Abstract
Polymer and lipid nanoparticles have been extensively used as carriers to address the biological barriers encountered in siRNA and mRNA delivery. In this review, we summarize the critical role of nanoparticle’s charges and ionizability in complexing RNAs, binding biological components, escaping from the endosome, and releasing RNAs to the cytoplasm. We highlight the significant impact of the apparent pKa of nanoparticles on the efficacy and toxicity of the nanoparticles and the importance of optimizing pKa in the development of lead formulations for RNAs. Also, we discuss the feasibility of fine-tuning of pKa in nanoparticles and the applications of using this approach in the optimization of delivery systems for RNAs.
Keywords: Lipid, polymer, RNA delivery, nanoparticles, pKa, endosomal escape, vaccine
Delivery Systems for RNA-Based Therapeutics
RNAs are rapidly expanding as a new class of therapeutics for novel druggable molecular targets including proteins, RNAs, and genomes [1]. Particularly, small interfering RNAs (siRNAs) and messenger RNA (mRNAs) have attracted the most attention as RNA therapeutics for a wide variety of diseases. siRNAs are 21-25 bp double-stranded non-coding RNAs that specifically cleave their target mRNAs in the cytoplasm [2]. A number of siRNA therapeutics have entered clinical trials, and three of them (Onpattro, Givlaari, and Oxlumo) have been approved by the Food and Drug Administration (FDA) for the treatment of genetic diseases. While siRNAs are relatively small with a molecular weight of ~14 kDa, mRNAs are long single-stranded RNAs with a wide range of molecular weight from 300 kDa to 5,000 kDa [3]. siRNAs are chemically synthesized, but mRNAs are synthesized by in vitro transcription (IVT) (See Glossary). After entering the cells, mRNAs express encoded proteins to produce a rapid and transient therapeutic effect [4]. mRNA-based therapies have been investigated in clinical trials as cancer therapies, treatment of genetic diseases, protein replacement therapies, and vaccines [5]. Very recently, two mRNA-based vaccines against COVID-19 have been approved by the FDA, and the breakthrough has spurred global interest in mRNA-based therapies. Antigen produced by mRNA vaccines exhibit more natural representation to the immune system, which leads to better T cell responses compared to viral vectored-based conventional vaccines [6].
RNA-based therapeutics are very promising because they do not require nuclear transportation and have a low risk for mutagenesis compare to DNA-based medicine [7]. The intact form of siRNAs and mRNAs should be delivered into target cells to exert their therapeutic effect [8]. However, the negative charge and the large size of RNAs prevent their cellular entry [9]. In addition, naked RNAs are highly susceptible to nuclease degradation as well as renal clearance in the human body. Exogenous RNAs can also be detected by the innate immune system and trigger immune responses [10]. A robust and safe delivery system is, therefore, essential to translate RNA-based therapeutics from bench to bedside.
Chemical modification and encapsulation in nanoparticles are commonly used to overcome the biological barriers of RNA therapeutics in vivo. Chemical modification of RNAs was extensively reviewed elsewhere[11], and this review will focus on nanoparticles, which have been widely used for plasmid DNA, oligonucleotides, and RNAs. Positively charged nanomaterials form nanoparticles with anionic RNAs to protect RNAs from nucleases and help in binding and penetrating the negatively charged cell membrane [12]. However, endosomal escape and dissociation of RNAs from the nanoparticles in the cytoplasm are the two major challenges that need to be overcome to achieve the maximum therapeutic index [13, 14].
Among different types of nanomaterials, lipids and polymers stand out as the most commonly used nanomaterials to deliver RNAs because of their safety, flexibility, and efficiency [12, 15]. A great variety of lipids and polymers with different compositions and physicochemical properties have been developed to construct nanoparticles encapsulating RNAs [16, 17]. Particle size, shape, surface charge, surface area, ionization constant, and aggregation of nanoparticles are the key characteristics that determine the efficacy and safety of the RNA nanoparticles [18–20]. Optimization of these characteristics is, therefore, essential to develop a successful nanoparticle formulation for RNAs.
Nanoparticles bearing ionizable amine headgroups represent a promising platform in RNA delivery [21, 22]. The acid dissociation constant (pKa) is one of the most important physicochemical properties of the ionizable headgroups of the nanoparticles. pKa determines the ionization behavior and surface charge of the nanoparticles, which substantially influences the stability, potency, and toxicity of the nanoparticles [8]. The biological performance depends on the interactions of cationic nanoparticles with negatively charged blood proteins and cell membranes. These interactions are strongly influenced by the charge state of the nanoparticles’ amine headgroups [23]. In addition, surface charge affects cellular uptake, endosomal release, and biodistribution of nanoparticles [24, 25]. Although a wide variety of lipids and polymers have been developed for RNA delivery, very few of them are optimized using pKa as a tool to achieve the highest therapeutic index. In this review, we summarize the importance, applications, tuning, and measurement techniques of pKa of nanoparticles encapsulating RNAs. We hope the information presented in this review will facilitate the designing and optimization of nanoparticles for RNA-based therapeutics.
Apparent pKa - Basics and Measurement Techniques
Apparent pKa is an experimentally determined value of molecules or nanoparticles. This value is the pH at which the numbers of ionized (protonated) and deionized groups are equal in the systems. The surface charge and ionic interaction of assembled nanomaterials in nanoparticles can be estimated according to apparent pKa. The apparent pKa of a nanoparticle is the result of the average ratio of all the ionized to deionized groups in the nanoparticle. Thus, apparent pKa is not the intrinsic pKa value of any individual molecule [26, 27]. These ionizable amine groups of nanoparticles transform from deprotonated to protonated states as pH decreases. This transition occurs very rapidly near the apparent pKa value (Box 1). The graph of apparent pKa study provides information about the charge state of nanoparticles at various pH.
Box 1: Measurement of Apparent pKa.
The apparent pKa of nanoparticles can be measured by different techniques. Acid-base titration and 2-(p-toluidino)-6-naphthalene sulfonic acid (TNS) fluorescent methods are widely used in the determination of apparent pKa of blank nanoparticles. In the acid-base titration (also known as potentiometric titration method), the blank nanoparticles are first suspended in HCl solution, and titration is carried out by adding 0.1 M NaOH or 0.5 M KOH solution. The pKa value is determined as the pH in the midpoint of the two equivalence points of the titration curve [85]. Figure I–A shows the schematic illustration of the titration curve for ionizable amine-bearing nanoparticles. Automatic instruments, such as SiriusT3, are available to carry out the acid-base titration experiment at a small scale [23, 86].
TNS fluorescent method is very sensitive and has been extensively used to measure the pKa of LNPs [32]. TNS is non-fluorescent in aqueous solutions but exhibits strong fluorescence after binds with cationic lipids or polymers [23, 87]. As pH decreases, the interaction between TNS and the cationic surface increases, leading to a steady increment in fluorescence. The fluorescence reaches a maximum when all the ionizable groups are charged at a specific pH [49]. To carry out the TNS fluorescent assay, a series of buffer solutions are prepared in the pH range of 3 to 10 using 150 mM NaCl, 10 mM borate, 10 mM Phosphate, and 10 mM of Citrate. Blank nanoparticles are prepared and diluted in each buffer solution. TNS is dissolved in DMSO at 300 μM, and 2 μL of the TNS solution is mixed with 100 μL of blank nanoparticles. Then, the fluorescence is measured at excitation and emission wavelength of 325 nm and 425 nm, respectively [8]. Figure I–B shows the schematic of the fluorescence as a function of pH.
The fluorescence of TNS relies on the binding of TNS to positively charged lipids. TNS binding to the lipids is limited by the accessibility of the positively charged lipid headgroups, which are relatively smaller compared to TNS. TNS fluorescence increases steadily with the concentration of lipids before it reaches a plateau [23]. It is noted that it is challenging to observe lower pKa for the lipids bearing multiple amines due to steric hindrance of TNS [23]. However, if TNS is not completely utilized in the first inflection we can see a small inflection in fluorescence at lower pKa for lipids with multiple amines [48]. Similarly, the double sigmoidal ionization curve of lipids with two ionizable amines was normalized to the net charges of lipid against pH range using the single cationic DOTAP and uncharged DOPC nanoparticles. The net charge values are higher than one, suggesting that both the ionizable amines of the lipid respond to the TNS [88].
Figure I: Schematic diagram of the methods for pKa measurement:
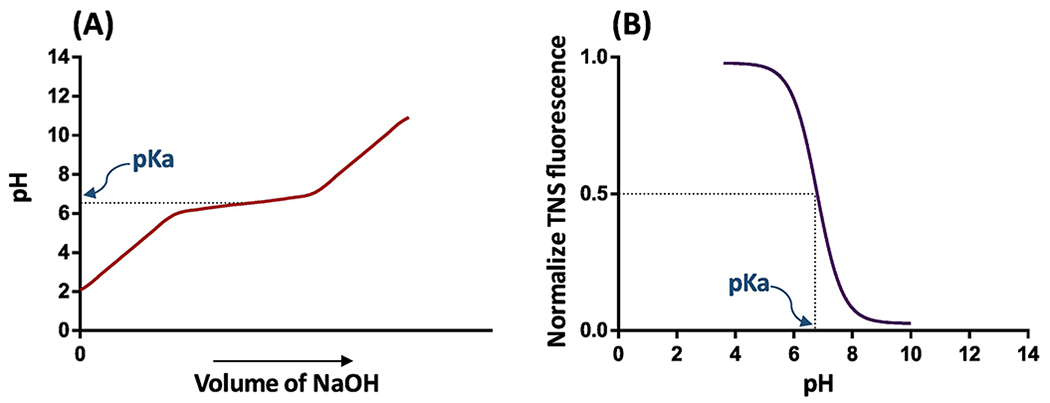
A) Potentiometric titration of nanoparticles (in an acidic solution) with base to increase the pH. The deionization of the nanoparticles inflects the titration curve by producing buffer action. The middle of the two equivalence points represents the pKa of nanoparticles. B) TNS fluorescent measurement of nanoparticles at various pH shows the fluorescence corresponding to the ratio of ionized to deionized amines. TNS interacts with positively charged amines and produces fluorescence signal. The pH value at the half maximum value of fluorescence represents the pKa of nanoparticles.
The measurement of apparent pKa can be strongly influenced by various noncovalent interactions and environmental parameters, such as ionic strength, dielectric constant, hydrophobic interactions, π–π stacking interactions, and the presence of neighboring charges [23, 28]. Additionally, the apparent pKa value of the ionizable ligand-modified nanoparticles changed with the particle size and shape of the nanoparticles [29]. These structural and environmental factors affect the actual ionization of nanoparticles. Thus, the apparent pKa of nanoparticles is relatively lower compared to the calculated pKa of the individual molecules or monomers in the nanoparticles.
Apparent pKa in Nanoparticle-Based RNA Delivery
Apparent pKa value reflects the charge interaction behavior of nanoparticles, which significantly affects their biological activity because the positively charged molecules of the nanoparticles can interact with negatively charged proteins and cells in the body [30]. Negatively charged RNAs condense with ionizable cationic nanomaterials to form nanoparticles by electrostatic interactions, thus protecting RNAs from nuclease degradation [31]. Nanoparticles with an optimum pKa carry negligible charges at physiological pH, thereby preventing nonspecific binding and toxicity in the body [32]. Also, the optimum pKa of nanoparticles plays important roles in endosomal escape and the release of RNAs in the cytosol to exert therapeutic effect (Figure 1) [33–36].
Figure 1: Delivery of RNAs into the cytoplasm through endosomal escape with ionizable nanoparticles.
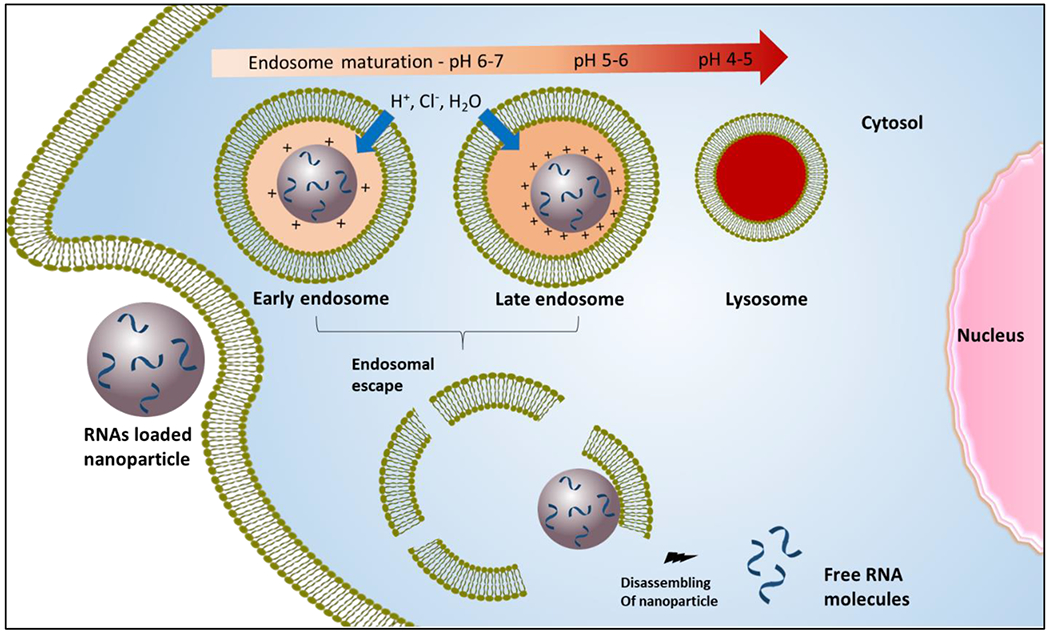
Once nanoparticles are taken up by the cells, charges of the nanoparticle increase as pH decrease below pKa during endosomal maturation (pH 7 to 5.5). The nanoparticles with pKa in this range are protonated due to the acceptance of protons by amine groups. The accumulation of protons with counter ions enhances the transportation of liquids from the cytosol to the endosome to counteract the osmotic pressure. The rapid ionization of nanoparticles near the pKa range creating a buffering capacity, which accounts for the proton sponge effect. The osmotic swelling, due to the buffering capacity of nanoparticles and/or membrane destabilization due to the interaction of negatively charged endosome bilayer with positively charged lipids or polymers of nanoparticle, leads to bursting of endosomes. The charges on nanoparticles decrease in the cytosol and weaken the binding interaction with RNAs. Finally, the nanoparticles are dissociated to release the RNAs to produce the desire activity.
Toxicity, inefficient endosomal escape, and inadequate dissociation of RNAs from nanoparticles in the cytoplasm are the major challenges that are correlated to the charge state of the nanoparticles, and these challenges can be overcome by tuning the apparent pKa. Therefore, the apparent pKa is the most important parameter to improve the efficacy of nanoparticles encapsulating RNAs [23, 32, 37].
a. Apparent pKa of lipid nanoparticles in siRNA delivery
Lipid nanoparticles (LNPs) are the most clinically advanced drug delivery systems for RNA delivery [38]. LNPs are composed of a mixture of ionizable or cationic lipid, phospholipid, PEG-lipid, and cholesterol[39]. Although ionizable lipid is the major and most important component of LNPs, other components also affect the stability and function of LNPs [40]. For example, Patisiran (ONPATTRO™) is the first-ever FDA-approved siRNA therapy,[41] and the transthyretin(TTR)-targeted siRNA is encapsulated in LNPs composed of the DLin-MC3-DMA (MC3) lipid [42].
The MC3 lipid was developed with rational design approaches by systematically varying the structure of lipid 1,2-dilinoleyloxy-3-dimethylaminopropane (DLinDMA). The two double bonds in each tail are responsible for the high gene silencing activity of DLinDMA. The degree of saturation of the lipids affects the phase transition temperature of LNPs. The double bond increases from 0 to 2 in the tail, decreases the phase transition temperature and enhances the fusogenicity of the lipids.[43]. Later, the DLin-K-DMA lipid was developed by introducing a ketal ring in the linker of DLinDMA, which increased the silencing activity by 2.5-fold. The activity was further enhanced by incorporating an additional methylene group between the ketal ring and ionizable amine, resulting in the DLin-KC2-DMA (KC2) lipid. KC2 LNPs with a pKa of 6.7 showed a potent in vivo activity with an ED50 of 0.1 mg/kg [35]. Finally, the MC3 lipid was discovered by screening 53 analogues of a highly potent KC2 lipid. These analogues were synthesized in the pKa range of 4.17 to 8.12 by varying the headgroup of the lipid KC2. A strong correlation was found between apparent pKa and in vivo activity [32]. Gene silencing activity of all the LNPs was evaluated in female C57BL/6 mice after intravenous (IV) administration. A plot of pKa versus ED50 shows a strong correlation between pKa and in vivo activity, and MC3 LNPs exhibit an ED50 of 0.03 mg/kg. The formulation was further optimized by varying the ratio of co-lipids, resulting in an ED50 of 0.005 mg/kg. According to the results, a pKa of 6.2 to 6.5 is the optimum range for maximum activity [32]. The MC3 lipid was further modified by adding an ester linkage at different sites on the aliphatic chain to improve biodegradability and safety. The modified lipids from MC3 with pKa values in the range of 6.2 to 6.4 demonstrated promising activity for hepatic gene silencing. The highly active biodegradable lipid L319 has an ED50 value less than 0.01 mg/kg [44]
Another approach to developing highly effective lipids is the screening of a large combinatorial library of lipid-like molecules [45, 46]. For example, a library containing 1400 lipids was screened using in vitro gene silencing study, and 96 lipids that showed more than 50 % silencing activity were further evaluated for in vivo activity. Fifteen lipids that showed effective gene silencing activity in mice had pKa values greater than 5.5. The lead compound 304O13 was identified based on its good liver biodistribution and gene silencing activity. 304O13 has a pKa value of 6.8. Other lead compounds, including 306O12 and 113O13, have pKa values of 6.8 and 6, respectively. However, their biodistribution profiles were relatively poor compared to 304O13 [47]. This combinatorial approach was also used to screen another lipid library to deliver siRNAs to leukocytes. Lipids with a piperazine head group accumulated more in the spleen compared to the liver. The ‘Lipid 10’ has a pKa in the range of 6.2 to 6.5 and showed significant gene silencing in animal studies [48].
To understand the effect of particle size, lipase sensitivity, and pKa on biodistribution, cell specificity, and gene silencing of LNPs in the liver, six novel lipids were developed by modifying the headgroup of a previously synthesized pH-sensitive lipid YSK05 [49]. The plot of ED50 against pKa showed a bell-shaped curve for FVII gene silencing in hepatocytes, and the lipid with the highest activity has a pKa of 6.45. By contrast, silencing of CD31 in liver sinusoidal endothelial cells exhibited a sigmoidal curve, showing that gene silencing activity increases with pKa up to 7 [49]. This result suggests that tuning the pKa of LNPs can be used to achieve the cell-specific activity.
In another study, multiple parameters (particle size, siRNA entrapment, stability, pKa, hemolysis, and cellular uptake) of LNPs were studied to understand how they affect the in vitro and in vivo silencing activity [8]. The results indicate that increased cellular uptake did not always correlate with increased silencing activity. LNPs should not dissociate before cellular entry, and they must escape the endosome to release siRNAs in an intact form to produce the silencing effect. The gene silencing activity is highly correlated with pKa as compared to particle size and siRNA entrapment. LNPs with pKa between 6 and 7 showed promising gene silencing, while LNPs with pKa between 3 and 6 exhibited poor stability and cellular uptake, resulting in poor silencing activity [8]. Another study showed the impact of structural changes on physicochemical parameters and in vivo activity. Nanoparticles with pKa (6 to 6.6) and calculated lipophilicity (10-14) showed good in vivo activity [50].
b. Apparent pKa of lipid nanoparticles in mRNA delivery
The recent success of two mRNA-based vaccines for COVID-19 has drawn increasing interest in mRNA therapy. Compare to conventional vaccines, it is much faster to develop mRNA-based vaccines, allowing us to promptly respond to virus outbreaks [4, 51]. Lipids SM-102 (Lipid H) and ALC-0315 are used in the formulation of Moderna’s COVID-19 vaccine (mRNA-1273) and Pfizer’s vaccine (BNT162b2 RNA), respectively [I, II].
Various routes of administration were evaluated for mRNA vaccines, and intramuscular (IM), intradermal and subcutaneous administrations produced robust protein expression at the site of injection [52, 53]. To develop a highly efficient delivery system for mRNA vaccines, Moderna synthesized 30 lipids and compared their efficacy to the MC3 lipid [6]. Among the 14 lipids that yielded higher α-H10 IgG titer than MC3, four lipids showed higher luciferase expression relative to MC3, but another four lipids exhibited lower luciferase expression than MC3. This interesting result suggests that protein expression upon IM administration cannot predict immunogenicity. By contrast, the pKa of LNPs is a strong determinant of the immunogenicity, and the optimum range of pKa is 6.6-6.9 for IM delivery. The ‘Lipid H’ (pKa 6.68) was finally selected as the best lipid to deliver mRNA vaccines because of its good biodegradability, tolerability, protein expression, and immunogenicity [6]. Later, the lipid H (SM-102) was used for the COVID-19 vaccine ‘mRNA -1273’.
LNPs that were originally designed for siRNA delivery can be modified to deliver mRNAs. For example, the C12-200 LNP designed for siRNAs was optimized by simultaneously varying the lipid ratios and structures. The optimized C12-200 LNP increased the potency of erythropoietin mRNA by 7-fold relative to the original LNP [54]. Both LNPs exhibit similar morphological characteristics except for the apparent pKa, which was changed from 7.25 to 6.96 for the optimized LNP. Interestingly, both LNPs showed similar potency in siRNA delivery, highlighting the differences in optimized formulation parameters for siRNA and mRNA [54]. Later, the same group synthesized a series of alkenyl amino alcohols (AAA) ionizable lipids for mRNA delivery by introducing different unsaturated fatty chains to the cKK-E12 lipid. The optimized lipid OF-02 lipid yielded a twofold increase in protein expression in vivo compared to cKK-E12 [55].
mRNA LNP was also used to express Chimeric Antigen Receptor (CAR) on human T cells to overcome the side effects associated with virus-based CAR T-cell therapy. Among 24 lipids, the best lipid C14-4 delivered the CD19 mRNA and produced similar amounts of CAR expression with less cytotoxicity compared to the electroporation method. Besides, The LNP from the pure C14-4 has a pKa of 6.5, and the mRNA-LNP-based CAR T-cell therapy elicited a potent antitumor activity in Nalm-6 cells [56].
Another lipid library was constructed by reacting the alkyl-amine 306 with alkyl-acrylate tail with different lengths [57]. The lipids with ten-carbon tails were highly effective and express encoded proteins in mice after IV injection. Moreover, the lipid ‘306i10’ with a branched tail showed a tenfold improvement in the efficiency compared to the lipid ‘30610’ which contains a straight tail. The improved efficacy of the lipid ‘306i10’ is owing to its high surface ionization in the late endosome [57].
c. Apparent pKa of polymeric nanoparticles in RNA delivery
Numerous polymer-based nanomedicines have been approved by the FDA [58]. A wide range of cationic, ionizable, biodegradable, and charge altering polymers have been used in the preparation of nanoparticles for effective delivery of RNAs [59–61]. The physicochemical properties (size, surface charges, stability, entrapment, and toxicity) of polymers can be optimized by modifying the ionizable groups or varying the size of polymers. Controlling these physicochemical characteristics allows the creation of an efficient polymeric delivery system for RNA therapeutics [62].
Polyethyleneimine (PEI) has been extensively used in DNA and RNA delivery because of its endosomal escape capability [63]. The protonation of amines produces a proton sponge effect in early endosomes [63]. However, PEI carries a positive charge in physiological conditions, and its interaction with biomolecules induces significant toxicity [64]. As a result, PEIs have been modified with various biomaterials to reduce their toxicity [65, 66]. For example, a definite number (1 to 4) of aminoethylene units, such as Ethylenediamine (EDA), Diethylenetriamine (DET), Triethylenetetramine (TET), and Tetraethylenepentamine (TEP), were introduced into polyaspartamides (Pasp) to form cationic copolymers with reduced toxicity [67]. Pasp(DET) and Pasp(TEP) have an amine region with pKa values of 6.2 and 6.3, respectively. Both polymers exhibit increased protonation in the endosome, leading to efficient endosomal escape. Subsequently, these polymers were used for the delivery of siRNA [68]. Moreover, these polymers were modified with polyethyleneglycol (PEG) and cholesterol (Chol) to form PEG-Pasp(DET) and PEG-Pasp(TEP)-Chol for mRNA delivery in CNS and treatment of pancreatic cancer, respectively [69, 70].
Chitosan is another well-characterized and extensively used polymer in RNA delivery [71]. The pKa of chitosan is in the range of 6.2 to 7.0 and can be influenced by the molecular weight and degree of deacetylation [72]. Thus, chitosan-based nanoparticles are safe and effective in delivering siRNAs and mRNAs [73–75]. Moreover, chitosan can be easily modified to meet the specific requirement of RNA delivery because each subunit of chitosan has two hydroxyl groups and one amine group [76].
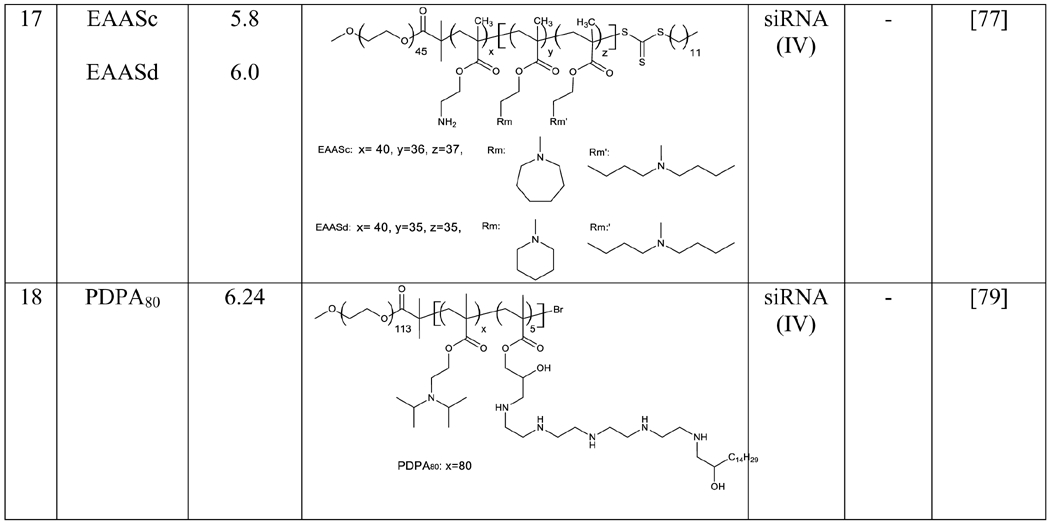
The relationship between pKa and siRNA delivery efficiency were studied in tri-block copolymers, which have been widely used for RNA delivery. A series of triblock copolymers with pKa ranging from 5.2 to 7.0 were synthesized by adjusting the number and the type of hydrophobic amine monomers. The copolymers with pKa of 5.8-6.2 showed better gene silencing effect. The lead copolymer, EAASc, exhibited high silencing effect in MDA-MB-231 (92.45%), HepG2 (89.94%), 293A (83.06%), and HeLa cells (80.27%). The polymer also showed silencing activity in tumors after peritumoral injection. [77].
pH-responsive polymers are another type of promising carriers for RNA delivery. The pH response is a result of the reversible deprotonation and protonation of ionizable groups. pKa is a critical parameter to reflect the ionization status of nanoparticles at various pH values. Nanoparticles with optimum pKa values exhibit maximum efficacy by responding to endosomal pH [78]. For example, a series of poly(2-(diisopropylamino)ethylmethacrylate) (PDPA)-based ultra pH-sensitive polymers were evaluated for siRNA delivery. Among them, PDPA80 has a pKa of 6.24 and showed excellent gene silencing through the proton sponge effect. siRNA nanoparticles made of the polymer exhibited excellent anti-cancer activity in vivo [79]. The preservation of pH sensitivity at optimum level in nanostructures is highly significant to generate maximum endosomal escape and RNAs efficacy.
Tuning of pKa in Nanoparticles
The apparent pKa of polymeric nanoparticles can be tuned by chemically modifying monomers, changing the molar ratio of different monomers in a copolymer, or changing the molecular weight of a polymer. For example, an ultra-pH sensitive nanoprobe library was developed using two randomly distributed monomer R1 and R2 in PEO-b-P(R1-r-R2). The polymer polyethyleneoxide-b-poly(2-(dibutylamino)ethylmethacrylate) (PEO-b-PDBA) and PEO-PDPA (propyl) have pKa of 5.3 and 6.2, respectively. The molar ratio of the monomers DBA and DPA was precisely controlled in the polymerization process to synthesize a series of PEO-b-P(DPA-r-DBA) polymers in the pKa range of 5.3 to 6.2. Similarly, another series of polymers were synthesized by controlling the polymerization of monomer DEA (ethyl) with DPA and D5A (pentyl) with DBA. The nanoprobe library covered the pH range of 4-74. The sharp pH transition of nanoprobes is strongly correlated with their apparent pKa. [80].
The apparent pKa of LNPs is dependent not only on the pKa of each lipid but also on the molar ratio of the lipids. Each lipid has a distinct pKa which can be changed by modifying its headgroup rather than hydrophobic tail [44]. Therefore, one strategy to adjust the apparent pKa of LNPs is to chemically modify the lipid. Another strategy is to use a mixture of two or more lipids with different pKa and adjust their ratio to achieve the desirable apparent pKa. For example, LNPs with pKa values between 5.64 and 6.93 were developed by mixing various ratios of two of the three structurally similar lipids, 15 (pKa 5.64), 16 (pKa 6.44), and 17 (pKa 6.93) [32]. Similarly, various ratios of lipids YSK05 (pKa 6.50) and YSK12-C4 (pKa 8.00) were mixed to form LNPs with apparent pKa values ranging between 6.50 to 8.00 [81].
Conclusion and Future Perspectives
The key requirement for a successful RNA therapy is to overcome the extracellular and intracellular barriers that may degrade RNAs before they reach the site of action. While nanoparticles have been extensively used to protect RANs from degradation, endosomal entrapment of the nanoparticles is a major bottleneck that limits the therapeutic effect. The mechanism of endosomal escape is not completely understood, and it varies from one type of cell to another [82, 83]. Significant efforts have been undertaken to understand the endosomal escape mechanisms of nanoparticles and overcome endosomal entrapment with different approaches.
It is generally believed that the structural and physicochemical properties of nanoparticles play important roles in delivering RNAs [8, 21, 35, 47, 67]. However, the structural requirements for effective RNA delivery are not conclusive, and the structural features of most active lipids are very different (Table 1). By contrast, the apparent pKa of nanoparticles stands out as a reliable criterion to predict the efficiency of nanoparticles encapsulating RNAs. The apparent pKa of nanoparticles has a high correlation with the efficacy and toxicity. Nanoparticles with apparent pKa in the optimum range (see Outstanding Questions) exhibit efficient endosomal escape and therapeutic effect (Figure 2). Incorporation of apparent pKa as a design criterion in the development of RNA nanoparticles will facilitate the discovery of effective and safe RNA therapies.
Table1:
List of the highly effective lipids and polymers for RNA delivery
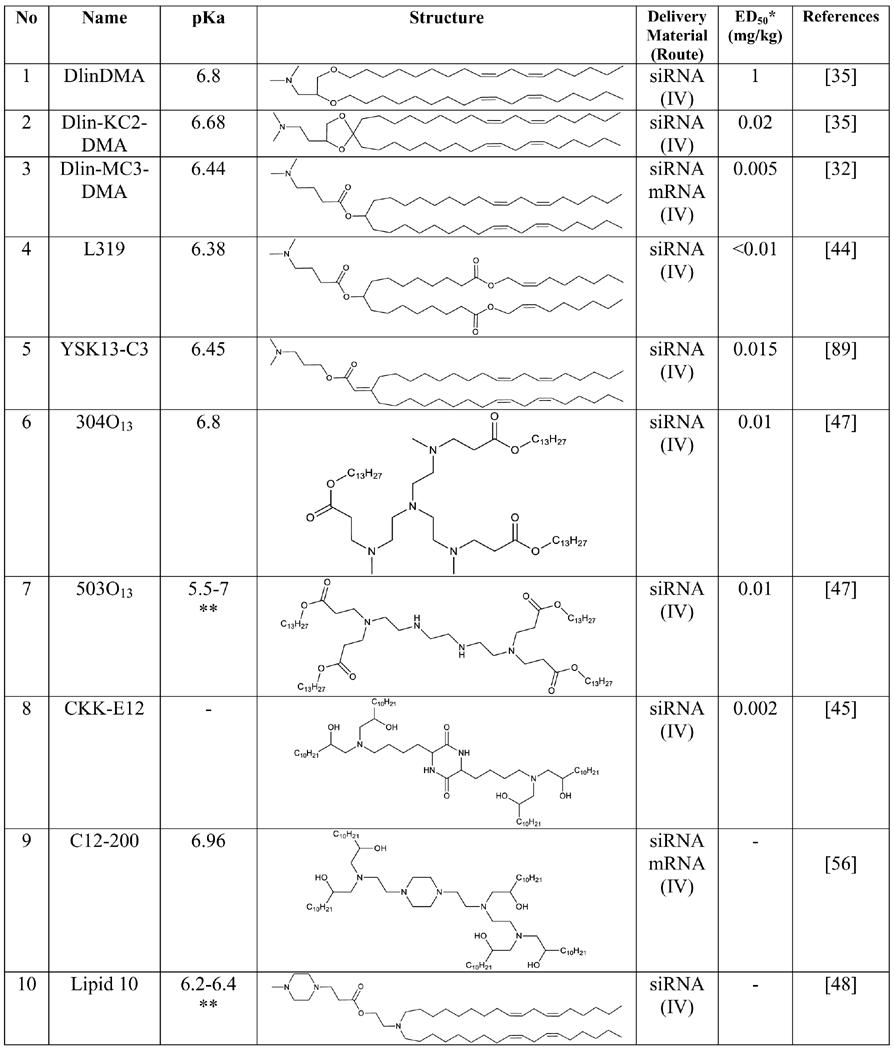 |
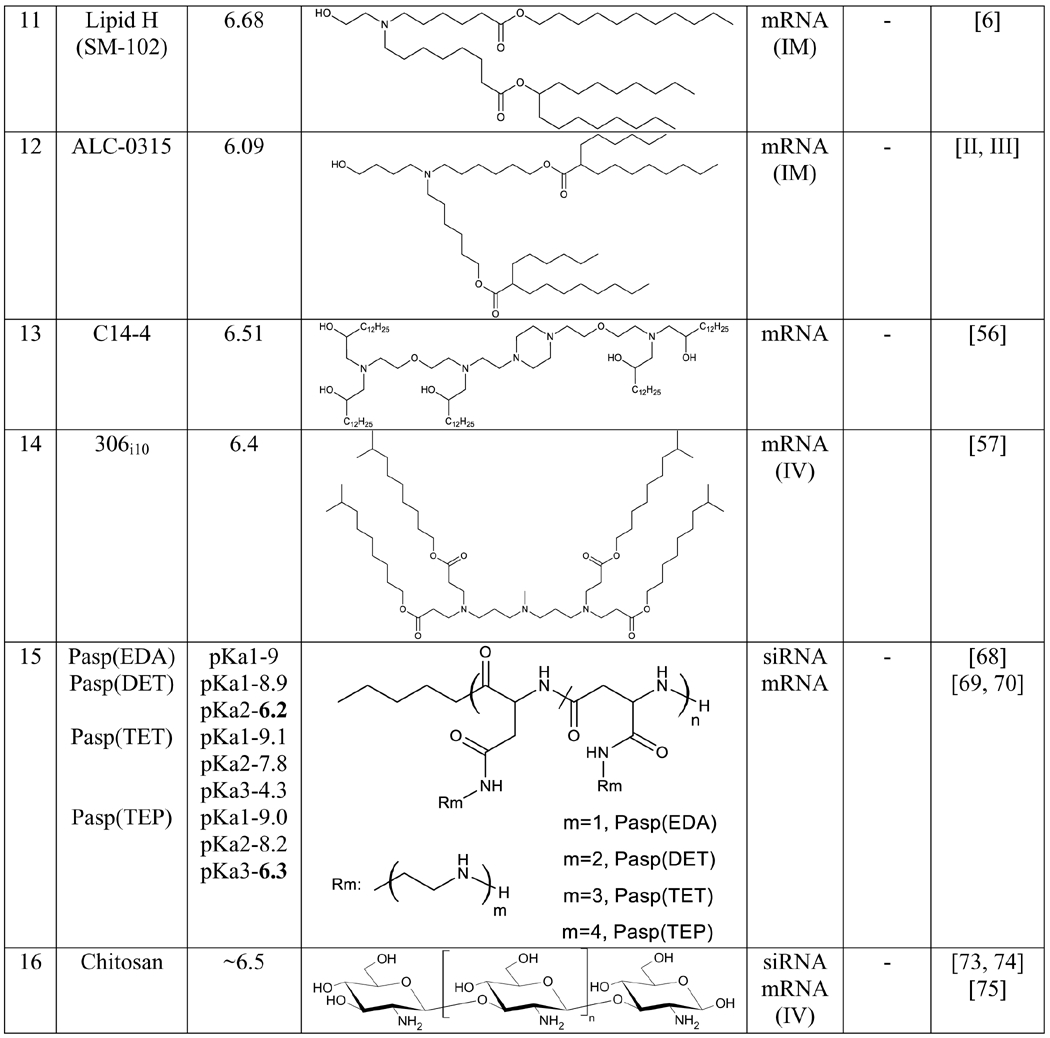 |
 |
The ED50 value is the dose of siRNA for 50% FVII gene silencing in mice with optimized LNPs formulation
The exact value of Lipid 503O13 and Lipid 10 have not been reported, however the measured value of pKa reported to be in between the given value.
Outstanding Questions:
What is the best strategy to develop or optimize siRNA- and mRNA-based formulations?
What are the ways to improve the therapeutic index of nanoparticle-based RNA Therapeutics?
What is the importance of measuring apparent pKa of nanoparticles?
Figure 2: Effects of pKa values on instability, potency, and toxicity of nanoparticles.
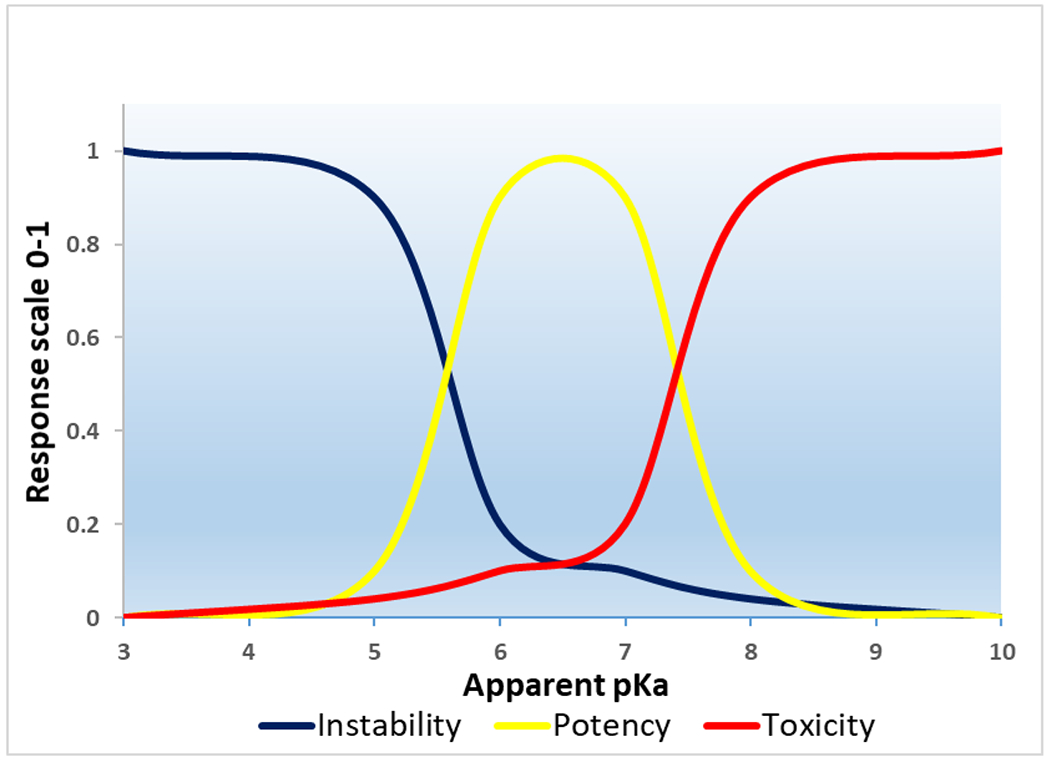
The graph represents the normal tendency of ionizable cationic nanoparticles with different pKa values in biological systems. An apparent pKa range of 6-7 is the optimum range for the development of highly efficient nanoparticles for RNA delivery. The nanoparticles with lower pKa values do not have enough ionic charges and polarity at neutral pH, thereby leading to the aggregation of nanoparticles as hydrophobic interactions are stronger between the inter-particles. Thus, they are less stable in biological systems. While the nanoparticles with higher pKa carry positive charges at physiological pH, which is the main reason for their toxicity. Most importantly, nanoparticles with lower and higher pKa outside the range of 6 to 7 do not ionize effectively during endosome maturation. As a result, they cannot efficiently release the cargo into the cytoplasm.
The most efficient lipid and polymer nanoparticles in RNA delivery have apparent pKa values between 6 and 7 (Table.1). LNPs with optimized pKa of 6.2-6.4 was found to be effective for hepatic delivery of siRNAs [32]. LNPs with optimal pKa in the rage of 6.6-6.9 showed very efficient immune responses after IM administration of mRNAs [6]. The endocytosis processes differ from cells to cells because the proliferation status of cells varies with their physiological roles [84]. The apparent pKa should be optimized according to the target tissue and disease condition. Overall, the optimum pKa value depends on a number of factors, including the structure of the carrier, target tissue, and the route of delivery. As a result, it is difficult to recommend a universal pKa value of nanoparticles for RNA delivery. Nevertheless, it would be appropriate to say that an optimum pKa range of 6 to 7 is the ideal range for the development of nanoparticles for RNA therapeutics.
Resources.
Safety and Immunogenicity Study of 2019-nCoV Vaccine (mRNA-1273) for Prophylaxis of SARS-CoV-2 Infection (COVID-19). ClinicalTrials.gov Identifier: NCT04283461, 2020.
Public Assessment Report: Authorisation for Temporary Supply COVID-19 mRNA Vaccine BNT162b2. Medicines and Healthcare products Regulatory Agency 2020. https://www.gov.uk/government/publications/regulatory-approval-of-pfizer-biontech-vaccine-for-covid-19/summary-public-assessment-report-for-pfizerbiontech-covid-19-vaccine
Ansell, S.M. and X. Du, Novel lipids and lipid nanoparticle formulation for delivery of nucleic acids. 2017, Google Patents / WO2017075531A1.
Highlights:
siRNA and mRNA (RNAs) therapeutics have a high potential in revolutionizing the field of medicine.
Clinical translation of RNAs is limited by their poor in vivo stability. The major challenge is the safe and efficient delivery of RNAs into the cytoplasm to produce the therapeutic effect.
Physiochemical optimization of RNA delivery systems is essential to enhance the effectiveness and eliminate the toxicity associated with the carriers.
The charged state of the amine groups in nanoparticles depends on the pH of the medium, and the changes can be predicted with their apparent pKa. The measurement of pKa help in understanding the advantages and problems of the nanoparticles in various biological processes distinct pH values.
Optimization of nanoparticles based on their apparent pKa dramatically increases the delivery efficiency of siRNA and Mrna
Acknowledgements
This work is supported by the National Institutes of Health (R01AA021510, R01CA23109, and R01GM121798).
Glossary:
- Acid dissociation constant (pKa)
The pKa of a drug is the pH at which molecules are half dissociated. pKa is of utmost importance to comprehend drug absorption and biodistribution in the systemic circulation. pKa measurements enable the determination of the proportion of the molecules in the ionized (charged) or deionized states.
- ED50 (Median effective dose)
The dose of siRNA for 50% FVII gene silencing in mice with optimized LNPs formulation
- In Vitro Transcription
It is a cell-free enzymatic transcription of mRNAs from a linearized DNA or a PCR template using RNA polymerase. The template contains a bacteriophage promoter, a 5’ UTR, an opening reading frame, a 3’ untranslated region, and optionally a poly(A) tail.
- Sharp pH transition
The sharp pH transition means the rapid protonation of all ionizable groups occurs within a narrow pH range.
- Therapeutic Index
Therapeutic index is the ratio of median lethal dose to median effective dose. The relative safety of the drug quantitatively measured with ‘Therapeutic index’.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Publisher's Disclaimer: Disclaimer Statement
None.
References
- 1.Yu A-M, et al. , RNA therapy: Are we using the right molecules? Pharmacology & Therapeutics, 2019. 196: p. 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng K and Mahato RI, Biological and therapeutic applications of small RNAs. Pharm Res, 2011. 28(12): p. 2961–5. [DOI] [PubMed] [Google Scholar]
- 3.Tang X, et al. , Therapeutic Prospects of mRNA-Based Gene Therapy for Glioblastoma. Front Oncol, 2019. 9: p. 1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong Z, et al. , mRNA therapeutics deliver a hopeful message. Nano Today, 2018. 23: p. 16–39. [Google Scholar]
- 5.Wadhwa A, et al. , Opportunities and Challenges in the Delivery of mRNA-based Vaccines. Pharmaceutics, 2020. 12(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hassett KJ, et al. , Optimization of Lipid Nanoparticles for Intramuscular Administration of mRNA Vaccines. Mol Ther Nucleic Acids, 2019. 15: p. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomez-Aguado I, et al. , Nanomedicines to Deliver mRNA: State of the Art and Future Perspectives. Nanomaterials (Basel), 2020. 10(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alabi CA, et al. , Multiparametric approach for the evaluation of lipid nanoparticles for siRNA delivery. Proc Natl Acad Sci U S A, 2013. 110(32): p. 12881–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tai W and Gao X, Noncovalent tagging of siRNA with steroids for transmembrane delivery. Biomaterials, 2018. 178: p. 720–727. [DOI] [PubMed] [Google Scholar]
- 10.Kaczmarek JC, Kowalski PS, and Anderson DG, Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome Med, 2017. 9(1): p. 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ku SH, et al. , Chemical and structural modifications of RNAi therapeutics. Adv Drug Deliv Rev, 2016. 104: p. 16–28. [DOI] [PubMed] [Google Scholar]
- 12.Chahal JS, et al. , Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc Natl Acad Sci U S A, 2016. 113(29): p. E4133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilleron J, et al. , Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol, 2013. 31(7): p. 638–46. [DOI] [PubMed] [Google Scholar]
- 14.Kwon YJ, Before and after endosomal escape: roles of stimuli-converting siRNA/polymer interactions in determining gene silencing efficiency. Acc Chem Res, 2012. 45(7): p. 1077–88. [DOI] [PubMed] [Google Scholar]
- 15.Kanasty R, et al. , Delivery materials for siRNA therapeutics. Nat Mater, 2013. 12(11): p. 967–77. [DOI] [PubMed] [Google Scholar]
- 16.Kulkarni JA, et al. , Lipid Nanoparticle Technology for Clinical Translation of siRNA Therapeutics. Acc Chem Res, 2019. 52(9): p. 2435–2444. [DOI] [PubMed] [Google Scholar]
- 17.Chenthamara D, et al. , Therapeutic efficacy of nanoparticles and routes of administration. Biomater Res, 2019. 23: p. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan I, Saeed K, and Khan I, Nanoparticles: Properties, applications and toxicities. Arabian Journal of Chemistry, 2019. 12(7): p. 908–931. [Google Scholar]
- 19.Jasinski DL, Li H, and Guo P, The Effect of Size and Shape of RNA Nanoparticles on Biodistribution. Mol Ther, 2018. 26(3): p. 784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi J, et al. , Cancer nanomedicine: progress, challenges and opportunities. Nat Rev Cancer, 2017. 17(1): p. 20–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akinc A, et al. , A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol, 2008. 26(5): p. 561–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin H, et al. , Non-viral vectors for gene-based therapy. Nat Rev Genet, 2014. 15(8): p. 541–55. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, et al. , Ionization behavior of amino lipids for siRNA delivery: determination of ionization constants, SAR, and the impact of lipid pKa on cationic lipid-biomembrane interactions. Langmuir, 2011. 27(5): p. 1907–14. [DOI] [PubMed] [Google Scholar]
- 24.Fromen CA, et al. , Nanoparticle surface charge impacts distribution, uptake and lymph node trafficking by pulmonary antigen-presenting cells. Nanomedicine, 2016. 12(3): p. 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kranz LM, et al. , Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature, 2016. 534(7607): p. 396–401. [DOI] [PubMed] [Google Scholar]
- 26.Tsui FC, Ojcius DM, and Hubbell WL, The intrinsic pKa values for phosphatidylserine and phosphatidylethanolamine in phosphatidylcholine host bilayers. Biophys J, 1986. 49(2): p. 459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cawley JJ, The determination of “apparent” pKa’s: An experiment for liberal arts or science students. Journal of Chemical Education, 1993. 70(7): p. 596. [Google Scholar]
- 28.Li Y, et al. , Non-covalent interactions in controlling pH-responsive behaviors of self-assembled nanosystems. Polym Chem, 2016. 7(38): p. 5949–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D, et al. , How and why nanoparticle’s curvature regulates the apparent pKa of the coating ligands. J Am Chem Soc, 2011. 133(7): p. 2192–7. [DOI] [PubMed] [Google Scholar]
- 30.Carnal F, Clavier A, and Stoll S, Polypeptide-Nanoparticle Interactions and Corona Formation Investigated by Monte Carlo Simulations. Polymers (Basel), 2016. 8(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johannes L and Lucchino M, Current Challenges in Delivery and Cytosolic Translocation of Therapeutic RNAs. Nucleic Acid Ther, 2018. 28(3): p. 178–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jayaraman M, et al. , Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew Chem Int Ed Engl, 2012. 51(34): p. 8529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gujrati M, et al. , Multifunctional cationic lipid-based nanoparticles facilitate endosomal escape and reduction-triggered cytosolic siRNA release. Mol Pharm, 2014. 11(8): p. 2734–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sahay G, et al. , Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat Biotechnol, 2013. 31(7): p. 653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Semple SC, et al. , Rational design of cationic lipids for siRNA delivery. Nat Biotechnol, 2010. 28(2): p. 172–6. [DOI] [PubMed] [Google Scholar]
- 36.Patel S, et al. , Brief update on endocytosis of nanomedicines. Adv Drug Deliv Rev, 2019. 144: p. 90–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maugeri M, et al. , Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat Commun, 2019. 10(1): p. 4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulkarni JA, Cullis PR, and van der Meel R, Lipid Nanoparticles Enabling Gene Therapies: From Concepts to Clinical Utility. Nucleic Acid Ther, 2018. 28(3): p. 146–157. [DOI] [PubMed] [Google Scholar]
- 39.Kulkarni JA, et al. , On the Formation and Morphology of Lipid Nanoparticles Containing Ionizable Cationic Lipids and siRNA. ACS Nano, 2018. 12(5): p. 4787–4795. [DOI] [PubMed] [Google Scholar]
- 40.Ramezanpour M, et al. , Ionizable amino lipid interactions with POPC: implications for lipid nanoparticle function. Nanoscale, 2019. 11(30): p. 14141–14146. [DOI] [PubMed] [Google Scholar]
- 41.Hoy SM, Patisiran: First Global Approval. Drugs, 2018. 78(15): p. 1625–1631. [DOI] [PubMed] [Google Scholar]
- 42.Titze-de-Almeida SS, et al. , Leading RNA Interference Therapeutics Part 1: Silencing Hereditary Transthyretin Amyloidosis, with a Focus on Patisiran. Mol Diagn Ther, 2020. 24(1): p. 49–59. [DOI] [PubMed] [Google Scholar]
- 43.Heyes J, et al. , Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J Control Release, 2005. 107(2): p. 276–87. [DOI] [PubMed] [Google Scholar]
- 44.Maier MA, et al. , Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol Ther, 2013. 21(8): p. 1570–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong Y, et al. , Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc Natl Acad Sci U S A, 2014. 111(11): p. 3955–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love KT, et al. , Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci U S A, 2010. 107(5): p. 1864–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitehead KA, et al. , Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat Commun, 2014. 5: p. 4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramishetti S, et al. , A Combinatorial Library of Lipid Nanoparticles for RNA Delivery to Leukocytes. Adv Mater, 2020. 32(12): p. e1906128. [DOI] [PubMed] [Google Scholar]
- 49.Sato Y, et al. , Relationship Between the Physicochemical Properties of Lipid Nanoparticles and the Quality of siRNA Delivery to Liver Cells. Mol Ther, 2016. 24(4): p. 788–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rajappan K, et al. , Property-Driven Design and Development of Lipids for Efficient Delivery of siRNA. J Med Chem, 2020. 63(21): p. 12992–13012. [DOI] [PubMed] [Google Scholar]
- 51.Pardi N, et al. , mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov, 2018. 17(4): p. 261–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pardi N, et al. , Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J Control Release, 2015. 217: p. 345–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bahl K, et al. , Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines against H10N8 and H7N9 Influenza Viruses. Mol Ther, 2017. 25(6): p. 1316–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kauffman KJ, et al. , Optimization of Lipid Nanoparticle Formulations for mRNA Delivery in Vivo with Fractional Factorial and Definitive Screening Designs. Nano Lett, 2015. 15(11): p. 7300–6. [DOI] [PubMed] [Google Scholar]
- 55.Fenton OS, et al. , Bioinspired Alkenyl Amino Alcohol Ionizable Lipid Materials for Highly Potent In Vivo mRNA Delivery. Adv Mater, 2016. 28(15): p. 2939–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Billingsley MM, et al. , Ionizable Lipid Nanoparticle-Mediated mRNA Delivery for Human CAR T Cell Engineering. Nano Lett, 2020. 20(3): p. 1578–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hajj KA, et al. , Branched-Tail Lipid Nanoparticles Potently Deliver mRNA In Vivo due to Enhanced Ionization at Endosomal pH. Small, 2019. 15(6): p. e1805097. [DOI] [PubMed] [Google Scholar]
- 58.Ventola CL, Progress in Nanomedicine: Approved and Investigational Nanodrugs. P T, 2017. 42(12): p. 742–755. [PMC free article] [PubMed] [Google Scholar]
- 59.Wu C, et al. , Rationally Designed Polycationic Carriers for Potent Polymeric siRNA-Mediated Gene Silencing. ACS Nano, 2018. 12(7): p. 6504–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McKinlay CJ, et al. , Charge-altering releasable transporters (CARTs) for the delivery and release of mRNA in living animals. Proc Natl Acad Sci U S A, 2017. 114(4): p. E448–E456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ben Djemaa S, et al. , Versatile electrostatically assembled polymeric siRNA nanovectors: Can they overcome the limits of siRNA tumor delivery? Int J Pharm, 2019. 567: p. 118432. [DOI] [PubMed] [Google Scholar]
- 62.Ulkoski D, et al. , Recent advances in polymeric materials for the delivery of RNA therapeutics. Expert Opin Drug Deliv, 2019. 16(11): p. 1149–1167. [DOI] [PubMed] [Google Scholar]
- 63.Akinc A, et al. , Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med, 2005. 7(5): p. 657–63. [DOI] [PubMed] [Google Scholar]
- 64.Kargaard A, Sluijter JPG, and Klumperman B, Polymeric siRNA gene delivery - transfection efficiency versus cytotoxicity. J Control Release, 2019. 316: p. 263–291. [DOI] [PubMed] [Google Scholar]
- 65.Chen S and Jin T, Poly-Cross-Linked PEI Through Aromatically Conjugated Imine Linkages as a New Class of pH-Responsive Nucleic Acids Packing Cationic Polymers. Frontiers in Pharmacology, 2016. 7(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao Z, et al. , Development of a Biocompatible Copolymer Nanocomplex to Deliver VEGF siRNA for Triple Negative Breast Cancer. Theranostics, 2019. 9(15): p. 4508–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Uchida H, et al. , Odd-even effect of repeating aminoethylene units in the side chain of N-substituted polyaspartamides on gene transfection profiles. J Am Chem Soc, 2011. 133(39): p. 15524–32. [DOI] [PubMed] [Google Scholar]
- 68.Suma T, et al. , Enhanced stability and gene silencing ability of siRNA-loaded polyion complexes formulated from polyaspartamide derivatives with a repetitive array of amino groups in the side chain. Biomaterials, 2012. 33(9): p. 2770–9. [DOI] [PubMed] [Google Scholar]
- 69.Uchida S, et al. , In vivo messenger RNA introduction into the central nervous system using polyplex nanomicelle. PLoS One, 2013. 8(2): p. e56220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Uchida S, et al. , Systemic delivery of messenger RNA for the treatment of pancreatic cancer using polyplex nanomicelles with a cholesterol moiety. Biomaterials, 2016. 82: p. 221–8. [DOI] [PubMed] [Google Scholar]
- 71.Molinaro R, et al. , Polyethylenimine and chitosan carriers for the delivery of RNA interference effectors. Expert Opinion on Drug Delivery, 2013. 10(12): p. 1653–1668. [DOI] [PubMed] [Google Scholar]
- 72.Wang QZ, et al. , Protonation constants of chitosan with different molecular weight and degree of deacetylation. Carbohydrate Polymers, 2006. 65(2): p. 194–201. [Google Scholar]
- 73.Vauthier C, Zandanel C, and Ramon AL, Chitosan-based nanoparticles for in vivo delivery of interfering agents including siRNA. Current Opinion in Colloid & Interface Science, 2013. 18(5): p. 406–418. [Google Scholar]
- 74.Song Y, Tang C, and Yin C, Combination antitumor immunotherapy with VEGF and PIGF siRNA via systemic delivery of multi-functionalized nanoparticles to tumor-associated macrophages and breast cancer cells. Biomaterials, 2018. 185: p. 117–132. [DOI] [PubMed] [Google Scholar]
- 75.Soliman OY, et al. , Efficiency of Chitosan/Hyaluronan-Based mRNA Delivery Systems In Vitro: Influence of Composition and Structure. J Pharm Sci, 2020. 109(4): p. 1581–1593. [DOI] [PubMed] [Google Scholar]
- 76.Cao Y, et al. , Recent Advances in Chitosan-Based Carriers for Gene Delivery. Mar Drugs, 2019. 17(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Du L, et al. , The study of relationships between pKa value and siRNA delivery efficiency based on tri-block copolymers. Biomaterials, 2018. 176: p. 84–93. [DOI] [PubMed] [Google Scholar]
- 78.Tang H, et al. , Recent Development of pH-Responsive Polymers for Cancer Nanomedicine. Molecules, 2018. 24(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu X, et al. , Ultra-pH-Responsive and Tumor-Penetrating Nanoplatform for Targeted siRNA Delivery with Robust Anti-Cancer Efficacy. Angew Chem Int Ed Engl, 2016. 55(25): p. 7091–7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ma X, et al. , Ultra-pH-sensitive nanoprobe library with broad pH tunability and fluorescence emissions. J Am Chem Soc, 2014. 136(31): p. 11085–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shobaki N, Sato Y, and Harashima H, Mixing lipids to manipulate the ionization status of lipid nanoparticles for specific tissue targeting. Int J Nanomedicine, 2018. 13: p. 8395–8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pei D and Buyanova M, Overcoming Endosomal Entrapment in Drug Delivery. Bioconjug Chem, 2019. 30(2): p. 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vermeulen LMP, et al. , Endosomal Size and Membrane Leakiness Influence Proton Sponge-Based Rupture of Endosomal Vesicles. ACS Nano, 2018. 12(3): p. 2332–2345. [DOI] [PubMed] [Google Scholar]
- 84.Hinze C and Boucrot E, Endocytosis in proliferating, quiescent and terminally differentiated cells. J Cell Sci, 2018. 131(23). [DOI] [PubMed] [Google Scholar]
- 85.Zhou K, et al. , Tunable, ultrasensitive pH-responsive nanoparticles targeting specific endocytic organelles in living cells. Angew Chem Int Ed Engl, 2011. 50(27): p. 6109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schonherr D, et al. , Characterisation of selected active agents regarding pKa values, solubility concentrations and pH profiles by SiriusT3. Eur J Pharm Biopharm, 2015. 92: p. 155–70. [DOI] [PubMed] [Google Scholar]
- 87.Tanaka H and Sakamoto Y, Polyelectrolyte titration using fluorescent indicator. I. Direct titration of anionic and cationic polyelectrolytes with 10–4N standard solutions. Journal of Polymer Science Part A: Polymer Chemistry, 1993. 31(11): p. 2687–2691. [Google Scholar]
- 88.Walsh CL, et al. , Synthesis, characterization, and evaluation of ionizable lysine-based lipids for siRNA delivery. Bioconjug Chem, 2013. 24(1): p. 36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yamamoto N, et al. , Novel pH-sensitive multifunctional envelope-type nanodevice for siRNA-based treatments for chronic HBV infection. J Hepatol, 2016. 64(3): p. 547–55. [DOI] [PubMed] [Google Scholar]


