Abstract
Background & aims
Obesity is associated with low grade systemic inflammation and insulin resistance. Although metabolic and immunological changes may contribute to the increased risk for COVID-19 mortality in obese, little is known about the impact of obesity in the lungs of patients with COVID-19.
Methods
We analyzed gene expression profiles of autopsy lungs of a cohort of 14 COVID-19 patients and 4 control individuals. Patients were divided into 3 groups according to their comorbidities: hypertension, type 2 diabetes (T2D) and obesity. We then identified the molecular alterations associated with these comorbidities in COVID-19 patients.
Results
Patients with only hypertension showed higher levels of inflammatory genes and B-cell related genes when compared to those with T2D and obesity. However, the levels of IFN-gamma, IL22, and CD274 (a ligand that binds to receptor PD1) were higher in COVID-19 patients with T2D and obesity. Several metabolic- and immune-associated genes such as G6PD, LCK and IL10 were significantly induced in the lungs of the obese group.
Conclusion
Our findings suggest that SARS-CoV-2 infection in the lungs may exacerbate the immune response and chronic condition in obese COVID-19 patients.
Keywords: COVID-19, Obesity, Leptin, Inflammation, Lung, Comorbidities
Abbreviations: BMI, Body Mass Index; COVID-19, Coronavirus Disease 2019; DEGs, Differentially Expressed Genes; FDR, False Discovery Rate; GEO, Gene Expression Omnibus; T2D, Type II diabetes; VL, Viral Load
1. Introduction
Chronic inflammation and dysfunction of immune response emerge as consequences of obesity, and lead to increase in the susceptibility to pulmonary diseases [1]. Although obesity is a key comorbidity associated with severe COVID-19 [2], little is known about its impact in the lungs of COVID-19 patients.
Here, we analyzed the transcriptional profile of autopsy lungs from obese and non-obese patients with COVID-19. The expression of several immune-, metabolic- and obesity-related genes was altered in obese patients compared to control individuals. We developed an online database to display all of the results. Our work highlights the impact of nutrition on the immune response to SARS-CoV-2 in the lungs.
2. Material & methods
2.1. Targeted transcriptome analysis of post-mortem lung samples
Publicly available data from 18 individuals were downloaded using the GEOquery R package (GEO accession number: GSE151764) [3]. The primary cause of death in all 18 individuals was respiratory failure, and sometimes multi-organ failure [3]. Laboratory results of COVID-19 patients showed high levels of circulating markers of inflammation: IL-6 (mean) = 5774 ng/l, C-reactive protein (mean) 216,36 mg/l [3]. Levels of circulating markers of glucose metabolism were not measured. The complete clinicopathological data of the 18 patients, including information about therapy, cause of death, thromboembolic events in the lungs, and disseminated intravascular coagulation can be found in Table S1 (which was adapted from [3]).
We summed the count expression of genes from technical replicates and performed differential expression analysis using the edgeR package. The cutoff for identifying the differentially expressed genes (DEGs) were |log2 fold-change| > 0.5849 and FDR < 0.05. We also correlated body mass index (BMI), SARS-CoV-2 viral load (in log10) and age of COVID-19 patients with gene expression. We then used the significant Pearson correlations (P < 0.05) to create a network using Cytoscape. The online database was created in python using streamlit, pandas and altair libraries for data manipulation and visualization.
3. Results
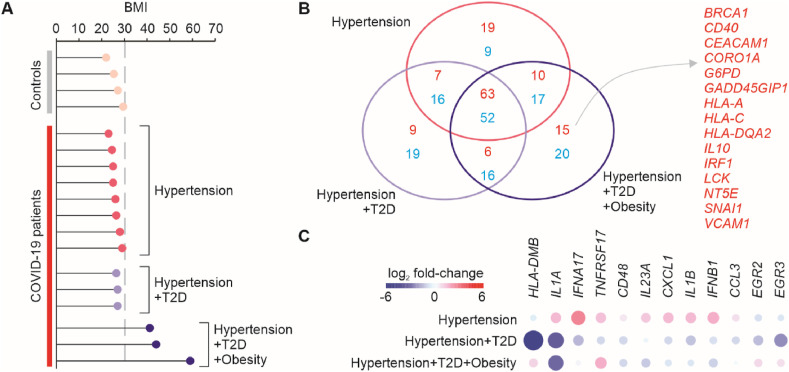
Recently, Nienhold et al. [3] described the immunopathological profiles in autopsy lungs of COVID-19 patients. However, they did not characterize the expression profiles according to the comorbidities associated with the severe form of the disease. We divided the COVID-19 patients into 3 groups: Hypertension (N = 8), Hypertension + T2D (N = 3), Hypertension + T2D + Obesity (N = 3) (Fig. 1 A). Given the limited number of samples, we considered overweight patients as non-obese patients. We then identified the differentially expressed genes between each one of these 3 groups and the control individuals not infected with SARS-CoV-2 (N = 4). To display the results, we developed an online database which is located at https://covid19.sysbio.tools/obesity. Most of the genes up- and down-regulated in COVID-19 when compared to controls were shared by all 3 groups (Fig. 1B). However, some of the genes, including IL1A, IL1B, IL23A, IFNA17, and IFNB1 were up-regulated in the hypertension group but down-regulated in either the Hypertension + T2D or Hypertension + T2D + Obesity groups (Fig. 1C). The expression of 15 genes was exclusively induced in patients with Hypertension + T2D + Obesity relative to controls, even including both normal weight and overweight controls (Fig. 1B). These included several genes associated with leptin (LCK, IRF1, VCAM1, SNAI1), MHC-class I and II (HLA-A, HLA-C, HLA-DQA2), glucose metabolism (G6PD), and immune regulatory functions (IL10, CD40, NT5E, CORO1A).
Fig. 1.
Differential gene expression analysis of the lungs of COVID-19 patients compared to controls. (A) Individuals utilized in the analysis grouped according to their comorbidities. The x-axis shows the body mass index (BMI) for each individual. Individuals with a BMI above 30 (grey dashed line) were considered obese. (B) Number of differentially expressed genes (DEGs) of each COVID-19 patient group compared to control. The Venn diagram shows the number of upregulated genes (red) and down-regulated genes (blue) that were shared or unique to each group. (C) DEGs with opposite expression among the groups. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
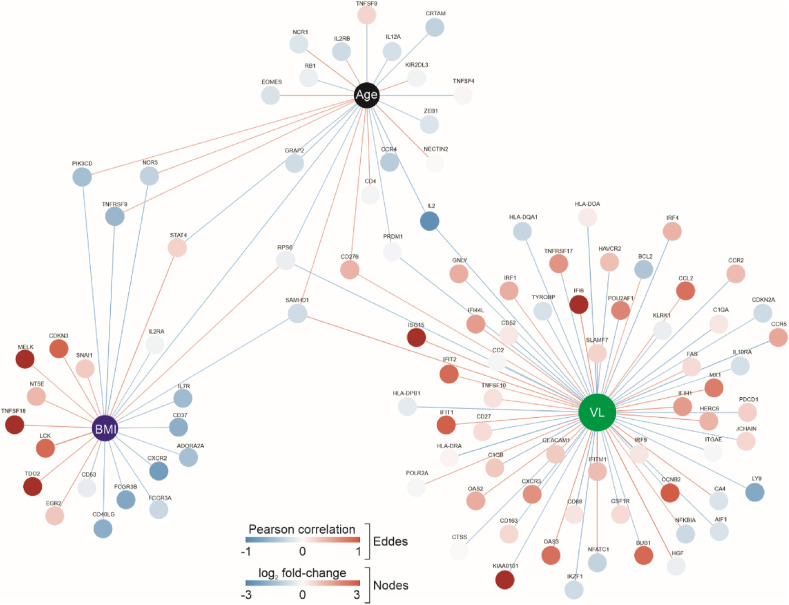
Using only the 14 COVID-19 patients, we then performed a correlation analysis between gene expression and age, SARS-CoV-2 viral load (VL) or BMI (Fig. 2 ). While genes associated with type I interferon were positively correlated with VL, genes related to Interleukin-2 signaling pathway (CD52, RPS6, AIF1, LY9, CTSS, IL2, CD2, KLRK1, GNLY, IRF4, CXCR3, CD27, CCR5, CD68, CCR2) were negatively correlated with VL (Fig. 2). There were 17 genes exclusively associated with BMI. Among them, genes involved with lipid metabolism, insulin signaling and cell cycle/maturation, such as LCK, EGR2, CDKN3 and MELK were positively correlated with BMI (Fig. 2). Some of the genes whose expression was negatively correlated only with BMI were associated with cytokine signaling and immune responses, such as CD40LG, CXCR2, ADORA2A, PIK3CD and CD37, as well as receptors for the Fc region of immunoglobulins gamma (FCGR3A and FCGR3B) (Fig. 2).
Fig. 2.
Network of genes associated with BMI, age and SARS-CoV-2 viral load. The network shows the genes whose expression was correlated with BMI, age and log10 viral load (VL) (P < 0.05). The edges colors represent the direction of correlation (negative correlation in blue and positive correlation in red). The nodes are colored with the log2 fold-change from Hypertension + T2D + Obesity group compared to controls. The size of the nodes is proportional to their degree. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Obesity is a key comorbidity associated with severe COVID-19 [2]. Here, we revealed significant alterations in gene expression in the lungs of obese patients that died from COVID-19 complications [3]. Type-I interferon and interleukin-1 genes were up-regulated only in non-obese and non-T2D patients, suggesting a lower capacity in inducing proper antiviral defense in the lungs of patients with T2D/obesity. Interestingly, genes up-regulated exclusively in the lungs of obese individuals were associated with energy metabolism, antigen presentation and immune regulatory functions, which might suggest an interplay between metabolism and immunity.
Obese people have high levels of leptin. We found that the expression of lymphocyte-specific kinase (LCK) gene and Early Growth Response 2 (EGR2) genes was exclusively up-regulated in obese patients and positively correlated with BMI. In kidney, it has been demonstrated that LCK plays an important role in leptin-induced inflammation [4]. Additionally, Jaedicke et al. showed that leptin up-regulates the expression of EGR2 in primary human monocytes [5]. Together with the fact that leptin dysregulation in the lungs is associated with abnormal ventilation, excessive remodeling, and immune dysregulation seen in pulmonary diseases [6], we think that leptin may play a key role in COVID-19 physiopathology in obese individuals.
Our findings also suggest that neutrophils may migrate less to the lungs of obese COVID-19 patients, compared to non-obese ones. This was supported by the fact that the expression of CXCR2 (a chemokine that mediates neutrophil migration to sites of inflammation), and FCGR3A and FCGR3B genes was negatively correlated with BMI. FCGR3A and FCGR3B are the two subunits of CD16 surface molecule, which is found in neutrophils, as well as in natural killer cells, monocytes, and macrophages. There is increase evidence that neutrophils and Neutrophil Extracellular Traps play an important role in the lungs of COVID-19 patients [7]. Nienhold et al. [3] have estimated the number of neutrophils per lung tissue section by immunohistochemical stains for CD15 and MPO. We found that in 83% of lung tissue sections of obese patients, few or no neutrophils were found, whereas in non-obese patients, the number of tissue sections with few or no neutrophils was around 60%.
Since obesity is associated with an increased risk of developing T2D and hypertension, we must also consider its impact as the driver of the signatures in the present data. However, our correlation analysis was performed taking into account these comorbidities. Also, our online database provides the scientific papers which are related to the genes and to the comorbidities shown in this work.
We recognize the study limitations, which mainly consist in a reduced number of samples and genes, absence of obese people in the healthy control group, and the presence of confounding factors [3], such as smoker status and the drug therapy taken by the COVID-19 patients before death. Indeed, these limitations prevent us to determine others key factors that may interfere to the observed phenomena. However, our results present novel insights about the mechanism and severity of COVID-19 in obese individuals.
5. Conclusion
The lungs of obese patients with COVID-19 display an altered metabolic and immunological gene profile that may explain the association between obesity and the increased risk for the severe COVID-19.
Declaration of competing interest
None of the authors have conflicts of interest to disclose.
Acknowledgements
We are thankful to the participants of the Systems Biology course (02.2020) and the members of Computational Systems Biology Laboratories (CSBL) for the discussions that refined this work, including Allan Rodrigo for his help with the online database.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.clnesp.2021.05.004.
Funding sources
This work was supported by Brazilian National Council for Scientific and Technological Development (grant numbers 313662/2017-7 to H.I.N.; 142058/2020-3 to IHYN); the São Paulo Research Foundation (grant numbers 2018/14933-2 to H.I.N.; 2019/27139-5 to JCSS; 2019/27146-1 to APV; 2020/04579-7 to PMMV); and CAPES (grant number 88882.180020/2018-01 to NYN).
Author contribution
Conceptualization, Investigation, Data Curation and Writing: JCSS, APV, IHYN, NYN, HIN. Software Programming, Formal analysis: JCSS, APV, HIN, AGCM, VS. Database was developed by RA, JG and LD. Resources, Writing Review & Editing, Supervision, Funding acquisition: PMMV, HIN.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Qi Y., Xie M., Wei L., Hou G. Insulin resistance exacerbates lung inflammation in obese patients via PI3K/Akt signaling pathway. Eur Respir J. 2019;54 [Google Scholar]
- 2.Simonnet A., Chetboun M., Poissy J., Raverdy V., Noulette J., Duhamel A., et al. High prevalence of obesity in severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) requiring invasive mechanical ventilation. Obesity. 2020;28(7):1195–1199. doi: 10.1002/oby.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nienhold R., Ciani Y., Koelzer V.H., Tzankov A., Haslbauer J.D., Menter T., et al. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat Commun. 2020;11(1):5086. doi: 10.1038/s41467-020-18854-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim D.H., Park J.W., Jeong H.O., Lee B., Chung K.W., Lee Y., et al. Novel role of lck in leptin-induced inflammation and implications for renal aging. Aging Dis. 2019;10(6):1174–1186. doi: 10.14336/AD.2019.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaedicke K.M., Roythorne A., Padget K., Todryk S., Preshaw P.M., Taylor J.J. Leptin up-regulates TLR2 in human monocytes. J Leukoc Biol. 2013;93(4):561–571. doi: 10.1189/jlb.1211606. [DOI] [PubMed] [Google Scholar]
- 6.Jutant E.M., Tu L., Humbert M., Guignabert C., Huertas A. The thousand faces of leptin in the lung. Chest. 2021;159(1):239–248. doi: 10.1016/j.chest.2020.07.075. [DOI] [PubMed] [Google Scholar]
- 7.Wang J., Li Q., Yin Y., Zhang Y., Cao Y, Lin X., et al. Excessive neutrophils and neutrophil extracellular Traps in COVID-19. Front Immunol. 2020;11:2063. doi: 10.3389/fimmu.2020.02063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.