Abstract
Simple Summary
Human bladder cancer, one of the most common cancers worldwide, is a molecularly heterogenous and complex disease. Identifying novel prognostic biomarkers and establishing new predictive signatures are important for personalized medicine and effective treatment of bladder cancer patients. Autophagy, a cell self-maintenance process that removes damaged organelles and misfolded proteins, displays both tumor promotion and suppression activities. The aim of our study is to investigate the function of autophagy-related genes in bladder cancer with the main focus on their contribution to prognostic outcome. By analyzing data obtained from The Cancer Genome Atlas (TCGA), we identified 32 autophagy-related genes that were highly associated with overall survival of bladder cancer patients. Further statistical assessment established an 11-autophagy-related-gene signature as an effective prognostic biomarker to predict the survival outcomes of bladder cancer patients.
Abstract
Background: Survival rates for highly invasive bladder cancer (BC) patients have been very low, with a 5-year survival rate of 6%. Accurate prediction of tumor progression and survival is important for diagnosis and therapeutic decisions for BC patients. Our study aims to develop an autophagy-related-gene (ARG) signature that helps to predict the survival of BC patients. Methods: RNA-seq data of 403 BC patients were retrieved from The Cancer Genome Atlas Urothelial Bladder Carcinoma (TCGA-BLCA) database. Univariate Cox regression analysis was performed to identify overall survival (OS)-related ARGs. The Lasso Cox regression model was applied to establish an ARG signature in the TCGA training cohort (N = 203). The performance of the 11-gene ARG signature was further evaluated in a training cohort and an independent validation cohort (N = 200) using Kaplan-Meier OS curve analysis, receiver operating characteristic (ROC) analysis, as well as univariate and multivariate Cox regression analysis. Results: Our study identified an 11-gene ARG signature that is significantly associated with OS, including APOL1, ATG4B, BAG1, CASP3, DRAM1, ITGA3, KLHL24, P4HB, PRKCD, ULK2, and WDR45. The ARGs-derived high-risk bladder cancer patients exhibited significantly poor OS in both training and validation cohorts. The prognostic model showed good predictive efficacy, with the area under the ROC curve (AUCs) for 1-year, 3-year, and 5-year overall survival of 0.702 (0.695), 0.744 (0.640), and 0.794 (0.658) in the training and validation cohorts, respectively. A prognostic nomogram, which included the ARGs-derived risk factor, age and stage for eventual clinical translation, was established. Conclusion: We identified a novel ARG signature for risk-stratification and robust prediction of overall survival for BC patients.
Keywords: bladder cancer, autophagy, gene signature, overall survival, The Cancer Genome Atlas
1. Introduction
Bladder cancer (BC) is one of the most common cancers worldwide, with an estimated 550,000 new cases (almost 425,000 in males and over 125,000 in females) in 2018 [1]. More than 60% of all bladder cancer cases and half of all the 165,000 bladder cancer deaths occur in the less developed regions of the world. Bladder cancer is a molecularly heterogeneous and complex disease [2,3]. Approximately 30% of cases of invasive bladder cancer are associated with occult distant metastasis at the time of diagnosis, leading to a disappointing 5-year survival rate in patients with muscle-invasive bladder cancer [1]. Environmental and occupational exposure to risk factors such as cigarette smoking, chemical carcinogens, arsenic in drinking water, and endemic chronic urinary infections with Schistosoma haematobium are the main causes of bladder cancer [4]. Among these, cigarette smoking is the top risk factor, accounting for 50–65% of male cases and 20–30% of female cases [5,6]. The prognosis of patients with invasive bladder cancer is still very poor. The most common histopathological prognostic variables are tumor stage and lymph node density [7]. Recently, the Cancer Genome Atlas (TCGA) classified bladder cancer into five molecular subtypes (luminal-papillary, luminal-infiltrated, luminal, neuronal, and basal-squamous), each of which has distinct differentiation patterns, oncogenic mechanisms, tumor microenvironments, and clinical and histological associations [8]. The luminal-papillary, luminal-infiltrated, and luminal subtypes highly express luminal markers, including GATA3, UPK1A, UPK2, KTT20, FGFR3, etc. The extracellular matrix (ECM) and smooth muscle genes are highly expressed in the luminal-infiltrated subtype. True to name, the neural subtype highly expresses neural differentiation genes. Basal squamous subtypes highly express squamous epithelium-like genes (GSDMC, TGM1, etc.), immune marker genes (L1CAM, CXCL11, etc.), and basal marker genes (CD44, KRT5, KRT6A, etc.). The molecular diversity of subtypes contributes to the different responses to conventional and targeted therapies [9]. Thus, biomarker identification is of importance for diagnosis and prognosis of bladder cancer in the genomic era [10].
Autophagy is an evolutionarily conserved “self-eating” process, in which aberrant intracellular proteins and damaged organelles are engulfed in double-membrane structures and subsequently delivered to lysosomes for degradation. Both natural and pathological processes, including nutrient deprivation, organelle damage, and chemotherapy or radiation therapy of cancer cells, are able to trigger autophagy [11,12,13]. In cancer, autophagy functions as a double-edged sword with both tumor-promoting and tumor-suppressing activities, depending on tumor type, stage, and genetic context [14]. In early tumor formation, autophagy acts as a tumor suppressor by maintaining genomic integrity, preventing cell proliferation and inflammation. However, once established, tumor cells utilize autophagy to survive [15,16].
Very few studies have reported the function of autophagy in human bladder cancer. Dickstein et al. reported that AZ7328, an AKT inhibitor, induced autophagy in human bladder cancer cell lines, which protected cancer cells from AZ7328-induced apoptosis [17]. In addition, maternally expressed gene 3 (MEG3) has been reported to be downregulated in bladder cancer tissues compared to normal controls, resulting in increased cell proliferation and autophagy activity [18]. In contrast to the tumor promotion activity shown by these two studies, autophagy has been recently reported to be able to inhibit growth and invasion of bladder cancer cells [19,20]. Downregulation of high-mobility group AT-hook1 (HMGA1) and miR-211 in bladder cancer cells induced cytotoxic autophagy and suppressed cell proliferation, migration, and invasion [19]. Treating bladder cancer cells with isorhapontogenin, a small stilbene derivative, induced autophagy and inhibited cell growth and invasion via the MAPK8-JUN-SESN2 axis [20]. However, the molecular pathways that mediate autophagy’s tumor promotion or tumor suppression activities in bladder cancers are not fully characterized. Gene expression signatures for survival stratification in bladder cancer patients have been proposed by various studies [21,22,23,24]. A recent study has reported a 7-autophagy-gene signature in bladder cancer patients [25]. Four out of seven genes were significantly downregulated in bladder cancer compared to the control group, and displayed high sensitivity and specificity in bladder cancer diagnosis [25]. However, a prognosis-related autophagy-related gene (ARG) signature has not been reported in bladder cancer. Thus, for this paper, we aimed to investigate the function of autophagy-related genes in bladder cancer and their contribution to prognostic outcomes using a data-based approach. By analyzing RNA-seq data of bladder cancer patients and their clinical follow-up information, obtained from TCGA, we identified the prognosis-related ARGs that were highly associated with overall survival. Then, we used multiple statistical analysis methods to identify the potential correlation of these genes and prognostic outcome. We eventually built a convenient nomogram to predict survival rate by combining the risk score of autophagy-related genes and major confounding factors.
2. Methods
2.1. Data Acquisition
A total of 213 autophagy-related genes were obtained from the HADB database (Human Autophagy Database, http://autophagy.lu/clustering/index.html, accessed on 1 May 2021), which provides a comprehensive and up-to-date list of autophagy-related genes and proteins (Table S1). The RNA sequencing (RNA-seq) data and corresponding clinical follow-up information were extracted from the Bladder Carcinoma (TCGA-BLCA) database (https://cancergenome.nih.gov, accessed on 1 May 2021) database, a project that was initiated in 2005 by the National Cancer Institute to identify genetic mutations implicated in cancer using genome sequencing and bioinformatics. Out of 414 bladder cancer patients with the clinic follow-up data (ranging from 0 to 5050 days) collected in the TCGA Urothelial Bladder Carcinoma (TCGA-BLCA) database, our study included 403 BC patients with a minimum follow-up duration of more than one month. Univariate Cox regression analyses were performed to identify ARGs associated with overall survival of patients for subsequent model construction.
2.2. Functional Analysis
To investigate a comprehensive set of functional annotation of prognosis-related ARGs, gene ontology (GO) term enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed using the “Clusterprofiler” package in R.
2.3. Construction of an ARGs-Related Prognostic Model
The prognostic value of autophagy-related genes of all samples was computed by a univariate Cox regression analysis. Then, samples were randomly divided into two cohorts—the training cohort (N = 203) and the validation cohort (N = 200) (Table S2). The least absolute shrinkage and selection operator (LASSO) method was used to identify gene signatures and obtain their respective coefficients value. After incorporating expression values for each particular gene, a risk score formula for each patient was constructed and weighted according to its estimated regression coefficients in a multivariate Cox regression analysis. The prognostic risk score for predicting overall survival was calculated as follows:
According to the risk scoring formula, the median risk score in the training cohort and the validation cohort was used as the cutoff value. Bladder cancer patients in each cohort were divided into low-risk and high-risk groups. The overall survival curves of the two groups were generated through the “survival” package in R. 1-, 3-, and 5-year receiver operating characteristic (ROC) curve analyses were performed to estimate the predictive value of the candidate genes. Sensitivity and specificity of the risk model of CESC were calculated by the AUC of the ROC curve with “survival ROC” R package. A p value < 0.05 was considered statistically significant in the prognostic signature analysis.
2.4. Statistical Analysis
Overall survival differences in the low- and high-risk groups in each cohort were evaluated by the Kaplan–Meier curve and compared by log-rank statistical methods. The median values were used as cut-off thresholds to plot the KM curves, and the statistical significance was evaluated by the log rank test. Both multivariate Cox regression analysis and stratification analysis were implemented to examine the role of risk scores in predicting patient outcomes. The ‘glmnet’ R package (version 2.0–16) was employed to perform the least absolute shrinkage and selection. The univariate and multivariate analysis was enforced using the Cox proportional hazard regression model. A nomogram for predicting the OS was built using the R library “rms” package. The survival probabilities were predicted by receiver operating characteristic (ROC) curve analysis for both 3- and 5-year survival. The validation of the nomogram-based prediction model was accessed via bootstrapped calibration curves and quantified as a C-index. The “B” and “m” parameters were set as 200 and 30, respectively. All statistical analyses were performed using the R language (version 3.6). A p-value of < 0.05 was identified as statistically significant.
3. Result
3.1. Identification of Prognostic Autophagy-Related Genes
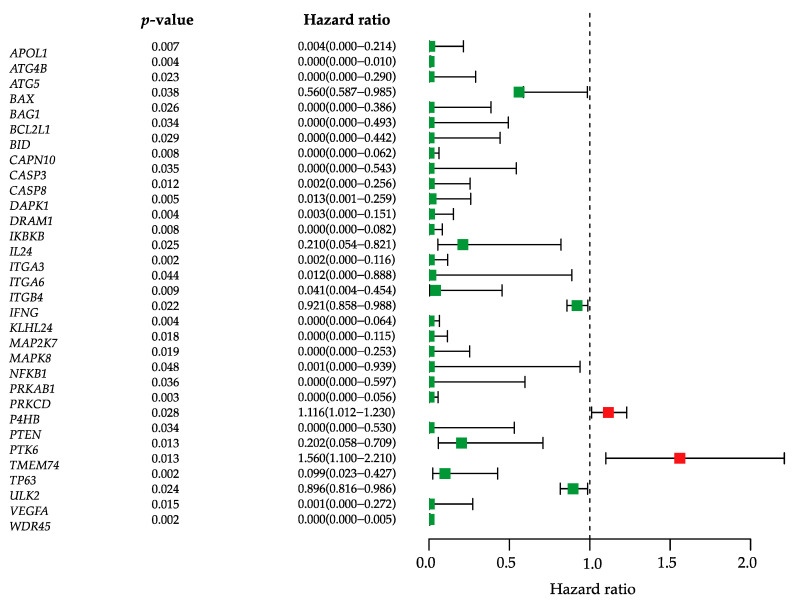
We obtained the 213 autophagy-related genes (ARGs) (Table S1) from the Human Autophagy Database (HADB, http://autophagy.lu/clustering/index.html, accessed on 1 May 2021) and merged them with RNA sequencing (RNA-seq) data of 403 BLCA samples on the Illumina HiSeq RNA-Seq platform from The Cancer Genome Atlas Urothelial Bladder Carcinoma (TCGA-BLCA) database. The demographic and clinical features of patients were listed in Table 1. Univariate Cox regression analysis was performed to identify prognostic ARG biomarkers, and yielded 32 genes that were significantly associated with the overall survival in bladder cancer (Figure 1). Most of these genes have hazard ratio below 1, which means that they are protective factors. The two genes with hazard ratio higher than 1 were P4HB and TMEM74. While P4HB has been recently identified as a novel prognostic biomarker associated with overall survival of bladder cancer patients [26], TMEM74 has been shown to promote tumor cell survival by inducing autophagy. High expression of TMEM74 significantly shortens the surviving periods of patients in several types of cancer [27].
Table 1.
Characteristics of patients in TCGA database.
| Variables | TCGA (N = 403) |
|---|---|
| Status | |
| Alive | 249 |
| Dead | 154 |
| Age | 68 ± 10.57 |
| Gender | |
| Female | 102 (26.2) |
| Male | 299 (73.8) |
| Race | |
| White | 332 (82.5) |
| Asian | 43 (10.6) |
| Black or African | 28 (6.9) |
| AJCC -T | |
| T0/Ta | 1 (0.2) |
| T1 | 3 (0.8) |
| T2 | 117 (28.9) |
| T3 | 193 (47.7) |
| T4 | 58 (14.3) |
| Unknown | 31 (8.1) |
| AJCC -N | |
| N0 | 235 (58.0) |
| N1 | 46 (11.4) |
| N2 | 75 (18.5) |
| N3 | 7 (1.7) |
| Unknown | 40 (10.4) |
| AJCC -M | |
| M0 | 195 (48.1) |
| M1 | 11 (2.7) |
| Mx | 197 (49.2) |
| Pathologic_stage | |
| I&II | 130 (32.1) |
| III&IV | 271 (67.4) |
| Unknown | 2 (0.5) |
| Tumor_grade | |
| G1/G2 | 20 (5.0) |
| G3/G4 | 380 (94.3) |
| Unknown | 3 (0.7) |
Figure 1.
Identification of prognostic autophagy-related genes. Univariate Cox regression analysis obtained 32 ARGs. The hazard ratio forest plot shows the prognostic value of each gene. Green squares represent genes with hazard ratio <1, while red squares represent genes with hazard ratio >1. Horizontal lines are 95% CIs.
3.2. Identification of Involved Signaling Pathways and Functional Annotation
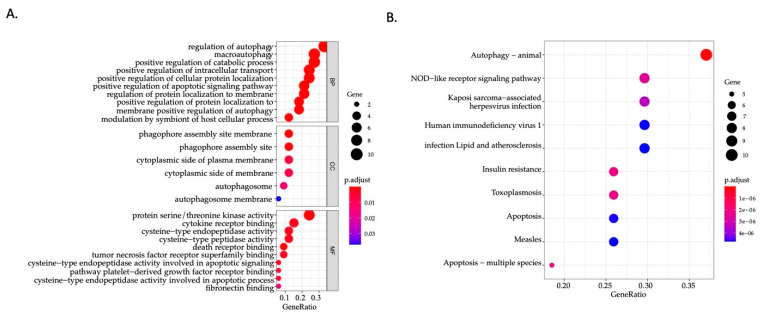
To investigate potential signaling pathways and potential function related to the 32 ARGs in bladder cancer (Table 2), we explored their biological characteristics and pathways using Gene Ontology (GO) enrichment analysis as well as the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. GO enrichment analysis of 32 prognostic ARGs provides a biological understanding of these genes. Biological function analysis (Figure 2A) showed that these genes were significantly associated with regulation of autophagy, macroautophagy, positive regulation of catabolic process, and positive regulation of apoptotic signaling pathway. According to cellular component enrichment, these genes were mainly located in the cytoplasmic side of plasma membrane, the phagophore assembly site membrane, and the autophagosome. Molecular function analysis suggested that the major functions of these genes were protein serine/threonine kinase activity, cytokine receptor binding, cysteine-type endopeptidase activity, cysteine-type peptidase activity, death receptor binding, and tumor necrosis factor receptor superfamily binding. KEGG analysis (Figure 2B) showed that pathways of prognostic autophagy-related genes were mostly enriched in autophagy, followed by the NOD-like receptor signaling pathway, the Kaposi sarcoma-associated herpesvirus infection, and the human immunodeficiency virus 1 infection.
Table 2.
Prognostic autophagy-related genes.
| Id | HR | HR.95 L | HR.95H | p-Value |
|---|---|---|---|---|
| APOL1 | 3.68 × 10−3 | 6.33 × 10−5 | 2.14 × 10−1 | 6.87 × 10−3 |
| ATG4B | 5.23 × 10−7 | 2.63 × 10−11 | 1.04 × 10−2 | 4.18 × 10−3 |
| ATG5 | 1.06 × 10−4 | 3.86 × 10−8 | 2.90 × 10−1 | 2.34 × 10−2 |
| BAX | 5.60 × 10−1 | 5.87 × 10−1 | 9.85 × 10−1 | 3.79 × 10−2 |
| BAG1 | 3.33 × 10−4 | 2.86 × 10−7 | 3.86 × 10−1 | 2.61 × 10−2 |
| BCL2L1 | 1.17 × 10−4 | 2.79 × 10−8 | 4.93 × 10−1 | 3.35 × 10−2 |
| BID | 3.72 × 10−4 | 3.14 × 10−7 | 4.42 × 10−1 | 2.88 × 10−2 |
| CAPN10 | 2.97 × 10−5 | 1.42 × 10−8 | 6.20 × 10−2 | 7.52 × 10−3 |
| CASP3 | 1.89 × 10−4 | 6.55 × 10−8 | 5.43 × 10−1 | 3.48 × 10−2 |
| CASP8 | 1.99 × 10−3 | 1.55 × 10−5 | 2.56 × 10−1 | 1.21 × 10−2 |
| DAPK1 | 1.27 × 10−2 | 6.27 × 10−4 | 2.59 × 10−1 | 4.52 × 10−3 |
| DRAM1 | 2.98 × 10−3 | 5.88 × 10−5 | 1.51 × 10−1 | 3.70 × 10−3 |
| IKBKB | 6.90 × 10−5 | 5.82 × 10−8 | 8.18 × 10−2 | 7.97 × 10−3 |
| IL24 | 2.10 × 10−1 | 5.38 × 10−2 | 8.21 × 10−1 | 2.48 × 10−2 |
| ITGA3 | 2.35 × 10−3 | 4.75 × 10−5 | 1.16 × 10−1 | 2.34 × 10−3 |
| ITGA6 | 1.16 × 10−2 | 1.51 × 10−4 | 8.88 × 10−1 | 4.40 × 10−2 |
| ITGB4 | 4.12 × 10−2 | 3.74 × 10−3 | 4.54 × 10−1 | 9.19 × 10−3 |
| IFNG | 9.21 × 10−1 | 8.58 × 10−1 | 9.88 × 10−1 | 2.22 × 10−2 |
| KLHL24 | 1.45 × 10−4 | 3.29 × 10−7 | 6.36 × 10−2 | 4.41 × 10−3 |
| MAP2K7 | 2.72 × 10−6 | 6.43 × 10−11 | 1.15 × 10−1 | 1.84 × 10−2 |
| MAPK8 | 1.91 × 10−4 | 1.44 × 10−7 | 2.53 × 10−1 | 1.95 × 10−2 |
| NFKB1 | 8.46 × 10−4 | 7.61 × 10−7 | 9.39 × 10−1 | 4.80 × 10−2 |
| PRKAB1 | 3.01 × 10−4 | 1.52 × 10−7 | 5.97 × 10−1 | 3.63 × 10−2 |
| PRKCD | 1.65 × 10−4 | 4.86 × 10−7 | 5.63 × 10−2 | 3.42 × 10−3 |
| P4HB | 1.12 × 10−1 | 1.01 × 10−1 | 1.23 × 10−1 | 2.80 × 10−2 |
| PTEN | 2.27 × 10−4 | 9.75 × 10−8 | 5.30 × 10−1 | 3.40 × 10−2 |
| PTK6 | 2.02 × 10−1 | 5.76 × 10−2 | 7.09 × 10−1 | 1.26 × 10−2 |
| TMEM74 | 1.56 × 10−1 | 1.10 × 10−1 | 2.21 × 10−1 | 1.32 × 10−2 |
| TP63 | 9.94 × 10−2 | 2.31 × 10−2 | 4.27 × 10−1 | 1.91 × 10−3 |
| ULK2 | 8.96 × 10−1 | 8.16 × 10−1 | 9.86 × 10−1 | 2.39 × 10−2 |
| VEGFA | 1.15 × 10−3 | 4.88 × 10−6 | 2.72 × 10−1 | 1.52 × 10−2 |
| WDR45 | 6.04 × 10−7 | 7.23 × 10−11 | 5.04 × 10−3 | 1.88 × 10−3 |
p < 0.05 was considered statistically significant. HR, hazard ratio.
Figure 2.
Functional annotation related to the 32 prognostic ARGs in bladder cancer. (A) GO analysis of 32 prognostic ARGs. (B) KEGG pathway enrichment of 32 prognostic ARGs. The size of the circle represents the number of genes. The color of the circle represents p-value. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; ARG, autophagy-related genes.
3.3. Identification of Autophagy-Related Gene Signatures
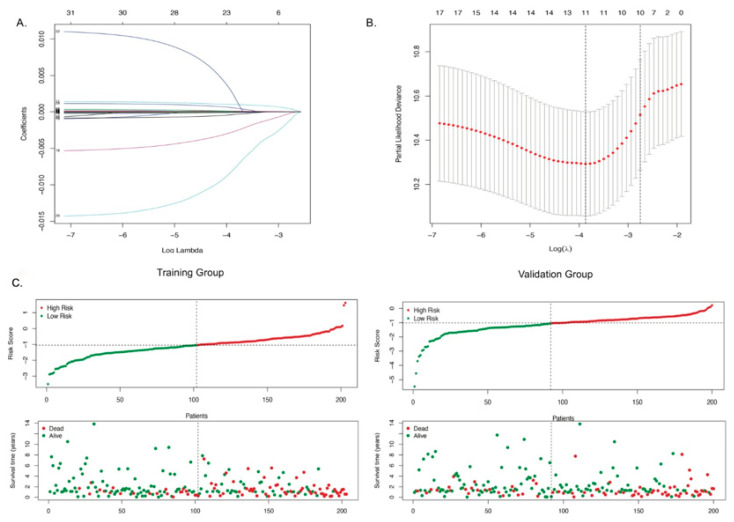
It has been reported that autophagy plays a vital role in bladder cancer progression [28,29]. To test whether autophagy-related genes can be used as prognostic biomarkers, we applied the Least Absolute Shrinkage and Selection Operator (LASSO) Cox regression model to the 32 genes extracted from RNA-seq data of bladder cancer patients. We randomly divided the 403 patients into two cohorts, a training cohort (N = 203) and a validation cohort (N = 200) (Table S2). A LASSO regression model was then used to select key prognosis-associated genes. In the LASSO-penalized Cox regression, the corresponding coefficients of certain genes were reduced to zero along with log λ (a tuning parameter) changed, indicating that they were shrinking parameters and their effects on the model could be omitted (Figure 3A). Following cross-validation, 11 genes achieved the minimum partial likelihood deviance (Figure 3B). Moreover, at this point, log λ was approximately -3.9, and the 11 genes displayed non-zero effects. These genes are APOL1, ATG4B, BAG1, CASP3, DRAM1, ITGA3, KLHL24, P4HB, PRKCD, ULK2, and WDR45. Using the multivariate Cox regression analysis, we established an autophagy-clinical prognostic index (API) based on the 11-gene prognostic model. Then we calculated the risk score for each individual patient using API and the mRNA expression of 11 genes (Table S2). Patients with a risk score higher than the median were classified into the high-risk group and those with a risk score lower than the median were classified into the low-risk group. Figure 3C showed the distribution of risk scores in patients with bladder cancer and the correlation between survival time and risk score in training cohorts. Patients with higher score were correlated with higher number of dead cases, which indicated that high-risk patients generally had poor overall survival. Similar trending was identified in validation cohorts (Figure 3C). Further analysis revealed that a significant correlation exists between the mRNA expression of 8 genes (APOL1, ITGA3, WDR45, CASP3, PRKCD, ATG4B, ULK2, and P4HB) and overall survival of bladder cancer patients (Figure S1). In addition, 10 out of 11 genes (except CASP3) exhibited a significant difference in mRNA expression levels between high- and low-risk groups (Figure S2).
Figure 3.
Expression profile and prognostic value of ARGs. (A) LASSO coefficient spectrum of 11 genes in BLCA. The coefficient distribution map was generated for a logarithmic (λ) sequence. (B) Selection of the best parameters for BLCA in the LASSO model (λ). (C) Distribution of prognostic index (upper panel) and survival status of patients (lower panel) in different groups.
3.4. The 11-ARG Signature as an Independent Risk Factor for Bladder Cancer
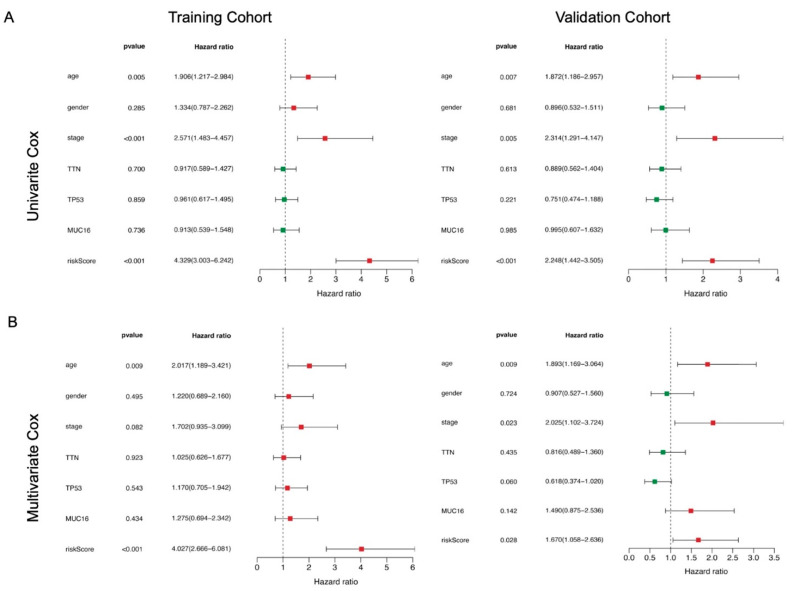
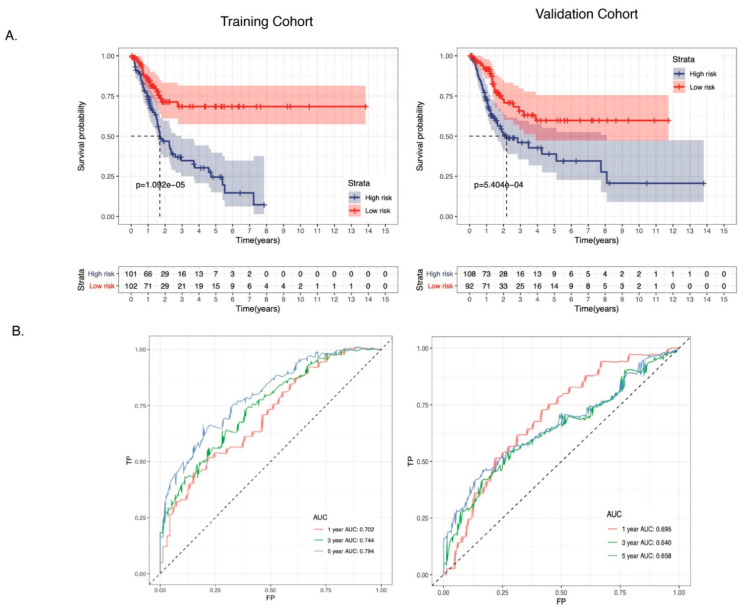
We performed univariate and multivariate Cox regression analyses on this 11-gene panel by including various clinical variables (e.g., age, gender, stage) of bladder cancer patients, as well as the mutation status of TNN, TP53, and MUC16, which are the top 3 mutated genes in bladder cancer (Figure S3). Univariate Cox regression analysis showed that API was significantly associated with patient prognosis in both training cohorts (p-value <0.001) and validation cohorts (p-value < 0.001) (Table 3, Figure 4A). Clinicopathological features including age, gender, stage, as well as mutation status of TTN, TP53, and MUC16 were further adjusted by multivariate Cox regression analysis. API remained an independent prognostic indicator for bladder cancer in training and validation cohorts with p-value < 0.01 and <0.05, respectively (Figure 4B). Kaplan–Meier overall survival curve analysis was plotted to determine the performance of the API in predicting clinical outcomes in bladder cancer patients. As shown in Figure 5A, the survival rate of patients in the high-risk group was significantly lower than that in the low-risk group in both training (p-value < 0.001) and validation (p-value < 0.001) cohorts (Figure 5A). Then, survival-dependent receiver operating characteristic (ROC) curves were performed to validate the accuracy of ARGs-based prognostic indicators. For the training group, the areas under the ROC curve (AUC) for 1-year, 3-year, and 5-year survival rates were 0.702, 0.744, and 0.794, respectively (Figure 5B). For the validation group, the AUC for 1-year, 3-year, and 5-year survival rates were 0.695, 0.640, and 0.658, respectively (Figure 5B). These results indicated that the ARGs-based prognostic indicators have a robust potential in survival prediction for bladder cancer patients.
Table 3.
Univariate and multivariate Cox regression analysis of clinical pathologic features for OS.
| Variables | Univariate Cox Regression | Multivariate Cox Regression | ||||||
|---|---|---|---|---|---|---|---|---|
| Training Group | Validation Group | Training Group | Validation Group | |||||
| HR | p-Value | HR | p-Value | HR | p-Value | HR | p-Value | |
| Age | 1.90599 | 0.00480 | 1.87244 | 0.00712 | 2.01716 | 0.00923 | 1.89282 | 0.00942 |
| Gender | 1.33404 | 0.28459 | 0.89624 | 0.68090 | 1.21995 | 0.49531 | 0.90684 | 0.72394 |
| Stage | 2.57129 | 0.00077 | 2.31364 | 0.00483 | 1.70227 | 0.08188 | 2.02544 | 0.02310 |
| TTN | 0.91655 | 0.69983 | 0.88862 | 0.61304 | 1.02461 | 0.92293 | 0.81568 | 0.43476 |
| TP53 | 0.96076 | 0.85924 | 0.75083 | 0.22132 | 1.17030 | 0.54285 | 0.61782 | 0.04981 |
| MUC16 | 0.91318 | 0.73605 | 0.99522 | 0.98484 | 1.27489 | 0.43391 | 1.48957 | 0.14201 |
| Risk Score | 4.32931 | 0.00000 | 2.24823 | 0.00035 | 4.02659 | 0.00000 | 1.67008 | 0.02760 |
p < 0.05 was considered statistically significant. HR, hazard ratio. Age (>65 vs. 65<). Gender (female vs. male). Stage (stage I&II vs. stage III&IV). TNN, TP53, MUC16 (mutant vs. wildtype).
Figure 4.
Prognostic indicators based on ARGs show good predictive performance and development of a prognostic index based on ARGs. A forest plot of (A) univariate and (B) multivariate Cox regression analysis in the training group and the validation group. Green squares represent genes with hazard ratio <1, while red squares represent genes with hazard ratio >1. Horizontal lines are 95% CIs.
Figure 5.
Evaluation of the 11-gene ARG signature in TCGA training and validation cohorts. (A) The Kaplan–Meier survival curve analysis revealed a shorter overall survival in the high-risk group. The blue line represents the high-risk group. The red line represents the low-risk group. (B) Survival-dependent ROC curves validated the prognostic significance of ARGs-based prognostic indicators. The red line represents the 1-year AUC; the green line represents the 3-year AUC; and the blue line represents the 5-year AUC. ROC, receiver operating characteristic; AUC, the area under the ROC curve.
3.5. Establishment of a Nomogram for Predicting Survival in Bladder Cancer Patients
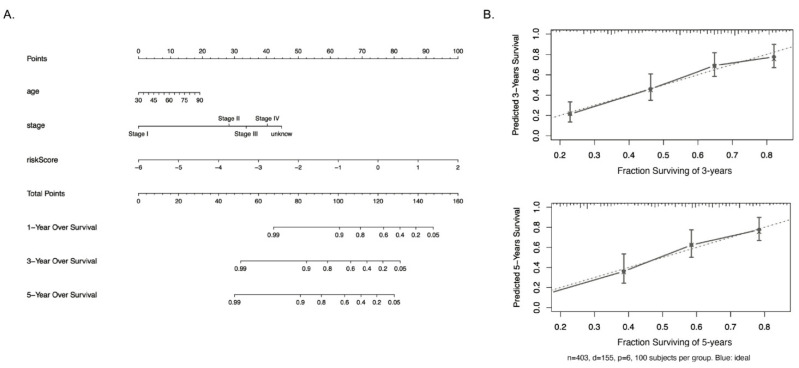
In order to further improve the prediction accuracy and practicability of API, we established a risk nomogram, which combined our 11-ARGs gene signature and other univariate significant features, for predicting the survival probability in bladder cancer patients. As shown in Figure 6A, a higher total score calculated by the sum of different factors in the nomogram was indicated with worse 1-year, 3-year, and 5-year overall survival rates. For example, a 60-year-old stage II BLCA patient with API of -2 would yield a total of 87.5 (10 points for age, 27.5 points for stage II, and 50 for -2 API score), with predicted 1-year, 3-year, and 5-year overall survival rates around 0.92, 0.80, and 0.73. Figure 6B showed calibration plots for the prediction of 3- and 5-year OS rates, which demonstrated a robust similarity between observed outcomes and predicted survival probabilities. These results strongly suggest our ARGs signature combined with age and bladder cancer stages have an overall superior clinical predictive power for determining survival outcomes in bladder cancer patients.
Figure 6.
Nomograms predicting survival in bladder cancer patients. (A) The nomogram to predict 1-, 3-, and 5-year overall survival of bladder cancer patients was created based on three independent factors. (B) The 403-patient bootstrapped calibration plots show the comparison of predicted and actual overall survival probabilities at 3- and 5-year follow-up. The dotted line represents the ideal fit; circles represent nomogram-predicted probabilities; stars represent the bootstrap-corrected estimates; and error bars represent the 95% CIs of these estimates.
4. Discussion
As the prognosis of bladder cancer is still poor, new strategies are urgently needed to be established to achieve better prognosis performance and increase overall survival of bladder cancer patients. Biomarkers are reliable prognostic methods for identifying the oncologic outcome including disease progression in patient and overall survival. In this study, we identified the autophagy-related genes that were significantly correlated to overall survival of bladder cancer patients. Among these prognostic autophagy-related genes, we discovered a novel 11-gene ARG signature that could classify bladder cancer patients into two distinct groups, high-risk and low-risk groups, with different clinical and molecular features. The prognostic power of the identified ARG signature was further validated in both training and validation groups. Furthermore, we combined this with patient clinical information, including age and bladder cancer stage, to establish an easy-to-use risk-assessment nomogram, which can predict 1-year, 3-year, and 5-year overall survivals of bladder cancer patients.
Several studies have highlighted the biomarker signatures associated with prognosis for bladder cancer patients. Chen et al. identified a four-gene signature from the gene expression profiles of 93 bladder cancer patients from the GEO database and 408 bladder tumor patients from TCGA database [30]. Dancik and Theodorescu performed a gene set enrichment analysis of 1968 patients with lung, bladder, or head and neck cancer, and identified 10–15 cell cycle-related genes as a predictive signature [31]. Mo et al. reported an 18-gene signature to characterize two major tumor differentiation subgroups, “basal” and “differentiated,” based on gene expression profiles from three bladder tumor datasets (TCGA, MDA, and LUND) [32]. In the context of cancer, dysfunction of autophagy may lead to increased free radicals and aggregated macromolecules, which may result in elevated DNA damage, mutation, metabolic deficiencies, and global cellular damage. On the other hand, abnormal autophagy could help cancer cells resistant to cell death. For these reasons, autophagy-related genes could be promising predictors in cancer. In the present study, we identified 32 ARGs that were significantly associated with the overall survival of bladder cancer patients. Among these genes, 11 ARGs (APOL1, ATG4B, BAG1, CASP3, DRAM1, ITGA3, KLHL24, P4HB, PRKCD, ULK2, and WDR45) displayed a high potential in predicting clinical outcome of bladder cancer patients. When dividing the patients into high- and low- risk groups based on median risk score calculated from these 11 ARGs, the high-risk bladder cancer patients exhibited significantly poor overall survival, while the low-risk patients generally have better overall survival. In addition, to further improve the prognostic accuracy of the 11-ARGs signature, we combined it with clinical features including age and stage. This method is much more potent than single factor prognosis methods.
Among genes identified in 11-ARG gene signature, ATG4B, WDR45, and DRAM1 play an important role in autophagy regulation. ATG4B is a core autophagy protein involved in formation of autophagosomes [33]. Recent studies suggest that ATG4B is a potential biomarker and vital in cancer therapy through the regulation of autophagy [33,34]. WDR45 has been shown to regulate autophagy via the AMPK and mTOR pathways and may also modulate autophagosome size [35]. DRAM1 is a p53 target gene and a regulator of the ATG5-independent autophagy pathway [36,37]. It has been reported to induce migration and invasion abilities of glioblastoma stem cells [38]. BAG1, PRKCD, and CASP3 are important regulators of apoptosis and recently were found to participate in autophagy. BAG1, a BCL2-associated athanogene, binds to oncogene BCL2 and enhances its anti-apoptotic effects. It has been reported that BAG1, associated with LC3-II, may participate in the induction of autophagy through interaction with Hsc70 [39]. Dysregulation of BAG1 has been reported in many human cancers, including breast cancer, non-small cell lung cancer, glioblastoma, etc. [40]. Several studies on breast cancer suggested that BAG1 is a prognostic marker in breast cancer [40,41]. Protein kinase C delta isoform (PRKCD) belongs to the protein kinase C (PKC) family, which consists of serine/threonine protein kinases and is a critical regulator of cell proliferation, survival, and cell death [42]. Recent studies have reported its potential role in modulating the crosstalk between apoptosis and autophagy. It has been reported that PRKCD activated GSK3αβ by phosphorylation and inhibited autophagy in cadmium-exposed NRK52E kidney cells [43]. Consistently, Rottlerin, an effective inhibitor of PRKCD, induced autophagy and caused apoptosis in breast cancer stem cells [44]. PRKCD has been shown to suppress breast cancer cell migration [45,46]. The potential role of BAG1 and PRKCD in bladder cancer has not been reported. In contrast, CASP3, caspase 3, has been recognized as a prognostic predictor in bladder cancer [47,48,49]. As an important effector caspase, CASP3 not only plays a crucial role in apoptosis [47] but also modulates autophagy through the cleavage of core autophagy proteins, such as Beclin-1, ATG5, and ATG4D [50,51,52]. APOL1, apolipoprotein L1, is a BH3-only protein and belongs to the APOL family of proteins [53]. APOLs are predominantly involved in the regulation of lipid transport and metabolism [54,55], and are expressed in papillary thyroid carcinomas [56]. APOL1 has been reported to induce autophagy and autophagy-associated cell death in a variety of cell types [57]. APOL1 genetic variants are reported to be a major drive of kidney diseases [58], but their function in bladder cancer has not been reported. ITGA3 and ULK2 have been studied in bladder cancer [59,60]. ITGA3, integrin alpha-3, was known to influence the development of various cancers including bladder cancer [60,61,62,63]. Upregulation of ITGA3 was observed in clinical specimens and bladder cancer cell lines [60]. Silencing ITGA3 inhibited tumor cell migration and invasion through regulating FAK/PI3K/AKT and FAK/Src signaling mechanisms [60]. ULK2 is a serine/threonine kinase that participates in autophagosome formation and initiation of autophagy [64]. ULK2 knockdown inhibited autophagy and apoptosis in bladder cancer cells [59]. ULK family proteins also have been used both as a direct or indirect target for tumor therapy [65,66,67]. KLHL24 belongs to the Kelch-like gene family, which contains a bric-a-brac, tramtrack, broad complex (BTB)/poxvirus and zinc finger (POZ) domain that can function in transcriptional repression through interacting with Cullin3 to form E3 ligase and mediating the ubiquitination of substrate proteins [68]. P4HB is one of the hub genes in the beta subunit of prolyl 4-hydroxylase and play an important function in inhibiting the aggregation of misfolded proteins [69]. It has recently been reported as a novel diagnosis and prognosis marker for kidney renal clear cell carcinoma [70]. P4HB was found to be significantly correlated with a high frequency of TP53 mutations in glioma cells [71]. It is worth noting that high TP53 mutations is a hallmark of muscle-invasive bladder cancer [72]. Additionally, P4HB has been utilized as an autophagy-related prognostic marker for cancer [70,73,74].
When tested as individual gene, 9 genes from the 11-ARG signature (APOL1, ATG4B, CASP3, ITGA3, P4HB, PRKCD, ULK2, and WDR45) exhibited a significant association between mRNA expression levels and overall survival of bladder cancer patients (Figure S1). While higher expression of APOL1, ATG4B, ITGA3, WDR45, CASP3, and PRKCD is associated with favorable survival in human bladder cancer patients, increased ULK2 and P4HB mRNA levels are negatively correlated to overall survival of bladder cancer patients. Despite the favorable survival rate observed in the high expression group of DRAM1, BAG1, and KLHL24, the correlations are less significant. Thus, our results suggest that survival analysis of individual genes might be insufficient in addressing the multilevel complexity in cancer patients. Consistent with the negative correlation to the overall survival of patients, the mRNA expression of ULK2 and P4HB are significantly higher in the high-risk group than the low-risk group (Figure S2). The other 8 genes, except for CASP3, were significantly higher in the low-risk group compared to the high-risk group, which supports their correlation to favorable survival in bladder cancer patients. Indeed, cancer diagnosis and prognosis are challenging due to the complexity of gene expression alteration in individual patients. Identification of a multi-gene signature from cancer patients as well as a combination of the gene signature-derived risk factor and other clinical parameters will provide more robust evaluation of cancer prognosis.
Our study identified a novel 11-gene ARG signature that was significantly correlated with the overall survival of human bladder cancer patients. Based on the expression value of 11 ARGs, an autophagy-related prognostic index was generated and successfully divided the patients into low-risk or high-risk subgroups. Moreover, a nomogram model combining the risk score values and the tumor stage and age of the patients exhibited good predictive efficacy on the overall survival of bladder cancer patients. However, our study solely relies on the TCGA dataset by dividing the patients randomly to the testing and validation group. Due to this limitation, our model has not been validated in an independent dataset or in cohorts with different TMN stages. In addition, AUCs in our analysis are relatively low. Thus, the model’s clinical accuracy needs to be further assessed with more datasets and different sample attributes.
5. Conclusions
Taken together, our work provides a comprehensive, accurate, and convenient prognosis of the survival outcomes of bladder cancer patients. These genotypic risk assessment markers may serve to be useful in assessing patients with higher survival rates. This can assist in the creation of patient-specific treatment regimens that will improve patient care. At the same time, our results show great potential of innovative molecular targets for bladder cancer treatment strategies. For further studies, clinical application of this signature for the prognosis of bladder cancer patients are needed for further evaluation.
Acknowledgments
The results published here are in whole based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga (accessed on 1 May 2020).
Supplementary Materials
The following is available online at: https://www.mdpi.com/article/10.3390/biology10050375/s1, Figure S1: The Kaplan–Meier survival curve analysis of individual genes of 11-gene ARG signature, Figure S2: Boxplot showing the mRNA expression levels of 11 genes in high-risk and low-risk groups, Figure S3: Summary of mutations in bladder cancer patients, Table S1: 213 autophagy-related genes (ARGs) obtained from the Human Autophagy Database, Table S2: Summary of patients’ survival information, 11-ARG signature genes expression and risk score in training and validation cohorts.
Author Contributions
Conceptualization, H.S.; Data curation, C.Z. and A.H.L.; Funding acquisition, H.S.; Supervision, H.S.; Writing—original draft, C.Z.; Writing—review & editing, A.H.L., S.L., and H.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by funding from the NIH/NCI, R01 CA217923, and NIH/NIEHS Center Grant P30 ES000260.
Institutional Review Board Statement
Data included in this study were obtained from the public database TCGA with no access to any identifiable personal information. Ethical review and approval were not required.
Informed Consent Statement
Not applicable.
Data Availability Statements
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ferlay J.E.M., Lam F., Colombet M., Mery L., Piñeros M., Znaor A., Soerjomataram I., Bray F. Global Cancer Observatory: Cancer Today. [(accessed on 20 May 2020)];2018 Available online: https://gco.iarc.fr/today.
- 2.Damrauer J.S., Hoadley K.A., Chism D.D., Fan C., Tiganelli C.J., Wobker S.E., Yeh J.J., Milowsky M.I., Iyer G., Parker J.S., et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc. Natl. Acad. Sci. USA. 2014;111:3110–3115. doi: 10.1073/pnas.1318376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richters A., Aben K.K.H., Kiemeney L.A.L.M. The global burden of urinary bladder cancer: An update. World J. Urol. 2020;38:1895–1904. doi: 10.1007/s00345-019-02984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cumberbatch M.G., Rota M., Catto J.W., La Vecchia C. The Role of Tobacco Smoke in Bladder and Kidney Carcinogenesis: A Comparison of Exposures and Meta-analysis of Incidence and Mortality Risks. Eur. Urol. 2016;70:458–466. doi: 10.1016/j.eururo.2015.06.042. [DOI] [PubMed] [Google Scholar]
- 6.Freedman N.D. Association between Smoking and Risk of Bladder Cancer among Men and Women. JAMA. 2011;306:737–745. doi: 10.1001/jama.2011.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witjes J.A., Bruins H.M., Cathomas R., Compérat E.M., Cowan N.C., Gakis G., Hernández V., Espinós E.L., Lorch A., Neuzillet Y., et al. European Association of Urology Guidelines on Muscle-invasive and Metastatic Bladder Cancer: Summary of the 2020 Guidelines. Eur. Urol. 2020;79:82–104. doi: 10.1016/j.eururo.2020.03.055. [DOI] [PubMed] [Google Scholar]
- 8.Inamura K. Bladder Cancer: New Insights into Its Molecular Pathology. Cancers. 2018;10:100. doi: 10.3390/cancers10040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi W., Porten S., Kim S., Willis D., Plimack E.R., Hoffman-Censits J., McConkey D.J. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152–165. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson A.G., Kim J., Al-Ahmadie H., Bellmunt J., Guo G., Cherniack A.D., Hinoue T., Laird P.W., Hoadley K.A., Akbani R., et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell. 2017;171:540–556.e25. doi: 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apel A., Herr I., Schwarz H., Rodemann H.P., Mayer A. Blocked Autophagy Sensitizes Resistant Carcinoma Cells to Radiation Therapy. Cancer Res. 2008;68:1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 12.Zachari M., Gudmundsson S.R., Li Z., Manifava M., Cugliandolo F., Shah R., Smith M., Stronge J., Karanasios E., Piunti C., et al. Selective Autophagy of Mitochondria on a Ubiquitin-Endoplasmic-Reticulum Platform. Dev. Cell. 2019;50:627–643.e5. doi: 10.1016/j.devcel.2019.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dower C.M., Wills C.A., Frisch S.M., Wang H.-G. Mechanisms and context underlying the role of autophagy in cancer metastasis. Autophagy. 2018;14:1110–1128. doi: 10.1080/15548627.2018.1450020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White E., DiPaola R.S. The Double-Edged Sword of Autophagy Modulation in Cancer. Clin. Cancer Res. 2009;15:5308–5316. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Apel A., Zentgraf H., Büchler M.W., Herr I. Autophagy-A double-edged sword in oncology. Int. J. Cancer. 2009;125:991–995. doi: 10.1002/ijc.24500. [DOI] [PubMed] [Google Scholar]
- 16.Mathew R., Karantza-Wadsworth V., White E. Role of autophagy in cancer. Nat. Rev. Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickstein R.J., Nitti G., Dinney C.P., Davies B.R., Kamat A.M., McConkey D.J. Autophagy limits the cytotoxic effects of the AKT inhibitor AZ7328 in human bladder cancer cells. Cancer Biol. Ther. 2012;13:1325–1338. doi: 10.4161/cbt.21793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ying L., Huang Y., Chen H., Wang Y., Xia L., Chen Y., Liu Y., Qiu F. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol. BioSyst. 2013;9:407–411. doi: 10.1039/c2mb25386k. [DOI] [PubMed] [Google Scholar]
- 19.Liu X., Zhou Z., Wang Y., Zhu K., Deng W., Li Y., Fu B. Downregulation of HMGA1 Mediates Autophagy and Inhibits Migration and Invasion in Bladder Cancer via miRNA-221/TP53INP1/p-ERK Axis. Front. Oncol. 2020;10:589. doi: 10.3389/fonc.2020.00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang Y., Zhu J., Huang H., Xiang D., Li Y., Zhang D., Li J., Wang Y., Jin H., Jiang G., et al. SESN2/sestrin 2 induction-mediated autophagy and inhibitory effect of isorhapontigenin (ISO) on human bladder cancers. Autophagy. 2016;12:1229–1239. doi: 10.1080/15548627.2016.1179403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dyrskjøt L., Zieger K., Real F.X., Malats N., Carrato A., Hurst C., Kotwal S., Knowles M., Malmström P.-U., De La Torre M., et al. Gene Expression Signatures Predict Outcome in Non–Muscle-Invasive Bladder Carcinoma: A Multicenter Validation Study. Clin. Cancer Res. 2007;13:3545–3551. doi: 10.1158/1078-0432.CCR-06-2940. [DOI] [PubMed] [Google Scholar]
- 22.von Rundstedt F.C., Rajapakshe K., Ma J., Arnold J.M., Gohlke J., Putluri V., Putluri N. Integrative Pathway Analysis of Metabolic Signature in Bladder Cancer: A Linkage to The Cancer Genome Atlas Project and Prediction of Survival. J. Urol. 2016;195:1911–1919. doi: 10.1016/j.juro.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abudurexiti M., Xie H., Jia Z., Zhu Y., Zhu Y., Shi G., Zhang H., Dai B., Wan F., Shen Y., et al. Development and External Validation of a Novel 12-Gene Signature for Prediction of Overall Survival in Muscle-Invasive Bladder Cancer. Front. Oncol. 2019;9:856. doi: 10.3389/fonc.2019.00856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao R., Yuan L., Ma B., Wang G., Qiu W., Tian Y. An EMT-related gene signature for the prognosis of human bladder cancer. J. Cell. Mol. Med. 2020;24:605–617. doi: 10.1111/jcmm.14767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eissa S., Matboli M., Awad N., Kotb Y. Identification and validation of a novel autophagy gene expression signature for human bladder cancer patients. Tumor Biol. 2017;39:1010428317698360. doi: 10.1177/1010428317698360. [DOI] [PubMed] [Google Scholar]
- 26.Wu Y., Peng Y., Guan B., He A., Yang K., He S., Gong Y., Li X., Zhou L. P4HB: A novel diagnostic and prognostic biomarker for bladder carcinoma. Oncol. Lett. 2020;21:1. doi: 10.3892/ol.2020.12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Y., Chen Y., Zhang J., Cao L., He M., Liu X., Zhao N., Yin A., Huang H., Wang L. TMEM74 promotes tumor cell survival by inducing autophagy via interactions with ATG16L1 and ATG9A. Cell Death Dis. 2017;8:e3031. doi: 10.1038/cddis.2017.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li F., Guo H., Yang Y., Feng M., Liu B., Ren X., Zhou H. Autophagy modulation in bladder cancer development and treatment (Review) Oncol. Rep. 2019;42:1647–1655. doi: 10.3892/or.2019.7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin J.-F., Hwang T.I.S. Autophagy regulation in bladder cancer as the novel therapeutic strategy. Transl. Cancer Res. 2017;6:S708–S719. doi: 10.21037/tcr.2017.06.26. [DOI] [Google Scholar]
- 30.Chen S., Zhang N., Shao J., Wang T., Wang X. A novel gene signature combination improves the prediction of overall survival in urinary bladder cancer. J. Cancer. 2019;10:5744–5753. doi: 10.7150/jca.30307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dancik G.M., Theodorescu D. Robust Prognostic Gene Expression Signatures in Bladder Cancer and Lung Adenocarcinoma Depend on Cell Cycle Related Genes. PLoS ONE. 2014;9:e85249. doi: 10.1371/journal.pone.0085249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mo Q., Nikolos F., Chen F., Tramel Z., Lee Y.-C., Hayashi K., Xiao J., Shen J., Chan K.S. Prognostic Power of a Tumor Differentiation Gene Signature for Bladder Urothelial Carcinomas. J. Natl. Cancer Inst. 2018;110:448–459. doi: 10.1093/jnci/djx243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akin D., Wang S.K., Habibzadegah-Tari P., Law B., Ostrov D., Li M., Yin X.-M., Kim J.-S., Horenstein N., William A.D., Jr. A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy. 2014;10:2021–2035. doi: 10.4161/auto.32229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y., Luo Q., Yuan L., Miao C., Mu X., Xiao W., Li J., Sun T., Ma E. JNK-dependent Atg4 upregulation mediates asperphenamate derivative BBP-induced autophagy in MCF-7 cells. Toxicol. Appl. Pharmacol. 2012;263:21–31. doi: 10.1016/j.taap.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 35.Bakula D., Müller A.J., Zuleger T., Takacs Z., Franz-Wachtel M., Thost A.K., Proikas-Cezanne T. WIPI3 and WIPI4 β-propellers are scaffolds for LKB1-AMPK-TSC signalling circuits in the control of autophagy. Nat. Commun. 2017;8:15637. doi: 10.1038/ncomms15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crighton D., Wilkinson S., Ryan K.M. DRAM Links Autophagy to p53 and Programmed Cell Death. Autophagy. 2007;3:72–74. doi: 10.4161/auto.3438. [DOI] [PubMed] [Google Scholar]
- 37.Nagata M., Arakawa S., Yamaguchi H., Torii S., Endo H., Tsujioka M., Honda S., Nishida Y., Konishi A., Shimizu S. Dram1 regulates DNA damage-induced alternative autophagy. Cell Stress. 2018;2:55–65. doi: 10.15698/cst2018.03.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen C., Liang Q., Chen H., Wu P., Feng Z., Ma X., Wu H., Zhou G. DRAM1 regulates the migration and invasion of hepatoblastoma cells via autophagy‑EMT pathway. Oncol. Lett. 2018;16:2427–2433. doi: 10.3892/ol.2018.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gurusamy N., Lekli I., Gherghiceanu M., Popescu L.M., Das D.K. BAG-1 induces autophagy for cardiac cell survival. Autophagy. 2009;5:120–121. doi: 10.4161/auto.5.1.7303. [DOI] [PubMed] [Google Scholar]
- 40.Cutress R.I., Townsend P.A., Brimmell M., Bateman A.C., Hague A., Packham G. BAG-1 expression and function in human cancer. Br. J. Cancer. 2002;87:834–839. doi: 10.1038/sj.bjc.6600538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner B.C., Krajewski S., Krajewska M., Takayama S., Gumbs A.A., Carter D., Rebbeck T.R., Haffty B.G., Reed J.C. BAG-1: A Novel Biomarker Predicting Long-Term Survival in Early-Stage Breast Cancer. J. Clin. Oncol. 2001;19:992–1000. doi: 10.1200/JCO.2001.19.4.992. [DOI] [PubMed] [Google Scholar]
- 42.Reyland M.E. Protein kinase C isoforms: Multi-functional regulators of cell life and death. Front. Biosci. 2009;14:2386–2399. doi: 10.2741/3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.So K.-Y., Oh S.-H. Cadmium-induced heme-oxygenase-1 expression plays dual roles in autophagy and apoptosis and is regulated by both PKC-δ and PKB/Akt activation in NRK52E kidney cells. Toxicology. 2016;370:49–59. doi: 10.1016/j.tox.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 44.Kumar D., Shankar S., Srivastava R.K. Rottlerin-induced autophagy leads to the apoptosis in breast cancer stem cells: Molecular mechanisms. Mol. Cancer. 2013;12:171. doi: 10.1186/1476-4598-12-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santiago-Walker A.E., Fikaris A.J., Kao G.D., Brown E.J., Kazanietz M.G., Meinkoth J.L. Protein kinase C delta stimulates apoptosis by initiating G1 phase cell cycle progression and S phase arrest. J. Biol. Chem. 2005;280:32107–32114. doi: 10.1074/jbc.M504432200. [DOI] [PubMed] [Google Scholar]
- 46.Jackson D., Zheng Y., Lyo D., Shen Y., Nakayama K., Nakayama K.I., Foster D.A. Suppression of cell migration by protein kinase Cdelta. Oncogene. 2005;24:3067–3072. doi: 10.1038/sj.onc.1208465. [DOI] [PubMed] [Google Scholar]
- 47.Johnson A., Bridgham J. Caspase-3 and -6 expression and enzyme activity in hen granulosa cells. Biol. Reprod. 2000;62:589–598. doi: 10.1095/biolreprod62.3.589. [DOI] [PubMed] [Google Scholar]
- 48.Wang J., Zhang X., Wei P., Zhang J., Niu Y., Kang N., Zhang Y., Zhang W., Xing N. Livin, Survivin and Caspase 3 as early recurrence markers in non-muscle-invasive bladder cancer. World J. Urol. 2014;32:1477–1484. doi: 10.1007/s00345-014-1246-0. [DOI] [PubMed] [Google Scholar]
- 49.Burton P.B., Anderson C.J., Corbishly C.M. Caspase 3 and p27 as Predictors of Invasive Bladder Cancer. N. Engl. J. Med. 2000;343:1418–1420. doi: 10.1056/NEJM200011093431915. [DOI] [PubMed] [Google Scholar]
- 50.Zhu Y., Zhao L., Liu L., Gao P., Tian W., Wang X., Jin H., Xu H., Chen Q. Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein Cell. 2010;1:468–477. doi: 10.1007/s13238-010-0048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Betin V.M., Lane J.D. Atg4D at the interface between autophagy and apoptosis. Autophagy. 2009;5:1057–1059. doi: 10.4161/auto.5.7.9684. [DOI] [PubMed] [Google Scholar]
- 52.Tsapras P., Nezis I.P. Caspase involvement in autophagy. Cell Death Differ. 2017;24:1369–1379. doi: 10.1038/cdd.2017.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhaorigetu S., Wan G., Kaini R., Jiang Z., Hu C.-A.A. ApoL1, a BH3-only lipid-binding protein, induces autophagic cell death. Autophagy. 2008;4:1079–1082. doi: 10.4161/auto.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duchateau P.N., Pullinger C.R., Cho M.H., Eng C., Kane J.P. Apolipoprotein L gene family: Tissue-specific expression, splicing, promoter regions; discovery of a new gene. J. Lipid Res. 2001;42:620–630. doi: 10.1016/S0022-2275(20)31171-8. [DOI] [PubMed] [Google Scholar]
- 55.Duchateau P.N., Pullinger C.R., Orellana R.E., Kunitake S.T., Naya-Vigne J., O’Connor P.M., Kane J.P. Apolipoprotein L, a new human high density lipoprotein apolipoprotein expressed by the pancreas Identification, cloning, characterization, and plasma distribution of apolipoprotein L. J. Biol. Chem. 1997;272:25576–25582. doi: 10.1074/jbc.272.41.25576. [DOI] [PubMed] [Google Scholar]
- 56.Chidiac M., Fayyad-Kazan M., Daher J., Poelvoorde P., Bar I., Maenhaut C., Delrée P., Badran B., Vanhamme L. ApolipoproteinL1 is expressed in papillary thyroid carcinomas. Pathol. Res. Pr. 2016;212:631–635. doi: 10.1016/j.prp.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 57.Hu C.-A.A., Klopfer E.I., Ray P.E. Human apolipoprotein L1 (ApoL1) in cancer and chronic kidney disease. FEBS Lett. 2012;586:947–955. doi: 10.1016/j.febslet.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Genovese G., Friedman D.J., Ross M.D., Lecordier L., Uzureau P., Freedman B.I., Bowden D.W., Langefeld C.D., Oleksyk T.K., Knob A.L.U., et al. Association of Trypanolytic ApoL1 Variants with Kidney Disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hao K., Zhao S., Cui D., Zhang Y., Jiang C., Jing Y., Xia S., Han B. Androgen receptor antagonist bicalutamide induces autophagy and apoptosis via ULK2 upregulation in human bladder cancer cells. Int. J. Clin. Exp. Pathol. 2017;10:7603–7615. [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J.-R., Liu B., Zhou L., Huang Y.-X. MicroRNA-124-3p suppresses cell migration and invasion by targeting ITGA3 signaling in bladder cancer. Cancer Biomark. 2019;24:159–172. doi: 10.3233/CBM-182000. [DOI] [PubMed] [Google Scholar]
- 61.Jiao Y., Li Y., Liu S., Chen Q., Liu Y. ITGA3 serves as a diagnostic and prognostic biomarker for pancreatic cancer. OncoTargets Ther. 2019;12:4141–4152. doi: 10.2147/OTT.S201675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang H., Cui X., Cao A., Li X., Li L. ITGA3 interacts with VASP to regulate stemness and epithelial-mesenchymal transition of breast cancer cells. Gene. 2020;734:144396. doi: 10.1016/j.gene.2020.144396. [DOI] [PubMed] [Google Scholar]
- 63.Sa K.-D., Zhang X., Li X.-F., Gu Z.-P., Yang A.-G., Zhang R., Li J.-P., Sun J.-Y. A miR-124/ITGA3 axis contributes to colorectal cancer metastasis by regulating anoikis susceptibility. Biochem. Biophys. Res. Commun. 2018;501:758–764. doi: 10.1016/j.bbrc.2018.05.062. [DOI] [PubMed] [Google Scholar]
- 64.Lee E.-J., Tournier C. The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy. Autophagy. 2011;7:689–695. doi: 10.4161/auto.7.7.15450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qiu L., Zhou G., Cao S. Targeted inhibition of ULK1 enhances daunorubicin sensitivity in acute myeloid leukemia. Life Sci. 2020;243:117234. doi: 10.1016/j.lfs.2019.117234. [DOI] [PubMed] [Google Scholar]
- 66.Lu J., Zhu L., Zheng L.-P., Cui Q., Zhu H.-H., Zhao H., Shen Z.-J., Dong H.-Y., Chen S.-S., Wu W.-Z., et al. Overexpression of ULK1 Represents a Potential Diagnostic Marker for Clear Cell Renal Carcinoma and the Antitumor Effects of SBI-0206965. EBioMedicine. 2018;34:85–93. doi: 10.1016/j.ebiom.2018.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang F., Hu P., Yang Z., Xue C., Gong J., Sun S., Shi L., Zhang S., Li Z., Yang C., et al. SBI0206965, a novel inhibitor of Ulk1, suppresses non-small cell lung cancer cell growth by modulating both autophagy and apoptosis pathways. Oncol. Rep. 2017;37:3449–3458. doi: 10.3892/or.2017.5635. [DOI] [PubMed] [Google Scholar]
- 68.Shi X., Xiang S., Cao J., Zhu H., Yang B., He Q., Ying M. Kelch-like proteins: Physiological functions and relationships with diseases. Pharmacol. Res. 2019;148:104404. doi: 10.1016/j.phrs.2019.104404. [DOI] [PubMed] [Google Scholar]
- 69.Zhang J., Guo S., Wu Y., Zheng Z.-C., Wang Y., Zhao Y. P4HB, a Novel Hypoxia Target Gene Related to Gastric Cancer Invasion and Metastasis. BioMed. Res. Int. 2019;2019:1–13. doi: 10.1155/2019/9749751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie L., Li H., Zhang L., Ma X., Dang Y., Guo J., Liu J., Ge L., Nan F., Dong H., et al. Autophagy-related gene P4HB: A novel diagnosis and prognosis marker for kidney renal clear cell carcinoma. Aging. 2020;12:1828–1842. doi: 10.18632/aging.102715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zou H., Wen C., Peng Z., Shao Y.-Y., ChunJie W., Li S., Li C., Zhou H.-H. P4HB and PDIA3 are associated with tumor progression and therapeutic outcome of diffuse gliomas. Oncol. Rep. 2017;39:501–510. doi: 10.3892/or.2017.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lindgren D., Frigyesi A., Gudjonsson S., Sjödahl G., Hallden C., Chebil G., Veerla S., Ryden T., Månsson W., Liedberg F., et al. Combined Gene Expression and Genomic Profiling Define Two Intrinsic Molecular Subtypes of Urothelial Carcinoma and Gene Signatures for Molecular Grading and Outcome. Cancer Res. 2010;70:3463–3472. doi: 10.1158/0008-5472.CAN-09-4213. [DOI] [PubMed] [Google Scholar]
- 73.Lyu L., Xiang W., Zheng F., Huang T., Feng Y., Yuan J., Zhang C. Significant Prognostic Value of the Autophagy-Related Gene P4HB in Bladder Urothelial Carcinoma. Front. Oncol. 2020;10:1613. doi: 10.3389/fonc.2020.01613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei X., Wang W., Wang H., Wang Y., Wang Y., Li G., Ji C., Ren X., Song N., Qin C. Identification of an independent autophagy-gene prognostic index for papillary renal cell carcinoma. Transl. Androl. Urol. 2020;9:1945–1956. doi: 10.21037/tau-20-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.