Abstract
The current health crisis caused by coronavirus 2019 (COVID-19) and associated pathogens emphasize the urgent need for vaccine systems that can generate protective and long-lasting immune responses. Vaccination, employing peptides, nucleic acids, and other molecules, or using pathogen-based strategies, in fact, is one of the most potent approaches in the management of viral diseases. However, the vaccine candidate requires protection from degradation and precise delivery to the target cells. This can be achieved by employing different types of drug and vaccine delivery strategies, among which, nanotechnology-based systems seem to be more promising. This entry aims to provide insight into major aspects of vaccine design and formulation to address different diseases, including the recent outbreak of SARS-CoV-2. Special emphasis of this review is on the technical and practical aspects of vaccine construction and theranostic approaches to precisely target and localize the active compounds.
Keywords: Corona, COVID-19, drug delivery, lipid particles, liposomes, nanoliposomes, SARS-CoV-2, theranostics, tocosome, virus
1. Introduction
Viruses are microscopic infectious agents, which replicate only inside the living cells of host organisms. They infect all types of life forms from microorganisms (including archaea and bacteria) to plants, animals, and humans [1]. There are mainly four groups of viruses, namely: (i) bacteriophages (viruses that infect bacterial cells); (ii) plant viruses (e.g., TMV: tobacco mosaic virus); (iii) animal viruses (e.g., DHV: duck hepatitis virus; SIV: simian immunodeficiency virus); and (iv) human viruses (e.g., coronaviruses) (see Figure 1). Bacterial viruses, known as bacteriophages, are one class of viruses that are identified as the most widespread biological organisms on the earth [2,3]. They bind to bacterial cells and perfuse the viral genome into the cell via lytic or lysogenic cycles [4,5]. The genetic material of these types of viruses consists of either DNA or RNA molecules [6].
Figure 1.
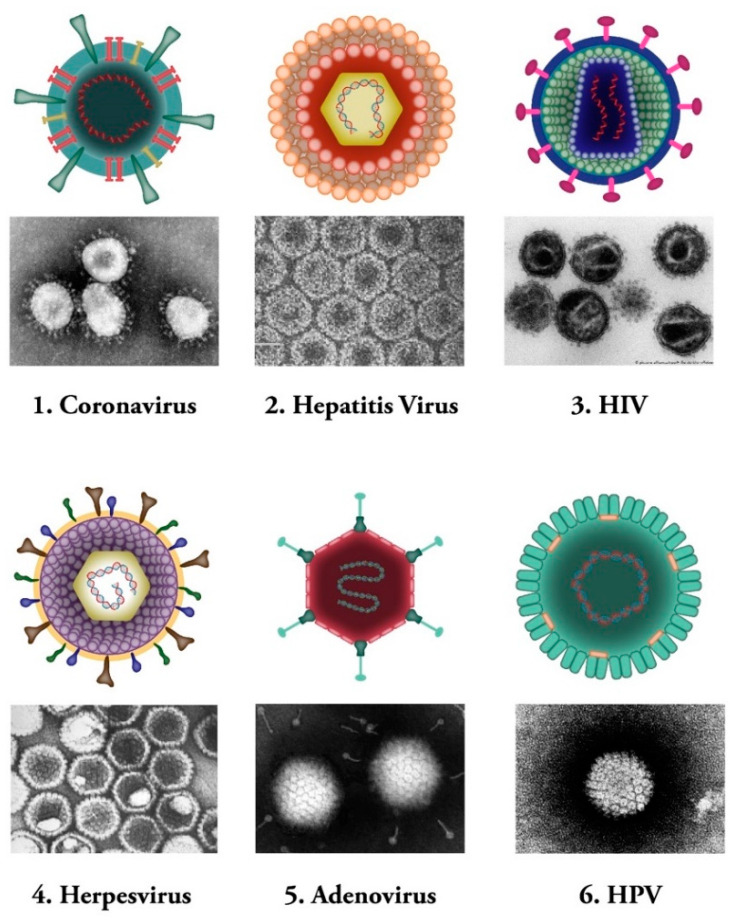
Common human viral pathogens. Six types of human infecting viruses, including coronavirus, hepatitis virus, human immunodeficiency virus (HIV), herpesvirus, adenovirus, human papilloma virus (HPV) are depicted schematically or as observed under electron microscopes.
Plant viruses, on the other hand, are parasitic organisms that use plant cells as their hosts. They rely on the host cellular resources and other factors essential for their replication cycle and movement [7]. Plant viruses utilize two types of movement, known as local (cell-to-cell movement in the infected leaves) and systemic (long-distance mobility through the plant vascular system). The genome of these viruses is made of ssDNA, dsRNA, positive-single-strand RNA, or negative-single-strand RNA [8,9].
Animal viruses are another group of viruses that cause infection and disease in animal cells. They need to identify a certain host cellular receptor for their entrance into the cell and the initiation of their life cycle [10]. The genome of animal viruses can be made of either DNA or RNA. Certain subgroups of animal viruses are classified as reverse transcribing viruses (also known as RT viruses), examples of which are simian immunodeficiency virus (SIV) and human immunodeficiency virus (HIV) [11].
Human viruses, on the other hand, are defined as specific pathogens that have to confiscate some of the host cellular pathways to survive [12]. These viruses can either cause disease or could be asymptomatic [13,14]. Human viruses manifest a broad range of various structures [15]. Same as other groups of viruses mentioned above, Human viruses contain either RNA or DNA as their genome. Their nucleic acid material can be single- or double-stranded molecule. Human viruses are classified into several subgroups. Some of the human viruses, such as the herpes virus, might stay hidden in the body and be activated under specific circumstances of stress and immune system compromise [16]. Hepatitis B and C viruses are examples of another type of human pathogens that maintain constant existence and virulency in the body, and people are permanent carriers of these pathogens [17,18] (Figure 1).
In the recent years we are facing novel viral pathogens that are defined as new etiological agents [19]. Emergence of these viral infections has notably affected human health and has caused concerns for the health care systems around the world [20]. Vaccination is one of the most potent approaches in the management of viral diseases. In many cases, the vaccine candidate requires protection from degradation and precise delivery to the target cells. This can be achieved by employing different types of drug and vaccine delivery strategies, among which, nanotechnology-based systems seem to be promising alternatives to the conventional vaccinations. The present narrative review aims to provide insight into major aspects of vaccine design and formulation to address different diseases, including the recent outbreak of SARS-CoV-2. This name was chosen based on the fact that the virus is genetically related to the coronavirus responsible for the SARS outbreak of 2003 and the Middle East Respiratory Syndrome (MERS) outbreak of 2012. On 11 February 2020, the International Committee on Taxonomy of Viruses (ICTV) announced “severe acute respiratory syndrome coronavirus 2” (SARS-CoV-2) as the official scientific name of this new pathogen [21]. Special emphasis of this entry is on the technical and practical aspects of vaccine construction and theranostic approaches to precisely localize the payload.
2. Vaccination Platforms
Vaccines are medical tools that cause pathogen-specific immune responses in the body [22]. They induce the immune system to produce efficient antibodies that can attach to the antigens strongly and prevent them from infecting cells. In the recent outbreak of SARS-CoV-2, we are facing an urgent need for an efficient vaccine to minimize the threats of the virus on the human health and global economy as soon as possible [23]. Table 1 lists different types of vaccine protocols currently available to tackle viral diseases. Some of different available vaccine protocols are explained in the following sections.
Table 1.
Different types of currently available protocols employed in vaccine construction.
| Vaccine Type | Target Pathogen/Disease | |
|---|---|---|
| Nucleic acid based vaccine | SARS-Cov-2 | |
| Peptide based vaccine | SARS-Cov-2, Hepatitis B | |
| Pathogen based vaccine | Live attenuated viral vaccine | Vaccina (smallpox), Measles, mumps, and rubella (MMR combined vaccine), Varicella (chickenpox), Influenza (nasal spray), Rotavirus, Zoster (shingles), Yellow fever |
| Inactivated viral vaccine | Polio (IPV), Hepatitis A, Rabies | |
| Viral subunit vaccine | Hepatitis B, Influenza (injection), Haemophilus influenzae type b (Hib), Pertussis (part of DTaP combined immunization), Pneumococcal, Meningococcal, Human papillomavirus (HPV) | |
| Adjuvant | Human papillomavirus (HPV) types 16 and 18, influenza (flu), Hepatitis B | |
2.1. Nucleic Acid-Based Strategies
The nucleic acid-based vaccines are generally selected from either an RNA molecule (e.g., mRNA) or a plasmid or a double-stranded DNA that encode antigens. These antigens provoke humoral and/or cell-mediated immune responses in the body [24,25]. The DNA/RNA -based vaccines are considered flexible, as they allow manipulating the antigens easily. The synthetic DNA/RNA stimulates the protein production, similar to the infection situation, in the target cells. The synthesized protein will be placed in the cytosolic plasma membrane (e.g., endoplasmic reticulum) and protein modification processes can precisely take place [26]. The important point towards this end is that DNA (or RNA) needs a delivery system to achieve its aims. In the context of nucleic acid-based COVID-19 vaccine formulation, liposomes and lipid nanoparticles (LNP), together with certain electrophysiological approaches (e.g., electroporation), in certain cases, appear to be among the most promising systems to deliver the nucleic acid construct to the target cell [27,28,29,30].
2.2. Peptide-Based Vaccines
Peptide-based vaccines have been designed against a variety of infectious and tumorigenic diseases [31]. Their design is based on the prediction and evaluation of specific epitopes of the B and T cells [32]. Epitopes are defined as antigenic regions, which are recognized by the immune cells and hold the immunogenic property for these cells [33]. To conduct an accurate cellular immune response in the body, antigenic segments should bind to a certain class of molecules known as Major Histocompatibility Complex (MHC). The antigenic fragments are processed by the MHC molecules, which will eventually present them to T cells. These peptidic epitopes should be linear in order to be able to bind MHC molecules. B cells possess receptors and are responsible for producing antibodies in the immune system [34]. Immunogenic carrier proteins, nanoparticle delivery systems, and potent adjuvants could be combined in the formulation in order to boost the immunogenicity of peptide-based vaccines. Several peptide-based vaccines (such as multi-epitope vaccines) against SARS-CoV-2 were formulated by interaction with T cells (CD4+ and CD8+) and B cells along with an adjuvant and assembling multi-epitope by using EAAAK linkers [35,36,37].
2.3. Pathogen-Based Strategies
2.3.1. Live Attenuated Viral Vaccines
Attenuated vaccines represent a weakened form of various pathogens, e.g., bacteria and viruses (including the yellow fever virus strain 17D) that lead to several different diseases. They induce potent and long-lasting cellular and humoral immunity in the body similar to the natural response, and are considered to be the oldest and the most efficient mode of vaccination [38,39,40]. Different approaches have been used for the production of the attenuated vaccines, such as employment of related non-human viruses (e.g., rotavirus, smallpox), administration at different body sites (adenovirus), and laboratory adaptation (polio and others) [41].
2.3.2. Inactivated Viral Vaccines
Whole virus vaccines utilize the complete particle of the virus that has been deactivated by processes employing certain chemical compounds, radiation, or heat. The immune responses of these type of vaccines qualitatively differ from those vaccines that are processed through intracellular pathways. In addition, inactivated viral vaccines often need extensive extra trials to confirm their safety because some studies have indicated increased infectivity following immunization [42,43,44].
2.3.3. Viral Subunit Vaccines
Subunit vaccines are specific antigenic segments (such as synthetic peptide or protein) of a pathogen, that are used to induce an immune response and provoke acquired immunity against the pathogen from which they are derived [45,46]. They possess high safety profiles, specifically targeting well-defined neutralizing epitopes [45,47]. There is no risk of incomplete inactivation, regain of virulence of the attenuated virus, or unfavorable host responses to viral subunit vectors [46,48]. Since the prevalence of SARS in 2002, subunit vaccines based on this virus have been studied and tested widely, demonstrating adequate efficacy and protection against SARS-CoV infections in several animal models [49,50,51,52].
2.4. Adjuvants
Adjuvants are pharmacological or immunological agents that are being used to improve immune responses. In other words, they are capable of increasing the biological half-life of vaccines and antigen uptake by the antigen-presenting cells (APCs). Currently, many adjuvants with different mechanisms of action are being utilized in vaccine constructs. Examples of adjuvants include mineral salts, saponins, cytokines, microbial components, microparticles, emulsions, and certain liposomes and nanoliposomes. They induce the production of immune regulatory cytokines, activate inflammations, local inflammation, cellular recruitment, and induce more rapid, broader, and stronger immune responses that are essential for good immunogenicity [53,54]. Nanoparticles and nanovesicles, due to their intrinsic adjuvanticity (by activating complement system, inducing autophagy and activation of inflammasome) are also considered as vaccine adjuvants nanosystems [55,56,57,58].
3. Nanotechnology in Drug and Vaccine Delivery
Nanotechnology techniques provide substantial advantages in the formulation of novel vaccines and have significantly enhanced their effectiveness [57]. A broad range of active moieties can be encapsulated and delivered by nanovesicles and nanocarriers. These nanosystems possess more surface area to volume ratio when compared to micrometric drug carriers and, as such, they can attach to/interact with the target cells more efficiently. Nanotechnology-based vaccines can be formulated employing nucleic acids, purified subunit proteins or polysaccharides, synthetic peptides, or recombinant proteins. These classes of vaccines may stand in need of adjuvants to become sufficiently immunogenic, as explained above. As explained in the previous section, nanovesicles are able to function as adjuvants for viral vaccines; therefore, they are regarded as nano-adjuvants (NAs) [55,58]. They can also increase immunogenicity, cellular uptake, and stability of vaccines by encapsulation or absorption of the vaccine antigen or DNA in a proper formulation. Moreover, it is postulated that various designs of nanoparticles are able to modulate the systemic release of a vaccine and its bio-distribution [58]. The recent SARS-CoV-2 pandemic has seriously put human health in danger and has resulted in an extensive magnitude of mortality and morbidity all around the world [59]. In the context of this outbreak, emerging nanotechnologies, including some mRNA-based vaccines carried by lipidic nanovesicles and viral vectors, have reached Phase II and III clinical trials and some of such formulations have even been approved for human use so far [60,61,62,63].
An important feature of nanotechnology applications in drug delivery is the possibility of elimination of adverse drug effects through a theranostic approach. Theranostic drug delivery employs the combined features of diagnosis and therapy, and hence, is extremely beneficial in the development of targeted drug delivery systems. Theranostic approach is utilized to design and formulate biocompatible and efficient protocols for stimuli-responsive, controlled vaccine, or drug delivery for various applications [61,62]. The success of theranostics depends on multidisciplinary collaborations, combined with our understanding of host responses to customized therapeutic or preventive agents and formulating individualized therapies.
Associating the right therapeutic moiety to the right nanocarrier or nanovesicle, aimed for a specific disease condition, is essential for the clinical success of nanomedicine against the SARS-CoV-2 and other viral infections. Nanotechnology and nanomedicine enable reformulating different drug candidates to improve their “therapeutic index”, mainly by addressing the limitations associated with the drug molecule and alleviating the conventional toxicity or side effects [59,60,61]. The following sections bring more insight into the micro- and nano-carrier systems for vaccine delivery.
4. Vaccine Delivery Techniques
A perfect vaccine delivery technique requires encompassing a mixture of high loading capacity, prolonged half-life, no premature leakage or release of the active compound, no intervention with the consistency of the therapeutic molecule, and a simple, scalable, and reproducible manufacturing procedure. Acceptable bio-compatibility, bio-degradability, and safety aspects of the formulation, in addition to a clear understanding of the mechanism of action of the vaccine delivery system, are also desirable factors. The current COVID-19 pandemic has increased the necessity of constructing safe and efficacious vaccine delivery systems [61]. Nanotechnology and nanomedicine provide substantial advantages for the formulation of vaccine protocols and enable either the targeted delivery or co-delivery of the therapeutic molecules and reinforce the delivery of high-value and high-tech bio-therapeutic agents, such as proteins or genetic material [62]. Some of the main vaccine encapsulation and targeting techniques are presented in the following sub-sections.
4.1. Polymeric Carriers
Polymer science possesses an efficient role in producing nanomaterials in a simple, cost-beneficial, and scrutinized way [64]. The design of polymeric therapeutic delivery technologies and vaccine carriers (especially biodegradable and biocompatible constructs) are considered as an essential part of the design and formulation of vaccines [65]. Certain features, such as size, charge, structure, hydrophobicity, aggregation, and natural or synthetic composition, can influence the immunogenicity of the polymeric nanostructures. In the context of vaccine construction, polymeric nanoparticles can be applied as controlled delivery systems of the immunomodulatory adjuvants and antigenic compounds. Antigens with an amphoteric nature can be attached by adsorption or surface immobilization to certain polymeric nanocarriers, such as chitosan or dextran sulfate-based nanoparticles. Release of the antigenic compound in such cases is predesigned based on the properties of the biological milieu (e.g., pH, temperature, ionic strength, etc.) [64,65].
Certain polymers, including dendrimers, polyamines, polyethylenimines, and copolymers, are functional materials capable of delivering mRNA and other vaccine candidates. Similar to other drug delivery systems, polymers can also protect encapsulated material from enzyme-mediated degradation and facilitate intracellular delivery. Nevertheless, the formulation of polymer-based vaccine nanoparticles tends to have high polydispersity [66]. To stabilize the formulation and improve the safety profile, structural modifications of polymer materials, such as incorporating hyperbranched groups, lipid chains, and biodegradable subunits, have been explored [67].
Despite the considerable potential of the polymeric nanocarriers and the magnitude of the bibliography on their therapeutic applications, a very limited number of polymeric constructs has reached clinical trials thus far [68].
4.2. Surfactant-Based Carriers
Non-ionic surfactant vesicles (NSVs), also known as niosomes, are relatively new drug delivery systems that are employed for managed and targeted delivery of drugs because of their ease of preparation on large scales and cost-effectiveness [69,70]. Moreover, their characteristics such as biocompatibility, biodegradability, and flexibility in structure have made them ideal carriers for loading hydrophobic and hydrophilic drugs [71]. These carriers have been applied in many areas, including gene transfer and vaccine delivery [72]. Despite all benefits of NSV delivery systems, their stability could be an issue with respect of the possibility of hydrolysis of the entrapped or encapsulated drug in an aqueous media. The problem of untimely and uncontrolled drug leakage from the non-ionic surfactant vesicles and aggregate formation of the niosomes may also occur. In order to alleviate these problems, incorporation of certain phospholipids and/or sterols (e.g., cholesterol—as a bilayer modulating agent) in the structure of niosomes is suggested [70,73,74,75].
4.3. Lipidic Delivery Systems
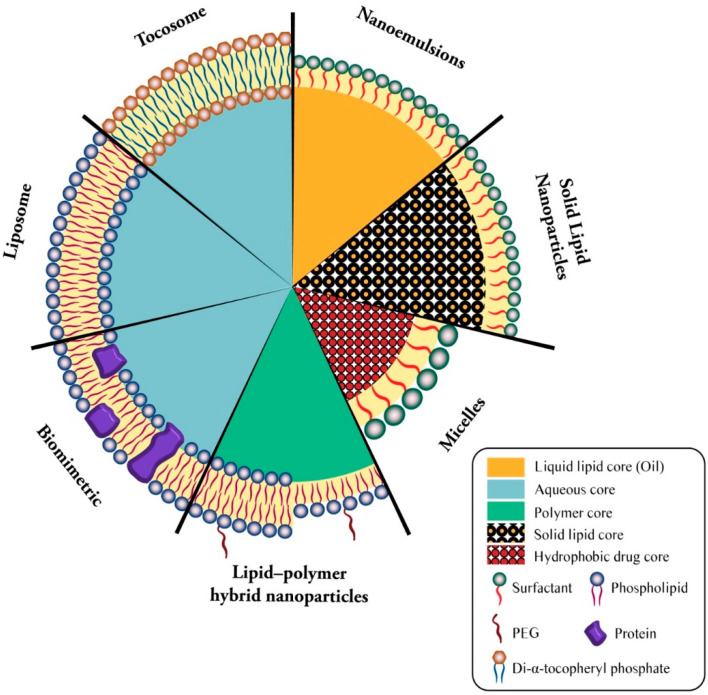
Lipid-based bioactive delivery systems are among the most promising technologies employed for the encapsulation and targeted release of drug and vaccine molecules. They include bilayer lipid vesicles (e.g., liposomes, nanoliposomes, archaeosomes, vesicular lipid gels, immunovesicles, lipospheres), solid lipid nanoparticles (SLN), tocosomes, and some other micro- and nanocarrier systems [76]. Figure 2 demonstrates the structure of available lipid-based drug and vaccine carriers.
Figure 2.
Simplified structure of main lipidic carrier systems. Seven types of lipidic vaccine/drug carriers consists of liposome (phospholipid bilayer + aqueous core), biomimetric (proteinated phospholipid bilayer + aqueous core), lipid-polymer hybrid nanoparticles (bi- or monolayer phospholipid + polymer core), micelles (surfactant monolayer + hydrophobic drug core), solid lipid nanoparticles (surfactant monolayer + solid lipid core), nanoemulsion (surfactant monolayer + liquid lipid core), and tocosome (Di-α-tocopheryl phosphate bilayer + aqueous core).
4.3.1. Liposomes as Drug and Vaccine Carriers
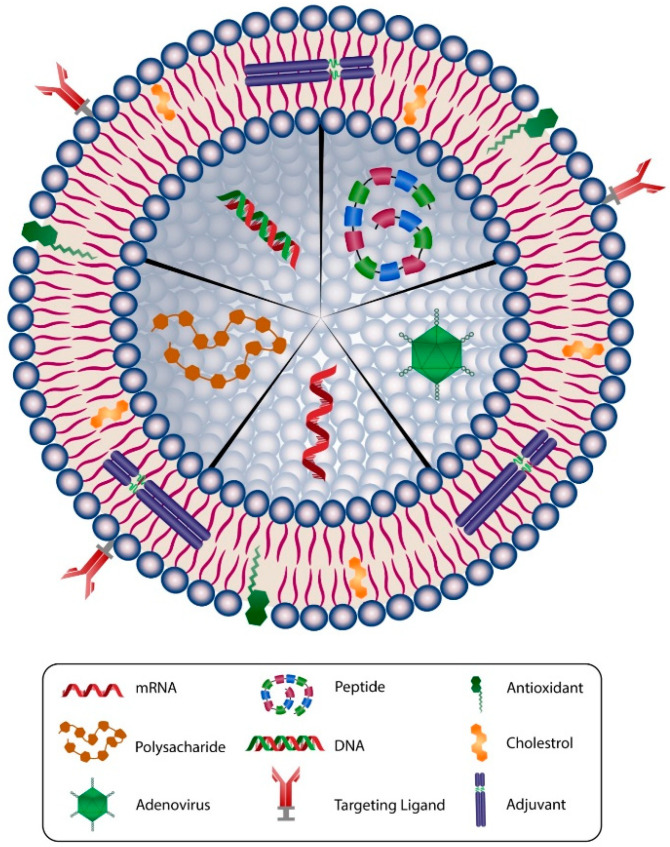
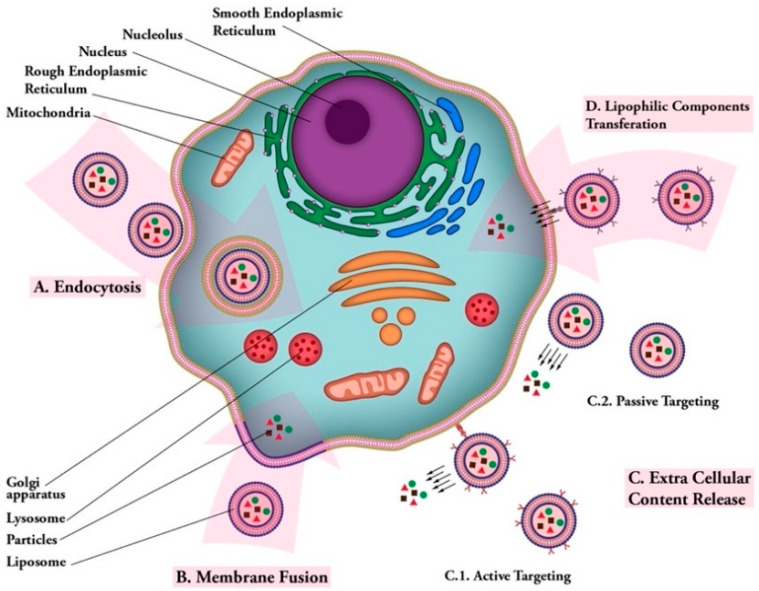
One of the first and most versatile drug delivery protocols is liposome, which is also known as a bilayer lipid or phospholipid vesicle. The word liposome refers to mesomorphic, nematic, and smectic structures that are mainly composed of lipid, phospholipid, and water molecules. The main ingredients of liposomes are amphiphilic lipid and phospholipid molecules [77]. They improve the stability and efficacy of bioactive compounds by entrapment and release of water-soluble, lipid-soluble, and amphiphilic materials, as well as targeting the encapsulated compounds to particular cells or tissues inside the body. Liposomes can be made on a small scale (e.g., for lab-scale research and development), pilot-scale, or industrial scales using natural ingredients, such as egg or soy lecithin. It is also possible to include other ingredients, such as sterols (cholesterol or plant-derived phytosterols), polypeptides (e.g., antigens), antioxidants (e.g., α-tocopherol), as well as polymers (such as poly-ethylene-glycol or chitosan) in the structure of the lipid vesicles. Figure 3 depicts main components used in the construction of a liposomal or nanoliposomal vaccine formulation. These additives assist in modulating the bilayer structure of liposomes, increase their tolerance towards reactive oxygen assaults, increase their blood circulation half-life, and provide a targeting strategy for the lipid vesicles [78]. Different types of liposomal vaccine uptake mechanisms are illustrated in Figure 4.
Figure 3.
Schematic representation of main components used in a typical liposomal/nanoliposomal vaccine formulation. Liposomes can carry different types of drug or vaccine core, such as mRNA, DNA, peptide, adenovirus, or polysacharide. The membranes of liposomal carriers are made of phospholipids, cholesterol, targeting ligands, antioxidants, and adjuvants.
Figure 4.
Liposomal vaccine uptake mechanisms. (A) Endocytosis: uptake of vaccine carrier by cell; (B) membrane fusion: fusion of vaccine carrier and cell membranes and release of vaccine particles into cytoplasm; (C) extra cellular content release: (C.1); active targeting: releasing vaccine particles near cell membrane after targeting via ligand-receptor connection. (C.2); passive targeting: release of vaccine particles near cell membrane; (D) lipophilic components transferation: transfer of lipophilic vaccine particles through cell membrane after carrier ligand-receptor binding.
4.3.2. Nanoliposomes
Nanoliposomes (nanometric versions of liposomes or lipidic nanovesicles) can be briefly defined as colloidal nanostructures composed of lipid or phospholipid molecules [79]. It should be noted that liposomes and their nano-versions possess the same physical, chemical, and thermodynamic properties, which are mainly governed by their ingredients and their suspension media. The difference between the two related drug carriers comes from their size range. It is known that the smaller the particle size, the larger the surface-to-volume ratio they will acquire. Therefore, in comparison with liposomes, nanoliposomes provide more surface area and have more potential to increase solubility, enhance bioavailability, improve the controlled release of the loaded material, and enable accurate targeting of the encapsulated drug or vaccine moieties. Manufacture of both liposomes and nanoliposomes involves input of energy (in the form of agitation, heat, ultrasound, etc.) to a dispersion of lipid and phospholipid molecules in an aqueous medium [77,78,79,80,81]. Although liposomes and nanoliposomes are initially prepared as a liquid product (aqueous suspension), they can be subsequently incorporated in a cream, lotion, aerosol, soft-gel, powder (via lyophilization or spray drying for instance), or other formulations and dosage forms [76]. These versatilities open significant opportunities for efficient vaccine design and formulation.
4.3.3. Solid Lipid Nanoparticles (SLNs)
Solid lipid nanoparticles can be defined as nano-sized colloidal carriers made up of accurate ratios of lipid, surfactants, and bioactive compounds. These relatively new drug delivery systems are superior to other available drug carriers, due to their small size and lipid core that provide some unbeatable properties. They also provide a suitable alternative to the existing drug delivery technologies, such as liposomes, emulsions, and polymeric nanocarriers. In these new generations of submicron-sized lipidic carriers, the liquid-phase lipid has been replaced by a solid-phase lipid, resulting in the lipidic particle that is solid at room temperature or body temperature [80]. Triglycerides, acetyl alcohol, emulsifying wax, beeswax, carnauba wax, cholesterol, and cholesterol butyrate are some of the solid lipids employed in the preparation of solid lipid nanoparticles [76,81].
4.3.4. Immunostimulatory Complexes (ISCOMs)
The immune stimulatory complexes, known as ISCOMs, are other vaccine delivery carriers with potent adjuvanticity, as attested clinically [82]. They size about 40–60 nm and are self-assembled in solution at well-defined ratios of saponin adjuvant Quil A and a protein antigen, cholesterol, and phospholipid. The neutral or positively charged dimethylaminoethane-carbamoyl (DC)-cholesterol, and zwitterionic phospholipids or the cationic dioleoyl-trimethyl-ammonium-propane (DOTAP) are such materials to be used for ISCOM formation. These cage-like particles with a hollow center are also used for trapping hydrophobic antigens [82,83].
ISCOMs-based vaccines possess the potentiality to boost both humoral and cellular immune responses. ISCOMATRIX is a vaccine delivery carrier and adjuvant, which does not contain antigen and operates similarly to ISCOMs. Nevertheless, it is more applicable than ISCOMs due to its potency to remove the limitation of hydrophobic antigens. Antigens added later to the preformed ISCOMATRIX particles can be employed to entrap hydrophilic antigens [84,85]. Several research groups have published the usage of different antigens for forming ISCOMs and ISCOMATRIX vaccines, including antigens derived from HIV [84,85], HPV, Newcastle disease, and influenza [86,87].
4.4. Tocosome as a Potential Vaccine Carrier
Tocosome is a vesicular and colloidal bioactive carrier system that is mainly composed of phosphate-group-bearing alpha tocopherols [88]. Tocosomes can also accommodate proteins, polymers, and sterols in their structures, as is the case with liposomes and nanoliposomes. Alpha-tocopherol phosphate (TP), a main ingredient of tocosome, is the phosphorylated form of alpha-tocopherol that is available naturally in human and some animal tissues and in certain food compounds (such as fruits, green vegetables, cereals, dairy products, as well as deferent nuts and seeds) [89,90]. TP is made up of a phosphate group binded to one hydrophobic chain (phytyl tail), composed of three isoprene units. Di-alpha-tocopherol phosphate (T2P), a molecule related to TP, is composed of two phytyl chains. The hydrophobic phytyl chains of T2P molecules cannot arrange in a parallel position because of the presence of bulky isoprene side-chains (unlike phosphatidylcholine and some other phospholipids). Accordingly, T2P possesses a conical geometric shape, while the TP has a cylindrical geometric shape, similar to PC [91]. Clinical studies have demonstrated that TP and T2P molecules have many advantages for human health, including atherosclerotic prevention as well as cardioprotective and anti-inflammatory properties [92,93]. In addition, the TP molecule has an inhibitory effect against tumor invasion [94]. Investigations have also reported that TP can protect primary cortical neuronal cells from glutamate-induced cytotoxicity in vitro and can decrease the amount of lipid peroxidation products in the plasma and liver of mice in vivo [95]. Formulations of tocosomes include various phospholipid molecules (in addition to TP and T2P components) and different combinations of cholesterol. They have recently been used for the entrapment and controlled release of the anticancer drug 5-fluorouracil successfully [88].
5. Lipidic Carrier-Based Vaccine Design and Formulation
Lipids, lipid-like compounds, tocopheryl-phosphates, and lipid derivatives have been widely used to formulate nanovesicles and nanoparticles for in vivo delivery of vaccines [96,97,98,99,100]. Lipidic nanocarriers are generally defined as nano-sized particulate or colloidal systems that are composed of natural, synthetic, or physiological lipid/phospholipid materials [101]. Lipidic nanocarriers are being developed for vaccine delivery for the following two main reasons. Firstly, they can encapsulate vaccine molecules and compounds, and hence, protect them from enzymatic degradation [96]; in the case of mRNA vaccines, the reported encapsulation efficiency by lipidic nanocarriers was usually high, designating the mRNA molecules were mostly internalized within the vesicles [102,103]. Secondly, the nanocarriers can effectively transport vaccine molecules into the cell cytosol through a series of endocytosis processes [104]; for example, it was reported that the surface adsorption of apolipoprotein E on the lipid-based nanoparticles might facilitate its intracellular delivery via low-density lipoprotein, receptor-mediated, clathrin-dependent endocytosis [105]. This endocytosis process delivered the vaccine-loaded nanocarriers to the cell membrane-bound vesicles, including endosomes and lysosomes [104,106]. Eventually, the lipid-based nanocarriers facilitated translocation of the vaccine cargos into the cytosol for protein expression [97].
The lipidic nanocarriers usually contain one or more of the functional lipid components, which are crucial for the intracellular vaccine delivery [66,97,107]. The cationic or ionizable lipid materials, such as 1,2-di-O-octadecenyl-3-trimethylammonium propane (DOTMA), N,N-Dimethyl-2,3-bis[(9Z,12Z)-octadeca-9,12-dienyloxy]propan-1-amine (DLinDMA), and N1,N3,N5-tris(3-(didodecylamino)propyl)benzene-1,3,5-tricarboxamide (TT3) usually contain one or more amino groups [108,109,110,111]. These lipid molecules can be positively charged at a certain pH to encapsulate the negatively charged molecules (e.g., DNA or RNA) via electrostatic interactions and help interact with the cell membrane on the target cells. Recent studies indicated that the final step of vaccine release from lipidic nanocarriers into the cytosol might involve the membrane disruption of endosomes. In this process, the ionizable cationic lipids were suggested to interact with anionic phospholipids on the endosomal membrane and form disruptive non-bilayer structures that finally released the encapsulated vaccine into the cell cytoplasm [112]. Moreover, the structure–activity relationship of the lipidic structures for vaccine delivery and endosomal escape was studied. Results of these studies indicated that the hydrophilic head group of the phospholipid molecules might determine the acid dissociation constant (pKa) and influence the vaccine delivery efficiency [113]. Besides, modification of fatty acids structures in the hydrophobic tails may also affect the vaccine delivery efficiency [113,114]. Even though the membrane disruptive features of the lipid molecules improve the delivery efficiency, certain synthetic materials may cause side effects in vivo [115]. The helper lipids, such as 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), and cholesterol, stabilize the structures of lipidic nanocarriers and facilitate endosomal escape [116]. The PEG-lipid conjugates could stabilize the nanocarriers during preparation and provide a hydrophilic outer layer that prolongs the circulation time after in vivo administration [66,116,117,118]. In addition to these functions, the engineered ionizable lipid molecules containing cyclic amino head groups, isocyanide linker, and two unsaturated alkyl tails were reported to provide adjuvant activities independent of the encapsulated vaccine. These cyclic amino head groups directly bound the STING (stimulator of interferon genes) protein and triggered the downstream signaling pathway, leading to an elevated innate response. After subcutaneous injection of an antigen-coding mRNA encapsulated by such lipidic nanocarriers into mice, the researchers observed the upregulation of antigen-specific T cells and inhibition of tumor growth [119].
The formulation methods of lipid-based vaccines mainly include thin-film hydration [120,121], direct mixing [122], ethanol injection, and continuous-flow microfluidic device [123,124]. Among these methods, the continuous-flow microfluidic device emerges as a prevalent method to prepare RNA encapsulated nanoparticle, especially lipid nanoparticles, for in vivo applications [66,125]. These chip-based microfluidic devices mix two laminar flows, the vaccine-containing aqueous phase, and the carriers-containing solvent phase, through a confined microchannel equipped with mixers at a controlled speed, leading to rapid diffusion, change of polarity, and self-assembly of vaccine-lipidic nanoparticles at the interface [126,127]. The resulting lipid nanoparticles are relatively homogeneous formulation and usually show spherical and multilamellar morphologies [127]. Compared to other preparation methods, the use of continuous-flow microfluidic devices increases reproducibility, improves molecular stability, reduces the chance of contamination, and is easily scaled up for preclinical and clinical studies [125,128]. Another safe, mild, and robust method of vaccine preparation on industrial scales is “Mozafari method” [129], which is based on a former technique known as heating method [130,131]. This patented process, and its modifications, does not require application of toxic solvents, detergents, high-shear forces, or extreme pH values [132,133]. Drug and vaccine formulations can be mass manufactured using a simple vessel at very low shear forces (e.g., maximum agitation power of 1000 rpm) and temperatures below 40 °C [134,135,136].
6. Vaccine Targeting Strategies
Covering the surface of lipidic nanocarriers with immune cell receptors may facilitate their uptake by the desired type of immune cells. For the immune system to be activated, or for an immune response to be provoked, specific APCs need to encounter an antigen and a risk signal. APCs are concentrated at high concentration in the lymph nodes. For lymph node targeting, the vaccine moiety can be injected directly in the nodes, or the nanocarriers can be designed to accumulate in the lymph nodes. The two most important parameters for lymph node accumulation are size and surface composition of the nanocarriers. Scientific reports indicate decreasing lymphatic uptake with increasing particle size. Only nanoparticles with diameters smaller than about 150 nm appear to enter the lymphatic capillaries, and are subsequently drained to the peripheral lymphatics [137,138,139]. On the other hand, larger lipid particles and vesicles are retained at the injection site [140,141]. Larger vesicles are believed to be recognized and cleared more rapidly by the complement system because they present a larger number of recognition sites on their surface [142]. Intravenously injected lipidic nanoparticles are mostly eliminated from the blood circulation, after opsonization, by the cells of the immune system, which are mainly present in the liver and spleen. Opsonization is the process by which particles are labeled for ingestion and elimination by the macrophages and phagocytes. Inclusion of certain polymers (e.g., polyethylene glycol (PEG), polyhydroxyethyl L-asparagine (PHEA)), or incorporation of glycolipids (e.g., monosialoganglioside) to the lipid vesicles, prevent opsonization and result in sterically stabilized vesicles. These carrier systems have several advantages over conventional formulations including reduced recognition and uptake by MPS/RES, extended circulation half-lives, and dose-independent pharmacokinetics (for a recent review see: [143]). It has been reported that anti-PEG antibody response following repeated intravenous (IV) administration of PEGylated vesicles dramatically accelerate blood clearance of the vesicles and leads to acute hypersensitivity [144,145]. This finding is a concern for immunotherapy applications, where multiple dosing may be required for long-lasting protection. A possible solution may be achieved by modifying the PEG molecule into a less immunogenic variant, using alternative molecules, such as PHEA, or by using different administration routes.
Another group of target cells for vaccination purposes are dendritic cells (DC). These cells are covered with different receptors, which include lectins that recognize carbohydrate moieties present on many pathogens, and are involved in antigen capture and presentation [146,147]. Other dendritic cell receptors are the mannose receptor, DC-SIGN, DEC-205, and Langerin [148,149,150,151]. Active targeting of dendritic cells has been extensively studied in recent years. The term active targeting is somewhat misleading in that the lipidic nanoparticles are not actually actively guided toward dendritic cells. In fact, uptake by dendritic cells is enhanced by modification of the surfaces of vesicles with suitable molecules. These molecules include antibodies (anti-CD11c, anti-DEC-205) or peptides (P-D2, RGD), where anti-CD11c antibody, P-D2, and RGD peptides target integrins and anti-DEC-205 antibody targets the c-type lectin receptor DEC-205 [152].
DC receptors have been characterized and utilized for targeted protein and protein–lipid-nanoparticle vaccines extensively. Initial studies employed mannose monosaccharides or disaccharides to target vaccines to dendritic cells, albeit with little success [153]. The binding affinity of such monosaccharaide molecules is very weak, typically in the mM range. However, the binding affinity can be enhanced by orders of magnitude by coupling the monosaccharaides to a scaffold that forms a multivalent cluster or by using multibranched saccharides [154,155]. Biodistribution and pharmacokinetics of such engineered nanoparticles is altered significantly when varying the density of the sugar moieties [155]. A high dendritic cell specificity was observed for LNPs containing 11% mannosylated lipids, while no specificity was observed for LNPs containing 3% mannosylated lipids [156]. In another study, mannosylated mRNA–lipid nanoparticles coding for MART-1 also showed higher vaccination rates compared with their nonmannosylated analogs [157]. It would be interesting to investigate if surface modifications with ligands for different DC subsets also increase the potency of liposomal mRNA vaccines, as has been shown in the case of peptide-based vaccines [158].
7. Synopsis
Recent progress in nanobiotechnology and advanced nanomanufacturing lay the foundation for rapid development of innovative vaccine technologies to make an impact during the COVID-19 pandemic. Towards this end, efficient vaccine encapsulation and delivery systems with the capability of mass manufacturing the vaccine formulation play a critical role. However, improvements are still required to optimize the safety profile and to increase the vaccination efficacy. The progress in the development of various delivery systems has enabled numerous preclinical studies and clinical trials. Among the all-available drug delivery systems, lipidic carriers represent one of the most advanced platforms for vaccine delivery and targeting in vivo. Compared with other delivery protocols, the lipidic carriers have the longest history of being under research and development worldwide as well as having the highest number of FDA approved pharmaceutical products on the market. As the delivery methods and the vaccine formulations further advance, theranostic lipidic vaccines will become an important class of medicine to effectively tackle diverse health issues, such as viral and other infectious diseases.
Abbreviations
| Ab | antibody |
| Ag | antigen |
| APC | antigen-presenting cell |
| ApoE | apolipoprotein E |
| DC | dendritic cells |
| DHV | duck hepatitis virus |
| DLinDMA | N,N-Dimethyl-2,3-bis[(9Z,12Z)-octadeca-9,12-dienyloxy]propan-1-amine |
| DOPE | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine |
| DOTMA | 1,2-di-O-octadecenyl-3-trimethylammonium propane |
| DSPC | 1,2-distearoyl-sn-glycero-3-phosphocholine |
| DTaP | a vaccine that helps children younger than age 7 develop immunity to three deadly diseases caused by bacteria: diphtheria, tetanus, and whooping cough (pertussis) |
| EAAAK | alpha helix-forming linkers with the sequence of (EAAAK)n, which are being applied to the construction of many recombinant fusion proteins |
| HIV | Human immunodeficiency virus |
| HPV | Human papillomavirus |
| ICTV | international committee on taxonomy of viruses |
| ISCOMs | immune stimulatory complexes |
| MERS | Middle-east respiratory syndrome |
| MHC | major histocompatibility complex |
| MPS | mononuclear phagocytic system |
| NA | Nano adjuvant |
| NSV | nonionic surfactant vesicle |
| PC | phosphatidylcholine |
| PEG | polyethylene glycol |
| PHEA | polyhydroxyethyl L-asparagine |
| RES | reticuloendothelial system |
| RT | reverse transcriptase |
| SIV | simian immunodeficiency virus |
| SLN | solid lipid nanoparticles |
| STING | stimulator of interferon genes |
| TMV | tobacco mosaic virus |
| TP | tocopheryl phosphate |
| T2P | di-tocopheryl phosphate |
| TT3 | N1,N3,N5-tris(3-(didodecylamino)propyl)benzene-1,3,5-tricarboxamide |
Author Contributions
In the preparation of this Review Article, all authors contributed equally to the compilation and editing of the text and table, as well as graphical designs of figures of this article. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable. This Review Article analyzed and discussed already published data and did not report any new data.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Smith A.E., Helenius A. How Viruses Enter Animal Cells. [(accessed on 1 March 2021)];Science. 2004 304:237–242. doi: 10.1126/science.1094823. Available online: https://science.sciencemag.org/content/304/5668/237. [DOI] [PubMed] [Google Scholar]
- 2.Breitbart M., Rohwer F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005;13:278–284. doi: 10.1016/j.tim.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Martel B., Moineau S. CRISPR-Cas: An Efficient Tool for Genome Engineering of Virulent Bacteriophages. [(accessed on 1 March 2021)];Nucleic Acids Res. 2014 2:9504–9513. doi: 10.1093/nar/gku628. Available online: https://academic.oup.com/nar/article/42/14/9504/1283784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Labrie S.J., Samson J.E., Moineau S. Bacteriophage Resistance Mechanisms. [(accessed on 1 March 2021)];Nat. Rev. Genet. 2010 8:317–327. doi: 10.1038/nrmicro2315. Available online: https://www.nature.com/articles/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 5.Vu N.T., Oh C.S. Bacteriophage usage for bacterial disease management and diagnosis in plants. [(accessed on 1 March 2021)];Plant Pathol. J. 2020 36:204–217. doi: 10.5423/PPJ.RW.04.2020.0074. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7272851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatfull G.F. Bacteriophage genomics. Curr. Opin. Microbiol. 2008;11:447–453. doi: 10.1016/j.mib.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Ruiz H. Host factors against plant viruses. Mol. Plant Pathol. 2019;20:1588–1601. doi: 10.1111/mpp.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Ruiz H. Susceptibility Genes to Plant Viruses. [(accessed on 1 March 2021)];Viruses. 2018 10:484. doi: 10.3390/v10090484. Available online: http://www.mdpi.com/1999-4915/10/9/484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plant Virology—Roger Hull—Google Books. [(accessed on 1 March 2021)]; Available online: https://books.google.com/books?hl=en&lr=&id=PYrZAAAAQBAJ&oi=fnd&pg=PP1&dq=Hull,+R.;+Plant+virology.+2013:+Academic+press&ots=NJHjP_M4WU&sig=Ybvvy1UVPPemv2i24LhWEFmVMFE#v=onepage&q=Hull%2C R.%2C Plant virology. 2013%3A Academic press&f=false.
- 10.Avaratnarajah C., Warrier R., Kuhn R. Encyclopedia of Virology. Elsevier Ltd.; Amsterdam, The Netherlands: 2008. Assembly of Viruses: Enveloped Particles; pp. 193–200. [Google Scholar]
- 11.Molecular Virology of Human Pathogenic Viruses—1st Edition. [(accessed on 1 March 2021)]; Available online: https://www.elsevier.com/books/molecular-virology-of-human-pathogenic-viruses/ryu/978-0-12-800838-6.
- 12.Greco A. Involvement of the nucleolus in replication of human viruses. Rev. Med. Virol. 2009;19:201–214. doi: 10.1002/rmv.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pride D.T., Salzman J., Haynes M., Rohwer F., Davis-Long C., White R.A., Loomer P., Armitage G.C., Relman D.A. Evidence of a Robust Resident Bacteriophage Population Revealed through Analysis of the Human Salivary Virome. [(accessed on 1 March 2021)];ISME J. 2011 6:915–926. doi: 10.1038/ismej.2011.169. Available online: www.nature.com/ismej. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wylie K.M., Weinstock G.M., Storch G.A. Emerging view of the human virome. Transl. Res. 2012;160:283–290. doi: 10.1016/j.trsl.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bushman F.D., McCormick K., Sherrill-Mix S. Virus Structures Constrain Transmission Modes. [(accessed on 1 March 2021)];Nat. Microbiol. 2019 4:1778–1780. doi: 10.1038/s41564-019-0523-5. Available online: https://www.nature.com/articles/s41564-019-0523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen C.-H., Chiu Y.-L., Wei F.-C., Koong F.-J., Liu H.-C., Shaw C.-K., Hwu H.-G., Hsiao K.-J. High Seroprevalence of Borna Virus Infection in Schizophrenic Patients, Family Members and Mental Health Workers in Taiwan. [(accessed on 1 March 2021)];Mol. Psychiatry. 1999 4:33–38. doi: 10.1038/sj.mp.4000484. Available online: https://www.nature.com/articles/4000484. [DOI] [PubMed] [Google Scholar]
- 17.Lauer G.M., Walker B.D. Hepatitis C Virus Infection. N. Engl. J. Med. 2001;345:41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 18.Lee W.M. Hepatitis B Virus Infection. N. Engl. J. Med. 1997;337:1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 19.Parvez M.K., Parveen S. Evolution and Emergence of Pathogenic Viruses: Past, Present, and Future. [(accessed on 1 March 2021)];Intervirology. 2017 60:1–7. doi: 10.1159/000478729. Available online: https://www.karger.com/Article/FullText/478729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krause R.M. The Origin of Plagues: Old and New. [(accessed on 1 March 2021)];Science. 1992 257:1073–1078. doi: 10.1126/science.257.5073.1073. Available online: https://science.sciencemag.org/content/257/5073/1073. [DOI] [PubMed] [Google Scholar]
- 21.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., et al. Severe acute respiratory syndrome-related coronavirus: The species and its viruses—A statement of the Coronavirus Study Group. bioRxiv. 2020 [Google Scholar]
- 22.Pandolfi F., Franza L., Todi L., Carusi V., Centrone M., Buonomo A., Chini R., Newton E.E., Schiavino D., Nucera E. The Importance of Complying with Vaccination Protocols in Developed Countries: “Anti-Vax” Hysteria and the Spread of Severe Preventable Diseases. Curr. Med. Chem. 2019;25:6070–6081. doi: 10.2174/0929867325666180518072730. [DOI] [PubMed] [Google Scholar]
- 23.Ganti R.S., Chakraborty. A.K. Mechanisms underlying vaccination protocols that may optimally elicit broadly neutralizing antibodies against highly mutable pathogens. bioRxiv. 2020:330340. doi: 10.1101/2020.10.07.330340. [DOI] [PubMed] [Google Scholar]
- 24.Geall A.J., Mandl C.W., Ulmer J.B. RNA: The new revolution in nucleic acid vaccines. Semin. Immunol. 2013;25:152–159. doi: 10.1016/j.smim.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Piyush R., Rajarshi K., Chatterjee A., Khan R., Ray S. Nucleic acid-based therapy for coronavirus disease 2019. Heliyon. 2020;6:e05007. doi: 10.1016/j.heliyon.2020.e05007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Restifo N.P., Ying H., Hwang L., Leitner W.W. The Promise of Nucleic Acid Vaccines. [(accessed on 1 March 2021)];Gene Ther. 2000 7:89–92. doi: 10.1038/sj.gt.3301117. Available online: www.nature.com/gt. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Vrieze J., Louage B., Deswarte K., Zhong Z., De Coen R., Van Herck S., Nuhn L., Frich C.K., Zelikin A.N., Lienenklaus S., et al. Potent Lymphatic Translocation and Spatial Control Over Innate Immune Activation by Polymer–Lipid Amphiphile Conjugates of Small-Molecule TLR7/8 Agonists. Angew. Chem. Int. Ed. 2019;58:15390–15395. doi: 10.1002/anie.201905687. [DOI] [PubMed] [Google Scholar]
- 28.Wadhwa A., Aljabbari A., Lokras A., Foged C., Thakur A. Opportunities and Challenges in the Delivery of mRNA-based Vaccines. [(accessed on 1 March 2021)];Pharmaceutics. 2020 12:102. doi: 10.3390/pharmaceutics12020102. Available online: https://www.mdpi.com/1999-4923/12/2/102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bookstaver M.L., Tsai S.J., Bromberg J.S., Jewell C.M. Improving Vaccine and Immunotherapy Design Using Biomaterials. Trends Immunol. 2018;39:135–150. doi: 10.1016/j.it.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye T., Zhong Z., García-Sastre A., Schotsaert M., De Geest B.G. Current Status of COVID-19 (Pre)Clinical Vaccine Development. Angew. Chem. Int. Ed. 2020;59:18885–18897. doi: 10.1002/anie.202008319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabatino D. Medicinal Chemistry and Methodological Advances in the Development of Peptide-Based Vaccines. J. Med. Chem. 2020;63:14184–14196. doi: 10.1021/acs.jmedchem.0c00848. [DOI] [PubMed] [Google Scholar]
- 32.Hemmati M., Raoufi E., Fallahi H. Advances in Ebola Control. InTech; London, UK: 2018. [(accessed on 1 March 2021)]. Predicting Candidate Epitopes on Ebola Virus for Possible Vaccine Development. Available online: http://www.intechopen.com/books/advances-in-ebola-control/predicting-candidate-epitopes-on-ebola-virus-for-possible-vaccine-development. [Google Scholar]
- 33.Raoufi E., Hemmati M., Eftekhari S., Khaksaran K., Mahmodi Z., Farajollahi M.M., Mohsenzadegan M. Epitope Prediction by Novel Immunoinformatics Approach: A State-of-the-art Review. [(accessed on 1 March 2021)];Int. J. Pept. Res. Ther. 2020 26:1155–1163. doi: 10.1007/s10989-019-09918-z. Available online: https://pubmed.ncbi.nlm.nih.gov/32435171/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raoufi E., Hemmati M., Einabadi H., Fallahi H. Predicting candidate epitopes on Ebolaviruse for possible vaccine development; Proceedings of the 2015 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining 2015; Paris, France. 25–28 August 2015; New York, NY, USA: Association for Computing Machinery, Inc.; 2015. pp. 1083–1088. [DOI] [Google Scholar]
- 35.Lim H.X., Lim J., Jazayeri S.D., Poppema S., Poh C.L. Development of multi-epitope peptide-based vaccines against SARS-CoV-2. Biomed. J. 2020;44:18–30. doi: 10.1016/j.bj.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kar T., Narsaria U., Basak S., Deb D., Castiglione F., Mueller D.M., Srivastava A.P. A candidate multi-epitope vaccine against SARS-CoV-2. Sci. Rep. 2020;10:1–24. doi: 10.1038/s41598-020-67749-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalita P., Padhi A.K., Zhang K.Y.J., Tripathi T. Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microb. Pathog. 2020;145:104236. doi: 10.1016/j.micpath.2020.104236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss C.M., Liu H., Riemersma K.K., Ball E.E., Coffey L.L. Engineering a fidelity-variant live-attenuated vaccine for chikungunya virus. Vaccines. 2020;5:1–13. doi: 10.1038/s41541-020-00241-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salmona M., Gazaignes S., Mercier-Delarue S., Garnier F., Korimbocus J., De Verdière N.C., LeGoff J., Roques P., Simon F., Gazaigne S. Molecular characterization of the 17D-204 yellow fever vaccine. Vaccine. 2015;33:5432–5436. doi: 10.1016/j.vaccine.2015.08.055. [DOI] [PubMed] [Google Scholar]
- 40.Vignuzzi M., Wendt E., Andino R. Engineering Attenuated Virus Vaccines by Controlling Replication Fidelity. [(accessed on 1 March 2021)];Nat. Med. 2008 14:154–161. doi: 10.1038/nm1726. Available online: http://www.nature.com/naturemedicine. [DOI] [PubMed] [Google Scholar]
- 41.Powers D.C., Murphy B.R., Fries L.F., Adler W.H., Clements M.L. Reduced Infectivity of Cold-Adapted Influenza A H1N1 Viruses in the Elderly: Correlation with Serum and Local Antibodies. J. Am. Geriatr. Soc. 1992;40:163–167. doi: 10.1111/j.1532-5415.1992.tb01938.x. [DOI] [PubMed] [Google Scholar]
- 42.Chen W.-H., Strych U., Hotez P.J., Bottazzi M.E. The SARS-CoV-2 Vaccine Pipeline: An Overview. Curr. Trop. Med. Rep. 2020;7:61–64. doi: 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eckels K.H., Putnak R. Advances in Virus Research. Academic Press Inc.; Cambridge, MA, USA: 2003. Formalin-inactivated Whole Virus and Recombinant Subunit Flavivirus Vaccines; pp. 395–418. [DOI] [PubMed] [Google Scholar]
- 44.Jiang S., Bottazzi M.E., Du L., Lustigman S., Tseng C.-T.K., Curti E., Jones K., Zhan B., Hotez P.J. Roadmap to developing a recombinant coronavirus S protein receptor-binding domain vaccine for severe acute respiratory syndrome. Expert Rev. Vaccines. 2012;11:1405–1413. doi: 10.1586/erv.12.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du L., He Y., Jiang S., Zheng B.-J. Development of subunit vaccines against severe acute respiratory syndrome. Drugs Today. 2008;44:63–73. doi: 10.1358/dot.2008.44.1.1131830. [DOI] [PubMed] [Google Scholar]
- 46.Deng M.-P., Hua-Lin W., Wang H.-L., Deng F. Developments of subunit and VLP vaccines against influenza a virus. Virol. Sin. 2012;27:145–153. doi: 10.1007/s12250-012-3241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang N., Jiang S., Du L. Current advancements and potential strategies in the development of MERS-CoV vaccines. Expert Rev. Vaccines. 2014;13:761–774. doi: 10.1586/14760584.2014.912134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang N., Channappanavar R., Ma C., Wang L., Tang J., Garron T., Tao X., Tasneem S., Lu L., Tseng C.-T.K., et al. Identification of an Ideal Adjuvant for Receptor-Binding Domain-Based Subunit Vaccines against Middle East Respiratory Syndrome Coronavirus. [(accessed on 1 March 2021)];Cell. Mol. Immunol. 2016 13:180–190. doi: 10.1038/cmi.2015.03. Available online: http://www.fda.gov/BiologicsBloodVaccines/Safety. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang N., Shang J., Jiang S., Du L. Subunit Vaccines Against Emerging Pathogenic Human Coronaviruses. [(accessed on 1 March 2021)];Front. Microbiol. 2020 11:29. doi: 10.3389/fmicb.2020.00298. Available online: www.frontiersin.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du L., Zhao G., Chan C.C.S., Sun S., Chen M., Liu Z., Guo H., He Y., Zhou Y., Zheng B.J., et al. Recombinant receptor-binding domain of SARS-CoV spike protein expressed in mammalian, insect and E. coli cells elicits potent neutralizing antibody and protective immunity. Virology. 2009;393:144–150. doi: 10.1016/j.virol.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zakhartchouk A.N., Sharon C., Satkunarajah M., Auperin T., Viswanathan S., Mutwiri G., Petric M., See R.H., Brunham R.C., Finlay B.B., et al. Immunogenicity of a receptor-binding domain of SARS coronavirus spike protein in mice: Implications for a subunit vaccine. Vaccine. 2007;25:136–143. doi: 10.1016/j.vaccine.2006.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang N., Tang J., Lu L., Jiang S., Du L. Receptor-binding domain-based subunit vaccines against MERS-CoV. Virus Res. 2015;202:151–159. doi: 10.1016/j.virusres.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hajizade A., Ebrahimi F., Salmanian A.H., Arpanaei A., Amani J. Nanoparticles in vaccine development. J. Appl. Biotechnol. Rep. 2014;1:125–134. [Google Scholar]
- 54.Di Pasquale A., Preiss S., Silva F.M.D.O.E., Garçon N. Vaccine Adjuvants: From 1920 to 2015 and Beyond. [(accessed on 1 March 2021)];Vaccines. 2015 3:320–343. doi: 10.3390/vaccines3020320. Available online: http://www.mdpi.com/2076-393X/3/2/320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laupèze B., Hervé C., Di Pasquale A., Da Silva F.T. Adjuvant Systems for vaccines: 13 years of post-licensure experience in diverse populations have progressed the way adjuvanted vaccine safety is investigated and understood. Vaccine. 2019;37:5670–5680. doi: 10.1016/j.vaccine.2019.07.098. [DOI] [PubMed] [Google Scholar]
- 56.Lakshmi P., Kumar S., Pawar S., Sudheesh M., Pawar R.S. Plant-based Adjuvant in Vaccine Immunogenicity: A Review. Curr. Tradit. Med. 2018;4:215–236. doi: 10.2174/2215083804666180830142648. [DOI] [Google Scholar]
- 57.Fries C.N., Curvino E.J., Chen J.-L., Permar S.R., Fouda G.G., Collier J.H. Advances in nanomaterial vaccine strategies to address infectious diseases impacting global health. Nat. Nanotechnol. 2020:1–14. doi: 10.1038/s41565-020-0739-9. [DOI] [PubMed] [Google Scholar]
- 58.Garg A., Dewangan H.K. Nanoparticles as Adjuvants in Vaccine Delivery. [(accessed on 1 March 2021)];Crit. Rev. Ther. Drug Carrier Syst. 2020 37:183–204. doi: 10.1615/CritRevTherDrugCarrierSyst.2020033273. Available online: http://www.dl.begellhouse.com/journals/3667c4ae6e8fd136,303ea22275400937,447d4b2056c2708f.html. [DOI] [PubMed] [Google Scholar]
- 59.Nasrollahzadeh M., Sajjadi M., Soufi G.J., Iravani S., Varma R.S. Nanomaterials and Nanotechnology-Associated Innovations against Viral Infections with a Focus on Coronaviruses. [(accessed on 1 March 2021)];Nanomatererials. 2020 10:1072. doi: 10.3390/nano10061072. Available online: https://www.mdpi.com/2079-4991/10/6/1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shin M.D., Shukla S., Chung Y.H., Beiss V., Chan S.K., Ortega-Rivera O.A., Wirth D.M., Chen A., Sack M., Pokorski J.K., et al. COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 2020;15:646–655. doi: 10.1038/s41565-020-0737-y. [DOI] [PubMed] [Google Scholar]
- 61.Theobald N. Emerging vaccine delivery systems for COVID-19: Functionalised Silica Nanoparticles Offer a Potentially Safe and Effective Alternative Delivery System for DNA/RNA Vaccines and May Be Useful in the Hunt for a COVID-19 Vaccine. [(accessed on 1 March 2021)];Drug Discov. Today. 2020 25:1556–1558. doi: 10.1016/j.drudis.2020.06.020. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7313501/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramsey J.M., McCloskey A., Gaul R., Fernandez E.F., Sweeney L., Greene C.M., Macloughlin R., Cryan S.-A. AAPS Advances in the Pharmaceutical Sciences Series. Springer; Cham, Switzerland: 2020. Respiratory Drug/Vaccine Delivery Using Nanoparticles; pp. 125–154. [DOI] [Google Scholar]
- 63.Krammer F., Srivastava K., Alshammary H., Amoako A.A., Awawda M.H., Beach K.F., Bermúdez-González M.C., Bielak D.A., Carreño J.M., Chernet R.L., et al. Antibody responses in seropositive persons after a single dose of SARS-CoV-2 mRNA vaccine. N. Engl. J. Med. 2021;384:1372–1374. doi: 10.1056/NEJMc2101667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nevagi R.J., Skwarczynski M., Toth I. Polymers for subunit vaccine delivery. Eur. Polym. J. 2019;114:397–410. doi: 10.1016/j.eurpolymj.2019.03.009. [DOI] [Google Scholar]
- 65.Shae D., Postma A., Wilson J.T. Vaccine delivery: Where Polymer Chemistry Meets Immunology. [(accessed on 1 March 2021)];Ther. Deliv. 2016 7:193–196. doi: 10.4155/tde-2016-0008. Available online: www.future-science.com. [DOI] [PubMed] [Google Scholar]
- 66.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol. Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeng C., Zhang C., Walker P.G., Dong Y. Formulation and Delivery Technologies for mRNA Vaccines. Springer; Berlin/Heidelberg, Germany: 2020. pp. 1–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simón-Vázquez R., Peleteiro M., González-Fernández Á. Polymeric nanostructure vaccines: Applications and challenges. Expert Opin. Drug Deliv. 2020;17:1007–1023. doi: 10.1080/17425247.2020.1776259. [DOI] [PubMed] [Google Scholar]
- 69.Kaur D., Kumar S. Niosomes: Present scenario and future aspects. J. Drug Deliv. Ther. 2018;8:35–43. doi: 10.22270/jddt.v8i5.1886. [DOI] [Google Scholar]
- 70.Bhardwaj P., Tripathi P., Gupta R., Pandey S. Niosomes: A review on niosomal research in the last decade. J. Drug Deliv. Sci. Technol. 2020;56:101581. doi: 10.1016/j.jddst.2020.101581. [DOI] [Google Scholar]
- 71.Durak S., Rad M.E., Yetisgin A.A., Sutova H.E., Kutlu O., Cetinel S., Zarrabi A. Niosomal Drug Delivery Systems for Ocular Disease—Recent Advances and Future Prospects. [(accessed on 1 March 2021)];Nanomaterials. 2020 10:1191. doi: 10.3390/nano10061191. Available online: https://www.mdpi.com/2079-4991/10/6/1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pardakhty A., Moazeni E. Nano-Niosomes in Drug, Vaccine and Gene Delivery: A Rapid Overview. [(accessed on 1 March 2021)];2013 Available online: http://nmj.mums.ac.ir/article_697.html.
- 73.Moghassemi S., Hadjizadeh A. Nano-niosomes as nanoscale drug delivery systems: An illustrated review. J. Control. Release. 2014;185:22–36. doi: 10.1016/j.jconrel.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 74.Pola Chandu V., Arunachalam A., Jeganath S., Yamini K., Tharangini K., Chaitanya G. Niosomes: A Novel Drug Delivery System. [(accessed on 1 March 2021)];Int. J. Nov. Trends Pharm. Sci. 2012 2:25–31. Available online: www.ijntps.org. [Google Scholar]
- 75.Gharbavi M., Amani J., Kheiri-Manjili H., Danafar H., Sharafi A. Niosome: A Promising Nanocarrier for Natural Drug Delivery through Blood-Brain Barrier. Adv. Pharmacol. Sci. 2018;2018:6847971. doi: 10.1155/2018/6847971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Danaei M., Dehghankhold M., Ataei S., Davarani F.H., Javanmard R., Dokhani A., Khorasani S., Mozafari M.R. Impact of Particle Size and Polydispersity Index on the Clinical Applications of Lipidic Nanocarrier Systems. [(accessed on 1 March 2021)];Pharmaceutics. 2018 10:57. doi: 10.3390/pharmaceutics10020057. Available online: http://www.mdpi.com/1999-4923/10/2/57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mozafari M.R. Liposomes: An Overview of Manufacturing Techniques. [(accessed on 1 March 2021)];Cell. Mol. Biol. Lett. 2005 10:711–719. Available online: https://research.monash.edu/en/publications/liposomes-an-overview-of-manufacturing-techniques. [PubMed] [Google Scholar]
- 78.Maherani B., Arab-Tehrany E.R., Mozafari M., Gaiani C., Linder M. Liposomes: A Review of Manufacturing Techniques and Targeting Strategies. Curr. Nanosci. 2011;7:436–452. doi: 10.2174/157341311795542453. [DOI] [Google Scholar]
- 79.Mozafari M.R. Nanoliposomes: Preparation and Analysis. Methods Mol. Biol. 2010;605:29–50. doi: 10.1007/978-1-60327-360-2_2. [DOI] [PubMed] [Google Scholar]
- 80.Sharma D., Ali A.A.E., Trivedi L.R. An Updated Review On: Liposomes as Drug Delivery System. PharmaTutor. 2018;6:50. doi: 10.29161/PT.v6.i2.2018.50. [DOI] [Google Scholar]
- 81.Amoabediny G., Haghiralsadat F., Naderinezhad S., Helder M.N., Kharanaghi E.A., Arough J.M., Zandieh-Doulabi B. Overview of preparation methods of polymeric and lipid-based (niosome, solid lipid, liposome) nanoparticles: A comprehensive review. Int. J. Polym. Mater. 2017;67:383–400. doi: 10.1080/00914037.2017.1332623. [DOI] [Google Scholar]
- 82.Morein B., Sundquist B., Höglund S., Dalsgaard K., Osterhaus A. Iscom, a Novel Structure for Antigenic Presentation of Membrane Proteins from Enveloped Viruses. [(accessed on 1 March 2021)];Nat. Cell Biol. 1984 308:457–460. doi: 10.1038/308457a0. Available online: https://www.nature.com/articles/308457a0. [DOI] [PubMed] [Google Scholar]
- 83.Alexyuk P.G., Bogoyavlenskiy A.P., Alexyuk M.S., Turmagambetova A.S., Zaitseva I.A., Omirtaeva E.S., Berezin V.E. Adjuvant activity of multimolecular complexes based on Glycyrrhiza glabra saponins, lipids, and influenza virus glycoproteins. Arch. Virol. 2019;164:1793–1803. doi: 10.1007/s00705-019-04273-2. [DOI] [PubMed] [Google Scholar]
- 84.Buglione-Corbett R., Pouliot K., Marty-Roix R., Li W., West K., Wang S., Morelli A.B., Lien E., Lu S. Reduced MyD88 Dependency of ISCOMATRIX™ Adjuvant in a DNA Prime-Protein Boost HIV Vaccine. [(accessed on 1 March 2021)];Hum. Vaccines Immunother. 2014 10:1078–1090. doi: 10.4161/hv.27907. Available online: http://www.tandfonline.com/doi/abs/10.4161/hv.27907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kheirollahpour M., Mehrabi M., Dounighi N.M., Mohammadi M., Masoudi A. Nanoparticles and Vaccine Development. [(accessed on 1 March 2021)];Pharm. Nanotechnol. 2020 8:6–21. doi: 10.2174/2211738507666191024162042. Available online: http://www.eurekaselect.com/176139/article. [DOI] [PubMed] [Google Scholar]
- 86.Pearse M.J., Drane D. ISCOMATRIX® adjuvant for antigen delivery. Adv. Drug Deliv. Rev. 2005;57:465–474. doi: 10.1016/j.addr.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 87.Peek L.J., Middaugh C.R., Berkland C. Nanotechnology in vaccine delivery. Adv. Drug Deliv. Rev. 2008;60:915–928. doi: 10.1016/j.addr.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mozafari M., Javanmard R., Raji M. Tocosome: Novel drug delivery system containing phospholipids and tocopheryl phosphates. Int. J. Pharm. 2017;528:381–382. doi: 10.1016/j.ijpharm.2017.06.037. [DOI] [PubMed] [Google Scholar]
- 89.Azzi A. Tocopheryl phosphate, a novel natural form of vitamin E: In vitro and in vivo studies. FASEB J. 2006;20:LB79–LB80. doi: 10.1096/fasebj.20.5.LB79-d. [DOI] [Google Scholar]
- 90.Gianello R., Libinaki R., Azzi A., Gavin P.D., Negis Y., Zingg J.-M., Holt P., Keah H.-H., Griffey A., Smallridge A., et al. α-Tocopheryl phosphate: A novel, natural form of vitamin E. Free Radic. Biol. Med. 2005;39:970–976. doi: 10.1016/j.freeradbiomed.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 91.Zarrabi A., Alipoor A.A.M., Khorasani S., Mohammadabadi M.R., Jamshidi A., Torkaman S., Taghavi E., Mozafari M.R., Rasti B. Nanoliposomes and Tocosomes as Multifunctional Nanocarriers for the Encapsulation of Nutraceutical and Dietary Molecules. [(accessed on 1 March 2021)];Molecules. 2020 25:638. doi: 10.3390/molecules25030638. Available online: https://www.mdpi.com/1420-3049/25/3/638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Munteanu A., Zingg J.M., Ogru E., Libinaki R., Gianello R., West S., Negis Y., Azzi A. Modulation of cell proliferation and gene expression by α-tocopheryl phosphates: Relevance to atherosclerosis and inflammation. Biochem. Biophys. Res. Commun. 2004;318:311–316. doi: 10.1016/j.bbrc.2004.04.028. [DOI] [PubMed] [Google Scholar]
- 93.Libinaki R., Tesanovic S., Heal A., Nikolovski B., Vinh A., Widdop R.E., Gaspari T.A., Devaraj S., Ogru E. Effect of tocopheryl phosphate on key biomarkers of inflammation: Implication in the reduction of atherosclerosis progression in a hypercholesterolaemic rabbit model. Clin. Exp. Pharmacol. Physiol. 2010;37:587–592. doi: 10.1111/j.1440-1681.2010.05356.x. [DOI] [PubMed] [Google Scholar]
- 94.Saitoh Y., Yumoto A., Miwa N. α-tocopheryl phosphate suppresses tumor invasion concurrently with dynamic morphological changes and delocalization of cortactin from invadopodia. Int. J. Oncol. 2009;35:1277–1288. doi: 10.3892/ijo_00000445/abstract. [DOI] [PubMed] [Google Scholar]
- 95.Nishio K., Ishida N., Saito Y., Ogawa-Akazawa Y., Shichiri M., Yoshida Y., Hagihara Y., Noguchi N., Chirico J., Atkinson J., et al. α-Tocopheryl phosphate: Uptake, hydrolysis, and antioxidant action in cultured cells and mouse. Free Radic. Biol. Med. 2011;50:1794–1800. doi: 10.1016/j.freeradbiomed.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 96.Reichmuth A.M., Oberli M.A., Jaklenec A., Langer R., Blankschtein D. mRNA vaccine delivery using lipid nanoparticles. Ther. Deliv. 2016;7:319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Midoux P., Pichon C. Lipid-based mRNA vaccine delivery systems. Expert Rev. Vaccines. 2014;14:221–234. doi: 10.1586/14760584.2015.986104. [DOI] [PubMed] [Google Scholar]
- 98.Corthésy B., Bioley G. Lipid-Based Particles: Versatile Delivery Systems for Mucosal Vaccination against Infection. [(accessed on 1 March 2021)];Front. Immunol. 2018 9 doi: 10.3389/fimmu.2018.00431. Available online: www.frontiersin.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pardi N., Hogan M.J., Porter F.W., Weissman D. Vaccines Prevent Many Millions of Illnesses and Save Numerous Lives Every Year. [(accessed on 1 March 2021)];Nat. Publ. Gr. 2018 17:261. Available online: www.nature.com/nrd. [Google Scholar]
- 100.Li B., Zhang X., Dong Y. Nanoscale platforms for messenger RNA delivery. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2019;11:e1530. doi: 10.1002/wnan.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ganesan P., Narayanasamy D. Lipid nanoparticles: Different preparation techniques, characterization, hurdles, and strategies for the production of solid lipid nanoparticles and nanostructured lipid carriers for oral drug delivery. Sustain. Chem. Pharm. 2017;6:37–56. doi: 10.1016/j.scp.2017.07.002. [DOI] [Google Scholar]
- 102.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C., et al. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell. 2017;168:1114–1125.e10. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Richner J.M., Jagger B.W., Shan C., Fontes C.R., Dowd K.A., Cao B., Himansu S., Caine E.A., Nunes B.T., Medeiros D.B., et al. Vaccine Mediated Protection Against Zika Virus-Induced Congenital Disease. Cell. 2017;170:273–283.e12. doi: 10.1016/j.cell.2017.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sahay G., Alakhova D.Y., Kabanov A.V. Endocytosis of nanomedicines. J. Control. Release. 2010;145:182–195. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Basak J.M., Verghese P.B., Yoon H., Kim J., Holtzman D.M. Low-density Lipoprotein Receptor Represents an Apolipoprotein E-independent Pathway of Aβ Uptake and Degradation by Astrocytes. J. Biol. Chem. 2012;287:13959–13971. doi: 10.1074/jbc.M111.288746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Patel S., Kim J., Herrera M., Mukherjee A., Kabanov A.V., Sahay G. Brief update on endocytosis of nanomedicines. Adv. Drug Deliv. Rev. 2019;144:90–111. doi: 10.1016/j.addr.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Verbeke R., Lentacker I., De Smedt S.C., Dewitte H. Three decades of messenger RNA vaccine development. Nano Today. 2019;28:100766. doi: 10.1016/j.nantod.2019.100766. [DOI] [Google Scholar]
- 108.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., Sah D.W.Y., Stebbing D., Crosley E.J., Yaworski E., et al. Rational Design of Cationic Lipids for siRNA Delivery. [(accessed on 1 March 2021)];Nat. Biotechnol. 2010 28:172–176. doi: 10.1038/nbt.1602. Available online: https://www.nature.com/articles/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 109.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., Butler D., Eltepu L., Matsuda S., Narayanannair J.K., et al. Maximizing the Potency of siRNA Lipid Nanoparticles for Hepatic Gene Silencing In Vivo. Angew. Chem. 2012;124:8657–8661. doi: 10.1002/ange.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Billingsley M.M., Singh N., Ravikumar P., Zhang R., June C.H., Mitchell M.J. Ionizable Lipid Nanoparticle-Mediated mRNA Delivery for Human CAR T Cell Engineering. Nano Lett. 2020;20:1578–1589. doi: 10.1021/acs.nanolett.9b04246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zeng C., Hou X., Yan J., Zhang C., Li W., Zhao W., Du S., Dong Y. Leveraging mRNAs sequences to express SARS-CoV-2 antigens in vivo. bioRxiv Prepr. Serv. Biol. 2020 doi: 10.1101/2020.04.01.019877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cullis P.R., Hope M.J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017;25:1467–1475. doi: 10.1016/j.ymthe.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sato Y., Hashiba K., Sasaki K., Maeki M., Tokeshi M., Harashima H. Understanding structure-activity relationships of pH-sensitive cationic lipids facilitates the rational identification of promising lipid nanoparticles for delivering siRNAs in vivo. J. Control Release. 2019;295:140–152. doi: 10.1016/j.jconrel.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 114.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J., et al. A Novel Amino Lipid Series for mRNA Delivery: Improved Endosomal Escape and Sustained Pharmacology and Safety in Non-human Primates. Mol. Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sedic M., Senn J.J., Lynn A., Laska M., Smith M., Platz S.J., Bolen J., Hoge S., Bulychev A., Jacquinet E., et al. Safety Evaluation of Lipid Nanoparticle–Formulated Modified mRNA in the Sprague-Dawley Rat and Cynomolgus Monkey. Veter- Pathol. 2017;55:341–354. doi: 10.1177/0300985817738095. [DOI] [PubMed] [Google Scholar]
- 116.Cheng X., Lee R.J. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug Deliv. Rev. 2016;99:129–137. doi: 10.1016/j.addr.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 117.Ambegia E., Ansell S., Cullis P., Heyes J., Palmer L., MacLachlan I. Stabilized plasmid–lipid particles containing PEG-diacylglycerols exhibit extended circulation lifetimes and tumor selective gene expression. Biochim. Biophys. Acta BBA Biomembr. 2005;1669:155–163. doi: 10.1016/j.bbamem.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 118.Heyes J., Palmer L., Bremner K., MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 119.Miao L., Li L., Huang Y., Delcassian D., Chahal J., Han J., Shi Y., Sadtler K., Gao W., Lin J., et al. Delivery of mRNA Vaccines with Heterocyclic Lipids Increases Anti-Tumor Efficacy by STING-Mediated Immune cell Activation. [(accessed on 1 March 2021)];Nat. Biotechnol. 2019 37:1174–1185. doi: 10.1038/s41587-019-0247-3. Available online: https://www.nature.com/articles/s41587-019-0247-3. [DOI] [PubMed] [Google Scholar]
- 120.Akbarzadeh A., Rezaei-Sadabady R., Davaran S., Joo S.W., Zarghami N., Hanifehpour Y., Samiei M., Kouhi M., Nejati-Koshki K. Liposome: Classification, Preparation, and Applications. [(accessed on 1 March 2021)];Nanoscale Res. Lett. 2013 8:102. doi: 10.1186/1556-276X-8-102. Available online: https://nanoscalereslett.springeropen.com/articles/10.1186/1556-276X-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., Meng M., Fritz D., Vascotto F., Hefesha H., et al. Systemic RNA Delivery to Dendritic Cells Exploits Antiviral Defence for Cancer Immunotherapy. [(accessed on 1 March 2021)];Nature. 2016 534:396–401. doi: 10.1038/nature18300. Available online: https://www.nature.com/articles/nature18300. [DOI] [PubMed] [Google Scholar]
- 122.Borrego B., Blanco E., Pulido M.R., Mateos F., Lorenzo G., Cardillo S., Smitsaart E., Sobrino F., Sáiz M. Combined administration of synthetic RNA and a conventional vaccine improves immune responses and protection against foot-and-mouth disease virus in swine. Antivir. Res. 2017;142:30–36. doi: 10.1016/j.antiviral.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 123.Chen D., Love K.T., Chen Y., Eltoukhy A.A., Kastrup C., Sahay G., Jeon A., Dong Y., Whitehead K.A., Anderson D.G. Rapid Discovery of Potent siRNA-Containing Lipid Nanoparticles Enabled by Controlled Microfluidic Formulation. [(accessed on 1 March 2021)];J. Am. Chem. Soc. 2012 134 doi: 10.1021/ja301621z. Available online: https://pubs.acs.org/doi/abs/10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- 124.Kose N., Fox J.M., Sapparapu G., Bombardi R., Tennekoon R.N., De Silva A.D., Elbashir S.M., Theisen M.A., Humphris-Narayanan E., Ciaramella G., et al. A Lipid-Encapsulated mRNA Encoding a Potently Neutralizing Human Monoclonal Antibody Protects against Chikungunya Infection. [(accessed on 1 March 2021)];Sci. Immunol. 2019 4:eaaw6647. doi: 10.1126/sciimmunol.aaw6647. Available online: http://immunology.sciencemag.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu C., Feng Q., Sun J. Lipid Nanovesicles by Microfluidics: Manipulation, Synthesis, and Drug Delivery. Adv. Mater. 2019;31:e1804788. doi: 10.1002/adma.201804788. [DOI] [PubMed] [Google Scholar]
- 126.Belliveau N.M., Huft J., Lin P.J., Chen S., Leung A.K., Leaver T.J., Wild A.W., Lee J.B., Taylor R.J., Tam Y.K., et al. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol. Ther. Nucleic Acids. 2012:e37. doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Arteta M.Y., Kjellman T., Bartesaghi S., Wallin S., Wu X., Kvist A.J., Dabkowska A., Székely N., Radulescu A., Bergenholtz J., et al. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc. Natl. Acad. Sci. USA. 2018;115:E3351–E3360. doi: 10.1073/pnas.1720542115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Damiati S., Kompella U., Damiati S., Kodzius R. Microfluidic Devices for Drug Delivery Systems and Drug Screening. [(accessed on 1 March 2021)];Genes. 2018 9:103. doi: 10.3390/genes9020103. Available online: http://www.mdpi.com/2073-4425/9/2/103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Colas J.C., Shi W., Rao V.M., Omri A., Mozafari M.R., Singh H. Microscopical investigations of nisin-loaded nanoliposomes prepared by Mozafari method and their bacterial targeting. Micron. 2007;38:841–847. doi: 10.1016/j.micron.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 130.Mozafari M.R., Reed C.J., Rostron C., Kocum C., Piskin E. Formation and characterisation of non-toxic anionic liposomes for delivery of therapeutic agents to the pulmonary airways. Cell. Mol. Biol. Lett. 2002;7:243–244. [PubMed] [Google Scholar]
- 131.Mozafari M.R., Reed C.J., Rostron C. Development of non-toxic liposomal formulations for gene and drug delivery to the lung. Technol. Health Care. 2002;10:342–344. [Google Scholar]
- 132.Mozafari M.R. Method and Apparatus for Producing Carrier Complexes. GB0404993,8. Patent Application. 2005 Sep 15;
- 133.Mozafari M.R. Method for the Preparation of Micro- and Nano-Sized Carrier Systems for the Encapsulation of Bioactive Substances. 12/790,991. U.S. Patent Application. 2010 Sep 23;
- 134.ElMeshad A.N., Mortazavi S.M., Mozafari M.R. Formulation and characterization of nanoliposomal 5-fluorouracil for cancer nanotherapy. J. Liposome Res. 2013;24:1–9. doi: 10.3109/08982104.2013.810644. [DOI] [PubMed] [Google Scholar]
- 135.Savaghebi D., Barzegar M., Mozafari M.R. Manufacturing of nanoliposomal extract from Sargassum boveanum algae and investigating its release behavior and antioxidant activity. Food Sci. Nutr. 2020;8:299–310. doi: 10.1002/fsn3.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rasti B., Jinap S., Mozafari M.R., Abd-Manap M.Y. Optimization on preparation condition of polyunsaturated fatty acids nanoliposome prepared by Mozafari method. J. Liposome Res. 2013;24:99–105. doi: 10.3109/08982104.2013.839702. [DOI] [PubMed] [Google Scholar]
- 137.Allen T.M., Hansen C.B., Guo L.S.S. Subcutaneous administration of liposomes: A comparison with the intravenous and intraperitoneal routes of injection. Biochim. Biophys. Acta BBA Biomembr. 1993;1150:9–16. doi: 10.1016/0005-2736(93)90115-G. [DOI] [PubMed] [Google Scholar]
- 138.Swartz M.A. The physiology of the lymphatic system. Adv. Drug Deliv. Rev. 2001;50:3–20. doi: 10.1016/S0169-409X(01)00150-8. [DOI] [PubMed] [Google Scholar]
- 139.Oussoren C., Velinova M., Scherphof G., van der Want J.J., van Rooijen N., Storm G. Lymphatic uptake and biodistribution of liposomes after subcutaneous injection: IV. Fate of liposomes in regional lymph nodes. Biochim. Biophys. Acta BBA Biomembr. 1998;1370:259–272. doi: 10.1016/S0005-2736(97)00275-7. [DOI] [PubMed] [Google Scholar]
- 140.Henriksen-Lacey M., Bramwell V.W., Christensen D., Agger E.-M., Andersen P., Perrie Y. Liposomes based on dimethyldioctadecylammonium promote a depot effect and enhance immunogenicity of soluble antigen. J. Control. Release. 2010;142:180–186. doi: 10.1016/j.jconrel.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 141.Henriksen-Lacey M., Christensen D., Bramwell V.W., Lindenstrøm T., Agger E.M., Andersen P., Perrie Y. Comparison of the depot effect and immunogenicity of liposomes based on DDA, DC-Chol and DOTAP: Prolonged liposome retention mediates stronger Th1 responses. Mol Pharm. 2011;8:153–161. doi: 10.1021/mp100208f. [DOI] [PubMed] [Google Scholar]
- 142.Gabizon A., Papahadjopoulos D. The role of surface charge and hydrophilic groups on liposome clearance in vivo. Biochim. Biophys. Acta BBA Biomembr. 1992;1103:94–100. doi: 10.1016/0005-2736(92)90061-P. [DOI] [PubMed] [Google Scholar]
- 143.Mozafari M., Torkaman S., Karamouzian F.M., Rasti B., Baral B. Antimicrobial Applications of Nanoliposome Encapsulated Silver Nanoparticles: A Potential Strategy to Overcome Bacterial Resistance. Curr. Nanosci. 2021;17:26–40. doi: 10.2174/1573413716999200712184148. [DOI] [Google Scholar]
- 144.Judge A., McClintock K., Phelps J.R., MacLachlan I. Hypersensitivity and loss of disease site targeting caused by antibody responses to PEGylated liposomes. Mol. Ther. 2006;13:328–337. doi: 10.1016/j.ymthe.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 145.Yang Q., Lai S.K. Anti-PEG immunity: Emergence, characteristics, and unaddressed questions. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2015;7:655–677. doi: 10.1002/wnan.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Caminschi I., Proietto A.I., Ahmet F., Kitsoulis S., Teh J.S., Lo J.C.Y., Rizzitelli A., Wu L., Vremec D., Van Dommelen S.L.H., et al. The dendritic cell subtype-restricted C-type lectin Clec9A is a target for vaccine enhancement. Blood. 2008;112:3264–3273. doi: 10.1182/blood-2008-05-155176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Alavi M., Asare-Addo K., Nokhodchi A. Lectin Protein as a Promising Component to Functionalize Micelles, Liposomes and Lipid NPs against Coronavirus. Biomedicine. 2020;8:580. doi: 10.3390/biomedicines8120580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Martinez-Pomares L. The mannose receptor. J. Leukoc. Biol. 2012;92:1177–1186. doi: 10.1189/jlb.0512231. [DOI] [PubMed] [Google Scholar]
- 149.Geijtenbeek T.B., Torensma R., van Vliet S.J., van Duijnhoven G.C., Adema G.J., van Kooyk Y., Figdor C.G. Identification of DC-SIGN, a Novel Dendritic Cell–Specific ICAM-3 Receptor that Supports Primary Immune Responses. Cell. 2000;100:575–585. doi: 10.1016/S0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- 150.Jiang W., Swiggard W.J., Heufler C., Peng M., Mirza A., Steinman R.M., Nussenzweig M.C. The receptor DEC-205 expressed by dendritic cells and thymic epithelial cells is involved in antigen processing. Nature. 1995;375:151–155. doi: 10.1038/375151a0. [DOI] [PubMed] [Google Scholar]
- 151.Valladeau J., Ravel O., Dezutter-Dambuyant C., Moore K., Kleijmeer M., Liu Y., Duvert-Frances V., Vincent C., Schmitt D., Davoust J., et al. Langerin, a Novel C-Type Lectin Specific to Langerhans Cells, Is an Endocytic Receptor that Induces the Formation of Birbeck Granules. Immunology. 2000;12:71–81. doi: 10.1016/S1074-7613(00)80160-0. [DOI] [PubMed] [Google Scholar]
- 152.Lewis J.S., Zaveri T.D., Crooks C.P., II, Keselowsky B.G. Microparticle surface modifications targeting dendritic cells for non-activating applications. Biomaterials. 2012;33:7221–7232. doi: 10.1016/j.biomaterials.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Karanikas V., Hwang L.A., Pearson J., Ong C.S., Apostolopoulos V., Vaughan H., Xing P.X., Jamieson G., Pietersz G., Tait B., et al. Antibody and T cell responses of patients with adenocarcinoma immunized with mannan-MUC1 fusion protein. J. Clin. Investig. 1997;100:2783–2792. doi: 10.1172/JCI119825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Espuelas S., Thumann C., Heurtault B., Schuber F., Frisch B. Influence of Ligand Valency on the Targeting of Immature Human Dendritic Cells by Mannosylated Liposomes. Bioconj. Chem. 2008;19:2385–2393. doi: 10.1021/bc8002524. [DOI] [PubMed] [Google Scholar]
- 155.Reina J.J., Rojo J. Glycodendritic structures: Tools to interact with DC-SIGN. Braz. J. Pharm. Sci. 2013;49:109–124. doi: 10.1590/S1984-82502013000700009. [DOI] [Google Scholar]
- 156.Perche F., Gosset D., Mével M., Miramon M.-L., Yaouanc J.-J., Pichon C., Benvegnu T., Jaffrès P.-A., Midoux P. Selective gene delivery in dendritic cells with mannosylated and histidylated lipopolyplexes. J. Drug Target. 2010;19:315–325. doi: 10.3109/1061186X.2010.504262. [DOI] [PubMed] [Google Scholar]
- 157.Perche F., Benvegnu T., Berchel M., Lebegue L., Pichon C., Jaffrès P.-A., Midoux P. Enhancement of dendritic cells transfection in vivo and of vaccination against B16F10 melanoma with mannosylated histidylated lipopolyplexes loaded with tumor antigen messenger RNA. Nanomed. Nanotechnol. Biol. Med. 2011;7:445–453. doi: 10.1016/j.nano.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 158.Dudziak D., Kamphorst A.O., Heidkamp G.F., Buchholz V.R., Trumpfheller C., Yamazaki S., Cheong C., Liu K., Lee H.-W., Park C.G., et al. Differential Antigen Processing by Dendritic Cell Subsets in Vivo. Science. 2007;315:107–111. doi: 10.1126/science.1136080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable. This Review Article analyzed and discussed already published data and did not report any new data.