Graphical abstract
Keywords: Crystal structure, Glycoside hydrolase, Pulp and paper, Xylose, Xylooligosaccharides
Abstract
Endoxylanases active under extreme conditions of temperature and alkalinity can replace the use of highly pollutant chemicals in the pulp and paper industry. Searching for enzymes with these properties, we carried out a comprehensive bioinformatics study of the GH10 family. The phylogenetic analysis allowed the construction of a radial cladogram in which protein sequences putatively ascribed as thermophilic and alkaliphilic appeared grouped in a well-defined region of the cladogram, designated TAK Cluster. One among five TAK sequences selected for experimental analysis (Xyn11) showed extraordinary xylanolytic activity under simultaneous conditions of high temperature (90 °C) and alkalinity (pH 10.5). Addition of a carbohydrate binding domain (CBM2) at the C-terminus of the protein sequence further improved the activity of the enzyme at high pH. Xyn11 structure, which has been solved at 1.8 Å resolution by X-ray crystallography, reveals an unusually high number of hydrophobic, ionic and hydrogen bond atomic interactions that could account for the enzyme’s extremophilic nature.
1. Introduction
Enzymes play a fundamental role in the transformation of the traditional chemical industry into a future, more sustainable, green alternative. The great diversity of enzymes existing in Nature can supply suitable catalysts to any type of biochemical reaction. Although most enzymes are labile material, unable to stand the harsh physicochemical conditions used in industry, life shows a surprising plasticity, being able to generate organisms that thrive under harsh conditions of temperature, pH, pressure, salinity, etc., which a priori would be unimaginable. Extremophilic organisms, mainly archaea and bacteria, adapted to such extreme conditions become a source of enzymes that widen the range of processes and operational conditions in which enzymes can be applied [1], [2].
Use of xylanases by the paper industry, for pulp bleaching, offer a challenging case for stretching the enzyme capability to the limit. Xylanases facilitate the removal of residual lignin that causes the dark color of the pulp and can be a total or partial replacement to the use of hazardous chemical agents [3], [4]. The predominant method used by the paper industry, the kraft process, requires harsh conditions. Therefore, the availability of xylanases active under extreme environments of temperature and alkalinity represents an economic bonus.
Enzymes with xylanolytic activity appear in at least nine families of the CAZY database [5]. However, most true xylanases (EC 3.2.1.8), considered as such because of their high substrate specificity, are classified in families GH10 and GH11 [6], [7]. Family GH10 enzymes are characterized by the presence of a catalytic domain of ca. 40 kD, with (α/β)8-barrel structure. Family GH11 enzymes contain a smaller (ca. 30 k Da) catalytic domain with β-jelly roll structure. There is abundant published information about enzymes from both families. Up to the present the number of annotated protein sequences and resolved atomic structures is about 5000 and 50 for GH10, respectively and 2000 and 30 for GH11 (CAZY database, accessed on January 2021). Although only a relatively small fraction of the sequences corresponds to enzymes whose activity has been characterized experimentally, the available data provide very useful information to predict the enzymatic properties of an uncharacterized sequence. Several bioinformatic approaches to this end have been reported.
Conventional strategies used to identify enzymes with extremophilic properties relied on their isolation from microorganisms with such properties, for instance, Thermotoga and Thermoascus [8]. However, this approach is too limited to yield enzymes with optimal physicochemical and catalytic properties that fulfill industry. Protein engineering techniques provide a complementary tool for obtaining enzymes with enhanced thermostability or alkalinity [9], [10], [11]. In silico screening of protein sequences from databases, often derived from genomic and metagenomic analysis, has been shown to be a powerful methodology that has been successfully applied to identify extremophilic xylanases in GH11 family [12] and for discovery of hidden properties of carbohydrate active enzymes [13], [14], [15].
In this communication we describe an in silico analysis of GH10 family, searching for xylanases with extreme thermophilic and alkalophilic properties, and the functional and structural characterization of the enzyme with best characteristics. Additionally, we show that the modification of the protein structure by the addition of carbohydrate binding domains improves the enzyme performance, which is relevant for biotechnological applications.
2. Materials and methods
2.1. Phylogenetic analysis of GH10 sequences
Accession numbers of GH10 protein sequences were obtained from the CAZy database [5]. The amino acid sequences were obtained from the NCBI database, using the Batch Entrez Tool (https://www.ncbi.nlm.nih.gov/sites/batchentrez). Pfam [16] provided protein domain composition and coordinates for each sequence.
The linear composition of domains of a given sequence in N-terminal to C-terminal order, defined what has been called domain architecture (DA). The assignment of specific DA from the sequences of the GH10 family was performed following the methodology previously described for the GH2 family [13]. Only sequences containing a catalytic domain Glyco_hydro_10 (GH10) matching at least 80% with the Pfam consensus were included in this study.
Sequence alignment of GH10 catalytic domains was performed with CLC sequence viewer (Quiagen), using ClustalO MSA algorithm [17]. Trees were built using W-IQ-TREE Maximum Likehood algorithm with JTT matrix [15] and a bootstrap of 1000 replicates. Results were analyzed on Dendroscope Software [18] and represented using FigTree (http://tree.bio.ed.ac.uk/software/figtree/) and MESQUITE software (https://www.mesquiteproject.org).
2.2. Molecular biology
DNA sequences and amino acid selected for experimental analysis were edited before cloning. Signal peptide sequences were detected using the Phobius Tool [19] and removed. The coding sequences were optimized for E. coli expression by using the Integrated DNA Technologies (IDT) Codon Optimization Tool (www.idtdna.com). Native restriction sites were eliminated and SacI and SalI restriction sites were added in 5′ and 3′ respectively, to facilitate the cloning in vector pQE80L (Quiagen).
Synthetic genes (codon-optimized) of sequences encoding five selected putative xylanases (Xyn10-14) were purchased from IDT (Supplementary Table S3). The DNA fragments (except Xyn10 and Xyn14) were digested with endonucleases SacI and SalI and cloned into pQE80L plasmid cut with the same enzymes. Xyn10 and Xyn14 could not be synthesized as a single piece and were therefore assembled by joining two fragments (F1 and F2). Xyn10-F1 and Xyn14-F1 were cut with SacI and EcoRI, whereas Xyn10-F2 and Xyn14-F2 were cut with EcoRI and SalI. The corresponding fragments were then cloned as one piece in pQE80L. Fast Digest enzymes and T4 ligase were purchased from ThermoScientific.
The xylanase encoding plasmids were used for transforming E. coli XL1 Blue. Selection of E. coli clones expressing xylanase genes was performed as described in (Talens-Perales et al., 2020). The cloned sequences were confirmed by sequencing. Once checked, the plasmids were transferred to E. coli Rosetta (Stratagene) for protein production. Hybrid enzymes Xyn11-CBM2 and Xyn11-CBM9 were constructed from plasmids Xyn5-CBM2 PQE80L and Xyn5-CBM9, respectively [12], replacing the Xyn5 gene by the Xyn11 gene.
2.3. Protein purification
Cell crude extracts were obtained from E. coli cultures grown at 37 °C until reaching OD600 of 0.6 and then were induced with 1 mM IPTG, either at 37 °C for 5 h or 16 °C overnight. Buffer A (20 mM phosphate buffer, pH 7.4, 10 mM imidazole, 500 mM NaCl) was used to disrupt the cells by sonication. Protein extracts were obtained by centrifugation at 12,000 g during 25 min. Protein purification was performed using nickel affinity chromatography with 1 mL HisTrap FF crude column (GE Healthcare) mounted in AKTA-Purifier (GE). Buffer B (20 mM phosphate buffer, pH 7.4, 500 mM imidazole, 500 mM NaCl) was used for elution. Eluted fractions with xylanase activity were dialyzed against buffer C (20 mM Tris-HCl, pH 7.0, 50 mM NaCl). Protein concentration in the eluted fractions was determined in a NanoDrop spectrophotometer (Thermo Fisher). The purity of the protein recovered was analyzed by SDS-PAGE, using Blue Safe staining (Nzytech). An image of the gel was taken with a Proxima AQ-4 gel documentation system (Isogen) and the amount of protein in the gel bands was quantified using FIJI software [20].
2.4. Evaluation of xylanase activity at different conditions of temperature and pH
Xylanase activity of purified enzymes was assayed at range of temperature and pH. Enzyme reactions for temperature assays were prepared by mixing 180 μL of substrate (1% oat spelt xylan, in Tris-HCl 50 mM buffer pH 9.0) and 20 μL of purified protein (protein concentrations were adjusted to assure linear enzyme response) and then incubated at 60 °C, 70 °C, 80 °C or 90 °C, for 10 min. The reaction was stopped by putting the tubes on ice.
Activity at different pH was determined using 50 mM buffered solutions, at the following pH values: 5.0 (acetate), 6.0 and 7.0 (phosphate), 8.0, 9.0 and 10.0 (Tris-HCl). The enzyme reactions were prepared as above described, using the appropriate buffer. The reactions were incubated at 65 °C during 10 min and then stopped on ice. Control reactions, without enzyme, were carried out for each assay condition.
Measurement of reducing sugars was carried out by adding 100 μL of DNS reactive to the reaction tubes that were then boiled for 10 min. Next, 900 μL of miliQ H2O were added and the tubes were centrifuged. Measurements were done at OD540 in 96-well plates by transferring 300 μL of the supernatant using PowerWave HT equipment, from BioTek Instruments (Winooski, VT, USA).
Cleavage pattern of xylooligosaccharides was analyzed by using 500 μL of 2 mM of the corresponding substrate, from 2 to 6 units (Megazyme), dissolved in 50 mM Tris-HCl buffer pH 9.0. To each tube, 20 μL of enzyme solution were added. The amount of enzyme used was estimated to obtain 1 μmol of reducing sugars min−1 mg−1 of enzyme. The reactions were carried for 5 h at 90 °C and stopped on ice. Analysis of reaction products was carried out by ion exchange chromatography using a Dionex (Thermo Fisher Scientific) with CarbonPac PA100 column and a pulsed amperometric detector.
2.5. Protein crystallization, data collection and structure determination
Firstly, different crystallization screens were explored, using the sitting-drop vapor-diffusion method. A wide variety of crystals grew in several commercial kits such as Index (Hamptom Research), JBScreen JCSG++ (Jena Bioscience), JBScreen Classic (Jena Bioscience). Different manuals grids were used in order to optimize the quality of the crystals. Finally, the best crystals were obtained with 10% PEG 3350, 0.2 M Proline, 0.1 M Hepes pH 7.5. Each drop presented the same proportion of protein (6.47 mg/mL) and reservoir solution (250 nl). Mother liquor solution was supplemented with 25% of glycerol to cryoprotect the crystal during the data collection. Diffraction data were collected at the ALBA synchrotron station of Barcelona (Spain). The X-ray images were processed with XDS [21], and merged using Aimless from CCP4 suite package [22]. P 21 21 21 space group was obtained with cell parameters 91.45 × 95.27 × 100.53 Å3 and two molecules in the asymmetric unit.
The structure was solved by molecular replacement using MOLREP from CCP4 [23]. The xylanase CbXyn10C from Caldicellulosiruptor bescii [24], with the Protein Data Bank code 5OFJ (56.5% sequence homology) was selected as a template. Restraint refinement was carried out using REFMAC5 from CCP4 with local non-crystallographic symmetry (NCS) [25]. This was combined with several rounds of model building in COOT [26]. Figures were prepared with PyMOL [27]. Final crystallography results are given in Supplementary Table 4.
2.6. Analysis of protein stability
Different web servers were employed to analyze the structure and thermostability of the protein. Protein Blast [28] and ENDscript server [29] were used to compare protein sequence and the tridimensional structures were inspected in Dali server [30]. ProtParam tool (ExPASy) [31] was used to compute amino acid composition. Intraprotein hydrophobic and ionic interactions, hydrogen bonds, aromatic and cation-π interactions were determined by PIC server (Protein Interactions Calculator) [32]. The accessible surfaced area (ASA) and the exposed charged accessible area were both calculated with VADAR server [33]. The formula for compactness was: ASA/number of total residues, and for % exposed charged accessible surface area was: exposed charged accessible area /ASA *100.
3. Results and discussion
3.1. In silico screening of GH10 sequences for thermophilic alkaliphilic xylanases
The nearly 5000 entries currently available in the CAZy database were expurgated discarding repetitions, incomplete sequences and those having less than 20% matching with the Pfam Glyco_Hydro_10 domain (GH10). This yielded a total of 2309 sequences that were further processed (Supplementary material, Table S1). About 80% of these sequences (1848) corresponded to bacteria whereas the remaining 20% (461) were from eukaryotes. No archaeal sequences matched the established requirements. GH10 protein sequences showed a great variety of domain architectures. Up to 197 different domain architectures (DA) could be distinguished (Supplementary material, Table S2), in contrast with the lesser diversity (69 DA) described for functionally related GH11 family [12]. About one half of the bacterial sequences and one tenth of the eukaryotic, showed the simplest possible DA, consisted of just the catalytic GH10 domain. Regarding more complex DA, the presence of CBM4-9, which binds amorphous cellulose and soluble oligosaccharides [34], [35], was ubiquitous and detected in 14% of the sequences, in some cases in tandem repeats at N-terminal position. CBM2 which binds cellulose and chitin [36], or Ricin-B-lectin domains that mediate sugar recognition [37], were quite abundant, present in more than 5% of the sequences. A small fraction of the sequences (1–2%) contained C-terminal extensions not corresponding with identified motifs. These tails were labeled by their length, as Ct1 (50–150), Ct2 (150–200), Ct3 (200–300), Ct4 (300–400) and Ct5 (400–500) (Supplementary material, Table S2).
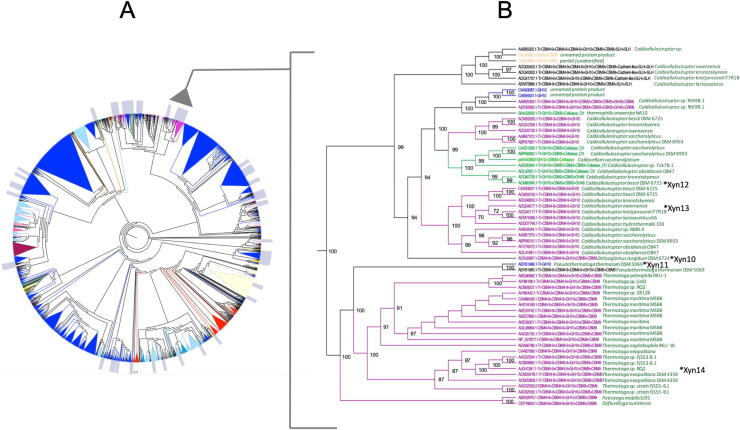
Fig. 1A shows a schematic representation of the cladogram resulting from the phylogenetic analysis of the Glyco_Hydro_10 domain of the 2309 selected sequences. A detailed version of the cladogram is presented as supplementary material (Fig. S1). In Fig. 1 and S1 and in Table S2, different DA with common domains are represented by the same color code. In the cladogram, eukaryotic sequences appear in three groups, generated by different evolutionary events, at a late stage in the evolutionary process. The simplest DA, a single GH10 domain, appears in some instances mingled with composed DA, but most sequences with additional, non-catalytic domains, emerge from separate nodes. Sequences putatively identified as thermoresistant, are marked by a gray label in Fig. 1A. Among these, a cluster (hereinafter designated as TAK) marked by a triangle, corresponds to sequences that putatively are thermostable and alkaliphilic. The sequences included in the TAK cluster, belong to genera the Caldicellulosiruptor, Thermotoga, Pseudothermotoga, Petrotoga and Defluviitoga, and are listed in Fig. 1B. Most of these sequences contain a multidomain DA, with CBM4-9 present in ca. 70% of them. Another domains present are CBM9, CBM3 and the catalytic domain GH48 with cellulase activity [38]. CBM9 and CBM3 have been described to have xylan-binding activity [38], [39]. Five of these sequences, representing different DA, were selected for experimental analysis: Xyn10 from Dyctioglomus turgidum, Xyn11 from Pseudothermotoga thermarum, Xyn12 and Xyn13 from Caldicellulosiruptor bescii and Caldicellulosiruptor owensensis, respectively and Xyn14 from Thermotoga sp.
Fig. 1.
(A) Cladogram of the GH10 family. The gray triangle marks the TAK cluster. (B) Phylogenetic tree of the TAK cluster sequences. Asterisks mark the sequences that were selected for experimental analysis.
3.2. Production, purification and functional characterization of selected enzymes
Synthetic, codon-optimized gene sequences encoding the selected putative xylanases were cloned and expressed in E. coli. Synthetic DNA coding sequences could be obtained as a single fragment, except for Xyn10 and Xyn14, which had to be reconstructed from two fragments (Xyn10-F1, Xyn14-F1 and Xyn10-F2, Xyn14-F2). Proteins were purified from bacterial crude cell extracts by thermal treatment and nickel affinity chromatography. Xyn12 was discarded at this point because no substantial amount of protein could be recovered from the recombinant E. coli cultures. Semiquantitative evaluation of Xyn14 showed very low level of xylanase activity (results not shown) and therefore was also discarded. The other three enzymes were purified and analyzed by SDS-PAGE (Fig. 2). In all three cases, electrophoretical mobility was in accordance with the expected molecular mass of the polypeptides, predicted by ProtParam [40].
Fig. 2.
SDS-PAGE analysis and domain architecture of selected enzymes. Protein ladder mass in kDa.
Oat spelt xylan hydrolysis by Xyn10, Xyn11, and Xyn13 was measured at different pH and temperature. Assays at different values of pH were carried out at 90 °C (Fig. 3A). Xyn10 and Xyn11 showed the expected profile for alkaliphilic enzymes, with optimal activity at pH between 8.0 and 9.0. Xyn13 showed maximal activity at pH between 6.0 and 7.0, but retained high activity at pH 9.0. In buffered solution at pH 9.0, the three xylanases showed a thermophilic profile, with optimal temperature 70 °C and 90 °C. The highest activity measured corresponded to Xyn11 at 90 °C (Fig. 3B). These results are remarkably different from a report by Shi et al., [41], describing the properties of a xylanase encoded by the Xyn10B gene of T. thermarum (now Pseudothermotoga thermarum), same sequence as Xyn11, but optimal activity at 80 °C, and weak activity at pH > 8.0. A different xylanase from the GH10 family from P. thermarum has also been described. This one displays a complex DA, which three tandem CBM4-9 at N-terminal position and two CBM9 at C-terminal position. Despite of the differences in DA, the catalytic domain of this enzyme, named Xyn10A, is very similar to Xyn11 being located very close in the phylogenetic tree (Figs. 1 and S1). Xyn10A, was reported to be optimally active at 95 °C and pH 7.0 [8]. Other thermostable xylanases (from GH10 and GH11 families) have been characterized but their optimal temperatures are around 70 °C and their optimum pH ranged from 6.5 to 9.0. Only Bacillus halodurans TSEV xylanase combines thermostability and alkaliphility, with optimal values of pH of 9.0 and temperature 80 °C [42], but still below the score of Xyn11. Xylanases from Geobacillus, Caldicellusiruptor [43] and Thermotoga, showed high optimum temperatures. Remarkably, Thermotoga naphthophila RKU-10 and Thermotoga petrophila RKU-1 (95 °C) or Thermotoga maritima MSB8, (90 °C), but optimum pH values for these enzymes were between 5.0 and 6.0 [44], [45], [46]. Xyn11 represented an extraordinary performance under simultaneous conditions of high temperature and alkalinity, showing at pH 10.5 and 90 °C, an activity value of 600 µmol min−1 mg−1 of enzyme.
Fig. 3.
Activity of selected enzymes at different values of pH (A) and temperatures (B). Activity values correspond to μmol of reducing sugars min−1 mg-1 of enzyme.
A previous study carried out with GH11 xylanases showed that in some instances, the addition of a Carbohydrate Binding Domain (CBM2, CBM9) to the protein structure has a positive effect on enzyme performance [12]. Therefore, we undertook the construction of hybrid enzymes derived from Xyn11, by fusing this enzyme with CBM2 and CBM9. The SDS-PAGE profile and DA of the hybrids are shown in Fig. 4A. Addition of CBM9 caused a decrease of Xyn11 activity at different pH (Fig. 4B). This result was unexpected as it is the opposite to what was observed by the addition of CBM9 to a GH11 xylanase [12]. However, the addition of CBM2 improved the activity of the enzyme at high pH. Hybrid Xyn11-CBM2 showed an activity two times higher than Xyn11 at 90 °C and pH 10.5 (Fig. 4B).
Fig. 4.
(A) Physical characterization (DA and SDS-PAGE analysis) of hybrid xylanases. (B) Activity of hybrid xylanases assayed at 90 °C and different pH. Displayed values correspond to activity expressed as μmol of reducing sugars min−1 mmol−1 of enzyme. (C) Activity of hybrid xylanases at pH 9.0 and different temperatures.
The cleavage pattern of xylanases was analyzed using as substrate oligoxylosides from 2 to 6 units. Enzyme reactions were carried out for 5 h at pH 9.0 and 90 °C. The products released from the reactions were analyzed chromatographically. As expected, the enzymes showed characteristic behavior of endoxylanases, but with some interesting differences. Xyn10 and Xyn13 were unable to hydrolyze xylobiose and xylotriose. Xyn10 hydrolyzed xylotetraose, xylopentaose and xylohexaose, yielding mainly xylobiose and xylotriose (Fig. 5A). Xyn13 was inactive towards xylotetraose, but excised xylopentaose and xylohexose, yielding mainly xylobiose and xylotriose (Fig. 5C). Xyn11, as well as its hybrids Xyn11-CBM9 and Xyn11-CBM2, were unable to cut xylobiose, but were active against xylotriose, xylotetraose, xylopentaose and xylohexaose yielding mainly xylose and xylobiose (Fig. 5B). These results agree with previous studies on GH10 xylanases [42]. The observed differences in the cleavage pattern of Xyn10, Xyn11 and Xyn13 is relevant from a biotechnological point of view, as it can be used to produce short-chain oligoxylosides from xylan. These oligosaccharides are compounds with recognized value as prebiotics, whose administration confers a proven heath benefit [47], [48].
Fig. 5.
Cromatographic profile of reaction products obtained from the action of xylanases on xylooligosaccharides of different degree of polymerization.
3.3. Crystallographic structure of Xyn11
The structure of Xyn11 was solved at 1.8 Å resolution by X-ray crystallography. The protein folds into an (β/α)8 barrel (TIM-barrel) architecture (Fig. 6A), typical of GH10 xylanases. From the loops linking the C-terminal of each β-strand to the succeeding α-helix, the long L4, L7 and L8 loops protrude markedly and define an extended groove to accommodate the substrate. An additional β-hairpin motif is present in the loop L2, linking β2 to α2.
Fig. 6.
Crystal structure of Xyn11. (A) Overall folding as an (β/α)8 barrel (TIM-barrel) architecture, represented in green (helices) and violet (strands). The most relevant loops that participate in the substrate binding and the catalytic residues are colored in orange. (B) The active site tunnel with a heptaxylose molecule, modelled from structural superimposition with its complex from Caldicellulosiruptor bescii Xyn10C (PDB code 5OFK). (C) Proposed atomic interactions of heptaxylose at Xyn11 active site. The catalytic residues are colored in orange, while the non-conserved residues are colored in yellow. Polar interactions are marked in dashed lines. (D) Detail of the reducing-end moiety of the active site, showing two glycerol molecules (blue) trapped in the crystals, and the three aromatic residues shaping a pocket in the tunnel. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
According to its primary structure, the closest homologues of Xyn11 are thermoresistant multidomain proteins from Caldicellulosiruptor species (N-terminal domain of CbXyn10C-Cel48B [24], PDB code 5OFJ, and GH10 domain of WP_045175321 [49], PDB code 6D5C, showing 55% sequence identity, both presenting the same β-hairpin observed in Xyn11. Next homologues are two intracellular xylanases, IXT6 from Geobacillus stearothermophilus [50], PDB code 1 N82, with 50% identity, and the mesophilic Xyn10B from Paenibacillus barcinonensis [51], PDB code 3EMC, showing 44% identity. These enzymes show very similar topology, with equivalent L4, L7 and L8 loops both in length and position.
Despite fairly conserved topology, loops L7 and L8 are highly variable in sequence (Fig. 7). Therefore, these loops shape very different active site crevices. To fully depict the Xyn11 active site, the binding mode of an heptaxylose has been inferred from structural superimposition of its complex from Caldicellulosiruptor bescii Xyn10C [43], PDB code 5OFK, onto the Xyn11 coordinates (Fig. 6B–D). As it is observed in Fig. 6, Xyn11 seems able to accommodate at least seven units of xylose spanning its catalytic tunnel. The most peculiar feature of Xyn11 is the presence at the non-reducing end of three consecutive Phe residues (Phe312, Phe313 and Phe314) at L8 (Fig. 7) that define a narrow entrance to the catalytic channel. Only the central Phe313 is conserved in homologues, which have Ser/Thr and Pro at the other two positions. This feature places Phe313 in a more prominent position within a narrower cavity and suggests a potential subsite −3, by hydrophobic interaction to Phe312, not seen in the Xyn11 analogues. Apart from this hydrophobic wall, subsite −2 is well-characterized by the presence of a great amount of hydrogen bonds at the opposite wall with a number of very conserved residues. Thus, and according to the complex model, O2 for the xylose would interact with Trp301 and Glu50, O3 with Glu50 and Asn51, O4 would be stabilized by Trp301 and Lys54, which also form a hydrogen bond with O5. Furthermore, the xylose bound at subsite −1 would also present polar interactions to conserved residues at the same wall of the active site, i.e. to Lys54 (O3), His91 (O2 and O3), and Asn143 and Gln220, (O2).
Fig. 7.
Sequence variability of loops L7 and L8. Sequences are of xylanases CbXynb10C from C. bescii (5OFJ), GH10 module from C. danielli (6D5C), IXT6 from G. stearothermophilus (1N82), and Xyn10B from P. barcinonensis (3EMC). (ESPript - http://espript.ibcp.fr).
The xylose bound at subsite + 1 is mainly fixed by stacking to Tyr189 and hydrophobic interaction to Trp309 and Phe313, all conserved residues, and lacking direct polar interactions as in other xylanases. However, in Xyn11, the presence of the pair Phe313-Phe314 narrow the cavity at this subsite + 1 and, more important, fixes the conformation of the Arg260 side-chain in a very restrained position pointing to the inner part of the cavity, in which it could be hydrogen linked to O2 and O3 of xylose bound at subsite + 2. As a consequence, Xyn11 does not present at this subsite + 2 the pocket observed in its homologues that seems able to allocate a putative xylan decoration [51]. Furthermore, Ser259, a position in which most analogues present an aromatic residue with a protruding side-chain that makes a more restricted subsite + 3, precedes Arg260, unique to Xyn11 at loop L7 (Fig. 7). Thus, the reducing-end of the Xyn11 tunnel exhibit rather open subsites from + 3 that are markedly different to that observed in its homologues. Only the bifurcated polar link from Asn190 to the xyloses bound at subsite + 2 (O5) and + 3 (O3) is conserved.
A last interesting feature of the channel is the presence of a small pocket created by a cluster of three aromatic residues located at the end of the tunnel, Phe192, Phe193 and Trp231, only Phe193 being conserved (Fig. 6D). Interestingly, a glycerol molecule from the cryoprotectant has been trapped within this pocket in our crystals, suggesting a putative role in accommodating a potential O3 substituent at the succeeding xylose unit, which would represent a putative subsite + 5. As it is shown in Fig. 6D, an additional glycerol molecule has been trapped in a groove close to the free O2 and O3 hydroxyls of xylose at subsite + 3. The particular shape of both cavities might reflect a specific pattern of xylan decorations that would be recognized and bound at subsite + 3 and + 5.
In summary, Xyn11, as its closest thermoresistant homologues, is able to degrade xylan, and presents 6–7 subsites (from −2 to + 4/+5) for binding this polysaccharide. However, inspection of its active site crevice reveals that substitutions are not allowed at −2 and −1, nor at + 2 and + 4. Therefore, two consecutive unsubstituted xylose units are required to bind at positions −2, −1, for hydrolysis to occur, and a subsequent sequence of alternate substituted/unsubstituted xyloses is envisaged from subsite + 1. This result is in accordance with the cleavage pattern observed in Fig. 5. The especial shape of the cavities described above may match the chemical structure of its natural substrate.
3.4. Structural basis of Xyn11 extremophilic properties
A great effort has been directed to disclose the molecular basis of protein adaptability to extreme conditions [52], [53], [54], [55]. The general agreement is that this ability results from the cumulative effect of multiple stabilizing factors that may be present to a different degree in each extremophilic protein. Nevertheless, despite the ample variation observed, some trends can be observed between the sequences and structures of extremophilic vs their mesophilic orthologues.
In order to explain the extremophilic properties of Xyn11, its primary and tertiary structure was compared to other family GH10 xylanases, including thermoresistant CbXyn10C (PDB code 5OFJ) [49] and three mesophiles: PbXynB from Paenibacillus barcinonensis; (3EMC) [51], CmXyn10B from Cellvibrio mixtus; (2CNC) [53] and SoXyn10A from Streptomyces olivaceoviridis (1V6Y) [56]. Several features analyzed in Xyn11 and its homologues are summarized in Tables 1 and 2.
Table 1.
Analysis of amino acid composition. Percentage of Asn, Gln, Ser, Thr, Pro, Met, Cys, Glu, Arg and Lys in Xyn11, CbXyn10C from Caldicellulosiruptor bescii (5OFJ), XynB from Paenibacillus barcinonensi (3EMC), CmXyn10B from Cellvibrio mixtus (2CNC) and SoXyn10A from Streptomyces olivaceoviridis (1V6Y). In each column, the two first-ranked proteins are shown in bold type, underlined number indicate the most favorable value.
| AA
Composition (%) |
|||||
|---|---|---|---|---|---|
| Asn + Gln | Ser + Thr | Pro | Met + Cys | Glu + Arg + Lys | |
| Xyn11 | 7.4 | 10.0 | 4.9 | 3.0 | 19.0 |
| CbXyn10C | 10.0 | 13.0 | 4.1 | 3.3 | 17.1 |
| PbXynB | 9.0 | 10.8 | 3.3 | 3.0 | 18.4 |
| CmXyn10B | 7.2 | 9.6 | 3.4 | 3.1 | 18.6 |
| SoXyn10A | 10.8 | 11.8 | 2.8 | 4 | 13.9 |
Table 2.
Analysis of the atomic interactions and packing. Number of hydrophobic and aromatic interactions, main chain-main chain hydrogen bonds and ionic interactions in Xyn11, CbXyn10C from Caldicellulosiruptor bescii (5OFJ), XynB from Paenibacillus barcinonensi (3EMC), CmXyn10B from Cellvibrio mixtus (2CNC) and SoXyn10A from Streptomyces olivaceoviridis (1V6Y). Compactness and percentage of exposed residues in the accessible surface area are also computed and shown. In each column, the two first-ranked proteins are shown in bold type, underlined number indicate the most favorable value.
| Atomic
Interactions and Packing |
||||||
|---|---|---|---|---|---|---|
| HYDROPHOBIC | AROMATIC | HYDROGEN BONDS (mch-mch) | IONIC | COMPACTNESS | EXPOSED CHARGED ASA (%) | |
| Xyn11 | 381 | 32 | 449 | 47 | 42.4 | 21.8 |
| CbXyn10C | 361 | 21 | 497 | 39 | 40.6 | 19.1 |
| PbXynB | 323 | 24 | 428 | 38 | 42.7 | 25.1 |
| CmXyn10B | 347 | 27 | 419 | 56 | 43.1 | 21.4 |
| SoXyn10A | 274 | 18 | 439 | 34 | 37.9 | 16.2 |
As a general rule, extremophilic proteins contain low number of residues that decompose easily, such as hydroxy amino acids: Ser and Thr [57], amino acids with an amide terminal group: Asn, Gln [58] or sulfur containing amino acids: Cys, Met [57]. Accordingly, Xyn11 complies this rule (Table 1). On the other hand, the relative abundance of Pro observed in Xyn11, compared with its homologues, can been related to increased stabilization provided by a higher chain rigidity that reduces fluctuation of the secondary structure and helps to keep the proper folding at high temperature [59]. Finally, an increased number of charged residues, meaning an increment in the ion pair interactions, derive in thermostability [58], [60]. Thus, Xyn11 presents the highest value of Lys, Arg and Glu compared with the homologues.
Considering the tertiary structure (Table 2), hydrophobic interactions have been reported to play a key role in protein folding [53]. Thus, the buried hydrophobic interactions in the core of the protein increase the van der Waals contacts, reducing the exposure of hydrophobic residues to solvent and promoting rigidity [53]. In addition, aromatic clusters have also been commonly considered to contribute to the extreme stability of the proteins, resulting in internal stacking interactions that enhance the stability of the protein [61]. In this respect, Xyn11 presents the highest number of both, hydrophobic and aromatic interactions. Whereas Xyn11 compactness is similar to its mesophilic analogues, its accessible surface area (ASA) is higher. Hydrogen bonding is correlated with an increase in rigidity in the core of the protein [62] and is considered one of the major contributors to extreme-stability, especially when considering the main chain-main chain interactions [53], [63]. Theoretical studies show the stabilizing effect of electrostatic interactions and salt-bridges at high temperature [64]. Again, Xyn11 presents a high number of hydrogen bonds and ionic interactions.
It is worth noting that, even when Xyn11 is first-ranked in only 5 out of 11 stabilizing parameters that we have analyzed (Table 1, Table 2), it is one of the two best proteins in all cases, revealing that its molecular structure is well suited to stand denaturing conditions. All the atomic interactions considered in this work have been previously identified as molecular mechanism underlying resistance in both, thermophilic and also alkaliphilic proteins [53], which explains not only Xyn11 thermal resistance but also its activity at high pH. Furthermore, the increased number of atomic interactions observed in Xyn11 must derive in protein rigidity, which is the key to high-temperature adaptation by preventing unfolding. In this context, Xyn11 presents a markedly low chain flexibility, as deduced from the analysis of the atomic B factors values along the polypeptide chain that is shown in Fig. 8A.
Fig. 8.
Putative structural elements of Xyn11 thermoresistance. (A) Crystallographic atomic B factors in the polypeptide chain shown in rainbow code, from low (blue) to high (red) values. (B) Cation- π interactions stabilizing the β-hairpin present in loop L2. A relevant hydrogen bond is represented as dashed line. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Lastly, an interesting feature in Xyn11 structure is the long loop L2 (Fig. 6A), connecting β2 to α2, also present in close homologues from Caldicellulosiruptor species (xylanases from C. bescii, PDB code 5OFK and C. danielli, PDB code 6D5C). This loop is making a β-hairpin which is protruding from the (α/β)8 barrel that, nevertheless is stabilized by two cation- π interactions keeping a close packing of L2 (Fig. 8B). Thus, Arg61 is stacking to Trp102, located at L3, while Arg131, at the end of helix α3, is stacking to Tyr69. Only the last interaction is conserved in the other thermoresistant homologues. Cation-π interactions have been described as another form of electrostatic interaction that make an important contribution to protein stability, which appears to increase at higher temperatures [65]. Thus, Xyn11 might have developed additional local molecular mechanisms that combined to the above parameters make a very resistant protein.
4. Conclusions
The comprehensive bioinformatics screening of the GH10 family reported in this study represents a powerful methodological approach that allowed the identification and characterization of xylanases active at extreme conditions of pH and temperature. The enzyme with best performance, Xyn11, corresponds to a xylanase from the bacterium Pseudothermotoga thermarum. The enzyme shows an exceptional level of activity at 90 °C and pH 10.5. Fusion of a carbohydrate binding module (CBM2) to Xyn11 further increased its activity at extreme conditions. In addition to Xyn11, two other xylanases, Xyn10 and Xyn13 showed activity high at extreme conditions. Interestingly, these enzymes differ in their cleavage pattern on xylan derived substrates, yielding different proportions of xylose, xylobiose and xylotriose as final products, which may have practical consequences from a biotechnological point of view. The crystallographic resolution at 1.8 Å of Xyn11 structure provides an explanation of its function at extreme conditions. Not surprisingly, qualitatively, the atomic interactions responsible for Xyn11 resistance are the same known to sustains protein stability, namely hydrogen bonds, ion pairs, hydrophobic and aromatic interactions. However, the number and distribution in which these interactions appear and its extremophilic enzyme properties makes Xyn11 an outstanding case for study.
5. Accesion code
PDB 7NL2
CRediT authorship contribution statement
David Talens-Perales: Formal analysis, Investigation. Elena Jiménez-Ortega: Investigation. Paloma Sánchez-Torres: Investigation. Julia Sanz-Aparicio: Supervision, Funding acquisition. Julio Polaina: Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded as part of the European Project WOODZYMES, by the Bio Based Industries Joint Undertaking, under the European Union’s Horizon 2020 research and innovation program (Grant Agreement H2020-BBI-JU-792070) and by grants BIO2016-76601-C3-3-R and PID2019-105838RB-C33 from the Spanish Government.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.05.004.
Contributor Information
Julia Sanz-Aparicio, Email: xjulia@iqfr.csic.es.
Julio Polaina, Email: jpolaina@iata.csic.es.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Kohli I., Joshi N.C., Mohapatra S., Varma A. Extremophile - an adaptive strategy for extreme conditions and applications. Curr Genomics. 2020;21(2):96–110. doi: 10.2174/1389202921666200401105908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krüger A., Schäfers C., Schröder C., Antranikian G. Towards a sustainable biobased industry - highlighting the impact of extremophiles. N Biotechnol. 2018;40(Pt A):144–153. doi: 10.1016/j.nbt.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Walia A., Guleria S., Mehta P., Chauhan A., Parkash J. Microbial xylanases and their industrial application in pulp and paper biobleaching: a review. 3 Biotech. 2017;7(1):11. doi: 10.1007/s13205-016-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar V., Marín-Navarro J., Shukla P. Thermostable microbial xylanases for pulp and paper industries: trends, applications and further perspectives. World J Microbiol Biotechnol. 2016;32(2):34. doi: 10.1007/s11274-015-2005-0. [DOI] [PubMed] [Google Scholar]
- 5.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. The Carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(D1):D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verma D., Satyanarayana T. Cloning, expression and applicability of thermo-alkali-stable xylanase of Geobacillus thermoleovorans in generating xylooligosaccharides from agro-residues. Bioresour Technol. 2012;107:333–338. doi: 10.1016/j.biortech.2011.12.055. [DOI] [PubMed] [Google Scholar]
- 7.Paës G., Berrin J.-G., Beaugrand J. GH11 xylanases: structure/function/properties relationships and applications. Biotechnol Adv. 2012;30(3):564–592. doi: 10.1016/j.biotechadv.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Shi H., Zhang Y.u., Li X., Huang Y., Wang L., Wang Y.e. A novel highly thermostable xylanase stimulated by Ca2+ from Thermotoga thermarum: cloning, expression and characterization. Biotechnol Biofuels. 2013;6(1):26. doi: 10.1186/1754-6834-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paës G., O’Donohue M.J. Engineering increased thermostability in the thermostable GH-11 xylanase from Thermobacillus xylanilyticus. J Biotechnol. 2006;125(3):338–350. doi: 10.1016/j.jbiotec.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 10.Li H.e., Kankaanpää A., Xiong H., Hummel M., Sixta H., Ojamo H. Thermostabilization of extremophilic Dictyoglomus thermophilum GH11 xylanase by an N-terminal disulfide bridge and the effect of ionic liquid [emim]OAc on the enzymatic performance. Enzyme Microb Technol. 2013;53(6-7):414–419. doi: 10.1016/j.enzmictec.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Bai W., Cao Y., Liu J., Wang Q., Jia Z. Improvement of alkalophilicity of an alkaline xylanase Xyn11A-LC from Bacillus sp. SN5 by random mutation and Glu135 saturation mutagenesis. BMC Biotech. 2016;16(1) doi: 10.1186/s12896-016-0310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talens-Perales D., Sánchez-Torres P., Marín-Navarro J., Polaina J. In silico screening and experimental analysis of family GH11 xylanases for applications under conditions of alkaline pH and high temperature. Biotechnol Biofuels. 2020;13(1):1–15. doi: 10.1186/s13068-020-01842-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talens-Perales D., Górska A., Huson D.H., Polaina J., Marín-Navarro J., Mallela K. Analysis of domain architecture and phylogenetics of family 2 glycoside hydrolases (GH2) PLoS ONE. 2016;11(12):e0168035. doi: 10.1371/journal.pone.0168035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones D.R., Thomas D., Alger N., Ghavidel A., Inglis G.D., Abbott D.W. SACCHARIS: an automated pipeline to streamline discovery of carbohydrate active enzyme activities within polyspecific families and de novo sequence datasets. Biotechnol Biofuels. 2018;11(1):1–15. doi: 10.1186/s13068-018-1027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015;32:268–274. [DOI] [PMC free article] [PubMed]
- 16.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer E, Hirsh L, Paladin L, Piovesan D, Tosatto S, Finn RD. The Pfam protein families database in 2019. Nucleic Acids Res 2019;47(D1):D427–D432. [DOI] [PMC free article] [PubMed]
- 17.Sievers F., Higgins D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27(1):135–145. doi: 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huson DH, Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol 2012;61:1061–1067. [DOI] [PubMed]
- 19.Käll L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server, Nucleic Acids Res 2007;35(Web Server issue):W429–W432. [DOI] [PMC free article] [PubMed]
- 20.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabsch W. Xds. Acta Crystallogr Sect D Biol Crystallogr 2010;66(Pt 2):125–132. [DOI] [PMC free article] [PubMed]
- 22.Evans P.R. An introduction to data reduction: Space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr. 2011;67(4):282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lebedev A.A., Vagin A.A., Murshudov G.N. Model preparation in MOLREP and examples of model improvement using X-ray data. Acta Crystallogr D Biol Crystallogr. 2007;64(1):33–39. doi: 10.1107/S0907444907049839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu Y., Tu T., Penttinen L., Xue X., Wang X., Yi Z. Insights into the roles of non-catalytic residues in the active site of a GH10 xylanase with activity on cellulose. J Biol Chem. 2017;292(47):19315–19327. doi: 10.1074/jbc.M117.807768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(12 I):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.DeLano W.L. Pymol: an open-source molecular graphics tool. CCP4 Newslett Protein Crystallogr. 2002;40(1):82–92. [Google Scholar]
- 28.Madden T. The NCBI Handbook [Internet] 2nd ed. National Center for Biotechnology Information (US); 2013. The BLAST sequence analysis tool. [Google Scholar]
- 29.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42(W1):W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holm L., Laakso L.M. Dali server update. Nucleic Acids Res. 2016;44(W1):W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilkins M.R., Gasteiger E., Bairoch A., Sanchez J.C., Williams K.L., Appel R.D. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol (Clifton N.J.) 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 32.Tina K.G., Bhadra R., Srinivasan N. PIC: protein interactions calculator. Nucleic Acids Res. 2007;35(SUPPL.2):473–476. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willard L., Ranjan A., Zhang H., Monzavi H., Boyko R.F., Sykes B.D. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 2003;31(13):3316–3319. doi: 10.1093/nar/gkg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson P.E., Joshi M.D., Tomme P., Kilburn D.G., McIntosh L.P. Structure of the N-terminal cellulose-binding domain of Cellulomonas fimi CenC determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1996;35(45):14381–14394. doi: 10.1021/bi961612s. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura T., Mine S., Hagihara Y., Ishikawa K., Ikegami T., Uegaki K. Tertiary structure and carbohydrate recognition by the chitin-binding domain of a hyperthermophilic chitinase from Pyrococcus furiosus. J Mol Biol. 2008;381(3):670–680. doi: 10.1016/j.jmb.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Hirabayashi J., Arai R. Lectin engineering: the possible and the actual. Interface Focus. 2019;9(2):20180068. doi: 10.1098/rsfs.2018.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chu Y., Hao Z., Wang K., Tu T., Huang H., Wang Y. The GH10 and GH48 dual-functional catalytic domains from a multimodular glycoside hydrolase synergize in hydrolyzing both cellulose and xylan. Biotechnol Biofuels. 2019;12(1) doi: 10.1186/s13068-019-1617-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Notenboom V., Boraston A.B., Kilburn D.G., Rose D.R. Crystal structures of the family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A in native and ligand-bound forms. Biochemistry. 2001;40(21):6248–6256. doi: 10.1021/bi0101704. [DOI] [PubMed] [Google Scholar]
- 39.Sakka M., Higashi Y., Kimura T., Ratanakhanokchai K., Sakka K. Characterization of Paenibacillus curdlanolyticus B-6 Xyn10D, a xylanase that contains a family 3 carbohydrate-binding module. Appl Environ Microbiol. 2011;77(12):4260–4263. doi: 10.1128/AEM.00226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: John M. Walker (Ed.), The Proteomics Protocols Handbook, Humana Press; 2005, p. 571–607.
- 41.Shi H., Zhang Y.u., Zhong H., Huang Y., Li X., Wang F. Cloning, over-expression and characterization of a thermo-tolerant xylanase from Thermotoga thermarum. Biotechnol Lett. 2014;36(3):587–593. doi: 10.1007/s10529-013-1392-2. [DOI] [PubMed] [Google Scholar]
- 42.Wang K., Cao R., Wang M., Lin Q., Zhan R., Xu H. A novel thermostable GH10 xylanase with activities on a wide variety of cellulosic substrates from a xylanolytic Bacillus strain exhibiting significant synergy with commercial Celluclast 1.5 L in pretreated corn stover hydrolysis. Biotechnol Biofuels. 2019;12(1):1–13. doi: 10.1186/s13068-019-1389-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.An J., Xie Y., Zhang Y., Tian D., Wang S., Yang G. Characterization of a thermostable, specific GH10 xylanase from Caldicellulosiruptor bescii with high catalytic activity. J Mol Catal B: Enzymatic. 2015;117:13–20. [Google Scholar]
- 44.ul Haq I., Akram F. Insight into kinetics and thermodynamics of a novel hyperstable GH family 10 endo-1, 4-β-xylanase (TnXynB) with broad substrates specificity cloned from Thermotoga naphthophila RKU-10T. Enzyme Microb Technol. 2019;127:32–42. doi: 10.1016/j.enzmictec.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 45.ul Haq I., Hussain Z., Khan M.A., Muneer B., Afzal S., Majeed S., Akram F. Kinetic and thermodynamic study of cloned thermostable endo-1,4-β-xylanase from Thermotoga petrophila in mesophilic host. Mol Biol Rep. 2012;39(7):7251–7261. doi: 10.1007/s11033-012-1555-6. [DOI] [PubMed] [Google Scholar]
- 46.Yu Tianyi, Anbarasan Sasikala, Wang Yawei, Telli Kübra, Aslan Aşkın Sevinç, Su Zhengding. Hyperthermostable Thermotoga maritima xylanase XYN10B shows high activity at high temperatures in the presence of biomass-dissolving hydrophilic ionic liquids. Extremophiles: life under extreme conditions. Extremophiles. 2016;20(4):515–524. doi: 10.1007/s00792-016-0841-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amorim Cláudia, Silvério Sara C., Prather Kristala L.J., Rodrigues Lígia R. From lignocellulosic residues to market: production and commercial potential of xylooligosaccharides. Biotechnol Adv. 2019;37(7):107397. doi: 10.1016/j.biotechadv.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 48.Santibáñez Luciana, Henríquez Constanza, Corro-Tejeda Romina, Bernal Sebastián, Armijo Benjamín, Salazar Oriana. Xylooligosaccharides from lignocellulosic biomass: a comprehensive review. Carbohydr Polym. 2021;251:117118. doi: 10.1016/j.carbpol.2020.117118. [DOI] [PubMed] [Google Scholar]
- 49.Conway Jonathan M., Crosby James R., Hren Andrew P., Southerland Robert T., Lee Laura L., Lunin Vladimir V. Novel multidomain, multifunctional glycoside hydrolases from highly lignocellulolytic Caldicellulosiruptor species. AIChE J. 2018;64(12):4218–4228. [Google Scholar]
- 50.Solomon V., Teplitsky A., Shulami S., Zolotnitsky G., Shoham Y., Shoham G. Structure-specificity relationships of an intracellular xylanase from Geobacillus stearothermophilus. Acta Crystallogr D Biol Crystallogr. 2007;63(8):845–859. doi: 10.1107/S0907444907024845. [DOI] [PubMed] [Google Scholar]
- 51.Gallardo Óscar, Pastor F.I. Javier, Polaina Julio, Diaz Pilar, Łysek Robert, Vogel Pierre. Structural insights into the specificity of Xyn10B from Paenibacillus barcinonensis and its improved stability by forced protein evolution. J Biol Chem. 2010;285(4):2721–2733. doi: 10.1074/jbc.M109.064394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.González-Blasco Gracia, Sanz-Aparicio Juliana, González Beatriz, Hermoso Juan A., Polaina Julio. Directed evolution of β-glucosidase A from Paenibacillus polymyxa to thermal resistance. J Biol Chem. 2000;275(18):13708–13712. doi: 10.1074/jbc.275.18.13708. [DOI] [PubMed] [Google Scholar]
- 53.Chakravorty Debamitra, Khan Mohd Faheem, Patra Sanjukta. Multifactorial level of extremostability of proteins: can they be exploited for protein engineering? Extremophiles. 2017;21(3):419–444. doi: 10.1007/s00792-016-0908-9. [DOI] [PubMed] [Google Scholar]
- 54.Panja Anindya S., Maiti S., Bandyopadhyay B. Protein stability governed by its structural plasticity is inferred by physicochemical factors and salt bridges. Sci Rep. 2020;10(1):1822. doi: 10.1038/s41598-020-58825-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie Hefang, Flint James, Vardakou Maria, Lakey Jeremy H., Lewis Richard J., Gilbert Harry J. Probing the structural basis for the difference in thermostability displayed by family 10 xylanases. J Mol Biol. 2006;360(1):157–167. doi: 10.1016/j.jmb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Kaneko Satoshi, Ichinose Hitomi, Fujimoto Zui, Kuno Atsushi, Yura Kei, Go Mitiko. Structure and function of a family 10 β-xylanase chimera of Streptomyces olivaceoviridis E-86 FXYN and Cellulomonas fimi cex. J Biol Chem. 2004;279(25):26619–26626. doi: 10.1074/jbc.M308899200. [DOI] [PubMed] [Google Scholar]
- 57.Ihsanawati, Kumasaka Takashi, Kaneko Tomonori, Morokuma Chihiro, Yatsunami Rie, Sato Takao. Structural basis of the substrate subsite and the highly thermal stability of Xylanase 10B from Thermotoga maritima MSB8. Proteins Struct Funct Genet. 2005;61(4):999–1009. doi: 10.1002/prot.20700. [DOI] [PubMed] [Google Scholar]
- 58.Cambillau Christian, Claverie Jean-Michel. Structural and genomic correlates of hyperthermostability. J Biol Chem. 2000;275(42):32383–32386. doi: 10.1074/jbc.C000497200. [DOI] [PubMed] [Google Scholar]
- 59.Watanabe K., Hata Y., Kizaki H., Katsube Y., Suzuki Y. The refined crystal structure of Bacillus cereus oligo-1, 6-glucosidase at 2.0 Å resolution: structural characterization of proline-substitution sites for protein thermostabilization. J Mol Biol. 1997;269(1):142–153. doi: 10.1006/jmbi.1997.1018. [DOI] [PubMed] [Google Scholar]
- 60.Arrizubieta María Jesús, Polaina Julio. Increased thermal resistance and modification of the catalytic properties of a β-glucosidase by random mutagenesis and in vitro recombination. J Biol Chem. 2000;275(37):28843–28848. doi: 10.1074/jbc.M003036200. [DOI] [PubMed] [Google Scholar]
- 61.Feller G., Gerday C. Psychrophilic enzymes: molecular basis of cold adaptation. Cel Mol Life Sci. 1997;53(10):830–841. doi: 10.1007/s000180050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Auerbach G., Ostendorp R., Prade L., Korndörfer I., Dams T., Huber R. Lactate dehydrogenase from the hyperthermophilic bacterium Thermotoga maritima: The crystal structure at 2.1 Å resolution reveals strategies for intrinsic protein stabilization. Structure. 1998;6(6):769–781. doi: 10.1016/s0969-2126(98)00078-1. [DOI] [PubMed] [Google Scholar]
- 63.Kollman Peter A., Allen Leland C. Theory of the hydrogen bond. Chem Rev. 1972;72(3):283–303. [Google Scholar]
- 64.Panja Anindya Sundar, Bandopadhyay Bidyut, Maiti Smarajit, Bhattacharjya Surajit. Protein thermostability is owing to their preferences to non-polar smaller volume amino acids, variations in residual physico-chemical properties and more salt-bridges. PLoS ONE. 2015;10(7):e0131495. doi: 10.1371/journal.pone.0131495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prajapati R.S., Sirajuddin M., Durani V., Sreeramulu S., Varadarajan R. Contribution of cation-pi interactions to protein stability. Biochemistry. 2006 Dec 19;45(50):15000–15010. doi: 10.1021/bi061275f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.