Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is a major cause of nosocomial infections because of its high resistance. Here, we study the antibiotic resistance in MRSA clinical isolates and their relation to integron I occurrence. A total of 88 clinical Staphylococcus aureus isolates were collected. MRSA were identified by the disk diffusion method (DDM) and confirmed by PCR, and antibiogram was determined by DDM. Integron I, II and the aacA4 gene were investigated by PCR. Integrase-positive strains were analyzed for the presence of resistance gene cassettes by sequencing. All isolates were identified as MRSA by DDM and confirmed by PCR. All isolates were resistant to ampicillin and cefoxitin. Concerning aminoglycosides, the frequency of resistance was reported for streptomycin (60.7%), tobramycin (37.1%) gentamicin (36%), and for amikacin (15.9%). Integron I was detected in 41 isolates (46.6%), while integron II was detected in three isolates (3.4%). Sequencing of the integron I-cassette indicated the exclusive prevalence of addA gene variants mediating aminoglycoside resistance. The aacA4 gene was found in DNA of 31 isolates (35.22%). This study revealed the high existence of MRSA. Furthermore, the AacA4 gene and class I integron harboring aadA gene were predominant in MRSA isolates.
Keywords: MRSA, integron I, aminoglycoside resistance, sequencing, aadA gene variants, aacA4 gene
1. Introduction
Staphylococcus aureus is a major human pathogen responsible for a wide range of infections including superficial and fatal systemic infections [1]. S. aureus is identified as one of the world’s leading causes of nosocomial infections [2]. In the past, infections caused by S. aureus were well-controlled by penicillin [3]. With the misuse of penicillin, penicillin-resistant S. aureus and methicillin-resistant S. aureus (MRSA) have appeared; MRSA resistance is mediated by the mecA gene [4,5]. MRSA has rapidly become the most frequently occurring resistant pathogen that leads to high rates of morbidity and mortality [6,7]. In the last few decades, multidrug resistant (MDR)-MRSA isolates have become prominent in Egypt due to several factors including carriage between healthcare workers, persistence in the hospital environment and the misuse of antibiotics [8,9].
Several mechanisms involving mobile genetic elements, such as plasmids, transposons and integrons, have been shown to contribute to the spread of antibiotic resistant genes [10]. Integrons are mobile-DNA elements that play an important role in the spread of antibiotics and biocides resistance among bacteria [11,12]. Integrons I and II are the classes of integrons that are common in Gram-positive bacteria, including S. aureus [13,14].
A complete functional integron consists of three elements: the integrase gene (intI) encoding an integrase protein, a recombination site (attI) and a promoter (Pc) gene that directs transcription of the gene cassettes. The integrase protein catalyzes recombination between incoming gene cassettes and the recombination site [15]. Integrons usually carry antibiotic resistance genes that contribute to resistance against aminoglycosides, macrolides, β-lactams, chloramphenicol, and sulfonamides [16].
Aminoglycosides are bactericidal agents that play an important role in the anti-staphylococcal therapies. They act by binding specifically to 16S rRNA of the 30S ribosomal subunit, thus interfering with protein synthesis [17]. Gentamicin, tobramycin and amikacin are the most active aminoglycosides against staphylococci and are often used in combination with other groups such as β-lactams [18]. The main mechanism of aminoglycoside resistance is drug inactivation by aminoglycoside-modifying enzymes (AMEs) that decrease aminoglycosides’ affinity to their natural target (16S rRNA).
The most important AMEs are adenylyltransferase (ANT), acetyltransferase (AAC), and phosphotransferase (APH) enzymes [19]. The AACs acetylate an amino group, while the ANTs and APHs adenylate and phosphorylate a hydroxyl group of the antibiotic, respectively. Some AACs are found as fusion proteins located adjacent to other AMEs such as APH and ANT [17,20].
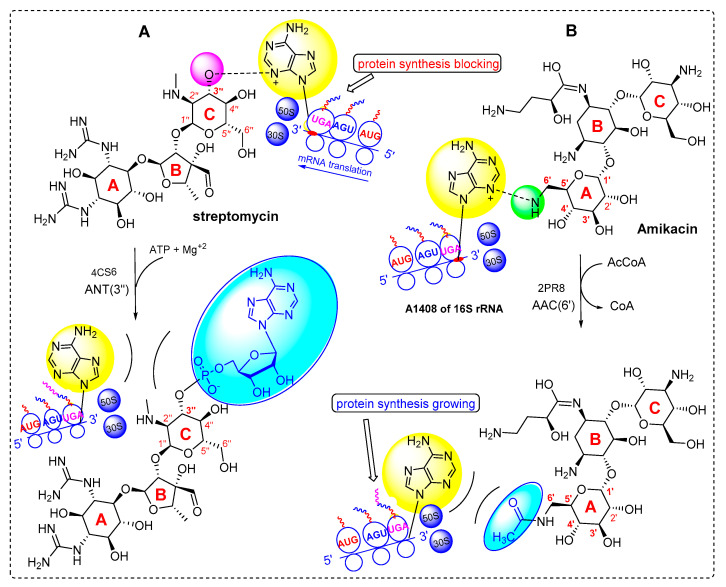
Four types of adenylyltransferases have been identified, including: ANT (2″), ANT (3″), ANT (4′), and ANT (6′). The ANT (3″) are the most common ANT enzymes, they specify resistance to streptomycin, and their coding genes are commonly named aadA; at least 22 highly related aadA gene variants are found in GenBank. The aadA genes exist as gene cassettes and are part of integrons, plasmids or transposons [17,21]. The ANT (3″) adenylates streptomycin at 3″-position (labelled pink) give rise to the cleavage of drug/16S-rRNA complex, restoring the function of ribosomes (Figure 1A).
Figure 1.
Mechanism of action of AMEs, (A) the adenyltransferase aadA (PDB; 4CS6) adenylate streptomycin at 3″ position, (B) acetyltransferase AAC (6′)-Ib (PDB; 2PR) acetylate amikacin at the 6′ position.
The AACs are subdivided into several groups based on the position where the acetyl group is inserted in the acceptor aminoglycoside drug. Known AACs catalyze acetylation at the 1 (AAC (1)), 3 (AAC (3)), 2′ (AAC (2′)), or 6′ (AAC (6′)) positions. AAC (6′) enzymes are the most common group of AACs that contain more than 40 enzymes [22]. A plasmid-mediated acetyltransferase AAC(6′)-Ib, also known as aacA4, specify resistance to several aminoglycosides including amikacin [20]. AAC (6′) modifies the aminoglycosides (gentamicin, tobramycin and amikacin) by the acetylation of the amino group at 6’-position (labelled green), setting the 16S-rRNA receptor free and turning the ligand into an inactive drug (Figure 1B).
The main purpose of the current study is to investigate the resistance profile of clinical MRSA isolates, to determine the frequency of class I and II integrons in these isolates, and to investigate the pattern of resistance linked to integron I-positive/cassette-positive isolates especially resistant against aminoglycosides. Furthermore, aminoglycoside resistance due to the aacA4 gene was investigated.
2. Results
2.1. Identification of S. aureus Isolates
A total of 88 non-duplicate clinical S. aureus isolates originated from wounds (46%), urine (30%), sputum (13%), and blood (11%) were identified in this study. The majority of cases (57.95%) were males, 32.95% of the total number of cases were <19 years old, 37.5% of cases were between 19 to 40 years, 11.36% of cases were between 41 and 60 years, and 18.18% of them were >60 years. All S. aureus isolates showed golden yellow colonies on nutrient agar, and yellow fermentation on mannitol salt agar. Microscopically, all isolates were positive in Gram staining with the arrangement in grapes clusters. Biochemically, all isolates were mannitol fermenters. All the isolates were identified phenotypically as MRSA by the disk diffusion method using a cefoxitin disk.
2.2. Antimicrobial Susceptibility Testing
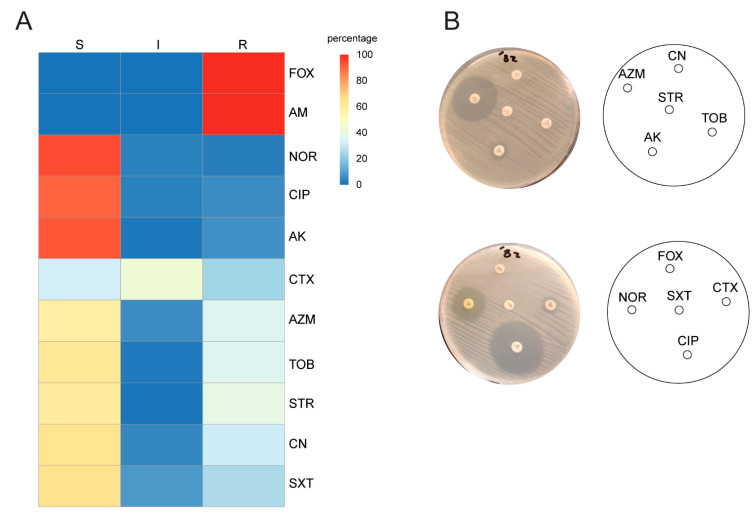
The antimicrobial susceptibility test revealed that all the isolates were resistant to ampicillin and cefoxitin (100%). Frequency of resistance was commonly reported with cefotaxime (67.41%) and streptomycin (60.67%), while intermediate resistance was detected in azithromycin (43.82%), tobramycin (37.1%), gentamicin (35.95%), and sulfamethoxazole/trimethoprim (35.33%). Resistance was rarely observed with amikacin (15.9%), ciprofloxacin (9%), and norfloxacin (5.68%), (Figure 2). Forty-nine of the isolates (55.7%) were multidrug resistant (MDR).
Figure 2.
Antimicrobial susceptibility of MRSA isolates: (A) representative heatmap depicting the observed antibiogram for the S. aureus isolates to different antimicrobial chemotherapeutics. Cefoxitin (FOX), ampicillin (AM), cefotaxime (CTX), ciprofloxacin (CIP), norfloxacin (NOR), amikacin (AK), tobramycin (TOB), gentamicin (CN), streptomycin (STR), azithromycin (AZM), and sulfamethoxazole/trimethoprim (SXT). Sensitive (S), intermediate resistant (I), resistant (R). The heatmaps were created using R version 4.0.1 with the package ggplot2 version 3.3.1 [23]; the heatmap matrix was retrieved from scored susceptibility identified by DDM in (B). (B) Antimicrobial susceptibility of S. aureus isolates against different antibiotics by DDM.
2.3. Confirmation of MRSA Identification, Screening of Integron I, II and aacA4 Genes by Polymerase Chain Reaction (PCR)
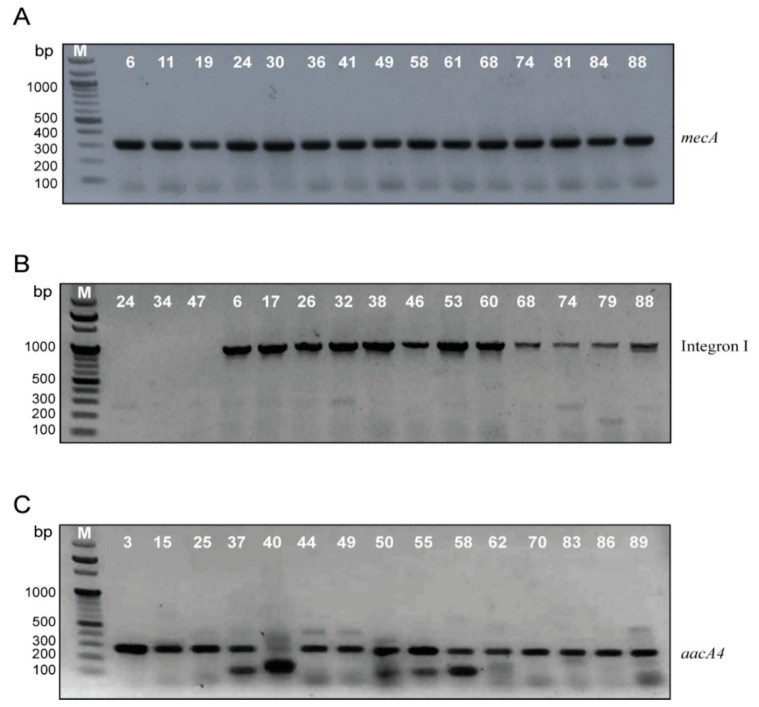
PCR was used for the amplification of the mecA gene to confirm MRSA identification. All the isolates gave a single band at 310 bp matched to the mecA gene (Figure 3A).
Figure 3.
Electrophoretic graph of PCR products on 1.5% agarose gel stained with ethidium bromide for (A) mecA gene among S. aureus isolates, positive isolates gave a single band of 310 bp. (B) Integron I amplicon among MRSA, positive isolates gave a single band of either 250 bp or 1000 bp. (C) aacA4 gene among MRSA, positive isolates gave a single band of 200 bp.
PCR was also used for integron detection using a primer specific for class I integron (intI1). Integron I was detected in 41 isolates (46.6%) with a variable amplicon size, 35 isolates (39.8%) with a single band of approximately 1000 bp (Figure 3B), and only six isolates (6.8%) produced a single band of 250 bp. Integron II with 200 bp amplicon was detected in only three isolates (3.4%).
The aacA4 gene encoding for aminoglycoside resistance was detected in DNA of 31 isolates (35.22%) with an amplicon size of 300 bp (Figure 3C)
2.4. Sequencing of Integron I-Positive Isolates
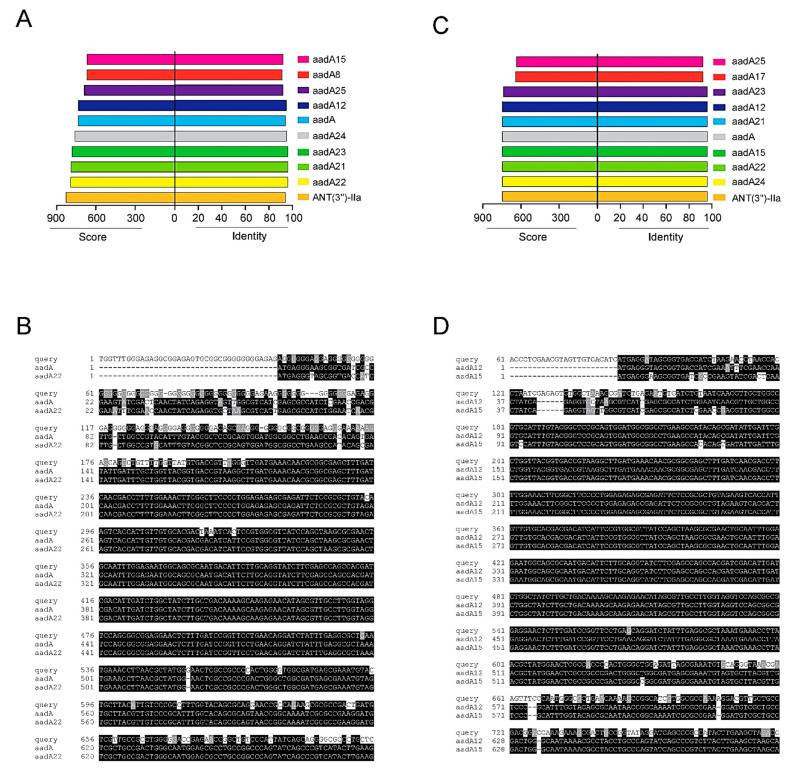
Integron-borne resistance was investigated by PCR amplification for the integrase gene followed by sequencing the variable region and 3′-conserved regions. The results are shown in Figure 4. Multiple sequence alignment showed that integron I of 1 kb size has a sequence similarity with aadA gene variants (aadA22, aadA12, and aadA15), while integron I with 250 bp does not contain any gene (empty) cassette (Table 1). Unique sequences analyzed by the Comprehensive Antibiotic Resistance Database (CARD) detected two major categories of cassette arrays coding aminoglycosides resistance genes with a high degree of inferred sequence homology. The two frequently detected AMR genes both belonging to the aminoglycoside nucleotidyltransferase are usually responsible for nucleotidylylation of streptomycin at the hydroxyl group at position 3″(ANT(3″)). One of the two subfamilies shows high identity (95 to 98%) to aadA15, identified in the resistome of Pseudomonas aeruginosa and aadA12, identified in the resistome of Escherichia coli, Yersinia enterocolitica and Salmonella enterica. The other subfamily shows a 95% identity to aadA, the aminoglycoside nucleotidyltransferase gene encoded by plasmids, transposons, and integrons in Enterobacteriaceae, A. baumannii, P. aeruginosa and Vibrio cholerae, and a 95% identity to aadA22 in S. enterica and E. coli. Other closely related resistomes with sequence variants for the two AMR gene variants were detected by the sequence recognition analysis against a library of identified aminoglycosides resistomes annotated in the resistome database (Figure 4A,C).
Figure 4.
(A) Bar blots indicating the identity percentage and covered score of the sequenced query 1 against the most likelihood AMR genes recognized by the Comprehensive Antibiotic Resistance Database (CARD). (B) Multiple sequence alignment of one category of the identified AMR genes (query 1) with the sequences of aadA and aadA22 genes retrieved from banked microbial resistome that give a high identity to the queried sequence analyzed via CARD; the available sequences are aligned via ClustalW and BOXSHADE and were employed to highlight the multiple alignment. Black shading indicates that the residue is identical to the column consensus; gray shading indicates that the residue is not identical but at least similar to the column consensus; and no shading indicates that the residue is neither identical nor similar to the consensus [24]. (C) Bar blots indicating the identity percentage and covered score of the sequenced query 2 against the most likelihood AMR genes retrieved from CARD. (D) CLUSTAL W multiple sequence alignment of the other category of the identified AMR genes (query 2) with the sequences of aadA12 and aadA15 genes retrieved from banked microbial resistomes that give a high identity to the queried sequence using CARD; the available sequences are aligned and shaded via ClustalW and BOXSHADE, as previously described.
Table 1.
Resistance profile and characters of integron I-positive MRSA isolates.
| Isolate No | Integron I (bp) | Sequence Profile of Integron I |
aacA4 Gene |
Antibiotic Resistance Profile |
|---|---|---|---|---|
| 6 | 1000 | query 2 (aadA12,15) | − | FOX, AM, CTX (I), STR, SXT |
| 8 | 1000 | query 1 (aadA22,23) | − | FOX, AM, STR, CTX(I), SXT(I) |
| 11 | 1000 | query 1 (aadA22,23) | − | FOX, AM, CTX (I), STR |
| 13 | 1000 | query 1 (aadA22,23) | − | FOX, AM, CTX, STR AZM, |
| 15 | 1000 | query 2 (aadA12,15) | + | FOX, AM, CTX(I), STR, TOB (I), CN (I), AK(I) AZM |
| 17 | 1000 | query 1 (aadA22,23) | − | FOX, AM, CTX, STR, SXT(I), AZM |
| 19 | 1000 | query 1 (aadA22,23) | − | FOX, AM, CTX, STR, SXT |
| 20 | 1000 | query 1 (aadA22,23) | − | FOX, AM, CTX(I), STR, SXT |
| 21 | 1000 | query 1 (aadA22,23) | + | FOX, AM, CTX(I), STR, CN, TOB, AK, AZM |
| 24 | 250 | Empty cassette | − | FOX, AM |
| 25 | 1000 | query 2 (aadA12,15) | + | FOX, AM, CTX(I), STR, CN, TOB, AK, AZM(I) |
| 26 | 1000 | query 1 (aadA22,23) | − | FOX, AM, CTX, STR, SXT |
| 27 | 1000 | query 1 (aadA22,23) | − | FOX, AM, CTX (I), STR, AZM |
| 28 | 1000 | query 2 (aadA12,15) | − | FOX, AM, STR |
| 29 | 250 | Empty cassette | − | FOX, AM |
| 30 | 1000 | query 1 (aadA22,23) | − | FOX, AM, STR |
| 31 | 250 | Empty cassette | − | FOX, AM |
| 32 | 1000 | query 2 (aadA12,15) | − | FOX, AM, CTX (I), STR, SXT |
| 34 | 250 | Empty cassette | − | FOX, AM |
| 35 | 1000 | query 1 (aadA22,23) | − | FOX, AM, STR, SXT |
| 36 | 1000 | query 1 (aadA22,23) | − | FOX, AM, STR, AZM |
| 37 | 1000 | query 2 (aadA12,15) | + | FOX, AM, STR, AK, CN, TOB, AZM, SXT |
| 38 | 1000 | query 1 (aadA22,23) | − | FOX, AM, STR, AZM |
| 41 | 250 | Empty cassette | − | FOX, AM |
| 42 | 1000 | query 1 (aadA22,23) | + | FOX, AM, CTX (I), STR, AK, CN, TOB, AZM, SXT |
| 46 | 1000 | query 2 (aadA12,15) | + | FOX, AM, CTX (I), STR, AK, CN, TOB, AZM |
| 47 | 250 | Empty cassette | − | FOX, AM |
| 49 | 1000 | query 2 (aadA12,15) | + | FOX, AM, CTX, STR, AK, CN, TOB, NOR, CIP, SXT, AZM |
| 53 | 1000 | query 1 (aadA22,23) | − | FOX, AM, CTX, STR, AZM |
| 58 | 1000 | query 1 (aadA22,23) | + | FOX, AM, CTX(I), STR, CN, TOB, AK, AZM |
| 60 | 1000 | query 1 (aadA22,23) | − | FOX, AM, CTX(I), CIP, NOR(I), STR, SXT(I) |
| 61 | 1000 | query 2 (aadA12,15) | + | FOX, AM, CTX(I), STR, CN, TOB, AK(I) |
| 65 | 1000 | query 1 (aadA22,23) | − | FOX, AM, CTX, STR, AZM |
| 68 | 1000 | query 1 (aadA22,23) | − | FOX, AM, CTX, STR, SXT |
| 71 | 1000 | query 2 (aadA12,15) | + | FOX, AM, CTX, STR, CN, TOB, AK(I), AZM |
| 74 | 1000 | query 1 (aadA22,23) | − | FOX, AM, CTX, STR, SXT |
| 78 | 1000 | query 2 (aadA12,15) | − | FOX, AM, CTX(I), STR, SXT |
| 79 | 1000 | query 1 (aadA22,23) | − | FOX, AM, STR, SXT |
| 81 | 1000 | query 1 (aadA22,23) | + | FOX, AM, STR, AK, CN, TOB, AZM, SXT |
| 84 | 1000 | query 1 (aadA22,23) | + | FOX, AM, CTX(I), STR, AK, CN, TOB, AZM, SXT |
| 88 | 1000 | query 2 (aadA12,15) | + | FOX, AM, CTX, STR, CN, TOB, AK, AZM |
Cefoxitin (FOX), ampicillin (Am), cefotaxime (CTX), ciprofloxacin (CIP), norfloxacin (NOR), amikacin (AK), tobramycin (TOB), gentamicin (CN), streptomycin (STR), azithromycin (AZM), and sulfamethoxazole/trimethoprim (SXT).
2.5. Classification of Isolates According to Resistance Profile, Integron I and aacA4 Presence
Merging the results retrieved from the molecular analysis of the most probable elements for resistance, the integron I-positive isolates can be divided into three groups. Group one includes 13 isolates which are integron I-positive (cassette-positive) and aacA4-positive; all of these isolates were resistant to all aminoglycosides including amikacin. Group two includes 22 isolates that are integron I-positive (cassette-positive) and aacA4-negative; these isolates were resistant to streptomycin only, while Group three includes six isolates only integron-positive (cassette-negative) and aacA4-negative; these isolates are sensitive to all aminoglycosides as shown in Table 1.
Table 2 contain the characters of the integron I-negative, aacA4-positive isolates (18 isolates). A total of 17 of these isolates were resistant to all aminoglycosides except amikacin, while only one isolate was resistant to all aminoglycosides including amikacin.
Table 2.
Resistance profile of integron-negative /aacA4-positive MRSA isolates.
| Isolate No. | Integron I | aacA4 Gene | Antibiotic Resistance Profile |
|---|---|---|---|
| 3 | − | + | FOX, AM, CTX (I), CN, TOB, STR, |
| 4 | − | + | FOX, AM, CTX, CN (I), TOB, STR, SXT |
| 5 | − | + | FOX, AM, CTX (I), CN, TOB, STR, AZM |
| 23 | − | + | FOX, AM, CTX, CN, TOB, STR, AZM |
| 40 | − | + | FOX, AM, CTX (I), CN, TOB, STR, AZM (I) |
| 43 | − | + | FOX, AM, CN, TOB, STR, AZM (I) |
| 44 | − | + | FOX, AM, CTX, CIP (I), CN, TOB, STR, AZM |
| 48 | − | + | FOX, AM, CTX (I), CN (I), TOB, STR, AZM |
| 50 | − | + | FOX, AM, CTX, CN, TOB, STR, AZM |
| 51 | − | + | FOX, AM, CTX (I), CN, TOB, STR, AZM |
| 55 | − | + | FOX, AM, CTX (I), CN, TOB, STR, AZM |
| 57 | − | + | FOX, AM, CTX (I), CN, TOB, STR, AZM |
| 62 | − | + | FOX, AM, CTX, CN (I), TOB, STR, SXT |
| 70 | − | + | FOX, AM, CTX (I), CN, TOB, STR, AZM |
| 85 | − | + | FOX, AM, CN, TOB, STR, AZM (I) |
| 86 | − | + | FOX, AM, CTX, CIP (I), CN, TOB, STR, AZM |
| 87 | − | + | FOX, AM, CTX(I), CIP, NOR, AK, CN, TOB, STR, AZM, SXT |
| 89 | − | + | FOX, AM, CTX (I), CN, TOB, STR, AZM |
Cefoxitin (FOX), ampicillin (Am), cefotaxime (CTX), ciprofloxacin (CIP), norfloxacin (NOR), amikacin (AK), tobramycin (TOB), gentamicin (CN), streptomycin (STR), azithromycin (AZM), and sulfamethoxazole/trimethoprim (SXT).
3. Discussion
The screening of antibiotic resistance in pathogenic bacteria, especially S. aureus, is very important for infection control practice. MRSA has been labelled as a ‘super bug’ due to its widespread resistance to commonly used antibiotics [25]. The present study was performed to investigate the resistance of MRSA isolates from Egyptian hospitals and their relation to integron I occurrence.
Phenotypic MRSA identification was confirmed by the disk diffusion method using a cefoxitin disk; all of the isolates were resistant to cefoxitin (100%). The usefulness of the cefoxitin disk in predicting methicillin resistance has been reported, with sensitivity values reaching 100% [26,27]. However, identification of the mecA gene is the most reliable method of detecting MRSA isolates [28]. Hence, mecA amplification by PCR was performed to confirm MRSA identification. All of the isolates were mecA-positive, and a high prevalence (75%) of MRSA was reported in Nepal [29] and 72% in Eritrea [30], while lower rates were reported in Europe, ranging from 0.9% in the Netherlands to 56% in Romania [6]. The observed existence of MRSA in this study may be related to several factors, including: carriage between healthcare workers, persistence in the hospital environment and the misuse of antibiotics [9,31].
The rise of MDR S. aureus strains, particularly MRSA, is a serious problem in the treatment and control of staphylococcal infections [32]. Approximately 55% of the MRSA isolates were MDR. A quite similar percentage of MDR (47%) was reported recently in S. aureus isolates from dairy cattle in China [33], while a higher percentage of MDR (78%) was reported among clinical S. aureus isolates in Iran [13].
All of the isolates in this study were resistant to ampicillin and cefoxitin (100%). Similarly, Gurung and his colleagues reported that all MRSA isolates were resistant to penicillin and cefoxitin [29]. Regarding aminoglycoside resistance, resistance was often reported with streptomycin (60.67%), intermediate resistance with tobramycin (37.1%), and gentamicin (35.95%), while resistance was less frequently observed for amikacin (15.9%). However, slightly lower resistance to gentamicin (10.2%) was previously reported by Gurung and his colleagues [29]. The low resistance detected for ciprofloxacin (9%), and norfloxacin (5.68%), makes these drugs the best treatment options for MRSA infections in our hospitals.
The occurrence of integrons in clinical isolates and their role in antimicrobial resistance have been widely studied in the past few decades [11,12,34]. In the current study, integron I (with variable amplicon) was detected in 41 isolates (46.6%), while integron II (with 200 bp amplicon) was detected in only three isolates (3.4%). Similarly, class I integron was found in 42.5% of nosocomial MRSA isolates (76/179) in China [25]. Furthermore, another study reported that the rate of detection of integron I (72.6%) was more than integron II (35.2%), but with higher percentages than our study [13]. In contrast, a recent study in Iran reported a lower prevalence of class I integron (24.8%), and complete absence of class II in their isolates at all [16].
Cassette genes encoding resistance to aminoglycosides were found to be predominant in the class I integron. The aadB and aadA genes encoding aminoglycoside resistance were most commonly found. aadB confer resistance to gentamicin, tobramycin, and kanamycin, while aadA confer resistance to streptomycin [13]. In this study, integron I cassette gene sequencing revealed the presence of different alleles of the aadA gene (aadA12, 15, 22, 23) in 35 isolates (39.8%). Similarly, the aadA gene was frequently detected in MRSA isolates harboring class I integron from China [25]. In fact, the aadA gene cassettes were found to be the most often reported cassettes in different bacterial isolates harboring class I integron [35]. Furthermore, similar allelic diversity of the aadA gene has been reported among Gram-negative isolates, including clinical Klebsiella pneumoniae strains [36,37].
In the current study, only six of the integron I-positive isolates with 250 bp amplicon (6.8%) were found to have empty cassette. Similarly, it was previously reported by Li and Zhao) (2018) in China [33] that the class I integron with a 153 bp size does not usually harbor any gene cassette, but this empty integron was found in 21.5% of their isolates.
Previous studies reported the occurrence of the aacA4 gene among integron cassettes but with lower existence than aadA genes [13,33]. However, none of the current isolates contain aacA4 gene within integron I; however, the aacA4 gene was detected in the genomic DNA of 31 isolates. In our study, 13 of the aacA4-positive isolates were also integron I/cassette-positive (carrying the aadA gene variant); those isolates were resistant to all tested aminoglycosides including amikacin. Other isolates that are integron I-positive (carrying the aadA gene) but negative for the aacA4 gene, were resistant to streptomycin only. The results obtained are in consistence with the literature stating that aadA variants confer resistance to streptomycin and spectinomycin only [38,39].
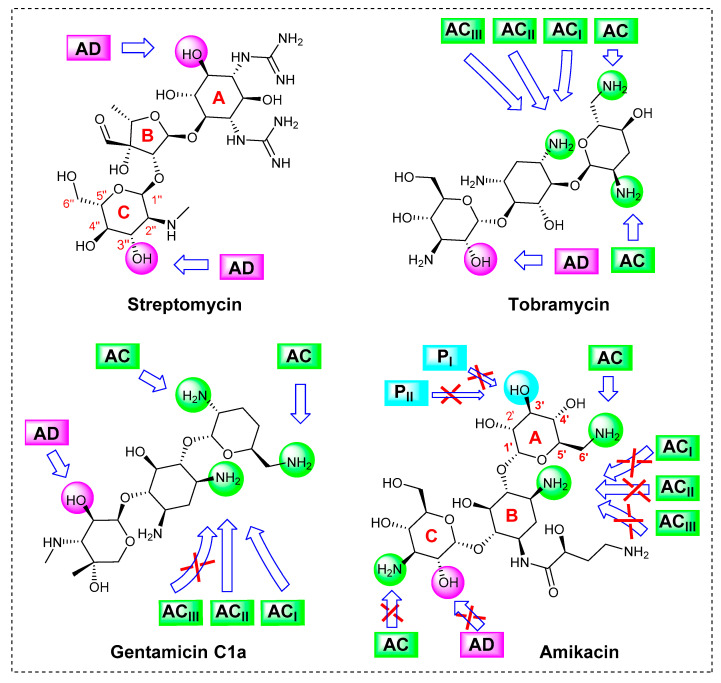
Almost all the integron I-negative but aacA4-positive isolates (17/18) were resistant to all aminoglycoside except amikacin. Overall, it seems that the presence of both adenylyltransferase (aadA) and acetyltransferase (aacA4) is essential for amikacin resistance. Previous studies related amikacin resistance in E. coli to the presence of aacA4 and aacA7 together [40]; along the same lines with our finding, it was reported that the ANT(3″) enzymes confer more resistance against amikacin in Serratia marcescens by increasing the affinity of the aminoglycoside to bind AAC(6′) enzyme [41]. Only one isolate (1/18) was integron I-negative and aacA4-positive, and it showed resistance to amikacin; in this isolate, other mechanisms could play a role in such resistance. For example, reduced drug uptake was reported as a mechanism of resistance to amikacin. Interestingly, amikacin is considered the most resistant drug to the action of AMEs, as shown in Figure 5 [20].
Figure 5.
Chemical structures of aminoglycosides under investigation showing positions targeted by modifying enzymes: acetyltransferase (AC), adenylyltransferase (AD) and phosphotransferase (P). Gentamicin and amikacin show resistance to some AMEs. Adapted from [42] with some modifications.
Furthermore, integron I-positive isolates showed resistance to other classes of antimicrobials (beta-lactam, macrolide, quinolone, sulphonamides); however, related resistance gene cassettes were not found on integron I, suggesting non-integron sources of resistance to these antimicrobials. Similarly, the absence of any gene cassette conferring resistance to erythromycin, clindamycin, tetracycline, penicillin, and ampicillin was previously reported by Li and Zhao (2018) in China [33] although the isolates were resistant to these antibiotics, which suggest other sources of such resistance by other different mobile DNA elements, such as plasmid and transposon, or other resistance genes under chromosomal control [13,33].
4. Materials and Methods
4.1. Bacteria Isolation and Identifications
A total of eighty-eight S. aureus isolates were collected from clinical laboratories in Mansoura University Hospital, Egypt, from different clinical samples (wounds, urine, sputum, and blood). All isolates were identified using standard microbiological tests, including Gram stain and growth on mannitol salt agar [43].
4.2. Antimicrobial Susceptibility
Antimicrobial susceptibility was determined by the disk diffusion method according to the Clinical and Laboratory Standard Institute [44]. Susceptibility testing was performed using antibiotics from 5 classes including: ampicillin (Am, 10 µg), cefoxitin (FOX, 30 µg), cefotaxime (CTX, 30 µg), ciprofloxacin (CIP, 5 µg), norfloxacin (NOR, 10 µg), tobramycin (TOB, 10 µg), gentamicin (CN, 10 µg), streptomycin (STR, 10 µg), amikacin (AK, 30 µg), azithromycin (AZM, 15 µg), and sulfamethoxazole/trimethoprim (SXT, 1.25/23.75 µg). Antibiotic disks were obtained from Oxoid, (Hampshire, England). The results were interpreted according to the criteria indicated in the CLSI guidelines [44].
4.3. Amplification of Integron I, Integron II, mecA and aacA4 Genes Using PCR
Total DNAs from different samples were extracted using the boiling method as described previously [45]. Each PCR reaction was performed using a Biometra T-personal thermocycler (Goettingen, Germany), in 25 reaction mixtures containing 2.5 µL of DNA, 12.5 µL of MyTaq™ red mix (Bioline Co., London, UK), 1 µL of forward primer (10 µM), 1 µL of reverse primer (10 µM) and nuclease free water to 25 µL.
For confirmation of MRSA identification, mecA genes were detected by polymerase chain reaction (PCR) using the primers listed in Table 3 [46]. the cycling conditions include heating at 94 °C for 10 min, then 30 cycles of 94 °C for 30 s, annealing at 52 °C for 30 s and 72 °C for 45 s and finally heating at 72 °C for 5 min.
Table 3.
Primers and amplicon size for mecA gene, integron class I, class II and aacA4 genes.
| Primer | Sequence (5′-3′) | Amplicon Size | Target | Annealing Temperature |
Reference |
|---|---|---|---|---|---|
| mecA-F | GTAGAAATGACTGAACGTCCGATAA | 310 bp | mecA gene | 52 °C | [46] |
| mecA-R | CCAATTCCACATTGTTCGGTCTAA | ||||
| 5′-CS | GGCATACAAGCAGCAAGC | Variable | Intg I gene cassette(s) | 49 °C | [47] |
| 3′-CS | AAGCAGACTTGACCTGAT | ||||
| Ti-F | ACCTTTTTGTCGCATATCCGTG | Variable | Intg II gene cassette(s) | 52 °C | |
| TI-B | CTAACGCTTGAGTTAAGCC | ||||
| aacA4F | GCTCTTGGAAGCGGGGACGG | 300 bp | aacA4 gene | 54 °C | [48] |
| aacA4R | TCGCTCGAATGCCTGGCGTG |
Different classes of integrons (I and II) were also detected by PCR using the primers listed in Table 1 [47]. The cycling conditions include heating at 94 °C for 5 min, then 35 cycles of 94 °C for 30 s, 49 °C and 52 °C for 20 s and 72 °C for 30 s and finally heating at 72 °C for 3 min.
Furthermore, the aacA4 gene (mediating aminoglycoside resistance) was detected in the total DNA of S. aureus isolates by PCR as previously described [48]. The cycling conditions include heating at 94 °C for 5 min, then 35 cycles of 94 °C for 30 s, 54 °C for 45 s and 72 °C for1 min, and then finally heating at 72 °C for 10 min. The PCR products as well as GeneRuler 100 bp plus DNA ladder (Thermo scientific, Waltham, MA, USA) were separated on 1.5% agarose gel, stained with ethidium bromide, and visualized by UV transilluminator.
4.4. Sequencing of Amplified Integrons
PCR amplicons comprising integrons were cleaned up using QIAquick PCR Purification (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. DNA was eluted using the elution buffer, and the concentration of eluted DNA was measured using a NanoDropTM 2000/2000c Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) including the quantification of 260/280 and 260/230 nm ratios with 1 µL of sample.
Purified PCR samples were used for sequencing according to service requirements; 5 µL of template DNA (20–80 ng) were mixed with 5 µL of the specific primers (5 pmol/µL). PCR samples were sequenced using the Illumina HiSeq platform using 300 PE chemistry (GATC-Biotech, Konstanz, Germany, now part of Eurofins Genomics Germany GmbH).
Multiple sequence alignment of the identified AMR genes (query 1 and 2) with the sequences of the aadA and aadA22 genes retrieved from banked microbial resistomes that give high identity to the queried sequence were analyzed via the Comprehensive Antibiotic Resistance Database (CARD) (https://card.mcmaster.ca/analyze/blast), access date 15 January 2021. The available sequences aligned via ClustalW and BOXSHADE were employed to highlight the multiple alignment.
5. Conclusions
The current study revealed the alarming existence of MRSA isolates. Notable levels of resistance to penicillin, cephalosporins and aminoglycosides were observed. Both the AacA4 gene and class I integrons harboring the aadA gene (mediating resistance to aminoglycosides) were predominant in MRSA isolates. The link between integrons and aminoglycoside resistance in MRSA isolates is notable and could be clinically important.
Acknowledgments
The authors would like to acknowledge the clinical laboratory medical staff at Zagazig University Hospitals and Mansoura University Hospital for providing the clinical specimens. G.Y. is a Georg Forster Research Fellow, awarded from the Alexander von Humboldt Foundation.
Author Contributions
Conceptualization, A.M.E.-B. and M.M.A.E.-S.; methodology, A.M.E.-B., G.Y., and A.M.E.-G.; software, G.Y. and B.M.; formal analysis, A.M.E.-B., G.Y., M.M.A.E.-S. and A.M.E.-G.; investigation, A.M.E.-B., G.Y., B.M. and A.M.E.-G.; data curation, G.Y., R.A. and A.A.; writing—original draft preparation, A.M.E.-G.; writing—review and editing, A.M.E.-B., G.Y., B.M., M.M.A.E.-S., R.A., A.A. and A.M.E.-G.; supervision, A.M.E.-G.; resource, A.M.E.-B., M.M.A.E.-S., R.A., A.A. and A.M.E.-G. All authors have read and agreed to the published version of the manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pollitt E.J., Szkuta P.T., Burns N., Foster S.J. Staphylococcus aureus infection dynamics. PLoS Pathog. 2018;14:e1007112. doi: 10.1371/journal.ppat.1007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tong S.Y., Davis J.S., Eichenberger E., Holland T.L., Fowler V.G. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbial. Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein E.Y., Mojica N., Jiang W., Cosgrove S.E., Septimus E., Morgan D.J., Laxminarayan R. Trends in methicillin-resistant Staphylococcus aureus hospitalizations in the United States, 2010–2014. Clin. Infect. Dis. 2017;65:1921–1923. doi: 10.1093/cid/cix640. [DOI] [PubMed] [Google Scholar]
- 4.Pichereau S., Rose W.E. Invasive community-associated MRSA infections: Epidemiology and antimicrobial management. Expert Opin. Pharmacother. 2010;11:3009–3025. doi: 10.1517/14656566.2010.511614. [DOI] [PubMed] [Google Scholar]
- 5.Otto M. Community-associated MRSA: What makes them special? Int. J. Med. Microbiol. 2013;303:324–330. doi: 10.1016/j.ijmm.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hassoun A., Linden P.K., Friedman B. Incidence, prevalence, and management of MRSA bacteremia across patient populations—A review of recent developments in MRSA management and treatment. Crit. Care. 2017;21:1–10. doi: 10.1186/s13054-017-1801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lakhundi S., Zhang K. Methicillin-resistant Staphylococcus aureus: Molecular characterization, evolution, and epidemiology. Clin. Microbial. Rev. 2018;31:e00020-18. doi: 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daef E.A., Elsherbiny N.M., Ibrahim M.A., Ahmed E.H. Decolonization of methicillin resistant Staphylococcus aureus nasal carriage among health care workers. Life Sci. J. 2012;9:223–227. [Google Scholar]
- 9.Abdel-Maksoud M., El-Shokry M., Ismail G., Hafez S., El-Kholy A., Attia E., Talaat M. Methicillin-resistant Staphylococcus aureus recovered from healthcare-and community-associated infections in Egypt. Int. J Bact. 2016;2016:5751785. doi: 10.1155/2016/5751785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stokes H.W., Gillings M.R. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbial. Rev. 2011;35:790–819. doi: 10.1111/j.1574-6976.2011.00273.x. [DOI] [PubMed] [Google Scholar]
- 11.Mazel D. Integrons: Agents of bacterial evolution. Nat. Rev. Microbiol. 2006;4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 12.Kadry A.A., Serry F.M., El-Ganiny A.M., El-Baz A.M. Integron occurrence is linked to reduced biocide susceptibility in multidrug resistant Pseudomonas aeruginosa. Br. J Biomed. Sci. 2017;74:78–84. doi: 10.1080/09674845.2017.1278884. [DOI] [PubMed] [Google Scholar]
- 13.Mostafa M., Siadat S.D., Shahcheraghi F., Vaziri F., Japoni-Nejad A., Yousefi J.V., Rajaei B., Mood E.H., Moshiri A., Siamdoust S.A.S. Variability in gene cassette patterns of class 1 and 2 integrons associated with multi drug resistance patterns in Staphylococcus aureus clinical isolates in Tehran-Iran. BMC Microbial. 2015;15:1–9. doi: 10.1186/s12866-015-0488-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren C., Zhao Y., Shen Y. Analysis of the effect of integrons on drug-resistant Staphylococcus aureus by multiplex PCR detection. Mol. Med. Rep. 2013;7:719–724. doi: 10.3892/mmr.2013.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillings M.R. Integrons: Past, Present, and Future. Microbiol. Mol. Biol. Rev. 2014;78:257–277. doi: 10.1128/MMBR.00056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohammadi M., Bahrami N., Khajavian M., Faghri J. The Occurrence of Type I, II, and III Integrons in Multi-drug Resistance and Methicillin-Resistant Staphylococcus aureus Isolates in Iran. Curr. Microbial. 2020;77:1653–1659. doi: 10.1007/s00284-020-01956-x. [DOI] [PubMed] [Google Scholar]
- 17.Zárate S.G., De la Cruz Claure M.L., Benito-Arenas R., Revuelta J., Santana A.G., Bastida A. Overcoming aminoglycoside enzymatic resistance: Design of novel antibiotics and inhibitors. Molecules. 2018;23:284. doi: 10.3390/molecules23020284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitz F.-J., Fluit A.C., Gondolf M., Beyrau R., Lindenlauf E., Verhoef J., Heinz H.-P., Jones M.E. The prevalence of aminoglycoside resistance and corresponding resistance genes in clinical isolates of staphylococci from 19 European hospitals. J. Antimicrob. Chemother. 1999;43:253–259. doi: 10.1093/jac/43.2.253. [DOI] [PubMed] [Google Scholar]
- 19.Serio A., Magalhães M., Blanchard J., Connolly L. Antimicrobial Drug Resistance. National Institute of Allergy and Infectious Diseases; Bethesda, MD, USA: 2017. Aminoglycosides: Mechanisms of Action and Resistance; pp. 213–229. [Google Scholar]
- 20.Ramirez M.S., Tolmasky M.E. Amikacin: Uses, resistance, and prospects for inhibition. Molecules. 2017;22:2267. doi: 10.3390/molecules22122267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramirez M.S., Tolmasky M.E. Aminoglycoside modifying enzymes. Drug Resist. Updat. 2010;13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramirez M.S., Nikolaidis N., Tolmasky M. Rise and dissemination of aminoglycoside resistance: The aac (6′)-Ib paradigm. Front. Microbiol. 2013;4:121. doi: 10.3389/fmicb.2013.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wickham H., Chang W. Computer Software. [(accessed on 15 January 2021)];2012 Available online: http://ggplot2.org.
- 24.Hofmann K., Baron M.B. Version 3.21 of BOXSHADE. [(accessed on 15 January 2021)]; Available online: http://sourceforge.net/projects/boxshade/
- 25.Xu Z., Li L., Shirtliff M., Peters B., Li B., Peng Y., Alam M.J., Yamasaki S., Shi L. Resistance Class 1 integron in clinical methicillin-resistant Staphylococcus aureus strains in southern China, 2001–2006. Clin. Microbiol. Infect. 2011;17:714–718. doi: 10.1111/j.1469-0691.2010.03379.x. [DOI] [PubMed] [Google Scholar]
- 26.Velasco D., del Mar Tomas M., Cartelle M., Beceiro A., Perez A., Molina F., Moure R., Villanueva R., Bou G. Evaluation of different methods for detecting methicillin (oxacillin) resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 2005;55:379–382. doi: 10.1093/jac/dki017. [DOI] [PubMed] [Google Scholar]
- 27.Sousa Júnior F.C.d., Néri G.d.S., Silva A.K., Araújo B.P.R.C.d., Guerra M.J.d.P.D., Fernandes M.J.d.B.C., Milan E.P., Melo M.C.N.d. Evaluation of different methods for detecting methicillin resistance in Staphylococcus aureus isolates in a university hospital located in the Northeast of Brazil. Braz. J. Microbiol. 2010;41:316–320. doi: 10.1590/S1517-83822010000200009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadry A., Shaker G., El-Ganiny A., Youssef C. Phenotypic and Genotypic detection of local MRSA isolates. ZJPS. 2016;25:39–46. doi: 10.21608/zjps.2016.38164. [DOI] [Google Scholar]
- 29.Gurung R.R., Maharjan P., Chhetri G.G. Antibiotic resistance pattern of Staphylococcus aureus with reference to MRSA isolates from pediatric patients. Future Sci. OA. 2020;6:FSO464. doi: 10.2144/fsoa-2019-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garoy E.Y., Gebreab Y.B., Achila O.O., Tekeste D.G., Kesete R., Ghirmay R., Kiflay R., Tesfu T. Methicillin-resistant Staphylococcus aureus (MRSA): Prevalence and antimicrobial sensitivity pattern among patients—A multicenter study in Asmara, Eritrea. Can. J. Infect. Dis. Med. Microbiol. 2019;2019:8321834. doi: 10.1155/2019/8321834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El Aila N.A., Al Laham N.A., Ayesh B.M. Nasal carriage of methicillin resistant Staphylococcus aureus among health care workers at Al Shifa hospital in Gaza Strip. BMC Infect. Dis. 2017;17:1–7. doi: 10.1186/s12879-016-2139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaur D.C., Chate S.S. Study of antibiotic resistance pattern in methicillin resistant Staphylococcus aureus with special reference to newer antibiotic. J. Glob. Infect. Dis. 2015;7:78. doi: 10.4103/0974-777X.157245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L., Zhao X. Characterization of the resistance class 1 integrons in Staphylococcus aureus isolates from milk of lactating dairy cattle in Northwestern China. BMC Vet. Res. 2018;14:1–7. doi: 10.1186/s12917-018-1376-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng Y., Bao X., Ji L., Chen L., Liu J., Miao J., Chen D., Bian H., Li Y., Yu G. Resistance integrons: Class 1, 2 and 3 integrons. Ann. Clin. Microbiol. Antimicrob. 2015;14:1–11. doi: 10.1186/s12941-015-0100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun G., Yi M., Shao C., Ma J., Zhang Q., Shao S. Novel class 1 integrons in multi-drug resistant isolates from Eastern China. Indian J. Microbiol. 2014;54:227–231. doi: 10.1007/s12088-013-0441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grape M., Farra A., Kronvall G., Sundström L. Integrons and gene cassettes in clinical isolates of co-trimoxazole-resistant Gram-negative bacteria. Clin. Microbiol. Infect. 2005;11:185–192. doi: 10.1111/j.1469-0691.2004.01059.x. [DOI] [PubMed] [Google Scholar]
- 37.Firoozeh F., Mahluji Z., Khorshidi A., Zibaei M. Molecular characterization of class 1, 2 and 3 integrons in clinical multi-drug resistant Klebsiella pneumoniae isolates. Antimicrob. Resist. Infect. Control. 2019;8:1–7. doi: 10.1186/s13756-019-0509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kehrenberg C., Catry B., Haesebrouck F., de Kruif A., Schwarz S. Novel spectinomycin/streptomycin resistance gene, aadA14, from Pasteurella multocida. Antimicrob. Agents Chemother. 2005;49:3046–3049. doi: 10.1128/AAC.49.7.3046-3049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cameron A., Klima C.L., Ha R., Gruninger R.J., Zaheer R., McAllister T.A. A novel aadA aminoglycoside resistance gene in bovine and porcine pathogens. mSphere. 2018;3:e00568-17. doi: 10.1128/mSphere.00568-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruiz J., Bertran S., Sauca G., Julià A., Vila X., Gómez E., Jiménez de Anta M., Vila J. Isolation of an amikacin-resistant Escherichia coli strain after tobramycin treatment of previous recurrent episodes of respiratory tract infections caused by Pseudomonas aeruginosa. Clin. Microbiol. Infect. 2005;11:71–73. doi: 10.1111/j.1469-0691.2004.01039.x. [DOI] [PubMed] [Google Scholar]
- 41.Green K.D., Garneau-Tsodikova S. Domain dissection and characterization of the aminoglycoside resistance enzyme ANT (3″)-Ii/AAC (6′)-IId from Serratia marcescens. Biochimie. 2013;95:1319–1325. doi: 10.1016/j.biochi.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hilal-Dandan R., Brunton L. Aminoglycosides. Goodman and Gilman’s Manual of Pharmacology and Therapeutics. 2nd ed. McGraw-Hill Education; New York, NY, USA: 2016. [Google Scholar]
- 43.Boerlin P., Kuhnert P., Hüssy D., Schaellibaum M. Methods for identification of Staphylococcus aureus isolates in cases of bovine mastitis. J. Clin. Microbiol. 2003;41:767–771. doi: 10.1128/JCM.41.2.767-771.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinstein M.P., Lewis J.S. The clinical and laboratory standards institute subcommittee on antimicrobial susceptibility testing: Background, organization, functions, and processes. J. Clin. Microbial. 2020;58:e01864-19. doi: 10.1128/JCM.01864-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dilhari A., Sampath A., Gunasekara C., Fernando N., Weerasekara D., Sissons C., McBain A., Weerasekera M. Evaluation of the impact of six different DNA extraction methods for the representation of the microbial community associated with human chronic wound infections using a gel-based DNA profiling method. AMB Express. 2017;7:1–11. doi: 10.1186/s13568-017-0477-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun J., Yang M., Sreevatsan S., Davies P.R. Prevalence and characterization of Staphylococcus aureus in growing pigs in the USA. PLoS ONE. 2015;10:e0143670. doi: 10.1371/journal.pone.0143670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su J., Shi L., Yang L., Xiao Z., Li X., Yamasaki S. Analysis of integrons in clinical isolates of Escherichia coli in China during the last six years. FEMS Microbiol. Lett. 2006;254:75–80. doi: 10.1111/j.1574-6968.2005.00025.x. [DOI] [PubMed] [Google Scholar]
- 48.Sacha P., Jaworowska J., Ojdana D., Wieczorek P., Czaban S., Tryniszewska E. Occurrence of the aacA4 gene among multidrug resistant strains of Pseudomonas aeruginosa isolated from bronchial secretions obtained from the intensive therapy unit at University hospital in Bialystok, Poland. Folia Histochem. Cytobiol. 2012;50:322–324. doi: 10.5603/FHC.2012.0043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.