Abstract
In response to the ongoing coronavirus disease 2019 (COVID-19) pandemic, a panel of assays has been developed and applied to screen collections of approved and investigational drugs for anti-severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) activity in a quantitative high-throughput screening (qHTS) format. In this review, we applied data-driven approaches to evaluate the ability of each assay to identify potential anti-SARS-CoV-2 leads. Multitarget assays were found to show advantages in terms of accuracy and efficiency over single-target assays, whereas target-specific assays were more suitable for investigating compound mechanisms of action. Moreover, strict filtering with counter screens might be more detrimental than beneficial in identifying true positives. Thus, developing novel HTS assays acting simultaneously against multiple targets in the SARS-CoV-2 life cycle will benefit anti-COVID-19 drug discovery.
Keywords: High-throughput screening, SARS-CoV-2, COVID-19, In vitro assay, Cytopathic effect reduction assay
The ongoing COVID-19 pandemic is a serious infectious disease caused by SARS-CoV-2, which is an enveloped virus with a single-strand, positive-sense RNA genome.1, 2 The clinical spectrum of COVID-19 is highly variable, ranging from asymptomatic to multiorgan failure and ultimately death.3 Virus-specific vaccines and antiviral drugs are two common strategies to combat viral diseases. The WHO had listed >200 COVID-19 vaccines (including 51 candidate vaccines in clinical evaluation, and 163 candidate vaccines in preclinical evaluation) as under development as of December 2, 2020.4 Since December 2020, two COVID-19 mRNA vaccines and one adenovirus vector vaccine have been approved by the US Food and Drug Administration (FDA) and other regulatory agencies for Emergency Use Authorization. There are still several crucial issues regarding COVID-19 vaccines that need to be addressed, such as the correlates of protective immunity after natural infection or vaccination, the duration of vaccine-mediated immunity, and the potential risk of vaccine-associated enhanced disease.5, 6 Although some antiviral drugs (e.g., remdesivir7 and favipiravir8) have been used in the clinical treatment of patients with COVID-19, it is clear that the currently available drugs are not sufficient to fight the global COVID-19 pandemic. Moreover, the effectiveness of some antiviral drugs is also controversial. Remdesivir, the first FDA-approved drug for COVID-19 treatment, did not show any significant association with clinical benefits in reducing the recovery time for patients with severe COVID-19, based on a double-blind, placebo-controlled randomized clinical trial.9 Given the high infectivity of SARS-CoV-2, the number of confirmed cases of COVID-19 worldwide has excessed 130 million according to data released by the WHO as of April 5, 2021,10 and the number is increasing rapidly. Therefore, there is still an urgent need to identify new compounds with potent anti-SARS-CoV-2 activity.
HTS has been used as an efficient tool in identifying new lead compounds for antiviral drug development.11, 12, 13 For example, HTS assays successfully identified several potential drug leads (e.g,. emricasan, and niclosamide) against Ebola virus infection in 201414 and the Zika virus infection in South American regions in 2015.11 During the early COVID-19 outbreak, several high-throughput assays were developed and applied to screen collections of approved and investigational drugs to identify potential anti-SARS-CoV-2 compounds at the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health (NIH), which cover a wide spectrum of the SARS-CoV-2 life cycle, including viral entry, viral replication, in vitro infectivity, and live virus infectivity.15, 16, 17, 18, 19 Several potential anti-SARS-CoV-2 compounds targeting different steps in the SARS-CoV-2 life cycle were identified. For example, cepharanthine, a natural product with anti-inflammatory activities, was reported to rescue the cytopathic effects (CPEs) of SARS-CoV-2 to full efficacy, probably because of the inhibition of spike-mediated cell entry.15 Corilagin, a polyphenolic natural product, showed activity against the angiotensin-converting enzyme (ACE2) receptor-binding domain (RBD) with an IC50 of 5.5 μM.16 Walrycin B, an analog of toxoflavin, which potently inhibits bacteria growth, inhibited the replication of SARS-CoV-2 via 3-chymotrypsin-like protease (3CL) inhibition (IC50 = 0.27 μM).19
In this review, we applied computational approaches to analyze the data from the above compound screens using the SARS-CoV-2-related assays and compared the compound screening results from the target-specific assays with those from phenotypic assays. We will discuss the advantages and disadvantages of this panel of anti-SARS-CoV-2 HTS assays. The experience gained from the current screens can be used to design new assays for future compound screens for anti-COVID-19 drug development.
HTS assays used for the identification of potential anti-SARS-CoV-2 compounds
To date, NCATS has developed five HTS assays that have been used for compound screening to identify potential anti-SARS-CoV-2 drugs. These assays can be divided into three groups: single-target assays [ACE2 activity assay (ACE2 assay), and 3CL protease activity assay (3CL assay)], multitarget assays [Spike-ACE2 protein–protein interaction assay (Spike-ACE2 assay), and pseudotyped particle entry assay (PP assay)], and phenotypic antiviral efficacy assay [CPE reduction (CPE assay)] (Table 1 ).17
Table 1.
Overview of HTS assays for the identification of anti-SARS-CoV-2 compounds.
| Assay name | Abbreviated name | Assay type | Target category | Cell line | Refs |
|---|---|---|---|---|---|
| ACE2 activity assay | ACE2 assay | Biochemical | Viral entry | N/A | 17 |
| Spike-ACE2 protein–protein interaction assay | Spike-ACE2 | Proximity | Viral entry | N/A | 16 |
| 3CL protease activity assay | 3CL assay | Biochemical | Viral replication | N/A | 19 |
| Pseudotyped particle entry assay | PP entry | Cell based | In vitro infectivity | Vero E6 | 15 |
| Cytopathic effect reduction assay | CPE assay | Cell viability | Live virus infectivity | Vero E6 | 21 |
The ACE2 and 3CL assays are both fluorescence-based cell-free biochemical assays that measure the inhibitory effect of a test compound on the human ACE2 activity or SARS-CoV-2 3CL protease activity, respectively.17, 19 The Spike-ACE2 assay is a proximity assay that uses the AlphaLISA technology to identify compounds that can disrupt the interaction between the SARS-CoV-2 Spike protein and its cellular receptor ACE2.16 The PP entry and CPE assays are both cell-based assays with a luminescence readout.15, 20 The PP entry assay, which can be conducted in biosafety level 2 (BSL2) laboratories, facilitates the identification of viral cell entry inhibitors using pseudotyped viral particles that incorporate SARS-CoV-2 Spike proteins without the viral genome. Compared with the assays designed to screen potential anti-SARS-CoV-2 compounds acting via specific mechanisms, the CPE assay is suitable for assessing the general anti-SARS-CoV-2 efficacy of potential compounds. Here, we use the CPE assay as the benchmark assay to evaluate the robustness and reliability of the target-oriented assays.20, 21
The CPE assay measures the ability of compounds to prevent the live SARS-CoV-2-induced cytopathic effects in human host cells through various molecular mechanisms, such as inhibition of viral entry or replication, virus-induced apoptosis, and activation of host immune responses. However, the CPE assay has some inherent limitations. It requires a relatively long protocol time, mainly due to the 72-h incubation of SARS-CoV-2 and Vero-E6 cells in the presence of test compounds.21 As a method that indirectly measures the ability of compounds to inhibit viral infection-caused cell death, the CPE assay cannot identify specific anti-SARS-CoV-2 mechanisms, and it might not be as sensitive as assays that measure viral load directly.21 Furthermore, these five HTS assays cover multiple key stages of the SARS-CoV-2 life cycle, including viral entry into host cells (ACE2 and Spike-ACE2 assays), viral replication (3CL assay), in vitro infectivity (PP entry assay), and live virus infectivity (CPE assay) (Table 1). Fig. 1 shows the number of compounds screened using these in vitro assays. A detailed description of these HTS assays and all the screening data are publicly available through the NCATS/NIH open science data portal (OpenData, https://opendata.ncats.nih.gov/covid19/).
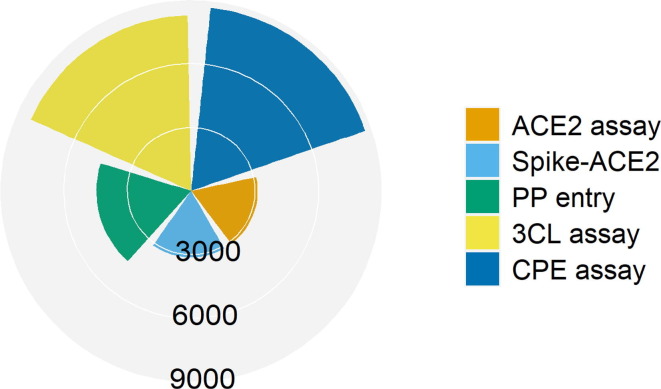
Figure 1.
Distribution of compounds screened in the anti-severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) high-throughput screening (HTS) assays. The radar plot shows the number of compounds screened in each of the five assays that cover different stages in the SARS-CoV-2 life cycle, such as viral entry into host cells [angiotensin-converting enzyme 2 (ACE2) assay and Spike-ACE2 assay], viral replication [3-chymotrypsin-like protease (3CL) assay], in vitro infectivity [pseudotyped particle (PP) entry assay], and live virus infectivity [cytopathic effect (CPE) assay]. In the plot, each slice represents an assay, and the radius of the slice is proportional to the number of compounds screened in the assay.
Activity profiles of compounds in the panel of anti-SARS-CoV-2 HTS assays
The primary screening data and concentration–response curves were analyzed using custom software developed internally at NCATS.22 Concentration–response titration points for each compound were fitted to a four-parameter Hill equation yielding half-maximal activity concentrations (AC50) and maximal response (efficacy) values.23 Compounds were further designated as class 1–4 based on the shape of the concentration–response curve.23 Compounds that showed activation were assigned class 1.1, 1.2, 1.3, 1.4, 2.1, 2.2, 2.3, 2.4, and 3 curves. Compounds that showed inhibition were assigned class −1.1, −1.2, −1.3, −1.4, −2.1, −2.2, −2.3, −2.4, and −3 curves. Compounds that showed no significant concentration response were considered inactive and assigned class 4. Curve classes were further combined with efficacy and converted to a numeric rank, such that more potent and efficacious compounds with higher quality curves were assigned a higher rank. The curve rank is a value ranging from −9 to +9, with −9 to −1 indicating inhibitory ability, 1–9 indicating activating ability, and 0 indicating inactive.23, 24 In this study, compounds with an absolute curve rank >0 were labeled as active, and inactive otherwise. That is, any compound that showed a significant concentration-dependent response was considered active. This definition of ‘active’ is not equivalent to what is traditionally considered a ‘hit’, which normally also has certain potency and/or efficacy requirements. Our active classification is used solely for the purpose of comparing activities from different assays.
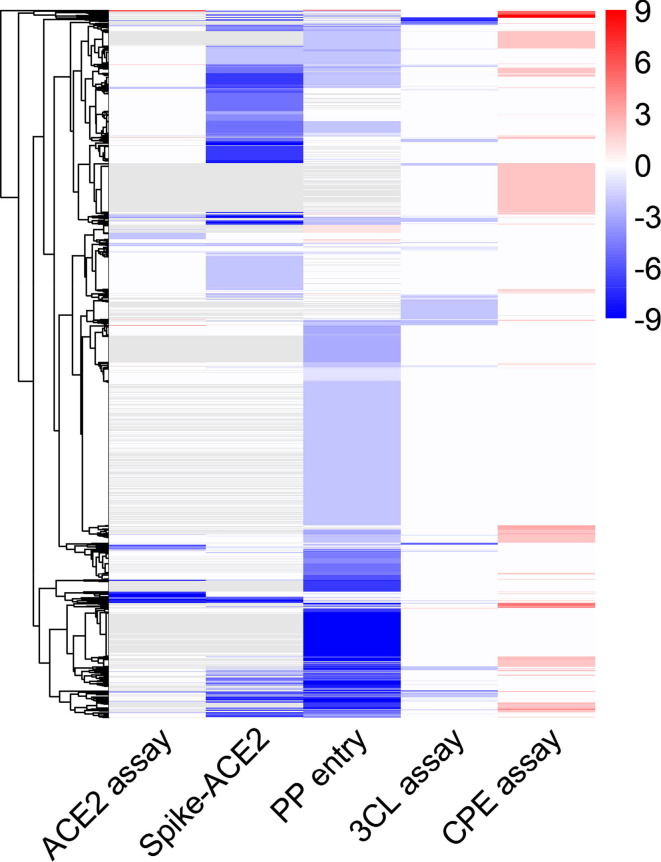
Overall, most compounds (>92%) exhibited some activity in at least one of the anti-SARS-CoV-2 assays (Fig. 2 ). The hit rates of the multitarget assays (31% for the Spike-ACE2 assay, and 46% for the PP entry assay) were approximately one order of magnitude higher than that of the single-target assays (5% for the 3CL assay, and 6% for the ACE2 assay) and the CPE assay (8%), suggesting that multitarget assays are more sensitive in identifying potential anti-SARS-CoV-2 compounds. Only three compounds (chloroxine, celastrol, and eltrombopag olamine) exhibited activities against all five anti-SARS-CoV-2 assays (Fig. 2). These three compounds were reported to have different pharmacological mechanisms of action (chloroxine is an antibacterial drug; celastrol is a heat shock protein 90 (hsp90) inhibitor,25 and eltrombopag olamine is a thrombopoietin receptor (TpoR) agonist26). Chloroxine has also been reported to bind with a strong predicted affinity to the SARS-CoV-2 3CL protease (−8.24 kcal/mol), suggesting that its anti-SARS-CoV-2 activity could be attributed to inhibition of the 3CL protease.27 Celastrol was known to exhibit antiviral activity against SARS-CoV-1 via inhibition of the viral 3CL protease; however, data showing its impact on the SARS-CoV-2 3CL protease remain lacking. In vitro molecular interaction experiments have demonstrated that Eltrombopag potently inhibited the binding between the SARS-CoV-2 Spike protein and human ACE2, indicating that it might inhibit SARS-CoV-2 by affecting the stability of the Spike-ACE2 protein complex.28
Figure 2.
Activity profiles of compounds in anti-severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) high-throughput screening (HTS) assays. In the heat map, each row is a compound, and each column is an assay readout. The colors of the heat map range from blue to red based on compound activity. Blue colors indicate inhibitors, and red colors indicate activators. Gray colors indicate missing values (not tested). Compounds are grouped into clusters of similar activity profiles. Compounds were assigned to different clusters based on the similarity of their activities using R package ‘pheatmap’ with its default clustering method (complete) and distance function (Euclidean). Abbreviations: 3CL, 3-chymotrypsin-like protease; ACE2, angiotensin-converting enzyme 2; CPE, cytopathic effect; PP, pseudotyped particle.
Correlations between the activity profiles of different HTS assays
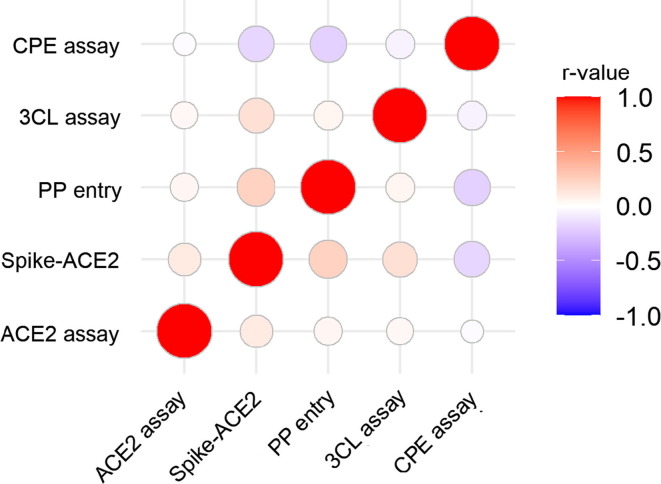
To further investigate the relationship between these HTS assays, Pearson's correlation coefficients (r) were computed between assay pairs using the compound activity profiles. The r-value is a number ranging from −1 (perfect negative correlation) to +1 (perfect positive correlation) with 0 representing no correlation. We found that the r-values between different assay pairs in this study ranged from −0.20 to 0.24, indicating overall low correlations among these assays (Fig. 3 ). Differences in targets could explain the poor correlations observed between the assays with different targeting mechanisms (r-values ranging from 0.04 to 0.24). The lack of correlation (r-values ranging from −0.02 to −0.2) between each target specific assay and the CPE assay suggests that the target specific assays did not fully reflect the anticytopathic effect or overall anti-SARS-CoV-2 effectiveness. That is, a single target cannot capture all the antiviral mechanisms captured by the CPE assay. By contrast, multitarget assays (Spike-ACE2 assay and PP entry assay) showed better correlations with the CPE assay when compared with the single-target assays (3CL and ACE2 assays), indicating that multitarget assays might be better suited for identifying anti-SARS-CoV-2 compounds more efficiently.29 For example, Z-FA-FMK exhibited higher potency and efficacy in the CPE assay than in the 3CL assay possibly because of its involvement in multiple steps/targets in the virus replication process.19 In addition, the 3CL and ACE2 assays are enzymatic assays without cells, but the other assays (PP entry and CPE) are cell-based assays, which could also contribute to the difference. Consistent with our findings, multitarget strategies have been widely used in the field of drug discovery, especially for various complex diseases, such as neuropsychiatric and neurodegenerative disorders, and cancers.30, 31, 32
Figure 3.
Correlations between compound activity profiles in different anti-severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) high-throughput screening (HTS) assays. The heat map is colored by the pairwise Pearson's correlation coefficient (r-value) between assays, such that red colors indicate positive correlations and blue colors indicate negative correlations. The diameter of each circle is proportional to the magnitude of the r-value. Abbreviations: 3CL, 3-chymotrypsin-like protease; ACE2, angiotensin-converting enzyme 2; CPE, cytopathic effect; PP, pseudotyped particle.
Efficiency of target-specific assays in identifying anti-SARS-CoV-2 compounds measured by the CPE assay
The CPE assay has been used extensively as the benchmark to evaluate hits identified from target-specific assays to confirm their potential anti-SARS-CoV-2 activities. Here, we compared the compound activities in the CPE assay with other assays using data-driven approaches. Briefly, each compound identified in any assay was assigned as positive (anti-SARS-CoV-2 effect) or negative (no anti-SARS-CoV-2 effect) based on its curve rank and efficacy. For example, compounds with curve rank < 0 and efficacy < −50% in the 3CL assay were considered positive, whereas other compounds were considered negative. A few counter-screen assays were used in parallel with the above assays to eliminate potential false positive compounds. The Spike-ACE2 TruHits assay directly measures the streptavidin donor beads bound to biotinylated acceptor beads without intermediary molecules (i.e., SARS-CoV-2 Spike protein and human ACE2) and, thus, can be used to eliminate the false positive compounds that interfere with the AlphaLISA readout. Given that the PP entry assay is a cell-based assay, cytotoxic compounds could also show up as inhibitors in the assay, resulting in false positives. The efficiency of target-specific assays in identifying anti-SARS-CoV-2 compounds was assessed by calculating the true positive (TP), true negative (TN), false positive (FP), false negative (FN), sensitivity [TP/(TP + FN)], specificity [TN/(FP + TN)], balanced accuracy (BA, the average of sensitivity and specificity), and positive predictive value [PPV, TP/(TP + FP)] compared with the CPE assay. TP is the number of positive compounds in a target assay (e.g., 3CL assay) that are also classified as positive in the CPE assay. TN is the number of negative compounds in a target assay (e.g., 3CL assay) that are also classified as negative in the CPE assay. FN is the number of negative compounds in a target assay (e.g., 3CL assay) classified as positive in CPE assay. FP is the number of positive compounds in a target assay (e.g., 3CL assay) classified as negative in CPE assay. The efficiency in identifying anti-SARS-CoV-2 compounds varied among target-specific assays.
We first looked at the effect of activity filtering using counter-screen assays. Filtering using the cytotoxicity counter screen only had a minor effect on the PP entry assay, with the assay PPV increasing slightly from 0.23 to 0.24. (Table 2 ). As for the Spike-ACE2 assay, the assay sensitivity and PPV in detecting anti-SARS-CoV-2 compounds suffered one order of magnitude reduction after filtering with its counter-screen assay (Spike-ACE2 TruHits assay) (sensitivity dropped from 0.54 to <0.01, and PPV decreased from 0.2 to 0.02) (Table 2), suggesting that the counter-screen assay used for the Spike-ACE2 assay is too strict, resulting in a large number of FNs (TPs decreased from 122 to 1).
Table 2.
Comparison of target assay efficiency in identifying anti-SARS-CoV-2 compounds as measured by the CPE assay.
| Assay name | TP | TN | FP | FN | Sensitivity | Specificity | PPV | BA |
|---|---|---|---|---|---|---|---|---|
| ACE2 assay | 12 | 2694 | 102 | 245 | 0.05 | 0.96 | 0.11 | 0.51 |
| Spike-ACE2 | 122 | 2044 | 494 | 106 | 0.54 | 0.81 | 0.20 | 0.67 |
| Spike-ACE2_Filtera | 1 | 2495 | 43 | 227 | <0.01 | 0.98 | 0.02 | 0.49 |
| PP entry | 319 | 2333 | 1063 | 84 | 0.79 | 0.69 | 0.23 | 0.74 |
| PP entry_Filterb | 138 | 2965 | 431 | 265 | 0.34 | 0.87 | 0.24 | 0.61 |
| 3CL assay | 21 | 7173 | 111 | 615 | 0.03 | 0.98 | 0.16 | 0.51 |
Inclusion criteria: curve rank in the Spike-ACE2 TruHits assay > 0, or efficacy in the Spike-ACE2 TruHits assay > −40%, or AC50 (Spike-ACE2 assay)/AC50 (Spike-ACE2 TruHits assay) greater than sixfold.
Inclusion criteria: curve rank in the cytotoxicity assay > 0, or efficacy in the cytotoxicity assay > −40%, or AC50 (PP entry assay)/AC50 (cytotoxicity assay) greater than sixfold.
In addition, multitarget cell-based assays (e.g., the PP entry assay) might be more efficient in identifying potential anti-SARS-CoV-2 compounds compared with multitarget cell-free assays (e.g., the Spike-ACE2 assay). The single-target assays (ACE2 and 3CL assays) showed very low sensitivity (≤0.05) and PPV (≤0.16) (Table 2), suggesting that such assays are more suitable for anti-SARS-CoV-2 mechanism deconvolution rather than serving as initial screens for compound identification. In addition, the low PPV of a single-target assay means that the assay has a high FP rate. This further implies that the target/mechanism measured by the assay is not very druggable or not sufficient to suppress SARS-CoV-2 infection. In addition, the other possibilities are that the compounds identified by the enzymatic assays cannot enter cells or be metabolized in cells, or that the CPE assay is not efficient at identifying compounds acting through this mechanism. In terms of BA and PPV, the PP entry assay showed the best performance, the Spike-ACE2 assay came up second, and the ACE2 and 3CL assays performed the worst (Table 2). These results suggest that multitarget assays are superior to single-target assays serving as initial compound screens for SARS-CoV-2.
HTS assays moving forward
Thus far, five types of in vitro HTS assay for identifying anti-SARS-CoV-2 compounds according to different targets and design strategies have been established and applied to drug-repurposing screens at NCATS. Compared with single-target assays (ACE2 and 3CL assays), multitarget assays (PP entry and Spike-ACE2 assays) showed advantages in terms of accuracy and efficiency using the CPE assay as a benchmark. Consistent with other studies,33 strict filtering using counter-screens can result in the loss of a large number of true hits. In our study, when a counter-screen assay was applied to filter out potential artifacts from the Spike-ACE2 assay hits using strict filtering criteria, the TP rates dropped significantly. We used the CPE assay, a general antiviral efficacy assay, as the benchmark to evaluate the other mechanism-based anti-SARS-CoV-2 assays; however, the CPE assay itself also has inherent limitations, such as BSL-3 lab requirements, time-consuming, insufficient coverage of virus life cycle in host cells, lack of information and limited coverage regarding anti-SARS-CoV-2 mechanisms, and slight underestimation of potency and efficacy of the anti-SARS-CoV-2 compounds identified (i.e., is less sensitive). The results from this study show that there is a lot of room, as well as need, for improvement in current HTS assays for the identification of compounds against SARS-CoV-2.
Several paths can be considered for new assay design and implementation for SARS-CoV-2 drug development: (i) novel phenotypic HTS assays are needed for the identification of antiviral compounds that act simultaneously against multiple targets in the SARS-CoV-2 life cycle. Drug combination therapy has been demonstrated as a necessary therapeutic strategy for the successful treatment of HIV and hepatitis C.34 Compounds with polypharmacology could have higher efficacy against SARS-CoV-2 infection. Furthermore, multitargeted compounds might improve the existing therapeutic strategies and clinical outcomes for patients with COVID-1929; (ii) cell-based enzyme assays are needed to replace biochemical enzyme assays for SARS-CoV-2 drug development. Although biochemical enzyme assays can directly measure the potencies of inhibitory compounds, the recombinant enzymes might miss cofactors and subunits in cell-free assays. In addition, compound membrane permeability and drug metabolism in cells might affect the final antiviral activities of the compounds found active in biochemical assays.35 Recently, a cell-based SARS-CoV-2 main protease assay was reported36 that provides a direction for the development of cell-based assays for other SARS-CoV-2 targets; (iii) use of disease-relevant human cell lines might improve the efficiency of the SARS-CoV-2 screening assays. Given the lack of ACE2 receptors required for SARS-CoV-2 entry in many human cell lines, Vero E6 cells (originally isolated from kidney epithelial cells extracted from an African green monkey) have been used extensively in the SARS-CoV-2 pseudotyped particle entry and live virus assays. The use of human cell lines, such as Caco2 cells and A549 cells transfected with ACE2, might improve the disease relevance of cell-based SARS-CoV-2 screening assays37; (iv) application of human induced pluripotent stem cell (hiPSC)-derived organoids and 3D tissue models in SARS-CoV-2 assays. Han et al. developed a lung organoid model using hPSCs and applied this model to perform a HTS of FDA-approved drugs.38 The screen identified three entry inhibitors of SARS-CoV-2: imatinib, mycophenolic acid (MPA), and quinacrine dihydrochloride (QNHC).38 Consistent with these results, the three compounds were also found to be active in our CPE and PP entry assays. Compared with the lung organoid model results, imatinib and MPA were less potent, whereas QNHC was more potent in the CPE assay. In addition, all three compounds were less potent in our PP entry assay, and MPA was filtered out by the cytotoxicity counter-screen. Nevertheless, the lung organoid model assay has low-screening throughput and is not suitable for HTS of large compound collections. Compared with phenotypic assays such as the CPE assay, target-specific assays might be more suitable for investigating the mechanisms of action of anti-SARS-CoV-2 compounds; and (v) given the limitation of SARS-CoV-2 live virus assays in the BSL-3 lab settings, new cell-based assays in addition to the CPE assay are needed. The cytopathic effects of SARS-CoV-2 infection do not occur in all host cells because not all host cells are killed by the virus infection. Recently, an antibody-based AlphaLISA assay measuring the SARS-CoV-2 N-protein was developed that can be used for compound screening in BSL-3 labs. This assay can be miniaturized and has the homogenous assay format suitable for HTS. This type of assays can measure the activity of compounds against SARS-CoV-2 infection and replication in host cells without the involvement of cell killing. Machine-learning classification models are highly efficient computer algorithms capable of predicting categories for new unseen data based on annotated training data sets and have been widely applied to improve the speed and efficiency of therapeutic drug candidate development processes.39, 40 The existing HTS assay data provide good training sets for the implementation of machine-learning models to improve the hit rates of anti-SARS-CoV-2 drug screens.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the teams at NCATS/DPI for generating and making publicly available the COVID-19 screening data used in this study. This work was supported by the Intramural Research Programs of the National Center for Advancing Translational Sciences, National Institutes of Health.
References
- 1.Yang Y., Peng F., Wang R., Guan K., Jiang T., Xu G., et al. The deadly coronaviruses: The 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Draft landscape and tracker of COVID-19 candidate vaccines. www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines [accessed May 18, 2021].
- 5.Haynes B.F., Corey L., Fernandes P., Gilbert P.B., Hotez P.J., Rao S., et al. Prospects for a safe COVID-19 vaccine. Sci Translat Med. 2020;12(568):eabe0948. doi: 10.1126/scitranslmed.abe0948. [DOI] [PubMed] [Google Scholar]
- 6.Castells M.C., Phillips E.J. Maintaining safety with SARS-CoV-2 vaccines. New Engl J Med. 2020;384(7):643–649. doi: 10.1056/NEJMra2035343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., et al. Compassionate use of remdesivir for patients with severe Covid-19. New Engl J Med. 2020;382(24):2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kocayiğit H., Özmen Süner K., Tomak Y., Demir G., Yaylacı S., Dheir H., et al. Observational study of the effects of Favipiravir vs Lopinavir/Ritonavir on clinical outcomes in critically Ill patients with COVID-19. J Clin Pharm Therap. 2021;46:454–459. doi: 10.1111/jcpt.13305. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395(10236):1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/ [accessed May 18, 2021].
- 11.Xu M., Lee E.M., Wen Z., Cheng Y., Huang W.K., Qian X., et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med. 2016;22(10):1101–1107. doi: 10.1038/nm.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee E.M., Titus S.A., Xu M., Tang H., Zheng W. High-throughput Zika viral titer assay for rapid screening of antiviral drugs. Assay Drug Develop Technol. 2019;17(3):128–139. doi: 10.1089/adt.2018.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rumlová M., Ruml T. In vitro methods for testing antiviral drugs. Biotechnol Adv. 2018;36(3):557–576. doi: 10.1016/j.biotechadv.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kouznetsova J., Sun W., Martinez-Romero C., Tawa G., Shinn P., Chen C.Z., et al. Identification of 53 compounds that block Ebola virus-like particle entry via a repurposing screen of approved drugs. Emerg Microbes Infect. 2014;3(12) doi: 10.1038/emi.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C.Z., Xu M., Pradhan M., Gorshkov K., Petersen J.D., Straus M.R., et al. Identifying SARS-CoV-2 entry inhibitors through drug repurposing screens of SARS-S and MERS-S pseudotyped particles. ACS Pharmacol Transl Sci. 2021;3:1165–1175. doi: 10.1021/acsptsci.0c00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanson Q.M., Wilson K.M., Shen M., Itkin Z., Eastman R.T., Shinn P., et al. Targeting ACE2–RBD interaction as a platform for COVID-19 therapeutics: development and drug-repurposing screen of an AlphaLISA proximity assay. ACS Pharmacol Transl Sci. 2020;3(6):1352–1360. doi: 10.1021/acsptsci.0c00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brimacombe KR, Zhao T, Eastman RT, Hu X, Wang K, Backus M, et al. An OpenData portal to share COVID-19 drug repurposing data in real time. bioRxiv http://dx.doi.org/10.1101/2020.06.04.135046 [Published online June 5, 2020].
- 18.Gorshkov K, Chen CZ, Bostwick R, Rasmussen L, Xu M, Pradhan M, et al. The SARS-CoV-2 cytopathic effect is blocked with autophagy modulators. bioRxiv http://dx.doi.org/10.1101/2020.05.16.091520 [published online May 28, 2020].
- 19.Zhu W., Xu M., Chen C.Z., Guo H., Shen M., Hu X., et al. Identification of SARS-CoV-2 3CL protease inhibitors by a quantitative high-throughput screening. ACS Pharmacol Transl Sci. 2020;3(5):1008–1016. doi: 10.1021/acsptsci.0c00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z.R., Zhang Y.N., Li X.D., Zhang H.Q., Xiao S.Q., Deng F., et al. A cell-based large-scale screening of natural compounds for inhibitors of SARS-CoV-2. Signal Transd Targeted Therapy. 2020;5(1):1–3. doi: 10.1038/s41392-020-00343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C.Z., Shinn P., Itkin Z., Eastman R.T., Bostwick R., Rasmussen L., et al. Drug repurposing screen for compounds inhibiting the cytopathic effect of SARS-CoV-2. Front Pharmacol. 2021;11 doi: 10.3389/fphar.2020.592737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Jadhav A., Southal N., Huang R., Nguyen D.-T. A grid algorithm for high throughput fitting of dose-response curve data. Curr Chem Genom. 2010;4:57–66. doi: 10.2174/1875397301004010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang R. A quantitative high-throughput screening data analysis pipeline for activity profiling. Methods Mol Biol. 2016;1473:111–122. doi: 10.1007/978-1-4939-6346-1_12. [DOI] [PubMed] [Google Scholar]
- 24.Huang R., Xia M., Cho M.H., Sakamuru S., Shinn P., Houck K.A., et al. Chemical genomics profiling of environmental chemical modulation of human nuclear receptors. Environ Health Perspect. 2011;119(8):1142–1148. doi: 10.1289/ehp.1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang T., Hamza A., Cao X., Wang B., Yu S., Zhan C.G., et al. A novel Hsp90 inhibitor to disrupt Hsp90/Cdc37 complex against pancreatic cancer cells. Mol Cancer Therap. 2008;7(1):162–170. doi: 10.1158/1535-7163.MCT-07-0484. [DOI] [PubMed] [Google Scholar]
- 26.Corman S.L., Mohammad R.A. Eltrombopag: a novel oral thrombopoietin receptor agonist. Ann Pharmacotherapy. 2010;44(6):1072–1079. doi: 10.1345/aph.1P042. [DOI] [PubMed] [Google Scholar]
- 27.Gao K., Nguyen D.D., Chen J., Wang R., Wei G.-W. Repositioning of 8565 existing drugs for COVID-19. J Phys Chem Lett. 2020;11(13):5373–5382. doi: 10.1021/acs.jpclett.0c01579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng S., Luan X., Wang Y., Wang H., Zhang Z., Wang Y., et al. Eltrombopag is a potential target for drug intervention in SARS-CoV-2 spike protein. Infect Genet Evol. 2020;85 doi: 10.1016/j.meegid.2020.104419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shyr Z.A., Gorshkov K., Chen C.Z., Zheng W. Drug discovery strategies for SARS-CoV-2. J Pharmacol Exp Therap. 2020;375(1):127–138. doi: 10.1124/jpet.120.000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bawa P., Pradeep P., Kumar P., Choonara Y.E., Modi G., Pillay V. Multi-target therapeutics for neuropsychiatric and neurodegenerative disorders. Drug Discov Today. 2016;21(12):1886–1914. doi: 10.1016/j.drudis.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Csermely P., Agoston V., Pongor S. The efficiency of multi-target drugs: the network approach might help drug design. Trends Pharmacol Sci. 2005;26(4):178–182. doi: 10.1016/j.tips.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 32.Ramsay R.R., Popovic-Nikolic M.R., Nikolic K., Uliassi E., Bolognesi M.L. A perspective on multi-target drug discovery and design for complex diseases. Clin Transl Med. 2018;7(1):1–14. doi: 10.1186/s40169-017-0181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian J., Fu Y., Li Q., Xu Y., Xi X., Zheng Y., et al. Differential expression and bioinformatics analysis of circRNA in PDGF-BB-induced vascular smooth muscle cells. Front Genet. 2020;11:530. doi: 10.3389/fgene.2020.00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meganck R.M., Baric R.S. Developing therapeutic approaches for twenty-first-century emerging infectious viral diseases. Nat Med. 2021;27(3):401–410. doi: 10.1038/s41591-021-01282-0. [DOI] [PubMed] [Google Scholar]
- 35.Zheng W., Thorne N., McKew J.C. Phenotypic screens as a renewed approach for drug discovery. Drug Discov Today. 2013;18(21–22):1067–1073. doi: 10.1016/j.drudis.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rawson J.M.O., Duchon A., Nikolaitchik O.A., Pathak V.K., Hu W.S. Development of a cell-based luciferase complementation assay for identification of SARS-CoV–2 3CL(pro) inhibitors. Viruses. 2021;13(2):173. doi: 10.3390/v13020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wurtz N., Penant G., Jardot P., Duclos N., La Scola B. Culture of SARS-CoV-2 in a panel of laboratory cell lines, permissivity, and differences in growth profile. Eur J Clin Microbiol Infect Dis. 2021;40(3):477–484. doi: 10.1007/s10096-020-04106-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han Y., Duan X., Yang L., Nilsson-Payant B.E., Wang P., Duan F., et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589(7841):270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J., Kumar S., Lee S.-Y., Park S.J., Kim M.-H. Development of predictive models for identifying potential S100A9 inhibitors based on machine learning methods. Front Chem. 2019;7:779. doi: 10.3389/fchem.2019.00779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maharao N., Antontsev V., Wright M., Varshney J. Entering the era of computationally driven drug development. Drug Metabolism Rev. 2020;52(2):283–298. doi: 10.1080/03602532.2020.1726944. [DOI] [PubMed] [Google Scholar]