Abstract
In 2017, a 560-ha area of hybrid poplar plantation in northern Poland showed symptoms of tree decline. The leaves appeared smaller, yellow-brown, and were shed prematurely. Twigs and smaller branches died without distinct cankers. Trunks decayed from the base. The phloem and xylem showed brown necrosis. Ten percent of the trees died 1–2 months after the first appearance of the symptoms. None of these symptoms were typical for known poplar diseases. The trees’ mycobiota were analysed using Illumina sequencing. A total of 69 467 and 70 218 operational taxonomic units (OTUs) were obtained from the soil and wood. Blastocladiomycota and Chytridiomycota occurred only in the soil, with very low frequencies (0.005% and 0.008%). Two taxa of Glomeromycota, with frequencies of 0.001%, occurred in the wood. In the soil and wood, the frequencies of Zygomycota were 3.631% and 0.006%, the frequencies of Ascomycota were 45.299% and 68.697%, and the frequencies of Basidiomycota were 4.119% and 2.076%. At least 400 taxa of fungi were present. The identifiable Zygomycota, Ascomycota, and Basidiomycota were represented by at least 18, 263 and 81 taxa, respectively. Many fungi were common to the soil and wood, but 160 taxa occurred only in soil and 73 occurred only in wood. The root pathogens included species of Oomycota. The vascular and parenchymal pathogens included species of Ascomycota and of Basidiomycota. The initial endophytic character of the fungi is emphasized. Soil, and possibly planting material, may be the sources of the pathogen inoculum, and climate warming is likely to be a predisposing factor. A water deficit may increase the trees’ susceptibility. The epidemiology of poplar vascular wilt reminds grapevine trunk diseases (GTD), including esca, black foot disease and Petri disease.
Keywords: fungi, pathogens, plantation, poplar hybrids, vascular wilt
1. Introduction
Populus is a genus of deciduous trees in the family Salicaceae, native to most of the Northern Hemisphere. They are among the fastest-growing trees, and the most efficient in terms of sustainability. Poplar is significant because of: (i) its rapid production of wood (in Europe, 1 m3 of lumber can be produced on average in 15 years, six times faster than with oak); (ii) its very versatile wood, with an excellent ratio between specific weight and mechanical features, making it suitable for furniture, plywood and the paper industry; (iii) its excellent capacity for purifying the air by capturing CO2 and storing it in the biomass (1 ha can capture 11 t CO2/year); (iv) its capacity for purifying water while acting as a green filter, absorbing nitrates and sediments; (v) its potential for biofuel production using the coppicing method; (vi) the possibility for its cultivation on abandoned and degraded land, thus optimizing land use.
Poplar is an important source of wood for pulp and paper products, but mostly paper, for which worldwide production reaches 420 Mt, including 5 Mt in Poland [1]. Its wood is also suitable for use as a renewable energy source. The development of renewable sources for energy purposes has been substantially supported and promoted by a European Union Directive. Poland is obliged to obtain at least 30% of its energy from renewable sources by 2030 (Directive (EU) 2018/2001). Wood that is suitable for renewable energy includes that derived from trees grown in short- and medium-rotation plantations, often on agricultural land or non-forested areas. Plantations based on varieties of Acacia and Eucalyptus have been particularly effective in tropical countries with favourable climate and soil conditions for faster growth; Eucalyptus has produced 25 m3 of wood per ha annually, compared with 7–8 m3 in the temperate climate zone (1). Plantations of fast-growing trees are now also being established in the temperate zone. The most promising genus in Poland is poplar (Populus spp.), with plantations usually in short- (up to 10 years) or medium-rotation (up to 15–25 years) coppice systems [2,3,4].
Hybrid poplar trees are often the progeny of crosses between cottonwood (Populus deltoides W. Bartram ex Marshall) and black poplar (Populus nigra L. ‘Italica’). They have the advantages of: (i) rapid growth (1.5–2.5 m per year), (ii) a large range of hardiness zones (3–9), (iii) high productivity resulting from a prolonged vegetation period, and (iv) better resistance to pests and diseases [5].
Poplars are frequently attacked by microorganisms that cause discolorations, necrosis, depressions, deformations (thickening of the trunk and branches, the abnormal proliferation of the underlying phloem, the formation of the corky ridges or woody galls). Stresses predispose trees to infection by phytopathogens. Attacks on the trunk and branches of younger trees often kill the main shoot.
The bark necrosis of poplars can be caused by Discosporium populeum (Sacc.) B. Sutton (=Chondroplea populea (Sacc.) Kleb. = Dothichiza populea Sacc. Sacc. & Briard, anamorph of Cryptodiaporthe populea (Sacc.) Butin). Necrosis and cankers are often caused by Cytospora spp. (C. populina (Pers.) Rabenh. = C. ambiens Sacc., teleomorph Valsa ambiens (Pers.) Fr., and C. nivea Fuckel, teleomorph V. nivea (Hoffm.) Fr.). Cankers can be caused by Entoleuca mammata (Wahlenb.) Rogers and Ju (=Hypoxylon mammatum (Wahl.: Fr.) Karst.). Sooty-bark canker is caused by Sclerencoelia pruinosa (Ellis and Everh.) Pärtel and Baral (=Encoelia pruinosa (Ell. and Ev.) Torkelsen and Eckblad). Black or target canker can be caused by Ceratocystis fimbriata Ellis and Halst. Other agents of necrosis and cankers or wood rots and bark alterations, of which the incidence is more local and/or secondary, include Boeremia populi (Gruyter and Scheer) Jayawardena, Jayasiri and Hyde (=Phoma exigua var. populi Gruyter and Scheer), Botryodiplodia populea Zhong, Diplodia tumefaciens (Shear) Zalasky (the anamorph of Keissleriella emergens (Karst.) Bose), Fusarium spp., Neofusicoccum ribis (Slippers, Crous and M.J. Wingf.) Crous, Slippers and Phillips (=Dothiorella gregaria Sacc., the anamorph of Botryosphaeria dothidea (Moug.) Ces. and De Not), Neonectria ditissima (Tul. and C. Tul.) Samuels and Rossman (with anamorph Cylindrocarpon mali (Allesch.) Wollenw.), Phomopsis spp., Rhytidiella moriformis Zalasky, Rhytidiella baranyayi Funk and Zalasky, and basidiomycetous Erythricium salmonicolor (Berk. and Broome) Burds. (=Corticium salmonicolor Berk. and Broome). Damage to heartwood can be caused by bacteria (Erwinia nimipressuralis). Disease of the leaves are usually caused by Melampsora medusae Thüm. (rust), Venturia tremulae Aderh. (scab, shoot blight), Sphaerulina musiva (Peck) Quaedvl., Verkley and Crous (=Septoria musiva Peck), and Marssonina spp. Most infections of woody tissues are initiated by wind-borne ascospores, which are forcibly ejected from perithecia during periods of damp weather. Fungi infect trees through wounds and invade the inner bark and cambium.
In 2017, a 560 ha plantation of hybrid poplar (P. deltoides × P. nigra) in northern Poland showed symptoms of tree decline. The leaves of the diseased trees appeared smaller, turned yellow-brown, and were shed prematurely. Twigs and smaller branches died without definite cankers. The bark of the entire trunk was sunken and discolored, often loosened and split. It often fell off, exposing wet wood. The trunks decayed from the base. The phloem showed brown necrosis. Ten percent of the trees died in 1–2 months (in June) after the first appearance of the symptoms. None of the observed symptoms were typical for known poplar diseases.
The objectives of the study on the structure of the fungal communities present in the rotten wood of poplar trunks and in the soil were to: (i) determine the abundance and diversity of pathogens and other fungi; (ii) identify interactions among fungi that may contribute to the disease progress; (iii) assess associations between the disease and global warming, with consequences for host and pathogen physiology, reproduction, survival, spatial and temporal distribution, resource availability and competition.
2. Materials and Metods
2.1. Site and Sampling
The study was carried out in the Łoża, Czarne District, Człuchów County, Pomeranian Voivodeship, northern Poland (53°41′29″ N 17°04′19″ E), in a 560 ha plantation of 5–6-year-old hybrid poplar (P. deltoides × P. nigra, cultivar AF2, from Italy) showing symptoms of crown decline, trunk-base decay (520 ha) and tree death (40 ha) (Figure 1 and Figure 2). The plantation was so intensively affected that the inclusion of a control (healthy plantation) from the same area with the same conditions of climate and soil was impossible.
Figure 1.
Poplar plantation with diseased trees.
Figure 2.
Necrosis and decay at the base of the trunk of a diseased poplar.
The trees were grown at a density of 425 trees/ha (4 m × 4m spacing), and had a mean diameter of 9–10 cm at breast height. The post-agricultural soil was sandy loam, consisting of sand (60%), silt (20%) and clay (20%), with a low humus level. The former crop was rye (Secale cereale L.). The average temperature is 7.9 °C and the rainfall is 680 mm.
The understorey vegetation included Achillea millefolium L., Agrostis stolonifera L., Artemisia absinthium L., Artemisia vulgaris L., Cichorium intybus L., Elymus repens (L.) Gould, Lamium purpureum L., Lolium perenne L., Papaver rhoeas L., Poa annua L., Poa pratensis L., Poa trivialis L., Polygonum aviculare L., Polypodium vulgare L., Polytrichum commune Hedw., Stellaria media Hist. Pl. Dauphiné, Taraxacum officinale F.H. Wigg., and Trifolium arvense L.
Five wood cores; 10 cm long and 3 cm in diam., each including bark, phloem and xylem, were sampled from the bases of the necrotic trunks of five symptomatic trees, 0 cm and 50 cm above the ground, with a Pressler borer. The core samples were surface-sterilized and ground to sawdust with a cordless SPARKY BUR2 15E drill. Additionally, five subsamples of soil were taken as cylindrical cores, 10 cm long and 5 cm in diam., from the surroundings of roots of five symptomatic trees. They were placed in sterile glass containers and refrigerated for 48 h.
2.2. DNA Extraction, Amplification and Illumina Sequencing
Five samples of sawdust were prepared from five wood cores in the SPEXTM SamplePrepTM Freezer/MillTM cryogenic mill. The wood’s genomic DNA was extracted from each of five 30 mg heavy sawdust samples using a Plant Genomic DNA Purification Kit (Thermo Scientific, Carlsbad, California, USA). The soil’s genomic DNA was extracted from each 300 mg soil subsample using a Power SoilM DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, USA).
The rDNA was amplified with fungi specific primers ITS1 FI2 (5′-GAACCWGCGGARGGATCA-3′) [6] and 5.8 S (5′-CGCTGCGTT CTTCATCG-3′) [7].
The PCR reaction mixture consisted of 12.5 μL of 2 × Mix PCR (A & A Biotechnology, Gdańsk, Poland), 0.2 μM of each primer, 1.5 μL purified and diluted DNA, and 10.6 μL water. The DNA amplification was performed under the following conditions: denaturation at 94 °C for 5 min followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 56 °C for 30 s, elongation at 72 °C for 30 s, and a final elongation at 72 °C for 7 min. The visualization of 5-μL amplicons was performed in 1.0% agarose gel dyed with Midori Green Advance DNA (Genetics). The pooled PCR products were purified using a MinElute PCR Purification Kit (Qiagen, Hilden, Germany). The concentration of PCR products was quantified using a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA), and an equimolar mix of PCR products from each sample was prepared. The amplicons were sequenced using the Illumina system in the Genomic Laboratory, DNA Research Center, Rubież 46, Poznań, Poland.
2.3. Bioinformatics Analysis
A table of Operational Taxonomic Units (OTUs) was prepared by PIPITS, version 1.2.0 [8]. The read-pairs were joined with PEAR, version 0.9.6 [9], filtered with a quality threshold of q = 30 by FASTX-toolkit, version 0.0.13 (http:hannonlab.cshl.edu/fastx_toolkit/index.html, accessed on 26 April 2012) converted to the Fasta format, and merged into a single file. The prepared sequences were de-replicated, and subregions of ITS were selected with the use of ITSx, version 1.0.11 [10]. Unique sequences and those shorter than 100 bp were removed. The remaining sequences were clustered with 97% sequence identity. The resulting representative sequences for each cluster were subjected to chimera detection and removal using the UNITE UCHIME reference dataset, version 6.0 (https://unite.ut.ee/index.php (accessed on 26 April 2012)). The input sequences were then mapped onto the representative sequences, and taxonomy was assigned using RDP Classifier, version 2.10.2 [11] against the UNITE fungal ITS reference database, version 11.2 [12]. This process resulted in the creation of a table of OTUs. The sequences were identified by comparison with reference sequences from the National Center for Biotechnology Information (NCBI) database.
The abundance of fungi was defined as the average number of OTUs from five subsamples. The frequency of an individual taxon was defined as the percentage (%) of OTUs in the total number of OTUs. The similarity and relationships between the fungal communities from the soil and wood is shown by a heat map.
2.4. Statistical Analyses
The differences in the abundance of microfungi in the soil and wood were analysed with chi-squared tests (χ2). The diversity between the communities of microfungi was compared with Margalef’s diversity index (DMg), Shannon’s diversity index (H), Simpson’s diversity index (D), Shannon’s evenness index (E) and Berger–Parker’s index (d) [13].
3. Results
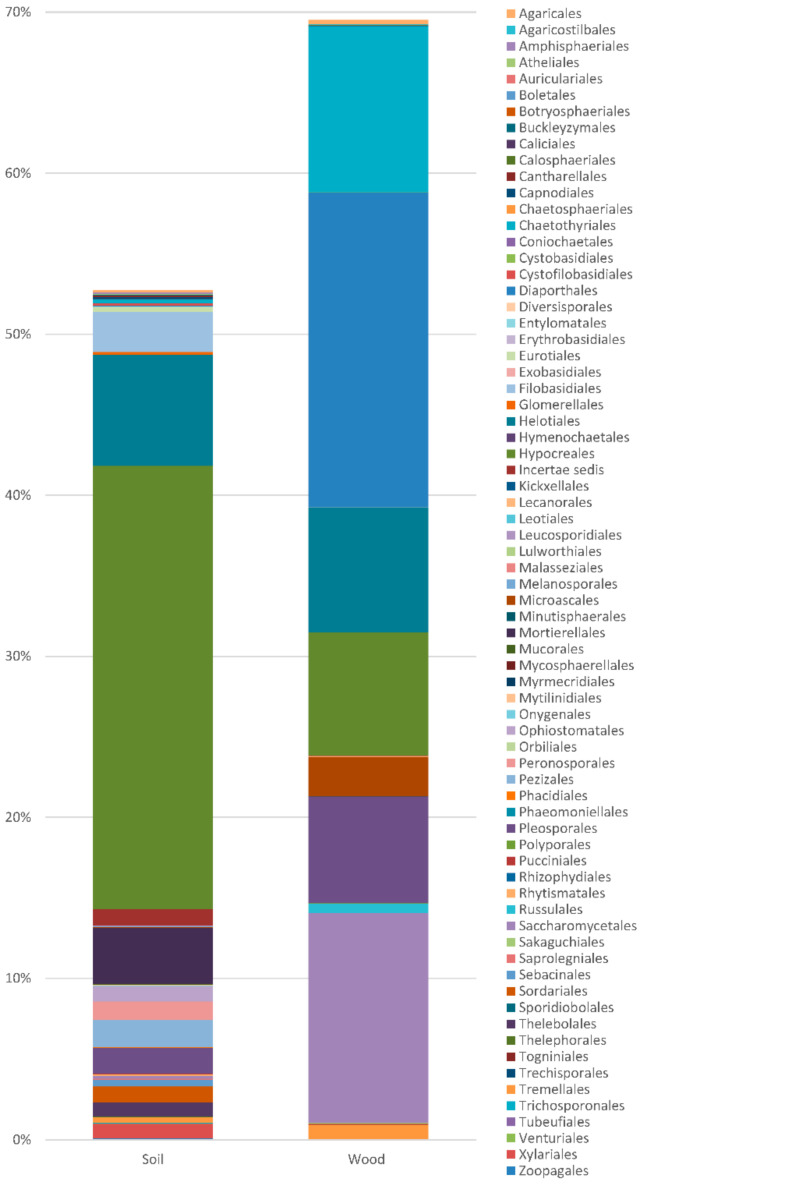
Totals of 69 467 and 70 218 OTUs were obtained, respectively, from the soil and wood of the Populus hybrid using the Illumina sequencing technique (Table 1, Figure 3). Of these, 44 506 (64%) and 53 592 (76%) were of fungi known from culture, and 24 961 (36%) and 16,628 (24%) were unidentified fungi and other organisms. Fungi from Blastocladiomycota, Chytridiomycota, Glomeromycota, Zygomycota, Ascomycota and Basidiomycota were detected. Blastocladiomycota and Chytridiomycota occurred only in the soil, with very low frequencies of 0.005% and 0.008%. Two taxa of Glomeromycota with a frequency of 0.001% occurred in the wood. The frequencies of Zygomycota in the soil and wood were 3.631% and 0.006%, the frequencies of Ascomycota were 45.299% and 68.697%, and the frequencies of Basidiomycota were 4.119% and 2.076%. The samples were colonized by at least 400 taxa of fungi. Identifiable Zygomycota, Ascomycota, and Basidiomycota were represented by at least 18, 263 and 81 taxa, respectively. Many fungi were common to the soil and wood, but 160 taxa occurred only in the soil, and 73 occurred only in the wood.
Table 1.
Microbiota present in the soil and wood of the diseased poplar.
| No. | Taxon | Order | Soil | Wood | Trophic Group |
|---|---|---|---|---|---|
| Chromista | |||||
| Oomycota | |||||
| 1. | Aphanomyces spp. | Saprolegniales | 0.042 | Pathogens | |
| 2. | Elongisporangium anandrum (Drechsler) Uzuhasi, Tojo & Kakish | Peronosporales | 0.004 | Pathogen | |
| 3. | Globisporangium apiculatum (B. Paul) Uzuhashi, Tojo & Kakish. + G. heterothallicum W.A. Campb. & F.F. Hendrix + G. intermedium (de Bary) Uzuhashi, Tojo & Kakish. + G. macrosporum (Vaartaja & Plaäts-Nit.) Uzuhashi, Tojo & Kakish. + G. mamillatum (Meurs) Uzuhashi, Tojo & Kakish. + G. pleroticum (Takesi Itô) Uzuhashi, Tojo & Kakish. + G. sylvaticum (W.A. Campb. & F.F. Hendrix) Uzuhashi, Tojo & Kakish. + G. ultimum (Trow) Uzuhashi, Tojo & Kakish | Peronosporales | 1.010 | 0.001 | Pathogens |
| 4. | Hyaloperonospora cochleariae (Gäum.) Göker, Riethm., Voglmayr, Weiss & Oberw | Peronosporales | 0.017 | Pathogen | |
| 5. | Isoachlya intermedia (Coker & J.V. Harv.) Coker | Saprolegniales | 0.007 | Saprotroph | |
| 6. | Myzocytiopsis sp. | Peronosporales | 0.005 | Nematopathogenic | |
| 7. | Phytophthora brassicae De Cock & Man in ‘t Veld + P. citricola Sawada + P. clandestina P.A. Taylor, Pascoe & F.C. Greenh | Peronosporales | 0.040 | Pathogens | |
| 8. | Pythium conidiophorum Jokl. + P. oligandrum Drechsler + P. pachycaule Ali-Shtayeh + P. selbyi M.L. Ellis, Broders & Dorrance + P. vanterpoolii V. Kouyeas & H. Kouyeas + P. volutum Vanterp. & Truscott + Pythium spp. | Peronosporales | 0.053 | 0.001 | Pathogens |
| 9. | Thraustotheca clavata (de Bary) Humphrey | Saprolegniales | 0.021 | Saprotroph | |
| Frequency Oomycota | 1.199 | 0.002 | |||
| Number of taxa Oomycota | 26 | 2 | |||
| Fungi | |||||
| Blastocladiomycota | |||||
| Frequency Blastocladiomycota | 0.005 | ||||
| Number of taxa Blastocladiomycota | 1 | ||||
| Chytridiomycota | |||||
| 1. | Chytridiomycota | 0.004 | |||
| 2. | Rhizophydium sp. | Rhizophydiales | 0.004 | Pathogen | |
| Frequency Chytridiomycota | 0.008 | ||||
| Number of taxa Chytridiomycota | 2 | ||||
| Glomeromycota | |||||
| 1. | Entrophospora sp. | Diversisporales | 0.001 | ||
| Frequency Glomeromycota | 0.001 | Mycorrhizal | |||
| Number of taxa Glomeromycota | 2 | ||||
| Zygomycota | |||||
| 1. | Mortierella alpina Peyronel + M. amoeboidea W. Gams + M. antarctica Linnem. + M. elongata Linnem. + M. epicladia W. Gams & Emden + M. exigua Linnem. + M. fatshederae Linnem. + M. gamsii Milko + M. horticola Linnem. + M. humilis Linnem. + M. hyalina (Harz) W. Gams + Mortierella spp. | Mortierellales | 3.483 | 0.006 | Saprotrophs |
| 2. | Mortierellales | Mortierellales | 0.006 | ||
| 3. | Mucor racemosus Bull. | Mucorales | 0.012 | Saprotrophs | |
| 4. | Ramicandelaber sp. | Kickxellales | 0.004 | ||
| 5. | Rhizopus arrhizus A. Fisch. + R. oryzae Went & Prins. Geerl. | Mucorales | 0.019 | ||
| 6. | Syncephalis sp. | Zoopagales | 0.107 | Mycoparasite | |
| Frequency Zygomycota | 3.631 | 0.006 | |||
| Number of taxa Zygomycota | 18 | 3 | |||
| Ascomycota | |||||
| 1. | Acaulium retardatum (Udagawa & T. Muroi) Lei Su | Microascales | 0.004 | Saprotroph | |
| 2. | Acericola italica Wanas., Camporesi, E.B.G. Jones & K.D. Hyde | Pleosporales | 0.001 | ||
| 3. | Acremonium persicinum (Nicot) W. Gams + A. rutilum W. Gams | Hypocreales | 0.001 | 0.002 | Saprotrophs |
| 4. | Acrodontium crateriforme (J.F.H. Beyma) de Hoog | Incertae sedis | 0.013 | ||
| 5. | Alatospora acuminata Ingold + Alatospora sp. | Helotiales | 0.113 | 0.026 | |
| 6. | Alternaria alternata (Fr.) Keissl. + A. botrytis (Preuss) Woudenb. & Crous + A. infectoria E.G. Simmons + A. tenuissima (Kunze) Wiltshire + Alternaria sp. | Pleosporales | 0.065 | 0.039 | Pathogens |
| 7. | Amesia nigricolor (L.M. Ames) X. Wei Wang & Samson | Sordariales | 0.001 | Saprotroph | |
| 8. | Angustimassarina acerina Jayasiri, Thambug., R.K. Schumach. & K.D. Hyde + A. populi Thambug. & K.D. Hyde | Pleosporales | 0.354 | Mycoparasite | |
| 9. | Arthoniomycetes | 0.001 | 0.001 | ||
| 10. | Ascobolus sp. | Pezizales | 0.005 | Saprotroph, coprophilous | |
| 11. | Ascochyta skagwayensis (R. Sprague) Punith. | Pleosporales | 0.001 | Saprotroph, pathogen | |
| 12. | Ascomycete | 0.027 | |||
| 13. | Ascomycota | 1.123 | 0.215 | ||
| 14. | Aspergillus conicus Blochwitz + A. niger Tiegh. + A. penicillioides Speg. + A. versicolor (Vuill.) Tirab. | Eurotiales | 0.008 | 0.003 | Saprotrophs |
| 15. | Atrocalyx lignicola (Ying Zhang, J. Fourn. & K.D. Hyde) A. Hashim. & Kaz. Tanaka | Pleosporales | 0.009 | Saprotroph | |
| 16. | Aureobasidium melanogenum (Herm.-Nijh.) Zalar, Gostinčar & Gunde-Cim. + A. pullulans (de Bary & Löwenthal) G. Arnaud + Aureobasidium sp. | Dothideales | 0.003 | 0.013 | Saprotrophs, often aquatic |
| 17. | Bacidina sp. | Lecanorales | 0.018 | Lichenicolous | |
| 18. | Beauveria bassiana (Bals.-Criv.) Vuill. + Beauveria sp. | Hypocreales | 0.049 | 0.002 | Entomopathogenic |
| 19. | Blastobotrys malaysiensis Kurtzman + Blastobotrys sp. | Saccharomycetales | 0.009 | 0.013 | Saprotrophs |
| 20. | Boeremia exigua (Desm.) Aveskamp, Gruyter & Verkley + B. noackiana (Allesch.) Gruyter & Verkley | Pleosporales | 0.006 | 0.017 | Pathogens |
| 21. | Cadophora luteo-olivacea (J.F.H. Beyma) T.C. Harr. & McNew + C. spadicis Travadon, D.P. Lawr., Roon.-Lath., Gubler, W.F. Wilcox, Rolsh. & K. Baumgartner + Cadophora sp. | Helotiales | 0.114 | 1.435 | Pathogens |
| 22. | Candida sake (Saito & M. Ota) Uden & H.R. Buckley ex S.A. Mey. & Ahearn + C. subhashii M. Groenew., Sigler & S.E. Richardson + C. vartiovaarae (Capr.) Uden & H.R. Buckley + Candida sp. | Saccharomycetales | 0.093 | 0.012 | Saprotrophs |
| 23. | Capnobotryella renispora Sugiy | Capnodiales | 0.005 | Saprotroph | |
| 24. | Capnodiales | Capnodiales | 0.017 | ||
| 25. | Cenococcum geophilum Fr. | Mytilinidiales | 0.039 | Ectomycorrhizal | |
| 26. | Cephalothecaceae | Sordariales | 0.003 | Saprotrophs, mycoparasites | |
| 27. | Ceratostomataceae | Melanosporales | 0.004 | Saprotrophs, mycoparasite | |
| 28. | Cercophora sp. | Sordariales | 0.014 | Coprophilous | |
| 29. | Cercosporabeticola Sacc. | Capnodiales | 0.012 | Pathogen | |
| 30. | Chaetomiaceae | Sordariales | 0.085 | Saprotrophs | |
| 31. | Chaetomium globosum Kunze + Ch. piluliferum J. Daniels + Chaetomium sp. | Sordariales | 0.062 | 0.002 | Saprotrophs, endophytes |
| 32. | Chaetosphaeria vermicularioides (Sacc. & Roum.) W. Gams & Hol.-Jech. | Chaetosphaeriales | 0.005 | Saprotroph | |
| 33. | Chaetothyriales | Chaetothyriales | 0.104 | Parasites of humans and cold-blooded animals | |
| 34. | Chalara microspora (Corda) S. Hughes + Chalara sp. | Helotiales | 0.007 | 0.001 | Saprotroph |
| 35. | Chloridium paucisporum C.J.K. Wang & H.E. Wilcox | Helotiales | 0.001 | Ectendomycorrhizal | |
| 36. | Chrysosporium pseudomerdarium Oorschot | Onygenales | 0.004 | Endophyte | |
| 37. | Cistella albidolutea (Feltgen) Baral | Helotiales | 0.003 | Saprotroph | |
| 38. | Cladophialophora minutissima M.L. Davey & Currah + Cladophialophora sp. | Chaetothyriales | 0.002 | Saprotrophs, human pathogens | |
| 39. | Cladorrhinum flexuosum Madrid, Cano, Gené & Guarro | Sordariales | 0.008 | Saprotroph | |
| 40. | Cladosporium allicinum (Fr.) Bensch, U. Braun & Crous + C. cladosporioides (Fresen.) G.A. de Vries + C. colocasiae Sawada | Capnodiales | 0.096 | 0.015 | Saprotrophs, facultative plant pathogens, mycoparasites |
| 41. | Clonostachys divergens Schroers + C. parva (Schroers) Rossman, L. Lombard & Crous + C. rosea (Link) Schroers, Samuels + Clonostachys sp. | Hypocreales | 0.187 | 0.033 | Endophytes, mycoparasites |
| 42. | Coleophoma cylindrospora (Desm.) Höhn | Helotiales | 0.010 | Saprotroph | |
| 43. | Collophorina sp. | Leotiales | 0.001 | Saprotroph | |
| 44. | Coniochaeta sp. | Coniochaetales | 0.015 | 0.002 | Pathogens, saprotrophs, endophytes, coprophilous, mycoparasite, human pathogens |
| 45. | Cordyceps bassiana Z.Z. Li, C.R. Li, B. Huang & M.Z. Fan + C. brongniartii Shimazu | Hypocreales | 0.047 | Enthomopathogenic, mycoparasite | |
| 46. | Cosmospora berkeleyana (P. Karst.) Gräfenhan, Seifert & Schroers | Hypocreales | 0.027 | Saprotroph, pathogen, mycoparasite | |
| 47. | Crocicreas sp. | Helotiales | 0.005 | Saprotrophs | |
| 48. | Cucurbitariaceae | Pleosporales | 0.076 | Saprotrophs, pathogens | |
| 49. | Cudoniella indica J. Webster, Eicker & Spooner | Helotiales | 0.002 | Saprotroph | |
| 50. | Cyathicula cyathoidea (Bull.) Thüm | Helotiales | 0.006 | Saprotrophs | |
| 51. | Cyphellophora sessilis (de Hoog) Réblová & Unter | Chaetothyriales | 0.001 | Pathogen | |
| 52. | Cytospora davidiana Y.L. Wang & X.Y. Zhang + C. leucostoma (Pers.) Sacc. + C. paratranslucens Norphanph., Bulgakov, T.C. Wen & K.D. Hyde + Cytospora sp. | Diaporthales | 0.012 | 13.720 | Pathogens |
| 53. | Dactylaria dimorphospora Veenb.-Rijks | Helotiales | 0.016 | Saprotroph | |
| 54. | Dactylonectria torresensis (A. Cabral, Rego & Crous) L. Lombard & Crous | Hypocreales | 0.008 | Pathogen | |
| 55. | Debaryomyces hansenii (Zopf) Lodder & Kreger-van Rij | Saccharomycetales | 0.023 | Pathogen | |
| 56. | Dendryphion europaeum Crous & R.K. Schumach. + D. nanum (Nees) S. Hughes | Pleosporales | 0.268 | 0.006 | Saprotroph |
| 57. | Dermateaceae | Helotiales | 0.002 | ||
| 58. | Desmazierella acicola Lib. | Pezizales | 0.001 | Saprotroph | |
| 59. | Diaporthe cynaroidis Marinc., M.J. Wingf. & Crous + D. foeniculina (Sacc.) Udayanga & Castl. + D. helicis Niessl + D. novem J.M. Santos, Vrandečić & A.J.L. Phillips + D. rudis (Fr.) Nitschke + Diaporthe sp. | Diaporthales | 0.017 | 3.327 | Pathogens, endophytes |
| 60. | Didymella macrostoma (Mont.) Qian Chen & L. C + D. pedeiae (Aveskamp, Gruyter & Verkley) Qian Chen & L. Cai + D. pinodes (Berk. & A. Bloxam) Petr. + D. pomorum (Thüm.) Qian Chen & L. Cai | Pleosporales | 0.039 | 0.036 | Pathogens |
| 61. | Didymosphaeria futilis (Berk. & Broome) Rehm | Pleosporales | 0.005 | Saprotroph | |
| 62. | Dissoconium eucalypti Crous & Carnegie | Capnodiales | 0.001 | Commensalist, mycoparasite | |
| 63. | Dothideomycetes | 0.018 | 0.014 | ||
| 64. | Emericellopsis glabra (J.F.H. Beyma) Backus & Orpurt + E. minima Stolk | Hypocreales | 0.179 | Endophytes | |
| 65. | Endophoma elongata Tsuneda & M.L. Dave | Incertae sedis | 0.005 | ||
| 66. | Epicoccum nigrum Link | Pleosporales | 0.002 | 0.001 | Endophyte, saprotroph, pathogen |
| 67. | Eurotiales | Eurotiales | 0.001 | ||
| 68. | Eurotiomycetes | 0.002 | 0.020 | ||
| 69. | Exophiala capensis Crous + E. equina (Pollacci) de Hoog, V.A. Vicente, Najafz., Harrak, Badali & Seyedm. + E. opportunistica de Hoog, V.A. Vicente, Najafz., Harrak, Badali & Seyedm. + Exophiala sp. | Chaetothyriales | 0.129 | 0.031 | Saprotrophs, human pathogens |
| 70. | Fusarium avenaceum (Fr.) Sacc. + F. equiseti (Corda) Sacc. + F. fujikuroi Nirenberg + F. oxysporum Schltdl. + F. petersiae L. Lombard + F. redolens Wollenw. + F. solani (Mart.) Sacc. + F. torulosum (Berk. & M.A. Curtis) Gruyter & J.H.M. Schneid. + Fusarium sp. + Neocosmospora solani (Mart.) L. Lombard & Crous | Hypocreales | 0.890 | 0.104 | Pathogens |
| 71. | Fusicolla aquaeductuum (Radlk. & Rabenh.) Gräfenhan, Seifert & Schroers + F. merismoides (Corda) Gräfenhan, Seifert & Schroers | Hypocreales | 0.096 | Pathogens | |
| 72. | Gibellulopsis nigrescens (Pethybr.) Zare, W. Gams & Summerb | Glomerellales | 0.009 | Saprotroph | |
| 73. | Gliomastix murorum var. felina (Marchal) S. Hughes | Hypocreales | 0.023 | Saprotroph | |
| 74. | Graphium basitruncatum (Matsush.) Seifert & G.Okada + G. penicillioides Corda | Microascales | 0.007 | 2.451 | Saprotrophs, plant and human pathogens |
| 75. | Gaphostroma platystomum (Schwein.) Piroz. | Xylariales | 0.004 | Saprotroph | |
| 76. | Halenospora varia (Anastasiou) E.B.G. Jones + Halenospora sp. | Helotiales | 0.443 | Saprotrophs, aquatic | |
| 77. | Halokirschsteiniothelia maritima (Linder) Boonmee & K.D. Hyde | Mytilinidiales | 0.023 | Saprotroph | |
| 78. | Halosphaeria quadri-remis (Höhnk) Kohlm | Microascales | 0.007 | Saprotroph | |
| 79. | Halosphaeriaceae | Microascales | 0.008 | ||
| 80. | Harzia acremonioides (Harz) Costantin + H. sphaerospora (Matsush.) D.W. Li & N.P. Schultes | Melanosporales | 0.028 | Saprotrophs | |
| 81. | Helicodendron luteoalbum Glen Bott + H. westerdijkiae Beverw | Helotiales | 0.009 | Saprotrophs | |
| 82. | Helicosporium sp. | Tubeufiales | 0.006 | Saprotrophs | |
| 83. | Helotiaceae | Helotiales | 0.005 | ||
| 84. | Helotiales | Helotiales | 3.087 | 4.565 | |
| 85. | Hemibeltrania sp. | Amphisphaeriales | 0.007 | Pathogen | |
| 86. | Herpotrichia pinetorum (Fuckel) G. Winter + Herpotrichia sp. | Pleosporales | 0.183 | 0.002 | Pathogens |
| 87. | Herpotrichiellaceae | Chaetothyriales | 0.004 | ||
| 88. | Hyalodendriella betulae Crous | Helotiales | 0.012 | 0.001 | Saprotroph, pathogen |
| 89. | Hyalopeziza sp. | Helotiales | 0.014 | Saprotroph | |
| 90. | Hyaloscypha bicolor (Hambl. & Sigler) Vohník, Fehrer & Réblová | Helotiales | 0.012 | Endophyte, saprotroph | |
| 91. | Hyaloscyphaceae | Helotiales | 0.003 | 0.040 | |
| 92. | Hymenoscyphus caudatus (P. Karst.) Dennis + H. imberbis (Bull.) Dennis | Helotiales | 0.007 | 0.017 | Pathogens, saprotrophs |
| 93. | Hypocreales | Hypocreales | 2.979 | ||
| 94. | Hypoxylon fragiforme (Pers.) J. Kickx f. | Xylariales | 0.469 | 0.002 | Saprotroph, pathogen |
| 95. | Ilyonectria crassa (Wollenw.) A. Cabral & Crous + I. cyclaminicola A. Cabral & Crous + I. destructans (Zinssm.) Rossman, L. Lombard & Crous + I. europaea A. Cabral, Rego & Crous + I. mors-panacis (A.A. Hildebr.) A. Cabral & Crous + I. robusta (A.A. Hildebr.) A. Cabral & Crous + Ilyonectria sp. + Cylindrocarpon sp. | Hypocreales | 2.031 | 6.710 | Saprotrophs, pathogens |
| 96. | Infundichalara microchona (W. Gams) Réblová & W. Gams + I. minuta Koukol | Helotiales | 0.014 | 0.001 | Saprotrophs, patogens, mycoparasitic |
| 97. | Jattaea taediosa (Sacc.) Réblová & Jaklitsch | Calosphaeriales | 0.005 | Endophyte | |
| 98. | Juxtiphoma eupyrena Sacc. | Pleosporales | 0.001 | Pathogen | |
| 99. | Knufia cryptophialidica L.J. Hutchison & Unter. + K. peltigerae (Fuckel) Réblová & Unter | Incertae sedis | 0.006 | 0.015 | Pathogens, lichenicolous |
| 100. | Lambertella tubulosa Abdullah & J. Webster | Helotiales | 1.445 | Saprotroph | |
| 101. | Lasiosphaeriaceae | Sordariales | 0.095 | 0.005 | |
| 102. | Lecania cyrtella (Ach.) Th. Fr. + L. naegelii (Hepp) Diederich & van den Boom | Lecanorales | 0.001 | 0.034 | Lichenicolous |
| 103. | Lecanorales | Lecanorales | 0.001 | ||
| 104. | Lemonniera terrestris Tubaki | Helotiales | 0.014 | Saprotroph, aquatic | |
| 105. | Leohumicola minima (de Hoog & Grinb.) Seifert & Hambl | Helotiales | 0.002 | Saprotroph | |
| 106. | Leotiomycetes | 0.003 | 0.876 | ||
| 107. | Lepraria caesiella R.C. Harris | Lecanorales | 0.002 | Lichenicolous | |
| 108. | Leptodontidium sp. | Helotiales | 0.011 | 0.254 | Endophyte, mycorrhizal |
| 109. | Leptosphaeria sp. | Pleosporales | 0.023 | Endophytes, saprotrophs, pathogens | |
| 110. | Leptosphaerulina australis McAlpine | Pleosporales | 0.014 | Endophyte | |
| 111. | Lophiostoma corticola (Fuckel) E.C.Y. Liew, Aptroot & K.D. Hyde + Lophiostoma sp. | Pleosporales | 0.788 | Pathogens | |
| 112. | Lophodermium pinastri (Schrad.) Chevall. + L. seditiosum Minter, Staley & Millar + Lophodermium sp. | Rhytismatales | 0.107 | 0.003 | Pathogens |
| 113. | Lophotrichus sp. | Microascales | 0.017 | Patogen, coprophilus, human pathogen | |
| 114. | Macroconia sphaeriae (Fuckel) Gräfenhan & Schroers | Hypocreales | 0.013 | Saprotroph, mycoparasitic | |
| 115. | Magnohelicospora fuscospora (Linder) R.F. Castañeda, Hern.-Restr. & Gené | Incertae sedis | 0.269 | Saprotroph | |
| 116. | Massarina sp. | Pleosporales | 0.002 | Saprotroph | |
| 117. | Megacapitula villosa J.L. Chen & Tzean | Incertae sedis | 0.001 | Saprotroph | |
| 118. | Melanospora kurssanoviana (Beliakova) Czerepan | Melanosporales | 0.009 | Saprotroph, mycoparasitic | |
| 119. | Metarhizium marquandii (Massee) Kepler, S.A. Rehner & Humber | Hypocreales | 0.495 | Endophyte | |
| 120. | Meyerozyma guilliermondii (Wick.) Kurtzman & M. Suzuki | Saccharomycetales | 0.003 | 0.022 | Coprophilous, human pathogen |
| 121. | Micarea adnata Coppins | Lecanorales | 0.006 | Lichenicolous | |
| 122. | Microascaceae | Microascales | 0.002 | ||
| 123. | Microdochium sp. | Amphisphaeriales | 0.063 | 0.001 | Pathogen |
| 124. | Microthecium fimicola (E.C. Hansen) Y. Marín, Stchigel, Guarro & Cano + M. quadrangulare (Dania García, Stchigel & Guarro) Y. Marín, Stchigel, Guarro & Cano | Melanosporales | 0.012 | 0.002 | Saprotrophs |
| 125. | Minutisphaera parafimbriatispora Raja, Oberlies, Shearer & A.N. Mill | Minutisphaerales | 0.017 | Saprotroph, aquatic | |
| 126. | Mollisia sp. | Helotiales | 0.021 | Saprotroph | |
| 127. | Monographella nivalis (Schaffnit) E. Müll | Amphisphaeriales | 0.004 | Pathogen | |
| 128. | Montagnulaceae | Pleosporales | 0.005 | Saprotrophs, endophytes, pathogens | |
| 129. | Mycofalcella calcarata Marvanová, Om-Kalth. & J. Webster | Helotiales | 0.002 | Saprotroph, aquatic | |
| 130. | Myco sphaerella tassiana (De Not.) Johanson | Capnodiales | 0.008 | Pathogen, saprotroph | |
| 131. | Myrmecridium schulzeri (Sacc.) Arzanlou, W. Gams & Crous | Myrmecridiales | 0.010 | Saprotroph | |
| 132. | Naevala perexigua (Roberge ex Desm.) K. Holm & L. Holm | Helotiales | 0.001 | Saprotroph | |
| 133. | Nakazawaea anatomiae (Zwillenb.) Kurtzman & Robnett + N. populi (Hagler, Mend.-Hagler & Phaff) Kurtzman & Robnett | Saccharomycetales | 0.016 | 12.941 | Saprotrophs |
| 134. | Nectria sp. | Hypocreales | 0.032 | Pathogens, saprotrophs | |
| 135. | Nectriaceae | Hypocreales | 0.432 | ||
| 136. | Neoascochytaexitialis (Morini) Qian Chen & L. Cai | Pleosporales | 0.012 | Pathogen | |
| 137. | Neobulgaria premnophila Roll-Hansen & H. Roll-Hansen + N. pura (Pers.) Petr. + Neobulgaria sp. | Helotiales | 0.684 | Saprotrophs | |
| 138. | Neocatenulostroma germanicum (Crous & U. Braun) Quaedvl. & Crous | Capnodiales | 0.001 | Pathogen | |
| 139. | Neocucurbitaria cava (Schulzer) Gruyter, Aveskamp & Verkley | Pleosporales | 0.002 | Saprotroph | |
| 140. | Neofabraea perennans Kienholz | Helotiales | 0.009 | Pathogen | |
| 141. | Neolepto sphaeria rubefaciens (Togliani) Gruyter, Aveskamp & Verkley | Pleosporales | 0.003 | Pathogen | |
| 142. | Neonectria candida (Ehrenb.) Rossman, L. Lombard & Crous + Neonectria sp. | Hypocreales | 0.560 | 0.763 | Pathogen |
| 143. | Neopyrenochaeta acicola ((Moug. & Lév.) Valenz.-Lopez, Crous, Stchigel, Guarro & Cano + N. inflorescentiae (Crous, Marinc. & M.J. Wingf.) Valenz.-Lopez, Crous, Stchigel, Guarro & Cano | Pleosporales | 0.014 | 0.058 | Pathogens, saprotrophs |
| 144. | Neosetophoma clematidis Wijayaw., Camporesi & K.D. Hyde | Pleosporales | 0.046 | Saprotroph | |
| 145. | Neurospora terricola Goch. & Backus | Sordariales | 0.004 | Saprotroph | |
| 146. | Niesslia mucida (W. Gams) W. Gams & Stielow | Hypocreales | 0.004 | Saprotroph | |
| 147. | Nigrograna mycophila Jaklitsch, Friebes & Voglmayr | Pleosporales | 0.007 | Saprotroph, mycoparasitic | |
| 148. | Nigro spora oryzae (Berk. & Broome) Petch | Incertae sedis | 0.535 | Saprotroph, pathogen | |
| 149. | Ochrocladosporium elatum (Harz) Crous & U. Braun | Pleosporales | 0.022 | 0.084 | Endophyte |
| 150. | Oedocephalum nayoroense Ts. Watan | Pezizales | 0.049 | Saprotroph | |
| 151. | Onygenales | Onygenales | 0.005 | ||
| 152. | Ophiostomataceae | Ophiostomatales | 0.790 | Pathogens | |
| 153. | Orbilia auricolor (A. Bloxam) Sacc. | Orbiliales | 0.026 | Saprotroph | |
| 154. | Orbiliaceae | Orbiliales | 0.006 | ||
| 155. | Pachyramichloridium pini (de Hoog & Rahman) C. Nakash., Videira & Crous | Capnodiales | 0.017 | Pathogen | |
| 156. | Papulaspora pisicola J.F.H. Beyma | Incertae sedis | 0.019 | Saprotroph | |
| 157. | Paraphoma chrysanthemicola (Hollós) Gruyter, Aveskamp & Verkley + P. radicina (McAlpine) Morgan-Jones & J.F. White + Paraphoma sp. | Pleosporales | 4.852 | Saprotrophs, pathogens | |
| 158. | Penicillium citreonigrum Dierckx + P. citreosulfuratum Biourge + P. georgiense S.W. Peterson & B.W. Horn + P. glandicola (Oudem.) Seifert & Samson + P. halotolerans Frisvad, Houbraken & Samson + P. lapidosum Raper & Fennell + P. nothofagi Houbraken, Frisvad & Samson + P. raphiae Houbraken, Frisvad & Samson + P. roseomaculatum Biourge + P. sacculum E. Dale + P. unicum Tzean, J.L. Chen & Shiu + P. virgatum Nirenberg & Kwaśna + Penicillium sp. + Talaromyces luteus C.R. Benj. | Eurotiales | 0.295 | 0.001 | Saprotrophs |
| 159. | Periconia sp. | Pleosporales | 0.012 | Endophyte | |
| 160. | Petriella sordida (Zukal) G.L. Barron & J.C. Gilman | Microascales | 0.001 | Coprophilous | |
| 161. | Phacidium lacerum Fr. + Phacidium sp. | Phacidiales | 0.027 | Saprotroph | |
| 162. | Phaeoacremonium cinereum Gramaje, Mohammadi, Banihash., Armengol & L. Mostert + P. hungaricum Essakhi, Mugnai, Surico & Crous | Togniniales | 0.044 | Pathogens | |
| 163. | Phaeoisaria loranthacearum Crous & R.K. Schumach. + P. sparsa B. Sutton | Xylariales | 0.347 | Saprotrophs, coprophilous | |
| 164. | Phaeomoniella sp. | Phaeomoniellales | 0.001 | ||
| 165. | Phaeosphaeria sp. | Pleosporales | 0.007 | Pathogens | |
| 166. | Phaeosphaeriaceae | Pleosporales | 0.013 | ||
| 167. | Phaeosphaeriopsis sp. | Pleosporales | 0.032 | Pathogens, saprotrophs | |
| 168. | Phialocephala sp. | Helotiales | 0.004 | Saprotrophs | |
| 169. | Phialophora sp. | Chaetothyriales | 10.291 | Saprotrophs, pathogens | |
| 170. | Phoma boeremae Gruyter + Phoma sp. | Pleosporales | 0.010 | 0.007 | Saprotrophs, pathogens |
| 171. | Phomopsis phaseoli (Desm.) Sacc. + P. velata (Sacc.) Traverso + Phomopsis sp. | Diaporthales | 1.186 | Pathogens, saprothrophs endophytes | |
| 172. | Physcia tenella (Scop.) DC. | Caliciales | 0.001 | Lichenicolous | |
| 173. | Pilophorus strumaticus Nyl. ex Cromb | Lecanorales | 0.001 | Lichenicolous | |
| 174. | Plagiostoma jonesii Senan. & K.D. Hyde | Diaporthales | 0.031 | Saprotroph, endophyte | |
| 175. | Plectosphaerella cucumerina (Lindf.) W. Gams + P. niemeijerarum L. Lombard | Glomerellales | 0.140 | 0.014 | Pathogens |
| 176. | Pleosporaceae | Pleosporales | 0.003 | ||
| 177. | Pleosporales | Pleosporales | 0.161 | 0.504 | |
| 178. | Pleotrichocladium opacum (Corda) Hern.-Restr., R.F. Castañeda & Gené | Pleosporales | 0.307 | 0.013 | Saprotroph, aquatic |
| 179. | Pleurophoma ossicola Crous, Krawczynski & H.-G. Wagner + Pleurophoma sp. | Xylariales | 0.016 | 0.005 | Saprotroph |
| 180. | Podospora appendiculata (Auersw. ex Niessl) Niessl + P. bulbillosa (W. Gams & Mouch.) X. Wei Wang & Houbraken. + P. leporina (Cain) Cain + Podospora sp. | Sordariales | 0.074 | Saprotroph, coprophilous | |
| 181. | Preussia flanaganii Boylan + P. typharum (Sacc.) Cain | Pleosporales | 0.058 | Saprotrophs, endophytes, coprophilous | |
| 182. | Pseudeurotium hygrophilum (Sogonov, W. Gams, Summerb. & Schroers) Minnis & D.L. Lindner + P. ovale Stolk + P. zonatum J.F.H. Beyma | Thelebolales | 0.804 | Saprotrophs, human pathogens | |
| 183. | Pseudocercospora angolensis (T. Carvalho & O. Mendes) Crous & U. Braun | Mycosphaerellales | 0.004 | Pathogen | |
| 184. | Pseudogymnoascus pannorum (Link) Minnis & D.L. Lindner + P. roseus Raillo | Thelebolales | 0.068 | Saprotrophs | |
| 185. | Pyrenochaeta sp. | Incertae sedis | 0.105 | 0.005 | Pathogen, saprotroph |
| 186. | Pyrenochaetopsis leptospora (Sacc. & Briard) Gruyter, Aveskamp & Verkley + P. microspora (Gruyter & Boerema) Gruyter, Aveskamp & Verkley | Pleosporales | 0.007 | 0.001 | Pathogens, saprotrophs, endophytes |
| 187. | Pyronemataceae | Pezizales | 0.081 | ||
| 188. | Saccharomyces cerevisiae (Desm.) Meyen | Saccharomycetales | 0.001 | Saprotroph | |
| 189. | Schizothecium glutinans (Cain) N. Lundq | Sordariales | 0.015 | Saprotroph, coprophilous | |
| 190. | Scolecobasidium constrictum E.V. Abbott + S. umbrinum (Ach.) Arnold | Incertae sedis | 0.016 | 0.002 | Saprotrophs, endophytes |
| 191. | Scutellinia scutellata (L.) Lambotte | Pezizales | 0.005 | Saprotroph | |
| 192. | Scytalidium lignicola Pesante + S. multiseptatum Hol.-Jech | Helotiales | 0.055 | 0.001 | Pathogens, saprotrophs, mycoparasitic |
| 193. | Sordariales | 0.008 | |||
| 194. | Sordariomycetes | 0.211 | 0.003 | ||
| 195. | Sphaeropsis sapinea (Fr.) Dyko & B. Sutton | Botryosphaeriales | 0.003 | Pathogen | |
| 196. | Sporormiaceae | Pleosporales | 0.003 | ||
| 197. | Sporothrix dentifunda Aghayeva & M.J. Wingf. + S. stenoceras (Robak) Z.W. de Beer, T.A. Duong & M.J. Wingf. + S. narcissi (Limber) Z.W. de Beer, T.A. Duong & M.J. Wingf | Ophiostomatales | 0.161 | 0.001 | Pathogens, saprotrophs |
| 198. | Stemphylium herbarum E.G. Simmons + S. majusculum E.G. Simmons + S. vesicarium (Wallr.) E.G. Simmons | Pleosporales | 0.027 | Pathogens | |
| 199. | Subramaniula flavipila X. Wei Wang & Samson | Sordariales | 0.014 | Saprotroph | |
| 200. | Sydowia polyspora (Bref. & Tavel) E. Müll | Dothideales | 0.004 | 1.028 | Pathogen, endophyte, saprotroph |
| 201. | Tetracladium furcatum Descals + T. setigerum (Grove) Ingold + Tetracladium sp. | Helotiales | 1.171 | 0.862 | Saprotrophs |
| 202. | Thelonectria blackeriella + T. olida (Wollenw.) Wollenw. + T. nodosa Salgado & P. Chaverri | Hypocreales | 0.012 | 0.006 | Pathogens |
| 203. | Tricharina sp. | Pezizales | 1.55 | Saprotrophs | |
| 204. | Trichocladium asperum Harz + T. griseum (Traaen) X. Wei Wang & Houbraken | Sordariales | 0.593 | Saprotrophs | |
| 205. | Trichoderma aerugineum Jaklitsch + T. hamatum (Bonord.) Bainier + T. koningiopsis Samuels, Carm. Suárez & H.C. Evans + T. martiale Samuels + T. neokoningii Samuels & Soberanis + T. piluliferum J. Webster & Rifai + T. Polysporum (Link) Rifai + T. pubescens Bissett + T. stilbohypoxyli Samuels & Schroers + T. viride Pers. + Trichoderma sp. | Hypocreales | 19.464 | 0.001 | Saprotrophs |
| 206. | Tricladium splendens Ingold | Helotiales | 0.040 | 0.057 | Saprotroph, acquatic |
| 207. | Truncatella an gustata (Pers.) S. Hughes + T. restionacearum S.J. Lee & Crous | Amphisphaeriales | 0.003 | 0.001 | Pathogens |
| 208. | Valsa malicola Z. Urb. + V. sordida Sacc. + V. leucostoma (Pers.) Fr. | Diaporthales | 0.012 | 0.214 | Pathogens |
| 209. | Valsaceae | Diaporthales | 0.003 | ||
| 210. | Venturia hystrioides (Dugan, R.G. Roberts & Hanlin) Crous & U. Braun | Venturiales | 0.018 | Pathogen | |
| 211. | Venturiaceae sp. | Venturiales | 0.001 | ||
| 212. | Verticillium dahliae Kleb. + V. longisporum (C. Stark) Karapapa, Bainbr. & Heale | Glomerellales | 0.029 | Pathogens, saprotrophs | |
| 213. | Volutella ciliata (Alb. & Schwein.) Fr. + Volutella sp. | Hypocreales | 0.009 | 0.009 | Saprotrophs, pathogen |
| 214. | Xanthoparmelia subchalybaeizans (Hale) G. Amo, A. Crespo, Elix & Lumbsch | Lecanorales | 0.005 | Lichenicolous | |
| 215. | Xenochalara sp. | Helotiales | 0.033 | Saprotroph | |
| 216. | Xenopolyscytalum pinea Crous + Xenopolyscytalum sp. | Helotiales | 0.001 | 0.001 | Saprotrophs |
| 217. | Xenoramularia arxii Videira, Crous & U. Braun | Capnodiales | 0.001 | Pathogen | |
| 218. | Xylariales | Xylariales | 0.061 | ||
| 219. | Yamadazyma mexicana (M. Miranda, Holzschu, Phaff & Starmer) Billon-Grand | Saccharomycetales | 0.039 | Saprotroph | |
| 220. | Yarrowia lipolytica (Wick., Kurtzman & Herman) Van der Walt & Arx | Saccharomycetales | 0.001 | Saprotroph | |
| 221. | Zalerion sp. | Lulworthiales | 0.001 | Saprotroph, aquatic | |
| 222. | Zopfiella marina Furuya & Udagawa + Z. pilifera Udagawa & Furuya | Sordariales | 0.027 | Saprotrophs, aquatic | |
| Frequency of Ascomycota | 45.299 | 68.697 | |||
| Number of taxa Ascomycota | 263 | 178 | |||
| Basidiomycota | |||||
| 1. | Aecidium sp. | Pucciniales | 0.034 | Pathogen | |
| 2. | Agaricales | 0.054 | |||
| 3. | Agaricomycetes | 0.008 | 0.074 | ||
| 4. | Agaricostilbomycetes | 0.001 | |||
| 5. | Apiotrichum dulcitum (Berkhout) Yurkov & Boekhout + A. gracile (Weigmann & A. Wolff) Yurkov & Boekhout | Trichosporonales | 0.047 | Saprotrophs | |
| 6. | Armillaria mellea (Vahl) P. Kumm | Agaricales | 0.025 | Pathogen | |
| 7. | Athelia acrospora Jülich | Atheliales | 0.001 | Saprotroph | |
| 8. | Atheliaceae | Atheliales | 0.023 | ||
| 9. | Aurantiporus fissilis (Berk. & M.A. Curtis) H. Jahn ex Ryvarden | Polyporales | 0.002 | Saprotroph, pathogen | |
| 10. | Auriculariales | 0.004 | |||
| 11. | Basidiomycota | 0.031 | 0.038 | ||
| 12. | Bensingtonia sp. | Agaricostilbales | 0.001 | Saprotroph | |
| 13. | Bjerkandera adusta (Willd.) P. Karst | Polyporales | 0.002 | Saprotroph, pathogen | |
| 14. | Buckleyzyma aurantiaca (Saito) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout | Buckleyzymales | 0.048 | 0.007 | Saprotroph |
| 15. | Bullera crocea Buhagiar | Tremellales | 0.008 | 0.001 | Saprotroph |
| 16. | Bulleromyces albus Boekhout & Á. Fonseca | Tremellales | 0.001 | 0.001 | Saprotroph |
| 17. | Burgoa anomala (Hotson) Goid | Cantharellales | 0.009 | Saprotroph | |
| 18. | Camarophyllus sp. | Agaricales | 0.001 | Mycorrhizal | |
| 19. | Cantharellales | 0.002 | |||
| 20. | Chondrostereum purpureum (Pers.) Pouzar | Agaricales | 0.018 | Pathogen, saprotroph | |
| 21. | Coprinellus disseminatus (Pers.) J.E. Lange | Agaricales | 0.230 | Saprotroph | |
| 22. | Cryptococcus tephrensis Vishniac + Cryptococcus sp. | Tremellales | 0.220 | 0.406 | Saprotrophs, endophytes |
| 23. | Curvibasidium pallidicorallinum Golubev, Fell & N.W. Golubev | Incertae sedis | 0.001 | Mycocinogenic | |
| 24. | Cystobasidiomycetes | 0.003 | |||
| 25. | Cystobasidium pinicola (F.Y. Bai, L.D. Guo & J.H. Zhao) Yurkov, Kachalkin, H.M. Daniel, M. Groenew., Libkind, V. de Garcia, Zalar, Gouliam., Boekhout & Begerow + C. psychroaquaticum A.M. Yurkov, Kachalkin, H.M. Daniel, M. Groenew., Libkind, V. de Garcia, Zalar, Gouliamova, Boekhout & Begerow | Cystobasidiales | 0.002 | 0.016 | Saprotrophs, mycoparasitic |
| 26. | Cystofilobasidiales | Cystofilobasidiales | 0.004 | 0.001 | |
| 27. | Cystofilobasidium infirmominiatum (Fell, I.L. Hunter & Tallman) Hamam., Sugiy. & Komag. + C. macerans J.P. Samp. | Cystofilobasidiales | 0.012 | 0.001 | Saprotrophs, acquatic |
| 28. | Daedaleopsis confragosa (Bolton) J. Schröt | Polyporales | 0.001 | Saprotroph | |
| 29. | Efibulobasidium sp. | Sebacinales | 0.020 | Mycorrhizal | |
| 30. | Entyloma gaillardianum Vánky + E. polysporum (Peck) Farl. | Entylomatales | 0.044 | Pathogens | |
| 31. | Erythrobasidiales | Erythrobasidiales | 0.001 | 0.001 | |
| 32. | Erythrobasidium hasegawae (Y. Yamada & Komag.) Hamam., Sugiy. & Komag | Erythrobasidiales | 0.008 | Saprotroph | |
| 33. | Exidiopsis sp. | Auriculariales | 0.001 | Saprotroph | |
| 34. | Exobasidium arescens Nannf. + Exobasidium sp. | Exobasidiales | 0.001 | 0.001 | Pathogen |
| 35. | Fellomyces sp. | Tremellales | 0.001 | Saprotroph | |
| 36. | Fellozyma inositophila (Nakase & M. Suzuki) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout | Incertae sedis | 0.007 | Saprotroph | |
| 37. | Fibulobasidium inconspicuum Bandoni | Tremellales | 0.004 | 0.379 | Saprotroph |
| 38. | Filobasidium wieringae (Á. Fonseca, Scorzetti & Fell) Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout | Filobasidiales | 0.008 | Saprotroph | |
| 39. | Fomitopsis pinicola (Sw.) P. Karst | Polyporales | 0.005 | Pathogen, saprotroph | |
| 40. | Geotrichopsis mycoparasitica Tzean & Estey | Incertae sedis | 0.033 | Mycoparasitic | |
| 41. | Gymnopus androsaceus (L.) Della Magg. & Trassin | Agaricales | 0.001 | Saprotroph, mycoparasitic | |
| 42. | Hannaella zeae (O. Molnár & Prillinger) F.Y. Bai & Q.M. Wang | Tremellales | 0.047 | Saprotroph, endophyte | |
| 43. | Hebeloma mesophaeum (Pers.) Quél | Agaricales | 0.007 | Mycorrhizal | |
| 44. | Hydnaceae | Cantharellales | 0.004 | ||
| 45. | Hygrophoraceae | Agaricales | 0.008 | ||
| 46. | Hymenogaster arenarius Tul. & C. Tul. | Agaricales | 0.005 | Ectomycorrhizal | |
| 47. | Hyphodontia pallidula (Bres.) J. Erikss | Hymenochaetales | 0.003 | Saprotroph | |
| 48. | Hypochnicium lundellii (Bourdot) J. Erikss | Polyporales | 0.012 | Saprotroph | |
| 49. | Inocybe curvipes P. Karst | Agaricales | 0.043 | Ectomycorrhizal | |
| 50. | Itersonilia perplexans Derx | Cystofilobasidiales | 0.001 | Pathogen | |
| 51. | Kockovaella machilophila Cañ.-Gib., M. Takash., Sugita & Nakase | Tremellales | 0.001 | ||
| 52. | Kondoa yuccicola (Nakase & M. Suzuki) Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout | Agaricostilbales | 0.012 | Saprotroph | |
| 53. | Kwoniella newhampshirensis K. Sylvester, Q.M. Wang & Hittinger + K. pini (Golubev & I. Pfeiff.) Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout | Tremellales | 0.016 | 0.003 | Entomopathogenic |
| 54. | Laccaria sp. | Agaricales | 0.001 | Ectomycorrhizal | |
| 55. | Lachnella alboviolascens (Alb. & Schwein.) Fr. | Agaricales | 0.007 | Saprotroph | |
| 56. | Leptosporomyces galzinii (Bourdot) Jülich | Atheliales | 0.054 | Saprotroph | |
| 57. | Leucosporidiales | Leucosporidiales | 0.007 | ||
| 58. | Malassezia globosa Midgley, E. Guého & J. Guillot + M. restricta E. Guého, J. Guillot & Midgley + | Malasseziales | 0.016 | 0.001 | Human pathogens |
| 59. | Marasmius cohaerens (Pers.) Cooke & Quél | Agaricales | 0.008 | Saprotroph | |
| 60. | Microbotryomycetes | 0.042 | |||
| 61. | Minimedusa polyspora (Hotson) Weresub & P.M. LeClair | Cantharellales | 0.069 | Saprotroph, mycoparasitic | |
| 62. | Mrakia frigida (Fell, Statzell, I.L. Hunter & Phaff) Y. Yamada & Komag. + Mrakia sp. | Cystofilobasidiales | 0.012 | 0.001 | Saprotroph |
| 63. | Mycena aurantiomarginata (Fr.) Quél. + M. galericulata (Scop.) Gray | Agaricales | 0.003 | 0.001 | Saprotroph |
| 64. | Naganishia cerealis (Passoth, A.-C. Andersson, Olstorpe, Theelen, Boekhout & Schnürer) Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout + N. diffluens (Zach) Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout | Tremellales | 0.021 | 0.001 | Saprotroph |
| 65. | Oberwinklerozyma silvestris Golubev & Scorzetti ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout | Incertae sedis | 0.012 | ||
| 66. | Oliveonia sp. | Auriculariales | 0.008 | Saprotroph | |
| 67. | Peniophora sp. | Russulales | 0.593 | Pathogen, saprotroph | |
| 68. | Phaeotremella frondosa (Fr.) Spirin & V. Malysheva + P. roseotincta (Lloyd) V. Malysheva | Tremellales | 0.001 | 0.123 | Saprotrophs, mycoparasites |
| 69. | Phloeomana speirea (Fr.) Redhead | Agaricales | 0.024 | Saprotroph, aquatic | |
| 70. | Piskurozyma sp. | Filobasidiales | 0.024 | Saprotroph | |
| 71. | Psathyrella squamosa (P. Karst.) A.H. Sm. | Agaricales | 0.004 | Saprotroph | |
| 72. | Rhodotorula glutinis (Fresen.) F.C. Harrison + Rhodotorula sp. | Sporidiobolales | 0.003 | 0.001 | Saprotrophs |
| 73. | Saitozyma podzolica (Babeva & Reshetova) Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout | Tremellales | 0.001 | Saprotroph | |
| 74. | Sakaguchia lamellibrachiae (Nagah., Hamam., Nakase & Horikoshi) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout | Sakaguchiales | 0.027 | Saprotroph | |
| 75. | Sebacinales | Sebacinales | 0.392 | 0.001 | |
| 76. | Serendipita vermifera Oberw | Sebacinales | 0.017 | Endophyte, mycorrhizal | |
| 77. | Serpula himantioides (Fr.) P. Karst | Boletales | 0.001 | Saprotroph, pathogen | |
| 78. | Sirotrema translucens (H.D. Gordon) Bandoni | Tremellales | 0.001 | Saprotroph | |
| 79. | Sistotremastrum sp. | Trechisporales | 0.001 | Saprotroph | |
| 80. | Slooffia pilatii (F.H. Jacob, Faure-Reayn. & Berton) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout | Incertae sedis | 0.001 | Saprotroph | |
| 81. | Solicoccozyma fuscescens (Golubev) Yurkov + S. phenolica (Á. Fonseca, Scorzetti & Fell) A.M. Yurkov + S. terrea (Di Menna) A.M. Yurkov + S. terricola (T.A. Pedersen) Yurkov | Filobasidiales | 2.451 | 0.004 | Saprotrophs |
| 82. | Sporobolomyces roseus Kluyver & C.B. Niel + Sporobolomyces sp. | 0.008 | 0.001 | ||
| 83. | Stilbum sp. | Agaricostilbales | 0.018 | Saprotroph | |
| 84. | Symmetrospora coprosmae (Hamam. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout + S. gracilis (Derx) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout | Incertae sedis | 0.005 | 0.001 | Saprotrophs |
| 85. | Tausonia pullulans (Lindner) Xin Zhan Liu, F.Y. Bai, J.Z. Groenew. & Boekhout | Cystofilobasidiales | 0.094 | 0.012 | Saprotrophs |
| 86. | Thelephoraceae | Thelephorales | 0.058 | Pathogens | |
| 87. | Tomentella sp. | Thelephorales | 0.001 | Ectomycorrhizal | |
| 88. | Tremella encephala Pers. | Tremellales | 0.003 | Saprotroph | |
| 89. | Tremellales | 0.014 | 0.001 | Saprotrophs | |
| 90. | Tremellomycetes | 0.003 | |||
| 91. | Tricholomataceae | Agaricales | 0.004 | ||
| 92. | Trichosporon otae Sugita, Takshima & Kikuchi | Trichosporonales | 0.003 | Human pathogen | |
| 93. | Tulasnellaceae | Cantharellales | 0.005 | ||
| 94. | Typhula incarnata Lasch | Agaricales | 0.004 | Pathogen | |
| 95. | Pappia fissilis (Berk. & M.A. Curtis) Zmitr | Polyporales | 0.004 | Saprotroph | |
| 96. | Vishniacozyma carnescens (Verona & Luchetti) Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout + V. globispora (B.N. Johri & Bandoni) Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout + V. victoriae (M.J. Montes, Belloch, Galiana, M.D. García, C. Andrés, S. Ferrer, Torr.-Rodr. & J. Guinea) Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout | Tremellales | 0.007 | 0.005 | Pathogens, saprotrophs |
| Frequency Basidiomycota | 4.119 | 2.076 | |||
| Number of Basidiomycota taxa | 81 | 59 | |||
| Frequency | |||||
| Oomycota | 1.199 | 0.002 | |||
| Culturable fungi | 53.062 | 70.780 | |||
| Non-culturable fungi | 25.645 | 17.435 | |||
| Other Kingdoms | 15.822 | 11.728 | |||
| No sequence in NCBI database | 4.272 | 0.055 | |||
| Number | |||||
| Total OTUs | 69,467 a | 70,218 a | |||
| Culturable fungal OTUs | 44,506 a | 53,592 a | |||
| Taxa | 474 a | 309 a | |||
| Fungal taxa | 364 a | 242 a | |||
| Margalef’s diversity index–DMg | 65.54 | 21.72 | |||
| Shannon’s diveristy index–H | 2.55 | 0.77 | |||
| Simpson’s diversity index–D | 0.21 | 0.74 | |||
| Shannon’s evenness index–E | 0.39 | 0.17 | |||
| Berger-Parker’s dominance index–d | 0.20 | 0.46 | |||

| |||||
Percentage of variation. Pathogens are in bold. a Indicates a statistically significant difference according to a χ2-test, p < 0.001.
Figure 3.
Frequency of the fungi in taxonomic orders.
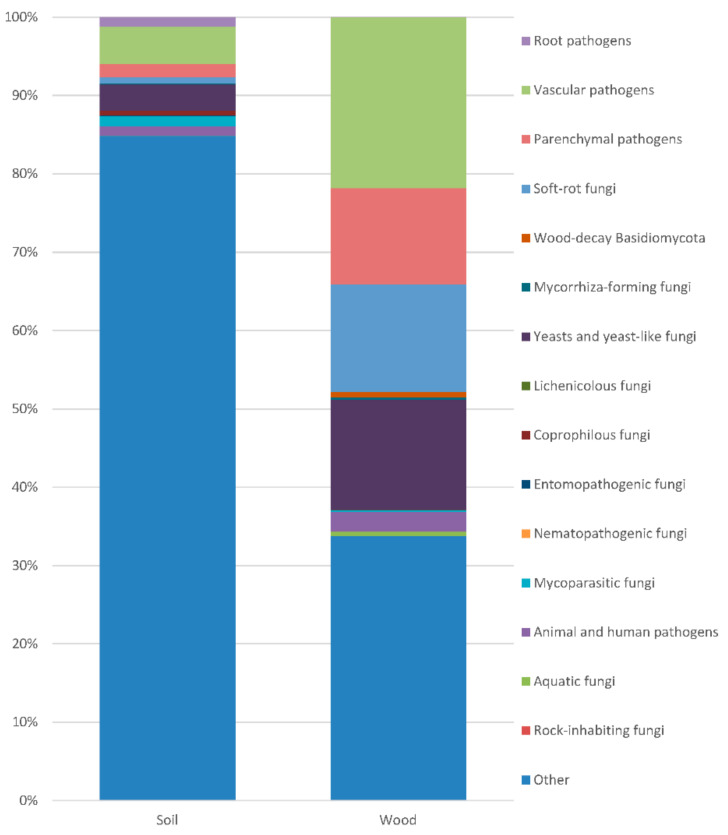
Saprotrophs were the most abundant (Figure 4). In the soil, their frequency exceeded 80%. In the soil, the most common (with frequency > 0.1%) were species of Mortierella (Zygomycota), Alatospora, Clonostachys, Dendryphion, Emericellopsis, Exophiala, Halenospora, Lambertella, Leptodontidium, Magnohelicospora, Metarhizium, Neobulgaria, Nigrospora, Penicillium, Petriella, Pleotrichocladium, Pseudeurotium, Tetracladium, Tricharina and Trichoderma (Ascomycota), Coprinellus, Cryptococcus, Fibulobasidium, Phaeotremella and Solicoccozyma (Basidiomycota).
Figure 4.
Frequency of the fungi in specific trophic groups.
Individual taxa of obligate or facultative phytopathogens were more or less frequent.
The root pathogens included species of Aphanomyces, Globisporangium, Phytophthora and Pythium (Oomycota: 1.17%), and Truncatella (Ascomycota: 0.003% in the soil, 0. 001% in the wood).
Vascular pathogens included species of Cadophora, Dactylonectria, Debaryomyces, Fusarium, Fusicolla, Graphium, Hymenoscyphus, Ilyonectria, Microdochium, Neonectria, Ophiostomataceae, Phaeoacremonium, Phaeomoniella, Phialophora, Sporothrix, Thelonectria and Verticillium (Ascomycota: 4.783% in soil, 21.831% in the wood).
The parenchymal pathogens included species of Alternaria, Boeremia, Cladosporium, Coniochaeta, Cosmospora, Cytospora, Diaporthe, Didymella, Epicoccum, Herpotrichia, Hypoxylon, Lophiostoma, Mycosphaerella, Neoascochyta, Neocatenulostroma, Neofabraea, Neoleptosphaeria, Neopyrenochaeta, Paraphoma, Phaeoisaria, Phaeosphaeria, Phaeosphaeriopsis, Phoma, Phomopsis, Plectosphaerella, Pseudocercospora, Pyrenochaeta, Pyrenochaetopsis, Scytalidium, Sphaeropsis, Stemphylium, Sydowia, Valsa, Volutella and Xenoramularia (Ascomcota: 1.647% in the soil, 11.645% in the wood), and Armillaria, Aurantiporus, Chondrostereum, Fomitopsis, Peniophora and Serpula (Basidiomycota: 0.026% in the soil, 0.618% in the wood).
The soft-rot fungi included species of Alatospora, Alternaria, Cadophora, Chaetomium, Cladosporium, Clonostachys, Exophiala, Halenospora, Leptodontidium, Neosetophoma, Orbilia, Phialophora, Plagiostoma, Sydowia and Tricladium (Ascomycota: 0.821% in the soil, 13.757% in the wood).
The wood-decay Basidiomycota included the white rot fungi Armillaria mellea, Aurantiporus fissilis, Bjerkandera adusta, Chondrostereum purpureum, Hyphodontia pallidula and Peniophora, and the brown rot fungus Fomitopsis piniola. They occurred with frequencies of 0.028% in the soil and 0.62% in the wood.
The mycorrhiza-forming fungi present in the soil and wood included 12 taxa: arbuscular Entrophospora (Glomeromycota: 0.001% in the wood); ectomycorrhizal Cenococcum geophilum (Ascomycota; 0.039% in the soil), Hymenogaster arenarius, Inocybe curvipes, Laccaria sp., Serendipita vermifera and Tomentella (Basidiomycota: 0.048% in the soil, 0.019% in the wood); ectendomycorrhizal Chloridium paucisporum and Leptodontidium sp. (Ascomycota), and Camarophyllus sp., Efibulobasidium sp. and Hebeloma mesophaeum (Basidiomycota: 0.039% in the soil, 0.254% in the wood).
The yeasts and yeast-like fungi present in the soil and wood included 52 taxa: Aureobasidium melanogenum, Blastobotrys spp., Candida spp., Capnobotryella renispora, Cladophialophora spp., Cyphellophora sessilis, Debaryomyces hansenii, Exophiala spp., Meyerozyma guilliermondii, Micarea agnata, Nakazawaea spp., Saccharomyces cerevisiae, Yamadazyma mexicana, Yarrowia lipolytica and Xanthoparmelia subchalybaeizans (Ascomycota: 0.296% in the soil, 13.072% in the wood); Apiotrichum dulcitum, Bensingtonia spp., Buckleyzyma aurantiaca, Bullera croce, Bulleromyces albus, Cryptococcus spp., Curvibasidium pallidicorallinum, Cystobasidium spp., Cystofilobasidium spp., Erythrobasidium hasegawianum, Fellomyces spp., Fellozyma inositophila, Fibulobasidium inconspicuum, Filobasidium wieringae, Hannaella zeae, Itersonilia perplexans, Kockovaella machilophila, Kondoa yuccicola, Kwoniella newhampshirensis, Malassezia spp., Mrakia frigida, Naganishia cerealis, Phaeotremella spp., Piskurozyma sp., Rhodotorula spp., Saitozyma podzolica, Sakaguchia lamellibrachiae, Sirotrema translucens, Slooffia pilatii, Solicoccozyma spp., Sporobolomyces spp., Symmetrospora coprosmae, Tausonia pullulans, Tremella encephala, Trichosporon otae and Vishniacozyma carnescens (Basidiomycota: 3.061% in the soil, 1.017% in the wood).
The lichenicolous fungi present in the soil and wood included eight taxa: Bacidina sp., Knufia peltigerae, Lecania cyrtella, Lepraria caesiella, Micarea agnata, Physcia tenella, Pilophorus strumaticusa and Xanthoparmelia subchalybaeizans (Ascomycota: 0.02% in the soil, 0.068% in the wood).
The coprophilous fungi present in the soil and wood included 10 taxa: Ascobolus sp., Cercophora sp. Coniochaeta sp., Lophotrichus sp., Meyerozyma guilliermondii, Petriella sordida, Phaeoisaria, Podospora appendiculata (forest specific), Preussia spp. and Schizothecium glutinans (Ascomycota: 0.548% in the soil, 0.002% in the wood). The entomopathogenic fungi present in the soil and wood included three taxa: Beauveria bassiana and Cordyceps spp. (Ascomycota: 0.096% in the soil, 0.023% in the wood), and Kwoniella spp. (Basidiomycota: 0.016% in the soil, 0.003% in the wood).
The nematopathogenic fungi included one species, Myzocytiopsis sp. (Oomycota: 0.005% in the soil).
The mycoparasitic fungi present in the soil and wood included 18 taxa: Syncephalis sp. (Zygomycota: 0.107% in the soil), Angustimassarina spp., Cladosporium spp., Clonostachys spp., Coniochaeta sp., Cordyceps spp., Cosmospora sp., Dissoconium eucalypti, Infundichalara microchona, Macroconia sphaeriae, Melanospora kurssanoviana, Nigrograna mycophila and Scytalidium lignicola (Ascomycota: 1.063% in the soil, 0.056% in the wood), Cystobasidium spp., Geotrichopsis mycoparasitica, Gymnopus androsaceus, Minimedusa polyspora and Phaeotremella frondosa (Basidiomycota: 0.16% in the soil, 0.139% in the wood).
The animal and human pathogens included Coniochaeta, Exophiala, Graphium spp., Lophotrichus sp., Meyerozyma guilliermondii and Pseudeurotium ovale (Ascomycota: 0.975% in the soil, 2.504% in the wood), and Malassezia spp. (Basidiomycota: 0.16% in the soil, 0.001% in the wood).
The aquatic fungi present in the soil and wood included 11 taxa: Aureobasidium melanogenum, Halenospora spp., Lemonniera terrestris, Minutisphaera parafimbriatispora, Mycofalcella calcarata, Pleotrichocladium opacum, Tricladium splendens, Zalerion sp. and Zopfiella spp. (Ascomycota: 0.041% in the soil, 0.527% in the wood), Cystofilobasidium spp. and Phloeomana speirea (Basidiomycota: 0.012% in the soil, 0.025% in the wood).
The rock-inhabiting fungi included one taxon, Capnobotryella renispora (Ascomycota: 0.005% in the soil).
The individual fungi often belonged to more than one trophic group.
Margalef’s index (DMg), Shannon’s diversity index (H) and Simpson’s diversity index (D) indicated greater diversity in the soil than in the wood. Shannon’s evenness index (E) showed more evenness in the soil and, conversely, Berger-Parker’s dominance index (d) showed more dominance of individual taxa in the wood.
4. Discussion
4.1. Disease Characteristics
The vascular wilt of hybrid poplar appeared locally in Poland in 2017. The symptoms appeared suddenly in 5–6-year-old trees, and the disease developed very quickly, in less than 2 months. The activity of the pathogens, either already known or previously unrecognized, apparently circumvented any resistance in the host and led to the failure of the plantations. The disease was asymptomatic in its initial stage. Diagnosis at the final stage was not possible because of either: (i) the immaturity of the pathogen, or (ii) the absence of the distinctive morphological elements essential for the identification of causal fungi. Poplar diseases have a serious economic impact on wood production worldwide, and so the development of effective management strategies depends on the clear identification of the pathogens involved. The affected tissues were therefore analyzed by DNA sequencing.
The symptomatology of poplar wilt can be compared with that of some grapevine diseases, notably grapevine trunk diseases (GTD), including the esca and black foot diseases, and Petri disease [14,15]. Grapevine trunk disease symptoms include the sectorial and/or central necrosis of the trunk wood, brown streaking of the wood, cankers, and the discoloration and wilting of the foliage, which can occur suddenly [15,16]. Petri disease is a vascular disease associated with the decline and dieback of young grapevines. Typical black foot disease symptoms include stunted growth, reduced vigour, retarded or absent sprouting, sparse and chlorotic foliage with necrotic margins, wilting, dieback and death. Characteristic sunken necrotic root lesions with a reduction in root biomass and root hairs may also occur.
Grapevine trunk disease is caused by fungi in the Botryosphaeriaceae [17,18], Phomopsis viticola [17,19], Eutypa lata [20] and Truncatella [21]. Petri disease and esca are caused by six species of Cadophora, including C. luteo-olivacea, 29 species of Phaeoacremonium (particularly P. cinereum), Phaeomoniella chlamydospora (Gams, Crous, Wingf. and Mugnai) Crous and Gams, Pleurostoma richardsiae (Nannf.) Réblová and Jaklitsch (=Phialophora richardsiae (Nannf.) Conant), and basidiomycetous Fomitiporia mediterranea (Fisch.) and Stereum hirsutum (Willd.) Pers. [15,22,23,24,25]. Black foot disease is caused by species of Campylocarpon, Cylindrocladiella, Dactylonectria, Ilyonectria, Neonectria and Thelonectria [26]. The fungal species associated with grapevine diseases, mentioned above, have also been reported from a broad range of woody and herbaceous host plants [23,27,28,29,30]. In Italy, Cadophora, Coniochaeta (in its Lecythophora anamorphic stage) and Phaeoacremonium have been isolated from the wood of kiwifruit plants suffering from elephantiasis, which had trunk necrosis, hypertrophy and longitudinal bark cracks [31].
4.2. Pathogens in Diseased Poplar Trunk
According to EN 350:2016, poplar wood is non-durable, and some studies have shown that it is highly susceptible to wood-rotting fungi [32,33].
The dominant taxonomic group of poplar-associated fungi was Ascomycota. Those fungi are often cosmopolitan species known from the above- and below-ground parts of Populus species. Many species found in the wood of diseased trees are, however, known from diseased grapevine: Botryosphaeriaceae, C. luteo-olivacea, Dactylonectria spp., Ilyonectria spp., Neonectria spp., P. cinereum, Phaeomoniella spp., Phialophora spp., Phomopsis spp., Thelonectria spp. and Truncatella spp. Other vascular and parenchymal fungi, frequently necrotrophic species, were also found: Angustimassarina, Aureobasidium, Boeremia, Chaetomium, Chaetosphaeria, Cyathicula, Cudoniella, Dendryphion, Didymella, Fusarium, Graphium, Helicodendron, Helicosporium, Hymenoscyphus, Hypoxylon, Knufia, Leptodontidium, Leptosphaeria, Lophiostoma, Massarina, Megacapitula, Mollisia, Neocatenulostroma, Neoleptosphaeria, Neosetophoma, Niesslia, Ophiostomatacea (with its anamorphs), Phoma, Plagiostoma, Pleurophoma, Podospora, Pyrenochaeta, Scutellinia, Scytalidium, Sporothrix, Tricharina, Xenopolyscytalum, Verticillium, and basidiomycetous Burgoa. These fungi were also often in the surrounding soil. Some of them seem likely to have contributed to the disease-causing species complex. The fungi associated with the diseased poplars, and which had been found previously in the wood of poplar or other deciduous trees, included: Angustimassarina on the wood of grapevine and poplar [34], Chaetosphaeria on the necrotic wood of Prunus [35], Graphium penicillioides in a wood core of Populus nigra in the Czech Republic 200 years ago [36], Graphostroma platystomum on the bark of oak [37], Helicodendron luteoalbum on poplar roots [38], Helicosporium on a wilted chestnut tree [39], and Hymenoscyphus caudatus on the rotten leaves of Populus nigra [40]. The last species is related to Hymenoscyphus fraxineus (T. Kowalski) Baral, Queloz and Hosoya, which causes a very destructive wilt disease of ash, ash dieback—with similar trunk symptoms to those observed in the hybrid poplar [41,42]. Infundichalara microchona occurred in conifers [43,44]; Knufia in black galls on the stems and branches of Populus tremuloides Michx. in Canada [45]; Leptodontidium on the roots of healthy Populus deltoides [46]; Lophiostoma corticola on the above-ground organs of dying oaks in Poland [47]; Megacapitula on fallen, decaying petioles of broad-leaves trees [48]. Mollisia occurred on decaying plant tissues throughout the Northern Hemisphere; Neocatenulostroma germanicum in oak-wood debris [49]; Neoleptosphaeria rubefaciens occurred on the wood, bark and fruits of herbaceous or woody plants in terrestrial habitats [50,51,52]. Neosetophoma clematidis occurred on the branches of Clematis vitalba L. [53] and Niesslia mucida on the bark of diverse plants, especially conifers [54]. Ophiostomataceae have been associated with wounds on hardwood trees in Poland [55]. Phaeoacremonium species occurred on European olive, quince and willow [27]; Phialocephala on rotten deciduous wood [56]; Phoma on the decaying wood of oak and pine [57]; Plagiostoma in the stems, twigs, and branches of woody and herbaceous plants from a wide range of plants in temperate regions of the Northern Hemisphere [58,59]. Pleurophoma ossicola occurred in Scots pine [60], and Pyrenochaeta occurred in oak [57]. Scytalidium lignicola causes diseases in Citrus and Manihot [58,61,62]. Sporothrix occurred in eucalyptus, pine and rosebush [63], and Xenopolyscytalum pinea in pine stumps [64].
Basidiomycetous Burgoa anomala was found in pine wood and litter [65].
Some of the fungi are, surprisingly, often common on wood in water, including sea water. This group includes Didymosphaeria futilis, Halenospora varia, Halosphaeria quadriemis, Paraphoma radicina, Trichocladium and basidiomycetous Cystobasidium [66,67,68,69,70,71,72]. Fusarium spp. were not abundant in the poplar wood, but occurred frequently in the soil. Various Fusarium spp. have been reported in Poland as causing swellings, necrosis, bark fray, reddish-purple discoloration, and ultimately the characteristic cankers in poplar [73]. Fusarium avenaceum is perhaps the most important species, first reported in the 1950s on Euramerican poplar clones in France. Since then it has spread in Europe, from central and eastern areas with a continental climate to sub-mediterranean areas, and recently to Portugal, with its oceanic climate. Neocosmospora solani (=Fusarium solani (Mart.) Sacc. (found mostly on Aigeiros and Tacamahaca poplars and intersectional hybrids) seemed to be confined to North America until it was reported in Poland [74]. Species with sporadic occurrence and of limited importance include F. lateritium Nees, observed in France and in the USA on Populus trichocarpa Torr. and A. Gray, and F. sporotrichioides Sherb., observed in eastern Europe and central Italy on Populus × euramericana. Fusarium spp., constituting a threat to young trees. Colonized trunks are susceptible to breakage, and to attacks by other bark parasites which are also active during a plantation’s early years. The symptoms are not immediately visible, and mostly take the form of the disorganization of the cortical tissues in part of the trunk.
Fungi which are more frequent and perhaps more significant than Fusarium spp. in diseased poplar wood include Cytospora, Diaporthe (with its Phomopsis anamorph), Graphium, Ilyonectria, Paraphoma, Phaeoisaria and Phialophora.
Cytospora species are cosmopolitan, facultative parasites, and appear in tree stands subjected to some form or stress, with poor agronomic management or infected by other pathogens. Infection occurs in late autumn or winter, when the host is dormant, usually behaving as a distinctly secondary parasite. The initial symptoms include brown-blackish discolorations, necrosis, depressions in the bark and underlying wood, callus production and withering. Older, sturdier tissues may develop resistance to further invasion. The disease then appears as small brown depressions bounded by distinct calluses. In the advanced stage, the bark tissues may peel away to reveal underlying stained wood [75]. Cytospora ambiens, C. chrysosperma and C. nivea (Hoffm.) Sacc., which are usually present on/in poplar wood worldwide, with their highest incidence in central and southern Italy, eastern Europe, the Near East, northern India, southern Africa (mainly in plantations) and the west-central USA (especially in Colorado), were not detected in the diseased hybrid poplars.
Species of Diaporthe and its Phomopsis anamorph comprise a phytopathologically important group, with diverse host associations and worldwide distribution. They cause leaf spots, blights, decay, wilt, root rots, dieback and cankers. Phomopsis pathogens are hemibiotrophs, i.e., first latent endophytes requiring living plants as a nutrient source, then sometimes becoming necrotrophic in the latent phase of colonization, or saprotrophic, their nutrients provided by tissue they have killed [76,77]. They occur in both temperate and tropical regions, and are especially common in the sapwood of angiosperms [78,79,80,81,82,83,84,85,86,87,88,89,90,91,92]. Endophytic and saprotrophic strains of Phomopsis produce similar degrading enzymes, supporting the thesis that endophytes become saprotrophs at the plant’s senescence [87,93]. Graphium basitruncatum has been reported from the gallery of the ambrosia beetle in poplar in South America [94]. Graphium penicillioides has been detected in the fully functional, wet sapwood of poplars [36] Baobab. Although the teleomorph of G. penicillioides is unknown, the genus is believed to have ophiostomatoid affinities [95,96,97].
Paraphoma is root-associated on Populus, although P. chrysanthemicola has so far been reported only from Juniperus, Malus and herbaceous plants [97,98].The fungus can infect the leaves of certain plant species and provoke disease [99]. On poplar, it caused foliar blight [100]. The fungus can also live benignly in asymptomatic plant tissues, and has been detected or isolated from the roots of healthy plants [101].
Phaeoisaria loranthacearum has so far been reported from twigs of Loranthus europaeus in Germany [102].
Phialophora species, found very abundantly, may include P. richardsiae, a serious pathogen implicated in the Petri disease of grapevine. The significance of other Phialophora spp. potentially occurring in the diseased poplar wood should also be emphasized. They are mostly saprotrophic and common in soil and wood, in which they cause soft rot. Growth at the hyphal tip and the secretion of lignolytic enzymes (pectinase, amylase, xylanase, cellulase and mannanase) causes widened cavities in sapwood and the degradation of the wood [103,104]. They can also cause cavities in the wood and plants via an erosion-type attack [105]. The degradation of Populus tremuloides wood has been known to affect sales of commercial aspen timber. The blue staining of wood by Phialophora has also been reported [106]. The fungus is psychrotolerant (able to grow at a low temperature).
Many of the taxa recorded, especially in the soil, may not be poplar-specific. They would originate from nearby vegetation, litter and decaying organic matter. Ascomycetous Boeremia spp., Desmazierella acicola, Dissoconium eucalypti, Entyloma gaillardianum, Lambertella tubulosa, Leptosphaerulina australis, Microdochium sp., Monographella nivalis, Neosetophoma clematidis, Periconia sp., Phacidium spp., Phaeosphaeria sp., Phaeosphaeriopsis sp., Phialocephala sp., Pyrenochaetopsis spp., Schizothecium glutinans, Xenochalara sp., Xenopolyscytalum spp., Xenoramularia arxii, and basidiomycetous Aecidium sp., Entyloma spp. and Itersonilia perplexans possibly spread from weeds, grass roots, leaf litter and woody debris [107,108,109,110,111,112,113,114,115,116,117,118,119,120,121]. Neocatenulostroma germanicum, recently found in Europe, seems to spread from pine needles or oak wood debris [49,122].
The cosmopolitan Cenococcum geophilum, one of the most frequently encountered ectomycorrhizal fungi in nature, is well recognized for its extremely wide host and habitat range [123].
Fungi of the genera Alternaria, Epicoccum, Fusarium, Cladosporium, Penicillium and Trichoderma are highly robust and ubiquitous, with an almost global distribution, occurring in the Americas, Asia, and Europe [103]. Their spores have been found in a variety of habitats, predominantly in soil of various types and in sand, often in extreme conditions. Epicoccum can grow on leaves submerged in water, even at 0 °C; hyphal growth can resume within an hour of exposure to water [104,124].
Some fungi were recorded for the first time on wood, or have been found rarely on wood. Ascomycetous Neocatenulostroma germanicum is known from pine needles, and is known to cause needle blight on Pinus mugo Turra, P. nigra Arn. ssp. pallasiania and P. sylvestris L. in Lithuania, Poland and Ukraine [44,122], but has also occurred in the soil in Poland [125]. Sydowia polyspora is so far known from the foliage of Abies spp., Pinus spp. and Pseudotsuga menziesii (Mirb.), and litter [126]. Research suggests that some of these hosts can be primary inoculum sources when located near poplar plantations [127].
Some more- or less-frequent colonizers are untypical and dubious. Acaulium retardatum has so far been recorded from rice-field soil [128], Acrodontium crateriforme from trap-liquid of pitcher plant Nepenthes khasiana Hook f. A.L.P.P. de Candolle, Prodr. in India [129], Alatospora has been recorded from aquatic habitats [130], Amesia nigricolor has been recorded from an indoor habitat in India [131], Cercospora beticola from sugar beet leaves, Desmazierella acicola from pine needle litter [132,133], Dissoconium eucalypti from Eucalyptus leaf [134], Halokirschsteiniothelia maritima from decaying wood in Thailand [135], Nigrospora oryzae from tropical plants [136], Pleurophoma ossicola from bone [102], Pseudocercospora angolensis from leaf spot on Citrus in Africa [137], Sakaguchia lamellibrachiae (Nagah., Hamam., Nakase and Horikoshi) Wang, Bai, Groenew. and Boekhout from a deep-sea tubeworm in Japan [138], and the basidiomycetous yeast Erythrobasidium hasegawianum has been recorded from old beer yeast culture in USA [139].
Some can occur at the extreme of their host ranges. Graphium basitruncatum has been isolated from wood and soil, even in the Solomon Islands and Japan, and from a leukemic patient [140,141]. Scytalidium lignicola and Sporothrix are recognized as saprotrophic opportunists of which the lifestyle can change from plant to human or animal pathogenicity.
Oomycota with eight species of Globisporangium, two species of Phytophthora and eight species of Pythium were mostly in the soil, and were not very common. Their contribution to the development of the disease cannot be excluded. All of them are plant pathogens, which cause root rot and damping off in a multitude of species. Phytophthora plurivora Jung and Burgess, followed by P. pini Leonian, P. polonica Belbahri, E. Moralejo, Calmin and Oszako, P. lacustris Brasier, Cacciola, Nechw., Jung and Bakonyi, P. cactorum (Lebert and Cohn) Schröt, and P. gonapodyides (Petersen) Buisman. were common in three declining and three healthy poplar plantations in Serbia [142].
4.3. Yeasts in Diseased Poplar Trunks
Yeasts are now identified and classified almost exclusively by DNA sequence analysis, which has resulted in the discovery of many new species and taxonomic revisions.
Filamentous fungi have a key role in the decomposition of plant material because of their ability to produce a wide range of extracellular enzymes that efficiently attack the recalcitrant lignocellulose matrix. However, the presence of yeasts during the different stages of wood breakdown highlights the ecological role of these microorganisms. Yeasts have been found to produce enzymes acting on cellulose, hemicelluloses and pectin [143]. They can therefore degrade plant material. They can also be transient fungi, using products released during decomposition by other organisms. Many yeast species found in live or decaying plant parts are associated with insects that also use these habitats as feeding or breeding sites.
The general opinion is that the most abundant yeast taxa associated with decayed wood are basidiomycetous (Agaricomycotina) and xylose-assimilating species. The present data do not support this thesis. Some ascomycetous yeasts were particularly abundant in the wood, where basidiomycetous yeasts were much less frequent.
Ascomycetous Aureobasidium pullulans and Candida spp., and basidiomycetous species of Apiotrichum, Cystofilobasidium, Naganishia, Saitozyma, Solicoccozyma, Tausonia, Tremella, Trichosporon and Vishniacozyma are frequently found in decaying plant material [143]. However, variations in their abundance and diversity reflect the environment, and also correlate with the natural abundance and distribution of basidiomycetous fungi in the study areas [144]; Apiotrichum, for example, was reported as being abundant in wood decayed by Armillaria. The abundance of ascomycetous yeasts in the wood resulted from the high frequency of Nakazawaea spp., especially N. populi, which was previously found in exudates of Populus species [145].
4.4. Mycorrhiza-Forming Fungi
Mycorrhiza-forming fungi were rare, especially in the soil. Basidiomycetous species occurred, surprisingly, more often in the wood, probably as: (i) facultative biotrophic encounters that either formed mycorrhizal structures or colonized the tissues as endophytes (i.e., grew within living plant tissues, without apparent infection, but not forming true mycorrhizae or causing any disease symptoms), or (ii) saprotrophs. Transition from saprotrophy to mycorrhizal status is common in fungal development [146], and other unexpected trophic conversions within the mycobiota may be possible.
4.5. The Endophytic State/Habit/Lifestyle of Fungi
As with grapevine diseases, it is assumed that the causal fungi are endophytic, living for a time asymptomatically in the plant. Then, at some point, in association with plant stress, they modify their behaviour and become pathogenic, which leads to the expression of disease symptoms [147]. As endophytes, they would often have key positive roles in plant function and fitness [148,149]. As parasites, they are cryptic, often opportunistic pathogens, which in special conditions induce disease [150]. Their virulence may be dictated by multi-partner interactions and environmental conditions. The most favoured conditions include: (i) the presence of very vigorous plants with succulent tissues; (ii) prolonged periods of damp and wet weather; (iii) free-standing water on the leaves; (iv) injuries such as pruning and leaf wounds; (v) the presence of senescent tissues, especially older, lower leaves; (vi) frost damage; and (vii) excessive crowding. Tissues are invaded by enzyme action, and roots and stems are gradually enveloped until the vessels are eventually reached, and wilting and desiccation occur. Different lifestyles and functions may occur depending on the situation. Phoma may at first be a plant-growth–promoting fungus [151].The lifestyles of Phaeoisaria and Pyrenochaetopsis depend on secreted peptidases [121,152]. Plectosphaerella (mostly P. populi) damages poplar stems [102,152], but simultaneously induces the formation of antifungal phenolic metabolites that protect poplar against foliar pathogens [153]. Some, such as Pyrenochaeta, are weak pathogens [154], but their adaptability to different climates allows them to infect many hosts and to survive in a broad range of pH, temperature and aeration conditions and soil types. Fungi such as Ilyonectria may survive in the roots of apparently healthy (asymptomatic) poplars, where they may suppress other fungal root pathogens and help maintain tree health [27,30]. These examples show that caution is necessary in classifying fungi according to function. There is no indication that other species, uncommon on Populus or so far not detected, might be pathogenic.
4.6. Interactions among Fungi
Trichoderma spp. occurred at a high natural frequency in the plantation soil. They are well known for their antagonistic activity, hyperparasitism and ability to induce defensive systems in plants to other microorganisms (specifically soil microorganisms). They are used in the biological control of several pathogens. Trichoderma harzianum Rifai and T. atroviride Karst. have shown promise in controlling Botryosphaeria dieback and esca disease in vineyards and other common trunk diseases [155]. Trichoderma significantly improved grapevine root growth and decreased the incidence of fungi involved in diseases when tested in vitro or in nurseries [24,156]. Grapevine defence systems have also been induced by Oomycota. The necrosis of root systems of vine cuttings was reduced by 50% after colonization by Pythium oligandrum [157,158,159]. Other biological control agents (Aureobasidium pullulans, Cladosporium herbarum, Fusarium lateritium and Rhodotorula rubra) have been reported to be effective against grapevine trunk disease pathogens, alone or in combination with fungicides, although some were tested only in vitro or in nurseries [160]. Arbuscular mycorrhizal fungi have been shown to increase the tolerance of grapevine rootstocks to Ilyonectria spp. [161]; Glomus intraradices was the most effective [162]. Aureobasidium pullulans, P. oligandrum, Trichoderma spp. and two species of Glomeromycota, present in the poplar plantation soil, may naturally decrease the incidence of pathogens involved in disease. Mortierella elongata, also detected, has been found to manipulate poplar defenses while promoting plant growth [30].This response was particularly beneficial because it was independent of cultivars.
4.7. Soil and Planting Material as the Source of the Inoculum
The soil origin was shown to be a significant factor affecting the composition of the fungal communities and networks in Populus [149,163].
The soil was here shown to be a natural source of many vascular and parenchymal pathogens found in the affected hybrid poplars, i.e., species of ascomycetous Alternaria, Cadophora, Cladosporium, Fusarium, Ilyonectria, Nectria, Neonectria, Neopyrenochaeta Ophiostomataceae, Phoma, Pyrenochaeta, Sporothrix, Thelonectria and Verticillium, and of basidiomycetous Armillaria and Entyloma. Their presence in the soil has been associated with their occurrence on plant debris and plant roots [164]. Soil was also the main source of pathogenic Oomycota (Aphanomyces, Elongisporangium, Globisporangium, Phytophthora and Pythium), which can, generally, cause extensive and devastating root rot. The destruction of roots can lead to minor or severe wilting caused by impeded root functioning or further biotrophic infections that can become necrotrophic in response to infection pressure or environmental stress. Oomycota tend to be very generalistic and non-specific, with a wide range of susceptible host roots, including poplar [142]. The wilt results from root degradation by Oomycota and a lack of oxygen, followed by disrupted water transport. A moist habitat and low pH in forest soils favour the growth, propagation, and dispersal of Oomycota spores. At optimal temperatures (28–30 °C), some species of Globisporangium grow very fast, i.e., 2.7 cm in 24-h.
Fungi such as Collophorina, Hyalodendriella and Hyaloscypha bicolor, which occurred sporadically in the soil, whilst being biotrophic parasites, may contribute to the final wilt [165,166].
The planting material may, however, already have been infected, either systemically from infected mother poplars or by contamination during the propagation process.
4.8. Colonization
As in grapevine disease, poplar wilt may be a complex disease in which symptoms result from the concomitant action of several factors.
The initial stage of the disease seems to be accomplished by highly specialized vascular fungi in the plant’s phloem. Their presence in the soil suggests that the infection can be soil-borne. Hyphae from established mycelia, and germ tubes developing from spores, perceive signals from root exudates. The hyphae secrete cell-wall–degrading enzymes and enter roots through wounds, at branching points, or directly through root tips. The mycelium spreads between root cortex cells to reach phloem and xylem vessels, from which the fungus travels as conidia in the sap stream, mostly upwards. The phloem and xylem become obstructed by mycelium and spores, and by plant-produced gels, gums and tyloses. Water transport to the leaves fails, and the plant wilts and dies. The fungus then invades all of the plant tissues and obtains nutrition by decomposing them. The response to the degradation of hemicellulose or lignin by the pathogen is usually the accumulation of tylose, polysaccharides and phenolic compounds (gummosis), tannins and phytoalexins. It is likely that at least a part of the external and internal symptoms are caused by phytotoxic fungal metabolites produced in decayed wood, or by the oxidation of some host-response substances. Some chemicals produced in grapevine in response to fungal infection are toxic, notably α-glucans and two naphthalenone pentaketides, scytalone and isosclerone [22]. A similar situation may be expected in poplar.
The final stage of the disease is apparently accomplished by parenchymal fungi. The spores released from reproductive structures produced in dead wood in the presence of water are dispersed by wind, potentially infecting fresh new wounds. Among the parenchymal fungi, bracket fungi (Polyporales, Basidiomycota) were, surprisingly, found only sporadically; they usually dominate communities of wood-rotting organisms. In grapevine, the phytoalexin resveratrol showed a direct antifungal effect, inhibiting the in vitro growth of two bracket species, Fomitiporia mediterranea and Stereum hirsutum. It is possible that the accumulation of certain compounds produced by poplar suppresses the colonization of wood by bracket fungi.
4.9. Effects of Climate
Up to 133 fungal species of 34 genera have so far been associated with grapevine trunk diseases worldwide [127]. The incidence of particular taxa differs between regions. All known grapevine trunk pathogens have been encountered in all grape-cultivation regions, mainly between latitudes of 30° to 50°, where annual mean temperatures are generally 10–20 °C [127,167]. There are conflicting reports on the effects of temperature and water stress on the incidence of grapevine trunk disease [127]. Therefore, it is not possible to assume a straightforward relationship between poplar disease and climatic conditions, particularly concerning water stress. Water stress is likely, however, to increase susceptibility. In recent years, precipitation in central Europe has often been characterized by extreme events (fog, hailstorms, thunderhails, heat waves, heavy rains, floods, winds), followed by drought. Increased humidity favours disease development. Infection by ascospores or conidia released from perithecia or pycnidia embedded in the bark or wood will be promoted by high humidity, often associated with higher temperatures; such conditions encourage the release and spread of spores, and favour spore germination [168,169,170,171]. The inoculum potential is consequently increased.
An extremely hot and dry summer (particularly August and September) occurred across Poland in 2015. The climate projections for Poland and central Europe predict further warming and the continuation of the changes already observed, including decreased precipitation and drought, especially in summer [172]. Such conditions may be expected to affect the health of poplar and other trees.
4.10. Control and Mitigation
Fungicides such as sodium arsenite or 8-hydroxyquinoline, used against esca and with the potential to control the wilt of poplar, are banned in Europe. No other highly effective treatments are available. Other chemical products and biological stimulators used in vineyards are not curative, and so only preventive methods are available in poplar plantations. Infections in grapevine from propagating materials can increase from 40% before cuttings are taken up to 70% after nursery processing [172]. Detection prior to planting is therefore critical to assure the longevity of newly established plantations [173]. A healthy poplar at planting is fundamental to the establishment and sustainability of a plantation. Good hygiene and wound protection are of the utmost importance. The disinfection of propagating materials with fungicides or hot water treatment (50 °C for 30 min), applied correctly to avoid plant stress and death, is advisable. Where soil constitutes the main source of the inoculum, disease management practices based on soil disinfestation and amendments, plant-based resistance to infection, and prophylactic cultural practices should be applied. Infected plant parts and infected dead wood on the soil should be removed, pruning wounds should be chemically protected, and the elimination of plant-stress factors should be taken into account.
5. Conclusions
-
1.
Populus hybrids may be subjected to various, thus far unidentified pathogenic agents.
-
2.
New diseases may be asymptomatic, at least in the initial phase.
-
3.
The indigenous microbiota can be involved in the development of the disease, but can also have an important role in limiting or preventing the development of pathogens.
-
4.
The development of new diseases is related to climate change. It can lead to the near-total disappearance of some diseases, the sudden emergence of a new pathogens, or to the fungi already present becoming pathogenic.
-
5.
Poplar wilt symptoms may be a consequence of various factors, the most important being climate and its effects on fungal development and the host–pathogen relationship.
-
6.
Fungal diseases can spread from the soil or from introduced plant material, with the latter potentially introducing them into new areas.
Author Contributions
Conceptualization, W.S. and J.B.-B.; methodology, J.B.-B.; formal analysis, E.G. and M.W.; investigation, M.B.; resources, W.S.; writing H.K., writing, review and editing, H.K., visualization, J.B.-B. and H.K., supervision, J.B.-B.; project administration, W.S., funding acquisition, J.B.-B.; M.B.; W.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research did not obtain any external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board and Ethics Committee of Poznan University of Life Sciences.
Informed Consent Statement
Informed Consent Statement was obtained from all subjects involved in the study.
Data Availability Statement
Data supporting reported results can be found at https://figshare.com/s/2c89719675a6859ee8a6 (accessed on 11 April 2021).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Przybysz K., Przybysz P. Poplar wood as a raw material for the paper industry in the twenty-first century. Ann. Warsaw Univ. Life Sci. 2013;84:56–59. [Google Scholar]
- 2.Herve C., Ceulemans R. Short-rotation coppiced vs. non-coppiced poplar: A comparative study at two different field sites. Biomass Bioenergy. 1996;11:139–150. doi: 10.1016/0961-9534(96)00028-1. [DOI] [Google Scholar]
- 3.Zajączkowski K. Hodowla Lasu: Plantacje Drzew Szybko Rosnących. Powszechne Wydawnictwo Rolnicze i Leśne; Warszawa, Poland: 2013. pp. 1–168. [Google Scholar]
- 4.Benetka V., Novotná K., Štochlová P. Biomass production of Populus nigra L. clones grown in short rotation coppice systems in three different environments over four rotations. iFor. Biogeosci. For. 2014;7:233–239. doi: 10.3832/ifor1162-007. [DOI] [Google Scholar]
- 5.Yu Q., Tigerstedt P., Haapanen M. Growth and phenology of hybrid aspen clones (Populus tremula L. × Populus tremuloides Michx.) Silva Fenn. 2001;35:15–25. doi: 10.14214/sf.600. [DOI] [Google Scholar]
- 6.Schmidt P.-A., Bálint M., Greshake B., Bandow C., Römbke J., Schmitt I. Illumina metabarcoding of a soil fungal community. Soil Biol. Biochem. 2013;65:128–132. doi: 10.1016/j.soilbio.2013.05.014. [DOI] [Google Scholar]
- 7.Vilgalys R., Gonzalez D. Organization of ribosomal DNA in the basidiomycete Thanatephorus praticola. Curr. Genet. 1990;18:277–280. doi: 10.1007/BF00318394. [DOI] [PubMed] [Google Scholar]
- 8.Gweon H.S., Oliver A., Taylor J., Booth T., Gibbs M., Read D.S., Griffiths R.I., Schonrogge K. PIPITS: An automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods Ecol. Evol. 2015;6:973–980. doi: 10.1111/2041-210X.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J., Kobert K., Flouri T., Stamatakis A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bengtsson-Palme J., Ryberg M., Hartmann M., Branco S., Wang Z., Godhe A., De Wit P.J.G.M., Sánchez-García M., Ebersberger I., De Sousa F., et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013;4:914–919. doi: 10.1111/2041-210X.12073. [DOI] [Google Scholar]
- 11.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian Classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole J.R., Wang Q., Fish J.A., Chai B., Mc Garrell D.M., Sun Y., Tiedje J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magurran A.E. Ecological Diversity and Its Measurement. Springer Science and Business Media, L.L.C.; Berlin/Heidelberg, Germany: 1988. pp. 1–180. [Google Scholar]
- 14.Mugnai L., Graniti A., Surico G. Esca (Black Measles) and Brown Wood-Streaking: Two old and elusive diseases of grapevines. Plant Dis. 1999;83:404–418. doi: 10.1094/PDIS.1999.83.5.404. [DOI] [PubMed] [Google Scholar]
- 15.Carlucci A., Lops F., Mostert L., Halleen F., Raimondo M. Occurrence fungi causing black foot on young grapevines and nursery rootstock plants in Italy. Phytopathol. Mediterr. 2017;56:10–39. [Google Scholar]
- 16.Larignon P., Fontaine F., Farine S., Clément C. Esca et Black Dead Arm: Deux acteurs majeurs des mala-dies du bois chez la Vigne. Comptes Rendus Biol. 2009;332:765–783. doi: 10.1016/j.crvi.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Van Niekerk J.M., Groenewald J.Z., Farr D.F., Fourie P.H., Halleen F., Crous P.W. Reassessment of Phomopsis species on grapevine. Austral. Plant Pathol. 2005;34:27–39. doi: 10.1071/AP04072. [DOI] [Google Scholar]
- 18.Úrbez-Torres J.R., Leavitt G.M., Voegel T.M., Gubler W.D. Identification and distribution of Botryosphaeria spp. associated with grapevine cankers in California. Plant Dis. 2006;90:1490–1503. doi: 10.1094/PD-90-1490. [DOI] [PubMed] [Google Scholar]
- 19.De Guido M.A., Pollastro S., Carlucci A., De Miccolis Angelini R.M., Faretra F. Phomopsis viticola is easily transformed with Hph and Bmlr genes. J. Plant Pathol. 2013;85:43–52. [Google Scholar]
- 20.Larignon P., Dubos B. Fungi associated with esca disease in grapevine. Eur. J. Plant Pathol. 1997;103:147–157. doi: 10.1023/A:1008638409410. [DOI] [Google Scholar]
- 21.Arzanlou M., Narmani A., Moshari S., Khodaei S., Babai-Ahari A. Truncatella angustata associated with grapevine trunk disease in northern Iran. Arch. Phytopathol. Plant Prot. 2013;46:1168–1181. doi: 10.1080/03235408.2012.761417. [DOI] [Google Scholar]
- 22.Bruno G., Sparapano L. Effects of three esca-associated fungi on Vitis vinifera L.: V. Changes in the chemical and biological profile of xylem sap from diseased cv. Sangiovese vines. Physiol. Mol. Plant Pathol. 2007;71:210–229. doi: 10.1016/j.pmpp.2008.02.005. [DOI] [Google Scholar]
- 23.Gramaje D., Armengol J., Mohammadi H., Banihashemi Z., Mostert L. Novel Phaeoacremonium species associated with Petri disease and esca of grapevine in Iran and Spain. Mycologia. 2009;101:920–929. doi: 10.3852/08-222. [DOI] [PubMed] [Google Scholar]
- 24.Agusti-Brisach C., Armengol J. Black-foot disease of grapevine: An update on taxonomy, epidemiology and management strategies. Phytopathol. Mediterr. 2013;52:245–261. [Google Scholar]
- 25.Carlucci A., Cibelli F., Lops F., Phillips A., Ciccarone C., Raimondo M. Pleurostomophora richardsiae asso-ciated with trunk diseases of grapevines in southern Italy. Phytopathol. Mediterr. 2015;54:109–123. [Google Scholar]
- 26.Lombard L., Van Der Merwe N.A., Groenewald J.Z., Crous P.W. Lineages in Nectriaceae: Re-evaluating the generic status of Ilyonectria and allied genera. Phytopathol. Mediterr. 2014;53:515–532. [Google Scholar]
- 27.White N.H., Chilvers G.A., Evans G. Antifungal activity of Cylindrocarpon radicicola Wr. Nat. Cell Biol. 1962;195:406–407. doi: 10.1038/195406a0. [DOI] [Google Scholar]
- 28.Spies C., Moyo P., Halleen F., Mostert L. Phaeoacremonium species diversity on woody hosts in the Western Cape Province of South Africa. Persoonia. 2018;40:26–62. doi: 10.3767/persoonia.2018.40.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cabral A., Groenewald J.Z., Rego C., Oliveira H., Crous P.W. Cylindrocarpon root rot: Multi-gene analysis reveals novel species within the Ilyonectria radicicola species complex. Mycol. Prog. 2011;11:655–688. doi: 10.1007/s11557-011-0777-7. [DOI] [Google Scholar]
- 30.Liao H.-L., Bonito G., Rojas J.A., Hameed K., Wu S., Schadt C.W., Labbé J., Tuskan G.A., Martin F., Grigoriev I.V., et al. Fungal endophytes of Populus trichocarpa alter host phenotype, gene expression, and rhizobiome composition. Mol. Plant-Microbe Interact. 2019;32:853–864. doi: 10.1094/MPMI-05-18-0133-R. [DOI] [PubMed] [Google Scholar]
- 31.Prodi A., Sandalo S., Tonti S., Pisi A. Phialophora-like fungi associated with kiwi fruit elephantiasis. J. Plan. Pathol. 2008;90:487–494. [Google Scholar]
- 32.Diaz B., Murace M., Peri P., Keil G., Luna L., Otaño M.Y. Natural and preservative-treated durability of Populus nigra cv Italica timber grown in Santa Cruz Province, Argentina. Int. Biodeterior. Biodegrad. 2003;52:43–47. doi: 10.1016/S0964-8305(03)00034-9. [DOI] [Google Scholar]
- 33.Xing J.-Q., Ikuo M., Wakako O. Natural resistance of two plantation woods Populus × canadensis cv. and Cunninghamia lanceolata to decay fungi and termites. For. Stud. China. 2005;7:36–39. doi: 10.1007/s11632-005-0055-3. [DOI] [Google Scholar]
- 34.Del Frari G., Gobbi A., Aggerbeck M.R., Oliveira H., Hansen L.H., Ferreira R.B. Characterization of the wood mycobiome of Vitis vinifera in a vineyard affected by esca. Spatial distribution of fungal communities and their putative relation with leaf symptoms. Front. Plant Sci. 2019;10:910. doi: 10.3389/fpls.2019.00910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Réblová M., Seifert K.A. A new species of Chaetosphaeria with Menispora ciliata and phialophora-like anamorphs. Fungal Divers. 2008;29:99–105. [Google Scholar]
- 36.Corda A.K.J. Leones fungorum hucusque cognitorum. Praha Tomus. 1837;1:1–32. [Google Scholar]
- 37.Pirozynski K.A. Xenotypa Petrak and Graphostroma gen. nov., segregates from Diatrypaceae. Can. J. Bot. 1974;52:2129–2135. doi: 10.1139/b74-274. [DOI] [Google Scholar]
- 38.Zhao G., Liu X., Wu W. Helicosporous hyphomycetes from China. Fungal Divers. 2007;26:313–524. [Google Scholar]
- 39.Choi Y.W. A Novel Helicosporium isolate and its antimicrobial and cytotoxic pigment. J. Microbiol. Biotechnol. 2012;22:1214–1217. doi: 10.4014/jmb.1204.04063. [DOI] [PubMed] [Google Scholar]
- 40.Hengstmengel J. Notes on Hymenoscyphus—II. On three non-fructicolous species of the ‘fructi-genus-group’ with croziers. Persoonia. 1996;16:191–207. [Google Scholar]
- 41.Kowalski T. Chalara fraxinea sp. nov. associated with dieback of ash (Fraxinus excelsior) in Poland. For. Pathol. 2006;36:264–270. doi: 10.1111/j.1439-0329.2006.00453.x. [DOI] [Google Scholar]
- 42.Kowalski T., Holdenrieder O. The teleomorph of Chalara fraxinea, the causal agent of ash dieback. For. Pathol. 2009;39:304–308. doi: 10.1111/j.1439-0329.2008.00589.x. [DOI] [Google Scholar]
- 43.Réblová M., Gams W., Štěpánek V. The new hyphomycete genera Brachyalara and Infundichalara, the similar Exochalara and species of ‘Phialophora sect. Catenulatae’ (Leotiomycetes) Fungal Divers. 2011;46:67–86. doi: 10.1007/s13225-010-0077-6. [DOI] [Google Scholar]
- 44.Behnke-Borowczyk J., Kwaśna H., Kulawinek B. Fungi associated with Cyclaneusma needle cast in Scots pine in the west of Poland. For. Pathol. 2019;49:e12487. doi: 10.1111/efp.12487. [DOI] [Google Scholar]
- 45.Hutchison L.J., Untereiner W.A., Hiratsuka Y. Knufia cryptophialidica gen. et sp. nov., a Dematiaceous Hyphomycete isolated from black galls of trembling aspen (Populus tremuloides) Mycologia. 1995;87:902. doi: 10.1080/00275514.1995.12026613. [DOI] [Google Scholar]
- 46.Fernando A., Currah R. A comparative study of the effects of the root endophytes Leptodontidium orchi-dicola and Phialocephala fortinii (Fungi Imperfecti) on the growth of some subalpine. Can. J. Bot. 1996;74:1071–1078. doi: 10.1139/b96-131. [DOI] [Google Scholar]
- 47.Wit M., Sierota Z., Osako T., Mirzwa-Mróz E., Wakuliński W. Fusarium spp. na nadziemnych organach zamierających dębów—Nowe zagrożenie? (Fusarium spp. on the above–ground organs of dying—A new threat?) Sylwan. 2015;159:403–410. [Google Scholar]
- 48.Chen J.L., Tzean S.S. Megacapitula villosa gen. et sp. nov. from Taiwan. Mycol. Res. 1993;97:347–350. doi: 10.1016/S0953-7562(09)81134-9. [DOI] [Google Scholar]
- 49.Behnke-Borowczyk J., Kwaśna H., Kokot K., Hałuszczak M., Łakomy P. Abundance and diversity of fungi in oak wood. Dendrobiology. 2018;80:143–160. doi: 10.12657/denbio.080.014. [DOI] [Google Scholar]
- 50.Rooney S.N., Eskalen A., Gubler W.D. Recovery of Phaeomoniella chlamydospora and Phaeoacremonium in-flatipes from soil and grapevine tissues. Phytopathol. Mediterr. 2001;40:S351–S356. [Google Scholar]
- 51.Ariyawansa H.A., Phukhamsakda C., Thambugala K.M., Bulgakov T.S., Wanasinghe D.N., Perera R.H., Mapook A., Camporesi E., Kang J.-C., Jones E.B.G., et al. Revision and phylogeny of Leptosphaeriaceae. Fungal Divers. 2015;74:19–51. doi: 10.1007/s13225-015-0349-2. [DOI] [Google Scholar]
- 52.El-Demerdash A., El-Demerdash A. Chemical diversity and biological activities of Phaeosphaeria fungi genus: A systematic review. J. Fungi. 2018;4:130. doi: 10.3390/jof4040130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J.K., Hyde K.D., Jones E.B.G., Ariyawansa H.A., Bhat D.J., Boonmee S., Maharachchikumbura S.S.N., McKenzie E.H.C., Phookamsak R., Phukhamsakda C., et al. Fungal diversity notes 1–110, Taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 2015;72:1–197. doi: 10.1007/s13225-015-0324-y. [DOI] [Google Scholar]
- 54.Gams W., Stielow B., Gräfenhan T., Schroers H.-J. The ascomycete genus Niesslia and associated Monocillium-like anamorphs. Mycol. Prog. 2019;18:5–76. doi: 10.1007/s11557-018-1459-5. [DOI] [Google Scholar]
- 55.Jankowiak R., Bilański P., Ostafińska A., Linnakoski R. Ophiostomatales associated with wounds on hardwood trees in Poland. Plant Pathol. 2019;68:1407–1424. doi: 10.1111/ppa.13061. [DOI] [Google Scholar]
- 56.Kendrick W.B. The Leptographium complex Verticicladiella Hughes. Can. J. Bot. 1961;39:1079–1085. doi: 10.1139/b61-094. [DOI] [Google Scholar]
- 57.Kwaśna H., Mazur A., Łabędzki A., Kuźmiński R., Łakomy P. Zbiorowiska grzybów w rozkładającym się drewnie dębu i sosny (Communities of fungi in decomposed wood of oak and pine) For. Res. Pap. 2016;77:261–275. [Google Scholar]
- 58.Sogonov M.V., Castlebury L.A., Rossman A.Y., Mejía L.C., White J.F., Jr. Leaf-inhabiting genera of the Gnomoniaceae, Diaporthales. Stud. Mycol. 2008;62:1–79. doi: 10.3114/sim.2008.62.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanney J., Seifert K. Mollisiaceae: An overlooked lineage of diverse endophytes. Stud. Mycol. 2020;95:293–380. doi: 10.1016/j.simyco.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Behnke-Borowczyk J., Kwaśna H., Kartawik N., Sijka B., Bełka M., Łakomy P. Effect of management on fungal communities in dead wood of Scots pine. For. Ecol. Manag. 2021;479:118528. doi: 10.1016/j.foreco.2020.118528. [DOI] [Google Scholar]
- 61.Oren Y., Sadowsky A., Gefen D., Solel Z., Kimchy M. Scytalidium wilt of citrus. Eur. J. Plant Pathol. 2001;107:467–470. doi: 10.1023/A:1011283318617. [DOI] [Google Scholar]
- 62.Machado A.R., Pinho D.B., De Oliveira S.A.S., Pereira O.L. New occurrences of Botryosphaeriaceae causing black root rot of cassava in Brazil. Trop. Plant Pathol. 2014;39:464–470. doi: 10.1590/S1982-56762014000600008. [DOI] [Google Scholar]
- 63.De Meyer E.M., De Beer Z.W., Summerbell R.C., Moharram A., De Hoog G.S., Vismer H.F., Wingfield M.J. Taxonomy and phylogeny of new wood- and soil-inhabiting Sporothrix species in the Ophiostoma stenoceras-Sporothrix schenckii complex. Mycologia. 2008;100:647–661. doi: 10.3852/07-157R. [DOI] [PubMed] [Google Scholar]
- 64.Wrzosek M., Sierota Z., Sikora K., Małecka M., Pawłowska J. Bogactwo grzybów zasiedlających drewno pniaków świerkowych po roku od sztucznego zakażenia Phlebiopsis gigantea. (The diversity of fungi present in the wood of spruce stumps one year after artificial infection by Phlebiopsis gigantea. Stud. Mater. CEPL. 2014;16:202–211. [Google Scholar]
- 65.Koukol O., Kubátová A. New European records of basidiomycete Burgoa anomala from coniferous litter and sediment in underground tunnel. Czech Mycol. 2015;67:241–247. doi: 10.33585/cmy.67207. [DOI] [Google Scholar]
- 66.Goh T.K., Hyde K.D. A synopsis of Trichocladium species, based on the literature. Fungal Divers. 1999;2:101–118. [Google Scholar]
- 67.Nagahama T. Yeast biodiversity in freshwater, marine and deep-sea environments. In: Rosa C.A., Peter G., editors. Biodiversity and ecophysiology of yeasts. Springer; Berlin/Heidelberg, Germany: 2006. pp. 241–263. The Yeast Handbook. [Google Scholar]
- 68.Alias S.A., Jones E.B.G. Marine Fungi from Mangroves of Malaysia. Institute Ocean and Earth Sciences, University of Malaya; Kuala Lumpur, Malaysia: 2009. pp. 1–108. [Google Scholar]
- 69.Pang K.L., Jheng J.S., Jones E.B.G. Marine Mangrove Fungi of Taiwan. National Taiwan Ocean University Press; Keelung, Taiwan: 2011. pp. 1–131. [Google Scholar]
- 70.El-Elimat T., Raja H.A., Figueroa M., Falkinham J.O., Oberlies N.H. Isochromenones, isobenzofuranone, and tetrahydronaphthalenes produced by Paraphoma radicina, a fungus isolated from a freshwater habitat. Phytochemistry. 2014;104:114–120. doi: 10.1016/j.phytochem.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 71.Hafellner J. Distributional and other data for some species of Didymocyrtis (Dothideomycetes, Pleo-sporales, Phaeosphaeriaceae), including their Phoma-type anamorphs. Fritschiana. 2015;80:43–88. [Google Scholar]
- 72.Tibell S., Tibell L., Pang K.-L., Jones E.G. A conspectus of the filamentous marine fungi of Sweden. Bot. Mar. 2019;63:141–153. doi: 10.1515/bot-2018-0114. [DOI] [Google Scholar]
- 73.Kwaśna H. Zdrowotność topoli na plantacjach w strefach ochronnych hut miedzi Legnica i Głogów. (Health of poplars in plantations in the sanitary protection zones of Legnica and Glogów copper mills) Sylwan. 2017;161:639–647. [Google Scholar]
- 74.Cellerino G.P. Review of Fungal Diseases in Poplar. Food and Agriculture Organization of the United Nations; Rome, Italy: 1999. [Google Scholar]
- 75.Kepley J.B., Jacobi W.R. Pathogenicity of Cytospora fungi on six hardwood species. J. Arboric. 2000;26:326–332. [Google Scholar]
- 76.Rosskopf E.N., Charudattan R., DeValerio J.T., Stall W.M. Field evaluation of Phomopsis amaranthicola, a biological control agent of Amaranthus spp. Plant Dis. 2000;84:1225–1230. doi: 10.1094/PDIS.2000.84.11.1225. [DOI] [PubMed] [Google Scholar]
- 77.Van Kan J.A.L. Licensed to kill: The lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 2006;11:247–253. doi: 10.1016/j.tplants.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 78.Bussaban B., Lumyong L., Lumyong P., McKenzie E.H.C., Hyde K.D. Endophytic fungi from Amomum siamense. Can. J. Microbiol. 2001;47:943–948. doi: 10.1139/w01-098. [DOI] [PubMed] [Google Scholar]
- 79.Kumaresan V., Suryanarayanan T.S. Endophyte assemblages in young, mature and senescent leaves of Rhizophora apiculata: Evidence for the role of endophytes in mangrove litter degradation. Fungal Divers. 2002;9:81–91. [Google Scholar]
- 80.Osono T., Takeda H. Comparison of litter decomposing ability among diverse fungi in cool temperate deciduous forest in Japan. Mycologia. 2002;94:421–427. doi: 10.1080/15572536.2003.11833207. [DOI] [PubMed] [Google Scholar]
- 81.Suryanarayanan T.S., Murali T.S., Venkatesan G. Occurrence and distribution of fungal endophytes in tropical forests across a rainfall gradient. Can. J. Bot. 2002;80:818–826. doi: 10.1139/b02-069. [DOI] [Google Scholar]
- 82.Yanna Ho W.H., Hyde K.D. Fungal succession on fronds of Phoenix hanceana in Hong Kong. Fungal Divers. 2002;10:185–211. [Google Scholar]
- 83.Tomita F. Endophytes in Southeast Asia and Japan: Their taxonomic diversity and potential applications. Fungal Divers. 2003;14:187–204. [Google Scholar]
- 84.Murali T.S., Suryanarayanan T.S., Geeta R. Endophytic Phomopsis species: Host range and implications for diversity estimates. Can. J. Microbiol. 2006;52:673–680. doi: 10.1139/w06-020. [DOI] [PubMed] [Google Scholar]
- 85.Hyde K.D., Bussaban B., Paulus B., Crous P.W., Lee S., McKenzie E.H.C., Photita W., Lumyong S. Biodiversity of saprobic fungi. Biodivers. Conserv. 2007;16:17–35. [Google Scholar]
- 86.Rossman A.Y., Farr D.F., Castlebury L.A. A review of the phylogeny and biology of the Diaporthales. Mycoscience. 2007;48:135–144. doi: 10.1007/S10267-007-0347-7. [DOI] [Google Scholar]
- 87.Promputtha I., Hyde K.D., McKenzie E.H.C., Peberdy J.F., Lumyong S. Can leaf degrading enzymes provide evidence that endophytic fungi becoming saprobes? Fungal Divers. 2010;41:89–99. doi: 10.1007/s13225-010-0024-6. [DOI] [Google Scholar]
- 88.Promputtha I., Lumyong S., Vijaykrishna D., McKenzie E.H.C., Hyde K.D., Jeewon R. A phylogenetic evaluation of whether endophytes become saprotrophs at host senescence. Microb. Ecol. 2007;53:579. doi: 10.1007/s00248-006-9117-x. [DOI] [PubMed] [Google Scholar]
- 89.Kodsueb R., McKenzie E.H.C., Lumyong S., Hyde K.D. Diversity of saprobic fungi on Magnoliaceae. Fungal Divers. 2008;30:37–53. [Google Scholar]
- 90.Kodsueb R., McKenzie E.H.C., Lumyong S., Hyde K.D. Fungal succession on woody litter of Magnolia liliiflora (Magnoliaceae) Fungal Divers. 2008;30:55–72. [Google Scholar]
- 91.Botella L., Diez J.J. Phylogenic diversity of fungal endophytes in Spanish stands of Pinus halepensis. Fungal Divers. 2010;47:9–18. doi: 10.1007/s13225-010-0061-1. [DOI] [Google Scholar]
- 92.González V., Tello M.L. The endophytic mycota associated with Vitis vinifera in central Spain. Fungal Divers. 2011;47:29–42. doi: 10.1007/s13225-010-0073-x. [DOI] [Google Scholar]
- 93.Dai C.C., Chen Y., Tian L., Sh Y. Correlation between invasion by endophytic fungus Phomopsis sp. and enzyme production. Afr. J. Agric. Res. 2010;5:1324–1340. [Google Scholar]
- 94.Ceriani-Nakamurakare E., Slodowicz M., Gonzalez-Audino P., Dolinko A., Carmarán C. Mycobiota associated with the ambrosia beetle Megaplatypus mutatus: Threat to poplar plantations. Forests. 2016;89:191–200. doi: 10.1093/forestry/cpw001. [DOI] [Google Scholar]
- 95.Goidànich G. Schema di una classificazione delle Stilbaceaeche erano riunite fin’ora nel genere Graphium Corda. Ann. Bot. 1935;21:40–50. [Google Scholar]
- 96.Upadhyay H.P. A monograph of Ceratocyslis and Ceratocystiopsi. University of Georgia Press; Athens, GA, USA: 1981. pp. 1–176. [Google Scholar]
- 97.Seifert K.A., Okada G. Graphium anamorphs of Ophiostoma species and similar anamorphs of other as-comycetes. In: Wingfield M.J., Seifert K.A., Webber J.F., editors. Ceratocyslis and Ophiostoma: Taxonomy, Ecology, and Pathology. American Phytopathological Society Press; St. Paul, MN, USA: 1993. pp. 27–41. [Google Scholar]
- 98.De Gruyter J., Woudenberg J.H., Aveskamp M.M., Verkley G.J., Groenewald J.Z., Crous P.W. Systematic reappraisal of species in Phoma section Paraphoma, Pyrenochaeta and Pleurophoma. Mycologia. 2010;102:1066–1081. doi: 10.3852/09-240. [DOI] [PubMed] [Google Scholar]
- 99.Ge X., Zhou R., Yuan Y., Xu H., Fu J., Li H. Identification and characterization of Paraphoma chrysanthemicola causing leaf spot disease on Atractylodes japonica in China. J. Phytopathol. 2016;164:372–377. doi: 10.1111/jph.12463. [DOI] [Google Scholar]
- 100.Dhillon G., Sandhu J.S., Singh P. Variation among poplar (Populus deltoides Bartr.) clones for growth, wood traits and tolerance to leaf spot diseases. Curr. Agric. Res. J. 2020;8:128–136. doi: 10.12944/CARJ.8.2.08. [DOI] [Google Scholar]
- 101.Yokoya K., Postel S., Fang R., Sarasan V. Endophytic fungal diversity of Fragaria vesca, a crop wild relative of strawberry, along environmental gradients within a small geographical area. PeerJ. 2017;5:e2860. doi: 10.7717/peerj.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Crous P.W., Schumacher R.K., Wingfield M.J., Lombard L., Giraldo A., Christensen M., Gardiennet A., Nakashima C., Pereira O., Smith A.J., et al. Fungal systematics and evolution: FUSE 1. Sydowia. 2015;67:81–118. [Google Scholar]
- 103.Domsch K.H., Gams W., Anderson T.-H. Compendium of Soil Fungi. TAXON. 1982;31:600. doi: 10.2307/1220704. [DOI] [Google Scholar]
- 104.Hale M.D., Eaton R.A. Oscillatory growth of fungal hyphae in wood cell walls. Trans. Br. Mycol. Soc. 1985;84:277–288. doi: 10.1016/S0007-1536(85)80079-6. [DOI] [Google Scholar]
- 105.Nilsson T. Microscopic studies on the degradation of cellophane and various cellulosic fibres by wood-attacking microfungi. Stud. For. Suec. 1947;117:1–27. [Google Scholar]
- 106.Hallaksela A.M., Niemistö P. Stem discoloration of planted silver birch. Scand. J. For. Res. 2008;13:169–176. doi: 10.1080/02827589809382973. [DOI] [Google Scholar]
- 107.Coetsee C., Wingfield M.J., Crous P.W., Wingfield B.D. Xenochalara, a new genus of dematiaceous hypho-mycetes for Chalara-like fungi with apical wall building conidial development. S. Afr. J. Bot. 2000;66:99–103. doi: 10.1016/S0254-6299(15)31069-3. [DOI] [Google Scholar]
- 108.Fonseca A., Scorzetti G., Fell J.W. Diversity in the yeast Cryptoccocus albidus and related species as re-vealed by ribosomal DNA sequence analysis. Can. J. Microbiol. 2000;46:7–27. doi: 10.1139/cjm-46-1-7. [DOI] [PubMed] [Google Scholar]
- 109.Cai L., Jeewon R., Hyde K., Hyde R. Phylogenetic evaluation and taxonomic revision of Schizothecium based on ribosomal DNA and protein coding genes. Fungal Divers. 2005;19:1–21. [Google Scholar]
- 110.Arenz B.E., Held B.W., Jurgens J.A., Farrell R.L., Blanchette R.A. Fungal diversity in soils and historic wood from the Ross Sea Region of Antarctica. Soil Biol. Biochem. 2006;38:3057–3064. doi: 10.1016/j.soilbio.2006.01.016. [DOI] [Google Scholar]
- 111.McGovern R.J., Horita H., Stiles C.M., Seijo T.E. Host range of Itersonilia perplexans and management of Itersonilia petal blight of China Aster. Plant Health Prog. 2006;7:7. doi: 10.1094/PHP-2006-1018-02-RS. [DOI] [Google Scholar]
- 112.Aveskamp M.M., de Gruyter J., Woudenberg J.H.C., Verkley G.J.M., Crous P.W. Highlights of the Didymellaceae: A polyphasic approach to characterise Phoma and related pleosporalean genera. Stud Mycol. 2010;65:1–60. doi: 10.3114/sim.2010.65.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Branda E., Turchetti B., Diolaiuti G., Pecci M., Smiraglia C., Buzzini P. Yeast and yeast-like diversity in the southernmost glacier of Europe (Calderone Glacier, Apennines, Italy) FEMS Microbiol. Ecol. 2010;72:354–369. doi: 10.1111/j.1574-6941.2010.00864.x. [DOI] [PubMed] [Google Scholar]
- 114.Crous P.W., Groenewald J.Z., Diederich P. Xenopolyscytalum pinae. Fungal Planet 55. Persoonia. 2010;5:130–131. [Google Scholar]
- 115.Kowalski T., Kehr R.D. Two new species of Phialocephala occurring on Picea and Alnus. Can. J. Bot. 1995;73:26–32. doi: 10.1139/b95-004. [DOI] [Google Scholar]
- 116.Markovskaja S. Aero-aquatic fungi colonizing decaying leaves in woodland swampy pools of Aukštadvaris Regional Park (Lithuania) Bot. Lithuania. 2012;18:123–132. [Google Scholar]
- 117.Savchenko K.G., Heluta V.P. Smut fungi of Ukraine, a checklist. Sydowia. 2012;64:281–300. [Google Scholar]
- 118.Hyde K.D., Jones E.B.G., Liu J.-K., Ariyawansa H., Boehm E.W., Boonmee S., Braun U., Chomnunti P., Crous P.W., Dai D.-Q., et al. Families of Dothideomycetes. Fungal Divers. 2013;63:1–313. doi: 10.1007/s13225-013-0263-4. [DOI] [Google Scholar]
- 119.Quaedvlieg W., Verkley G., Shin H.-D., Barreto R., Alfenas A., Swart W., Groenewald J., Crous P. Sizing up Septoria. Stud. Mycol. 2013;75:307–390. doi: 10.3114/sim0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Marin-Felix Y., Groenewald J., Cai L., Chen Q., Marincowitz S., Barnes I., Bensch K., Braun U., Camporesi E., Damm U., et al. Genera of phytopathogenic fungi: GOPHY 1. Stud. Mycol. 2017;86:99–216. doi: 10.1016/j.simyco.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Valenzuela-Lopez N., Cano-Lira J., Guarro J., Sutton D., Wiederhold N., Crous P., Stchigel A. Coelomycetous Dothideomycetes with emphasis on the families Cucurbitariaceae and Didymellaceae. Stud. Mycol. 2017;90:1–69. doi: 10.1016/j.simyco.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Markovskaja S., Kačergius A., Davydenko K., Fraser S. First record of Neocatenulostroma germanicum on pines in Lithuania and Ukraine and its co-occurrence with Dothistroma spp. and other pathogens. For. Pathol. 2016;46:522–533. doi: 10.1111/efp.12308. [DOI] [Google Scholar]
- 123.LoBuglio K.F. Cenococcum. In: Cairney J.W.G., Chambers S.M., editors. Ectomycorrhizal Fungi Key Genera in Profile. Springer; Berlin/Heidelberg, Germany: 1999. [DOI] [Google Scholar]
- 124.Cole G.T., Kendrick B., editors. Biology of Conidial Fungi. Academic Press; New York, NY, USA: 1981. pp. 1–680. [Google Scholar]
- 125.Kwaśna H., Behnke-Borowczyk J., Gornowicz R., Łakomy P. Effects of preparation of clear-cut forest sites on the soil mycobiota with consequences for Scots pine growth and health. For. Pathol. 2019;49:e12494. doi: 10.1111/efp.12494. [DOI] [Google Scholar]
- 126.Boberg J.B., Ihrmark K., Lindahl B.D. Decomposing capacity of fungi commonly detected in Pinus sylvestris needle litter. Fungal. Ecol. 2011;4:110–114. doi: 10.1016/j.funeco.2010.09.002. [DOI] [Google Scholar]
- 127.Gramaje D., Úrbez-Torres J.R., Sosnowski M.R. Managing grapevine trunk diseases with respect to etiology and epidemiology: Current strategies and future prospects. Plant Dis. 2018;102:12–39. doi: 10.1094/PDIS-04-17-0512-FE. [DOI] [PubMed] [Google Scholar]
- 128.Su L., Zhu H., Niu Y., Guo Y., Du X., Guo J., Zhang L., Qin C. Phylogeny and taxonomic revision of Kernia and Acaulium. Sci. Rep. 2020;10:1–11. doi: 10.1038/s41598-020-67347-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Prabhugaonkar A., Jalmi P. Isolation of Acrodontium crateriforme as a pitcher trap inquiline. Curr. Res. Environ. Appl. Mycol. 2017;7:203–207. doi: 10.5943/cream/7/3/7. [DOI] [Google Scholar]
- 130.Ingold C. Aquatic hyphomycetes of decaying alder leaves. Trans. Br. Mycol. Soc. 1942;25:339–417. doi: 10.1016/S0007-1536(42)80001-7. [DOI] [Google Scholar]
- 131.Wang X., Houbraken J., Groenewald J., Meijer M., Andersen B., Nielsen K., Crous P., Samson R. Diversity and taxonomy of Chaetomium and chaetomium-like fungi from indoor environments. Stud. Mycol. 2016;84:145–224. doi: 10.1016/j.simyco.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kendrick W.B., Burges A. Biological aspects of the decay of Pinus sylvestris leaf litter. Nova Hedwigia. 1962;4:313–359. [Google Scholar]
- 133.Martinović T., Koukol O., Hirose D. Distinct phylogeographic structure recognized within Desmazierella acicola. Mycologia. 2016;108:20–30. doi: 10.3852/14-291. [DOI] [PubMed] [Google Scholar]
- 134.Li H., Sun G., Zhai X., Batzer J., Mayfield D., Crous P., Groenewald J., Gleason M. Dissoconiaceae associated with sooty blotch and flyspeck on fruits in China and the United States. Pers. Mol. Phylogeny Evol. Fungi. 2012;28:113–125. doi: 10.3767/003158512X651157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Boonmee S., Ko T.W.K., Chukeatirote E., Hyde K.D., Chen H., Cai L., McKenzie E.H., Jones E.B.G., Kodsueb R., Hassan B.A. Two new Kirschsteiniothelia species with Dendryphiopsis anamorphs cluster in Kirschsteiniotheliaceae fam. nov. Mycologia. 2012;104:698–714. doi: 10.3852/11-089. [DOI] [PubMed] [Google Scholar]
- 136.Abass M., Hussein N. Morphological, molecular and pathological study on Nigrospora oryzae and Nigrospora sphaerica, the leaf spot fungi of date palm. Basra J. Date Palm Res. 2014;13:1313. [Google Scholar]
- 137.Brentu F.C., Cornelius E.W., Lawson L.E.V., Oduro K.A., Vicent A. First Report of Pseudocercospora angolensis causing fruit and leaf spot of Citrus in Ghana. Plant Dis. 2013;97:1661. doi: 10.1094/PDIS-06-13-0615-PDN. [DOI] [PubMed] [Google Scholar]
- 138.Nagahama T., Hamamoto M., Nakase T., Horikoshi K. Rhodotorula lamellibrachii sp. nov., a new yeast species from a tubeworm collected at the deep-sea floor in Sagami bay and its phylogenetic analysis. Antonie Leeuwenhoek. 2001;80:317–323. doi: 10.1023/A:1013043301388. [DOI] [PubMed] [Google Scholar]
- 139.Hamamoto M. Erythrobasidium Hamamoto, Sugiyama & Komagata. In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The Yeasts, a Taxonomic Study. 5th ed. Volume 3. Elsevier; London, UK: 2011. pp. 1433–1435. [Google Scholar]
- 140.Kumar D., Sigler L., Gibas C.F.C., Mohan S., Schuh A., Medeiros B.C., Peckham K., Humar A. Graphium basitruncatum fungemia in a patient with acute leukemia. J. Clin. Microbiol. 2007;45:1644–1647. doi: 10.1128/JCM.02466-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lackner M., de Hoog G.S. Parascedosporium and its relatives: Phylogeny and ecological trends. IMA Fungus. 2011;2:39–48. doi: 10.5598/imafungus.2011.02.01.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Milenković I., Keča N., Karadžić D., Radulović Z., Nowakowska J.A., Oszako T., Sikora K., Corcobado T., Jung T. Isolation and pathogenicity of Phytophthora species from poplar plantations in Serbia. Forests. 2018;9:330. doi: 10.3390/f9060330. [DOI] [Google Scholar]
- 143.Cadete R.M., Lopes M.R., Rosa C.A. Yeasts associated with decomposing plant material and rotting wood. In: Buzzini P., Lachance M.-A., Yurkov A., editors. Yeasts in Natural Ecosystems: Diversity. Springer International Publishing; Cham, Switzerland: 2017. pp. 265–292. [DOI] [Google Scholar]
- 144.González A., Martínez A.T., Almendros G., Grinbergs J. A study of yeasts during the delignification and fungal transformation of wood into cattle feed in Chilean rain forest. Antonie Leeuwenhoek. 1989;55:221–236. doi: 10.1007/BF00393851. [DOI] [PubMed] [Google Scholar]
- 145.Hagler A.N., Mendonca-Hagler L.C., Phaff H.J. Candida populi, a new species of yeast occurring in exudates of Populus and Betula species. Int. J. Syst. Bacteriol. 1989;39:97–99. doi: 10.1099/00207713-39-2-97. [DOI] [Google Scholar]
- 146.Selosse M.A., Martos F., Perry B., Maj P., Roy M., Pailler T. Saprotrophic fungal symbionts in tropical achlorophyllous orchids. Plant Signal. Behav. 2010;5:349–353. doi: 10.4161/psb.5.4.10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hofstetter V., Buyck B., Croll D., Viret O., Couloux A., Gindro K. What if esca disease of grapevine were not a fungal disease? Fungal Divers. 2012;54:51–67. doi: 10.1007/s13225-012-0171-z. [DOI] [Google Scholar]
- 148.Shakya M., Gottel N., Castro H., Yang Z.K., Gunter L., Labbe J., Muchero W., Bonito G., Vilgalys R., Tuskan G., et al. A multifactor analysis of fungal and bacterial community structure in the root microbiome of mature Populus deltoides trees. PLoS ONE. 2013;8:e76382. doi: 10.1371/journal.pone.0076382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cregger M.A., Veach A.M., Yang Z.K., Crouch M.J., Vilgalys R., Tuskan G.A., Schadt C.W. The Populus holobiont: Dissecting the effects of plant niches and genotype on the microbiome. Microbiome. 2018;6:31. doi: 10.1186/s40168-018-0413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ridout M., Newcombe G. Sydowia polyspora is both a foliar endophyte and a pre-emergent seed pathogen in Pinus ponderosa. Plant Dis. 2018;102:640–644. doi: 10.1094/PDIS-07-17-1074-RE. [DOI] [PubMed] [Google Scholar]
- 151.Hamayun M., Khan S.A., Khan A.L., Rehman G., Sohn E.-Y., Shah A.A., Kim S.-K., Joo G.-J., Lee I.-J. Phoma herbarum as a new gibberellin-producing and plant growth-promoting fungus. J. Microbiol. Biotechnol. 2009;19:1244–1249. [PubMed] [Google Scholar]
- 152.Da Silva R.R., Da Rosa N.G., De Oliveira L.C.G., Juliano M.A., Juliano L., Rosa J.C., Cabral H. Biochemical properties and catalytic specificity of a novel neutral serine peptidase secreted by fungus Pyrenochaetopsis sp. Appl. Biochem. Biotechnol. 2018;187:1158–1172. doi: 10.1007/s12010-018-2875-3. [DOI] [PubMed] [Google Scholar]
- 153.Ullah C., Unsicker S.B., Reichelt M., Gershenzon J., Hammerbacher A. Accumulation of catechin and proanthocyanidins in black poplar stems after infection by Plectosphaerella populi: Hormonal regulation, biosynthesis and antifungal activity. Front. Plant Sci. 2019;10:1441. doi: 10.3389/fpls.2019.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Schwartz H.F., Mohan S.K. Compendium of Onion and Garlic Diseases and Pests. 2nd ed. The American Phytopathological Society; St. Paul, MN, USA: 2016. pp. 8–86. [DOI] [Google Scholar]
- 155.Larignon P. Réflexions sur l’esca: Ce que l’on sait déjà montre qu’il en reste beaucoup à apprendre: Vigne. Phytoma-La Défense des Végétaux. 2004;576:28–31. [Google Scholar]
- 156.Fourie P.H., Halleen F., van der Vyver J., Schreuder W. Effects of Trichoderma treatments on the occurrence of decline pathogens in the roots and rootstocks of nursery grapevines. Phytopathol. Mediterr. 2001;40:473–478. [Google Scholar]
- 157.Gerbore J. Ph.D. Thesis. Punjab Agricultural University; Punjab, India: 2013. Lutte Biologique Contre un Champignon Pathogène Impliqué dans L’esca de la Vigne, par Utilisation de L’oomycète Pythium oligandrum; pp. 1–270. [Google Scholar]
- 158.Yacoub A., Gerbore J., Magnin N., Chambon P., Dufour M.C., Corio-Costet M.F., Guyoneaud R., Rey P. Ability of Pythium oligandrum strains to protect Vitis vinifera against Phaeomoniella chlamydsopora, a pathogen involved in Esca, by inducing plant resistance. Biol. Control. 2016;92:7–16. [Google Scholar]
- 159.Yacoub A., Gerbore J., Magnin N., Vallance J., Grizard D., Guyoneaud R., Rey P. Induction of grapevine defence systems using the oomycete Pythium oligandrum against a pathogenic fungus involved in Esca. Phytopathol. Mediterr. 2014;53:574–575. [Google Scholar]
- 160.Bertsch C., Ramírez-Suero M., Magninrobert M., Larignon P., Chong J., Aboumansour E., Spagnolo A., Clément C., Fontaine F. Grapevine trunk diseases: Complex and still poorly understood. Plant Pathol. 2012;62:243–265. doi: 10.1111/j.1365-3059.2012.02674.x. [DOI] [Google Scholar]
- 161.Jones E.E., Hammond S., Blond C., Brown D.S., Ridgway H.J. Interaction between arbuscular mycorrhizal fungi and rootstock cultivar on the susceptibility to infection by Ilyonectria species. Phytopathol. Mediterr. 2014;53:582–583. [Google Scholar]
- 162.Petit E., Gubler W.D. Influence of Glomus intraradices on black foot disease caused by Cylindrocarpon macrodidymum on Vitis rupestris under controlled conditions. Plant Dis. 2006;90:1481–1484. doi: 10.1094/PD-90-1481. [DOI] [PubMed] [Google Scholar]
- 163.Bonito G., Reynolds H., Robeson M.S., Nelson J., Hodkinson B.P., Tuskan G., Schadt C.W., Vilgalys R. Plant host and soil origin influence fungal and bacterial assemblages in the roots of woody plants. Mol. Ecol. 2014;23:3356–3370. doi: 10.1111/mec.12821. [DOI] [PubMed] [Google Scholar]
- 164.Zhu Y.J., Yu X.Y., Wang B.T., Jin L., Jin F.J. Description of Fusarium solani isolated from the soil of a poplar plantation in China. Int. J. Agric. Biol. 2020;24:663–670. [Google Scholar]
- 165.Crous P., Braun U., Schubert K., Groenewald J. Delimiting Cladosporium from morphologically similar genera. Stud. Mycol. 2007;58:33–56. doi: 10.3114/sim.2007.58.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Baral H.-O., De Sloover J.R., Huhtinen S., Laukka T., Stenroos S. An emendation of the genus Hyaloscypha to include Fuscoscypha (Hyaloscyphaceae, Helotiales, Ascomycotina) Karstenia. 2009;49:1–17. doi: 10.29203/ka.2009.430. [DOI] [Google Scholar]
- 167.Van Niekerk J.M., Bester W., Halleen F., Crous P.W., Fourie P.H. The distribution and symptomatology of grapevine trunk disease pathogens are influenced by climate. Phytopathol. Mediterr. 2011;50:98–111. [Google Scholar]
- 168.Úrbez-Torres J.R., Battany M., Bettiga L.J., Gispert C., McGourty G., Roncoroni J., Smith R.J., Verdegaal P., Gubler W.D. Botryosphaeriaceae species spore-trapping studies in California vineyards. Plant Dis. 2010;94:717–724. doi: 10.1094/PDIS-94-6-0717. [DOI] [PubMed] [Google Scholar]
- 169.Úrbez-Torres J., Bruez E., Hurtado J., Gubler W.D. Effect of temperature on conidial germination of Botryosphaeriaceae species infecting grapevines. Plant Dis. 2010;94:1476–1484. doi: 10.1094/PDIS-06-10-0423. [DOI] [PubMed] [Google Scholar]
- 170.Van Niekerk J.M., Calitz F.J., Halleen F., Fourie P.H. Temporal spore dispersal patterns of grapevine trunk pathogens in South Africa. Eur. J. Plant Pathol. 2010;127:375–390. doi: 10.1007/s10658-010-9604-2. [DOI] [Google Scholar]
- 171.Szwed M. Variability of precipitation in Poland under climate change. Theor. Appl. Clim. 2018;135:1003–1015. doi: 10.1007/s00704-018-2408-6. [DOI] [Google Scholar]
- 172.Gramaje D., Armengol J. Fungal trunk pathogens in the grapevine propagation process: Potential inoculum sources, detection, identification, and management strategies. Plant Dis. 2011;95:1040–1055. doi: 10.1094/PDIS-01-11-0025. [DOI] [PubMed] [Google Scholar]
- 173.Úrbez-Torres J.R., Haag P., Bowen P., Lowery T., O’Gorman D. Development of a DNA macroarray for the detection and identification of fungal pathogens causing decline of young grapevines. Phytopathology. 2015;105:1373–1388. doi: 10.1094/PHYTO-03-15-0069-R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data supporting reported results can be found at https://figshare.com/s/2c89719675a6859ee8a6 (accessed on 11 April 2021).