Abstract
Conjugation of polyethylene glycol (PEGylation) is a well-known strategy for extending the serum half-life of protein drugs and for increasing their resistance to proteolysis and aggregation. We previously showed that PEGylation can increase protein conformational stability; the extent of PEG-based stabilization depends on the PEGylation site, the structure of the PEG-protein linker, and the ability of PEG to release water molecules from the surrounding protein surface to bulk solvent. The strength of a non-covalent interaction within a protein depends highly on its microenvironment, with salt-bridge and hydrogen-bond strength increasing in non-polar vs. aqueous environments. Accordingly, we wondered whether partial desolvation by PEG of the surrounding protein surface might result in measurable increases in the strength of a salt bridge near a PEGylation site. Here we explore this possibility using triple mutant box analysis to assess the impact of PEGylation on the strength of nearby salt bridges at specific locations within three peptide model systems. The results indicate that PEG can increase nearby salt bridge strength, though this effect is not universal, and its precise structural prerequisites are not a simple function of secondary structural context, of orientation and distance between PEGylation site and salt bridge, or of salt-bridge residue identity. We obtained high-resolution x-ray diffraction data for a PEGylated peptide in which PEG enhances the strength of a nearby salt bridge. Comparing the electron density map of this PEGylated peptide vs. that of its non-PEGylated counterpart provide evidences for localized protein surface desolvation as a mechanism for the PEG-based salt-bridge stabilization.
Graphical Abstract
Conjugation of polyethylene glycol (PEGylation) to proteins is a well-known strategy for enhancing the pharmacokinetic properties of protein pharmaceuticals.1–4 The large hydrodynamic radius of the PEG conjugate shields the protein from proteases, neutralizes antibodies and decreases its clearance from serum via renal filtration.5, 6 First-generation PEG-protein conjugates were prepared by reacting a non-selective PEG-electrophile with surface nucleophiles, resulting in a heterogeneous mixture of PEGylated proteins differing in both number and location of the attached PEGs.3, 4 The resulting population of non-specifically PEGylated protein isoforms often exhibits decreased in vitro activity, presumably because PEG also interferes with the binding events that are essential for protein-protein interactions and/or catalysis.
Advances in chemoselective biorthogonal reactions now allow chemists to install PEG site-specifically at any arbitrary position within a proteins.7–19 Using these reactions, chemists can now choose surface PEGylation sites that are far from enzyme active sites and from substrate binding interfaces. The resulting second-generation PEG-protein conjugates often retain more in vitro activity than did their first-generation counterparts, relative to the non-PEGylated protein. However, aside from these simple and intuitive guidelines, finding a PEGylation site that provides an optimal balance between increased proteolytic stability, lengthened serum half-life, and retained biological activity is often an empirical matter of trial and error.
We have previously hypothesized that such optimal PEGylation sites will be characterized by the increased conformational stability of the PEG-protein conjugate relative to its non-PEGylated counterpart.20, 21 The rationale for this hypothesis is that proteolysis, aggregation, and recognition by neutralizing antibodies are generally more severe for unfolded proteins than folded proteins.22 Consequently, we have explored the impact of PEGylation on the conformational stability of peptide and protein model systems in search of structure- or sequence-based criteria for identifying conformationally stabilizing PEGylation sites. We found that PEG can increase conformational stability, but the extent of stabilization depends strongly the location of the PEGylation site21 and the chemistry used to connect PEG and protein.23, 24 Detailed thermodynamic analyses and computational simulations suggest that the stabilizing impact of PEGylation derives from an entropic effect, likely because PEG allows some first-shell water molecules to be released to bulk solvent.21 We wondered whether such localized desolvation might increase the strength of non-covalent interactions in the immediate vicinity of the PEGylation site; this possibility seemed reasonable in light of several recent reports linking the strength of a salt bridge or of a hydrogen bond to the polarity of its immediate microenvironment and its exposure to solvent.25–29
Results and Discussion
We explored this possibility in the context of dimeric helix-bundle peptide GCN4-p1 (Figure 1A).30 As with other helix bundles, the primary sequence of GCN4-p1 is characterized by a seven-residue repeating pattern in which non-polar residues occupy positions a and d of an abcdefg heptad, with polar and/or charged residues predominantly occupying the other positions. Such an arrangement aligns the non-polar a- and d-position along the same side of the peptide in the α-helical conformation; burial of these residues at the interface between the two helices provides the driving force for helix-bundle formation.31 In contrast, residues at the other five positions are solvent-exposed and are therefore ideal locations for probing the impact of PEG-based desolvation on the strength of non-covalent interactions at the protein surface. Accordingly, we prepared GCN4-p1 variants p2α1, p2α3, p2α4, p2α6, p2α7, p2α10, p2α14, p2α18, p2α21, p2α25, and p2α28, in which we incorporated an asparagine-linked monomethoxy-PEG (i.e., AsnPEG, comprised of four ethylene oxide units; Figure 1B) at solvent exposed positions 1, 3, 4, 6, 7, 10, 14, 18, 21, 25, and 28, respectively (Figure 1, Table 1). We also prepared their Asn-containing sequence-matched non-PEGylated counterparts 2α1, 2α3, 2α4, 2α6, 2α7, 2α10, 2α14, 2α18, 2α21, 2α25, and 2α28. We used variable temperature CD experiments to obtain the apparent melting temperatures (Tm) and folding free energies (ΔGf) of each variant listed in Table 1, along with the impact of PEGylation on the conformational stability of each PEGylated variant relative to its non-PEGylated counterpart (ΔΔGf).
Figure 1.
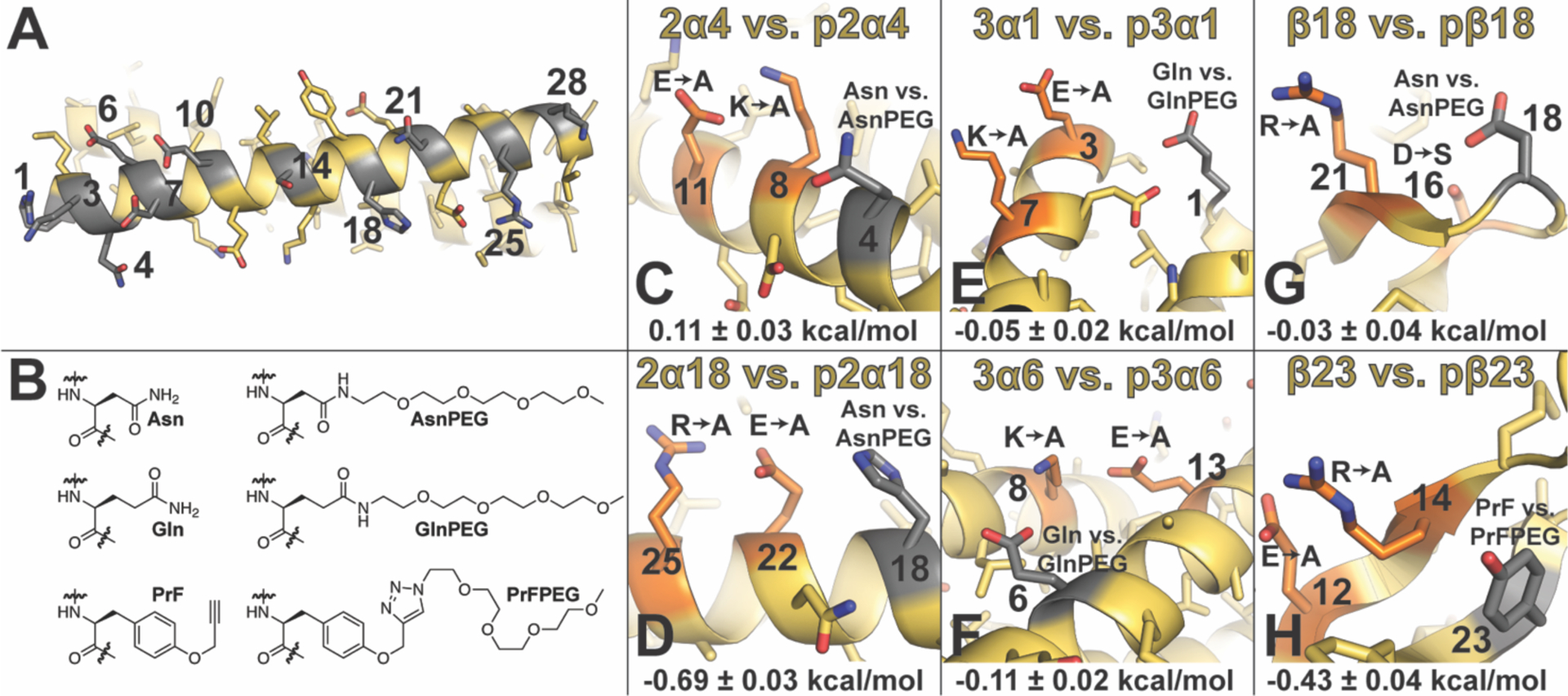
(A) Ribbon diagram of dimeric helix bundle formed by peptide GCN4-p1 (PDB ID: 2ZTA) with side chains shown as sticks. Positions where we incorporated Asn vs. AsnPEG are numbered and highlighted in gray. (B) Structures of Asn vs. AsnPEG; Gln vs. GlnPEG; and PrF vs. PrFPEG. (C)–(H) Summary of triple mutant box analysis of PEGylation sites and nearby salt bridges (highlighted in orange and gray, respectively) within peptides based on α-helical coiled-coil dimer GCN4 (PDB ID: 2ZTA): (C) 2α4 vs. p2α4 and (D) 2α18 vs. p2α18; α-helical coiled-coil trimer 1CW (PDB ID: 1COI): (E) 3α1 vs p3α1 and (F) 3α6 vs. p3α6; and two closely related variants of β-sheet protein WW (PDB IDs: 2F21 and 1PIN): (G) β18 vs. pβ18 and (H) β23 vs. pβ23. Impact of PEGylation on salt bridge strength (ΔΔΔΔGf) is indicated below each figure ± standard error in kcal/mol.
Table 1.
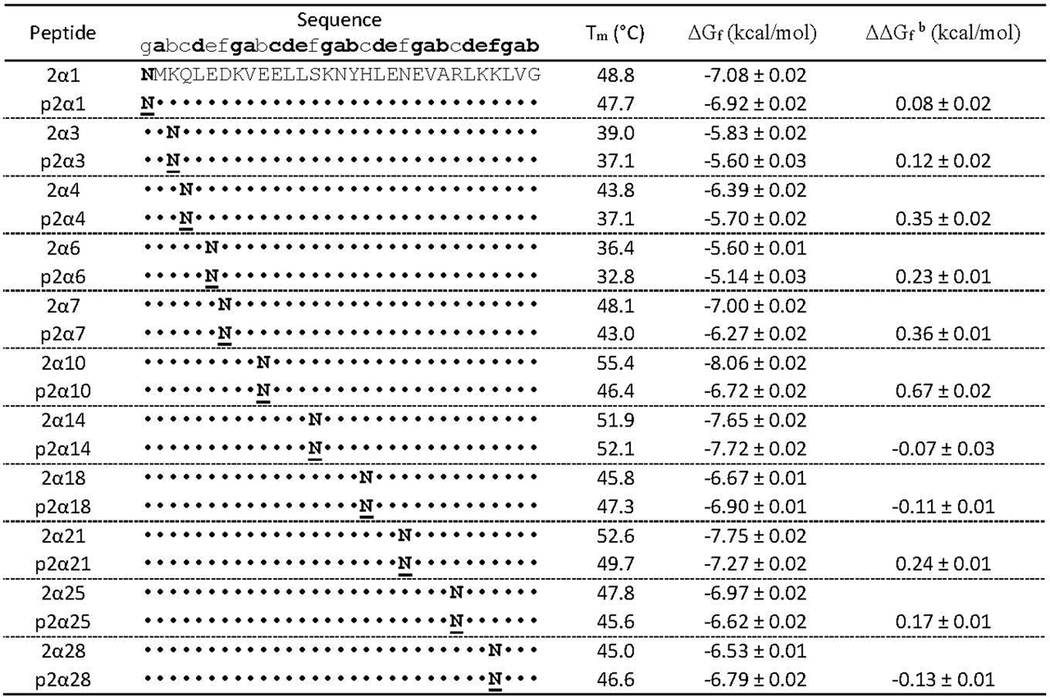
Sequences, melting temperatures and folding free energies of GCN4-p1 variants 2α1, 2α3, 2α4, 2α6, 2α7, 2α10, 2α14, 2α18, 2α21, 2α25, 2α28 and their PEGylated counterparts.a
|
Folding free energies are given ± standard error in kcal/mol at 30 μM protein concentration in 20 mM sodium phosphate buffer (pH 7) at the average melting temperature of variants 2α1, 2α3, 2α4, 2α6, 2α7, 2α10, 2α14, 2α18, 2α21, 2α25, 2α28 and their PEGylated counterparts (318.5 K). ΔΔGf values represent the energetic contribution of a single AsnPEGylation event to helix-bundle conformational stability.
Asn-PEGylation has a minimal effect at positions 1, 3, 14 within GCN4-p1; is destabilizing at positions 4, 6, 7, 10, 21, and 25; and is slightly stabilizing at positions 18 and 28 (Table 1). We examined the structural context of each of these PEGylation sites to explore whether their proximity to any nearby non-covalent interactions was related to the overall impact of PEGylation on the helix bundle. Interestingly, positions 4 and 18 share a similar structural context: they each occupy the i–4 position relative to an i to i+3 salt bridge: Lys8–Glu11 in p2α4 (Figure 1C) and Glu22–Arg25 in p2α18 (Figure 1D). However, PEGylation has opposite effects at these positions: p2α4 is less stable than 2α4, whereas p2α18 is more stable than 2α18. We wondered whether these differences might be reflected in the impact of PEG on the strength of the Lys8–Glu11 salt bridge in p2α4 vs. the Glu22–Arg25 salt bridge in p2α18.
We explored this possibility using triple mutant cycle analysis as we have done previously.13 We prepared six additional variants of 2α4 and p2α4 in which we replaced Lys8 and Glu11 with Ala, individually (2α4-ΚΑ, 2α4-AE, p2α4-ΚΑ, p2α4-AE) and in combination (2α4-AΑ, p2α4-AA). For the non-PEGylated variants, comparing the impact of the Ala8 to Lys8 mutation in the presence of Ala11 (2α4-KΑ vs. 2α4-AΑ, ΔΔGf = 0.03 ± 0.02 kcal/mol) vs. Glu11 (2α4 vs. 2α4-AE, ΔΔGf = −0.03 ± 0.02 kcal/mol) reveals that the Lys8–Glu11 salt bridge does not contribute substantially to the stability of 2α4 (ΔΔΔGf = −0.06 ± 0.02 kcal/mol). For the PEGylated variants, comparing analogous variants (p2α4-KA vs. p2α4-AA, ΔΔGf = 0.36 ± 0.01 kcal/mol; 2α4 vs. 2α4-AE, ΔΔGf = 0.41 ± 0.01 kcal/mol) reveals that the Lys8-Glu11 salt bridge has a similarly small impact on the stability of p2α4 (ΔΔΔGf = 0.05 ± 0.02 kcal/mol), indicating that PEG has a minimal impact on the strength of the Lys8–Glu11 salt bridge (ΔΔΔΔGf = 0.11 ± 0.03 kcal/mol).
We performed analogous experiments with 2α18 and p2α18 and their six sequence mutants (2α18-EΑ, 2α18-AR, 2α18-AA, p2α18-EΑ, p2α18-AR, p2α18-AA). When Asn occupies position 18, the Glu22–Arg25 salt bridge contributes −0.45 ± 0.02 kcal/mol to the stability of 2α18; when AsnPEG occupies position 18, the Glu22–Arg25 salt bridge contributes −1.14 ± 0.03 kcal/mol to the stability of p2α18, indicating that PEG stabilizes the Glu22–Arg25 salt bridge by −0.69 ± 0.03 kcal/mol. We wondered whether other stabilizing PEGylation sites might be similarly characterized by their ability to increase the strength of nearby salt bridges. To test this hypothesis, we explored PEGylation sites near salt bridges within a second α-helical model system (i.e the trimeric helix bundle 1CW)32–34 and within a β-sheet model protein (the WW domain of the human protein Pin1).35, 36 The variants and positions we explored are summarized in Figure 1E–H and in Table 2.
Table 2.
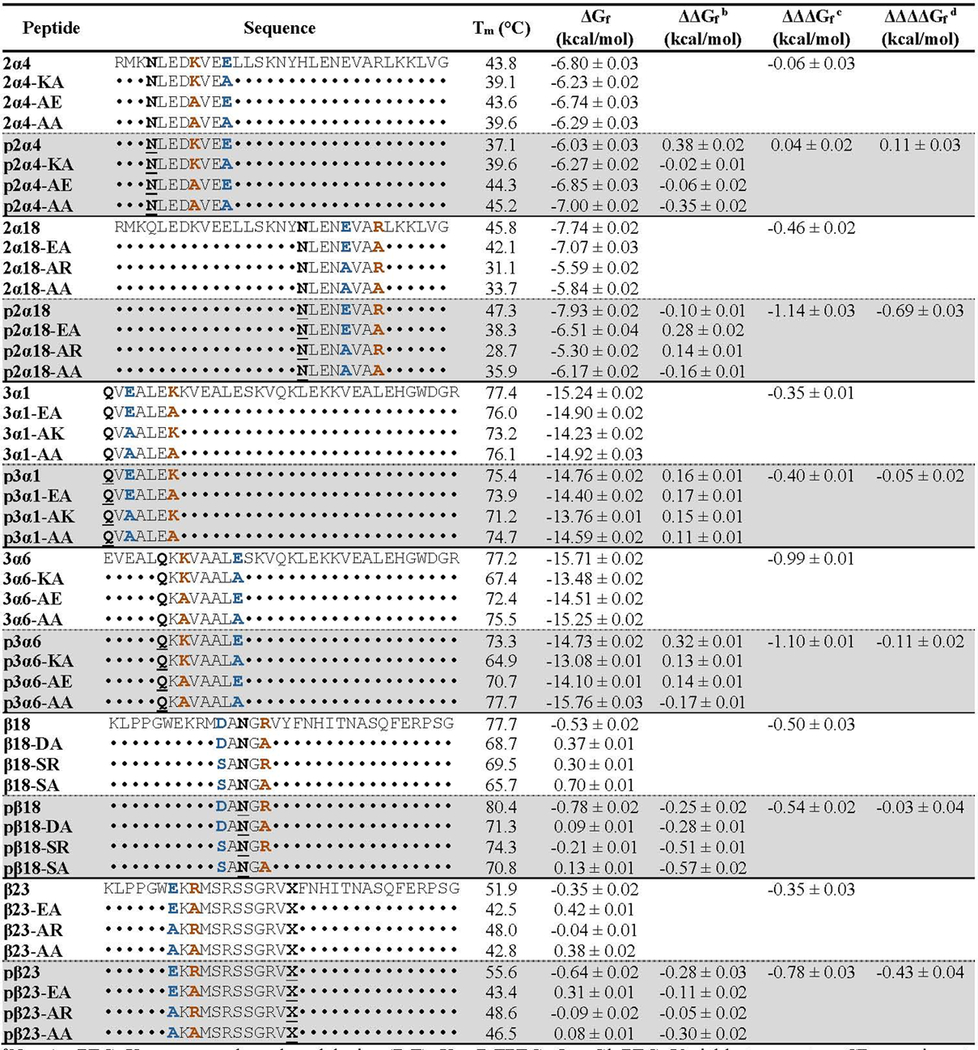
Triple mutant box analysis of the impact of PEGylation on salt-bridge strength within peptides p2α4, p2α18, p3α1, p3α6, pβ18 and pβ23.a
|
N = AsnPEG; X = propargyloxyphenylalanine (PrF); X = PrFPEG; Q = GlnPEG. Variable temperature CD experiments were performed in 20 mM phosphate buffer (pH 7) at 30 μM protein concentration except for the WW variants β18 and β23 and their derivatives, which were characterized at 50 μΜ. ΔGf values are given ± standard error. ΔGf values for each of the eight peptides within the same triple mutant cycle were calculated at their average melting temperature: 314.7 K for 2α4 and its derivatives; 311.0 K for 2α18 and its derivatives; 347.9 K for 3α1 and its derivatives; 345.5 K for 3α6 and its derivatives; 345.4 K for β18 and its derivatives; 320.6 K for β23 and its derivatives.
Impact of PEGylation on peptide/protein conformational stability.
Strength of salt-bridge interaction.
Impact of PEGylation on salt-bridge strength.
Position 1 within 1CW is oriented toward a nearby i to i+4 salt bridge comprised of Glu3 and Lys7 on an adjacent helix within the trimer (Figure 1E). We generated variant 3α1 and its PEGylated counterpart p3α1 by incorporating Gln vs. GlnPEG (Figure 1B) at position 1 within 1CW (Table 2). We chose to use Gln vs. GlnPEG because they more closely resemble the residue that occupies position 1 (i.e. Glu) within the parent 1CW peptide from which these variants were derived. GlnPEGylation slightly destabilizes p3α1 and relative to 3α1. Comparing ΔGf values for p3α1 and 3α1 with those of their sequence variants (3α1-EA, 3α1-AK, 3α1-AA, p3α1-EA, p3α1- AK, and p3α1-AA) reveals that PEGylation at Gln1 does not substantially change the strength of the Glu3–Lys7 salt bridge (ΔΔΔΔGf = −0.05 ± 0.02 kcal/mol), which contributes favorably to helix bundle stability whether or not PEG is present (Table 2).
Position 6 within 1CW is oriented toward a salt bridge between Lys8 on an adjacent helix and Glu13 within the same helix (Figure 1F). Accordingly, we prepared 1CW variant 3α6 and its PEGylated counterpart p3α6, in which we incorporated Gln vs. GlnPEG, respectively at position 6 (as with position 1 above, the residue that occupies this position in the parent 1CW is Glu). GlnPEGylation destabilizes p3α6 by 0.32 ± 0.01 kcal/mol relative to 3α6 (Table 2). Triple mutant cycle analysis of p3α6, 3α6, and their sequence variants at positions 8 and 13 (p3α6-KA, p3α6-AE, p3α6-AA, 3α6-KA, 3α6-AE, and 3α6-AA) indicates that the Lys8–Glu13 salt bridge contributes strongly to helix bundle conformational stability (ΔΔΔGf = −0.99 ± 0.01 kcal/mol) with PEGylation of Gln6 only slightly increasing salt bridge strength (ΔΔΔΔGf = −0.11 ± 0.02 kcal/mol).
Position 18 within WW (normally occupied by Asp) occupies the same reverse turn as nearby positions 16 and 21, which are normally occupied by Ser and Arg, respectively (Figure 1G). Side chains at these three positions occupy the same face of the reverse turn. We envisioned that incorporating AsnPEG vs. Asn at position 18 and Asp at position 16 might allow PEG to stabilize a salt bridge between Asp16 and Arg21. Accordingly, we prepared WW variants β18 and pβ18, with Asp at position 16, Asn vs. AsnPEG at position 18, respectively, and Arg at position 21. PEGylation modestly stabilizes pβ18 relative to β18 (ΔΔGf = −0.25 ± 0.02 kcal/mol). Triple mutant cycle analysis of pβ18, β18, and their sequence variants (pβ18-DA, pβ18-SR, pβ18-SA, β18-DA, β18-SR, and β18-SA) indicates that the Asp16-Arg21 salt bridge is stabilizing (ΔΔΔGf = −0.50 ± 0.03 kcal/mol), but PEGylation at position 18 does not affect its (ΔΔΔΔGf = −0.03 ± 0.04 kcal/mol).
Finally, we recently found that conjugating an azido-functionalized four-unit PEG to a propargyloxyphenylalanine residue (PrF, Figure 1B) at position 23 within WW increases the conformational stability of the PEGylated protein (pβ23) relative to its non-PEGylated counterpart β23 (Table 2; ΔΔGf = −0.28 ± 0.03 kcal/mol). Position 23 within WW (normally occupied by Tyr) is oriented toward a salt bridge between Glu12 and Arg14 on an adjacent β-strand (Figure 1H), and we wondered whether PEG-based stabilization of the Glu12–Arg14 salt bridge might partially account for the increased stability of pβ23 relative to β23. Triple mutant cycle analysis of pβ23 and β23 and their sequence variants pβ23-EA, pβ23-AR, pβ23-AA, β23-EA, β23-AR, and β23-AA indicates that the Glu12–Arg14 salt bridge contributes −0.35 ± 0.03 kcal/mol to WW conformational stability, with PEGylation increasing the strength of the salt bridge by an additional −0.43 ± 0.04 kcal/mol.
These results are summarized in Figure 2, which plots salt bridge strength (ΔΔΔGf) within non-PEGylated variants 2α4, 2α18, 3α1, 3α6, β18, and β23 (x-axis) vs. salt bridge strength (ΔΔΔGf) within PEGylated variants p2α4, p2α18, p3α1, p3α6, pβ18, and pβ23 (y-axis). PEGylation data points for 2α4, 3α1, 3α6, and β18 fit readily to a line with slope = 1.2 ± 0.01 (p = 0.001), indicating that PEG has no substantial impact on salt bridge strength for these variants. In contrast, data points for 2α18 and β23 are exceptions to this trend: the salt bridges we investigated are stronger within PEGylated p2α18 and pβ23 than in non-PEGylated 2α18 and β23. We wondered whether p2α18 and pβ23 had any common features that might explain this observation. We noticed that both p2α18 and pβ23 contained a Glu–Arg salt bridge; whereas, p2α4, p3α1, p3α6 contain a Glu–Lys salt bridge and pβ18 contains an Asp–Arg salt bridge. To explore whether Glu–Arg salt bridges are uniquely subject to PEG-based stabilization, we prepared 1CW variants 3α1-ER, 3α1-AR, p3α1-ER, p3α1-AR, in which Arg occupies position 7 rather than Lys. Comparing folding free energies of these variants (see supporting information) vs. those of previously characterized 3α1-EA, 3α1-AA, p3α1-EA, and p3α1-AA reveals that PEGylation does not substantially change the strength of the Glu3-Arg7 salt bridge (although the Glu3-Arg7 salt bridge in 3α1-ER is stronger than the corresponding Glu3-Lys7 salt bridge in 3α1). It appears from this observation that salt-bridge residue identity alone is not a sufficient predictor of the impact of PEGylation on salt bridge strength.
Figure 2.
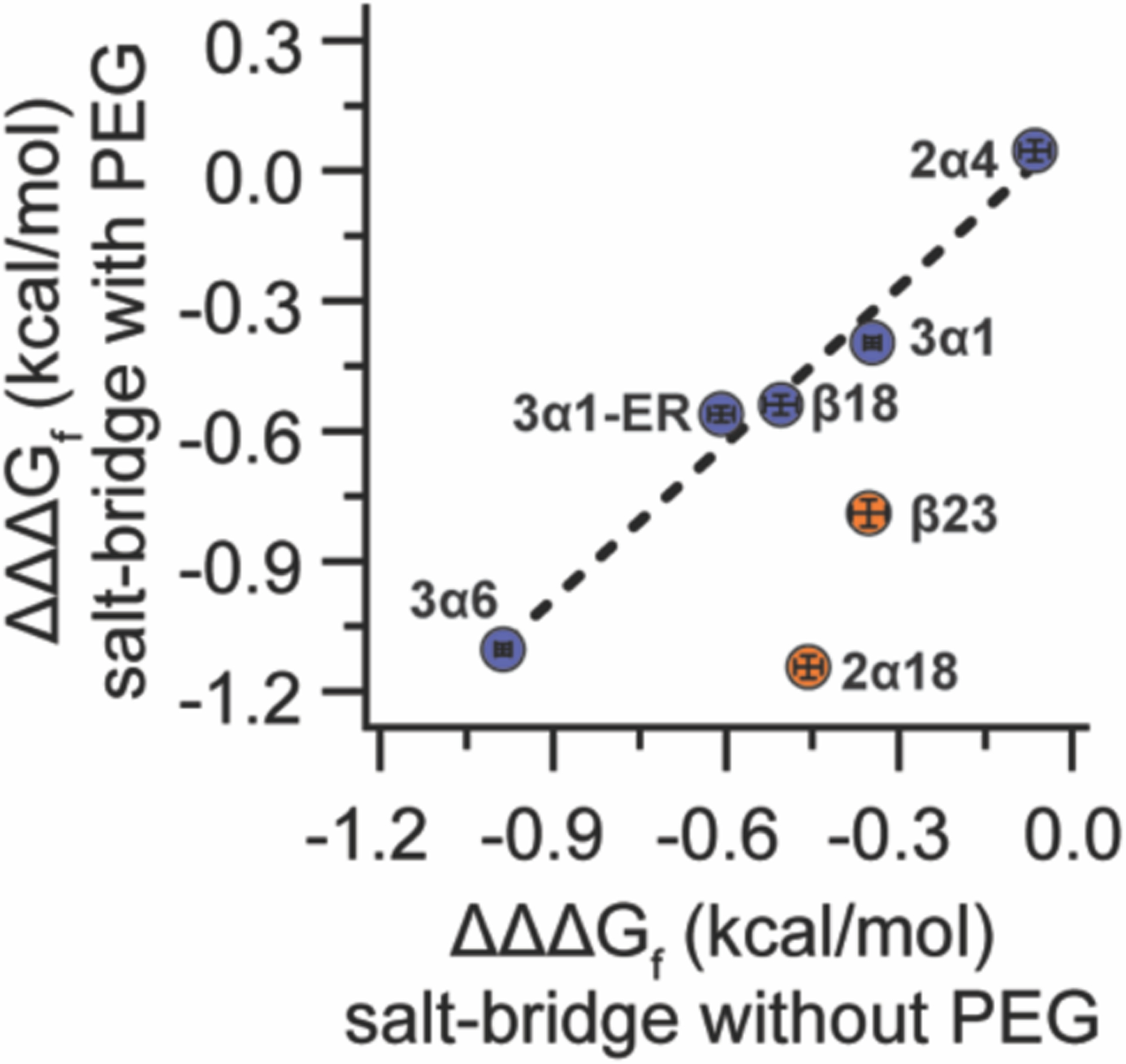
Plot of salt-bridge strength (ΔΔΔGf) within PEGylated variants p2α4, p2α18, p3α1, p3α1-ER, p3α6, pβ18, and pβ23 vs. salt-bridge strength (ΔΔΔGf) within the corresponding non-PEGylated variants 2α4, 2α18, 3α1, p3α1-ER, 3α6, β18, and β23. Data for p2α18 vs. 2α18 and pβ23 vs. β23 are highlighted in orange; at these positions salt bridges are stronger in the presence of PEG than without PEG. Dotted line represents linear regression of the data for p2α4 vs. 2α4, p3α1 vs. 3α1, p3α1-ER vs. 3α1-ER, p3α6 vs. 3α6, and pβ18 vs. β18 (points highlighted in blue), in which PEGylation does not substantially change the salt-bridge strength.
To gain structural insight into how PEG affects the Glu22–Arg25 salt bridge in 2α18 vs p2α18, we crystallized these variants, obtained x-ray diffraction data, and solved their structures via molecular replacement (Figure 3A–B, respectively). The structures for 2α18 (PDB ID: 6O2E) and p2α18 (PDB ID: 6O2F) are extremely similar overall (rmsd = 0.113 Å) with identical distances between Glu22 and Arg25, indicating that the increased strength of the Glu22–Arg25 salt bridge in the presence of PEG does not come from increased proximity of these charged groups. However, differences in the crystallographic data become more apparent upon close inspection of the electron density maps of 2α18 vs. p2α18: the area immediately surrounding Asn18, Glu22 and Arg25 in PEGylated p2α18 contains substantially more electron density than the corresponding area in non-PEGylated 2α18 (Figure 3A–B). Indeed, electron density for the Asn18 side chain in p2α18 continues to extent outward from the side-chain amide nitrogen, which is consistent with the presence of AsnPEG at position 18 within p2α18 (as confirmed by mass spectrometry; see supporting information). Including an explicit Asn-linked PEG at position 18 in our crystallographic model for p2α18 allowed us to account for some of this extra electron density and led to modest improvements in the statistical quality of the model. However, no single conformation of PEG accounted for all of the observed extra electron density; similar improvements in the statistical quality of the model resulted from including explicit crystallographic water molecules in the place of PEG. Based on these observations, we speculate that the Asn-linked PEG at position 18 in the crystalline state of p2α18 is disordered but likely occupies the space between Asn18, Glu22 and Arg25. Though these observations apply specifically to the crystalline state, it is tempting to extrapolate them to the solution behavior of p2α18; it is possible that PEG occupies space in the immediate vicinity of the Glu22–Arg25 salt bridge and partially desolvates it, thereby increasing its strength.
Figure 3.
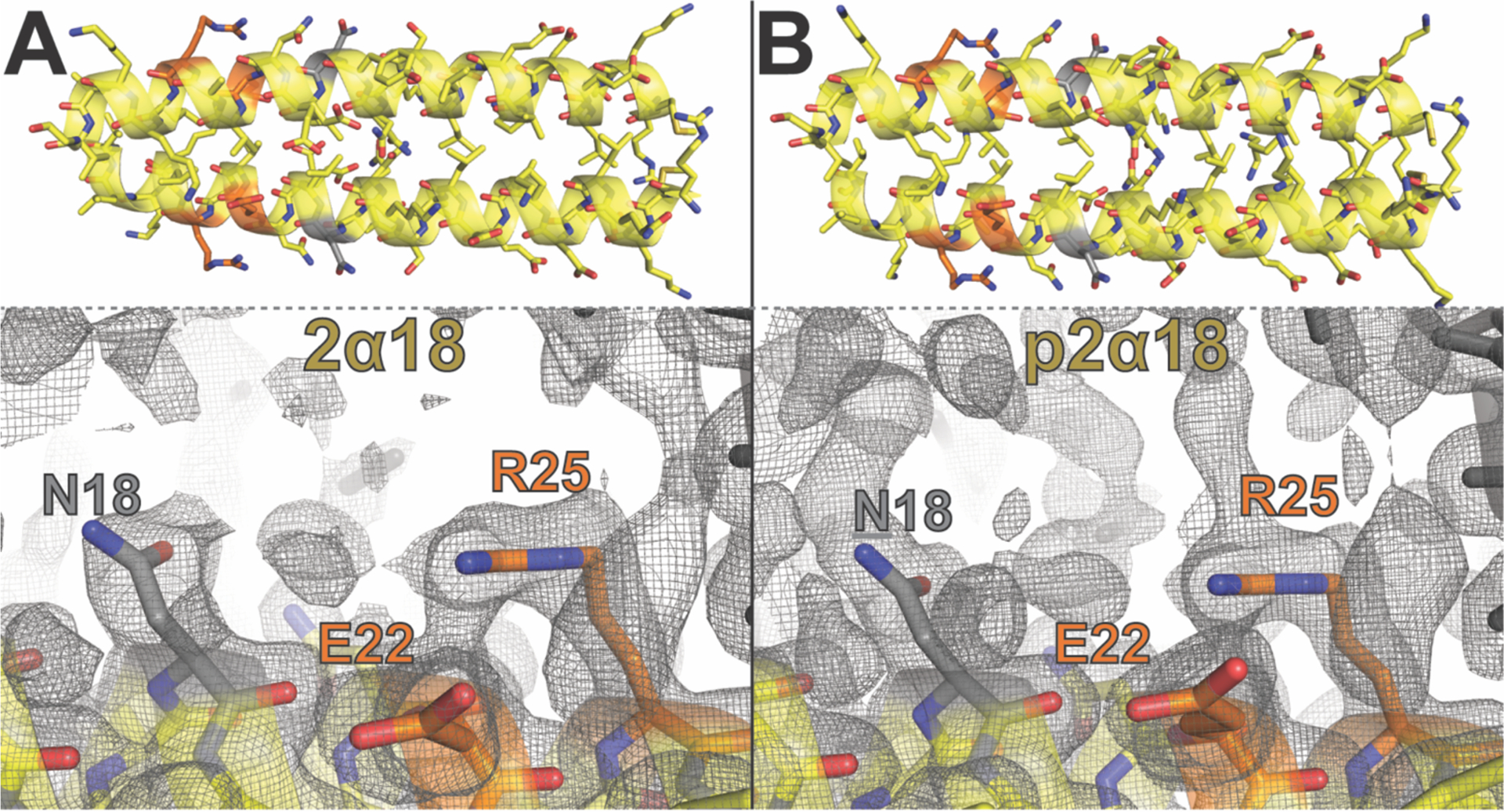
(A)–(B) Ribbon diagram of α-helical coiled-coil dimers formed by GCN4-p1 variant 2α18 (PDB ID: 6O2E) and its PEGylated counterpart p2α18 (PDB ID: 6O2F), respectively, with side chains shown as sticks. Asn18 is highlighted in gray, with the Glu22-Arg25 salt bridge highlighted in orange. Also shown are close up views of Asn18, Glu22, and Arg25 within a single helix (yellow) within the crystalline lattices formed by variants 2α18 and p2α18, respectively. Gray mesh represents electron density contoured at 0.5 σ.
Here we have shown that a short PEG oligomer can increase protein conformational stability by increasing the strength of a nearby salt bridge in two distinct secondary structural contexts, including an i to i+4 Glu-Arg salt bridge within an α-helix and an intra-strand i to i+2 Glu-Arg salt bridge within a β-sheet. High-resolution crystallography data supports the possibility that PEG adopts a disordered conformation near the Glu22–Arg25 salt bridge within GCN4 variant p2α18, occupying space that would normally be occupied by water. It is possible that this effect is partially responsible for the PEG-based stabilization of the Glu22–Arg25 salt bridge. However, the structural prerequisites for this effect are not a simple function of secondary structural context, orientation and distance of the PEGylation site with respect to the salt bridge, or of salt-bridge residue identity; PEGylation did not increase the strength of some salt-bridges, for reasons that remain unclear. Previous work showed that the structure of the PEG-protein linkage is a major determinant of PEG-based stabilization at a given PEGylation site.23, 24 Moreover, a short PEG oligomer comprised of three ethylene oxide units can almost entirely recapitulate the PEG-based stabilization associated with a longer 45-unit PEG.20, 21 These observations suggest that the impact of PEG-based protein stabilization derives primarily from the PEG-protein linker and the first few atoms of the attached PEG, which perturb the microenvironment in the immediate vicinity of the PEGylation site, including nearby side-chain and backbone groups, along with water molecules in the first solvent shell). If so, the minimal impact of PEG on the stability of nearby salt bridges within p2α4, p3α1, p3α6, and pβ18 could reflect our choice of suboptimal linkers at these positions (AsnPEG, GlnPEG, GlnPEG, and AsnPEG, respectively) vs. serendipitously optimal linkers at corresponding positions within p2α18 and pβ23 (AsnPEG and PrFPEG, respectively). More broadly, our results also highlight the importance of considering the extent to which side-chain functionalization (e.g. with a fluorophore, an affinity tag, a post-translational modification, etc.) perturbs interactions with and among water molecules and side-chain or backbone groups in the localized microenvironment and hydration sphere of the conjugation site.
Methods
Peptide Synthesis
Peptides were synthesized via solid phase synthesis using a standard Fmoc strategy, described in detail in the Supporting Information. Fmoc-protected and PEGylated derivatives of l-asparagine and l -glutamine were synthesized as reported previously.21, 23 Fmoc-protected propargyloxyphenylalanine (PrF) was synthesized as reported previously.37 A previously described PEG-azide was conjugated to PrF via copper(I)-catalyzed azide-alkyne cycloaddition as we described recently.21, 23 Following cleaving from resin, peptides were purified by HPLC with a reverse phase C18 column and a linear gradient of water and acetonitrile with 0.1% v/v TFA. The mass of each peptide was confirmed by matrix-assisted laser desorption/ionization time-of-flight spectrometry, and purity was evaluated by analytical HPLC. Mass spectra and analytical HPLC chromatograms for all peptides are available in the supporting information.
CD Measurements
CD spectra and variable temperature CD data were collected using an Aviv 420 spectropolarimeter. We used non-linear regression to fit the variable temperature CD data to equations derived from a two-state folding model for each variant to obtain Tm and ΔGf values for each peptide, as described in detail the Supporting Information.
Supplementary Material
References
- 1.Veronese FM, (Ed.) (2009) PEGylated Protein Drugs: Basic Sience and Clinical Applications, Birkhauser Verlag, Basel. [Google Scholar]
- 2.Frokjaer S, and Otzen DE (2005) Protein drug stability: a formulation challenge, Nat. Rev. Drug. Discov 4, 298–306. [DOI] [PubMed] [Google Scholar]
- 3.Abuchowski A, Mccoy JR, Palczuk NC, Vanes T, and Davis FF (1977) Effect of Covalent Attachment of Polyethylene-Glycol on Immunogenicity and Circulating Life of Bovine Liver Catalase, J. Biol. Chem 252, 3582–3586. [PubMed] [Google Scholar]
- 4.Abuchowski A, Vanes T, Palczuk NC, and Davis FF (1977) Alteration of Immunological Properties of Bovine Serum-Albumin by Covalent Attachment of Polyethylene-Glycol, J. Biol. Chem 252, 3578–3581. [PubMed] [Google Scholar]
- 5.Harris JM, Martin NE, and Modi M (2001) Pegylation - A novel process for modifying pharmacokinetics, Clin. Pharmacokinet 40, 539–551. [DOI] [PubMed] [Google Scholar]
- 6.Harris JM, and Chess RB (2003) Effect of pegylation on pharmaceuticals, Nat. Rev. Drug Discov 2, 214–221. [DOI] [PubMed] [Google Scholar]
- 7.Hermanson GT (1996) Bioconjugate Techniques, Academic Press, San Diego. [Google Scholar]
- 8.Nischan N, and Hackenberger CPR (2014) Site-specific PEGylation of Proteins: Recent Developments, J. Org. Chem 79, 10727–10733. [DOI] [PubMed] [Google Scholar]
- 9.Jan C. M. v. H., Kristi LK, and David AT. (2000) Efficient Incorporation of Unsaturated Methionine Analogues into Proteins in Vivo, J Am Chem Soc 122. [Google Scholar]
- 10.Wang L, Zhang ZW, Brock A, and Schultz PG (2003) Addition of the keto functional group to the genetic code of Escherichia coli, P Natl Acad Sci USA 100, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deiters A, Cropp TA, Summerer D, Mukherji M, and Schultz PG (2004) Site-specific PEGylation of proteins containing unnatural amino acids, Bioorg. Med. Chem. Lett 14, 5743–5745. [DOI] [PubMed] [Google Scholar]
- 12.Doherty D, Rosendahl M, Smith D, Hughes J, Chlipala E, and Cox G (2005) Site-specific PEGylation of engineered cysteine analogues of recombinant human granulocyte-macrophage colony-stimulating factor, Bioconjugate Chem 16, 1291–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosendahl MS, Doherty DH, Smith DJ, Carlson SJ, Chlipala EA, and Cox GN (2005) A Long-Acting, Highly Potent Interferon α-2 Conjugate Created Using Site-Specific PEGylation, Bioconjugate Chem. 16, 200–207. [DOI] [PubMed] [Google Scholar]
- 14.Soderquist RG, Milligan ED, Sloane EM, Harrison JA, Douvas KK, Potter JM, Hughes TS, Chavez RA, Johnson K, Watkins LR, and Mahoney MJ (2009) PEGylation of brain-derived neurotrophic factor for preserved biological activity and enhanced spinal cord distribution, J. Biomed. Mater. Res. A 91, 719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shozen N, Iijima I, and Hohsaka T (2009) Site-specific incorporation of PEGylated amino acids into proteins using nonnatural amino acid mutagenesis, Bioorg. Med. Chem. Lett 19, 4909–4911. [DOI] [PubMed] [Google Scholar]
- 16.Cho H, Daniel T, Buechler YJ, Litzinger DC, Maio Z, Putnam AM, Kraynov VS, Sim BC, Bussell S, Javahishvili T, Kaphle S, Viramontes G, Ong M, Chu S, Becky GC, Lieu R, Knudsen N, Castiglioni P, Norman TC, Axelrod DW, Hoffman AR, Schultz PG, DiMarchi RD, and Kimmel BE (2011) Optimized clinical performance of growth hormone with an expanded genetic code, Proc. Natl. Acad. Sci. USA 108, 9060–9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, Lu YF, Fang ZZ, Liu JX, Tian H, Gao XD, and Yao WB (2011) High-level production of uricase containing keto functional groups for site-specific PEGylation, Biochem Eng J 58–59, 25–32. [Google Scholar]
- 18.Chalker JM, Lercher L, Rose NR, Schofield CJ, and Davis BG (2012) Conversion of Cysteine into Dehydroalanine Enables Access to Synthetic Histones Bearing Diverse Post-Translational Modifications, Angew. Chem. Int. Ed 51, 1835–1839. [DOI] [PubMed] [Google Scholar]
- 19.Tada S, Andou T, Suzuki T, Dohmae N, Kobatake E, and Ito Y (2012) Genetic PEGylation, Plos One 7, e49235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandey BK, Smith MS, Torgerson C, Lawrence PB, Matthews SS, Watkins E, Groves ML, Prigozhin MB, and Price JL (2013) Impact of Site-Specific PEGylation on the Conformational Stability and Folding Rate of the Pin WW Domain Depends Strongly on PEG Oligomer Length, Bioconjugate Chem. 24, 796–802. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence PB, Gavrilov Y, Matthews SS, Langlois MI, Shental-Bechor D, Greenblatt HM, Pandey BK, Smith MS, Paxman R, Torgerson CD, Merrell JP, Ritz CC, Prigozhin MB, Levy Y, and Price JL (2014) Criteria for Selecting PEGylation Sites on Proteins for Higher Thermodynamic and Proteolytic Stability, J. Am. Chem. Soc 136, 17547–17560. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence PB, and Price JL (2016) How PEGylation influences protein conformational stability, Curr. Opin. Chem. Biol 34, 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence PB, Billings WM, Miller MB, Pandey BK, Stephens AR, Langlois MI, and Price JL (2016) Conjugation Strategy Strongly Impacts the Conformational Stability of a PEG-Protein Conjugate, ACS Chem. Biol 11, 1805–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Draper SRE, Lawrence PB, Billings WM, Xiao Q, Brown NP, Becar NA, Matheson DJ, Stephens AR, and Price JL (2017) Polyethylene Glycol Based Changes to beta-Sheet Protein Conformational and Proteolytic Stability Depend on Conjugation Strategy and Location, Bioconjugate Chem. 28, 2507–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pluharova E, Marsalek O, Schmidt B, and Jungwirth P (2012) Peptide salt bridge stability: From gas phase via microhydration to bulk water simulations, J. Chem. Phys 137, 185101. [DOI] [PubMed] [Google Scholar]
- 26.Hendsch ZS, and Tidor B (1994) Do Salt Bridges Stabilize Proteins - a Continuum Electrostatic Analysis, Protein Sci. 3, 211–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takano K, Tsuchimori K, Yamagata Y, and Yutani K (2000) Contribution of salt bridges near the surface of a protein to the conformational stability, Biochemistry 39, 12375–12381. [DOI] [PubMed] [Google Scholar]
- 28.Efimov AV, and Brazhnikov EV (2003) Relationship between intramolecular hydrogen bonding and solvent accessibility of side-chain donors and acceptors in proteins, FEBS Lett. 554, 389–393. [DOI] [PubMed] [Google Scholar]
- 29.Gao J, Bosco DA, Powers ET, and Kelly JW (2009) Localized thermodynamic coupling between hydrogen bonding and microenvironment polarity substantially stabilizes proteins, Nat. Struct. Mol. Biol 16, 684–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Shea EK, Klemm JD, Kim PS, and Alber T (1991) X-Ray Structure of the Gcn4 Leucine Zipper, a 2-Stranded, Parallel Coiled Coil, Science 254, 539–544. [DOI] [PubMed] [Google Scholar]
- 31.Woolfson DN (2005) The design of coiled-coil structures and assemblies, Adv. Protein Chem 70, 79–112. [DOI] [PubMed] [Google Scholar]
- 32.Ogihara NL, Weiss MS, Degrado WF, and Eisenberg D (1997) The crystal structure of the designed trimeric coiled coil coil-V(a)L(d): Implications for engineering crystals and supramolecular assemblies, Protein Sci. 6, 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandey BK, Smith MS, and Price JL (2014) Cys(i)-Lys(i+3)-Lys(i+4) Triad: A General Approach for PEG-Based Stabilization of alpha-Helical Proteins, Biomacromolecules 15, 4643–4647. [DOI] [PubMed] [Google Scholar]
- 34.Smith MS, Billings WM, Whitby FG, Miller MB, and Price JL (2017) Enhancing a long-range salt bridge with intermediate aromatic and nonpolar amino acids, Org. Biomol. Chem 15, 5882–5886. [DOI] [PubMed] [Google Scholar]
- 35.Ranganathan R, Lu KP, Hunter T, and Noel JP (1997) Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent, Cell 89, 875–886. [DOI] [PubMed] [Google Scholar]
- 36.Jäger M, Zhang Y, Bieschke J, Nguyen H, Dendle M, Bowman ME, Noel JP, Gruebele M, and Kelly JW (2006) Structure-function-folding relationship in a WW domain, Proc. Natl. Acad. Sci. USA 103, 10648–10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deiters A, Cropp TA, Mukherji M, Chin JW, Anderson JC, and Schultz PG (2003) Adding Amino Acids with Novel Reactivity to the Genetic Code of Saccharomyces Cerevisiae, J. Am. Chem. Soc 125, 11782–11783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


