Abstract
The fission yeast Schizosaccharomyces japonicus, comprising S. japonicus var. japonicus and S. japonicus var. versatilis varieties, has unique characteristics such as striking hyphal growth not seen in other Schizosaccharomyces species; however, information on its diversity and evolution, in particular mating and sporulation, remains limited. Here we compared the growth and mating phenotypes of 17 wild strains of S. japonicus, including eight S. japonicus var. japonicus strains newly isolated from an insect (Drosophila). Unlike existing wild strains isolated from fruits/plants, the strains isolated from Drosophila sporulated at high frequency even under nitrogen-abundant conditions. In addition, one of the strains from Drosophila was stained by iodine vapor, although the type strain of S. japonicus var. japonicus is not stained. Sequence analysis further showed that the nucleotide and amino acid sequences of pheromone-related genes have diversified among the eight strains from Drosophila, suggesting crossing between S. japonicus cells of different genetic backgrounds occurs frequently in this insect. Much of yeast ecology remains unclear, but our findings suggest that insects such as Drosophila might be a good niche for mating and sporulation, and will provide a basis for the understanding of sporulation mechanisms via signal transduction, as well as the ecology and evolution of yeast.
Keywords: fission yeast, Schizosaccharomyces japonicus, mating, sporulation, pheromone, Drosophila
1. Introduction
The fission yeast Schizosaccharomyces japonicus, which was first isolated from strawberries at a farm of Kyusyu University in Japan [1], is a close relative of the well-studied model organism Schizosaccharomyces pombe [2]. However, S. japonicus differs from other fission yeast species with respect to several features. First, the size of S. japonicus haploid cells is bigger, facilitating observation of chromosome behavior such as condensed mitotic chromosomes [3,4,5]. Second, similar to many fungal species, S. japonicus is dimorphic and can exist in the form of both yeast and filamentous (hyphae) cells [6]. Fungal dimorphism is usually associated with pathogenic activity towards animals and plants [7] because a hyphae cell can penetrate into a host cell; however, S. japonicus is nonpathogenic to humans. Hyphal growth of S. japonicus is triggered by environmental stresses as nutritional starvation [8,9,10] and DNA damage [11]. This response is thought to enable hyphae cells of S. japonicus to escape more quickly from harmful stimuli. S. japonicus also has unique physiological features such as semi-open mitosis [4], light response [12], and growth via fermentation under anaerobic conditions [13]. Despite these interesting properties, relatively few studies on S. japonicus have been reported to date.
More than a decade since S. japonicus was isolated by Yukawa and Maki, a variant termed S. japonicus var. versatilis was isolated from home-canned grape juice in the United States [14]. S. japonicus var. japonicus and S. japonicus var. versatilis show some differences in characteristics and phenotypes; for example, sporulated colonies of the former are not stained by iodine vapor [15], whereas S. japonicus var. versatilis sporulated colonies are stained by iodine vapor [16]. Nevertheless, S. japonicus var. japonicus and S. japonicus var. versatilis have been demonstrated to be genetically compatible and can mate with each other following sporulation [17]. Notably, sporulated cells of the cross exhibit an iodine-negative phenotype, suggesting that the negative iodine-staining genotype of S. japonicus var. japonicus is dominant [17]. The difference in iodine staining phenotype has been attributed to variations in at least two unlinked genes between the two strains, but the mechanism remains unclear [17].
In recent years, genetic methods for studying S. japonicus (e.g., transformation [18,19] and spore dissection [20]) and bioresources (e.g., heterothallic strains [15]) have become available. In addition, whole-genome sequences of S. japonicus were published by the Broad Institute in 2011 [21]. S. japonicus is gradually becoming a popular experimental microorganism in addition to S. pombe. However, because only a few S. japonicus wild strains have been isolated, mainly from fruits and plants such as grapes and from slime fluxes, information on its diversity and evolution, especially mating and sporulation, remains limited.
Although key aspects of yeast ecology remain poorly elucidated, it has been proposed that insects allow yeasts to disperse [22]. Many yeasts and insects depend similarly on a source of sugar, such as rotting fruit or floral nectar; thus, insects such as fruit flies, which are known to accumulate yeast and bacteria in their crop, may provide a means for immobile yeasts to access sugar sources and transfer between sources [23]. Furthermore, insects may also increase outbreeding between different yeast strains: experimental data have suggested that the gut environment of insects, including fruit flies, might facilitate outbreeding [24,25]. This might also assist in adaptive outbreeding among diverse yeasts in the wild.
In this study, we successfully isolated, for the first time to our knowledge, eight S. japonicus var. japonicus strains possessing different genetic backgrounds from Drosophila species sampled at two distinct locations in Japan. Interestingly, most of the isolated strains were found to have a high-temperature tolerance and to sporulate at high frequency on nitrogen-abundant medium, although the type strain of S. japonicus var. japonicus does not generally mate under this condition. Sequence analysis subsequently revealed that pheromone-related genes were diversified among the eight strains. Further genetic characterization is needed to understand the mating responses of S. japonicus under various culture conditions, but we believe that the information and resources presented in this study will facilitate future studies on signal transduction and the regulation of sporulation, as well as unraveling yeast ecology and evolution in nature.
2. Materials and Methods
2.1. Yeast Strains, Media, and Culture Conditions
The wild strains of S. japonicus examined used in this study are listed in Table 1. Three strains (FY16936, FY32181*, and FY33672) were provided by the National Bio-Resource Project (NBRP), Japan (https://yeast.nig.ac.jp/yeast/top.xhtml (accessed on 16 February 2021)). Six strains (NBRC1646, NBRC1712, NBRC1713, NBRC10527*, NBRC10528*, and NBRC10529*) were obtained from the Biological Resource Center, NITE (NBRC), Japan (https://www.nite.go.jp/en/index.html (accessed on 16 February 2021)). The other strains were isolated by Taisuke Seike and Natsue Sakata from Drosophila captured inside RIKEN Center for Biosystems Dynamics Research or Suita Campus at Osaka University, Japan. Strain TS16 (h− sxa2::kanMX6) constructed in this study was a derivative of NIG5872 (h−) [15].
Table 1.
S. japonicus wild strains used in this study.
| Strain | Variety | Origin | Source | Reference |
|---|---|---|---|---|
| FY16936 | S. japonicus var. japonicus | Japan, Kyushu University | strawberry | NBRP |
| FY32181 * | S. japonicus var. versatilis | USA | home-canned grape juice | NBRP |
| FY33672 | S. japonicus var. japonicus | Japan, Suzuka College | NBRP | |
| NBRC1646 | S. japonicus var. japonicus | Japan | slime flux of tree | NBRC |
| NBRC1712 | S. japonicus var. japonicus | Unknown | NBRC | |
| NBRC1713 | S. japonicus var. japonicus | Unknown | NBRC | |
| NBRC10527 * | S. japonicus var. versatilis | Unknown | grape juice | NBRC |
| NBRC10528 * | S. japonicus var. versatilis | Portugal | white wine | NBRC |
| NBRC10529 * | S. japonicus var. versatilis | Unknown | slime flux, Ulmus carpinifolia | NBRC |
| TN33 | S. japonicus var. japonicus | Japan, RIKEN | Drosophila | This study |
| TN40 | S. japonicus var. japonicus | Japan, RIKEN | Drosophila | This study |
| TN82 | S. japonicus var. japonicus | Japan, RIKEN | Drosophila | This study |
| TN83 | S. japonicus var. japonicus | Japan, RIKEN | Drosophila | This study |
| TN84 | S. japonicus var. japonicus | Japan, RIKEN | Drosophila | This study |
| TN104 | S. japonicus var. japonicus | Japan, RIKEN | Drosophila | This study |
| TN108 | S. japonicus var. japonicus | Japan, RIKEN | Drosophila | This study |
| TN313 | S. japonicus var. japonicus | Japan, Osaka University | Drosophila | This study |
In this study, to distinguish between the two subspecies more easily, S. japonicus var. versatilis strains were marked with an asterisk (*).
Cells were typically grown in YE medium (5 g/L of Bacto Yeast Extract [BD Bioscience, Sparks, MD, USA] and 30 g/L of D-glucose [Wako Pure Chemicals, Osaka, Japan], supplemented with adenine sulfate [200 mg/L], uracil [200 mg/L], and leucine [100 mg/L]). For the solid medium, 15 g/L of agar [Wako Pure Chemicals] was added to the YE medium to form the YEA medium. Where appropriate, geneticin (G418) disulfate (Nacalai Tesque, Kyoto, Japan) was added to YEA medium at a final concentration of 100 μg/mL. For hyphal induction, YMoA medium (35 g/L of DifcoTM yeast morphology agar [BD Bioscience]) was used. For mating and sporulation, the MEA medium (30 g/L of extract malt [Nacalai Tesque] and 15 g/L of agar, pH 5.5) and Edinburgh minimal medium without nitrogen (EMM2−N; 26.77 g/L of EMM−Nitrogen [Sunrise Science Products, Knoxville, TN, USA]) were used. For solid medium, 20 g/L of agar was added to EMM2−N medium to form EMM2−N agar medium. For vegetative growth, hyphal growth, mating, and sporulation, cells were incubated at 30 °C, unless stated otherwise.
2.2. Isolation of S. japonicus from Drosophila
To collect large numbers of Drosophila, we used commercially available bananas. We placed a piece of ripe bananas in a milk carton and then hung it with a rope on tree branches located approximately 2 m from the ground. After a few days, we covered the milk carton with an aseptic plastic bag and captured Drosophila that escaped toward the bag by tapping the carton. Next, in the laboratory, we transferred the obtained Drosophila (about ten) in a clean 1.5-mL tube, and then washed their body surface with sterilized water. We homogenized Drosophila to obtain samples, including their crops, using a homogenizer pestle (INA•OPTIKA, Osaka, Japan). The samples were diluted with sterilized water and spread onto YEA plates containing 100 µg/mL ampicillin and 100 µg/mL chloramphenicol (to prevent the growth of any bacteria) at 30 °C for 2 days. The colonies that appeared on YEA plates were classified based on colony color (white, yellowish, and red) and surface (diaphanous or opaque), and their morphology was inspected using a microscope. Consequently, we collected several wild yeasts. Most of the isolated strains were budding-type yeasts, such as those from the genera Candida, Pichia, and Hanseniaspora. However, some strains were judged to be the fission yeast S. japonicus based on their fission proliferation pattern, larger cell size, and formation of eight spores.
2.3. Sequence Analysis of S. japonicus Wild Strains
Genomic DNA was extracted from cultures grown overnight in YE medium by using a Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA). The D1/D2 divergent domains of the large subunit ribosomal DNA (rDNA) were determined by sequencing using the primers oTS1250/oTS1251 [26] and were used to identify the varieties of S. japonicus (Table 1). Each of the DNA fragments containing map2 (P-factor gene; SJAG_00781), map3 (M-factor receptor gene; SJAG_01345), and mam2 (P-factor receptor gene; SJAG_01928) was amplified by using the respective primer sets: oTS1254/1255, oTS1258/1259, and oTS1262/1263 (all primers are listed in Table S1). The PCR products were sequenced by using internally specific primers: oTS1256 (map2), oTS1260/oTS1261 (mam2), and oTS1264/oTS1265 (map3). The sequences obtained were compared with those of the FY16936 strain; the differences are listed in Table 2 and Table S2.
Table 2.
Polymorphisms of pheromone-associated genes in the 17 S. japonicus strains investigated.
| Gene | Amino acid Substitution | No. of Strains | Strain |
|---|---|---|---|
|
map2 (SJAG_00781) |
WT | 15 | FY16936, FY32181 *, FY33672, NBRC1646, NBRC1712, NBRC1713, NBRC10527 *, NBRC10529 *, TN33, TN40, TN82, TN83, TN84, TN104, TN108 |
| S22N/N32V | 1 | NBRC10528 * | |
| A29S | 1 | TN313 | |
|
map3 (SJAG_01345) |
WT | 11 | FY16936, FY33672, NBRC1646, NBRC1713, TN40, TN82, TN83, TN84, TN104, TN108, TN313 |
| S3P/V4E/M27L/V43L/I54V/I59V/F117L/I121V/I125V/I179V/T222S/M223L/M261L | 4 | FY32181 *, NBRC10527 *, NBRC10528 *, NBRC10529 * | |
| A44S/L169V | 1 | NBRC1712 | |
| I328V | 1 | TN33 | |
|
mam2 (SJAG_01928) |
WT | 10 | FY16936, FY32181 *, TN33, TN40, TN82, TN83, TN84, TN104, TN108, TN313 |
| A13T/T28V/V170I | 1 | FY33672 | |
| A185G | 1 | NBRC1646 | |
| V170I | 1 | NBRC1712 | |
| A13T/V170I | 2 | NBRC1713, NBRC10527 * | |
| A13T/V170I/R319G | 1 | NBRC10528 * | |
| A13T/V170I/S277W/L283F R319G/S330F | 1 | NBRC10529 * |
S. japonicus var. versatilis strains were marked with an asterisk (*).
2.4. Growth Assay at Various Temperatures
Cells were pre-cultured on YEA medium at 30 °C overnight, harvested in sterilized water, and inoculated at an optical density at 600 nm of 0.1 (OD600 = 0.1) into 1 mL of YE medium. Next, 200-µL of cell culture was added to each well of 96-well plates (CELLSTAR 96-Well Plates Polystyrene flat bottom well plates; Greiner Bio-One, Kremsmünster, Austria). During 24 hours of cultivation with continuous shaking in orbital mode at 355 cpm (4 mm) at various temperatures (30, 37, 40, and 42 °C), the OD600 of each well containing S. japonicus cells was measured every 30 min automatically by using a microplate reader (SynergyTM HTX Multi Mode Microplate Reader; BioTek Instruments, Winooski, VT, USA). The growth assay was performed in quadruplicate for each sample (n = 4), and the mean ± standard deviation (SD) of growth rate (per hour) was calculated as described previously [27].
2.5. Spotting Assay
Cells were pre-cultured on YEA medium at 30 °C overnight and then diluted to an OD600 of 10. The diluted cultures were added to sterile 96-well plates and four fivefold serial dilutions (OD600 of 2.0, 0.4, 0.08, and 0.016) were prepared by adding sterilized water. A 6-µL aliquot of the resulting cell suspensions was spotted onto YEA plates and incubated at 42 °C for 3 days. After incubation, each plate was photographed.
2.6. Hyphal Formation
Cells were pre-cultured on YEA medium at 30 °C overnight, harvested in sterilized water, and diluted to an OD600 of 0.1. A 5-µL aliquot of the diluted culture was spotted in the center of a YMoA plate, and incubated at 30 °C for 10 days, as described previously [9]. Colonies with hyphal zones were photographed.
2.7. Mating Frequency
Cells grown on YEA plates overnight were resuspended in sterilized water at an OD600 of 5.0. A 30-μL aliquot of the resulting suspension was spotted onto EMM2−N or YEA plates (including a nitrogen source) and incubated for 48 hours at 30 °C as described previously [20]. The number of cells was counted under a differential interference contrast (DIC) microscope (BX53; Olympus, Tokyo, Japan). Cell types were classified into four groups: vegetative cells (V), zygotes (Z), asci (A), and spores (S). Mating was calculated by using the following equation [28]:
| Mating (%) = 100 × (2Z + 2A + S/4)/(V + 2Z + 2A + S/4) |
In all cases, S. japonicus cells were counted and classified in nine randomly photographed digital images of each strain (>400 cells counted in total), and the mean ± SD was calculated.
2.8. Iodine Staining
Cells grown on YEA plates overnight were resuspended in sterilized water at an OD600 of 0.1. A 6-μl aliquot of the resulting suspension was spotted onto MEA plates and then incubated at 30 °C for 2 days. The colonies of S. japonicus strains on MEA plates were stained by iodine vapor (Wako Pure Chemicals) for 1 min. The stained colonies were photographed.
2.9. Plasmid Construction and Transformation
To delete the sxa2+ gene, encoding P-factor-degrading enzyme (SJAG_00975), from S. japonicus cells, the 5’-upstream sequence (500 bp) of sxa2+ was first amplified from FY16936 genomic DNA by using the primers oTS9/oTS10 (Table S1), and the BamHI/PacI fragment was cloned into pFA6a-kanMX6 to create pTS2 [pFA6a-kanMX6(Sjsxa2UP)]. Next, the 3’-downstream sequence (1 kb) of the sxa2+ gene was amplified from FY16936 genomic DNA by using the primers oTS61/oTS62, and the DNA fragment was fused to a linearized vector derived from pTS2, which was prepared by inverse PCR using KOD FX Neo (TOYOBO, Osaka, Japan) with primers oTS37/oTS38 by Gibson Assembly (New England Biolabs, Ipswich, MA, USA). The resultant plasmid was called pTS7 [pFA6a-kanMX6(Sisxa2UPDOWN)]. A 3-kb fragment was amplified from pTS7 and purified by using the QIAquick PCR Purification Kit (Qiagen, Tokyo, Japan). The DNA fragment was transformed in a heterothallic M-strain, NIG5872, by electroporation as described previously [18], and transformants were selected on YEA containing G418. Primers oTS9/oTS38 were used to confirm that the ORF of sxa2+ had been deleted.
2.10. Shmoo Formation by Pheromone Treatment
P-factor peptides were chemically synthesized (Eurofins Genomics, Tokyo, Japan) [29]. The purity of the preparations was confirmed to be more than 95% by HPLC. Each P-factor was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 500 µM. For the assay, a heterothallic haploid S. japonicus strain lacking the endogenous sxa2+ gene was used. The cells were grown in YE medium overnight, washed with sterilized water three times, and then resuspended in EMM2−N medium at OD600 of 2.0. The cells were incubated with each P-factor peptide (5 µM) for exactly 24 hours with gentle shaking and then observed by DIC microscopy.
2.11. Data Availability
All relevant data are included in the paper and the Supporting Information file (Table S3).
3. Results
3.1. Isolation of the Fission Yeast S. japonicus var. japonicus from Drosophila
To investigate the diversity of the fission yeast S. japonicus in sexual reproduction, we obtained the following wild strains from NBRP and NBRC: for S. japonicus var. japonicus, the type strain FY16936 (also known as ATCC10660/IFO1609) and four other strains; for S. japonicus var. versatilis, the type strain FY32181* (also known as ATTC9987/IFO1607) and three other strains (Table 1). These nine strains were isolated mainly from fruits and plants. We, therefore, attempted to isolate additional S. japonicus species from an insect source. Because fruit flies feed on immobile yeast and are known to accumulate yeast and bacteria in their crop [1], we attempted to isolate novel S. japonicus strains from Drosophila. By trapping hundreds of flies with a banana, we successfully isolated eight S. japonicus strains from two locations: the campus of RIKEN and Osaka University in Japan (Table 1). We determined the sequences of the D1/D2 domains of rDNA genes in these strains, which confirmed that all of the strains isolated were S. japonicus var. japonicus. Below, we analyzed similarities and differences in various features of the 17 wild S. japonicus strains (Table 1).
3.2. Heat Tolerance in Most of the Strains Isolated from Drosophila
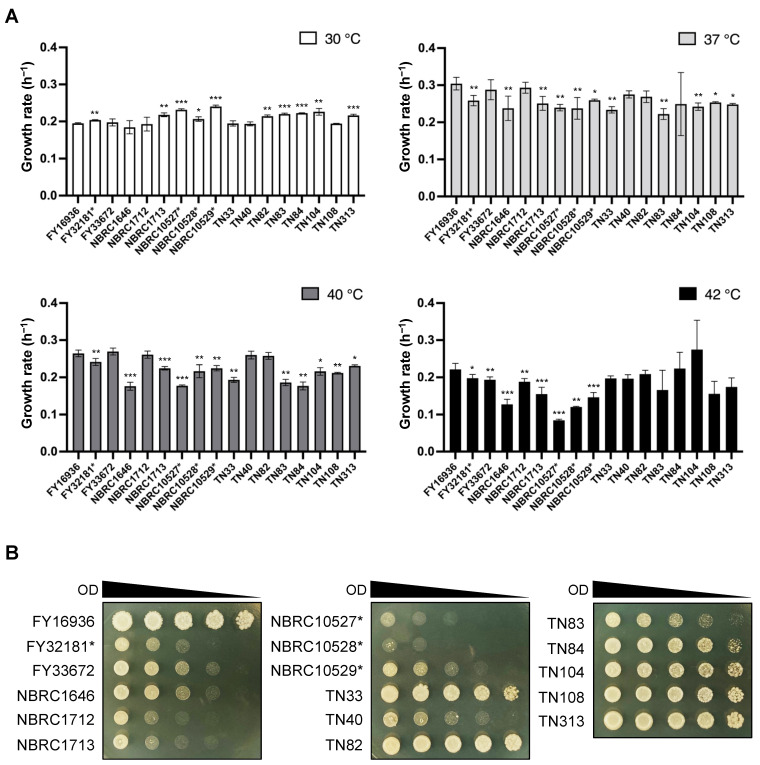
In the genus Schizosaccharomyces, S. japonicus is the only species that can grow at high temperatures (e.g., 40 °C) [13]. We, therefore, compared the growth rate of the 17 S. japonicus wild strains under high-temperature conditions. Cells were incubated in YEL medium at 30, 37, 40, and 42 °C, and the OD600 was measured with a photo recorder. At 30 °C, S. japonicus var. versatilis strains grew slightly faster than S. japonicus var. japonicus strains (e.g., the growth rate of NBRC10529* vs FY16936 was 0.241 ± 0.004 h−1 vs 0.195 ± 0.002 h−1, respectively); by contrast, most S. japonicus var. versatilis strains showed slower growth at 42 °C (Figure 1A). Almost all strains of S. japonicus grew fastest at 37 °C, consistent with findings from a previous study [6].
Figure 1.
Effects of high temperature on growth. (A) Growth rate per hour (h−1) of 17 S. japonicus strains incubated in YEL medium at various temperatures (30 °C, 37 °C, 40 °C, and 42 °C). Data are the mean ± SD of quadruplicate samples. t-test: * p < 0.05; ** p < 0.01; *** p < 0.001. (B) Heat tolerance of the strains. Samples were withdrawn from cultures, and the OD600 was adjusted to 10. Fivefold serial dilutions (from 5−1 to 5−4) were prepared in sterilized water, and 6-μL of each sample (the culture at OD600=10 and its dilutions) spotted onto YEA plates. The plates were incubated at 42 °C for 3 days and photographed.
To further evaluate tolerance to high temperature, we carried out spot assays of S. japonicus cells on YEA medium at 42 °C. These assays indicated that the growth of S. japonicus var. versatilis strains was severely inhibited at 42 °C (Figure 1B). Better growth was observed for the S. japonicus var. japonicus strains TN33, TN82–TN84, TN104, TN108, and TN313, as well as FY16936 (Figure 1B). Thus, most of the strains that were isolated from Drosophila showed tolerance to high-temperature stress.
3.3. Differences in Hyphal Formation among S. japonicus Strains
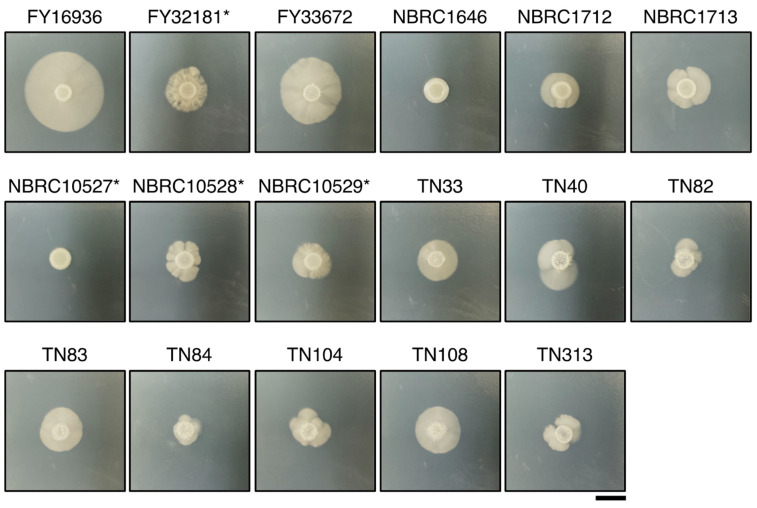
Unlike other fission yeast species, S. japonicus is a dimorphic yeast that can transit from unicellular yeast to filamentous hyphae [6]. Therefore, we examined whether the S. japonicus strains could undergo hyphal transition dependent on the culture medium. A conventional medium, yeast morphology agar (YMoA), has been reported to induce a transition to hyphal cells in S. japonicus due to nutritional deficiencies [9]; therefore, we spotted S. japonicus cells on YMoA and incubated them at 30 °C for 10 days. Among the 17 strains, FY16936, the type strain of S. japonicus var. japonicus, showed the biggest hyphal zone on the plate, and FY33672 also showed strong induction of hyphae (Figure 2). By contrast, NBRC1646 and NBRC10527* did not show any hyphal zones under this condition (Figure 2). Among the eight strains isolated from flies, the size of the hyphal zone differed to some extent (Figure 2), implying that S. japonicus var. japonicus strains are diversified in Drosophila.
Figure 2.
Differences in hyphal formation. Hyphal growth of 17 S. japonicus strains incubated on YMoA medium at 30 °C for 10 days. Two strains, NBRC1646 and NBRC10527*, did not form hypha under these conditions. S. japonicus var. versatilis strains were marked with an asterisk (*). Scale bar, 1 cm.
3.4. Elevated Sporulation Efficiency in S. japonicus Strains Isolated from Drosophila
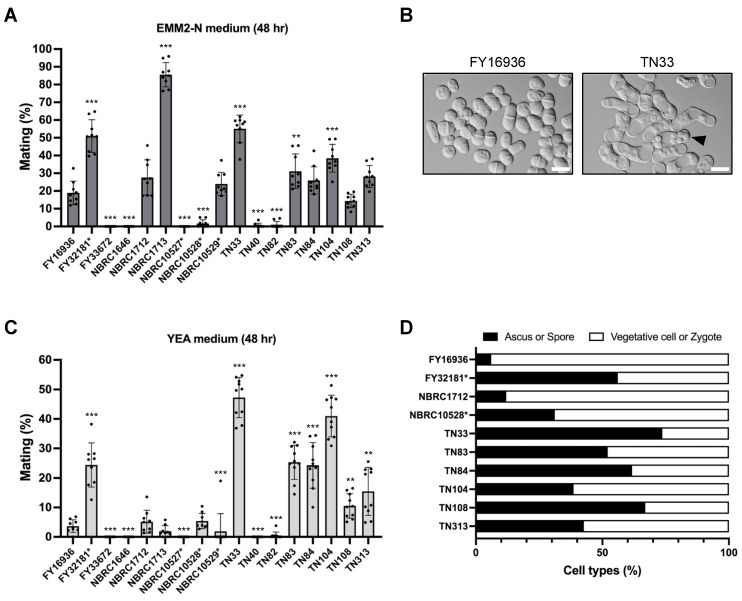
Next, we compared the mating efficiency of the 17 S. japonicus wild strains on a standard sporulation medium, EMM2−N. After 48 hours of incubation at 30 °C, some S. japonicus strains mated and formed asci with eight spores; however, the mating frequency varied greatly (Figure 3A). For example, cells mated most efficiently in the NBRC1713 strain (85.6% ± 6.8%; Figure 3A), whereas the FY33672, NBRC1646, and NBRC10527* strains were completely sterile (0%). Six of the eight strains isolated from Drosophila mated at a relatively high frequency (ranging from 14.4% ± 3.8% to 55.0% ± 7.7%), but the remaining two strains, TN40 and TN82, hardly mated on EMM2−N agar medium (Figure 3A).
Figure 3.
Mating response on different media. Mating frequency of 17 S. japonicus strains incubated on EMM2−N (A) or YEA (C) medium at 30 °C for 48 hours. At least 400 cells were examined for each strain. Data are the mean ± SD of nine samples. (B) Micrographs showing FY16936 (wild strain) and TN33 (a strain isolated from flies) incubated on YEA medium at 30 °C for 1 day. Scale bar, 10 µm. (D) Cell type proportion of some strains. Black bars indicate ascus or spore; white bars indicate vegetative cell or zygote. t-test: * p < 0.05; ** p < 0.01; *** p < 0.001.
We noticed that the strains isolated from Drosophila mated and sporulated even on YEA medium containing an abundant nitrogen source (Figure 3B); therefore, we also examined mating efficiency on YEA medium in the same way. Notably, strains TN33, TN83, TN84, TN104, TN108, and TN313 showed markedly high mating frequency under this condition (Figure 3C). In particular, approximately half of the cells of the TN33 strain mated on YEA medium after incubation at 30 °C for 48 hours (48.3% ± 6.3%). This tendency was also seen in FY32181*, the type strain of S. japonicus var. versatilis (Figure 3C). Interestingly, these strains that showed high mating frequency on the YEA medium also formed spores at high frequency after mating (Figure 3D). As far as we know, the mating response is not induced in laboratory strains of other fission yeast species under this condition. Thus, these data, showing that most of the strains isolated from Drosophila can sporulate even on YEA medium, may reflect a specific environment in Drosophila.
3.5. The TN33 Strain of S. japonicus var. japonicus Shows Iodine-Positivity
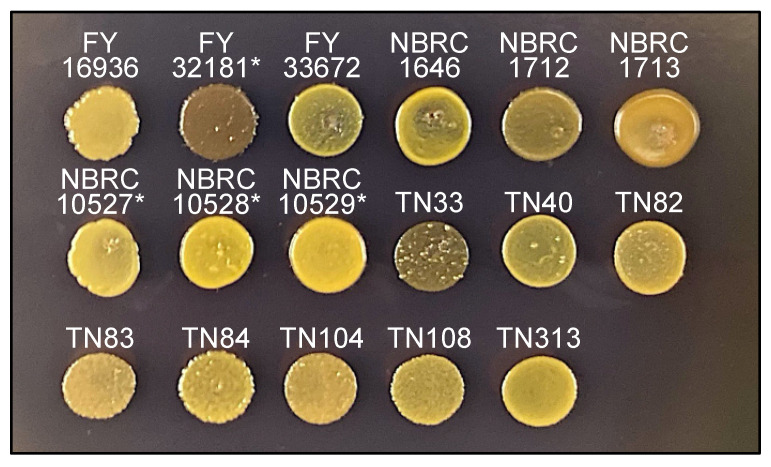
Sporulated colonies of S. pombe and S. octosporus are stained by iodine vapors [28] because the spores consist of a starch-like compound. Spores of the type strain of S. japonicus var. versatilis are also stained with iodine vapor; however, those of S. japonicus var. japonicus are not [15,16], although the underlying reason is unknown.
Next, therefore, we examined whether the 17 S. japonicus wild strains are stained by iodine vapors. As expected, after treatment of the strains on sporulation medium (MEA) with iodine vapor, colonies of the FY32181* strain were clearly stained, but those of the FY16936 strain were not (Figure 4). Unexpectedly, among the strains that were isolated from Drosophila, the TN33 strain was strongly stained dark brown by iodine vapor after 1 minute (Figure 4). Possibly, S. japonicus cells might mate repeatedly with each other in Drosophila, leading to the occurrence of a strain such as TN33 with a genetic background showing positive iodine staining.
Figure 4.
Iodine staining. Sporulating cells of 17 S. japonicus strains incubated on MEA medium at 30 °C for 2 days were stained by iodine vapor for 1 min. As reported previously, the type stain (FY16936) of S. japonicus var. japonicus was negatively stained, whereas the type stain (FY32181*) of S. japonicus var. versatilis was positively stained. Under this condition, TN33 was strongly positive; the others stains were negative (or weakly positive).
3.6. Differences in Pheromone-Related Genes among S. japonicus Strains
The S. japonicus strains that we investigated showed diverse phenotypes in sexual reproduction (Figure 2, Figure 3 and Figure 4); therefore, we surmised that the mating pheromones and their corresponding receptor genes were diversified among the strains. Fission yeast has two mating types, Plus and Minus, which each secrete the respective mating pheromones p-factor and M-factor for mating [30,31]. Because the M-factor gene has not been identified in S. japonicus, we sequenced the P-factor gene (map2+), M-factor receptor gene (map3+), and P-factor receptor gene (mam2+) of all 17 strains and compared the nucleotide sequences with those of the FY16936 strain. Many nucleotide differences in the three genes were found among the strains (Table S2). As summarized in Table 2, the amino acid sequences of Map3 and Mam2 were relatively diversified; for example, the map3 gene was identical in all four strains of S. japonicus var. versatilis, whereas there were many nucleotide differences and 14 amino acid differences between the Map3 proteins of S. japonicus var. japonicus and versatilis (Table S2). In addition, some mutations in the mam2 genes were common to strains isolated in certain regions and countries (Table S2).
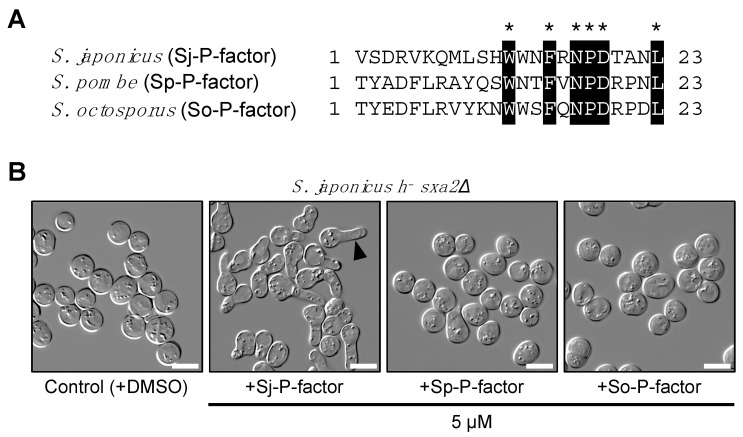
In contrast, sequencing indicated that the map2 gene of all 17 strains produced four copies of an identical mature P-factor peptide, VSDRVKQMLSHWWNFRNPDTANL (Figure 5A), although some missense and silent mutations were found in the pro sequences and mature pheromone sequences (Table S2). Therefore, we chemically synthesized this simple peptide and used it to treat S. japonicus cells. In S. pombe, P-factor is degraded outside the cell by the carboxyl peptidase Sxa2 [32,33,34]. Here, therefore, we constructed a heterothallic M-type strain (TS16) lacking the sxa2+ gene (SJAG_00975) and examined whether the corresponding P-factor peptide of the three fission yeast species can be recognized by Mam2 of S. japonicus in the absence of Sxa2. The TS16 strain was treated with synthetic P-factors at 5 µM in EMM2−N medium and the formation of a shmoo – the mating projection that is elongated when yeast cells sense pheromones – was monitored. When treated with Sj-P-factor, S. japonicus cells showed sufficient shmoo formation, whereas the other two P-factor peptides, Sp-P-factor and So-P-factor, were not recognized at all (Figure 5B). These data indicate that Mam2 of S. japonicus specifically recognizes Sj-P-factor.
Figure 5.
Shmooing assay using synthetic P-factor peptides. (A) Amino acid alignment of P-factor peptide from three Schizosaccharomyces species. Numerals to the right of each sequence indicate the length of the peptide product. Identical amino acids in the three peptides are highlighted by asterisks (*). (B) Shmoo formation in S. japonicus M cells lacking the sxa2+ gene (h− sxa2Δ). Cells were treated with each of the synthetic P-factors at 5 µM and incubated in EMM2−N medium with gentle shaking for 24 hours. Arrow indicates a representative shmooing cell. Scale bar, 10 μm.
3.7. Diversify of S. japonicus Wild Strains in Nature
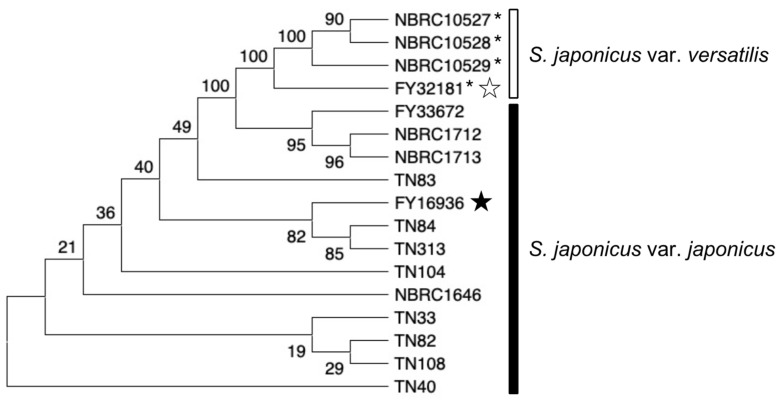
We constructed a phylogenetic tree based on the sequences of three genes (map2, map3, and mam2) in the 17 S. japonicus strains (Figure 6). In the tree, strains of each subspecies were clustered, while the evolutionary distance among the strains was not necessarily consistent with sampling locations. For example, the TN84 strain (isolated at RIKEN) is evolutionarily closer to the TN313 strain (isolated at Osaka University) than to the TN40 strain (isolated at RIKEN). These data suggest that Drosophila consume yeast while flying around the neighborhood, and accumulate many strains of yeast in their crop for a while. A strategy for isolating yeasts from highly mobile flies might be effective for the collection of yeasts showing various phenotypes.
Figure 6.
Evolutionary relationship of S. japonicus strains. Phylogenetic tree of the 17 S. japonicus wild strains investigated in this study. The analysis combined multiple sequence alignments for the map2, map3, and mam2 genes. The evolutionary history was inferred by using neighbor-joining [35] and the Kimura 2-parameter method [36], and the number of base substitutions per site is shown. All ambiguous positions were removed for each sequence pair. In total, there were 2766 positions in the final data set. Evolutionary analyses were conducted in MEGA X [37,38]. Black and white stars indicate the type strain of S. japonicus var. japonicus and S. japonicus var. versatilis, respectively. S. japonicus var. versatilis strains were marked with an asterisk (*).
4. Discussion
Yeasts are widely dispersed in nature across a variety of habitats and are commonly found on fruits and plants, in alcohol, as well as on the surface of the skin of animals and humans. In this study, we isolated eight strains from fruit flies sampled at two distinct locations in Japan, indicating that the fission yeast S. japonicus species may live symbiotically (or temporally) in Drosophila. As far as we know, this is the first report of the isolation of this species from insects.
It is well documented that Drosophila species in the wild use yeast as a food source [39,40,41]. Laboratory studies on the budding yeast Saccharomyces cerevisiae and Drosophila melanogaster have demonstrated that almost all vegetative cells are killed by passage through the gut in D. melanogaster, but spores show enhanced survival [24,42]. Spores are quiescent cells that have resistance to various environmental stimuli, probably due to their two-layer outer wall, which comprises chitosan [43] and a di-tyrosine (in S. cerevisiae [44]) or isp3 (in S. pombe [45]) polymer. Accordingly, spores have resistance to stresses such as ethanol, low and high pH, and high temperatures (>50 °C) [45,46,47]. These experimental data might explain our finding that S. japonicus strains isolated from Drosophila show elevated sporulation efficiency (Figure 3). Our sequencing analysis further showed that the strains highlighted from Drosophila have different genetic backgrounds (Table 2 and Table S2). Interestingly, a previous study reported that outbreeding of S. cerevisiae can be enhanced by the degradation of interspore bridges by Drosophila [24]. Therefore, we speculate that sporulation followed by mating is facilitated in Drosophila, where crossing between closely related species may frequently occur.
In many immobile organisms such as plants and fungi, the sexual reproduction phase (pollen and spores) is the mobile phase of the life cycle. A strategy to migrate to a new environment via predation by flies (or other animals and humans) is probably convenient because fries give yeast not only the ability to disperse but also a higher chance of migrating to a viable environment (e.g., the host’s preferred food) rather than just dispersing. Insects can increase the probability of outbreeding by flying from flower to flower in search of nectar, collecting yeasts and their spores, and then dropping them all in one place.
Isolation of yeasts from insects such as Drosophila will be beneficial for the collection of yeasts that have various phenotypes due to genetic diversification, such as the iodine-positive S. japonicus var. japonicus strain TN33 identified herein (Figure 4). Further detailed analysis of other phenotypes of these strains, such as nutritional requirement, mating type, and the effects of mutations in pheromone-related genes, should be conducted in the future. We hope that the information and resources presented in this study will provide a basis for future studies on the signal transduction mechanisms of sporulation, as well as the ecology and evolution of yeast.
Acknowledgments
We thank the National Bio-Resource Project (NBRP), Japan, and the Biological Resource Center, NITE (NBRC) for providing S. japonicus strains and plasmids. We are also grateful to Y. Ishikawa and R. Tanaka (Nagoya University, Japan), H. Takekata (University of the Ryukyus, Japan) for technical support in catching Drosophila samples, H. Niki (National Institute of Genetics) for helpful comments, and all members of our laboratory for useful suggestions.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jof7050350/s1, Table S1: Primers used in this study. Table S2: Nucleotide sequences of three pheromone-associated genes (map2, map3, and mam3) in 17 wild S. japonicus strains, Table S3: Numerical data for all of the graphs presented in this study.
Author Contributions
Conceptualization: T.S.; Data curation: T.S.; Formal analysis: T.S.; Funding acquisition: T.S.; Investigation: T.S. and N.S.; Methodology: T.S. and N.S.; Project administration: T.S.; Resources: T.S., N.S., F.M., and C.F.; Visualization: T.S.; Writing—original draft: T.S.; Writing—review and editing: F.M. and C.F. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Japan Society for the Promotion of Science KAKENHI https://kaken.nii.ac.jp/en/index/ (accessed on 16 February 2021) (Grant Number JP19K16197, JP20H04790, and JP21K05342) to TS. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All relevant data are included in the paper and its Supplementary Materials.
Conflicts of Interest
The authors have declared that no competing interests exist.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mrak E.M. The fate of yeast in the digestive tract of drosophila. Am. Nat. 1951;85:381–383. doi: 10.1086/281692. [DOI] [Google Scholar]
- 2.Sipiczki M. Where does fission yeast sit on the tree of life? Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-2-reviews1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinow C., Hyams J. Molecular Biology of the Fission Yeast. Elsevier; Amsterdam, The Netherlands: 1989. General cytology of fission yeasts; pp. 273–330. [Google Scholar]
- 4.Aoki K., Hayashi H., Furuya K., Sato M., Takagi T., Osumi M., Kimura A., Niki H. Breakage of the nuclear envelope by an extending mitotic nucleus occurs during anaphase in Schizosaccharomyces japonicus. Genes Cells. 2011;16:911–926. doi: 10.1111/j.1365-2443.2011.01540.x. [DOI] [PubMed] [Google Scholar]
- 5.Yam C., He Y., Zhang D., Chiam K.-H., Oliferenko S. Divergent strategies for controlling the nuclear membrane satisfy geometric constraints during nuclear division. Curr. Biol. 2011;21:1314–1319. doi: 10.1016/j.cub.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 6.Niki H. Schizosaccharomyces japonicus: The fission yeast is a fusion of yeast and hyphae. Yeast. 2013;31:83–90. doi: 10.1002/yea.2996. [DOI] [PubMed] [Google Scholar]
- 7.Nemecek J.C. Global control of dimorphism and virulence in fungi. Science. 2006;312:583–588. doi: 10.1126/science.1124105. [DOI] [PubMed] [Google Scholar]
- 8.Sipiczki M., Takeo K., Yamaguchi M., Yoshida S., Miklos I. Environmentally controlled dimorphic cycle in a fission yeast. Microbiology. 1998;144:1319–1330. doi: 10.1099/00221287-144-5-1319. [DOI] [PubMed] [Google Scholar]
- 9.Nozaki S., Furuya K., Niki H. The Ras1-Cdc42 pathway is involved in hyphal development of Schizosaccharomyces japonicus. FEMS Yeast Res. 2018;18 doi: 10.1093/femsyr/foy031. [DOI] [PubMed] [Google Scholar]
- 10.Kinnaer C., Dudin O., Martin S.G. Yeast-to-hypha transition of Schizosaccharomyces japonicus in response to environmental stimuli. Mol. Biol. Cell. 2019;30:975–991. doi: 10.1091/mbc.E18-12-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furuya K., Niki H. The DNA damage checkpoint regulates a transition between yeast and hyphal growth in Schizosaccharomyces japonicus. Mol. Cell. Biol. 2010;30:2909–2917. doi: 10.1128/MCB.00049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okamoto S., Furuya K., Nozaki S., Aoki K., Niki H. Synchronous activation of cell division by light or temperature stimuli in the dimorphic yeast Schizosaccharomyces japonicus. Eukaryot. Cell. 2013;12:1235–1243. doi: 10.1128/EC.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaino T., Tonoko K., Mochizuki S., Takashima Y., Kawamukai M. Schizosaccharomyces japonicus has low levels of CoQ10 synthesis, respiration deficiency, and efficient ethanol production. Biosci. Biotechnol. Biochem. 2018;82:1031–1042. doi: 10.1080/09168451.2017.1401914. [DOI] [PubMed] [Google Scholar]
- 14.Wickerham L.J., Duprat E. A Remarkable fission yeast, Schizosaccharomyces versatilis nov. sp. J. Bacteriol. 1945;50:597–607. doi: 10.1128/JB.50.5.597-607.1945. [DOI] [PubMed] [Google Scholar]
- 15.Furuya K., Niki H. Isolation of heterothallic haploid and auxotrophic mutants of Schizosaccharomyces japonicus. Yeast. 2009;26:221–233. doi: 10.1002/yea.1662. [DOI] [PubMed] [Google Scholar]
- 16.Yu C., Bonaduce M.J., Klar A.J.S. Defining the epigenetic mechanism of asymmetric cell division of Schizosaccharomyces japonicus Yeast. Genetics. 2013;193:85–94. doi: 10.1534/genetics.112.146233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klar A.J.S. Schizosaccharomyces japonicus yeast poised to become a favorite experimental organism for eukaryotic research. Genes Genomes Genet. 2013;3:1869–1873. doi: 10.1534/g3.113.007187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aoki K., Niki H. Transformation of Schizosaccharomyces japonicus. Cold Spring Harb. Protoc. 2017;2017 doi: 10.1101/pdb.prot091850. [DOI] [PubMed] [Google Scholar]
- 19.Aoki K., Nakajima R., Furuya K., Niki H. Novel episomal vectors and a highly efficient transformation procedure for the fission yeast Schizosaccharomyces japonicus. Yeast. 2010;27:1049–1060. doi: 10.1002/yea.1815. [DOI] [PubMed] [Google Scholar]
- 20.Furuya K., Niki H. Mating, spore dissection, and selection of diploid cells in Schizosaccharomyces japonicus. Cold Spring Harb. Protoc. 2017;2017 doi: 10.1101/pdb.prot091843. [DOI] [PubMed] [Google Scholar]
- 21.Rhind N., Chen Z., Yassour M., Thompson D.A., Haas B.J., Habib N., Wapinski I., Roy S., Lin M.F., Heiman D.I., et al. Comparative Functional Genomics of the Fission Yeasts. Science. 2011;332:930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madden A.A., Epps M.J., Fukami T., Irwin R.E., Sheppard J., Sorger D.M., Dunn R.R. The ecology of insect—Yeast rela-tionships and its relevance to human industry. Proc. R. Soc. B Biol. Sci. 2018;285:20172733. doi: 10.1098/rspb.2017.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buzzini P., Lachance M.A., Yurkov A. Yeasts in Natural Ecosystems: Diversity. Springer; Cham, Switzerland: 2017. [Google Scholar]
- 24.Reuter M., Bell G., Greig D. Increased outbreeding in yeast in response to dispersal by an insect vector. Curr. Biol. 2007;17:R81–R83. doi: 10.1016/j.cub.2006.11.059. [DOI] [PubMed] [Google Scholar]
- 25.Stefanini I., Dapporto L., Berná L., Polsinelli M., Turillazzi S., Cavalieri D. Social wasps are a Saccharomyces mating nest. Proc. Natl. Acad. Sci. USA. 2016;113:2247–2251. doi: 10.1073/pnas.1516453113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spencer J., Rawling S., Stratford M., Steels H., Novodvorska M., Archer D., Chandra S. Yeast identification: Reassessment of assimilation tests as sole universal identifiers. Lett. Appl. Microbiol. 2011;53:503–508. doi: 10.1111/j.1472-765X.2011.03130.x. [DOI] [PubMed] [Google Scholar]
- 27.Moriya H., Shimizu-Yoshida Y., Kitano H. In vivo robustness analysis of cell division cycle genes in Saccharomyces cerevisiae. PLoS Genet. 2006;2:e111. doi: 10.1371/journal.pgen.0020111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seike T., Niki H. Mating response and construction of heterothallic strains of the fission yeast Schizosaccharomyces octosporus. FEMS Yeast Res. 2017;17:1–14. doi: 10.1093/femsyr/fox045. [DOI] [PubMed] [Google Scholar]
- 29.Seike T., Shimoda C., Niki H. Asymmetric diversification of mating pheromones in fission yeast. PLoS Biol. 2019;17:e3000101. doi: 10.1371/journal.pbio.3000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutz H., Heslot H., Leupold U.L.N. In: Handbook of Genetics. King R., editor. Plenum Press; New York, NY, USA: 1974. [Google Scholar]
- 31.Richard E. The Molecular Biology of Schizosaccharomyces Pombe. Springer; Berlin/Heidelberg, Germany: 1974. Fission yeast in general genetics; pp. 1–12. [Google Scholar]
- 32.Ladds G., Rasmussen M., Davey J. Characterisation of Sxa2, a protease involved in pheromone communication in fission yeast. Biochem. Soc. Trans. 1995;23:565S. doi: 10.1042/bst023565s. [DOI] [PubMed] [Google Scholar]
- 33.Ladds G., Rasmussen E.M., Young T., Nielsen O., Davey J. The sxa2-dependent inactivation of the P-factor mating pheromone in the fission yeast Schizosaccharomyces pombe. Mol. Microbiol. 1996;20:35–42. doi: 10.1111/j.1365-2958.1996.tb02486.x. [DOI] [PubMed] [Google Scholar]
- 34.Imai Y., Yamamoto M. Schizosaccharomyces pombe sxa1+ and sxa2+ encode putative proteases involved in the mating response. Mol. Cell. Biol. 1992;12:1827–1834. doi: 10.1128/MCB.12.4.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 36.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 37.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stecher G., Tamura K., Kumar S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 2020;37:1237–1239. doi: 10.1093/molbev/msz312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamby K.A., Hernández A., Boundy-Mills K., Zalom F.G. Associations of yeasts with spotted-wing drosophila (drosophila suzukii; diptera: Drosophilidae) in cherries and raspberries. Appl. Environ. Microbiol. 2012;78:4869–4873. doi: 10.1128/AEM.00841-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chandler J.A., Eisen J.A., Kopp A. Yeast communities of diverse drosophila species: Comparison of two symbiont groups in the same hosts. Appl. Environ. Microbiol. 2012;78:7327–7336. doi: 10.1128/AEM.01741-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koerte S., Keesey I.W., Easson M.L.A.E., Gershenzon J., Hansson B.S., Knaden M. Variable dependency on associated yeast communities influences host range in drosophila species. Oikos. 2020;129:964–982. doi: 10.1111/oik.07180. [DOI] [Google Scholar]
- 42.Coluccio A.E., Rodriguez R.K., Kernan M.J., Neiman A.M. The yeast spore wall enables spores to survive passage through the digestive tract of drosophila. PLoS ONE. 2008;3:e2873. doi: 10.1371/journal.pone.0002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Briza P., Ellinger A., Winkler G., Breitenbach M. Chemical composition of the yeast ascospore wall. The second outer layer consists of chitosan. J. Biol. Chem. 1988;263:11569–11574. doi: 10.1016/S0021-9258(18)37997-3. [DOI] [PubMed] [Google Scholar]
- 44.Briza P., Winkler G., Kalchhauser H., Breitenbach M. Dityrosine is a prominent component of the yeast ascospore wall. A proof of its structure. J. Biol. Chem. 1986;261:4288–4294. doi: 10.1016/S0021-9258(17)35659-4. [DOI] [PubMed] [Google Scholar]
- 45.Fukunishi K., Miyakubi K., Hatanaka M., Otsuru N., Hirata A., Shimoda C., Nakamura T. The fission yeast spore is coated by a proteinaceous surface layer comprising mainly Isp3. Mol. Biol. Cell. 2014;25:1549–1559. doi: 10.1091/mbc.e13-12-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dawes I.W., Hardie I.D. Selective killing of vegetative cells in sporulated yeast cultures by exposure to diethyl ether. Mol. Genet. Genom. 1974;131:281–289. doi: 10.1007/BF00264859. [DOI] [PubMed] [Google Scholar]
- 47.Briza P., Breitenbach M., Ellinger A., Segall J. Isolation of two developmentally regulated genes involved in spore wall maturation in Saccharomyces cerevisiae. Genes Dev. 1990;4:1775–1789. doi: 10.1101/gad.4.10.1775. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are included in the paper and its Supplementary Materials.