Abstract
INTRODUCTION:
Effective colorectal cancer (CRC) prevention and screening requires sensitive detection of all advanced neoplasias (CRC and advanced adenomas [AA]). However, existing noninvasive screening approaches cannot accurately detect adenomas with high sensitivity.
METHODS:
Here, we describe a multifactor assay (RNA-FIT test) that combines 8 stool-derived eukaryotic RNA biomarkers, patient demographic information (smoking status), and a fecal immunochemical test (FIT) to sensitively detect advanced colorectal neoplasias and other non-advanced adenomas in a 1,305-patient, average-risk, prospective cohort. This cohort was supplemented with a 22-patient retrospective cohort consisting of stool samples obtained from patients diagnosed with AA or CRC before treatment or resection. Participants within these cohorts were evaluated with the RNA-FIT assay and an optical colonoscopy. RNA-FIT test results were compared with colonoscopy findings.
RESULTS:
Model performance was assessed through 5-fold internal cross-validation of the training set (n = 939) and by using the model on a hold out testing set (n = 388). When used on the hold out testing set, the RNA-FIT test attained a 95% sensitivity for CRC (n = 22), 62% sensitivity for AA (n = 52), 25% sensitivity for other non-AA (n = 139), 80% specificity for hyperplastic polyps (n = 74), and 85% specificity for no findings on a colonoscopy (n = 101).
DISCUSSION:
The RNA-FIT assay demonstrated clinically relevant detection of all grades of colorectal neoplasia, including carcinomas, AAs, and ONAs. This assay could represent a noninvasive option to screen for both CRC and precancerous adenomas.
INTRODUCTION
Colorectal cancer (CRC) is the second leading cause of cancer related deaths in the United States. If detected early, CRC has a 5-year survival rate of 92%. However, 63% of newly diagnosed patients have advanced disease, with an associated 5-year survival rate as low as 14% (1–3). Disease onset is typically insidious, starting as a small polyp which can take several years to further accrue somatic mutations and develop into an invasive carcinoma (4–6). For this reason, the detection and resection of adenomas through colonoscopy significantly reduces morbidity and mortality associated with CRC (7,8). One study estimates that for every 1% increase in the rate of adenoma detection through screening, there is a 3% decrease in the rate of cancer development (9).
The annual malignant transformation rate of any adenoma is approximately 0.25% (10). However, advanced adenomas (AA), which are the most severe category of adenomas, can have an annual malignant transformation rate between 5% and 50% depending on pathological characteristics (10,11). AA are defined by size (>10 mm) and/or histology on surgical pathology (villous/tubulovillous growth pattern, high-grade dysplasia, and/or carcinoma in situ). Although the importance of AA detection has been extensively recognized, the US Multi-Society Task Force on CRC recently determined that non-AA/polyps (e.g., 3–4 adenomas <10 mm and hyperplastic polyps >10 mm) are clinically relevant and alter the recommended screening interval if discovered via colonoscopy (12). Therefore, detection of all adenomas is an important aspect of CRC screening programs.
Although invasive screening methods (e.g., colonoscopy or sigmoidoscopy) can detect up to 95% of advanced colorectal neoplasias (CRC and AA) (13), CRC screening compliance by colonoscopy has remained stagnant at approximately 60% for decades (1,14) and nearly a quarter of adults in average-risk populations have never been screened (3). To combat low compliance, noninvasive screening alternatives have been developed that evaluate stool- and blood-derived biomarkers.
The fecal immunochemical test (FIT)-DNA test (Cologuard; Exact Sciences Corporation, Madison, WI) is considered to be the most accurate of all noninvasive diagnostic tests with 92% sensitivity for CRC and 42% sensitivity for AA, at an 87% specificity (15). However, recent studies indicate that the accuracy profile of the FIT-DNA test might not significantly differ from modern FITs performed alone (16). In addition, the FIT-DNA test classifies non-AA as negative findings, despite these lesions having known malignant potential that impact the patient's risk-stratified screening interval recommendation if detected through colonoscopy (12). Other Food and Drug Administration–approved or –cleared diagnostics (fecal occult blood test, FIT, and the plasma Septin9 test [Epi proColon; Epigenomics, San Diego, CA]) demonstrate comparable or inferior metrics regarding advanced neoplasia detection (16,17).
Blood-based diagnostics, which evaluate circulating tumor cells (18), cell-free DNA (19,20), and/or immune markers, are also being developed to address CRC screening noncompliance, but existing evidence for these tests highlights a number of challenges. For example, clinical utility assessments of these tests are limited to retrospective or case-control cohorts (18,19,21). Scalability of these diagnostics is unknown because huge feature pools (whole genome or >100,000 methylome sites) increase the risk of false discovery and limit reproducibility in prospective cohorts (19,20). Furthermore, study results typically report on fewer than 50% of enrolled participants (19,21), implying either selection bias or inability to scalably evaluate all participants. Blood-based diagnostics face the additional challenge of determining tumor of origin and demonstrating high sensitivity for early-stage carcinomas (19). A recent study attempting to address these concerns showed low sensitivity for early-stage cancer detection (stage I = 28%), which is compounded by imperfect tumor of origin predictions (19). To date, most blood-based diagnostics have not assessed sensitivity for adenomas, likely due to biological and technical barriers, which significantly limits their utility in the colorectal cancer screening setting (22).
It has been proposed that stool-derived eukaryotic RNA (seRNA) biomarkers are the ideal platform for detection of colorectal neoplasms because of the transcriptome's ability to provides a real-time snapshot of cellular activity using signals that are exponentially amplified relative to DNA markers (23). In previous studies, seRNA has been leveraged to sensitively detect CRC and non-AA (24,25). Here, we present novel data from a new cohort that further supports the ability for RNA biomarkers to accurately identify colorectal lesions in an average-risk asymptomatic population. Using a prospective cohort from a multicenter clinical trial, the RNA-FIT test demonstrated accurate detection of colorectal neoplasias when compared with a screening colonoscopy. A retrospective cohort was used to supplement the accuracy of the RNA-FIT test on patients with advanced neoplasias. The results suggest that the RNA-FIT test could improve access to sensitive and specific noninvasive screening tests for CRC and AA.
METHODS
Study design
This study required prospective collection of stool samples from participants prior to undergoing average-risk CRC screening via colonoscopy. Samples were collected up to 60 days before the colonoscopy procedure. In addition, stool samples were retrospectively obtained from participants who had been diagnosed with CRC or AA through colonoscopy, but had not yet been treated for disease. The protocol and research procedures were approved by the Advarra Institutional Review Board (IRB #00032825) and the Washington University School of Medicine Institutional Review Board (IRB #20111107). The primary objective of the study was to assess the ability for the RNA-FIT test to detect advanced neoplasias (CRC and AA), with an emphasis on sensitivity for highly aggressive precancerous adenomas (AAs with high-grade dysplasia, carcinoma in situ, or villous/tubulovillous architecture). The secondary objective was to assess RNA-FIT test sensitivity for other non-AA (ONA).
Patient eligibility criteria (prospective cohort)
Individuals who were prospectively recruited for this study were identified by using online engagement platforms. Initial eligibility was determined through an online survey, and final eligibility was subsequently confirmed verbally using predetermined criteria (see Supplementary Methods, Supplementary Digital Content 1, http://links.lww.com/CTG/A618). For a patient to be considered for the analysis, it was required that the patient (i) submit a stool sample, (ii) undergo a screening colonoscopy, and (iii) provide colonoscopy/histopathology reports, as applicable. If any of these requirements were not completed, the patient was considered ineligible. Reports provided by physicians after colonoscopy procedures (e.g., colonoscopy report, histopathology, and/or history and physical) were also reviewed to ensure that eligibility criteria were met.
Patient eligibility criteria (retrospective cohort)
Individuals who were retrospectively recruited for the study were identified by the Digestive Disease Research Core Center at the Washington University School of Medicine. Stool samples from this cohort were subject to the same inclusion criteria as those identified from the prospective cohort (see Supplementary Methods, Supplementary Digital Content 1, http://links.lww.com/CTG/A618).
Demographic characteristics
Demographics, including age, smoking status, sex, family history of CRC, ethnic background, average income, insurance, and screening history, were captured using a combination of the online survey and verbal communication. Some data were verified using clinical information from the physician-provided colonoscopy notes and/or history and physical reports. Geographic designations were based on linking a patient's zip code to the US core-based statistical areas (26,27). Rural areas were defined as containing an urban core of fewer than 10,000 residents. If a zip code was on the border between rural and urban, the residential ratio was used to classify the participant as either rural or urban.
Acquisition of stool samples
Once a patient was enrolled into the study, a stool sample collection kit was sent to the patient's residence. Participants produced a stool sample acquired from a single bowel movement, collected a stool swab for FIT analysis (OC-Light S FIT; Polymedco, Cortlandt Manor, NY) in accordance with manufacturer instructions, and shipped the stool sample in a stabilization buffer at ambient temperature to a centralized lab (Geneoscopy, St. Louis, MO). If a sample was produced >96 hours before receipt, then the sample was considered ineligible.
seRNA biomarker extraction and quantification
Once a stool sample was received, seRNA was extracted based on previously described methods (24). Briefly, isolation and purification of seRNA required differential centrifugation, automated nucleic acid extraction (EMAG; bioMérieux, Marcy-l'Étoile, France), and DNAse treatment (Baseline-ZERO DNase, Middleton, WI). Quantification of seRNA used a digital droplet polymerase chain reaction system (QXDx AutoDG ddPCR System; Bio-Rad Laboratories, Hercules, CA).
FIT analysis
The OC-Light S FIT (Polymedco; Cortlandt Manor, NY) was processed in accordance with manufacturer instructions. FIT assessments were performed by researchers within 30 days of sample collection by inserting the lateral flow test strip into the sample bottle and visualizing the experimental line after 5 minutes. If the experimental line was visible and the control line was visible, then the test was positive. If the experimental line was not visible and the control line was visible, then the test was negative. If the control line was not visible, then the test was considered ineligible. Faint experimental lines and experimental lines that did not entirely cross the test strip were considered negative, as described in internal standard operating procedures.
Risk-stratification of lesions based on optical colonoscopy and histopathology
Patient classifications were based on risk-stratified disease severity (Table 1) and were generated from colonoscopy and histopathology findings (12). Lesion classifications directly reflected those required by the Food and Drug Administration for other approved CRC screening diagnostics (15) with updates based on the most recent US Multi-Society Task Force guidelines (12). Lesion assessment was performed by 2 blinded independent reviewers, and lesion determinations were compared for consistency. As needed, a third independent blinded pathologist was used to obtain agreement across discordant lesion determinations. Patient classification was performed centrally, with the same reviewers assessing all colonoscopy/histopathology reports associated with eligible participants.
Table 1.
Patient lesion categories for the 1,305-patient prospective cohort and 22-patient retrospective cohort
| Lesion categories | Description | Total study (n = 1,327) | Prospective training set (n = 939) | Prospective testing set (n = 366) | Retrospective testing set (n = 22) |
| CRC (1.0) | Stage I–IV CRC | 25 (1.7%) | 3 (0.3%) | 2 (0.6%) | 20 (91.0%) |
| AA (2.1) | >10 adenomas (TA + SSA + VA + TVA); adenoma with high-grade dysplasia; adenoma with carcinoma in situ | 7 (0.5%) | 3 (0.3%) | 3 (0.8%) | 1 (4.5%) |
| AA (2.2) | Villous adenoma; tubulovillous adenoma | 39 (2.9%) | 23 (2.5%) | 15 (4.1%) | 1 (4.5%) |
| AA (2.3) | Tubular adenoma ≧ 10 mm; 5–10 adenomas (TA only) | 50 (3.8%) | 28 (3.0%) | 22 (6.0%) | 0 (0.0%) |
| AA (2.4) | SSA ≧ 10 mm; 5–10 adenomas (TA + SSA) | 22 (1.7%) | 12 (1.3%) | 10 (2.7%) | 0 (0.0%) |
| ONA (3.0) | 1–2 adenomas (TA + SSA) between 5 and 10 mm; >20 hyperplastic polyps | 123 (9.3%) | 81 (8.6%) | 42 (11.5%) | 0 (0.0%) |
| ONA (4.0) | 3–4 adenomas (TA + SSA) < 10 mm | 62 (4.7%) | 35 (3.7%) | 27 (7.4%) | 0 (0.0%) |
| ONA (5.0) | 1–2 adenomas (TA + SSA) ≦5 mm | 233 (17.6%) | 163 (17.4%) | 70 (19.1%) | 0 (0.0%) |
| Hyperplastic polyp (6.1) | ≦20 hyperplastic polyps | 229 (17.3%) | 155 (16.5%) | 74 (20.2%) | 0 (0.0%) |
| No findings (6.2) | No findings on colonoscopy; No histopathological abnormalities | 537 (40.5%) | 436 (46.4%) | 101 (27.6%) | 0 (0.0%) |
| Total | 1,327 | 939 | 366 | 22 |
This table enumerates lesion categories for the total study, the prospective training set, the prospective testing set, and the retrospective testing set.
AA, advanced adenoma; CRC, colorectal cancer; ONA, other non-AA; SSA, sessile serrated adenoma/sessile serrated polyp; TA, tubular adenoma; TVA, tubulovillous adenoma; VA, villous adenoma.
Data normalization and quality assessment
seRNA quantification data were processed before model input. All markers were evaluated against internal controls, and experimental markers were normalized to the housekeeping gene GAPDH. Samples with a GAPDH ≤ no template control + 0.05 copies/μL were considered as having failed quantification and were not eligible for model development or assessment.
Machine learning model development and assessment
All eligible features, including seRNA biomarkers, are provided in Supplementary Table 1, Supplementary Digital Content 1, http://links.lww.com/CTG/A618. Primer/probe sequences for all seRNA biomarkers were previously described (24). Model parameter and biomarker selections were performed using bootstrapped grid selection (24). Model performance was initially assessed using 5-fold internal cross-validation (ICV). This required splitting the training cohort into 5 even folds with stratified lesion categories, thereby creating 5 ICV-training cohorts (80% of samples) and 5 ICV-testing cohorts (20% of samples). The ICV-testing cohorts were designed such that each patient in the training cohort was only assessed in a single ICV-testing cohort. The model for each ICV-training set was built using the parameters and features listed in Supplementary Table 1, Supplementary Digital Content 1, http://links.lww.com/CTG/A618 (24). An optimal threshold for a positive outcome was determined for each ICV model in the training cohort by maximizing the Youden J statistic for the constrained specificity and sensitivity (28). ICV performance was assessed by evaluating point sensitivities for the ICV-testing fold with the median area under the curve (AUC) (median accuracy) and by evaluating predictions of all 5 models across all folds (concatenated accuracy) (see Supplementary Methods, Supplementary Digital Content 1, http://links.lww.com/CTG/A618).
Hold out testing set assessment
A final model was trained using all individuals in the training set and the parameters/features defined in Supplementary Table 1, Supplementary Digital Content 1, http://links.lww.com/CTG/A618. The ultimate binarization threshold for predicting positive findings was calculated by maximizing Youden J statistic (28) within the training set. The model performance was assessed using a hold out testing set. Point sensitivities, 95% confidence intervals, and AUC were determined for participants with CRCs, AA, and ONA.
RESULTS
Study population
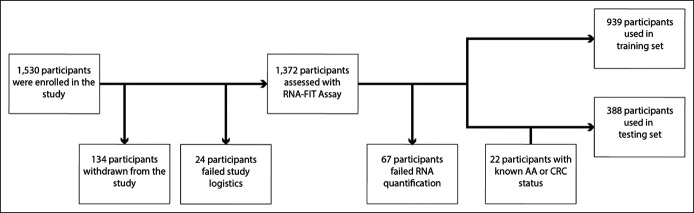
Participants were recruited for the prospective cohort using virtual enrollment strategies. A total of 1,530 participants were enrolled from the 48 contiguous states over the course of 131 days (October 8, 2019, to February 15, 2020). Of the total participants, 158 (10.3%) did not complete study requirements. Of the remaining 1,372 evaluable participants, 67 (4.9%) failed the quality metrics when evaluated by the RNA-FIT test. The remaining 1,305 participants were split into a 939-patient training set and a 366-patient hold out testing set. The study was performed chronologically, such that the training set comprised the first 939 participants to complete all requirements, and the hold out testing set comprised all remaining participants. Participants were recruited for the retrospective cohort through the Digestive Disease Research Core Center at the Washington University School of Medicine. In total, 22 samples were obtained from patients after diagnosis of CRC or AA but before surgical resection or chemotherapy. All samples from the retrospective cohort (n = 22) were used in the hold out testing set (Table 1 and Figure 1).
Figure 1.
Flow chart showing patient recruitment. In total, 1,530 patients were prospectively enrolled into the study. Of the 1,530 enrolled participants, 134 were withdrawn from the study because of lack of eligibility (see Supplementary Methods, Supplementary Digital Content 1, http://links.lww.com/CTG/A618). An additional 24 participants completed the study but were considered ineligible because of failed study logistics (e.g., collection kit was lost in the mail, collection kit was used improperly). Of the 1,372 participants assessed with the RNA-FIT assay, 67 failed RNA quantification (4.9%). No participants failed FIT assessment. The study was performed chronologically, such that the training set comprised the first 939 participants to complete all requirements, and the hold out testing set comprised all remaining participants. An additional retrospective cohort of 22 samples were collected from patients with known disease status (CRC or advanced adenoma) before surgical resection or treatment. AA, advanced adenomas; CRC, colorectal cancer.
Participant demographics
Demographics for the 1,305 participants in the prospective cohort are provided in Supplementary Table 2, Supplementary Digital Content 1, http://links.lww.com/CTG/A618. The average age of participants was 55 years (range = 44–80 years), and 63% were female (37% male). Thirteen percent of participants were African American, 2.2% were Asian American, 5.6% were Hispanic, 75.4% were white, and 3.1% were of other ethnic backgrounds; 6.5% of all participants had a family history of CRC. Participant accrual was from all 48 contiguous states, which corresponded to 1,180 zip codes. Approximately 4.3% of participants lived in rural areas (26,27). Finally, 21% of the study population was low income (<$29,999 per year), and 24% of eligible participants were on public insurance (Medicaid and Medicare).
Internal cross-validation of the training set
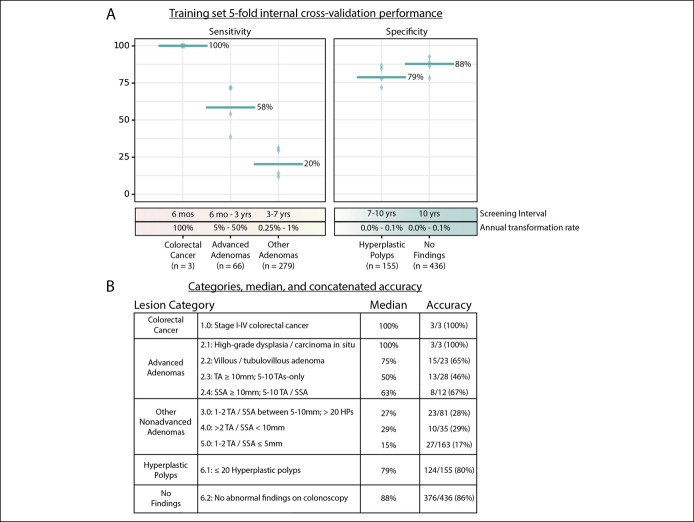
There were 348 participants (37% of total) in the training set who were considered to have positive colonoscopy findings; CRC (n = 3), AA (n = 66), or ONA (n = 279). Among the participants with AA, 3 were classified as having high-grade dysplasia or carcinoma in situ, 23 were classified as having tubulovillous or villous architecture, 28 had large (≥10-mm) tubular adenomas, and 12 had large (≥10-mm) sessile serrated adenomas/sessile serrated polyps. There were 591 participants (63% of total) in the training set who were considered to have negative findings; hyperplastic polyps (n = 155) or no findings on a colonoscopy (n = 436) (Table 1).
The performance of 5-fold internal cross-validation in the training set is shown in Figure 2A. The median accuracy showed 100% sensitivity for CRC, 58% sensitivity for AA, 20% sensitivity for ONA, 79% specificity for hyperplastic polyps, and 88% specificity for no findings on a colonoscopy. When evaluating the aggregated performance of all 5 ICV folds (concatenated accuracy), the RNA-FIT assay detected 3 of 3 participants with CRC (100% sensitivity; CI95% 29%–100%), 39 of 66 participants with AA (59% sensitivity; CI95%, 46%–71%), and 60 of 279 participants with ONA (22% sensitivity; CI95%, 17%–26%) (Figure 2B). Among participants with AA, 100% of participants with high-grade dysplasia (n = 3) and 65% of participants with villous or tubulovillous adenomas (n = 23) were detected as positive using the RNA-FIT test. This accuracy profile was achieved with 80% specificity (CI95%, 73%–86%) for hyperplastic polyps (n = 155) and 86% specificity (CI95%, 82%–89%) for no findings on colonoscopy (n = 436) (Figure 2B).
Figure 2.
Model performance on the training set. (a) 5-fold internal cross-validation was performed for the 939-patient training set. Sensitivity was reported for 3 lesion categories: CRC, advanced adenomas, and other non-AA. Specificity was reported for 2 lesion categories: hyperplastic polyps and no findings on a colonoscopy. For each lesion category, the accuracy metric for the ICV-testing cohort in each fold is reported using a green diamond and the accuracy metric for the median fold is reported with a green bar. The sensitivity/specificity for all folds was determined based on a threshold identified in the ICV-training cohort using the Youden J statistic (28). Recommended screening intervals (12) and malignant transformation rates (10,11) for the 5 lesion categories are also reported. (b) The index provides lesion categories (Table 1), median accuracy, and concatenated accuracy for each lesion category in the testing set (see Supplementary Methods, Supplementary Digital Content 1, http://links.lww.com/CTG/A618). TA, tubular adenoma; TVA, tubulovillous adenoma; SSA, sessile serrated adenoma/sessile serrated polyp; VA, villous adenoma.
Hold out testing set performance
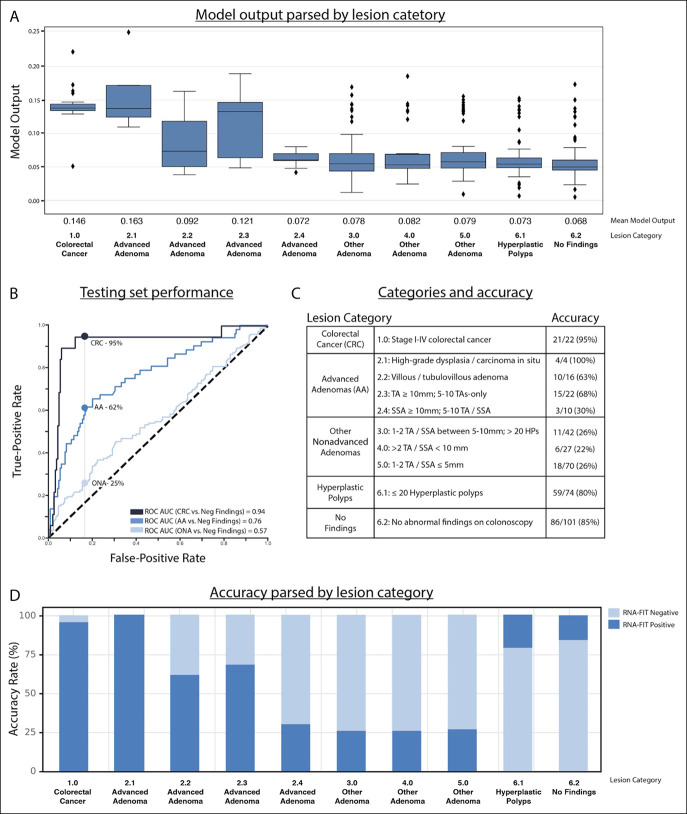
There were 213 participants (55% of total) in the testing set who were considered to have positive colonoscopy findings; CRC (n = 22), AA (n = 52), or non-AA (n = 139). Among the 52 participants with AA, 4 were classified as having high-grade dysplasia or carcinoma in situ, 16 were classified as having tubulovillous or villous architecture, 22 had large (≥10-mm) tubular adenomas, and 10 had large (≥10-mm) sessile serrated adenomas. There were 175 participants (45% of total) in the hold out testing set who were considered to have negative findings; hyperplastic polyps (n = 74) or no findings on a colonoscopy (n = 101) (Table 1).
A final model was built using the 939-participant training set and used on the 388-patient hold out testing set. The hold out testing set model predictions were correlated with disease severity (Figure 3A). Receiver operator characteristic AUC for CRC, AA, and ONA was 0.94, 0.76, and 0.57, respectively, when compared with negative findings (hyperplastic polyps and no findings on colonoscopy) (Figure 3B). The RNA-FIT test detected 21 of the 22 participants with CRC (95%; CI95%, 77%–100%); 32 of the 52 participants with AA (62%; CI95%, 47%–75%); and 35 of the 139 participants with ONA (25%; CI95%, 18%–33%). Of participants with CRC, the RNA-FIT test detected 100% of patients with local disease (stage I–II) (n = 6). Of participants with AA, the RNA-FIT test detected 100% of participants with high-grade dysplasia (n = 4) and 63% of participants with villous or tubulovillous adenomas (n = 16) (Figure 3C). This accuracy profile was achieved with 80% specificity (CI95%, 67%–88%) for hyperplastic polyps (n = 74) and 85% specificity (CI95%, 77%–91%) for no findings on a colonoscopy (n = 101) (Figure 3c, d).
Figure 3.
Model performance on the 388-patient hold out testing set. (a) The box plot shows model output parsed by lesion category. The box encases the first/third quartile, the bar represents the median, whiskers represent 1.5 times the interquartile range, and diamonds represent outliers. (b) The ROC curve shows model performance for CRC, advanced adenomas, and other nonadvanced adenomas. Hyperplastic polyps and no findings on a colonoscopy were considered negative. (c) The index provides lesion categories (Table 1) and total accuracy for each lesion category. (d) Bar plot shows model accuracy parsed by lesion category. AA, advanced adenomas; AUC, area under the curve; CRC, colorectal cancer; ONA, other nonadvanced adenoma; ROC, receiver operator characteristic; SSA, sessile serrated adenoma; TA, tubular adenoma; TVA, tubulovillous adenoma; VA, villous adenoma.
Anecdotal performance compared with other noninvasive screening tests
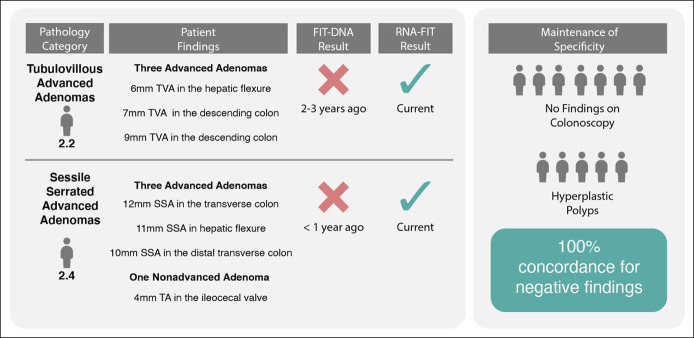
There were 25 participants who had received a negative FIT-DNA test within the 3 years before enrollment in this study. Colonoscopy results from this study identified 2 participants as having AA, 11 as having non-AA, and the remaining 12 participants as having negative findings (hyperplastic polyps or no findings on a colonoscopy). The RNA-FIT test detected both participants with AA (100%) as positive and 2 of the 11 participants with non-AA (18%) as positive. All 12 participants with negative findings (hyperplastic polyps and no findings on colonoscopy) were detected as negative (100%) using the RNA-FIT test (Figure 4).
Figure 4.
Evaluation of the RNA-FIT assay on patients with a previous negative FIT-DNA test within the past 3 years. Pathology category corresponds to the pathology category defined in Table 1. Patient findings define the number and type of lesions that were observed on colonoscopy. FIT-DNA and RNA-FIT results show the output of the assays and time of assay occurrence. Performance of the RNA-FIT was derived from either internal cross-validation of the training set or model performance on the hold out testing set. Specificity shows patients with a negative FIT-DNA result and a negative RNA-FIT result. Two of the 11 participants with other nonadvanced adenomas (not depicted here) were detected as positive using the RNA-FIT. SSA, sessile serrated adenoma/sessile serrated polyp; TA, tubular adenoma; TVA, tubulovillous adenoma.
DISCUSSION
The RNA-FIT test presented here represents the first prospective development study aimed at accurately detecting CRC, AA, and other non-AAs with high sensitivity and specificity. The conclusions from this study are strengthened by the use of a pragmatic study design, which included collecting stool before screening colonoscopy; using a patient cohort that matched the intended use population; using numerous/geographically diverse test collection and endoscopy sites; using varied reagent lots and test systems; limiting features used in model development; and assessing the model performance by setting thresholds within the folds of the various training cohorts. Unlike many previous development studies that use large feature pools, use a highly controlled design, and leverage retrospective/case-control patient cohorts (29), the accuracy profile presented here will likely be maintained as the RNA-FIT diagnostic progresses toward clinical application.
Virtual enrollment permitted significant advantages relative to traditional enrollment strategies. Study enrollment was completed on an expedited timeline (131 days) with high patient compliance (90% completed study requirements). The patient population for this study included participants across all 48 contiguous states, and colonoscopy results were derived from >600 different endoscopy centers. In addition, virtual enrollment assisted with overcoming bias associated with traditional clinical trials by including participants from rural areas, participants with low income, and participants with varied insurance (30,31). Use of this method for enrollment more closely approximates the clinical setting, which further supports reproducibility and clinical utility of the accuracy profile.
The RNA-FIT assay has increased ability to detect AAs that have a high malignant transformation rate. In the hold out testing set, the RNA-FIT assay detected 62% of all AAs, which represents a significant improvement relative to existing alternatives (OC FIT-CHECK = 24% sensitivity (15), FIT-DNA test = 42% sensitivity (15), and plasma septin-9 test = 22% sensitivity (17)). The importance of this improvement is evidenced by evaluating the 1,680,000 patients who were screened using a noninvasive FIT-DNA test in 2019 (32). Relative to the FIT-DNA test, which is currently the most sensitive noninvasive screening alternative, the RNA-FIT test would have returned positive results on 26,880 additional patients with AA. Given an estimated annual transformation rate of 5%, the RNA-FIT assay could prevent cancer development in up to 4,032 additional patients over a 3-year screening interval (11). Even in this modestly sized study of 1,305 prospective participants, the RNA-FIT test identified 2 patients with multiple aggressive AAs and a negative FIT-DNA test within the past 3 years. The positive result of the RNA-FIT assay for these 2 patients would have resulted in a recommendation for a colonoscopy to have these lesions removed (Figure 4).
Ultimately, implementation of the RNA-FIT test might improve the efficacy of CRC screening on several fronts. Noninvasive stool-based screening options can increase patient compliance through a patient-friendly, at-home sample collection (33). In addition, for individuals who require a colonoscopy to remove lesions, positive noninvasive tests before a colonoscopy have been shown to increase adenoma detection rates (34). Most importantly, using the highly sensitive RNA-FIT assay presented here, of the 28% of participants who would have been referred for a colonoscopy, more than 68% would have had an adenoma or a carcinoma removed. More importantly, the 72% of participants with a negative RNA-FIT assay could have been followed noninvasively without missing the vast majority of participants with CRC, carcinoma in situ, or high-grade dysplasia. In a clinical setting where noninvasive tests are not used for CRC screening, up to 40% of patients will elect to forgo the recommended colonoscopy, and of those who do complete the procedure, only 41% would have had a malignant or premalignant lesion removed.
There were several limitations of this study that can be addressed through follow on research. First, the use of virtual enrollment includes increased noise from using hundreds of disparate sites with differing standard protocols for colonoscopy procedures, varying methods for colonoscopy reporting, and varying adenoma detection rates (35). An additional limitation of this study includes the limited number of patients with CRC and the use of retrospective cancer samples to ascertain cancer sensitivity. Finally, it was observed that specificity for no findings on a colonoscopy was slightly reduced relative to existing screening alternatives (86% specificity in the training set and 85% specificity in the testing set). Given the increased sensitivity of the RNA-FIT assay for both AA and non-AA, it is possible that this reduction in specificity was partially attributable to missed adenomas on colonoscopy (13). This is supported by the observation that false-positive RNA-FIT results had a 28% reduction in colonoscopy withdrawal times when compared with true-negative RNA-FIT results (8.1 vs 10.4 minutes; 1-sided t test P value = 0.085) (36).
The RNA-FIT assay presented here provides a robust method to noninvasively detect colorectal neoplasms with high sensitivity and specificity. Using ICV of a 939-patient training set and validation on a 388-patient hold out testing set, we demonstrated significant improvement relative to existing noninvasive diagnostics across all cancerous and precancerous lesions: 95% sensitivity for CRC, 62% sensitivity for AA (100% of AA with high-grade dysplasia and/or carcinoma in situ), and 25% sensitivity for ONA. Accurate detection of precancerous lesions in a prospective screening population demonstrates the potential for an RNA-FIT diagnostic to prevent CRC development through adenoma detection. Analytical and clinical validation of the RNA-FIT diagnostic is currently underway to further substantiate that this noninvasive test could serve as a valuable tool to prevent and detect cancer as part of routine CRC screening.
CONFLICTS OF INTEREST
Guarantor of the article: Erica K. Barnell, PhD.
Specific author contributions: E.K.B. and Y.K. drafted the manuscript; E.K.B., Y.K., A.R.B., and E.M.W. provided analysis and interpretation of data; E.K.B., Y.K., A.R.B., K.R.K., J.F., Z.P., F.H., T.H., M.G., O.L.G., A.A.C., and E.M.W. provided critical revision of the manuscript for important intellectual content; E.K.B, Y.K., A.R.B., K.R.K., J.R., Z.P., and E.M.W. contributed to acquisition of data; E.K.B, F.H., T.H., and E.M.W. contributed to pathology review; F.H. and E.M.W. provided study supervision; M.G., O.L.G., and A.A.C. advised on study development.
Financial support: This research was funded by Geneoscopy Inc. (St. Louis, MO), a molecular diagnostics company. A. Chaudhuri is supported by the V Foundation V Scholar Award, the Alvin Siteman Cancer Research Fund, and the NCI K08CA238711 Career Development Award. M. Griffith is supported by the National Human Genome Research Institute under award number R00HG007940. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Potential competing interests: These authors disclose the following: Erica Barnell, Yiming Kang, Andrew Barnell, and Elizabeth Wurtzler are inventors of the intellectual property owned by Geneoscopy Inc. Erica Barnell, Yiming Kang, and Andrew Barnell are owners of Geneoscopy Inc. Aadel Chaudhuri is a scientific advisor for Geneoscopy Inc. Andrew Barnell, Elizabeth Wurtzler, Erica Barnell, Yiming Kang, Kimberly Kruse, and Jared Fiske are employees of Geneoscopy Inc. Aadel Chaudhuri has received speaker honoraria and travel support from Roche, research support from Roche, and has served as a consultant for Tempus, Roche, Guidepoint, and Fenix Group International. Aadel Chaudhuri has stock options in Geneoscopy, and ownership interests in Droplet Biosciences. The remaining authors disclose no conflicts.
Study sponsor: Geneoscopy Inc. was the sponsor for this study and provided funding for all study-related costs.
Study Highlights.
WHAT IS KNOWN
✓ Colorectal cancer (CRC) is the second leading cause of cancer-related deaths in the United States; however, CRC screening compliance by colonoscopy has remained stagnant at approximately 60% for decades. Noninvasive screening alternatives have a high sensitivity for CRC (up to 92%); however, there is a significant reduction in sensitivity for small CRCs, advanced adenomas, and other precancerous lesions relative to colonoscopies.
WHAT IS NEW HERE
✓ Use of a decentralized enrollment strategy permitted appropriate representation of women, minorities, participants with low income, and participants in rural areas, which reduced recruitment bias in this study. Using a prospectively collected hold out test set, the multifactor assay (RNA-FIT test) detected 21 of 22 participants with CRC (95%), 32 of 52 participants with advanced adenomas (62%), and 35 of 139 participants with other precancerous adenomas (25%).
✓ For all malignant and premalignant lesions, the RNA-FIT test demonstrated the highest accuracy profile ever reported in a prospective cohort for a noninvasive CRC screening test.
TRANSLATIONAL IMPACT
✓ Detection of precancerous adenomas using the RNA-FIT test could reduce CRC morbidity and mortality through cancer prevention. Due to its sensitivity for high-risk adenoma and carcinoma detection, use of the RNA-FIT test could increase adenoma detection rate and enhance productivity of colonoscopies performed.
Supplementary Material
Acknowledgements
We thank the participants and their families for their selfless and generous contributions to science. We greatly appreciate the Geneoscopy Quality Team (Ian Rappold and Matthew Schnell) and the Geneoscopy Science/Software Team (Ben Wedeking, Brett Gleason, and Beekey Cheung) for their contribution to the maintenance of high data integrity. Finally, this research was made possible by the individuals at TynanDx (Katherine Tynan), at Elligo Health Research Team (Tyler Payne, Kellie Esinhart, Paul Couey, Beverly Cerda, Kacey Christian, Jackie Bodmer, Hicham Fayad, Rob Shepler, Matt Tibby, Pablo Alvarez, Patrick Riordan, Dan Hedges, Kristen Amarone, Anita Suri, Deborah Svochak, Valeria Apodaca, and Leviette Sigala), and at 83Bar (Karen Wallingford, Alex Hilderbrand, Mike Zangrilli, Amanda Westcott, Meredyth Glass, and Sara Shissler). We also thank our advisors for their contribution to this work (Liz Lison, Mike Wood, and Jon Harrington).
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A618.
Contributor Information
Yiming Kang, Email: yiming.kang@geneoscopy.com.
Andrew R. Barnell, Email: andrew.barnell@geneoscopy.com.
Kimberly R. Kruse, Email: kimberly.kruse@geneoscopy.com.
Jared Fiske, Email: jared.fiske@geneoscopy.com.
Zachary R. Pittz, Email: zach.pittz@slu.edu.
Thomas A. Huebner, Email: tom.a.huebner@gmail.com.
Faith L. Holmes, Email: faith.holmes@elligodirect.com.
Malachi Griffith, Email: mgriffit@wustl.edu.
Obi L. Griffith, Email: obigriffith@wustl.edu.
Aadel A. Chaudhuri, Email: aadel@wustl.edu.
Elizabeth M. Wurtzler, Email: elizabeth.wurtzler@geneoscopy.com.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975-2013. National Cancer Institute; April 2016: Bethesda, MD, 2017. [Google Scholar]
- 3.Montminy EM, Karlitz JJ, Landreneau SW. Progress of colorectal cancer screening in United States: Past achievements and future challenges. Prev Med 2019;120:78. [DOI] [PubMed] [Google Scholar]
- 4.Haggar FA, Boushey RP. Colorectal cancer epidemiology: Incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg 2009;22:191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med 1988;319:525–32. [DOI] [PubMed] [Google Scholar]
- 6.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759–67. [DOI] [PubMed] [Google Scholar]
- 7.Kaminski MF, Wieszczy P, Rupinski M, et al. Increased rate of adenoma detection associates with reduced risk of colorectal cancer and death. Gastroenterology 2017;153:98–105. [DOI] [PubMed] [Google Scholar]
- 8.Shaukat A, Mongin SJ, Geisser MS, et al. Long-term mortality after screening for colorectal cancer. N Engl J Med 2013;369:1106–14. [DOI] [PubMed] [Google Scholar]
- 9.Corley DA, Levin TR, Doubeni CA. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med 2014;370:2541–1306. [DOI] [PubMed] [Google Scholar]
- 10.Conteduca V, Sansonno D, Russi S, et al. Precancerous colorectal lesions (Review). Int J Oncol 2013;43:973–84. [DOI] [PubMed] [Google Scholar]
- 11.Brenner H, Hoffmeister M, Stegmaier C, et al. Risk of progression of advanced adenomas to colorectal cancer by age and sex: Estimates based on 840,149 screening colonoscopies. Gut 2007;56:1585–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta S, Lieberman D, Anderson JC, et al. Recommendations for follow-up after colonoscopy and polypectomy: A consensus update by the US multi-society Task Force on colorectal cancer. Gastroenterology 2020;158:1131.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn SB, Han DS, Bae JH, et al. The miss rate for colorectal adenoma determined by quality-adjusted, back-to-back colonoscopies. Gut Liver 2012;6:64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meester RG, Doubeni CA, Zauber AG, et al. Public health impact of achieving 80% colorectal cancer screening rates in the United States by 2018. Cancer 2015;121:2281–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 2014;370:1287–97. [DOI] [PubMed] [Google Scholar]
- 16.Imperiale TF, Gruber RN, Stump TE, et al. Performance characteristics of fecal immunochemical tests for colorectal cancer and advanced adenomatous polyps: A systematic review and meta-analysis. Ann Intern Med 2019;170:319–29. [DOI] [PubMed] [Google Scholar]
- 17.Johnson DA, Barclay RL, Mergener K, et al. Plasma Septin9 versus fecal immunochemical testing for colorectal cancer screening: A prospective multicenter study. PLoS One 2014;9:e98238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta P, Gulzar Z, Hsieh B, et al. Analytical validation of the CellMax platform for early detection of cancer by enumeration of rare circulating tumor cells. J Circ Biomark 2019;8:1849454419899214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu MC, Oxnard GR, Klein EA, et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol 2020;31:745–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan N, Weinberg D, Liu TY, et al. Machine learning enables detection of early-stage colorectal cancer by whole-genome sequencing of plasma cell-free DNA. BMC Cancer 2019;19:832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai WS, You JF, Hung HY, et al. Novel circulating tumor cell assay for detection of colorectal adenomas and cancer. Clin Transl Gastroenterol 2019;10:e00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbosh C, Birkbak NJ, Swanton C. Early stage NSCLC - challenges to implementing ctDNA-based screening and MRD detection. Nat Rev Clin Oncol 2018;15:577–86. [DOI] [PubMed] [Google Scholar]
- 23.Dickinson BT, Kisiel J, Ahlquist DA, et al. Molecular markers for colorectal cancer screening. Gut 2015;64:1485–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barnell EK, Kang Y, Wurtzler EM, et al. Noninvasive detection of high-risk adenomas using stool-derived eukaryotic RNA sequences as biomarkers. Gastroenterology 2019;157:884–e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barnell EK, Kang Y, Barnell A, et al. Stool-derived eukaryotic RNA biomarkers for detection of high-risk adenomas. bioRxiv 2019:534412 (doi: 10.1101/534412). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.HUD USPS ZIP Code Crosswalk Files | HUD USER. (https://www.huduser.gov/portal/datasets/usps_crosswalk.html). Accessed May 10, 2021. [Google Scholar]
- 27.Core Based Statistical Areas (National) - Data.Gov. (https://catalog.data.gov/dataset/core-based-statistical-areas-national). Accessed May 10, 2021. [Google Scholar]
- 28.Youden WJ. Index for rating diagnostic tests. Cancer 1950;3:32–5. [DOI] [PubMed] [Google Scholar]
- 29.Lidgard GP, Domanico MJ, Bruinsma JJ, et al. Clinical performance of an automated stool DNA assay for detection of colorectal neoplasia. Clin Gastroenterol Hepatol 2013;11:1313–8. [DOI] [PubMed] [Google Scholar]
- 30.Mills EJ, Seely D, Rachlis B, et al. Barriers to participation in clinical trials of cancer: A meta-analysis and systematic review of patient-reported factors. Lancet Oncol 2006;7:141–8. [DOI] [PubMed] [Google Scholar]
- 31.Niranjan SJ, Martin MY, Fouad MN, et al. Bias and stereotyping among research and clinical professionals: Perspectives on minority recruitment for oncology clinical trials. J Clin Oncol 2019;37:152. [DOI] [PubMed] [Google Scholar]
- 32.Exact Sciences Announces Preliminary Fourth Quarter 2019 Results (http://investor.exactsciences.com/investor-relations/press-releases/press-release-details/2020/Exact-Sciences-Announces-Preliminary-Fourth-Quarter-2019-Results/default.aspx). Accessed May 10, 2021. [Google Scholar]
- 33.Prince M, Lester L, Chiniwala R, et al. Multitarget stool DNA tests increases colorectal cancer screening among previously noncompliant Medicare patients. World J Gastroenterol 2017;23:464–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eckmann JD, Ebner DW, Kisiel JB. Multi-target stool DNA testing for colorectal cancer screening: Emerging learning on real-world performanc. Curr Treat Options Gastro 2020;18:109–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rees CJ, Bevan R, Zimmermann-Fraedrich K, et al. Expert opinions and scientific evidence for colonoscopy key performance indicators. Gut 2016;65:2045–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butterly L, Robinson CM, Anderson JC, et al. Serrated and adenomatous polyp detection increases with longer withdrawal time: Results from the New Hampshire colonoscopy registry. Am J Gastroenterol 2014;109:417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.