Abstract
Ca2+-activated Cl- channel A2 (CLCA2), a tumor suppressor, is associated with the development of several cancers. However, little is known about CLCA2 in human cervical cancer. Therefore, the aim of the present study was to investigate the effects of CLCA2 on cervical cancer. Reverse transcription-quantitative (RT-q)PCR was used to examine the mRNA expression levels of CLCA2 in eight pairs of cervical cancer tissues. Immunohistochemistry was used to investigate CLCA2 protein expression in 144 archived cervical cancer specimens. The association of the CLCA2 with clinicopathological parameters was statistically evaluated. Cell proliferation and invasion capability were examined by MTT and Transwell assays, respectively. RT-qPCR analysis revealed that CLCA2 expression was decreased in cervical cancer compared with that in adjacent normal tissues. The expression levels of CLCA2 in patients were correlated with tumor stage (P=0.028), tumor size (P=0.009), and human papillomavirus (HPV) infection status (P=0.041). In addition, CLCA2 upregulation was associated with longer overall and recurrence-free survival time after surgery (P=0.016 and P=0.009, respectively). Multivariate Cox regression analysis demonstrated that CLCA2 expression had a predictive value for overall survival of patients with cervical cancer (P=0.017 and P=0.025, respectively). Knockdown of CLCA2 by small interfering RNA suppressed tumor cell proliferation and migration. Mechanistically, CLCA2 was involved in Wnt/β-catenin signaling. In conclusion, the results of the present study demonstrated that CLCA2 suppressed the proliferation, migration and invasion of cervical cancer cells, and that CLCA2 may be a potential therapeutic target of cervical cancer.
Keywords: Ca2+-activated Cl- channel A2, cervical cancer, proliferation, metastasis, Wnt
Introduction
Cervical carcinoma (CaCx) is one of the most common cancers and one of the leading causes of cancer death among females, worldwide (1). Despite the recent advances in the understanding and treatment of this distressing disease, the prognosis of patients with CaCx remains unsatisfactory due to the advanced stage of the disease at diagnosis (2). Thus, there is an urgent need for the identification of biological factors in the tumorigenesis and development of CaCx in order to improve prognosis of advanced CaCx.
The Ca2+-activated Cl- channel A2 (CLCA2) is a member of the Ca2+-activated Cl- channel (CACC) family, characterized by a single COOH-terminal transmembrane segment followed by a 16 amino acid cytoplasmic tail, a structure incompatible with function as a channel (3). CLCA2 serves vital roles in a variety of biological processes, including cell proliferation, DNA damage and other stressors (4). In human stratified epithelium, the expression of CLCA2 has been reported to be strongly associated with basal cell adhesion molecules, suggesting its role in basal cell detachment (5). Several studies have focused on the function of CLCA2 in tumorigenesis and tumor development. For example, as a p53-inducible growth inhibitor, CLCA2 induced the expression of p53, cyclin-dependent kinase inhibitors p21 and p27, and cell cycle arrest in breast cancer cells (4). In addition, CLCA2 interacts with the cell junctional protein eva-1 homolog A to mediate homophilic cell-cell adhesion and epithelial-to-mesenchymal transition in breast cancer cells (6). Additionally, an in vivo study reported that CLCA2 is an ultraviolet (UV) B target gene that may serve a role in epidermal differentiation and UV-dependent skin malignancies (7). However, the role of CLCA2 in CaCx has not been well elucidated. The present study focused on the expression of CLCA2 in CaCx and the role of CLCA2 in the prognosis of patients with CaCx.
Materials and methods
Patients and tissue specimens
The samples were collected from 144 cervical cancer patients (mean age, 43.7 years; age range, 25-68 years) who underwent radical hysterectomy from January 2007 to December 2013 at Guangzhou Women and Children's Medical Center (Guangdong, China). The tumor stage of patients was classified according to the 2018 International Federation of Gynecology and Obstetrics staging system (8). The overall survival (OS) and recurrence-free survival (RFS) was calculated as the time from radical hysterectomy to the date of death and recurrence, respectively. The resected specimens were promptly laid out and fixed in 10% phosphate-buffered formalin at room temperature, and then processed for pathological assessment. Additionally, eight pairs of snap-frozen cervical cancer and paired adjacent non-cancerous tissues (>5 cm from the tumor) were collected for reverse transcription-quantitative (RT-q)PCR in 2018 at Guangzhou Women and Children's Medical Center. The tissues were carefully excised, frozen at -20˚C and evaluated to exclude dysplasia and the presence of inflammatory cells.
This study was approved by the Research Ethics Committee of Guangzhou Women and Children's Medical Center (approval no. GWCMC201224) and written informed consent was obtained from each patient.
Cell Lines
Cervical cancer cell lines, SiHa, C33A, HeLa, CaSki, and ME-180 were cultured in DMEM medium (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.). Primary normal cervical epithelial cells (NCEC) were obtained from the patient's fresh tissues and cultured in keratinocyte serum-free medium (Invitrogen; Thermo Fisher Scientific, Inc.) (9).
Small interfering RNA (siRNA) transfection
CLCA2 knockdown was performed by transfecting the synthetic siRNA duplexes in SiHa and CaSki cells (3x105 cells/well) for 24-96 h. The CLCA2 siRNA sequences were designed and synthesized by RiboBio Co., Ltd. The sequences of the sense strand of siRNA duplexes were as follows: SiControl, 5'-CATTAATGTCGGACAAC-3'; CLCA2-Ri1, 5'-CGAAGTTCTGTTACCCATA-3'; CLCA2-Ri2, 5'-GGTCGTTGTATAAGTCGAA-3'. Cells were transfected with siRNA using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Briefly, a total of 5 µl Lipofectamine® 2000 and 20 µM siRNA were added into 120 µl 1X RiboFECT™ CP Buffer (Guangzhou RiboBio Co., Ltd.), and the mixture incubated for 5 min at 37˚C. Subsequently, 12 µl RiboFECT™ CP Reagent (Guangzhou RiboBio Co., Ltd.) was added into the mixture and incubated for 15 min at 37˚C. The riboFECT™ CP mixture was diluted with 1,863 µl RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) to a total volume of 2 ml. The cells were cultured for another 48 h, at which point the subsequent experiments were performed. The non-specific siRNA vector was used as a control.
RT-qPCR
Total RNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The resulting RNA was subsequently reversed transcribed into cDNA using PrimeScript™ RT Master Mix (Takara Biotechnology Co., Ltd.). qPCR was performed using the SYBR-Green mix kit (Bio-Rad Laboratories, Inc.). The thermocycling conditions of PCR amplification consisted of an initial denaturation at 95˚C for 10 min, followed by 40 cycles of denaturation at 95˚C for 10 sec and annealing at 60˚C for 30 sec. The primers used were as follows: CLCA2 forward, 5'-ACAGGCTGGTGACAAAGTGGTC-3' and reverse, 5'-ACTGGCAATGCCCACGAAGGTA-3'; β-actin forward, 5'-TCATGAAGTGTGACGTTGACATCCGT-3' and reverse 5'-CCTAGAAGCATTTGCGGTGCACGATG-3'. The 2-ΔΔCq method was used to calculate relative expression of CLCA2 mRNA (10).
Immunohistochemistry (IHC)
Tissues were fixed in 4% paraformaldehyde solution at room temperature for 24 h, embedded in paraffin and sliced into sections (4 µM thick). Sections were then dewaxed and rehydrated in an ethanol series (100, 95, 75 and 50%, each for 5 min). Endogenous peroxidase activity was blocked by immersing the sections in 3% H2O2 in methanol for 10 min at room temperature. The sections were blocked with 1% BSA for 15 min at room temperature. and then incubated overnight at 4˚C with rabbit anti-CLCA2 antibody (1:200; cat. no. HPA047192; Sigma-Aldrich; Merck KGaA). The following day, the slides were incubated with biotinylated anti-rabbit secondary antibody (1:1,000; cat. no. 66467-1-Ig; Proteintech Group, Inc.) for 30 min at room temperature, followed by further incubation with streptavidin-horseradish peroxidase (HRP) at 37˚C for 30 min. Slides were observed under a light microscope (magnification, x40).
The CLCA2 IHC results were assessed by two pathologists using a blind protocol design. The analysis was assessed according to both the intensity of staining and the proportion of positively stained tumor cells. The area of staining was stratified into five scoring groups: 0, not detected; 1, <10% positive cells; 2, 11-50% positive cells; 3, 51-80% positive cells; 4, >80% positive cells. Intensity of stained cells was graded into four levels: 0, no staining; 1, weak staining; 2, moderate staining; 3, strong staining. Using this method, the expression of CLCA2 was evaluated by the staining index (SI; multiplying values of the above two scores). The SI score of ≥6 was considered as high CLCA2 expression and SI <6 was considered as low CLCA2 expression.
Western blotting
Western blotting was performed according to standard methods as previously described (11). Cells and tissues were placed in RIPA lysis buffer (Beyotime Institute of Biotechnology) at 4˚C for 1 h. A Pierce BCA Protein assay kit (Thermo Fisher Scientific, Inc.) was used to quantify protein expression. Protein samples (30 µg/lane) were separated using 8-15% sodium dodecyl sulfate-polyacrylamide gels and transferred to PVDF membranes. After the blots were blocked in 5% milk for 1 h at room temperature, the membranes were incubated overnight at 4˚C. With primary antibodies against CLCA2 (1:5,000, cat. no. HPA047192; Sigma-Aldrich; Merck KGaA), β-catenin (1:4,000; cat. no. ab22656; Abcam). The membranes were then incubated with HRP-conjugated anti-rabbit secondary antibody (1:4,000; cat. no. ab6112; Abcam) for 2 h at room temperature. A membrane-enhanced chemiluminescence reagent kit (Amersham; Cytiva) was used to detect the immunoreactive bands. Nuclear extracts were prepared using the Nuclear Extraction kit (Active Motif, Inc.), according to the manufacturer's instructions. Anti-GAPDH (1:5,000; cat. no. ab9485; Abcam) and anti-P84 (1:10,000; cat. no. ab131268; Abcam) antibodies were used as loading controls.
MTT assay
Cell viability was measured using MTT assay. After CLCA2 knockdown, growing cells (2x103/well) were seeded in 96-well plates. At each time point (day 1, 2, 3, 4 and 5), cells were stained with 100 µl sterile MTT dye (0.5 mg/ml, Sigma-Aldrich; Merck KGaA), followed by removal of the culture medium and addition of 150 µl of dimethyl sulfoxide to dissolve the purple formazan (Sigma-Aldrich; Merck KGaA). Absorbance was determined at 570 nm immediately. Experiments were repeated three times, and data were represented as the mean ± SEM.
Cell migration and invasion assay
Cell migration and invasion abilities were analyzed by Transwell chamber assays. A total of 200 µl cell medium (1x105 cells/100 µl in RPMI 1640 medium) were added to the upper chamber of Transwell equipment, and medium containing 10% FBS (HyClone; GE Healthcare Life Sciences) was added to the lower chamber. For invasion assays, Matrigel-coated Transwell cell culture chambers (1:5; BD Biosciences) were introduced. After 24 h, SiHa and CaSki cells on the upper side of the filter were removed with a cotton swab. Migrated and invaded cells on the lower membrane surface were fixed in 4% paraformaldehyde for 30 min at room temperature, stained with 0.1% crystal violet for 15 min at 25˚C and visualized using a light microscope (Olympus Corp.; magnification, x20).
Gene set enrichment analysis (GSEA)
GSEA was performed using the GSEA platform (version 2.0.9; http://www.broadinstitute.org/gsea/) to analyze the correlation between CLCA2 expression and Wnt pathway.
Statistical analysis
All statistical analyses were performed with SPSS 18.0 software (SPSS, Inc.). Comparison of CLCA2 expression between different groups was assessed with paired Student's t-tests or one-way analysis of variance (ANOVA) followed by a Student-Newman-Keuls post-hoc test. Continuous variables were expressed as mean ± SD. A standard χ2 test was performed to assess the association between CLCA2 expression and the clinicopathological characteristics. OS and RFS curves were obtained by the Kaplan-Meier method and were compared with the log-rank test. Multivariate analysis was performed using the Cox regression model. P<0.05 was considered to indicate a statistically significant difference.
Results
CLCA2 is downregulated in cervical cancer
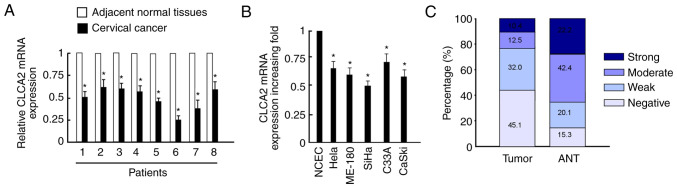
RT-qPCR revealed that CLCA2 mRNA expression levels were significantly downregulated in cervical cancer tissues compared with those in adjacent normal tissues (Fig. 1A). Additionally, CLCA2 mRNA expression levels were significantly decreased in cervical cancer cell lines compared with those in NCEC (Fig. 1B). CaSki and SiHa cells were selected for further analysis as their expression levels were decreased more obviously in cervical cancer lines compared with NCEC. Additionally, CLCA2 staining intensity was scored as strong or moderate intensity in 22.9% of tumor tissues, whereas 64.6% of corresponding adjacent normal cervical tissues were scored as strong or moderate intensity (Fig. 1C).
Figure 1.
Expression pattern of CLCA2 in cervical cancer. (A) CLCA2 mRNA expression in eight pairs of cervical cancer and adjacent normal cervical tissues. *P<0.05 vs. cervical cancer. (B) CLCA2 mRNA expression in cervical cancer cells and normal cervical epithelial cells. *P<0.05 vs. NCEC. (C) Bar graph demonstrating the % of different staining intensity for CLCA2 in 144 patients with cervical cancer. CLCA2, Ca2+-activated Cl- channel A2; ANT, adjacent normal tissues; NCEC, normal cervical epithelial cells.
Correlation of CLCA2 expression and clinicopathological features
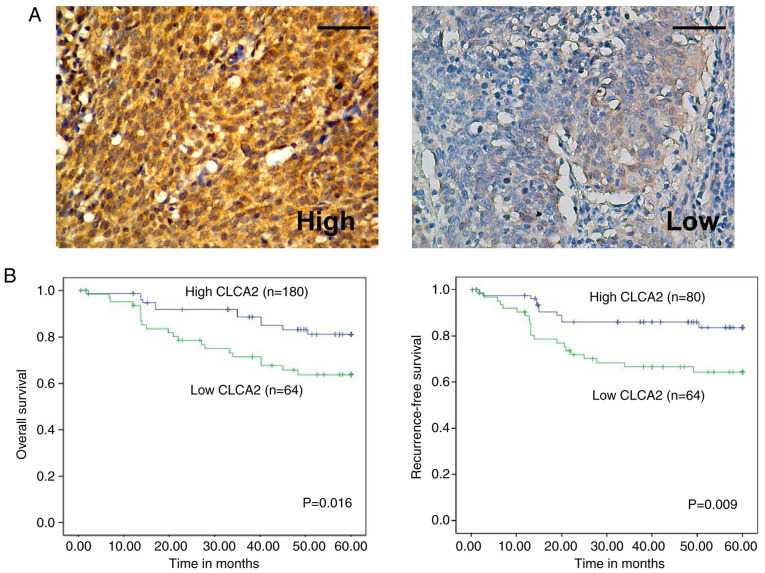
The expression of CLCA2 in 144 cervical cancer specimens was examined by IHC. In Table I, patient age was divided at 45 years on the basis of several previous studies (12). The representative immunostaining of high and low CLCA2 expression in cervical cancer tissues was demonstrated in Fig. 2A. Statistical analysis further revealed a positive correlation between CLCA2 expression and tumor size (P=0.009), tumor stage (P=0.028) and HPV status (P=0.041; Table I). In addition, no significant differences were observed between high and low expression levels of CLCA2 in terms of patient's age, tumor differentiation, histological type or lymph node involvement (Table I).
Table I.
The association between clinical parameters with Ca2+-activated Cl- channel A2.
| CLCA2 | ||||
|---|---|---|---|---|
| Features | Total | High | Low | P-value |
| Age, years | ||||
| ≤45 y | 90 | 46 | 44 | 0.166 |
| >45 y | 54 | 34 | 20 | |
| Tumor stage | ||||
| IB | 95 | 59 | 36 | 0.028a |
| IIA | 49 | 21 | 28 | |
| Tumor size, cm | ||||
| ≤4 | 87 | 56 | 31 | 0.009a |
| >4 | 57 | 24 | 33 | |
| Differentiation | ||||
| 1/2 | 108 | 43 | 65 | 0.174 |
| 3 | 36 | 19 | 17 | |
| Histological type | ||||
| Squamous cell cancer | 89 | 49 | 40 | 0.878 |
| Adenocarcinoma | 55 | 31 | 24 | |
| Human papillomavirus 16/18 | ||||
| + | 115 | 59 | 56 | 0.041a |
| - | 29 | 21 | 8 | |
| LN metastasis | ||||
| No | 110 | 64 | 46 | 0.254 |
| Yes | 34 | 16 | 18 | |
| Total | 144 | 80 | 64 | |
aP<0.05. CALC2, Ca2+-activated Cl- channel A2. LN, lymph nodes.
Figure 2.
CLCA2 expression is correlated with the clinical prognosis of patients with cervical cancer. (A) Representative immunostaining images of CLCA2 in cervical cancer. Magnification, x400; scale bar, 50 µm. (B) Kaplan-Meier curves for overall survival and recurrence-free survival based on CLCA2 expression in patients with cervical cancer. CLCA2, Ca2+-activated Cl- channel A2.
CLCA2 expression predicts favorable prognosis in cervical cancer
Statistical analysis also revealed a correlation between CLCA2 expression and patients' clinical prognosis. Kaplan-Meier analysis indicated that higher expression of CLCA2 was correlated with more favorable OS (P=0.016) and RFS (P=0.009; Fig. 2B). Multivariate Cox regression analysis revealed that CLCA2 expression was an independent predictor for OS (P=0.017) and RFS (P=0.025) in patients with CaCx (Table II).
Table II.
Multivariate Cox proportional hazards regression models for estimating overall survival and recurrence-free survival.
| Overall survival | Recurrence-free survival | |||
|---|---|---|---|---|
| Prognostic variables | Hazard ratio (95% CI) | P-values | Hazard ratio (95% CI) | P-values |
| Age, years | ||||
| ≤45 vs. >45 y | - | - | ||
| Tumor stage | ||||
| IB vs. IIA | 2.769 (0.401-7.158) | 0.022a | - | |
| Tumor size, cm | ||||
| ≤4 vs. >4 | 2.293 (0.097-8.340) | 0.034a | - | |
| Histological type | ||||
| SCC vs. AC | - | - | ||
| LN metastasis | ||||
| + vs. - | - | 33.278 (1.071-10.432) | 0.026a | |
| HPV 16/18 | ||||
| + vs. - | 2.178 (0.242-5.219) | 0.015a | - | |
| CLCA2 expression | ||||
| Low vs. high | 2.838 (0.651-6.519) | 0.017a | 3.265 (1.282-7.984) | 0.025a |
aP<0.05. CI, confidence interval; SCC, squamous cell cancer; AC, adenocarcinoma; LN, lymph nodes; HPV, human papillomavirus; CLCA2, Ca2+-activated Cl- channel A2.
CLCA2 inhibition promotes CaCx cell proliferation, migration, and invasion
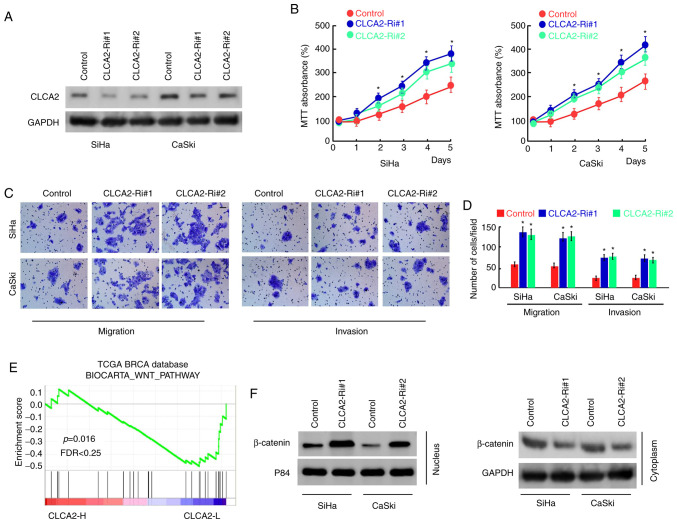
To further examine the role of CLCA2 in CaCx cells, CLCA2 expression was inhibited using siRNAs in SiHa and CaSki cells (Fig. 3A). The results of MTT assays demonstrated that the silencing of CLCA2 expression significantly increased the proliferation of SiHa and CaSki cells compared with that of the control group (Fig. 3B). In addition, Transwell assays demonstrated that knockdown of CLCA2 effectively promoted CaCx cell migration and invasion (Fig. 3C and D). These results indicated that CLCA2 inhibits the proliferation, migration and invasion of CaCx cells.
Figure 3.
Depletion of CLCA2 increases cervical cancer cell proliferation and migration. (A) Western blot analysis of CLCA2 expression in vector and in cervical cancer cell lines stably silencing CLCA2. (B) Inhibition of CLCA2 increases growth rate of cervical cancer cells, determined by MTT assay. *P<0.05 vs. control group; (C) Representative micrographs and (D) quantification of Transwell assay in SiHa and CaSki cell lines demonstrating knockdown of CLCA2 promotes migration and invasion of cancer cells. Scale bar, 50 µm. *P<0.05 vs. control group; (E) Gene set enrichment analysis plot demonstrating that CLCA2 expression negatively correlated with Wnt/β-catenin-activated gene signatures in published cervical cancer gene expression profiles. (F) Western blotting analysis of β-catenin expression in the cytoplasm and nucleus of the indicated cells. GAPDH and P84 were used as loading controls for the cytoplasmic and nuclear fractions, respectively. CLCA2, CLCA2, Ca2+-activated Cl- channel A2; Ri, small interfering RNA; FDR, false discover rate.
CLCA2 suppresses Wnt/β-catenin signaling in cervical cancer cells
In order to further explore the mechanism of CLCA2 regulating cell functions, the publicly available gene expression array data for CaCx using GSEA was analyzed. The results demonstrated that CLCA2 expression was negatively correlated with the activation of the Wnt/β-catenin pathway (Fig. 3E). In addition, the effect of CLCA2 on the subcellular localization of β-catenin was assessed. The results demonstrated that knockdown of CLCA2 increased the translocation of β-catenin into the nucleus (Fig. 3F), indicating that CLCA2 suppressed the activation of Wnt/β-catenin signaling in cervical cancer cells.
Discussion
This study demonstrated that CLCA2 functions as a tumor suppressor to inhibit CaCx progression for the very first-time, to the best of our knowledge. This study also demonstrated that the expression of CLCA2 is negatively correlated with tumor stage, tumor size, HPV status and the prognosis of patients. In the in vitro assays, decreased expression of CLCA2 promoted CaCx cell proliferation, migration and invasion. These findings suggested that CLCA2 may function as a tumor-suppressive protein in CaCx and may be regarded as a potential prognostic marker of CaCx.
The CLCA proteins, originally named Cl- channels, have various members expressed throughout epithelia, endothelia, and other cell types, and mediate a constitutively expressed endogenous Cl- current (13). The CLCAs are transmembrane proteins and distinguished by the juxtaposition of metalloprotease and VWA domains and the capacity to self-cleave (14). Overexpression of CLCA proteins produce a novel Cl- current that can be activated by Ca2+ and blocked by typical Cl- channel inhibitors, which results in their designation as Cl- channels (15). As a type I integral transmembrane protein, CLCA2 is induced by p53 in response to cell detachment, DNA damage and other stressors (4). CLCA2 was originally identified as a tumor suppressor in breast cancer, and its ectopic expression retarded tumor formation (16). A subsequent study revealed that CLCA2 was downregulated in cervical and ovarian cancer cells, suggesting its potential tumor-suppressing role in these malignancies (17). The results of the present study demonstrated that CLCA2 expression was decreased in CaCx compared with that in adjacent normal cervix tissues. This study also demonstrated significant association between CLCA2 and tumor size, tumor stage and HPV status, indicating the specific involvement of CLCA2 in CaCx development.
CLCA2 has been reported to negatively regulate cancer cell migration and invasion (18), however, the underlying mechanism has not been defined yet. Qiang et al (19) demonstrated that CLCA2 inhibits growth and metastasis of nasopharyngeal carcinoma cells via inhibition of focal adhesion kinase (FAK)/extracellular signal-regulated kinases signaling. In corroboration with their study, Sasaki et al (18) revealed that CLCA2 function as a target of P53 to inhibit cancer cell migration and invasion through suppression of the FAK signaling pathway. The results of the present study demonstrated that attenuated expression of CLCA2 increased β-catenin nuclear translocation in CaCx cells, suggesting that the tumor-suppressing role of CLCA2 in CaCx is related to the inactivation of Wnt/β-catenin signaling. To the best of our knowledge, this study is the first study to suggest that CLCA2 may contribute to the development of CaCx. As CLCA2 is a CACC regulator, the underlying mechanisms of CLCA2 mediated cancer cell migration and invasion need further investigation.
The clinical significance of CLCA2 has been shown in previous studies. Jiang et al (20) investigated the expression of acute myeloid leukemia 1 protein (AML1) fusion genes and demonstrated that AML1-CLCA2 can be potential molecular markers for diagnostics and prognostic evaluation of acute myeloid leukemia. Man et al (21) suggested that the combination of the four markers cytokine 7, CLCA2, hyaluronan mediated motility receptor or telomerase reverse transcriptase has great value in lung adenocarcinoma prognosis evaluation. However, the role of CLCA2 in predicting the survival of cancer patients has never been reported. This study demonstrated that patients with lower levels of CLCA2 had a significantly poorer OS and RFS compared with patients with higher levels of CLCA2, suggesting that CLCA2 may be used as a prognostic marker for predicting clinical outcome of patients with CaCx.
In conclusion, this is the first study that explored the role of CLCA2 in CaCx and demonstrated that CLCA2 may act as a tumor suppressor in CaCx, to the best of our knowledge. Downregulation of CLCA2 in CaCx, which is strongly correlated with tumor progression and clinical prognosis, merits further development as a prognostic marker.
Acknowledgements
Not applicable.
Funding Statement
Funding: The present study was supported by the Innovation Fund of Guangzhou Women and Children's Medical Center (grant no. 201513521).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
PZ participated in data collection and drafted the manuscript. YLin performed the statistical analysis. YLiu participated in the design of the study. All authors analyzed and interpreted the patient data. PZ and YLiu confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Research Ethics Committee of Guangzhou Women and Children's Medical Center (approval no. GWCMC201224). Written informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Peiretti M, Zapardiel I, Zanagnolo V, Landoni F, Morrow CP, Maggioni A. Management of recurrent cervical cancer: A review of the literature. Surg Oncol. 2012;21:e59–e66. doi: 10.1016/j.suronc.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Cerrotta A, Gardan G, Cavina R, Raspagliesi F, Stefanon B, Garassino I, Musumeci R, Tana S, De Palo G. Concurrent radiotherapy and weekly paclitaxel for locally advanced or recurrent squamous cell carcinoma of the uterine cervix. A pilot study with intensification of dose. Eur J Gynaecol Oncol. 2002;23:115–119. [PubMed] [Google Scholar]
- 3.Elble RC, Walia V, Cheng HC, Connon CJ, Mundhenk L, Gruber AD, Pauli BU. The putative chloride channel hCLCA2 has a single C-terminal transmembrane segment. J Biol Chem. 2006;281:29448–29454. doi: 10.1074/jbc.M605919200. [DOI] [PubMed] [Google Scholar]
- 4.Walia V, Ding M, Kumar S, Nie D, Premkumar LS, Elble RC. hCLCA2 is a p53-inducible inhibitor of breast cancer cell proliferation. Cancer Res. 2009;69:6624–6632. doi: 10.1158/0008-5472.CAN-08-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connon CJ, Kawasaki S, Yamasaki K, Quantock AJ, Kinoshita S. The quantification of hCLCA2 and colocalisation with integrin beta4 in stratified human epithelia. Acta Histochem. 2005;106:421–425. doi: 10.1016/j.acthis.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Ramena G, Yin Y, Yu Y, Walia V, Elble RC. CLCA2 interactor EVA1 is required for mammary epithelial cell differentiation. PLoS One. 2016;11(e0147489) doi: 10.1371/journal.pone.0147489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bart G, Hämäläinen L, Rauhala L, Salonen P, Kokkonen M, Dunlop TW, Pehkonen P, Kumlin T, Tammi MI, Pasonen-Seppänen S, Tammi RH. rClca2 is associated with epidermal differentiation and is strongly downregulated by ultraviolet radiation. Br J Dermatol. 2014;171:376–387. doi: 10.1111/bjd.13038. [DOI] [PubMed] [Google Scholar]
- 8.Grigsby PW, Massad LS, Mutch DG, Powell MA, Thaker PH, McCourt C, Hagemann A, Fuh K, Kuroki L, Schwarz JK, et al. FIGO 2018 staging criteria for cervical cancer: Impact on stage migration and survival. Gynecol Oncol. 2020;157:639–643. doi: 10.1016/j.ygyno.2020.03.027. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Ou J, Guo Y, Dai T, Li X, Liu J, Xia M, Liu L, He M. TBLR1 is a novel prognostic marker and promotes epithelial-mesenchymal transition in cervical cancer. Br J Cancer. 2014;111:112–124. doi: 10.1038/bjc.2014.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 11.Wang B, Zhang L, Zhao L, Zhou R, Ding Y, Li G, Zhao L. LASP2 suppresses colorectal cancer progression through JNK/p38 MAPK pathway meditated epithelial-mesenchymal transition. Cell Commun Signal. 2017;15(21) doi: 10.1186/s12964-017-0179-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park S, Kim J, Eom K, Oh S, Kim S, Kim G, Ahn S, Park KH, Chung D, Lee H. microRNA-944 overexpression is a biomarker for poor prognosis of advanced cervical cancer. BMC Cancer. 2019;19(419) doi: 10.1186/s12885-019-5620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pauli BU, Abdel-Ghany M, Cheng HC, Gruber AD, Archibald HA, Elble RC. Molecular characteristics and functional diversity of CLCA family members. Clin Exp Pharmacol Physiol. 2000;27:901–905. doi: 10.1046/j.1440-1681.2000.03358.x. [DOI] [PubMed] [Google Scholar]
- 14.Pawlowski K, Lepisto M, Meinander N, Sivars U, Varga M, Wieslander E. Novel conserved hydrolase domain in the CLCA family of alleged calcium-activated chloride channels. Proteins. 2006;63:424–439. doi: 10.1002/prot.20887. [DOI] [PubMed] [Google Scholar]
- 15.Fuller CM, Ji HL, Tousson A, Elble RC, Pauli BU, Benos DJ. Ca(2+)-activated Cl(-) channels: A newly emerging anion transport family. Pflugers Archiv. 2001;443 (Suppl 1):S107–S110. doi: 10.1007/s004240100655. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Cowell JK, Sossey-Alaoui K. CLCA2 tumour suppressor gene in 1p31 is epigenetically regulated in breast cancer. Oncogene. 2004;23:1474–1480. doi: 10.1038/sj.onc.1207249. [DOI] [PubMed] [Google Scholar]
- 17.Zhao G, Chen J, Deng Y, Gao F, Zhu J, Feng Z, Lv X, Zhao Z. Identification of NDRG1-regulated genes associated with invasive potential in cervical and ovarian cancer cells. Biochem Biophys Res Commun. 2011;408:154–159. doi: 10.1016/j.bbrc.2011.03.140. [DOI] [PubMed] [Google Scholar]
- 18.Sasaki Y, Koyama R, Maruyama R, Hirano T, Tamura M, Sugisaka J, Suzuki H, Idogawa M, Shinomura Y, Tokino T. CLCA2, a target of the p53 family, negatively regulates cancer cell migration and invasion. Cancer Biol Ther. 2012;13:1512–1521. doi: 10.4161/cbt.22280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiang YY, Li CZ, Sun R, Zheng LS, Peng LX, Yang JP, Meng DF, Lang YH, Mei Y, Xie P, et al. Along with its favorable prognostic role, CLCA2 inhibits growth and metastasis of nasopharyngeal carcinoma cells via inhibition of FAK/ERK signaling. J Exp Clin Cancer Res. 2018;37(34) doi: 10.1186/s13046-018-0692-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang MM, Gao L, Jing Y, Ding Y, Xu YY, Zhou MH, Ma C, Wang N, Wang W, Han XP, et al. Rapid detection of AML1 associated fusion genes in patients with adult acute myeloid leukemia and its clinical significance. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2013;21:821–829. doi: 10.7534/j.issn.1009-2137.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Man Y, Cao J, Jin S, Xu G, Pan B, Shang L, Che D, Yu Q, Yu Y. Newly identified biomarkers for detecting circulating tumor cells in lung adenocarcinoma. Tohoku J Exp Med. 2014;234:29–40. doi: 10.1620/tjem.234.29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.