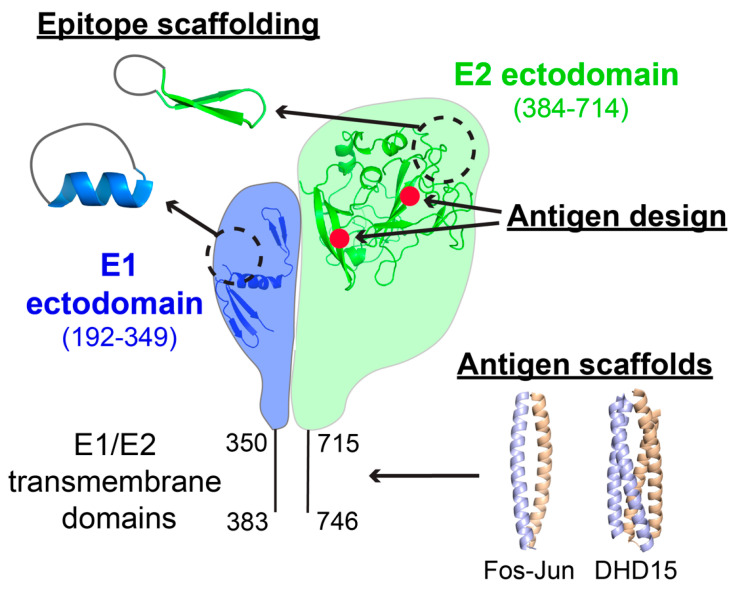
Figure 1.
Structure-based approaches for HCV E1E2 vaccine design. Approaches shown represent: (1) scaffolding of epitopes from E1 and E2, (2) design of the E2 antigen through truncation or residue substitutions to alter antigenicity, immunogenicity, or epitope exposure, and (3) scaffolding of E1E2 to generate stable, secreted glycoproteins. These design strategies are labeled as “Epitope scaffolding”, “Antigen design”, and “Antigen scaffolds”, respectively. Molecular structures shown are from PDB codes 4N0Y (E1 epitope) [61], 5KZP (E2 epitope) [77], 4UOI (E1 N-terminal ectodomain) [79], 6MEI (E2 ectodomain core) [80], 1FOS (Fos-Jun E1E2 scaffold) [81] and 6DMA (DHD15 E1E2 scaffold) [82], and were rendered in PyMOL v. 1.8 (Schrödinger, LLC). Red points on E2 represent rationally selected modifications of E2 (residue substitutions or loop truncations). Residue ranges for ectodomains and transmembrane domains are labeled according to H77 numbering.