Abstract
Inorganic polyphosphate (polyP) is a linear polymer composed of up to a few hundred orthophosphates linked together by high-energy phosphoanhydride bonds, identical with those found in ATP. In mammalian mitochondria, polyP has been implicated in multiple processes, including energy metabolism, ion channels function, and the regulation of calcium signaling. However, the specific mechanisms of all these effects of polyP within the organelle remain poorly understood. The central goal of this study was to investigate how mitochondrial polyP participates in the regulation of the mammalian cellular energy metabolism. To accomplish this, we created HEK293 cells depleted of mitochondrial polyP, through the stable expression of the polyP hydrolyzing enzyme (scPPX). We found that these cells have significantly reduced rates of oxidative phosphorylation (OXPHOS), while their rates of glycolysis were elevated. Consistent with this, metabolomics assays confirmed increased levels of metabolites involved in glycolysis in these cells, compared with the wild-type samples. At the same time, key respiratory parameters of the isolated mitochondria were unchanged, suggesting that respiratory chain activity is not affected by the lack of mitochondrial polyP. However, we detected that mitochondria from cells that lack mitochondrial polyP are more fragmented when compared with those from wild-type cells. Based on these results, we propose that mitochondrial polyP plays an important role as a regulator of the metabolic switch between OXPHOS and glycolysis.
Introduction
Inorganic polyphosphate (polyP) is an ancient molecule that has been well-conserved throughout evolution and is present in all studied organisms [1]. It is composed of tens to hundreds of units of orthophosphate linked together by high-energy phosphoanhydride bonds. In mammalian cells and tissues, polyP is a ubiquitous molecule, present in many different intracellular locations, including mitochondria [1]. In fact, in these organisms, mitochondria appear to be one of the preferred locations of polyP [1–3].
PolyP, which bonds are isoenergetic to those found in ATP [4], has been proposed to play a crucial role in cellular energy storage, allowing for the stored energy to be easily available under conditions of cellular stress [5,6]. In fact, the role of this polymer as a key energy metabolite has already been demonstrated in a wide-variety of organisms and models [2,3,7–14]. Consistent with its high energy feature and its preferred mitochondrial localization, polyP was recently proposed of being produced and hydrolyzed by the mammalian mitochondrial ATP synthase [15]. It has been further hypothesized that polyP might regulate the mitochondrial energy metabolism in mammalian cells, either through its direct participation in the ATP production or through the regulation of the rates of key enzymatic activities [6,11]. As a result, in mammalian models, amorphous polyP has been shown to have a protective effect when cells experience decreased levels of ATP as a result of the presence of stressors, such as amyloid peptides [12].
PolyP and other polyphosphate compounds have also been proposed to play a role in energy metabolism in other organisms. Specifically, it has been shown that inositol pyrophosphate represses mitochondrial functionality and enhances glycolysis in yeast [16]. The authors hypothesized that the ability of inositol pyrophosphate to regulate the cellular energetic metabolism is the result of the interaction between inositol pyrophosphates, ATP, and polyP. Moreover, accumulation of polyP has also found to play a key role in the energy metabolism of Dictyostelium discouideum, which show a 2.5-fold decrease in their ATP levels when the polyphosphate synthesizing enzyme PPK1 is deleted [17]. In addition to its potentially direct involvement into cellular energy metabolism, mitochondrial polyP has also been implied in the regulation of the mitochondrial permeability transition pore (mPTP) [18–21] and in the buffering of mitochondrial calcium [3,7,14]. Both mPTP and mitochondrial calcium homeostasis are closely related to the bioenergetic status of the cells.
Dysfunctional cellular bioenergetics is a crucial component in the etiopathology of many diseases and dysfunctions, ranging from diabetes to many neurodegenerative disorders, and cancer [22–24]. Interestingly, polyP has been shown to protect cells against stressors commonly associated with dysfunctional bioenergetics, such as increased oxidative stress or the presence of amyloids [4,12,25–29]. However, it is unclear whether these effects are a direct consequence of polyP’s role in regulating bioenergetics and/or are related to other functions of this multifaceted polymer.
The central aim of the current study was to elucidate the relationship between mitochondrial polyP levels and the status of the cellular energy metabolism in mammalian cells. To do this, we compared ATP levels and mitochondrial bioenergetics parameters in Wild-type (Wt) HEK 293 cells and cells lacking mitochondrial polyP (MitoPPX). Our studies demonstrated that MitoPPX cells have decreased ATP levels, lower rates of OXPHOS and increased levels of glycolysis, compared with Wt cells. Yet mitochondria isolated from the two cell lines showed no significant functional differences. Subsequent morphological and biochemical analysis revealed that cells lacking mitochondrial polyP show significant mitochondrial fragmentation, compatible with increased mitochondrial fission, even if this point should be further investigated. Based on these data, we propose that mitochondrial polyP is a previously unrecognized regulator of the metabolic balance between OXPHOS and glycolysis.
Material and methods
Reagents
Dulbecco’s Modified Eagle medium (DMEM), penicillin-streptomycin, Hank’s Balanced Salt Solution (HBSS), G418, trypsin, and heat-inactivated fetal bovine serum (FBS) were purchased from Gibco-Invitrogen (Carlsbad, California, U.S.A.); Phosphate-Buffered Saline (PBS), poly-L-lysine, Tris-buffered saline (TBS), Tris (hydroxymethyl)-1,3-propanediol hydrochloride (TRIS-HCl), Glycerol, Bovine Serum Albumina (BSA), Phenylmethylsulfonyl fluoride (PMSF), Tween-20, potassium chloride, pyruvate solution, L-malate, MOPS (3-(N-Morpholino) propanesulfonic acid), magnesium chloride, EGTA (ethylene glycol tetraacetic acid) ammonium acetate, mannitol, sucrose, dipotassium phosphate and HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) from Sigma–Aldrich (San Luis, Missouri, U.S.A.); tetramethylrhodamine methyl ester (TMRM), Alkaline Phosphatase (1 U/μl), 4′−6-diamino-2-phenylindole (DAPI), Pierce BCA Protein Assay kit and Pierce ECL Western Blotting Substrate form ThermoFisher Scientific (Waltham, Massachusetts, U.S.A.); Calcein-AM from BD Pharmingen (San Jose, California, U.S.A.); microscopy glasses and the Optima Grade reagents for the metabolomics assays (acetonitrile, methanol and acetic acid) from ThermoFisher Scientific (Waltham, Massachusetts, U.S.A.); secondary antibodies, protein ladders and polyacrylamide precast gels from Bio-Rad (Hercules, California, U.S.A.), and anti-OXPHOS, anti-TOMM20, and anti-pDrp1 antibodies and ATP luminescent measurement kit from AbCam (Cambridge, Cambridge, U.K.). All the materials, drugs and reagents used on the Seahorse experiments were purchased from Agilent Technologies (Santa Clara, California, U.S.A.) and all the materials and reagents used in the Western Blotting experiments were obtained from Bio-Rad (Hercules, California, U.S.A.). PolyP medium chain was a gift from Dr. Toshikazu Shiba, from Kitasato University, Tokyo, Japan.
Cell cultures
Wt HEK 293 cells were purchased from the American Type Culture Collection (ATCC; Manassas, Virginia, U.S.A.) and grown following the supplier’s recommendations. MitoPPX cells are stably transfected cells, grown under the same conditions as the Wt cells, with the only difference being the addition of 40 μl/ml G418 to the medium, as the selection antibiotic. Both cell types were grown in a humidified cell culture incubator, under a 5% CO2 atmosphere, at 37°C.
MitoPPX cell line generation
These cells were generated following the protocol published in [7]. Briefly, we transfected HEK 293 cells with the plasmid containing the sequence for the mitochondrial expression of the yeast exopolyphosphatase enzyme (PPX), GFP and the resistance to geneticin. We selected the cells positive for the expression of this construct by treating our samples with geneticin.
Seahorse assays
Cells grown to the confluence of 80–90% in 75 cm2 flasks were trypsinized and centrifuged. Thereafter, the supernatant was discarded, and the pellet was re-suspended in 1 ml of medium. 10 μl of the cellular suspension were transferred to the Hemocytometer Neubauer Counting Chamber. Cells were counted and diluted with growing medium, prior to being plated on the poly-L-lysine-coated Seahorse XFe24 Cell culture 24-wells microplates at a density of 50 000 cells per well. Subsequently, cells were incubated under normal conditions for 48 h.
The night before the experiment, the cartridge containing the sensors was hydrated with 1 ml of XF Calibrant Solution and was kept overnight in an incubator containing 0% CO2. The day of the experiment, cells were washed three times with Seahorse XF DMEM medium that contained 1 mM pyruvate, 2 mM glutamine, and 10 mM glucose. The medium for the Glycolytic Rate Assay was supplemented with 0.5 mM HEPES and afterwards, the plates that contained the cells were incubated for 1 h in the same CO2-free incubator (hypoxia), while the drugs from the specific Agilent XF kits were loaded onto the cartridge. Specifically, we added 0.5 μM oligomycin, different concentrations of FCCP (see figures) and 0.5 μM rotenone/antimycin A (Rot/AA) to the XF MitoStress kit and 50 μM Rot/AA and 50 mM 2-deoxy-D-glucose (2-DG), to the XF Glycolytic Rate Assay kit. All the drugs were dissolved to the working concentrations using the Seahorse XF DMEM medium. The cartridge containing the drugs and the sensors were introduced in the Seahorse XFe24 Analyzer for calibration. Subsequently, the cells were loaded onto the analyzer and the measurements were conducted. The obtained data was exported and analyzed using the Seahorse Wave Desktop Software, which was downloaded from the Agilent Technologies’ website.
Once the Seahorse measurements were concluded, the medium was carefully removed and cells were incubated with 5 μM Calcein-AM in PBS, in a regular incubator, for 30 min. Thereafter, fluorescence and cell number were measured, using a Cell Insight CX7 LZR reader, with a 4× magnification objective lens (ThermoFisher Scientific, Waltham, Massachusetts, U.S.A.). The data obtained from the Seahorse measurements were normalized with the number of cells calculated using this technology.
Image acquisition (Seahorse experiments)
Before and after conducting the Seahorse experiments, cells were imaged using an EVOS FL Auto Cell Imaging System, with a 20× magnification objective lens (ThermoFisher Scientific, Waltham, Massachusetts, U.S.A.). Specifically, the cells on the Seahorse plates were placed on the microscope just before the hypoxia incubation and just after the Seahorse assay. Images were taken from cells from the exact same wells before and after the experiments.
Fluorescence and confocal microscopy
Cells were plated on 25-mm borosilicate poly-L-lysine-coated glass coverslips at a 50% confluence. 24 h later, cells were washed twice using sterile HBSS and mounted on Sykes-Moore microscopy chambers (BellCo, Vineland, New Jersey, U.S.A.). Sterile HBSS was added to the glasses on the chambers and cells were imaged on a Nikon Fluorescence microscope, using a 20× magnification objective lens (Chiyoda, Tokyo, Japan); or on a Leica SP8 Confocal microscope, using a 40× oil-immersion magnification objective lens (Wetzlar, Germany).
ATP luminescence measurement
ATP was measured per the instructions that were provided by the manufacturer. Briefly, 10 000 cells per well were plated on a flat bottom, white, 96-well plate. We prepared triplicates of each condition. The day after, cells were incubated for 5 min with the detergent contained in the kit, at room temperature and under constant agitation. Subsequently, the substrate solution, also provided in the kit, was added and the cells were incubated under the same conditions as in the previous incubation. Cells were then incubated for another 10 min in the dark, in order to dark-adapt the plate. Lastly, luminescence was measured, at 500 ms, from the bottom.
Mitochondrial length measurement
Mitochondria from Wt and MitoPPX cells were loaded with TMRM, following the same protocol published in [30]. Subsequently, cells were mounted on microscopy chambers and images were acquired through the protocol previously described above. The mitochondrial length was then measured, using ImageJ software (National Institutes of Health, Bethesda, Maryland, U.S.A.) and following the protocol published in [31].
Western blotting
Western blotting assays were conducted as previously described in [32,33] through the use of the specific antibodies for the proteins of interest. Briefly, cells were scraped on ice and lysed. Protein was assayed using the Pierce BCA Protein Assay kit. Thereafter, equal amounts of protein were loaded on 12% polyacrylamide gels and electrophoresis was conducted at 100 V. The proteins were then transferred from the gel to a PVDF membrane, blocked and incubated with a 1 : 1000 dilution of the specific antibody overnight. The day after, the membrane was washed three times with TBS-Tween, incubated with the specific secondary antibodies, washed three times and the signal was visualized using Pierce ECL Western Blotting Substrate. Subsequently, membranes were stripped, blocked and Western blotting for actin (loading control) was conducted, following the same protocol.
Targeted metabolomics assays
LC–MS/MS-based metabolomics targeting a list of 310 metabolites was performed on a system consisting of Shimadzu Nexera XR LC-20AD pumps (Kyoto, Kyoto, Japan) coupled to a Sciex 6500+ triple quadrupole spectrometer (Framingham, Massachusetts, U.S.A.) operating in MRM detection mode through Sciex Analyst 1.6.3 software. The system includes a dual chromatography column setup with dedicated columns for positive and negative ionization modes. The samples were separated on a Waters Xbridge BEH amide column (2.5 μm, 130 Å, 2.1 × 150 mm) (Milford, Massachusetts, U.S.A.) operated in a HILIC regime at 40°C. Solvent A consisted of 95% water, 3% acetonitrile, 2% methanol, 0.2% acetic acid (v/v/v/v) 10 mM ammonium acetate, with a pH of ∼4.2. Solvent B consisted of 93% acetonitrile, 5% water, 2% methanol, 0.2% acetic acid and 10 mM ammonium acetate. 18.2 MOhm water was obtained from a Synergy UV system (MilliporeSigma, Burlington, Massachusetts, U.S.A.). Gradient elution at a flow rate of 0.300 ml/min was as follows: 0–3 min 95% B, 3–8 min 95–50% B, 8–12 min 50% B, 12–13 min 50–95% B, 13–18.1 min 95% B with linear ramps. During the injection on columns of opposite polarity, the solvent continued at 95% B, giving each column ∼23 min of equilibration time. Each sample was injected twice, 5 μl for positive mode and 10 μl for negative. Relative metabolite levels were quantified using Sciex Multiquant 3.0 software (Framingham, Massachusetts, U.S.A.).
Mitochondrial isolation and high-resolution respirometry
Wt and MitoPPX cells were grown until 90% confluence, after which they were trypsinized and spun down. The cellular pellet was then resuspended in 25 ml of isolation buffer, containing 200 mM mannitol, 50 mM sucrose, 5 mM dipotassium phosphate, and 0.1% v/v BSA, at pH = 7.15. Homogenization was done using an Omni International Handled homogenizer (Kennesaw, Georgia, U.S.A.) at 18.000 rpm for 20 s. Mitochondria were then recovered by gradient centrifugation in isolation buffer, at 4°C. Mitochondrial protein was quantified with the Pierce BCA Protein Assay kit and BSA standards, using an Olis DM-245 spectrofluorometer (Bogart, Georgia, U.S.A.). The mitochondrial stock solution was then diluted to a working concentration of 10 mg/ml.
Oxygen consumption data was obtained at 37°C, using Oxygraph 2k (Oroboros Instruments, Graz, Austria). Each respiratory chamber was loaded with respiration buffer, consisting of 130 mM potassium chloride, 5 mM dipotassium phosphate, 20 mM MOPS, 1 mM magnesium chloride, 1 mM EGTA, and 0.1% w/v BSA at pH 7.1. Before the addition of mitochondria, 5 mM pyruvate and 1 mM L-malate were added to each chamber. Mitochondria were then injected into each chamber at a concentration of 0.1 mg/ml. After the leak state respiratory rate stabilized (∼5 min), 1 mM of ADP was injected into each chamber, to achieve the OXPHOS respiratory state. The OXPHOS respiratory state reached a steady-state after an additional 5 min. The respiratory control ratio was calculated from the steady-state OXPHOS rate divided by the steady-state leak respiration rate. In the calcium retention experiments, EGTA was not added to the buffer.
Polyp degradation measurement
Wt and MitoPPX cells were grown under regular conditions on Petri Dishes. When the confluence was ∼90%, they were washed three times in ice-cold PBS, scraped on ice using the same PBS and collected into a 1 ml tube. Cellular suspensions were then centrifuged at 1000 rpm for 5 min, at 4°C. Thereafter, supernatants were discarded and the pellets, containing the cells were re-suspended in 500 μl of lysis buffer. The composition of this buffer and the protocol for the assay were modified from [2]. Specifically, our solution contained 20 mM TRIS-HCl, 10% glycerol, 1% Triton X100 and 1 mM PMSF. Cellular suspensions were then sonicated four times for 25 s, at 100% amplitude, with 45 s of cooling on ice between sonication cycles. BCA Protein Assay was conducted on the samples and appropriate amounts of the buffer were added to the samples, in order to have the same concentration of protein in Wt and MitoPPX cells. Subsequently, the incubation buffer, which contained 0.1% BSA (w/v), 10% glycerol, 20 mM TRIS-HCl (pH = 7.5) and 50 mM potassium chloride, was prepared and kept in an ice bucket. Samples were diluted 1 : 1 in this buffer.
A solution containing 40 μM DAPI was prepared in incubation buffer and kept in the dark. DAPI has been proven to be a useful probe to analyze the presence of polyP in biological samples [34]. Medium chain polyP was diluted to 0.5 M phosphate (51 mg/ml polyP), in purified water to prepare the stock solution. Thereafter, a small aliquot of this solution was diluted 1 : 100 in the incubation buffer, to obtain our working solution.
An amount of 10 μl of the cell suspension were added to different wells of a 96-wells plate. All the conditions were tested in triplicate at a minimum. On top of the cell suspension, we added 10 μl of the working solution of polyP and 1 μl of the DAPI/polyP solution. Plates were then incubated at room temperature and fluorescent readings were performed at the indicated times, using a FlexStation Microplate reader (Molecular Devices, San Jose, California, U.S.A.) at the excitation/emission wavelengths of 415/500 nm, without any cut off filter. 1 U of alkaline phosphatase was added as a positive control for degradation of polyP [35].
Statistical analysis of the experimental data
Statistical significance of differences between groups was determined by the Student’s test or the two-tailed Student’s test. The level of statistical significance was set at α = 0.05 (* P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001). For the statistical analysis and graphical representation, Origins Lab software (Northampton, Massachusetts, U.S.A.) was used.
Results
MitoPPX cells show decreased levels of oxidative phosphorylation
To assess the metabolic consequences of mitochondrial polyP depletion, we first compared OXPHOS-linked ATP production in Wt and MitoPPX cells. We used HEK 293 cells for these studies since their glucose metabolism has been extensively characterized [36]. Specifically, it has been shown that 15% of the consumed glucose is channeled through the pentose phosphate pathway (PPP) whereas 77% of metabolized glucose enters glycolysis. About 22% of the pyruvate generated in this process is then metabolized into acetyl-CoA for energy production and lipid biosynthesis [36].
Stable HEK 293 MitoPPX cells were previously generated and characterized [3,7]. These cells express GFP-labeled yeast exopolyphosphatase (PPX) selectively in the mitochondria, and appear to contain substantially decreased levels of mitochondrial polyP compared with Wt HEK 293 cells. To independently verify the functional activity of PPX specifically in MitoPPX cells, we prepared cell lysates from both cell lines, added 5 mM polyP50 (concentration given in inorganic phosphate — Pi — units) and monitored phosphate production over time (Supplementary Figure S1A). Our data revealed a significantly increased polyP-hydrolyzing activity in the lysate of MitoPPX cells, compared with Wt samples. Moreover, analysis of the subcellular localization of GFP-PPX revealed a clear GFP signal in the mitochondria of MitoPPX cells, whereas no such signal was detected in the Wt cells (Supplementary Figure S1B). Based on these results, we conclude that MitoPPX cells express functional yeast PPX specifically in the mitochondria, leading to decreased levels of mitochondrial polyP.
To assess the metabolic consequences of mitochondrial polyP depletion, we monitored ATP production in Wt and MitoPPX cells, using the Seahorse technology [37,38]. Specifically, we employed the MitoStress Kit to assay the Oxygen Consumption Rate (OCR) in both cell lines. This kit includes the following components: oligomycin, which serves as an inhibitor of the ATP synthase; FCCP, an uncoupling agent that collapses the proton gradient; and rot/AA, which inhibits Complex I and III of the Electron Transfer Chain (ETC) and hence shuts down mitochondrial respiration. We focused our studies initially on select parameters of mitochondrial respiration. Specifically, we measured ATP-linked respiration by determining the difference between basal respiration (the basal signal) and the signal after addition of oligomycin, as well as the spare capacity by determining the difference between basal respiration and maximal respiration upon addition of FCCP.
The observed oligomycin response in Wt and MitoPPX cells indicated that ATP production was dramatically reduced in MitoPPX cells, compared with Wt cells (Figure 1A,B). Furthermore, and in striking contrast with Wt cells, MitoPPX cells exhibited a significantly altered response to FCCP addition. In Wt cells, addition of FCCP elicited the expected stimulation of mitochondrial respiration, with a maximal stimulation being achieved at 0.5 μM of FCCP. In stark contrast, addition of the same concentration of FCCP to MitoPPX cells reduced respiration from ∼200 pmolO2/min/cells to 50 pmol O2/min/cells. (Figure 1A,B). Lower concentrations (i.e. 0.25 μM) of FCCP were still able to stimulate respiration in MitoPPX cells, indicating that the respiratory chain was generally functional in MitoPPX cells (Figure 1C). However, MitoPPX cells appear to be more susceptible to the membrane depolarization that is triggered by higher concentrations of FCCP (Supplementary Figure S3). Microscopy images that were taken before and after the Seahorse experiments indicated that similar number of cells were present under both conditions and that no major cell detachment occurred during the treatment (Supplementary Figure S4A). These results confirmed that the observed differences were not caused by differences in cell number.
Figure 1. Mitochondrial polyP depletion decreases OXPHOS in mammalian cells.
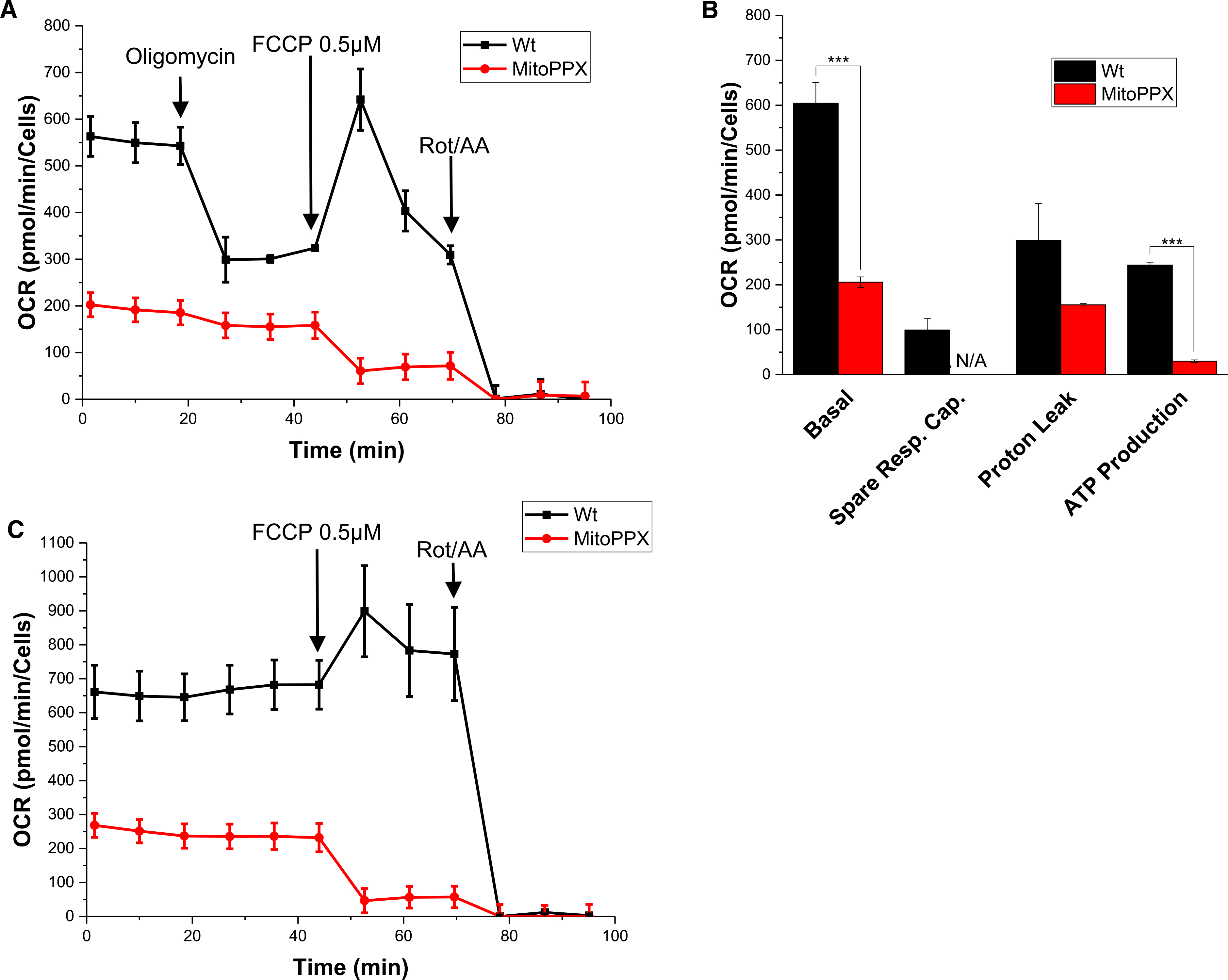
Wt and MitoPPX HEK 293 cells were cultivated and the oxygen consumption rate (OCR) was assayed using the Seahorse technology. Each experiment was conducted in biological triplicates. (A) A representative graph showing decreased OXPHOS in MitoPPX cells compared with Wt cells after treatment with 0.5 μM FCCP. (B) Quantification of the differences in basal respiration, spare respiratory capacity, proton leak, and ATP production between Wt and MitoPPX cells. (C) Response of Wt and MitoPPX cells to the treatment with 0.5 μM FCCP and 0.5 μM rot/AA. In this case, oligomycin was not added to the samples to avoid the inhibition of the ATP synthase. Data in the graphs is shown as average ± SEM.
MitoPPX cells show increased rates of glycolysis
The decrease in ATP generation can be either an indication of the overall slower energy metabolism or of a shift of ATP production towards the less effective glycolysis. To explore these possibilities, we measured glycolytic rates in Wt and MitoPPX cells utilizing the Glycolytic Rate Assay kit. This assay measures the protons extruded into the extracellular media as a consequence of the breakdown of glucose into lactate during glycolysis (ECAR, Extracellular Acidification Rate). Since the Krebs cycle also acidifies the media through the generation of CO2, we inhibited oxygen consumption during the assay using rot/AA. By using this approach, the rate of proton efflux from respiration can be calculated and subtracted from the total proton efflux rate, yielding in the glycolytic proton efflux rate (glycoPER). As control, we administered 2-deoxy-D-glucose (2-DG), which serves as a glucose analog that competitively inhibits glycolysis [39].
As shown in Figure 2A,B, MitoPPX cells showed a 1.5-fold increase in ECAR and an almost 2-fold increase in glycoPER compared with Wt cells. Inhibition of mitochondrial OXPHOS by the addition of rot/AA revealed the same maximal rates of glycolysis, and addition of 2-DG triggered the same abrupt signal decrease in both Wt and MitoPPX cells (Figure 2A). These experiments suggest that cells lacking mitochondrial polyP undergo a metabolic shift from OXPHOS to glycolysis. As before, we confirmed that both Wt and MitoPPX samples show a similar cell density (Supplementary Figure S4B). Notably, increased glycolysis was not able to completely compensate for the loss of aerobic ATP production. Indeed, measurements of total endogenous levels of ATP using a luminescence assay revealed decreased levels of ATP in the MitoPPX cells, compared with the Wt cells (Figure 2C).
Figure 2. MitoPPX cells show increased glycolysis compared with Wt cells.
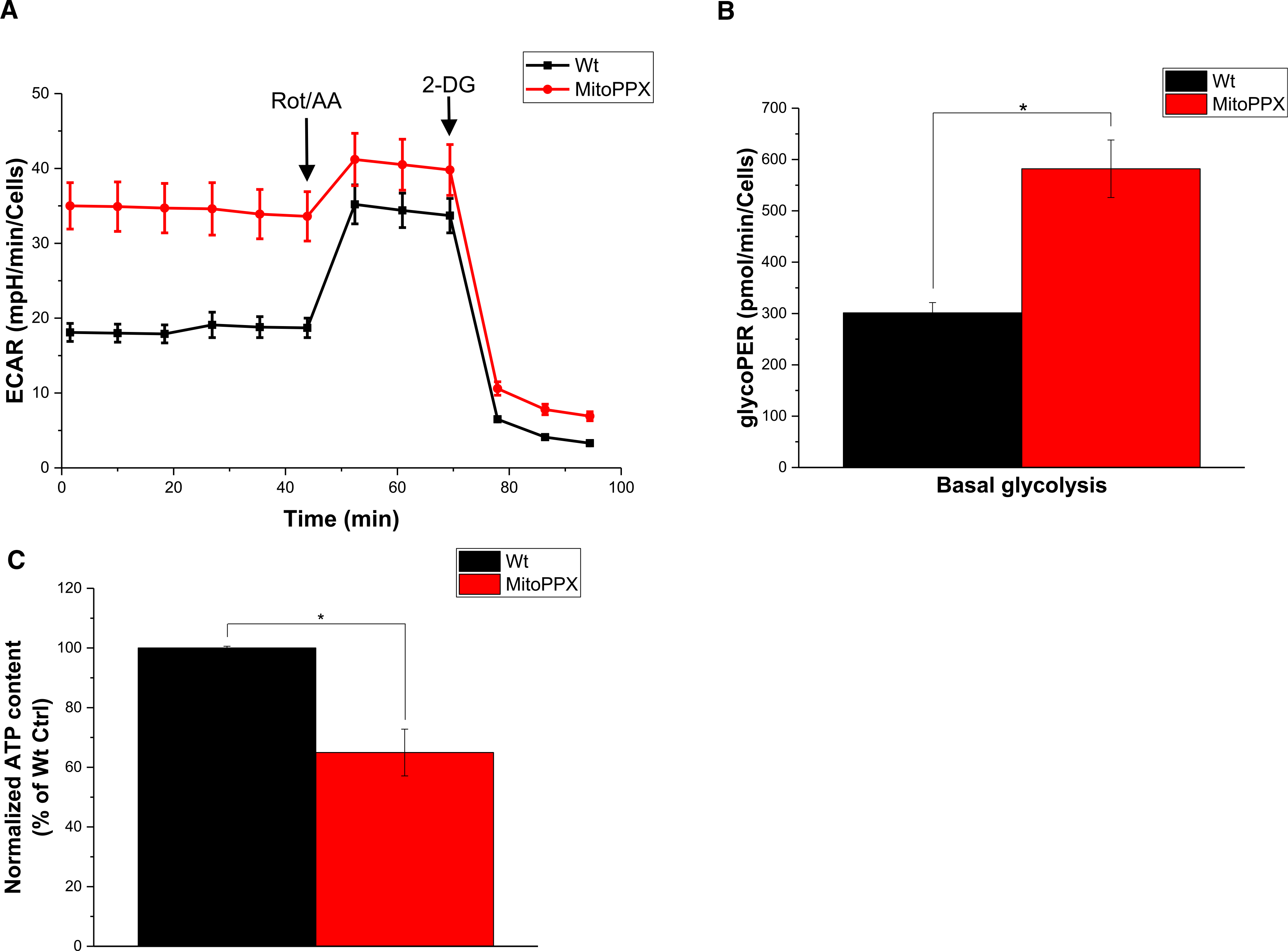
(A) Analysis of the glycolysis rate in Wt and MitoPPX HEK 293 cells using the Glycolytic Rate Assay Kit and Seahorse technology. Each experiment was conducted in biological triplicates and a representative graph is shown. (B) Quantification of the data shown in A. (C) Quantification of cellular ATP levels in Wt and MitoPPX cells. Data in the graphs is shown as average ± SEM.
Metabolomics data indicate changes in glycolysis, and energy and nucleotide metabolism
To independently assess the functional consequences of mitochondrial polyP depletion on cellular bioenergetics, we conducted metabolomic analysis in our Wt and MitoPPX samples. We used a LC–MS/MS-based metabolomics approach targeting a list of 310 metabolites, many of them related to cellular bioenergetics. To conduct these experiments, we prepared six biological duplicates of each cell type (Wt and MitoPPX). Out of the 310 metabolites targeted, we consistently detected 175 in our cell samples. Of these, 45 had P-values <0.05, and 15 remained significant after FDR correction using a Benjamini–Hochberg approach with q < 0.05 (see Supplementary Table S1).
The metabolomics analysis showed a large and highly significant reduction (>16 fold) in sugar monophosphates in MitoPPX cells, compared with the results in the Wt samples (Table 1). Moreover, the ratio (glucose/lactate) in MitoPPX cells did not change significantly, compared with the same ratio in the Wt samples (P = 0.023, Supplementary Table S1). Furthermore, we observed significant differences in the abundance of the energy-related coenzymes, NADP and NADPH, as well as of ADP and AMP. All of these compounds were significantly increased in MitoPPX cells, when compared with Wt samples. In fact, AMP was one of the most highly altered metabolites in MitoPPX cells compared with Wt samples. The levels of two other energy-related metabolites, succinylcarnitine and isovalerylcarnitine, were also significantly increased in MitoPPX cells compared with Wt cells. In addition to these changes, we found that MitoPPX cells contain higher levels of GMP, IMP, CMP, DTMP, GDP, DCMP and UDP, and lower levels of uracil. Finally, levels in selected methylated nucleosides were significantly different between the cell lines, as were changes in a number of amino acid metabolism-related metabolites. Pathway analysis of the metabolomics data confirmed our Seahorse measurements, and were indicative of increased rates of glycolysis and glycolysis-specific metabolites in MitoPPX cells compared with Wt cells (Figure 3). Notably, as stated, in addition to changes in glycolysis, our data also indicate alterations in purine, pyrimidine, glutathione and sugar metabolism in cells lacking mitochondrial polyP.
Table 1.
Major metabolites involved in biogenetics analyzed in the Wt and MitoPPX samples
| Metabolite | LogFC | P-value | FDR | HMDB ID | Metabolic pathway |
|---|---|---|---|---|---|
| G1P/G6P/F6P/F1P | −4.335 | 0.000 | 0.002 | HMDB01401 | Glycolosis/PPP |
| 2/3-Phosphoglyceric Acid | −0.918 | 0.024 | 0.120 | HMDB00362 | Glycolosis |
| NADP | 1.216 | 0.018 | 0.114 | HMDB00217 | Coenzyme |
| NADPH | 2.864 | 0.001 | 0.025 | HMDB00221 | Coenzyme |
| Valine | 0.796 | 0.012 | 0.085 | HMDB00883 | Amino Acid |
| Serine | −0.576 | 0.049 | 0.189 | HMDB00187 | Amino Acid |
| Pentothenate | 2.257 | 0.003 | 0.036 | HMDB00210 | AA metabolism/alanine, CoA |
| 3-hydroxy-3-methylglutarate | −1.333 | 0.005 | 0.050 | HMDB00355 | AA metabolism/Leu |
| N-formylmethionine | 1.586 | 0.011 | 0.081 | HMDB01015 | AA metabolism/Met |
| Taurine | 1.317 | 0.029 | 0.138 | HMDB00251 | AA metabolism/Sulfur metabolism |
| Phosphoserine | −1.758 | 0.037 | 0.163 | HMDB00272 | AA metabolism/Ser |
| UDP-GlcNAc | 1.133 | 0.038 | 0.165 | HMDB00290 | Amino sugar and nucleotide sugar metabolism |
| 2-Hydroxyglutarate | −0.858 | 0.045 | 0.186 | HMDB59655 | AA metabolism & Gly/Ser/Thr metabolism |
| 1/3-Methylhistidine | 1.129 | 0.048 | 0.189 | HMDB00001 | Amino acid metabolism/His |
| AMP | 3.319 | 0.000 | 0.000 | HMDB00045 | Nucleotide |
| GMP | 3.235 | 0.000 | 0.017 | HMDB01397 | Nucleotide/Purine metabolism |
| IMP | 3.526 | 0.001 | 0.025 | HMDB00175 | Nucleotide/Purine metabolism |
| CMP | 3.103 | 0.001 | 0.025 | HMDB00095 | Nucleotide/Pyrimidine metabolism |
| Orotate | −4.184 | 0.002 | 0.030 | HMDB00226 | Nucleotide/Pyrimidine metabolism |
| ADP | 1.690 | 0.002 | 0.030 | HMDB01341 | Nucleotide/Purine metabolism |
| DTMP | 2.482 | 0.004 | 0.048 | HMDB01227 | Nucleic Acid |
| GDP | 1.783 | 0.008 | 0.068 | HMDB01201 | Nucleotide/Purine metabolism |
| Uracil | −0.951 | 0.015 | 0.102 | HMDB00300 | Nucleotide/Pyrimidine metabolism |
| DCMP | 0.930 | 0.022 | 0.120 | HMDB01202 | Nucleotide/pyrimidine |
| UDP | 2.007 | 0.043 | 0.183 | HMDB00295 | Nucleotide/Pyrimidine metabolism |
| 5-Hydroxymethyl-2’-deoxyuridine | 1.757 | 0.003 | 0.041 | N/A | Modified nucleoside |
| 5-Methylcytidine | 3.168 | 0.006 | 0.053 | HMDB00982 | Modified nucleoside |
| 5-Methyluridine | 2.743 | 0.009 | 0.074 | HMDB00884 | Methylated nucleoside |
| 5’-Methylthioadenosine | −0.733 | 0.022 | 0.120 | HMDB01173 | AA metabolism/Cys, Met |
| 2-Aminoisobutyric acid | 1.458 | 0.030 | 0.140 | HMDB01906 | Nucleotide/Pyrimidine metabolism |
| Pseudouridine | 1.573 | 0.027 | 0.130 | HMDB00767 | Nucleotide/Pyrimidine metabolism |
| Succinylcarnitine | 4.220 | 0.002 | 0.030 | HMDB61717 | Acyl carnitine |
| Isovalerylcarnitine | 1.954 | 0.023 | 0.120 | HMDB00688 | Acyl carnitine |
| Arachidonate | −1.221 | 0.001 | 0.025 | HMDB60102 | Lipids/phospholipids, ligand |
| o-phosphoethanolamine | 1.483 | 0.001 | 0.025 | HMDB00224 | Glycerophospholipid modification/metabolism |
| ethanolamine | 1.647 | 0.011 | 0.081 | HMDB00149 | Glycerophospholipid modification/metabolism |
| Glycerol-3-P | 1.276 | 0.016 | 0.104 | HMDB00126 | Lipids/Glycerollipid |
| Reduced glutathione | 0.926 | 0.025 | 0.123 | HMDB00125 | Oxidative Damage |
| Putrescine | −2.930 | 0.001 | 0.025 | HMDB01414 | Polyamine metabolism |
| Sorbitol | −1.055 | 0.022 | 0.120 | HMDB00247 | Sugar |
| Riboflavin | −1.220 | 0.005 | 0.051 | HMDB00244 | Vitamin |
| Suberic acid | −0.650 | 0.049 | 0.189 | HMDB00893 | Fatty acid metabolism |
| Inositol | 1.689 | 0.007 | 0.062 | N/A | Glucose/inositol metabolism |
All major metabolites analyzed in the samples are listed and arranged by the pathways they are involved in.
Figure 3. Metabolomic comparison of Wt and MitoPPX cells.
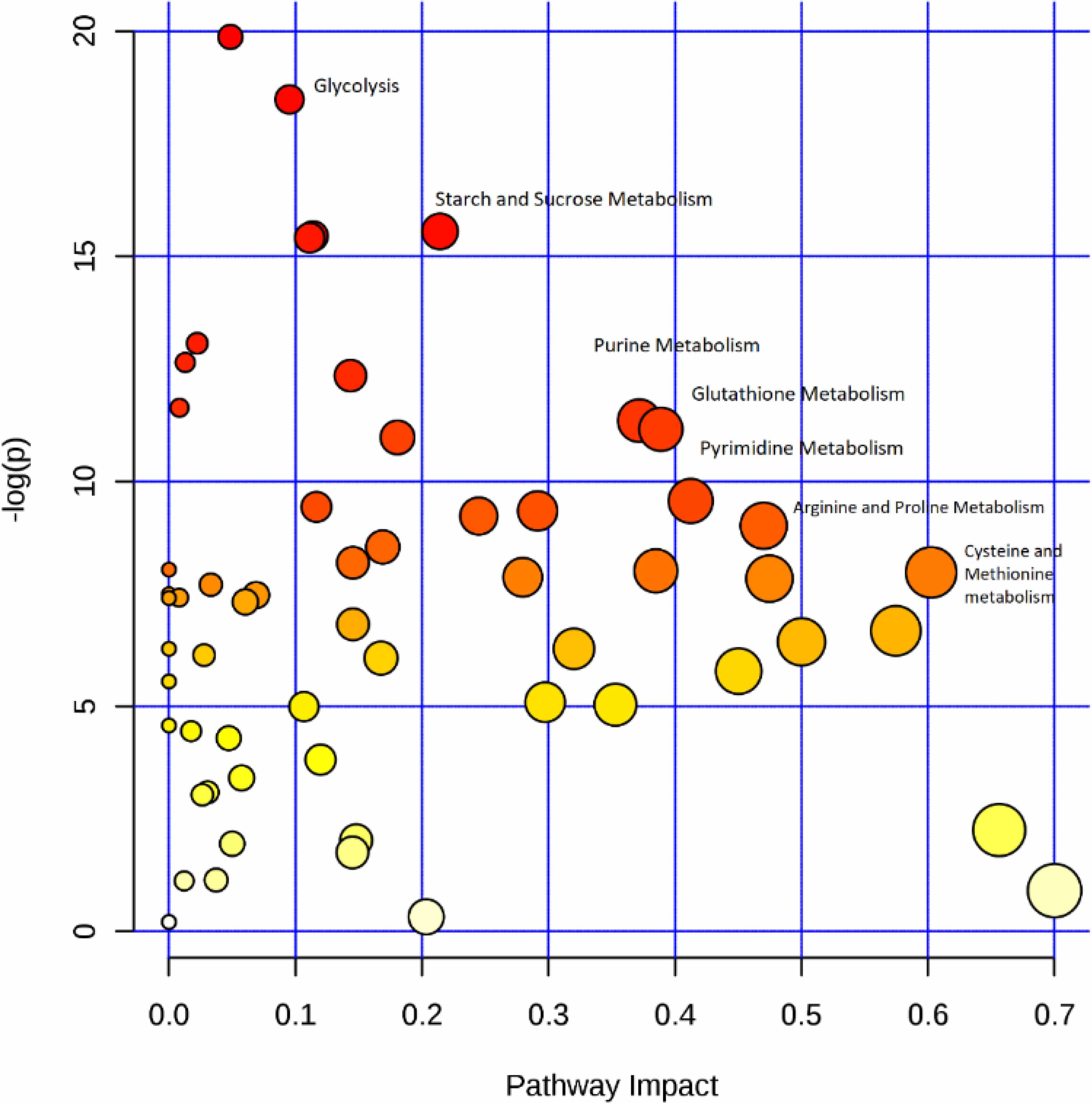
Pathway analysis illustrating the primary differences between Wt and MitoPPX cells. The analysis was arranged by scores from pathway enrichment (y axis) and topology analysis (x axis). The size and the color of each circle is based on P-values and pathway impact values, respectively (Xia and Wishart 2011).
Isolated MitoPPX and Wt mitochondria have similar activity
The shift in energy metabolism towards glycolysis that we observed in MitoPPX cells might be caused by a loss in mitochondrial function. To test this possibility, we measured the rates of respiration in isolated mitochondria, using resolution respirometry. This technique, which is based on the amperometric measurement of O2, allows us to assay OCR in isolated mitochondria, providing minimal O2 leak and maximum sensitivity to O2 Hutter, [40–42].
To our surprise, we found that Wt and MitoPPX cells behave very similarly in this assay. As shown in Figure 4A,B, the leak respiration and ADP-coupled respiration rates from Wt and MitoPPX cells were not significantly different. Since ADP-coupled respiration, which drives OXPHOS of ADP to ATP, is mediated by proton pumps across the inner mitochondrial membrane [41], this result indicates that both mitochondrial inner membrane and the ETC are fully functional in MitoPPX cells. The same is true for leak respiration, which is defined as the electron flow, coupled to proton pumping to compensate for proton leak [41]. Accordingly, when we calculated the Respiratory Control Ratios for Wt and MitoPPX isolated mitochondria, we did not find significant differences between the two cell types. Analysis of the steady-state levels of selected protein components of the ETC complexes did not reveal any significant differences between Wt and MitoPPX cells, consistent with our results showing that mitochondria from MitoPPX cells are fully functional ex vivo (Figure 4C). These results suggest however that the differences between Wt and MitoPPX cells that we observe in regards to the in vivo ATP production are more likely due to changes in the regulation of the energy metabolic pathways than due to structural or functional defects in the ETC. We cannot eliminate the possibility that mitochondria isolated from MitoPPX cells will also present increased rates of ATP hydrolysis. This could be also affecting the levels of mitochondrial respiration in our samples.
Figure 4. Isolated mitochondria from MitoPPX cells show Wt activity.
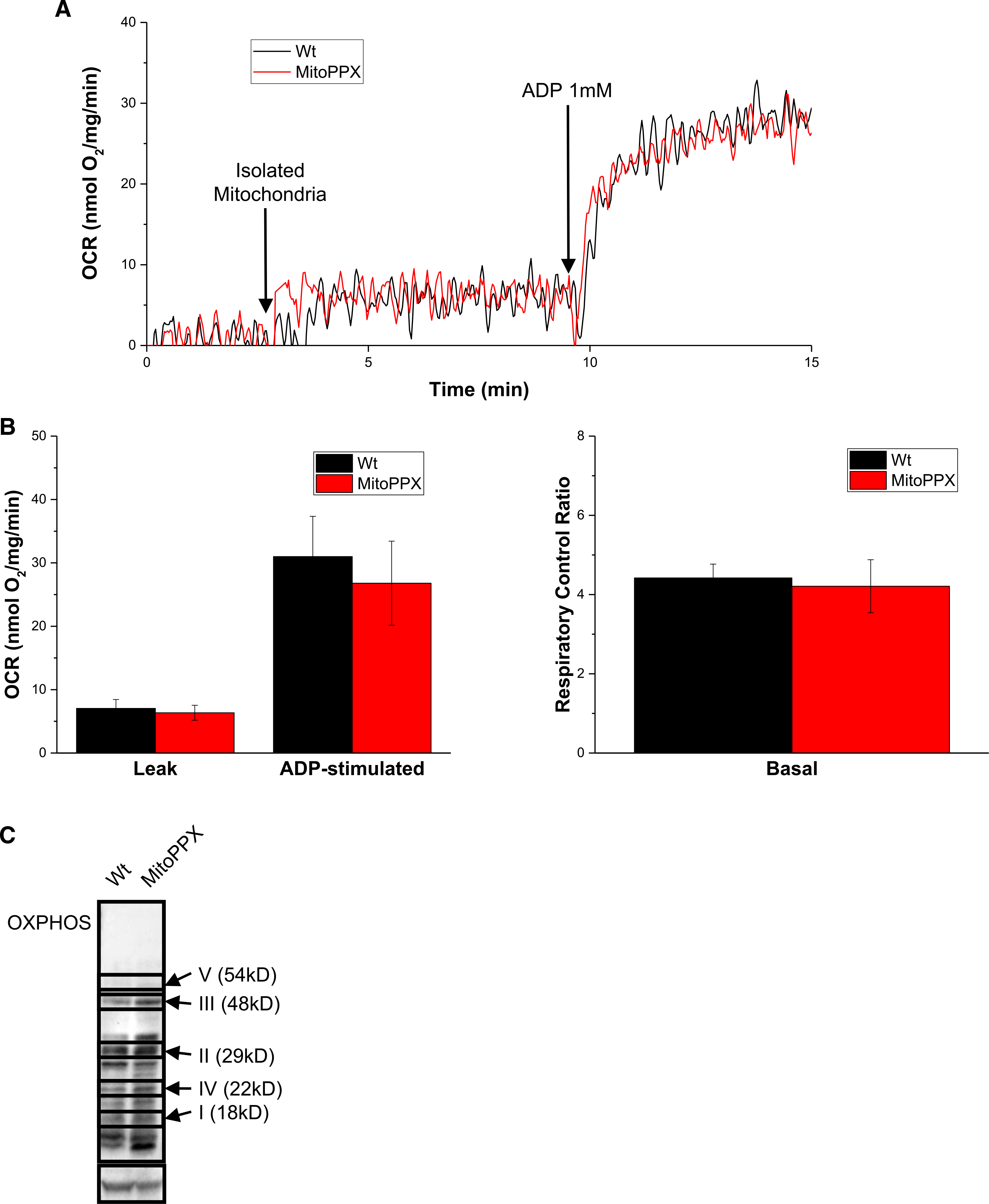
(A) OCR measurements of mitochondria isolated from either Wt or MItoPPX cells. (B) Analysis of OCR and the respiratory control rate of Wt and MitoPPX cells during the respirometry experiment. (C) Steady-state levels of different components of the respiratory chain in from Wt and MitoPPX HEK 293 cells, using Western blotting. Immunoblot was conducted using a commercially available cocktail antibody, containing all the components of the ETC. Bands not labeled are unspecific. Actin was used as loading control. Data in the graphs are shown as average ± SEM.
MitoPPX cells show increased fragmentation
It has recently been proposed that increased mitochondrial fission might trigger the metabolic shift from OXPHOS to glycolysis [43]. Increased mitochondrial fission, which has been broadly reported in different pathologies, leads to higher levels of mitochondrial mass but with a defective ability to produce ATP. However, mitochondrial fission is also a physiological process, naturally present in cells and crucial to maintain the physiology or the organelle [44].
To investigate the effect of the depletion of mitochondrial polyP in the fission of the organelle, we measured mitochondrial length in Wt and MitoPPX cells by microscopy. Our analysis revealed that mitochondria of MitoPPX cells are significantly shorter compared with mitochondria of Wt cells (Figure 5A,B). Shorter and more abundant mitochondria have been widely associated with the over-activation of mitochondrial fission [45]. To further assess the state of mitochondrial fission in our cells, we assayed the levels of Drp1, which is the main protein involved in mitochondrial fission in mammalian cells [46] Consistent with our microscopy data, we found that MitoPPX cells contain substantially increased levels of Drp1 (Figure 5C). However, when we addressed the levels of phosphorylated Drp1 (serine 616), which is a clear marker of Drp1-dependent mitochondrial fission [47], our data showed similar levels of the presence of this protein in both Wt and MitoPPX samples (Supplementary Figure S5B). Accordingly, no increased levels of Drp1 were found on mitochondria isolated from MitoPPX cells, when compared with mitochondria isolated from the Wt samples (Supplementary Figure S5A). These results suggest that in our model mitochondrial fission might occur by a mechanism not directly linked to the Drp1 pathway. Furthermore, no differences were observed in the maximal respiration rate between our Wt and MitoPPX mitochondria. These data align with two works showing that the treatment of cellular samples with increasing concentrations of Mdivi-1, which is a well-known inhibitor of mitochondrial fission in mammals, does not affect the maximal respiration, measured by Seahorse technology, when the cells were treated with concentrations of up to 20 μM of the drug (10 μM is usually enough to prevent increased fission in response to increased oxidative stress in cells, as we demonstrated) [48–50].
Figure 5. MitoPPX cells reveal increased mitochondrial fragmentation.
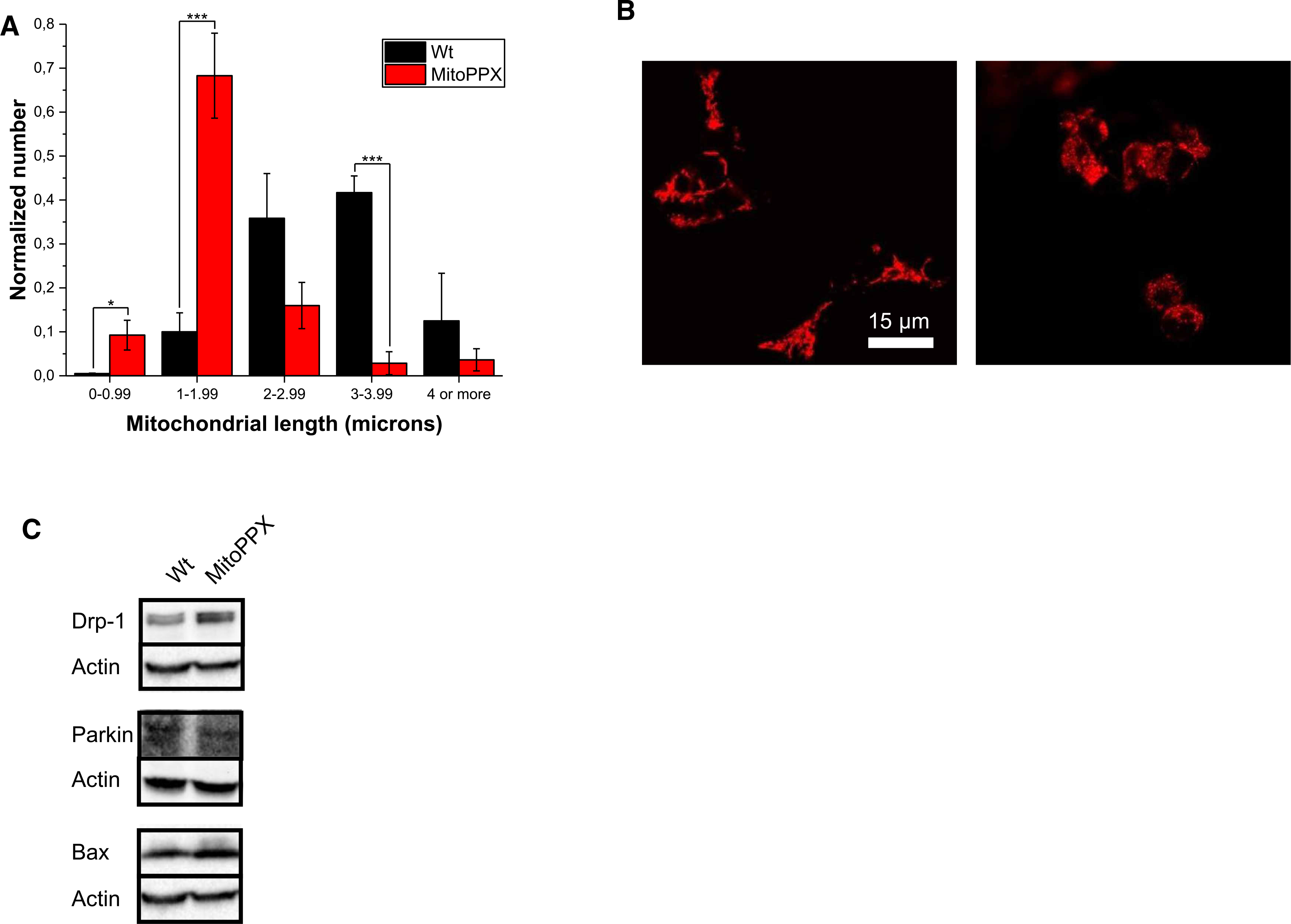
(A) Analysis of mitochondrial length in Wt and MitoPPX cells. Data was normalized with the total number of mitochondria counted in each picture. (B) Significant pictures of Wt and MitoPPX HEK 293 cells showing shorter mitochondria in MitoPPX cells, compared with Wt samples. Shorter mitochondria are a classical feature of increased mitochondrial fission. Mitochondria were labeled using TMRM. (C) Steady state levels of Drp1, Parkin and Bax in cell lysates of Wt and MitoPPX HEK 293 cells. Actin was used as loading control. Data in the graphs are shown as average of six pictures ± SEM.
Importantly, we did not detect any differences in the activation of the mitochondrial-specific authophagy (mitophagy) or the intrinsic pathway of apoptosis, two processes that can act downstream of Drp1-mediated mitochondrial fission. This result became evident when we assayed for Parkin, one of the main proteins involved in mitophagy [51] or Bax, a key regulator of the intrinsic pathway of apoptosis [52] (Figure 5C). Our data suggest that the direct effect of polyP in Drp1 does not activate other pathways within the organelle. These data could further support that the activation of mitochondrial fission in our samples, even if present, is still within the physiological range, as well as that the mechanism involved on this activation might not be the Drp1-mediated classical pathway.
Discussion
Here we present a comprehensive study of the effects of mitochondrial polyP depletion on the energy metabolism of mammalian cells. One of the key findings of our study is the observation that lack of mitochondrial polyP decreases OXPHOS, and concomitantly leads to an increase in glycolysis. Since mitochondria isolated from either Wt or MitoPPX cells showed very similar rates of OXPHOS and both cell types showed comparable expression levels of the key enzymes of the respiratory chain, we propose that mitochondrial polyP is not required for the process of OXPHOS per se, but it might play a regulatory role in energy metabolism.
The metabolomics analysis that we conducted shows a major up-regulation in the production of a number of nucleotides, including several energy coenzymes in cells lacking mitochondrial polyP. This increase could be a consequence of the PPX activity. Moreover, we found that the lactate to glucose ratio increases in MitoPPX cells, consistent with the increased glycolytic conversion of glucose to lactate. It remains to be investigated how lack of mitochondrial polyP triggers the observed changes in other glycolysis-related pathways, such as the pentose phosphate pathway, as well as the nucleotide synthesis. However, these results demonstrate that mitochondrial polyP plays a central metabolic role in eukaryotic cells.
It is possible that the observed metabolic shift from OXPHOS to glycolysis in MitoPPX cells is linked to the levels of mitochondrial fission, through a mechanism independent of Drp1. However, further studies will have to be conducted to unequivocally confirm the mechanistic relationship between these processes. The link between increased mitochondrial fission and a shift from OXPHOS to glycolysis has been demonstrated before in cancer models [43,53]. However, in our experiments the morphological changes in mitochondria, which are compatible with increase fission, did not appear to be linked to the classical Drp1-mediated mitochondrial fission pathway. This suggests that other mechanisms might be in play. Based on these results, we now consider the intriguing possibility that mitochondrial polyP serves as a regulator of the mitochondrial fission machinery, maybe through a Drp1-independent mechanism. It is of note that the AMP-activated Protein Kinase (AMPK) enzyme is regulated by the inositol polyphosphate multikinase (IPMK), which is involved in the catalysis of inositol polyP and inositol pyrophosphate [54–56]. Moreover, AMPK has been described as an energy sensor, involved in cellular bioenergetics, regulating mitochondrial fission, in response to bioenergetics stress [57,58]. Thus, decreased levels of polyP might result in decreased activity of IPMK, dysregulating AMPK and, hence stimulating mitochondria fission.
In summary, we report that enzymatic depletion of the mitochondrial polyP leads to the metabolic shift from OXPHOS to glycolysis which is linked to the increased mitochondrial fragmentation. Our data suggest that polyP is a previously unrecognized regulator of the cellular energy metabolism in mammalian cells.
Supplementary Material
Acknowledgements
We kindly thank the Northwest Metabolomics Research Center, at University of Washington, for the metabolomics analysis. Moreover, we thank the High Throughput Biology Laboratory, at New York University-Langone Health, especially Sabina Cook, Rebecca Lee and Chi Yun for their help with planning, preparing and conducting the Seahorse experiments, as well as to Kevin Bittman for his input on interpreting the obtained data on those experiments. We also thank Dr. Toshikazu Shiba, from Kitasato University, Tokyo, Japan for kindly provide us with medium chain PolyP. Lastly, we thank Mr. Mitch Maleki, Esq., for editing this manuscript.
Funding
This study was supported by the National Institutes of Health (1K99AG055701–01A1 and 4R00AG055701–03 to M.E.S., GM115570–01A1 to E.V.P., 1S10OD021562 to D.R., and GM122506 to U.J.), by Rutgers University (Start Up funds to M.E.S.), and by the American Heart Association, Transformation Project Award (18TPA34230060 to E.V.P.).
Abbreviations
- BSA
Bovine Serum Albumina
- DAPI
4′−6-diamino-2-phenylindole
- ECAR
extracellular acidification rate
- EGTA
ethylene glycol tetraacetic acid
- ETC
electron transfer chain
- HBSS
Hank’s Balanced Salt Solution
- IPMK
inositol polyphosphate multikinase
- MitoPPX
mitochondrial polyP
- OCR
oxygen consumption rate
- OXPHOS
oxidative phosphorylation
- PBS
Phosphate-Buffered Saline
- PMSF
Phenylmethylsulfonyl fluoride
- PPP
pentose phosphate pathway
- TBS
Tris-buffered saline
- TMRM
tetramethylrhodamine methyl ester
Footnotes
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Data Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Maria E. Solesio Torregrosa (m.solesio@rutgers.edu).
References
- 1.Kumble KD and Kornberg A (1995) Inorganic polyphosphate in mammalian cells and tissues. J. Biol. Chem. 270, 5818–5822 10.1074/jbc.270.11.5818 [DOI] [PubMed] [Google Scholar]
- 2.Abramov AY, Fraley C, Diao CT, Winkfein R, Colicos MA, Duchen MR et al. (2007) Targeted polyphosphatase expression alters mitochondrial metabolism and inhibits calcium-dependent cell death. Proc. Natl Acad. Sci. U.S.A. 104, 18091–6 10.1073/pnas.0708959104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solesio ME, Demirkhanyan L, Zakharian E and Pavlov EV (2016) Contribution of inorganic polyphosphate towards regulation of mitochondrial free calcium. Biochim. Biophys. Acta 1860, 1317–1325 10.1016/j.bbagen.2016.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kornberg A, Rao NN and Ault-Riche D (1999) Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68, 89–125 10.1146/annurev.biochem.68.1.89 [DOI] [PubMed] [Google Scholar]
- 5.Beauvoit B, Rigoulet M, Raffard G, Canioni P and Guerin B (1991) Differential sensitivity of the cellular compartments of saccharomyces cerevisiae to protonophoric uncoupler under fermentative and respiratory energy supply. Biochemistry 30, 11212–11220 10.1021/bi00111a004 [DOI] [PubMed] [Google Scholar]
- 6.Pavlov E, Aschar-Sobbi R, Campanella M, Turner RJ, Gomez-Garcia MR and Abramov AY (2010) Inorganic polyphosphate and energy metabolism in mammalian cells. J. Biol. Chem. 285, 9420–9428 10.1074/jbc.M109.013011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solesio ME, Garcia Del Molino LC, Elustondo PA, Diao C, Chang JC and Pavlov EV (2020) Inorganic polyphosphate is required for sustained free mitochondrial calcium elevation, following calcium uptake. Cell Calcium 86, 102127 10.1016/j.ceca.2019.102127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seidlmayer LK, Juettner VV, Kettlewell S, Pavlov EV, Blatter LA and Dedkova EN (2015) Distinct mPTP activation mechanisms in ischaemia-reperfusion: contributions of Ca2+, ROS, pH, and inorganic polyphosphate. Cardiovasc. Res. 106, 237–248 10.1093/cvr/cvv097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seidlmayer LK, Gomez-Garcia MR, Shiba T, Porter GA Jr, Pavlov EV, Bers DM et al. (2019) Dual role of inorganic polyphosphate in cardiac myocytes: The importance of polyP chain length for energy metabolism and mPTP activation. Arch. Biochem. Biophys. 662, 177–189 10.1016/j.abb.2018.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suess PM, Watson J, Chen W and Gomer RH (2017) Extracellular polyphosphate signals through Ras and Akt to prime dictyostelium discoideum cells for development. J. Cell Sci. 130, 2394–2404 10.1242/jcs.203372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muller WEG Wang S, Neufurth M, Kokkinopoulou M, Feng Q, Schroder HC et al. (2017) Polyphosphate as a donor of high-energy phosphate for the synthesis of ADP and ATP. J. Cell Sci. 130, 2747–2756 10.1242/jcs.204941 [DOI] [PubMed] [Google Scholar]
- 12.Muller WEG Wang S, Ackermann M, Neufurth M, Steffen R, Mecja E et al. (2017) Rebalancing beta-amyloid-induced decrease of ATP level by amorphous nano/micro polyphosphate: suppression of the neurotoxic effect of amyloid beta-protein fragment 25–35. Int. J. Mol. Sci. 18, 2154 10.3390/ijms18102154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freimoser FM, Hurlimann HC, Jakob CA, Werner TP and Amrhein N (2006) Systematic screening of polyphosphate ( poly P) levels in yeast mutant cells reveals strong interdependence with primary metabolism. Genome Biol. 7, R109 10.1186/gb-2006-7-11-r109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borden EA, Furey M, Gattone NJ, Hambardikar VD, Liang XH, Scoma ER et al. (2021) Is there a link between inorganic polyphosphate ( polyP), mitochondria, and neurodegeneration? Pharmacol. Res. 163, 105211 10.1016/j.phrs.2020.105211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bayev AY, Angelova PR and Abramov AY (2020) Inorganic polyphosphate is produced and hydrolysed in F0F1-ATP synthase of mammalian mitochondria. Biochem. J. 477, 1515–1524 10.1042/BCJ20200042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szijgyarto Z, Garedew A, Azevedo C and Saiardi A (2011) Influence of inositol pyrophosphates on cellular energy dynamics. Science 334, 802–805 10.1126/science.1211908 [DOI] [PubMed] [Google Scholar]
- 17.Livermore TM, Chubb JR and Saiardi A (2016) Developmental accumulation of inorganic polyphosphate affects germination and energetic metabolism in dictyostelium discoideum. Proc. Natl Acad. Sci. U.S.A. 113, 996–1001 10.1073/pnas.1519440113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seidlmayer LK, Blatter LA, Pavlov E and Dedkova EN (2012) Inorganic polyphosphate–an unusual suspect of the mitochondrial permeability transition mystery. Channels (Austin) 6, 463–467 10.4161/chan.21939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amodeo GF, Solesio ME and Pavlov EV (2017) From ATP synthase dimers to C-ring conformational changes: unified model of the mitochondrial permeability transition pore. Cell Death Dis. 8, 1 10.1038/s41419-017-0042-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solesio ME, Elustondo PA, Zakharian E and Pavlov EV (2016) Inorganic polyphosphate ( polyP) as an activator and structural component of the mitochondrial permeability transition pore. Biochem. Soc. Trans. 44, 7–12 10.1042/BST20150206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seidlmayer LK, Gomez-Garcia MR, Blatter LA, Pavlov E and Dedkova EN (2012) Inorganic polyphosphate is a potent activator of the mitochondrial permeability transition pore in cardiac myocytes. J. Gen. Physiol. 139, 321–331 10.1085/jgp.201210788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haythorne E, Rohm M, van de Bunt M, Brereton MF, Tarasov AI, Blacker TS et al. (2019) Diabetes causes marked inhibition of mitochondrial metabolism in pancreatic beta-cells. Nat. Commun. 10, 2474 10.1038/s41467-019-10189-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowling AC and Beal MF (1995) Bioenergetic and oxidative stress in neurodegenerative diseases. Life Sci. 56, 1151–1171 10.1016/0024-3205(95)00055-B [DOI] [PubMed] [Google Scholar]
- 24.Zheng J (2012) Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (Review). Oncol Lett. 4, 1151–1157 10.3892/ol.2012.928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray MJ and Jakob U (2015) Oxidative stress protection by polyphosphate–new roles for an old player. Curr. Opin. Microbiol. 24, 1–6 10.1016/j.mib.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dahl JU, Gray MJ and Jakob U (2015) Protein quality control under oxidative stress conditions. J. Mol. Biol. 427, 1549–1563 10.1016/j.jmb.2015.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cremers CM, Knoefler D, Gates S, Martin N, Dahl JU, Lempart J et al. (2016) Polyphosphate: A conserved modifier of amyloidogenic processes. Mol. Cell 63, 768–780 10.1016/j.molcel.2016.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lempart J, Tse E, Lauer JA, Ivanova MI, Sutter A, Yoo N et al. (2019) Mechanistic insights into the protective roles of polyphosphate against amyloid cytotoxicity. Life Sci. Alliance 2, e201900486 10.26508/lsa.201900486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lempart J and Jakob U (2019) Role of polyphosphate in amyloidogenic processes. Cold Spring Harb. Perspect. Biol. 11, a034041 10.1101/cshperspect.a034041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solesio ME, Peixoto PM, Debure L, Madamba SM, de Leon MJ, Wisniewski T et al. (2018) Carbonic anhydrase inhibition selectively prevents amyloid beta neurovascular mitochondrial toxicity. Aging Cell 17, e12787 10.1111/acel.12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pavlov E, Gutierrez RC, Zhang Y, Kertesz AC, Hung J, Espina FJ et al. (2010) Development of an in vitro model of neuronal activity induced excitotoxicity using photoconductive stimulation. Cell Calcium 47, 441–448 10.1016/j.ceca.2010.03.005 [DOI] [PubMed] [Google Scholar]
- 32.Solesio ME, Saez-Atienzar S, Jordan J and Galindo MF (2013) 3-Nitropropionic acid induces autophagy by forming mitochondrial permeability transition pores rather than activating the mitochondrial fission pathway. Br. J. Pharmacol. 168, 63–75 10.1111/j.1476-5381.2012.01994.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solesio ME, Saez-Atienzar S, Jordan J and Galindo MF (2012) Characterization of mitophagy in the 6-hydoxydopamine Parkinson’s disease model. Toxicol. Sci. 129, 411–420 10.1093/toxsci/kfs218 [DOI] [PubMed] [Google Scholar]
- 34.Aschar-Sobbi R, Abramov AY, Diao C, Kargacin ME, Kargacin GJ, French RJ et al. (2008) High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J. Fluoresc. 18, 859–866 10.1007/s10895-008-0315-4 [DOI] [PubMed] [Google Scholar]
- 35.Lorenz B and Schroder HC (2001) Mammalian intestinal alkaline phosphatase acts as highly active exopolyphosphatase. Biochim. Biophys. Acta 1547, 254–261 10.1016/S0167-4838(01)00193-5 [DOI] [PubMed] [Google Scholar]
- 36.Henry O, Jolicoeur M and Kamen A (2011) Unraveling the metabolism of HEK-293 cells using lactate isotopomer analysis. Bioprocess Biosyst. Eng. 34, 263–273 10.1007/s00449-010-0468-9 [DOI] [PubMed] [Google Scholar]
- 37.Brand MD and Nicholls DG (2011) Assessing mitochondrial dysfunction in cells. Biochem. J. 435, 297–312 10.1042/BJ20110162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salabei JK, Gibb AA and Hill BG (2014) Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat. Protoc. 9, 421–438 10.1038/nprot.2014.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wick AN, Drury DR, Nakada HI and Wolfe JB (1957) Localization of the primary metabolic block produced by 2-deoxyglucose. J. Biol. Chem. 224, 963–969 10.1016/S0021-9258(18)64988-9 [DOI] [PubMed] [Google Scholar]
- 40.Hutter E, Unterluggauer H, Garedew A, Jansen-Durr P and Gnaiger E (2006) High-resolution respirometry–a modern tool in aging research. Exp. Gerontol. 41, 103–109 10.1016/j.exger.2005.09.011 [DOI] [PubMed] [Google Scholar]
- 41.Pesta D and Gnaiger E (2012) High-resolution respirometry: OXPHOS protocols for human cells and permeabilized fibers from small biopsies of human muscle. Methods Mol. Biol. 810, 25–58 10.1007/978-1-61779-382-0_3 [DOI] [PubMed] [Google Scholar]
- 42.Perry CG, Kane DA, Lanza IR and Neufer PD (2013) Methods for assessing mitochondrial function in diabetes. Diabetes 62, 1041–1053 10.2337/db12-1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guido C, Whitaker-Menezes D, Lin Z, Pestell RG, Howell A, Zimmers TA et al. (2012) Mitochondrial fission induces glycolytic reprogramming in cancer-associated myofibroblasts, driving stromal lactate production, and early tumor growth. Oncotarget 3, 798–810 10.18632/oncotarget.574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho B, Choi SY, Cho HM, Kim HJ and Sun W (2013) Physiological and pathological significance of dynamin-related protein 1 (drp1)-dependent mitochondrial fission in the nervous system. Exp. Neurobiol. 22, 149–157 10.5607/en.2013.22.3.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sesaki H and Jensen RE (1999) Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147, 699–706 10.1083/jcb.147.4.699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imoto M, Tachibana I and Urrutia R (1998) Identification and functional characterization of a novel human protein highly related to the yeast dynamin-like GTPase Vps1p. J. Cell Sci. 111, 1341–1349 PMID:9570752 [DOI] [PubMed] [Google Scholar]
- 47.Taguchi N, Ishihara N, Jofuku A, Oka T and Mihara K (2007) Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 282, 11521–9 10.1074/jbc.M607279200 [DOI] [PubMed] [Google Scholar]
- 48.Alam S, Abdullah CS, Aishwarya R, Miriyala S, Panchatcharam M, Peretik JM et al. (2018) Aberrant mitochondrial fission Is maladaptive in desmin mutation-Induced cardiac proteotoxicity. J. Am. Heart Assoc. 7, e009289 10.1161/JAHA.118.009289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peiris-Pages M, Bonuccelli G, Sotgia F and Lisanti MP (2018) Mitochondrial fission as a driver of stemness in tumor cells: mDIVI1 inhibits mitochondrial function, cell migration and cancer stem cell (CSC) signalling. Oncotarget 9, 13254–13275 10.18632/oncotarget.24285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Solesio ME, Prime TA, Logan A, Murphy MP, Del Mar Arroyo-Jimenez M, Jordan J et al. (2013) The mitochondria-targeted anti-oxidant mitoQ reduces aspects of mitochondrial fission in the 6-OHDA cell model of Parkinson’s disease. Biochim. Biophys. Acta 1832, 174–182 10.1016/j.bbadis.2012.07.009 [DOI] [PubMed] [Google Scholar]
- 51.Narendra D, Tanaka A, Suen DF and Youle RJ (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pawlowski J and Kraft AS (2000) Bax-induced apoptotic cell death. Proc. Natl Acad. Sci. U.S.A. 97, 529–531 10.1073/pnas.97.2.529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hagenbuchner J, Kuznetsov AV, Obexer P and Ausserlechner MJ (2013) BIRC5/Survivin enhances aerobic glycolysis and drug resistance by altered regulation of the mitochondrial fusion/fission machinery. Oncogene 32, 4748–4757 10.1038/onc.2012.500 [DOI] [PubMed] [Google Scholar]
- 54.Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P and Snyder SH (1999) Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 9, 1323–1326 10.1016/S0960-9822(00)80055-X [DOI] [PubMed] [Google Scholar]
- 55.Resnick AC, Snowman AM, Kang BN, Hurt KJ, Snyder SH and Saiardi A (2005) Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc. Natl Acad. Sci. U.S.A. 102, 12783–8 10.1073/pnas.0506184102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bang S, Kim S, Dailey MJ, Chen Y, Moran TH, Snyder SH et al. (2012) AMP-activated protein kinase is physiologically regulated by inositol polyphosphate multikinase. Proc. Natl Acad. Sci. U.S.A. 109, 616–620 10.1073/pnas.1119751109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Long YC and Zierath JR (2006) AMP-activated protein kinase signaling in metabolic regulation. J. Clin. Invest. 116, 1776–1783 10.1172/JCI29044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toyama EQ, Herzig S, Courchet J, Lewis TL Jr, Loson OC, Hellberg K, et al. (2016) Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science 351, 275–281 10.1126/science.aab4138 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


