Abstract
Nervous necrosis virus (NNV), the causative agent of viral encephalopathy and retinopathy (VER), is one of the most threatening viruses affecting marine and freshwater fish species worldwide. Senegalese sole is a promising fish species in Mediterranean aquaculture but also highly susceptible to NNV and VER outbreaks, that puts its farming at risk. The development of vaccines for aquaculture is one of best tools to prevent viral spread and sudden outbreaks, and virus inactivation is the simplest and most cost-effective method available. In this work, we have designed two inactivated vaccines based on the use of formalin or binary ethylenimine (BEI) to inactivate a reassortant NNV strain. After vaccination, the BEI-inactivated vaccine triggered the production of specific IgM-NNV antibodies and stimulated innate and adaptive immune responses at transcriptional level (rtp3, mx, mhcii and tcrb coding genes). Moreover, it partially improved survival after an NNV in vivo challenge, reducing the mid-term viral load and avoiding the down-regulation of immune response post-challenge. On the other hand, the formalin-inactivated vaccine improved the survival of fish upon infection without inducing the production of IgM-NNV antibodies and only stimulating the expression of herc4 and mhcii genes (in head-kidney and brain, respectively) during the vaccination period; this suggests that other immune-related pathways may be involved in the partial protection provoked. Although these vaccines against NNV showed encouraging results, further studies are needed to improve sole protection and to fully understand the underlying immune mechanism.
Keywords: Senegalese sole, inactivated vaccine, BEI, formalin, nervous necrosis virus, immune response, antibodies, gene expression
1. Introduction
Aquaculture is one of the fastest growing sectors of the food industry worldwide. Nowadays, global aquaculture has surpassed marine catches in terms of fish protein production for human consumption [1]. As a result of this growth, the spread of viruses has also risen, due to the constant transport of eggs and larvae between fish farms all over the world and because survivors of an epizootic outbreak often become asymptomatic carriers.
One of most widespread and dangerous viruses among fish farms is the nervous necrosis virus (NNV; Genus Betanodavirus, Family Nodaviridae). NNV is the causative agent of viral encephalopathy and retinopathy (VER), a neuropathological condition which provokes mortality rates of up to 100%, particularly at early stages of development (larvae and juveniles), especially in marine fish [2]. NNV is an icosahedral non-enveloped virus composed of two positive sense single stranded RNA segments—RNA 1 coding for the non-structural RNA-dependent RNA-polymerase, and RNA2 coding for the capsid protein. NNV is grouped into four different genotypes: red-spotted grouper nervous necrosis virus (RGNNV), barfin flounder nervous necrosis virus, tiger puffer nervous necrosis virus and striped jack nervous necrosis virus (SJNNV) [2]. In addition, natural reassortants between RGNNV and SJNNV genotypes have been isolated as the causative agents of disease outbreaks in different species, including sole (Solea sp.) and gilthead seabream (Sparus aurata), the latter previously thought to be an asymptomatic NNV carrier [3,4,5,6].
To date, the most common preventive strategies regarding husbandry and NNV-free broodstock selection appear to be inefficient at avoiding NNV outbreaks on fish farms due to the stability of the virus in the aquatic environment [2]. Therefore, vaccination stands as a cost-effective, innocuous and sustainable strategy to prevent NNV infections and severe episodes. Different strategies have been followed to design NNV vaccines, including live or inactivated viruses, DNA, virus-like particles, virus subunits, viral synthetic peptides or recombinant protein using diverse administration methods [7,8,9,10,11,12,13,14]. Although the immune response was triggered by most of them, they were only partially protective. As a first approach to inactivated dead recombinant bacteria, a spinycterin system expressing a downsized NNV coat protein was reported to induce 100% of sea bass (Dicentrarchus labrax) survival upon NNV infection [15]. However, the existence of contradictory data and the limited number of fish species on which vaccines have been tested make it difficult to release an effective and wide-spectrum vaccine against NNV onto the market. In fact, only two commercial formalin-inactivated vaccines against the RGNNV genotype, ALPHA JECT micro® 1Noda (Pharmaq) and ICTHIOVAC® VNN (Hipra), are available for sea bass vaccination in the Mediterranean area.
Senegalese sole (Solea senegalensis) is a highly promising flatfish species for aquaculture in Southern-European countries. However, one of the main drawbacks for its intensive farming is its high susceptibility to reassortant RGNNV/SJNNV (referring to the RNA1/RNA2 of the donors) strains [2]. In this work, a reassortant, sole pathogenic NNV strain has been used to design two inactivated vaccine formulations (using either formalin or binary ethylenimine, BEI). Juvenile sole individuals were intraperitoneally vaccinated and the antibody response and expression profiles of immune-related genes of vaccinated fish was assessed. In addition, fish survival upon NNV infection and the immunostimulation of challenged sole was also analyzed.
2. Materials and Methods
2.1. Viruses and Cells
The strain used in this study, SpSs-IAusc160.03 (hereafter Ss160), a reassortant strain exhibiting an RNA1 typed as RGNNV and an SJNNV-type RNA2, was isolated from Senegalese sole (Solea senegalensis) during an acute disease outbreak in a rearing facility in Spain in 2003 [3]. E-11 cell line was cultured in L-15 Leibovitz (Lonza) medium supplemented with 5% fetal bovine serum (FBS), penicillin (100 IU/mL) and streptomycin (100 mg/mL) at 25 °C. For viral propagation, cells were inoculated at an MOI = 0.1 and incubated in L-15 with 2% FBS at 25 °C until the cytopathic effect (CPE) was extensive. The supernatant was harvested and centrifuged to eliminate cell debris. Virus stock was titrated in 96-well plates and expressed as the viral dilution infecting 50% of the cell cultures (TCID50), following a previously defined methodology [16].
2.2. Vaccine Inactivation
Either formalin or binary ethylenimine (BEI) was used to inactivate Ss160 (iSs160; 108.5 TCID50/mL). Formalin inactivation was accomplished by adding the reagent into the viral suspension to a final concentration of 0.2% during 7 or 9 days (d) at 25 °C in agitation. BEI inactivation was achieved by mixing the virus with freshly prepared 0.1 M BEI dissolved in 0.175 M NaOH to a final concentration of 0.1, 1 or 4 mM. Inactivation was tested after 1, 24, 48 or 72 h of inactivation at 25 °C in agitation. After incubation, 1 M sodium thiosulphate was added (1:10 in relation to BEI). Vaccines were confirmed to be completely inactivated by the absence of CPE and viral titer in the E-11 cell line, after three 10-day blind passages, as elsewhere [3]. In addition, in vivo toxicity was ruled out by intraperitoneal injection (ip) in sole juveniles.
2.3. Animals and Sampling Procedure
A total of 920 healthy Senegalese sole juveniles (2.9 ± 0.1 g body weight) from a local farm were transferred to the Universidade de Santiago de Compostela (Spain) aquarium facilities. Fish were randomly divided into 300 L running seawater tanks (33‰ salinity) at 19 °C and with a 12 h light: 12 h dark photoperiod and acclimatized for 15 days prior to the experiments. During acclimatization, fish were tested for the presence of NNV by RT-qPCR (see below). Twenty fish were reserved to be mock-vaccinated and -infected until the end of the trials.
Sampled fish were analyzed as follows: blood was collected from the caudal peduncles and serum samples were obtained by centrifugation at 10,000× g for 10 min at 4 °C and immediately stored at −20 °C until use. Afterwards fish were sacrificed with a tricaine methanesulfonate overdose (MS-222, Sigma-Aldrich, St. Louis, MO, USA). Head-kidney and brain were removed and immediately stored at −80 °C until later use for RT-qPCR assays and cell culture when applicable.
2.4. Fish Vaccination
Senegalese sole specimens were randomly divided into ten 100 L tanks (n = 90/tank), forming five experimental groups in duplicate. Fish were gently sedated with MS-222 and intraperitoneally (ip) vaccinated as follows: Control group (Control) was injected with PBS (100 μL/fish) while vaccinated groups received a single ip injection of Ss160 inactivated with either formalin (form-iSs160) or BEI (BEI-iSs160) with high and low dose, 105 (iSs160L) and 107 TCID50/mL (iSs160H), respectively. After vaccination, fish (n = 6 fish/group and time point) were sampled at 7-, 15-, and 30-days post-vaccination (dpv) as described above.
2.5. NNV Challenge
Thirty days after vaccination, the remaining fish were challenged by immersion and exposed to an Ss160 concentration of 105 TCID50/mL for 3 h with strong aeration (dissolved oxygen concentration 11 ± 1 mg/L) at 22 °C. Control fish were mock-infected with L-15 medium. Mortalities and clinical signs were recorded daily. Viral load quantification in brain tissue was performed on fish (n = 3 fish/group) sampled at 12-day intervals from the initial detection of VER signs (see below). After 45 days of infection, serum, head-kidney and brain samples from surviving fish (n = 6 fish/group) were analyzed.
2.6. Specific and Neutralizing Antibody Levels
The detection of specific antibodies against NNV (IgM-NNV) was performed by a previously described indirect ELISA with slight modifications [17]. Briefly, 20 μg of total proteins from serum samples were diluted in coating buffer [100 mM Bicarbonate/Carbonate, pH 9.6] and incubated overnight at 4 °C in 96 High Binding flat-bottomed plates (Sarsted, Newton, NC, USA). After washing with PBST [PBS with 0.2% of Tween-20], the samples were blocked with 5% skimmed milk in PBST for 1 h. Afterwards, incubation with a rabbit anti-NNV (Abcam, Inc., Hong Kong; 1:10,000) was performed for 1 h at room temperature. After three 5-min washes, the samples were incubated with the anti-rabbit IgG-HRP (Sigma Aldrich; 1:25,000) for 1 h at room temperature. The reaction was revealed with 100 μL per well of 3,3′,5,5′-tetramethylbenzidine single solution (ThermoFisher, Waltham, MA, USA) for 20 min and stopped with 50 μL of 2 M sulphuric acid. The absorbance was read at 450 nm with an iMark™ Microplate Absorbance Reader (BioRad, Heracles, CA, USA). All assays were performed in duplicate. Previously assayed positive serum was used as a positive control whilst the absence of sample, NNV, primary or secondary antibodies were used as negative controls.
Neutralizing antibodies were assayed as elsewhere [14], with few modifications. In brief, sera from fish obtained during vaccination and after 45 days post-challenge (dpc) were decomplemented at 56 °C during 30 min. Serial dilutions (from 10 to 640-fold) of decomplemented sera were incubated with equal volumes of 102.5 TCID50 mL of Ss160 for 1 h at 25 °C. After incubation, samples were assayed for NNV replication on E-11 cells as above, and serum dilution to provoke the absence of CPE was determined. A serum from an infected fish served as a positive control.
2.7. Gene Expression by Real-Time Polymerase Chain Reaction
Total RNA was isolated from head-kidney and brain using an EZNA Total RNA purification Kit (VWR) following the manufacturer’s instructions. The RNA samples were resuspended in 70 µL of nuclease-free water (VWR), quantified by absorbance at 260 nm in a Nanodrop ND-100 spectrophotometer (Nanodrop Technologies) and stored at −80 °C. The first strand of cDNA was synthetized using Superscript IV (ThermoFisher) with Random Hexameres (ThermoFisher) as previously described [18].
The expression of sole immune-related genes, namely the receptor transporter protein (rtp3), E3 ubiquitin-protein ligase (herc4), interferon-induced GTP-binding protein Mx (mx), major histocompatibility complex II (mhcii) and T-cell receptor beta (tcrb), were analyzed by RT-qPCR, in an iCycler iQ CFX96TM Real Time System (BioRad) following the manufacturer’s instructions using iQTM SYBR® Green Supermix (BioRad). Reaction mixtures (containing 20 μL of SYBR Green supermix with 0.2 μM of the specific primers and 2 μL of cDNA template) were incubated for 3 min at 95 °C as an activation/denaturation step, followed by 40 cycles of 15 s at 95 °C and 30 s at 55 (mhcii, mx and rtp3) or 58 °C (tcrb and herc4). The specific primers used are shown in Table 1. Negative controls with no template were always included in the reactions. The relative expression of all genes was calculated by the 2−ΔΔCt method [19] using the beta actin (actb) coding gene as the endogenous reference.
Table 1.
Primer sequences used for gene expression analysis.
| Protein Name | Gene Name | Accession Number, UniGen Name or Reference | Sequence (5′–3′) | ||
|---|---|---|---|---|---|
| Nodavirus | RNA-dependent RNA polymerase | RNA1 | FJ803911 | F | TCCAAAAGAAAGAAGCATAC |
| R | TGGCATGTACCACGGAAC | ||||
| Senegalese sole | Receptor transporter protein | rtp3 | [20] | F | GACGCCCCAATGGTGGAT |
| R | CCAGATTCTTCATGAGGATGGTGAT | ||||
| E3 ubiquitin-protein ligase | herc4 | [20] | F | GCCAAAACACTGGCATGGTT | |
| R | AACGCCAAACAGGAAGTACCT | ||||
| Interferon-induced GTP-binding protein Mx | mx | [20] | F | CCTCTCTCCTTCAGGATCCTCCTCCTGTGC | |
| R | CAAAACAAGAAACTATCTGCCTGGTGGTTC | ||||
| Major histocompatibility complex II | mhcii | [20] | F | CGCTGATGAAAATGATCCACCTTCT | |
| R | ACCAGTCACATGACAGATCAGAGT | ||||
| T-cell receptor beta chain | tcrb | solea_v4.1_unigene681812 * | F | CAGGAGGCACAGCTATGAAA | |
| R | TCTCCACCCAAATCTCCAAA | ||||
| Beta actin | actb | DQ485686 | F | GACGACATGGAGAAGATC | |
| R | GGTGTTGAAGGTCTCAAA | ||||
* NOTE: UniGen transcriptomic database in http://www.scbi.uma.es/soleadb (accessed on 21 February 2021).
2.8. Viral Quantification
Betanodavirus RNA1 extraction and amplification was accomplished as described above using SnodR1 primers (Table 1). The corresponding standard curve was prepared using 20-fold dilutions of a plasmid containing the full-length RNA1 of strain Ss160 in the range of 101 to 107 copies/μL. Viral load data were calculated as RNA1 copies per g of fish tissue. All samples were tested in triplicate.
2.9. Calculations and Statistics
All the data are shown as the mean ± standard error of the mean (SEM). The data corresponding to the mRNA transcriptional levels was expressed as the relative gene expression of the control or vaccinated group. IgM-NNV results are described as the optical density (OD) at 450 nanometers. The relative percent survival (RPS) was calculated from the cumulative mortality by the following formula:
| (1) |
Variations between different time points and groups were analyzed by a two-way ANOVA followed by Tukey’s post-hoc analysis. A non-parametric Kruskal-Wallis test, followed by a multiple comparison test, was used when data did not meet parametric assumptions. Survival rates were compared between groups using the Kaplan Meier test. Letters denote statistical differences between groups at a same time-point (p < 0.05), whilst asterisks indicate those differences between time-points in the same group (* p < 0.05; ** p < 0.01; **** p < 0.0001). All statistical analyses were conducted using GraphPad Prism 8.
3. Results
Prior to the vaccination assays, fish were tested by RT-qPCR and were confirmed to be free of NNV (data not shown). In addition, five fish per vaccine formulation were ip injected and monitored for 48 h before the beginning of the vaccination experiments to rule out the toxicity of the inactivated vaccines. A control group with mock-vaccinated and infected specimens was maintained until the end of the experiments as tank-effect control.
3.1. Formalin and BEI Effectively Inactivated Ss160
The effectivity of inactivation was tested using different chemical concentrations and incubation times (Table 2). Complete formalin inactivation was achieved after nine days of incubation since neither CPE nor viral titer were recorded after three blind passages (Table 2). In the case of BEI, inactivation was accomplished with 1 mM after 72 or 4 mM at any time point tested (Table 2). Based on these results, a form-iSs160 vaccine (0.2% formalin for nine days) and a BEI-iSs160 vaccine (1 mM BEI for 72 h) were chosen for in vivo vaccination assays. After 7, 15 and 30 dpv, NNV was not recovered in cell culture from the brain of vaccinated sole (three brains individually, data not shown) corroborating the complete inactivation of the vaccines chosen.
Table 2.
Safety test of form- and BEI-iSs160 vaccines after different concentration of chemical compounds treatments and incubation periods.
| Chemical Compound | Concentration | Time | CPE under Each Blind Passage | Titer after Virus Inactivation(Log10 TCID50/mL) | ||||
|---|---|---|---|---|---|---|---|---|
| % | mM | Days | Hours | 1 | 2 | 3 | ||
| Formalin | 0.2 | 7 | − | − | + | nt | ||
| 9 | − | − | − | <1.0 | ||||
| BEI | 0.1 | 1 | + | nt | nt | nt | ||
| 24 | + | nt | nt | nt | ||||
| 48 | + | nt | nt | nt | ||||
| 72 | + | nt | nt | nt | ||||
| 1 | 1 | + | + | nt | nt | |||
| 24 | + | + | nt | nt | ||||
| 48 | − | − | + | nt | ||||
| 72 | − | − | − | <1.0 | ||||
| 4 | 1 | − | − | − | <1.0 | |||
| 24 | − | − | − | <1.0 | ||||
| 48 | − | − | − | <1.0 | ||||
| 72 | − | − | − | <1.0 | ||||
NOTE: −: negative; +: positive; nt: non-tested.
3.2. BEI-iSs160 Vaccine Induces the Production of Specific NNV-IgM
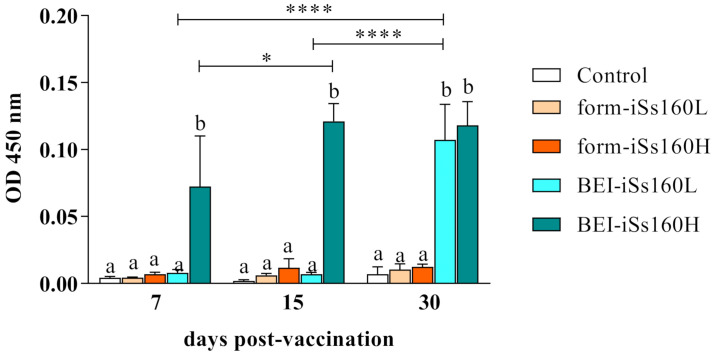
After 7, 15 and 30 dpv, the production of specific antibodies against NNV was studied (Figure 1). Only the BEI-iSs160 vaccine induced the synthesis of NNV-IgM from 7 dpv onwards or after 30 dpv (iSs160H or iSs160L, respectively). Moreover, this increase was significant with time in both BEI-iSs160 vaccinated groups, although no neutralizing activity was observed (data not shown). On the contrary, form-iSs160 vaccines did not provoke the production of specific antibodies at any time tested.
Figure 1.
BEI-iSs160 vaccine induces specific humoral immunity. Specific anti-NNV IgM levels in the serum of Senegalese sole specimens 7-, 15- and 30-days after intraperitoneal vaccination with low and high dosages (105 and 107 TCID50/mL, iSs160L and iSs160H, respectively) of BEI-iSs160 vaccine (inactivated with BEI) and form-iSs160 (formalin inactivated) or PBS (Control). Data represent the mean ± standard error of the mean (SEM; n = 6 fish/group and time point). Lower letters denote statistical differences between groups at a same time-point (p < 0.05), whilst asterisks indicate those between time-points in the same group (* p < 0.05; **** p < 0.0001).
3.3. Immune-Related Gene Transcription in Vaccinated Fish
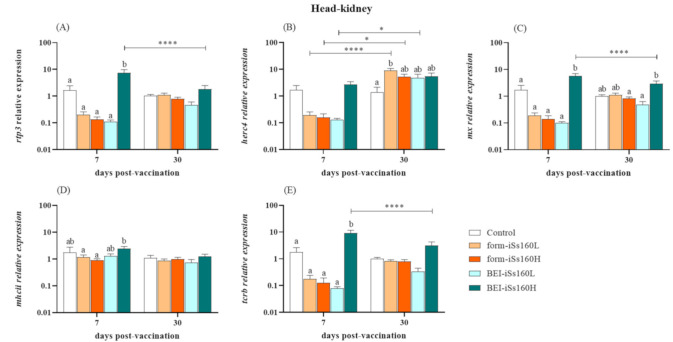
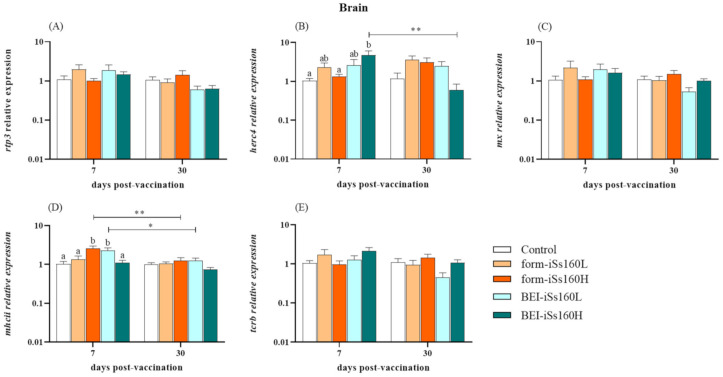
At the gene expression level, most of the studied immune-related markers were modulated in the head-kidney of vaccinated fish, whilst in the brain they were barely altered (Figure 2 and Figure 3). Thus, in head-kidney, an overexpression of rpt3, mx and tcrb genes was observed in the BEI-iSs160H group compared to the controls and the other vaccinated groups after 7 dpv. However, a significant decrease down to control levels was observed after 30 dpv in the transcription of the three genes (Figure 2A,C,E). Regarding the fish vaccinated with form-iSs160, only the form-iSs160L triggered increased transcriptional levels of herc4 at 30 dpv (Figure 2B). Finally, although mhcii gene expression was unaltered in vaccinated groups compared to the controls, at 7 dpv in the BEI-iSs160H group, up-regulated levels were observed when compared with formalin vaccinated fish (Figure 2D). In brain tissue, only a few changes were seen at the short time point (7 dpv; Figure 3). Thus, herc4 gene expression was increased in the BEI-iSs160H group whilst mhcii was increased in the form-iSs160H and BEI-iSs160L groups (Figure 3D). In all cases, gene expression returned to control levels after 30 dpv (Figure 3B,D).
Figure 2.
Vaccination produces the increment of immune response at transcriptional level in head-kidney. Expression of immune-related genes (A) rtp3, (B) herc4, (C) mx, (D) mhcii and (E) tcrb in the head-kidney of Senegalese sole specimens 7-, 15- and 3-days after intraperitoneal vaccination with low and high dosages (105 and 107 TCID50/mL, iSs160L and iSs160H, respectively) of BEI-iSs160 vaccine (inactivated with BEI) and form-iSs160 (formalin inactivated) or PBS (Control). Data represent the mean ± standard error of the mean (SEM; n = 6 fish/group and time). Lower letters denote statistical differences between groups at a same time-point (p < 0.05), whilst asterisks indicate those between time-points in the same group (* p < 0.05; **** p < 0.0001).
Figure 3.
Vaccination barely alters the immune response at transcriptional level in brain. Expression of immune-related genes (A) rtp3, (B) herc4, (C) mx, (D) mhcii and (E) tcrb in the brain of Senegalese sole specimens 7-, 15- and 30-days after intraperitoneal vaccination with low and high dosages (105 and107 TCID50/mL, iSs160L and iSs160H, respectively) of BEI-iSs160 vaccine (inactivated with BEI) and form-iSs160 (formalin inactivated) or PBS (Control). Data represent the mean ± standard error of the mean (SEM; n = 6 fish/group and time). Lower letters denote statistical differences between groups at a same time-point (p < 0.05), whilst asterisks indicate those between time-points in the same group (* p < 0.05; ** p < 0.01).
3.4. BEI-iSs160 Vaccine Improves Survival and Reduces Viral Load in Sole Brain after Challenge
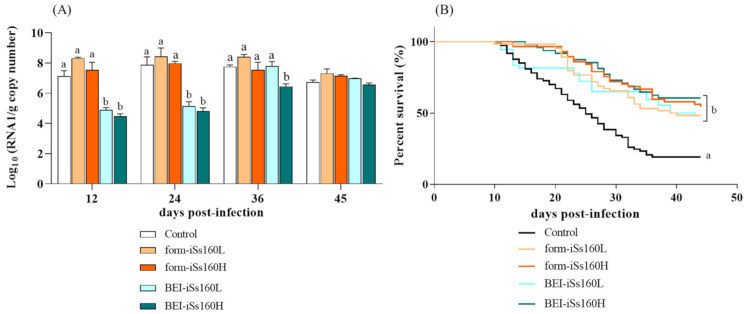
At 30 dpv, fish were bath-challenged with the Ss160 strain and at 12 dpc, control fish started to show typical symptoms of VER disease such as erratic swimming or changes in skin color (data not shown), whereas no external disease signs were observed in the vaccinated fish. Therefore, from this time point and at 12-day intervals the viral load in the fish brain tissue (n = 3 fish/group and time point) was individually assessed. From 12–24 dpc, in the BEI-vaccinated groups the viral load was significantly lower (8.82 × 104–1.98 × 105 and 3.46 × 104–7.99 × 104 RNA copies per gram in the BEI-iSs160L and BEI-iSs160H groups, respectively) than in the control fish (2.71 × 108 and 7.91 × 107 copies) (Figure 4A). However, at 36 dpc viral RNA copies only remained low in the BEI-iSs160H group (3.3 × 106 versus 6.2 × 107 in the control) and at the end of the challenge (45 dpc) no significant differences were found between vaccinated and control fish. No variations with the control group were observed in both form-iSs160 vaccinated groups at the time-point assayed (Figure 4A).
Figure 4.
All vaccines induce partial protection and BEI-iSs160 decreases the viral load after challenge. (A) viral load (n = 3 fish/group and time) in brain and (B) Percent of survival during 45 days of in vivo infection in intraperitoneally vaccinated Senegalese sole specimens vaccinated with low and high dosages (105 and 107 TCID50/mL, iSs160L and iSs160H, respectively) of BEI-iSs160 vaccine (inactivated with BEI) and form-iSs160 (formalin inactivated) or PBS (Control). Infection was performed by 3 h of immersion with 107 TCID50/mL. Survival rates were compared between groups using Kaplan Meier test. Lower letters denote statistical differences between groups of the same time point according to the two-way ANOVA test (p < 0.05).
After 45 dpc, a significant improvement of survival was observed with all vaccines tested (Figure 4B), although the best results were obtained with the high concentrations. Thus, whereas in the control group only 19.2% of the individuals survived, form- and BEI-iSs160H groups displayed survival values of 54.38 and 60.42%, respectively (RPS 43.7 and 51.0). Low vaccine concentrations showed very similar RPS values (35.8 and 36.2, BEI- and form-iSs160L, respectively).
3.5. NNV Infection Induces the Production of Specific NNV-IgM Similarly in All Fish
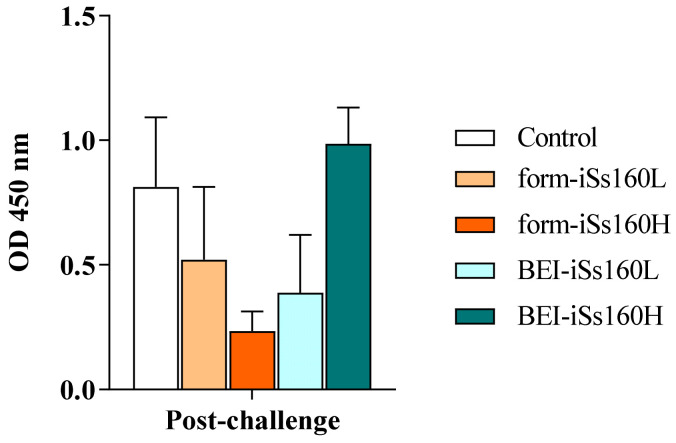
The level of specific NNV-IgM was analyzed 45 dpc (Figure 5). The in vivo infection elicited the production of specific antibodies in vaccinated and control fish with no statistical differences between groups. In addition, no NNV-neutralizing activity was recorded (data not shown).
Figure 5.
Vaccination does not alter specific humoral immunity upon NNV infection. Specific anti-NNV IgM levels in the serum of Senegalese sole specimens 45 days after 3 h of immersion with 105 TCID50/mL of Ss160 in previously intraperitoneally vaccinated Senegalese sole specimens vaccinated with low and high dosages (105 and 107 TCID50/mL, iSs160L and iSs160H, respectively) of BEI-iSs160 vaccine (inactivated with BEI) and form-iSs160 (formalin inactivated) or PBS (Control). Infection was performed by 3 h of immersion with 107 TCID50/mL. Data represent the mean ± standard error of the mean (SEM; n = 6 fish/group and time). Statistical assay was performed by the two-way ANOVA test (p < 0.05).
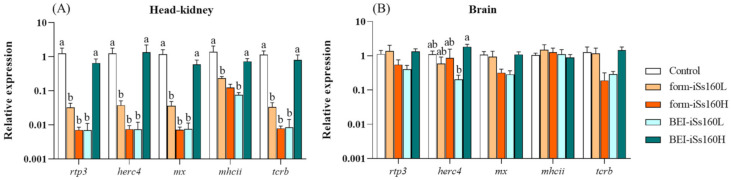
3.6. Immune-Related Genes in Head-Kidney after NNV Challenge
At 45 dpc, the analyzed genes were down-regulated in the head-kidney of all fish treated with the formalin-inactivated vaccine. This down-regulation was also observed in the BEI-iSs160L group, whereas it remained unaltered in the BEI-iSs160H vaccinated fish (Figure 6A). In the brain, the mRNA transcriptomic levels of the studied immune-related genes were unaltered in all vaccinated fish when compared with the control group (Figure 6B). However, comparison among vaccinated groups showed that herc4 gene expression was significantly higher in the BEI-iSs160H than in the BEI-iSs160L group (Figure 6B).
Figure 6.
Vaccination mainly decreases the immune response upon NNV infection at a transcriptional level in head-kidney but not in brain. Expression of immune-related genes in (A) head-kidney and (B) brain of Senegalese sole specimens 45 days after 3 h of immersion with 105 TCID50/mL of Ss160 in fish previously vaccinated with PBS (Control) or with low and high dosages (105 and 107 TCID50/mL, iSs160L and iSs160H, respectively) of BEI-iSs160 vaccine (inactivated with BEI) and form-iSs160 (formalin inactivated) or PBS (Control). Data represent the mean ± standard error of the mean (SEM; n = 6 fish/group and time). Lower letters denote statistical differences between groups according to the two-way ANOVA test (p < 0.05).
4. Discussion
Although Senegalese sole is a promising species for Mediterranean aquaculture that shows extraordinary susceptibility to NNV [18,21], there is only one study describing an experimental recombinant NNV vaccine for this fish species which induced a good antibody response but whose protective effect was not assessed [22]. Therefore, aiming to prevent VER disease in sole, and considering that the simplicity of the production method makes the inactivated vaccines one of the most practical, we have designed a BEI- and a formalin-inactivated vaccine using the sole-pathogenic reassortant strain, Ss160.
The main concept linked to the inactivation of viruses for vaccine production is to generate totally safe vaccines, with the complete blockage of viral replication, without damaging its antigenicity [23]. Our first observation was that the complete inactivation of NNV was achieved with 0.2% formalin or 1 mM BEI after 9 days or 72 h of incubation, respectively. Previously, NNV has shown a certain resistance to be inactivated by low concentrations of formalin (up to 0.16%), needing concentrations of at least 0.5% or higher [24,25,26,27]. In the case of BEI, a previous concentration used to block NNV replication was 4 mM [7,26,28,29]. Consumers today tend to be increasingly concerned about the origin and the composition of the food they eat, so it may be worth reducing any chemical compounds used in vaccination strategies, as done in this work, to avoid a generalized rejection of the product but ensuring, at the same time, that NNV is completely inactivated with no possibility of being spread into the wild.
Our post-vaccination results showed the abrupt induction of specific IgM-NNV production in BEI-iSs160 vaccinated fish, in a concentration dependent manner, although they were not neutralizing. Similarly, other BEI inactivated NNV vaccines for grouper strains triggered the synthesis of IgM-NNV, but they also possessed neutralizing activity [28,29]. On the contrary, fish immunized with the form-iSs160 vaccine did not elicit an antibody response. Most of the formalin inactivated vaccines against NNV reported in the literature reveal a significant increment in neutralizing antibodies, regardless of whether the sera are decomplemented or not [26,27,30,31,32,33,34,35,36]. However, formalin-inactivated vaccines appear to be dose dependent, since dosages of around 107 TCID50/fish are necessary to provoke an effective production of specific NNV antibodies in several fish species [27,31,32,33,34,35,36]. As a matter of fact, the minimum vaccine concentration to successfully generate neutralizing antibodies in groupers is 107 TCID50/fish [36]. Therefore, it is reasonable to assume that the lack of antibody production observed in sole is due to the low vaccine concentration used in this study (106 and 104 TCID50/fish).
To the best of our knowledge, this is the first study addressing the immune response at a transcriptional level of sole immunized with NNV inactivated vaccines. Interestingly, the transcription of rtp3, mx, herc4 and tcrb (all of them demonstrated to be up-regulated in NNV-infected S. sole [20]) was considerably induced in the head-kidney of fish vaccinated with the BEI-iSs160H, whereas in brain tissue it was barely altered. Thus, the gene expression of rtp3, a virus-responsive gene [37], was increased in the kidney soon after vaccination (7 dpv) although the values were similar to the control fish at 30 dpv. This gene was also greatly up-regulated in the head-kidney and brain of sole infected with the reassortant Ss160 strain [20], just like in the brain and in several organs of NNV-infected Atlantic salmon (Salmo salar) and Asian seabass (Lates calcarifer), respectively [37,38]. The rtp3 gene shows an extraordinary sensitivity to NNV since its up-regulation is highly stimulated soon after infection, appearing to be linked to the resistance to infection [38,39]. This information, taken together with our results, points to Rtp3 as a relevant protein in the fight against NNV. However, further studies are needed to clarify its specific function. In a similar manner, the mx gene expression peaked after 7 dpv, perfectly simulating a natural NNV infection. As a matter of fact, mx gene stimulation has usually been used as an indicator of the vaccine’s antiviral effectiveness due to the significant induction of the type I interferon (IFN) route—the most powerful antiviral pathway—in fish infected with NNV [20,40,41,42,43,44]. This induction seems to be regardless of the administration method, given that other tested BEI-vaccines showed a suitable mx gene up-regulation during the vaccination period, when administered either by bath or intraperitoneal injection [7,28]. Finally, the BEI-vaccine seems to organize complete adaptive cellular immunity in head-kidney because tcrb (a marker of cytotoxic T cells subsets) and mhcii (marker of T helper cells route activation via antigen presenting cells) genes were up-regulated 7 days after vaccination, as seen in groupers [7,28], concomitantly with the induction of specific IgM-NNV production observed. It has previously been described how T cells were activated upon in vivo infection with NNV [41,45,46,47] and even after vaccination with UV-inactivated, recombinant or DNA vaccines [8,14,48], as with the BEI-iSs160H vaccine. Exceptionally, the herc4 gene was the only gene up-regulated in the brain after BEI vaccination but not in head-kidney. Although its function in fish is still unclear, the herc4 gene is up-regulated in the head-kidney and brain of infected susceptible fish whilst inhibited in asymptomatic carriers of NNV [20,49]. In addition, this molecule seems to be relevant in antiviral responses, as the viral mimic poly (I:C) in macrophages induced an increment of its transcriptional levels [50]. Regarding the formalin-inactivated vaccines, only herc4 was overexpressed in the head-kidney of fish injected with the form-iSs160L, whereas mhcii transcriptional levels were increased in the brain of fish vaccinated with the form-iSs160H. Several formalin inactivated vaccines have been reported to confer protection against NNV in different fish species [26,27,31,32,33,34,51], but immune-related gene expression was only studied in European sea bass [30]. In agreement with our results, mx was not significantly overexpressed in kidneys although a significant increase was observed in the gut at 48 h post-vaccination [31].
Thirty days after vaccination, fish were bath-challenged. Although mortalities appeared approximately at the same time as in most of the groups, only control fish showed VER disease signs, such as erratic swimming and alterations in the skin color. The absence of symptomatology coincided with the improvement in the survival of vaccinated fish, especially those injected with BEI-iSs160H, which showed an RPS value of 51, whereas survival of control fish was only 19.2%. Our results agree with those found in groupers of different ages, where BEI-inactivated NNV triggered partial survival rates ranging between 52–67% in early juveniles when ip challenged [28]. However, no differences were shown in the production of specific antibodies at the end of the challenge, contrary to what happens in groupers immunized either with formalin- or BEI-inactivated vaccines [27,28,32,36]. In fact, in this fish species, a potent humoral response against NNV has been associated with the improvement in the survival of vaccinated fish [32]. Our results indicate, therefore, that cellular immunity can play an important role in the partial protection achieved in sole, and that the form-iSs160 vaccine could induce additional defense mechanisms to those analyzed in this study. Regarding survival rates provoked by formalin-inactivated vaccines against NNV, there are many controversial results in which survival rates range from 10.1–100% with an unclear common pattern [26,27,31,32,35,36]. Many variables may be affecting the differential effectivity of this type of vaccine, such as the viral strain used, the formalin dosage and inactivation period, the vaccine dosage or the virus load used in the challenge. However, the vaccine dosage appears to be the most plausible explanation as it is determinant for triggering neutralizing antibodies. Nevertheless, further studies might be necessary to fully understand the conditioning factors for the effectivity of this type of vaccine.
After the challenge, we observed a generalized down-regulation of all the genes studied in the head-kidney of fish vaccinated with form-iSs160 (both concentrations). However, the BEI-iSs160H was the only one maintaining transcriptional levels of immune-related genes with no inhibition. This may be relevant given that NNV has been reported to eradicate the innate immune response during the first moments of infection to freely spread and colonize target tissues [40,52]. Finally, and despite the similarities in the survival rates found in all groups, it is worth noting that BEI-iSs160H, in addition to maintaining the transcriptional levels of immune-related genes, provoked a significant decrease in the viral load in the brain tissue of challenged fish from 12–36 dpc, coinciding with the onset of disease signs (day 12 post challenge) and the period of higher mortalities in control fish (20–30 dpc).
5. Conclusions
NNV inactivated vaccines generated in this study conferred partial protection to Senegalese sole when administered by ip injection, although they induced a different immune response. On the one hand, BEI-iSs160 generates the innate (type I interferon pathway, rtp3) and adaptive (specific IgM-NNV production, T cell markers) immune responses which last for at least 30 days. This vaccine improves sole survival upon NNV infection by decreasing the viral load in brain tissue and maintaining their immune response unaltered until the end of the challenge. The consistency of the results obtained with BEI inactivation ([28] and this study) suggest that the reassortant strain Ss160 inactivated with BEI has the potential to be used for developing vaccines which generate a robust immune response in S. sole. However, further experimental approaches are needed to improve this promising vaccine, including either booster immunizations or adjuvants. On the other hand, form-iSs160 decreases the mortality rates upon NNV infection even when a scarce immune stimulation was observed, pointing to the fact that different immune-related pathways are involved.
Acknowledgments
Y.V. would like to thank the Ministerio de Ciencia, Innovación y Universidades (MICIU) for her Juan de la Cierva Formación contract. Authors also thank J.F.-A. and A.A.S.-M. for their technical support during all the experiments and to Stolt Sea Farm S.A. for providing the Senegalese sole fish.
Author Contributions
Y.V., J.G.O. and C.L.-V. performed the trials, the sampling, and the experimental analysis; Y.V., C.L.-V. and I.B. performed the data analysis and presentation; Y.V. and I.B. wrote the manuscript; I.B. and C.P.D. conceived and supported the study. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grant RTI2018-094687-B-C21 from MICIU (Spain) co-funded by FEDER.
Institutional Review Board Statement
Fish were handled in strict accordance with good animal practices as defined by EU guidelines (directive 2010/63/UE). The protocol was approved by the Bioethics and Experimental Animal Welfare Committees of the University of Santiago de Compostela and by the Xunta de Galicia (Permit Id. 15010/2020/004).
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.FAO . The State of World Fisheries and Aquaculture. Opportunities and Challenges. Food and Agriculture Organization of the United Nations; Rome, Italy: 2012. [Google Scholar]
- 2.Bandín I., Souto S. Betanodavirus and VER disease: A 30-year research review. Pathogens. 2020;9:106. doi: 10.3390/pathogens9020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olveira J.G., Souto S., Dopazo C.P., Thiéry R., Barja J.L., Bandín I. Comparative analysis of both genomic segments of betanodaviruses isolated from epizootic outbreaks in farmed fish species provides evidence for genetic reassortment. J. Gen. Virol. 2009;90:2940–2951. doi: 10.1099/vir.0.013912-0. [DOI] [PubMed] [Google Scholar]
- 4.Volpe E., Gustinelli A., Caffara M., Errani F., Quaglio F., Fioravanti M.L., Ciulli S. Viral nervous necrosis outbreaks caused by the RGNNV/SJNNV reassortant betanodavirus in gilthead sea bream (Sparus aurata) and European sea bass (Dicentrarchus labrax) Aquaculture. 2020;523:735155. doi: 10.1016/j.aquaculture.2020.735155. [DOI] [Google Scholar]
- 5.Panzarin V., Fusaro A., Monne I., Cappellozza E., Patarnello P., Bovo G., Capua I., Holmes E.C., Cattoli G. Molecular epidemiology and evolutionary dynamics of betanodavirus in southern Europe. Infect. Genet. Evol. 2012;12:63–70. doi: 10.1016/j.meegid.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Toffan A., Pascoli F., Pretto T., Panzarin V., Abbadi M., Buratin A., Quartesan R., Gijón D., Padros F. Viral nervous necrosis in gilthead sea bream (Sparus aurata) caused by reassortant betanodavirus RGNNV/SJNNV: An emerging threat for Mediterranean aquaculture. Sci. Rep. 2017;2:46755. doi: 10.1038/srep46755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kai Y.-H., Wu Y.-C., Chi S.-C. Immune gene expressions in grouper larvae (Epinephelus coioides) induced by bath and oral vaccinations with inactivated betanodavirus. Fish Shellfish Immunol. 2014;40:563–569. doi: 10.1016/j.fsi.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Valero Y., Mokrani D., Chaves-Pozo E., Arizcun M., Oumouna M., Meseguer J., Esteban M.Á., Cuesta A. Vaccination with UV-inactivated nodavirus partly protects European sea bass against infection, while inducing few changes in immunity. Dev. Comp. Immunol. 2018;86:171–179. doi: 10.1016/j.dci.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Chien M.-H., Wu S.-Y., Lin C.-H. Oral immunization with cell-free self-assembly virus-like particles against orange-spotted grouper nervous necrosis virus in grouper larvae, Epinephelus coioides. Vet. Immunol. Immunopathol. 2018;197:69–75. doi: 10.1016/j.vetimm.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Coeurdacier J.-L., Laporte F., Pepin J.-F. Preliminary approach to find synthetic peptides from nodavirus capsid potentially protective against sea bass viral encephalopathy and retinopathy. Fish Shellfish Immunol. 2003;14:435–447. doi: 10.1006/fsim.2002.0449. [DOI] [PubMed] [Google Scholar]
- 11.Kim J.-O., Kim W.-S., Oh M.-J. Development of a Recombinant Protein Vaccine Based on Cell-Free Protein Synthesis for Sevenband Grouper Epinephelus septemfasciatus Against Viral Nervous Necrosis. J. Microbiol. Biotechnol. 2015;25:1761–1767. doi: 10.4014/jmb.1507.07004. [DOI] [PubMed] [Google Scholar]
- 12.González-Silvera D., Guardiola F.A., Espinosa C., Chaves-Pozo E., Esteban M.Á., Cuesta A. Recombinant nodavirus vaccine produced in bacteria and administered without purification elicits humoral immunity and protects European sea bass against infection. Fish Shellfish Immunol. 2019;88:458–463. doi: 10.1016/j.fsi.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Lin C.-F., Jiang H.-K., Chen N.-C., Wang T.-Y., Chen T.-Y. Novel subunit vaccine with linear array epitope protect giant grouper against nervous necrosis virus infection. Fish Shellfish Immunol. 2018;74:551–558. doi: 10.1016/j.fsi.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 14.Valero Y., Awad E., Buonocore F., Arizcun M., Esteban M.Á., Meseguer J., Chaves-Pozo E., Cuesta A. An oral chitosan DNA vaccine against nodavirus improves transcription of cell-mediated cytotoxicity and interferon genes in the European sea bass juveniles gut and survival upon infection. Dev. Comp. Immunol. 2016;65:64–72. doi: 10.1016/j.dci.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 15.Lama R., Pereiro P., Novoa B., Coll J. Sea bass immunization to downsize the betanodavirus protein displayed in the surface of inactivated repair-less bacteria. Vaccines. 2019;7:19. doi: 10.3390/vaccines7030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reed L.J., Müench H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 17.Gye H.J., Nishizawa T. Reducing background optical density in enzyme-linked immunosorbent assay for detecting nervous necrosis virus (NNV)-specific IgM by immobilizing fish sera. Aquaculture. 2018;485:93–100. doi: 10.1016/j.aquaculture.2017.11.039. [DOI] [Google Scholar]
- 18.Souto S., Olveira J.G., Bandín I. Influence of temperature on Betanodavirus infection in Senegalese sole (Solea senegalensis) Vet. Microbiol. 2015;179:162–167. doi: 10.1016/j.vetmic.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gémez-Mata J., Labella A.M., Bandín I., Borrego J.J., García-Rosado E. Immunogene expression analysis in betanodavirus infected-Senegalese sole using an OpenArray® platform. Gene. 2021;774:145430. doi: 10.1016/j.gene.2021.145430. [DOI] [PubMed] [Google Scholar]
- 21.Souto S., López-Jimena B., Alonso M.C., García-Rosado E., Bandín I. Experimental susceptibility of European sea bass and Senegalese sole to different betanodavirus isolates. Vet. Microbiol. 2015;177:53–61. doi: 10.1016/j.vetmic.2015.02.030. [DOI] [PubMed] [Google Scholar]
- 22.Thwaite R., Berbel C., Aparicio M., Torrealba D., Pesarrodona M., Villaverde A., Borrego J.J., Manchado M., Roher N. Nanostructured recombinant protein particles raise specific antibodies against the nodavirus NNV coat protein in sole. Fish Shellfish Immunol. 2020;99:578–586. doi: 10.1016/j.fsi.2020.02.029. [DOI] [PubMed] [Google Scholar]
- 23.Delrue I., Verzele D., Madder A., Nauwynck H.J. Inactivated virus vaccines from chemistry to prophylaxis: Merits, risks and challenges. Expert Rev. Vaccines. 2014;11:695–719. doi: 10.1586/erv.12.38. [DOI] [PubMed] [Google Scholar]
- 24.Arimoto M., Sato J., Maruyama K., Mimura G., Furusawa I. Effect of chemical and physical treatments on the inactivation of striped jack nervous necrosis virus (SJNNV) Aquaculture. 1996;143:15–22. doi: 10.1016/0044-8486(96)01261-6. [DOI] [Google Scholar]
- 25.Frerichs G.N., Tweedie A., Starkey W.G., Richards R.H. Temperature, pH and electrolyte sensitivity, and heat, UV and disinfectant inactivation of sea bass (Dicentrarchus labrax) neuropathy nodavirus. Aquaculture. 2000;185:13–24. doi: 10.1016/S0044-8486(99)00337-3. [DOI] [Google Scholar]
- 26.Kai Y.H., Chi S.C. Efficacies of inactivated vaccines against Betanodavirus in grouper larvae (Epinephelus coioides) by bath immunization. Vaccine. 2008;26:1450–1457. doi: 10.1016/j.vaccine.2007.12.043. [DOI] [PubMed] [Google Scholar]
- 27.Yamashita H., Fujita Y., Kawakami H., Nakai T. The efficacy of inactivated virus vaccine against viral nervous necrosis (VNN) Fish Pathol. 2005;40:15–21. doi: 10.3147/jsfp.40.15. [DOI] [Google Scholar]
- 28.Cheng Y.K., Wu Y.C., Chi S.C. Humoral and cytokine responses in giant groupers after vaccination and challenge with betanodavirus. Dev. Comp. Immunol. 2017;67:385–394. doi: 10.1016/j.dci.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 29.Kai Y.H., Su H.M., Tai K.T., Chi S.C. Vaccination of grouper broodfish (Epinephelus tukula) reduces the risk of vertical transmission by nervous necrosis virus. Vaccine. 2010;28:996–1001. doi: 10.1016/j.vaccine.2009.10.132. [DOI] [PubMed] [Google Scholar]
- 30.Huang S.M., Cheng J.H., Tu C., Chen T.N., Lin C.T., Chang S.K. A bivalent inactivated vaccine of viral nervous necrosis virus and grouper iridovirus applied to grouper broodfish (Epinephelus coioides) reduces the risk of vertical transmission. Taiwan Vet. J. 2017;43:1–6. doi: 10.1142/S1682648517500032. [DOI] [Google Scholar]
- 31.Núñez-Ortiz N., Pascoli F., Picchietti S., Buonocore F., Bernini C., Toson M., Scapigliati G., Toffan A. A formalin-inactivated immunogen against viral encephalopathy and retinopathy (VER) disease in European sea bass (Dicentrarchus labrax): Immunological and protection effects. Vet. Res. 2016;47:89. doi: 10.1186/s13567-016-0376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pakingking R., Bautista N.B., de Jesus-Ayson E.G., Reyes O. Protective immunity against viral nervous necrosis (VNN) in brown-marbled grouper (Epinephelus fuscogutattus) following vaccination with inactivated betanodavirus. Fish Shellfish Immunol. 2010;28:525–533. doi: 10.1016/j.fsi.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 33.Pakingking R., de Jesus-Ayson E.G., Reyes O., Brian Bautista N. Immunization regimen in Asian sea bass (Lates calcarifer) broodfish: A practical strategy to control vertical transmission of nervous necrosis virus during seed production. Vaccine. 2018;36:5002–5009. doi: 10.1016/j.vaccine.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 34.Pakingking R., Seron R., dela Peña L., Mori K., Yamashita H., Nakai T. Immune responses of Asian sea bass, Lates calcarifer Bloch, against an inactivated betanodavirus vaccine. J. Fish Dis. 2009;32:457–463. doi: 10.1111/j.1365-2761.2009.01040.x. [DOI] [PubMed] [Google Scholar]
- 35.Pascoli F., Guazzo A., Buratin A., Toson M., Buonocore F., Scapigliati G., Toffan A. Lack of in vivo cross-protection of two different betanodavirus species RGNNV and SJNNV in European sea bass Dicentrachus Labrax. Fish Shellfish Immunol. 2019;85:85–89. doi: 10.1016/j.fsi.2017.10.033. [DOI] [PubMed] [Google Scholar]
- 36.Yamashita H., Mori K., Kuroda A., Nakai T. Neutralizing antibody levels for protection against betanodavirus infection in sevenband grouper, Epinephelus septemfasciatus (Thunberg), immunized with an inactivated virus vaccine. J. Fish Dis. 2009;32:767–775. doi: 10.1111/j.1365-2761.2009.01054.x. [DOI] [PubMed] [Google Scholar]
- 37.Krasnov A., Timmerhaus G., Schiøtz B.L., Torgersen J., Afanasyev S., Iliev D., Jørgensen J., Takle H., Jørgensen S.M. Genomic survey of early responses to viruses in Atlantic salmon, Salmo salar L. Mol. Immunol. 2011;49:163–174. doi: 10.1016/j.molimm.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Liu P., Wang L., Ye B.Q., Huang S., Wong S.-M., Yue G.H. Characterization of a novel disease resistance gene rtp3 and its association with VNN disease resistance in Asian seabass. Fish Shellfish Immunol. 2017;61:61–67. doi: 10.1016/j.fsi.2016.12.021. [DOI] [PubMed] [Google Scholar]
- 39.Liu P., Wang L., Kwang J., Yue G.H., Wong S.-M. Transcriptome analysis of genes responding to NNV infection in Asian seabass epithelial cells. Fish Shellfish Immunol. 2016;54:342–352. doi: 10.1016/j.fsi.2016.04.029. [DOI] [PubMed] [Google Scholar]
- 40.Valero Y., Morcillo P., Meseguer J., Buonocore F., Esteban M.Á., Chaves-Pozo E., Cuesta A. Characterization of the interferon pathway in the teleost fish gonad against the vertically transmitted viral nervous necrosis virus. J. Gen. Virol. 2015;96:2176–2187. doi: 10.1099/vir.0.000164. [DOI] [PubMed] [Google Scholar]
- 41.Valero Y., Boughlala B., Arizcun M., Patel S., Fiksdal I.U., Esteban M.Á., De Juan J., Meseguer J., Chaves-Pozo E., Cuesta A. Genes related to cell-mediated cytotoxicity and interferon response are induced in the retina of European sea bass upon intravitreal infection with nodavirus. Fish Shellfish Immunol. 2018;74:627–636. doi: 10.1016/j.fsi.2018.01.034. [DOI] [PubMed] [Google Scholar]
- 42.Carballo C., García-Rosado E., Borrego J.J., Alonso M.C. SJNNV down-regulates RGNNV replication in European sea bass by the induction of the type I interferon system. Vet. Res. 2016;47:6. doi: 10.1186/s13567-015-0304-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poisa-Beiro L., Dios S., Montes A., Aranguren R., Figueras A., Novoa B. Nodavirus increases the expression of Mx and inflammatory cytokines in fish brain. Mol. Immunol. 2008;45:218–225. doi: 10.1016/j.molimm.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 44.Álvarez-Torres D., Podadera A.M., Alonso M.C., Bandín I., Béjar J., García-Rosado E. Molecular characterization and expression analyses of the Solea senegalensis interferon-stimulated gene 15 (isg15) following NNV infections. Fish Shellfish Immunol. 2017;66:423–432. doi: 10.1016/j.fsi.2017.05.040. [DOI] [PubMed] [Google Scholar]
- 45.Øvergård A.C., Nerland A.H., Fiksdal I.U., Patel S. Atlantic halibut experimentally infected with nodavirus shows increased levels of T-cell marker and IFNgamma transcripts. Dev. Comp. Immunol. 2012;37:139–150. doi: 10.1016/j.dci.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 46.González-Fernández C., Esteban M.Á., Cuesta A. Molecular characterization of the T cell costimulatory receptors CD28 and CTLA4 in the European sea bass. Fish Shellfish Immunol. 2021;109:106–115. doi: 10.1016/j.fsi.2020.12.006. [DOI] [PubMed] [Google Scholar]
- 47.López-Muñoz A., Sepulcre M.P., García-Moreno D., Fuentes I., Béjar J., Manchado M., Álvarez M.C., Meseguer J., Mulero V. Viral nervous necrosis virus persistently replicates in the central nervous system of asymptomatic gilthead seabream and promotes a transient inflammatory response followed by the infiltration of IgM+ B lymphocytes. Dev. Comp. Immunol. 2012;37:429–437. doi: 10.1016/j.dci.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 48.Øvergård A.C., Patel S., Nøstbakken O.J., Nerland A.H. Atlantic halibut (Hippoglossus hippoglossus L.) T-cell and cytokine response after vaccination and challenge with nodavirus. Vaccine. 2013;31:2395–2402. doi: 10.1016/j.vaccine.2013.01.034. [DOI] [PubMed] [Google Scholar]
- 49.Rise M.L., Hall J.R., Rise M., Hori T.S., Browne M.J., Gamperl A.K., Hubert S., Kimball J., Bowman S., Johnson S.C. Impact of asymptomatic nodavirus carrier state and intraperitoneal viral mimic injection on brain transcript expression in Atlantic cod (Gadus morhua) Physiol. Genom. 2010;42:266–280. doi: 10.1152/physiolgenomics.00168.2009. [DOI] [PubMed] [Google Scholar]
- 50.Eslamloo K., Xue X., Booman M., Smith N.C., Rise M.L. Transcriptome profiling of the antiviral immune response in Atlantic cod macrophages. Dev. Comp. Immunol. 2016;63:187–205. doi: 10.1016/j.dci.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka S., Mori K., Arimoto M., Iwamoto T., Nakai T. Protective immunity of sevenband grouper, Epinephelus septemfasciatus Thunberg, against experimental viral nervous necrosis. J. Fish Dis. 2001;24:15–22. doi: 10.1046/j.1365-2761.2001.00259.x. [DOI] [Google Scholar]
- 52.Qin Y., Wang Y., Liu J., Lu Y., Liu X. Red-grouper nervous necrosis virus B2 protein negatively regulates fish interferon response by suppressing host transcription directed by RNA polymerase II. Aquaculture. 2021;536:736488. doi: 10.1016/j.aquaculture.2021.736488. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.