins from ubiquitination [59]. It has been described that lncRNAs are involved in signaling pathways and the cellular transport of molecules, including other lncRNAs [60], which may function in organelles [61].
As lncRNAs are crucial components of many cellular processes and their functional scope is broad, it is evident that their dysregulation could lead to pathological conditions and the development of various diseases, depending on the mechanism that was disrupted. LncRNAs are associated with neurological and neurodegenerative diseases [62], such as Alzheimer’s disease [63], diabetes mellitus and its complications [64], autoimmune [65], inflammatory [66], or cardiovascular diseases [67]. The most attention, however, has been given to tumor diseases. LncRNAs can act both as oncogenes and tumor suppressors. They regulate the expression of such genes [68] and modulate all the tumor hallmarks, according to Hanahan and Weinberg [69], such as the independent production of proliferation signals, insensitivity to antigrowth signals and programmed cell death escape [70,71], genomic instability, inflammation, disruption of energetic metabolism, and immune surveillance escape [72].
3. Role of PVT1 in Development of Renal Cell Carcinoma
Encoded by the human PVT1 (plasmacytoma variant translocation 1) gene of nine exons, PVT1 is 1957-bp-long lncRNA located at 8q24.21 [73]. PVT1 is generally known as an oncogene involved in tumorigenesis [74]. The artificial silencing of PVT1 represses EMT and affects the proliferation, apoptosis, migration, and cell cycle by regulating many prominent tumorigenesis players, such as cyclin D1, p21, and Myelocytomatosis (MYC) [75,76,77,78]. The latter mentioned originates in a famous proto-oncogene MYC localized close to the PVT1 gene. PVT1 and MYC are known to interact with each other and are commonly coamplified [79].
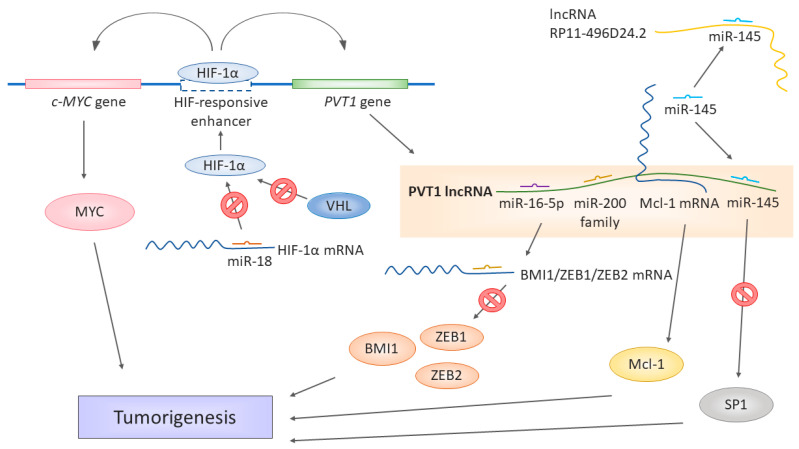
Corresponding with the dysregulation in RCC tumor samples, PVT1 is overexpressed in RCC cells as well. The results show that PVT1 affects apoptosis through myeloid leukemia cell differentiation protein 1 (Mcl-1), a member of the B-cell leukemia lymphoma protein (Bcl-2) family of proteins involved in regulating cell death and comprising both pro- and antiapoptotic factors. Mcl-1 is antiapoptotic; thus, its upregulation leads to cell death signal resistance and increased survival typical for human cancer cells [80]. PVT1 knockdown in RCC cells leads to the downregulation of Mcl-1 as PVT1 enhances the Mcl-1 mRNA stability, thus keeping its levels higher (Figure 1). Downregulating either member of the PVT1/Mcl-1 axis inhibits proliferation and colony formation and promotes apoptosis [81].
Figure 1.
Different ways of lncRNA PVT1 contributing to the development of renal cell carcinoma. miR-18 binds to HIF-1α mRNA and prevents it from translation. If downregulated, the miR-18 levels are not sufficient to keep the expression of HIF-1α in check. However, while the normal oxygen levels last, protein HIF-1α is targeted by VHL for degradation. In hypoxia, HIF-1α is stabilized and cannot be targeted by VHL and can be transferred to the nucleus. The PVT1 gene is under the HIF-responsive enhancer, along with cMYC. HIF-1α binding to this enhancer activates the expression of both c-MYC and PVT1. PVT1 itself serves as a miRNA sponge binding the miR-16-5p, miR-145, and miR-200 family members, thus liberating the target mRNA from miRNA-induced degradation, for example, BMI1, ZEB1, ZEB2, and SP1. miR-145 is a target of both PVT1 and RP11-496D24.2 in the ceRNA network. Moreover, PVT1 stabilizes larger mRNA molecules such as Mcl-1, which, in turn, can be translated into functional Mcl-1 proteins. The upregulation of PVT1 is thus associated or directly responsible for the upregulation of significant factors promoting tumorigeneses, such as MYC, Mcl-1, BMI1, ZEB1, ZEB2, and SP1.
PVT1 seems to be also tightly connected to VHL signaling and hypoxia sensing [82], a significant feature of RCC tumorigenesis. On a molecular level, the most frequent cause of either sporadic or hereditary RCC is the inactivation of the von Hippel-Lindau (VHL) gene [83]. Mutations causing the loss of heterozygosity of the 3p chromosome [83,84,85] or hypermethylation of the VHL promotor [86] are common in RCC patients. Defective protein VHL enables the stabilization of the hypoxia-induced factor (HIF) protein family, which would be otherwise degraded in a proteasome. When stabilized, the HIF protein family activates and regulates the cellular response to hypoxia, affecting the genes involved in angiogenesis (for example, VEGF—vascular endothelial growth factor), cell migration, or apoptosis. VHL thus functions as a tumor suppressor [87,88].
The results of Grampp et al. [89] tie the involvement of PVT1 in MYC and VHL regulation together (Figure 1), as their team identified a unique polymorphism at 8q24.21 associated with renal cancer susceptibility. The polymorphism influences the expression of MYC and PVT1, as it is localized in the HIF-binding enhancer, affecting both the cellular MYC (c-MYC) gene and PVT1 gene. The polymorphism’s effect is restricted to renal tubular cells, and the renal cancer susceptibility depends on the genotype of the respective polymorphism site in the enhancer, affecting its accessibility to HIF proteins. pVHL defects enhanced expression of c-MYC and PVT1, hence the notorious upregulation of PVT1 in RCC. The polymorphisms in the HIF-responding regulatory elements could affect renal tumorigenesis. As the authors pointed out, the currently known SNPs associated with RCC are all linked to modulation of the VHL/HIF pathway [89,90]. Therefore, the renal cancer-promoting or protective effect of a given SNP depends on its effect on the HIF expression and HIF-mediated regulation [89].
The regulation of oxygen sensing has been expanded for the role of miR-18a [82], which regulates the expression of the HIF-1α protein. The downregulation of miR-18a leads to higher levels of the HIF-1α protein, whose expression is then unrestricted and affects the expression of PVT1 (Figure 1). The miR-18a/HIF-1α/PVT1 regulatory pathway plays a crucial role in the development and prognosis of ccRCC, as miR-18a has been identified in this work as a significant biomarker, and prognosis and RCC development [82].
The complexity of gene expression regulation by non-coding RNAs was further illustrated by Yang et al. [73] in their work showing that PVT1 also serves as a ceRNA in the context of RCC. The PVT1 levels negatively correlate with the miR-200 family and miR-20a, miR-20b, and miR-203s. PVT1 shares the miRNA-binding “seed” sequence with the miR-200-regulated mRNAs of BMI1, ZEB1, and ZEB2, all three being major players in cancer [91,92,93] (Figure 1). The subsequent in vitro experiments showed that PVT1 upregulates the levels of BMI1, ZEB1, and ZEB2, while the overexpression of the miR-200s downregulated them. The authors further identified a new splicing variant of PVT1, whose levels were even higher in the tumor tissue and tumor cell lines than the levels of the full-length original splicing variant and had an even higher effect on the cell proliferation. The spliced region, exon 4, does not contain binding sites for the miR-200 family. Therefore, even this variant can act as ceRNA [73]. Ren et al. [77] identified the functional connection of PVT1 with miR-16-5p, as the silencing of this miRNA has led to the reversion of the regulatory effect on RCC cells induced by the downregulation of PVT1. An in-silico target prediction showed that miR-16-5p is a target of PVT1. Thus, its levels decrease as a result of binding to PVT1 (Figure 1). Moreover, the strong negative regulatory connection of PVT1 and miR-16-5p has been observed in colorectal cancer [94].
A similar ceRNA mechanism has been observed in PRCC by Huang et al. [95]. In their work, they found the interactions of miR-145 miR-211, miR-216a, mIR-133a, and miR-133b with PVT1. These interactions overlapped with lncRNA RP11-496D24.2, which also contained binding sites for some of these miRNAs (mir-145, mir-211, and mir-216a). The authors suggested a competition mechanism between PVT1 and RP11-496D24.2 for these common miRNAs, thus being critical regulators of pathogenesis in PRCC. However, the downstream targets profiting from RP11-496D24.2-mediated miRNA binding are not yet known. In PVT1, though, SP1 has been identified as a potential target of PVT1, miR-145, miR-133a, and miR-133b. SP1 is a versatile transcription factor [96] involved in many cellular processes as an activator or repressor, depending on the context. Suggested lncRNA-miRNA-mRNA ceRNA pathway regulating of such a potent regulator of gene expressions could be a significant driving force in the pathogenesis of PRCC, especially with the upregulation of PVT1, which was shown by Huang et al. [95] in PRCC patients.
The complex system of PVT1 tumorigenesis regulation via the regulation of miRNA was exceptionally summarized in the work of Wang et al. [97]. PVT1 functions as a miRNA “sponge”, inhibiting their activity by lowering the levels of more than 20 miRNAs. Should those miRNAs be involved in the regulation of other oncogenic mRNAs, the upregulation of PVT1 would lead to their decrease and, thus, the promotion of tumorigenesis. Moreover, PVT1 seems to be a precursor to other miRNAs with either oncogenic or tumor-suppressive effects [97].
4. PVT1 as a Diagnostic and Prognostic Biomarker
The prominent association of PVT1 with renal cell carcinoma has been outlined repeatedly in lncRNA profiling studies (Table 1). Posa et al. [98] analyzed over 700 samples of patients with different types of cancer, including RCC. Already here, the upregulation of PVT1 in RCC overshadows other types of tumors. This trend is promising for further uses in diagnostics, and luckily, subsequent papers seem to validate PVT1 as a specific marker of ccRCC. However, PVT1 was shown to also be significantly upregulated in PRCC [95]. Liu et al. [99] later identified a panel of four lncRNAs, including PVT1, specific for ccRCC, and developed a risk score for the ccRCC prognosis estimation. Wang et al. [100] created a model of independent prognostic lncRNAs, miRNAs, and mRNAs, including PVT1, associated with the overall survival. Moreover, lncRNAs PVT1, LINC00472, TCL6, WT-AS1, and mRNA of the COL4A4 protein were identified as independent prognostic factors.
Table 1.
PVT1 in lncRNA profiling studies in renal cell carcinoma. *—Papillary renal cell carcinoma; otherwise, clear cell renal cell carcinoma; **—Included patients with ccRCC, chromophobe RCC, and PRCC; and ***—Included both serum and tissue samples; otherwise, only tumor and nontumor tissues.
| Reference | Primary Endpoint | N of Patients/Controls (Source) | Diagnostic Properties | Prognostic Properties | |
|---|---|---|---|---|---|
| p-Value, AUC | p-Value | Risk Ratio, 95% CI | |||
| Studies focused on the diagnostic properties of PVT1 only | |||||
| Wu et al., 2016 ** [101] | - | 71/62 healthy + 8 benign (patient samples) *** | 0.007, 0.900 | - | - |
| Grampp et al., 2016 ** [89] | - | 453/453 (patient samples) 138/138 (TCGA) |
<0.05 | - | - |
| Studies focused on the prognostic properties of PVT1 only | |||||
| Posa et al., 2016 [98] | OS | -/- (TCGA) |
- | <0.0001 | - |
| Huang et al., 2017 * [95] | OS | 289/32 (TCGA) | - | 0.024 (panel of 15 lncRNAs) | - |
| Wu et al., 2017 [81] | OS | 55/55 (patient samples) 534/72 (TCGA) |
- | 0.00007 | - |
| Xu et al., 2017 [102] | OS | 530/72 (TCGA) 109/100 (microarray) |
- | 9.15 × 10−7 | - |
| Wang et al., 2018 [100] | OS | 539/72 (TCGA) | - | 0.0002 (panel of 11 lncRNAs) |
1.47, - |
| Liu et al., 2020 [99] | OS | 525/- (TCGA) 60/60 (patient samples) |
- | 00002 | 1.79, 1.32–2.43 |
| Studies showing both the diagnostic and prognostic properties of PVT1 | |||||
| Yang et al., 2017 [73] | OS DFS |
50/50 (patient samples), 534/72 (TCGA) |
<0.001 | 0.014 NS |
1.494, 1.081–2.063 1.469, 0.976–2.211 |
| Bao et al., 2018 [103] | OS DFS |
129/129 (patient samples) | <0.01 | 0.012 0.004 |
4.445, 1.515–8.392 3.553, - |
| Li et al., 2018 [75] | OS | 448/67 (TCGA) 40/40 (patient samples) |
<0.001 | <0.01 |
- |
| Ren et al., 2019 [77] | OS | 25/25 (patient samples) | <0.001 | 0.007 | - |
Interestingly, the association with the tumor stage seemed to be highest in stage III, while in stages I, II, and IV, there were lower proportions of patients with a high level of PVT1 [98]. Conversely, Wu et al. [81] used TCGA data as well; however, they observed an increase in the PVT1 expression with the histological and TNM stages.
The association of PVT1 upregulation with several clinicopathological parameters is popping up repeatedly in several works. The most prominent is the association with the TNM stage [73,77,81,103]. Li et al. [75] identified the association of PVT1 expression with the T, M, and AJCC stages but not the N stage and grade. Other works have frequently identified the association of PVT1 expression with the histological grade, Fuhrman stage, distant metastasis, overall survival, and disease-free survival, and the results typically overlap. In almost all of the reviewed publications, PVT1 upregulation correlates with a shorter survival in RCC patients, even though some exceptions need to be highlighted [102] where PVT1 was not among the most prominent candidates. Although this would seem promising, most of the papers utilized the same TCGA cohorts repeatedly; only some also recruited independent cohorts of RCC patients [73,75,81,89,99]. Typically, the TCGA cohort was used for exploration, while the patient samples served as a validation cohort. Only three papers utilized independently collected patient samples [77,101,103]. An independent cohort showed its importance for Wu et al. [81], who observed differences in the TCGA and independent cohorts, as PVT1 was more upregulated in the TCGA one.
Possibly, the highest aim for biomarker studies is to find a molecule feasible in so-called liquid biopsy— biomarkers detectable in body fluids with little to no burden for the patient, faster and simpler analyses, and precise results. The potential use of lncRNA in the form of liquid biopsy from bodily fluids stems from their stability, although the exact mechanism is still unclear [101]. The encapsulation of lncRNAs in vesicles such as apoptotic bodies, microvesicles, and exosomes or the formation of complexes with proteins might play a part [104,105,106,107]. So far, the only study aimed in this direction was provided by Wu et al. [101]. The authors measured 82 lncRNAs chosen from the literature in the tissues of ccRCC, chromophobe RCC, and PRCC.
Further validation in the tissues and serum of ccRCC patients and healthy controls led to a five lncRNA diagnostic model. Combining lncRNA-LET, PVT1, PANDAR, PTENP1, and linc00963 provided a diagnostic ability with the area under the curve of 0.90. Interestingly, PVT1 was downregulated in the serum ccRCC samples (p = 0.021), which did not correspond with the higher expression in the ccRCC tissues. The authors suggested that this discrepancy might be caused by the mechanism of lncRNA export from the cell. The recruitment of lncRNA into extracellular vesicles can be facilitated through ECV-bound proteins, limiting the capacity of transferable lncRNAs. Therefore, it might not reflect the levels in the tissues [101,108,109].
5. Conclusions
Many new lncRNAs have been described and studied in recent years thanks to the extensive development of sequencing technology and bioinformatics approaches primarily for data assessment. A quick search in PubMed returned more than 400 papers on lncRNAs in renal cell carcinoma. PVT1 has become a staple of the standard RCC lncRNA profile, as it has been repeatedly identified among the most dysregulated lncRNAs in RCC. Its role in regulating the cellular processes has been investigated in several papers, which show that PVT1 is a significant factor promoting tumorigenesis. Association with the development of a tumor creates a biomarker potential, which can be put to good use. Dysregulation of the PVT1 expression is easily detectable and correlates with a specific clinical behavior and RCC subtype.
Although so far, most of the studies used TCGA datasets, several groups also added their independent exploratory cohorts. However, more alarming is the lack of independent validation by qPCR, which belongs to the gold standard of study design when searching for potential diagnostic, prognostic, and predictive biomarkers. Stemming from the use of TCGA datasets, most of the studies were focused solely on ccRCC. Only two [69,82] also included other types of renal cancer, which supports the results in the context of day-to-day diagnostic conditions and expectations from a potential biomarker. The ability to differentiate ccRCC from PRCC, chRCC, and other rarer types is more than desirable.
Nevertheless, PVT1 showed a specific upregulation in ccRCC, which outlined its future use in clinical practices. Moreover, PVT1 seems to be a good marker of the worse prognosis and shorter survival of patients with higher PVT1 levels. Either singularly or in combination with other non-coding RNAs, PVT1 should have its place in therapy personalization in the future for its fundamental biomarker value and the undeniable role in RCC development.
Author Contributions
Conceptualization, O.S. and J.B.; methodology, J.B.; writing—original draft preparation, A.K.; writing—review and editing, J.B.; visualization, J.B.; and supervision and funding acquisition, O.S. All authors have read and agreed to the published version of the manuscript.
Funding
This publication was funded by the Ministry of Health of the Czech Republic, grant no. NV18-03-00554.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Capitanio U., Bensalah K., Bex A., Boorjian S.A., Bray F., Coleman J., Gore J.L., Sun M., Wood C., Russo P. Epidemiology of Renal Cell Carcinoma. Eur. Urol. 2019;75:74–84. doi: 10.1016/j.eururo.2018.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Athanazio D.A., Amorim L.S., da Cunha I.W., Leite K.R.M., da Paz A.R., de Paula Xavier Gomes R., Tavora F.R.F., Faraj S.F., Cavalcanti M.S., Bezerra S.M. Classification of Renal Cell Tumors–Current Concepts and Use of Ancillary Tests: Recommendations of the Brazilian Society of Pathology. Surg. Exp. Pathol. 2021;4:4. doi: 10.1186/s42047-020-00084-x. [DOI] [Google Scholar]
- 4.Warren A.Y., Harrison D. WHO/ISUP Classification, Grading and Pathological Staging of Renal Cell Carcinoma: Standards and Controversies. World J. Urol. 2018;36:1913–1926. doi: 10.1007/s00345-018-2447-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Escudier B., Porta C., Schmidinger M., Rioux-Leclercq N., Bex A., Khoo V., Grünwald V., Gillessen S., Horwich A. ESMO Guidelines Committee Renal Cell Carcinoma: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2019;30:706–720. doi: 10.1093/annonc/mdz056. [DOI] [PubMed] [Google Scholar]
- 6.Lopez-Beltran A., Henriques V., Cimadamore A., Santoni M., Cheng L., Gevaert T., Blanca A., Massari F., Scarpelli M., Montironi R. The Identification of Immunological Biomarkers in Kidney Cancers. Front. Oncol. 2018;8:456. doi: 10.3389/fonc.2018.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jabandziev P., Kakisaka T., Bohosova J., Pinkasova T., Kunovsky L., Slaby O., Goel A. MicroRNAs in Colon Tissue of Pediatric Ulcerative Pancolitis Patients Allow Detection and Prognostic Stratification. J. Clin. Med. 2021;10:1325. doi: 10.3390/jcm10061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brito G.C., Fachel A.A., Vettore A.L., Vignal G.M., Gimba E.R.P., Campos F.S., Barcinski M.A., Verjovski-Almeida S., Reis E.M. Identification of Protein-Coding and Intronic Noncoding RNAs down-Regulated in Clear Cell Renal Carcinoma. Mol. Carcinog. 2008;47:757–767. doi: 10.1002/mc.20433. [DOI] [PubMed] [Google Scholar]
- 9.Yu G., Yao W., Wang J., Ma X., Xiao W., Li H., Xia D., Yang Y., Deng K., Xiao H., et al. LncRNAs Expression Signatures of Renal Clear Cell Carcinoma Revealed by Microarray. PLoS ONE. 2012;7:e42377. doi: 10.1371/journal.pone.0042377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou B., Wang D., Xu K., Liu J., Yuan D., Meng Z., Zhang B. Prognostic Value of Long Non-Coding RNA Plasmacytoma Variant Translocation1 in Human Solid Tumors. Medicine. 2019;98:e16087. doi: 10.1097/MD.0000000000016087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.International Human Genome Sequencing Consortium Finishing the Euchromatic Sequence of the Human Genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 12.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., et al. GENCODE: The Reference Human Genome Annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., et al. Landscape of Transcription in Human Cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frith M.C., Pheasant M., Mattick J.S. The Amazing Complexity of the Human Transcriptome. Eur. J. Hum. Genet. 2005;13:894–897. doi: 10.1038/sj.ejhg.5201459. [DOI] [PubMed] [Google Scholar]
- 15.Fang S., Zhang L., Guo J., Niu Y., Wu Y., Li H., Zhao L., Li X., Teng X., Sun X., et al. NONCODEV5: A Comprehensive Annotation Database for Long Non-Coding RNAs. Nucleic Acids Res. 2018;46:D308–D314. doi: 10.1093/nar/gkx1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ravasi T., Suzuki H., Pang K.C., Katayama S., Furuno M., Okunishi R., Fukuda S., Ru K., Frith M.C., Gongora M.M., et al. Experimental Validation of the Regulated Expression of Large Numbers of Non-Coding RNAs from the Mouse Genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mercer T.R., Dinger M.E., Sunkin S.M., Mehler M.F., Mattick J.S. Specific Expression of Long Noncoding RNAs in the Mouse Brain. Proc. Natl. Acad. Sci. USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L. Integrative Annotation of Human Large Intergenic Noncoding RNAs Reveals Global Properties and Specific Subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., et al. The GENCODE v7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bánfai B., Jia H., Khatun J., Wood E., Risk B., Gundling W.E., Kundaje A., Gunawardena H.P., Yu Y., Xie L., et al. Long Noncoding RNAs Are Rarely Translated in Two Human Cell Lines. Genome Res. 2012;22:1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chooniedass-Kothari S., Emberley E., Hamedani M.K., Troup S., Wang X., Czosnek A., Hube F., Mutawe M., Watson P.H., Leygue E. The Steroid Receptor RNA Activator Is the First Functional RNA Encoding a Protein. FEBS Lett. 2004;566:43–47. doi: 10.1016/j.febslet.2004.03.104. [DOI] [PubMed] [Google Scholar]
- 22.Kondo T., Plaza S., Zanet J., Benrabah E., Valenti P., Hashimoto Y., Kobayashi S., Payre F., Kageyama Y. Small Peptides Switch the Transcriptional Activity of Shavenbaby during Drosophila Embryogenesis. Science. 2010;329:336–339. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- 23.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P., et al. Chromatin Signature Reveals over a Thousand Highly Conserved Large Non-Coding RNAs in Mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L., Duff M.O., Graveley B.R., Carmichael G.G., Chen L.-L. Genomewide Characterization of Non-Polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu H., Yang L., Chen L.-L. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet. 2017;33:540–552. doi: 10.1016/j.tig.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Wang J., Zhang J., Zheng H., Li J., Liu D., Li H., Samudrala R., Yu J., Wong G.K.-S. Mouse Transcriptome: Neutral Evolution of “non-Coding” Complementary DNAs. Nature. 2004;14:431. [PubMed] [Google Scholar]
- 27.Mercer T.R., Dinger M.E., Mattick J.S. Long Non-Coding RNAs: Insights into Functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 28.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The Transcriptional Landscape of the Mammalian Genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 29.Ponjavic J., Ponting C.P., Lunter G. Functionality or Transcriptional Noise? Evidence for Selection within Long Noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marques A.C., Ponting C.P. Catalogues of Mammalian Long Noncoding RNAs: Modest Conservation and Incompleteness. Genome Biol. 2009;10:R124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ørom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q., et al. Long Noncoding RNAs with Enhancer-like Function in Human Cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Statello L., Guo C.-J., Chen L.-L., Huarte M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2020;22:96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Oliveira J.C., Oliveira L.C., Mathias C., Pedroso G.A., Lemos D.S., Salviano-Silva A., Jucoski T.S., Lobo-Alves S.C., Zambalde E.P., Cipolla G.A., et al. Long Non-Coding RNAs in Cancer: Another Layer of Complexity. J. Gene Med. 2019;21:e3065. doi: 10.1002/jgm.3065. [DOI] [PubMed] [Google Scholar]
- 34.Wang K.C., Chang H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L.-L. Linking Long Noncoding RNA Localization and Function. Trends Biochem. Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Schmitz K.-M., Mayer C., Postepska A., Grummt I. Interaction of Noncoding RNA with the RDNA Promoter Mediates Recruitment of DNMT3b and Silencing of RRNA Genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Ruscio A., Ebralidze A.K., Benoukraf T., Amabile G., Goff L.A., Terragni J., Figueroa M.E., De Figueiredo Pontes L.L., Alberich-Jorda M., Zhang P., et al. DNMT1-Interacting RNAs Block Gene-Specific DNA Methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., Thomas K., Presser A., Bernstein B.E., van Oudenaarden A., et al. Many Human Large Intergenic Noncoding RNAs Associate with Chromatin-Modifying Complexes and Affect Gene Expression. Proc. Natl. Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Q., Hao Q., Prasanth K.V. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet. 2018;34:142–157. doi: 10.1016/j.tig.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lam M.T.Y., Li W., Rosenfeld M.G., Glass C.K. Enhancer RNAs and Regulated Transcriptional Programs. Trends Biochem. Sci. 2014;39:170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng J., Bi C., Clark B.S., Mady R., Shah P., Kohtz J.D. The Evf-2 Noncoding RNA Is Transcribed from the Dlx-5/6 Ultraconserved Region and Functions as a Dlx-2 Transcriptional Coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shamovsky I., Ivannikov M., Kandel E.S., Gershon D., Nudler E. RNA-Mediated Response to Heat Shock in Mammalian Cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 43.Yang L., Lin C., Jin C., Yang J.C., Tanasa B., Li W., Merkurjev D., Ohgi K.A., Meng D., Zhang J., et al. LncRNA-Dependent Mechanisms of Androgen-Receptor-Regulated Gene Activation Programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lai F., Orom U.A., Cesaroni M., Beringer M., Taatjes D.J., Blobel G.A., Shiekhattar R. Activating RNAs Associate with Mediator to Enhance Chromatin Architecture and Transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian D., Sun S., Lee J.T. The Long Noncoding RNA, Jpx, Is a Molecular Switch for X Chromosome Inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun S., Del Rosario B.C., Szanto A., Ogawa Y., Jeon Y., Lee J.T. Jpx RNA Activates Xist by Evicting CTCF. Cell. 2013;153:1537–1551. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X., Arai S., Song X., Reichart D., Du K., Pascual G., Tempst P., Rosenfeld M.G., Glass C.K., Kurokawa R. Induced NcRNAs Allosterically Modify RNA-Binding Proteins in Cis to Inhibit Transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martianov I., Ramadass A., Serra Barros A., Chow N., Akoulitchev A. Repression of the Human Dihydrofolate Reductase Gene by a Non-Coding Interfering Transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 49.Ramos A.D., Andersen R.E., Liu S.J., Nowakowski T.J., Hong S.J., Gertz C., Salinas R.D., Zarabi H., Kriegstein A.R., Lim D.A. The Long Noncoding RNA Pnky Regulates Neuronal Differentiation of Embryonic and Postnatal Neural Stem Cells. Cell Stem Cell. 2015;16:439–447. doi: 10.1016/j.stem.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonzalez I., Munita R., Agirre E., Dittmer T.A., Gysling K., Misteli T., Luco R.F. A LncRNA Regulates Alternative Splicing via Establishment of a Splicing-Specific Chromatin Signature. Nat. Struct. Mol. Biol. 2015;22:370–376. doi: 10.1038/nsmb.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gong C., Maquat L.E. LncRNAs Transactivate STAU1-Mediated MRNA Decay by Duplexing with 3’ UTRs via Alu Elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Damas N.D., Marcatti M., Côme C., Christensen L.L., Nielsen M.M., Baumgartner R., Gylling H.M., Maglieri G., Rundsten C.F., Seemann S.E., et al. SNHG5 Promotes Colorectal Cancer Cell Survival by Counteracting STAU1-Mediated MRNA Destabilization. Nat. Commun. 2016;7:13875. doi: 10.1038/ncomms13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu G., Lou Z., Gupta M. The Long Non-Coding RNA GAS5 Cooperates with the Eukaryotic Translation Initiation Factor 4E to Regulate c-Myc Translation. PLoS ONE. 2014;9:e107016. doi: 10.1371/journal.pone.0107016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carrieri C., Cimatti L., Biagioli M., Beugnet A., Zucchelli S., Fedele S., Pesce E., Ferrer I., Collavin L., Santoro C., et al. Long Non-Coding Antisense RNA Controls Uchl1 Translation through an Embedded SINEB2 Repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 55.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A CeRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kino T., Hurt D.E., Ichijo T., Nader N., Chrousos G.P. Noncoding RNA Gas5 Is a Growth Arrest- and Starvation-Associated Repressor of the Glucocorticoid Receptor. Sci. Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee S., Kopp F., Chang T.-C., Sataluri A., Chen B., Sivakumar S., Yu H., Xie Y., Mendell J.T. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell. 2016;164:69–80. doi: 10.1016/j.cell.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoon J.-H., Abdelmohsen K., Kim J., Yang X., Martindale J.L., Tominaga-Yamanaka K., White E.J., Orjalo A.V., Rinn J.L., Kreft S.G., et al. Scaffold Function of Long Non-Coding RNA HOTAIR in Protein Ubiquitination. Nat. Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang F., Zhang H., Mei Y., Wu M. Reciprocal Regulation of HIF-1α and LincRNA-P21 Modulates the Warburg Effect. Mol. Cell. 2014;53:88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 60.Willingham A.T., Orth A.P., Batalov S., Peters E.C., Wen B.G., Aza-Blanc P., Hogenesch J.B., Schultz P.G. A Strategy for Probing the Function of Noncoding RNAs Finds a Repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 61.Noh J.H., Kim K.M., Abdelmohsen K., Yoon J.-H., Panda A.C., Munk R., Kim J., Curtis J., Moad C.A., Wohler C.M., et al. HuR and GRSF1 Modulate the Nuclear Export and Mitochondrial Localization of the LncRNA RMRP. Genes Dev. 2016;30:1224–1239. doi: 10.1101/gad.276022.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vučićević D., Schrewe H., Orom U.A. Molecular Mechanisms of Long NcRNAs in Neurological Disorders. Front. Genet. 2014;5:48. doi: 10.3389/fgene.2014.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Faghihi M.A., Modarresi F., Khalil A.M., Wood D.E., Sahagan B.G., Morgan T.E., Finch C.E., St Laurent G., Kenny P.J., Wahlestedt C. Expression of a Noncoding RNA Is Elevated in Alzheimer’s Disease and Drives Rapid Feed-Forward Regulation of Beta-Secretase. Nat. Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goyal N., Kesharwani D., Datta M. Lnc-Ing Non-Coding RNAs with Metabolism and Diabetes: Roles of LncRNAs. Cell. Mol. Life Sci. 2018;75:1827–1837. doi: 10.1007/s00018-018-2760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu F., Jin L., Jin Y., Nie Z., Zheng H. Long Noncoding RNAs in Autoimmune Diseases. J. Biomed. Mater. Res. A. 2019;107:468–475. doi: 10.1002/jbm.a.36562. [DOI] [PubMed] [Google Scholar]
- 66.Jabandziev P., Bohosova J., Pinkasova T., Kunovsky L., Slaby O., Goel A. The Emerging Role of Noncoding RNAs in Pediatric Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2020;26:985–993. doi: 10.1093/ibd/izaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hennessy E.J. Cardiovascular Disease and Long Noncoding RNAs: Tools for Unraveling the Mystery Lnc-Ing RNA and Phenotype. Circ. Cardiovasc. Genet. 2017;10:e001556. doi: 10.1161/CIRCGENETICS.117.001556. [DOI] [PubMed] [Google Scholar]
- 68.Bhan A., Soleimani M., Mandal S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanahan D., Weinberg R.A. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 70.Gutschner T., Diederichs S. The Hallmarks of Cancer: A Long Non-Coding RNA Point of View. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schmitt A.M., Chang H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hanahan D., Weinberg R.A. Hallmarks of Cancer: The next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 73.Yang T., Zhou H., Liu P., Yan L., Yao W., Chen K., Zeng J., Li H., Hu J., Xu H., et al. LncRNA PVT1 and Its Splicing Variant Function as Competing Endogenous RNA to Regulate Clear Cell Renal Cell Carcinoma Progression. Oncotarget. 2017;8:85353–85367. doi: 10.18632/oncotarget.19743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu D., Luo P., Wang Q., Ye Y., Wang B. LncRNA PVT1 in Cancer: A Review and Meta-Analysis. Clin. Chim Acta. 2017;474:1–7. doi: 10.1016/j.cca.2017.08.038. [DOI] [PubMed] [Google Scholar]
- 75.Li W., Zheng Z., Chen H., Cai Y., Xie W. Knockdown of Long Non-Coding RNA PVT1 Induces Apoptosis and Cell Cycle Arrest in Clear Cell Renal Cell Carcinoma through the Epidermal Growth Factor Receptor Pathway. Oncol. Lett. 2018;15:7855–7863. doi: 10.3892/ol.2018.8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y., Tan Y., Wang H., Xu M., Xu L. Long Non-Coding RNA Plasmacytoma Variant Translocation 1 (PVT1) Enhances Proliferation, Migration, and Epithelial-Mesenchymal Transition (EMT) of Pituitary Adenoma Cells by Activating β-Catenin, c-Myc, and Cyclin D1 Expression. Med. Sci. Monit. 2019;25:7652–7659. doi: 10.12659/MSM.917110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 77.Ren Y., Huang W., Weng G., Cui P., Liang H., Li Y. LncRNA PVT1 Promotes Proliferation, Invasion and Epithelial-Mesenchymal Transition of Renal Cell Carcinoma Cells through Downregulation of MiR-16-5p. OncoTargets Ther. 2019;12:2563–2575. doi: 10.2147/OTT.S190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zeidler R., Joos S., Delecluse H.J., Klobeck G., Vuillaume M., Lenoir G.M., Bornkamm G.W., Lipp M. Breakpoints of Burkitt’s Lymphoma t(8;22) Translocations Map within a Distance of 300 Kb Downstream of MYC. Genes Chromosomes Cancer. 1994;9:282–287. doi: 10.1002/gcc.2870090408. [DOI] [PubMed] [Google Scholar]
- 79.Jin K., Wang S., Zhang Y., Xia M., Mo Y., Li X., Li G., Zeng Z., Xiong W., He Y. Long Non-Coding RNA PVT1 Interacts with MYC and Its Downstream Molecules to Synergistically Promote Tumorigenesis. Cell Mol. Life Sci. 2019;76:4275–4289. doi: 10.1007/s00018-019-03222-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perciavalle R.M., Opferman J.T. Delving Deeper: MCL-1’s Contributions to Normal and Cancer Biology. Trends Cell Biol. 2013;23:22–29. doi: 10.1016/j.tcb.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu Q., Yang F., Yang Z., Fang Z., Fu W., Chen W., Liu X., Zhao J., Wang Q., Hu X., et al. Long Noncoding RNA PVT1 Inhibits Renal Cancer Cell Apoptosis by Up-Regulating Mcl-1. Oncotarget. 2017;8:101865–101875. doi: 10.18632/oncotarget.21706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang H., Li Z.-Y., Xu Z.-H., Chen Y.-L., Lu Z.-Y., Shen D.-Y., Lu J.-Y., Zheng Q.-M., Wang L.-Y., Xu L.-W., et al. The Prognostic Value of MiRNA-18a-5p in Clear Cell Renal Cell Carcinoma and Its Function via the MiRNA-18a-5p/HIF1A/PVT1 Pathway. J. Cancer. 2020;11:2737–2748. doi: 10.7150/jca.36822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Latif F., Tory K., Gnarra J., Yao M., Duh F.M., Orcutt M.L., Stackhouse T., Kuzmin I., Modi W., Geil L. Identification of the von Hippel-Lindau Disease Tumor Suppressor Gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 84.Gnarra J.R., Tory K., Weng Y., Schmidt L., Wei M.H., Li H., Latif F., Liu S., Chen F., Duh F.M. Mutations of the VHL Tumour Suppressor Gene in Renal Carcinoma. Nat. Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 85.Shuin T., Kondo K., Torigoe S., Kishida T., Kubota Y., Hosaka M., Nagashima Y., Kitamura H., Latif F., Zbar B. Frequent Somatic Mutations and Loss of Heterozygosity of the von Hippel-Lindau Tumor Suppressor Gene in Primary Human Renal Cell Carcinomas. Cancer Res. 1994;54:2852–2855. [PubMed] [Google Scholar]
- 86.Herman J.G., Latif F., Weng Y., Lerman M.I., Zbar B., Liu S., Samid D., Duan D.S., Gnarra J.R., Linehan W.M. Silencing of the VHL Tumor-Suppressor Gene by DNA Methylation in Renal Carcinoma. Proc. Natl. Acad. Sci. USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kaelin W.G. Molecular Basis of the VHL Hereditary Cancer Syndrome. Nat. Rev. Cancer. 2002;2:673–682. doi: 10.1038/nrc885. [DOI] [PubMed] [Google Scholar]
- 88.Kaelin W.G. Von Hippel-Lindau Disease. Annu. Rev. Pathol. 2007;2:145–173. doi: 10.1146/annurev.pathol.2.010506.092049. [DOI] [PubMed] [Google Scholar]
- 89.Grampp S., Platt J.L., Lauer V., Salama R., Kranz F., Neumann V.K., Wach S., Stöhr C., Hartmann A., Eckardt K.-U., et al. Genetic Variation at the 8q24.21 Renal Cancer Susceptibility Locus Affects HIF Binding to a MYC Enhancer. Nat. Commun. 2016;7:1–11. doi: 10.1038/ncomms13183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schödel J., Bardella C., Sciesielski L.K., Brown J.M., Pugh C.W., Buckle V., Tomlinson I.P., Ratcliffe P.J., Mole D.R. Common Genetic Variants at the 11q13.3 Renal Cancer Susceptibility Locus Influence Binding of HIF to an Enhancer of Cyclin D1 Expression. Nat. Genet. 2012;44:420–425. doi: 10.1038/ng.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vaishnave S. BMI1 and PTEN Are Key Determinants of Breast Cancer Therapy: A Plausible Therapeutic Target in Breast Cancer. Gene. 2018;678:302–311. doi: 10.1016/j.gene.2018.08.022. [DOI] [PubMed] [Google Scholar]
- 92.Caramel J., Ligier M., Puisieux A. Pleiotropic Roles for ZEB1 in Cancer. Cancer Res. 2018;78:30–35. doi: 10.1158/0008-5472.CAN-17-2476. [DOI] [PubMed] [Google Scholar]
- 93.Yu X., Wang W., Lin X., Zheng X., Yang A. Roles of ZEB2 and RBM38 in Liver Cancer Stem Cell Proliferation. J. BUON. 2020;25:1390–1394. [PubMed] [Google Scholar]
- 94.Wu H., Wei M., Jiang X., Tan J., Xu W., Fan X., Zhang R., Ding C., Zhao F., Shao X., et al. LncRNA PVT1 Promotes Tumorigenesis of Colorectal Cancer by Stabilizing MiR-16-5p and Interacting with the VEGFA/VEGFR1/AKT Axis. Mol. Ther. Nucleic Acids. 2020;20:438–450. doi: 10.1016/j.omtn.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang C., Yuan N., Wu L., Wang X., Dai J., Song P., Li F., Xu C., Zhao X. An Integrated Analysis for Long Noncoding RNAs and MicroRNAs with the Mediated Competing Endogenous RNA Network in Papillary Renal Cell Carcinoma. OncoTargets Ther. 2017;10:4037–4050. doi: 10.2147/OTT.S141951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chu S. Transcriptional Regulation by Post-Transcriptional Modification--Role of Phosphorylation in Sp1 Transcriptional Activity. Gene. 2012;508:1–8. doi: 10.1016/j.gene.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 97.Wang W., Zhou R., Wu Y., Liu Y., Su W., Xiong W., Zeng Z. PVT1 Promotes Cancer Progression via MicroRNAs. Front. Oncol. 2019;9:609. doi: 10.3389/fonc.2019.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Posa I., Carvalho S., Tavares J., Grosso A.R. A Pan-Cancer Analysis of MYC-PVT1 Reveals CNV-Unmediated Deregulation and Poor Prognosis in Renal Carcinoma. Oncotarget. 2016;7:47033–47041. doi: 10.18632/oncotarget.9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu H., Ye T., Yang X., Lv P., Wu X., Zhou H., Zeng J., Tang K., Ye Z. A Panel of Four-LncRNA Signature as a Potential Biomarker for Predicting Survival in Clear Cell Renal Cell Carcinoma. J. Cancer. 2020;11:4274–4283. doi: 10.7150/jca.40421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang J., Zhang C., He W., Gou X. Construction and Comprehensive Analysis of Dysregulated Long Non-Coding RNA-Associated Competing Endogenous RNA Network in Clear Cell Renal Cell Carcinoma. J. Cell. Biochem. 2018;120:2576–2593. doi: 10.1002/jcb.27557. [DOI] [PubMed] [Google Scholar]
- 101.Wu Y., Wang Y.-Q., Weng W.-W., Zhang Q.-Y., Yang X.-Q., Gan H.-L., Yang Y.-S., Zhang P.-P., Sun M.-H., Xu M.-D., et al. A Serum-Circulating Long Noncoding RNA Signature Can Discriminate between Patients with Clear Cell Renal Cell Carcinoma and Healthy Controls. Oncogenesis. 2016;5:e192. doi: 10.1038/oncsis.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu X., Xu Y., Shi C., Wang B., Yu X., Zou Y., Hu T. A Genome-Wide Comprehensively Analyses of Long Noncoding RNA Profiling and Metastasis Associated LncRNAs in Renal Cell Carcinoma. Oncotarget. 2017;8:87773–87781. doi: 10.18632/oncotarget.21206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bao X., Duan J., Yan Y., Ma X., Zhang Y., Wang H., Ni D., Wu S., Peng C., Fan Y., et al. Upregulation of Long Noncoding RNA PVT1 Predicts Unfavorable Prognosis in Patients with Clear Cell Renal Cell Carcinoma. Cancer Biomark. 2017;21:55–63. doi: 10.3233/CBM-170251. [DOI] [PubMed] [Google Scholar]
- 104.Fachel A.A., Tahira A.C., Vilella-Arias S.A., Maracaja-Coutinho V., Gimba E.R.P., Vignal G.M., Campos F.S., Reis E.M., Verjovski-Almeida S. Expression Analysis and in Silico Characterization of Intronic Long Noncoding RNAs in Renal Cell Carcinoma: Emerging Functional Associations. Mol. Cancer. 2013;12:140. doi: 10.1186/1476-4598-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kogure T., Yan I.K., Lin W.-L., Patel T. Extracellular Vesicle–Mediated Transfer of a Novel Long Noncoding RNA TUC339. Genes Cancer. 2013;4:261–272. doi: 10.1177/1947601913499020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hasselmann D.O., Rappl G., Tilgen W., Reinhold U. Extracellular Tyrosinase MRNA within Apoptotic Bodies Is Protected from Degradation in Human Serum. Clin. Chem. 2001;47:1488–1489. doi: 10.1093/clinchem/47.8.1488. [DOI] [PubMed] [Google Scholar]
- 107.Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., Ochiya T. Secretory Mechanisms and Intercellular Transfer of MicroRNAs in Living Cells. J. Biol. Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Batagov A.O., Kuznetsov V.A., Kurochkin I.V. Identification of Nucleotide Patterns Enriched in Secreted RNAs as Putative Cis-Acting Elements Targeting Them to Exosome Nano-Vesicles. BMC Genom. 2011;12(Suppl. 3):S18. doi: 10.1186/1471-2164-12-S3-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Irion U., St Johnston D. Bicoid RNA Localization Requires Specific Binding of an Endosomal Sorting Complex. Nature. 2007;445:554–558. doi: 10.1038/nature05503. [DOI] [PMC free article] [PubMed] [Google Scholar]