Abstract
Mitophagy is a selective form of autophagy by which dysfunctional and damaged mitochondria are degraded in autolysosomes. Since defective mitophagy is closely related to various pathological processes, investigation on the detailed mitophagy process is of great importance. In this respect, disclosing the alterations of mitochondrial microenvironments is expected to be a promising way. However, an appropriate method for monitoring the fluctuations of mitochondrial polarity during mitophagy is still lacking. Here, we report a near-infrared hydroxyl-hemicyanine fluorescent probe that responds to polarity exclusively. Both the shift of emission maxima and the fluorescence intensity ratios at two different wavelengths of the probe can be applied to quantifying the polarity accurately. With ratiometric fluorescence imaging, the polarity differences of normal and cancer cells are clearly discriminated. Most importantly, the mitochondrial polarity variations during starvation and drug-induced mitophagy are determined for the first time. The observed decrease of mitochondrial polarity during mitophagy, together with the rationally designed probe, may facilitate the study on the vital role of mitophagy in physiological and pathological bioprocesses.
A near-infrared mitochondria-targeting fluorescent probe has been developed to monitor the mitophagy-specific polarity dynamics in living cells.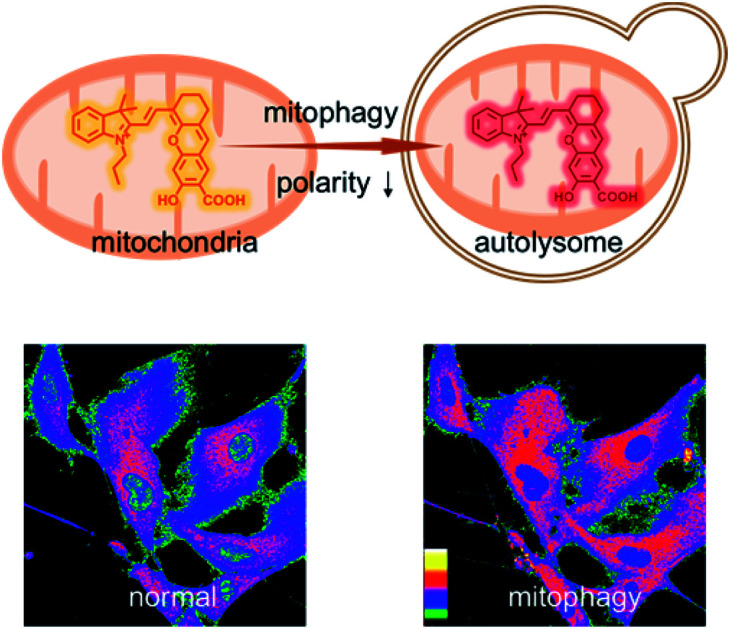
Introduction
Mitochondria are essential organelles that regulate cellular energy metabolism and cell death.1 Starvation, hypoxia or other stress stimulations may lead to mitochondrial oxidative damage and dysfunction, which results in several pathological processes, including aging, apoptosis, and cellular injury.2 The quality and quantity control of mitochondria through a process called mitophagy is thus critical for maintaining proper cellular functions.3 Indeed, mitophagy has been proposed to play critical roles in terminal differentiation of red blood cells,4 paternal mitochondrial degradation,5 neurodegenerative diseases,6 ischemia and drug-induced tissue injury.7 Therefore, monitoring the detailed mitophagy process is of great importance. Several methods have been developed for mitophagy-related detection, including transmission electron microscopy, immunoblot for mitochondrial proteins, citrate synthase activity assay, etc.8 However, these classical approaches are time-consuming, costly, and unable to provide real-time information regarding mitophagy in living cells. A simple and cost-effective method for the precise and vivid visualization of mitophagy dynamics is still urgently needed.
It is known that during mitophagy, mitochondria are first encircled into double-membrane vesicles called autophagosomes and then delivered into lysosomes to form autolysosomes for bulk degradation.1a Owing to the obvious differences of the microenvironments [e.g., polarity (ε), pH and viscosity (η)] between mitochondria and autophagosomes, detecting the microenvironment changes may be an efficient way for monitoring the mitophagy process. In this respect, small molecular fluorescent probes have attracted much attention due to their high sensitivity, good biocompatibility and incomparable spatiotemporal resolution.9 Several small molecular fluorescent probes have been designed for the detection of mitochondrial pH and viscosity changes, revealing a decreased pH and an increased viscosity during mitophagy.10 However, the polarity changes in this process are still unclear. Considering the massive formation of isolation membranes (autophagosomes, etc.),11 it may be reasonable to speculate that the mitochondrial polarity tends to decrease during mitophagy.
Herein, to test our hypothesis, we synthesized a mitochondria-targeting near-infrared fluorescent probe HXPI-P which displays red shifts in the fluorescence emission peaks as the medium polarity decreases (Fig. 1A). Though bearing a hydroxyl-hemicyanine skeleton, HXPI-P is insensitive to pH and viscosity variations. By analyzing the spectra of the probe, we show that the shift of emission maxima is linearly related to the polarity, and the fluorescence intensity ratios at the wavelengths of 730 nm and 670 nm versus the dielectric constant follow a Lorentz function. Therefore, the quantitative measurement of the media polarity values (ε) could be achieved accurately. HXPI-P is successfully utilized to discern the polarity difference between normal and cancer cells. Most importantly, the mitochondrial polarity fluctuations during mitophagy are monitored in real time, which has never been achieved before. This probe is thus expected to be helpful to visualize the mitophagy procedure by detecting the changes of mitochondrial polarity.
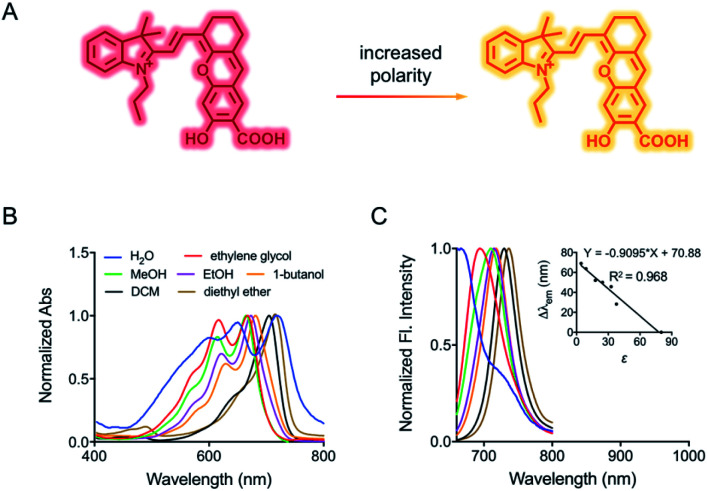
Fig. 1. (A) Fluorescence response of HXPI-P to polarity. (B) Normalized absorption and corresponding (C) fluorescence emission spectra of HXPI-P (10 μM) in solvents with different polarities. Inset: the linear fitting curve of Δλemvs. the dielectric constant. λex = 635 nm. Δλem = λem (in an organic solvent) − λem (in water). Data are expressed as the mean ± SD of three separate measurements.
Results and discussion
Solvent effects on the spectroscopic properties of HXPI-P
The spectroscopic behavior of HXPI-P was studied in various solvents covering a wide range of polarities such as water (ε = 80.1), ethylene glycol (ε = 37.7), methanol (MeOH, ε = 32.7), ethanol (EtOH, ε = 24.6), 1-butanol (ε = 17.8), dichloromethane (DCM, ε = 8.93) and diethyl ether (ε = 4.34). As shown in Fig. 1B and C, both the normalized absorption and emission maxima of HXPI-P display obvious polarity-dependent behavior. Moreover, the probe exhibits red shifts in the fluorescence emission wavelength as the solvent polarity decreases (Fig. 1C). Meanwhile, a good linear relationship between Δλem (shift of the maximum emission wavelength) and the dielectric constant over the range of 4.36–80.1 can be obtained (Fig. 1C), indicating the potential of the probe for quantitative measurements of polarity. Notably, HXPI-P shows a near-infrared emission in all solvents, which is beneficial to reduce the biological damage and the interference of autofluorescence from biological matrices. The detailed photo-physical properties of HXPI-P in various solvents are summarized in Table S1.†
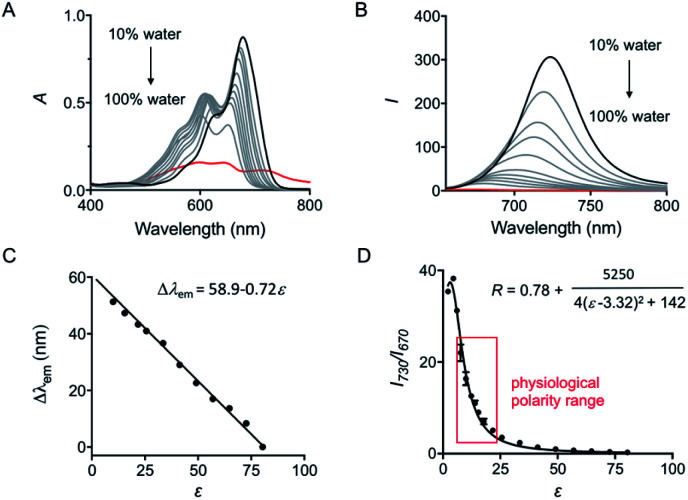
To accurately analyze the relationship between spectroscopic properties and solvent polarity, the absorption and emission spectra of HXPI-P were collected in the H2O–1,4-dioxane mixture system since 1,4-dioxane could be the best mimic of the structures of bio-membranes among other organic solvents. The polarity of the mixed solutions can be finely tuned by the volume fraction of H2O. As shown in Fig. 2A, the maximum absorption band at λ = 678 nm in 10% water media is blue-shifted to λ = 640 nm in pure water media accompanied by a color change from green to blue (Fig. S10†). Upon excitation at λ = 635 nm, the near-infrared emission wavelength of the probe undergoes a corresponding blue shift from λ = 724 nm in 10% water media to λ = 672 nm in pure water media accompanied by an obvious decrease in fluorescence intensity (Fig. 2B). A calibration curve was then obtained by plotting the shift of emission maximum against the dielectric constant (in the range of ε = 10.0 to ε = 80.1) with an excellent linearity (Fig. 2C; R2 = 0.991). Moreover, a good Lorentz function correlation between I730/I670 [the fluorescence intensity ratios at λ = 730 nm (approximate emission maxima in 1,4-dioxane) and λ = 670 nm (approximate emission maxima in water)] and the solvent polarity was obtained (Fig. 2D; R2 = 0.991), which is highly sensitive to weak polarity media, especially that of the physiological polarity range (from ε = 10.0 to ε = 25.6).12 These unique properties enable HXPI-P to be feasible to accurately measure the polarity changes in living cells based on changes in its emission maxima or the fluorescence intensity ratios I730/I670.
Fig. 2. (A) Absorption and (B) fluorescence emission spectra of HXPI-P (10 μM) in the mixture of H2O and 1,4-dioxane. The percentage indicates the volume fraction. (C) Plot of Δλemvs. the dielectric constant in the range of ε = 10.0 to ε = 80.1. Δλem = λem (in a mixed solvent) − λem (in water). (D) Plot of I730/I670 (R) vs. the dielectric constant in the range of ε = 2.21 (1.4-dioxane) to ε = 80.1 (water). λex = 635 nm. Data are expressed as the mean ± SD of three separate measurements.
It is interesting that HXPI-P shows a hypsochromic behavior in the solvents with higher polarity (Fig. 1 and 2), which is contrary to the behavior of most typical polarity-sensitive fluorophores, like neutral red,13 coumarin 343,14 and cyanine,15 whose emission maxima are red-shifted with increasing solvent polarity. The reason for this hypsochromic behavior should be that, according to the Lippert–Metaga equation (eqn (S1)†),16 the ground state dipole moment of HXPI-P is larger than that of the excited state. Moreover, a theoretical calculation for the dipole moments under the ground state and first excited state was conducted with the global hybrid B3LYP and basis set 6-31+G(d). As shown in Table S2,† in water, the ground state dipole moment of HXPI-P is 8.15 D, a bit larger than the excited state one which is 6.62 D. Similar calculation values could be observed in 1,4-dioxane. The final optimized structures of the ground state and first excited state are listed in Table S3 at the end of the ESI.† These estimated values are consistent with our experimental results that the excited state dipole moment is smaller than the ground state one and further, well explain the blue-shift behavior of the probe in solvents with higher polarity.
Selectivity of HXPI-P to pH, viscosity and biological active substances
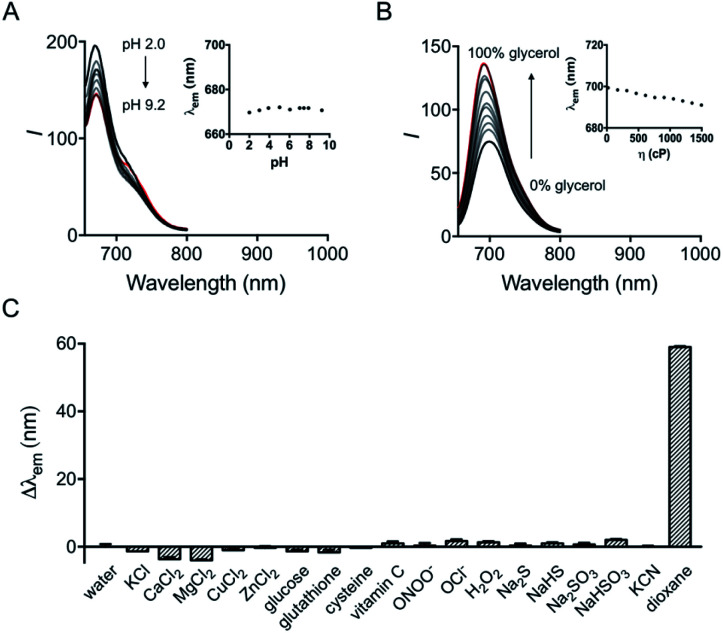
The influence of pH on the probe was first investigated. Although hydroxyl hemicyanine is a widely used pH-sensitive fluorophore,17 the emission maximum and the fluorescence intensity ratios I730/I670 of HXPI-P remain barely changed in 20 mM phosphate buffer solutions at pH values from 2.0 to 9.2 (Fig. 3A and S11A†). We speculate that this may be due to the formation of intramolecular hydrogen bonds which “lock” the hydroxyl group (Fig. S12A†), making it irresponsive to pH. To verify this hypothesis, two model compounds, HXPI-M with a carboxyl group at the meta-position of the hydroxyl group and HXPI-E with an esterified carboxyl group at the ortho-position of the hydroxyl group, were synthesized (Fig. S12B and C†), since both of them were unable to form intramolecular hydrogen bonds. As expected, upon excitation at λ = 635 nm, HXPI-M and HXPI-E display obvious changes of fluorescence intensity in different pH buffer solutions (Fig. S13†), which are ascribed to the protonation/deprotonation of the “unlocked” hydroxyl groups. From these explicit differences of spectroscopic behavior, we conclude that the formation of intramolecular hydrogen bonds in HXPI-P plays a vital role in its highly selective response to solvent polarity.
Fig. 3. (A) Fluorescence emission spectra of HXPI-P (10 μM) in phosphate buffer (20 mM) at different pH values. Inset: the plot of λemvs. pH in the range of pH 2.0 to pH 9.2. (B) Fluorescence emission spectra of HXPI-P (10 μM) in the mixture of glycol and glycerol. Inset: the plot of λemvs. viscosity in the range of η = 22.1 cP to η = 1495 cP. The percentage indicates the volume fraction. (C) Fluorescence responses of HXPI-P (10 μM) to various substances: water, KCl (150 mM), CaCl2 (2.0 mM), MgCl2 (2.0 mM), CuCl2 (100 μM), ZnCl2 (100 μM), glucose (10 mM), glutathione (1.0 mM), cysteine (100 μM), vitamin C (1.0 mM), ONOO− (100 μM), OCl− (100 μM), H2O2 (100 μM), Na2S (100 μM), NaHS (100 μM), Na2SO3 (100 μM), NaHSO3 (100 μM), KCN (100 μM) and 1,4-dioxane. λex = 635 nm. Δλem = λem (in an examed solution) − λem (in water). Data are expressed as the mean ± SD of three separate measurements.
Moreover, the response of HXPI-P to viscosity was studied. As the solvent viscosity changes from η = 22.1 cP (glycol) to 1495 cP (glycerol), both the emission maxima and the fluorescence intensity ratios I730/I670 display slight decreases (Fig. 3B and S11B†). Notably, during mitophagy, the increased mitochondrial viscosity10b,c might lead to a tiny blue shift of λem and a decrease of I730/I670 whereas the supposedly decreased mitochondrial polarity tends to cause a great red shift of λem and an increase of I730/I670, which could exclude the false positive signal derived from the variation of viscosity.
The selectivity of the probe was examined over various biological coexisting species as well. As shown in Fig. 3C and S11C,† the fluorescence emission maxima and the fluorescence intensity ratios I730/I670 remain almost unchanged in water in the presence of various common metal ions (K+, Ca2+, Mg2+, Cu2+, and Zn2+), glucose, redox substances (glutathione, cysteine, vitamin C, ONOO−, OCl−, and H2O2) and anions (S2−, HS−, SO32−, HSO3−, and CN−). These results indicate that HXPI-P could be applied to measure cellular polarity with minimum interference from pH, viscosity and other biologically related species.
Mitochondria-targeting ability of HXPI-P
Before intracellular experiments, a standard MTT assay was performed to determine the cytotoxicity of HXPI-P. As shown in Fig. S14,† HXPI-P displayed good biocompatibility since over 95% of HeLa cells survived even when the probe concentration reached 30 μM.
The photostability of the probe in water and in HeLa cells was then examined by comparison with that of Mito Green (a commercial mitochondria-specific dye). As shown in Fig. S15,† the fluorescence of Mito Green is bleached quickly in 10 min and decreases to about 25% of the initial intensity after 30 min. In contrast, HXPI-P is rather stable, and only a decrease of less than 5% in its fluorescence intensity occurs even after 30 min. These results are consistent with the observations in water (Fig. S16†), indicating high photostability of HXPI-P.
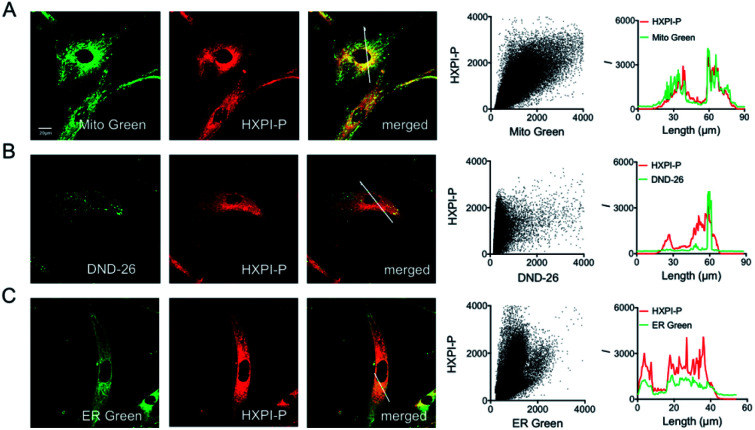
To evaluate the mitochondria-targeting ability, HeLa or HFL-1 cells were co-stained with HXPI-P and Mito Green. As shown in Fig. 4A, the fluorescence distribution of HXPI-P overlaps well with that of Mito Green (Pearson's coefficient: 0.97). In contrast, a poor overlap between the probe and DND-26 (a commercial lysosome-targeting dye) or ER Green (a commercial endoplasmic reticulum-targeting dye) is found (Pearson's coefficient: 0.65 and 0.82, respectively; Fig. 4B and C). Similar results are obtained for HFL-1 cells (Fig. S17†). These results confirm the superior mitochondria-targeting ability of HXPI-P.
Fig. 4. Mitochondria-targeting properties of HXPI-P in HeLa cells. (A) Colocalization images of HeLa cells stained with Mito Green (500 nM, green channel, λex = 488 nm, λem = 500–550 nm) and HXPI-P (2.0 μM, red channel, λex = 635 nm, λem = 650–750 nm), and the correlation of HXPI-P and Mito Green intensities as well as the intensity profiles within the ROI (region of interest; white line in the merged image of (A)). (B) Colocalization images of HeLa cells stained with DND-26 (1.0 μM, green channel, λex = 488 nm, λem = 500–550 nm) and HXPI-P [as in (A)], and the correlation of HXPI-P and DND-26 intensities as well as the intensity profiles within the ROI (white line in the merged image of (B)). (C) Colocalization images of HeLa cells stained with ER Green (1.0 μM, green channel, λex = 488 nm, λem = 500–550 nm) and HXPI-P [as in (A)], and the correlation of HXPI-P and ER Green intensities as well as the intensity profiles within the ROI (white line in the merged image of (C)). Scale bar = 20 μm. The fluorescence images were collected using a 100× oil immersion objective lens.
It has been reported that the mitochondria-targeting ability of the cationic hemicyanine fluorophore is highly dependent on the negative mitochondrial membrane potential (MMP).18 To assess whether HXPI-P could still stain mitochondria with decreased MMP, a mitochondrial uncoupling experiment was carried out. HFL-1 or HeLa cells were incubated with HXPI-P and rhodamine 123 (an MMP indicator whose fluorescence intensity would fade significantly when MMP decreased)19 for 10 min. Then, carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added to eliminate the MMP by rapid acidification of mitochondria.19 As can be seen from Fig. S18B and D,† after a 5 min incubation of CCCP, the fluorescence intensity from the rhodamine 123 channel reduces greatly, confirming the absence of MMP. However, after mitochondrial uncoupling, the fluorescence intensity of the HXPI-P channel only exhibits slight decreases (Fig. S18F and H†), which is still strong enough for intracellular imaging. This phenomenon indirectly confirms the mitochondria-targeting ability of HXPI-P and may be explained by the good lipid solubility of the probe (log Poct = 0.76). In conjunction with the data presented in Fig. 4 and S17, S18,† we conclude that HXPI-P is capable of monitoring the state of mitochondria in cell depolarization associated events.
Ratiometric fluorescence imaging of mitochondrial polarity in normal and cancer cells
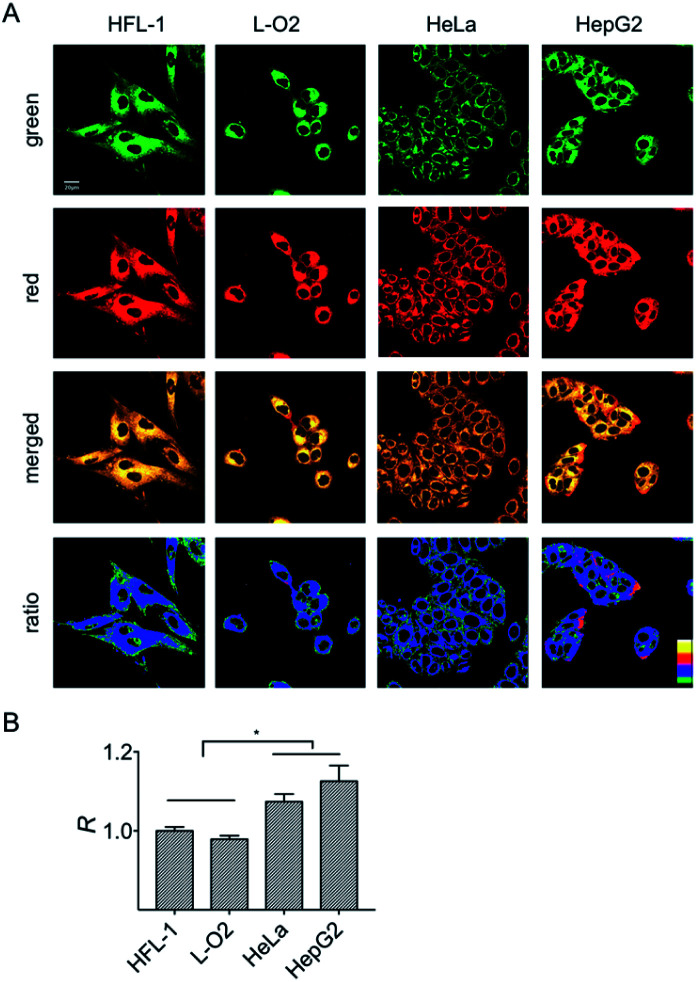
It is known that normal and cancer cells display different microenvironments.20 To exploit the biological feasibility of HXPI-P as a ratiometric fluorescence imaging probe, we intend to detect the variations of mitochondrial polarity among normal and cancer cells. Two kinds of normal (HFL-1 and L-O2 cells) and cancer cells (HeLa and HepG2 cells) were incubated with HXPI-P (5.0 μM) for 10 min, respectively, and then fluorescence images were collected in green and red channels. As illustrated in Fig. 5A, compared with the merged images of the HXPI-P-stained normal cells, those of the cancer ones are redder in color. In addition, the fluorescence intensity ratios (R) between the red and green channels of the cancer cells are higher than those of the normal ones (Fig. 5B), indicating a lower polarity in cancer cells. These results confirm the capability of HXPI-P in determining the mitochondrial polarity in living cells by ratiometric fluorescence imaging.
Fig. 5. Confocal fluorescence images of HFL-1, L-O2, HeLa and HepG2 cells stained with HXPI-P (5.0 μM). (A) Images collected in green (λem = 655–685 nm) and red (λem = 700–730 nm) channels at λex = 635 nm. The images in the third row are the merged ones of red and green channels. The fourth row shows the corresponding ratiometric images between red and green channels. Scale bar = 20 μm. (B) Polarity differences of mitochondria in different cells. The fluorescence intensity ratio of HFL-1 cells is defined as 1.0. The data are expressed as the mean ± SD of three measurements. Significant differences (*p < 0.05) are obtained by Student's t-test.
Monitoring the mitochondrial polarity changes during mitophagy
First, the MMP dependency on the fluorescence intensity ratios of HXPI-P was studied by using varied concentrations of CCCP. HeLa cells were incubated with HXPI-P (5.0 μM) for 10 min and then treated with different concentrations of CCCP for 5 min. Fluorescence images were collected in red, green and orange channels, respectively. As expected, the fluorescence intensity in HeLa cells gradually decreases with increasing concentrations of CCCP (Fig. S19A and B†). However, the fluorescence intensity ratios (R) between the orange and green channels remain barely changed (Fig. S19C†), suggesting that the decrease of MMP will not affect the polarity of mitochondria in a short period of time. These results indicate that HXPI-P could be applied to monitoring the polarity changes in uncoupled mitochondria by ratiometric fluorescence imaging.
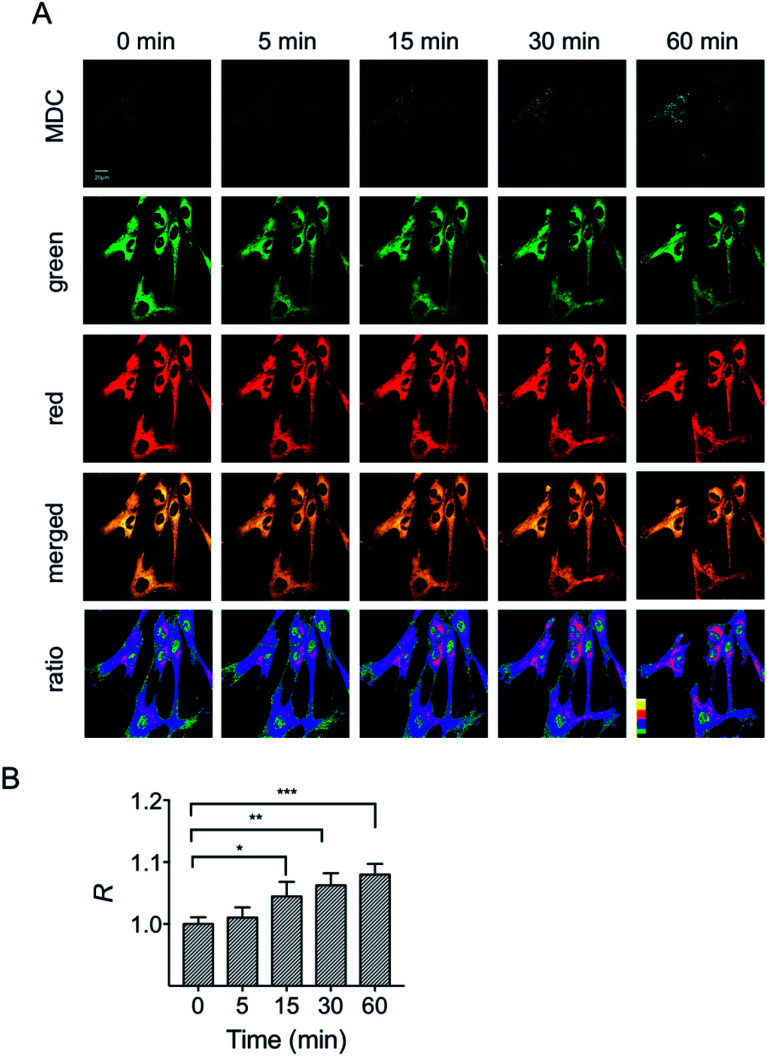
To investigate the variations of mitochondrial polarity during mitophagy, a starvation model was used. Nutrient deprivation could cause mitochondrial impairment through metabolic inhibition, further leading to mitophagy.21 For this purpose, HFL-1 cells were co-incubated with HXPI-P and MDC (dansylcadaverine, a commercial autophagy tracker)22 in a serum-free medium. Subsequently, fluorescence ratiometric images were recorded at different time points. As shown in Fig. 6A, the fluorescence intensity of the MDC channel enhances gradually over time, indicating an expanding degree of mitophagy ascribed to the nutrient deprivation. Moreover, the merged images turn redder in color (Fig. 6A) and the fluorescence intensity ratios between the red and green channels become higher as the time of mitophagy prolongs (Fig. 6B). These results confirm our hypothesis on the decreased polarity of mitochondria in the process of mitophagy. To further verify this conclusion, an autophagy inhibition assay was carried out. HFL-1 cells were treated with chloroquine (an autophagy inhibitor that prevents the fusion of lysosomes with autophagosomes), HXPI-P and MDC in a serum-free medium. As demonstrated in Fig. S20,† not only the fluorescence intensity of the MDC channel, but also the ratios between the red and green channels remain scarcely changed with time. This control study further supports our hypothesis that mitophagy is accompanied by a decreased polarity.
Fig. 6. Confocal fluorescence images of HFL-1 cells incubated with HXPI-P (5.0 μM) and MDC (1.0 μM). (A) Images collected in the MDC (λem = 450–550 nm) channel at λex = 405 nm; images in the green (λem = 655–685 nm) and red (λem = 700–730 nm) channels were collected at λex = 635 nm. The images in the fourth row are the merged ones of red and green channels. The fifth row shows the corresponding ratiometric images between red and green channels. Scale bar = 20 μm. (B) Polarity changes of mitochondria with time. The fluorescence intensity ratio at 0 min is defined as 1.0. The data are expressed as the mean ± SD of three measurements. Significant differences (*p < 0.05, **p < 0.01) are obtained by Student's t-test.
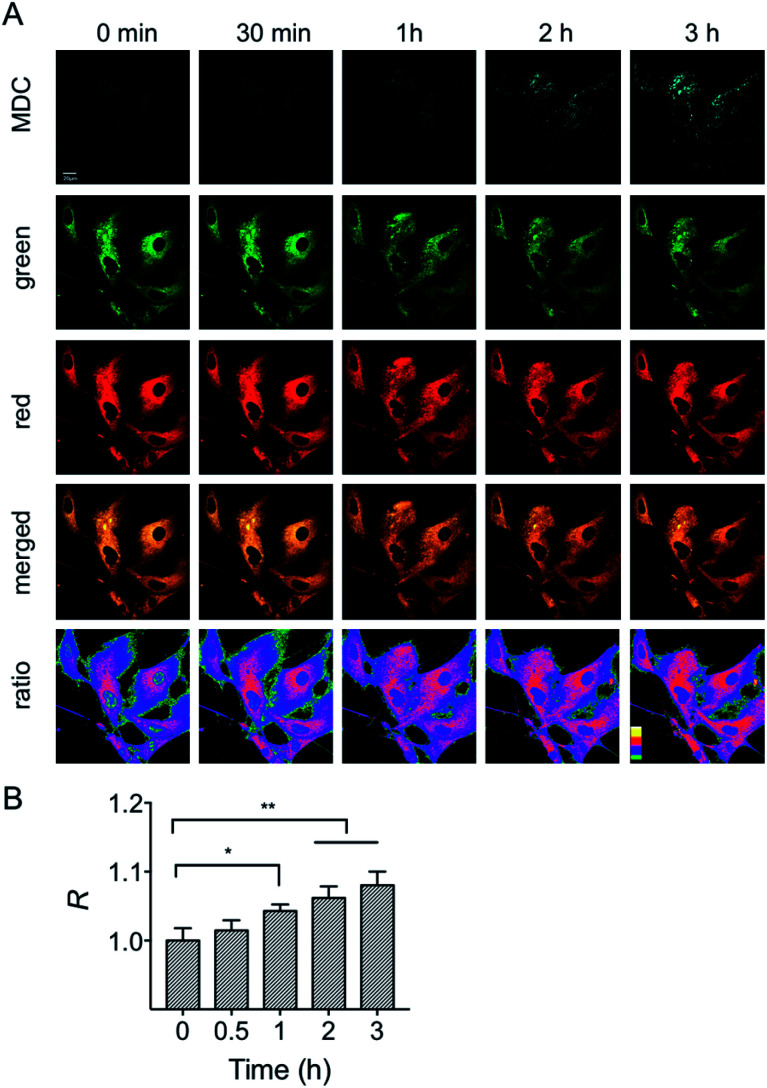
In addition to the starvation model, the mitochondrial polarity changes during drug-induced mitophagy were also studied. Rapamycin is a widely used macrolide antibiotic which induces mitophagy in a variety of cell types.23 In this experiment, HFL-1 cells were co-incubated with HXPI-P, MDC and rapamycin in a serum-free medium. Subsequently, fluorescence ratiometric images were recorded at different time points. As shown in Fig. 7, treatment of rapamycin leads to a significant increase in the fluorescence intensity ratios between the red and green channels within 30 min, which is more obvious than that in the starvation model (Fig. 6B), and is ascribed to the rapamycin-induced mitophagy. It is also seen from Fig. 7 that a larger decrease in polarity occurs at about 60 min, which may be attributed to the severer mitophagy caused by both starvation and rapamycin stimulation. Taken together, these results confirm the utility of HXPI-P for monitoring the mitophagy-specific polarity dynamics in mitochondria.
Fig. 7. Confocal fluorescence images of HFL-1 cells incubated with HXPI-P (5.0 μM), MDC (1.0 μM) and rapamycin (50 nM). (A) Images in the MDC (λem = 450–550 nm) channel was collected at λex = 405 nm; images in the green (λem = 655–685 nm) and red (λem = 700–730 nm) channels were collected at λex = 635 nm. The images in the fourth row are the merged ones of red and green channels. The fifth row shows the corresponding ratiometric images between red and green channels. Scale bar = 20 μm. (B) Polarity changes of mitochondria with time. The fluorescence intensity ratio at 0 min is defined as 1.0. The data are expressed as the mean ± SD of three measurements. Significant differences (*p < 0.05, **p < 0.01, ***p < 0.001) are obtained by Student's t-test.
Conclusions
In summary, by incorporating a carboxyl group into the hydroxyl hemicyanine skeleton, we have developed a near-infrared fluorescent probe, HXPI-P, for mitochondrial polarity measurements. By using HXPI-P with confocal fluorescence imaging, we show that the polarity of normal cells is usually higher than that of cancer cells. Moreover, a decrease in mitochondrial polarity during starvation or rapamycin-induced mitophagy was observed. We believe that, owing to its synthetic convenience, non-invasiveness, high selectivity and photostability, HXPI-P may serve as a promising tool for mitophagy-related fundamental and clinical research.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by grants from the NSF of China (No. 21775152, 21820102007, 21535009, and 21621062) and the Chinese Academy of Sciences (Grant XDB14030102).
Electronic supplementary information (ESI) available. See DOI: 10.1039/c9sc05505c
Notes and references
- (a) Ernster L. Schatz G. J. Cell Biol. 1981;91:227s–255s. doi: 10.1083/jcb.91.3.227s. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Nie G. Blood. 2005;105:2161–2167. doi: 10.1182/blood-2004-07-2722. [DOI] [PubMed] [Google Scholar]; (c) Nakagawa T. Guarente L. Aging. 2009;1:578–581. doi: 10.18632/aging.100062. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Logan C. V. Szabadkai G. Sharpe J. A. Parry D. A. Torelli S. Childs A.-M. Kriek M. Phadke R. Johnson C. A. Roberts N. Y. Bonthron D. T. Pysden K. A. Whyte T. Munteanu I. Foley A. R. Wheway G. Szymanska K. Natarajan S. Abdelhamed Z. A. Morgan J. E. Roper H. Santen G. W. E. Niks E. H. van der Pol W. L. Lindhout D. Raffaello A. De Stefani D. den Dunnen J. T. Sun Y. Ginjaar I. Sewry C. A. Hurles M. Rizzuto R. UK10K Consortium Duchen M. R. Muntoni F. Sheridan E. Nat. Genet. 2013;46:188–193. doi: 10.1038/ng.2851. [DOI] [PubMed] [Google Scholar]
- (a) Liochev S. I. Free Radical Biol. Med. 2013;60:1–4. doi: 10.1016/j.freeradbiomed.2013.02.011. [DOI] [PubMed] [Google Scholar]; (b) Trachootham D. Lu W. Ogasawara M. A. Nilsa R.-D. V. Huang P. Antioxid. Redox Signaling. 2008;10:1343–1374. doi: 10.1089/ars.2007.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R. J. Narendra D. P. Nat. Rev. Mol. Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval H. Thiagarajan P. Dasgupta S. K. Schumacher A. Prchal J. T. Chen M. Wang J. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen D.-F. Narendra D. P. Tanaka A. Manfredi G. Youle R. J. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11835–11840. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon R. A. Nat. Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- (a) Huang C. Andres A. M. Ratliff E. P. Hernandez G. Lee P. Gottlieb R. A. PLoS One. 2011;6:e20975. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hoshino A. Matoba S. Iwai-Kanai E. Nakamura H. Kimata M. Nakaoka M. Katamura M. Okawa Y. Ariyoshi M. Mita Y. Ikeda K. Ueyama T. Okigaki M. Matsubara H. J. Mol. Cell. Cardiol. 2012;52:175–184. doi: 10.1016/j.yjmcc.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Ding W. X. Yin X. M. Biol. Chem. 2012;393:547–564. doi: 10.1515/hsz-2012-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Li X. H. Gao X. H. Shi W. Ma H. M. Chem. Rev. 2014;114:590–659. doi: 10.1021/cr300508p. [DOI] [PubMed] [Google Scholar]; (b) Zhang Y. B. Xia S. Mikesell L. Whisman N. Fang M. X. Steenwinkel T. E. Chen K. Luck R. L. Werner T. Liu H. Y. ACS Appl. Bio Mater. 2019;2:4986–4997. doi: 10.1021/acsabm.9b00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Liu Y. C. Teng L. L. Chen L. L. Ma H. C. Liu H. W. Zhang X. B. Chem. Sci. 2018;9:5347–5353. doi: 10.1039/C8SC01684D. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zou Z. Yan Q. Ai S. X. Qi P. Yang H. Zhang Y. F. Qing Z. F. Zhang L. H. Feng F. Yang R. H. Anal. Chem. 2019;91:8574–8581. doi: 10.1021/acs.analchem.9b01861. [DOI] [PubMed] [Google Scholar]; (c) Hou L. L. Ning P. Feng Y. Ding Y. Q. Bai L. Li L. Yu H. Z. Meng X. M. Anal. Chem. 2018;90:7122–7126. doi: 10.1021/acs.analchem.8b01631. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Komatsu M. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- (a) Jiménez-Sánchez A. Lei E. K. Kelley S. O. Angew. Chem., Int. Ed. 2018;57:8891–8895. doi: 10.1002/anie.201802796. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Jiang N. Fan J. L. Xu F. Peng X. J. Mu H. Y. Wang J. Y. Xiong X. Q. Angew. Chem., Int. Ed. 2015;54:2510–2514. doi: 10.1002/anie.201410645. [DOI] [PubMed] [Google Scholar]
- Wang S. J. Wang X. C. Shi W. Wang K. Ma H. M. Biochim. Biophys. Acta. 2008;1784:415–422. doi: 10.1016/j.bbapap.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Liu X. G. Xu Z. C. Cole J. M. J. Phys. Chem. C. 2013;117:16584–16595. doi: 10.1021/jp404170w. [DOI] [Google Scholar]
- Xiao H. Li P. Zhang W. Tang B. Chem. Sci. 2016;7:1588–1593. doi: 10.1039/C5SC04099J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M. K. Pal H. Bhasikuttan A. C. Sapre A. V. Photochem. Photobiol. 1998;68:32–38. doi: 10.1111/j.1751-1097.1998.tb03249.x. [DOI] [Google Scholar]
- (a) Wan Q. Q. Chen S. M. Shi W. Li L. H. Ma H. M. Angew. Chem., Int. Ed. 2014;53:10916–10920. doi: 10.1002/anie.201405742. [DOI] [PubMed] [Google Scholar]; (b) Li X. Y. Hu Y. M. Li X. H. Ma H. M. Anal. Chem. 2019;91:11409–11416. doi: 10.1021/acs.analchem.9b02782. [DOI] [PubMed] [Google Scholar]
- (a) Liu Y. Zhou J. Wang L. L. Hu X. X. Liu X. J. Liu M. R. Cao Z. H. Shangguan D. H. Tan W. H. J. Am. Chem. Soc. 2016;138:12368–12374. doi: 10.1021/jacs.6b04048. [DOI] [PubMed] [Google Scholar]; (b) Cheng D. Peng J. J. Lv Y. Su D. D. Liu D. J. Chen M. Yuan L. Zhang X. B. J. Am. Chem. Soc. 2019;141:6352–6361. doi: 10.1021/jacs.9b01374. [DOI] [PubMed] [Google Scholar]
- Li H. Y. Li X. H. Wu X. F. Shi W. Ma H. M. Anal. Chem. 2017;89:5519–5525. doi: 10.1021/acs.analchem.7b00503. [DOI] [PubMed] [Google Scholar]
- (a) Hanahan D. Weinberg R. A. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]; (b) Allinen M. Beroukhim R. Cai L. Brennan C. Lahti-Domenici J. Huang H. Porter D. Hu M. Chin L. Richardson A. Schnitt S. Sellers W. Polyak K. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Hailey D. W. Rambold A. S. Satpute-Krishnan P. Mitra K. Sougrat R. Kim P. K. Lippincott-Schwartz J. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederbick A. Kern H. F. Elsässer H. P. Eur. J. Cell Biol. 1995;66:3–14. [PubMed] [Google Scholar]
- Sarkar S. Rubinsztein D. C. Mol. BioSyst. 2008;4:895–901. doi: 10.1039/B804606A. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.