Abstract
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-COV-2), which causes coronavirus disease-19 (COVID-19) has caused more than 2 million deaths around the globe. The high transmissibility rate of the disease is related to the strong interaction between the virus spike receptor-binding domain (RBD) and the human angiotensin-converting enzyme 2 (ACE2) as documented in several reports. In this study, using state-of-the-art computational methods, natural products were screened and their molecular mechanism to disrupt spike RBD-ACE2 recognition was evaluated. There is the sensitivity of results to receptor ensemble docking calculations. Binding free energy and MD simulation are important tools to evaluate the thermodynamics of binding stability and the capacity of top hits to disrupt RBD-ACE2 recognition. The free energy profiles provide a slight decrease in binding affinity of the virus-receptor interaction. Three flavonoids parvisoflavone B (3), alpinumisoflavone (5) and norisojamicin (2) were effective in blocking the viral entry by binding strongly at the spike RBD-ACE2 interface with the inhibition constant of 0.56, 0.78 and 0.93 μM, respectively. The same compounds demonstrated similar effect on free ACE2 protein. Compound (2), also demonstrated ability to bind strongly on free spike RBD. Well-tempered metadynamics established that parvisoflavone B (3) works by binding to three sites namely interface α, β and loop thereby inhibiting viral cell entry. Owing to their desirable pharmacokinetic properties, the presented top hit natural products are suggested for further SARS-COV-2 molecular targets and subsequent in vitro and in vivo evaluations.
Keywords: Natural products, Covid-19, SARS-CoV-2, Molecular docking, MD simulation
Graphical abstract
Highlights
-
•
The recent outbreak of SARS-CoV-2 is responsible for high morbidity and mortality rate across the global.
-
•
Natural products from African medicinal plants were screened against the spike RBD-ACE2.
-
•
One potent molecule with desirable pharmacokinetics properties was identified.
-
•
Stability studies ascertained the selected molecule to be potent inhibitor of SARS-CoV-2 viral entry.
The emergency of coronavirus disease (COVD-19) in December 2019 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in an unprecedented health crisis throughout the globe [1]. The structure of SARS-COV-2 is well described in the literature [2]. Based on its structure, therapies for SARS-COV-2 can be grouped into two categories, i.e those acting on the human immune system and those acting on the virus itself. The latter category can be divided into two groups, those preventing viral RNA synthesis and those blocking the entry to human cell receptors [2]. Today, efforts to develop drugs and/or vaccine to prevent the disease are going on around the globe. Recently, many vaccines have entered into clinical trials; however, there is still limited information on their long-term effectiveness, safety and efficacy to humans [1].
Natural products have a long history of serving human life for the treatment of many diseases including viral related infections. Several natural products have been investigated as SARS-COV-2 RNA viral replication inhibitors or cell entry inhibitors. The latter is achieved by preventing the interaction between the spike receptor-binding domain (RBD) and human angiotensin-converting enzyme 2 (ACE2) from recognition or by blocking the activities of transmembrane protease serine 2 (TMPRSS2). Various classes of natural products as inhibitors of TMPRSS2 and spike SARS-COV-2 RBD-ACE2 have been screened and reported [[3], [4], [5]]. On the other hands, natural product inhibitors of SARS-COV-2 main protease have also been reported using computational [[6], [7], [8], [9]] or a combination both computational and experimental approaches as detailed in several reviews [10,11]. For instance, escin and sodium aescinate are historically natural products for treatment of respiratory infection, currently registered in clinical trials targeting SARS-COV-2 inhibition [1]. These reports suggest that classes of natural products are excellent structural lead/hit molecules in discovering new therapies for viral related diseases, hence efforts to identify new natural products to fight viral related diseases is deemed a continuous process.
Advances of computer hardware and algorithms have accelerated drug discovery process. Some drugs discovered by computational approaches are in clinical trials and some are approved, a HIV protease inhibitor is a notable successful example [12]. The use of computer simulation in structure-based drug design (SBDD) has increasingly been reported in recent years [1,12,13]. Molecular docking is an effective and widely used tool in SBDD, for screening large libraries and predicting receptor-ligand complexes [1,13]. However, the accuracy of docking calculations are limited by scoring functions which lacks protein recognition and complete treatment of desolvation [13]. This results in screening ligand libraries containing a large number of false-positive binders [13]. Although some docking codes e.g autodock/vina are able to accommodate side chain residue flexibility, accommodating full protein flexibility in docking calculations remains a challenging task. Thus, top scores from docking calculations cannot be necessarily trusted, and hence requires further analysis [14]. Recently, numerous efforts to overcome the limitations of protein recognition have been established [14]. Relaxed complex scheme (RCS), is one of the approaches that was first suggested by the group of McCammon [12] to incorporate protein flexibility in docking calculations. RCS incorporates ensemble structures of a protein generated from molecular dynamic (MD) simulation [12,15]. The use of RCS to incorporate protein flexibility in SBDD has been considered by different research groups [[15], [16], [17], [18], [19], [20], [21]].
In this study, we report the effectiveness of combining RCS, pose clustering, end-point free energy methods based on LIE and metadynamics simulation to screen natural products as binders of the spike RBD-ACE2 protein. Pose clustering serves as a means of ranking binding energies and eliminating false positive binders from ensemble structures [14]. Herein, we demonstrate that probable poses can be re-scored by end-point free energy methods and metadynamics simulation to optimize the ligand binding affinity, orientation and unbinding processes which further eliminates inappropriate poses. Experience from our previous work [21] shows that RSC improves the ligand binding affinity to its receptor.
To provide plausible binding modes of the natural products, blind and restricted docking calculations at the interface were performed using autodock vina [22], which is effective in screening large libraries and accommodates ligand flexibility as described in our previous work [21]. To assess the stability and dynamical behavior of the complex, molecular dynamics (MD) simulation and free energy calculations based on linear interaction energy (LIE) and metadynamics simulation, an enhanced sampling method were performed as described in the SI.
Natural products screened in this study were obtained from NANPDB (Northern African Natural Products Database) [23], comprising of ≈4500 natural products from African medicinal plants. NANPDB database was considered as it contain several classes of natural products isolated from African medicinal plants, to our knowledge, compounds from this database have not yet explored as inhibitors of spike RBD-ACE2 cell entry. In addition, the database was recently updated and included new compounds; this motivated us to screen and identify compounds that could block the interaction and recognition of spike RBD-ACE2 protein. The details of the compounds including their chemical smiles and physicochemical properties are provided in the supplementary material as compounds1.csv and compounds2.csv
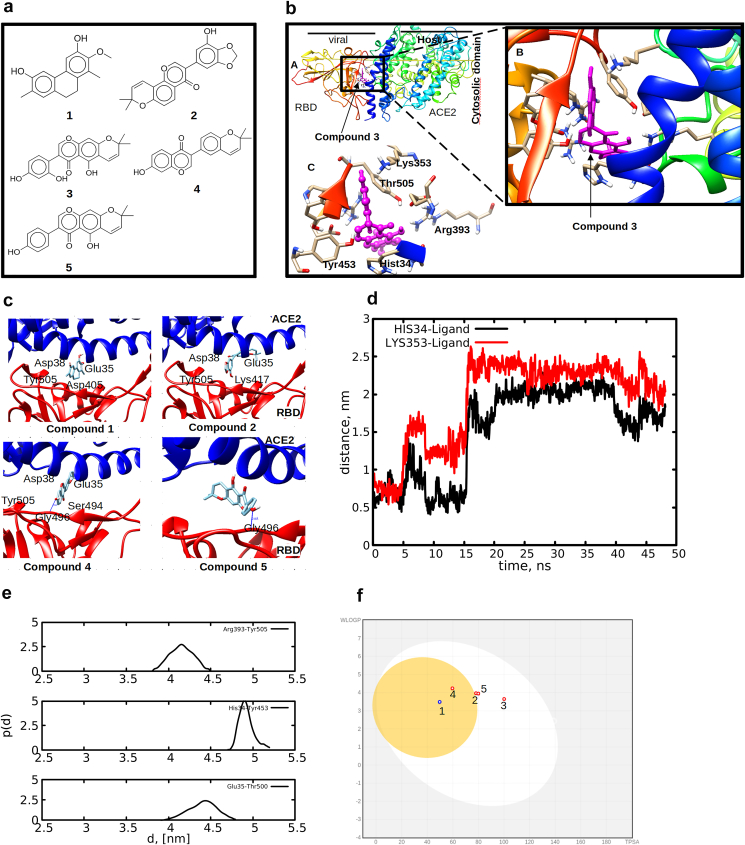
The SARS-COV-2 virion have many glycosylated spike proteins which extends from the envelope forming two subunits S1 and S2. The S1 comprises the RBD, is responsible for interaction with cell surfaces of the human ACE2. The S2 responsible for facilitating the fusion of the viral membrane into host cells. Thus, targeting inhibition of the ACE2 from recognition and interaction with spike RBD is considered a promising approach in finding therapies against COVID-19. Besides, there is accumulation of evidences supporting that spike protein is essential for the viral life cycle and is important target to block viral cell entry [2,24]. In this study, we focused on identifying natural products isolated from African medicinal plants as blockers of the spike RBD-ACE2 interface. The chemical structures and binding free energy values (BFE) of the best-docked compounds at the crystal structure of spike RBD-ACE2 interfaces are presented in Fig. 1a and Table 1. 5 compounds with BFE of −10.6 kcal/mol similar to inhibition constant of 0.93 μM were considered as potential spike RDB-ACE2 inhibitors. Selected compounds had best docking BFE than hesperidin, a known compound to bind at the interface with the BFE of −7.3 kcal/mol [2]. Compounds 1 and 2 favorably docked against the crystal structure with the BFE of −11 and −11.6 kcal/mol, respectively (Table 1). Compounds 3 and 5 had BFE of −10.7 and compound 4 had the BFE of −10.8 kcal/mol. Results obtained based on the crystal structure suggests potential disruption of RBD-ACE2 recognition and interaction, hence preventing viral fusion to human cell.
Fig. 1.
(a) 2D chemical structures of the top selected compounds from virtual screening, (b–c) binding mode and orientations preference of compounds 1–5 at the RBD-ACE2 interface, (d)distance measured between His34 and Lys353 and compound 3, the selected residues formed hydrogen bonds during docking calculations, (e) probability distribution for the holo spikeRBD-ACE2 separation distances for selected residues at the interface, (f) Boiled egg model showing the HIA and BBB distribution of the top compounds.
Table 1.
Binding free energy (ΔG (kcal/mol)) and predicted inhibition constant (K i) of the top compounds. K i was calculated from the relation ΔG = -RTlnK i, where, R is the universal gas constant, and T is Temperature (298 K).
| RBD-ACE2 |
ACE2 |
S-RBD |
|||
|---|---|---|---|---|---|
| Compounds | ΔG crystal | ΔG ensemble | Ki(ensemble) (μM) | ΔG crystal | ΔG crystal |
| 1 | −11 | −6.9 ± 0.72 | 8.39 | −6.8 | −6.3 |
| 2 | −11.6 | −8.2 ± 0.80 | 0.93 | −8.8 | −8.2 |
| 3 | −10.7 | −8.5 ± 1.17 | 0.56 | −10.7 | −7 |
| 4 | −10.8 | −8.1 ± 0.76 | 1.09 | −8.1 | −7.7 |
| 5 | −10.7 | −8.3 ± 1.24 | 0.78 | −10.7 | −6 |
Compound 1, neonthrene, belongs to a class of phenantherene and subclass of dihydrophenantherene was isolated for the first time from the leaves of Neoboutonia macrocalyx L. (Euphorbaceae). Traditionally, the plant is used for the treatment of headaches and fevers. The known biological activities of this compound includes antiplasmodial and cytotoxic activity [25]. Compound 2, norisojamicin, belongs to a class of flavonoid and subclass isoflavone was isolated for the first time from Millettia usaramensis (Leguminosae). This plant is locally used as a shade and ornamental tree [26]. Compounds 3, parvisoflavone B, is a flavonoid isolated from Erythrina schliebenii and was reported to posses moderate antitubercular and cytotoxic activity [27]. Traditionally, the plant is used for stomachache, diarrhea, for prevention of jaundice of newborn babies and as an abortive agent [27]. Compound 4, corylin, is a flavonoid belonging to a subclass of isoflavone isolated from Erythrina sacleuxii (Leguminosae). The compound was reported to possess antiplasmodial activity. Traditionally, the root and stem bark of Erythrina species are used in the treatment of malaria and microbial infections [28]. Compound 5, alpinumisoflavone, is a flavonoid isolated from Erythrina lysistemon cultivated in Egypt [29]. All compounds, except 1 shares a common class of flavonoids, the observed binding affinity of these compounds is also supported by other computational or experimental works where many flavonoids have demonstrated inhibition of SARS-COV-2 viral cell entry [30,31]. Some of the flavonoids such as hesperidin and diosimin have succeeded into clinical trial as blockers of spike RBD-ACE2. The reported flavonoinds may also possess similar activities in vitro and in vivo.
Docking calculations based on single crystal structure may results to generation of false positive hits. RCS was performed to establish the sensitivity of results to flexible receptor structures. A total of 40 ensemble structures were extracted at every 1 ns from the equilibrium MD simulation of the apo protein, and top selected compounds were docked to each ensemble structure. Interestingly, sensitivity of docking BFE was observed to different ensemble structures, for example, compound 1 which ranked higher (−11 kcal/mol) in the crystal structure, ranked least during the RCS docking with the binding free energy of −6.9 kcal/mol showing a difference of 4.1 kcal/mol (Table 1). Changes in BFE between the crystal and ensemble structures as well as wide distribution of binding free energy in the ensemble structures (Fig. 2), suggests the sensitivity of docking results to ensemble structures. Such sensitivity was also previously observed in the discovery of HIV protease inhibitors [12]. Compound 1 which ranked least and 3 which ranked higher during the ensemble based docking screening were selected for further discussion. As shown in Fig. 2 the BFE of compound 1 in the ensemble structures consistently ranked least compared to compound 3.
Fig. 2.
(a) Changes in BFE for compound 1 and 3 for different ensemble structures (b) WT- MetaD time dependent unbinding profile for compound 3 at the spike RBD-ACE2 interface.
It should be noted that the difference in binding free energy of ≈4.1 kcal/mol represents 1000-folds difference in dissociation constant [12]. It was further noted that the BFE difference between compound 1 and 3 in the ensemble structure was 1.6 kcal/mol. In other words, the dissociation constant of compound 1 is 390-folds lower than of compound 3. These findings suggest that incorporating ensemble structures in virtual screening has an advantage of eliminating false positive binders in drug design.
The ability of the selected compounds to bind to free spike RBD and ACE2 was evaluated. Table 1 shows that the binding of the compounds to ACE2 does not change significantly from the complex, two compounds; 3 and 5 were observed to have similar effects on the free ACE2 and RBD-ACE2 complex with binding affinity of −10.7 kcal/mol, however, compound 1 demonstrated a least binding affinity to free ACE2 with binding affinity of −6.8 kcal/mol. Since spike changes conformation upon binding with ACE2, the ability of compounds 3 and 5 to bind and inhibit ACE2 from changing its conformation represents them as potential candidates for the discovery of SARS-COV-2. Interestingly, when the compounds were targeted to bind free spike RBD, compound 2 showed strong binding affinity of −8.2 kcal/mol, which is comparable to the binding affinity of the flexible spike RBD-ACE2 complex. Compounds 4 and 3 also showed reasonable binding affinity of −7.7 and −7.0 kcal/mol, respectively. These results suggests that, compounds 2–5 can simultaneously bind to viral spike RBD, ACE2 and or spike RBD-ACE2 complex thereby preventing viral cell entry.
The selected compounds were further evaluated for their pharmacokinetic profiles using Swissadmet tool [32] and admetSAR [33]. The absorption, distribution, metabolism, excretion and toxicity (ADMET) profiles of the selected compounds are shown in Table 2. The Lipinski rule of five (RO5) [34] was used to evaluate the orally active and oral availability of the compounds. The RO5 requires that for an orally active drug should have (i) no more than 5 hydrogen bond donors, (ii) no more than 10 hydrogen bond acceptors, (iii) should have a molecular mass/weight (MW) less than 500 Da, (iv) should have partition coefficient (log P) range of −0.4 to +5.6, should have a topological polar surface area (TPSA) no greater than 140 Å2 [34]. As shown in Table 2, all compounds did not violate the RO5 and possessed favorable aqueous solubility, human intestinal absorption, and caco-2 permeability, except for compound 3 which demonstrated negative human oral bioavailability with a probability of 0.51. This problem could be solved by using nanotechnology to deliver the compounds to specific cells or site of action. The drug absorption percentage (%AB) was calculated using equation (1). It is interestingly to note that all compounds had the %AB of ≥65% which implies higher absorption. The calculated percentage is in agreement with the observed HIA for all compounds. For compounds to be predicated with negative HIA must have less than 30%, the selected compounds possessed a higher percentage signifying good absorption. Furthermore, these compounds showed less toxicity and did not inhibit human ether-a-go-go-related gene (hERG), suggesting them to be good drug candidates. It should be noted that, inhibition of hERG results into a potentially fetal disorder known as long QT syndrome.
| AB% = 109 − (0.345 × T P SA) | (1) |
Table 2.
ADMET profiles of the top selected compounds.
| AMEDT/ID | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| MW | 270.32 | 364.35 | 352.34 | 320.34 | 336.34 |
| TPSA | 49.69 | 78.13 | 100.13 | 59.67 | 79.9 |
| LogP o/w | 2.62 | 3.51 | 2.89 | 3.07 | 2.47 |
| LogS | −4.66 | −3.57 | −3.36 | −3.9 | −3.61 |
| HBA | 3 | 6 | 6 | 4 | 5 |
| HBD | 2 | 1 | 3 | 1 | 2 |
| RB | 1 | 1 | 1 | 1 | 1 |
| HIA | 0.99 | 0.95 | 0.97 | 0.97 | 0.97 |
| HOB | 0.61 | 0.55 | 0.51(−) | 0.51 | 0.58(−) |
| Caco-2 | 0.84 | 0.51 | 0.64 | 0.6 | 0.7 |
| %AB | 82.86 | 73.05 | 65.46 | 79.42 | 72.44 |
| BBB | 0.84 | 0.93 | 0.79 | 0.83 | 0.23(−) |
| PPB | 1.12 | 0.87 | 0.916 | 0.83 | 0.84 |
| h-ERG | 0.48(−) | 0.47(−) | 0.59(−) | 0.63(−) | 0.56(−) |
To capture detailed information on the HIA and BBB, a BIOLED Egg model which computes the lipophilicity and polarity of small molecules was used. Fig. 1f, show that all compounds were within the egg york or white egg which is an indication of good bioavailability.
Docking calculation provides a better way of understanding the binding orientation of a compound. However, they cannot give information on the stability and kinetics, and often poorly estimates the binding free energy of the complex. Although RCS is effective in accomodating receptor flexibility, it is still limited by docking algorithms. To evaluate the effectiveness of the RCS approach in discriminating false-positive binders and true binders as well as providing reasonable sampling, MD simulation was performed to compounds 3, which was predicted to be good binders with desirable pharmacokinetic profile. In addition, the selection of compound 3 was further based on the presence of many –OH and C O group and the polycyclic ring which are beneficial for drugs to bind at SARS-COV-2. It is important to highlight that the presence of many OH and C O groups and/or polycyclic ring were important for compounds to bind at the interface. For instance, although compound 3 possessed few ring than compound 2, the presence of OH and C O were important for stable interaction at the interface. The binding free energy was observed to decrease as the number of OH and C O decreased in the following order: 3 > 5 > 2 > 4 > 1.
It is worth noting that the binding of compound 3 at the RBD-AC2 interface identified three binding sites i.e interface-β, α and loop (Fig. S1). This prompted us to examine the time dynamical behavior of the title compound at the interface by computing the distance between specific residues that formed hydrogen bonds from the best docking pose. The distance presented in Fig. 1d, does not reflect ligand re-orientation rather, compound 3 changed from its initial docking pose and exhibited multiple binding poses as shown in the snapshot taken at 0 and 30 ns (Fig. S2). During the first 15 ns compound 3 bound at the interface-β then quickly moved to interface-α and then occupied a different pose where it stayed up to 50 ns simulation time. The ability of compound 3 to disrupt the recognition of spike RBD-ACE2 was investigated by measuring the distances of the residues responsible for forming hydrogen bond at the interface.
The fusion of SARS-COV-2 occurs through strong interaction between residues His34–Tyr453, Lys353–Gly502, Asp355–Thr500, Glu35–Thr500 which forms strong hydrogen bond [24] as observed in our work (see Fig. 1b). Weakening these hydrogen bond interactions result in increased protein-protein distances which causes weak interaction as well as poor recognition and hence failure of the virus to penetrate human cells. Figs. 1e and S3, shows separation distance between the spike RBD-ACE2 residues of the holo protein with respect to apo protein. Interestingly, when compound 3 bound near interface-β the separation distance between Glu35–Thr500 slightly increased over the simulation time (Fig. S3), suggesting the ability to disrupt RBD-ACE2 recognition. Surprisingly, the calculated binding free energy based on LIE yielded unexpected results. Fig. S3(a) shows the average LIE which suggests that when compound 3 bound to the regions other than interface-α and β, its binding free energy decreased. The decrease in LIE could be attributed to the weak contribution of electrostatic forces of interaction at the region near interface-β where it stayed for long time.
The unbinding process and molecular interaction of compound 3 at the RBD-ACE2 interface was further characterized by means of WT-MetaD. The time dependence for the unbinding process presented in Fig. 2b shows that the system diffused many times reflecting convergence of the system. The 1D free energy surface (FES) as a function of d1COM shows three distinct (un)binding states (Fig. 3b). As observed in the unbiased simulation (Fig. 1d), compound 3 exhibited multiple binding poses characterized by the measured distances from the initial docking pose. The biased simulation shows a local free energy minimum at d1COM = 0.3 and 0.7 nm corresponding to state I, the d1COM = 1.4 nm represents local free energy minimum corresponding to state II. The two states i.e I and II are separated by energy barrier of 5 kJ/mol. The d1COM = 0.7 and 1.45 nm suggests two binding states of compound 3 at the interface, which could be a way to disrupt the recognition of spike RBD-ACE2 protein-protein interaction. The ability of compound 3 to disrupt RBD-ACE2 protein-protein recognition was assessed by measuring the residues distances at the interface-α and β which are involved in recognition and fusion. 1D FES shown in Fig. 3b indicates three free energy minimum at d2 = 4.25, 4.6 and 5 nm, where the minimum at d2 = 4.25 and 5 nm are separated by the energy barrier of 15 kJ/mol. The minimum free energy at d2 = 4.6 and 5 nm suggests weak interaction between the two residues. Interestingly, when compound 3 bound at the interface-α, the RDB-ACE2 distance between His34–Tyr453 increased to 5 nm and when bound at interface-β the distance reduced to 4.25 nm, while increasing the distance between Glu35–Try500 (Fig. 3c). The ability of compound 3 to simultaneously bind at the two interfaces and increasing the RBD-ACE2 distances suggests its ability to weaken and disrupt the virus-receptor recognition.
Fig. 3.
Binding process of compound 3 at the RBD-ACE interface. (a) 1D-FES profile of the unbinding of compound 3 (b) spike RBD-ACE2 separation distance (c) 2D-FES shows the. minimum free energy and the associated structures.
The binding interaction analysis of compound 3 from biased simulation shows occupancy to all sites i.e interface-β and α (Fig. 3c) interacting with many residues from ACE2. The interaction with residues at the ACE2 interface represents an important option for treatment of SARS-COV-2 because ACE2 residues at the interface are susceptible for strong interaction and recognition of the RBD. Fig. 3c shows the 2D free energy and the associated structures for the different free energy minimum. It is observed that when bound at the interface-β it interacted and formed hydrogen bonds with Asn33 and Arg393 both from ACE2. Another important feature observed is that the RBD-ACE2 separation distance is slightly increased by ~1.5 Å. When bound at interface-α the separation distance decreased by ~1.0 Å and interacted with residues Glu37, Lys353, Gln409 and Lys417 from RBD and ACE2 at the interface. The slight increase of residues distances at the interface is also observed in the unbiased simulation (see Fig. 1c). We further noted that compound 3 could indirectly weaken the recognition of spike RBD-ACE2 through forming hydrogen bonds with many residues from the ACE2 domain as well as bridging water molecules that formed hydrogen bond with compound 3 at the interfaces. This provides an important rationally of blocking the spike RBD from recognizing the ACE2.
Although compound 3 presents a small noticeable increase in protein-protein distances, it is observed to occupy all sites, i.e loop and interfaces-α and β necessary for viral attachment. As observed from the equilibrium MD, our biased simulation further shows that the unbinding of compound 3 starts from interface-β to α (Fig. 3c). The occupancy to different binding sites at the ACE2 domain suggests that, compound 3 could be a promising agent for reducing and preventing viral infection.
In summary, the strong interaction of SARS-COV-2 spike RBD through ACE2 protein provides peculiarity for the virus transmissibility rate as compared to SARS-COV [[35], [36], [37]]. Although currently there is no approved medication, some promising vaccination are in clinical trial. Efforts to find drugs to treat the disease are going-on in different research groups. Some groups have focused on repositioning old drugs while others have focused on establishing new molecules from natural products. The latter approach has an advantage of introducing new molecules for further investigations, should they contain desirable pharmacokinetic properties. Due to the natural products potential in treating various diseases, the present study, by incorporating various computational protocols, screened and evaluated natural products as blockers of SARS-COV-2 spike from recognizing and binding to ACE2. This strategy has the advantage of weakening the interaction and hence preventing viral fusion. Furthermore, the methodological approach reported here provides an optimized and efficient multi scale protocol ranging from rigid and flexible molecular docking to enhanced sampling as well as free energy techniques, that enables us to quantify and assess the interactions between the human protein and virus. Provided with the fact that, these results need further in vitro and in vivo evaluation that could strengthen the results presented in this work, we want to share our findings with the scientific community as soon as possible and providing the basis for discovering new therapy against COVID-19 and viral related diseases.
Declaration of competing interest
I confirm that all there is no any conflict of interest to all author.
Acknowledgments
The study was sponsored by the University of Dodoma through the 2020 JAS funding. Authors are grateful to the Lengau CHPC for providing access to computational facilities.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2021.101024.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Huang J., Tao G., Liu J., Cai J., Huang Z., Chen J.-x. Current prevention of covid-19: natural products and herbal medicine. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.588508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sethi A., Sanam S., Munagalasetty S., Jayanthi S., Alvala M. Understanding the role of galectin inhibitors as potential candidates for sars-cov-2 spike protein: in silico studies. RSC Adv. 2020;10(50):29873–29884. doi: 10.1039/d0ra04795c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chikhale R.V., Gupta V.K., Eldesoky G.E., Wabaidur S.M., Patil S.A., Islam M.A. Identification of potential anti-tmprss2 natural products through homology modelling, vir- tual screening and molecular dynamics simulation studies. J. Biomol. Struct. Dynam. 2020:1–16. doi: 10.1080/07391102.2020.1784289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vivek-Ananth R., Rana A., Rajan N., Biswal H.S., Samal A. 2020. In Silico Identification of Potential Natural Product Inhibitors of Human Proteases Key to Sars-Cov-2 Infection. arXiv preprint arXiv:2006.00652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. Sars-cov-2 cell entry depends on Ace2 and Tmprss2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan M.T., Ali A., Wang Q., Irfan M., Khan A., Zeb M.T., Zhang Y.-J., Chinnasamy S., Wei D.-Q. Marine natural compounds as potents inhibitors against the main protease of sars-cov-2 a molecular dynamic study. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1769733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forrestall K.L., Burley D.E., Cash M.K., Pottie I.R., Darvesh S. 2-pyridone natural products as inhibitors of sars-cov-2 main protease. Chem. Biol. Interact. 2021;335 doi: 10.1016/j.cbi.2020.109348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sayed A.M., Alhadrami H.A., El-Gendy A.O., Shamikh Y.I., Belbahri L., Hassan H.M., Abdelmohsen U.R., Rateb M.E. Microbial natural products as potential inhibitors of sars-cov-2 main protease (mpro) Microorganisms. 2020;8(7):970. doi: 10.3390/microorganisms8070970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gentile D., Patamia V., Scala A., Sciortino M.T., Piperno A., Rescifina A. Putative inhibitors of sars-cov-2 main protease from a library of marine natural products: a virtual screening and molecular modeling study. Mar. Drugs. 2020;18(4):225. doi: 10.3390/md18040225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z., Yang L. Turning the tide: natural products and natural-product-inspired chem- icals as potential counters to sars-cov-2 infection. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verma S., Twilley D., Esmear T., Oosthuizen C.B., Reid A.-M., Nel M., Lall N. Anti- sars-cov natural products with the potential to inhibit sars-cov-2 (covid-19) Front. Pharmacol. 2020;11:1514. doi: 10.3389/fphar.2020.561334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin J.-H., Perryman A.L., Schames J.R., McCammon J.A. Computational drug design accommodating receptor flexibility: the relaxed complex scheme. J. Am. Chem. Soc. 2002;124(20):5632–5633. doi: 10.1021/ja0260162. [DOI] [PubMed] [Google Scholar]
- 13.Deng N., Forli S., He P., Perryman A., Wickstrom L., Vijayan R., Tiefenbrunn T., Stout D., Gallicchio E., Olson A.J. Distinguishing binders from false positives by free energy calculations: fragment screening against the flap site of hiv protease. J. Phys. Chem. B. 2015;119(3):976–988. doi: 10.1021/jp506376z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makeneni S., Thieker D.F., Woods R.J. Applying pose clustering and md simulations to eliminate false positives in molecular docking. J. Chem. Inf. Model. 2018;58(3):605–614. doi: 10.1021/acs.jcim.7b00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meagher K.L., Carlson H.A. Incorporating protein flexibility in structure-based drug discovery: using hiv-1 protease as a test case. J. Am. Chem. Soc. 2004;126(41):13276–13281. doi: 10.1021/ja0469378. [DOI] [PubMed] [Google Scholar]
- 16.Amaro R.E., Baron R., McCammon J.A. An improved relaxed complex scheme for receptor flexibility in computer-aided drug design. J. Comput. Aided Mol. Des. 2008;22(9):693–705. doi: 10.1007/s10822-007-9159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin J.-H., Perryman A.L., Schames J.R., McCammon J.A. The relaxed complex method: accommodating receptor flexibility for drug design with an improved scoring scheme. Biopolymers: Orig. Res. Biomol. 2003;68(1):47–62. doi: 10.1002/bip.10218. [DOI] [PubMed] [Google Scholar]
- 18.Chan A.H., Wereszczynski J., Amer B.R., Yi S.W., Jung M.E., McCammon J.A., Clubb R.T. Discovery of s taphylococcus aureus sortase a inhibitors using virtual screening and the relaxed complex scheme. Chem. Biol. Drug Des. 2013;82(4):418–428. doi: 10.1111/cbdd.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barakat K., Tuszynski J. Relaxed complex scheme suggests novel inhibitors for the lyase activity of dna polymerase beta. J. Mol. Graph. Model. 2011;29(5):702–716. doi: 10.1016/j.jmgm.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Cheng L.S., Amaro R.E., Xu D., Li W.W., Arzberger P.W., McCammon J.A. Ensemble- based virtual screening reveals potential novel antiviral compounds for avian influenza neuraminidase. J. Med. Chem. 2008;51(13):3878–3894. doi: 10.1021/jm8001197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shadrack D.M., Swai H.S., Hassanali A. A computational study on the role of water and conformational fluctuations in hsp90 in response to inhibitors. J. Mol. Graph. Model. 2020;96 doi: 10.1016/j.jmgm.2019.107510. [DOI] [PubMed] [Google Scholar]
- 22.Trott O., Olson A.J. Autodock vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ntie Kang F., Telukunta K.K., Döring K., Simoben C.V., Moumbock A.F.A., Malange Y.I., Njume L.E., Yong J.N., Sippl W., Günther S. Nanpdb: a resource for natural products from northern african sources. J. Nat. Prod. 2017;80(7):2067–2076. doi: 10.1021/acs.jnatprod.7b00283. [DOI] [PubMed] [Google Scholar]
- 24.Gangadevi S., Badavath V.N., Thakur A., Yin N., De Jonghe S., Acevedo O., Jochmans D., Leyssen P., Wang K., Neyts J. Kobophenol a inhibits binding of host ace2 receptor with spike rbd domain of sars-cov-2, a lead compound for blocking covid-19. J. Phys. Chem. Lett. 2021;12:1793–1802. doi: 10.1021/acs.jpclett.0c03119. [DOI] [PubMed] [Google Scholar]
- 25.Namukobe J., Kiremire B.T., Byamukama R., Kasenene J.M., Dumontet V., Guéritte F., Krief S., Florent I., Kabasa J.D. Cycloartane triterpenes from the leaves of neoboutonia macrocalyx l. Phytochemistry. 2014;102:189–196. doi: 10.1016/j.phytochem.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Yenesew A., Midiwo J.O., Waterman P.G. Rotenoids, isoflavones and chalcones from the stem bark of millettia usaramensis subspecies usaramensis. Phytochemistry. 1998;47(2):295–300. [Google Scholar]
- 27.Nyandoro S.S., Munissi J.J., Kombo M., Mgina C.A., Pan F., Gruhonjic A., Fitz- patrick P., Lu Y., Wang B., Rissanen K. Flavonoids from erythrina schliebenii. J. Nat. Prod. 2017;80(2):377–383. doi: 10.1021/acs.jnatprod.6b00839. [DOI] [PubMed] [Google Scholar]
- 28.Andayi A., Abiy Y., Solomon D., Jocob M.O., Peter G.M., Ogoche J.J.I., Hosea A., Pamela L., Julia W., Norman W.C., Matthias H., Martin P.G. Antiplasmodial flavonoids from erythrina Sacleuxii. Planta Medica. 2005:187–189. doi: 10.1055/s-2005-873200. [DOI] [PubMed] [Google Scholar]
- 29.El-Masry S., Amer M.E., Abdel-Kader M.S., Zaatout H.H. Prenylated flavonoids of erythrina lysistemon grown in Egypt. Phytochemistry. 2002;60(8):783–787. doi: 10.1016/s0031-9422(02)00202-9. [DOI] [PubMed] [Google Scholar]
- 30.Bhowmik D., Nandi R., Prakash A., Kumar D. Evaluation of flavonoids as 2019-ncov cell entry inhibitor through molecular docking and pharmacological analysis. Heliyon. 2021 doi: 10.1016/j.heliyon.2021.e06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muchtaridi M., Fauzi M., Khairul Ikram N.K., Mohd Gazzali A., Wahab H.A. Natu- ral flavonoids as potential angiotensin-converting enzyme 2 inhibitors for anti-sars-cov-2. Molecules. 2020;25(17):3980. doi: 10.3390/molecules25173980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daina A., Michielin O., Zoete V. Swissadme: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng F., Li W., Zhou Y., Shen J., Wu Z., Liu G., Lee P.W., Tang Y. 2012. Admetsar: a Comprehensive Source and Free Tool for Assessment of Chemical Admet Properties. [DOI] [PubMed] [Google Scholar]
- 34.Lipinski C.A. Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y. Structural and functional basis of sars-cov-2 entry by using human ace2. Cell. 2020;181(4):894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu J., Sun P.D. High affinity binding of sars-cov-2 spike protein enhances ace2 car- boxypeptidase activity. J. Biol. Chem. 2020;295(52):18579–18588. doi: 10.1074/jbc.RA120.015303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calcagnile M., Forgez P., Iannelli A., Bucci C., Alifano M., Alifano P. Molecular docking simulation reveals ace2 polymorphisms that may increase the affinity of ace2 with the sars-cov-2 spike protein. Biochimie. 2021;180:143–148. doi: 10.1016/j.biochi.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.