Abstract
Sarcopenia is the loss of both muscle mass and function with age. Although the molecular underpinnings of sarcopenia are not fully understood, numerous pathways are implicated, including autophagy, in which defective cargo is selectively identified and degraded at the lysosome. The specific tagging and degradation of mitochondria is termed mitophagy, a process important for the maintenance of an organelle pool that functions efficiently in energy production and with relatively low reactive oxygen species production. Emerging data, yet insufficient, have implicated various steps in this pathway as potential contributors to the aging muscle atrophy phenotype. Included in this is the lysosome, the end-stage organelle possessing a host of proteolytic and degradative enzymes, and a function devoted to the hydrolysis and breakdown of defective molecular complexes and organelles. This review provides a summary of our current understanding of how the autophagy-lysosome system is regulated in aging muscle, highlighting specific areas where knowledge gaps exist. Characterization of the autophagy pathway with a particular focus on the lysosome will undoubtedly pave the way for the development of novel therapeutic strategies to combat age-related muscle loss.
Keywords: skeletal muscle, aging, autophagy, mitophagy, lysosomes, sarcopenia
1. Introduction
Skeletal muscle comprises approximately 40% of total body mass and is integral for locomotion and metabolic control [1]. Mitochondria, the organelles responsible for energy provision through the electron transport chain, make up approximately 2 to 7% of muscle cell volume [2,3]. These organelles also play an essential role within muscle through the production of reactive oxygen species (ROS), in Ca2+ handling, and in apoptotic signaling. Muscle cells display a high degree of adaptive plasticity, and as such, respond to the stimuli placed upon them, as commonly seen with endurance exercise and periods of disuse/inactivity. The lability of this tissue is controlled by a balance of organelle and protein biosynthesis, and their degradation. Autophagy is a major catabolic system within the cell, principally responsible for the selective degradation of long-lived proteins, protein aggregates and organelles via lysosomes [4]. Autophagic breakdown is essential for the maintenance of cell health through the removal of damaged or dysfunctional cellular components, such as mitochondria via mitophagy. Importantly, inhibition of autophagy in skeletal muscle elicits atrophy, perturbations in sarcomere structure, neuromuscular junction decay, and weakness [5,6,7,8,9].
With aging there is a progressive decline in muscle mass [1] and quality [10], commonly referred to as sarcopenia [10,11,12]. Concomitantly, mitochondrial content and function are reduced [13,14,15] which contributes to the metabolic decrements observed in aged skeletal muscle. In the elderly population, the effects of sarcopenia can be exacerbated by prolonged periods of inactivity related to injury or hospitalization [16]. Fortunately, exercise is capable of restoring muscle health following periods of disuse, and slowing age-related muscle wasting [14,17].
Further understanding of the molecular differences between young and old muscle in regard to the autophagy-lysosome system will guide the development of strategies to enhance skeletal muscle health with increasing age. Such an understanding will likely extend beyond age-related sarcopenia, as dystrophic, cachectic and lysosome storage disease muscles all commonly display altered autophagy, mitochondrial defects and muscle atrophy [18,19,20,21,22,23,24,25,26]. The goal of this review is to summarize the current understanding of the autophagy-lysosome system in aging muscle.
2. Regulation of Autophagy in Muscle
In its basic form, macroautophagy (herein referred to as autophagy) is the engulfment of materials in a double-membraned autophagosome, followed by their delivery to lysosomes for degradation. The process can be broken down into a series of steps, including initiation/activation, nucleation, elongation/maturation, transport, fusion with lysosomes, and degradation. These steps are regulated by a family of autophagy-related genes (Atgs), the function of which has been elucidated primarily in yeast and confirmed in mammalian cells/tissue. The steps and the key players have been reviewed extensively in the past [4,27], and we highlight these as a framework and describe the research that has been undertaken in the context of muscle physiology.
2.1. Overview of the Autophagy Pathway
2.1.1. Transcriptional Control of the Autophagy Lysosome System
The transcription factor forkhead box O3 (FoxO3) and members of the MiTE/TFE family, particularly Tfeb and Tfe3, coordinate autophagy-lysosome gene expression pathways. FoxO3 is an integral transcriptional regulator of the muscle atrophy program through the ubiquitin-proteasome and autophagy systems [28,29,30,31]. FoxO3 also aids in the control of mitophagy through Bcl2 and adenovirus E1b 19-kDa-interacting protein 3 (Bnip3) and Bnip3-like (Nix) gene expression [31]. The importance of FoxO3 in muscle was shown in a series of seminal studies, whereby constitutively active FoxO3 promotes atrophy [28,30,31], whereas blunted FoxO3 activity prevents stimulus-induced atrophy of muscle fibers through perturbations in both proteasomal and autophagic pathways [28,32,33,34,35,36]. Tfeb and Tfe3 homodimerize and/or heterodimerize and interact with E-box and M-Box elements in promoters [37,38,39], and they regulate the expression of both lysosome and autophagy-related genes via their ability to activate the coordinated lysosome enhancement and regulation (CLEAR) network of genes [38,40,41,42,43].
2.1.2. The Autophagy Pathway from Activation to Execution
Autophagy begins with the induction of a pre-autophagosomal structure, mediated by the Ulk1 complex [44,45,46,47,48,49]. Ulk1 activation is negatively regulated by the mammalian target of rapamycin complex 1 (mTORC1), and positively regulated by AMP-activated protein kinase (AMPK) [50,51,52,53], as discussed further below. Activated Ulk1 stimulates a class III PI3K Complex I (PI3KC3-C1), consisting of Vps15, Vps34, Atg14, Beclin1 and Ambra1 [54,55,56] to generate phosphatidylinositol-2-phosphate [PI(3)P], which is utilized in the nucleation of autophagosomal membranes [57,58]. Thus, Ulk1 activity is essential for the activation of autophagy in muscle, and knockout of Ulk1 perturbs autophagy flux [59,60].
Phagophore expansion occurs via two ubiquitin-like conjugation systems. The first creates the Atg12-Atg5-Atg16L1 complex, which aids in the second conjugation system, ultimately forming pro-microtubule-associated protein 1A/1V chain 3 (LC3) I, and then LC3-II. These processes are mediated by Atg3, Atg7, and Atg10 [58,61]. LC3-II and its related protein family members, γ-aminobutyric acid receptor-associated protein (Gabarap), and Golgi-associated ATPase enhancer of 16kDa (Gate16), embed in the autophagosomal membrane and act as receptors for cargo selection [62,63,64].
Cargo selection can be Ub-dependent or -independent. Both mechanisms utilize receptors that bind to digestible material directly, or via Ub-chains that are tethered to cargo [62,65,66]. Ultimately, these receptors interact with the autophagosome via an LC3-interacting region (LIR) [67,68]. Autophagosomes are then transported across microtubule tracts to lysosomes [58,69,70], where the membranes of these two compartments fuse, forming an autolysosome. This is mediated by Ras-related protein 7 (Rab7), Snare proteins and lysosomal associated membrane proteins (Lamps) [69].
2.1.3. Important Considerations in Autophagic Measurements
Various studies investigating autophagy regulation highlight alterations in autophagic markers and make inferences about the activation or repression of the autophagy pathway. Although such studies are informative, they do not accurately encapsulate the dynamics of the autophagic process. Critical to this analysis is the ability to assess flux through the pathway, given its dynamic nature. In the presence of inhibitors, such as the microtubule destabilizer colchicine, which dissociates autophagosome formation from transport to the lysosomes for terminal degradation, or bafilomycin A, which inhibits autophagosome-lysosome fusion, a comparison of autophagosomal proteins (i.e., LC3-II or p62) between drug- and vehicle-treated conditions is a reliable measure of autophagy flux [71]. However, such methodologies come with limitations. For example, these assays fail to capture the role of the lysosomes in autophagic resolution, and may also create a backlog of autophagic substrates, thereby creating intracellular stress. As such, these agents may alter the phenotype of the tissue, and this can impose a confounding variable. Thus, other assays have also been developed which may be feasible to apply to the muscle biology field, including the use of fluorescent-labelled markers and pH-sensitive probes for lysosome function (i.e., Mitotimer or mtKeima) [72]. These methods are useful in pre-clinical animal models or in cell culture, but they cannot be applied to the assessment of autophagy in human muscle, where the reliance on autophagy markers which correlate with flux is required. This technicality currently limits our understanding of autophagy in human skeletal muscle physiology.
2.2. Upstream Regulation of the Autophagy Lysosome System
2.2.1. Coordinated Activation of the Autophagy Lysosome System by Nutrient Availability
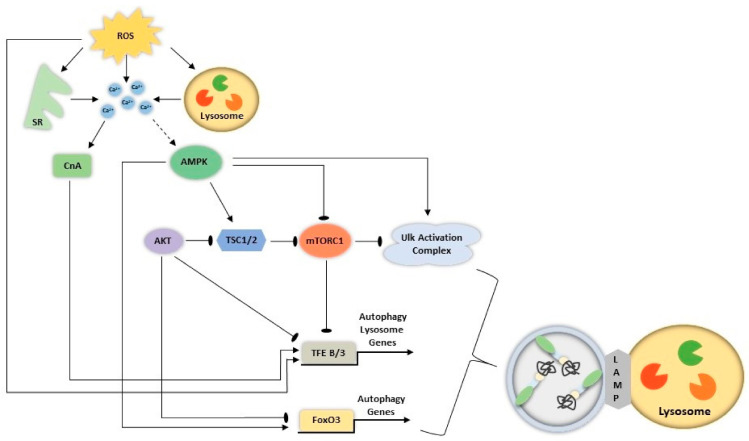
Autophagy-lysosome related genetic programs require sensing of the cellular milieu to coordinate the expression of essential gene-products. As such, FoxO3, Tfeb and Tfe3 undergo a series of post-translational modifications which dictate their nuclear localization and activity. Further, the Ulk1-complex, at the early stage of autophagy induction, is also tightly integrated with the signaling mechanisms that regulate these transcription factors, ultimately leading to a coordinated ALS activation. These pathways are summarized below and depicted in Figure 1.
Figure 1.
Proposed coordination of pathways that regulate the autophagy-lysosome System (ALS). ROS and cytosolic calcium ([Ca2+]IC) act as upstream signaling mechanisms that converge on the gene expression and protein activation of the ALS. AMPK is activated directly via compromised energy status (increase AMP:ADP), and indirectly through [Ca2+]IC via CaMKII (not shown). Elevated [Ca2+]IC may occur due to the damaging effects of elevated ROS, from mitochondria or other sources, on the sarcoplasmic reticulum and lysosomes. AMPK synchronously activates FoxO3, Tfeb and Tfe3 (Tfeb/3) through (1) direct phosphorylation of FoxO3, (2) activation of TSC1/2 and (3) inhibition of mTORC1. Simultaneously, AMPK activates autophagy induction through the Ulk1 complex and by alleviating mTORC1 inhibition on this complex. In a similar cellular milieu, FoxO3 repression is alleviated via AKT inhibition. Increases in [Ca2+]IC activate the phosphatase CnA, which activates Tfeb/3. ROS also activate Tfeb/3 through oxidation, an added layer of coordination in this autophagy-lysosome system.
Under fed conditions, FoxO3, Tfeb and Tfe3 localization is primarily cytosolic due to repression mediated by active AKT [73,74]. mTORC1, positioned downstream, and localized to the lysosome [52], phosphorylates Tfeb on Ser211 [75] and Tfe3 on Ser321 [41]. This promotes their interaction with 14-3-3 in the cytosol, rendering them inactive [41,75,76,77]. mTORC1 also inhibits Ulk1 through direct interaction and phosphorylation [78]. In this cellular milieu, AMPK activity is low, and thus its impact on activating FoxO3 and Ulk1-complex through phosphorylation is minimal [73,79,80,81]. These conditions promote a coordinated suppression of the autophagy-lysosome program.
In contrast, under energy stress, when AMPK is active, FoxO3 [73,79,80] and Ulk1 [81] activity are enhanced through direct phosphorylation. AMPK phosphorylates and inhibits mTORC1 [82] and the mTORC1 inhibitor Tsc1/2 [83] which will ultimately alleviate mTORC1′s repression on Ulk1, and Tfeb/Tfe3 to promote their nuclear localization. In this way, AMPK ultimately coordinates the activation and induction of the autophagy-lysosome system.
The importance of these signaling pathways has been shown, whereby Tsc1 knockout blocks autophagy induction and promotes muscle atrophy [84]. However, this effect can be prevented by the mTOR inhibitor rapamycin [84], highlighting the importance of mTORC1 in repressing autophagic induction. Further, AMPK knockout leads to an attenuation of atrophy [85,86], likely due to a failure to induce autophagy [87] and to promote protein degradation. Thus, this regulatory network, including both mTORC1 and AMPK are essential for the maintenance of muscle mass.
2.2.2. Other Mechanisms Coordinating Activation of the Autophagy Lysosome System
Other integral regulators of this coordinated response include reactive oxygen species (ROS) and Ca2+ [88,89,90,91,92]. Specifically, ROS activate AMPK [93] thereby enhancing FoxO3 and Ulk1 activity. ROS also inhibit AKT signaling [91,94] and ultimately the mTORC1 pathway [95], thereby alleviating its suppression on Ulk1 and FoxO3, further promoting autophagic induction. ROS can also potentiate CLEAR network expression via direct oxidation on Cys212 and Cys322 of Tfeb and Tfe3, respectively, which trigger their nuclear localization in muscle cells [95]. Finally, through oxidation of lysosomal transient receptor potential mucolipin 1 (Trpml1) on lysosomes [96] as well as Ryanodine Receptors (RyR) on the sarcoplasmic reticulum [97], ROS can promote the elevation of intracellular calcium ([Ca2+]IC) [89]. [Ca2+]IC may induce autophagy via the activation of CAMKKB and subsequently AMPK, which has implications for Ulk1 and FoxO3 phosphorylation, as described above [98]. Furthermore, elevations in cytosolic Ca2+ facilitate nuclear localization of Tfeb [99] and Tfe3 [100] via the phosphatase calcineurin (CnA), which dephosphorylates these proteins.
2.3. Autophagy in Aging Muscle
2.3.1. Upstream Regulation of Autophagy in Aging Muscle
A hallmark characteristic of sarcopenia is muscular atrophy, the result of a progressive loss of muscle protein over time. This could be achieved by an imbalance between muscle protein synthesis and the pathways that mediate protein degradation, including the ubiquitin proteasome pathway, apoptosis, and autophagy. Much remains to be elucidated regarding the signaling toward, and the activity of, autophagy-lysosome regulatory proteins. For example, there is evidence of upregulated FoxO3 [101,102,103] and Tfeb [102] content in aged muscle, supporting elevated expression of the ubiquitin-proteosome system and autophagy genes in old muscle. However, there are no clear indications of whether AKT-mediated FoxO3 phosphorylation [104,105] and localization [104,106] are enhanced or reduced with age. This may be explained, in part, by the relatively inconsistent measures of AKT activity that exist in the literature [104,106,107,108,109]. Further, deacetylation of FoxO3 by histone deacetylase 1 (HDAC1) [73,110,111] and co-activation by CARM1 [32,33] enhance FoxO3 function, and are implicated in disuse/denervation atrophy. Both mechanisms have not been investigated in aged muscle. On the other hand, AMPK signaling is lower in aged individuals [112,113], and although the ramifications of this for FoxO3 activity is yet to be uncovered, it has been shown that, the muscle of aged humans and rats contain lower basal AMPK-activating-Ulk1 phosphorylation [59]. Furthermore, mTORC1 activity is sustained in aged muscle [114,115,116], corresponding to increases in inhibitory Ulk1 phosphorylation [107,114,117]. This mTORC1-mediated inhibition may mitigate the extent of atrophy in aging muscle. However, no reports have investigated Tfeb or Tfe3 activity or localization in aged muscle. The lack of data on these signaling pathways suggest that more work is necessary to understand the influence of the autophagy-lysosome system in mediating the atrophy evident in aging muscle.
Both ROS [118,119,120] and [Ca2+]IC [97,118,121,122] are elevated in aged skeletal muscle, which may promote both autophagy-lysosome related gene expression and autophagic induction. In fact, scavenging of ROS via antioxidant and genetic models, in muscle disuse [94,123] and denervation [124,125,126] suppresses autophagic pathways and muscle loss, which may be true in aging muscle. Similarly, it would be essential to understand how age-related [Ca2+]IC elevations impact the autophagy lysosome system, as the literature has been primarily focused on Calpain activity in response to Ca2+ overload. Cumulatively, these limited data also point to the requirement of assessing the necessity of ROS- and Ca2+-induced autophagy signaling in aging muscle.
2.3.2. Autophagy Flux in Aging Skeletal Muscle
There are a variety of reports that support both enhanced, as well as reduced, autophagy in aged skeletal muscle, based on assessment of protein markers. Evidence of normally functioning autophagosome formation, with impairments in the processing and degradation of autophagosomes was observed based on accumulations in Beclin-1, LC3-I and LC3-II, without a change in upstream autophagy markers Atg7 and 9 or Lc3 transcript [127]. Our laboratory and others have similarly reported elevations in LC3-II, its processing protein Atg-7, LC3-II/I, and p62 [103,128,129,130,131]. In humans, Fry et al., measured elevations in autophagic proteins Atg7 and Beclin1 without changes in LC3-II/I, a result that is suggestive of perturbed autophagosomal breakdown with age [132].
In contrast, others have documented no change in Beclin1 or LC3-II/I in humans [133], and LC3-II/I or p62 protein expression in aged mice [107], findings that were interpreted as unaltered autophagy with age. These changes in autophagy execution may be fiber-type specific as some investigations have found that protein markers such as Atg5, LC3 and p62 are maintained in slow, but not fast muscle [134]. This fiber type difference is relevant in young, healthy muscle, whereby oxidative muscle has greater autophagy flux than predominantly fast muscle [135]. This fiber-type specific response requires further investigation.
Inconsistencies in these investigations may be explained by the “snapshots” measures of autophagosomal transcript and protein levels used to determine autophagy, which fail to capture the dynamic nature of the pathway. Recently, two studies utilized colchicine to assess autophagy flux in aged Fisher344BN rats. Baehr et al., measured an accumulation of p62 and LC3-II in the soleus and tibialis anterior muscles of 29mo rats [136], suggesting diminished autophagic degradation. However, autophagy flux was enhanced in the aged cohort [136]. Similarly, we have recently reported elevation in autophagy flux in 36mo rats [102]. Thus, it appears that autophagy flux in muscle is elevated with age, at least up until the point of cargo delivery to the lysosome.
2.3.3. Ramifications on Aging Skeletal Muscle
Although these basal changes in autophagy are important, understanding the extent to which muscle adapts to stimuli placed upon it is critical. In response to starvation, aged muscle fails to elicit the AKT and mTORC1 inhibition that is typical of young muscle [114]. Furthermore, there is also evidence of blunted stimulus-induced AMPK [137,138,139] and mTORC1 [140] activation, supported by lower starvation-induced Ulk1 activation [114]. Interestingly, caloric restriction [127], endurance exercise training [127], and chronic muscle activity [102] do improve autophagic degradation in aged muscle, however, the response is curtailed compared to a younger cohort. These data provide evidence that senescent muscle has a perturbed autophagy program. This has widespread ramifications for the ability of aged muscle to adapt to stimuli such as exercise and caloric restriction, which impact muscle quality.
3. Regulation of Mitophagy in Muscle
Mitochondrial health within muscle depends on organellar turnover that is a product of the balance between the synthesis of new, healthy organelles via biogenesis, and the recycling of damaged or dysfunctional mitochondria through mitophagy. This ensures the sustenance of a mitochondrial reticulum which is well coupled, thereby producing ATP efficiently and generating a level of ROS sufficient to signal events that maintain homeostasis. When segmental mitochondrial dysfunction occurs, evident form varying degrees of respiratory impairment, elevated ROS production, or loss of membrane potential, these organelles are digested at the lysosomes via the mitophagy pathway, as depicted in Figure 2, and as descried below. If balanced with a concurrent change in mitochondrial biogenesis, this turnover maintains the metabolic capacity of the muscle [13,14]. However, if these processes become unbalanced, the overall health of the mitochondrial pool will be disrupted, a phenomenon which is commonly observed in both aging and muscle disuse [13,14,119,120].
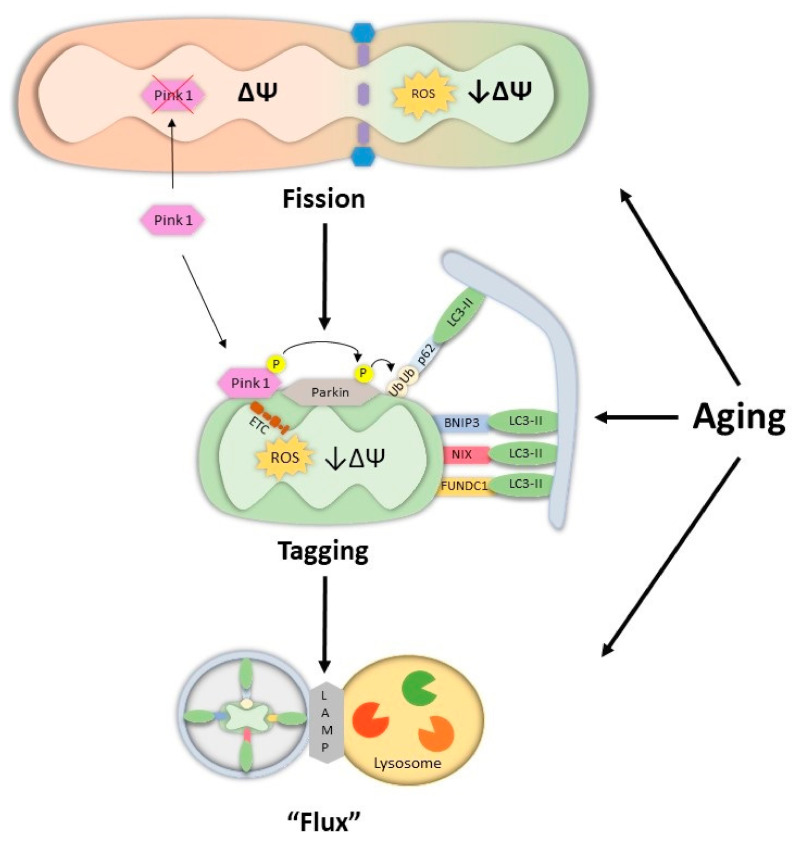
Figure 2.
The impact of age on mitophagy in skeletal muscle. When a portion of the mitochondrial reticulum becomes dysfunctional, with a low membrane potential (Ψ) and elevated ROS production, it will undergo fission, mediated by Drp1 and Fis1, depicted as green mitochondria. The fragmented mitochondrion will enter the mitophagy pathway. Pink1, which is normally imported and degraded in the mitochondria, will accumulate on the outer mitochondrial membrane (OMM), and will ultimately recruit and activate the E3-Ligase Parkin. Parkin subsequently poly-ubiquitinates OMM proteins which bind to adapt proteins such as p62. p62 will tether autophagosomal proteins, such as LC3-II. In Ub-independent mitophagy, receptors such as BNIP3, NIX and FUNDC1, found on the OMM, will anchor the mitochondria directly to autophagosomal proteins. Ultimately the autophagosome-bound mitochondria will be shuttled to lysosomes where they will be degraded. Aging muscle exhibits increased mitochondrial fission, autophagy tagging, and flux, to the point of the lysosomes. Evidence, somewhat limited, exists that point to lysosomal dysfunction with age.
3.1. Mitochondrial Dynamics in Muscle—Implications for Mitophagy
Skeletal muscle mitochondria exist in a reticular network which optimizes the metabolic capabilities of the tissue [2,141,142]. This network is maintained through the opposing processes of fusion and fission. As recently reviewed [143], mitochondrial dynamics are regulated via the pro-fusion proteins, mitofusin 1/2 (Mfn1/2) and optic atrophy protein 1 (Opa1), as well as the pro-fission dynamic-related protein 1 (Drp1) and mitochondrial fission protein 1(Fis1). When a portion of the network becomes impaired it is sequestered through mitochondrial fission, which primes this organelle segment for its ultimate degradative fate via the mitophagy pathway [144,145].
The vitality of these fusion and fission regulatory proteins has been revealed using genetic models. For example, knockdown of the pro-fission proteins Fis1 and Drp1 propagates muscle atrophy [146,147,148], promotes hyperfused mitochondria with impaired function [149] and perturbs mitophagy [148,149]. These data suggest that fission is required to maintain muscle organellar health via the provision of dysfunctional mitochondria through mitophagy pathway. With regard to fusion, Mfn2 [129,150] and Opa1 [151] deletion promotes mitochondrial fragmentation, excessive ROS production and muscle atrophy. In combination, these results suggest that a balance of fusion and fission is required to maintain a healthy mitochondrial pool, which has implications for muscle health with age.
Mitochondrial Dynamics in Aged Muscle
A number of studies have reported a greater abundance of fission relative to fusion proteins in aged muscle [113,129,130,131,152,153], although some inconsistent findings have been observed [154,155]. Elongated mitochondria in aged muscle have been noted, a finding the authors attributed to intrinsic mitochondrial defects that promote fusion as a compensatory mechanism [155]. Variability in the results may be explained by the aging model employed, species differences, or the degree of aging examined. However, as mitochondrial dysfunction is apparent in aged muscle, and as fission precedes mitophagy, it is not surprising that there is evidence of fragmented mitochondria in aged muscle, which may prime this organellar pool for mitophagy.
3.2. The Process of Mitophagy
3.2.1. Pink1-Parkin Mediated Mitophagy
The most well understood mitophagy system is termed ubiquitin-dependent mitophagy, which is controlled by the serine/threonine kinase, PTEN- induced putative kinase 1 (Pink1), and the E3-ubiquitin ligases Parkin. Pink1 is normally maintained at low levels within the cytosol under homeostatic conditions, as Pink1 is efficiently imported into functional mitochondria, processed by the mitochondrial protein peptidase (MPP) and subsequently degraded by the intramembrane protease presenilin-associated rhomboid-like protease (PARL) [156,157,158], or by Lon protease [159]. Following mitochondrial stress, particularly an accumulation of damaged intraorganellar proteins [160] or a loss of membrane potential [156,161,162,163], protein import is hindered, and Pink1 accumulates on the outer mitochondrial membrane (OMM). Here, Pink1 undergoes autophosphorylation and activation [164] to phosphorylate both Parkin [161,162,163,165] and ubiquitin [166,167,168,169]. The result is polyubiquitination of OMM proteins such as VDAC, Mfn1/2, mitochondrial translocase complex proteins (TOMs) and ubiquitin, which will feedforward to recruit more Parkin and ubiquitin to the membrane [161,162,163,170,171]. Ultimately, these ubiquitin chains act as a scaffold for adapter proteins such as p62 [172,173] optineurin [173,174] and Ndp52 [173], which interact with the autophagosomal proteins leading to organelle engulfment and subsequent degradation. An essential Parkin-regulated process is the ubiquitination and degradation of the fusion protein Mfn1/2, ensuring that re-fusion does not occur, and allowing for mitophagy to take place effectively [175].
The importance of Pink1/Parkin-mediated mitophagy in skeletal muscle has been demonstrated in lower organisms [176,177], cultured myotubes [178,179] and mice [180,181]. Specifically, Parkin knockout promotes muscle degeneration and cell death in drosophila [176], as well as insulin-insensitivity [178] and susceptibility to mitochondrial toxins [179] in myotubes. In the muscle of Parkin-null mice, there is evidence of mitochondrial fragmentation [182], impaired mitochondrial respiration [180,181,182] and reduced mitophagy flux [180,181]. Interestingly, Parkin-null mice fail to moderate mitophagy in response to acute exercise [180] and endurance training [181]. Parkin overexpression in mice prevents both aging- and sepsis-induced loss of muscle mass and strength [183,184] and extents lifespan through reduced proteotoxicity in drosophila [177]. Cumulatively, these reports highlight the importance of the Pink1-Parkin-mediated mitophagy system in skeletal muscle.
3.2.2. Receptor Mediated Mitophagy
Ubiquitin-independent mitophagy, also referred to as the receptor-mediated mitophagy system, operates more directly than the Parkin-driven pathway, and it is unclear if these mechanisms occur simultaneously or independently. Mitophagy receptors that have been characterized include Bnip3 [185,186,187], Bnip3-like protein (Nix) [188,189,190] and Fun14 domain containing protein (Fundc1) [191,192], which are normally localized to mitochondria [191,193,194,195]. Upon phosphorylation, these receptors interact with phagophore proteins such as LC3 and Gabarap [189,191,196]. In the early stages of mitophagy, Bnip/Nix also promote the mitochondrial localization of Drp1, which would enhance fission and aid in Parkin recruitment, leading to mitophagy [197,198,199].
The importance of these mitophagy mechanisms in muscle has recently been demonstrated. Knockout of Bnip3 in muscle cells prevented their differentiation, likely a result of an accumulation of dysfunctional mitochondria [200]. Similarly, knockout of both Nix and Fundc1 in cardiac progenitor cells led to cell death [201]. Further, skeletal muscle deletion of Fundc1 impaired mitochondrial function and exercise capacity, and this protein was required for both basal as well as exercise and contraction-induced mitophagy [202,203]. In contrast, the overexpression of the Bnip3 and Nix induced muscle atrophy [146], providing evidence that mitochondrial degradation is fine-tuned to the maintenance of muscle mass.
3.3. Mitophagy in Aging Skeletal Muscle
3.3.1. Mitophagy Targeting in Aged Muscle
As mitochondrial fragmentation and dysfunction are evident in senescent muscle, one would suspect that mitophagic signaling would be enhanced to remove these dysfunctional organelles. A variety of studies have reported the enhanced expression of the mitophagy proteins Bnip3, Nix, Pink1, and Parkin [102,129,180,204,205], but some studies have measured either no change and/or reduced levels of Bnip3 and Nix [130,133,206] in aged muscle. The phosphorylation status of these proteins would provide a clear indication if these mitophagy pathways are active, but this has not yet been explored in the context of aging muscle. We and others have reported decreases in Parkin protein [184] and mitochondrial Parkin [180] in 18 month old mice. In contrast, older rodents (i.e., 24mo mice and 35mo rats) exhibit elevations in Pink1, Parkin and Ubiquitin [130,131], suggesting an age-specific Pink1-Parkin mitophagic response.
3.3.2. Mitophagy Flux in Aged Muscle
Supportive of enhance targeting, our laboratory has reported elevated autophagosomal tethering to mitochondria in aged muscle, as measured by increased LC3-II [131] and p62 [102,131] in these isolated organelles. These data may suggest either (1) an enhanced rate of organellar targeting and degradation or (2) perturbed efficiency of mitophagic breakdown. Using colchicine, we have observed increased mitophagy flux in aged mouse [180] and rat [102] muscle, which occurs alongside deficits in mitochondrial biogenesis [13,14,15]. Although enhanced flux would implicate the accelerated removal of damaged mitochondria, the remaining presence of impaired organelles indicates that the rate may not be great enough to eliminate all dysfunctional cellular components, including mitochondria. This remaining organellar pool will not only have less metabolic capacity, but also contribute to oxidative damage within the muscle that can propagate further dysfunction. Thus, restoring mitophagy in aged muscle may hold therapeutic benefit in slowing age-related sarcopenia.
Aging is associated with both neuromuscular denervation and reduced physical activity. Since it is known that denervation [124,126,131,207,208,209,210,211,212,213] and muscle disuse [214,215,216] independently impact mitophagic pathways, work is required to determine the independent nature of these stimuli on mitophagy. Furthermore, acute endurance exercise acts as a stimulus that promotes mitophagy in young muscle, used to prune the mitochondrial pool and create a healthier tissue [13]. However, there is evidence for impaired acute exercise-induced mitophagy in aged muscle [180]. Further both the chronic contractile activity induced reduction of mitophagy flux [102] and the denervation-induced mitophagic upregulation [131] are impaired in aged muscle. Thus, it is becoming clear that aging alters the mitophagic phenotype of muscle, and the lack of appropriate stimulus-induced alterations in mitophagy may limit positive adaptations in the maintenance of a healthy organelle population.
4. The Lysosomes and Aging Muscle
Lysosomes are highly acidic organelles that contain an abundance of enzymes to aid in the degradation of autophagic substrates. Typically thought of as the digestive organelle, the literature has implicated the lysosome as a significant component in the monitoring and response to changes in the cellular milieu, such as nutrient availability [217,218]. Lysosomes are also an important intermediate in organellar communication, for example, between the nucleus and mitochondria [219,220].
Lysosomes are created through the integration of gene expression and protein trafficking with the endocytic pathway, processes that are not well characterized in skeletal muscle. Indeed, the role and importance of lysosomes have been elucidated in many cell types, but limited work has been conducted in the skeletal muscle field. Yet, the value of these organelles in muscle is clear when viewed in the context of Lysosomal Storage Diseases (LSDs). For example, Pompe [5,221,222,223,224,225,226] and Danon Disease [227,228,229,230,231,232] are both associated with an accumulation of non-digested autophagosomes and enlarged lysosomes, manifesting in diminished muscle mass and function.
4.1. Lysosomal Perturbations in Aging Muscle
Within aging tissue, there is a progressive increase in lipofuscin, commonly referred to as “the age pigment”, indicative of dysfunctional lysosomal clearance [233,234]. Skeletal muscle is not exempt from this phenomenon, as lipofuscin granules are visible in both rodent [131] and human [235] muscle. We recently reported increases in the lysosomal proteins Lamp2 and Cathepsin D in the tibialis anterior of aged rats, which may be due to the observed increase in Tfeb [102] and its function in controlling lysosomal synthesis. Others have reported similar findings in the extensor digitorum longus muscle of mice [134], although transcript levels may be reduced [127,236]. Presently there is a lack of investigation into lysosomal function in muscle, despite occasional measures of gene expression and enzyme activity. Disturbances in lysosome function in aged muscle would have widescale implications for autophagy and mitophagy. This could explain why dysfunctional mitochondria and oxidized proteins accumulate in the muscle of aged animals and humans. As the “Lysosomal Theory of Aging” would suggest, this blunted auto- and mitophagic capacity may in fact promote further damage within the tissue, which could potentiate myonuclear loss through apoptosis mediated by both mitochondria through the release of cytochrome c, and lysosomes via the release of cathepsins. In line with this, it is established that mitochondrially driven apoptosis is enhanced in aged skeletal muscle [237,238,239], but it remains unclear if lysosome-mediated apoptosis is a contributing factor in determining the degree of sarcopenia.
4.2. Future Work of Lysosomes in Aging Muscle
As noted above, there is virtually nothing known about lysosomal function in aging muscle. In other cell types, dysfunction is characterized by a loss in acidification due to membrane instability, organelle swelling, and the release of active cathepsins which can ultimately lead to cell death [240]. Thus, it seems wise to dedicate future investigations toward these types of analyses in young and old muscle. This is because the importance of enhancing lysosomal health is established in various experimental models. For example, the overexpression of Tfeb is able to reduce pathophysiology through the transcriptional induction of components of the autophagy-lysosome system in neuropathies [241,242,243]. Similarly, a direct Tfeb activator, curcumin analog C1, is able to prevent the pathology associated with Alzheimer’s disease [244]. In muscle, AAV-mediated TFEB overexpression attenuated the severe pathology evident in a murine model of Pompe Disease [221]. As aging muscle displays lysosome impairments, Tfeb activation and subsequent restoration of lysosome integrity may ultimately restore muscle health, at least in part. In addition, an assessment of the impact of ROS and Ca2+ on the lysosomes in aged muscle is warranted. This is because both are elevated in aging muscle and can impact Tfeb and Tfe3, and ultimately lysosomal biogenesis, as discussed above.
5. Conclusions
In recent years significant advances have been made to uncover the mechanisms that underly age-related sarcopenia. Although there are multiple mechanisms involved, this phenotypic change is mediated by alterations in protein and organelle homeostasis. One integral mediator of protein turnover is the activity of the autophagy-lysosomes system, and its interaction with mitochondrial turnover. Mitochondria are a predominant source of oxidative stress and are degraded via this pathway. Insufficient autophagic clearance can lead to a build-up of poorly functioning organelles that perpetuate intracellular damage. In aged muscle there is evidence of (1) oxidative damage, (2) mitochondrial dysfunction and (3) lysosome impairments. Thus, the activity of the complete autophagy-lysosome system, from organelle-tagging to destruction within lysosomes, may become insufficient with age. Moving forward, research has identified the key proteins within the autophagy pathway upstream of the lysosome with age, but current knowledge of lysosome function is lacking as it relates to age-related muscle atrophy. Once this is better appreciated, recognition may follow on how enhancing lysosome function pharmacologically can slow potentially slow the progress of sarcopenia.
Acknowledgments
David A. Hood is the holder of a Canada Research Chair in Cell Physiology.
Abbreviations
| AMP activated protein kinase | AMPK |
| Autophagy-related genes | Atg |
| Bcl2/adenovirus E1B 19-kDa protein-interacting protein 3 | Bnip3 |
| BNIP3-like protein | Nix |
| Calcium/calmodulin-dependent protein kinase kinase B | CAMKKB |
| Calcineurin | CnA |
| Class III PI3K Complex I | PI3KC3-C1 |
| Coactivator Associated Arginine Methyltransferase 1 | CARM1 |
| Coordinated lysosomal enhancement and regulation | CLEAR |
| Dynamic-related protein 1 | Drp1 |
| Fission protein 1 | Fis1 |
| Forkhead box O3 | FoxO3 |
| Fun14 domain containing protein | Fundc1 |
| γ-aminobutyric acid receptor-associated protein | Gabarap |
| Golgi-associated ATPase enhancer of 16kDa | Gate16 |
| Histone Deacetylase 1 | HDAC1 |
| LC3-II interacting region | LIR |
| Lysosomal-associated membrane protein | Lamp |
| Lysosomal storage disorder | LSD |
| Lysosome transient receptor potential mucolipin 1 | Trpml1 |
| Mammalian target of rapamycin | mTOR |
| Mitochondrial protein peptidase | MPP |
| Mitofusin 1/2 | Mfn1/2 |
| Optic atrophy protein 1 | Opa1 |
| Outer mitochondrial membrane | OMM |
| Phosphatidylethanolamine | PE |
| Presenilin-associated rhomboid-like protease | PARL |
| PTEN induced putative kinase | Pink1 |
| Ras-related protein | Rab |
| Reactive Oxygen Species | ROS |
| Ryanodine Receptors | RyR |
| Sequestosome 1 | Sqstm1/p62 |
| Trans-golgi network | TGN |
| Translocase of the outer membrane | TOM |
| Tuberous Sclerosis Complex 1/2 | Tsc1/2 |
| Transcription factor EB | Tfeb |
| Transcription factor E3 | Tfe3 |
| Ubiquitin | Ub |
Author Contributions
M.T. and D.A.H. drafted, edited, revised, and approved the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Science and Engineering Research Council of Canada [grant number 38462].
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Janssen I., Heymsfield S.B., Wang Z., Ross R. Skeletal Muscle Mass and Distribution in 468 Men and Women Aged 18–88 Yr. J. Appl. Physiol. 2000;89:81–88. doi: 10.1152/jappl.2000.89.1.81. [DOI] [PubMed] [Google Scholar]
- 2.Ogata T., Yamasaki Y. Scanning Electron-Microscopic Studies on the Three-Dimensional Structure of Mitochondria in the Mammalian Red, White and Intermediate Muscle Fibers. Cell Tissue Res. 1985;241:251–256. doi: 10.1007/BF00217168. [DOI] [PubMed] [Google Scholar]
- 3.Hoppeler H. Exercise-Induced Ultrastructural Changes in Skeletal Muscle. Int. J. Sports Med. 1986;7:187–204. doi: 10.1055/s-2008-1025758. [DOI] [PubMed] [Google Scholar]
- 4.Das G., Shravage B.V., Baehrecke E.H. Regulation and Function of Autophagy during Cell Survival and Cell Death. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a008813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raben N., Hill V., Shea L., Takikita S., Baum R., Mizushima N., Ralston E., Plotz P. Suppression of Autophagy in Skeletal Muscle Uncovers the Accumulation of Ubiquitinated Proteins and Their Potential Role in Muscle Damage in Pompe Disease. Hum. Mol. Genet. 2008;17:3897–3908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masiero E., Agatea L., Mammucari C., Blaauw B., Loro E., Komatsu M., Metzger D., Reggiani C., Schiaffino S., Sandri M. Autophagy Is Required to Maintain Muscle Mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 7.Carnio S., LoVerso F., Baraibar M.A., Longa E., Khan M.M., Maffei M., Reischl M., Canepari M., Loefler S., Kern H., et al. Autophagy Impairment in Muscle Induces Neuromuscular Junction Degeneration and Precocious Aging. Cell Rep. 2014;8:1509–1521. doi: 10.1016/j.celrep.2014.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paré M.F., Baechler B.L., Fajardo V.A., Earl E., Wong E., Campbell T.L., Tupling A.R., Quadrilatero J. Effect of Acute and Chronic Autophagy Deficiency on Skeletal Muscle Apoptotic Signaling, Morphology, and Function. Biochim. Biophys. Acta BBA Mol. Cell Res. 2017;1864:708–718. doi: 10.1016/j.bbamcr.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 9.Nemazanyy I., Blaauw B., Paolini C., Caillaud C., Protasi F., Mueller A., Proikas-Cezanne T., Russell R.C., Guan K.-L., Nishino I., et al. Defects of Vps15 in Skeletal Muscles Lead to Autophagic Vacuolar Myopathy and Lysosomal Disease. EMBO Mol. Med. 2013;5:870–890. doi: 10.1002/emmm.201202057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janssen I., Heymsfield S.B., Ross R. Low Relative Skeletal Muscle Mass (Sarcopenia) in Older Persons Is Associated with Functional Impairment and Physical Disability. J. Am. Geriatr. Soc. 2002;50:889–896. doi: 10.1046/j.1532-5415.2002.50216.x. [DOI] [PubMed] [Google Scholar]
- 11.Larsson L., Degens H., Li M., Salviati L., Lee Y.I., Thompson W., Kirkland J.L., Sandri M. Sarcopenia: Aging-Related Loss of Muscle Mass and Function. Physiol. Rev. 2018;99:427–511. doi: 10.1152/physrev.00061.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santilli V., Bernetti A., Mangone M., Paoloni M. Clinical Definition of Sarcopenia. Clin. Cases Miner. Bone Metab. 2014;11:177–180. doi: 10.11138/ccmbm/2014.11.3.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hood D.A., Memme J.M., Oliveira A.N., Triolo M. Maintenance of Skeletal Muscle Mitochondria in Health, Exercise, and Aging. Annu. Rev. Physiol. 2019;81:19–41. doi: 10.1146/annurev-physiol-020518-114310. [DOI] [PubMed] [Google Scholar]
- 14.Kim Y., Triolo M., Hood D.A. Impact of Aging and Exercise on Mitochondrial Quality Control in Skeletal Muscle. Oxidative Med. Cell. Longev. 2017;2017:3165396. doi: 10.1155/2017/3165396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tryon L.D., Vainshtein A., Memme J.M., Crilly M.J., Hood D.A. Recent Advances in Mitochondrial Turnover during Chronic Muscle Disuse. Integr. Med. Res. 2014;3:161–171. doi: 10.1016/j.imr.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welch C., K Hassan-Smith Z., A Greig C., M Lord J., A Jackson T. Acute Sarcopenia Secondary to Hospitalisation-An Emerging Condition Affecting Older Adults. Aging Dis. 2018;9:151–164. doi: 10.14336/AD.2017.0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodpaster B.H., Chomentowski P., Ward B.K., Rossi A., Glynn N.W., Delmonico M.J., Kritchevsky S.B., Pahor M., Newman A.B. Effects of Physical Activity on Strength and Skeletal Muscle Fat Infiltration in Older Adults: A Randomized Controlled Trial. J. Appl. Physiol. 2008;105:1498–1503. doi: 10.1152/japplphysiol.90425.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Triolo M., Hood D.A. Mitochondrial Breakdown in Skeletal Muscle and the Emerging Role of the Lysosomes. Arch. Biochem. Biophys. 2019;661:66–73. doi: 10.1016/j.abb.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 19.VanderVeen B.N., Fix D.K., Carson J.A. Disrupted Skeletal Muscle Mitochondrial Dynamics, Mitophagy, and Biogenesis during Cancer Cachexia: A Role for Inflammation. Oxidative Med. Cell. Longev. 2017;2017:1–13. doi: 10.1155/2017/3292087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Ende M., Grefte S., Plas R., Meijerink J., Witkamp R.F., Keijer J., van Norren K. Mitochondrial Dynamics in Cancer-Induced Cachexia. Biochim. Biophys. Acta BBA Rev. Cancer. 2018;1870:137–150. doi: 10.1016/j.bbcan.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 21.Moore T.M., Lin A.J., Strumwasser A.R., Cory K., Whitney K., Ho T., Ho T., Lee J.L., Rucker D.H., Nguyen C.Q., et al. Mitochondrial Dysfunction Is an Early Consequence of Partial or Complete Dystrophin Loss in Mdx Mice. Front. Physiol. 2020;11 doi: 10.3389/fphys.2020.00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes M.C., Ramos S.V., Turnbull P.C., Rebalka I.A., Cao A., Monaco C.M.F., Varah N.E., Edgett B.A., Huber J.S., Tadi P., et al. Early Myopathy in Duchenne Muscular Dystrophy Is Associated with Elevated Mitochondrial H2O2 Emission during Impaired Oxidative Phosphorylation. J. Cachexiasarcopenia Muscle. 2019;10:643–661. doi: 10.1002/jcsm.12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Penna F., Ballarò R., Martinez-Cristobal P., Sala D., Sebastian D., Busquets S., Muscaritoli M., Argilés J.M., Costelli P., Zorzano A. Autophagy Exacerbates Muscle Wasting in Cancer Cachexia and Impairs Mitochondrial Function. J. Mol. Biol. 2019;431:2674–2686. doi: 10.1016/j.jmb.2019.05.032. [DOI] [PubMed] [Google Scholar]
- 24.Ballarò R., Beltrà M., De Lucia S., Pin F., Ranjbar K., Hulmi J.J., Costelli P., Penna F. Moderate Exercise in Mice Improves Cancer plus Chemotherapy-Induced Muscle Wasting and Mitochondrial Alterations. FASEB J. 2019;33:5482–5494. doi: 10.1096/fj.201801862R. [DOI] [PubMed] [Google Scholar]
- 25.Pauly M., Daussin F., Burelle Y., Li T., Godin R., Fauconnier J., Koechlin-Ramonatxo C., Hugon G., Lacampagne A., Coisy-Quivy M., et al. AMPK Activation Stimulates Autophagy and Ameliorates Muscular Dystrophy in the Mdx Mouse Diaphragm. Am. J. Pathol. 2012;181:583–592. doi: 10.1016/j.ajpath.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Aversa Z., Pin F., Lucia S., Penna F., Verzaro R., Fazi M., Colasante G., Tirone A., Fanelli F.R., Ramaccini C., et al. Autophagy Is Induced in the Skeletal Muscle of Cachectic Cancer Patients. Sci. Rep. 2016;6:30340. doi: 10.1038/srep30340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandri M. Autophagy in Skeletal Muscle. FEBS Lett. 2010;584:1411–1416. doi: 10.1016/j.febslet.2010.01.056. [DOI] [PubMed] [Google Scholar]
- 28.Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S.H., Goldberg A.L. Foxo Transcription Factors Induce the Atrophy-Related Ubiquitin Ligase Atrogin-1 and Cause Skeletal Muscle Atrophy. Cell. 2004;117:399–412. doi: 10.1016/S0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bodine S.C., Latres E., Baumhueter S., Lai V.K.M., Nunez L., Clarke B.A., Poueymirou W.T., Panaro F.J., Erqian N., Dharmarajan K., et al. Identification of Ubiquitin Ligases Required for Skeletal Muscle Atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 30.Zhao J., Brault J.J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S.H., Goldberg A.L. FoxO3 Coordinately Activates Protein Degradation by the Autophagic/Lysosomal and Proteasomal Pathways in Atrophying Muscle Cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P., Burden S.J., Di Lisi R., Sandri C., Zhao J., et al. FoxO3 Controls Autophagy in Skeletal Muscle In Vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 32.Stouth D.W., van Lieshout T.L., Ng S.Y., Webb E.K., Manta A., Moll Z., Ljubicic V. CARM1 Regulates AMPK Signaling in Skeletal Muscle. iScience. 2020;23:101755. doi: 10.1016/j.isci.2020.101755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y., Li J., Shang Y., Guo Y., Li Z. CARM1 Contributes to Skeletal Muscle Wasting by Mediating FoxO3 Activity and Promoting Myofiber Autophagy. Exp. Cell Res. 2019;374:198–209. doi: 10.1016/j.yexcr.2018.11.024. [DOI] [PubMed] [Google Scholar]
- 34.Lee D., Goldberg A.L. SIRT1 Protein, by Blocking the Activities of Transcription Factors FoxO1 and FoxO3, Inhibits Muscle Atrophy and Promotes Muscle Growth. J. Biol. Chem. 2013;288:30515–30526. doi: 10.1074/jbc.M113.489716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang H., Inoki K., Lee M., Wright E., Khuong A., Khuong A., Sugiarto S., Garner M., Paik J., DePinho R.A., et al. MTORC1 Promotes Denervation-Induced Muscle Atrophy through a Mechanism Involving the Activation of FoxO and E3 Ubiquitin Ligases. Sci. Signal. 2014;7:ra18. doi: 10.1126/scisignal.2004809. [DOI] [PubMed] [Google Scholar]
- 36.Bertaggia E., Coletto L., Sandri M. Posttranslational Modifications Control FoxO3 Activity during Denervation. Am. J. Physiol. Cell Physiol. 2012;302:587–596. doi: 10.1152/ajpcell.00142.2011. [DOI] [PubMed] [Google Scholar]
- 37.Ploper D., De Robertis E.M. The MITF Family of Transcription Factors: Role in Endolysosomal Biogenesis, Wnt Signaling, and Oncogenesis. Pharmacol. Res. 2015;99:36–43. doi: 10.1016/j.phrs.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 38.Martina J.A., Diab H.I., Li H., Puertollano R. Novel Roles for the MiTF/TFE Family of Transcription Factors in Organelle Biogenesis, Nutrient Sensing, and Energy Homeostasis. Cell. Mol. Life Sci. 2014;71:2483–2497. doi: 10.1007/s00018-014-1565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slade L., Pulinilkunnil T. The MiTF/TFE Family of Transcription Factors: Master Regulators of Organelle Signaling, Metabolism, and Stress Adaptation. Mol. Cancer Res. 2017;15:1637–1643. doi: 10.1158/1541-7786.MCR-17-0320. [DOI] [PubMed] [Google Scholar]
- 40.Palmieri M., Impey S., Kang H., di Ronza A., Pelz C., Sardiello M., Ballabio A. Characterization of the CLEAR Network Reveals an Integrated Control of Cellular Clearance Pathways. Hum. Mol. Genet. 2011;20:3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 41.Martina J.A., Diab H.I., Lishu L., Jeong-A L., Patange S., Raben N., Puertollano R. The Nutrient-Responsive Transcription Factor TFE3 Promotes Autophagy, Lysosomal Biogenesis, and Clearance of Cellular Debris. Sci. Signal. 2014;7:ra9. doi: 10.1126/scisignal.2004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. TFEB Links Autophagy to Lysosomal Biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Malta C.D., Donaudy F., Embrione V., Polishchuk R.S., et al. A Gene Network Regulating Lysosomal Biogenesis and Function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 44.Hosokawa N., Sasaki T., Iemura S.I., Natsume T., Hara T., Mizushima N. Atg101, a Novel Mammalian Autophagy Protein Interacting with Atg13. Autophagy. 2009;5:973–979. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- 45.Mercer C.A., Kaliappan A., Dennis P.B. A Novel, Human Atg13 Binding Protein, Atg101, Interacts with ULK1 and Is Essential for Macroautophagy. Autophagy. 2009;5:649–662. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- 46.Noda N.N., Mizushima N. Atg101: Not Just an Accessory Subunit in the Autophagy-Initiation Complex. Cell Struct. Funct. 2016;41:13–20. doi: 10.1247/csf.15013. [DOI] [PubMed] [Google Scholar]
- 47.Hara T., Mizushima N. Role of ULK-FIP200 Complex in Mammalian Autophagy: FIP200, a Counterpart of Yeast Atg17? Autophagy. 2009;5:85–87. doi: 10.4161/auto.5.1.7180. [DOI] [PubMed] [Google Scholar]
- 48.Hara T., Takamura A., Kishi C., Iemura S.I., Natsume T., Guan J.L., Mizushima N. FIP200, a ULK-Interacting Protein, Is Required for Autophagosome Formation in Mammalian Cells. J. Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang Y.Y., Neufeld T.P. An Atg1/Atg13 Complex with Multiple Roles in TOR-Mediated Autophagy Regulation. Mol. Biol. Cell. 2009;20:2004–2014. doi: 10.1091/mbc.e08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alers S., Loffler A.S., Wesselborg S., Stork B. Role of AMPK-MTOR-Ulk1/2 in the Regulation of Autophagy: Cross Talk, Shortcuts, and Feedbacks. Mol. Cell. Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., Sabatini D.M. MTORC1 Senses Lysosomal Amino Acids through an Inside-out Mechanism That Requires the Vacuolar H+-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sancak Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., Sabatini D.M. Ragulator-Rag Complex Targets MTORC1 to the Lysosomal Surface and Is Necessary for Its Activation by Amino Acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Korolchuk V.I., Saiki S., Lichtenberg M., Siddiqi F.H., Roberts E.A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F.M., et al. Lysosomal Positioning Coordinates Cellular Nutrient Responses. Nat. Cell Biol. 2011;13:453–462. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Di Bartolomeo S., Corazzari M., Nazio F., Oliverio S., Lisi G., Antonioli M., Pagliarini V., Matteoni S., Fuoco C., Giunta L., et al. The Dynamic Interaction of AMBRA1 with the Dynein Motor Complex Regulates Mammalian Autophagy. J. Cell Biol. 2010;191:155–168. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Backer J.M. The Intricate Regulation and Complex Functions of the Class III Phosphoinositide 3-Kinase Vps34. Biochem. J. 2016;473:2251–2271. doi: 10.1042/BCJ20160170. [DOI] [PubMed] [Google Scholar]
- 56.Yue Z., Zhong Y. From a Global View to Focused Examination: Understanding Cellular Function of Lipid Kinase VPS34-Beclin 1 Complex in Autophagy. J. Mol. Cell Biol. 2010;2:305–307. doi: 10.1093/jmcb/mjq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wei Y., Liu M., Li X., Liu J., Li H. Origin of the Autophagosome Membrane in Mammals. BioMed Res. Int. 2018 doi: 10.1155/2018/1012789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parzych K.R., Klionsky D.J. An Overview of Autophagy: Morphology, Mechanism, and Regulation. Antioxid. Redox Signal. 2014;20:460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nichenko A.S., Sorensen J.R., Southern W.M., Qualls A.E., Schifino A.G., McFaline-Figueroa J., Blum J.E., Tehrani K.F., Yin H., Mortensen L.J., et al. Lifelong Ulk1-Mediated Autophagy Deficiency in Muscle Induces Mitochondrial Dysfunction and Contractile Weakness. Int. J. Mol. Sci. 2021;22:1937. doi: 10.3390/ijms22041937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fuqua J.D., Mere C.P., Kronemberger A., Blomme J., Bae D., Turner K.D., Harris M.P., Scudese E., Edwards M., Ebert S.M., et al. ULK2 Is Essential for Degradation of Ubiquitinated Protein Aggregates and Homeostasis in Skeletal Muscle. FASEB J. 2019;33:11735–11745. doi: 10.1096/fj.201900766R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a Mammalian Homologue of Yeast Apg8p, Is Localized in Autophagosome Membranes after Processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khaminets A., Behl C., Dikic I. Ubiquitin-Dependent and Independent Signals In Selective Autophagy. Trends Cell Biol. 2016;26:6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 63.Hemelaar J., Lelyveld V.S., Kessler B.M., Ploegh H.L. A Single Protease, Apg4B, Is Specific for the Autophagy-Related Ubiquitin-like Proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J. Biol. Chem. 2003;278:51841–51850. doi: 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- 64.Weidberg H., Shvets E., Shpilka T., Shimron F., Shinder V., Elazar Z. LC3 and GATE-16/GABARAP Subfamilies Are Both Essential yet Act Differently in Autophagosome Biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pohl C., Dikic I. Cellular Quality Control by the Ubiquitin-Proteasome System and Autophagy. Science. 2019;366:818–822. doi: 10.1126/science.aax3769. [DOI] [PubMed] [Google Scholar]
- 66.Lippai M., Lőw P. The Role of the Selective Adaptor P62 and Ubiquitin-Like Proteins in Autophagy. BioMed Res. Int. 2014 doi: 10.1155/2014/832704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Birgisdottir Å.B., Lamark T., Johansen T. The LIR Motif—Crucial for Selective Autophagy. J. Cell Sci. 2013;126:3237–3247. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- 68.Johansen T., Lamark T. Selective Autophagy Mediated by Autophagic Adapter Proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Noda T., Fujita N., Yoshimori T. The Late Stages of Autophagy: How Does the End Begin? Cell Death Differ. 2009;16:984–990. doi: 10.1038/cdd.2009.54. [DOI] [PubMed] [Google Scholar]
- 70.Monastyrska I., Rieter E., Klionsky D.J., Reggiori F. Multiple Roles of the Cytoskeleton in Autophagy. Biol. Rev. 2009;84:431–448. doi: 10.1111/j.1469-185X.2009.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ju J.-S., Varadhachary A.S., Miller S.E., Weihl C.C. Quantitation of “Autophagic Flux” in Mature Skeletal Muscle. Autophagy. 2010;6:929–935. doi: 10.4161/auto.6.7.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Klionsky D.J., Abdel-Aziz A.K., Abdelfatah S., Abdellatif M., Abdoli A., Abel S., Abeliovich H., Abildgaard M.H., Abudu Y.P., Acevedo-Arozena A., et al. Guidelines for the Use and Interpretation of Assays for Monitoring Autophagy (4th Edition) Autophagy. 2021:1–382. doi: 10.1080/15548627.2020.1797280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang X., Hu S., Liu L. Phosphorylation and Acetylation Modifications of FOXO3a: Independently or Synergistically? (Review) Oncol. Lett. 2017;13:2867–2872. doi: 10.3892/ol.2017.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Palmieri M., Pal R., Nelvagal H.R., Lotfi P., Stinnett G.R., Seymour M.L., Chaudhury A., Bajaj L., Bondar V.V., Bremner L., et al. MTORC1-Independent TFEB Activation via Akt Inhibition Promotes Cellular Clearance in Neurodegenerative Storage Diseases. Nat. Commun. 2017;8:14338. doi: 10.1038/ncomms14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martina J.A., Chen Y., Gucek M., Puertollano R. MTORC1 Functions as a Transcriptional Regulator of Autophagy by Preventing Nuclear Transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu Y., Ren J., He X., Chen H., Wei T., Feng W. YWHA/14-3-3 Proteins Recognize Phosphorylated TFEB by a Noncanonical Mode for Controlling TFEB Cytoplasmic Localization. Autophagy. 2019;15:1017–1030. doi: 10.1080/15548627.2019.1569928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., et al. A Lysosome-to-Nucleus Signalling Mechanism Senses and Regulates the Lysosome via MTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., et al. Nutrient-Dependent MTORC1 Association with the ULK1–Atg13–FIP200 Complex Required for Autophagy. Mol. Biol. Cell. 2009;20:1981–1991. doi: 10.1091/mbc.e08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Greer E.L., Oskoui P.R., Banko M.R., Maniar J.M., Gygi M.P., Gygi S.P., Brunet A. The Energy Sensor AMP-Activated Protein Kinase Directly Regulates the Mammalian FOXO3 Transcription Factor. J. Biol. Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 80.Sanchez A.M., Csibi A., Raibon A., Cornille K., Gay S., Bernardi H., Candau R. AMPK Promotes Skeletal Muscle Autophagy through Activation of Forkhead FoxO3a and Interaction with Ulk1. J. Cell. Biochem. 2012;113:695–710. doi: 10.1002/jcb.23399. [DOI] [PubMed] [Google Scholar]
- 81.Roach P.J. AMPK -> ULK1 -> Autophagy. Mol. Cell. Biol. 2011;31:3082–3084. doi: 10.1128/MCB.05565-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim J., Kundu M., Viollet B., Guan K.L. AMPK and MTOR Regulate Autophagy through Direct Phosphorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Inoki K., Zhu T., Guan K.L. TSC2 Mediates Cellular Energy Response to Control Cell Growth and Survival. Cell. 2003;115:577–590. doi: 10.1016/S0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 84.Castets P., Lin S., Rion N., Di Fulvio S., Romanino K., Guridi M., Frank S., Tintignac L.A., Sinnreich M., Rüegg M.A. Sustained Activation of MTORC1 in Skeletal Muscle Inhibits Constitutive and Starvation-Induced Autophagy and Causes a Severe, Late-Onset Myopathy. Cell Metab. 2013;17:731–744. doi: 10.1016/j.cmet.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 85.Guo Y., Meng J., Tang Y., Wang T., Wei B., Feng R., Gong B., Wang H., Ji G., Lu Z. AMP-Activated Kinase A2 Deficiency Protects Mice from Denervation-Induced Skeletal Muscle Atrophy. Arch. Biochem. Biophys. 2016;600:56–60. doi: 10.1016/j.abb.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 86.Egawa T., Goto A., Ohno Y., Yokoyama S., Ikuta A., Suzuki M., Sugiura T., Ohira Y., Yoshioka T., Hayashi T., et al. Involvement of AMPK in Regulating Slow-Twitch Muscle Atrophy during Hindlimb Unloading in Mice. Am. J. Physiol. Endocrinol. Metab. 2015;309:E651–E662. doi: 10.1152/ajpendo.00165.2015. [DOI] [PubMed] [Google Scholar]
- 87.Bujak A.L., Crane J.D., Lally J.S., Ford R.J., Kang S.J., Rebalka I.A., Green A.E., Kemp B.E., Hawke T.J., Schertzer J.D., et al. AMPK Activation of Muscle Autophagy Prevents Fasting-Induced Hypoglycemia and Myopathy during Aging. Cell Metab. 2015;21:883–890. doi: 10.1016/j.cmet.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dargelos E., Brulé C., Stuelsatz P., Mouly V., Veschambre P., Cottin P., Poussard S. Up-Regulation of Calcium-Dependent Proteolysis in Human Myoblasts under Acute Oxidative Stress. Exp. Cell Res. 2010;316:115–125. doi: 10.1016/j.yexcr.2009.07.025. [DOI] [PubMed] [Google Scholar]
- 89.Kourie J.I. Interaction of Reactive Oxygen Species with Ion Transport Mechanisms. Am. J. Physiol. Cell Physiol. 1998;275 doi: 10.1152/ajpcell.1998.275.1.C1. [DOI] [PubMed] [Google Scholar]
- 90.Dobrowolny G., Aucello M., Rizzuto E., Beccafico S., Mammucari C., Bonconpagni S., Belia S., Wannenes F., Nicoletti C., Del Prete Z., et al. Skeletal Muscle Is a Primary Target of SOD1G93A-Mediated Toxicity. Cell Metab. 2008;8:425–436. doi: 10.1016/j.cmet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 91.Rahman M., Mofarrahi M., Kristof A.S., Nkengfac B., Harel S., Hussain S.N.A. Reactive Oxygen Species Regulation of Autophagy in Skeletal Muscles. Antioxid. Redox Signal. 2013;20:443–459. doi: 10.1089/ars.2013.5410. [DOI] [PubMed] [Google Scholar]
- 92.Sun F., Xu X., Wang X., Zhang B. Regulation of Autophagy by Ca2+ Tumour Biol. 2016;37:15467. doi: 10.1007/s13277-016-5353-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Irrcher I., Ljubicic V., Hood D.A. Interactions between ROS and AMP Kinase Activity in the Regulation of PGC-1α Transcription in Skeletal Muscle Cells. Am. J. Physiol. Cell Physiol. 2009;296:C116–C123. doi: 10.1152/ajpcell.00267.2007. [DOI] [PubMed] [Google Scholar]
- 94.Talbert E.E., Smuder A.J., Min K., Kwon O.S., Szeto H.H., Powers S.K. Immobilization-Induced Activation of Key Proteolytic Systems in Skeletal Muscles Is Prevented by a Mitochondria-Targeted Antioxidant. J. Appl. Physiol. 2013;115:529–538. doi: 10.1152/japplphysiol.00471.2013. [DOI] [PubMed] [Google Scholar]
- 95.Wang H., Wang N., Xu D., Ma Q., Chen Y., Xu S., Xia Q., Zhang Y., Prehn J.H.M., Wang G., et al. Oxidation of Multiple MiT/TFE Transcription Factors Links Oxidative Stress to Transcriptional Control of Autophagy and Lysosome Biogenesis. Autophagy. 2020;16:1683–1696. doi: 10.1080/15548627.2019.1704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang X., Cheng X., Yu L., Yang J., Calvo R., Patnaik S., Hu X., Gao Q., Yang M., Lawas M., et al. MCOLN1 Is a ROS Sensor in Lysosomes That Regulates Autophagy. Nat. Commun. 2016;7:12109. doi: 10.1038/ncomms12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Andersson D.C., Betzenhauser M.J., Reiken S., Meli A.C., Umanskaya A., Xie W., Shiomi T., Zalk R., Lacampagne A., Marks A.R. Ryanodine Receptor Oxidation Causes Intracellular Calcium Leak and Muscle Weakness in Aging. Cell Metab. 2011;14:196–207. doi: 10.1016/j.cmet.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scotto Rosato A., Montefusco S., Soldati C., Di Paola S., Capuozzo A., Monfregola J., Polishchuk E., Amabile A., Grimm C., Lombardo A., et al. TRPML1 Links Lysosomal Calcium to Autophagosome Biogenesis through the Activation of the CaMKKβ/VPS34 Pathway. Nat. Commun. 2019;10:1–16. doi: 10.1038/s41467-019-13572-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Medina D.L., Di Paola S., Peluso I., Armani A., De Stefani D., Venditti R., Montefusco S., Scotto-Rosato A., Prezioso C., Forrester A., et al. Lysosomal Calcium Signalling Regulates Autophagy through Calcineurin and TFEB. Nat. Cell Biol. 2015;17:288–299. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Martina J.A., Diab H.I., Brady O.A., Puertollano R. TFEB and TFE3 Are Novel Components of the Integrated Stress Response. EMBO J. 2016;35:479–495. doi: 10.15252/embj.201593428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mikkelsen U.R., Agergaard J., Couppé C., Grosset J.F., Karlsen A., Magnusson S.P., Schjerling P., Kjaer M., Mackey A.L. Skeletal Muscle Morphology and Regulatory Signalling in Endurance-Trained and Sedentary Individuals: The Influence of Ageing. Exp. Gerontol. 2017;93:54–67. doi: 10.1016/j.exger.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 102.Carter H.N., Kim Y., Erlich A.T., Zarrin-khat D., Hood D.A. Autophagy and Mitophagy Flux in Young and Aged Skeletal Muscle Following Chronic Contractile Activity. J. Physiol. 2018;596:3567–3584. doi: 10.1113/JP275998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fritzen A.M., Frøsig C., Jeppesen J., Jensen T.E., Lundsgaard A.-M., Serup A.K., Schjerling P., Proud C.G., Richter E.A., Kiens B. Role of AMPK in Regulation of LC3 Lipidation as a Marker of Autophagy in Skeletal Muscle. Cell. Signal. 2016;28:663–674. doi: 10.1016/j.cellsig.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 104.Wagatsuma A., Shiozuka M., Takayama Y., Hoshino T., Mabuchi K., Matsuda R. Effects of Ageing on Expression of the Muscle-Specific E3 Ubiquitin Ligases and Akt-Dependent Regulation of Foxo Transcription Factors in Skeletal Muscle. Mol. Cell. Biochem. 2016;412:59–72. doi: 10.1007/s11010-015-2608-7. [DOI] [PubMed] [Google Scholar]
- 105.Stefanetti R.J., Zacharewicz E., Gatta P.D., Garnham A., Russell A.P., Lamon S. Ageing Has No Effect on the Regulation of the Ubiquitin Proteasome-Related Genes and Proteins Following Resistance Exercise. Front. Physiol. 2014;5 doi: 10.3389/fphys.2014.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Léger B., Derave W., De Bock K., Hespel P., Russell A.P. Human Sarcopenia Reveals an Increase in SOCS-3 and Myostatin and a Reduced Efficiency of Akt Phosphorylation. Rejuvenation Res. 2008;11:163B–175B. doi: 10.1089/rej.2007.0588. [DOI] [PubMed] [Google Scholar]
- 107.White Z., Terrill J., White R.B., McMahon C., Sheard P., Grounds M.D., Shavlakadze T. Voluntary Resistance Wheel Exercise from Mid-Life Prevents Sarcopenia and Increases Markers of Mitochondrial Function and Autophagy in Muscles of Old Male and Female C57BL/6J Mice. Skelet. Muscle. 2016;6 doi: 10.1186/s13395-016-0117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu M., Katta A., Gadde M.K., Liu H., Kakarla S.K., Fannin J., Paturi S., Arvapalli R.K., Rice K.M., Wang Y., et al. Aging-Associated Dysfunction of Akt/Protein Kinase B: S-Nitrosylation and Acetaminophen Intervention. PLoS ONE. 2009;4:e6430. doi: 10.1371/journal.pone.0006430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Paturi S., Gutta A.K., Katta A., Kakarla S.K., Arvapalli R.K., Gadde M.K., Nalabotu S.K., Rice K.M., Wu M., Blough E. Effects of Aging and Gender on Muscle Mass and Regulation of Akt-MTOR-P70s6k Related Signaling in the F344BN Rat Model. Mech. Ageing Dev. 2010;131:202–209. doi: 10.1016/j.mad.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 110.Stefanetti R.J., Voisin S., Russell A., Lamon S. Recent Advances in Understanding the Role of FOXO3. F1000Research. 2018;7 doi: 10.12688/f1000research.15258.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Beharry A.W., Sandesara P.B., Roberts B.M., Ferreira L.F., Senf S.M., Judge A.R. HDAC1 Activates FoxO and Is Both Sufficient and Required for Skeletal Muscle Atrophy. J. Cell Sci. 2014;127:1441–1453. doi: 10.1242/jcs.136390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kim K.W., Cho H.-J., Khaliq S.A., Son K.H., Yoon M.-S. Comparative Analyses of MTOR/Akt and Muscle Atrophy-Related Signaling in Aged Respiratory and Gastrocnemius Muscles. Int. J. Mol. Sci. 2020;21:2862. doi: 10.3390/ijms21082862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Koltai E., Hart N., Taylor A.W., Goto S., Ngo J.K., Davies K.J.A., Radak Z. Age-Associated Declines in Mitochondrial Biogenesis and Protein Quality Control Factors Are Minimized by Exercise Training. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012;303:R127–R134. doi: 10.1152/ajpregu.00337.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.White Z., White R.B., McMahon C., Grounds M.D., Shavlakadze T. High MTORC1 Signaling Is Maintained, While Protein Degradation Pathways Are Perturbed in Old Murine Skeletal Muscles in the Fasted State. Int. J. Biochem. Cell Biol. 2016;78:10–21. doi: 10.1016/j.biocel.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 115.Sandri M., Barberi L., Bijlsma A.Y., Blaauw B., Dyar K.A., Milan G., Mammucari C., Meskers C.G.M., Pallafacchina G., Paoli A., et al. Signalling Pathways Regulating Muscle Mass in Ageing Skeletal Muscle. The Role of the IGF1-Akt-MTOR-FoxO Pathway. Biogerontology. 2013;14:303–323. doi: 10.1007/s10522-013-9432-9. [DOI] [PubMed] [Google Scholar]
- 116.Parkington J.D., LeBrasseur N.K., Siebert A.P., Fielding R.A. Contraction-Mediated MTOR, P70S6k, and ERK1/2 Phosphorylation in Aged Skeletal Muscle. J. Appl. Physiol. 2004;97:243–248. doi: 10.1152/japplphysiol.01383.2003. [DOI] [PubMed] [Google Scholar]
- 117.Segalés J., Perdiguero E., Serrano A.L., Sousa-Victor P., Ortet L., Jardí M., Budanov A.V., Garcia-Prat L., Sandri M., Thomson D.M., et al. Sestrin Prevents Atrophy of Disused and Aging Muscles by Integrating Anabolic and Catabolic Signals. Nat. Commun. 2020;11:1–13. doi: 10.1038/s41467-019-13832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fodor J., Al-Gaadi D., Czirják T., Oláh T., Dienes B., Csernoch L., Szentesi P. Improved Calcium Homeostasis and Force by Selenium Treatment and Training in Aged Mouse Skeletal Muscle. Sci. Rep. 2020;10:1–14. doi: 10.1038/s41598-020-58500-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ziaaldini M.M., Marzetti E., Picca A., Murlasits Z. Biochemical Pathways of Sarcopenia and Their Modulation by Physical Exercise: A Narrative Review. Front. Med. 2017;4 doi: 10.3389/fmed.2017.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Marzetti E., Calvani R., Cesari M., Buford T.W., Lorenzi M., Behnke B.J., Leeuwenburgh C. Mitochondrial Dysfunction and Sarcopenia of Aging: From Signaling Pathways to Clinical Trials. Int. J. Biochem. Cell Biol. 2013;45:2288–2301. doi: 10.1016/j.biocel.2013.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fraysse B., Desaphy J.F., Rolland J.F., Pierno S., Liantonio A., Giannuzzi V., Camerino C., Didonna M.P., Cocchi D., De Luca A., et al. Fiber Type-Related Changes in Rat Skeletal Muscle Calcium Homeostasis during Aging and Restoration by Growth Hormone. Neurobiol. Dis. 2006;21:372–380. doi: 10.1016/j.nbd.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 122.Damiano S., Muscariello E., La Rosa G., Di Maro M., Mondola P., Santillo M. Dual Role of Reactive Oxygen Species in Muscle Function: Can Antioxidant Dietary Supplements Counteract Age-Related Sarcopenia? Int. J. Mol. Sci. 2019;20:3815. doi: 10.3390/ijms20153815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Smuder A.J., Sollanek K.J., Nelson W.B., Min K., Talbert E.E., Kavazis A.N., Hudson M.B., Sandri M., Szeto H.H., Powers S.K. Crosstalk between Autophagy and Oxidative Stress Regulates Proteolysis in the Diaphragm during Mechanical Ventilation. Free Radic. Biol. Med. 2018;115:179–190. doi: 10.1016/j.freeradbiomed.2017.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huang Z., Fang Q., Ma W., Zhang Q., Qiu J., Gu X., Yang H., Sun H. Skeletal Muscle Atrophy Was Alleviated by Salidroside through Suppressing Oxidative Stress and Inflammation during Denervation. Front. Pharmacol. 2019;10:997. doi: 10.3389/fphar.2019.00997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Qiu J., Fang Q., Xu T., Wu C., Xu L., Wang L., Yang X., Yu S., Zhang Q., Ding F., et al. Mechanistic Role of Reactive Oxygen Species and Therapeutic Potential of Antioxidants in Denervation- or Fasting-Induced Skeletal Muscle Atrophy. Front. Physiol. 2018;9 doi: 10.3389/fphys.2018.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shen Y., Zhang Q., Huang Z., Zhu J., Qiu J., Ma W., Yang X., Ding F., Sun H. Isoquercitrin Delays Denervated Soleus Muscle Atrophy by Inhibiting Oxidative Stress and Inflammation. Front. Physiol. 2020;11 doi: 10.3389/fphys.2020.00988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wohlgemuth S.E., Seo A.Y., Marzetti E., Lees H.A., Leeuwenburgh C. Skeletal Muscle Autophagy and Apoptosis during Aging: Effects of Calorie Restriction and Life-Long Exercise. Exp. Gerontol. 2010;45:138–148. doi: 10.1016/j.exger.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sakuma K., Kinoshita M., Ito Y., Aizawa M., Aoi W., Yamaguchi A. P62/SQSTM1 but Not LC3 Is Accumulated in Sarcopenic Muscle of Mice. J. Cachexiasarcopenia Muscle. 2016;7:204–212. doi: 10.1002/jcsm.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sebastián D., Sorianello E., Segalés J., Irazoki A., Ruiz-Bonilla V., Sala D., Planet E., Berenguer-Llergo A., Muñoz J.P., Sánchez-Feutrie M., et al. Mfn2 Deficiency Links Age-related Sarcopenia and Impaired Autophagy to Activation of an Adaptive Mitophagy Pathway. EMBO J. 2016;35:1677–1693. doi: 10.15252/embj.201593084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yeo D., Kang C., Gomez-Cabrera M.C., Vina J., Ji L.L. Intensified Mitophagy in Skeletal Muscle with Aging Is Downregulated by PGC-1alpha Overexpression in Vivo. Free Radic. Biol. Med. 2019;130:361–368. doi: 10.1016/j.freeradbiomed.2018.10.456. [DOI] [PubMed] [Google Scholar]
- 131.O’Leary M.F., Vainshtein A., Iqbal S., Ostojic O., Hood D.A. Adaptive Plasticity of Autophagic Proteins to Denervation in Aging Skeletal Muscle. Am. J. Physiol. Cell Physiol. 2013;304:C422–C430. doi: 10.1152/ajpcell.00240.2012. [DOI] [PubMed] [Google Scholar]
- 132.Fry C.S., Drummond M.J., Glynn E.L., Dickinson J.M., Gundermann D.M., Timmerman K.L., Walker D.K., Volpi E., Rasmussen B.B. Skeletal Muscle Autophagy and Protein Breakdown Following Resistance Exercise Are Similar in Younger and Older Adults. J. Gerontol. A Biol. Sci. Med. Sci. 2013;68:599–607. doi: 10.1093/gerona/gls209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Distefano G., Standley R.A., Dubé J.J., Carnero E.A., Ritov V.B., Stefanovic-Racic M., Toledo F.G.S., Piva S.R., Goodpaster B.H., Coen P.M. Chronological Age Does Not Influence Ex-Vivo Mitochondrial Respiration and Quality Control in Skeletal Muscle. J. Gerontol. A Biol. Sci. Med. Sci. 2017;72:535–542. doi: 10.1093/gerona/glw102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fernando R., Castro J.P., Flore T., Deubel S., Grune T., Ott C. Age-Related Maintenance of the Autophagy-Lysosomal System Is Dependent on Skeletal Muscle Type. Oxid. Med. Cell Longev. 2020;2020 doi: 10.1155/2020/4908162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lira V.A., Okutsu M., Zhang M., Greene N.P., Laker R.C., Breen D.S., Hoehn K.L., Yan Z. Autophagy Is Required for Exercise Training-Induced Skeletal Muscle Adaptation and Improvement of Physical Performance. FASEB J. 2013;27:4184–4193. doi: 10.1096/fj.13-228486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Baehr L.M., West D.W.D., Marcotte G., Marshall A.G., De Sousa L.G., Baar K., Bodine S.C. Age-Related Deficits in Skeletal Muscle Recovery Following Disuse Are Associated with Neuromuscular Junction Instability and ER Stress, Not Impaired Protein Synthesis. Aging. 2016;8:127–146. doi: 10.18632/aging.100879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ljubicic V., Hood D.A. Diminished Contraction-Induced Intracellular Signaling towards Mitochondrial Biogenesis in Aged Skeletal Muscle. Aging Cell. 2009;8:394–404. doi: 10.1111/j.1474-9726.2009.00483.x. [DOI] [PubMed] [Google Scholar]
- 138.Hardman S.E., Hall D.E., Cabrera A.J., Hancock C.R., Thomson D.M. The Effects of Age and Muscle Contraction on AMPK Activity and Heterotrimer Composition. Exp. Gerontol. 2014;55:120–128. doi: 10.1016/j.exger.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Reznick R.M., Zong H., Li J., Morino K., Moore I.K., Yu H.J., Liu Z.X., Dong J., Mustard K.J., Hawley S.A., et al. Aging-Associated Reductions in AMP-Activated Protein Kinase Activity and Mitochondrial Biogenesis. Cell Metab. 2007;5:151–156. doi: 10.1016/j.cmet.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Funai K., Parkington J.D., Carambula S., Fielding R.A. Age-Associated Decrease in Contraction-Induced Activation of Downstream Targets of Akt/MTor Signaling in Skeletal Muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006;290:R1080–R1086. doi: 10.1152/ajpregu.00277.2005. [DOI] [PubMed] [Google Scholar]
- 141.Glancy B., Hartnell L.M., Malide D., Yu Z.-X., Combs C.A., Connelly P.S., Subramaniam S., Balaban R.S. Mitochondrial Reticulum for Cellular Energy Distribution in Muscle. Nature. 2015;523:617–620. doi: 10.1038/nature14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Glancy B., Hartnell L.M., Combs C.A., Fenmou A., Sun J., Murphy E., Subramaniam S., Balaban R.S. Power Grid Protection of the Muscle Mitochondrial Reticulum. Cell Rep. 2017;19:487–496. doi: 10.1016/j.celrep.2017.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Romanello V., Sandri M. The Connection between the Dynamic Remodeling of the Mitochondrial Network and the Regulation of Muscle Mass. Cell. Mol. Life Sci. 2020 doi: 10.1007/s00018-020-03662-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Iqbal S., Hood D.A. The Role of Mitochondrial Fusion and Fission in Skeletal Muscle Function and Dysfunction. Front. Biosci. 2015;20:157–172. doi: 10.2741/4303. [DOI] [PubMed] [Google Scholar]
- 145.Twig G., Elorza A., Molina A.J.A., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., et al. Fission and Selective Fusion Govern Mitochondrial Segregation and Elimination by Autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Romanello V., Guadagnin E., Gomes L., Roder I., Sandri C., Petersen Y., Milan G., Masiero E., Del Piccolo P., Foretz M., et al. Mitochondrial Fission and Remodelling Contributes to Muscle Atrophy. EMBO J. 2010;29:1774–1785. doi: 10.1038/emboj.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dulac M., Leduc-Gaudet J.-P., Reynaud O., Ayoub M.-B., Guérin A., Finkelchtein M., Hussain S.N., Gouspillou G. Drp1 Knockdown Induces Severe Muscle Atrophy and Remodelling, Mitochondrial Dysfunction, Autophagy Impairment and Denervation. J. Physiol. 2020;598:3691–3710. doi: 10.1113/JP279802. [DOI] [PubMed] [Google Scholar]
- 148.Favaro G., Romanello V., Varanita T., Andrea Desbats M., Morbidoni V., Tezze C., Albiero M., Canato M., Gherardi G., De Stefani D., et al. DRP1-Mediated Mitochondrial Shape Controls Calcium Homeostasis and Muscle Mass. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-10226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang Z., Sliter D.A., Bleck C.K.E., Ding S. Fis1 Deficiencies Differentially Affect Mitochondrial Quality in Skeletal Muscle. Mitochondrion. 2019;49:217–226. doi: 10.1016/j.mito.2019.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sebastián D., Hernández-Alvarez M.I., Segalés J., Sorianello E., Muñoz J.P., Sala D., Waget A., Liesa M., Paz J.C., Gopalacharyulu P., et al. Mitofusin 2 (Mfn2) Links Mitochondrial and Endoplasmic Reticulum Function with Insulin Signaling and Is Essential for Normal Glucose Homeostasis. Proc. Natl. Acad. Sci. USA. 2012;109:5523–5528. doi: 10.1073/pnas.1108220109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tezze C., Romanello V., Desbats M.A., Fadini G.P., Albiero M., Favaro G., Ciciliot S., Soriano M.E., Morbidoni V., Cerqua C., et al. Age-Associated Loss of OPA1 in Muscle Impacts Muscle Mass, Metabolic Homeostasis, Systemic Inflammation, and Epithelial Senescence. Cell Metab. 2017;25:1374–1389.e6. doi: 10.1016/j.cmet.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Joseph A.M., Adhihetty P.J., Buford T.W., Wohlgemuth S.E., Lees H.A., Nguyen L.M.D., Aranda J.M., Sandesara B.D., Pahor M., Manini T.M., et al. The Impact of Aging on Mitochondrial Function and Biogenesis Pathways in Skeletal Muscle of Sedentary High- and Low-Functioning Elderly Individuals. Aging Cell. 2012 doi: 10.1111/j.1474-9726.2012.00844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Iqbal S., Ostojic O., Singh K., Joseph A.-M., Hood D.A. Expression of Mitochondrial Fission and Fusion Regulatory Proteins in Skeletal Muscle during Chronic Use and Disuse. Muscle Nerve. 2013;48:963–970. doi: 10.1002/mus.23838. [DOI] [PubMed] [Google Scholar]
- 154.Konopka A.R., Suer M.K., Wolff C.A., Harber M.P. Markers of Human Skeletal Muscle Mitochondrial Biogenesis and Quality Control: Effects of Age and Aerobic Exercise Training. J. Gerontol. A Biol. Sci. Med. Sci. 2014;69:371–378. doi: 10.1093/gerona/glt107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Leduc-Gaudet J.-P., Picard M., Pelletier F.S.-J., Sgarioto N., Auger M.-J., Vallée J., Robitaille R., St-Pierre D.H., Gouspillou G. Mitochondrial Morphology Is Altered in Atrophied Skeletal Muscle of Aged Mice. Oncotarget. 2015;6:17923–17937. doi: 10.18632/oncotarget.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jin S.M., Lazarou M., Wang C., Kane L.A., Narendra D.P., Youle R.J. Mitochondrial Membrane Potential Regulates PINK1 Import and Proteolytic Destabilization by PARL. J. Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pickles S., Vigié P., Youle R.J. Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr. Biol. 2018;28:R170–R185. doi: 10.1016/j.cub.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yamano K., Youle R.J. PINK1 Is Degraded through the N-End Rule Pathway. Autophagy. 2013;9:1758–1769. doi: 10.4161/auto.24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Thomas R.E., Andrews L.A., Burman J.L., Lin W.-Y., Pallanck L.J. PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix. PLoS Genet. 2014;10:e1004279. doi: 10.1371/journal.pgen.1004279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jin S.M., Youle R.J. The Accumulation of Misfolded Proteins in the Mitochondrial Matrix Is Sensed by PINK1 to Induce PARK2/Parkin-Mediated Mitophagy of Polarized Mitochondria. Autophagy. 2013;9:1750–1757. doi: 10.4161/auto.26122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Narendra D.P., Jin S.M., Tanaka A., Suen D.-F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. PINK1 Is Selectively Stabilized on Impaired Mitochondria to Activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kondapalli C., Kazlauskaite A., Zhang N., Woodroof H.I., Campbell D.G., Gourlay R., Burchell L., Walden H., Macartney T.J., Deak M., et al. PINK1 Is Activated by Mitochondrial Membrane Potential Depolarization and Stimulates Parkin E3 Ligase Activity by Phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.-S., Saiki S., Kawajiri S., Sato F., et al. PINK1 Stabilized by Mitochondrial Depolarization Recruits Parkin to Damaged Mitochondria and Activates Latent Parkin for Mitophagy. J. Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Okatsu K., Oka T., Iguchi M., Imamura K., Kosako H., Tani N., Kimura M., Go E., Koyano F., Funayama M., et al. PINK1 Autophosphorylation upon Membrane Potential Dissipation Is Essential for Parkin Recruitment to Damaged Mitochondria. Nat. Commun. 2012;3:1–10. doi: 10.1038/ncomms2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shiba-Fukushima K., Imai Y., Yoshida S., Ishihama Y., Kanao T., Sato S., Hattori N. PINK1-Mediated Phosphorylation of the Parkin Ubiquitin-like Domain Primes Mitochondrial Translocation of Parkin and Regulates Mitophagy. Sci. Rep. 2012;2:1002. doi: 10.1038/srep01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kane L.A., Lazarou M., Fogel A.I., Li Y., Yamano K., Sarraf S.A., Banerjee S., Youle R.J. PINK1 Phosphorylates Ubiquitin to Activate Parkin E3 Ubiquitin Ligase Activity. J. Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kazlauskaite A., Kondapalli C., Gourlay R., Campbell D.G., Ritorto M.S., Hofmann K., Alessi D.R., Knebel A., Trost M., Muqit M.M.K. Parkin Is Activated by PINK1-Dependent Phosphorylation of Ubiquitin at Ser65. Biochem. J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., Kimura Y., Tsuchiya H., Yoshihara H., Hirokawa T., et al. Ubiquitin Is Phosphorylated by PINK1 to Activate Parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 169.Okatsu K., Koyano F., Kimura M., Kosako H., Saeki Y., Tanaka K., Matsuda N. Phosphorylated Ubiquitin Chain Is the Genuine Parkin Receptor. J. Cell Biol. 2015;209:111–128. doi: 10.1083/jcb.201410050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sarraf S.A., Raman M., Guarani-Pereira V., Sowa M.E., Huttlin E.L., Gygi S.P., Harper J.W. Landscape of the PARKIN-Dependent Ubiquitylome in Response to Mitochondrial Depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gegg M.E., Cooper J.M., Chau K.Y., Rojo M., Schapira A.H.V., Taanman J.W. Mitofusin 1 and Mitofusin 2 Are Ubiquitinated in a PINK1/Parkin-Dependent Manner upon Induction of Mitophagy. Hum. Mol. Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Geisler S., Holmström K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. PINK1/Parkin-Mediated Mitophagy Is Dependent on VDAC1 and P62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 173.Lazarou M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L., Sideris D.P., Fogel A.I., Youle R.J. The Ubiquitin Kinase PINK1 Recruits Autophagy Receptors to Induce Mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wong Y.C., Holzbaur E.L.F. Optineurin Is an Autophagy Receptor for Damaged Mitochondria in Parkin-Mediated Mitophagy That Is Disrupted by an ALS-Linked Mutation. Proc. Natl. Acad. Sci. USA. 2014;111:E4439–E4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tanaka A., Cleland M.M., Xu S., Narendra D.P., Suen D.-F., Karbowski M., Youle R.J. Proteasome and P97 Mediate Mitophagy and Degradation of Mitofusins Induced by Parkin. J. Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Greene J.C., Whitworth A.J., Kuo I., Andrews L.A., Feany M.B., Pallanck L.J. Mitochondrial Pathology and Apoptotic Muscle Degeneration in Drosophila Parkin Mutants. Proc. Natl. Acad. Sci. USA. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rana A., Rera M., Walker D.W. Parkin Overexpression during Aging Reduces Proteotoxicity, Alters Mitochondrial Dynamics, and Extends Lifespan. Proc. Natl. Acad. Sci. USA. 2013;110:8638–8643. doi: 10.1073/pnas.1216197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Drew B.G., Ribas V., Le J.A., Henstridge D.C., Phun J., Zhou Z., Soleymani T., Daraei P., Sitz D., Vergnes L., et al. HSP72 Is a Mitochondrial Stress Sensor Critical for Parkin Action, Oxidative Metabolism, and Insulin Sensitivity in Skeletal Muscle. Diabetes. 2014;63:1488–1505. doi: 10.2337/db13-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rosen K.M., Veereshwarayya V., Moussa C.E.-H., Fu Q., Goldberg M.S., Schlossmacher M.G., Shen J., Querfurth H.W. Parkin Protects against Mitochondrial Toxins and β-Amyloid Accumulation in Skeletal Muscle Cells. J. Biol. Chem. 2006;281:12809–12816. doi: 10.1074/jbc.M512649200. [DOI] [PubMed] [Google Scholar]
- 180.Chen C.C.W., Erlich A.T., Crilly M.J., Hood D.A. Parkin Is Required for Exercise-Induced Mitophagy in Muscle: Impact of Aging. Am. J. Physiol. Endocrinol. Metab. 2018;315:E404–E415. doi: 10.1152/ajpendo.00391.2017. [DOI] [PubMed] [Google Scholar]
- 181.Chen C.C.W., Erlich A.T., Hood D.A. Role of Parkin and Endurance Training on Mitochondrial Turnover in Skeletal Muscle. Skelet. Muscle. 2018 doi: 10.1186/s13395-018-0157-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gouspillou G., Godin R., Piquereau J., Picard M., Mofarrahi M., Mathew J., Purves-Smith F.M., Sgarioto N., Hepple R.T., Burelle Y., et al. Protective Role of Parkin in Skeletal Muscle Contractile and Mitochondrial Function. J. Physiol. 2018;596:2565–2579. doi: 10.1113/JP275604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Leduc-Gaudet J.-P., Mayaki D., Reynaud O., Broering F.E., Chaffer T.J., Hussain S.N.A., Gouspillou G. Parkin Overexpression Attenuates Sepsis-Induced Muscle Wasting. Cells. 2020;9:1454. doi: 10.3390/cells9061454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Leduc-Gaudet J., Reynaud O., Hussain S.N., Gouspillou G. Parkin Overexpression Protects from Ageing-related Loss of Muscle Mass and Strength. J. Physiol. 2019;597:1975–1991. doi: 10.1113/JP277157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hanna R.A., Quinsay M.N., Orogo A.M., Giang K., Rikka S., Gustafsson Å.B. Microtubule-Associated Protein 1 Light Chain 3 (LC3) Interacts with Bnip3 Protein to Selectively Remove Endoplasmic Reticulum and Mitochondria via Autophagy. J. Biol. Chem. 2012;287:19094–19104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhang H., Bosch-Marce M., Shimoda L.A., Tan Y.S., Baek J.H., Wesley J.B., Gonzalez F.J., Semenza G.L. Mitochondrial Autophagy Is an HIF-1-Dependent Adaptive Metabolic Response to Hypoxia. J. Biol. Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 187.Rikka S., Quinsay M.N., Thomas R.L., Kubli D.A., Zhang X., Murphy A.N., Gustafsson Å.B. Bnip3 Impairs Mitochondrial Bioenergetics and Stimulates Mitochondrial Turnover. Cell Death Differ. 2011;18:721–731. doi: 10.1038/cdd.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Novak I., Kirkin V., McEwan D.G., Zhang J., Wild P., Rozenknop A., Rogov V., Löhr F., Popovic D., Occhipinti A., et al. Nix Is a Selective Autophagy Receptor for Mitochondrial Clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Rogov V.V., Suzuki H., Marinković M., Lang V., Kato R., Kawasaki M., Buljubašić M., Šprung M., Rogova N., Wakatsuki S., et al. Phosphorylation of the Mitochondrial Autophagy Receptor Nix Enhances Its Interaction with LC3 Proteins. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-01258-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Schweers R.L., Zhang J., Randall M.S., Loyd M.R., Li W., Dorsey F.C., Kundu M., Opferman J.T., Cleveland J.L., Miller J.L., et al. NIX Is Required for Programmed Mitochondrial Clearance during Reticulocyte Maturation. Proc. Natl. Acad. Sci. USA. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Liu L., Feng D., Chen G., Chen M., Zheng Q., Song P., Ma Q., Zhu C., Wang R., Qi W., et al. Mitochondrial Outer-Membrane Protein FUNDC1 Mediates Hypoxia-Induced Mitophagy in Mammalian Cells. Nat. Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 192.Wu W., Tian W., Hu Z., Chen G., Huang L., Li W., Zhang X., Xue P., Zhou C., Liu L., et al. ULK1 Translocates to Mitochondria and Phosphorylates FUNDC1 to Regulate Mitophagy. EMBO Rep. 2014;15:566–575. doi: 10.1002/embr.201438501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Quinsay M.N., Thomas R.L., Lee Y., Gustafsson A.B. Bnip3-Mediated Mitochondrial Autophagy Is Independent of the Mitochondrial Permeability Transition Pore. Autophagy. 2010;6:855–862. doi: 10.4161/auto.6.7.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hamacher-Brady A., Brady N.R., Logue S.E., Sayen M.R., Jinno M., Kirshenbaum L.A., Gottlieb R.A., Gustafsson Å.B. Response to Myocardial Ischemia/Reperfusion Injury Involves Bnip3 and Autophagy. Cell Death Differ. 2007;14:146–157. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- 195.Chen G., Cizeau J., Vande Velde C., Park J.H., Bozek G., Bolton J., Shi L., Dubik D., Greenberg A. Nix and Nip3 Form a Subfamily of Pro-Apoptotic Mitochondrial Proteins. J. Biol. Chem. 1999;274:7–10. doi: 10.1074/jbc.274.1.7. [DOI] [PubMed] [Google Scholar]
- 196.Zhu Y., Massen S., Terenzio M., Lang V., Chen-Lindner S., Eils R., Novak I., Dikic I., Hamacher-Brady A., Brady N.R. Modulation of Serines 17 and 24 in the LC3-Interacting Region of Bnip3 Determines pro-Survival Mitophagy versus Apoptosis. J. Biol. Chem. 2013;288:1099–1113. doi: 10.1074/jbc.M112.399345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lee Y., Lee H.-Y., Hanna R.A., Gustafsson Å.B. Mitochondrial Autophagy by Bnip3 Involves Drp1-Mediated Mitochondrial Fission and Recruitment of Parkin in Cardiac Myocytes. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H1924–H1931. doi: 10.1152/ajpheart.00368.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Simpson C.L., Tokito M.K., Uppala R., Sarkar M.K., Gudjonsson J.E., Holzbaur E.L.F. NIX Initiates Mitochondrial Fragmentation via DRP1 to Drive Epidermal Differentiation. Cell Rep. 2021;34:108689. doi: 10.1016/j.celrep.2021.108689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ding W.-X., Ni H.-M., Li M., Liao Y., Chen X., Stolz D.B., Dorn G.W., Yin X.-M., Yin X.-M. Nix Is Critical to Two Distinct Phases of Mitophagy, Reactive Oxygen Species-Mediated Autophagy Induction and Parkin-Ubiquitin-P62-Mediated Mitochondrial Priming. J. Biol. Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Baechler B.L., Bloemberg D., Quadrilatero J. Mitophagy Regulates Mitochondrial Network Signaling, Oxidative Stress, and Apoptosis during Myoblast Differentiation. Autophagy. 2019;15:1606–1619. doi: 10.1080/15548627.2019.1591672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lampert M.A., Orogo A.M., Najor R.H., Hammerling B.C., Leon L.J., Wang B.J., Kim T., Sussman M.A., Gustafsson Å.B. BNIP3L/NIX and FUNDC1-Mediated Mitophagy Is Required for Mitochondrial Network Remodeling during Cardiac Progenitor Cell Differentiation. Autophagy. 2019;15:1182–1198. doi: 10.1080/15548627.2019.1580095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Fu T., Xu Z., Liu L., Guo Q., Wu H., Liang X., Zhou D., Xiao L., Liu L., Liu Y., et al. Mitophagy Directs Muscle-Adipose Crosstalk to Alleviate Dietary Obesity. Cell Rep. 2018;23:1357–1372. doi: 10.1016/j.celrep.2018.03.127. [DOI] [PubMed] [Google Scholar]
- 203.Gao J., Yu L., Wang Z., Wang R., Liu X. Induction of Mitophagy in C2C12 Cells by Electrical Pulse Stimulation Involves Increasing the Level of the Mitochondrial Receptor FUNDC1 through the AMPK-ULK1 Pathway. Am. J. Transl. Res. 2020;12:6879–6894. [PMC free article] [PubMed] [Google Scholar]
- 204.Zampieri S., Pietrangelo L., Loefler S., Fruhmann H., Vogelauer M., Burggraf S., Pond A., Grim-Stieger M., Cvecka J., Sedliak M., et al. Lifelong Physical Exercise Delays Age-Associated Skeletal Muscle Decline. J. Gerontol. A Biol. Sci. Med. Sci. 2015;70:163–173. doi: 10.1093/gerona/glu006. [DOI] [PubMed] [Google Scholar]
- 205.Ko F., Abadir P., Marx R., Westbrook R., Cooke C., Yang H., Walston J. Impaired Mitochondrial Degradation by Autophagy in the Skeletal Muscle of the Aged Female Interleukin 10 Null Mouse. Exp. Gerontol. 2016;73:23–27. doi: 10.1016/j.exger.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ogborn D.I., McKay B.R., Crane J.D., Safdar A., Akhtar M., Parise G., Tarnopolsky M.A. Effects of Age and Unaccustomed Resistance Exercise on Mitochondrial Transcript and Protein Abundance in Skeletal Muscle of Men. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015;308:R734–R741. doi: 10.1152/ajpregu.00005.2014. [DOI] [PubMed] [Google Scholar]
- 207.Vainshtein A., Desjardins E.M., Armani A., Sandri M., Hood D.A. PGC-1α Modulates Denervation-Induced Mitophagy in Skeletal Muscle. Skelet. Muscle. 2015;5:9. doi: 10.1186/s13395-015-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Tamura Y., Kitaoka Y., Matsunaga Y., Hoshino D., Hatta H. Daily Heat Stress Treatment Rescues Denervation-Activated Mitochondrial Clearance and Atrophy in Skeletal Muscle. J. Physiol. 2015;593:2707–2720. doi: 10.1113/JP270093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Yang X., Xue P., Chen H., Yuan M., Kang Y., Duscher D., Machens H.-G., Chen Z. Denervation Drives Skeletal Muscle Atrophy and Induces Mitochondrial Dysfunction, Mitophagy and Apoptosis via MiR-142a-5p/MFN1 Axis. Theranostics. 2020;10:1415–1432. doi: 10.7150/thno.40857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Graham Z.A., Harlow L., Bauman W.A., Cardozo C.P. Alterations in Mitochondrial Fission, Fusion, and Mitophagic Protein Expression in the Gastrocnemius of Mice after a Sciatic Nerve Transection. Muscle Nerve. 2018;58:592–599. doi: 10.1002/mus.26197. [DOI] [PubMed] [Google Scholar]
- 211.Furuya N., Ikeda S.-I., Sato S., Soma S., Ezaki J., Oliva Trejo J.A., Takeda-Ezaki M., Fujimura T., Arikawa-Hirasawa E., Tada N., et al. PARK2/Parkin-Mediated Mitochondrial Clearance Contributes to Proteasome Activation during Slow-Twitch Muscle Atrophy via NFE2L1 Nuclear Translocation. Autophagy. 2014;10:631–641. doi: 10.4161/auto.27785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Huang Z., Zhong L., Zhu J., Xu H., Ma W., Zhang L., Shen Y., Law B.Y.-K., Ding F., Gu X., et al. Inhibition of IL-6/JAK/STAT3 Pathway Rescues Denervation-Induced Skeletal Muscle Atrophy. Ann. Transl. Med. 2020;8 doi: 10.21037/atm-20-7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wan Q., Zhang L., Huang Z., Zhang H., Gu J., Xu H., Yang X., Shen Y., Law B.Y.-K., Zhu J., et al. Aspirin Alleviates Denervation-Induced Muscle Atrophy via Regulating the Sirt1/PGC-1α Axis and STAT3 Signaling. Ann. Transl. Med. 2020;8 doi: 10.21037/atm-20-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kang C., Yeo D., Ji L.L. Muscle Immobilization Activates Mitophagy and Disrupts Mitochondrial Dynamics in Mice. Acta Physiol. 2016;218:188–197. doi: 10.1111/apha.12690. [DOI] [PubMed] [Google Scholar]
- 215.Leermakers P.A., Kneppers A.E.M., Schols A.M.W.J., Kelders M.C.J.M., de Theije C.C., Verdijk L.B., van Loon L.J.C., Langen R.C.J., Gosker H.R. Skeletal Muscle Unloading Results in Increased Mitophagy and Decreased Mitochondrial Biogenesis Regulation. Muscle Nerve. 2019;60:769–778. doi: 10.1002/mus.26702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Deval C., Calonne J., Coudy-Gandilhon C., Vazeille E., Bechet D., Polge C., Taillandier D., Attaix D., Combaret L. Mitophagy and Mitochondria Biogenesis Are Differentially Induced in Rat Skeletal Muscles during Immobilization and/or Remobilization. Int. J. Mol. Sci. 2020;21:3691. doi: 10.3390/ijms21103691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ballabio A., Bonifacino J.S. Lysosomes as Dynamic Regulators of Cell and Organismal Homeostasis. Nat. Rev. Mol. Cell Biol. 2020;21:101–118. doi: 10.1038/s41580-019-0185-4. [DOI] [PubMed] [Google Scholar]
- 218.Lawrence R.E., Zoncu R. The Lysosome as a Cellular Centre for Signalling, Metabolism and Quality Control. Nat. Cell Biol. 2019;21:133–142. doi: 10.1038/s41556-018-0244-7. [DOI] [PubMed] [Google Scholar]
- 219.Ramachandran P.V., Savini M., Folick A.K., Hu K., Masand R., Graham B.H., Wang M.C. Lysosomal Signaling Promotes Longevity by Adjusting Mitochondrial Activity. Dev. Cell. 2019;48:685–696. doi: 10.1016/j.devcel.2018.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Todkar K., Ilamathi H.S., Germain M. Mitochondria and Lysosomes: Discovering Bonds. Front. Cell Dev. Biol. 2017;5 doi: 10.3389/fcell.2017.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Gatto F., Rossi B., Tarallo A., Polishchuk E., Polishchuk R., Carrella A., Nusco E., Alvino F.G., Iacobellis F., De Leonibus E., et al. AAV-Mediated Transcription Factor EB (TFEB) Gene Delivery Ameliorates Muscle Pathology and Function in the Murine Model of Pompe Disease. Sci. Rep. 2017;7:15089. doi: 10.1038/s41598-017-15352-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Spampanato C., Feeney E., Li L., Cardone M., Lim J.-A., Annunziata F., Zare H., Polishchuk R., Puertollano R., Parenti G., et al. Transcription Factor EB (TFEB) Is a New Therapeutic Target for Pompe Disease. EMBO Mol. Med. 2013;5:691–706. doi: 10.1002/emmm.201202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Manganelli F., Ruggiero L. Clinical Features of Pompe Disease. Acta Myol. 2013;32:82–84. [PMC free article] [PubMed] [Google Scholar]
- 224.Xu S., Galperin M., Melvin G., Horowits R., Raben N., Plotz P., Yu L. Impaired Organization and Function of Myofilaments in Single Muscle Fibers from a Mouse Model of Pompe Disease. J. Appl. Physiol. 2010;108:1383–1388. doi: 10.1152/japplphysiol.01253.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lim J.-A., Li L., Kakhlon O., Myerowitz R., Raben N. Defects in Calcium Homeostasis and Mitochondria Can Be Reversed in Pompe Disease. Autophagy. 2015;11:385–402. doi: 10.1080/15548627.2015.1009779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Feeney E.J., Austin S., Chien Y.-H., Mandel H., Schoser B., Prater S., Hwu W.-L., Ralston E., Kishnani P.S., Raben N. The Value of Muscle Biopsies in Pompe Disease: Identifying Lipofuscin Inclusions in Juvenile- and Adult-Onset Patients. Acta Neuropathol. Commun. 2014;2:2. doi: 10.1186/2051-5960-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Endo Y., Furuta A., Nishino I. Danon Disease: A Phenotypic Expression of LAMP-2 Deficiency. Acta Neuropathol. 2015;129:391–398. doi: 10.1007/s00401-015-1385-4. [DOI] [PubMed] [Google Scholar]
- 228.Stevens-Lapsley J.E., Kramer L.R., Balter J.E., Jirikowic J., Boucek D., Taylor M. Functional Performance and Muscle Strength Phenotypes in Men and Women with Danon Disease. Muscle Nerve. 2010;42:908–914. doi: 10.1002/mus.21811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Nascimbeni A.C., Fanin M., Angelini C., Sandri M. Autophagy Dysregulation in Danon Disease. Cell Death Dis. 2017;8:e2565. doi: 10.1038/cddis.2016.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Hashem S.I., Murphy A.N., Divakaruni A.S., Klos M.L., Nelson B.C., Gault E.C., Rowland T.J., Perry C.N., Gu Y., Dalton N.D., et al. Impaired Mitophagy Facilitates Mitochondrial Damage in Danon Disease. J. Mol. Cell. Cardiol. 2017;108:86–94. doi: 10.1016/j.yjmcc.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 231.Sugie K., Noguchi S., Kozuka Y., Arikawa-Hirasawa E., Tanaka M., Yan C., Saftig P., von Figura K., Hirano M., Ueno S., et al. Autophagic Vacuoles with Sarcolemmal Features Delineate Danon Disease and Related Myopathies. J. Neuropathol. Exp. Neurol. 2005;64:513–522. doi: 10.1093/jnen/64.6.513. [DOI] [PubMed] [Google Scholar]
- 232.Ma S., Zhang M., Zhang S., Wang J., Zhou X., Guo G., Wang L., Wang M., Peng Z., Guo C., et al. Characterisation of Lamp2-Deficient Rats for Potential New Animal Model of Danon Disease. Sci. Rep. 2018;8:6932. doi: 10.1038/s41598-018-24351-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Brunk U.T., Terman A. The Mitochondrial-Lysosomal Axis Theory of Aging. Eur. J. Biochem. 2002;269:1996–2002. doi: 10.1046/j.1432-1033.2002.02869.x. [DOI] [PubMed] [Google Scholar]
- 234.Terman A., Kurz T., Navratil M., Arriaga E.A., Brunk U.T. Mitochondrial Turnover and Aging of Long-Lived Postmitotic Cells: The Mitochondrial-Lysosomal Axis Theory of Aging. Antioxid. Redox Signal. 2010;12:503–535. doi: 10.1089/ars.2009.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hütter E., Skovbro M., Lener B., Prats C., Rabøl R., Dela F., Jansen-Dürr P. Oxidative Stress and Mitochondrial Impairment Can Be Separated from Lipofuscin Accumulation in Aged Human Skeletal Muscle. Aging Cell. 2007;6:245–256. doi: 10.1111/j.1474-9726.2007.00282.x. [DOI] [PubMed] [Google Scholar]
- 236.Kim Y.A., Kim Y.S., Oh S.L., Kim H.J., Song W. Autophagic Response to Exercise Training in Skeletal Muscle with Age. J. Physiol. Biochem. 2013;69:697–705. doi: 10.1007/s13105-013-0246-7. [DOI] [PubMed] [Google Scholar]
- 237.Dirks A., Leeuwenburgh C. Apoptosis in Skeletal Muscle with Aging. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002;282:R519–R527. doi: 10.1152/ajpregu.00458.2001. [DOI] [PubMed] [Google Scholar]
- 238.Cheema N., Herbst A., Mckenzie D., Aiken J.M. Apoptosis and Necrosis Mediate Skeletal Muscle Fiber Loss in Age-Induced Mitochondrial Enzymatic Abnormalities. Aging Cell. 2015;14:1085–1093. doi: 10.1111/acel.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Marzetti E., Leeuwenburgh C. Skeletal Muscle Apoptosis, Sarcopenia and Frailty at Old Age. Exp. Gerontol. 2006;41:1234–1238. doi: 10.1016/j.exger.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 240.Wang F., Gómez-Sintes R., Boya P. Lysosomal Membrane Permeabilization and Cell Death. Traffic. 2018;19:918–931. doi: 10.1111/tra.12613. [DOI] [PubMed] [Google Scholar]
- 241.Arotcarena M.-L., Bourdenx M., Dutheil N., Thiolat M.-L., Doudnikoff E., Dovero S., Ballabio A., Fernagut P.-O., Meissner W.G., Bezard E., et al. Transcription Factor EB Overexpression Prevents Neurodegeneration in Experimental Synucleinopathies. JCI Insight. 2019;4 doi: 10.1172/jci.insight.129719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Decressac M., Mattsson B., Weikop P., Lundblad M., Jakobsson J., Björklund A. TFEB-Mediated Autophagy Rescues Midbrain Dopamine Neurons from α-Synuclein Toxicity. Proc. Natl. Acad. Sci. USA. 2013;110:E1817–E1826. doi: 10.1073/pnas.1305623110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Xiao Q., Yan P., Ma X., Liu H., Perez R., Zhu A., Gonzales E., Tripoli D.L., Czerniewski L., Ballabio A., et al. Neuronal-Targeted TFEB Accelerates Lysosomal Degradation of APP, Reducing Aβ Generation and Amyloid Plaque Pathogenesis. J. Neurosci. 2015;35:12137–12151. doi: 10.1523/JNEUROSCI.0705-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Song J.-X., Malampati S., Zeng Y., Durairajan S.S.K., Yang C.-B., Tong B.C.-K., Iyaswamy A., Shang W.-B., Sreenivasmurthy S.G., Zhu Z., et al. A Small Molecule Transcription Factor EB Activator Ameliorates Beta-Amyloid Precursor Protein and Tau Pathology in Alzheimer’s Disease Models. Aging Cell. 2020;19:e13069. doi: 10.1111/acel.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]