Abstract
Background
Alzheimer’s disease (AD) is a progressive and irreversible brain disorder. Hippocampus is one of the involved regions and its atrophy is a widely used biomarker for AD diagnosis. We have recently developed DenseCNN, a lightweight 3D deep convolutional network model, for AD classification based on hippocampus magnetic resonance imaging (MRI) segments. In addition to the visual features of the hippocampus segments, the global shape representations of the hippocampus are also important for AD diagnosis. In this study, we propose DenseCNN2, a deep convolutional network model for AD classification by incorporating global shape representations along with hippocampus segmentations.
Methods
The data was obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) and was T1-weighted structural MRI from initial screening or baseline, including ADNI 1,2/GO and 3. DenseCNN2 was trained and evaluated with 326 AD subjects and 607 CN hippocampus MRI using 5-fold cross-validation strategy. DenseCNN2 was compared with other state-of-the-art machine learning approaches for the task of AD classification.
Results
We showed that DenseCNN2 with combined visual and global shape features performed better than deep learning models with visual or global shape features alone. DenseCNN2 achieved an average accuracy of 0.925, sensitivity of 0.882, specificity of 0.949, and area under curve (AUC) of 0.978, which are better than or comparable to the state-of-the-art methods in AD classification. Data visualization analysis through 2D embedding of UMAP confirmed that global shape features improved class discrimination between AD and normal.
Conclusion
DenseCNN2, a lightweight 3D deep convolutional network model based on combined hippocampus segmentations and global shape features, achieved high performance and has potential as an efficient diagnostic tool for AD classification.
Keywords: Alzheimer’s disease, Hippocampus, Magnetic resonance imaging, 3D Convolutional neural network, Classification
Introduction
Alzheimer’s disease (AD) is a chronic neurological brain disorder characterized by memory loss and cognitive impairment [1]. Currently, there are no effective drug treatments available to cure AD, and the existing medicines can only ease symptoms or slow down its progression [2]. An early diagnosis of AD can help in determining its progression and also improve the quality of life of AD patients [3].
The current diagnosis of AD is made by clinical, neuropsychological, and neuroimaging assessments [4–9]. More recently, a variety of imaging modalities, including structural and functional magnetic resonance imaging (MRI) and positron emission tomography (PET) studies of cerebral metabolism, have shown characteristic changes in the brains of patients with AD [10–12]. MRI is considered the preferred neuroimaging examination for AD as it allows for high tissue contrast and accurate measurement of the 3-dimensional (3D) volume of brain structures, especially the size of the hippocampus and related regions [13]. Also, MRI has an exceptional spatial resolution, high accessibility, and good contrast. In recent years, extensive efforts have been done to identify biomarkers for structural changes and disease states of the brain with structural MRIs [12, 14].
Hippocampal atrophy measures from MRI are powerful biomarkers for monitoring AD progression [15, 16]. Features from the hippocampus have been studied for AD diagnosis-based structural MRIs [16–19]. Hippocampal visual features have been used in Support Vector Machines and Bayesian classifiers for AD diagnosis [17]. Hippocampal volume changes based on MRI were used as biomarkers and features for AD diagnosis [18]. However, methods based on volumetric analysis alone provide limited information about morphological changes that characterize the appearance and progression of AD. Recently, shape description/modeling methods have been proposed to analyze the development of AD [19]. Shape descriptors can be broadly divided into three categories: (1) histogram-based, (2) graph-based, and (3) transform-based shape descriptors. Some of the histogram-based descriptors are the shape spectrum, generalized shape distributions, probability density-based descriptors, 3D shape contexts, etc. [20]. The challenges here are to select discriminating shape functions and to robustly compute the dissimilarity between probability distributions [21]. The graph-based descriptors like medial axis, Reeb graph, skeletal graph, etc., capture geometrical and topological shape properties, but are more complex and difficult to be constructed and derived [22]. Transform-based descriptors mainly include Fourier transform descriptors, spherical space transform descriptor, spherical harmonics, etc. [23], and have been widely applied in the neuroimaging fields [24–26]. Gutman et al. performed shape analysis for the hippocampus using the spherical harmonic shape description [24]. Spherical harmonics requires spherical parameterization, a smooth mapping from the surface to a unit sphere [27]. But the shape descriptors based on Laplace Beltrami (LB) spectrum can be modeled for any Riemannian manifold, is isometry-invariant, and avoids pre-processing steps like mapping, registration, and alignment [28]. In [25], shape changes of MRI brain regions were studied using LB eigen value-based features.
Recently, deep learning (DL) techniques have been developed/used to find links between different parts of images and to identify disease-related patterns [29], including AD classification based on detailed hippocampus analysis using structural MRIs [30–33]. Deep learning models can extract the features directly from medical images to discover hidden representations. These models often outperform other machine learning approaches with the best results on image classification tasks. However, recent researches suggest that these models have limitations in recognizing objects by their global shapes or shape surfaces [34, 35]. And, it is well-known that the hippocampal shape changes are important biomarker in AD [15]. In this study, we hypothesize that integration of global shape representations with visual features of the hippocampus in a deep learning framework will improve the performance of AD classification.
We have recently developed a densely connected 3D convolution neural network (CNN) model (DenseCNN) for classifying AD from normal based on hippocampus segmentations [36]. DenseCNN is a lightweight model with fewer convolutional kernels, relatively simple structure, and fewer total parameters than other state-of-the-art deep learning models for AD classification. Recent studies demonstrated that deep convolution neural networks (DCNN) often did not capture global object shape features [34, 35]. DCNNs are able to encode the local shape features including local edge segments and relations. But, it is sensitive to how these local features fit together as a whole to represent global shape features. DCNNs trained for object recognition do not appear to represent global shape at all [34]. Thus, in this study, we propose DenseCNN2, a lightweight DenseCNN model with combined global shape and visual hippocampus segmentation features of hippocampus, to improve AD classification. Different from DenseCNN, DenseCNN2 is built not only on hippocampus segmentations but also their global shape representations. We demonstrated that DenseCNN2 performed better than DenseCNN and other state-of-the-art methods.
Materials and method
Data
The MRI Data was obtained from the ADNI (http://adni.loni.usc.edu) and the data is T1-weighted structural from initial screening or baseline, including ADNI 1,2/GO and 3. Hippocampus segmentation was performed on this MRI data. After the segmentation, data contained 326 AD subjects and 607 control normal (CN) subjects, totaling 933 hippocampus segmentations. This work is aimed at classifying AD vs. CN. The demographic information of the subjects is provided in Table 1.
Table 1.
Demographic characteristics of the subjects from ADNI database (age and years of education are given as mean (standard deviation))
| AD (N=326) | CN (N=607) | P value | |
|---|---|---|---|
| Gender (% female) | 47.54 | 54.36 | 0.054 |
| Age | 74.9 (7.6) | 74.4 (7.3) | 0.008 |
| Years of education | 15.0 (2.9) | 16.5 (2.5) | < 0.001 |
| APOE ε4 (% at least one allele) | 67.27 | 28.62 | < 0.001 |
There are two hippocampi (left and right) in the brain. We segmented left and right hippocampi using the very recent segmentation tool Hippmapp3r, which is based on 3D CNNs and robust for MRI images with brain atrophy and lesions associated with aging and neurodegeneration [37]. Hippmapp3r has shown to be producing accurate and fast hippocampal segmentations when compared to the existing segmentation algorithms [37]. Figure 1 shows examples of hippocampal segmentation results from the two groups AD and CN.
Fig. 1.
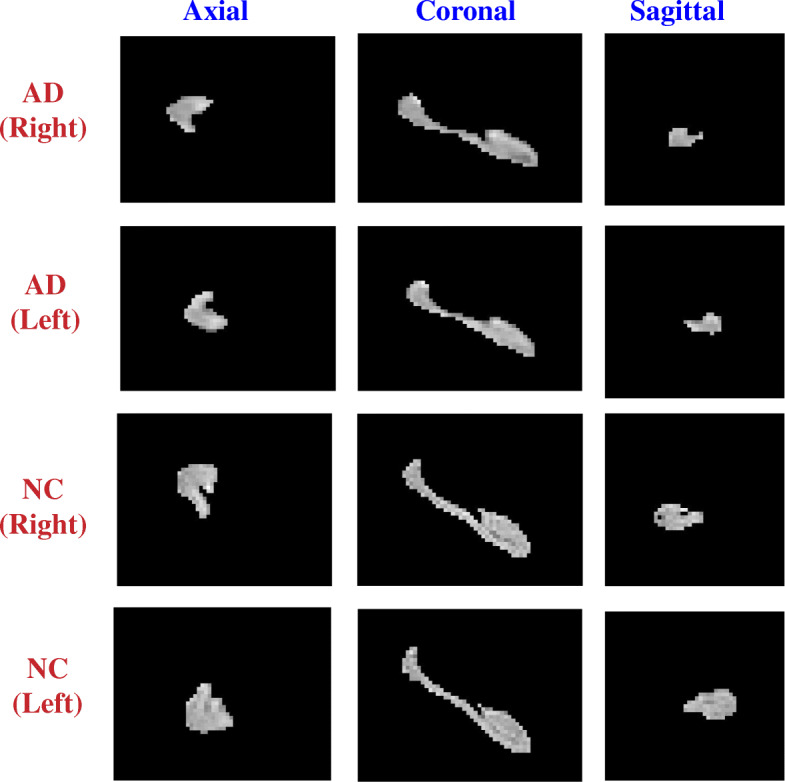
Examples of hippocampal segmentations (both left and right) from the AD and NC
Global shape representations
Global shape features, for each left (L) and right (R) hippocampus, were obtained using LB spectrum [25]. Compared to other shape descriptors such as spherical harmonics [24], LB spectrum is isometry-invariant and avoids pre-processing steps like mapping, registration, and alignment. Also, LB spectrum works for any Riemannian manifold [28], whereas spherical harmonics requires spherical parameterization, a smooth mapping from the surface to a unit sphere [27]. The shape and geometrical information of volumetric data can be discovered and extracted by taking the eigenvalues (i.e., the spectrum) of its LB operator. The LB spectrum can be considered as the set of squared frequencies that are associated with the eigen modes of a generalized oscillating membrane defined on the manifold [25]. Also, eigen values are invariant to isometric transformations and rely on the deformation applied to the boundary of the object [27]. Since, LB spectrum is isometric invariant; it is one of the most powerful ways to represent shape [25]. A brief detail of the LB spectrum is provided below.
Spectrum of the LB operator is defined for real-valued functions on Riemannian manifolds. For a real-valued function f defined on a Riemannian manifold M, the LB operator Δ is given as:
| 1 |
with grad “f” being the gradient of “f” and the div being the divergence on the manifold. The LB operator is a linear differential operator and it can be calculated in local coordinates. LB operator is self-adjoint and semi-positive definite. It follows that the operator Δ admits an orthonormal eigensystem. It consists of eigenvalues λi ϵ ℝ and eigen function fi pairs. More details of LB operator can be found in [28, 38]. The eigen values are called LB the spectrum and contain intrinsic geometrical information of the segmented data. Generally, normalized eigen values are utilized to obtain a scale-invariant global shape representation. This spectrum was calculated through finite element computations [38].
Deep visual features of the hippocampus segments
We obtained deep visual features of the hippocampus segments from DenseCNN, a deep convolution neural network model for AD classification that we have recently developed based on hippocampus segments [36]. DenseCNN has 3 dense layers, with each layer consisting of 2 convolutional layers, combined with Batch normalization (BN) layers and Relu activation layers. Transition layers end with a max pooling layer to decrease the size of input data. DenseCNN has two streams for left and right hippocampus segments correspondingly (Fig. 2). Each stream has an initial 3D convolutional layer followed by a BN layer and a Relu activation layer, extracting low-level image features. Then a max pooling was used to ignore 0 voxels on the edges of the input data and reduce the data size. Two dense blocks and a transition layer were stocked in each stream, using 8 and 16 filters correspondingly. At the end of each stream is a global average pooling (GAP) layer, which compresses high dimensional image features to 1-dimensional features. After the GAP layer, two streams were merged followed by a dropout layer. Finally, a fully connected layer and a SoftMax layer were used for generating prediction. The architecture of this model is shown in Fig. 2. The output of the last GAP layer is the CNN features considered here. For each left and right hippocampus, deep visual features were obtained after the last GAP layer of the DenseCNN.
Fig. 2.
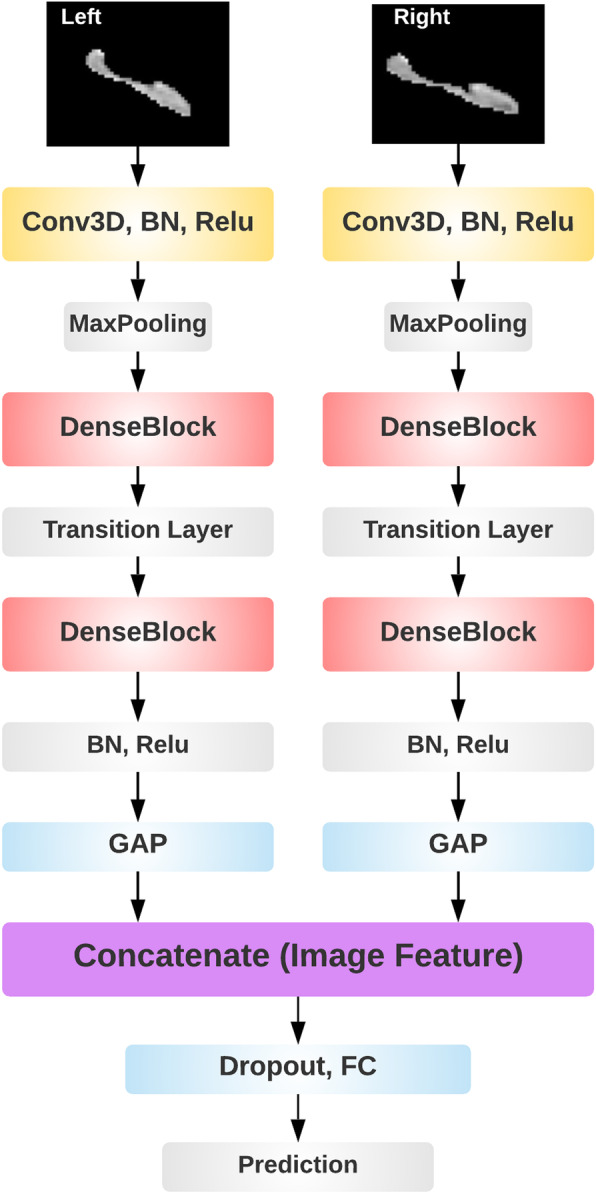
The architecture of DenseCNN
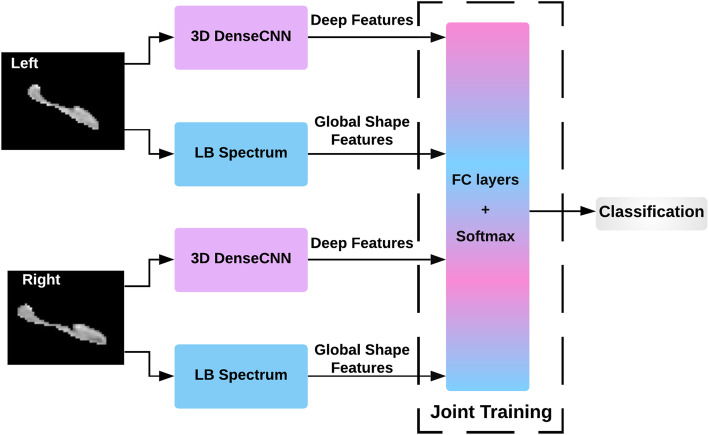
Joint training DenseCNN2 using DenseCNN features and global shape features
Using the abovementioned methods, both shape and deep features were extracted using hippocampus segments. Global shape description for each of the left and right hippocampus is calculated using the LB spectrum (see the “Global shape representations” section) where “f” is the output of the segmentation tool Hippmapp3r. These two types of (shape and DenseCNN features) were expanded and connected together. The two parts of features re-trained in a neural network using a joint training strategy. The learned features from DenseCNN and shape features were combined by a network architecture with fully connected layers followed by softmax layer for AD classification. This joint training strategy can make better global optimization for two models. The performance of this joint model was analyzed by varying the number of fully connected layers and also varying the number of neurons in each layer. Both training and testing data were normalized using a z-score to combine deep features and shape features on the same scale. Figure 3 illustrates the architecture of direct combination of CNN features and shape features.
Fig. 3.
The structure of the joint model
Evaluation
To train and test the classification performance of the proposed DenseCNN2 model, a total of 326 AD subjects and 607 CN subjects were used. DenseCNN2 was implemented with python and Keras library on the Tensorflow backend. Fivefold cross-validation with validation and test set was used for model evaluation. Each time, 1-fold of the data set was used for testing, and 4 folds were further split into training and validation with the validation part consisting of 10% of the training data. This process was repeated 10 times and the average result was reported. In this experiment, as the spectrum of the LB operator contains intrinsic shape information, we considered the top 30 LB eigen values. Normalized eigen values were used and the type of normalization used here was λn → λn/λ1 with logarithmic scale.
To evaluate the performance of DenseCNN2, five measures were computed for evaluation: overall accuracy, sensitivity, specificity, receiver operating characteristic (ROC) curve, and the area under ROC curve (AUC).
Comparison with other models
The performance of DenseCNN2 with combined visual and shape features was compared with deep learning models with shape features alone and visual features alone (DenseCNN). We also compared DenseCNN2 with existing traditional and deep learning methods such as hippocampus volumes, CHF features [17], ResNet [39], 3D CNN [32], hybrid CNN-RNN [33], multi-model CNN [40], and 3D DenseNet [41]. For existing deep learning methods, the reported results are considered here. Overall accuracy, sensitivity, specificity, and AUC were compared.
Data visualization using UMAP
We performed data visualization to demonstrate how combined visual and shape features contributed in separating AD from normal. Dimension reduction plots were used to visualize the data by placing similar data points in close proximity in a low-dimensional space. UMAP (Uniform Manifold Approximation and Projection), an effective tool for visualizing clusters or groups of data points and their relative proximities with nonlinear mapping [42], were used for data visualization. The quality of the separation was computed using three commonly used probabilistic class separability measures, namely, Jeffries-Matusita distance, Bhattacharya distance, and the transformed divergence [43]. The greater the values of these measures indicate better separation between the classes.
Results
Comparison of DenseCNN2 with combined visual and shape features with deep models with shape or visual features alone
DenseCNN2 was compared with deep learning model using shape features alone (DL_shape), and with DenseCNN with visual features. DL_shape was constructed using fully connected layers network with softmax layer classification. For training DenseCNN2 model, the network parameters were randomly initialized at the beginning. For learning deep features, batch-normalization has been performed in order to stop learning irrelevant features at convolutional layers and for faster training. Also, to avoid overfitting, the dropout and L2 regularization were used in the network. Optimal performance of the model is found with dropout factor of 0.5 and L2 weight decay of 0.02. The performance of the joint training with both (deep and shape) features was analyzed by varying the number of fully connected layers and also varying the number of neurons. The stochastic gradient descent optimizer was used with the initial learning rate of 1e−3. The momentum was set to 0.9 and the cross-entropy loss function was used. DenseCNN2 has achieved an accuracy of 92.52%, sensitivity of 88.20%, specificity of 94.95%, and AUC of 97.89%, which are better than DL_shape and DenseCNN (Table 2). DCNNs are able to encode the local shape features including local edge segments and relations. But, it is sensitive to how these local features fit together as a whole to represent global shape features. Thus, our combined model DenseCNN2 has better performance over DenseCNN.
Table 2.
Performance comparison of DL_shape, DenseCNN, and DenseCNN2
| Method | Accuracy | Sensitivity | Specificity | AUC |
|---|---|---|---|---|
| DL_shape | 70.89 | 54.31 | 75.42 | 76.15 |
| DenseCNN | 89.91 | 84.91 | 94.01 | 96.42 |
| DenseCNN2 | 92.52 | 88.20 | 94.95 | 97.89 |
Figure 4 illustrates the ROC curves of the shape and DenseCNN and DenseCNN2 for classifying AD vs. NC. These results demonstrate that combining CNN features with shape features in DenseCNN2 improved the performance of AD classification compared with CNN features or shape features alone. These results indicate that the model has learned complementary features for AD classification based on global shape and visual features of hippocampus segments. Thus, DenseCNN2 is better than DenseCNN by modeling the global shape features along with visual features.
Fig. 4.
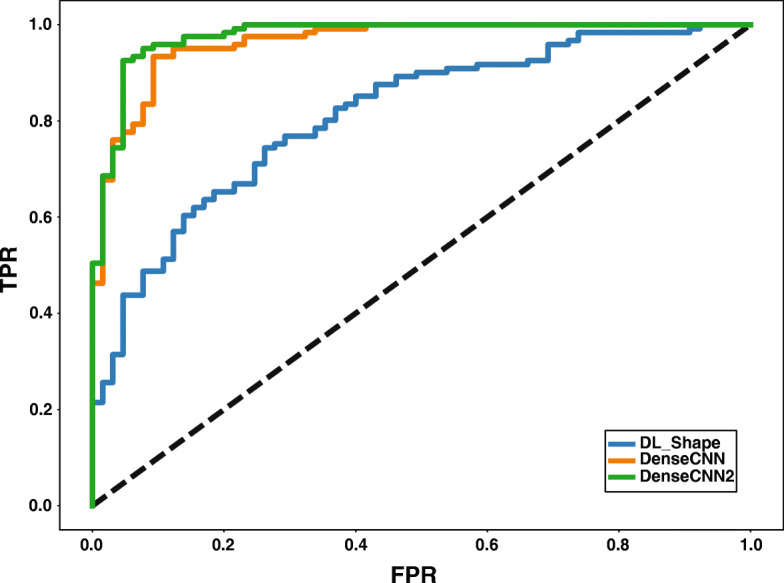
ROC curves of shape, DenseCNN, and DenseCNN2
Comparison with other methods
The performance of the proposed model was compared with seven traditional and deep learning methods. Table 3 shows the comparison performances between DenseCNN2 and the existing methods. DenseCNN2 achieved a classification accuracy of 92.52%, a specificity of 94.85%, and an AUC of 97.89 for AD vs. NC classification, which is higher than both traditional and deep learning methods. Although we reported higher performance, the number of subjects and partitions of training, testing data are different with the existing methods. Compared to other models, DenseCNN2 is a lightweight 3D deep convolutional network model based on DenseCNN, with no particular feature engineering needed. DenseCNN2 does not heavily rely on data augmentation and is fast in training and prediction and has potential to be used for practical and clinical purposes. It integrates additional global shape information into the model for improved performance.
Table 3.
Performance comparison of DenseCNN2 with other methods
| Method | Accuracy | Sensitivity | Specificity | AUC |
|---|---|---|---|---|
| Hippo volumes | 50.54 | 42.12 | 58.40 | 53.54 |
| Hippo CHF features | 85.12 | 76.31 | 81.40 | – |
| 3D CNN | 86.94 | 79.36 | 93.21 | 86.40 |
| ResNet | 90.00 | – | – | 95.60 |
| Hybrid CNN-RNN | 89.17 | 84.64 | 93.16 | 91.00 |
| Multi-model CNN | 88.90 | 86.62 | 90.81 | 92.50 |
| 3D DenseNet | 92.29 | 90.63 | 93.72 | 96.95 |
| DenseCNN2 | 92.52 | 88.20 | 94.95 | 97.89 |
Data visualization using UMAP with DenseCNN features and with DenseCNN2 features
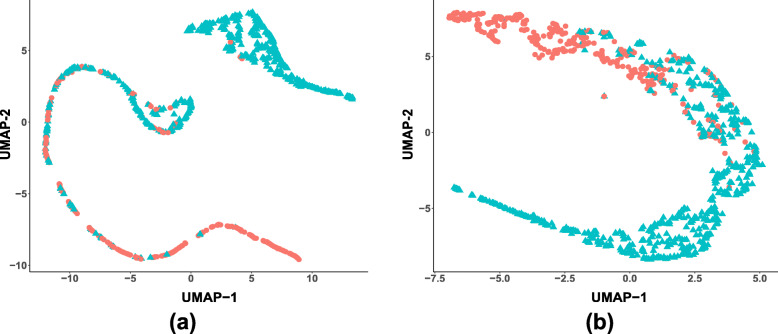
We used data visualization to get insights into the data and the discrimination information between classes. We demonstrated through 2D embedding of UMAP that the class discrimination was improved when shape features were added. Figure 5a, b provides the 2D embedding UMAP of hippocampus data with visual features and with combined visual and shape features respectively. Visually, the separation between the two classes with combined features is greater than the separation with visual features alone. In Fig. 5a, UMAP provided two well separated clusters, but many data points from different classes are overlapping and mis-classified. On the other hand, while there is overlap with combined features, but greater amount of the data is well separated (Fig. 5b).
Fig. 5.
Two dimensional UMAP embedding with visual features (left) and with combined visual and global shape features (right). The data points in color red represent AD subjects and in color blue represent NC subjects
Table 4 provides quantitative measures of separations for the UMAP with visual features and with combined features. It is clear that the values of separations increased for combined features compared with deep visual features alone. This indicates that the classes with combined features have more separation than those with deep visual features alone. These results further validated that the integration of shape features in deep learning model is necessary for AD classification based on MRI data.
Table 4.
Comparison of class separability indices
| Method | Jeffries-Matusita | Bhattacharryya | Divergence |
|---|---|---|---|
| Visual features | 1.74 | 2.04 | 281.30 |
| Combined visual and shape features | 1.88 | 2.85 | 324.33 |
Discussions and conclusion
We proposed a lightweight 3D deep convolutional network model, DenseCNN2, for AD classification using combined hippocampus segmentations and global shape representations. We demonstrated that the combination of deep features and global shape features improved the performance of classifying AD from normal. Also, it is observed through 2D embedding of UMAP that the class discrimination is improved when shape features are added. Also, DenseCNN2 is compared with existing traditional and deep learning-based methods. It is performing better or comparable with all the existing methods.
Future works are warranted to further improve and extend the current study. First, the performances were heavily relied on high-quality hippocampus segments. However, current hippocampus segmentation tools are not ideal. We will develop more robust segmentation algorithms for the hippocampus. We can also avoid this problem by building deep learning models on other relevant regions or whole brain MRI, since brain segments on average have much higher quality than hippocampal segments. Second, while DenseCNN2 is a lightweight model without the need of particular feature engineering and data augmentation and is fast in training and prediction, the data preprocessing step was extensive including segmentation, normalization, and multiple extractions, which needed a well understanding of the dataset and expertise in image processing. In our future study, we will investigate the possibility of building an end-to-end deep learning model, to simplify the whole process of data preprocessing, model training and testing, and data visualization. Third, our model is based on only MRI data. It is known that AD risk is also affected by genetic variants, demographics, comorbidities, medications, socio-economic determinants among others. So, our future study would be to develop multi-modal prediction models for AD classification by combining the MRI data with other data available in ADNI. We will also investigate how patient genetics, demographics, medications, and comorbidities are involved in AD etiology by examining visual and shape changes in the hippocampus. Studying such correlations or interactions provides important insights for AD initiation and progression. Fourth, though our dataset consisted of 326 AD subjects and 607 control normal (CN) subjects from the ADNI database, it still may not be large enough to guarantee generalizability. Further study could utilize more samples from ADNI, or other datasets such as Oasis (http://www.oasis-brains.org). And we believe more new samples should be collected, since new MRI samples can test the generality of existing AD detection approaches.
Limitation
Limitations include the following: (1) current model was trained solely on hippocampus regions. Other brain regions could be used for further improvements; (2) current model used only MRI data. Other data types including clinical, genetic, and genomics will be incorporated in our future studies; (3) only ADNI data was used for both model training and testing. Other independent datasets could be used for both model improvement and independent testing; (4) this study focused on classifying AD versus normal control. We will adopt the “light” model approach demonstrated in this study to classify AD versus MCI versus normal, which is a more challenging task than classifying AD versus normal.
Data availability
Features and code for DenseCNN2 are publicly available at http://nlp.case.edu/public/data/DenseCNN2.
Acknowledgements
We thank the Alzheimer’s Disease Neuroimaging Initiative (ADNI) for generously sharing clinical, imaging, genetic, and biochemical biomarkers for the early detection and tracking of Alzheimer’s disease (AD).
Abbreviations
- MRI
Magnetic resonance imaging
- DCNN
Deep Convolution Neural Networks
- LB
Laplace Beltrami
- AD
Alzheimer’s disease
- NC
Normal control
- UMAP
Uniform Manifold Approximation and Projection
Authors’ contributions
Study conception and supervision: RX. Study design: SK, RX; data preprocessing: QYW; analysis and evaluation: SK; paper writing: SK; paper review and supervision: RX. The authors read and approved the final manuscript.
Funding
S.K., Q.Y.W., and R.X. are supported by NIH National Institute on Aging R01 AG057557, R01 AG061388, R56 AG062272, National Institute on Drug Addiction UG1DA049435, American Cancer Society Research Scholar Grant RSG-16-049-01 – MPC, The Clinical and Translational Science Collaborative (CTSC) of Cleveland 1UL1TR002548-01.
The funder of the study had no role in study design, data collection, data analysis, data interpretation, and writing of the report. The corresponding author (R.X) had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Availability of data and materials
The MRI data used in this study was from Alzheimer’s Disease Neuroimaging Initiative (ADNI).
Declarations
Ethics approval and consent to participate
Ethics approval and informed consents were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.de la Torre JC. Is Alzheimer’s disease a neurodegenerative or a vascular disorder? Data, dogma, and dialectics. Lancet Neurol. 2004;3(3):184–190. doi: 10.1016/S1474-4422(04)00683-0. [DOI] [PubMed] [Google Scholar]
- 2.Bature F, Guinn BA, Pang D, Pappas Y. Signs and symptoms preceding the diagnosis of Alzheimer's disease: a systematic scoping review of literature from 1937 to 2016. BMJ Open. 2017;7(8):e015746. 10.1136/bmjopen-2016-015746. [DOI] [PMC free article] [PubMed]
- 3.Chong MS, Sahadevan S. Preclinical Alzheimer’s disease: diagnosis and prediction of progression. Lancet Neurol. 2005;4(9):576–579. doi: 10.1016/S1474-4422(05)70168-X. [DOI] [PubMed] [Google Scholar]
- 4.Rentz DM, Rodriguez MA, Amariglio R, Stern Y, Sperling R, Ferris S. Promising developments in neuropsychological approaches for the detection of preclinical Alzheimer’s disease: a selective review. Alzheimer’s Res Ther. 2013;5(6):1–0. doi: 10.1186/alzrt222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selkoe DJ. Alzheimer’s disease--genotypes, phenotype, and treatments. Science. 1997;275(5300):630–631. doi: 10.1126/science.275.5300.630. [DOI] [PubMed] [Google Scholar]
- 6.Aël Chetelat G, Baron JC. Early diagnosis of Alzheimer’s disease: contribution of structural neuroimaging. Neuroimage. 2003;18(2):525–541. doi: 10.1016/S1053-8119(02)00026-5. [DOI] [PubMed] [Google Scholar]
- 7.Boss MA. Diagnostic approaches to Alzheimer’s disease. Biochim Biophys Acta. 2000;1502(1):188–200. doi: 10.1016/S0925-4439(00)00044-2. [DOI] [PubMed] [Google Scholar]
- 8.Vieira S, Pinaya WH, Mechelli A. Using deep learning to investigate the neuroimaging correlates of psychiatric and neurological disorders: methods and applications. Neurosci Biobehav Rev. 2017;74(Pt A):58–75. doi: 10.1016/j.neubiorev.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Blennow K, Hampel H, Weiner M, Zetterberg H. Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat Rev Neurol. 2010;6(3):131–144. doi: 10.1038/nrneurol.2010.4. [DOI] [PubMed] [Google Scholar]
- 10.Lu D, Popuri K, Ding GW, Balachandar R, Beg MF. Multimodal and multiscale deep neural networks for the early diagnosis of Alzheimer’s disease using structural MR and FDG-PET images. Sci Rep. 2018;8(1):1–3. doi: 10.1038/s41598-018-22871-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, Liang M, Wang L, Tian L, Zhang X, Li K, Jiang T. Altered functional connectivity in early Alzheimer's disease: A resting-state fMRI study. Hum Brain Mapp. 2007;28(10):967–978. doi: 10.1002/hbm.20324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jack CR, Jr, Bernstein MA, et al. The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27(4):685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritchie K, Lovestone S. The dementias. Lancet. 2002;360(9347):1759–1766. doi: 10.1016/S0140-6736(02)11667-9. [DOI] [PubMed] [Google Scholar]
- 14.Falahati F, Westman E, Simmons A. Multivariate data analysis and machine learning in Alzheimer’s disease with a focus on structural magnetic resonance imaging. J Alzheimer's Dis. 2014;41(3):685–708. doi: 10.3233/JAD-131928. [DOI] [PubMed] [Google Scholar]
- 15.Sabuncu MR, Desikan RS, Sepulcre J, Yeo BT, Liu H, Schmansky NJ, Reuter M, Weiner MW, Buckner RL, Sperling RA, Fischl B, Alzheimer's Disease Neuroimaging Initiative The dynamics of cortical and hippocampal atrophy in Alzheimer disease. Arch Neurol. 2011;68(8):1040–1048. doi: 10.1001/archneurol.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Epifanio I, Ventura-Campos N. Hippocampal shape analysis in Alzheimer’s disease using functional data analysis. Stat Med. 2014;33(5):867–880. doi: 10.1002/sim.5968. [DOI] [PubMed] [Google Scholar]
- 17.Ahmed OB, Benois-Pineau J, Allard M, Amar CB, Catheline G. Alzheimer’s Disease Neuroimaging Initiative. Classification of Alzheimer’s disease subjects from MRI using hippocampal visual features. Multimedia Tools Appl. 2015;74(4):1249–1266. doi: 10.1007/s11042-014-2123-y. [DOI] [Google Scholar]
- 18.Chincarini A, Sensi F, Rei L, Gemme G, Squarcia S, Longo R, Brun F, Tangaro S, Bellotti R, Amoroso N, Bocchetta M, Redolfi A, Bosco P, Boccardi M, Frisoni GB, Nobili F, Alzheimer's Disease Neuroimaging Initiative Integrating longitudinal information in hippocampal volume measurements for the early detection of Alzheimer’s disease. NeuroImage. 2016;125:834–847. doi: 10.1016/j.neuroimage.2015.10.065. [DOI] [PubMed] [Google Scholar]
- 19.Shen K, Bourgeat P, Fripp J, Meriaudeau F, Salvado O. Detecting hippocampal shape changes in Alzheimer’s disease using statistical shape models. Med Imaging. 2011;7962:796243. [Google Scholar]
- 20.Shilane P, Funkhouser T. Selecting distinctive 3D shape descriptors for similarity retrieval. In IEEE International Conference on Shape Modeling and Applications 2006 (SMI'06) 2006 Jun 14 (pp. 18-18). IEEE.
- 21.Osada R, Funkhouser T, Chazelle B, Dobkin D. Matching 3D models with shape distributions. In Proceedings International Conference on Shape Modeling and Applications 2001 May 7 (pp. 154-166). IEEE.
- 22.Kazmi IK, You L, Zhang JJ. A survey of 2D and 3D shape descriptors. In2013 10th International Conference Computer Graphics, Imaging and Visualization 2013 Aug 6 (pp. 1-10). IEEE.
- 23.Dutagaci H, Sankur B, Yemez Y. Transform-based methods for indexing and retrieval of 3d objects. In Fifth International Conference on 3-D Digital Imaging and Modeling (3DIM'05) 2005 Jun 13 (pp. 188-195). IEEE.
- 24.Gutman B, Wang Y, Lui LM, Chan TE, Thompson PM. Hippocampal surface analysis using spherical harmonic function applied to surface conformal mapping. 18th International Conference on Pattern Recognition. 2006 Aug 20; 3:964-967.
- 25.Ramaniharan AK, Manoharan SC, Swaminathan R. Laplace Beltrami eigen value based classification of normal and Alzheimer MR images using parametric and non-parametric classifiers. Expert Syst Appl. 2016;59:208–216. doi: 10.1016/j.eswa.2016.04.029. [DOI] [Google Scholar]
- 26.Liu H, Rashid T, Habes M. Cerebral Microbleed Detection Via Fourier Descriptor with Dual Domain Distribution Modeling. IEEE 17th International Symposium on Biomedical Imaging Workshops. 2020 Apr 4 (pp. 1-4). IEEE.
- 27.Bajaj CL, Xu G. Anisotropic diffusion of surfaces and functions on surfaces. ACM Transact Graphics. 2003;22(1):4–32. doi: 10.1145/588272.588276. [DOI] [Google Scholar]
- 28.Seo S, Chung MK. Laplace-Beltrami eigen function expansion of cortical manifolds. IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 2011 Mar 30; 372-375.
- 29.Razzak MI, Naz S, Zaib A. Deep learning for medical image processing: Overview, challenges and the future. Classif BioApps. 2018:323–50. 10.1007/978-3-319-65981-7_12.
- 30.Payan A, Montana G. Predicting Alzheimer’s disease: a neuroimaging study with 3D convolutional neural networks. arXiv:1502.02506. 2015
- 31.Cao L, Li L, Zheng J, Fan X, Yin F, Shen H, Zhang J. Multi-task neural networks for joint hippocampus segmentation and clinical score regression. Multimedia Tools Appl. 2018;77(22):29669–29686. doi: 10.1007/s11042-017-5581-1. [DOI] [Google Scholar]
- 32.Cui R, Liu M. Hippocampus analysis based on 3D CNN for Alzheimer’s disease diagnosis. Tenth Int Conf Digital Image Process. 2018;10806:1080650. [Google Scholar]
- 33.Li F. Manhua Liu, and Alzheimer's Disease Neuroimaging Initiative. A hybrid convolutional and recurrent neural network for hippocampus analysis in Alzheimer’s disease. J Neurosci Methods. 2019;323:108–118. doi: 10.1016/j.jneumeth.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 34.Baker N, Lu H, Erlikhman G, Kellman PJ. Deep convolutional networks do not classify based on global object shape. Plos Comput Biol. 2018;14(12):e1006613. doi: 10.1371/journal.pcbi.1006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinha A, Bai J, Ramani K. Deep learning 3D shape surfaces using geometry images. InEuropean conference on computer vision. Cham: Springer; 2016. p. 223-240.
- 36.Wang Q, Li Y, Zheng C, Xu R. DenseCNN: A Densely Connected CNN Model for Alzheimer's Disease Classification Based on Hippocampus MRI Data. InAMIA Annual Symposium Proceedings 2020 (Vol. 2020, p. 1277). American Medical Informatics Association. [PMC free article] [PubMed]
- 37.Goubran M, Ntiri EE, Akhavein H, Holmes M, Nestor S, Ramirez J, Adamo S, Ozzoude M, Scott C, Gao F, Martel A, Swardfager W, Masellis M, Swartz R, MacIntosh B, Black SE. Hippocampal segmentation for brains with extensive atrophy using three-dimensional convolutional neural networks. Hum Brain Mapp. 2020;41(2):291–308. doi: 10.1002/hbm.24811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chung MK, Taylor J. 2004 2nd IEEE International Symposium on Biomedical Imaging: Nano to Macro. 2004. Diffusion smoothing on brain surface via finite element method; pp. 432–435. [Google Scholar]
- 39.Li H, Habes M, Wolk DA, Fan Y. Alzheimer's Disease Neuroimaging Initiative. A deep learning model for early prediction of Alzheimer’s disease dementia based on hippocampal magnetic resonance imaging data. Alzheimer’s Dement. 2019;15(8):1059–1070. doi: 10.1016/j.jalz.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu M, Li F, Yan H, Wang K, Ma Y, Shen L, et al. A multi-model deep convolutional neural network for automatic hippocampus segmentation and classification in Alzheimer’s disease. NeuroImage. 2020;208:116459. doi: 10.1016/j.neuroimage.2019.116459. [DOI] [PubMed] [Google Scholar]
- 41.Cui R, Liu M. Hippocampus Analysis by Combination of 3-D DenseNet and Shapes for Alzheimer’s Disease Diagnosis. IEEE J Biomed Health Inform. 2019;23(5):2099–2107. doi: 10.1109/JBHI.2018.2882392. [DOI] [PubMed] [Google Scholar]
- 42.McInnes L, Healy J, Melville J. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv:1802.03426. 2018
- 43.Swain PH, King RC. Two effective feature selection criteria for multispectral remote sensing. LARS technical reports. 1973:39.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Features and code for DenseCNN2 are publicly available at http://nlp.case.edu/public/data/DenseCNN2.
The MRI data used in this study was from Alzheimer’s Disease Neuroimaging Initiative (ADNI).