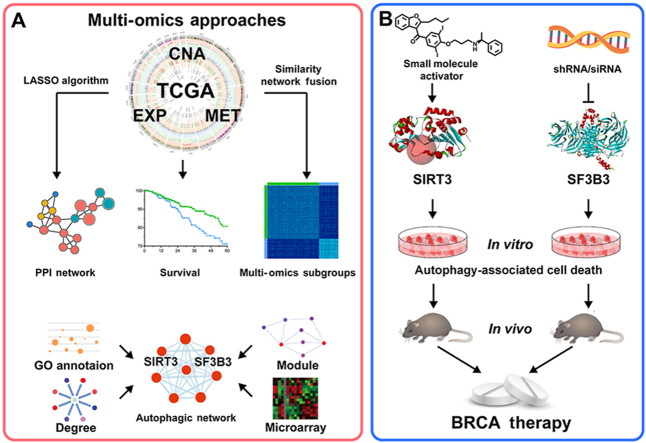
Abstract
Autophagy is a critical cellular homeostatic mechanism, and its dysfunction is linked to invasive breast carcinoma (BRCA). Recently, several omics methods have been applied to explore autophagic regulators in BRCA; however, more reliable and robust approaches for identifying crucial regulators and druggable targets remain to be discovered. Thus, we report here the results of multi-omics approaches to identify potential autophagic regulators in BRCA, including gene expression (EXP), DNA methylation (MET) and copy number alterations (CNAs) from The Cancer Genome Atlas (TCGA). Newly identified candidate genes, such as SF3B3, TRAPPC10, SIRT3, MTERFD1, and FBXO5, were confirmed to be involved in the positive or negative regulation of autophagy in BRCA. SF3B3 was identified firstly as a negative autophagic regulator, and siRNA/shRNA-SF3B3 were shown to induce autophagy-associated cell death in in vitro and in vivo breast cancer models. Moreover, a novel small-molecule activator of SIRT3, 1-methylbenzylamino amiodarone, was discovered to induce autophagy in vitro and in vivo. Together, these results provide multi-omics approaches to identify some key candidate autophagic regulators, such as the negative regulator SF3B3 and positive regulator SIRT3 in BRCA, and highlight SF3B3 and SIRT3 as new druggable targets that could be used to fill the gap between autophagy and cancer drug development.
KEY WORDS: Invasive breast carcinoma, Multi-omics approach, SIRT3, SF3B3, Autophagic regulator, Anti-proliferation, Migration, Druggable target
Abbreviations: ATG, autophagy-related gene; BRCA, invasive breast carcinoma; CNA, copy number alteration; EXP, gene expression; GO, Gene Ontology; LASSO, least absolute shrinkage and selection operator; MET, DNA methylation; PFS, progression-free survival; SNF, similarity network fusion; TCGA, The Cancer Genome Atlas; TNBC, triple-negative breast cancer
Graphical abstract
This study provides multi-omics approaches to identify some key candidate autophagic regulators, such as the negative regulator SF3B3 and positive regulator SIRT3 in invasive breast carcinoma, and highlight them as new druggable targets.
1. Introduction
Autophagy, an evolutionarily conserved process by which the cell degrades aged and damaged cellular components, plays context-dependent roles in various types of human cancers, including breast cancer1,2. It appears to be quite logical for autophagy to play an essential role in protecting cells from neoplastic transformation3. In early stages of cancer initiation, autophagy exhibits primarily an anti-tumor function. Studies indicated that loss of beclin-1, a protein instrumental in the induction and progression of autophagy, or its low expression promoted tumorigenesis in mammary epithelial cells, resulting in the formation of tumors with characteristics of triple negative breast cancer4,5. Although autophagy is closely associated with the occurrence, development and metastasis of breast cancer, the exploration of crucial autophagic regulators for potential targeted therapy remains limited6. Breast cancer continues to be the leading cause of cancer-related deaths in women, with BRCA ranking as the top breast cancer subtypes in incidence7. Although the survival rate of BRCA patients has greatly increased with the improvements in surgery, targeted therapy and chemotherapy in recent years8, the recurrence and metastasis of breast cancer cells remains an obstacle to satisfactory prognosis9. Since autophagy is closely associated with breast cancer, it might be beneficial to identify key autophagy-associated susceptibility genes and critical autophagic regulators to provide potential druggable targets for BRCA therapy10. For instance, the autophagic initiator ULK1 has recently been reported to be significantly downregulated in triple negative breast cancer11. In addition, the expression of beclin-1, the homolog of ATG6, has been confirmed to inversely correlate with growth factor signaling in breast cancer12. Therefore, targeting some key autophagic regulators such as ULK1 and beclin-1 may be a promising strategy for breast cancer therapy. Correspondingly, identification of the key autophagic regulators in BRCA would provide additional potential druggable targets for therapeutics.
Several investigations have identified autophagic regulators, including possible biomarkers or targets, by one-dimensional analyses of somatic mutations13, CNAs12, MET14, and EXP15. Unfortunately, these one-dimensional omics approaches often result in incomplete exploitation of the cancer genome. Thus, multidimensional searches for potential biomarkers or targets seem to be powerful alternatives to accelerate integrative analyses of cancer genomics data16. With the developing of large-scale technologies, TCGA has collected multiple sources of genomics data on more than 30 types of human cancers17. Some multi-omics methods, such as the graph diffusion-based method18, network-propagation algorithms and artificial neural networks19,20, have been used to identify cancer-related biomarkers and targets in many types of human cancers. The application of multi-omics approaches to specific biological process, such as autophagy, is considered a powerful method for exploring the relationships between autophagy and breast cancer21.
Thus, in this study, we developed multi-omics approaches that integrate omics data, including EXP, MET and CNA data from TCGA. We used some established bioinformatics methods and algorithms, such as the similarity network fusion (SNF) method and the least absolute shrinkage and selection operator (LASSO) algorithm, to identify potential autophagic regulators, such as SIRT3 and SF3B3, in the autophagic network of BRCA. Subsequently, we found siRNA/shRNA-SF3B3 could induce autophagy-associated cell death in in vitro and in vivo breast cancer models. In addition, we designed a novel small-molecule activator of SIRT3, 1-methylbenzylamino amiodarone, that was able to induce autophagy-associated cell death in the established breast cancer models. Our multi-omics approaches identified SF3B3 and SIRT3 as candidate autophagic regulators and druggable targets for BRCA. Thus, these candidates may provide the first comprehensive resource for further exploring the intricate relationships between autophagy and breast cancer treatment.
2. Material and methods
2.1. Data preprocessing
BRCA and 12 other types of cancers were screened from the TCGA dataset, which included more than 150 samples containing CNA, EXP, and MET data. The genomic and clinical data were downloaded from the Broad GDAC Firehose website (http://gdac.broadinstitute.org/)22.
For CNAs, we used data calculated by the genome-wide SNP6 array platform. The gain and loss levels of the copy number for a given gene were segmented by regions per sample using the GISTIC2.0 algorithm23. For MET, we used data from the JHU-USC-Human Methylation 450 platform. We extracted the beta values calculated as [M/(M+U)], which were scores derived from values of methylated (M) and unmethylated (U) signals that were used to measure the levels of MET. If multiple CpG islands existed for a single gene, we used the mean beta values as the overall methylation values. For EXP, we used RNASeqV2 normalized counts. For BRCA, ER, PR, and HER2 status were measured by immunohistochemistry. Patients with breast cancer were classified into four subtypes (luminal A, luminal B, HER2-enriched, and basal-like) based on the RNA-Seq measurements of the 50-gene PAM50 signature24.
2.2. SNF and spectral clustering by multi-omics profiles of autophagic genes
We used the SNF method to aggregate the different omics data (CNA, EXP, and MET data) of 34 core autophagic genes; this method can utilize biological evidence from different sources25. These 34 core autophagic genes, which participate in different autophagy signaling pathways (e.g., the ULK complex, the beclin-1 interactome, mTOR signaling and ATG4–LC3 signaling) were selected from the literature26. Due to a lack of data, CNA and MET of ATG4A were not considered. The best cluster number of spectral clustering was determined by eigengap, a component function of SNF packages. Briefly, SNF was used to calculate the patient similarity networks that were obtained separately from all data types. These networks were then fused into a single similarity network by keeping the high-weight edges in one or more networks and removing the low-weight edges not presented in all networks, which resulted in a fused network with shared and complementary information captured from different data sources.
2.3. Identification of the CNAs and MET alterations associated with SNF clustering subgroups
CNAs and MET alterations associated with SNF clustering subgroups were identified using Fisher's exact test, which was used to test the statistical significance of the differences in frequencies of amplifications and deletions or in the frequencies of hypermethylation and hypomethylation. Samples in one of the SNF clustering subgroups were compared to those in another subgroup. The upper 33% and lower 33% of the MET level of each gene in each sample were classified as hypermethylation and hypomethylation, respectively. The –lgP values corrected by the Benjamini Hochberg (BH) method for multiple testing were reported.
2.4. LASSO
LASSO is a penalized feature selection algorithm that has been described in a previous study27. LASSO tends to select a small subset of important features by generating coefficients that are equal to zero. For the linear regression model, suppose that we have response variables y = (y1, …, yn)t and predictor variables x = (x1j, …, xnj)t, where j = 1, …, p. Letting is defined as Eq. (1):
| (1) |
where s is a tuning parameter that controls the amount of shrinkage that is applied to the estimates. The tuning parameter was chosen by 10-fold cross-validation. The LASSO method described above was implemented in the R package and was applied to our data, which was calculated along a regularization path via a cyclical coordinate descent.
2.5. Construction of the multi-omics network from integrated data and the autophagic network
The overall network construction procedure can be divided into three main stages. In the first stage, known as the sample selecting stage, tumor samples with their CNA, methylation and EXP profile simultaneously available in TCGA were selected. In the second stage, known as the feature filtering stage, we aimed to select the CNA or methylation feature that is responsible for mRNA expression changes. To guarantee the successful running of the following procedure and to increase the precision of the calculation, we first reduced the number of input genes by applying the following filtering steps. First, genes with data on CNA, methylation and mRNA expression in more than 90% of the samples were retained. Second, genes that simultaneously had records of CNA, MET and EXP data were retained. After the above steps, approximately 17,263 genes with records of CNA, MET and EXP data remained for further analysis. To avoid irrelevant information that might increase the complexity and decrease the accuracy of model, we selected CNA and MET as features by using the following steps:
Step 1: We calculated the Pearson correlation coefficients between the CNA data or MET data and the corresponding EXP data. Then, we retained the CNA/MET features if the absolute value of Pearson's correlation coefficient was greater than 0.5 or less than −0.5, with significant P-values corrected by the BH method for multiple testing (FDR<0.05). We used a nonconservative threshold for cut-off to preserve the positive/negative correlation of CNA/MET with the EXP change. This step resulted in a feature pool that consisted of 9237 CNA features and 4628 MET features.
Step 2: (a) We calculated the absolute value of the Pearson correlation coefficient between the expression of each gene and the expression of all the other genes that belong to the same feature pool. If the absolute value of the Pearson correlation coefficient was greater than 0.5 with a significant P-value corrected by the BH method for multiple testing (FDR<0.05), we treated the genes as possible features for further consideration. This step was performed based on the assumption that genetic and epigenetic alterations should exert their influences through changes in mRNA expression. This step filtered out the irrelevant features of mRNA expression level. (b) We combined the genetic and epigenetic features that may affect EXP and incorporated them into the lasso linear regression model (details are described above).
Step 3: Step 2 was repeated until all 17,263 genes that simultaneously have records of CNA, MET and EXP data have been calculated.
Finally, the core autophagy network was constructed based on the computation results of the three types of molecular data (MET, CNA and mRNA expression) by using the method of lasso linear regression. In the created autophagy network of BRCA cancer datasets, the arrows indicate the direction of the putative regulation of features (MET or CNA) by response variables (mRNA expression). If the regression coefficient between a feature and the response variable was nonzero, we connected the feature and variable in the network, with an arrow depicting up- or down-regulation depending on whether the Pearson correlation coefficient was positive or negative, respectively.
2.6. Cell culture and reagents
MCF-7 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and used to conduct functional assays. The cells were cultured in DMEM medium supplemented with 10% FBS, 100 μg/mL streptomycin, 100 U/mL penicillin, and 0.03% l-glutamine and maintained at 37 °C with 5% CO2 at a humidified atmosphere. SBI-0206965, 3-MA, bafilomycin A1 and chloroquine were purchased from Sigma–Aldrich [the detail information are as follows: SBI-0206965 (SML1540), 3-MA (189490), bafilomycin A1 (196000) and chloroquine (C6628)]. The primary antibodies used for Western blot are as follows: LC3 B (Abcam, ab192890), beclin-1 (Abcam, ab207612), SF3B3 (Abcam, ab96683), ULK1 (8054, CST, MA, USA), p-ULK1Ser317 (12753, CST), p-ULK1Ser555 (5869, CST), mATG13 (13273, CST), p-mATG13Ser318 (PAB19948, Abnova, Taiwan), ATG101 (13492, CST), FIP200 (12436, CST), SQSTM1/P62 (5114, CST), ATG4B (13507, CST), ATG4C (5262, CST), ATG5 (12994, CST), ATG16L1 (8089, CST), SIRT3 (2627, CST), ac-MnSOD2 K68 (Abcam, ab137037), ac-MnSOD2 K122 (Abcam, ab214675), MMP-2(40994, CST), MMP-9 (13667, CST), E-cadherin (14472, CST), and β-actin (66009-1-Ig, Proteintech, IL, USA). In addition, the secondary infrared antibodies goat anti-rabbit IgG (Cell Signaling Technology #4410, 1:5000) and goat anti mouse IgG (Cell Signaling Technology #4414, 1:5000) were added.
2.7. RNA knockdown or cDNA overexpression of candidate genes
The MCF-7 cells were tested for mycoplasma contamination, and no contamination was found. Five siRNAs designed for SF3B3, TRAPPC10, FBXO5, METRFD1 and SIRT3 and one cDNA designed for SIRT3 were obtained from GeneChem. shRNA oligonucleotides for SF3B3 were annealed and ligated to pLKO-TRC vector digested with AgeI and EcoRI and purified with Qiaquick gel extraction kit to construct a recombinant plasmid. Then 293T cells were co-transfected with recombinant plasmids together with pHelper1.0 and pHelper2.0 plasmids to package lentivirus, and the virus supernatant was collected to infect MCF-7 target cells. qRT-PCR was used to monitor RNAi knockdown and cDNA overexpression efficiency. The primer sequences used for qRT-PCR are listed in Supporting Information Table S1. The RNAi knockdown or cDNA overexpression of candidate genes was conducted following previously reported methods28. The MCF-7 cells were transfected with 100 nmol/L siRNA or cDNA mixtures in 6-well plates and incubated for 48 h, followed by washing with phosphate-buffered saline buffer, and then collected for qRT-PCR and Western blot assays.
2.8. Western blot
For the Western blot assay, the cell pellets were re-suspended in lysis buffer consisting of 50 mmol/L HEPES, pH 7.4; 1% Triton-X100; 2 mmol/L sodium orthovanadate; 100 mmol/L sodium fluoride; 1 mmol/L edetic acid; 1 mmol/L PMSF; 10 mg/L aprotinin (Sigma, MO, USA) and 10 mg/L leupeptin (Sigma) and were lysed at 4 °C for 1 h. After centrifugation at 12,000 rpm (Legend Micro 17R, Thermo Fisher Scientific, Waltham, USA) for 15 min, the protein content of the supernatant was determined by the Bio-Rad DC protein assay (Bio-Rad Laboratories, Hercules, CA, USA). Equal amounts of total protein were separated by 10%–12% SDS-PAGE and were transferred to PVDF membranes, which were soaked in blocking buffer (5% skimmed milk or BSA). Proteins were detected by applying primary antibodies followed by HRP-conjugated secondary antibody and were visualized using ECL as the HRP substrate.
2.9. Immunofluorescence
Cells seeded on coverslips coated with FN1/fibronectin (MP Biomedicals, 02150025) were treated with complete medium and then treated according to demand. After three times of PBS washes, the cells were fixed with 4% paraformaldehyde for 20 min and then permeabilized with PBS containing 0.1% Triton-X100 (Sigma–Aldrich, T9284) for 10 min. Samples were blocked with PBS containing 3% BSA (Sigma–Aldrich, A1933) for 30 min and then incubated with primary LC3B, E-cadherin, MMP-2, SQSTM1/P62 and Ki-67 antibody solution at 4 °C overnight. The cells were then washed with PBS containing 0.05% Tween-20 three times and incubated with secondary antibody for 1 h at room temperature. Then, the cells were stained with DAPI (Sigma–Aldrich, 32670) for 5 min. Images were obtained with a Zeiss 710 Duo confocal microscope.
2.10. Autophagy, apoptosis and cell cycle assays
For the autophagy assay, MCF-7 cells were transfected with GFP/mRFP-LC3 for 24 h and then treated with 20 μmol/L MA for 24 h. Then, the cells were observed under a fluorescence microscope. For the apoptosis assay, apoptosis ratios were determined by flow cytometry analysis of Annexin-V/PI double staining. For cell cycle detection, cells were ethyl alcohol-fixed at 4 °C for 24 h, and the cell cycle distribution was determined by flow cytometry analysis of PI staining.
2.11. Colony formation assay
The stable knockdown of SF3B3 or overexpression of SIRT3 on the proliferation of MCF-7 cells were analyzed with a colony formation assay. Briefly, 200 treated cells per well were seeded into 6-well plates. After incubation for 2 weeks, the cells were fixed with 4% paraformaldehyde for 30 min and then stained with crystal violet.
2.12. Cell-migration assay
Wound healing and Transwell assays were performed to determine the cell migration ability. For the wound healing assay, cells were seeded in 6-well plates. After the cells reached confluency, wounds were created by the manual scraping of the cell monolayer with a pipette tip. After 24 h incubation, the wells were replenished with PBS, and the cells were photographed using a phase-contrast microscope. For the Transwell assay, cells were seeded into the upper chamber of a Transwell insert (6.5 mm diameter, 8 mm pores; Corning, New York, USA) at a density of 1.5 × 104 cells per well. DMEM (500 mL) with 10% FBS was added to the lower chamber, and the 24-well plate was incubated for 24 h. The non-migrated cells were scraped off the upper surface of the membrane with a cotton swab, and the migrated cells were dyed with crystal violet. After being washed with PBS, the migrated cells were photographed using a phase-contrast microscope.
2.13. Cellular SIRT3 activity assay
The cellular SIRT3 activity in an immunoprecipitate was determined using the fluorometric SIRT3 Activity Assay Kit (ab156067) according to the manufacturer's instructions. The cells were treated by adding fresh media containing the test compound for the desired time. To harvest cells under nondenaturing conditions, the media was removed, and the cells were rinsed once with ice-cold PBS. After removing the PBS, 0.5 mL 1× ice-cold cell lysis buffer was added to each plate (10 cm diameter), and the plate was incubated on ice for 5 min. The cells were then scraped off the plate and transferred to a microcentrifuge tube. After sonication for 5 s 4 times on ice, the tube was micro-centrifuged for 10 min at 4 °C, and the supernatant was transferred to a new tube to obtain the cell lysate. A total of 200 μL cell lysate was then incubated with anti-SIRT3 antibody or anti-IgG according to the manufacturer's instructions. After adding protein A agarose beads (20 μL of 50% bead slurry), the lysate was incubated with gentle rocking for 1–3 h at 4 °C and then micro-centrifuged for 30 s at 4 °C. The pellet was washed 3 times with 500 μL of 1× cell lysis buffer and once with 500 μL of SIRT3 assay buffer [50 mmol/L Tris-HCl (pH 8.8), 0.5 mmol/L DTT]. After immunoprecipitation, a reaction mixture containing Fluoro-Substrate peptide solution was added to protein A agarose beads as an “Enzyme Sample”, and NAD-dependent deacetylase activity was measured.
2.14. Xenograft breast cancer model
Thirty female BALB/c mice (6–8 weeks, 20–22 g) were obtained from Huafukang Biotechnology Co., Ltd. (Certificate No. SCXK [Jing] 2019-0008). Animal welfare and the experimental procedures were in accordance with the Ethical Regulations on the Care and Use of Laboratory Animals of Sichuan University (Chengdu, China) and were approved by the Animal Experimental Ethics Committee for all animal experiments. Thirty female BALB/c mice (6–8 weeks, 20–22 g) were injected subcutaneously with MCF-7WT cells (n = 20) or MCF-7shSF3B3 cells (n = 10) (5 × 106 cells/mouse). When the tumor volume reached approximately 0.2 cm3, the MCF-7WT mice were randomly assigned into two groups (n = 10): a WT group and an MA-treated group (50 mg/kg). Animals were weighed three times per week during the treatment period, and tumor size was measured three times per week by electronic calipers during the treatment period. All mice were sacrificed, and the tumor tissues were harvested, weighed, and photographed. Then, the tumor tissues were frozen in liquid nitrogen or fixed in formalin immediately for further investigation.
2.15. Immunohistochemistry analysis (IHC)
Tumor sections were submerged in EDTA antigenic retrieval buffer (pH 8.0) or citrate buffer (pH 6.0) and microwaved for antigenic retrieval. The slides were then incubated with polyclonal antibody (1:400) for 30–40 min at 37 °C. Normal rabbit/mouse IgG was used as a negative control. The slides were then treated with HRP polymer conjugated secondary antibody for 30 min and developed with diamino-benzidine solution. Meyer's hematoxylin was used as a counterstain.
2.16. Statistical analysis
Significantly differentially expressed genes between the SNF clustering subgroups in our network were identified by the Wilcoxon rank sum test. Differentially expressed genes were defined by median fold change >1.2 or <0.8 and P < 0.05. Significantly different MET/CNA were defined by FDR only. The upper 33% and lower 33% of EXP were defined as high and low expression, respectively. Kaplan–Meier survival analysis (Log-rank test) was used to evaluate the association of EXP with the OS of patients. All the experiments were performed independently at least three times. The data are expressed as mean ± standard error of mean (SEM), and were analyzed with GraphPad Prism 7.0. Statistical comparisons were made by one-way ANOVA and Student's t-test. P < 0.05 was considered statistically significant.
3. Results
3.1. Associations of autophagic subgroups with clinical features and multi-omics patterns
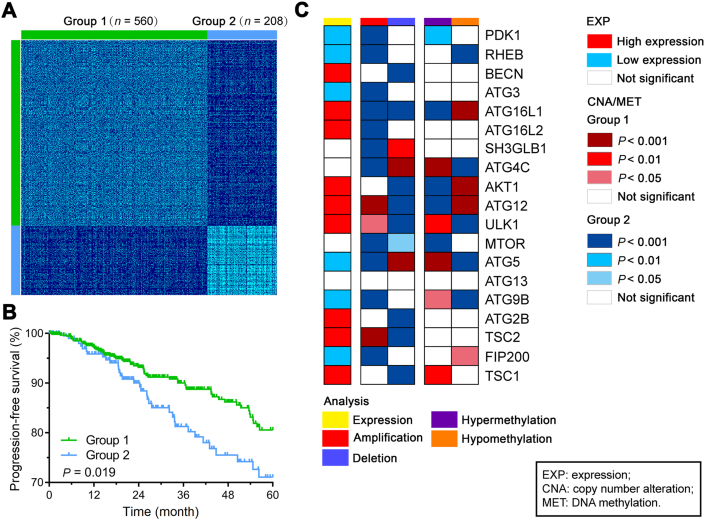
To identify distinct autophagic subgroups, we performed unsupervised clustering by using the SNF method against 34 known autophagy-related gene (ATG) profiles (CNA, EXP, and MET; Supporting Information Table S2) from TCGA. The BRCA patients were separated into 2 clusters with the optimal solution (n = 560 for group 1 and n = 208 for group 2, Fig. 1A). Then, a heatmap of 34 ATG gene profiles was obtained, which revealed the distinct molecular patterns between the subgroups in the three profiles (Supporting Information Fig. S1). For instance, copy number loss consistently induced low mRNA expression of BECN1/ULK1 in group 2, which has been demonstrated to be essential for autophagy and to serve as a tumor suppressor. The correlation between CNA and EXP is significant, such as BECN1, ULK1, ATG4B (Supporting Information Fig. S2). These results provide an overview of the multi-omics patterns of ATG genes in the BRCA subgroups and suggest the possibility of developing a manipulation technique for abnormally expressed genes.
Figure 1.
Clustering and molecular alterations of autophagic genes between patient groups identified by the SNF method. (A) Patient similarity matrix of two groups of BRCA patients obtained by the SNF method (n = 560 for group 1 and n = 208 for group 2). (B) Kaplan–Meier curves of PFS for the two groups clustered by SNF. (C) Summary of molecular alterations of core autophagic genes. The summary includes all the significant alterations of MET, CNA, and EXP. The rows present genes with significant alterations in at least one analysis. The columns are patient groups identified by the SNF method.
To assess the clinical relevance of the autophagic genes, we performed an association analysis of progression-free survival (PFS) with the autophagic subgroups identified by the SNF method. Patients in group 2 had significantly poorer PFS than patients in group 1 (P = 0.019; Fig. 1B). Moreover, the clinical features of BRCA patients were compared between the autophagic subgroups. The patients in the group 2 were associated with aggressive intrinsic subtypes, hormone receptor-negative cancers and histological types (Supporting Information Table S3). In particular, patients in the poor survival group were enriched in basal subtypes (Fisher's exact test; P = 1.62 × 10−78; odds ratio 58.11 [95% CI 33.33–105.91]) and triple-negative breast cancer (TNBC) (Fisher's exact test; P = 4.01 × 10−34; odds ratio 28.34 [95% CI 14.08–63.28]). Next, we investigated the differentially expressed ATG genes between the two subgroups, and genes with the most significant difference between the two groups was considered the top candidates. Among them, BECN1, ULK1, ATG2B and ATG12 had lower expression levels in group 2 than in group 1, while RHEB, RB1CC1, ATG5, and ATG9B had higher expression in group 2 than in group 1 (Fig. 1C, Table S2). Interestingly, ULK1 and BECN1 showed a favorable influence of autophagy on BRCA, consistent with previous reports of their roles as autophagic inducers12,29. The impacts of CNA and MET on the autophagic subgroups were consistent with the impact of EXP. We compared the frequencies of copy number amplification and deletion and the frequencies of hypermethylation and hypomethylation in the two subgroups (Supporting Information Tables S4 and S5). Significant enrichment of PDK1, ATG3, ATG5, RHEB, ATG9B, and RB1CC1 amplification and ATG16L1, ATG12, TSC1, ULK1, TSC2, ATG2B, and BECN1 deletion were observed in group 2 (Fig. 1C). In general, the differently expressed ATG genes showed significant differences in the frequencies of CNA gains and losses or in the frequencies of DNA hyper- and hypomethylation status between the autophagic subgroups (Supporting Information Fig. S3). These results indicate that differences in ATG genes between subgroups might reflect disease changes in breast cancer patients.
3.2. Multi-omics network-based identification of candidate autophagic regulators in BRCA
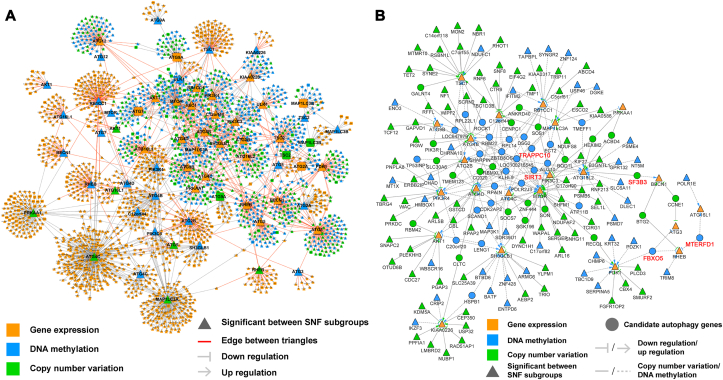
To identify the interactions between candidate autophagic regulators and ATG genes at the multi-omics level, an autophagic network for BRCA was established based upon TCGA data by using the LASSO algorithm (Fig. 2A). CNAs and methylations mostly exhibit their effects through the regulation of EXP. Thus, in our study, we focused on the cross-interactions of CNA-EXP and MET-EXP with CNA/MET as the upstream and EXP as the downstream. In total, 2271 nodes (i.e., 1354 of EXP, 680 of CNA, and 541 of MET) and 2812 interactions were identified (Supporting Information Table S6). Not surprisingly, all the CNA and MET of ATG genes were shown to regulate their own EXP. Moreover, 1168 EXP (86.3%), 662 CNA (97.4%), and 418 MET (77.3%) nodes exhibited significant differences between the two autophagic subgroups (indicated by triangles in Fig. 2A). Interestingly, we found that some positive autophagic regulators in the network that showed low expression levels in group 2 possessed the ability to serve as hub genes. For example, the number of interactions for ULK1, BECN1 and ATG2B was 45, 97, and 123, respectively, which were higher than the average (2.396).
Figure 2.
Construction of the multi-omics network for BRCA using the lasso algorithm with TCGA data. (A) The direction of edges in the multi-omics network is consistently from MET/CNA to EXP. The arrows between nodes depict up- or down-regulation depending on the sign of the Pearson correlation coefficient of MET/CNA with EXP. Details are provided in Supporting Information Table S6. Nodes with significant differences in DNA MET/CNA frequencies and EXP between autophagic subgroups are denoted by triangles, and red edges between triangles indicate that the edges may be key interactions in the cancer-related core autophagy network. (B) Each node of the network is a statistically significant node. Details are provided in Supporting Information Table S7. Nodes linked with multiple core autophagic genes may have key roles in the regulation of the autophagy process and were identified as candidate autophagy targets. Five genes validated experimentally are highlighted in red.
To identify the genes that can potentially regulate key autophagic process, we extracted the regulator network from the autophagic network. This regulator network contained only significantly different nodes between the subgroups, including MET and CNA nodes, with significantly different frequencies and differential expression downstream. The network contained 178 nodes and 248 interactions (Fig. 2B, Supporting Information Table S7). Notably, several genes (i.e., SIRT3, SF3B3, MTERFD1, TRAPPC10, and FBXO5) were connected by more than one core autophagic gene and were thus considered novel autophagic regulators (Supporting Information Table S8). For example, hypomethylation and amplification of SF3B3 might downregulate the expression of ATG16L2 and BECN1, respectively. Hypomethylation of MTERFD1 could downregulate the expression of RHEB and upregulate the expression of ATG16L1. Hypermethylation of SIRT3 could upregulate the expression of ATG4C and downregulate the expression of ATG4B, ATG4D, and ATG16L2. The prognostic potential of these candidate autophagic regulators was estimated (Supporting Information Fig. S4). As mentioned above, lower expression of SIRT3 predicted a significantly poor prognosis, whereas lower expression of SF3B3 was strongly associated with better outcomes. Interestingly, when analyzing the group 1 and group 2, separately, the prognostic correlation of SIRT3 and SF3B3 are opposite (Supporting Information Fig. S5).
3.3. Validation of SF3B3 and SIRT3 as potential autophagic regulators in BRCA
We hypothesized that these candidate autophagic regulators may affect autophagosome formation by regulating ATG genes. To investigate this possibility, the top candidates (e.g., TRPPC10, FBXO5, METRFD1, and SF3B3) were knocked down in human breast carcinoma MCF-7 cells. Consistent with our findings, the modification of these genes altered their predicted druggable targets. For instance, silencing both FBXO5 and METRFD1 down-regulated RHEB, which is a positive regulator of MTOR signaling (Supporting Information Fig. S6A–S6D). However, no obvious transformation of LC3 I to LC3 II was observed after METRFD1 knockdown (Supporting Information Fig. S7B), and LC3 intensity was also not changed obviously (Fig. S7C). Silencing of TRAPPC10 suppressed the expression of ATG4C and MAPLC3A, and promoted the expression of MTOR (Fig. S6E–S6H). The knockdown of SF3B3 resulted in the upregulation of BECN1 and ATG5 (Fig. S6I–S6K) and promoted the expression of beclin-1, the accumulation of LC3 II and an increase in LC3 B fluorescence intensity (Fig. S7A–S7C). These results indicate that SF3B3 might be a negative regulator of autophagy. In addition, SIRT3 could affect the autophagosome formation by regulating ATG4, ATG5, BECN1 and MAPLC3A, which were also identified in the autophagic network. No significant expression change was observed in these target genes after knocking down of SIRT3, while overexpression of SIRT3 significantly enhanced their expression, indicating that the overexpression of SIRT3 could promote autophagy (Supporting Information Fig. S8A–S8F). Based on these results, SF3B3 and SIRT3 were selected for further investigations as to their druggable target potential in breast cancer therapy.
3.4. SF3B3 is a negative autophagic regulator and druggable target by RNA silencing in vitro and in vivo
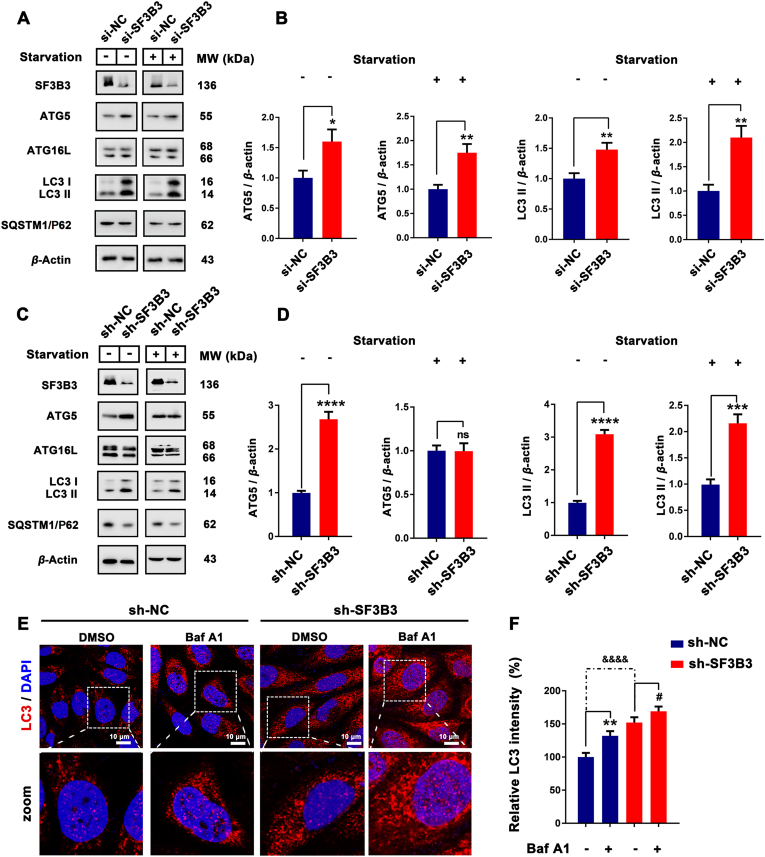
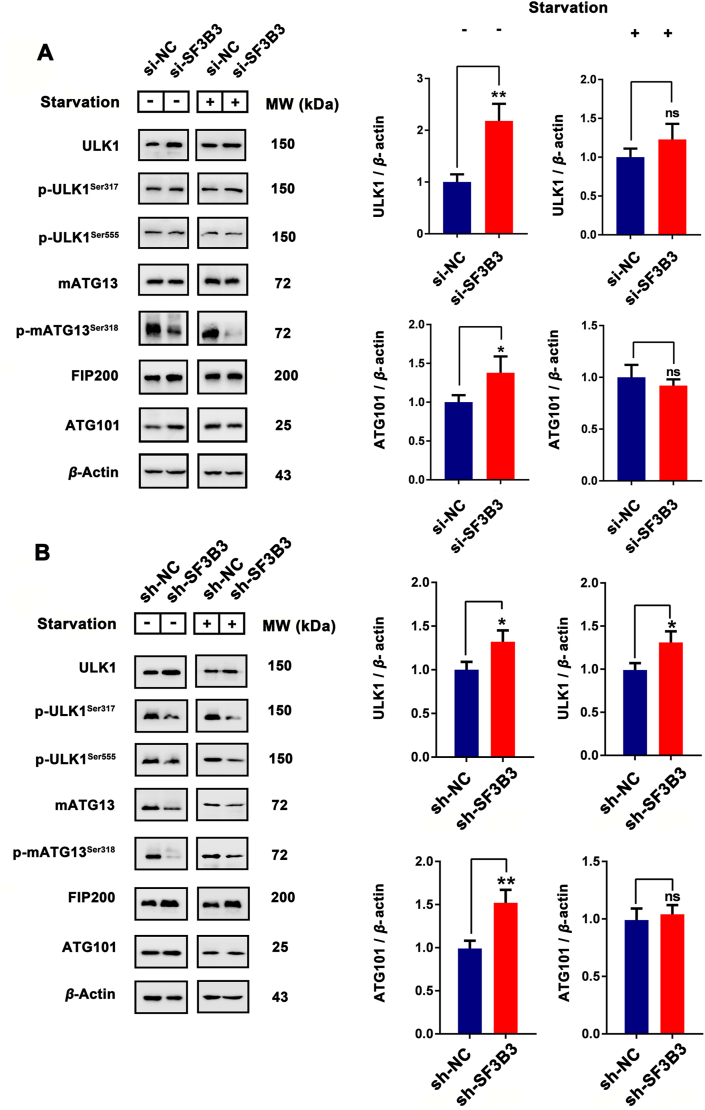
The network prediction results described above indicated that SF3B3 may mainly affect the autophagy process by affecting beclin-1 and ATG16L. Therefore, we detected the expression of ATG5, ATG16L, SQSTM1/P62 and LC3, the main proteins involved in the formation of autophagosome and autophagosome membrane, after the knockdown of SF3B3. The transient knockdown of SF3B3 with siRNA promoted the expression of ATG5 and the up-regulation of LC3 II under both normal and starvation conditions (Fig. 3A and B). Stable knockdown of SF3B3 promoted the expression of ATG5 and the up-regulation of LC3 II under normal conditions; however, under starvation conditions, it promoted the up-regulation of LC3 II but had no obvious effect on the expression of ATG5. It suggests that, in the process of autophagy that has already been carried out, SF3B3 might not regulate autophagy by affecting ATG5 expression (Fig. 3C and D). Additionally, transient knockdown of SF3B3 had little effect on the expression of ATG16L and SQSTM1/P62 under both normal and starvation conditions, but stable knockdown of SF3B3 promoted the degradation of SQSTM1/P62 (Fig. 3A, C, and Supporting Information Fig. S9A). These results indicate that knockdown of SF3B3 had a significant positive effect on LC3 and could activate autophagy. To confirm the effect of knockdown of SF3B3 on LC3 and autophagy, immunofluorescence was applied to detect the expression of LC3 after stable knockdown of SF3B3 together with Baf A1 (lysosome-mediated proteolysis inhibitor). LC3 fluorescence intensity of the stable SF3B3 knockdown group was significantly higher than that of the control group, which demonstrated the activation of LC3 and the occurrence of autophagy in the former. After Baf A1 treatment, the LC3 fluorescence intensity was increased and this demonstrated that SF3B3 knockdown induced autophagy was a continual process (Fig. 3E and F).
Figure 3.
SF3B3 silencing induces autophagy in vitro. (A)–(D) The MCF-7 cells transfected with si-NC, si-SF3B3 or long-term knockout SF3B3 and the expression of ATG5, ATG16L, SQSTM1/P62 and LC3 under normal or starvation conditions quantified by Western blot. ns, no significance; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. (E) Differential expression of LC3 in sh-NC/sh-SF3B3 and Baf A1 treated MCF-7 cells. Cell nucleus were stained with DAPI. Scale bar = 20 μm. ∗∗P < 0.01, sh-NC + Baf A1 vs. sh-NC; #P < 0.01, sh-SF3B3 + Baf A1 vs. sh-SF3B3; &&&&P < 0.0001, sh-SF3B3 vs. sh-NC.
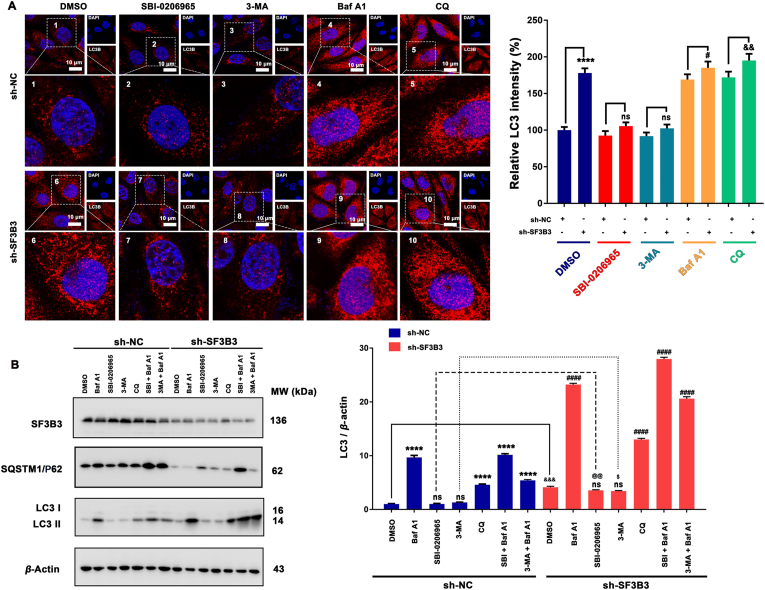
To further explore the effect of SF3B3 on autophagy, we investigated the effect of SF3B3 on the ULK complex, which is the core component of autophagy. The experimental results showed that under normal culture conditions, the expression of ULK1, FIP200 and ATG101 was up-regulated by both transient knockdown and stable knockdown of SF3B3, whereas the effect of transient SF3B3 knockdown on mATG13 expression was not significant and for stable knockdown of SF3B3, mATG13 expression was decreased. Interestingly, under starvation, transient knockdown of SF3B3 had no obvious effect on ULK1 or ATG101. Stable knockdown of SF3B3 up-regulated ULK1 but had no obvious effect on ATG101. Under normal and starvation conditions, the transient knockdown of SF3B3 had no significant effect on the phosphorylation of ULK1, and the phosphorylation of mATG13 was down-regulated. Under normal and starvation conditions, the stable knockdown of SF3B3 inhibited the phosphorylation of ULK1 and mATG13 (Fig. 4 and Fig. S9B–S9F). Since the ULK complex is activated after canonical autophagy initiation, and the main outcome is the phosphorylation of mATG13Ser318 by ULK1. While knockdown of SF3B3 inhibited the phosphorylation of mATG13Ser318, this indicated that knockdown of SF3B3 inhibited ULK1 activity. This is interesting, some of the ULK complex member were up-regulated after SF3B3 knockdown, but ULK1 activity were decreased. In addition, SF3B3 knockdown could also induced autophagy in other two breast cancer cells (MDA-MB-231 and MDA-MB-468), but has little effect on MCF-10A cells (Supporting Information Fig. S10). To further investigate how SF3B3 affect autophagy, we employed different stages of autophagy inhibitors [ULK1 inhibitor SBI-0206965; PI3K inhibitor 3-MA; lysosome-mediated proteolysis inhibitors bafilomycin A1 (Baf A1) and chloroquine (CQ)]. As show in Fig. 5A, ULK1 inhibitor SBI-0206965 and PI3K inhibitor 3-MA could inhibit the up-regulation of LC3 after SF3B3 knockdown compared with sh-SF3B3 control. This suggest ULK1 and PI3K were involved in SF3B3 knockdown induced autophagy. In addition, the autophagy level of SBI-0206965 and 3-MA treated sh-SF3B3 cells was higher than SBI-0206965 and 3-MA treated sh-NC cells, this indicated that ULK1 inhibition or PI3K inhibition could not totally block SF3B3 knockdown induced autophagy. These results suggest that canonical autophagy was contribute to SF3B3 knockdown induced autophagy. While Baf A1 and CQ can increase the accumulation of LC3, which confirmed SF3B3 knockdown induced autophagy was a continuous process. Interestingly, when cells were treated with SBI-0206965/3-MA together with Baf A1, we could observe obviously accumulation of LC3 compared with the SBI-0206965/3-MA treated alone group from the WB results (Fig. 5B), and this suggested some non-canonical autophagy pathways (independent on ULK1) were also involved in SF3B3 knockdown induced autophagy. Therefore, our results indicate that SF3B3 knockdown-induced autophagy maybe a form of canonical autophagy together with non-canonical autophagy in breast cancer cells.
Figure 4.
ULK complex was involved in SF3B3 silencing induces autophagy. MCF-7 cells transfected with (A) si-NC, si-SF3B3 or (B) long-term knockout SF3B3 and the expression of ULK1, p-ULK1Ser317, p-ULK1Ser555, mATG13, p-mATG13Ser318, FIP200 and ATG101 under normal or starvation conditions were quantified by Western blot. ns, no significance; ∗P < 0.05, ∗∗P < 0.01.
Figure 5.
Canonical and non-canonical autophagy both contribute to SF3B3 silencing induced autophagy. (A) MCF-7 cells transfected with sh-NC or sh-SF3B3 and then treated with different stages of autophagy inhibitors (ULK1 inhibitor SBI-0206965 (10 μmol/L); PI3K inhibitor 3-MA (1 mmol/L); lysosome-mediated proteolysis inhibitors Baf A1 (25 nmol/L) and CQ (5 μmol/L)) to investigate the expression and location of LC3 by immunofluorescence. Cell nucleus were stained with DAPI. Scale bar = 20 μm. ∗∗∗∗P < 0.0001, sh-SF3B3 vs. sh-NC; #P < 0.05, sh-SF3B3+Baf A1 vs. sh-NC+Baf A1; &&P < 0.01, sh-SF3B3+CQ vs. sh-NC+CQ. ns, no significance; (B) MCF-7 cells transfected with sh-NC or sh-SF3B3 and then treated with different stages of autophagy inhibitors (ULK1 inhibitor SBI-0206965 (10 μmol/L); PI3K inhibitor 3-MA (1 mmol/L); lysosome-mediated proteolysis inhibitors Baf A1 (25 nmol/L) and CQ (5 μmol/L); SBI-0206965+Baf A1; 3-MA+Baf A1) and the expression of SF3B3, SQSTM1/P62 and LC3 were quantified by Western blot. ns, no significance; ∗∗∗∗P < 0.0001 vs. sh-NC; ####P < 0.0001 vs. sh-SF3B3.
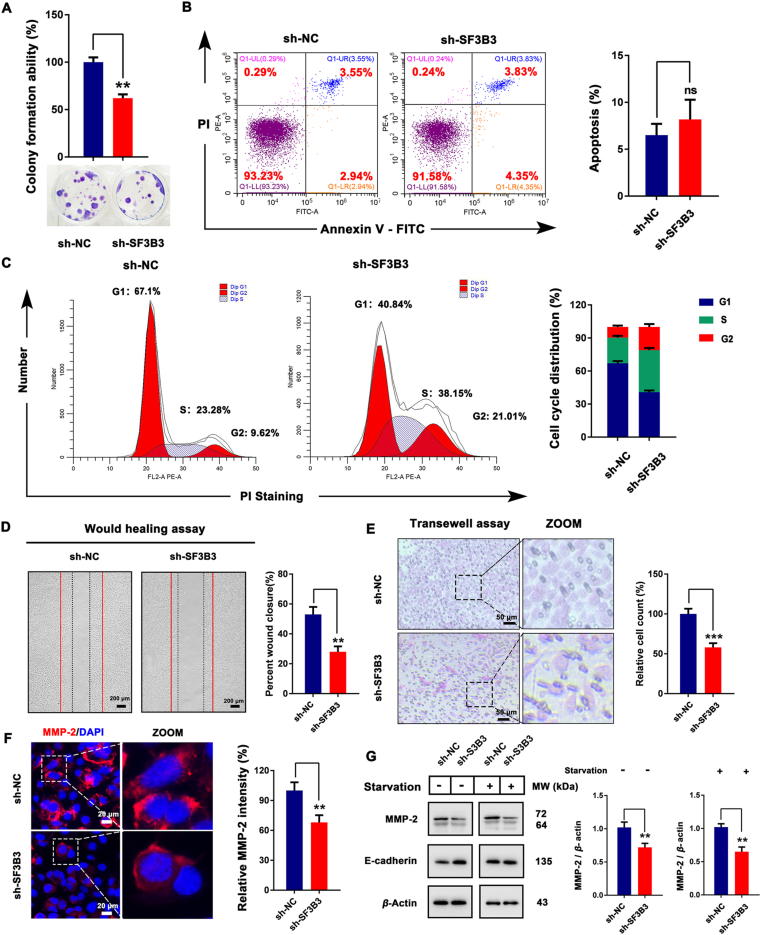
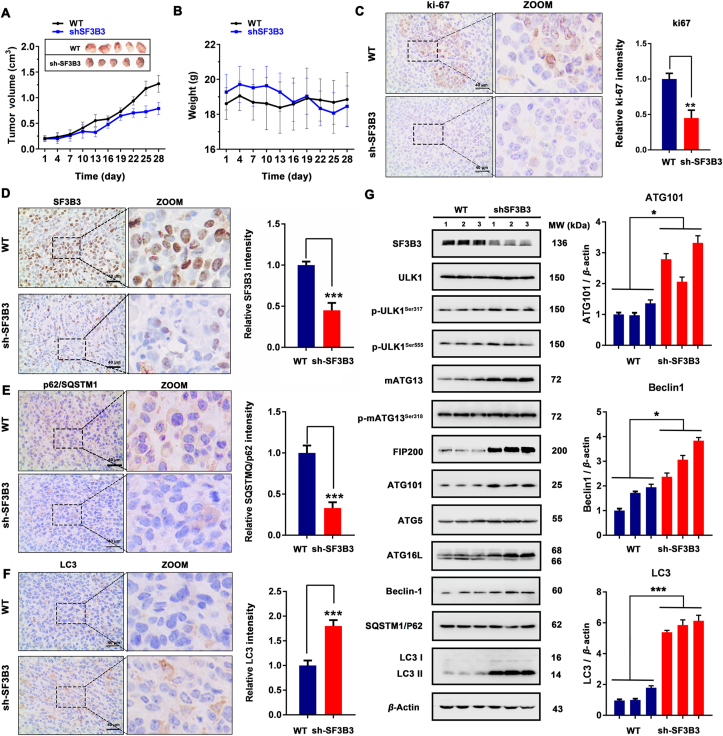
To investigate whether SF3B3 might be a potential drug target for the treatment of breast cancer, we evaluated the effects of SF3B3 knockdown on cell proliferation, apoptosis, cell cycle, and migration. We conducted colony formation experiments, which showed that SF3B3 knockdown inhibited the colony formation ability of MCF-7 cells (Fig. 6A). Subsequently, we examined the effects of SF3B3 knockdown on apoptosis and cell cycle and found that SF3B3 knockdown did not affect apoptosis (Fig. 6B) but significantly promoted MCF-7 cell cycle distribution change (Fig. 6C). Next, we investigated the effect of SF3B3 knockdown on cell migration by wound healing assay and Transwell assay. We found that SF3B3 knockdown inhibited cell migration (Fig. 6D and E). In addition, we detected the expression of MMP-2 and E-cadherin, two major proteins related to tumor migration and invasion. We found that SF3B3 knockdown significantly reduced the fluorescence intensity of MMP-2 and inhibited the expression of MMP-2 (Fig. 6F and G), suggesting that SF3B3 may positively regulate tumor migration and invasion. Interestingly, under normal conditions, the knockdown of SF3B3 increased the expression of E-cadherin, but under starvation conditions, it had no significant effect on E-cadherin expression. Taken together, these results suggest that SF3B3 knockdown inhibits the malignant progression of MCF-7 breast cancer cells. To further detect the relationship between SF3B3 knockdown induced autophagy and phenotypic change (proliferation, apoptosis, cell cycle and migration), we also employed these autophagy inhibitors. The results demonstrated that SBI-0206965, 3-MA, Baf A1 and CQ treated sh-SF3B3 cells had no significant effect on cell viability compared with sh-NC cells, but improved the colony formation ability compared with sh-SF3B3 cells (Supporting Information Figs. S11A, S11B, and S13A); SBI-0206965, 3-MA, Baf A1 and CQ treatment had no obvious effect on apoptosis and SF3B3 knockdown induced cell cycle distribution change (Figs. S11C, S11D, and S13B). Of note, SF3B3 knockdown induced obvious cell cycle distribution change, and this may contribute a lot to SF3B3 knockdown induced cell death. In addition, SBI-0206965, 3-MA and CQ could enhance cell migration after SF3B3 knockdown (Supporting Information Figs. S12 and S14). These results suggest that SF3B3 knockdown induced autophagy inhibited cell proliferation and migration, but has little effect on cell apoptosis and cell cycle distribution change. Subsequently, we evaluated the effect of SF3B3 knockdown on autophagy and proliferation in vivo in SF3B3-knockdown MCF-7 xenograft mice. After 28 days of growth, the mice were sacrificed and the tumor were removed for index detection. Based on the results of tumor volume (Fig. 7A), we found that SF3B3-knockdown could significantly inhibit the growth of xenograft MCF-7 breast cancer cells. The body weights of mice were relative stable, which indicated SF3B3-knockdown had no obvious side effects (Fig. 7B). In addition, SF3B3-knockdown could decrease the expression of Ki-67, and this also demonstrated its tumor growth inhibition ability (Fig. 7C). To detect whether SF3B3-knockdown in xenograft MCF-7 breast cancer cells could induce autophagy, we next evaluated the expression of some autophagy markers with immunohistochemical. From the results of SQSTM1/P62 and LC3 staining, we found that SF3B3-knockdown induced obvious autophagy in xenograft MCF-7 breast cancer cells, with significant degradation of SQSTM1/P62 and up-regulation of LC3 (Fig. 7D–F). Next, we carried out Western blot analysis to clarify the autophagy induction mechanism of SF3B3-knockdown in vivo. Similar as the in vitro results, SF3B3-knockdown up-regulated the expression of ATG101, FIP200 and LC3 II. But the expression and phosphorylation of ULK1 were not changed obviously. The expression of mATG13 was increased and its phosphorylation was not affected. beclin-1 and ATG16L were up-regulated, but had little effect on ATG5 and SQSTM1/P62. Interestingly, SF3B3 knockdown appears to be more beneficial to the autophagy process in vivo than in vitro (Fig. 7G). Thus, these results suggest that SF3B3 knock down has a positive regulatory effect on the progression of autophagy and negative effect on tumor growth. Together, these findings provide the first evidence that SF3B3 is a negative regulator of autophagy. Thus, SF3B3 may be a promising target in breast cancer.
Figure 6.
SF3B3 is a positive regulator of breast cancer progression in vitro. (A) Colony formation assay of MCF-7 cells after SF3B3 knockdown. ∗∗P < 0.01. (B) Apoptosis assay of MCF-7 cells after SF3B3 knockdown detected by Annexin V-PI double staining. (C) Cell cycle analysis of MCF-7 cells after SF3B3 knockdown detected by PI staining. (D) Would healing assay of MCF-7 cells after SF3B3 knockdown. Percent wound closure quantified by Image J software; ∗∗P < 0.01. (E) Transwell assay of MCF-7 cells after SF3B3 knockdown. Scale bar = 50 μm. (F) Differential expression of MMP-2 in sh-NC- or sh-SF3B3-treated MCF-7 cells. Cell nucleus were stained with DAPI. Relative MMP-2 intensity was quantified by Image J software; ∗∗∗P < 0.001. Scale bar = 20 μm. (G) Western blot analysis of MMP-2 and E-cadherin in MCF-7 cells after SF3B3 knockdown. Relative MMP-2 expression level was quantified by normalization to β-actin. ∗∗P < 0.01 vs. the NC group.
Figure 7.
SF3B3 knockdown induces autophagy in vivo. (A) Tumor volume and (B) body weight of WT and SF3B3-knockdown tumor tissue (n = 10); (C)–(F) The expression of Ki-67, SF3B3, SQSTM1/P62 and LC3B determined by immunohistochemistry in representative tumor sections of mice after SF3B3 knockdown. Scale bar = 40 μm. Relative Ki-67, SF3B3, SQSTM1/P62 and LC3B expression was quantified by normalization to the WT group; ∗∗P < 0.01, ∗∗∗P < 0.001. (G) Western blot analysis of SF3B3, ULK1, p-ULK1Ser317, p-ULK1Ser555, mATG13, p-mATG13Ser318, FIP200, ATG101, ATG5, ATG16L, beclin-1, SQSTM1/P62 and LC3 II. Relative ATG101, beclin-1 and LC3 II expression was quantified by normalization to β-actin expression. ∗P < 0.05, ∗∗∗P < 0.001.
3.5. SIRT3 is a positive autophagic regulator and a druggable target
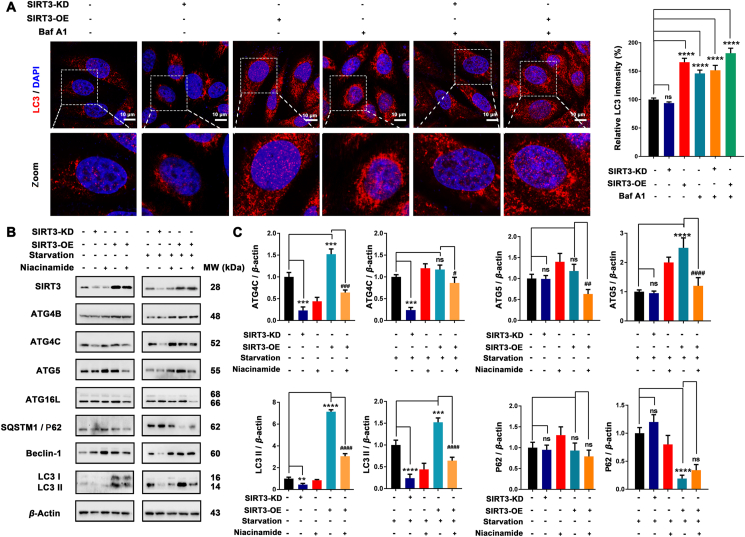
The effect of SIRT3 on autophagy was examined by knockdown or amplification. We found that the LC3 B puncta was a little reduced in SIRT3 knockdown cells compared with the control cells, while over-expression of SIRT3 markedly enhanced the expression of LC3, indicating that SIRT3 may positively regulate autophagy. In addition, Baf A1 plus SIRT3 overexpression could increase the accumulation of LC3 compared with SIRT3 overexpression alone (Fig. 8A), which confirmed that SIRT3 overexpression induced autophagy was a continuous process. Of note, SIRT3 is a deacetylase, which functions primarily by deacetylation of the corresponding substrate. Therefore, we explored whether the effect of SIRT3 on autophagy is dependent on its deacetylation function by introducing nicotinamide, which can inhibit the deacetylation function of SIRT3. Based on the predicted SIRT3 autophagy network, we investigated the effects of SIRT3 on ATG4, ATG5, ATG16, SQSTM1/P62, beclin-1 and LC3. Interestingly, knockdown of SIRT3 resulted in decreased expression of ATG4C and LC3-II in both normal and starvation conditions but inhibited the expression of beclin-1 only after starvation. However, overexpression of SIRT3 slightly increased the expression of ATG4B (with no significant effect under starvation condition), ATG5, ATG16L, beclin-1, and LC3 II and promoted the degradation of SQSTM1/P62. Notably, the expression-promoting effects of SIRT3 on ATG4C, ATG5, ATG16L and LC3 II were compromised (Fig. 8B and C) after niacinamide treatment. These results indicate that SIRT3 can significantly promote the expression of key proteins involved in autophagosome formation and that this ability is strongly dependent on deacetylation.
Figure 8.
SIRT3 activation induces deacetylation-dependent autophagy in vitro. (A) Differential expression of LC3B in MCF-7 cells after 48 h post-transfection with the indicated oligos and co-treated with Baf A1. Cell nucleus were stained with DAPI. Scale bar = 10 μm. Relative LC3 intensity of each group was quantified by Image J software; ns, no significance; ∗∗∗∗P < 0.0001. (B) Western blot analysis of SIRT3, ATG4B, ATG4C, ATG5, ATG16L, SQSTM1/P62, beclin-1 and LC3. (C) Relative ATG4C, ATG5, LC3 II and SQSTM1/P62 expression were quantified by normalization to β-actin expression. ns, no significance; ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 vs. the NC group; #P < 0.05, ###P < 0.001, and ####P < 0.0001 vs. the SIRT3-OE group.
Subsequently, we examined the effect of SIRT3 on the ULK complex. We evaluated the protein levels of ULK1, FIP200, mATG13, and ATG101, and we examined the phosphorylation levels of ULK1 and mATG13 to characterize the activation of autophagy. SIRT3 overexpression showed no significant effect on the expression levels of ULK1, FIP200, mATG13 and ATG101. However, the phosphorylation of ULK1 and mATG13 was significantly activated, and niacinamide blocked this phosphorylation. Intriguingly, under the starvation condition, the inhibitory effect of niacinamide on the phosphorylation of ULK1 and mATG13 induced by SIRT3 overexpression was not as strong as it was under normal conditions (Supporting Information Fig. S15). Thus, we speculate that SIRT3 deacetylation may not contribute significantly to the activation of the ULK complex when autophagy is already in progress. Since we noticed that SIRT3 overexpression promoted the phosphorylation of ULK1 at Ser317 and Ser555, which are two AMPK activated sites, so we knockdown of AMPK with si-RNA to identify the role of AMPK in SIRT3 overexpression induced autophagy. From the results we found that AMPK knockdown significantly block SIRT3 overexpression induced autophagy (Supporting Information Fig. S16). Therefore, we speculated that SIRT3 overexpression induced autophagy is highly dependent on AMPK regulated pathways. In addition, SIRT3 overexpression could also induced autophagy in other two breast cancer cells (MDA-MB-231 and MDA-MB-468), and promoted MCF-10A cells autophagy (Supporting Information Fig. S17).
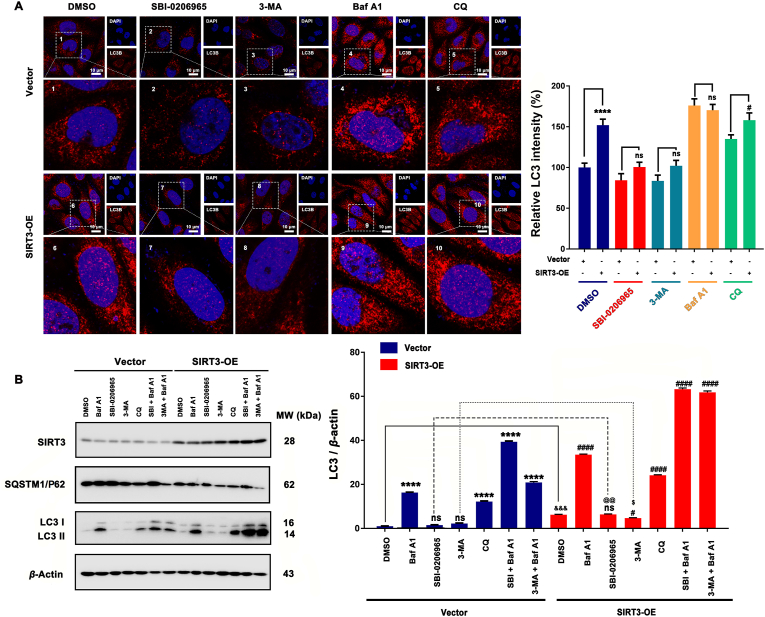
To further investigate how SIRT3 affect autophagy, we employed different stages of autophagy inhibitors (SBI-0206965; 3-MA; bafilomycin A1 and chloroquine). From the LC3 immunofluorescence staining results in Fig. 9A, ULK1 inhibitor SBI-0206965 and PI3K inhibitor 3-MA could inhibit the up-regulation of LC3 after SIRT3 overexpression compared with SIRT3-OE control. This suggest ULK1 and PI3K were involved in SIRT3 overexpression induced autophagy. In addition, the autophagy level of SBI-0206965 and 3-MA treated SIRT3-OE cells was higher than SBI-0206965 and 3-MA treated vector cells, this indicated that ULK1 inhibition or PI3K inhibition could not totally block SIRT3 overexpression induced autophagy. These results suggest canonical autophagy was contribute to SIRT3 overexpression induced autophagy. While Baf A1 and CQ can increase the accumulation of LC3, which confirmed SIRT3 overexpression induced autophagy was a continuous process. Interestingly, when cells were treated with SBI-0206965/3-MA together with Baf A1, we could observe obviously accumulation of LC3 compared with the SBI-0206965/3-MA treated alone group from the WB results (Fig. 9B), and this suggested some non-canonical autophagy pathways (Independent on ULK1) were also involved in SIRT3 overexpression induced autophagy.
Figure 9.
Canonical and non-canonical autophagy both contribute to SIRT3 overexpression induced autophagy. (A) MCF-7 cells transfected with vector or SIRT3 and then treated with different stages of autophagy inhibitors (ULK1 inhibitor SBI-0206965 (10 μmol/L); PI3K inhibitor 3-MA (1 mmol/L); lysosome-mediated proteolysis inhibitors Baf A1 (25 nmol/L) and CQ (5 μmol/L)) to investigate the expression and location of LC3 by immunofluorescence. Cell nucleus were stained with DAPI. Scale bar = 10 μm. ∗∗∗∗P < 0.0001, SIRT3-OE vs. Vector; #P < 0.05, SIRT3-OE+CQ vs. Vector+CQ; ns, no significance. (B) MCF-7 cells transfected with vector or SIRT3 and then treated with different stages of autophagy inhibitors (ULK1 inhibitor SBI-0206965 (10 μmol/L); PI3K inhibitor 3-MA (1 mmol/L); lysosome-mediated proteolysis inhibitors Baf A1 (25 nmol/L) and CQ (5 μmol/L); SBI-0206965+Baf A1; 3-MA+Baf A1 and the expression of SIRT3, SQSTM1/P62 and LC3 were quantified by Western blot. ns, no significance; ∗∗∗∗P < 0.0001 vs. Vector; ####P < 0.0001 vs. SIRT3-OE.
To investigate whether SIRT3 might be a potential drug target for the treatment of breast cancer, we examined the growth effects of SIRT3 on MCF-7 cells. We detected the effect of SIRT3 on cell proliferation through colony formation assay. SIRT3 overexpression inhibited the colony formation of MCF-7 cells (Supporting Information Fig. S18A). In addition, overexpression of SIRT3 significantly decreased the fluorescence intensity of ki-67, consistent with the previous result (Fig. S18B). Interestingly, SIRT3 knockdown had no significant effect on colony formation or ki-67 expression of MCF-7 cells, suggesting that SIRT3 may not be necessary for MCF-7 cell proliferation but that overexpression of SIRT3 inhibits MCF-7 cell proliferation. Subsequently, we investigated the effects of SIRT3 on key tumor migration and invasion proteins. SIRT3 knockdown inhibited the expression of E-cadherin, while SIRT3 overexpression significantly increased it (Fig. S18C). In addition, knockdown of SIRT3 promoted the expression of MMP-2 and MMP-9, while overexpression of SIRT3 had the opposite effect (Fig. S18D). These results suggest that SIRT3 overexpression may have a potential to inhibit the migration and invasion of MCF-7 cells. To further detect the relationship between SIRT3 overexpression induced autophagy and migration, we also employed these autophagy inhibitors. The results demonstrated SBI-0206965, 3-MA, Baf A1 and CQ treatment had no significant effect on cell viability but improved colony formation ability in SIRT3 overexpressed cells; and SBI-0206965, 3-MA and Baf A1 could enhance cell migration in SIRT3 overexpressed cells (Supporting Information Figs. S19 and S20). This suggest autophagy contribute to SIRT3 overexpression induced proliferation and migration suppression.
The above experimental results reveal that SF3B3 and SIRT3 can be used as negative or positive regulators of autophagy and that their regulatory effects on autophagy are a little different. The network prediction and experimental results reveal that canonical and non-canonical autophagy pathways were both contribute to SF3B3 knockdown and SIRT3 overexpression induced autophagy. And the knockdown of SF3B3 and the overexpression of SIRT3 were identified as beneficial for the treatment of breast cancer. Therefore, the development of SF3B3 inhibitor and SIRT3 activator may be a new strategy for the treatment of breast cancer with autophagy induction.
3.6. MA is a new SIRT3 activator with pronounced antiproliferative activity achieved by enhancing autophagy
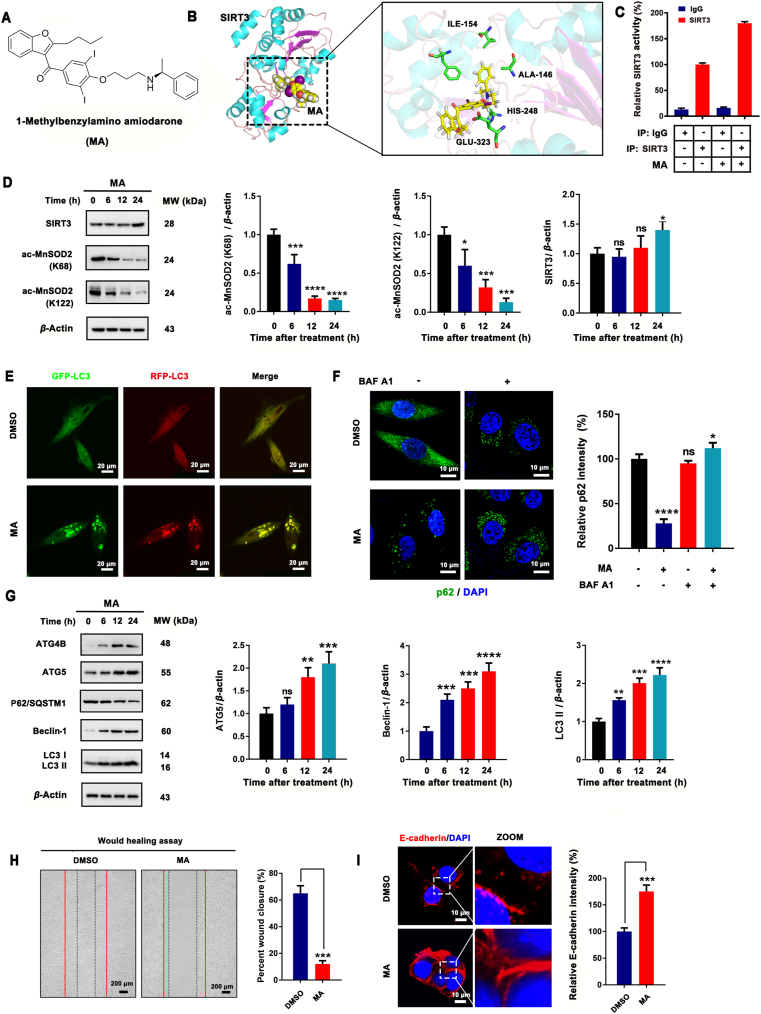
In this study, we utilized a multiple docking strategy to discover the lead activator of SIRT3. A total of 212,255 compounds in the SPECS library were filtered by the Lipinski's rule of five, yielding 97,360 retained compounds. Then, structure-based molecular docking was performed using LibDock and CDOCKER (Supporting Information Fig. S21). We then selected a new SIRT3 activator, 1-methylbenzylamino amiodarone (MA), based on the results of autophagic and SIRT3 activation experiments (Fig. 10A and B). MA was able to activate SIRT3 in MCF-7 cells, which stimulated the deacetylation ability of SIRT3 (Fig. 10C and D). The acetylation levels of MnSOD2 at K68 and K122, the substrates of SIRT3, were significantly downregulated in a time-dependent manner, as demonstrated by Western blot. In addition, the effect of MA on MCF-7 cell autophagy was examined. MA obviously promoted the aggregation of LC3 puncta (Fig. 10E) and enhanced the degradation of SQSTM1/P62. In addition, Baf A1 could increase the aggregation of SQSTM1/P62 after MA treatment (Fig. 10F), which proved that MA-induced autophagy was a continuous progress. Next, we evaluated the expression of ATG4B, ATG5, SQSTM1/P62, beclin1 and LC3 with Western blot and found MA up-regulated ATG4B, ATG5 and beclin-1 in a time-dependent manner. Furthermore, MA promoted the degradation of SQSTM1/P62 and the up-regulation of LC3 II (Fig. 10G) and inhibited the migration of MCF-7 cells via the up-regulation of E-cadherin (Fig. 10H and I). Taken together, these results suggest that MA, a small-molecule compound, harbors SIRT3-activation ability and potential anti-tumor activity.
Figure 10.
MA is a novel SIRT3 activator that induces MCF-7 cell autophagy. (A) Chemical structure of a new SIRT3 activator, 1-methylbenzylamino amiodarone (MA). (B) Molecular docking of MA and SIRT3. (C) The cellular SIRT3 deacetylase activity of MA was determined by SIRT3 activity assay kit using cellular extracts obtained from MCF-7 cells. (D) The expression levels of SIRT3 as well as acetylated MnSOD2 at K68 and K122 were determined by Western blot. The relative expression level of SIRT3, acetylated MnSOD2 at K68 and K122 was quantified by normalization to β-actin expression. ∗P < 0.05, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001 vs. control. (E) Representative immunofluorescence images of LC3 puncta in MCF-7 cells transiently expressing GFP-mRFP-LC3 plasmid followed by treatment with 20 μmol/L MA. Scale bar = 20 μm. (F) Representative immunofluorescence images of SQSTM1/P62 expression and location in MCF-7 cells treated with 20 μmol/L MA. Scale bar = 10 μm. (G) Western blot analysis of ATG4B, ATG5, SQSTM1/P62, beclin-1 and LC3 in MCF-7 cells treated with 20 μmol/L MA for 6, 12, or 24 h. Relative ATG5, beclin-1 and LC3 II expression were quantified by normalization to β-actin expression; ns, no significance; ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. (G) Would healing assay of MCF-7 cells after MA treatment. Percent wound closure was quantified by Image J software; ∗∗∗P < 0.001. (H) Differential expression of E-cadherin in MA-treated MCF-7 cells. Cells were counterstained with DAPI. Relative E-cadherin intensity was quantified by Image J software; ∗∗∗P < 0.01. Scale bar = 10 μm.
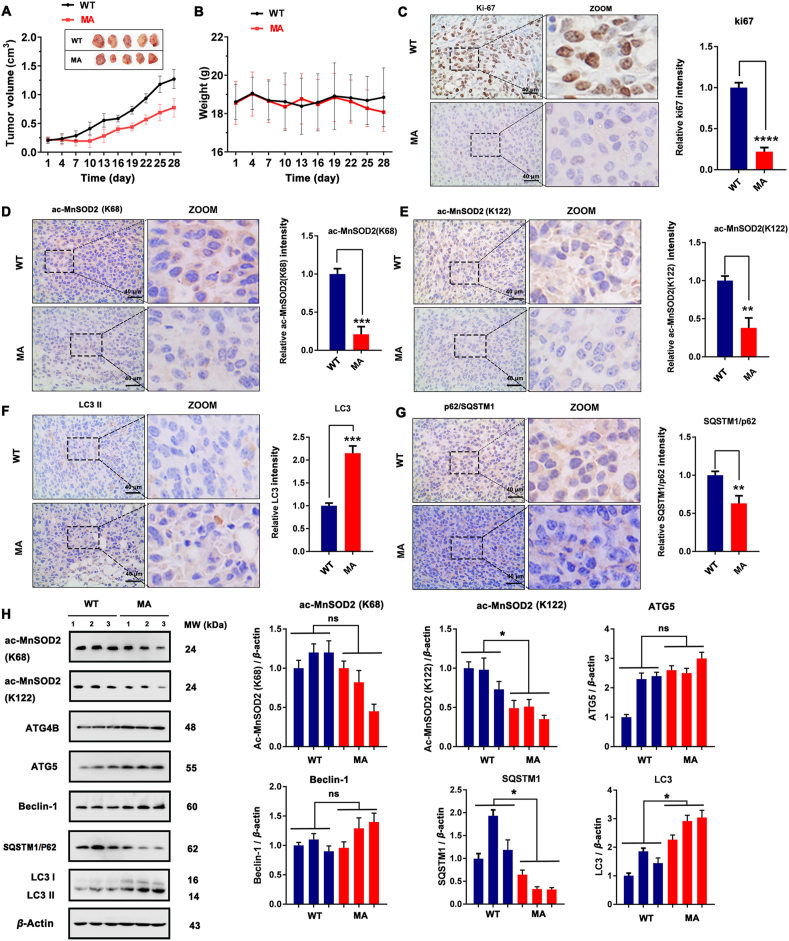
Intriguingly, MA exhibited significant SIRT3-activation ability and autophagy-induction activity in vivo as well as good anti-tumor activity. Based on the results of tumor volume (Fig. 11A), we found that MA could significantly inhibit the growth of xenograft MCF-7 breast cancer cells. The body weights of mice were relative stable, which indicated MA had no obvious side effects (Fig. 11B). In addition, MA decreased the expression of Ki-67, and this also proved it inhibited tumor growth (Fig. 11C). Next, we evaluated the expression of ac-MnSOD2 (K68 and K122) and autophagy markers (LC3 and SQSTM1/P62) with immunohistochemical. The results indicate that compared with the WT group, the MA group exhibited a significantly reduced acetylation level of MnSOD2 at K68 and K122 (Fig. 11D and E), increased LC3 II expression (Fig. 11F), and downregulated expression of SQSTM1/P62 (Fig. 11G). However, the activation of ATG4B, ATG5 and beclin1 in vivo was not obvious (Fig. 11H). Notably, MA exhibited apoptosis-induction ability and potential inhibitory activity of migration (Supporting Information Fig. S22). These results indicate that MA is an autophagy inducer with SIRT3-activation function and might be a candidate compound for the treatment of breast cancer.
Figure 11.
MA is a novel SIRT3 activator that induces MCF-7 cell autophagy in vivo. (A) Tumor volume and (B) body weight of WT and MA-treated tumor tissue; the MA-treated group shared the same control (WT) with the MCF7shSF3B3 group (n = 10). (C)–(G) The expression of Ki-67, ac-MnSOD2 (K68), ac-MnSOD2 (K122), SQSTM1/P62 and LC3 determined by immunohistochemistry in representative tumor sections of mice after MA treatment. Scale bar = 40 μm. Relative expressions of Ki-67, ac-MnSOD2 (K68), ac-MnSOD2 (K122), SQSTM1/P62 and LC3 were quantified by normalization to the WT group; ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. (H) Western blot analysis of the expressions of ac-MnSOD2 (K68), ac-MnSOD2 (K122), ATG4B, ATG5, beclin-1, SQSTM1/P62 and LC3. The relative expressions of ac-MnSOD2 (K68), ac-MnSOD2 (K122), ATG5, beclin-1, SQSTM1/P62 and LC3 II were quantified by normalization to β-actin expression. ns, no significance; ∗P < 0.05.
4. Discussion
To date, a few key autophagic regulators in breast cancer have been intensively explored by a range of elegant experimental methods; however, adequate autophagy-based therapeutic interventions have yet to be developed15,30. In silico autophagy-related methods, especially omics-based approaches, have been approved as crucial supplements to existing experimental methods for potential autophagy-targeted therapy31. With the development of next-generation sequencing, abundant multi-omics datasets of various cancer types have been gradually released. To decipher the rules of autophagy for breast cancer pharmaceutical applications and therapeutics, multi-omics approaches with experimental validation appear to be among the most promising therapeutic strategies.
Moreover, the detection of different types of autophagy-related omics alterations in breast cancer is becoming increasingly possible. As one example in genomics, copy number loss of BECN1 was reported to decrease BECN1 expression and thereby reduce the levels of autophagy in breast cancer12. As another example in epigenomics, alterations of MET have been recognized as a key component of cancer development. More recently, methylation alterations of the autophagy gene GABARAPL1 have been reported to be correlated with breast cancer grade32. Additionally, transcriptomics alterations based upon TCGA data, including differentially expressed genes, long noncoding RNAs33, and microRNAs34, have been validated as important information for identifying novel autophagic regulators in BRCA. However, these single-omics approaches cannot capture the entire biological complexity of autophagy in breast cancer35. Thus, the integration of multiple omics technologies has been emerging as a new approach to provide a more comprehensive view of breast cancer and autophagy. In this study, we subjected different types of omics data, i.e., EXP, MET and CNA data, from TCGA, to multi-omics approaches, including the SNF method and the LASSO algorithm, with the goal of discovering candidate autophagic regulators and druggable targets in BRCA. To our knowledge, our multi-omics approaches may provide the first comprehensive resource for identifying new autophagic regulators and druggable targets of BRCA for potential drug development. Some comprehensive resources of breast cancer exist, such as ActivePathways, which identified 192 significantly enriched GO biological processes and reactome pathways across four subtypes of breast cancer, providing prognostic signatures involved in pathways of immune response and apoptosis and other pathways36. Compared with our study, such resources have inherent limitations and lack experimental validation.
As mentioned above, in addition to employing multi-omics approaches, we experimentally elucidated some candidate autophagic regulators and relevant targets in breast cancer. We identified two key autophagic regulators, SF3B3 and SIRT3, and subsequently, further in vitro and in vivo experimental validations could support our results the context of BRCA (Supporting Information Fig. S23). SF3B3 has been reported to be associated with high tumor stage and poor overall survival in breast cancer37; however, to our knowledge, its correlation to autophagy has not been reported, indicating that SF3B3 might be a new autophagic regulator. Our study provides the first report that SF3B3 is a negative autophagic regulator in BRCA. To verify it target potential, we used SF3B3 silencing (siRNA/shRNA technology) to induce autophagy in BRCA. Interestingly, no SF3B3 inhibitor has been reported to date. More importantly, we found that SF3B3 silencing induced obvious autophagy, and autophagy was involved in oppose the cell proliferation and migration induced by SF3B3. This result suggests that SF3B3 is not only a negative regulator of autophagy but also a potential druggable target of BRCA in vitro and in vivo. In breast cancer, the development of SF3B3 inhibitors may become a promising treatment strategy. Considering its crosstalk with autophagy, autophagy activators might also be helpful in the treatment of SF3B3 highly expressed breast cancer. In our future studies, we aim to screen downstream splicing target genes of SF3B3 and elucidate its possible autophagy-regulated mechanisms in BRCA.
SIRT3 has been recently been reported to induce mitochondrial autophagy (mitophagy) and to act as both oncogene and tumor suppressor in different types of human cancers38,39; however, the conditions that determine which role SIRT3 plays remains to be clarified. Whereas SIRT3 can confer resistance to chemotherapy and tamoxifen40,41, lower SIRT3 expression is associated with poor outcome42. Interestingly, SIRT3 has not yet been included in Gene Ontology (GO) annotation of autophagy [GO:0016238], indicating that the role of SIRT3 in autophagy has not been totally confirmed. In this study, we identified that overexpression of SIRT3 could induce canonical and non-canonical autophagy, which demonstrated it might be a positive regulator of autophagy in breast cancer. In addition, we designed a novel small-molecule activator of SIRT3, 1-methylbenzylamino amiodarone, which could induce autophagy-associated cell death in breast cancer. By employing this activator, we demonstrated that SIRT3 is not only a positive regulator of autophagy but also a possible druggable target of BRCA in vitro and in vivo.
5. Conclusions
In summary, we used multi-omics approaches to identify candidate autophagic regulators in BRCA, such as genes with a prognosis-associated mutation (SF3B3 and TRAPPC10) and prognosis/survival-correlated expression (SIRT3, MTERFD1, and FBXO5), by integrating the SNF method and the LASSO algorithm. Moreover, we identified the positive regulator of autophagy SIRT3 and the negative regulator of autophagy SF3B3 as potential druggable targets by employing a new small-molecule activator of siRNA/shRNA in in vitro and in vivo breast cancer models. Taken together, the findings of this study identify SF3B3 and SIRT3 as candidate autophagic regulators and druggable targets and provide insight into the use of multi-omics approaches for developing autophagy-targeted therapeutic strategies and future breast cancer drug discovery.
Acknowledgments
We are grateful to Prof. Canhua Huang (Sichuan University), Prof. Haoyang Cai (Sichuan University), Dr. Lan Zhang (Southwest Jiaotong University) and Dr. Xin Wen (University of Michigan, Ann Arbor, USA) for their critical reviews of the manuscript. This work was supported by grants from National Science and Technology Major Project of the Ministry of Science and Technology of the People's Republic of China (No. 2018ZX09735005), National Natural Science Foundation of China (Grant Nos. 81522028, 81673452, 81673455, 81873939, 81803365 and 81602953). Post-Doctor Research Project (2018M643510, China) and Post-Doctor Research Project of West China Hospital, Sichuan University (Grant No. 2018HXBH065, China). Prof. Heng Xu was supported by the grant from “The Recruitment Program of Global Young Experts” (known as “the Thousand Young Talents Plan”, China).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2020.12.013.
Contributor Information
Heng Xu, Email: xuheng81916@scu.edu.cn.
Bo Liu, Email: liubo2400@163.com.
Author contributions
Heng Xu and Bo Liu designed the experiment; Shouyue Zhang and Ziyi Qin carried out the data processing and information generation analysis; Yang An, Xiaoxi Zeng, Jin Zhang and Yuqian Zhao carried out the biological experiment; Shouyue Zhang, Jin Zhang and Yang An wrote the manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Levine B., Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rybstein M.D., Bravo-San Pedro J.M., Kroemer G., Galluzzi L. The autophagic network and cancer. Nat Cell Biol. 2018;20:243–251. doi: 10.1038/s41556-018-0042-2. [DOI] [PubMed] [Google Scholar]
- 3.Ashrafi G., Schwarz T.L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei Y., Zou Z., Becker N., Anderson M., Sumpter R., Xiao G. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154:1269–1284. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cicchini M., Chakrabarti R., Kongara S., Price S., Nahar R., Lozy F. Autophagy regulator BECN1 suppresses mammary tumorigenesis driven by WNT1 activation and following parity. Autophagy. 2014;10:2036–2052. doi: 10.4161/auto.34398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalby K.N., Tekedereli I., Lopez-Berestein G., Ozpolat B. Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy. 2010;6:322–329. doi: 10.4161/auto.6.3.11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waks A.G., Winer E.P. Breast cancer treatment: a review. J Am Med Assoc. 2019;321:288–300. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- 8.Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esteva F.J., Hubbard-Lucey V.M., Tang J., Pusztai L. Immunotherapy and targeted therapy combinations in metastatic breast cancer. Lancet Oncol. 2019;20:e175–e186. doi: 10.1016/S1470-2045(19)30026-9. [DOI] [PubMed] [Google Scholar]
- 10.Xiang H., Zhang J., Lin C., Zhang L., Liu B., Ouyang L. Targeting autophagy-related protein kinases for potential therapeutic purpose. Acta Pharm Sin B. 2020;10:569–581. doi: 10.1016/j.apsb.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L., Fu L., Zhang S., Zhang J., Zhao Y., Zheng Y. Discovery of a small molecule targeting ULK1-modulated cell death of triple negative breast cancer and. Chem Sci. 2017;8:2687–2701. doi: 10.1039/c6sc05368h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang H., Sebti S., Titone R., Zhou Y., Isidoro C., Ross T.S. Decreased mRNA expression in human breast cancer is associated with estrogen receptor-negative subtypes and poor prognosis. EBioMedicine. 2015;2:255–263. doi: 10.1016/j.ebiom.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Li N., Jiang W., Deng W., Ye R., Xu C. Mutant LKB1 confers enhanced radiosensitization in combination with trametinib in KRAS-mutant non-small cell lung cancer. Clin Cancer Res. 2018;24:5744–5756. doi: 10.1158/1078-0432.CCR-18-1489. [DOI] [PubMed] [Google Scholar]
- 14.Muhammad J.S., Nanjo S., Ando T., Yamashita S., Maekita T., Ushijima T. Autophagy impairment by Helicobacter pylori-induced methylation silencing of MAP1LC3Av1 promotes gastric carcinogenesis. Int J Cancer. 2017;140:2272–2283. doi: 10.1002/ijc.30657. [DOI] [PubMed] [Google Scholar]
- 15.Shinde A., Hardy S.D., Kim D., Akhand S.S., Jolly M.K., Wang W.H. Spleen tyrosine kinase-mediated autophagy is required for epithelial-mesenchymal plasticity and metastasis in breast cancer. Cancer Res. 2019;79:1831–1843. doi: 10.1158/0008-5472.CAN-18-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pineda S., Real F.X., Kogevinas M., Carrato A., Chanock S.J., Malats N. Integration analysis of three omics data using penalized regression methods: an application to bladder cancer. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hudson T.J., Anderson W., Artez A., Barker A.D., Bell C., Bernabé R.R. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dimitrakopoulos C., Hindupur S.K., Häfliger L., Behr J., Montazeri H., Hall M.N. Network-based integration of multi-omics data for prioritizing cancer genes. Bioinformatics. 2018;34:2441–2448. doi: 10.1093/bioinformatics/bty148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruffalo M., Koyutürk M., Sharan R. Network-based integration of disparate omic data to identify “silent players” in cancer. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ching T., Zhu X., Garmire L.X. Cox-nnet: an artificial neural network method for prognosis prediction of high-throughput omics data. PLoS Comput Biol. 2018;14 doi: 10.1371/journal.pcbi.1006076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lebovitz C.B., Robertson A.G., Goya R., Jones S.J., Morin R.D., Marra M.A. Cross-cancer profiling of molecular alterations within the human autophagy interaction network. Autophagy. 2015;11:1668–1687. doi: 10.1080/15548627.2015.1067362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R.M., Ozenberger B.A., Ellrott K. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker J.S., Mullins M., Cheang M.C.U., Leung S., Voduc D., Vickery T. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang B., Mezlini A.M., Demir F., Fiume M., Tu Z., Brudno M. Similarity network fusion for aggregating data types on a genomic scale. Nat Methods. 2014;11:333–337. doi: 10.1038/nmeth.2810. [DOI] [PubMed] [Google Scholar]
- 26.Liang X.H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 27.Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc B. 1996;58:267–288. [Google Scholar]
- 28.Yang Y., Chen L., Gu J., Zhang H., Yuan J., Lian Q. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat Commun. 2017;8:14421. doi: 10.1038/ncomms14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang J., Deng R., Luo R.Z., Shen G.P., Cai M.Y., Du Z.M. Low expression of ULK1 is associated with operable breast cancer progression and is an adverse prognostic marker of survival for patients. Breast Cancer Res Treat. 2012;134:549–560. doi: 10.1007/s10549-012-2080-y. [DOI] [PubMed] [Google Scholar]
- 30.Lee M.H., Koh D., Na H., Ka N.L., Kim S., Kim H.J. MTA1 is a novel regulator of autophagy that induces tamoxifen resistance in breast cancer cells. Autophagy. 2018;14:812–824. doi: 10.1080/15548627.2017.1388476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y., Wang G., Cai H., Sun Y., Ouyang L., Liu B. Deciphering the rules of in silico autophagy methods for expediting medicinal research. J Med Chem. 2019;62:6831–6842. doi: 10.1021/acs.jmedchem.8b01673. [DOI] [PubMed] [Google Scholar]
- 32.Hervouet E., Claude-Taupin A., Gauthier T., Perez V., Fraichard A., Adami P. The autophagy GABARAPL1 gene is epigenetically regulated in breast cancer models. BMC Cancer. 2015;15:729. doi: 10.1186/s12885-015-1761-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H., Zhang N., Liu Y., Su P., Liang Y., Li Y. Epigenetic regulation of by drives metastatic progression in triple-negative breast cancer. Cancer Res. 2019;79:3347–3359. doi: 10.1158/0008-5472.CAN-18-3418. [DOI] [PubMed] [Google Scholar]
- 34.Liu L., He J., Wei X., Wan G., Lao Y., Xu W. MicroRNA-20a-mediated loss of autophagy contributes to breast tumorigenesis by promoting genomic damage and instability. Oncogene. 2017;36:5874–5884. doi: 10.1038/onc.2017.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karczewski K.J., Snyder M.P. Integrative omics for health and disease. Nat Rev Genet. 2018;19:299–310. doi: 10.1038/nrg.2018.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paczkowska M., Barenboim J., Sintupisut N., Fox N.S., Zhu H., Abd-Rabbo D. Integrative pathway enrichment analysis of multivariate omics data. Nat Commun. 2020;11:735. doi: 10.1038/s41467-019-13983-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gökmen-Polar Y., Neelamraju Y., Goswami C.P., Gu X., Nallamothu G., Janga S.C. Expression levels of SF3B3 correlate with prognosis and endocrine resistance in estrogen receptor-positive breast cancer. Mod Pathol. 2015;28:677–685. doi: 10.1038/modpathol.2014.146. [DOI] [PubMed] [Google Scholar]
- 38.Torrens-Mas M., Oliver J., Roca P., Sastre-Serra J. SIRT3: oncogene and tumor suppressor in cancer. Cancers (Basel) 2017;9:90. doi: 10.3390/cancers9070090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai Q., Zheng Z., Xia F., Liu P., Li M. A one-step specific assay for continuous detection of sirtuin 2 activity. Acta Pharm Sin B. 2019;9:1183–1192. doi: 10.1016/j.apsb.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torrens-Mas M., Pons D.G., Sastre-Serra J., Oliver J., Roca P. SIRT3 silencing sensitizes breast cancer cells to cytotoxic treatments through an increment in ROS production. J Cell Biochem. 2017;118:397–406. doi: 10.1002/jcb.25653. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L., Ren X., Cheng Y., Huber-Keener K., Liu X., Zhang Y. Identification of Sirtuin 3, a mitochondrial protein deacetylase, as a new contributor to tamoxifen resistance in breast cancer cells. Biochem Pharmacol. 2013;86:726–733. doi: 10.1016/j.bcp.2013.06.032. [DOI] [PubMed] [Google Scholar]
- 42.Desouki M.M., Doubinskaia I., Gius D., Abdulkadir S.A. Decreased mitochondrial SIRT3 expression is a potential molecular biomarker associated with poor outcome in breast cancer. Hum Pathol. 2014;45:1071–1077. doi: 10.1016/j.humpath.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.