Abstract
Hepatitis B virus infection affects over 250 million chronic carriers, causing more than 800,000 deaths annually, although a safe and effective vaccine is available. Currently used antiviral agents, pegylated interferon and nucleos(t)ide analogues, have major drawbacks and fail to completely eradicate the virus from infected cells. Thus, achieving a “functional cure” of the infection remains a real challenge. Recent findings concerning the viral replication cycle have led to development of novel therapeutic approaches including viral entry inhibitors, epigenetic control of cccDNA, immune modulators, RNA interference techniques, ribonuclease H inhibitors, and capsid assembly modulators. Promising preclinical results have been obtained, and the leading molecules under development have entered clinical evaluation. This review summarizes the key steps of the HBV life cycle, examines the currently approved anti-HBV drugs, and analyzes novel HBV treatment regimens.
Keywords: hepatitis B, hepatitis B virus, HBV, antiviral agents, HBV inhibitors, cccDNA, HBV life cycle, HBV treatment, nucleoside analogues, antiviral therapy
1. Introduction
Hepatitis B is a liver disease caused by the Hepatitis B Virus (HBV). HBV belongs to the Hepadnaviridae family and is classified into ten genotypes (A to J) [1]. It is transmitted by exposure to infectious blood or other body fluids (e.g., semen, vaginal secretions—sexual intercourse) as well as perinatally from infected mothers to infants [2]. The acute phase of the infection can be either symptomatic or asymptomatic. Acute infections can either spontaneously resolve or proceed to chronic infections. Chronic HBV infection is among the leading causes of hepatic cirrhosis and is the single largest cause of hepatocellular carcinoma (HCC). According to the World Health Organization (WHO), over 250 million people are chronically infected, and HBV caused 887,000 deaths in 2015 [3]. The highest epidemic prevalence is present in SE Asian, African, and Western Pacific countries [4].
The hepatitis B surface antigen (HBsAg), originally known as “Australia antigen” (AusAg), was firstly identified in the serum of indigenous Australians by Baruch Samuel Blumberg in 1965 [5]. This antigen was later related with viral hepatitis [6].
The goal of the current therapeutic development is a “functional cure” defined as sustained undetectable levels of HBsAg and HBV DNA in serum, with or without seroconversion to hepatitis B surface antibodies (anti-HBs) after the end of the treatment [7]. This reduction has been associated with an improved clinical condition and significantly decreased the chance of infection rebound. Other important HBV biomarkers include serum HBV DNA, hepatitis B core antigen (HBcAg), and its antibody anti-HBc, hepatitis B e antigen (HBeAg), and anti-HBe antibody [8,9,10]. HBeAg is a secreted variant of HBcAg, and viral infections are classified either as HbeAg-positive or HbeAg-negative, with HBeAg-positive patients having higher viral titers and a more frequent and rapid disease progression [11]. These biomarkers are used to guide treatment decisions following guidelines established by the major hepatology medical societies [12,13,14].
Despite the existence of a safe and effective vaccine, no therapeutic regimen that routinely induces a “functional cure” for chronic HBV has been identified yet. This review summarizes the HBV replication cycle, the existing treatment options and their significant disadvantages, and novel therapeutic approaches that are currently the subject of extensive scientific research, with the ultimate goal of achieving a “functional cure” of the disease.
2. HBV Replication Cycle
2.1. Virion Structure and Genome
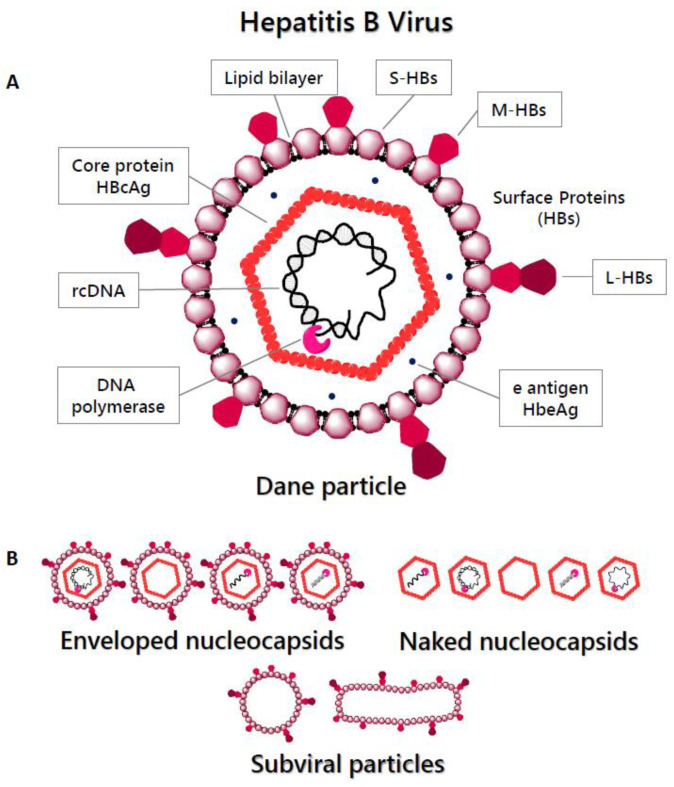
HBV particles, also known as Dane particles (Figure 1A), were firstly identified by Dane and colleagues in 1970 [15]. Their shape is spherical, with a diameter of ∼42 nm. They consist of an outer envelope, which is a host-derived lipid bilayer containing three different-sized HBV surface antigens (HBsAg or HBs)—large (L-HBs), middle (M-HBs) and small (S-HBs)—surrounding the viral nucleocapsid. The nucleocapsid (∼27 nm diameter) is icosahedral and comprises the HBV core protein (HBcAg), as well as the viral DNA genome and the viral DNA polymerase (P) [16,17]. The virus also secretes a wide range of defective particles (Figure 1B), including enveloped nucleocapsids that are empty or contain defective immature genomes and subviral lipid particles containing the viral surface antigens. The subviral particles are secreted along with the infectious virions at levels that are thousands of times higher, and they play an important role in suppressing antibody responses to the virus [18].
Figure 1.
Hepatitis B Virus particles. (A) Infectious HBV virion (Dane particle). The lipid envelope, bearing three types of surface proteins—small (S-HBs), middle (M-HBs) and large (L-HBs)—surrounds the nucleocapsid, consisting of HBV relaxed circular DNA (rcDNA), the viral DNA polymerase (P), and the core protein (HBcAg). (B) Non-infectious HBV particles; enveloped nucleocapsids containing immature or defective DNA/RNA, subviral particles, and naked nucleocapsids.
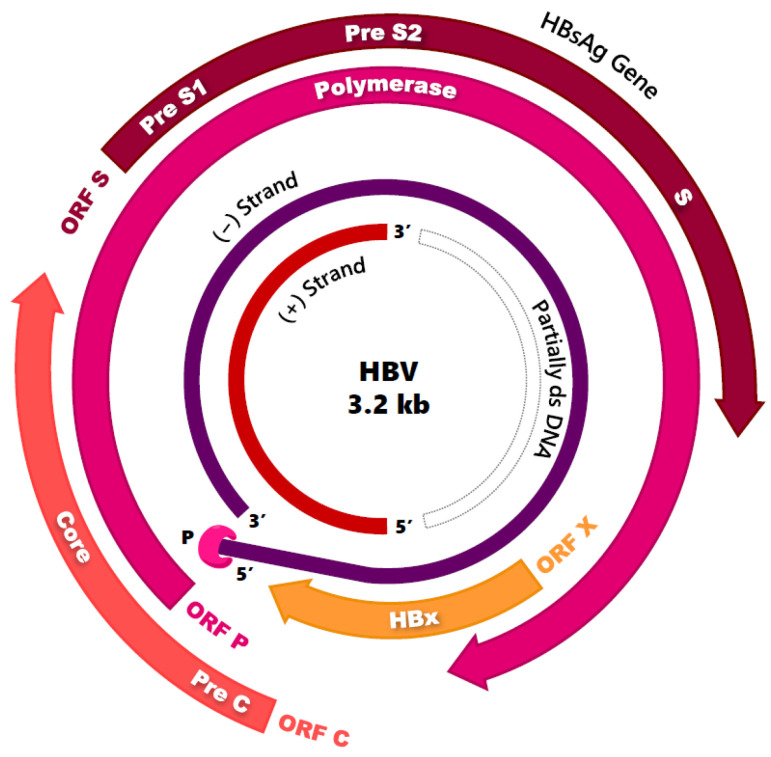
The HBV genome is a 3.2 kb circular, partially double-stranded DNA (relaxed circular DNA; rcDNA). The negative-sense, non-coding (−) DNA strand is complete and complementary to the mRNA transcripts, whereas the positive (+) DNA strand is incomplete and has a fixed 5′-end and a variable-size 3′-end [19,20,21]. The former contains four overlapping open reading frames (ORFs)—C, P, S, and X (Figure 2). These are transcribed into five RNA transcripts of varying lengths and are subsequently translated into seven functional proteins. HBcAg is produced from ORF-C, HBeAg is produced from ORF preC + C, DNA polymerase from ORF P, and HBV X protein (HBx) from ORF X. The ORF S, because of its multiple in-frame start codons, encodes the L-HBs, M-HBs, and S-HBs envelope proteins (pre-S1 + pre-S2 + S, pre-S2 + S, or S, respectively) [17,19]. This compact nature of the HBV genome results in approximately two thirds of nucleotides encoding more than one functional element [22,23]. The overlap of more than 1000 nucleotides between the P and S genes is the largest gene overlap of any known animal virus [24].
Figure 2.
Hepatitis B Virus genome. Partially double-stranded, relaxed circular DNA (rcDNA) with four overlapping open reading frames (ORFs).
2.2. Viral Entry
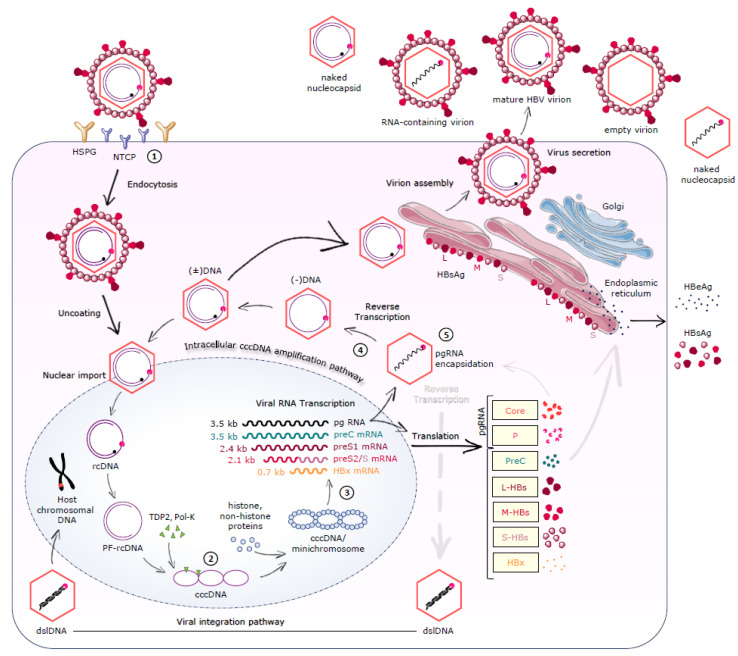
The HBV virion binds to the heparan sulfate proteoglycans (HSPGs) cell-surface receptors, via low-affinity and non-specific interactions. Afterwards, the Na(+)-taurocholate co-transporting polypeptide (NTCP) functions as a high affinity receptor for the recognition and attachment of the pre-S1 domain of L-HBsAg. NTCP is a liver-specific peptide that mediates the uptake of bile salts into hepatocytes, and it is also an entry receptor for Hepatitis D virus (HDV) [17,25]. Interactions between HBV and NTCP are responsible for the viral endocytosis (Figure 3). According to recent studies, a complex formed between NTCP and the epidermal growth factor receptor (EGFR) contributes to the HBV entry [26]. Due to its complicated structure, NTCP can be oligomerized, and this process seems to affect the viral internalization into the cell. Following entry, the nucleocapsid is released into the cytoplasm, followed by uncoating, and then the rcDNA is transported to the nucleus via the nuclear pore complexes [17,19,27].
Figure 3.
Main features of the hepatitis B virus replication cycle and potential therapeutic targets. (1) HBV entry inhibitors. Lipopeptides mimicking the pre-S1 region of HBV, monoclonal antibodies, and other small molecules under evaluation. (2) Targeting cccDNA. Damage and destruction of cccDNA via sequence-specific nucleases. Direct targeting of the HBx protein. (3) RNA interference. Small interfering RNAs (siRNAs), antisense oligonucleotides (ASOs). (4) HBV polymerase inhibitors. Reverse transcriptase inhibitors (nucleos(t)ide analogues) are part of the current treatment. RNaseH inhibitors are in preclinical evaluation. (5) Nucleocapsid assembly inhibitors or modulators can affect HBV capsid formation, reverse transcription, and pgRNA encapsidation. NTCP; Na(+) taurocholate co-transporting polypeptide, HSPG; heparan sulfate proteoglycan, rc-DNA; relaxed circular DNA, PF-rcDNA; protein-free rcDNA, cccDNA; covalently closed circular DNA, pgRNA; pregenomic RNA, preC; precore, mRNA; messenger RNA, P; polymerase, L-HBs; large hepatitis B surface protein, M-HBs; middle hepatitis B surface protein, S-HBs; small hepatitis B surface protein, HBx; hepatitis B X protein, HBsAg; hepatitis B surface antigen, HBeAg; hepatitis B e antigen, dslDNA; double-stranded linear DNA.
2.3. cccDNA Formation/Maintenance
Multiple cellular factors repair the HBV rcDNA to form the episomal covalently closed circular DNA (cccDNA) that is located in the nucleus (Figure 3). Both the viral P protein which is bound to the 5′-end of the minus-polarity DNA strand [28] and the RNA primer attached to the 5′-end of the plus DNA strand are removed to leave a protein-free rcDNA (PF-rcDNA). The gaps in both strands are filled and circularized to form the cccDNA. Cellular factors believed to be involved in this process include the DNA repair enzyme tyrosyl-DNA phosphodiesterase 2 (TDP2) by presumably breaking the phosphodiesterase bond between the HBV P and rcDNA [29,30]. Another enzyme that breaks down the RNA primer at the 5′-end of the minus strand is the flap endonuclease 1 (FEN1) [31]. After removal of the proteins, the fill-in of the strands, DNA ligation, and DNA repair are conducted by other host enzymes such as DNA polymerases (κ and α), DNA ligases (LIG1 and LIG3), and topoisomerases I and II (TOP1 and TOP2) [17]. However, there is some functional redundancy among these factors, and it is not fully clear which of them function in an infected liver. The cccDNA is the template for transcription of viral RNAs. The stability of cccDNA is regulated by several cellular factors, such as the APOBEC3 protein family, that triggers cccDNA degradation [32,33]. The cccDNA is rather stable during antiviral therapy, declining by only ~1 log10 after more than a year of nucleos(t)ide analogue therapy [34]. However, the half-life of cccDNA has been measured at only approximately 40 days in HepG2 cells [17,35], and studies of cccDNA replacement during the reversion of nucleoside analogue resistance following the cessation of the therapy indicate that the cccDNA half-life in the liver is 16–28 weeks [36].
2.4. Transcription-Translation-Reverse Transcription-Nucleocapsid Assembly
Using the cccDNA as a template, the host RNA polymerase II transcribes five RNAs of different lengths: three subgenomic mRNAs of 0.7, 2.1, and 2.4 kb and two longer than genomic mRNAs of 3.5 kb (Figure 3). All of them are heterogenous, positively orientated, and have a 5′-cap and a 3′-polyadenylated tail [21]. The 3.5 kb pregenomic RNA (pgRNA) has two functions: it is the template for reverse transcription to generate the minus DNA strand and also the mRNA for the translation of the core protein and HBV P. The preC mRNA is slightly longer than the pgRNA. It contains an open reading frame that starts about 90 nucleotides upstream of the HBc ORF on the 3.5 kb pre-C mRNA and encodes the precore protein, which is converted to HBeAg upon post-translational processing in the endoplasmic reticulum (ER). The 2.4 kb mRNA encodes the L-HBs, whilst both M-HBs and S-HBs are encoded by the 2.1 kb transcript. The shortest transcript encodes the HBx protein [37,38,39]. The transcription process is regulated by four promoters (precore/core, pre-S1, pre-S2, and X) and two enhancers (Enh1 and Enh2), as well as several cis-acting negative regulatory elements [19,40,41,42,43].
HBx is the only purely regulatory protein encoded by HBV and has a multifunctional role. HBx promotes the degradation of the Smc5/6 complex (structural maintenance of chromosomes) host factor, thus enhancing the transcription of cccDNA [44,45,46,47]. Moreover, HBx represses development of the immune response to HBV infection, protecting the infected hepatocytes from immune-mediated apoptosis, and interferes with the host gene expression, facilitating the development of HCC [48,49,50].
HBV P contains four domains: the terminal protein (TP), the spacer, the reverse transcriptase (RT) domain, and the ribonuclease H (RNaseH) domain [51,52]. The pgRNA binds to P via the ε-stem loop located close to its 5′-end with specific motifs in the TP, spacer, RT, and RNaseH domains to form a pgRNA-P ribonucleoprotein (RNP) complex [53,54,55,56,57]. This interaction is of great importance, as it is essential for the RNA packaging into nucleocapsids and initiation of reverse transcription [58,59]. Specifically, the RNP complex is packaged within HBcAg to form immature nucleocapsids, where reverse transcription occurs, producing either rcDNA, or less often, double-stranded linear DNA (dslDNA) forms (Figure 3).
HBV replicates by reverse transcription. The RT activity of the P protein primes DNA synthesis using a tyrosine in the TP domain, covalently linking the enzyme to the product DNA. P then catalyzes the synthesis of the (−) DNA strand, which is the pattern for the (+) DNA strand synthesis, to form double-stranded rcDNA and mature DNA nucleocapsids [19,60]. During the (−) DNA strand synthesis, the RNaseH domain degrades the pgRNA template inside the capsids after it is copied into the minus-polarity DNA [61,62]. Either the newly formed mature nucleocapsids are surrounded by HBsAg and secreted non-cytolytically as virions that can infect new hepatocytes or they can re-enter the nucleus to maintain the cccDNA reservoir (Figure 3). This intracellular cccDNA recycling is likely one factor that makes the complete elimination of the HBV infection in a patient so difficult. Smaller, non-infectious subviral particles (∼22 nm in diameter) are also released from the hepatocytes in vast excess over infectious virions [17]. These include empty envelopes of HBsAg (subviral particles), virions containing RNA, or a defective DNA genome, as well as naked nucleocapsids (lacking envelope) (Figure 1) [63]. These defective particles do not participate in viral replication, although the subviral HBsAg particles help suppress antiviral immunity [18].
The dslDNA is an aberrant reverse transcription product that is able to integrate into the cellular genome early after the initial HBV infection, and it has been associated with promoting the development of HCC. The integrated HBV DNA does not replicate, but it contributes to HBsAg expression, which contributes to HBV pathogenesis and modulates the immune response [64].
3. Current Therapies
Two types of treatment are currently available against hepatitis B viral infection, interferon α derivatives (IFNs), and nucleos(t)ide analogues (NAs).
Interferon α (IFN-α) was first approved for the treatment of HBV infection in 1991 [65]. However, the addition of a polyethylene glycol chain to IFN-α led to significantly improved pharmacological properties. Thus, IFN-α was replaced by its pegylated counterpart, PEG-IFN-α, in 2005. There are two forms of PEG-IFN-α available today, PEG-IFN-α2a (Pegasys©, Roche) and PEG-IFN-α2b (Pegintron©, Merck). They have improved pharmacokinetics and allowed for a longer half-life, enabling a weekly administration [66]. PEG-IFN-α is administered subcutaneously and has direct antiviral as well as immunomodulatory activity [67,68,69,70]. One year of PEG-IFN-α treatment in HbeAg-positive patients led to HbeAg seroconversion in 29–32% of the patients and sustainable reduced HbsAg levels in 3–7% of the patients, 24 weeks after the end of the treatment, highlighting the effectiveness of PEG-IFN-α against HBV [2,71]. Nevertheless, the PEG-IFN-α treatment causes adverse reactions including flu-like symptoms, bone marrow suppression, fatigue, and depression, and is contraindicated for patients suffering from hepatic failure or cirrhosis [2,72]. Patient compliance is also low due to the subcutaneous administration.
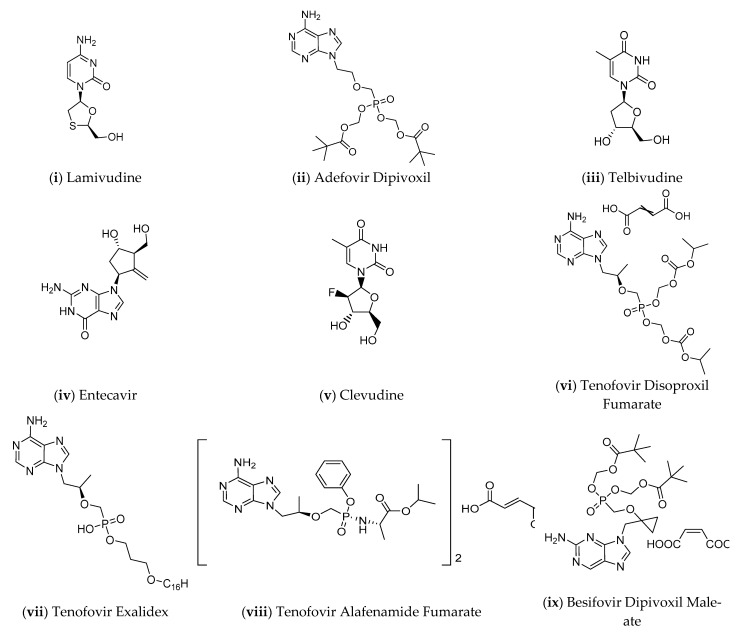
Nucleoside analogues (Figure 4) inhibit the HBV reverse transcriptase activity and therefore block HBV DNA replication. The active form of most of these drugs is the triphosphate that results from their phosphorylation by hepatocyte kinases. Nucleoside triphosphate analogues are substrates for the RT. During reverse transcription, they act as immediate or delayed transcriptional terminators and prevent the synthesis of both (−) and (+) HBV DNA strands. They are administered per os, having acceptable pharmacokinetics and limited drug-drug interactions. NAs suppress viremia at clinically undetectable levels in up to 76% of HBeAg (+) and 93% of HBeAg (−) patients after one year of treatment. Efficacy can vary in patients with different HBV genotypes [73,74]. Although some HBeAg (−) patients can discontinue treatment with NAs, their use is essentially life-long for the large majority of patients. However, virological relapse almost always occurs. Eight NAs have been approved against the HBV, of which the current recommended ones are entecavir and the two tenofovir prodrugs, disoproxil and alafenamide [73].
Figure 4.
Nucleos(t)ide Analogues (NAs) approved for the treatment of hepatitis B [28,73].
The first approved NA which was effective against HBV was lamivudine (3TC, LMV, Epivir©, Zeffix©, Heptodin©, Hepitec©). It was approved in the United States of America in 1998 [73], and it is administered once daily, with few side effects. It is no longer widely used because it is less potent than newer drugs and most patients develop resistance within one to five years [65]. Data from a randomized controlled trial showed that treatment with LMV for a median duration of approximately 32 months reduced the frequency of HCC occurrence [75]. As shown in another study, receiving LMV reduced the risk of HCC even in patients with liver cirrhosis [76]. The long-term use of LMV is limited by the development of resistance associated with mutations in the YMDD (tyrosine-methionine-aspartic acid-aspartic acid) motif in the viral RT active site. A study carried out by Kwon et al. in 2013 [77] suggests that treatment in patients without mutations in this region may be continued for more than five years until the complete loss of HBsAg is achieved [77]. However, the sustained viral response obtained with LMV for more than five years showed no further decrease in the incidence of HCC [75]. In the opposite direction, Eun et al. [78] found that long-term LMV administration and subsequent prolonged viral suppression had a beneficial impact on the risk of HCC [75]. Lamivudine therapy has been confirmed to reduce liver-related mortality in patients with HBV and even in patients with co-infection with human immunodeficiency virus (HIV), especially along with other NA as combination therapy [75,79].
The next approved ΝΑ was adefovir dipivoxil (bis(POM) PMEA, ADV, Hepsera© or Preveon©) in 2002 [73]. Given once daily, it has shown only few side effects; however, the renal function should be monitored to avoid the development of renal impairment. It is considered a second-line treatment option, except for the case of LMV resistance, where it is used as the drug of choice [65]. Although ADV monotherapy is effective in HBV patients and its long-term use reduced the rate of liver fibrosis, resistant mutations conferred decreased susceptibility to ADV [75].
Entecavir (BMS-200475-01, ETV, Baraclude©) was approved in 2005 [73]. It is taken once daily and causes few side effects. It is a first-line treatment with exceptional resistance profile [65], and it has been proved that it reduces the incidence of HCC [80]. The monitoring of serum alanine aminotransferase (ALT), an enzyme released by dead hepatocytes, is recommended at 6 and 12 months of treatment with ETV, since normal ALT levels are related to a reduced risk of developing HCC. Furthermore, the follow-up monitoring of serum alpha-fetoprotein as a biomarker for HCC is suggested [75].
Telbivudine (LdT, TBV, Tyzeka© or Sebivo©) was approved in 2006 as a second-line treatment option [65]. Randomized clinical trials revealed that TBV is superior to lamivudine and adefovir in the treatment of patients with chronic HBV, regardless of the HBeAg detection [81,82,83]. In 2013, Tsai et al. [84] found that the cumulative incidence of HCC in patients who had received telbivudine was 2.5% and 4.1% at two and three years, respectively, rates similar to that of entecavir administration (3.1% and 7.5% in two and three years, respectively). TBV is associated with few side effects, including muscle toxicity and peripheral neuropathy [85,86]. Renal function should be considered when choosing between NAs, and it is worth noting that TBV can prevent nephrotoxicity [75]. At the same year, clevudine (L-FMAU, CLV, Levovir© or Revovir©) was approved in South Korea and the Philippines. It was soon recalled due to the induction of skeletal myopathy caused by mitochondrial dysfunction [73].
Tenofovir Disoproxil Fumarate (bis(POC) PMPA Fumarate, TDF, Viread©) was first released in 2008. It is taken once daily and has few serious adverse effects, including dose-limiting renal toxicity. Although being a first-line treatment, TDF is also effective as a second-line rescue treatment after therapy with other nucleos(t)ide analogues has failed due to resistance evolution [65,75]. To a great extent, TDF is not susceptible to resistance development, and thus its use provides sufficient virological suppression [87]. Some studies demonstrate that patients receiving TDF have a lower incidence of HCC compared with patients receiving entecavir [75,88,89,90]. Contrarily, other studies indicate that both tenofovir and entecavir monotherapies display a comparable risk for HCC [91,92]. Tenofovir alafenamide fumarate (GS-7340-03, TAF, Vemlidy©) was first released in 2016, and it was developed to tackle the dose-limiting renal toxicity of TDF. The primary purpose of another analogue, tenofovir exalidex (a prodrug which is in early clinical development) is to improve the safety compared to formulations of TDF [93]. In 2017 another analogue was discovered, besifovir dipivoxil maleate (ANA-380/LB80380 maleate, BSV dipivoxil maleate, Besivo©), showing significantly reduced bone and kidney toxicity, compared to tenofovir [73,93].
Overall, NAs which are administered per os, require long-term duration of therapy, achieve better control of HBV replication, and show many fewer side effects compared to PEG-IFN-α. Treatment with PEG-IFN-α is shorter in duration and can lead to stable, off-treatment multi-log10 reductions in viral titers in about 30% of patients [2], but it is not well tolerated in many patients because of severe side effects [7,94].
4. Novel Therapeutic Strategies
Major scientific breakthroughs, such as the identification of the NTCP cell surface receptor, detailed knowledge gained about cccDNA formation, regulation and its epigenetic control, the mechanism of cccDNA and pgRNA degradation, and the determination of the HBx protein’s role in viral transcription, have enabled an in-depth understanding of the HBV life cycle. In addition, innovative cell and animal models have improved the in vitro and in vivo assessment of the antiviral activity and potential toxicity of novel compounds. All of the above have paved the way for investigating multiple new therapeutic targets that will lead to substantial progress toward achieving a functional HBV cure [7,95].
4.1. HBV Entry Inhibitors
The discovery of NTCP as the entry receptor for HBV provided key knowledge on the viral entry mechanism, thus facilitating the identification of a variety of compounds that block the viral entry into the host hepatocytes [96]. As mentioned in Section 2.2, interactions between the pre-S1 domain of L-HBsAg and NTCP are the key process for viral entry [97].
Various strategies have been proposed for the inhibition of HBV entry into hepatocytes. Small, acetylated peptides derived from the pre-S1 domain of L-HBs can effectively inhibit viral entry, exhibiting promising results both in vitro and in vivo [98,99,100]. In a recent report, novel cyclic peptides led to a significant HBsAg loss in vivo, with IC50 values between 0.66 and 2.54 μΜ, without affecting the physiological function of the NTCP receptor [101]. Another study revealed that peptide 4B10 was able to inhibit HBV infection in a human hepatocyte culture, with IC50 values in the nM range and no observed cytotoxicity [102]. The most important compound of this category is Myrcludex B (also known as Bulevirtide). Myrcludex B is a synthetic myristoylated lipopeptide consisting of 47 amino acids of the pre-S1 region. It strongly inhibits the HBV entry in the cell culture (IC50 = 80 pM) and in a uPA/SCID humanized mouse model of HBV infection [103,104,105]. Moreover, Myrcludex B inhibits bile salt uptake only at much higher concentrations (IC50 = 52.5 nM) [106]. The safety and efficacy results from clinical trials IIb are also excellent [107,108]. A liposomal formulation of Myrcludex B is under development for per os administration with an excellent pharmacological drug-drug interaction profile [109,110].
Several FDA-approved compounds have recently been identified as efficient inhibitors of the NTCP-L-HBs interaction. Those include the immunosuppressant cyclosporin A and its derivatives [106,111,112], the antihyperlipidemic ezetimibe [113], the angiotensin II receptor antagonist irbesartan [114], and the immunosuppressant rapamycin [115], among many drugs already in clinical use [116,117,118]. Another study identified that the green tea flavonoid epigallocatechin-3-gallate can efficiently block the NTCP-mediated viral entry [119]. Other recently identified HBV entry inhibitors include zafirlukast, vanitaracin A, proanthocyanidin, and its analogues, betulin derivatives, and novel synthetic compounds like B7 (Table 1) [117,120,121,122,123,124].
Table 1.
Compounds that inhibit HBV entry in hepatocytes and their in vitro IC50 values for HBV infection, measured in HepG2 cell cultures.
| Name | Structure | Class | IC50 1 | Ref. |
|---|---|---|---|---|
| Ezetimibe |
|
Cholesterol absorption inhibitor | 18 μΜ 2 | [113] |
| (−)-epigallocatechin-3-gallate |
|
Flavonoid in green tea extract | ≈10 μM | [119] |
| Proanthocyanidin |
|
Oligomeric flavonoid, derived from extract of grape seed | 7.8 μM | [122] |
| Oolonghomobisflavan C |
|
Proanthocyanidin analogue | 4.3 μM | [122] |
| Temsirolimus |
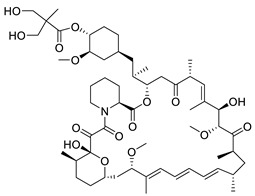
|
Rapamycin derivative and prodrug | 7.86 μM | [115] |
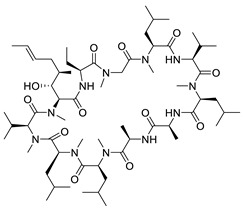
| Compound 2 |
|
Betulin derivative | 4 μM | [123] |
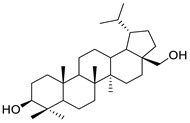
| Cyclosporin A |
|
Immunosuppressant | 1.17 μM | [111] |
| Rosiglitazone |
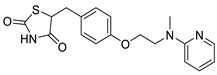
|
Thiazolidinedione (PPAR-γ agonist) | 5.1 μM | [117] |
| Ciglitazone |
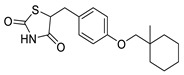
|
Thiazolidinedione (PPAR-γ agonist) | 4.7 μM | [116] |
| Irbesartan |
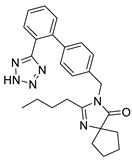
|
Angiotensin II receptor antagonist | 3.3 μM | [114] |
| Zafirlukast |
|
Leukotriene receptor antagonist | 6.5 μM | [117] |
| TRIAC |
|
Thyroid hormone analogue | 6.9 μM | [117] |
| B7 |
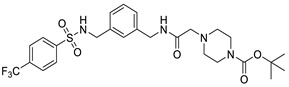
|
Novel synthetic compound | 7.36 μM | [124] |
| Vanitaracin A |
|
Isolated from Talaromyces sp. | 0.61 μM | [121] |
1 Inhibitor concentration required for 50% inhibition; 2 EC50 value (half-maximal effective concentration).
Monoclonal antibodies are also efficient HBV entry inhibitors [125,126,127,128]. Studies have proven that monoclonal antibodies not only inhibit viral entry but also block the secretion of new infectious virions from hepatocytes [129]. The combination of two monoclonal antibodies, HBV-Ab17 and HBV-Ab19, has been evaluated in phase I clinical trials, demonstrating great safety and efficacy against HBV infection [130].
4.2. Directly Targeting cccDNA
The ability to eliminate or inactivate HBV cccDNA has been considered as the “holy grail” of HBV treatments [93] because achieving a “functional cure” for chronic HBV infections requires the permanent inactivation or degradation of the cccDNA [131]. Steps toward disrupting cccDNA have been enabled by understanding the roles of host enzymes such as TDP2, FEN1, and Pol-K in cccDNA formation. Similarly, the identification of host cell nuclear histones and chromatin-modifying enzymes that are essential for viral minichromosome formation and function has also enabled novel drug discovery avenues. Thus, targeting the cccDNA formation has led to many novel candidates in the pipeline of HBV chemotherapy [132,133]. As referred in Section 2.3, APOBEC3 proteins can trigger cccDNA degradation. Several studies have proven that IFN-α administration induces APOBEC3 expression, resulting in the elimination of cccDNA in infected hepatocytes [134].
A potentially useful tool for the complete inactivation of cccDNA is the CRISPR-Cas9 endonuclease system to perform RNA-guided disruption and mutagenesis of cccDNA. The CRISPR-Cas9 endonuclease is complexed with a synthetic guide RNA (gRNA) that perfectly matches with the target sequence of cccDNA, resulting in the cleavage of the selected region. Therefore, to inactivate cccDNA, several gRNAs, targeting multiple different sites in the HBV genome, are required [133,135,136]. Integrated HBV DNA is also sensitive in CRISPR-Cas9-mediated inactivation, which could cause alterations in the host genome and subsequent gene malfunction [132]. Before using this genome editing approach in clinical practice, a number of serious potential issues need to be addressed. These include the incomplete cccDNA degradation, the need of a delivery system that will transfer the CRISPR-Cas9 system to all infected hepatic and extrahepatic cells, potential off-target effects, an immune response induced against the bacterial enzyme that could provoke serious toxicity, and the unpredictable effects of editing the integrated chromosomal HBV DNA [133,137].
Zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) are other methods that can destroy HBV cccDNA [133,138]. This treatment could slow down the growth of resistant HBV strains and increase the probability of a prolonged viral response [139,140,141], yet off-target activity, limited efficacy, effects on integrated HBV DNAs, and the potential induction of immune responses remain serious obstacles [140]. TALENs are newly developed nucleases that cleave selected DNA sequences, thus leading to gene disruptions. Several types of TALENs have been developed that target conserved regions of viral DNA among different HBV genotypes. Overall, TALENs can target and inactivate the HBV genome with a higher specificity than ZFNs and ameliorate the antiviral activity in synergy with IFN-α. Thus, a potential therapeutic strategy for the treatment of chronic hepatitis B infection is provided [140,141,142].
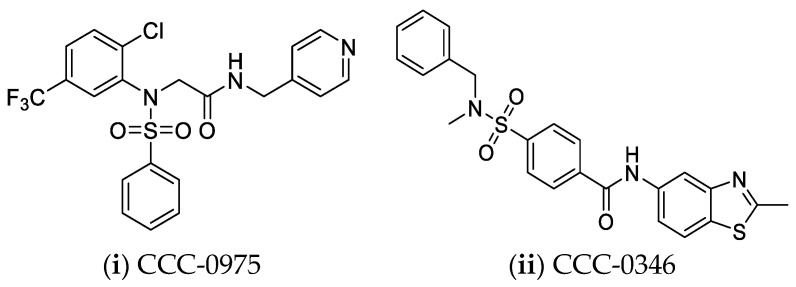
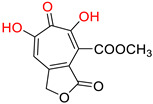
Another method that could contribute to the transcriptional control of cccDNA is direct targeting of the HBV X protein. HBx induces the proteasomal degradation of the Smc5/6 complex, that normally suppresses cccDNA transcription. Consequently, HBx protein inhibition will prevent the expression of all HBV transcripts from existing cccDNA molecules and suppress the formation of new cccDNA molecules [132,143]. cccDNA formation can also be directly targeted with substituted sulfonamides that interfere with the conversion of rcDNA to cccDNA (Figure 5) [144,145].
Figure 5.
Disubstituted Sulfonamides (DSS) [144].
4.3. Immune Therapy
4.3.1. Targeting Innate Immunity
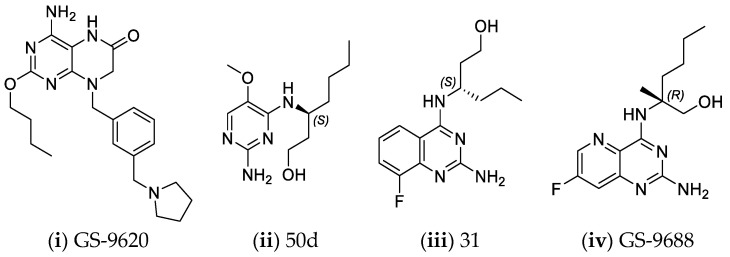
The innate immune responses constitute the first line of defense against pathogens. These systems include membrane and cytoplasmic pattern recognition receptors (PRRs). PRRs interact with specific components, essential for pathogens’ survival, called pathogen-associated molecular patterns (PAMPs), and trigger the production of pro-inflammatory factors, like cytokines from immune cells [146,147,148,149]. Thus, TLR, and RIG-I agonists can stimulate the immune response against HBV infection and contribute significantly to its “functional cure”. Several TLR7, TLR8, and TLR9 agonists are being evaluated in clinical trials [150,151,152,153]. Phase I clinical trials for TLR7 agonists RO7020531, RG7795 (ANA773), and RG7854 (Roche©) are currently underway. TLR7 agonist JNJ-64794964 (Janssen©) demonstrated an excellent safety and tolerability profile in healthy adults during a double-blinded, randomized phase I trial [154]. Phase II clinical trial results for TLR7 agonist GS-9620 (also known as vesatolimod) revealed that it is safe and well-tolerated in chronic hepatitis B patients receiving NAs, although no significant HBsAg loss was observed after 24 weeks of treatment [155]. The same compound also caused no significant HBsAg decline in combination with tenofovir in treatment-naïve patients [156]. Pyrimidine analogues were recently identified as potent dual TLR7/8 modulators (Figure 6ii) [157]. Structural modifications led to novel 2,4-diaminoquinazoline dual TLR7/8 agonists with increased potency and proved that changing the stereochemistry in one single stereocenter leads to TLR8 selectivity (Figure 6iii) [158]. TLR8 agonist GS-9688 (also known as selgantolimod) is under phase II clinical trial evaluation [150,159]. Finally, RIG-I agonist SB-9200 (also known as Inarigrivir) showed promising results in a woodchuck model of HBV infection [160,161], and phase II clinical trials demonstrated the increased benefit of combining classic antiviral treatment with immune therapy [162].
Figure 6.
Innate immunity modulators; (i) Selective TLR7 agonist [163] (ii) TLR7/8 dual agonist [157] (iii) Dual TLR7/8 agonist. (R) isomer results in selective TLR8 agonist [158] (iv) Selective TLR8 agonist [159].
4.3.2. Targeting Adaptive Immunity
The PD-1 (programmed death-1) receptor is expressed on HBV-specific T cells, and compounds that block the interactions with its physiological ligand, PD-L1, can increase the number and response of HBV-specific T cells, resulting in increased cytotoxic T cell activity against HBV-infected cells’ anti-HBV-antigen production by B cells [164,165,166]. Ex vivo studies have shown that blocking PD-1/PD-L1 interactions in chronically infected patients can partially restore the HBV-specific T and B cells’ function [167,168,169]. PD-1 antagonists have been associated with a high risk of hepatic failure [170]. On the other hand, the anti-PD-1:PD-L1 monoclonal antibody nivolumab has already been evaluated in phase I and II clinical trials in over 100 patients with advanced HCC and no hepatotoxicity incidents were observed [171,172].
4.4. RNA Interference—Post-Transcriptional Control
The inhibition of HBV replication by targeting mRNA production and stability is an innovative method for the therapy of chronic hepatitis B, whilst several inhibitors have made it into phase II clinical trials [7,28,93,173]. Inhibitors should bind to HBV mRNA with high specificity and therefore disrupt HBV protein expression by suppressing mRNA translation or inducing mRNA degradation. Such compounds are either small RNA interference (RNAi) molecules, antisense oligonucleotides (ASOs), or possibly even specific ribonucleic acid enzymes (riboenzymes) [133]. RNA interference is mediated by a sequence of 20–30 nucleotides, known as small interfering RNAs (siRNAs) [174]. An advantage stemming from HBV’s transcriptional profile is that multiple mRNA copies of HBV can be targeted selectively at the same time by selecting siRNAs that bind within the overlapping coding regions [133]. To date, three types of siRNAs with different modes of administration are under preclinical evaluation and/or in early-phase clinical trials [7].
An early RNAi drug against HBV, tested in human clinical trials, is ARC-520. The injection consists of two cholesterol-conjugated siRNAs, along with N-acetylgalactosamine (NAG) to achieve hepatocyte-specific delivery via the asialoglycoprotein receptor [39,93]. Potential use limitations are the intravenous administration, the contingent hepatotoxicity, and the off-target binding, as well as the risk of immune activation by PRRs [7]. Despite the barriers mentioned, ARC-520 seems to be very efficient in reducing HBV DNA, HBeAg, and HBsAg levels after experiments on chimpanzees [39,93]. Having passed through phase I with only few hypersensitivity reactions, it proceeded to phase II trials [28]. The co-administration of antihistamines is also recommended [28]. The next step in siRNA evolution came with JNJ-3989 (formerly ARO-HBV, phase II clinical trials), designed to target different HBV genome sites. It is administered subcutaneously, affecting transcripts from both cccDNA and integrated HBV DNA, is safe, and has not shown serious drug-drug interactions [39,93,175].
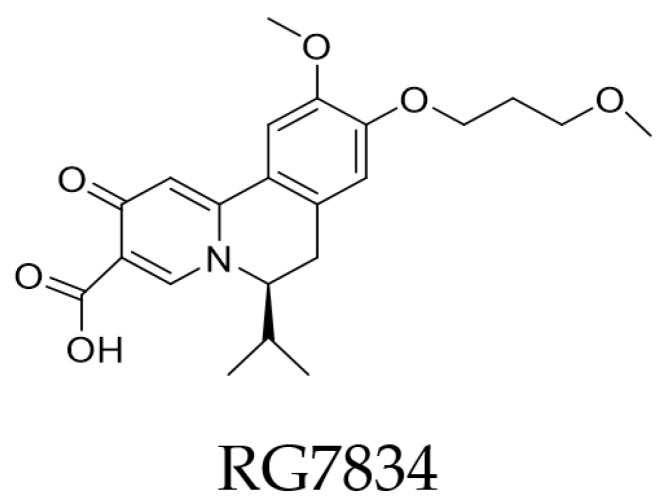
The HBV inhibitor RG7834 (Figure 7), has been studied for its complicity in hepatitis B and has been confirmed not to act as an RNAi molecule, but as an HBV transcription suppressor, in a specific, unknown manner [28,176]. Furthermore, the combination of RG7834, entecavir, and PEG-IFN-α significantly decreases HBV DNA and HBsAg levels [133]. AB-729 is also an siRNA molecule which is administered subcutaneously conjugated with NAG. This compound showed an important suppression of HBsAg in mice models infected by HBV [93].
Figure 7.
HBV transcription inhibitor [176].
Antisense oligonucleotides are short, single-stranded fragments of nucleic acids, either DNA or RNA, that bind to the complementary sequence of viral mRNAs through base pairing. As a result, when binding to RNA, they form hybrids of DNA:RNA (antisense DNA) and duplexes of RNA:RNA (antisense RNA), respectively. The subsequent degradation of the transcribed RNA and the silencing of protein expression occur via a host RNase H-dependent mechanism [7,177,178].
4.5. Ribonuclease H Inhibitors
Ribonucleases H are endonuclease enzymes that catalyze cleavage of RNA sequences in DNA:RNA hybrids [179,180]. The HBV RNaseH degrades the viral pgRNA during minus-polarity DNA strand synthesis by reverse transcriptase within immature nucleocapsids [181]. Inhibiting HBV RNaseH activity results in the accumulation of long DNA:RNA hybrids and halts the reverse transcription process [62]. Consequently, newly synthesized virions are non-infectious since they contain a defective genome [182]. Thus, compounds that inhibit RNaseH are promising antiviral candidates against HBV infection.
The RNaseH catalytic site includes a ‘DEDD’ (aspartic acid-glutamic acid-aspartic acid-aspartic acid) motif that coordinates two Mg2+ ions. Both Mg2+ are essential during the RNA hydrolysis process [183,184]. All known HBV RNaseH inhibitors contain three electron donors (O or N) that chelate these two cations [185]. RNaseH inhibitors primarily belong to two chemical classes: α-hydroxytropolones (α-HTs) and N-hydroxyimides. The latter include N-hydroxyisoquinolinediones (HIDs), N-hydroxynapthyrydinones (HNOs), N-hydroxypyridinediones (HPDs), and N-hydroxypyrimidinediones [185,186,187,188].
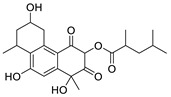
One of the first identified HBV RNaseH inhibitors was β-thujaplicinol (compound 46, Table 2), an α-HT isolated from the heartwood of western red cedar. β-thujaplicinol blocks the RNaseH of HBV genotypes D and H with EC50 values of 5.9 and 2.3 μΜ, respectively [189]. This finding led to the design and synthesis of several novel hydroxylated tropolone analogues that suppress HBV replication in EC50 values as low as 0.34 μM (compound 110) [185,190,191], and with CC50 values up to 100 μM. Therapeutic index values were up to 200 [191]. Compound 110 has also been found to inhibit the HBV RNaseH activity, in a molecular beacon assay, and to suppress viremia in animal models [192]. Further structure-activity relationship studies on the hydroxylated tropolone ring revealed that the α-OH substitution is essential for the HBV RNaseH inhibition. Bulky substitution in positions R1, R2 and R3 leads to a decreased inhibitory activity, indicating that sulfonyl- or lactone substituents can increase the efficacy [190,191,193]. These findings have been validated by recent studies, which also highlighted the increased efficacy of amide-substituted α-HTs [194,195,196].
Table 2.
HBV Ribonuclease inhibitors. The highlighted atoms chelate the two Mg2+ ions in the enzyme’s catalytic site. HID; N-hydroxyisoquinolinedione, HNO; N-hydroxynaphthyridinone, HPD; N-hydroxypyridinedione, EC50; half-maximal effective concentration, CC50; 50% cytotoxic concentration, TI; therapeutic index (TI = CC50/EC50).
| α-hydroxytropolones | |||||
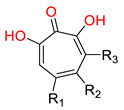 α-hydroxytropolones |
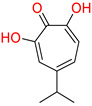 46 [190] EC50 = 1.0 μM CC50 = 25 μM TI = 25 |
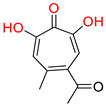 110 [190] EC50 = 0.34 μM CC50 = 32 μM TI = 94 |
 280 [191] EC50 = 0.5 μM CC50 = >77 M TI = >154 |
||
| N-hydroxyimides | |||||
 86 [188] HID EC50 = 1.4 μM CC50 = 99 μM TI = 71 |
 12 [185] HNO EC50 = 3.4 μM CC50 = 7.1 μM TI = 2 |
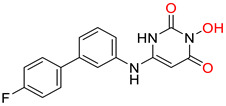 17 [185] N-hydroxypyrimidinedione EC50 = 5.5 μM CC50 = >100 μM TI = >18 |
|||
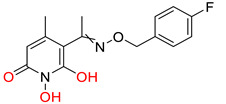 A23 [187] HPD EC50 = 0.11 μM CC50 = 33 μM TI = 300 |
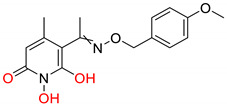 A24 [187] HPD EC50 = 0.29 μM CC50 = 102 μM TI = 352 |
||||
All four mentioned classes of N-hydroxyimides contain N or O atoms in suitable positions in order to chelate the two Mg2+ ions and inhibit RNaseH in the same way as the α-HTs [185]. Several analogues have been synthesized and assessed pharmacologically for their HBV RNaseH inhibitory activity, exhibiting low EC50 values (as low as 110 nM), limited cytotoxicity (most CC50 values between 25 and 100 μΜ), and TI values >300 [187,188,197,198]. One compound from this class has also been evaluated in vivo, and the results verified N-hydroxyimides as being effective HBV RNaseH inhibitors [192]. Finally, RNaseH inhibition is unlikely to be affected by HBV’s large genetic diversity, and RNaseH inhibitors have demonstrated great synergistic activity with antiviral compounds with a different mechanism of action, indicating that they can potentially be used in effective, combination therapeutic schemes against HBV infection [199,200]. The structures of several HBV RNaseH inhibitors identified up to date are shown in Table 2.
4.6. Nucleocapsid Assembly Inhibitors or Modulators
The HBV core particle is actively involved in the HBV replication cycle. It is required for the transfer of the viral genome to and from the nucleus of the infected hepatocyte, as well as for a successful reverse transcription [39]. Thus, it is a promising target for antiviral drugs [11]. New regulators or inhibitors of nucleocapsid assembly can affect various stages of the HBV replication cycle, including capsid formation, reverse transcription, and pgRNA encapsidation [28]. Based on the three-dimensional structure of capsids when they interact with a ligand, two categories of analogues have been developed [28].
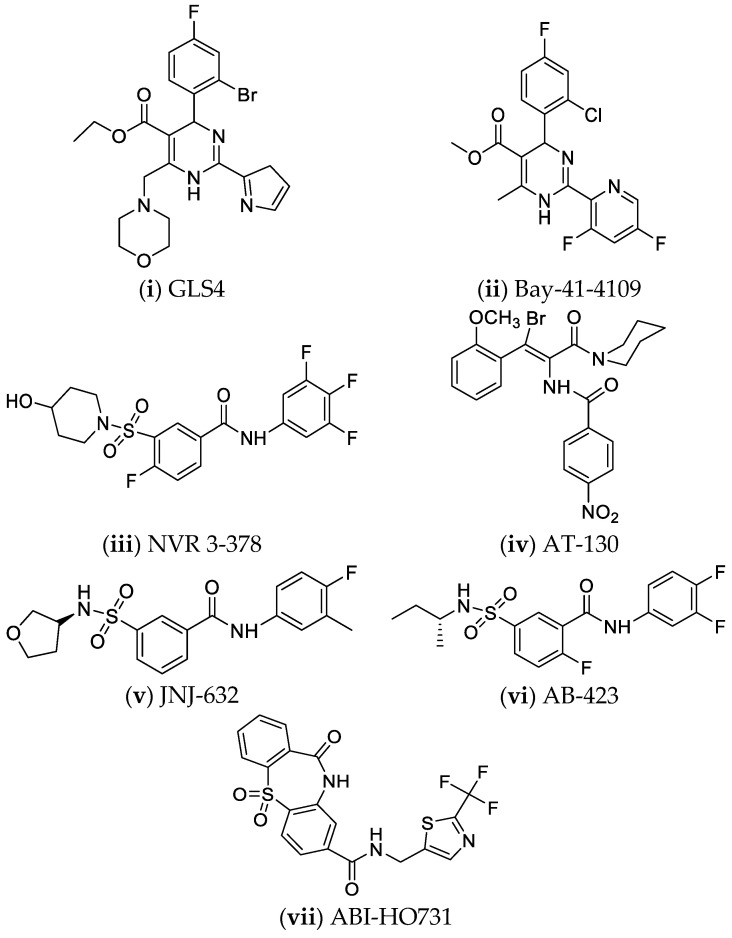
The first category is the Class I core protein allosteric modulators (CpAMs), represented by heteroaryldihydropyrimidines (HAPs) such as GLS4, RO7049389, and Bay41-4109 [28,201]. Class I CpAMs induce the formation of deformed nucleocapsids [11,39,133,145]. The other category is that of Class II CpAMs, such as phenylpropenamides (PP) or sulfamoylbenzamides (SBA), its main representatives being: AT-130, NVR-3778, JNJ6379, JNJ0440, JNJ-632, JNJ56136379, AB-423. These compounds accelerate the assembly of morphologically normal HBV capsids that lack the viral genome [28,133,201,202]. NVR 3-778 is the first SBA derivative developed in the USA, administered per os, and is under phase Ia clinical trial (NCT02112799, NCT02401737), exhibiting synergistic effects in combination with PEG-IFN-α, after evaluation in HBV-infected mice with a humanized liver [7,28,39,175]. Recent in vitro studies in primary human hepatocytes have shown that JNJ-632 (SBA) and Bay41-4109 (HAP) inhibit cccDNA formation and decrease both intracellular HBV RNA and HBeAg and HBsAg levels [28]. Further in vitro studies have proved that phenylpropenamide derivatives demonstrate improved antiviral activity when combined with NAs [145]. JNJ-6379 binds to the HBV core protein and disrupts the encapsidation of pgRNA. It also blocks the cccDNA formation. This drug seems to have a very long half-life, of approximately 120–140 h [175,203,204]. ABI-H0731 marked the beginning of a new category of compounds. It is a dibenzo-thiazepine-2-carboxamide derivative, and it has been shown to cause a significant reduction in HBV DNA and RNA levels in phase I clinical trials, as a core protein modulator [39,205].
Both classes of CpAMs inhibit the release of viral particles. Thus, the amount of HBV DNA and RNA leaving the hepatocyte is reduced. They also prevent de novo cccDNA formation due to blocking the formation of functional capsids, and hence viral replication [133]. The structures of the abovementioned compounds are shown in Figure 8.
Figure 8.
Nucleocapsid assembly modulators or inhibitors [201,205,206,207,208].
5. Perspectives
Hepatitis B vaccines are very effective in preventing infection, and antiviral drugs are partially effective in reducing disease progression and death from the infection. However, access to both the vaccine and the drugs remains a challenge for a large percentage of the world’s population because the majority of chronically infected patients live in developing countries, most often in sub-Saharan Africa or southeast Asia [209], with varying degrees of access to medical care. Therefore, developing an affordable and readily deliverable cure for chronic hepatitis B is urgent. It is widely believed that achieving a broadly applicable “functional cure” for chronic HBV infection will require a combination therapy using agents that target multiple different viral targets plus immune modulators that harness the power of the patients’ defenses against the virus [132,210]. Drugs to improve control and eliminate HBV will have to tackle the unique features of this infection, particularly the durability of the cccDNA during current therapies, which is the reason why the existing drugs so rarely induce a “functional cure”. Fortunately, there is a very wide range of drugs in preclinical and clinical development, so chances are high that combinations of these strategies may be found that substantially improve treatment for HBV patients.
Author Contributions
G.-M.P. writing—original draft, D.M. writing—original draft, E.G. writing—review & editing and preparing the original manuscript figures, V.P. writing—review & editing, J.E.T. review & editing G.Z. conceptualization, methodology, review & editing, supervision, funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
The APC was funded by the Special Account for Research Grants and the National and Kapodistrian University of Athens.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lin C.-L., Kao J.-H. Natural History of Acute and Chronic Hepatitis B: The Role of HBV Genotypes and Mutants. Best Pract. Res. Clin. Gastroenterol. 2017;31:249–255. doi: 10.1016/j.bpg.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Trépo C., Chan H.L.Y., Lok A. Hepatitis B Virus Infection. Lancet. 2014;384:2053–2063. doi: 10.1016/S0140-6736(14)60220-8. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization Hepatitis B-Key Facts. [(accessed on 14 March 2021)]; Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b.
- 4.Yuen M.-F., Chen D.-S., Dusheiko G.M., Janssen H.L.A., Lau D.T.Y., Locarnini S.A., Peters M.G., Lai C.-L. Hepatitis B Virus Infection. Nat. Rev. Dis. Primers. 2018;4:18035. doi: 10.1038/nrdp.2018.35. [DOI] [PubMed] [Google Scholar]
- 5.Suk-Fong Lok A. Hepatitis B: 50 Years after the Discovery of Australia Antigen. J. Viral Hepat. 2016;23:5–14. doi: 10.1111/jvh.12444. [DOI] [PubMed] [Google Scholar]
- 6.Trepo C. A Brief History of Hepatitis Milestones. Liver Int. 2014;34:29–37. doi: 10.1111/liv.12409. [DOI] [PubMed] [Google Scholar]
- 7.Lok A.S., Zoulim F., Dusheiko G., Ghany M.G. Hepatitis B Cure: From Discovery to Regulatory Approval. J. Hepatol. 2017;67:847–861. doi: 10.1016/j.jhep.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Hadziyannis E., Laras A. Viral Biomarkers in Chronic HBeAg Negative HBV Infection. Genes. 2018;9:469. doi: 10.3390/genes9100469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue T., Tanaka Y. Novel Biomarkers for the Management of Chronic Hepatitis B. Clin. Mol. Hepatol. 2020;26:261–279. doi: 10.3350/cmh.2020.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baudi I., Inoue T., Tanaka Y. Novel Biomarkers of Hepatitis B and Hepatocellular Carcinoma: Clinical Significance of HBcrAg and M2BPGi. Int. J. Mol. Sci. 2020;21:949. doi: 10.3390/ijms21030949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ko C., Michler T., Protzer U. Novel Viral and Host Targets to Cure Hepatitis B. Curr. Opin. Virol. 2017;24:38–45. doi: 10.1016/j.coviro.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 12.Lampertico P. EASL 2017 Clinical Practice Guidelines on the Management of Hepatitis B Virus Infection. J. Hepatol. 2017;67:370–398. doi: 10.1016/j.jhep.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 13.Terrault N.A., Lok A.S.F., McMahon B.J., Chang K.-M., Hwang J.P., Jonas M.M., Brown R.S., Bzowej N.H., Wong J.B. Update on Prevention, Diagnosis, and Treatment of Chronic Hepatitis B: AASLD 2018 Hepatitis B Guidance. Hepatology. 2018;67:1560–1599. doi: 10.1002/hep.29800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarin S.K., Kumar M., Lau G.K., Abbas Z., Chan H.L.Y., Chen C.J., Chen D.S., Chen H.L., Chen P.J., Chien R.N., et al. Asian-Pacific Clinical Practice Guidelines on the Management of Hepatitis B: A 2015 Update. Hepatol. Int. 2016;10:1–98. doi: 10.1007/s12072-015-9675-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karayiannis P. Hepatitis B Virus: Virology, Molecular Biology, Life Cycle and Intrahepatic Spread. Hepatol. Int. 2017;11:500–508. doi: 10.1007/s12072-017-9829-7. [DOI] [PubMed] [Google Scholar]
- 16.Dryden K.A., Wieland S.F., Whitten-Bauer C., Gerin J.L., Chisari F.V., Yeager M. Native Hepatitis B Virions and Capsids Visualized by Electron Cryomicroscopy. Mol. Cell. 2006;22:843–850. doi: 10.1016/j.molcel.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 17.Tsukuda S., Watashi K. Hepatitis B Virus Biology and Life Cycle. Antivir. Res. 2020;182:104925. doi: 10.1016/j.antiviral.2020.104925. [DOI] [PubMed] [Google Scholar]
- 18.Kuipery A., Gehring A.J., Isogawa M. Mechanisms of HBV Immune Evasion. Antivir. Res. 2020;179:104816. doi: 10.1016/j.antiviral.2020.104816. [DOI] [PubMed] [Google Scholar]
- 19.Tang H., editor. Hepatitis B Virus Infection: Molecular Virology to Antiviral Drugs. Volume 1179. Springer; Singapore: 2020. Advances in Experimental Medicine and Biology. [Google Scholar]
- 20.Glebe D., Bremer C. The Molecular Virology of Hepatitis B Virus. Semin. Liver Dis. 2013;33:103–112. doi: 10.1055/s-0033-1345717. [DOI] [PubMed] [Google Scholar]
- 21.Lamontagne R.J., Bagga S., Bouchard M.J. Hepatitis B Virus Molecular Biology and Pathogenesis. Hepatoma Res. 2016;2:163. doi: 10.20517/2394-5079.2016.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNaughton A.L., D’Arienzo V., Ansari M.A., Lumley S.F., Littlejohn M., Revill P., McKeating J.A., Matthews P.C. Insights From Deep Sequencing of the HBV Genome—Unique, Tiny, and Misunderstood. Gastroenterology. 2019;156:384–399. doi: 10.1053/j.gastro.2018.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Locarnini S., Zoulim F. Molecular Genetics of HBV Infection. Antivir. Ther. 2010;15:3–14. doi: 10.3851/IMP1619. [DOI] [PubMed] [Google Scholar]
- 24.Pavesi A. Different Patterns of Codon Usage in the Overlapping Polymerase and Surface Genes of Hepatitis B Virus Suggest a de Novo Origin by Modular Evolution. J. Gen. Virol. 2015;96:3577–3586. doi: 10.1099/jgv.0.000307. [DOI] [PubMed] [Google Scholar]
- 25.Yan H., Zhong G., Xu G., He W., Jing Z., Gao Z., Huang Y., Qi Y., Peng B., Wang H., et al. Sodium Taurocholate Cotransporting Polypeptide Is a Functional Receptor for Human Hepatitis B and D Virus. eLife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwamoto M., Saso W., Nishioka K., Ohashi H., Sugiyama R., Ryo A., Ohki M., Yun J.-H., Park S.-Y., Ohshima T., et al. The Machinery for Endocytosis of Epidermal Growth Factor Receptor Coordinates the Transport of Incoming Hepatitis B Virus to the Endosomal Network. J. Biol. Chem. 2020;295:800–807. doi: 10.1016/S0021-9258(17)49936-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herrscher C., Pastor F., Burlaud-Gaillard J., Dumans A., Seigneuret F., Moreau A., Patient R., Eymieux S., Rocquigny H., Hourioux C., et al. Hepatitis B Virus Entry into HepG2-NTCP Cells Requires Clathrin-mediated Endocytosis. Cell. Microbiol. 2020;22 doi: 10.1111/cmi.13205. [DOI] [PubMed] [Google Scholar]
- 28.Schinazi R.F., Ehteshami M., Bassit L., Asselah T. Towards HBV Curative Therapies. Liver Int. 2018;38:102–114. doi: 10.1111/liv.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui X., McAllister R., Boregowda R., Sohn J.A., Ledesma F.C., Caldecott K.W., Seeger C., Hu J. Does Tyrosyl DNA Phosphodiesterase-2 Play a Role in Hepatitis B Virus Genome Repair? PLoS ONE. 2015;10:e0128401. doi: 10.1371/journal.pone.0128401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Königer C., Wingert I., Marsmann M., Rösler C., Beck J., Nassal M. Involvement of the Host DNA-Repair Enzyme TDP2 in Formation of the Covalently Closed Circular DNA Persistence Reservoir of Hepatitis B Viruses. Proc. Natl. Acad. Sci. USA. 2014;111:E4244–E4253. doi: 10.1073/pnas.1409986111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei L., Ploss A. Hepatitis B Virus CccDNA Is Formed through Distinct Repair Processes of Each Strand. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-21850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lau K.C.K., Joshi S.S., Mahoney D.J., Mason A.L., van Marle G., Osiowy C., Coffin C.S. Differences in HBV Replication, APOBEC3 Family Expression, and Inflammatory Cytokine Levels Between Wild-Type HBV and Pre-Core (G1896A) or Basal Core Promoter (A1762T/G1764A) Mutants. Front. Microbiol. 2020;11:1653. doi: 10.3389/fmicb.2020.01653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo X., Huang Y., Chen Y., Tu Z., Hu J., Tavis J.E., Huang A., Hu Y. Association of Hepatitis B Virus Covalently Closed Circular DNA and Human APOBEC3B in Hepatitis B Virus-Related Hepatocellular Carcinoma. PLoS ONE. 2016;11:e0157708. doi: 10.1371/journal.pone.0157708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Werle–Lapostolle B., Bowden S., Locarnini S., Wursthorn K., Petersen J., Lau G., Trepo C., Marcellin P., Goodman Z., Delaney IV W.E. Persistence of CccDNA during the Natural History of Chronic Hepatitis B and Decline during Adefovir Dipivoxil Therapy1. Gastroenterology. 2004;126:1750–1758. doi: 10.1053/j.gastro.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 35.Xia Y., Guo H. Hepatitis B Virus CccDNA: Formation, Regulation and Therapeutic Potential. Antivir. Res. 2020;180:104824. doi: 10.1016/j.antiviral.2020.104824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Q., Zhou B., Cai D., Zong Y., Wu Y., Liu S., Mercier A., Guo H., Hou J., Colonno R., et al. Rapid Turnover of Hepatitis B Virus Covalently Closed Circular DNA Indicated by Monitoring Emergence and Reversion of Signature-Mutation in Treated Chronic Hepatitis B Patients. Hepatology. 2021;73:41–52. doi: 10.1002/hep.31240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seeger C., Mason W.S. Molecular Biology of Hepatitis B Virus Infection. Virology. 2015;479–480:672–686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kao J.-H., Chen D.-S., editors. Hepatitis B Virus and Liver Disease. Springer; Singapore: 2018. [Google Scholar]
- 39.Tang L., Kottilil S., Wilson E. Strategies to Eliminate HBV Infection: An Update. Future Virol. 2020;15:35–51. doi: 10.2217/fvl-2019-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moolla N., Kew M., Arbuthnot P. Regulatory Elements of Hepatitis B Virus Transcription. J. Viral Hepat. 2002;9:323–331. doi: 10.1046/j.1365-2893.2002.00381.x. [DOI] [PubMed] [Google Scholar]
- 41.Ren J.-H., Tao Y., Zhang Z.-Z., Chen W.-X., Cai X.-F., Chen K., Ko B.C.B., Song C.-L., Ran L.-K., Li W.-Y., et al. Sirtuin 1 Regulates Hepatitis B Virus Transcription and Replication by Targeting Transcription Factor AP-1. J. Virol. 2014;88:2442–2451. doi: 10.1128/JVI.02861-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’Arienzo V., Ferguson J., Giraud G., Chapus F., Harris J.M., Wing P.A.C., Claydon A., Begum S., Zhuang X., Balfe P., et al. The CCCTC-binding Factor CTCF Represses Hepatitis B Virus Enhancer I and Regulates Viral Transcription. Cell. Microbiol. 2021;23 doi: 10.1111/cmi.13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quasdorff M., Protzer U. Control of Hepatitis B Virus at the Level of Transcription: Control of Hepatitis B Virus. J. Viral Hepat. 2010;17:527–536. doi: 10.1111/j.1365-2893.2010.01315.x. [DOI] [PubMed] [Google Scholar]
- 44.Decorsière A., Mueller H., van Breugel P.C., Abdul F., Gerossier L., Beran R.K., Livingston C.M., Niu C., Fletcher S.P., Hantz O., et al. Hepatitis B Virus X Protein Identifies the Smc5/6 Complex as a Host Restriction Factor. Nature. 2016;531:386–389. doi: 10.1038/nature17170. [DOI] [PubMed] [Google Scholar]
- 45.Mitra B., Guo H. Hepatitis B Virus X Protein Crosses out Smc5/6 Complex to Maintain Covalently Closed Circular DNA Transcription. Hepatology. 2016;64:2246–2249. doi: 10.1002/hep.28834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy C.M., Xu Y., Li F., Nio K., Reszka-Blanco N., Li X., Wu Y., Yu Y., Xiong Y., Su L. Hepatitis B Virus X Protein Promotes Degradation of SMC5/6 to Enhance HBV Replication. Cell Rep. 2016;16:2846–2854. doi: 10.1016/j.celrep.2016.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Livingston C., Ramakrishnan D., Strubin M., Fletcher S., Beran R. Identifying and Characterizing Interplay between Hepatitis B Virus X Protein and Smc5/6. Viruses. 2017;9:69. doi: 10.3390/v9040069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lucifora J., Arzberger S., Durantel D., Belloni L., Strubin M., Levrero M., Zoulim F., Hantz O., Protzer U. Hepatitis B Virus X Protein Is Essential to Initiate and Maintain Virus Replication after Infection. J. Hepatol. 2011;55:996–1003. doi: 10.1016/j.jhep.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 49.Feitelson M.A., Bonamassa B., Arzumanyan A. The Roles of Hepatitis B Virus-Encoded X Protein in Virus Replication and the Pathogenesis of Chronic Liver Disease. Expert Opin. Ther. Targets. 2014;18:293–306. doi: 10.1517/14728222.2014.867947. [DOI] [PubMed] [Google Scholar]
- 50.Slagle B.L., Bouchard M.J. Hepatitis B Virus X and Regulation of Viral Gene Expression. Cold Spring Harb. Perspect. Med. 2016;6:a021402. doi: 10.1101/cshperspect.a021402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clark D.N., Hu J. Unveiling the Roles of HBV Polymerase for New Antiviral Strategies. Future Virol. 2015;10:283–295. doi: 10.2217/fvl.14.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clark D.N., Hu J. Hepatitis B Virus Reverse Transcriptase—Target of Current Antiviral Therapy and Future Drug Development. Antivir. Res. 2015;123:132–137. doi: 10.1016/j.antiviral.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu J. Hepatitis B and D Protocols. Humana Press; Totowa, NJ, USA: 2004. Studying DHBV Polymerase by In Vitro Transcription and Translation; pp. 259–270. [DOI] [PubMed] [Google Scholar]
- 54.Hu J., Boyer M. Hepatitis B Virus Reverse Transcriptase and ε RNA Sequences Required for Specific Interaction In Vitro. J. Virol. 2006;80:2141–2150. doi: 10.1128/JVI.80.5.2141-2150.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones S.A., Clark D.N., Cao F., Tavis J.E., Hu J. Comparative Analysis of Hepatitis B Virus Polymerase Sequences Required for Viral RNA Binding, RNA Packaging, and Protein Priming. J. Virol. 2014;88:1564–1572. doi: 10.1128/JVI.02852-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao F., Jones S., Li W., Cheng X., Hu Y., Hu J., Tavis J.E. Sequences in the Terminal Protein and Reverse Transcriptase Domains of the Hepatitis B Virus Polymerase Contribute to RNA Binding and Encapsidation. J. Viral Hepat. 2014;21:882–893. doi: 10.1111/jvh.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buhlig T.S., Bowersox A.F., Braun D.L., Owsley D.N., James K.D., Aranda A.J., Kendrick C.D., Skalka N.A., Clark D.N. Molecular, Evolutionary, and Structural Analysis of the Terminal Protein Domain of Hepatitis B Virus Polymerase, a Potential Drug Target. Viruses. 2020;12:570. doi: 10.3390/v12050570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones S.A., Hu J. Hepatitis B Virus Reverse Transcriptase: Diverse Functions as Classical and Emerging Targets for Antiviral Intervention. Emerg. Microbes Infect. 2013;2:1–11. doi: 10.1038/emi.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Badtke M.P., Khan I., Cao F., Hu J., Tavis J.E. An Interdomain RNA Binding Site on the Hepadnaviral Polymerase That Is Essential for Reverse Transcription. Virology. 2009;390:130–138. doi: 10.1016/j.virol.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Datta S., Chatterjee S., Veer V., Chakravarty R. Molecular Biology of the Hepatitis B Virus for Clinicians. J. Clin. Exp. Hepatol. 2012;2:353–365. doi: 10.1016/j.jceh.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Villa J.A., Pike D.P., Patel K.B., Lomonosova E., Lu G., Abdulqader R., Tavis J.E. Purification and Enzymatic Characterization of the Hepatitis B Virus Ribonuclease H, a New Target for Antiviral Inhibitors. Antivir. Res. 2016;132:186–195. doi: 10.1016/j.antiviral.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tavis J.E., Lomonosova E. The Hepatitis B Virus Ribonuclease H as a Drug Target. Antivir. Res. 2015;118:132–138. doi: 10.1016/j.antiviral.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu J., Liu K. Complete and Incomplete Hepatitis B Virus Particles: Formation, Function, and Application. Viruses. 2017;9:56. doi: 10.3390/v9030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tu T., Budzinska M.A., Vondran F.W.R., Shackel N.A., Urban S. Hepatitis B Virus DNA Integration Occurs Early in the Viral Life Cycle in an In Vitro Infection Model via Sodium Taurocholate Cotransporting Polypeptide-Dependent Uptake of Enveloped Virus Particles. J. Virol. 2018;92:e02007-17. doi: 10.1128/JVI.02007-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hepatitis B Foundation: Approved Drugs for Adults. [(accessed on 6 March 2021)]; Available online: https://www.hepb.org/treatment-and-management/treatment/approved-drugs-for-adults/
- 66.Woo A.S.J., Kwok R., Ahmed T. Alpha-Interferon Treatment in Hepatitis B. Ann. Transl. Med. 2017;5:159. doi: 10.21037/atm.2017.03.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kwon H., Lok A.S. Hepatitis B Therapy. Nat. Rev. Gastroenterol. Hepatol. 2011;8:275–284. doi: 10.1038/nrgastro.2011.33. [DOI] [PubMed] [Google Scholar]
- 68.Belloni L., Allweiss L., Guerrieri F., Pediconi N., Volz T., Pollicino T., Petersen J., Raimondo G., Dandri M., Levrero M. IFN-α Inhibits HBV Transcription and Replication in Cell Culture and in Humanized Mice by Targeting the Epigenetic Regulation of the Nuclear CccDNA Minichromosome. J. Clin. Investig. 2012;122:529–537. doi: 10.1172/JCI58847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lucifora J., Xia Y., Reisinger F., Zhang K., Stadler D., Cheng X., Sprinzl M.F., Koppensteiner H., Makowska Z., Volz T., et al. Specific and Nonhepatotoxic Degradation of Nuclear Hepatitis B Virus CccDNA. Science. 2014;343:1221–1228. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan Y., Yuan H., Yang G., Yun H., Zhao M., Liu Z., Zhao L., Geng Y., Liu L., Wang J., et al. IFN-α Confers Epigenetic Regulation of HBV CccDNA Minichromosome by Modulating GCN5-Mediated Succinylation of Histone H3K79 to Clear HBV CccDNA. Clin. Epigenetics. 2020;12:135. doi: 10.1186/s13148-020-00928-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Janssen H.L.A., van Zonneveld M., Senturk H., Zeuzem S., Akarca U.S., Cakaloglu Y., Simon C., So T.M.K., Gerken G., de Man R.A., et al. Pegylated Interferon Alfa-2b Alone or in Combination with Lamivudine for HBeAg-Positive Chronic Hepatitis B: A Randomised Trial. Lancet. 2005;365:123–129. doi: 10.1016/S0140-6736(05)17701-0. [DOI] [PubMed] [Google Scholar]
- 72.Zoulim F., Lebossé F., Levrero M. Current Treatments for Chronic Hepatitis B Virus Infections. Curr. Opin. Virol. 2016;18:109–116. doi: 10.1016/j.coviro.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 73.Pierra Rouviere C., Dousson C.B., Tavis J.E. HBV Replication Inhibitors. Antivir. Res. 2020;179:104815. doi: 10.1016/j.antiviral.2020.104815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bayliss J., Yuen L., Rosenberg G., Wong D., Littlejohn M., Jackson K., Gaggar A., Kitrinos K.M., Subramanian G.M., Marcellin P., et al. Deep Sequencing Shows That HBV Basal Core Promoter and Precore Variants Reduce the Likelihood of HBsAg Loss Following Tenofovir Disoproxil Fumarate Therapy in HBeAg-Positive Chronic Hepatitis B. Gut. 2017;66:2013–2023. doi: 10.1136/gutjnl-2015-309300. [DOI] [PubMed] [Google Scholar]
- 75.Abd El Aziz M.A., Sacco R., Facciorusso A. Nucleos(t)Ide Analogues and Hepatitis B Virus-Related Hepatocellular Carcinoma: A Literature Review. Antivir. Chem. Chemother. 2020;28:204020662092133. doi: 10.1177/2040206620921331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Su M.-H. Long-Term Lamivudine for Chronic Hepatitis B and Cirrhosis: A Real-Life Cohort Study. World J. Gastroenterol. 2015;21:13087. doi: 10.3748/wjg.v21.i46.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kwon J.H., Jang J.W., Choi J.Y., Park C.-H., Yoo S.H., Bae S.H., Yoon S.K. Should Lamivudine Monotherapy Be Stopped or Continued in Patients Infected with Hepatitis B with Favorable Responses after More than 5 Years of Treatment? J. Med. Virol. 2013;85:34–42. doi: 10.1002/jmv.23421. [DOI] [PubMed] [Google Scholar]
- 78.Eun J.R., Lee H.J., Kim T.N., Lee K.S. Risk Assessment for the Development of Hepatocellular Carcinoma: According to on-Treatment Viral Response during Long-Term Lamivudine Therapy in Hepatitis B Virus-Related Liver Disease. J. Hepatol. 2010;53:118–125. doi: 10.1016/j.jhep.2010.02.026. [DOI] [PubMed] [Google Scholar]
- 79.Luo A., Jiang X., Ren H. Lamivudine plus Tenofovir Combination Therapy versus Lamivudine Monotherapy for HBV/HIV Coinfection: A Meta-Analysis. Virol. J. 2018;15:139. doi: 10.1186/s12985-018-1050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hou J.-L., Zhao W., Lee C., Hann H.-W., Peng C.-Y., Tanwandee T., Morozov V., Klinker H., Sollano J.D., Streinu-Cercel A., et al. Outcomes of Long-Term Treatment of Chronic HBV Infection With Entecavir or Other Agents From a Randomized Trial in 24 Countries. Clin. Gastroenterol. Hepatol. 2020;18:457–467.e21. doi: 10.1016/j.cgh.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 81.Liaw Y., Gane E., Leung N., Zeuzem S., Wang Y., Lai C.L., Heathcote E.J., Manns M., Bzowej N., Niu J., et al. 2-Year GLOBE Trial Results: Telbivudine Is Superior to Lamivudine in Patients With Chronic Hepatitis B. Gastroenterology. 2009;136:486–495. doi: 10.1053/j.gastro.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 82.Hou J., Yin Y.-K., Xu D., Tan D., Niu J., Zhou X., Wang Y., Zhu L., He Y., Ren H., et al. Telbivudine versus Lamivudine in Chinese Patients with Chronic Hepatitis B: Results at 1 Year of a Randomized, Double-Blind Trial. Hepatology. 2007;47:447–454. doi: 10.1002/hep.22075. [DOI] [PubMed] [Google Scholar]
- 83.Lai C.-L., Hsu C.-W., Chen Y., Bzowej N., Bisceglie A.M.D., Moon Y.M., Chao G., Brown N.A. Telbivudine versus Lamivudine in Patients with Chronic Hepatitis B. N. Engl. J. Med. 2007;357:2576–2588. doi: 10.1056/NEJMoa066422. [DOI] [PubMed] [Google Scholar]
- 84.Tsai M.-C., Chen C.-H., Hung C.-H., Lee C.-M., Chiu K.-W., Wang J.-H., Lu S.-N., Tseng P.-L., Chang K.-C., Yen Y.-H., et al. A Comparison of Efficacy and Safety of 2-Year Telbivudine and Entecavir Treatment in Patients with Chronic Hepatitis B: A Match–Control Study. Clin. Microbiol. Infect. 2014;20:O90–O100. doi: 10.1111/1469-0691.12220. [DOI] [PubMed] [Google Scholar]
- 85.Wong G.L.-H., Seto W.-K., Wong V.W.-S., Yuen M.-F., Chan H.L.-Y. Review Article: Long-Term Safety of Oral Anti-Viral Treatment for Chronic Hepatitis B. Aliment. Pharmacol. Ther. 2018;47:730–737. doi: 10.1111/apt.14497. [DOI] [PubMed] [Google Scholar]
- 86.Wu X., Cai S., Li Z., Zheng C., Xue X., Zeng J., Peng J. Potential Effects of Telbivudine and Entecavir on Renal Function: A Systematic Review and Meta-Analysis. Virol. J. 2016;13:64. doi: 10.1186/s12985-016-0522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Y., Corsa A.C., Buti M., Cathcart A.L., Flaherty J.F., Miller M.D., Kitrinos K.M., Marcellin P., Gane E.J. No Detectable Resistance to Tenofovir Disoproxil Fumarate in HBeAg+ and HBeAg− Patients with Chronic Hepatitis B after 8 Years of Treatment. J. Viral Hepat. 2017;24:68–74. doi: 10.1111/jvh.12613. [DOI] [PubMed] [Google Scholar]
- 88.Zhang Z., Zhou Y., Yang J., Hu K., Huang Y. The Effectiveness of TDF versus ETV on Incidence of HCC in CHB Patients: A Meta Analysis. BMC Cancer. 2019;19:511. doi: 10.1186/s12885-019-5735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Choi W.-M., Choi J., Lim Y.-S. Effects of Tenofovir vs Entecavir on Risk of Hepatocellular Carcinoma in Patients With Chronic HBV Infection: A Systematic Review and Meta-Analysis. Clin. Gastroenterol. Hepatol. 2021;19:246–258.e9. doi: 10.1016/j.cgh.2020.05.008. [DOI] [PubMed] [Google Scholar]
- 90.Li M., Lv T., Wu S., Wei W., Wu X., Ou X., Ma H., Chow S.-C., Kong Y., You H., et al. Tenofovir versus Entecavir in Lowering the Risk of Hepatocellular Carcinoma Development in Patients with Chronic Hepatitis B: A Critical Systematic Review and Meta-Analysis. Hepatol. Int. 2020;14:105–114. doi: 10.1007/s12072-019-10005-0. [DOI] [PubMed] [Google Scholar]
- 91.Oh H., Yoon E.L., Jun D.W., Ahn S.B., Lee H.-Y., Jeong J.Y., Kim H.S., Jeong S.W., Kim S.E., Shim J.-J., et al. No Difference in Incidence of Hepatocellular Carcinoma in Patients With Chronic Hepatitis B Virus Infection Treated With Entecavir vs Tenofovir. Clin. Gastroenterol. Hepatol. 2020;18:2793–2802.e6. doi: 10.1016/j.cgh.2020.02.046. [DOI] [PubMed] [Google Scholar]
- 92.Hsu Y.-C., Wong G.L.-H., Chen C.-H., Peng C.-Y., Yeh M.-L., Cheung K.-S., Toyoda H., Huang C.-F., Trinh H., Xie Q., et al. Tenofovir Versus Entecavir for Hepatocellular Carcinoma Prevention in an International Consortium of Chronic Hepatitis B. Am. J. Gastroenterol. 2020;115:271–280. doi: 10.14309/ajg.0000000000000428. [DOI] [PubMed] [Google Scholar]
- 93.Lopatin U. Drugs in the Pipeline for HBV. Clin. Liver Dis. 2019;23:535–555. doi: 10.1016/j.cld.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 94.Rijckborst V., Sonneveld M.J., Janssen H.L.A. Review Article: Chronic Hepatitis B—Anti-Viral or Immunomodulatory Therapy?: Review: Therapy for Chronic Hepatitis B. Aliment. Pharmacol. Ther. 2011;33:501–513. doi: 10.1111/j.1365-2036.2010.04555.x. [DOI] [PubMed] [Google Scholar]
- 95.Revill P., Testoni B., Locarnini S., Zoulim F. Global Strategies Are Required to Cure and Eliminate HBV Infection. Nat. Rev. Gastroenterol. Hepatol. 2016;13:239–248. doi: 10.1038/nrgastro.2016.7. [DOI] [PubMed] [Google Scholar]
- 96.Fukano K., Tsukuda S., Watashi K., Wakita T. Concept of Viral Inhibitors via NTCP. Semin. Liver Dis. 2019;39:078–085. doi: 10.1055/s-0038-1676804. [DOI] [PubMed] [Google Scholar]
- 97.Dawson P.A., Lan T., Rao A. Bile Acid Transporters. J. Lipid Res. 2009;50:2340–2357. doi: 10.1194/jlr.R900012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gripon P., Cannie I., Urban S. Efficient Inhibition of Hepatitis B Virus Infection by Acylated Peptides Derived from the Large Viral Surface Protein. J. Virol. 2005;79:10. doi: 10.1128/JVI.79.3.1613-1622.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Glebe D., Urban S., Knoop E.V., Çaǧ N., Krass P., Grün S., Bulavaite A., Sasnauskas K., Gerlich W.H. Mapping of the Hepatitis B Virus Attachment Site by Use of Infection-Inhibiting PreS1 Lipopeptides and Tupaia Hepatocytes. Gastroenterology. 2005;129:234–245. doi: 10.1053/j.gastro.2005.03.090. [DOI] [PubMed] [Google Scholar]
- 100.Petersen J., Dandri M., Mier W., Lütgehetmann M., Volz T., von Weizsäcker F., Haberkorn U., Fischer L., Pollok J.-M., Erbes B., et al. Prevention of Hepatitis B Virus Infection in Vivo by Entry Inhibitors Derived from the Large Envelope Protein. Nat. Biotechnol. 2008;26:335–341. doi: 10.1038/nbt1389. [DOI] [PubMed] [Google Scholar]
- 101.Passioura T., Watashi K., Fukano K., Shimura S., Saso W., Morishita R., Ogasawara Y., Tanaka Y., Mizokami M., Sureau C., et al. De Novo Macrocyclic Peptide Inhibitors of Hepatitis B Virus Cellular Entry. Cell Chem. Biol. 2018;25:906–915.e5. doi: 10.1016/j.chembiol.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 102.Ye X., Zhou M., He Y., Wan Y., Bai W., Tao S., Ren Y., Zhang X., Xu J., Liu J., et al. Efficient Inhibition of Hepatitis B Virus Infection by a PreS1-Binding Peptide. Sci. Rep. 2016;6:29391. doi: 10.1038/srep29391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schulze A., Schieck A., Ni Y., Mier W., Urban S. Fine Mapping of Pre-S Sequence Requirements for Hepatitis B Virus Large Envelope Protein-Mediated Receptor Interaction. J. Virol. 2010;84:1989–2000. doi: 10.1128/JVI.01902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Volz T., Allweiss L., MBarek M.B., Warlich M., Lohse A.W., Pollok J.M., Alexandrov A., Urban S., Petersen J., Lütgehetmann M., et al. The Entry Inhibitor Myrcludex-B Efficiently Blocks Intrahepatic Virus Spreading in Humanized Mice Previously Infected with Hepatitis B Virus. J. Hepatol. 2013;58:861–867. doi: 10.1016/j.jhep.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 105.Zhao K., Liu S., Chen Y., Yao Y., Zhou M., Yuan Y., Wang Y., Pei R., Chen J., Hu X., et al. Upregulation of HBV Transcription by Sodium Taurocholate Cotransporting Polypeptide at the Postentry Step Is Inhibited by the Entry Inhibitor Myrcludex B. Emerg. Microbes Infect. 2018;7:1–14. doi: 10.1038/s41426-018-0189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nkongolo S., Ni Y., Lempp F.A., Kaufman C., Lindner T., Esser-Nobis K., Lohmann V., Mier W., Mehrle S., Urban S. Cyclosporin A Inhibits Hepatitis B and Hepatitis D Virus Entry by Cyclophilin-Independent Interference with the NTCP Receptor. J. Hepatol. 2014;60:723–731. doi: 10.1016/j.jhep.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 107.Blank A., Markert C., Hohmann N., Carls A., Mikus G., Lehr T., Alexandrov A., Haag M., Schwab M., Urban S., et al. First-in-Human Application of the Novel Hepatitis B and Hepatitis D Virus Entry Inhibitor Myrcludex B. J. Hepatol. 2016;65:483–489. doi: 10.1016/j.jhep.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 108.Kang C., Syed Y.Y. Bulevirtide: First Approval. Drugs. 2020;80:1601–1605. doi: 10.1007/s40265-020-01400-1. [DOI] [PubMed] [Google Scholar]
- 109.Uhl P., Helm F., Hofhaus G., Brings S., Kaufman C., Leotta K., Urban S., Haberkorn U., Mier W., Fricker G. A Liposomal Formulation for the Oral Application of the Investigational Hepatitis B Drug Myrcludex B. Eur. J. Pharm. Biopharm. 2016;103:159–166. doi: 10.1016/j.ejpb.2016.03.031. [DOI] [PubMed] [Google Scholar]
- 110.Blank A., Meier K., Urban S., Haefeli W.E., Weiss J. Drug–Drug Interaction Potential of the Hepatitis B and Hepatitis D Virus Entry Inhibitor Myrcludex B Assessed in Vitro. Antivir. Ther. 2017;23:267–275. doi: 10.3851/IMP3206. [DOI] [PubMed] [Google Scholar]
- 111.Watashi K., Sluder A., Daito T., Matsunaga S., Ryo A., Nagamori S., Iwamoto M., Nakajima S., Tsukuda S., Borroto-Esoda K., et al. Cyclosporin A and Its Analogs Inhibit Hepatitis B Virus Entry into Cultured Hepatocytes through Targeting a Membrane Transporter, Sodium Taurocholate Cotransporting Polypeptide (NTCP) Hepatology. 2014;59:1726–1737. doi: 10.1002/hep.26982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shimura S., Watashi K., Fukano K., Peel M., Sluder A., Kawai F., Iwamoto M., Tsukuda S., Takeuchi J.S., Miyake T., et al. Cyclosporin Derivatives Inhibit Hepatitis B Virus Entry without Interfering with NTCP Transporter Activity. J. Hepatol. 2017;66:685–692. doi: 10.1016/j.jhep.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lucifora J., Esser K., Protzer U. Ezetimibe Blocks Hepatitis B Virus Infection after Virus Uptake into Hepatocytes. Antivir. Res. 2013;97:195–197. doi: 10.1016/j.antiviral.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 114.Wang X., Hu W., Zhang T., Mao Y., Liu N., Wang S. Irbesartan, an FDA Approved Drug for Hypertension and Diabetic Nephropathy, Is a Potent Inhibitor for Hepatitis B Virus Entry by Disturbing Na+-Dependent Taurocholate Cotransporting Polypeptide Activity. Antivir. Res. 2015;120:140–146. doi: 10.1016/j.antiviral.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 115.Saso W., Tsukuda S., Ohashi H., Fukano K., Morishita R., Matsunaga S., Ohki M., Ryo A., Park S.-Y., Suzuki R., et al. A New Strategy to Identify Hepatitis B Virus Entry Inhibitors by AlphaScreen Technology Targeting the Envelope-Receptor Interaction. Biochem. Biophys. Res. Commun. 2018;501:374–379. doi: 10.1016/j.bbrc.2018.04.187. [DOI] [PubMed] [Google Scholar]
- 116.Fukano K., Tsukuda S., Oshima M., Suzuki R., Aizaki H., Ohki M., Park S.-Y., Muramatsu M., Wakita T., Sureau C., et al. Troglitazone Impedes the Oligomerization of Sodium Taurocholate Cotransporting Polypeptide and Entry of Hepatitis B Virus Into Hepatocytes. Front. Microbiol. 2019;9:3257. doi: 10.3389/fmicb.2018.03257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Donkers J.M., Zehnder B., van Westen G.J.P., Kwakkenbos M.J., IJzerman A.P., Oude Elferink R.P.J., Beuers U., Urban S., van de Graaf S.F.J. Reduced Hepatitis B and D Viral Entry Using Clinically Applied Drugs as Novel Inhibitors of the Bile Acid Transporter NTCP. Sci. Rep. 2017;7:15307. doi: 10.1038/s41598-017-15338-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Iwamoto M., Watashi K., Tsukuda S., Aly H.H., Fukasawa M., Fujimoto A., Suzuki R., Aizaki H., Ito T., Koiwai O., et al. Evaluation and Identification of Hepatitis B Virus Entry Inhibitors Using HepG2 Cells Overexpressing a Membrane Transporter NTCP. Biochem. Biophys. Res. Commun. 2014;443:808–813. doi: 10.1016/j.bbrc.2013.12.052. [DOI] [PubMed] [Google Scholar]
- 119.Huang H.-C., Tao M.-H., Hung T.-M., Chen J.-C., Lin Z.-J., Huang C. (−)-Epigallocatechin-3-Gallate Inhibits Entry of Hepatitis B Virus into Hepatocytes. Antivir. Res. 2014;111:100–111. doi: 10.1016/j.antiviral.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 120.Matsunaga H., Kamisuki S., Kaneko M., Yamaguchi Y., Takeuchi T., Watashi K., Sugawara F. Isolation and Structure of Vanitaracin A, a Novel Anti-Hepatitis B Virus Compound from Talaromyces sp. Bioorg. Med. Chem. Lett. 2015;25:4325–4328. doi: 10.1016/j.bmcl.2015.07.067. [DOI] [PubMed] [Google Scholar]
- 121.Kaneko M., Watashi K., Kamisuki S., Matsunaga H., Iwamoto M., Kawai F., Ohashi H., Tsukuda S., Shimura S., Suzuki R., et al. A Novel Tricyclic Polyketide, Vanitaracin A, Specifically Inhibits the Entry of Hepatitis B and D Viruses by Targeting Sodium Taurocholate Cotransporting Polypeptide. J. Virol. 2015;89:11945–11953. doi: 10.1128/JVI.01855-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tsukuda S., Watashi K., Hojima T., Isogawa M., Iwamoto M., Omagari K., Suzuki R., Aizaki H., Kojima S., Sugiyama M., et al. A New Class of Hepatitis B and D Virus Entry Inhibitors, Proanthocyanidin and Its Analogs, That Directly Act on the Viral Large Surface Proteins. Hepatology. 2017;65:1104–1116. doi: 10.1002/hep.28952. [DOI] [PubMed] [Google Scholar]
- 123.Kirstgen M., Lowjaga K.A.A.T., Müller S.F., Goldmann N., Lehmann F., Alakurtti S., Yli-Kauhaluoma J., Glebe D., Geyer J. Selective Hepatitis B and D Virus Entry Inhibitors from the Group of Pentacyclic Lupane-Type Betulin-Derived Triterpenoids. Sci. Rep. 2020;10:21772. doi: 10.1038/s41598-020-78618-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xiang H., Chen Y., Zhang J., Zhang J., Pan D., Liu B., Ouyang L. Discovery of a Novel Sodium Taurocholate Cotransporting Polypeptide (NTCP) Inhibitor: Design, Synthesis, and Anti-Proliferative Activities. Chin. Chem. Lett. 2020;31:1422–1426. doi: 10.1016/j.cclet.2020.03.017. [DOI] [Google Scholar]
- 125.Wi J., Jeong M.S., Hong H.J. Construction and Characterization of an Anti-Hepatitis B Virus PreS1 Humanized Antibody That Binds to the Essential Receptor Binding Site. J. Microbiol. Biotechnol. 2017;27:1336–1344. doi: 10.4014/jmb.1703.03066. [DOI] [PubMed] [Google Scholar]
- 126.Hong H.J., Ryu C.J., Hur H., Kim S., Oh H.K., Oh M.S., Park S.Y. In Vivo Neutralization of Hepatitis B Virus Infection by an Anti-PreS1 Humanized Antibody in Chimpanzees. Virology. 2004;318:134–141. doi: 10.1016/j.virol.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 127.Zhang T.-Y., Yuan Q., Zhao J.-H., Zhang Y.-L., Yuan L.-Z., Lan Y., Lo Y.-C., Sun C.-P., Wu C.-R., Zhang J.-F., et al. Prolonged Suppression of HBV in Mice by a Novel Antibody That Targets a Unique Epitope on Hepatitis B Surface Antigen. Gut. 2016;65:658–671. doi: 10.1136/gutjnl-2014-308964. [DOI] [PubMed] [Google Scholar]
- 128.Li D., He W., Liu X., Zheng S., Qi Y., Li H., Mao F., Liu J., Sun Y., Pan L., et al. A Potent Human Neutralizing Antibody Fc-Dependently Reduces Established HBV Infections. eLife. 2017;6:e26738. doi: 10.7554/eLife.26738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Neumann A.U., Phillips S., Levine I., Ijaz S., Dahari H., Eren R., Dagan S., Naoumov N.V. Novel Mechanism of Antibodies to Hepatitis B Virus in Blocking Viral Particle Release from Cells. Hepatology. 2010;52:875–885. doi: 10.1002/hep.23778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Galun E., Eren R., Safadi R., Ashour Y., Terrault N., Keeffe E.B., Matot E., Mizrachi S., Terkieltaub D., Zohar M., et al. Clinical Evaluation (Phase I) of a Combination of Two Human Monoclonal Antibodies to HBV: Safety and Antiviral Properties. Hepatology. 2002;35:673–679. doi: 10.1053/jhep.2002.31867. [DOI] [PubMed] [Google Scholar]
- 131.Zhao Q., Guo J. Have the Starting Lineup of Five for Hepatitis B Virus Covalently Closed Circular DNA Synthesis Been Identified? Hepatology. 2020;72:1142–1144. doi: 10.1002/hep.31408. [DOI] [PubMed] [Google Scholar]
- 132.Revill P.A., Chisari F.V., Block J.M., Dandri M., Gehring A.J., Guo H., Hu J., Kramvis A., Lampertico P., Janssen H.L.A., et al. A Global Scientific Strategy to Cure Hepatitis B. Lancet Gastroenterol. Hepatol. 2019;4:545–558. doi: 10.1016/S2468-1253(19)30119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fanning G.C., Zoulim F., Hou J., Bertoletti A. Therapeutic Strategies for Hepatitis B Virus Infection: Towards a Cure. Nat. Rev. Drug Discov. 2019;18:827–844. doi: 10.1038/s41573-019-0037-0. [DOI] [PubMed] [Google Scholar]
- 134.Bockmann J.-H., Stadler D., Xia Y., Ko C., Wettengel J.M., Schulze zur Wiesch J., Dandri M., Protzer U. Comparative Analysis of the Antiviral Effects Mediated by Type I and III Interferons in Hepatitis B Virus–Infected Hepatocytes. J. Infect. Dis. 2019;220:567–577. doi: 10.1093/infdis/jiz143. [DOI] [PubMed] [Google Scholar]
- 135.Seeger C., Sohn J.A. Complete Spectrum of CRISPR/Cas9-Induced Mutations on HBV CccDNA. Mol. Ther. 2016;24:1258–1266. doi: 10.1038/mt.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kostyushev D., Brezgin S., Kostyusheva A., Zarifyan D., Goptar I., Chulanov V. Orthologous CRISPR/Cas9 Systems for Specific and Efficient Degradation of Covalently Closed Circular DNA of Hepatitis B Virus. Cell. Mol. Life Sci. 2019;76:1779–1794. doi: 10.1007/s00018-019-03021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Uddin F., Rudin C.M., Sen T. CRISPR Gene Therapy: Applications, Limitations, and Implications for the Future. Front. Oncol. 2020;10:1387. doi: 10.3389/fonc.2020.01387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mohd-Ismail; Lim; Gunaratne; Tan Mapping the Interactions of HBV CccDNA with Host Factors. Int. J. Mol. Sci. 2019;20:4276. doi: 10.3390/ijms20174276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cradick T.J., Keck K., Bradshaw S., Jamieson A.C., McCaffrey A.P. Zinc-Finger Nucleases as a Novel Therapeutic Strategy for Targeting Hepatitis B Virus DNAs. Mol. Ther. 2010;18:947–954. doi: 10.1038/mt.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ely A., Moyo B., Arbuthnot P. Progress With Developing Use of Gene Editing To Cure Chronic Infection With Hepatitis B Virus. Mol. Ther. 2016;24:671–677. doi: 10.1038/mt.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ahmed M., Wang F., Levin A., Le C., Eltayebi Y., Houghton M., Tyrrell L., Barakat K. Targeting the Achilles Heel of the Hepatitis B Virus: A Review of Current Treatments against Covalently Closed Circular DNA. Drug Discov. Today. 2015;20:548–561. doi: 10.1016/j.drudis.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 142.Chen J., Zhang W., Lin J., Wang F., Wu M., Chen C., Zheng Y., Peng X., Li J., Yuan Z. An Efficient Antiviral Strategy for Targeting Hepatitis B Virus Genome Using Transcription Activator-Like Effector Nucleases. Mol. Ther. 2014;22:303–311. doi: 10.1038/mt.2013.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Seeger C. Control of Viral Transcripts as a Concept for Future HBV Therapies. Curr. Opin. Virol. 2018;30:18–23. doi: 10.1016/j.coviro.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 144.Cai D., Mills C., Yu W., Yan R., Aldrich C.E., Saputelli J.R., Mason W.S., Xu X., Guo J.-T., Block T.M., et al. Identification of Disubstituted Sulfonamide Compounds as Specific Inhibitors of Hepatitis B Virus Covalently Closed Circular DNA Formation. Antimicrob. Agents Chemother. 2012;56:4277–4288. doi: 10.1128/AAC.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Morikawa K., Suda G., Sakamoto N. Viral Life Cycle of Hepatitis B Virus: Host Factors and Druggable Targets: HBV Antivirals and Viral Life Cycle. Hepatol. Res. 2016;46:871–877. doi: 10.1111/hepr.12650. [DOI] [PubMed] [Google Scholar]
- 146.Bertoletti A., Gehring A.J. The Immune Response during Hepatitis B Virus Infection. J. Gen. Virol. 2006;87:1439–1449. doi: 10.1099/vir.0.81920-0. [DOI] [PubMed] [Google Scholar]
- 147.Akira S., Uematsu S., Takeuchi O. Pathogen Recognition and Innate Immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 148.Du K., Liu J., Broering R., Zhang X., Yang D., Dittmer U., Lu M. Recent Advances in the Discovery and Development of TLR Ligands as Novel Therapeutics for Chronic HBV and HIV Infections. Expert Opin. Drug Discov. 2018;13:661–670. doi: 10.1080/17460441.2018.1473372. [DOI] [PubMed] [Google Scholar]
- 149.Suslov A., Wieland S., Menne S. Modulators of Innate Immunity as Novel Therapeutics for Treatment of Chronic Hepatitis B. Curr. Opin. Virol. 2018;30:9–17. doi: 10.1016/j.coviro.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Meng Z., Chen Y., Lu M. Advances in Targeting the Innate and Adaptive Immune Systems to Cure Chronic Hepatitis B Virus Infection. Front. Immunol. 2020;10:3127. doi: 10.3389/fimmu.2019.03127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gehring A.J., Protzer U. Targeting Innate and Adaptive Immune Responses to Cure Chronic HBV Infection. Gastroenterology. 2019;156:325–337. doi: 10.1053/j.gastro.2018.10.032. [DOI] [PubMed] [Google Scholar]
- 152.Ma Z., Cao Q., Xiong Y., Zhang E., Lu M. Interaction between Hepatitis B Virus and Toll-Like Receptors: Current Status and Potential Therapeutic Use for Chronic Hepatitis B. Vaccines. 2018;6:6. doi: 10.3390/vaccines6010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lucifora J., Bonnin M., Aillot L., Fusil F., Maadadi S., Dimier L., Michelet M., Floriot O., Ollivier A., Rivoire M., et al. Direct Antiviral Properties of TLR Ligands against HBV Replication in Immune-Competent Hepatocytes. Sci. Rep. 2018;8:5390. doi: 10.1038/s41598-018-23525-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gane E., Pastagia M. Pharmacodynamics of Oral JNJ-64794964, a Toll-like Receptor-7 Agonist, in Healthy Adults. J. Hepatol. 2019;70:478. doi: 10.1016/S0618-8278(19)30943-0. [DOI] [Google Scholar]
- 155.Janssen H.L.A., Brunetto M.R., Kim Y.J., Ferrari C., Massetto B., Nguyen A.-H., Joshi A., Woo J., Lau A.H., Gaggar A., et al. Safety, Efficacy and Pharmacodynamics of Vesatolimod (GS-9620) in Virally Suppressed Patients with Chronic Hepatitis B. J. Hepatol. 2018;68:431–440. doi: 10.1016/j.jhep.2017.10.027. [DOI] [PubMed] [Google Scholar]
- 156.Agarwal K., Ahn S.H., Elkhashab M., Lau A.H., Gaggar A., Bulusu A., Tian X., Cathcart A.L., Woo J., Subramanian G.M., et al. Safety and Efficacy of Vesatolimod (GS-9620) in Patients with Chronic Hepatitis B Who Are Not Currently on Antiviral Treatment. J. Viral Hepat. 2018;25:1331–1340. doi: 10.1111/jvh.12942. [DOI] [PubMed] [Google Scholar]
- 157.McGowan D., Herschke F., Pauwels F., Stoops B., Last S., Pieters S., Scholliers A., Thoné T., Van Schoubroeck B., De Pooter D., et al. Novel Pyrimidine Toll-like Receptor 7 and 8 Dual Agonists to Treat Hepatitis B Virus. J. Med. Chem. 2016;59:7936–7949. doi: 10.1021/acs.jmedchem.6b00747. [DOI] [PubMed] [Google Scholar]
- 158.Embrechts W., Herschke F., Pauwels F., Stoops B., Last S., Pieters S., Pande V., Pille G., Amssoms K., Smyej I., et al. 2,4-Diaminoquinazolines as Dual Toll-like Receptor (TLR) 7/8 Modulators for the Treatment of Hepatitis B Virus. J. Med. Chem. 2018;61:6236–6246. doi: 10.1021/acs.jmedchem.8b00643. [DOI] [PubMed] [Google Scholar]
- 159.Mackman R.L., Mish M., Chin G., Perry J.K., Appleby T., Aktoudianakis V., Metobo S., Pyun P., Niu C., Daffis S., et al. Discovery of GS-9688 (Selgantolimod) as a Potent and Selective Oral Toll-Like Receptor 8 Agonist for the Treatment of Chronic Hepatitis B. J. Med. Chem. 2020;63:10188–10203. doi: 10.1021/acs.jmedchem.0c00100. [DOI] [PubMed] [Google Scholar]
- 160.Korolowicz K.E., Iyer R.P., Czerwinski S., Suresh M., Yang J., Padmanabhan S., Sheri A., Pandey R.K., Skell J., Marquis J.K., et al. Antiviral Efficacy and Host Innate Immunity Associated with SB 9200 Treatment in the Woodchuck Model of Chronic Hepatitis B. PLoS ONE. 2016;11:e0161313. doi: 10.1371/journal.pone.0161313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Suresh M., Korolowicz K.E., Balarezo M., Iyer R.P., Padmanabhan S., Cleary D., Gimi R., Sheri A., Yon C., Kallakury B.V., et al. Antiviral Efficacy and Host Immune Response Induction during Sequential Treatment with SB 9200 Followed by Entecavir in Woodchucks. PLoS ONE. 2017;12:e0169631. doi: 10.1371/journal.pone.0169631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Locarnini S., Wong D., Jackson K., Walsh R., Edwards R., Hammond R., Macfarlane C., Iyer R.P., Afdhal N., Yuen M.-F. Novel Anti-Viral Activity of SB 9200, a RIG-I Agonist; Results from Cohort 1 of the Achieve Trial. Hepatology. 2017;66:1269–1270. [Google Scholar]
- 163.Roethle P.A., McFadden R.M., Yang H., Hrvatin P., Hui H., Graupe M., Gallagher B., Chao J., Hesselgesser J., Duatschek P., et al. Identification and Optimization of Pteridinone Toll-like Receptor 7 (TLR7) Agonists for the Oral Treatment of Viral Hepatitis. J. Med. Chem. 2013;56:7324–7333. doi: 10.1021/jm400815m. [DOI] [PubMed] [Google Scholar]
- 164.Burton A.R., Pallett L.J., McCoy L.E., Suveizdyte K., Amin O.E., Swadling L., Alberts E., Davidson B.R., Kennedy P.T.F., Gill U.S., et al. Circulating and Intrahepatic Antiviral B Cells Are Defective in Hepatitis B. J. Clin. Investig. 2018;128:4588–4603. doi: 10.1172/JCI121960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fisicaro P., Valdatta C., Massari M., Loggi E., Biasini E., Sacchelli L., Cavallo M.C., Silini E.M., Andreone P., Missale G., et al. Antiviral Intrahepatic T-Cell Responses Can Be Restored by Blocking Programmed Death-1 Pathway in Chronic Hepatitis B. Gastroenterology. 2010;138:682–693.e4. doi: 10.1053/j.gastro.2009.09.052. [DOI] [PubMed] [Google Scholar]
- 166.Liu J., Zhang E., Ma Z., Wu W., Kosinska A., Zhang X., Möller I., Seiz P., Glebe D., Wang B., et al. Enhancing Virus-Specific Immunity In Vivo by Combining Therapeutic Vaccination and PD-L1 Blockade in Chronic Hepadnaviral Infection. PLoS Pathog. 2014;10:e1003856. doi: 10.1371/journal.ppat.1003856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ezzikouri S., Hoque Kayesh M.E., Benjelloun S., Kohara M., Tsukiyama-Kohara K. Targeting Host Innate and Adaptive Immunity to Achieve the Functional Cure of Chronic Hepatitis B. Vaccines. 2020;8:216. doi: 10.3390/vaccines8020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhang E., Zhang X., Liu J., Wang B., Tian Y., Kosinska A.D., Ma Z., Xu Y., Dittmer U., Roggendorf M., et al. The Expression of PD-1 Ligands and Their Involvement in Regulation of T Cell Functions in Acute and Chronic Woodchuck Hepatitis Virus Infection. PLoS ONE. 2011;6:e26196. doi: 10.1371/journal.pone.0026196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Salimzadeh L., Le Bert N., Dutertre C.-A., Gill U.S., Newell E.W., Frey C., Hung M., Novikov N., Fletcher S., Kennedy P.T.F., et al. PD-1 Blockade Partially Recovers Dysfunctional Virus–Specific B Cells in Chronic Hepatitis B Infection. J. Clin. Investig. 2018;128:4573–4587. doi: 10.1172/JCI121957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Dong H., Zhu G., Tamada K., Flies D.B., van Deursen J.M.A., Chen L. B7-H1 Determines Accumulation and Deletion of Intrahepatic CD8+ T Lymphocytes. Immunity. 2004;20:327–336. doi: 10.1016/S1074-7613(04)00050-0. [DOI] [PubMed] [Google Scholar]
- 171.El-Khoueiry A.B., Sangro B., Yau T., Crocenzi T.S., Kudo M., Hsu C., Kim T.-Y., Choo S.-P., Trojan J., Welling T.H., et al. Nivolumab in Patients with Advanced Hepatocellular Carcinoma (CheckMate 040): An Open-Label, Non-Comparative, Phase 1/2 Dose Escalation and Expansion Trial. Lancet. 2017;389:2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Scheiner B., Kirstein M.M., Hucke F., Finkelmeier F., Schulze K., Hinrichs J.B., Waneck F., Waidmann O., Reiberger T., Müller C., et al. Programmed Cell Death Protein-1 (PD-1)-targeted Immunotherapy in Advanced Hepatocellular Carcinoma: Efficacy and Safety Data from an International Multicentre Real-world Cohort. Aliment. Pharmacol. Ther. 2019;49:1323–1333. doi: 10.1111/apt.15245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gish R.G., Yuen M.-F., Chan H.L.Y., Given B.D., Lai C.-L., Locarnini S.A., Lau J.Y.N., Wooddell C.I., Schluep T., Lewis D.L. Synthetic RNAi Triggers and Their Use in Chronic Hepatitis B Therapies with Curative Intent. Antivir. Res. 2015;121:97–108. doi: 10.1016/j.antiviral.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 174.Grimm D., Thimme R., Blum H.E. HBV Life Cycle and Novel Drug Targets. Hepatol. Int. 2011;5:644–653. doi: 10.1007/s12072-011-9261-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Soriano V., Barreiro P., Cachay E., Kottilil S., Fernandez-Montero J.V., de Mendoza C. Advances in Hepatitis B Therapeutics. Ther. Adv. Infect. Dis. 2020;7:204993612096502. doi: 10.1177/2049936120965027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Han X., Zhou C., Jiang M., Wang Y., Wang J., Cheng Z., Wang M., Liu Y., Liang C., Wang J., et al. Discovery of RG7834: The First-in-Class Selective and Orally Available Small Molecule Hepatitis B Virus Expression Inhibitor with Novel Mechanism of Action. J. Med. Chem. 2018;61:10619–10634. doi: 10.1021/acs.jmedchem.8b01245. [DOI] [PubMed] [Google Scholar]
- 177.Chi X., Gatti P., Papoian T. Safety of Antisense Oligonucleotide and SiRNA-Based Therapeutics. Drug Discov. Today. 2017;22:823–833. doi: 10.1016/j.drudis.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 178.Billioud G., Kruse R.L., Carrillo M., Whitten-Bauer C., Gao D., Kim A., Chen L., McCaleb M.L., Crosby J.R., Hamatake R., et al. In Vivo Reduction of Hepatitis B Virus Antigenemia and Viremia by Antisense Oligonucleotides. J. Hepatol. 2016;64:781–789. doi: 10.1016/j.jhep.2015.11.032. [DOI] [PubMed] [Google Scholar]
- 179.Hausen P., Stein H. Ribonuclease H. An Enzyme Degrading the RNA Moiety of DNA-RNA Hybrids. Eur. J. Biochem. 1970;14:278–283. doi: 10.1111/j.1432-1033.1970.tb00287.x. [DOI] [PubMed] [Google Scholar]
- 180.Keller W., Crouch R. Degradation of DNA RNA Hybrids by Ribonuclease H and DNA Polymerases of Cellular and Viral Origin. Proc. Natl. Acad. Sci. USA. 1972;69:3360–3364. doi: 10.1073/pnas.69.11.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Edwards T.C., Ponzar N.L., Tavis J.E. Shedding Light on RNaseH: A Promising Target for Hepatitis B Virus (HBV) Expert Opin. Ther. Targets. 2019;23:559–563. doi: 10.1080/14728222.2019.1619697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gerelsaikhan T., Tavis J.E., Bruss V. Hepatitis B Virus Nucleocapsid Envelopment Does Not Occur without Genomic DNA Synthesis. J. Virol. 1996;70:4269–4274. doi: 10.1128/JVI.70.7.4269-4274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nowotny M., Gaidamakov S.A., Crouch R.J., Yang W. Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-Dependent Catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 184.Nowotny M., Yang W. Stepwise Analyses of Metal Ions in RNase H Catalysis from Substrate Destabilization to Product Release. EMBO J. 2006;25:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tavis J.E., Zoidis G., Meyers M.J., Murelli R.P. Chemical Approaches to Inhibiting the Hepatitis B Virus Ribonuclease H. ACS Infect. Dis. 2019;5:655–658. doi: 10.1021/acsinfecdis.8b00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Tramontano E., Corona A., Menéndez-Arias L. Ribonuclease H, an Unexploited Target for Antiviral Intervention against HIV and Hepatitis B Virus. Antivir. Res. 2019;171:104613. doi: 10.1016/j.antiviral.2019.104613. [DOI] [PubMed] [Google Scholar]
- 187.Edwards T.C., Mani N., Dorsey B., Kakarla R., Rijnbrand R., Sofia M.J., Tavis J.E. Inhibition of HBV Replication by N-Hydroxyisoquinolinedione and N-Hydroxypyridinedione Ribonuclease H Inhibitors. Antivir. Res. 2019;164:70–80. doi: 10.1016/j.antiviral.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Edwards T.C., Lomonosova E., Patel J.A., Li Q., Villa J.A., Gupta A.K., Morrison L.A., Bailly F., Cotelle P., Giannakopoulou E., et al. Inhibition of Hepatitis B Virus Replication by N -Hydroxyisoquinolinediones and Related Polyoxygenated Heterocycles. Antivir. Res. 2017;143:205–217. doi: 10.1016/j.antiviral.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hu Y., Cheng X., Cao F., Huang A., Tavis J.E. β-Thujaplicinol Inhibits Hepatitis B Virus Replication by Blocking the Viral Ribonuclease H Activity. Antivir. Res. 2013;99:221–229. doi: 10.1016/j.antiviral.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 190.Lu G., Lomonosova E., Cheng X., Moran E.A., Meyers M.J., Le Grice S.F.J., Thomas C.J., Jiang J., Meck C., Hirsch D.R., et al. Hydroxylated Tropolones Inhibit Hepatitis B Virus Replication by Blocking Viral Ribonuclease H Activity. Antimicrob. Agents Chemother. 2015;59:1070–1079. doi: 10.1128/AAC.04617-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lomonosova E., Daw J., Garimallaprabhakaran A.K., Agyemang N.B., Ashani Y., Murelli R.P., Tavis J.E. Efficacy and Cytotoxicity in Cell Culture of Novel α-Hydroxytropolone Inhibitors of Hepatitis B Virus Ribonuclease H. Antivir. Res. 2017;144:164–172. doi: 10.1016/j.antiviral.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Long K.R., Lomonosova E., Li Q., Ponzar N.L., Villa J.A., Touchette E., Rapp S., Liley R.M., Murelli R.P., Grigoryan A., et al. Efficacy of Hepatitis B Virus Ribonuclease H Inhibitors, a New Class of Replication Antagonists, in FRG Human Liver Chimeric Mice. Antivir. Res. 2018;149:41–47. doi: 10.1016/j.antiviral.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Agyemang N.B., Kukla C.R., Edwards T.C., Li Q., Langen M.K., Schaal A., Franson A.D., Casals A.G., Donald K.A., Yu A.J., et al. Divergent Synthesis of a Thiolate-Based α-Hydroxytropolone Library with a Dynamic Bioactivity Profile. RSC Adv. 2019;9:34227–34234. doi: 10.1039/C9RA06383H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Berkowitz A.J., Abdelmessih R.G., Murelli R.P. Amidation Strategy for Final-Step α-Hydroxytropolone Diversification. Tetrahedron Lett. 2018;59:3026–3028. doi: 10.1016/j.tetlet.2018.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Li Q., Lomonosova E., Donlin M.J., Cao F., O’Dea A., Milleson B., Berkowitz A.J., Baucom J.-C., Stasiak J.P., Schiavone D.V., et al. Amide-Containing α-Hydroxytropolones as Inhibitors of Hepatitis B Virus Replication. Antivir. Res. 2020;177:104777. doi: 10.1016/j.antiviral.2020.104777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Bak E., Miller J.T., Noronha A., Tavis J., Gallicchio E., Murelli R.P., Le Grice S.F.J. 3,7-Dihydroxytropolones Inhibit Initiation of Hepatitis B Virus Minus-Strand DNA Synthesis. Molecules. 2020;25:4434. doi: 10.3390/molecules25194434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cai C.W., Lomonosova E., Moran E.A., Cheng X., Patel K.B., Bailly F., Cotelle P., Meyers M.J., Tavis J.E. Hepatitis B Virus Replication Is Blocked by a 2-Hydroxyisoquinoline-1,3(2H,4H)-Dione (HID) Inhibitor of the Viral Ribonuclease H Activity. Antivir. Res. 2014;108:48–55. doi: 10.1016/j.antiviral.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Tavis J.E., Cheng X., Hu Y., Totten M., Cao F., Michailidis E., Aurora R., Meyers M.J., Jacobsen E.J., Parniak M.A., et al. The Hepatitis B Virus Ribonuclease H Is Sensitive to Inhibitors of the Human Immunodeficiency Virus Ribonuclease H and Integrase Enzymes. PLoS Pathog. 2013;9:e1003125. doi: 10.1371/journal.ppat.1003125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lu G., Villa J.A., Donlin M.J., Edwards T.C., Cheng X., Heier R.F., Meyers M.J., Tavis J.E. Hepatitis B Virus Genetic Diversity Has Minimal Impact on Sensitivity of the Viral Ribonuclease H to Inhibitors. Antivir. Res. 2016;135:24–30. doi: 10.1016/j.antiviral.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lomonosova E., Zlotnick A., Tavis J.E. Synergistic Interactions between Hepatitis B Virus RNase H Antagonists and Other Inhibitors. Antimicrob. Agents Chemother. 2017;61:e02441-16. doi: 10.1128/AAC.02441-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Yang L., Liu F., Tong X., Hoffmann D., Zuo J., Lu M. Treatment of Chronic Hepatitis B Virus Infection Using Small Molecule Modulators of Nucleocapsid Assembly: Recent Advances and Perspectives. ACS Infect. Dis. 2019;5:713–724. doi: 10.1021/acsinfecdis.8b00337. [DOI] [PubMed] [Google Scholar]
- 202.Pei Y., Wang C., Yan S.F., Liu G. Past, Current, and Future Developments of Therapeutic Agents for Treatment of Chronic Hepatitis B Virus Infection. J. Med. Chem. 2017;60:6461–6479. doi: 10.1021/acs.jmedchem.6b01442. [DOI] [PubMed] [Google Scholar]
- 203.Smolders E.J., Burger D.M., Feld J.J., Kiser J.J. Review Article: Clinical Pharmacology of Current and Investigational Hepatitis B Virus Therapies. Aliment. Pharmacol. Ther. 2020;51:231–243. doi: 10.1111/apt.15581. [DOI] [PubMed] [Google Scholar]
- 204.Zoulim F., Lenz O., Vandenbossche J.J., Talloen W., Verbinnen T., Moscalu I., Streinu-Cercel A., Bourgeois S., Buti M., Crespo J., et al. JNJ-56136379, an HBV Capsid Assembly Modulator, Is Well-Tolerated and Has Antiviral Activity in a Phase 1 Study of Patients With Chronic Infection. Gastroenterology. 2020;159:521–533.e9. doi: 10.1053/j.gastro.2020.04.036. [DOI] [PubMed] [Google Scholar]
- 205.Huang Q., Cai D., Yan R., Li L., Zong Y., Guo L., Mercier A., Zhou Y., Tang A., Henne K., et al. Preclinical Profile and Characterization of the Hepatitis B Virus Core Protein Inhibitor ABI-H0731. Antimicrob. Agents Chemother. 2020;64:e01463-20. doi: 10.1128/AAC.01463-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Mani N., Cole A.G., Phelps J.R., Ardzinski A., Cobarrubias K.D., Cuconati A., Dorsey B.D., Evangelista E., Fan K., Guo F., et al. Preclinical Profile of AB-423, an Inhibitor of Hepatitis B Virus Pregenomic RNA Encapsidation. Antimicrob. Agents Chemother. 2018;62:e00082-18. doi: 10.1128/AAC.00082-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wu G., Liu B., Zhang Y., Li J., Arzumanyan A., Clayton M.M., Schinazi R.F., Wang Z., Goldmann S., Ren Q., et al. Preclinical Characterization of GLS4, an Inhibitor of Hepatitis B Virus Core Particle Assembly. Antimicrob. Agents Chemother. 2013;57:5344–5354. doi: 10.1128/AAC.01091-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Berke J.M., Dehertogh P., Vergauwen K., Van Damme E., Mostmans W., Vandyck K., Pauwels F. Capsid Assembly Modulators Have a Dual Mechanism of Action in Primary Human Hepatocytes Infected with Hepatitis B Virus. Antimicrob. Agents Chemother. 2017;61:e00560-17. doi: 10.1128/AAC.00560-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Razavi-Shearer D., Gamkrelidze I., Nguyen M.H., Chen D.-S., Van Damme P., Abbas Z., Abdulla M., Abou Rached A., Adda D., Aho I., et al. Global Prevalence, Treatment, and Prevention of Hepatitis B Virus Infection in 2016: A Modelling Study. Lancet Gastroenterol. Hepatol. 2018;3:383–403. doi: 10.1016/S2468-1253(18)30056-6. [DOI] [PubMed] [Google Scholar]
- 210.Alter H., Block T., Brown N., Brownstein A., Brosgart C., Chang K.-M., Chen P.-J., Chisari F.V., Cohen C., El-Serag H., et al. A Research Agenda for Curing Chronic Hepatitis B Virus Infection. Hepatology. 2018;67:1127–1131. doi: 10.1002/hep.29509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.