Abstract
Pseudomonas aeruginosa is one of the most critical bacterial pathogens associated with chronic infections in cystic fibrosis patients. Here we show the phenotypic and genotypic characterization of five consecutive multidrug-resistant isolates of P. aeruginosa collected during a month from a CF patient with end-stage lung disease and fatal outcome. The isolates exhibited distinct colony morphologies and pigmentation and differences in their capacity to produce biofilm and virulence potential evaluated in larvae of Galleria mellonella. Whole genome-sequencing showed that isolates belonged to a novel sequence type ST3449 and serotype O6. Analysis of their resistome demonstrated the presence of genes blaOXA-396, blaPAO, aph(3’)-IIb, catB, crpP and fosA and new mutations in chromosomal genes conferring resistance to different antipseudomonal antibiotics. Genes exoS, exoT, exoY, toxA, lasI, rhlI and tse1 were among the 220 virulence genes detected. The different phenotypic and genotypic features found reveal the adaptation of clone ST3449 to the CF lung environment by a number of mutations affecting genes related with biofilm formation, quorum sensing and antimicrobial resistance. Most of these mutations are commonly found in CF isolates, which may give us important clues for future development of new drug targets to combat P. aeruginosa chronic infections.
Keywords: Pseudomonas aeruginosa, sequence type ST3449, multi-drug resistance, cystic fibrosis, biofilm, virulence
1. Introduction
Pseudomonas aeruginosa is among the most dreaded opportunistic human pathogens that affect the airways of cystic fibrosis (CF) individuals. This bacterium is notorious for its metabolic versatility and innate resistance to many antibiotics due to the permeability barrier conferred by its Gram-negative outer membrane. P. aeruginosa also has a tendency to form specialized bacterial communities called biofilms, which make bacterial cells impenetrable to therapeutic concentrations of antibiotics [1].
Respiratory tract infection in CF patient usually initiates with persistent infections by P. aeruginosa that are frequently eradicated by antibiotic treatments. Recolonization may take place, either by the same recalcitrant strain or other different strains, which can establish permanently in the airways leading to chronic infections in the respiratory tract of CF patients. Long-term colonization of the CF lung by P. aeruginosa generally occurs by single lineages, which are clonal to the strain acquired during initial colonization and can persist in the airway of a patient most of their lives [2]. P. aeruginosa displays high virulence at initial stages of infection and it is relatively susceptible to antibiotics, but during the transition to a chronic way of survival it undergoes several adaptations to overcome the hostile environment of the lung. Among the characteristics P. aeruginosa exhibits at chronic phases of infection the presence of a mucoid phenotype, reduced expression of virulence factors, reduced motility, enhanced biofilm formation ability, high mutation rates, appearance of small colony variants (SCV), pigmentation changes and resistance to antimicrobial agents [3,4].
Development of multidrug resistance (MDR) in P. aeruginosa during persistent infections is induced either by the accumulation of pathoadaptive chromosomal mutations or the acquisition of antimicrobial resistance determinants by horizontal gene transfer [2]. Increased ratios of resistance to beta-lactams, aminoglycosides and fluoroquinolones are usually detected among CF isolates. Moreover, some estimates point out that up to 40% of isolates from CF patients are MDR [5,6].
A deeper analysis in order to understand the role of mutations in the adaptive process of P. aeruginosa during CF, in particular of those related with the development of resistance to antimicrobial agents, is crucial for future development of new drug targets and the implementation of more efficient antibiotic treatments. In that sense, the advent of high-throughput sequencing technologies in combination with bioinformatics analysis has provided broad insights into bacterial evolution and pathogenesis, including polymorphisms associated with increased antimicrobial resistance and adaptation to specific niches. Some recent studies have employed whole genome sequencing (WGS) to examine the genetic variations of P. aeruginosa population during long-term infection of CF individuals [7,8,9,10,11,12].
With this work we aim to characterize five consecutive multidrug-resistant P. aeruginosa isolates belonging to a novel sequence type (ST3449) collected from a CF patient with end-stage pulmonary function. This study is a descriptive analysis where isolates were whole-genome sequenced by means of the Illumina Miseq platform to identify the antimicrobial resistance and virulence genes inventory. Our data provide new information about mutations in genes conferring antimicrobial resistance and in virulence factors that will need further investigation for a better understanding about the evolution of this clone.
2. Results and Discussion
2.1. Isolation, Phenotypic Features and Antimicrobial Susceptibility of the CF P. aeruginosa Strains
P. aeruginosa isolates MS6000, MS6002, MS6003, MS6004 and MS6005 were sequentially collected during a month (three first isolates from sputum, and the latter two from tracheal aspirate) from a CF patient with end-stage pulmonary disease who died before lung transplantation. Isolate numbering was assigned according to the consecutive date of isolation, that is to say MS6000 was the first strain isolated and MS6005 the last one. Isolates were identified as P. aeruginosa by means of the MALDI-TOF/Vitek MS instrument. The isolates displayed different colony morphology and pigmentation and all of them produced hemolysis (Table 1).
Table 1.
Phenotypic characteristics of the CF P. aeruginosa isolates.
| Isolate * | Source | Colony Morphology | Pigmentation | Hemolysis |
|---|---|---|---|---|
| MS6000 | Sputum | Small, mucoid | Brown | + |
| MS6002 | Sputum | Large, irregular | Blue-green opaque | + |
| MS6003 | Sputum | Small, circular | Green brilliant | + |
| MS6004 | Tracheal aspirate | Small, wrinkled | Green opaque | + |
| MS6005 | Tracheal aspirate | Large, irregular | Brown | + |
* Isolate numbering was assigned according to isolation date.
All the isolates were non-susceptible to aztreonam, cefepime, ceftazidime, piperacillin, piperacillin/tazobactam, imipenem, ciprofloxacin and levofloxacin. Conversely, all of them were susceptible to ceftazidime/avibactam and ceftolozane/tazobactam combinations. Except MS6002 that was resistant to meropenem and intermediate to doripenem, the rest of isolates were susceptible to these agents. All isolates were non-susceptible to gentamicin with the exception of MS6004. In the case of tobramycin, isolates MS6000, MS6003 and MS6005 were resistant to this drug. Amikacin was the most effective aminoglycoside antibiotic, as only isolate MS6003 showed resistance to this agent. Isolates MS6002 and MS6003 display resistance to colistin, but the rest remained susceptible. In the case of fosfomycin only MS6003 and MS6004 retained its susceptibility to this antibiotic. All isolates were categorized as XDR, except MS6004 that was considered as MDR (Table 2).
Table 2.
Antimicrobial susceptibility of P. aeruginosa ST3449 isolates.
| Isolate | CMI (µg/mL) | Resistance Phenotype | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AZT | FEP | CAZ | C/A * | C/T * | PIP | TZP | IMP | MER | DOR | GEN ** | TOB | AK | CIP | LEV | COL | FOS *** | ||
| MS6000 | 64 (R) | 16 (R) | 64 (R) | 4 (S) | 0.5 (S) | 256 (R) | 256 (R) | 1 (I) | 0.25 (S) | 0.25 (S) | 8 (I) | 4 (R) | 16 (S) | 0.5 (I) | 2 (R) | 2 (S) | 256 (R) | XDR |
| MS6002 | 64 (R) | 8 (I) | 16 (R) | 2 (S) | 0.38 (S) | 64 (R) | 128 (R) | 32 (R) | 16 (R) | 8 (I) | 8 (I) | 0.5 (S) | 8 (S) | 1 (R) | 4 (R) | 8 (R) | 512 (R) | XDR |
| MS6003 | 8 (I) | 16 (R) | 16 (R) | 1 (S) | 0.75 (S) | 128 (R) | 4 (I) | 8 (R) | 1 (S) | 2 (S) | 16 (R) | 1 (S) | 32 (R) | >32 (R) | >32 (R) | 4 (R) | 128 (S) | XDR |
| MS6004 | 0.25 (I) | 0.5 (I) | 1 (I) | 0.125 (S) | 0.125 (S) | 0.5 (I) | 1 (I) | 1 (I) | 0.06 (S) | 0.125 (S) | 4 (S) | 1 (S) | 8 (S) | 2 (R) | 4 (R) | 2 (S) | 64 (S) | MDR |
| MS6005 | >128 (R) | 128 (R) | 128 (R) | 3 (S) | 0.25 (S) | >256 (R) | 256 (R) | 4 (I) | 2 (S) | 2 (S) | 8 (I) | 4 (R) | 16 (S) | 4 (R) | 16 (R) | 2 (S) | 512 (R) | XDR |
AZT, aztreonam; FEP, cefepime; CAZ, ceftazidime; C/A, ceftazidime/avibactam; C/T, ceftolozane/tazobactam; PIP, piperacillin; TZP, piperacillin/tazobactam; IPM, imipenem; MEM, meropenem; DOR, doripenem; GEN, gentamicin, TOB, tobramycin; AK, amikacin; CIP, ciprofloxacin; LEV, levofloxacin; COL, colistin; FOS, fosfomycin.; * MICs of these antibiotics were determined by Etest; ** MIC interpretations were according to CLSI breakpoints; *** ECOFF used by EUCAST was applied for fosfomycin MIC interpretations: susceptible, ≤ 128 µg/mL; and non-susceptible or resistant, > 128 µg/mL; (S), susceptible; (I), intermediate; (R), resistant. MDR, multidrug-resistant; XDR, extensively drug-resistant.
2.2. Genome Analysis of the CF P. aeruginosa Isolates
Whole genome sequencing (WGS) of the five CF P. aeruginosa isolates was conducted in order to examine the molecular basis of their antimicrobial resistance and clonal relatedness, and their virulence genes repertoire. Summary and statistics for the genome sequencing of the P. aeruginosa isolates using the in-house bioinformatics pipeline TORMES version 1.0 [13] are detailed in Table 3.
Table 3.
Genome assembly summary of the CF P. aeruginosa isolates.
| Sample | Reads | Average Read Length (bp) | Contigs | Genome Length (bp) | Average Contig Length (bp) | N50 (bp) | ORF | GC Content |
Depth | Species | Percent ID |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MS6000 | 629064 | 278 | 119 | 6,326,892 | 576,053 | 262,138 | 5815 | 66.37 | 27X | P. aeruginosa | 99.29 |
| MS6002 | 876966 | 279 | 109 | 6,324,360 | 926,907 | 375,329 | 5830 | 66.38 | 38X | P. aeruginosa | 99.35 |
| MS6003 | 877052 | 277 | 123 | 6,339,440 | 920,338 | 351,508 | 5836 | 66.35 | 38X | P. aeruginosa | 99.28 |
| MS6004 | 915080 | 278 | 105 | 6,451,152 | 777,263 | 281,904 | 5953 | 66.37 | 39X | P. aeruginosa | 98.61 |
| MS6005 | 773800 | 278 | 122 | 6,334,587 | 777,177 | 351,513 | 5826 | 66.35 | 33X | P. aeruginosa | 99.30 |
| Average | 814392 | 278 | 116 | 6,355,286 | 795,548 | 324,478 | 5852 | 66.36 | 35X | - | 99.37 |
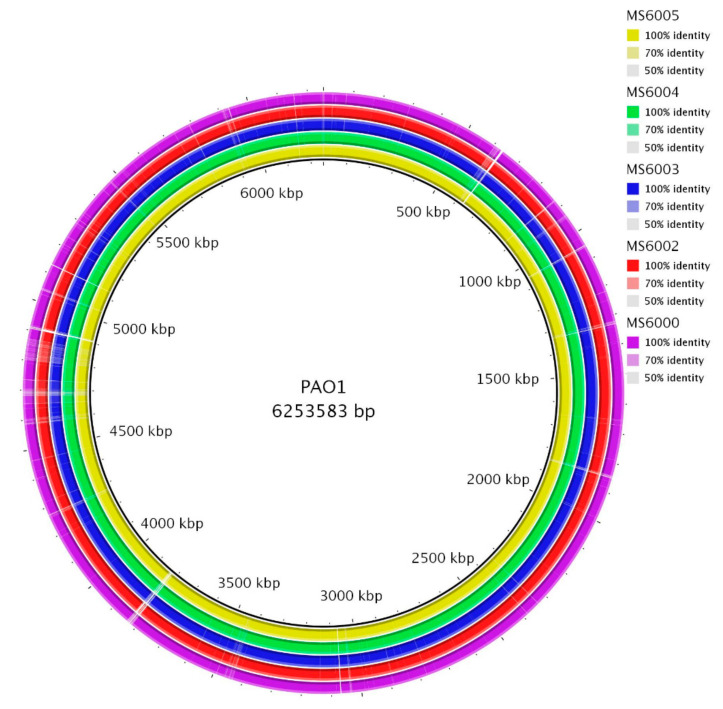
Genomic differences between CF isolates were determined by aligning the genomes using BRIG software [14] (Figure 1). Alignment of the assembled contig sequences of draft genomes of CF isolates with PAO1 showed a high level of conservation along their chromosomes. Based on the average genome length of the five CF isolates (6,355,286 bp) and the size of the PAO1 genome (6,253,583 bp, GenBank: JIEO00000000.1) used as a reference, we estimated that the average percentage of genome covered for the 5 CF isolates was 98.4%. MS6000 has an estimated genome size of 6,326,892 bp (5815 ORFs, GC% of 66.37), MS6002 of 6,324,360 bp (5830 ORFs, GC% of 66.38), MS6003 of 6,339,440 bp (5836 ORFs, GC% of 66.35), MS6004 of 6,451,152 bp (5953 ORFs, GC% of 66.37) and MS6005 of 6,334,587 bp (5826 ORFs, GC% of 66.36) (Table 3). The pangenome consisted of 5994 genes, with a core genome of 5753 (96%) genes and a shell genome of 241 (4%) genes. Core genome represents a pool of conserved genes, which are present in all genomes included in the analysis. The shell genes are those moderately common in the pangenome, including genes that are flexibly gained and lost, reflecting the evolution of a lineage and adaptation process of an organism to a particular niche. Soft-core category represents those genes present in 95% of genomes analyzed and cloud cluster is formed by genes present in less than 15% of genomes analyzed. In our analysis these two latter categories of genes were not represented. No plasmids were detected in any of the isolates.
Figure 1.
Circular representation of the CF P. aeruginosa isolates genomes. Comparison was performed using the blast ring image generator (BRIG) software [14]. Each genome is represented by a ring with different colors shown in the legend at the upper right panels. The genome of P. aeruginosa PAO1 (GenBank: JIEO00000000.1) was used as a reference sequence, represented in the innermost ring. The second innermost ring shows MS6005 genome and followed by isolates MS6004, MS6003, MS6002 and MS6000.
The MLST analysis showed that all CF P. aeruginosa isolates represented a novel sequence type ST3449 with the following housekeeping gene allele combination: acsA (28), aroE (150), guaA (11), mutL (7), nuoD (27), ppsA (6) and trpE (7). To the best of our knowledge this is the first report of ST3449 in isolates of P. aeruginosa from a CF patient.
The P. aeruginosa serotyper (PAst) program [15] was employed to perform the in silico serotyping of the isolates based on the sequences of the O-specific antigen (OSA) gene cluster. Typeability of P. aeruginosa is often lost in those isolates recovered from CF patients due to the adaptive process they undergo during chronic infections. The CF isolates analyzed here belonged to serotype O6. Earlier studies have also shown the presence of serotype O6 among CF P. aeruginosa isolates [16,17], which is often associated with poor clinical outcomes [6]. A recent Spanish national survey demonstrated that O6 was among the most prevalent serotypes in the population studied [18].
2.2.1. Resistome Analysis
Analysis of the horizontally acquired genes using the ResFinder database [19] revealed that all the isolates harbored resistance genes to beta-lactams (blaOXA-396 and blaPAO), aminoglycosides (aph(3’)-IIb), fluoroquinolones (crpP), phenicols (catB7) and fosfomycin (fosA). The presence of missense and nonsense mutations in genes known to confer resistance to the main groups of antipseudomonal agents such as beta-lactams, aminoglycosides, fluoroquinolones, polymixins and fosfomycin is shown in Table 4. A comprehensive list including all the mutations predicted by the breseq program [20] in the whole-genome sequences of the CF isolates analyzed in this work is available in the Supplementary Table S1.
Table 4.
Alterations in proteins/genes related with antimicrobial resistance found in the CF P. aeruginosa isolates of ST3449.
| Protein/Gene | MS6000 | MS6002 | MS6003 | MS6004 | MS6005 |
|---|---|---|---|---|---|
| AmpC | T105A | T105A | T105A | T105A | T105A |
| AmpR | +T at nt 860, stop TGA at 874 | +T at nt 860, stop TGA at 874 | +T at nt 860, stop TGA at 874 | +T at nt 860, stop TGA at 874 | +T at nt 860 stop TGA at 874 |
| AmpD | G148A, D183Y | G148A, D183Y | D183Y, +G at 443 | G148A, D183Y | G148A, D183Y |
| AmpDh2 | wt | wt | wt | wt | wt |
| AmpDh3 | A219T | A219T | A219T | A219T | A219T |
| mpl | Del. 14 bp (1120–1133) | Del. 14 bp (1120–1133) | wt | G133D | Del. 14 bp (1120–1133) |
| PBP1a (ponA) | D329G, +CCG at nt 1845 | D329G, +CCG at nt 1845 | D329G, +CCG at nt 1845 | D329G, +CCG at nt 1845 | D329G, +CCG at nt 1845 |
| PBP1b (mrcB) | L353Q | L353Q | L353Q | L353Q | L353Q |
| PBP3a (pbpC) | Not present | Not present | Not present | A104P | Not present |
| PBP7 (pbpG) | S250N | S250N | S250N | S250N | S250N |
| MexA | wt | wt | wt | wt | |
| MexB | L672R | L672R | L684P | L684P | L672R |
| OprM | wt | wt | wt | wt | wt |
| MexR | wt | wt | wt | wt | wt |
| NalC | G71E, S209R | G71E, S209R | G71E, S209R | G71E, S209R | G71E, S209R |
| NalD | wt | wt | wt | wt | wt |
| OprD | D43N, S57E, S59R, E202Q, I210A, E230K, S240T, N262T, A267S, A281G, K296Q, Q301E, R310G, V359L, M372V, S373D, D374S, N375S, N376S, V377S, Del.G378, Del.Y379, K380Y, N381A, Y382G, G383L | Stop at nt 18 (TGG→TGA) W6 * | D43N, S57E, S59R, E202Q, I210A, E230K, S240T, N262T, A267S, A281G, K296Q, Q301E, R310G, V359L, M372V, S373D, D374S, N375S, N376S, V377S, Del.G378, Del.Y379, K380Y, N381A, Y382G, G383L | D43N, S57E, S59R, E202Q, I210A, E230K, S240T, N262T, A267S, A281G, K296Q, Q301E, R310G, V359L, M372V, S373D, D374S, N375S, N376S, V377S, Del.G378, Del.Y379, K380Y, N381A, Y382G, G383L | D43N, S57E, S59R, E202Q, I210A, E230K, S240T, N262T, A267S, A281G, K296Q, Q301E, R310G, V359L, M372V, S373D, D374S, N375S, N376S, V377S, Del.G378, Del.Y379, K380Y, N381A, Y382G, G383L |
| MexX | K329Q, L331V, W358R | K329Q, L331V, W358R | K329Q, L331V, W358R | K329Q, L331V, W358R | K329Q, L331V, W358R |
| MexY | T543A, A992T | T543A, A992T | T543A | T543A | T543A, A992T |
| MexZ | wt | wt | wt | wt | wt |
| AmgS | wt | wt | wt | wt | wt |
| Fmt | I181V, E277K | I181V, E277K | I181V, E277K | I181V, E277K | I181V, E277K |
| FolD | T189A | T189A | T189A | T189A | T189A |
| FusA2 | S176A, G695A | S176A, G695A | S176A, G695A | S176A, G695A | S176A, G695A |
| NuoG | T484A | T484A | T484A | T484A | T484A |
| MexC | S330A | S330A | S330A | S330A | S330A |
| MexD | E257Q, S845A | E257Q, S845A | E257Q, S845A | E257Q, S845A | E257Q, S845A |
| OprJ | M69V | M69V | M69V | M69V | M69V |
| NfxB | wt | wt | wt | wt | wt |
| MexT | Del. GCCGGCCA (240–247), F172I | Del. GCCGGCCA (240–247), F172I | Del. GCCGGCCA (240–247), F172I | Del. GCCGGCCA (240–247), F172I | Del. GCCGGCCA (240–247), F172I |
| MexE | wt | wt | wt | wt | wt |
| MexF | wt | wt | wt | wt | wt |
| OprN | wt | wt | wt | wt | wt |
| MexS | D249N | D249N | D249N | D249N | D249N |
| GyrA | D87N | D87N | T83I | T83I | D87N |
| GyrB | S618L | S618L | wt | wt | S618L |
| ParC | V646L | V646L | E91K, V646L | E91K, V646L | E91K, V646L |
| ParE | G285S | G285S | G285S | G285S | G285S |
| CrpP | K4R, G7D | K4R, G7D | K4R, G7D | K4R, G7D | K4R, G7D |
| PmrA | L71R | L71R | L71R | L71R | L71R |
| PmrB | Y345H, V386M | Y345H, V386M | Y345H, V386M | Y345H, V386M | Y345H, V386M |
| ParR | L153R, S170N | L153R, S170N | L153R, S170N | L153R, S170N | L153R, S170N |
| ParS | H398R | H398R | Stop at nt 459 (TGG→TGA) W153 * | H398R | H398R |
| ArnB | V302A, E376D | V302A, E376D | V302A, E376D | V302A, E376D | V302A, E376D |
| ArnT | I509V | I509V | I509V | I509V | I509V |
| ArnF | T106I | T106I | T106I | T106I | T106I |
| PA3559 (ugd) | V101A | V101A | V101A | V101A | V101A |
| OprP | A98V | A98V | A98V | A98V | A98V |
The plus (+) sign indicates addition of a base at a certain position in the DNA sequence of the gene. In bold are represented those mutations/polymorphisms that to the best of our knowledge are described here for the first time. Del. (deletion). (*) an asterisk indicates that the protein is truncated from the amino acid residue due to the presence of a premature stop codon at a certain position of the sequence of the gene.
Resistance to beta-lactams in P. aeruginosa is primarily due to a contribution of several mechanisms such as overexpression of intrinsic beta-lactamase AmpC, horizontally acquired beta-lactamases carried by conjugative plasmids, low outer membrane permeability (porin mediated) and/or overproduction of RND (resistance-nodulation-division) efflux pumps, mainly MexAB-OprM and MexXY-OprM [21]. Role of penicillin binding proteins (PBPs) in beta-lactams resistance has also been demonstrated [22].
As mentioned before, the resistome analysis revealed that the five isolates harbored the beta-lactamase genes blaOXA-396 and blaPAO. Previous studies demonstrated the high prevalence of these genes in P. aeruginosa isolates from different sources and their contribution to antimicrobial resistance in this species [23,24,25,26,27]. According to the ResFinder tool on WGS from the five CF isolates, no carbapenemase genes were detected.
A missense mutation in the ampC gene leading to a T105A substitution in the beta-lactamase AmpC was found in all the isolates. It was formerly suggested that this mutation was related with reduced susceptibility to oxyiminocephalosporins and imipenem [28], but it did not affect ceftazidime [29]. Other studies have shown this is a common polymorphism in bela-lactam-resistant P. aeruginosa isolates [30,31,32,33,34]. However, its relationship with beta-lactam resistance is rather uncertain [31]. Overexpression of ampC is generally due to selected mutations in its regulator genes ampR, ampD and dacB. All our isolates presented a stop codon at nucleotide 874 in ampR due to the insertion of a thymine at position 860. Polymorphisms G148A, D183Y in AmpD were also detected, which were previously demonstrated not to be involved in ampC overexpression [30]. Isolate MS6003 showed an alteration in ampD consisting in the addition of a guanine at position 443 of the gene, resulting in a frameshift mutation (Table 4). AmpD homologues, AmpDh2 and AmpDh3 were previously described as negative regulators of AmpC [35]. No alterations were detected in AmpDh2, while mutation A219T in AmpDh3 was identified in all the isolates. This amino acid change in AmpDh3 was described before in multidrug-resistant P. aeruginosa isolates from ST175 and ST664 [36,37]. No mutations in the AmpC regulator dacB (PBP4) were found in our isolates.
A frameshift mutation was found in MS6000, MS60002 and MS6005 in the mpl gene, coding for a UDP-muramic acid/peptideligase, due to a deletion of 14 nucleotides. As far as we know, this deletion in mpl is described here for the first time. Inactivation of mpl has been shown to be responsible for increased activity of AmpC in P. aeruginosa [38,39]. Isolate MS6004 showed a missense mutation G133D in Mpl, previously observed in a CF isolate resistant to beta-lactams [33], but its implication in AmpC activation is unknown. Mutations in mpl are frequently found in P. aeruginosa isolates from CF individuals as a consequence of long-term colonization [40,41,42].
Mutations in some PBPs were documented in this study. All the isolates showed a missense mutation D329G in PonA (PBP1a) and the addition of the triplet CCG at position 1845 of ponA and the amino acid substitution L353Q in MrcB (PBP1b). PBP3a (pbpC) was absent in all except one, MS6004 that presented the mutation A104P. The substitution in residue S250N was found in PBP7 (pbpG) in all the isolates. Most of these mutations in PBP genes have been previously described, but their contribution to beta-lactams resistance remains to be demonstrated [33,36,43]. The rest of PBPs did not show any alteration.
The multidrug resistance efflux system MexAB-OprM is responsible for the extrusion of several antibiotics, among them, beta-lactams. Mutations accumulated in transcriptional regulators mexR, nalC and nalD lead to overexpression of this efflux pump. Of these three regulators, only NalC was found to display non-synonymous mutations G71E and S209R in all the isolates. These mutations seem to be frequent in multidrug resistant P. aeruginosa isolates, however their role in inducing overexpression of MexAB-OprM is unclear [33,36,44,45]. Two missense mutation not previously described (L627R and L684P) were detected in MexB (Table 4). However, we do not know their effect on the expression of this efflux system.
Porin OprD is a primary route of entry of imipenem in P. aeruginosa. OprD deficiency or loss from the outer membranes confers certain level of resistance to carbapenems [46]. Several polymorphisms were found in OprD (Table 4), affecting loops 1, 5, 6 and 7 of the porin. Some of these alterations in OprD had been described before with no contribution to carbapenem resistance [45,47,48,49]. Moreover, shortening of the loop 7 by deletion of residues G378 and Y379, here detected, has been associated with increased susceptibility to meropenem [50]. Isolate MS6002 presented a premature stop codon in oprD at position 18, thus generating a nonfunctional protein. This non-sense mutation possibly correlates with the resistance phenotype to imipenem displayed by MS6002 with the highest MIC value of this antibiotic among the isolates (Table 2).
Overexpression of MexXY-OprM is one of the major mechanisms of aminoglycoside resistance in P. aeruginosa, though this multidrug efflux system has a broad substrate specificity that also comprises quinolones, macrolides and some beta-lactams [51]. Overexpression of this pump is mainly due to mutations in its negative repressor MexZ and the activator AmgS. However, no mutations were detected in our isolates in any of these regulator genes. On the other hand, all the isolates presented the amino acid substitutions W358R, L331V and K329Q in MexX and T543A in MexY. All these mutations in MexXY have been related with resistance to beta-lactams, aminoglycosides and quinolones in a previous study conducted on CF P. aeruginosa isolates from the CC274 [33]. Nevertheless, an earlier study showed that these polymorphisms had no implication in resistance to aminoglycosides, as they were also found in strains susceptible to these agents [52]. MS6000, MS6002 and MS6005 presented a new amino acid substitution, A992T in MexY, which contribution to antimicrobial resistance needs to be investigated. Of note, all isolates displayed the amino acid substitutions I181V and E277K in Fmt, a methionyl-tRNA formyltransferase, and T189A in the methylene-tetrahydrofolate dehydrogenase, FolD. These two proteins were formerly determined to be involved in the regulation of MexXY expression [53]. The non-synonymous mutations mentioned above have been described before in CF P. aeruginosa isolates [33].
All the isolates displayed two missense mutations, S176A and G695A in FusA2. A previous study demonstrated that mutations in the elongation factor G encoding genes, fusA1 and fusA2, were responsible for high level of aminoglycoside resistance in CF isolates [33]. Mutation T484A was observed in NuoG in all the isolates. This mutation was previously documented in CF P. aeruginosa isolates [33]. The nuoG gene codes for a type I NADH dehydrogenase, whose inactivation leads to a decrease in aminoglycosides uptake [54].
High aminoglycoside resistance in P. aeruginosa is conferred by the presence of acquired aminoglycoside-modifying enzymes (AMEs). As mentioned above in the text, the aminoglycoside phosphotransferase gene, aph(3′)-IIb was found in the genomes of all the isolates. This enzyme confers resistance to kanamycin and neomycin but not to gentamicin, tobramycin or amikacin. Therefore, its contribution to the aminoglycoside resistant phenotype of the isolates is not relevant, suggesting that involvement of efflux activity and other mechanisms mentioned above should be playing a role in their aminoglycoside resistance phenotype.
Resistance to fluoroquinolones in P. aeruginosa involves a number of mechanisms. Fluroquinolones are substrates of the RND efflux pumps MexAB-OprM, MexXY-OprM, MexCD-OprJ and MexEF-OprN. However, some data suggested that the contribution of the overexpression of these efflux systems to high-level resistance to fluoroquinolones is rather limited [33]. Alterations in the MexAB-OprM and MexXY-OprM efflux systems and their respective regulator have been discussed above in the text. Some missense mutations were observed in all the structural components of MexCD-OprJ (Table 4). These mutations were previously described in multidrug-resistance P. aeruginosa isolates from different sources [33,55]. No sequence alterations were detected in the MexCD-OprJ repressor NfxB in any of the isolates, which lead us to think that MexCD-OprJ could be expressing in our isolates.
The positive regulator of the MexEF-OprN, MexT presented a missense mutation F172I and a deletion of eight nucleotides in mexT (Table 4), compared to the mexT of PAO1. The mexT gene missing the 8-bp insert have been described in prior studies and is related with the switch to an active MexT variant [47,56,57,58]. These mutations have been found before in isolates showing either basal or increased activity of MexEF-OprN [47,59]. All the isolates presented the amino acid change D249N in MexS, the negative regulator of MexEF-OprN. According to an earlier study the N249 is considered the wild type functional variant of MexS, present in strains PA14 and PAK [60]. Therefore, our isolates harbor an active MexS variant. It is known that MexS is a negative regulator of MexT. Despite not conducting any gene expression experiment to detect the activity of MexEF-OprN, we could think that as our isolates present a functional MexS, it would be expected that MexT were repressed and consequently activity of MexEF-OprN suppressed. However, overexpression of MexE have been demonstrated before in P. aeruginosa isolates with active variants of MexS and MexT, suggesting the existence of other regulatory mechanisms that may affect the expression of MexEF-OprN [45]. Search for mutations in additional regulators of MexEF-OprN such as MvaT and the two-component system (TCS) ParR/S were also carried out. Our isolates did not show any mutations in the mvaT gene. Conversely, several alterations were observed in ParR and ParS (Table 4). A premature stop codon (TGA) at position 459 in the parS gene found in isolate MS6003, leading to a non-functional two-component sensor ParS. A recent work proposed that mutations in the ParR/S lead to a reduction of mexS transcription, thus activating the expression of MexEF-OprN [61].
Resistance to fluoroquinolones also arises as a result of mutations in the quinolone resistance-determining regions (QRDRs) of gyrA, gyrB, parC and parE. The typical non-synonymous mutations T83I and D87N were found in GyrA in all the isolates. These mutations are clinically relevant and are often detected in fluoroquinolone-resistant P. aeruginosa [33,36,62]. Amino acid substitution S681L in GyrB was found in three of the isolates (Table 4). Missense mutations were also detected in the topoisomerase IV subunits ParC (E91K and V646L) and ParE (G285S) (Table 4). To our knowledge, mutations S681L, V646L and G285S accumulated in the topoisomerases II and IV, respectively, have not been described before. Additional analysis should be carried out to demonstrate whether these mutations are implicated in quinolone resistance or are just randomly selected during chronic infection in the lung of the CF patient. Interestingly, MS6003 was the isolate that accumulated the greatest number of mutations associated to fluoroquinolone resistance, which may correlate with its elevated ciprofloxacin and levofloxacin resistant profiles compared to the rest of isolates (Table 2).
The acquisition of the crpP gene, coding for a ciprofloxacin-modifying enzyme [63] was detected in our strains, all of them bearing the non-synonymous mutations K4R and G7D in CrpP, when compared with the CrpP (GenBank accession: NG_062203). A very recent study conducted on a large number of P. aeruginosa isolates from Portugal and Spain showed that CrpP proteins with the amino acid substitutions K4R and G7D were the most predominant variants among the population analyzed [64]. A reduced activity of CrpP on ciprofloxacin was previously observed when missense mutations occurred in the amino acid position G7, indicating that this is an essential residue for the enzymatic activity of CrpP [65]. Consequently, the presence of this CrpP variant in our isolates may not contribute to the ciprofloxacin resistance phenotype. On the other hand, based on our information, the implication of the missense mutation K4R on the CrpP activity is yet unknown.
Polymyxin resistance in P. aeruginosa is mostly attributable to mutations in the TCSs PhoP/Q, PmrA/B, ParR/S, ColR/S and CprR/S [66]. Several mutations in pmrA/pmrB and phoP/phoQ can induce upregulation of the arnBCADTEF-ugd (arn) operon, resulting in modifications of the lipopolysaccharide (LPS) that reduce the interaction of polymyxins with fatty acids and phosphates groups of the LPS [67,68]. When examining the amino acid sequences these TCSs in our isolates we only detected non-synonymous mutations in PmrA (L71R), PmrB (Y345H, V386M), ParR (L153R, S170N) and ParS (H398R) (Table 4). A thorough search in the literature demonstrated that these mutations were described before by other authors in colistin-susceptible strains [69,70], except for the amino acid substitution V386M observed in PmrB in all the isolates and the W153* in ParS found in isolate MS6003 (Table 4). Mutations in ArnB (V302A, E376D) and in other components of the arn operon were detected in all the isolates (Table 4 and Table S1), nonetheless the implication of this mutations on the arn operon activity and resistance to polymyxins needs to be demonstrated.
With regards to fosfomycin resistance, we carried out the sequence analysis of murA, glpT, oprO and oprP, involved in resistance to this agent. The entry of fosfomycin into the bacterium is still not very well understood. Three proteins are known so far to facilitate the entry of fosfomycin in P. aeruginosa, the glycerol-3-phosphate transporter, GlpT and porins OprO and OprP. Once in the periplasmic space, fosfomycin inhibits MurA, which initiates peptidoglycan biosynthesis. We did not observe any alteration in murA, glpT and oprO. However, all the isolates possessed the amino acid substitution A98V in OprP. This mutation was reported before, though its relation with fosfomycin resistance is yet unknown [43]. Resistance to fosfomycin is also conferred by FosA, a glutathione S-transferase that modifies enzymatically the antibiotic rendering it ineffective. As previously mentioned, all our isolates harbored the gene fosA, whose activity may have been the main contributor to fosfomycin resistance phenotypes showed by some of the isolates.
Altogether, the mutations found in antimicrobial resistance-related genes in this whole-genome sequence analysis may contribute to the antibiotic resistance displayed by our CF P. aeruginosa isolates, even when some phenotypic resistance could not be demonstrated in vitro. Therefore, additional in vivo analysis should be carried out to determine the actual role of these mutations developed during the course of chronic pulmonary diseases in CF patients.
2.2.2. Detection of Virulence Related Genes in the CF P. aeruginosa Isolates
A total of 220 virulence genes were found among our isolates by comparing the WGS data with the virulence factor database (VFDB), using as reference the P. aeruginosa strain PAO1. Supplementary Table S2 contains a list of the virulence factors found in each isolate, according to the VFDB. One of the most represented groups of virulence genes identified were those belonging to the Type III Secretion System (T3SS) machinery (17.7% of the genes), including its secreted exotoxin-coding genes exoS, exoT and exoY. Generally, isolates expressing exoS and exoT demonstrate an invasive phenotype while those isolates containing exoU, have a cytotoxic nature. Our strains did not contain the exoU gene. Generally, ExoS and ExoU are mutually exclusive, although some studies have reported the occurrence of isolates harboring both exotoxins [71,72,73,74]. Genes related with flagellar protein biosynthesis were also among the most represented virulence determinants (17%). Genes coding for Type IV pili related functions and twitching motility and those responsible for alginate biosynthesis and regulation represented the 14.5% and 12% of virulence genes, respectively. Several new polymorphisms were found in some genes involved in alginate production such as algC, algE, algG, algI, algX, mucD and mucK (Table S1). Of note, isolate MS6000 presented a new inactivating mutation in mucA, due to a substitution of five nucleotides at position 246 (Table S1), generating an anomalous MucA protein. Mutations in mucA are related with the transition from the nonmucoid to mucoid condition in P. aeruginosa [75,76], which correlates with the mucoid phenotype observed in isolate MS6000 (Table 1). Type VI secretion system (T6SS) genes from Hcp secretion island-1 (H1-T6SS) were also in all five genomes, among them the effector genes tse1, tse2 and tse3. The gene toxA, coding for exotoxin A, one of the most potent virulence factors produced by P. aeruginosa, was observed in all the isolates. Other virulence genes found included those related with biosynthesis of phenazines, pyoverdine and pyochelin, Type II Secretion System, LPS biosynthesis and quorum sensing (QS) genes rhlI, rhlC, lasS, lasB and lasI (Table S2). All the isolates presented the amino acid substitution S62G and D83E in RhlI (Table S1). These mutations have been described in P. aeruginosa isolates from different origins and do not seem to affect the protein function [77]. A frameshift mutation in the master QS signal receptor gene lasR due to a deletion of one nucleotide at position 605, was detected in all the isolates. It is known that LasR controls the expression of several genes, including lasI, rhlI and rhlR in laboratory adapted strains, such as PAO1. However, inactivating mutations in lasR are commonly found in P. aeruginosa isolates from chronic lung infections of CF individuals with an active RhlI/R QS system. This indicates that alternative pathways controlling the expression of RhlI/R system exist and that the las system is not entirely essential for virulence of P. aeruginosa, as was demonstrated before [77,78].
2.2.3. Analysis of Genes Related with P. aeruginosa Hypermutability
The hypermutator phenotype in P. aeruginosa appears as a result of mutations in genes related with in DNA repair processes, especially mutS, mutL and uvrD [33]. In this study we found a number of mutations in uvrD and mutL but not in mutS (Table S1). According to former studies, mutations in mutL seem to be the most frequent cause of hypermutability [79]. Mutations in other genes, including those involved in recombination functions (recB, recC, recD, recJ, recN, recR, recX, rdgC, rarA, uvrB and uvrC) and stress response-related polA and polB were also detected in our isolates (Table S1). The hypermutator phenotype is commonly found in P. aeruginosa isolates from CF patients and it is associated with reduced pulmonary function [80]. Hypermutation in specific genome regions is considered a beneficial mechanism in terms of bacterial fitness and adaptation of P. aeruginosa to persist in the CF lung environment. The hypermutator phenotype is usually linked to development of high levels of resistance to antibiotics in P. aeruginosa isolates from CF patients [42].
2.3. Biofilm Formation Capacities of CF P. aeruginosa Isolates
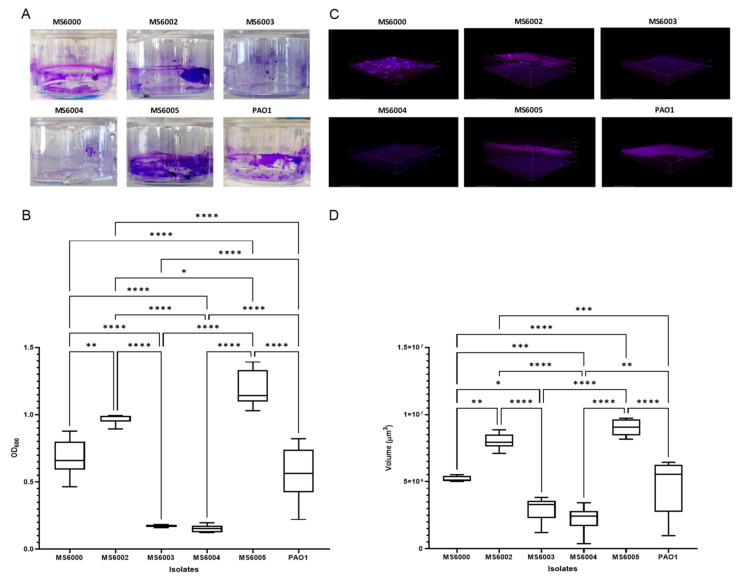
The ability of the CF P. aeruginosa isolates to form biofilm was visualized and quantified by using both the crystal violet (CV) and the combination of fluorescent dye SYPRO Ruby/DAPI (Figure 2). All the isolates grew well and were able to produce biofilm. According to the CV biofilm quantification results isolates MS6003 (O.D = 0.17 ± 0.008) and MS6004 (O.D = 0.15 ± 0.02) were classified as moderate biofilm producers, while isolates MS6000 (O.D = 0.68 ± 0.14), MS6002 (O.D = 0.97 ± 0.04) and MS6005 (O.D = 1.19 ± 0.13) were considered as strong biofilm producers. Biofilm production by the CV staining was compared among the isolates and P. aeruginosa PAO1 was used as a reference strain (O.D = 0.56 ± 0.21) (Figure 2B). We used confocal laser scanning microscopy (CSLM) to visualize the five CF P. aeruginosa isolates biofilm production and architecture (Figure 2C). SYPRO Ruby/DAPI fluorescent staining illustrated significant differences in total biofilm matrix quantity among isolates (Figure 2C). Isolates MS6005 (9.03 ± 0.61 × 106 µm3) and MS6002 (7.99 ± 0.59 × 106 µm3) displayed the largest volume of biofilm matrix, followed by isolate MS6000 (5.20 ± 0.21 × 106 µm3), which showed similar values to PAO1 (4.68 ± 2.12 × 106 µm3), while isolates MS6003 (2.96 ± 0.94 × 106 µm3) and MS6004 (2.24 ± 1.01 × 106 µm3) showed less matrix substance production (Figure 2D). There was a substantial correlation between the two quantification methods (R2 = 0.782) (Figure S1).
Figure 2.
Biofilm formation of the CF P. aeruginosa isolates of ST3449. (A) Biofilm were developed in 24-well plastic plates and quantified after crystal violet-staining. (C) Representative three-dimensional (3D) reconstructions of confocal laser scanning microscopy (CLSM) of P. aeruginosa biofilm developed on coated 4-well chamber slides and stained with SYPRO Ruby and DAPI. Representative extracted z images and their respective xy and xz planes are shown. (B,D) Corresponding levels of biofilm production represent mean values and standard deviations obtained from six independent experiments. Dunnett’s multiple-comparison test was performed and *, **, *** and **** refer to p < 0.05, p < 0.01, p < 0.001 and p < 0.0001 significant difference, respectively.
Biofilm production is of particularly concern because of its association with resistance to antimicrobial agents and evasion to host immune response mechanisms. When P. aeruginosa adopts the biofilm form it can easily adapt to the severe inflammatory conditions and antibiotic selective pressure during long periods without eradication [81].
Exopolysaccharides are known to play an important role in the biofilm formation and maintenance. Most of P. aeruginosa strains, including PAO1, rely on Pel and Psl polysaccharides to form biofilm and interact with host cells and there is a complex gene network involved in regulation and production of biofilm [82].
Biofilm production is of particular concern because of its association with resistance to antimicrobial agents and evasion to host immune response mechanisms. When P. aeruginosa adopts the biofilm form it can easily adapt to the severe inflammatory conditions and antibiotic selective pressure during long periods without eradication [81].
Exopolysaccharides are known to play an important role in the biofilm formation and maintenance. Most of P. aeruginosa strains, including PAO1, rely on Pel and Psl polysaccharides to form biofilm and interact with host cells and there is a complex gene network involved in regulation and production of this structure [82].
Similar to other studies, we observed that our CF isolates were able to produce different levels of biofilm in vitro. The WGS analysis revealed several polymorphisms and mutations in genes related with production the aforementioned exopolysaccharides and other key genes involved in biofilm formation such as lasR, ladS, pqsA, pqsD, rhlI and retS (Table S1). Interestingly, MS6000, MS6002 and MS6005, the isolates showing the most robust biofilm production, presented a premature stop codon in the gen pelD generated by a nucleotide substitution at position 731 (CAG by TAG), thus originating a truncated peptide at residue Q247. PelD is a cyclic diguanylate effector considered essential for biofilm production [83]. This observation led us to believe that alternative mechanism for biofilm production should exist in P. aeruginosa, as other plausible explanations are difficult to suggest from these experiments. Despite the large amount of information existing on biofilm production in P. aeruginosa, there are still many aspects to decipher as it is a multifarious mechanism, which depends not only on bacterial specific gene features but also on physiological and environmental conditions. In our case, more extensive experiments must be conducted to determine the actual contribution of those mutations to the differences observed in biofilm production in the CF P. aeruginosa isolates.
2.4. Virulence in Galleria mellonella
Virulence of the CF P. aeruginosa isolates was evaluated in the greater wax moth G. mellonella. Several studies have employed G. mellonella as a useful model host for investigating virulence traits of different human microbial pathogens [84].
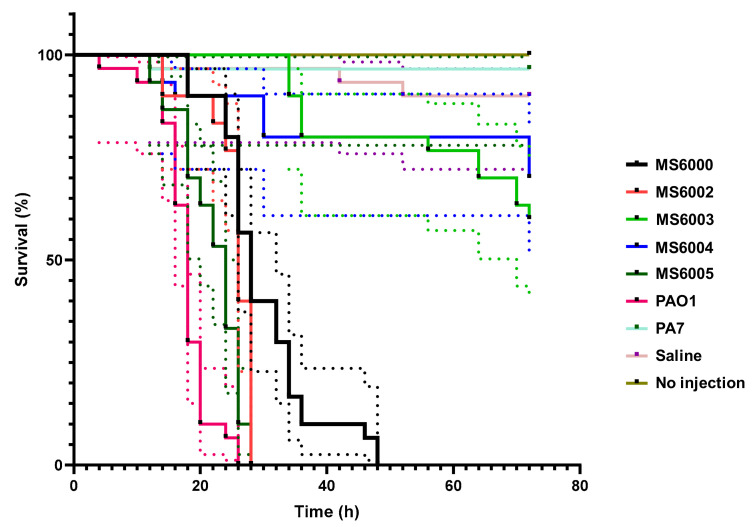
P. aeruginosa PAO1 and PA7 were employed as high and low virulence control strains, respectively. The survival of isolates was monitored for 72 h after infection in order to determine their pathogenic potential. We found significant differences in larvae mortality among different groups (p < 0.05) (Figure 3). The most virulent isolates were MS6005, MS6002 and MS6000, showing median survival times of 24, 26 and 28 h, respectively and for control strain PAO1 18 h. In contrast, isolates MS6003 and MS6004 presented a very low virulence capacity, as at 72 h post inoculation more than 50% of larvae in both groups remained still alive.
Figure 3.
Kaplan–Meier survival curves of G. mellonella larvae after injection with 10 µL of 1 × 103 CFUs of CF P. aeruginosa isolates of ST3449. Dead larvae were monitored every 2 h after inoculation up to 72 h. Uninoculated larvae and those inoculated with saline solution (0.85% NaCl), and P. aeruginosa strains PAO1 and PA7 were employed as control groups. The data shown represent three independent experiments with 95% confidence intervals, represented as dotted lines The isolates MS6002 and MS6005 showed high virulence capacity (killed 100% of the larvae at 28 h, similar to PAO1 that killed 100% of the larvae at 26 h) compared to that of MS6000 (killed 100% of the larvae at 48 h). Isolates MS6003 and MS6004 demonstrated very low virulence capacity.
Even when all our isolates presented similar number of virulence factors, the differences found in virulence patterns in G. mellonella reflects that virulence may be subjected to transcriptional regulation, depending on each isolate genetic background. Production of pigments are also associated with virulence in P. aeruginosa. As mentioned before our isolates displayed also differences in pigmentation, but we could not directly associate this phenotypic feature with larvae mortality observed in each group. Therefore, we corroborate that differences observed in the virulence patterns of the five isolates is not restricted to a certain number of virulence genes but it is a result of combinatorial mechanisms, which has been stated previously [85].
3. Materials and Methods
3.1. Bacterial Isolates and Antimicrobial Susceptibility Testing
Five consecutive P. aeruginosa isolates were collected during a month from respiratory samples (three from sputum and two from tracheal aspirate) from a cystic fibrosis patient with end-stage respiratory failure who died before lung transplantation.
The isolates were routinely grown on blood and/or Mueller–Hinton agar plates at 37 °C. Identification was performed by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF/Vitek-MS with SARAMIS MS-IVD v2) [86]. Phenotypic traits such as colony morphology, hemolysis and pigment production were also examined.
The minimum inhibitory concentrations (MICs) of aztreonam, cefepime, ceftazidime, piperacillin, piperacillin-tazobactam, imipenem, meropenem, doripenem, amikacin, gentamicin, tobramycin, ciprofloxacin, levofloxacin and colistin were determined by broth microdilution according to CLSI guidelines, except for fosfomycin that was performed by agar dilution and the combinations of ceftolozane-tazobactam and ceftazidime-avibactam that were determined by Etest according to the manufacturer’s recommendations (bioMérieux, Marcy-I’Etoile, France). All antibiotics were purchased from Sigma-Aldrich. MIC breakpoints for all agents were those established by EUCAST 2021 v_11.0 (http://www.eucast.org/clinical_breakpoints/ (accessed on January 2021), except for fosfomycin that was done using an ECOFF (epidemiological cutoff) value of ≤128 µ/mL, and gentamicin for which CLSI breakpoints were applied. Escherichia coli ATCC 25,922 and P. aeruginosa ATCC 27,853 were employed as quality control strains. Susceptibility categories of multidrug resistant (MDR) and extensively drug resistance (XDR) were assigned to the strains according to the expert consensus recommendations previously reported [87].
3.2. DNA Extraction and Sequencing
Total DNA from the P. aeruginosa isolates was purified with the DNeasy Blood and Tissue Kit (Qiagen) and sequenced on a MiSeq device using reagents kit v3 for 2 × 300 paired-end libraries (Illumina) as previously described [88].
3.3. Bioinformatics Analysis
Raw reads from the sequencing platform were directly analyzed using the in-house bioinformatics pipeline TORMES® [13]. P. aeruginosa PAO1 was used as a reference strain. The options used in this study included quality control and filtering of the reads by using Trimmomatic [89], Prinseq [90] and Kraken [91]. Genome assembly was performed with SPAdes [92] and Quast [93] and genome annotation with Prokka [94]. Multilocus sequence typing was performed using an open source tool (MLST, T. Seemann, https://github.com/tseemann/mlst (accessed on September 2020). Search of antibiotic resistance genes and plasmid replicons was done using BLAST [95] and ABRicate (https://github.com/tseemann/abricate (accessed on September 2020)) against ResFinder [19] and PlasmidFinder [96] databases, respectively. Search for single nucleotide polymorphisms (SNPs), insertion and deletions (indels) of nucleotides was performed with breseq [20]. Pangenome was created with Roary [97] and FastTree [98]. In silico serotyping was performed using the P. aeruginosa serotyper (PAst) program [15]. Plotting was carried out in R environment [99] by using ggplot2 [100], RColorBrewer [101] and reshape2 [102] packages. Blast Ring Image Generator (BRIG) [14] was used to show a genome wide visualization.
3.4. Biofilm Formation Assay
Biofilm formation was evaluated by means of the crystal violet staining assay as described before [103], with slight modifications. Briefly, P. aeruginosa overnight cultures were adjusted to an optical density at 600 nm (O.D600) of 0.2 in MHB. Biofilms were developed in 24 well plates (Nunc, Thermo Fisher Scientific). Subsequently, 100 µL of each bacterial suspension were placed in each well containing 900 μL of MHB and statically incubated at 37 °C for 24 h. Planktonic cells were removed and biofilms were washed three times with distilled water and air-dried for 20 min. Biofilms were stained with 1.5 mL/well of 0.7% crystal violet (wt/vol) solution (Sigma-Aldrich) for 12 min. The amount of dye (proportional to the density of adherent cells) was determined at 590 nm using a plate reader (Infinite® 200 PRO, Tecan). Results were corrected for background staining by subtracting the value for crystal violet bound to uninoculated MHB control wells. The OD590/OD600 ratio was determined in order to normalize the amount of biofilm produced with respect to the total cell content in each sample to avoid variations due to differences in bacterial growth under different experimental conditions. The biofilm assay was performed six independent times. For the interpretation of biofilm results isolates were classified as non-biofilm producer (OD ≤ 0.05), weak-biofilm producer (OD > 0.05–0.1), moderate-biofilm producer (OD > 0.1–0.3) and strong-biofilm producer (OD > 0.3).
3.5. Confocal Laser Scanning Microscopy (CLSM)
Biofilm architecture was investigated by CLSM. Biofilms were developed in 4-well µ-slides (Ibidi, Martinsried, Germany) as previously described [104], but with few modifications. In brief, slides containing 500 µL of bacterial cultures (adjusted to an optical density OD600 = 0.2 in MHB), were placed inclined (45°) into an incubator at 37 °C in order to form a liquid–air interface. Planktonic cells were removed after 24 h, by rinsing three times with saline solution (0.85% NaCl), and then biofilms were stained with the FilmTracerTM SYPRO Ruby Biofilm Matrix Stain (ThermoFisher Scientific) and 4′,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich) to reveal the biofilm protein components and nucleic acids, respectively. Both staining procedures were performed according to the manufacturer’s instructions. The stained biofilms were visualized under a Nikon A1R confocal laser scanning microscope (Nikon Instruments Inc., Melville, NY, USA). Six surfaces of six independent biological replicates were observed in each CLSM experiment, and representative images were selected. The excitation wavelengths were 405 nm and emission filters of 425–475 nm and 570–620 nm were used for DAPI and SYPRO, respectively. Measurements of biofilms 3D images were performed with the NIS-Element AR NiKon instrument software, version 3.2 (Nikon Instruments Inc., Melville, NY, USA).
3.6. Galleria mellonella Killing Assays
To assess the virulence of the P. aeruginosa isolates in vivo, the greater wax moth G. mellonella was used as a model of infection. Larvae of G. mellonella were obtained from Animal Center Valencia S.L (Valencia, Spain), stored in the dark on wood chips at 8–10 °C and used within 10 days of receipt. Larvae were selected to be 20–25 mm in length, presenting a healthy creamy color without speckles or grey markings.
Previous to inoculation, bacteria were grown overnight in MHB at 37 °C and the bacterial cultures were normalized using optical density (OD600) and the colony forming units (CFU/mL) were confirmed by the viable count assay. Bacterial inocula were then adjusted to 1 × 103 CFUs with saline solution (0.85% NaCl) and 10 μL were injected into the hemocoel via the left proleg of each larva using a BD Micro-Fine sterile insulin syringe (0.33 mm (29G) × 12.7 mm, 500 µL capacity) (Becton Dickinson, Franklin Lakes, NJ, USA). The syringes were changed between treatments with different bacterial inocula. Two negative control groups were included; one group of 10 larvae were injected with 10 μL of saline solution in order to measure any lethal effect associated to the injection process and a second group of not injected larvae were used to measure the effects of the incubation procedure on larvae mortality. Two P. aeruginosa strains PAO1 [105] and PA7 [106] were used as controls for fast and slow killing assays, respectively. Three biological replicates of 10 larvae were used for each experimental condition. Larvae were then incubated in petri dishes at 37 °C in the dark under aerobic conditions and survival was recorded at 2 h intervals for 72 h. Larvae were considered dead when they displayed complete melanization and/or no response was observed following touch.
3.7. Statistical Analyses
All statistical analyses were carried out using GraphPad Prism version 9.0.2 (GraphPad Software, San Diego, CA, USA). All experiments were performed at least six times and the results were shown as means ± standard deviation. Differences between mean values were tested for significance by performing one-way ANOVA analysis followed by Tukey’s multiple-comparison test, when appropriate. Survival curves were plotted using the Kaplan–Meier method, and differences in survival were calculated using the log-rank (Mantel–Cox) test for multiple comparisons. In all cases differences were considered statistically significant at p < 0.05.
3.8. Nucleotide Accession Numbers
The whole-genome sequencing reads and annotated assembly of the five P. aeruginosa isolates of ST3449 are available under the BioProject ID PRJNA695127.
4. Conclusions
This work is relevant as it identified and characterized five consecutive clonally related multidrug-resistant P. aeruginosa isolates (four of them XDR) that belong to a novel sequence type ST3449 and serotype O6, from a CF patient with end-stage pulmonary disease, who died before lung transplantation.
Our principal aim was the whole-genome sequence analysis of these isolates showing diverse phenotypic features such as changes in pigmentation, colony morphology, biofilm production and virulence capacity. This study demonstrates that the isolates analyzed here are probably the ones that presented a better fitness to subsist in the CF lung environment at the end-stage patient’s disease. Other isolates with less capacity of adaptation or reduced fitness might have coexisted at the same spatial time, but we could not isolate those ones from early-phase (primo-colonization) or mid-term colonization. Our isolates were collected during a month, and the patient had died already when the isolates were examined. Analysis of earlier isolates would have allowed us to compare the evolution of this novel P. aeruginosa clone ST3449 or the possible presence of other clones coexisting within a defined period of time.
Our observations corroborate previous studies regarding the coexistence of clonally related P. aeruginosa strains showing different phenotypic and genotypic characteristics within the lung of CF patients, which is a reflect of the enormous capacity of adaptation of this pathogen to the hostile environment in the CF lung. Due to the WGS we could describe new mutation in several genes related with antimicrobial resistance, virulence and other cell functions, which reflects the high pressure P. aeruginosa experiences in the lung of CF patients to generate new allelic variants of these genes in order to persist and survive in such harsh environment. On the other hand we could observe that some mutations in antimicrobial resistance related genes are possible a distinctive feature of P. aeruginosa isolates from CF as they repeatedly occur in these isolates.
Despite not performing any experiment to demonstrate mutation frequencies, all our isolates are very likely hypermutable, as a number of mutations were described in some genes known to be responsible for the hypermutator phenotype.
We believe that our results provide some new valuable information about the evolution and dynamics of antimicrobial resistance and virulence mechanisms seen through the repertoire of mutational events accumulated by some P. aeruginosa clones as is the case of this novel ST3449, described here for the first time in a CF patient. We consider it is important to continue characterizing and monitoring this new clone due to the potential risk it may represent for human health.
Acknowledgments
We would like to thank Jens Klockgether and Burkhard Tümmler for providing the P. aeruginosa strain PA7 and Fidel Madrazo (Electron Microscopy Unit, Technology Support Services, IDIVAL) for helping with confocal microscopy experiments. We would also like to express our gratitude to the Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica (SEIMC) for supporting some of the work performed by C.D.-R.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10050491/s1, Table S1: Mutations found in the WGS analysis of the P. aeruginosa isolates from ST3449. Table S2: Virulence genes of P. aeruginosa isolates from ST3449. Figure S1: Correlation between biofilm formation and quantification methods.
Author Contributions
C.D.-R. and M.H. performed most experiments, analyzed the results and participated in draft preparation; D.A. performed genome assembly and bioinformatics analysis; L.Á.-M. Performed susceptibility tests and biofilm experiments; A.V. performed the statistical and sequence analysis; D.I. Formal analysis and edited the paper; J.C. Isolated the strains, preformed the previous antimicrobial susceptibility test and edited the paper; A.A.O.-S. acquired funds, conceived the study, designed the experiments, analyzed the results, wrote, reviewed and edited the last version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Instituto de Salud Carlos III, Subdirección General de Evaluación y Fomento de la Investigación, Ministerio de Economía, Industria y Competitividad (Grants PI15/00009 and PI18/00380) and by Plan Nacional de I + D + and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Ciencia, Innovación y Universidades, Spanish Network for Research in Infectious Diseases (REIPI RD12/0015 and RD16/0016)-cofinanced by European Development Regional Fund “A way to achieve Europe” A.A.O.-S. was supported by Miguel Servet research contracts from the Instituto de Salud Carlos III. CP12-03149 and CPII17-00011).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are in Supplementary Tables S1 and S2. Genome sequences information is available in https://www.ncbi.nlm.nih.gov/bioproject/ (accessed on 26 January 2021) under the BioProject ID PRJNA695127.
Conflicts of Interest
The authors have no conflict of interest to disclose.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Häußler S., Ziegler I., Löttel A., Götz F.V., Rohde M., Wehmhöhner D., Saravanamuthu S., Tümmler B., Steinmetz I. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J. Med. Microbiol. 2003;52:295–301. doi: 10.1099/jmm.0.05069-0. [DOI] [PubMed] [Google Scholar]
- 2.Bianconi I., D’Arcangelo S., Esposito A., Benedet M., Piffer E., Dinnella G., Gualdi P., Schinella M., Baldo E., Donati C., et al. Persistence and microevolution of pseudomonas aeruginosa in the cystic fibrosis lung: A single-patient longitudinal genomic study. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2018.03242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sousa A.M., Pereira M.O. Pseudomonas Aeruginosa diversification during infection development in cystic fibrosis Lungs—A review. Pathogens. 2014;3:680–703. doi: 10.3390/pathogens3030680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winstanley C., O’brien S., Brockhurst M.A. Pseudomonas aeruginosa Evolutionary Adaptation and Diversification in Cystic Fibrosis Chronic Lung Infections. Trends Microbiol. 2016;24:327–337. doi: 10.1016/j.tim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lechtzin N., John M., Irizarry R., Merlo C., Diette G.B., Boyle M.P. Outcomes of adults with cystic fibrosis infected with antibiotic-resistant Pseudomonas aeruginosa. Respiration. 2006;73:27–33. doi: 10.1159/000087686. [DOI] [PubMed] [Google Scholar]
- 6.Parkins M.D., Somayaji R., Waters V.J. Epidemiology, Biology, and Impact of Clonal Pseudomonas aeruginosa Infections in Cystic Fibrosis. Clin. Microbiol. Rev. 2018;31:e00019-18. doi: 10.1128/CMR.00019-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith E.E., Buckley D.G., Wu Z., Saenphimmachak C., Hoffman L.R., D’Argenio D.A., Miller S.I., Ramsey B.W., Speert D.P., Moskowitz S.M., et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feliziani S., Marvig R.L., Luján A.M., Moyano A.J., Di Rienzo J.A., Krogh Johansen H., Molin S., Smania A.M. Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable Pseudomonas aeruginosa in Long-term Cystic Fibrosis Infections. PLoS Genet. 2014;10:e1004651. doi: 10.1371/journal.pgen.1004651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Mansfeld R., De Been M., Paganelli F., Yang L., Bonten M., Willems R. Within-Host evolution of the Dutch high-prevalent pseudomonas aeruginosa clone ST406 during Chronic colonization of a patient with cystic fibrosis. PLoS ONE. 2016;11:e0158106. doi: 10.1371/journal.pone.0158106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hilliam Y., Moore M.P., Lamont I.L., Bilton D., Haworth C.S., Foweraker J., Walshaw M.J., Williams D., Fothergill J.L., De Soyza A., et al. Pseudomonas aeruginosa adaptation and diversification in the non-cystic fibrosis bronchiectasis lung. Eur. Respir. J. 2017;49:1602108. doi: 10.1183/13993003.02108-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klockgether J., Cramer N., Fischer S., Wiehlmann L., Tümmler B. Long-term microevolution of pseudomonas aeruginosa differs between mildly and severely affected cystic fibrosis lungs. Am. J. Respir. Cell Mol. Biol. 2018;59:246–256. doi: 10.1165/rcmb.2017-0356OC. [DOI] [PubMed] [Google Scholar]
- 12.Dettman J.R., Kassen R. Evolutionary Genomics of Niche-Specific Adaptation to the Cystic Fibrosis Lung in Pseudomonas aeruginosa. Mol. Biol. Evol. 2021;38:663–675. doi: 10.1093/molbev/msaa226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quijada N.M., Rodríguez-Lázaro D., Eiros J.M., Hernández M. TORMES: An automated pipeline for whole bacterial genome analysis. Bioinformatics. 2019;35:4207–4212. doi: 10.1093/bioinformatics/btz220. [DOI] [PubMed] [Google Scholar]
- 14.Alikhan N.F., Petty N.K., Zakour N.L.B., Beatson S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011;12 doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thrane S.W., Taylor V.L., Lund O., Lam J.S., Jelsbak L. Application of whole-genome sequencing data for o-specific antigen analysis and in silico serotyping of pseudomonas aeruginosa isolates. J. Clin. Microbiol. 2016;54:1782–1788. doi: 10.1128/JCM.00349-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavallo J.D., Leblanc F., Fabre R., Avril J.L., Bebear C., Cavallo J.D., Etienne J., Leclercq R., Marty N., Nguyen J., et al. Surveillance de la sensibilite de pseudomonas aeruginosa aux antibiotiques en france et distribution des mecanismes de resistance aux β- lactamines: Etude GERPB 1998. Pathol. Biol. 2001;49:534–539. doi: 10.1016/S0369-8114(01)00213-9. [DOI] [PubMed] [Google Scholar]
- 17.Leone I., Chirillo M.G., Raso T., Zucca M., Savoia D. Phenotypic and genotypic characterization of Pseudomonas aeruginosa from cystic fibrosis patients. Eur. J. Clin. Microbiol. Infect. Dis. 2008;27:1093–1099. doi: 10.1007/s10096-008-0551-1. [DOI] [PubMed] [Google Scholar]
- 18.Del Barrio-Tofiño E., Sánchez-Diener I., Zamorano L., Cortes-Lara S., López-Causapé C., Cabot G., Bou G., Martínez-Martínez L., Oliver A. Association between Pseudomonas aeruginosa O-antigen serotypes, resistance profiles and high-risk clones: Results from a Spanish nationwide survey. J. Antimicrob. Chemother. 2019;74 doi: 10.1093/jac/dkz346. [DOI] [PubMed] [Google Scholar]
- 19.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deatherage D.E., Barrick J.E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolter D.J., Lister P.D. Mechanisms of β-lactam resistance among Pseudomonas aeruginosa. Curr. Pharm. Des. 2013;19:209–222. doi: 10.2174/138161213804070311. [DOI] [PubMed] [Google Scholar]
- 22.Ropy A., Cabot G., Sánchez-Diener I., Aguilera C., Moya B., Ayala J.A., Oliver A. Role of Pseudomonas aeruginosa low-molecular-mass penicillin-binding proteins in AmpC expression, β-lactam resistance, and peptidoglycan structure. Antimicrob. Agents Chemother. 2015;59:3925–3934. doi: 10.1128/AAC.05150-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galetti R., Andrade L.N., Varani A.M., Darini A.L.C. SPM-1-producing Pseudomonas aeruginosa ST277 carries a chromosomal pack of acquired resistance genes: An example of high-risk clone associated with ‘intrinsic resistome. J. Glob. Antimicrob. Resist. 2019;16:183–186. doi: 10.1016/j.jgar.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 24.Hussain M., Suliman M., Ahmed A., Altayb H., Elneima E. Draft genome sequence of a multidrug-resistant Pseudomonas aeruginosa strain isolated from a patient with a urinary tract infection in khartoum, Sudan. Genome Announc. 2017;5 doi: 10.1128/genomeA.00203-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grandjean T., Le Guern R., Duployez C., Faure K., Kipnis E., Dessein R. Draft Genome Sequences of Two Pseudomonas aeruginosa Multidrug-Resistant Clinical Isolates, PAL0.1 and PAL1.1. Microbiol. Resour. Announc. 2018;7 doi: 10.1128/MRA.00940-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madaha E.L., Mienie C., Gonsu H.K., Bughe R.N., Fonkoua M.C., Mbacham W.F., Alayande K.A., Bezuidenhout C.C., Ateba C.N. Whole-genome sequence of multi-drug resistant Pseudomonas aeruginosa strains UY1PSABAL and UY1PSABAL2 isolated from human broncho-alveolar lavage, Yaoundé, Cameroon. PLoS ONE. 2020;15:e0238390. doi: 10.1371/journal.pone.0238390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan M., Stapleton F., Summers S., Rice S.A., Willcox M.D.P. Antibiotic resistance characteristics of pseudomonas aeruginosa isolated from keratitis in Australia and India. Antibiotics. 2020;9:600. doi: 10.3390/antibiotics9090600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodríguez-Martínez J.M., Poirel L., Nordmann P. Extended-spectrum cephalosporinases in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2009;53:1766–1771. doi: 10.1128/AAC.01410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tam V.H., Schilling A.N., LaRocco M.T., Gentry L.O., Lolans K., Quinn J.P., Garey K.W. Prevalence of AmpC over-expression in bloodstream isolates of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 2007;13:413–418. doi: 10.1111/j.1469-0691.2006.01674.x. [DOI] [PubMed] [Google Scholar]
- 30.Cabot G., Ocampo-Sosa A.A., Domínguez M.A., Gago J.F., Juan C., Tubau F., Rodríguez C., Moyà B., Peña C., Martínez-Martínez L., et al. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob. Agents Chemother. 2012;56:6349–6357. doi: 10.1128/AAC.01388-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berrazeg M., Jeannot K., Ntsogo Enguéné V.Y., Broutin I., Loeffert S., Fournier D., Plésiat P. Mutations in β-lactamase AmpC increase resistance of Pseudomonas aeruginosa isolates to antipseudomonal cephalosporins. Antimicrob. Agents Chemother. 2015;59:6248–6255. doi: 10.1128/AAC.00825-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kao C.Y., Chen S.S., Hung K.H., Wu H.M., Hsueh P.R., Yan J.J., Wu J.J. Overproduction of active efflux pump and variations of OprD dominate in imipenem-resistant Pseudomonas aeruginosa isolated from patients with bloodstream infections in Taiwan. BMC Microbiol. 2016;16:107. doi: 10.1186/s12866-016-0719-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.López-Causapé C., Sommer L.M., Cabot G., Rubio R., Ocampo-Sosa A.A., Johansen H.K., Figuerola J., Cantón R., Kidd T.J., Molin S., et al. Evolution of the Pseudomonas aeruginosa mutational resistome in an international Cystic Fibrosis clone. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-05621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zamudio R., Hijazi K., Joshi C., Aitken E., Oggioni M.R., Gould I.M. Phylogenetic analysis of resistance to ceftazidime/avibactam, ceftolozane/tazobactam and carbapenems in piperacillin/tazobactam-resistant Pseudomonas aeruginosa from cystic fibrosis patients. Int. J. Antimicrob. Agents. 2019;53:774–780. doi: 10.1016/j.ijantimicag.2019.02.022. [DOI] [PubMed] [Google Scholar]
- 35.Juan C., Moyá B., Pérez J.L., Oliver A. Stepwise upregulation of the Pseudomonas aeruginosa chromosomal cephalosporinase conferring high-level β-lactam resistance involves three AmpD homologues. Antimicrob. Agents Chemother. 2006;50:1780–1787. doi: 10.1128/AAC.50.5.1780-1787.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cabot G., López-Causapé C., Ocampo-Sosa A.A., Sommer L.M., Domínguez M.Á., Zamorano L., Juan C., Tubau F., Rodríguez C., Moyà B., et al. Deciphering the Resistome of the Widespread Pseudomonas aeruginosa Sequence Type 175 International High-Risk Clone through Whole-Genome Sequencing. Antimicrob. Agents Chemother. 2016;60:7415–7423. doi: 10.1128/AAC.01720-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z., Cai Z., Cai Z., Zhang Y., Fu T., Jin Y., Cheng Z., Jin S., Wu W., Yang L., et al. Molecular genetic analysis of an XDR Pseudomonas aeruginosa ST664 clone carrying multiple conjugal plasmids. J. Antimicrob. Chemother. 2020;75:1443–1452. doi: 10.1093/jac/dkaa063. [DOI] [PubMed] [Google Scholar]
- 38.Tsutsumi Y., Tomita H., Tanimoto K. Identification of novel genes responsible for overexpression of ampC in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 2013;57:5987–5993. doi: 10.1128/AAC.01291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calvopiña K., Avisona M.B. Disruption of mpl activates -lactamase production in stenotrophomonas maltophilia and pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.00638-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caballero J.D., Clark S.T., Coburn B., Zhang Y., Wang P.W., Donaldson S.L., Elizabeth Tullis D., Yau Y.C.W., Waters V.J., Hwang D.M., et al. Selective sweeps and parallel pathoadaptation drive Pseudomonas aeruginosa evolution in the cystic fibrosis lung. MBio. 2015;6:e00981-15. doi: 10.1128/mBio.00981-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams D., Evans B., Haldenby S., Walshaw M.J., Brockhurst M.A., Winstanley C., Paterson S. Divergent, coexisting Pseudomonas aeruginosa lineages in chronic cystic fibrosis lung infections. Am. J. Respir. Crit. Care Med. 2015;191:775–785. doi: 10.1164/rccm.201409-1646OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colque C.A., Albarracín Orio A.G., Feliziani S., Marvig R.L., Tobares A.R., Johansen H.K., Molin S., Smania A.M. Hypermutator pseudomonas aeruginosa exploits multiple genetic pathways to develop multidrug resistance during long-term infections in the airways of cystic fibrosis patients. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.02142-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papp-Wallace K.M., Zeiser E.T., Becka S.A., Park S., Wilson B.M., Winkler M.L., D’Souza R., Singh I., Sutton G., Fouts D.E., et al. Ceftazidime-Avibactam in Combination With Fosfomycin: A Novel Therapeutic Strategy Against Multidrug-Resistant Pseudomonas aeruginosa. J. Infect. Dis. 2019;220:666–676. doi: 10.1093/infdis/jiz149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Domitrovic T.N., Hujer A.M., Perez F., Marshall S.H., Hujer K.M., Woc-Colburn L.E., Parta M., Bonomo R.A. Multidrug resistant Pseudomonas aeruginosa causing prosthetic valve endocarditis: A genetic-based chronicle of evolving antibiotic resistance. Open Forum Infect. Dis. 2016;3:ofw188. doi: 10.1093/ofid/ofw188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horna G., López M., Guerra H., Saénz Y., Ruiz J. Interplay between MexAB-OprM and MexEF-OprN in clinical isolates of Pseudomonas aeruginosa. Sci. Rep. 2018 doi: 10.1038/s41598-018-34694-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H., Luo Y.F., Williams B.J., Blackwell T.S., Xie C.M. Structure and function of OprD protein in Pseudomonas aeruginosa: From antibiotic resistance to novel therapies. Int. J. Med. Microbiol. 2012;302:63–68. doi: 10.1016/j.ijmm.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ocampo-Sosa A.A., Cabot G., Rodríguez C., Roman E., Tubau F., Macia M.D., Moya B., Zamorano L., Suárez C., Peña C., et al. Alterations of OprD in carbapenem-intermediate and -susceptible strains of Pseudomonas aeruginosa isolated from patients with bacteremia in a spanish multicenter study. Antimicrob. Agents Chemother. 2012;56:1703–1713. doi: 10.1128/AAC.05451-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.López-García A., Rocha-Gracia R.D.C., Bello-López E., Juárez-Zelocualtecalt C., Sáenz Y., Castañeda-Lucio M., López-Pliego L., González-Vázquez M.C., Torres C., Ayala-Nuñez T., et al. Characterization of antimicrobial resistance mechanisms in carbapenem-resistant pseudomonas aeruginosa carrying IMP variants recovered from a Mexican hospital. Infect. Drug Resist. 2018;11:1523–1536. doi: 10.2147/IDR.S173455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruiz-Roldán L., Bellés A., Bueno J., Azcona-Gutiérrez J.M., Rojo-Bezares B., Torres C., Castillo F.J., Sáenz Y., Seral C. Pseudomonas aeruginosa Isolates from Spanish Children: Occurrence in Faecal Samples, Antimicrobial Resistance, Virulence, and Molecular Typing. Biomed Res. Int. 2018:8060178. doi: 10.1155/2018/8060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Epp S.F., Köhler T., Plésiat P., Michéa-Hamzehpour M., Frey J., Pechère J.C. C-terminal region of Pseudomonas aeruginosa outer membrane porin OprD modulates susceptibility to meropenem. Antimicrob. Agents Chemother. 2001;45:1780–1787. doi: 10.1128/AAC.45.6.1780-1787.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morita Y., Tomida J., Kawamura Y. Mexxy multidrug efflux system of Pseudomonas aeruginosa. Front. Microbiol. 2012;3:408. doi: 10.3389/fmicb.2012.00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vettoretti L., Plésiat P., Muller C., El Garch F., Phan G., Attrée I., Ducruix A., Llanes C. Efflux unbalance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 2009;53:1987–1997. doi: 10.1128/AAC.01024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caughlan R.E., Sriram S., Daigle D.M., Woods A.L., Buco J., Peterson R.L., Dzink-Fox J.A., Walker S., Dean C.R. Fmt bypass in Pseudomonas aeruginosa causes induction of MexXY efflux pump expression. Antimicrob. Agents Chemother. 2009;53:5015–5021. doi: 10.1128/AAC.00253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poonsuk K., Chuanchuen R. Contribution of the MexXY multidrug efflux pump and other chromosomal mechanisms on aminoglycoside resistance in Pseudomonas aeruginosa isolates from canine and feline infections. J. Vet. Med. Sci. 2012;74:1575–1582. doi: 10.1292/jvms.12-0239. [DOI] [PubMed] [Google Scholar]
- 55.Olsson A., Wistrand-Yuen P., Nielsen E.I., Friberg L.E., Sandegren L., Lagerbäck P., Tängdén T. Efficacy of antibiotic combinations against multidrug-resistant pseudomonas aeruginosa in automated time-lapse microscopy and static time-kill experiments. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.02111-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maseda H., Saito K., Nakajima A., Nakae T. Variation of the mexT gene, a regulator of the MexEF-OprN efflux pump expression in wild-type strains of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2000;192:107–112. doi: 10.1111/j.1574-6968.2000.tb09367.x. [DOI] [PubMed] [Google Scholar]
- 57.Llanes C., Köhler T., Patry I., Dehecq B., Van Delden C., Plésiat P. Role of the MexEF-OprN efflux system in low-level resistance of Pseudomonas aeruginosa to ciprofloxacin. Antimicrob. Agents Chemother. 2011;55 doi: 10.1128/AAC.00101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Köhler T., Van Delden C., Curty L.K., Hamzehpour M.M., Pechere J.C. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 2001;183:5213–5222. doi: 10.1128/JB.183.18.5213-5222.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cabot G., Zamorano L., Moyà B., Juan C., Navas A., Blázquez J., Oliver A. Evolution of Pseudomonas aeruginosa antimicrobial resistance and fitness under low and high mutation rates. Antimicrob. Agents Chemother. 2016;60 doi: 10.1128/AAC.02676-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Richardot C., Juarez P., Jeannot K., Patry I., Plésiat P., Llanes C. Amino acid substitutions account for most mexS alterations in clinical nfxC mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016;60:2302–2310. doi: 10.1128/AAC.02622-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang D., Seeve C., Pierson L.S., Pierson E.A. Transcriptome profiling reveals links between ParS/ParR, MexEF-OprN, and quorum sensing in the regulation of adaptation and virulence in Pseudomonas aeruginosa. BMC Genom. 2013;14:618. doi: 10.1186/1471-2164-14-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bruchmann S., Dötsch A., Nouri B., Chaberny I.F., Häussler S. Quantitative contributions of target alteration and decreased drug accumulation to pseudomonas aeruginosa fluoroquinolone resistance. Antimicrob. Agents Chemother. 2013;57 doi: 10.1128/AAC.01581-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chávez-Jacobo V.M., Hernández-Ramírez K.C., Romo-Rodríguez P., Pérez-Gallardo R.V., Campos-García J., Félix Gutiérrez-Corona J., García-Merinos J.P., Meza-Carmen V., Silva-Sánchez J., Ramírez-Díaz M.I. CrpP is a novel ciprofloxacin-modifying enzyme encoded by the pseudomonas aeruginosa pUM505 plasmid. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.02629-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hernández-García M., García-Castillo M., García-Fernández S., López-Mendoza D., Díaz-Regañón J., Romano J., Pássaro L., Paixão L., Cantón R. Presence of Chromosomal crpP-like Genes Is Not Always Associated with Ciprofloxacin Resistance in Pseudomonas aeruginosa Clinical Isolates Recovered in ICU Patients from Portugal and Spain. Microorganisms. 2021;9:388. doi: 10.3390/microorganisms9020388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chávez-Jacobo V.M., Merinos J.P.G., López Y., Meza-Carmen V., Ramírez-Díaz M.I. Identification of essential residues for ciprofloxacin resistance of ciprofloxacin-modifying enzyme (CrpP) of pUM505. Microbiology. 2020;166:367–374. doi: 10.1099/mic.0.000889. [DOI] [PubMed] [Google Scholar]
- 66.Olaitan A.O., Morand S., Rolain J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014;5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Romano K.P., Warrier T., Poulsen B.E., Nguyen P.H., Loftis A.R., Saebi A., Pentelute B.L., Hung D.T. Mutations in pmrB Confer Cross-Resistance between the LptD Inhibitor POL7080 and Colistin in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.00511-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Puja H., Bolard A., Noguès A., Plésiat P., Jeannot K. The efflux pump MexXY/OprM contributes to the tolerance and acquired resistance of Pseudomonas aeruginosa to colistin. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.02033-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee J.Y., Chung E.S., Na I.Y., Kim H., Shin D., Ko K.S. Development of colistin resistance in pmrA-, phoP-, parR- and cprR-inactivated mutants of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2014;69:2966–2971. doi: 10.1093/jac/dku238. [DOI] [PubMed] [Google Scholar]
- 70.Lee J.Y., Ko K.S. Mutations and expression of PmrAB and PhoPQ related with colistin resistance in Pseudomonas aeruginosa clinical isolates. Diagn. Microbiol. Infect. Dis. 2014;78:271–276. doi: 10.1016/j.diagmicrobio.2013.11.027. [DOI] [PubMed] [Google Scholar]
- 71.Park H., Hong M., Hwang S., Park Y., Kwon K., Yoon J., Shin S., Kim J., Park Y. Characterisation of Pseudomonas aeruginosa related to bovine mastitis. Acta Vet. Hung. 2014;62:1–12. doi: 10.1556/avet.2013.054. [DOI] [PubMed] [Google Scholar]
- 72.Rodrigues Y.C., Furlaneto I.P., Pinto Maciel A.H., Garcia Quaresma A.J.P., de Matos E.C.O., Conceição M.L., da Silva Vieira M.C., da Cunha Brabo G.L., Falcão Sarges E.S.N., Costa Lima L.N.G., et al. High prevalence of atypical virulotype and genetically diverse background among Pseudomonas aeruginosa isolates from a referral hospital in the Brazilian Amazon. PLoS ONE. 2020;15:e0238741. doi: 10.1371/journal.pone.0238741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sarges E.D.S.N.F., Rodrigues Y.C., Furlaneto I.P., de Melo M.V.H., Brabo G.D.L.C., Lopes K.C.M., Quaresma A.J.P.G., Lima L.N.G.C., Lima K.V.B. Pseudomonas aeruginosa type iii secretion system virulotypes and their association with clinical features of cystic fibrosis patients. Infect. Drug Resist. 2020;13:3771–3781. doi: 10.2147/IDR.S273759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bel Hadj Ahmed A., Salah Abbassi M., Rojo-Bezares B., Ruiz-Roldán L., Dhahri R., Mehri I., Sáenz Y., Hassen A. Characterization of Pseudomonas aeruginosa isolated from various environmental niches: New STs and occurrence of antibiotic susceptible “high-risk clones”. Int. J. Environ. Health Res. 2020;30:643–652. doi: 10.1080/09603123.2019.1616080. [DOI] [PubMed] [Google Scholar]
- 75.Martin D.W., Schurr M.J., Mudd M.H., Govan J.R.W., Holloway B.W., Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl. Acad. Sci. USA. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bragonzi A., Wiehlmann L., Klockgether J., Cramer N., Worlitzsch D., Döning G., Tümmler B. Sequence diversity of the mucABD locus in Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Microbiology. 2006;152:3261–3269. doi: 10.1099/mic.0.29175-0. [DOI] [PubMed] [Google Scholar]
- 77.Wang Y., Gao L., Rao X., Wang J., Yu H., Jiang J., Zhou W., Wang J., Xiao Y., Li M., et al. Characterization of lasR-deficient clinical isolates of Pseudomonas aeruginosa. Sci. Rep. 2018;8:13344. doi: 10.1038/s41598-018-30813-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kostylev M., Kim D.Y., Smalley N.E., Salukhe I., Peter Greenberg E., Dandekar A.A. Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy. Proc. Natl. Acad. Sci. USA. 2019;116:7027–7032. doi: 10.1073/pnas.1819796116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rees V.E., Deveson Lucas D.S., López-Causapé C., Huang Y., Kotsimbos T., Bulitta J.B., Rees M.C., Barugahare A., Peleg A.Y., Nation R.L., et al. Characterization of Hypermutator Pseudomonas aeruginosa Isolates from Patients with Cystic Fibrosis in Australia. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.02538-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ferroni A., Guillemot D., Moumile K., Bernede C., Le Bourgeois M., Waernessyckle S., Descamps P., Sermet-Gaudelus I., Lenoir G., Berche P., et al. Effect of mutator P. aeruginosa on antibiotic resistance acquisition and respiratory function in cystic fibrosis. Pediatr. Pulmonol. 2009;44:820–825. doi: 10.1002/ppul.21076. [DOI] [PubMed] [Google Scholar]
- 81.Walker T.S., Tomlin K.L., Worthen G.S., Poch K.R., Lieber J.G., Saavedra M.T., Fessler M.B., Malcolm K.C., Vasil M.L., Nick J.A. Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infect. Immun. 2005;73:3693–3701. doi: 10.1128/IAI.73.6.3693-3701.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thi M.T.T., Wibowo D., Rehm B.H.A. Pseudomonas aeruginosa biofilms. Int. J. Mol. Sci. 2020;21:8671. doi: 10.3390/ijms21228671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li Z., Chen J.H., Hao Y., Nair S.K. Structures of the PelD cyclic diguanylate effector involved in pellicle formation in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 2012;287:30191–30204. doi: 10.1074/jbc.M112.378273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kavanagh K., Reeves E.P. Exploiting the potential of insects for in vivo pathogenicity testing of microbial pathogens. FEMS Microbiol. Rev. 2004;28:101–112. doi: 10.1016/j.femsre.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 85.Lee D.G., Urbach J.M., Wu G., Liberati N.T., Feinbaum R.L., Miyata S., Diggins L.T., He J., Saucier M., Déziel E., et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006;7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wattal C., Oberoi J.K., Goel N., Raveendran R., Khanna S. Matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) for rapid identification of micro-organisms in the routine clinical microbiology laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 2017;36:807–812. doi: 10.1007/s10096-016-2864-9. [DOI] [PubMed] [Google Scholar]
- 87.Magiorakos A.-P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 88.Hernández M., Iglesias M.R., Rodríguez-Lázaro D., Gallardo A., Quijada N.M., Miguela-Villoldo P., Campos M.J., Píriz S., López-Orozco G., de Frutos C., et al. Co-occurrence of colistin-resistance genes mcr-1 and mcr-3 among multidrug-resistant Escherichia coli isolated from cattle, Spain, September 2015. Eurosurveillance. 2017;22:30586. doi: 10.2807/1560-7917.ES.2017.22.31.30586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wood D.E., Salzberg S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 95.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Carattoli A., Zankari E., Garciá-Fernández A., Larsen M.V., Lund O., Villa L., Aarestrup F.M., Hasman H. In Silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., Holden M.T.G., Fookes M., Falush D., Keane J.A., Parkhill J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Price M.N., Dehal P.S., Arkin A.P. Fasttree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.R Development Core Team . R: A Language and Environment for Statistical Computing. R Core Team; Vienna, Austria: 2008. [Google Scholar]
- 100.Wickham H., Wickham H. ggplot2. Springer; New York, NY, USA: 2009. ggplot2: Elegant graphics for data analysis; pp. 1–7. [Google Scholar]
- 101.Neuwirth E. ColorBrewer Palettes [R package RColorBrewer Version 1.1-2]; 2014. [(accessed on 23 April 2021)]; Available online: https://cran.r-project.org/web/packages/RColorBrewer/index.html.
- 102.Wickham H. Reshaping data with the reshape package. J. Stat. Softw. 2007;21:1–20. doi: 10.18637/jss.v021.i12. [DOI] [Google Scholar]
- 103.Ramos-Vivas J., Chapartegui-González I., Fernández-Martínez M., González-Rico C., Fortún J., Escudero R., Marco F., Linares L., Montejo M., Aranzamendi M., et al. Biofilm formation by multidrug resistant Enterobacteriaceae strains isolated from solid organ transplant recipients. Sci. Rep. 2019;10:7452. doi: 10.1038/s41598-020-60496-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Remuzgo-Martínez S., Lázaro-Díez M., Mayer C., Aranzamendi-Zaldumbide M., Padilla D., Calvo J., Marco F., Martínez-Martínez L., Icardo J.M., Otero A., et al. Biofilm formation and quorum-sensing-molecule production by clinical isolates of Serratia liquefaciens. Appl. Environ. Microbiol. 2015;81:3306–3315. doi: 10.1128/AEM.00088-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stover C.K., Pham X.Q., Erwin A.L., Mizoguchi S.D., Warrener P., Hickey M.J., Brinkman F.S.L., Hufnagle W.O., Kowallk D.J., Lagrou M., et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 106.Roy P.H., Tetu S.G., Larouche A., Elbourne L., Tremblay S., Ren Q., Dodson R., Harkins D., Shay R., Watkins K., et al. Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS ONE. 2010;5:e0008842. doi: 10.1371/journal.pone.0008842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are in Supplementary Tables S1 and S2. Genome sequences information is available in https://www.ncbi.nlm.nih.gov/bioproject/ (accessed on 26 January 2021) under the BioProject ID PRJNA695127.