Abstract
The recent discovery of activator compounds binding to an allosteric site on the NAD+-dependent protein lysine deacetylase, sirtuin 6 (SIRT6) has attracted interest and presents a pharmaceutical target for aging-related and cancer diseases. However, the mechanism underlying allosteric activation of SIRT6 by the activator MDL-801 remains largely elusive because no major conformational changes are observed upon activator binding. By combining molecular dynamics simulations with biochemical and kinetic analyses of wild-type SIRT6 and its variant M136A, we show that conformational rotation of 2-methyl-4-fluoro-5-bromo substituent on the right phenyl ring (R-ring) of MDL-801, which uncovers previously unseen hydrophobic interactions, contributes to increased activating deacetylation activity of SIRT6. This hypothesis is further supported by the two newly synthesized MDL-801 derivatives through the removal of the 5-Br atom on the R-ring (MDL-801-D1) or the restraint of the rotation of the R-ring (MDL-801-D2). We further propose that the 5-Br atom serves as an allosteric driver that controls the ligand allosteric efficacy. Our study highlights the effect of allosteric enzyme catalytic activity by activator binding and provides a rational approach for enhancing deacetylation activity.
KEY WORDS: Allosteric driver, Allosteric sites, Allosteric mechanisms, Drug design, Enzyme catalysis, Protein dynamics
Abbreviations: ADPR, ADP-ribose; EC50, Effective concentration; FDL, Fluor de Lys; H3K9, histone 3 lysine 9; H3K56, histone 3 lysine 56; HPLC, high-performance liquid chromatography; MD, molecular dynamics; MST, microscale thermophoresis; Myr-H3K9, myristoyl H3K9; NAM, nicotinamide; PCA, principal component analysis; RMSD, root-mean-square deviation; SIRT6, sirtuin 6
Graphical abstract
Sirtuin 6 (SIRT6), a NAD+-dependent protein lysine deacetylase, has attracted interest and presents a pharmaceutical target for aging-related and cancer diseases. By combining molecular dynamics simulations, compound synthesis, and biochemical and kinetic analyses of wild-type SIRT6 and its variants, we elucidate the allosteric activation mechanism of SIRT6 by small-molecule compounds.
1. Introduction
Allostery is a key biological process in which two spatially and topographically distinct orthosteric and allosteric sites within a protein are functionally coupled1,2. Allosteric drugs, by targeting much more diversified allosteric sites, have significant pharmacological advantages such as higher selectivity and lower toxicity compared to orthosetric drugs that are bound to highly conserved orthosteric sites. Allostery can occur in an individual protein, or through protein–protein interactions that transmit allosteric signals to a long distance3, 4, 5, 6. Allosterism in drug discovery has, therefore, established as a novel avenue to develop safe therapeutic agents7, 8, 9, 10, 11, 12, 13. Allosteric regulation is an important feature of protein function that is rooted in its dynamics14, 15, 16, 17, 18 and dynamics-based regulation of allosteric proteins has been regarded as an effective mechanism for modulating function19,20. Static crystal structures represent snapshots of proteins that may ignore the roles of conformational dynamics in allosteric regulation21, 22, 23, 24. That is indeed the case for sirtuin 6 (SIRT6)—an NAD+-dependent protein lysine deacetylase—that catalyzes the transfer of an acetyl moiety on histone 3 (H3) lysine 9 (H3K9ac) and lysine 56 (H3K56ac) to the cofactor NAD+, thus yielding the products nicotinamide (NAM), a mixture of 2′- and 3′-O-acetyl-ADP ribose (2′-O-acetyl-ADPR and 3′-O-acetyl-ADPR) along with the deacetylated substrate (Fig. 1)25. Decreased SIRT6 deacetylase activity has been linked to shortened lifespan in male mice and promotes aging phenotypes and cancer progression26,27. As such, pharmacological activation of SIRT6 represents a promising strategy for the treatment of aging-related diseases and cancer28.
Figure 1.
Catalytic mechanism for sirtuin-catalyzed deacetylation reactions.
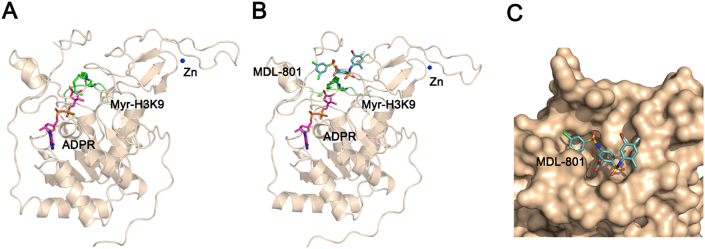
SIRT6 has a two-domain architecture composed of a large Rossmann fold and a small zinc-binding domain (Fig. 2A)29. This enzyme forms a hydrophobic-channel pocket that binds the substrate acyl group and ADP-ribose (ADPR) between the two domains. Using in silico, crystallographic, in vitro, and mouse studies, we have recently identified an SIRT6 allosteric activator, MDL-800 (Fig. 3), which significantly increased the SIRT6 deacetylase activity by ∼20 fold and suppressed the proliferation of human hepatocellular carcinoma30. A co-crystal structure of SIRT6 in complex with MDL-801 (Fig. 3), an MDL-800 derivative, and its substrate, the H3K9 myristoyl peptide, and cofactor, ADPR, clearly indicates that MDL-801 occupies an allosteric site at the back of the substrate site, nonoverlapping with the cofactor and substrate sites (Fig. 2B). Notably, the overall structures are nearly identical [root-mean-square deviation (RMSD) over equivalent Cα atoms is only 0.32 Å] in terms of the MDL-801-bound (PDB code 5Y2F)30 and -unbound (PDB code 5X16)30 enzyme and do not thoroughly decipher the dramatic enhancement in activating SIRT6 deacetylation by MDL compounds. In particular, the right phenyl ring of MDL-801 equipped with a 2-methyl-4-fluoro-5-bromo substituent (hereafter referred to as the R-ring) has no direct contact with the enzyme, as revealed by the co-crystal structure of the SIRT6‒MDL-801 complex (Fig. 2B and C). As such, the role of this moiety in the activation of SIRT6 deacetylation remains enigmatic without analysis of the enzyme dynamics.
Figure 2.
(A) Overall structure of SIRT6 in complex with myristoyl H3K9 (Myr-H3K9) peptide and ADP-ribose (ADPR) (PDB code 3ZG6). (B) Overall structure of SIRT6 in complex with Myr-H3K9 peptide, ADPR, and MDL-801 (PDB code 5Y2F). (C) Surface representation of MDL-801 in the SIRT6 allosteric site. Zinc ion is shown as a blue sphere. ADPR, the Myr-Lys9, and MDL-801 are depicted by stick representations.
Figure 3.

Chemical structures of MDL-800 and MDL-801 as well as two derivatives of MDL-801, MDL-801-D1 and MDL-801-D2. The right phenyl ring (R-ring) of MDL801 with a 2-methyl-4-fluoro-5-bromo substituent is marked by a red dotted ellipse.
2. Results and discussion
To examine allosteric activation of SIRT6, we first performed atomistic molecular dynamics (MD) simulations of SIRT6‒NAD+ and the acetylated peptide substrate in the presence or absence of MDL-801 over 500 ns in explicit water (Supporting Information Section 2). The extent of correlated SIRT6 fluctuations in response to activator binding was explored by calculating the magnitude of the pairwise cross-correlation coefficients cij of the Cα atoms31. The positive value of cij represents correlated motions, whereas the negative value represents anticorrelated motions. In both apo (Supporting Information Fig. S1A) and activator-bound (Fig. S1B) SIRT6, the patterns of fluctuations in the enzyme are very similar with strong anticorrelation motions between residues. However, the intrachain motions between residues are less anticorrelated in the presence of an allosteric activator, as revealed by the smaller cij in the activator-bound enzyme compared to its unbound form. This implies that when bound to MDL-801, a significant part of the regions that allow anticorrelation between residues is restricted, thus leading to a more active conformation that elevates the catalytic efficiency of SIRT6.
To further explore the altered conformational landscape of SIRT6 triggered by MDL-801 binding, a principal component analysis (PCA) of the fluctuations of the cartesian coordinates of Cα atoms was determined by projecting the MD trajectories onto the 2D space spanned by the first two principal components (PC1 and PC2)32, 33, 34. A comparison between the free-energy landscapes of activator-bound and apo SIRT6 reveals that MDL-801-bound SIRT6 spans smaller ranges of PC1 and PC2 and exhibits a more confined conformational space than the apo enzyme (Supporting Information Fig. S2). This observation provides further evidence for the vital role of the enzyme conformational dynamics in the enhancement of enzyme catalysis35, 36, 37, 38, through stabilization of the catalytically competent conformation of the enzyme–substrate complex exerted by MDL-801.
In the solved co-crystal complex of SIRT6‒MDL-801 (Fig. 2B and C)30, the R-ring of MDL-801 with the 2-methyl-4-fluoro-5-bromo substituent protrudes into the solvent. We were curious whether the R-ring of MDL-801 undergoes conformational rearrangements during the simulations. To address this question, we extracted the major representative structure of the SIRT6‒MDL-801 complex obtained by clustering analysis of the simulated trajectory based on the largest free-energy basin located in regions with PC1 and PC2 values of 5.5–8.0 and −4 to 1.7, respectively (Fig. S2B). This representative structural complex obtained from the simulations was superimposed onto the co-crystal structure 5Y2F, and we largely focus on the conformational changes of MDL-801 in the two structures. A remarkable conformational difference was found at the R-ring of MDL-801, where it underwent nearly 90° anticlockwise rotation in the simulations, which is highly distinct from its original position in the co-crystal structure (Supporting Information Fig. S3). This novel R-ring conformation has not been previously observed by X-ray crystallography. The resulting rotation of the R-ring enables it to interact with allosteric site residues. Dramatically, the 5-Br atom of the R-ring protrudes deep into the cavity of the allosteric site and establishes additional van der Waals interactions with Met136 at the bottom of the allosteric site (Fig. 4). This situation is reminiscent of the role of halogenated aromatic groups of peptidomimetics in the improvement of their inhibitory abilities towards MDM239. We thus hypothesize that this novel R-ring conformation of MDL-801 may play a role in the catalytic activities of SIRT6.
Figure 4.
Surface representation of the dominant conformation of MDL-801 in the SIRT6 allosteric site obtained from molecular dynamics simulations. Met136 at the bottom of the allosteric site is highlighted with sulfur, oxygen, and carbon atoms that are colored yellow, red, and green, respectively.
Accumulating evidence indicates that perturbations by ligand binding at the allosteric sites tweak protein activity of functional sites40,41. To further uncover an allosteric communication from the allosteric MDL-801 site to the functional NAD+ site, the web server AlloSigMA42,43 was used to estimate free-energy change of the NAD+ site upon MDL-801 binding. AlloSigMA that was developed based on structure-based statistical-mechanical model of allostery40,44 has been widely used to estimate the causality and energetics of allosteric effects triggered by ligand binding and/or mutations in biomacromolecules21,45,46. The analysis exhibited a free-energy change (ΔG = −0.71 kcal/mol) at the NAD+ site in response to MDL-801 binding, revealing the existence of allosteric coupling between the NAD+ and MDL-801 sites and suggesting that MDL-801 binding may induce the stabilization of the catalytic site enhancing the catalysis, which is consistent with the experimentally observed enhancement of SIRT6 deacetylation activity by MDL-80130.
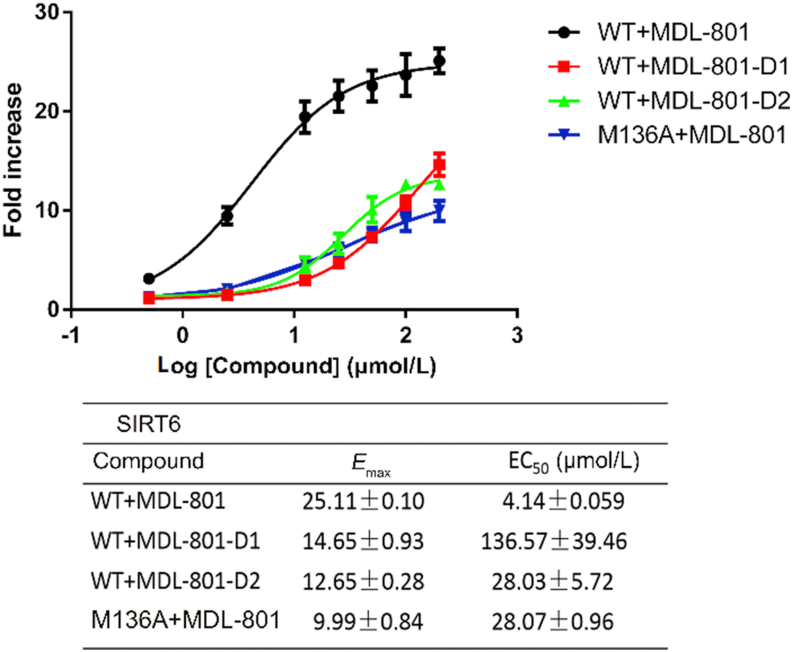
To assess the differential ability of MDL-801 to activate the wild-type and its M136A variant, we analyzed the in vitro deacetylase activity in the presence of MDL-801 using the Fluor de Lys (FDL) assay30. The wild-type SIRT6 and its M136A variant were expressed and purified (Supporting Information Section 1). Both enzymes were reacted with 75 μmol/L H3K9ac (RHKK-ac-AMC) peptide, which represents a physiological SIRT6 deacetylation site, and 2.5 mmol/L NAD+ in the presence of varied concentrations of MDL-801. The fold change in activity of the M136A variant in the presence of MDL-801 was compared to that of the wild-type enzyme. The FDL assay showed that MDL-801 enhanced the deacetylation activity of SIRT6, with a half-maximal effective concentration (EC50) of 4.14 ± 0.059 μmol/L (Fig. 5), which significantly increased SIRT6 deacetylation activity by ∼24-fold at 100 μmol/L. These observations are in good agreement with our previous results30. In contrast, the M136A variant showed a significantly decreased EC50 (28.07 ± 0.96 μmol/L) for MDL-801, resulting in a ∼7-fold decrease in activity compared to the wild-type enzyme (Fig. 5). Therefore, as expected, M136A mutation led to a marked reduction in MDL-801-mediated SIRT6 deacetylation activity.
Figure 5.
Determination of EC50 (μmol/L) values for compounds MDL-801, MDL-801-D1, and MDL-801-D2 toward wild-type SIRT6 or M136A deacetylation reaction. Dose-dependent effects of compounds on the activation of SIRT6 deacetylation, determined with the SIRT6 deacetylation assay. Data are presented as mean ± SD from three independent experiments.
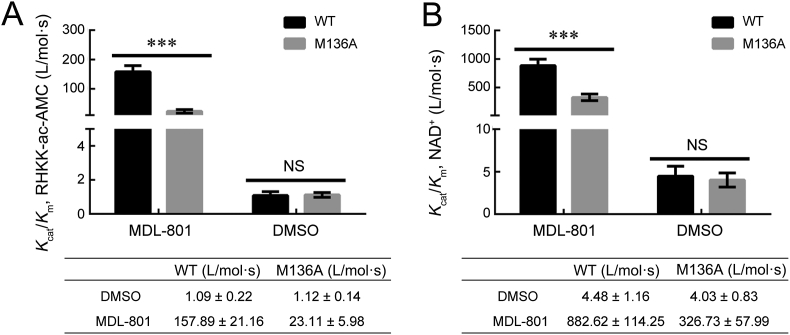
In the SIRT6 structure, Met136 is located in the zinc-binding domain and is ∼10 Å from both the NAM and ribose moieties of NAD+ and ∼11 Å from the acetyl moiety of K9ac from the substrate peptide (Supporting Information Fig. S4), highlighting the lack of direct contact between Met136 and either the NAD+ binding pocket or the substrate binding site. To further explore how a mutation of Met136 to alanine affects catalytic efficiency of SIRT6, the enzymatic kinetics of the deacetylation activity of wild-type SIRT6 and the M136A variant were measured in the presence of 25 μmol/L MDL-801 using high-performance liquid chromatography (HPLC) with 75 μmol/L RHKK-ac-AMC as substrate and 2.5 mmol/L NAD+ as cofactor (Supporting Information Fig. S5). The data were subjected to Michaelis–Menten analysis, and the kinetic constants were compared between wild-type SIRT6 and its mutant. In the presence of 25 μmol/L MDL-801, as shown in Fig. 6, the catalytic efficiency (Kcat/Km) of the deacetylation activity of wild-type SIRT6 for the acetylated peptide and NAD+ was 157.89 ± 21.86 and 882.62 ± 114.25 L/mol·s, respectively, which was ∼7- and ∼2.7-fold higher than that of SIRT6 M136A for the acetylated peptide (23.11 ± 5.98 L/mol·s) and NAD+ (326.73 ± 57.99 L/mol·s), respectively. Therefore, the dramatic decrease in the activating deacetylation activity of the M136A variant by the inhibitor MDL-801 is due to a decrease in the Kcat and an increase in the Km of the acetylated peptide, as well as a decrease in the Kcat and a decrease in the Km for NAD+. Indeed, the Kcat for the acetylated peptide decreased ∼4.7 fold [(248.08 ± 24.79) × 10−4 versus (1169.2 ± 55.64) × 10−4 s−1], and the Km increased ∼1.5 fold (1124 ± 240.9 versus 750 ± 87.03 μmol/L) for the M136A variant relative to wild-type SIRT6 (Supporting Information Fig. S6). The Kcat for NAD+ decreased ∼5.5 fold [(178.8 ± 7.18) × 10−4 versus (985.1 ± 32.67) × 10−4 s−1], and the Km for NAD+ decreased ∼2 fold (56.14 ± 9.82 versus 113.1 ± 14.09 μmol/L) relative to wild-type SIRT6 (Supporting Information Fig. S7). Together, these results indicate that the M136A mutation disrupts both the affinity of SIRT6 for the substrate peptide and catalysis, leading to a dramatic loss of MDL-801-enhanced SIRT6-dependent deacetylation.
Figure 6.
Stead-state kinetics for MDL-801 toward wild-type SIRT6 and M136A deacetylation reaction. (A). Kcat/Km, RHKK-ac-AMC value was determined for wild-type SIRT6 and M136A mutant with MDL-801 in the presence of different concentrations of RHKK-ac-AMC peptide. (B) Kcat/Km, NAD+ value was determined for wild-type SIRT6 and M136A with MDL-801 in the presence of different concentrations of NAD+. Calculated values were determined from nonlinear regression and fit to the Michaelis–Menten equation. Data are presented as mean ± SD from three independent experiments (∗∗∗P < 0.001).
To further determine the importance of the 5-Br atom at the R-ring in activating deacetylation activity of SIRT6, we first synthesized a new MDL-801 derivative without bromine substitution at the 5-position of the R-ring, designated as MDL-801-D1 (Fig. 3, Supporting Information). The hydrogen atom at the 5-position of the R-ring of MDL-801-D1 is therefore less capable of participating in van der Waals interactions compared with the bromine atom at the same position in MDL-801. Therefore, replacement of the 5-Br atom by a hydrogen atom is predicted to have a highly detrimental effect on the ability of MDL-801-D1 to catalyze deacetylation of SIRT6. Subsequently, the influence of MDL-801-D1 on SIRT6-dependent H3K9ac peptide deacetylation was evaluated using the FDL assay as described above. As expected, we observed that MDL-801-D1 showed a weak effect on activating SIRT6 deacetylation with an EC50 of 136.57 ± 39.46 μmol/L (Fig. 3), which showed a ∼33-fold decrease in the deacetylation potency compared to that with MDL-801, suggesting the critical role of bromination. To further elucidate the significance of hydrophobic interactions between the 5-Br atom and the Met136 at the base of the allosteric site, we performed two additional MD simulations of M136A SIRT6‒MDL-801 and wild-type SIRT6‒MDL-801-D1 systems. As shown in Supporting Information Fig. S8, backbone superimpositions of wild-type and M136A SIRT6‒MDL-801 as well as wild-type SIRT6‒MDL-801 and wild-type SIRT6‒MDL-801-D1 show that the R-ring in both the M136A SIRT6‒MDL-801 and wild-type SIRT6‒MDL-801-D1 underwent conformational changes compared to that in the wild-type SIRT6‒MDL-801, leading to the lack of hydrophobic contacts between the 5-Br atom and the Met136.
Next, we synthesized another MDL-801 derivative where the bromine substitution is at the 3-position of the R-ring instead of the 5-position, designated as MDL-801-E1 (Supporting Information). The FDL assay confirmed that MDL-801-E increased the SIRT6 deacetylation activity only by ∼4-fold (Supporting Information Fig. S9), which exhibited a markedly reduced activity compared to the MDL-801. Taken together, these data suggest that the engagement of the 5-Br atom of the R-ring with the Met136 at the bottom of the allosteric site, owing to the rotation of the R-ring, contributes to the enhanced deacetylation potency of the MDL-801.
It is recognized that a subtle chemical change of MDL-801—the removal of the 5-Br atom at the R-ring—significantly reduces the activation of SIRT6 deacetylation. To explore whether the reduced deacetylase activity of SIRT6 by MDL-801-D1 is due to reduced potency (binding affinity) and/or reduced efficacy, the binding affinities of MDL-801 and MDL-801-D1 to SIRT6 were assessed using microscale thermophoresis (MST) assays (Supporting Information). MST data showed that the binding affinity of MDL-801-D1 towards SIRT6 (Kd = 277.7 ± 1.40 μmol/L) was only 2-fold lower than that observed for MDL-801 (Kd = 129.15 ± 28.17 μmol/L, Supporting Information Fig. S10). Given the observations that the 5-Br atom at the R-ring has a marked effect on the deacetylase activity of SIRT6 (decrease by ∼33-fold), but has a minor effect on the binding affinity towards SIRT6 (decrease by ∼2-fold), it is suggested that the 5-Br atom of MDL-801 acts as an allosteric driver47, which controls the ligand allosteric efficacy40.
To further examine the role of the rotation of the R-ring in the improvement of enzyme activity, we synthesized another MDL-801 derivative in which the sulfonamide group was fused to the carboxyl group at the ortho-position of the central benzene ring, generating the saccharin scaffold in the central ring (Supporting Information); this derivative was designated as MDL-801-D2 (Fig. 3). MDL-801-D2 was designed to quench the rotation of the R-ring to the interior of the allosteric site. To test whether MDL-801-D2 occupied the same binding site of MDL-801, we individually mutated the residues F82 and F86 involved in binding of MDL-801 at the allosteric site. As shown in Supporting Information Fig. S11, compared to the wild-type SIRT6, F82A and F86A mutants showed a decreased EC50 for MDL-801-D2 by 3.52- and 3.16-fold, respectively. These data suggest that MDL-801-D2 binds to the same allosteric MDL-801 site of SIRT6. The FDL assay was then used to assess the influence of MDL-801-D2 on SIRT6-dependent H3K9ac peptide deacetylation. As expected, the activating SIRT6 deacetylation was compromised by MDL-801-D2, with an EC50 of 28.03 ± 5.72 μmol/L (Fig. 5)—a ∼7-fold lower potency against MDL-801. Collectively, these results indicate the importance of conformational rotation of the R-ring of MDL-801 in the activating deacetylation activity of SIRT6.
3. Conclusions
In summary, using MD simulations, we have investigated the binding mechanism of MDL-801 that allosterically activates SIRT6 through the rotation of the R-ring of MDL-801 to the interior of the allosteric site, generating additional hydrophobic interactions between the 5-Br atom of the R-ring and Met136 at the base of the allosteric site. This protein-ligand conformation was previously invisible in the crystal structure of SIRT6‒MDL-801 complex, thus arguing for conformational rotation of the R-ring of the MDL-801 as an important contribution to the binding and activation mechanism. Biochemical data combined with kinetic analysis of the M136A mutant as well as newly designed MDL-801 derivatives, created by the removal of the 5-Br atom on the R-ring or restraining the rotation of the R-ring, show that all these modifications markedly decrease the activating deacetylation activity of SIRT6 stimulated by MDL-801, strongly supporting our hypothesis. Specially, the 5-Br atom serves as an allosteric driver that is responsible for the ligand allosteric efficacy. Taken together, our data decipher the mechanism of the SIRT6 allosteric activation, which lays the foundation for the design of more potent activators as therapeutic agents or as tool compounds to advance understanding of SIRT6 biology.
Acknowledgments
This work was supported in part by grants from the National Natural Science Foundation of China (21778037, 81901423, 81903458, 81925034, 91753117 and 81721004, 22077082), China Postdoctoral Science Foundation (2019M660090), the Innovation Program of Shanghai Municipal Education Commission (2019-01-07-00-01-E00036, China), the Shanghai Science and Technology Innovation (19431901600, China), and the Shanghai Health and Family Planning System Excellent Subject Leader and Excellent Young Medical Talents Training Program (2018BR12, China).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting information to this article can be found online at https://doi.org/10.1016/j.apsb.2020.09.010.
Contributor Information
Shaoyong Lu, Email: lushaoyong@yeah.net.
Jian Zhang, Email: jian.zhang@sjtu.edu.cn.
Author contributions
Shaoyong Lu and Jian Zhang conceived and designed the study. Shaoyong Lu, Xinheng He and Duan Ni performed molecular dynamics simulations. Yingyi Chen and Mingzhu Zhao synthesized the compounds. Jiacheng Wei conducted biological assays. Shaoyong Lu, Yingyi Chen, Jiacheng Wei and Mingzhu Zhao wrote the paper.
Conflicts of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Lu S., Shen Q., Zhang J. Allosteric methods and their applications: facilitating the discovery of allosteric drugs and the investigation of allosteric mechanisms. Acc Chem Res. 2019;52:492–500. doi: 10.1021/acs.accounts.8b00570. [DOI] [PubMed] [Google Scholar]
- 2.Nussinov R., Tsai C.J. Allostery in disease and in drug discovery. Cell. 2013;153:293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 3.Serapian S.A., Colombo G. Designing molecular spanners to throw in the protein networks. Chem-A Eur J. 2020;26:4656–4670. doi: 10.1002/chem.201904523. [DOI] [PubMed] [Google Scholar]
- 4.Qiu Y., Li X., He X., Pu J., Zhang J., Lu S. Computational methods-guided design of modulators targeting protein‒protein interactions (PPIs) Eur J Med Chem. 2020;207:112764. doi: 10.1016/j.ejmech.2020.112764. [DOI] [PubMed] [Google Scholar]
- 5.Ni D., Lu S., Zhang J. Emerging roles of allosteric modulators in the regulation of protein‒protein interactions (PPIs): a new paradigm for PPI drug discovery. Med Res Rev. 2019;39:2314–2342. doi: 10.1002/med.21585. [DOI] [PubMed] [Google Scholar]
- 6.Xu X., Ni D., Lu S., Chen Y., Li X., Fu Q. The chemical diversity and structure-based discovery of allosteric modulators for the PIF-pocket of protein kinase PDK1. J Enzym Inhib Med Chem. 2019;34:361–374. doi: 10.1080/14756366.2018.1553167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guarnera E., Berezovsky I.N. Allosteric drugs and mutations: chances, challenges, and necessity. Curr Opin Struct Biol. 2020;62:149–157. doi: 10.1016/j.sbi.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Lu S., He X., Ni D., Zhang J. Allosteric modulator discovery: from serendipity to structure-based design. J Med Chem. 2019;62:6405–6421. doi: 10.1021/acs.jmedchem.8b01749. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y., Wang Z., Ma X., Yang S., Hu X., Tao J. Glycyrrhetinic acid binds to the conserved P-loop region and interferes with the interaction of RAS-effector proteins. Acta Pharm Sin B. 2019;9:294–303. doi: 10.1016/j.apsb.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao M., Guo W., Wu Y., Yang C., Zhong L., Deng G. SHP2 inhibition triggers anti-tumor immunity and synergizes with PD-1 blockade. Acta Pharm Sin B. 2019;9:304–315. doi: 10.1016/j.apsb.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guarnera E., Berezovsky I.N. Allosteric sites: remote control in regulation of protein activity. Curr Opin Struct Biol. 2016;37:1–8. doi: 10.1016/j.sbi.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Lu S., Zhang J. Small molecule allosteric modulators of G-protein-coupled receptors: drug–target interactions. J Med Chem. 2019;62:24–45. doi: 10.1021/acs.jmedchem.7b01844. [DOI] [PubMed] [Google Scholar]
- 13.Ni D., Li X., He X., Zhang H., Zhang J., Lu S. Drugging K-RasG12C through covalent inhibitors: mission possible?. Pharmacol Ther. 2019;202:1–17. doi: 10.1016/j.pharmthera.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Romero-Rivera A., Garcia-Borràs M., Osuna S. Role of conformational dynamics in the evolution of retro-aldolase activity. ACS Catal. 2017;7:8524–8532. doi: 10.1021/acscatal.7b02954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pantouris G., Ho J., Shah D., Syed M.A., Leng L., Bhandari V. Nanosecond dynamics regulate the MIF-induced activity of CD74. Angew Chem Int Ed. 2018;57:7116–7119. doi: 10.1002/anie.201803191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahuja L.G., Aoto P.C., Kornev A.P., Veglia G., Taylor S.S. Dynamic allostery-based molecular workings of kinase:peptide complexes. Proc Natl Acad Sci U S A. 2019;116:15052–15061. doi: 10.1073/pnas.1900163116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell E., Kaltenbach M., Correy G.J., Carr P.D., Porebski B.T., Livingstone E.K. The role of protein dynamics in the evolution of new enzyme function. Nat Chem Biol. 2016;12:944–950. doi: 10.1038/nchembio.2175. [DOI] [PubMed] [Google Scholar]
- 18.Guarnera E., Berezovsky I.N. On the perturbation nature of allostery: sites, mutations, and signal modulation. Curr Opin Struct Biol. 2019;56:18–27. doi: 10.1016/j.sbi.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y., Doruker P., Kaynak B., Zhang S., Krieger J., Li H. Intrinsic dynamics is evolutionarily optimized to enable allosteric behavior. Curr Opin Struct Biol. 2020;62:14–21. doi: 10.1016/j.sbi.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez-Martin C., Moroni E., Ferraro M., Laquatra C., Cannino G., Masgras I. Rational design of allosteric and selective inhibitors of the molecular chaperone TRAP1. Cell Rep. 2020;31:107531. doi: 10.1016/j.celrep.2020.107531. [DOI] [PubMed] [Google Scholar]
- 21.Lu S., Ni D., Wang C., He X., Lin H., Wang Z. Deactivation pathway of Ras GTPase underlies conformational substates as targets for drug design. ACS Catal. 2019;9:7188–7196. [Google Scholar]
- 22.Jagtap P.K.A., Asami S., Sippel C., Kaila V.R.I., Hausch F., Sattler M. Selective inhibitors of FKBP51 employ conformational selection of dynamic invisible states. Angew Chem Int Ed. 2019;58:9429–9433. doi: 10.1002/anie.201902994. [DOI] [PubMed] [Google Scholar]
- 23.Ma B., Nussinov R. Protein dynamics: conformational footprints. Nat Chem Biol. 2016;12:890–891. doi: 10.1038/nchembio.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm Sin B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schiedel M., Robaa D., Rumpf T., Sippl W., Jung M. The current state of NAD+-dependent histone deacetylases (sirtuins) as novel therapeutic targets. Med Res Rev. 2018;38:147–200. doi: 10.1002/med.21436. [DOI] [PubMed] [Google Scholar]
- 26.Bonkowski M.S., Sinclair D.A. Slowing ageing by design: the rise of NAD+ and sirtuin-activating compounds. Nat Rev Mol Cell Biol. 2016;17:679–690. doi: 10.1038/nrm.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanfi Y., Naiman S., Amir G., Peshti V., Zinman G., Nahum L. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 28.Chang A.R., Ferrer C.M., Mostoslavsky R. Multitasking abilities SIRT6 in physiology and disease. Physiol Rev. 2020;39:145–169. doi: 10.1152/physrev.00030.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X., Khan S., Jiang H., Antonyak M.A., Chen X., Spiegelman N.A. Identifying the functional contribution of the defatty-acylase activity of SIRT6. Nat Chem Biol. 2016;12:614–620. doi: 10.1038/nchembio.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Z., Zhao J., Deng W., Chen Y., Shang J., Song K. Identification of a cellularly active SIRT6 allosteric activator. Nat Chem Biol. 2018;14:1118–1126. doi: 10.1038/s41589-018-0150-0. [DOI] [PubMed] [Google Scholar]
- 31.Salvi N., Papadopoulos E., Blackledge M., Wagner G. The role of dynamics and allostery in the inhibition of the eIF4E/eIF4G translation initiation factor complex. Angew Chem Int Ed. 2016;55:7176–7179. doi: 10.1002/anie.201603254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker C., Wang Y., Olivieri C., Karamafrooz A., Casby J., Bathon K. Cushing's syndrome driver mutation disrupts protein kinase A allosteric network, altering both regulation and substrate specificity. Sci Adv. 2019;5 doi: 10.1126/sciadv.aaw9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang M., Jang H., Nussinov R. The mechanism of PI3Kα activation at the atomic level. Chem Sci. 2019;10:3671–3680. doi: 10.1039/c8sc04498h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X., Dai J., Ni D., He X., Zhang H., Zhang J. Insight into the mechanism of allosteric activation of PI3Kα by oncoprotein K-Ras4B. Int J Biol Macromol. 2020;144:643–655. doi: 10.1016/j.ijbiomac.2019.12.020. [DOI] [PubMed] [Google Scholar]
- 35.Maria-Solano M.A., Serrano-Hervás E., Romero-Rivera A., Iglesias-Fernández J., Osuna S. Role of conformational dynamics in the evolution of novel enzyme function. Chem Commun. 2018;54:6622–6634. doi: 10.1039/c8cc02426j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu S., Jang H., Muratcioglu S., Gursoy A., Keskin O., Nussinov R. Ras conformational ensembles, allostery, and signaling. Chem Rev. 2016;116:6607–6665. doi: 10.1021/acs.chemrev.5b00542. [DOI] [PubMed] [Google Scholar]
- 37.Chen X., Yao H., Wang H., Mao Y., Liu D., Long D. Extending the lifetime of native GTP-bound Ras for site-resolved NMR measurements: quantifying the allosteric dynamics. Angew Chem Int Ed. 2019;58:2730–2733. doi: 10.1002/anie.201812902. [DOI] [PubMed] [Google Scholar]
- 38.Fan Y., Cembran A., Ma S., Gao J. Connecting protein conformational dynamics with catalytic function as illustrated in dihydrofolate reductase. Biochemistry. 2013;52:2036–2049. doi: 10.1021/bi301559q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakurai K., Schubert C., Kahne D. Crystallographic analysis of an 8-mer p53 peptide analogue complexed with MDM2. J Am Chem Soc. 2006;128:11000–11001. doi: 10.1021/ja063102j. [DOI] [PubMed] [Google Scholar]
- 40.Guarnera E., Berezovsky I.N. Toward comprehensive allosteric control over protein activity. Structure. 2019;27:866–878. doi: 10.1016/j.str.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 41.Ni D., Li Y., Qiu Y., Pu J., Lu S., Zhang J. Combining allosteric and orthosteric drugs to overcome drug resistance. Trends Pharmacol Sci. 2020;41:336–348. doi: 10.1016/j.tips.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Guarnera E., Tan Z.W., Zheng Z., Berezovsky I.N. AlloSigMA: allosteric signaling and mutation analysis server. Bioinformatics. 2017;33:3996–3998. doi: 10.1093/bioinformatics/btx430. [DOI] [PubMed] [Google Scholar]
- 43.Tan Z.W., Guarnera E., Tee W.V., Berezovsky I.N. AlloSigMA 2: paving the way to designing allosteric effectors and to exploring allosteric effects of mutations. Nucleic Acids Res. 2020;48:W116–W124. doi: 10.1093/nar/gkaa338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guarnera E., Berezovsky I.N. Structure-based statistical mechanical model accounts for the causality and energetics of allosteric communication. PLoS Comput Biol. 2016;12:1–27. doi: 10.1371/journal.pcbi.1004678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abis G., Charles R.L., Kopec J., Yue W.W., Atkinson R.A., Bui T.T.T. 15-deoxy-Δ12,14-prostaglandin J2 inhibits human soluble epoxide hydrolase by a dual orthosteric and allosteric mechanism. Commun Biol. 2019;2:1–14. doi: 10.1038/s42003-019-0426-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurochkin I.V., Guarnera E., Wong J.H., Eisenhaber F., Berezovsky I.N. Toward allosterically increased catalytic activity of insulin-degrading enzyme against amyloid peptides. Biochemistry. 2017;56:228–239. doi: 10.1021/acs.biochem.6b00783. [DOI] [PubMed] [Google Scholar]
- 47.Nussinov R., Tsai C.J. Unraveling structural mechanisms of allosteric drug action. Trends Pharmacol Sci. 2014;35:256–264. doi: 10.1016/j.tips.2014.03.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.