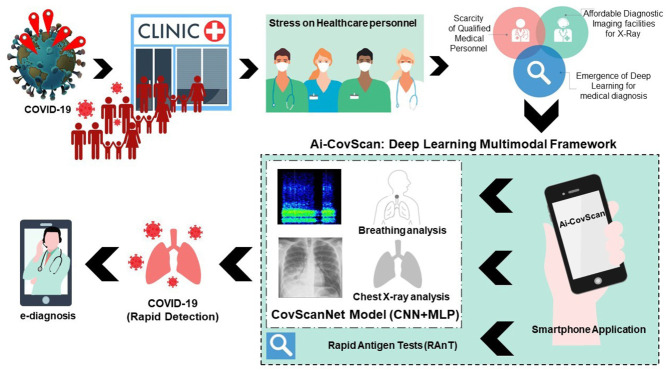
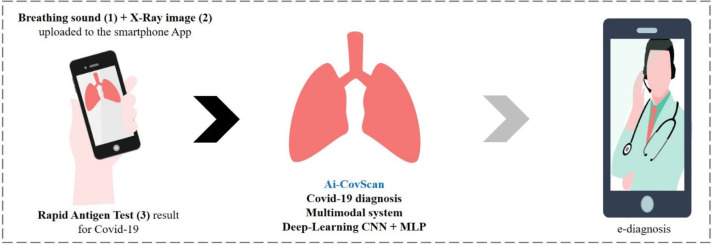
Abstract
Covid-19 has become a deadly pandemic claiming more than three million lives worldwide. SARS-CoV-2 causes distinct pathomorphological alterations in the respiratory system, thereby acting as a biomarker to aid its diagnosis. A multimodal framework (Ai-CovScan) for Covid-19 detection using breathing sounds, chest X-ray (CXR) images, and rapid antigen test (RAnT) is proposed. Transfer Learning approach using existing deep-learning Convolutional Neural Network (CNN) based on Inception-v3 is combined with Multi-Layered Perceptron (MLP) to develop the CovScanNet model for reducing false-negatives. This model reports a preliminary accuracy of 80% for the breathing sound analysis, and 99.66% Covid-19 detection accuracy for the curated CXR image dataset. Based on Ai-CovScan, a smartphone app is conceptualised as a mass-deployable screening tool, which could alter the course of this pandemic. This app’s deployment could minimise the number of people accessing the limited and expensive confirmatory tests, thereby reducing the burden on the severely stressed healthcare infrastructure.
Keywords: Covid-19, CNN, MLP, Chest X-ray images, Breathing sounds, Deep-learning
Graphical abstract
1. Introduction
Covid-19, declared as a global pandemic on 11th March 2020 by the World Health Organisation (WHO) [1], has severely affected the global population and brought the world to a standstill [2], [3], [4]. This airborne disease, known to spread from human to human via droplets and surface contamination, has affected millions from various age brackets. The Centre for Disease Prevention and Control (CDC) has estimated the fatality rate of the disease at 1.4% [5], where persons above the age of seventy-five are affected the most and those below the age of 19 contribute a lower percentage of the total cases [6]. As of 30th December 2020, this zoonotic virus has claimed over 1.8 million lives worldwide, with the US alone accounting for 341,000 deaths [7]. During the first wave of Covid-19, there were no licensed vaccines or therapeutics available. There are several therapeutics in phase III clinical trials and more than 20 vaccines in development against the SARS-CoV-2 [8], [9].
There are several testing methods for Covid-19— nasopharyngeal swab test, Rapid Antigen Test (RAnT), and RT-PCR (reverse transcription-polymerase chain reaction) [10], [11], [12]. An RT-PCR test is a well-established type of NAAT (Nucleic Acid Amplification Test), the current Gold standard for detecting Covid-19 [12]. This test directly detects the viral genome in the collected nasopharyngeal swab sample through a complex sequence of biochemical reactions to convert the RNA to DNA via Reverse Transcription and magnify the complementary DNA strands of the virus. This test indicates Covid-19 infection by detecting the specific genetic material of the virus. Though this test has the highest accuracy and precision, it is scarcely available, expensive, and time-consuming. It also requires expert lab technicians and expensive infrastructural facilities to safely carry out the sample testing protocol [11], [13]. These limitations restrict its use as a widely available confirmatory test, thus opening up an opportunity for other rapid tests to detect Covid-19.
During the pandemic, overwhelming Covid-19 positive cases have resulted in severe stress on existing healthcare facilities owing to a higher daily inflow of Covid-19 patients [14]. With a population of 7.8 billion, the world has witnessed a noticeable deficiency of Covid-19 testing kits, coupled with restricted access to healthcare [14]. Developing and underdeveloped nations worldwide are facing a crisis, where the urgent acquisition of cost-effective diagnostic solutions and novel portable testing mechanisms is required [13], [15].
A few studies have proposed technological tools like smartphone applications to provide e-diagnosis to patients [17], [18], [19]. 60% of the global population use the internet, and 67% possess smartphones [20], making smartphone applications a handy technological tool for the Covid-19 pandemic, as they can provide accessible healthcare and e-diagnosis to large populations from the comfort of their homes [21]. Many Covid-19 specific diagnostic applications are already available for smartphone users [22], [23], [24], [25], [26], [27], [28].
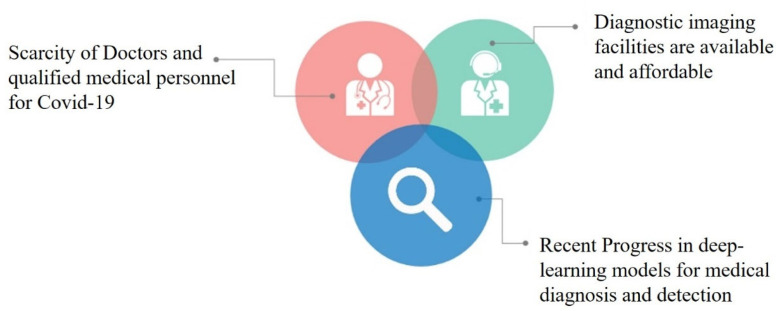
Given the recent progress in deep-learning models for medical diagnosis in the context of the opportunities and challenges posed by Covid-19 is depicted in Fig. 1. Medical image inspection was earlier performed manually by trained radiologists and physicians [16]. With access to organised, labelled, and clean images from available datasets online, these experts envisioned the long-term benefits of utilising computational technology and artificial intelligence for efficient image classification and preliminary medical diagnosis [16], [29]. Recent trends in artificial intelligence and computer-aided diagnosis have led to a proliferation of studies that recognise Deep-Learning (DL) as a useful tool in medical image analysis [29]. The Convolutional Neural Network (CNN), among several deep-learning algorithms, performs quickly and expertly in pattern recognition by eliminating the need for multiple training parameters for the network and pre-processing images. CNN is widely used for visual recognition tasks, as convolutions are performed using deep-learning architectures [30]. CNN has a self-learning capacity that may result in higher classification accuracy with techniques such as transfer learning [29]. Transfer learning further enhances image classification accuracy and efficiency, even while utilising a smaller dataset [31].
Fig. 1.
A depiction of opportunities and deficits in medical diagnosis.
Adapted from [16].
In the context of Covid-19, the use of deep-learning algorithms has led to various preliminary medical diagnostic methods using image analytics to identify abnormalities in chest X-rays, CT Scans, Ultrasounds, and cough sound patterns [31], [32], [33].
1.1. Chest X-ray
The use of X-rays in image analytics is gaining popularity and forms the basis of image classification for training neural networks [32]. Further, X-rays can identify patient attributes and specifics such as gender, bone-age, and diseases [31]. The last two decades have seen a growing trend towards medical image analysis for early detection and diagnosis utilising chest X-ray, CT Scans, and ultrasound [32]. Deep-learning has been chosen as the optimal tool for the classification of medical images obtained from these radiological apparatuses, given their high-resolution image acquisition [16], [31].
1.2. Sound based medical diagnosis
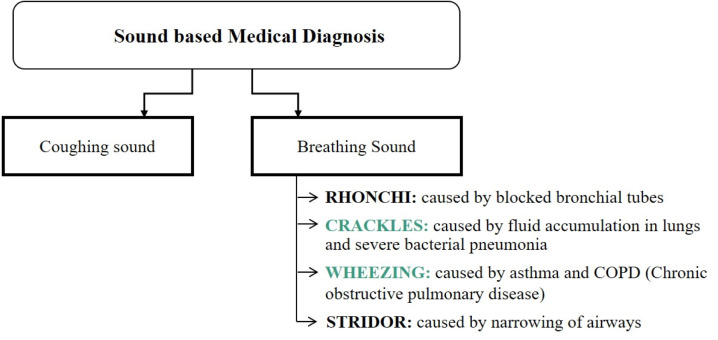
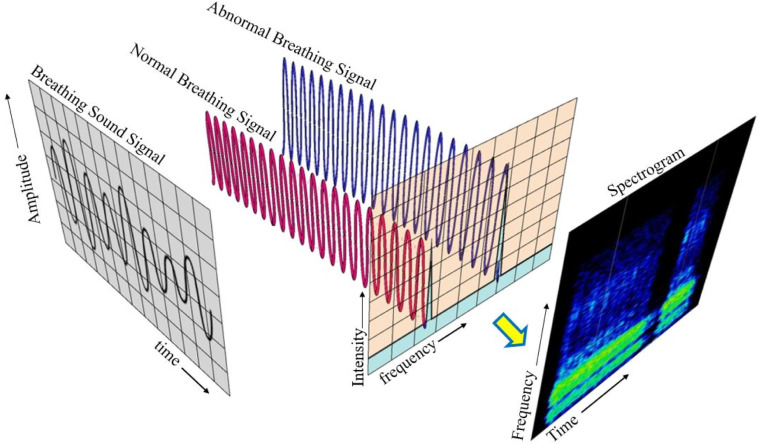
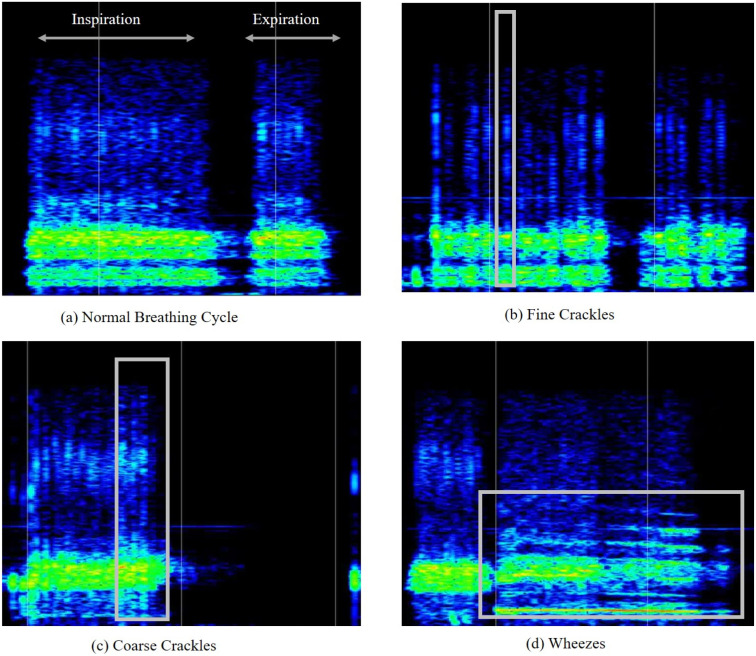
For the sound-based medical diagnosis, a few studies [33], [37], [38], [39], [40] have utilised cough sounds for Covid-19 diagnosis. Medical personnel follow manual methods of breathing sound capture and diagnosis [32]. In a pandemic scenario, manual testing for medical diagnosis is neither feasible nor safe given the considerable inflow of patients who can be the virus’s potential carriers [41]. In these situations, relying on self-diagnosis and telemedicine with preliminary symptom assessment is an alternative to manual testing [41], [42]. Breathing analysis utilises respiratory abnormalities like crackles (coarse and fine) and wheezes of patients as the basis for viral detection over other breathing sound abnormalities, as most Covid-19 patients exhibit pneumonia and asthma, which are linked to crackling and wheezing [43]. A brief overview of sound-based abnormalities utilised for medical diagnosis is listed in Fig. 2.
Fig. 2.
1.3. Rapid antigen test
Doctors recommend using the rapid antigen test for pre-screening purposes [11]; these tests are FDA approved—Food and Drug Administration is a federal organisation in the United States, which promotes health services by regulating pharmaceuticals, vaccines, cosmetics, diagnostic tests, food, and biomedical devices—and provide results in a few minutes, while also protecting others from potential exposure. Rapid antigen tests use nasopharyngeal swabs (from the nose and throat) to collect sample fluid, and immediately detect proteins of SARS-CoV-2 [11]. At 6.16 USD, the Rapid antigen test is economical and can cater to a large population at a given time [44]. The detection method can specifically identify infectious agents while being conducted from the comfort of one’s home. The rapid antigen testing kits, now in abundance, are being sold worldwide through online platforms [45].
Section 2 discusses the related works and the research highlights of this study. The methodology adopted in this paper is discussed in Section 3. Section 4 discusses the methodological framework for a deep-learning-based CovScanNet model. It also elucidates a multimodal decision-making framework named as Ai-CovScan. Section 5 presents the results and validation for the CovScanNet model on Covid-19 datasets. A smartphone app is developed to implement Ai-CovScan. Section 6 discusses the analysis and implementation of the multimodal framework, its advantages, and limitations. Finally, the conclusion and future scope are presented in Section 7.
2. Related works
Existing research recognises the critical role of cough sounds for Covid-19 screening and diagnosis [37]. A study was undertaken, where ten Covid-19 patients’ audio recordings were assessed using digital signal analysis [38]. It was revealed that signal analysis proved the reliability of common abnormalities in breathing sounds such as crackles, vocal resonance and murmurs for Covid-19 detection [38], [39]. The study aimed to fill the gap between medical data and scanning for viral presence [39]. Recently, researchers proposed a scalable screening tool with a three-tiered framework for Covid-19 detection using cough sounds [40]. This three-classifier approach used a deep-learning-based multi-class classifier using spectrogram [40]. The cough detection algorithm differentiates Covid-19 coughs from non-Covid-19 coughs with a reported accuracy of 97.91% [40]. A. Belkasem et al. proposed an early diagnosis tool that records users’ body temperature, coughing and airflow using sensors for symptom identification [45]. The recorded user data is converted into health data and fed into an ML module for further processing and validation. The AI framework identifies and classifies various respiratory illnesses linked to the Covid-19 with a smartphone application that also provides e-diagnosis [45]. Another study developed a model that identifies differences among Covid-19 coughs and non-Covid-19 coughs, and uses a deep-learning algorithm on a medical, demographic dataset of 150 patients’ cough sounds and audio segments. Using digital signal processing (DSP), cough features were re-boosted from natural cough sounds. The reported accuracy of the model was 96.83% [33]. A study collected Mel-Frequency Cepstral Coefficients (MFCC) [46], which used automatic speech recognition (ASR) and deep-learning algorithms [46] to obtain correlation coefficients of individual coughs, breathing sounds, and voices [46]. It is now well established from various studies that heavy cough sounds indicate respiratory illnesses such as Covid-19 that severely affects the lungs.
The primary cause of death due to Covid-19 was identified as pneumonia, a condition in which inflammation and fluid build-up in the lungs lead to difficulty in breathing [19], [47]. A collection of CT scans and chest X-rays could categorise Covid-19 patients’ respiratory abnormalities more accurately; and differentiate amongst mild, moderate and severe cases [48]. Recent trends indicate rapid progress in developing machine learning algorithms to recognise patterns from medical diagnostic images using image analytics. Studies have shown that healthy lungs usually show up dark in an X-ray or CT Scan, while Covid-19 X-ray images show a white haziness [49]. A few studies have used CNN and deep-learning to use chest X-ray analysis to categorise images into four classes—normal, viral-pneumonia, bacterial-pneumonia, and Covid-19 images [2]. Recently, there has been considerable literature [50], [51], [52] on the theme of using chest X-ray images for Covid-19 detection. It has been observed that ‘Deep Convolutional Neural Networks’ are well-recognised by researchers for Covid-19 detection [49]. H. Panwar et al. proposed a ‘nCOVnet’ model using a CNN-based approach for positive or negative detection under 5 s [50]. With a positive detection accuracy of 97% and sensitivity of 97.62%, the model was trained on 142 images of normal X-rays from a Kaggle dataset [53]. The network architecture of COVID-Net, a Deep Convolutional Neural Network model [2] by L. Wang, uses lightweight residual projection expansion [52]. The model recorded a Covid-19 sensitivity of 80% and a Covid-19 positive predictive value (PPV) of 80%. Studies of Inception-v3 models by A Abbas et al. [51] and AI Khan et al. [2] shows the importance of deep-learning and transfer learning for viral detection. The former proposed a CoroNet framework pre-trained on an ImageNet dataset based on ResNet50, Inception-v3 and InceptionResNet V2. An accuracy of 89.5% was reported with lower false negatives and higher recall values. The latter [2] uses a DCNN-based Inception-v3 model to classify chest X-ray images from the GitHub repository [2], [51]. This model has a classification accuracy of more than 98%. Several attempts have been made to detect Covid-19 using CT Scans [54]. A recent study used AI-based image analysis to generate a “corona score” with a 98.2 per cent sensitivity and 92.2 per cent specificity [55]. The findings have identified CT as a valuable tool in detecting and quantifying the disease [50], [51], [54]. [56] proposes the diagnosis of Covid-19 from CXR images using an optimised CNN architecture by automatic tuning of hyper-parameters yielding very high classification accuracy. Deep LSTM model proposed in [57] presents an alternative approach to detecting Covid-19 using MCWS images rather than raw images to obtain high accuracy for 3 class classification. A federated learning framework using VGG16 & ResNet50 as a decentralised data sharing option without compromising data privacy to improve data quality is proposed by Feki et al. [58].
Currently, one of the most significant discussions on Covid-19 detection is the feasibility of using multimodal frameworks. Researchers have followed a CNN-based multimodal approach through transfer learning to detect Covid-19 by pre-processing chest X-ray images [59], CT scans, and ultrasound images [51]. Along with unwanted noise removal, the images are filtered according to type. The study revealed that ultrasound images have a better prediction accuracy (100%) compared to X-ray (86%) and CT scans (84%) [32].
However, there has been little discussion about the procedure of reducing false negatives using transfer learning-based multimodal diagnostic methods. CT scans have been used as a method of viral detection in past studies [50], [51], [54]. However, chest X-rays are better suited towards preliminary diagnosis than CT-Scans considering radiation exposure [31]. CXR machinery is smaller, less complicated and low-cost, and has higher availability worldwide [31], [60]. Using a multimodal framework or combining multiple systems can increase any medical diagnostic method’s reliability compared to using a single framework. The majority of the studies [33], [37], [38], [39], [40] have used cough sounds to indicate the viral disease and compared its results with other cough samples [33]. A smartphone app named ResAppDx is proposed by Moschovis et al. for the detection of respiratory illnesses in children based on sound recordings obtained on the app with a proprietary algorithm named SMARTCOUGH-C 2 [18]. It has successfully been deployed in hospitals as an independent adjudicator for the disease [18]. Although one study compared cough sounds with breathing sounds [46], very few studies [46] have used breathing sounds as an indicator of Covid-19. A multimodal approach—utilising chest X-ray (CXR) images, breathing sound data, and antigen testing to detect Covid-19—has not yet been developed and validated.
This study set out to investigate the development of a multimodal framework for the rapid diagnosis of Covid-19—using CXR images, breathing sound data, and rapid antigen testing (RAnT)—that is deployed using a smartphone application. A deep-learning framework, named CovScanNet, based on Convolutional Neural Network (CNN) and Multi-Layer Perceptron (MLP) algorithm using the transfer-learning technique, is proposed for medical image analysis using CXR images and breathing sound spectrograms. Further, this paper aims to reduce the false negatives in the diagnosis of Covid-19, while also reducing the stress on healthcare infrastructure via e-diagnosis. The primary research highlights of this paper are presented below.
2.1. Research highlights
1. This paper proposes Ai-CovScan—a multimodal framework, including breathing sound analysis, chest X-ray image analysis, and antigen tests for detecting Covid-19—to reduce the false negatives and increase reliability.
2. The Ai-CovScan framework works on a deep-learning model named CovScanNet, which adopts a transfer-learning technique using CNN, where the output from CNN is fed into an MLP model.
3. A breathing sound analysis using spectrograms is proposed where the breathing sounds of patients at home or hospitals”- are recorded, and the percentage of breathing sound abnormalities are identified.
4. A chest X-ray image analysis is performed using a curated dataset, which is further validated.
5. The system is implemented using a smartphone application as a detection tool accessible to a large user base.
3. Materials and methods
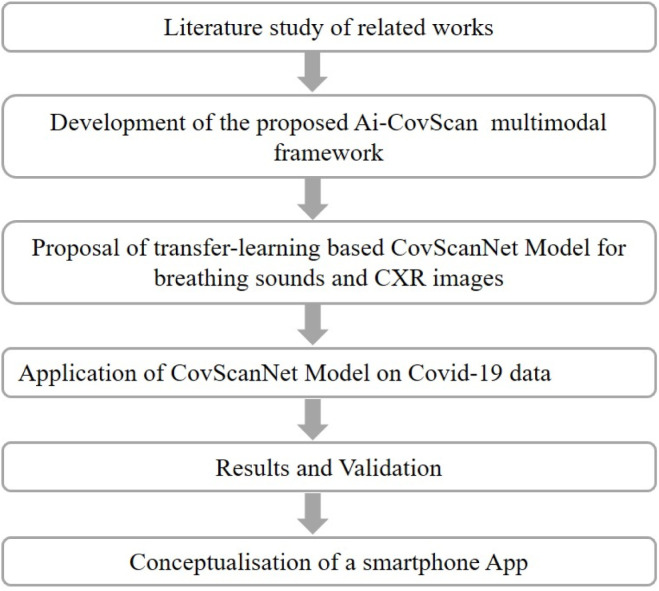
The methodology for this study is presented in Fig. 3. Initially, a comprehensive literature review is conducted to study the related works. A multimodal framework—Ai-CovScan—is developed, with the proposal of a transfer-learning-based CovScanNet model. The CovScanNet is applied to the collected/curated data for Covid-19. The model is validated, and the results are analysed for selected evaluation metrics. Finally, a smartphone app is envisaged to apply the Ai-CovScan framework.
Fig. 3.
Methodology for this study.
3.1. Ai-CovScan framework
3.1.1. Ai-CovScan: System description
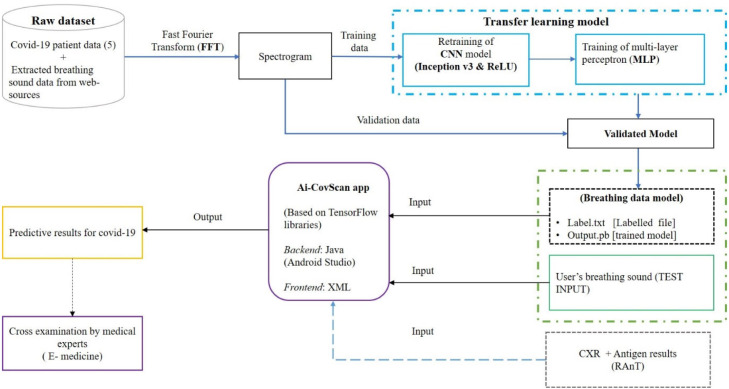
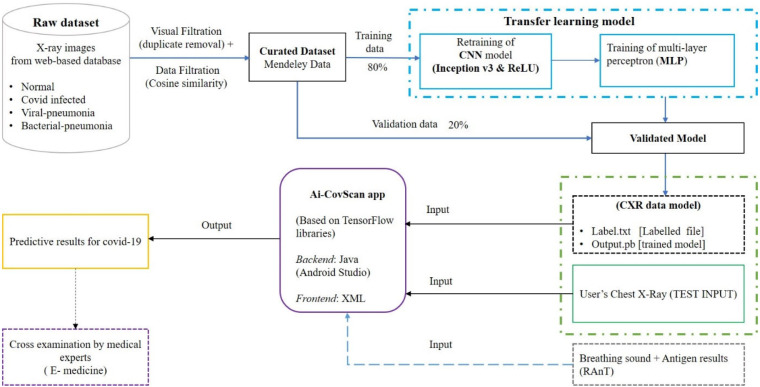
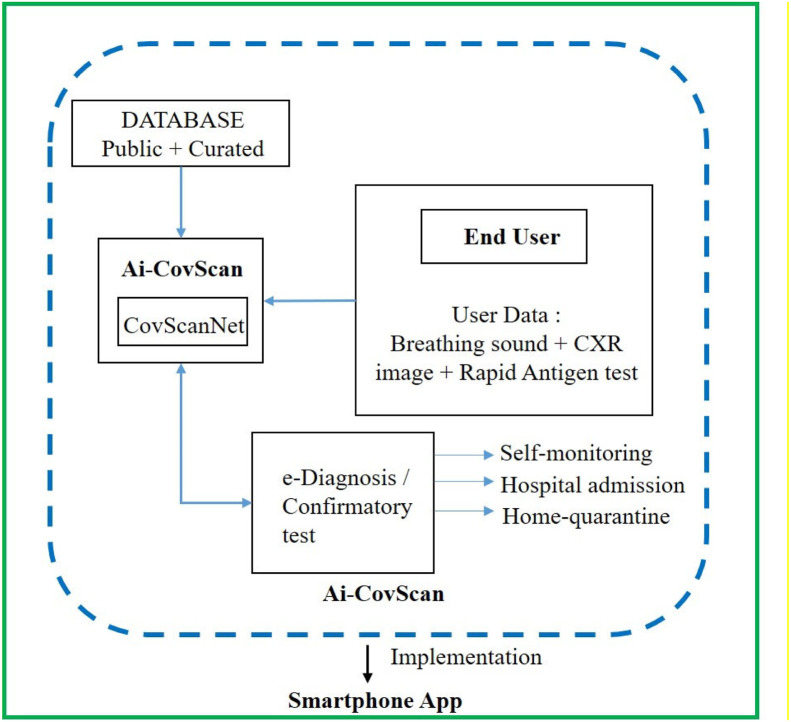
The Ai-CovScan framework is a multimodal approach to find a robust and reliable solution for rapid detection of Covid-19 using breathing sounds, CXR images, and combining it with the results from a disease-specific rapid antigen test. Fig. 4 presents the system description for the Ai-CovScan framework. Ai-CovScan framework has two components: the breathing sound analysis framework (Fig. 5) and the chest X-ray image-analysis framework (Fig. 6). These components exhibit the workflow for data collection, data processing, the development of the transfer learning model (CNN+MLP), validation, and implementation through a smartphone app.
Fig. 4.
System description of Ai-CovScan framework.
Fig. 5.
Breathing sound analysis component of Ai-CovScan framework.
Fig. 6.
X-ray image analysis component of Ai-CovScan framework.
3.1.2. Implementation of Ai-CovScan framework
The Ai-CovScan framework is implemented through a smartphone app named Ai-CovScan. The backend of this app is coded using Java, while the frontend is developed using XML with Android Studio. The prime focus of this app is to provide easy accessibility to any individual as an alternate mode of testing for respiratory diseases, such as the Covid-19, at the comfort of their living premises. For realising this vision, the app implements a three-tier detection model to be directly used by individual users, as shown in Fig. 4.
The data collection, curation (or pre-processing), and processing methodologies for breathing sound and CXR images are discussed in the following Sections 3.2, 3.3. Section 3.4 discusses the application of the rapid antigen test (RAnT) as an additional layer to the Ai-CovScan framework.
3.2. Breathing sound data collection
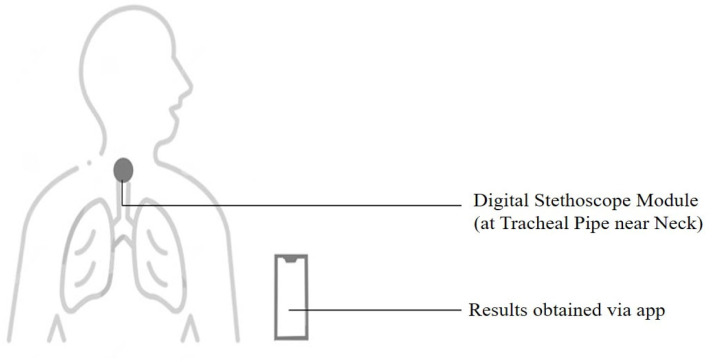
Breathing sounds are promising biomarkers that could indicate pathomorphological alterations in the respiratory system [59] arising due to Covid-19. Abnormal breathing sounds are often detected in patients with fluid-filled lungs or lung scarring, indicating pneumonia due to Covid-19 infection. An individual’s breathing sounds can be obtained with a digital stethoscope—developed using a standard stethoscope integrated with a Bluetooth module—or via a commercial digital stethoscope that can transfer data to a smartphone.
3.2.1. Data sourcing
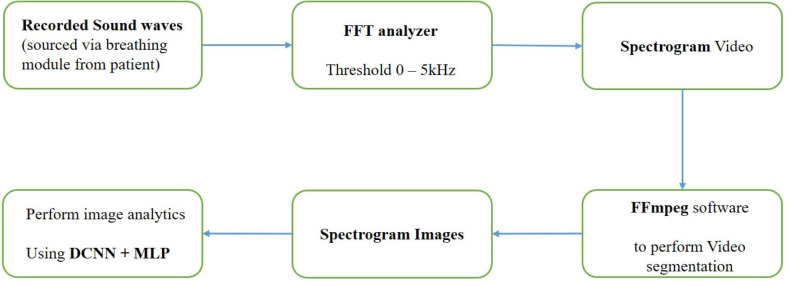
Although many datasets [53], [61], [62] relating to Chest radiology images are available online listed under various categories such as Normal, Pneumonia, Lung cancers, and other health conditions on public databases, access to Covid-19 related datasets for breathing sounds were limited. The breathing abnormality sounds available for detecting crackle and wheezes related to different respiratory diseases are obtained from an online source [63]. Audio files are extracted from this source, and each audio file is converted into a spectrogram video using the FFT analyzer. This spectrogram video is segmented into spectrogram images using FFmpeg software [64] to four seconds sample based on the breathing cycle. The resulting spectrogram images [65] are then used as a dataset for retraining the transfer learning model, as shown in Fig. 7. The significant limitations of the FFmpeg software are: — (a) the framerate and size limitations associated with different Codecs and containers, (b) Complex and tedious functions for the execution of the program, and (c) Lack of integrated Graphic User Interface (GUI), which limits debugging and troubleshooting.
Fig. 7.
Dataset preparation for breathing sound analysis.
A Preliminary breathing sound data collection of 10 individuals has been performed for validation. Of which, five individuals subsequently tested positive for Covid-19. Breathing sound data is collected for an interval of 10 to 30 s and is uploaded to an online database [65]. Table 1 presents the symptom analysis, diagnosis, vitals, and treatment protocol of the five covid-19 patients.
Table 1.
Covid-19 Patient details.
| Patient No. | Age | Gender | Symptoms | Vitals: SpO2, Pulse (BPM) average | Diagnosis Tests | Treatment Protocol | Date of collection | Survival | |
|---|---|---|---|---|---|---|---|---|---|
| Covid-19 Positive-Patient details | |||||||||
| 1 | 27 | M | Loss of smell and taste, Headache, throat pain | 96, 80 | RT-PCR, Antigen | Home quarantine | 29-sept-2020 | Yes | |
| 2 | 28 | M | Chest congestion, asymptomatic | 97, 72 | RT-PCR | Home quarantine | 29-sept-2020 | Yes | |
| 3 | 82 | F | Fever, Headache, fatigue, difficulty in breathing | 95, 70 | Antigen | Home quarantine | 27-sept-2020 | Yes | |
| 4 | 48 | F | Fever, Severe Headache, fatigue, loss of smell and taste | 96, 73 | Antibody | Home quarantine | 30-sept-2020 | Yes | |
| 5 | 56 | M | Asymptomatic | 96, 75 | RT-PCR | Home quarantine | 04-oct-2020 | Yes | |
The breathing sound recording device is a custom-made digital stethoscope assembly with an inbuilt Bluetooth module developed in an earlier study on pneumonia [44] to communicate with the smartphone for recording the breathing sounds. The stethoscope placement at the most appropriate part of the body is a critical factor [66]. The breathing sounds can be obtained from the following positions on the patient’s body—anterior, posterior, and tracheal. The tracheal position in the body is the ideal spot with the patient seated; this is due to the Tracheal fluid orientation inside the alveoli producing distinct pathomorphological sounds for analysis [37], [67], as shown in Fig. 8.
Fig. 8.
The positioning of the breathing sound recording module on the patient’s body.
3.2.2. Data pre-processing: Breathing sound
The breathing sounds contain a mixture of several frequencies along with noise. Performing Fourier transforms is an essential step in analysing the acquired breathing sounds, as analysis in the time domain is not feasible. Fourier transforms performed on the time-domain breathing sound signals convert them to the frequency domain. FFT spectrum analyser uses Fourier transformations to mathematically convert the time domain of spectrum to frequency domain. The input to FFT analyser software is the audio from the breathing sound module (microphone) connected via Bluetooth. The output includes a 2D-colour mapping of frequency in relation to time using digital signal processing.
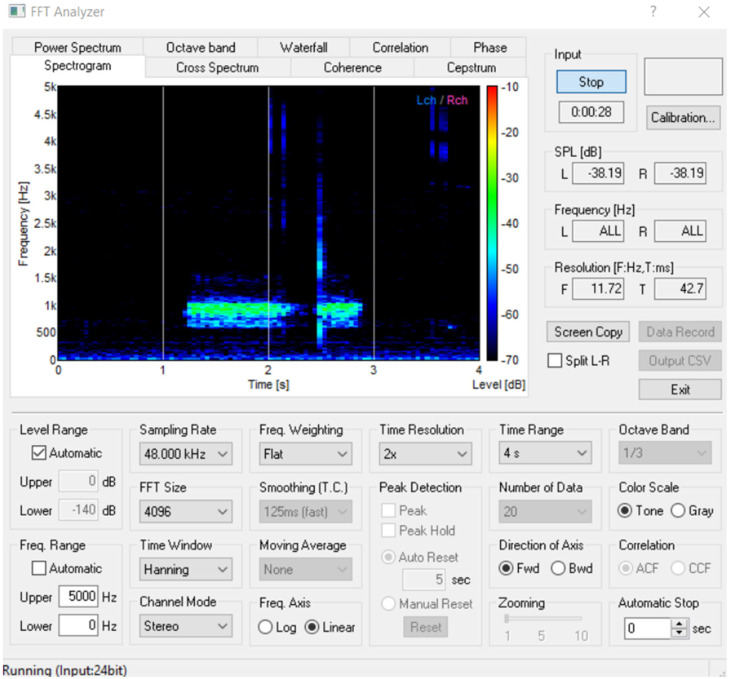
The Fast Fourier transform (FFT) is performed to analyse the energy distribution of the individual frames of signals in the frequency domain, as shown in Fig. 9. Then these frequencies are plotted to obtain a spectrogram where the differences in the frequencies are easily recognisable. Detailed settings of the ‘FFT analyzer’ software is presented in Fig. B.1 in Appendix B. Subsequently, deep-learning algorithms are employed to perform image processing for recognising patterns of abnormalities using deep CNN and MLP. Table 2 presents the frequency-, duration-, range (time and frequency for the abnormal and normal components of the breathing sound.
Fig. 9.
Conversion of breathing sound signals to spectrogram images using FFT.
Fig. B.1.
Snapshot of FFT Analyzer for converting the breathing sound into spectrogram.
Table 2.
Analysis of abnormal and normal breathing sound components in the study.
| Breathing Sound Component Analysis |
Frequency- | Duration- | Range |
||
|---|---|---|---|---|---|
| Abnormal | Normal | Time | Frequency | ||
| Crackles | 3462.24 Hz | 0.059 s | 0.1–0.3 s | 0–4500 Hz | |
| Wheezes | 1428.7 Hz | 1.49 s | 1.0–2.5 s | 0–2500 Hz | |
| Normal Sound | variable | variable | 0.0–4.0 s | 0–5000 Hz | |
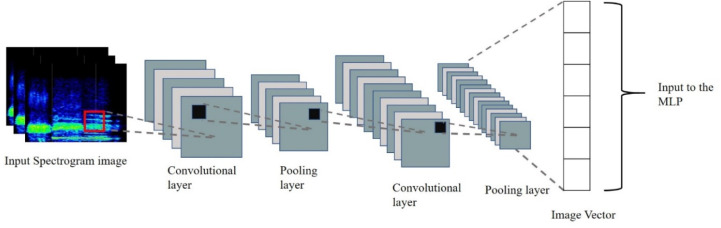
Breathing sound spectrograms obtained from [68] are provided as input to the Transfer learning model based on Deep Convolutional Neural Network (DCNN) for retraining. As mentioned in Section 3.2.1, a dataset of breathing sound patterns was uploaded on Mendeley data [69], which is used for further pre-processing. Breathing sound spectrograms are converted to a 2D-image vector using DCNN, which forms the input to the MLP to identify abnormalities due to Covid-19 (Fig. 10). This Transfer learning system is trained to recognise crackles (both coarse and fine) and wheezes in the range capped at a peak frequency of 5 kHz. The spectrogram is then analysed to predict the presence of breathing abnormalities due to Covid-19. All Spectrogram images are resized to a standard pixel dimension of 299*299, these resized images are used to obtain the 1D image vectors via transfer learning using Inception-v3.
Fig. 10.
Multi-class Image Classification Schematics.
3.3. Chest X-ray image collection
The CXR image dataset used in this study relies on multiple sources to collate a significant number of images; primarily, 15 datasets were collected from different sources [53], [61], [62]. Raw dataset is filtered for image defects, and good quality images are uploaded to an online database [62]. The composition of a combined dataset comprising Chest X-ray images of different abnormalities is presented in Table 3.
Table 3.
Combined dataset and the curated dataset.
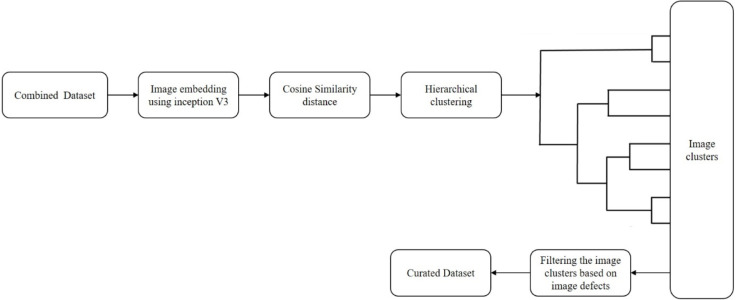
Duplicate X-ray images are removed from the combined dataset based on pixel-to-pixel image similarity [62]. After embedding the images using Inception-v3 architecture, the distances based on cosine similarity are computed. These images are then clustered and checked for defects—such as noise, pixelated, compressed, medical implants, and so on [62]—using an unsupervised learning algorithm, as shown in Fig. 12. During the curation process, the clusters with image defects are removed, and a curated dataset is derived [62]. This dataset is further split into two parts—where 80% is used to train the model, and the remaining 20% is used to validate the proposed model.
Fig. 12.
Curation process using unsupervised learning for finding image defects.
3.3.1. Data curation for CXR
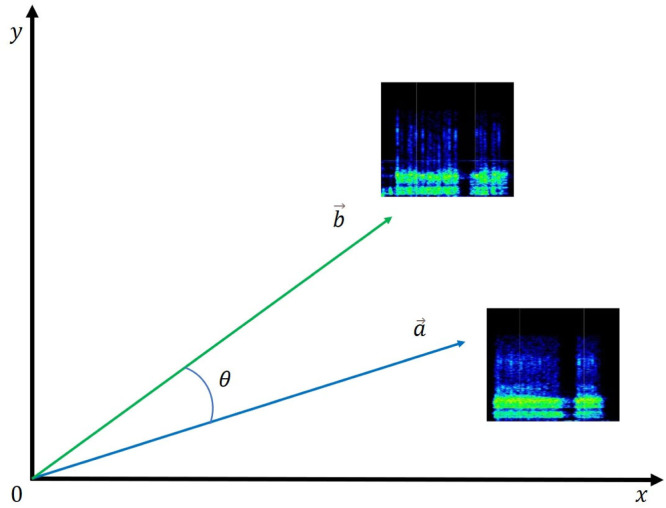
The embedded images contain an array of different image vector components, and their magnitude is computed. This image vector is computed for each image, and a cosine value is calculated between every combination of two different images using Eqs. (1), (2), and (3). Let a and b be the magnitudes of the resultant image vectors obtained for each image. These two image vectors, a and b, are compared using the cosine formula based on their dot product. The angle is the minimum for two similar image files, while angle reaches the maximum value for two different images. A diagrammatic description of cosine similarity distances between image vectors is shown in Fig. 11.
| (1) |
| (2) |
| (3) |
where ‘’ and ‘ ’ are resultant image vectors of any two different images—from the dataset—for which the cosine similarities are computed. , …, are the image vector components of the reference image while —are the image vectors of the image to be compared with the reference image.
Fig. 11.
A graph representing Cosine similarity distances () between image vectors.
3.4. Covid-19-specific antigen test
Rapid antigen testing (RAnT) is incorporated as a disease-specific chemical test to increase the proposed CovScanNet model’s reliability and accuracy. The RAnT could be available for home sample collection through the Ai-CovScan app. This test also has limitations, such as low accuracy, temperature sensitivity, and high false-negative rate. This test should not be used as a standalone test but could be highly beneficial with other supplementary testing facilities. Hence, RAnT has been used in the Ai-CovScan framework as an additional layer to the deep-learning diagnostic methodologies.
The following section discusses the framework developed for the CovScanNet model, followed by the development of the Ai-CovScan framework for decision making.
4. Framework development
4.1. Proposed covscannet model
4.1.1. The CNN component of covscannet
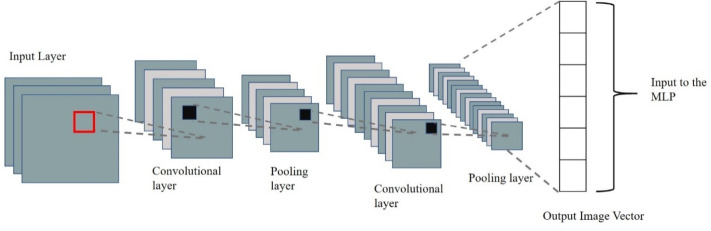
Medical image analysis using deep learning requires a large amount of training data that is challenging to acquire in the initial phases [59]. Previous studies have successfully used transfer learning techniques to retrain existing CNN models with high prediction accuracy [70], [71], [72], [73], [74]. In transfer learning, the training data and the classification task need not be in the same domain. Through this technique, highly accurate classification can be obtained using a relatively small dataset. A typical CNN architecture contains alternate layers of convolution and pooling, as shown in Fig. 13.
Fig. 13.
A typical Convolution Neural Network (CNN) Architecture.
In the proposed CovScanNet model, transfer learning is used for knowledge extraction from Inception-v3 trained on ImageNet dataset (containing 1.2 million images with 1000 classes) [76], [77], [78] and is applied to the target task of Covid-19 classification. The activation of the penultimate layer in Inception-v3 architecture is performed to obtain image embedding, where images are represented as vectors. The 882048 output map from Inception-v3 is flattened to 112048 image vector, which is then fed to the multi-layer perceptron (MLP). This would provide the possibility for optimising the model’s accuracy without using the conventional fully connected layer. The layer distribution of CovScanNet is—311 layers of Inception-v3, one ‘flatten’ layer, five layers of MLP—as presented in Table 4. In the Inception-v3 architecture, convolution layers are followed by ReLU (rectified linear unit) activations [79], and the max-pooling layers, successively. The convolution process is given by Eq. (4).
| (4) |
The output feature map is derived using the above Eq. (4) where represents local features from previous layers [59]. The components , f(.), , implies the input map selection, activation function, training bias, and variable kernels, respectively [59]. The non-linear ReLU function is used to activate the CNN layers for improving the ease-of-training and the performance of the model. The ReLU function definition is given in Eq. (5). A pooling layer is used to prevent the overfitting problem in the CNN model. As given in Eq. (6), the pooling layer reduces the number of computational nodes and further reduces the computational effort.
| (5) |
where x is the input activation and f(x) are the output activation of the node.
| (6) |
where down(.) represents the down-sampling, represents local features from previous layers, and represents the output activation of the subsequent layers.
Table 4.
Layer distribution of CovScanNet (See Ref. [75]).
| Sl. No. | Layer type | No. of layers | Part of: |
|---|---|---|---|
| 1 | Input Layer | 1 | 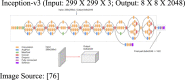 |
| 2 | Convolution Layer | 94 | |
| 3 | Batch Normalisation | 94 | |
| 4 | Activation | 94 | |
| 5 | Average Pooling | 9 | |
| 6 | Max Pooling | 4 | |
| 7 | Mixed | 13 | |
| 8 | Concatenate | 2 | |
| 9 | Flatten | 1 | (Input:8 X 8 X 2048; Output: 1 X 1 X 2048) |
| 10 | MLP Input | 1 | Multi Layered Perceptron (MLP) (Input: 1 X 1 X 2048; Output: 4) |
| 11 | Hidden MLP | 3 | |
| 12 | Output MLP | 1 | |
| Total | 317 | ||
4.1.2. The MLP component of CovScanNet
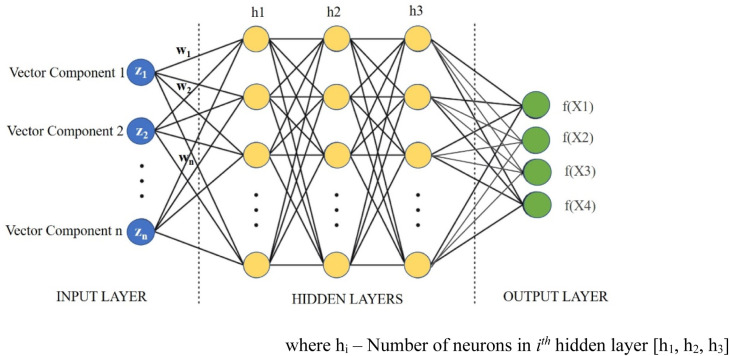
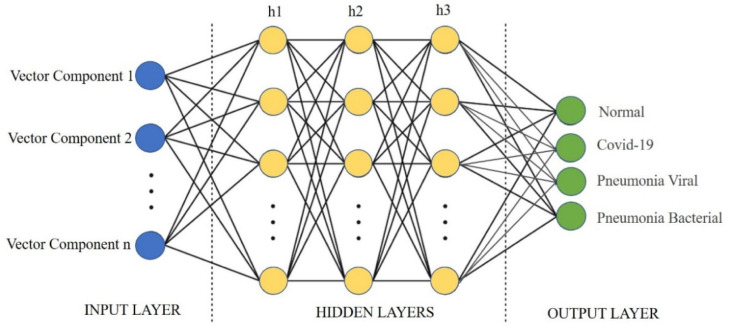
A Multi-Layer Perceptron (MLP)—a feedforward artificial neural network (ANN)—is implemented for classifying the categories from embedded images. The input to the MLP is the embedded images, and the output layer indicates the classes of the target labels. The hidden layers are modified to get the required accuracy for the MLP. The Scikit-Learn library is used to implement the MLP. Scikit Learn is an open-source library that includes various machine learning algorithms such as regression, classification, and clustering. It is built on SciPy library for data analysis and machine learning applications in the python programming language. It also contains the ‘MLPClassifier’ class, which utilises the MLP algorithm. The layers of MLP are—(a) the input layer, b() the hidden layers, and (c) the Output Layer [59], as shown in Figs. 14, 16, and 18. The activations of the hidden layer are calculated using ; where are the activations of the neurons in the input layer, and are the transformation weights applied to neurons in the input layers. Further, ReLU is applied for the activation of hidden layer neurons of the MLP, as given in Eq. (5).
Fig. 14.
Multi-Layer Perceptron for CovScanNet Model.
Fig. 16.
Multi-Layer Perceptron for the breathing sound component of CovScanNet.
Fig. 18.
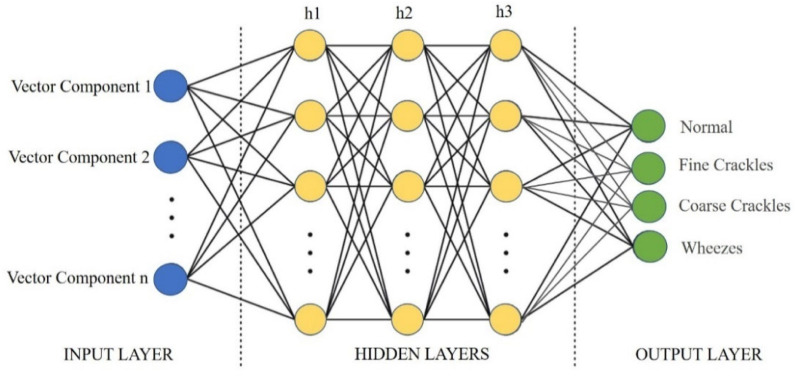
An MLP with input, output, and hidden layers for CXR CovScanNet Model.
A regularisation parameter ‘’ is used to penalise the higher magnitude weights, thereby avoiding overfitting. Increasing the regularisation parameter ‘’, decreases the accuracy of the model. The only exception being , which has slightly higher accuracy than (Refer to Table C.1 in Appendix C). In this study, is taken as 0.0001 to avoid overfitting in the training data, which might be the case at . Furthermore, a large value of ‘’ signifies a complex neural network that could be avoided by keeping ‘’ minimal.
Table C.1.
Model performance in relation to variation in Regularisation parameter .
| AUC | CA | F1-score | Precision | Recall | Specificity | |
|---|---|---|---|---|---|---|
| 0.0001 | 0.986 | 0.911 | 0.912 | 0.914 | 0.911 | 0.970 |
| 0.0005 | 0.986 | 0.911 | 0.912 | 0.914 | 0.911 | 0.971 |
| 0.001 | 0.986 | 0.906 | 0.907 | 0.910 | 0.906 | 0.969 |
| 0.005 | 0.986 | 0.909 | 0.909 | 0.910 | 0.909 | 0.969 |
| 0.05 | 0.983 | 0.900 | 0.902 | 0.907 | 0.900 | 0.969 |
| 0.1 | 0.985 | 0.907 | 0.908 | 0.909 | 0.907 | 0.968 |
| 0.5 | 0.986 | 0.914 | 0.914 | 0.916 | 0.914 | 0.971 |
| 1 | 0.985 | 0.907 | 0.908 | 0.911 | 0.907 | 0.970 |
| 5 | 0.973 | 0.881 | 0.882 | 0.884 | 0.881 | 0.959 |
| 50 | 0.929 | 0.772 | 0.704 | 0.690 | 0.772 | 0.892 |
| 500 | 0.500 | 0.356 | 0.187 | 0.126 | 0.356 | 0.644 |
The model is iterated for optimisation with a maximum epoch of 500. Epochs in the range of 400–500 are the early stopping point [80] for avoiding underfitting, overfitting, and improving the model’s learning. It is observed from previous studies [81], [82] that beyond this range, the generalisation error increases. The stochastic optimisation algorithm used in this model is Adaptive Moment Estimation (ADAM) [78]. It reduces the computational cost and uses less memory to solve complex and large-scale problem in an iterative process. Eq. (7) gives the expression for the ADAM stochastic optimisation algorithm.
| (7) |
where m moment and X is the random variable; E stands for the expectation of the random variable X to the power of n.
The softmax function ensures the maximum probability in the output layer classes, as given in Eq. (8).
| (8) |
where y is the ith element for the input component to the softmax and K is the number of classification categories.
4.1.3. Hyper-parameter Tuning (HT.)
The hyper-parameters for the MLP model are the number of neurons, hidden layers, and iterations. These hyper-parameters are tuned to improve the accuracy in the prediction of Covid-19. The hidden layer [h1, , ] and its structure are shown in Fig. 14. The time complexity of backpropagation (of the MLP) depends on the number of training samples (n), number of features (m), number of iterations (i), number of neurons(h) in the hidden layer (k), and number of output neurons (o). The big O denotes the time complexity function (Eq. (9)).
| (9) |
The tuning related to number of hidden layers started with a lower number of layers to minimise the time complexity and computational costs. The set of neurons and hidden layers are chosen from the power set of A {0, 10, 100, 200} limited to three hidden layers. A few combinations are selected from the power set for modelling. These selected combinations are represented by S(A) {[100, 0, 0], [200, 0, 0], [100, 10, 0], [100, 100, 0], [200, 10, 0], [200, 200, 0], [200,200,200]}.
The modification is such that the initial model was trained with a single hidden layer of [100, 0, 0] and then with [200, 0, 0]. Following which, the switching to double-layer [100, 10, 0], [100, 100, 0], [200, 10, 0] and, [200, 200, 0] is performed. After that, switching to three hidden layers with [200,200,200] neurons is done. This process is undertaken to avoid false negatives by identifying the optimal combination of hidden layers that gives the minimum number of false negatives, which increases the recall for Covid-19. This is crucial as any false-negative result could pose serious health concerns to the patients and their primary contacts.
4.2. CovScanNet for breathing sound
The model is trained on a dataset of spectrogram images for breathing sound, generated using the framework mentioned in Fig. 7. The input to the MLP is the image vector obtained from the CNN model based on Inception-v3, as shown in Fig. 15. The output layer of this MLP contains the classes of Normal, Fine Crackles, Coarse Crackles, and Wheezes, as shown in Fig. 16. Subsequently, validation is performed using breathing sound spectrograms acquired from Covid-19 patients and non-Covid-19 individuals. The evaluation metrics concerning total abnormalities, percentage abnormality, percentage normal, and minimum difference are given by Eqs. (10), (11), (12), and (13), respectively. The Minimum difference (Eq. (13)) is the difference between percentage abnormality in Covid-19 patients and percentage normality in non-Covid-19 patients. The Minimum difference indicates the efficiency of the model in predicting the results accurately.
| (10) |
| (11) |
| (12) |
| (13) |
where w is the total number of normal breathing sound, x is the total number of Coarse Crackles, y is the total number of Fine Crackles, and z is the total number of Wheezes.
Fig. 15.
CNN model for breathing sound spectrogram based on Inception-v3.
4.3. CovScanNet for CXR
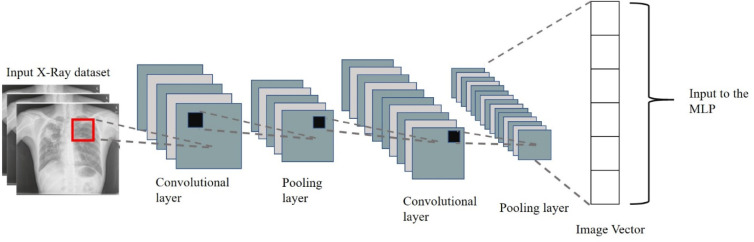
The model is trained on 80 per cent of the curated CXR images using the framework mentioned in Fig. 12. The input to the MLP is the image vector obtained from the CNN model based on Inception-v3, as shown in Fig. 17. The output layer of this MLP contains the classes of Covid-19, Normal, Pneumonia Bacterial, and Pneumonia Viral, as shown in Fig. 18. The validation is performed using 20 per cent of the total curated dataset.
| (14) |
| (15) |
| (16) |
| (17) |
Fig. 17.
Modified CNN framework for CXR CovScanNet.
Accuracy of a machine learning (ML) algorithm or model is used to measure the correct classification of data points out of total data points. It measures the correctness of the predicted data by the machine learning algorithm. Accuracy is not the only indicator to ensure the robustness of the ML algorithm. Recall—also known as sensitivity—gives the true-positive rate [83]. It is based on relevant instances out of the total instances retrieved. Precision—also known as positive predictive value—correctly classifies true positives amongst all actual positives. Recall compliments the type II error rate, while precision is related to the type I error rate [84]. F1 score is used to balance recall and precision and measure the model’s accuracy on a given dataset. Recall and precision and F1 score indicate the robustness of the machine learning model, which complements accuracy in reporting their performance. The evaluation metrics concerning total accuracy, recall, precision, and F1 score are given by Eqs. (14), (15), (16), and (17), respectively.
4.4. Ai-CovScan framework for decision making
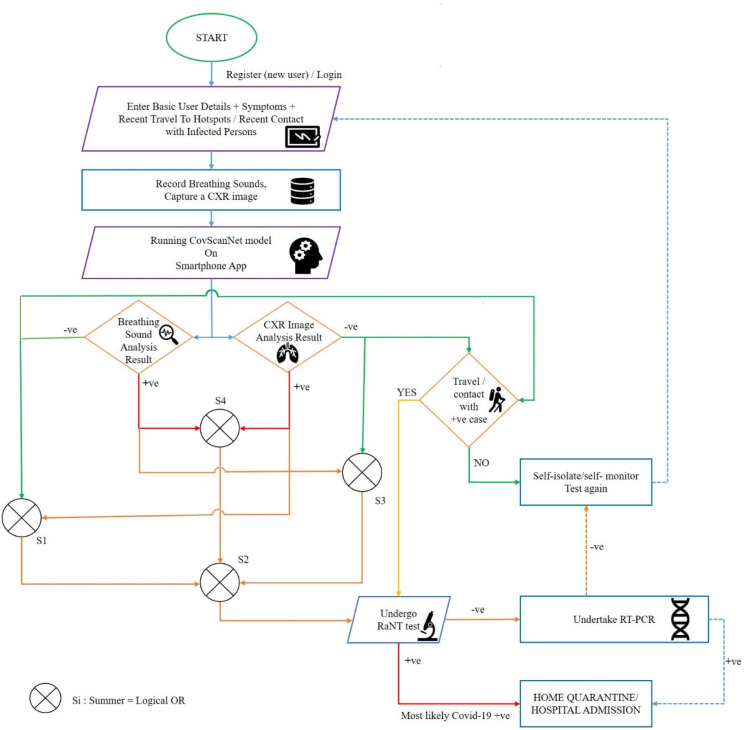
The Ai-CovScan framework—comprising the CovScanNet model—is devised to provide an effective decision-making tool for undergoing RT-PCR testing in low resource settings, based on the results from the analysis of CXR, breathing sound, and Rapid antigen test (RAnT) of the patient. The self-reported health data is procured from a susceptible person through a questionnaire in a mobile application. Based on the user’s response, they are directed to undergo a Rapid antigen test (RAnT), chest X-ray (CXR), and provide breathing sound data. The CXR is analysed using the proposed CovScanNet model to classify the X-ray image into Covid-19, Normal, Pneumonia-bacterial, and Pneumonia-viral categories. The proposed CovScanNet model is also used to analyse the spectrogram images of the recorded breathing sound data. The decision-making algorithm for RT-PCR is based on RAnT, CXR analysis, and breathing sound analysis, as shown in Fig. 19. The user can take help from medical experts via online consultation—an additional feature of the developed framework—for all other cases. Alternatively, the user can choose to go for an RT-PCR test for confirmation.
Fig. 19.
Ai-CovScan—decision-making framework.
5. Results
5.1. Breathing sound analysis
After converting the breathing sounds into spectrograms, distinguishable patterns across different abnormalities are observed. Abnormalities like fine crackles, coarse crackles, and wheezes are visually recognised (shown in Fig. 20) while inspection by a focused group comprising medical professionals, subject experts, biomedical researchers, and the authors. Subsequently, the CovScanNet model is trained to classify breathing abnormalities, as discussed above. Following this, the model is tested on the Covid-19 database, and the results are presented below.
Fig. 20.
An observed sample of spectrogram images for different types of breathing sounds.
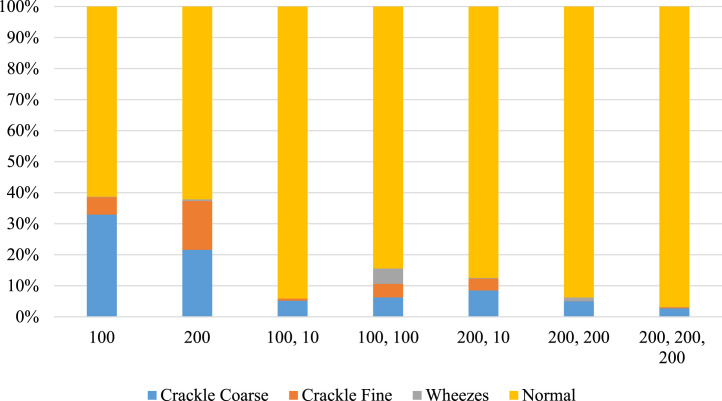
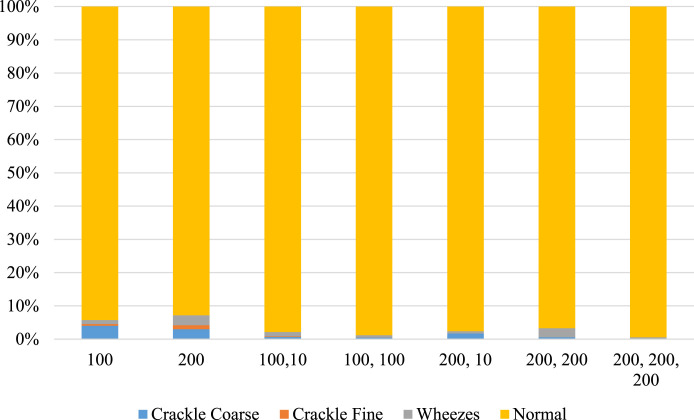
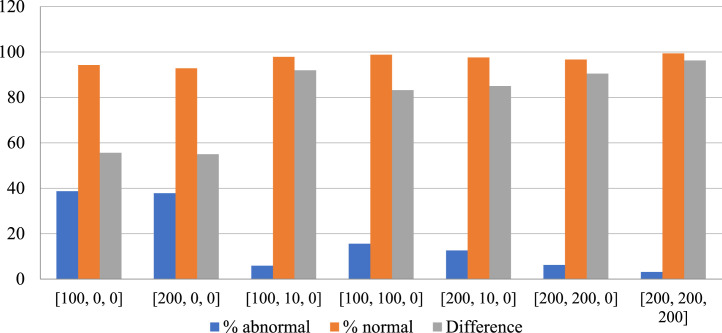
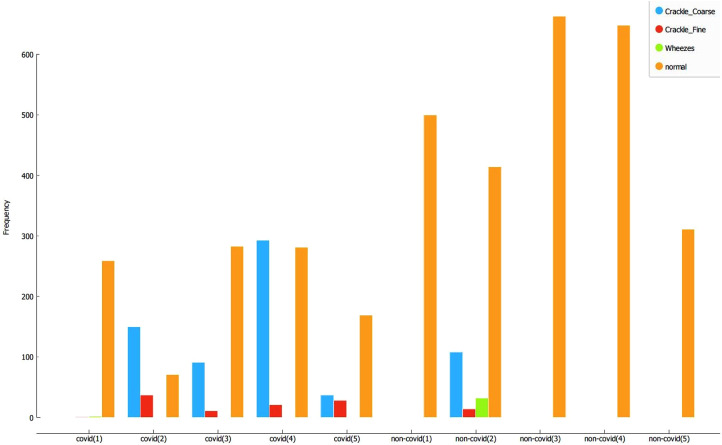
The predicted composition of the spectrogram—percentage abnormalities versus normal percentage—are calculated. These results are plotted in Fig. 21 for Covid-19 patients, and Fig. 22 for non-Covid-19 individuals across the selected hyper-parameters. Fig. 23 provides the selection criteria for selecting the hidden layer composition and the corresponding number of neurons.
Fig. 21.
Components of breathing sound for Covid-19 confirmed cases.
Fig. 22.
Components of breathing sound for Non-Covid-19 cases.
Fig. 23.
Accuracy in % of normal vs abnormal study for Covid-19 (Histogram plot).
It can be inferred from Fig. 23 that the hidden layer composition [100, 0, 0] is performing the best in classifying abnormalities in Covid-19 patients. This [100, 0, 0] layer also performs better in identifying normal individual’s breathing sound, and the minimum difference between percentage abnormality and percentage normality for the total dataset. Hence, the [100, 0, 0] is selected for pattern recognition of Covid-19.
The selected hyper-parameter [100, 0, 0] is employed to identify the composition of abnormalities (like fine crackles, coarse crackles, and wheezes) and normal breathing sound in each individual, both Covid-19 and non-Covid-19. The results are presented in Fig. 24. It can be observed that all the Covid-19 patients are showing significant breathing abnormalities, especially crackles. As an exception, one patient infected by Covid-19 is showing negligible breathing abnormalities. This observation may be explained because the patient is asymptomatic and mildly infected. The accuracy for the detection of Covid-19 is 80% for the dataset considered for validation. Furthermore, normal patients returned zero or minimal abnormalities except non-Covid-19 individual number 2 (NC-2). After consultation with medical experts, it is assumed that NC-2 had a pre-existing disorder. Nonetheless, more data and further testing is required to provide concrete results.
Fig. 24.
Components of breathing sound for Covid-19 and non-Covid-19 patient.
Though the model returns impressive results, it needs to be further tested on a more extensive and more diverse dataset, improving the classification accuracy. Also, the dataset of five covid patients is not robust enough to make highly accurate predictions; hence it should be treated as a preliminary methodological contribution. In future studies, as the number of data samples increases, the accuracy and performance of this model would increase too. The background noise while recording the breathing sound and the microphone’s sensitivity is the other limitations of this model. The model could potentially identify other respiratory diseases when trained with disease-specific datasets. A database of sound signatures for many respiratory diseases could also be created to identify diseases rapidly and conveniently in the early stages of a pandemic.
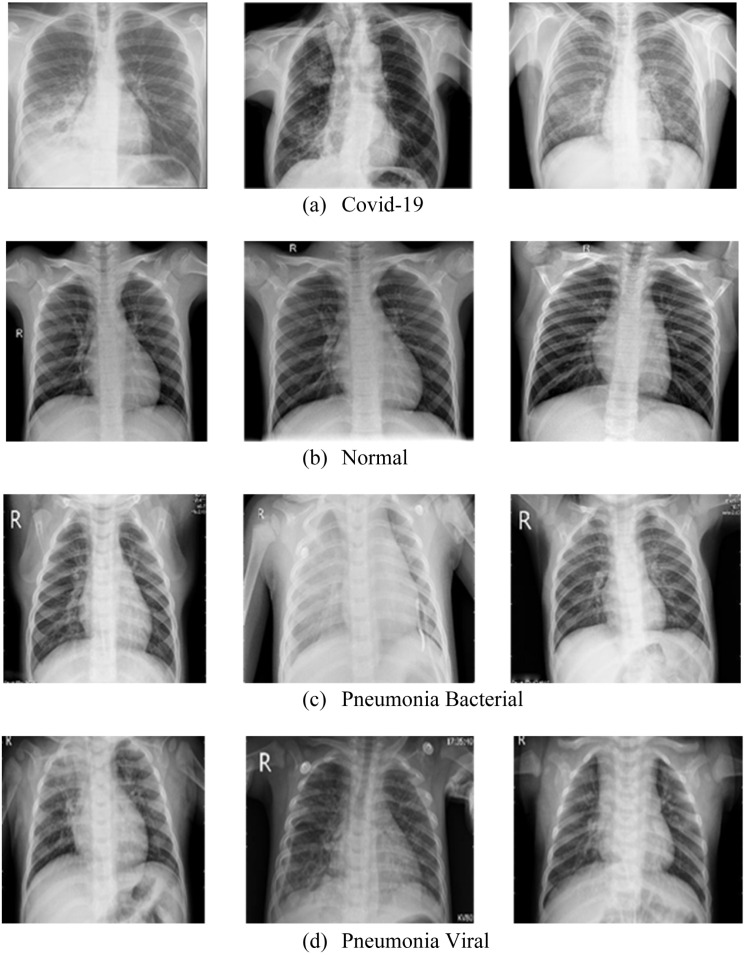
5.2. Chest X-ray image analysis
The curated data is segmented into four classes—(a) Covid-19, (b) Normal, (c) Pneumonia Bacterial, and (d) Pneumonia Viral, as shown in Fig. 25. Subsequently, the dataset is split into train and test data with a ratio of 80:20. The CovScanNet model is trained using the trained data, which classifies the test data with significant accuracy. The accuracy, recall, precision, and F1 score [59] is reported in Table 5. The area under the Receiver Operating Characteristic (ROC) is computed for selected layer compositions with different hyper-parameters.
Fig. 25.
Chest X-ray for (a) Covid-19, (b) Normal, (c) Pneumonia Bacterial, and (d) Pneumonia Viral.
Table 5.
Comparison matrix for models with different hidden layer composition.
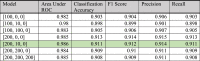 |
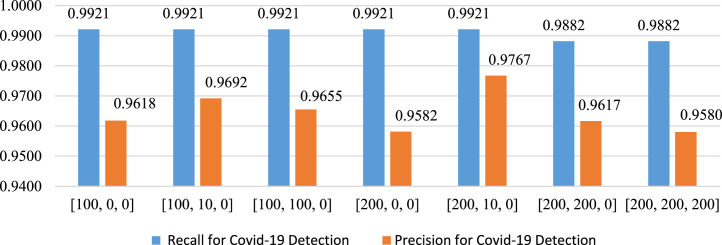
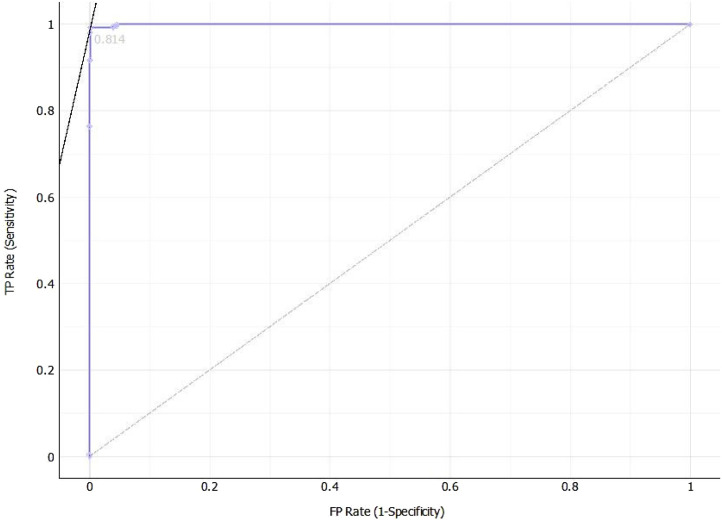
Moreover, precision and recall specific to Covid-19 are presented in Fig. 26; and the [200, 10, 0] layer composition returns the best results. It can be observed that the precision of this layer composition is 97.67%, while the recall is 99.21%. The ROC curve for the selected layer is presented in Fig. 27.
Fig. 26.
Recall and precision of different layer combinations for Covid-19.
Fig. 27.
ROC curve plotted for the transfer learning model with [200, 10, 0] hidden layer.
5.2.1. Confusion matrices
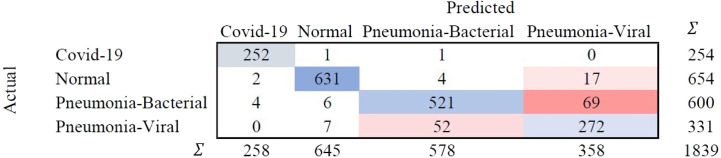
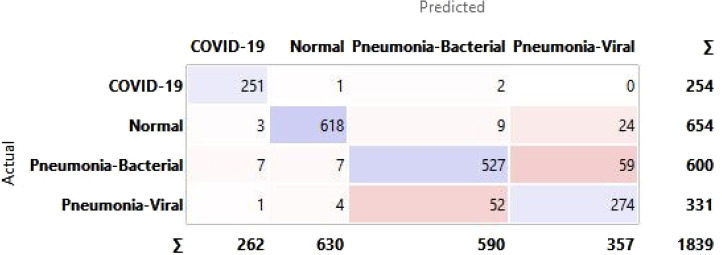
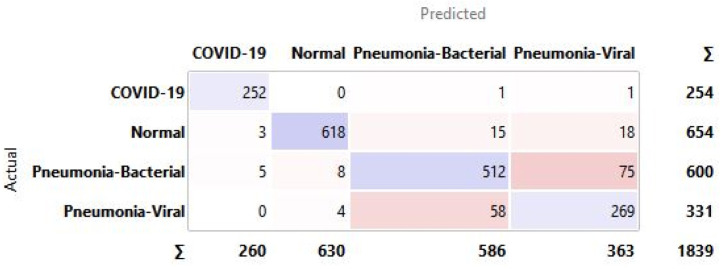
A confusion matrix is a tabular representation to summarise the training model’s success in accurately identifying the specific labels for the actual results. The labels identified in this training model are COVID-19, Normal, Pneumonia-Bacterial, and Pneumonia-viral. The rows represent actual figures, while the columns represent the predicted results of the neural network. The model is tested for a different number of hidden layers combinations incorporated in the training model to identify the suitable one with the best predictive outcome. The hidden layers are chosen FOR CXR, and the resulting outcomes are summarised in the confusion matrix as shown in Fig. 28.
Fig. 28.
Confusion matrix for selected hidden layer composition [200, 10, 0] for CXR.
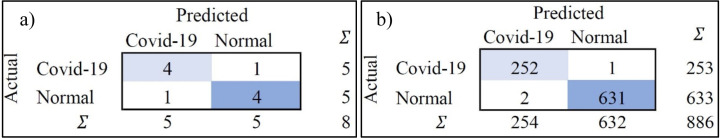
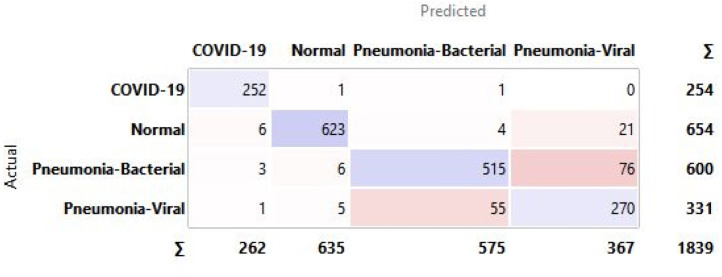
The confusion matrix highlights that the predicted false negatives are minimal, i.e., only two out of 254 Covid-19-positive cases are inaccurately reported for CXR. Also, the model has comparatively lesser accuracy in distinguishing between viral and bacterial pneumonia. The Covid-19-specific accuracy (comparing Covid-19 against the normal) is reported as 80% for breathing sound, and 99.66% for CXR, as calculated from confusion matrices (using Eq. (14)) shown in Figs. 29(a) and 29(b), respectively. The collected dataset is small and hence would need further testing and validation for a larger population. The noise due to CXR-image-capture while scanning it to the Ai-CovScan app can distort the results. Hence, a detailed manual to train the user for scanning images is required to increase the CovScanNet model’s resilience.
Fig. 29.
Confusion matrix for Covid-19-specific accuracy of—(a) Breathing Sound, and (b) CXR images.
5.3. Covid-19 specific antigen test
The methods adopted for Covid-19 diagnosis in the CovScanNet model are the breathing sound and the patient’s CXR, which are indirect diagnostics methods. The limited dataset available for training and testing breathing sound- and CXR-images is constrained in its specificity and sensitivity. Rapid antigen test is a direct indicator of the disease vector’s presence and could be the critical factor in identifying Covid-19. When the proposed model is augmented with improvement in the quantity and quality of datasets, ideally, there would not be a requirement for the antigen test to confirm the disease. In such a scenario, a two-tier testing model with breathing sounds and CXR could provide comparable results to the three-tier testing model proposed in this study.
5.4. Smartphone app development
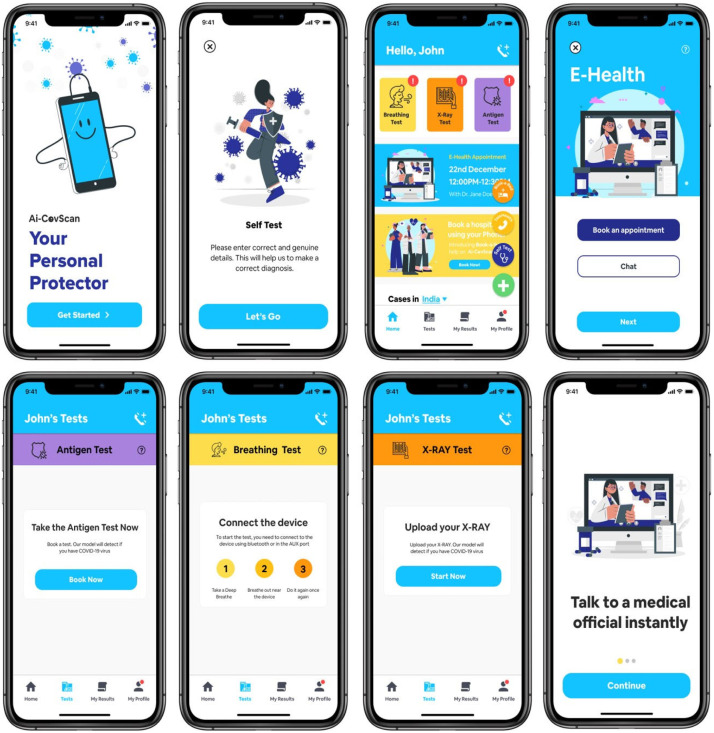
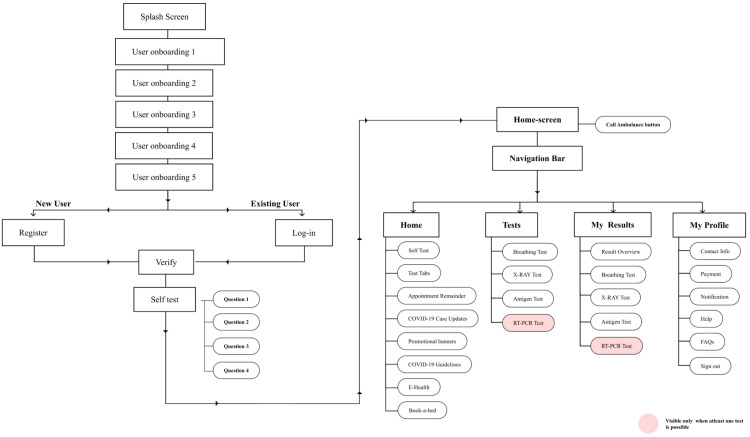
The smartphone app serves as the window to the Ai-CovScan framework to function in a real-world scenario. It incorporates the following significant functionalities—(a) get a questionnaire response from the user regarding the symptoms exhibited and their recent travel history; (b) accept user’s breathing sound via USB or Bluetooth connectivity between the module or digital stethoscope and the smartphone app; (c) accept user’s CXR image via direct upload from a file or smartphone camera; (d) accept rapid antigen test results of the user. The app feeds the user’s inputs into the CovScanNet model to predict the results for further evaluation, such as undertaking a confirmatory RT-PCR test or seeking expert medical advice. There are added features in the e-diagnosis facility where users can seek professional medical advice on their test results for Breathing sound, CXR, Antigen or a combination of these test results. An option to book for an Antigen test, CXR imaging, antibody test, or an RT-PCR test could also be provided with the active participation of other stakeholders involved in pandemic response. The app screens are presented in Fig. 30.
Fig. 30.
Ai-CovScan app screens.
The app is tested for three smartphone devices with low, medium, and high specifications, as given in Table 6. The app works satisfactorily, even for the low-end specifications of the tested smartphones.
Table 6.
Hardware specifications for the smartphone for testing Ai-CovScan .
| Phone specification | CPU | GPU | RAM | OS | Reference |
|---|---|---|---|---|---|
| Low | Octa-core 1.4 GHz Cortex-A53 | Adreno 505 | 2 GB | Android 6.0 | [85] |
| Medium | Octa-core (42.35 GHz Kryo and 41.9 GHz Kryo) | Adreno 540 | 4 GB | Android 8.0 | [86] |
| High | Octa-core (12.96 GHz Kryo 485 and 32.42 GHz Kryo 485 and 41.78 GHz Kryo 485) | Adreno 640 | 8 GB | Android 10.0 | [87] |
6. Discussion
6.1. Ai-covscan framework
Ai-CovScan has been developed as a self-screening tool for Covid-19, based on the analysis of breathing sound and CXR image (Fig. 31). If the model predicts any abnormalities in the analysis, then the user is advised to undertake an antigen test as the next course of action. This framework advises users to follow self-isolation and social distancing protocols, where users can test themselves at home with a smartphone app. Preliminary detection methods such as antigen tests and antibody tests are limited in their accuracy and prediction, requiring a confirmatory test for Covid-19. The confirmatory tests are, however, expensive and limited. CovScanNet, a novel approach based on CNN and MLP, is developed to identify the presence of Covid-19 that uses medical image analysis—chest X-ray or breathing sound, or both—with a disease-specific antigen test. Inception-v3 combined with MLP is retrained for recognising Covid-19 and pneumonia from a patient’s CXR images and breathing abnormalities.
Fig. 31.
An overview of the system catering to the end-user.
6.2. CovScanNet model
Breathing sound patterns may provide unique signatures indicating damage caused to the respiratory system due to Covid-19. CovScanNet is a preliminary model trained on the limited data relating to Covid-19 breathing abnormalities. Hence this model needs further improvements to enhance the prediction accuracy. Testing the Covid-19-positive patients for their breathing sound abnormalities yielded significantly promising outcomes. Pulmonary-related abnormalities such as pneumonia are commonly observed in Covid-19 infected individuals who may later develop severe complications. Therefore, the detection of pneumonia in the lungs can function as a tool for diagnosis. The CovScanNet model is trained on a curated dataset of several CXR images, and abnormalities in the breathing sound spectrogram images to identify Covid-19 with high accuracy and precision. In the Ai-CovScan framework, the RAnT test results are combined with the patients’ breathing sounds and CXR image analysis providing a higher prediction accuracy. This framework, when implemented through a smartphone app, reduces the demand for RT-PCR testing. The potential limitations while accepting the user’s breathing sound through the app are:—(a) sensitivity of the microphone, (b) noise filtration rate, (c) specification and computing capability of the smartphone, d) use of a lossless recording format, and (e) amplification of the breathing sound.
6.3. Smartphone app
Ai-CovScan app provides a means for testing and self- monitoring to reach a large section of the population at the comfort of their living spaces. When lockdowns and restrictions are in place to contain the pandemic, the free movement of people becomes restricted, and access to healthcare facilities are severely undermined. There is also a ‘safety’ factor that one must consider when they venture out to public spaces. This factor restricts individuals from accessing necessary diagnostic facilities for fear of contracting the disease. In the initial stages of a pandemic, the test facilities and methods are evolving, and skilled medical professionals involved in diagnostics are minimal. The limited healthcare workforce may be inadequately trained to follow standard testing protocols for proper sample collection and processing, resulting in several errors. The developed smartphone app can predict the likelihood of infection, further enhancing the usability and implementation of the Ai-CovScan framework.
7. Conclusion
Covid-19 is a major health concern for vulnerable populations worldwide, especially concerning the elderly and individuals with other underlying health conditions. A pandemic response demands swift actions involving accurate identification of infected individuals and their isolation to prevent further disease spread. As the number of people involved in a pandemic scenario outpaces the existing healthcare infrastructure’s capacity, denial of health services becomes seldom. Testing infrastructure is limited in its capacity to include a large proportion of the susceptible population in the pandemic’s initial phases, leading to further disease spread. The proposed multimodal diagnostic framework allows the user to test for Covid-19 using CXR images and breathing sounds. Chest X-ray images can be readily scanned using a smartphone. Covid-19, if predicted, can help the patient in decision-makingfor further confirmatory tests. Breathing sounds can be recorded using the sound recorder module to communicate with the smartphone. The breathing sound detection framework is trained to recognise crackles present in the spectrogram, where wheezing sounds are also identified and filtered. The model provided a tentative accuracy of 80% for breathing sound data analysis and a 99.66% Covid-19 detection accuracy for the curated CXR image dataset. Detection of Covid-19 using X-ray images and breathing sounds is a cost-effective and time-saving method for pre-screening. The framework is reliable for patients and their caretakers for frequent testing. This framework further aims at reducing our need for confirmatory tests that are not easily accessible, sometimes inaccurate, and expensive to deploy, hence saving critical resources during Covid-19—a global pandemic. The proposed framework is not a substitute for standard clinical-grade testing tools but merely a self-screening tool deployed in a smartphone app. Future work could improve the dataset for training and validating the model using advanced artificial intelligence algorithms.
CRediT authorship contribution statement
Unais Sait: Conceptualization, Modelling, Experimentation, Analysis, Writing. Gokul Lal K.V.: Writing, Structure, Figures, Experimentation. Sanjana Shivakumar: Writing, Review, Figures. Tarun Kumar: Supervision, Review and Editing, Analysis, Experimentation. Rahul Bhaumik: Analysis, Experimentation, Editing. Sunny Prajapati: Analysis, Experimentation, Editing. Kriti Bhalla: Writing, Review, Figures. Anaghaa Chakrapani: Application Development.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank Dr Subin Shaji (AIIMS, Rishikesh) for helping us to understand and validate the dataset. We acknowledge Mr Gautham Ravikiran (Student, PES University) for exploring the form of the breathing sound recording module, and Ms Bhavani S P (Student, PES University) for helping us designing the UX for the app. We also acknowledge Dr Neha Bharti (Patna Medical College) for helping us to identify and validate breathing sound data.
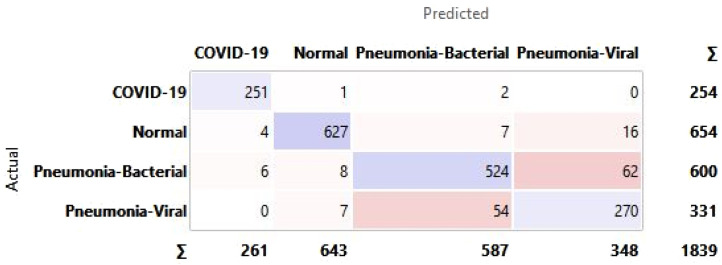
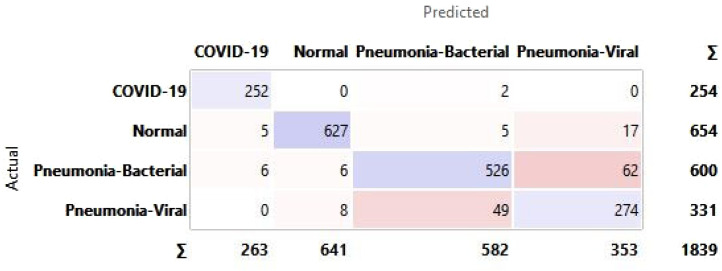
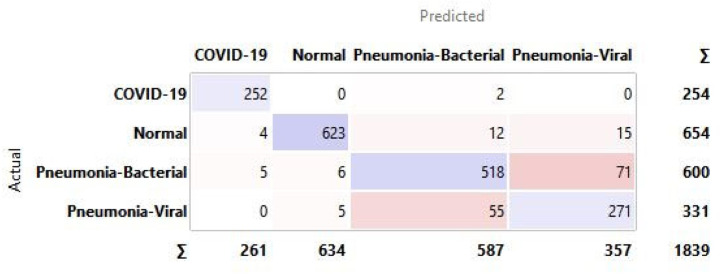
Appendix A. Confusion matrix for all hidden layer combinations for CXR
See Fig. A.1, Fig. A.2, Fig. A.3, Fig. A.4, Fig. A.5, Fig. A.6.
Fig. A.1.
Confusion matrix for selected hidden layer composition [200, 200, 200].
Fig. A.2.
Confusion matrix for selected hidden layer composition [200, 200, 0].
Fig. A.3.
Confusion matrix for selected hidden layer composition [200, 0, 0].
Fig. A.4.
Confusion matrix for selected hidden layer composition [100, 100, 0].
Fig. A.5.
Confusion matrix for selected hidden layer composition [100, 10, 0].
Fig. A.6.
Confusion matrix for selected hidden layer composition [100, 0, 0].
Appendix B. FFT analyzer software for breathing sound spectrogram generation
See Fig. B.1.
Appendix C. Model performance in relation to variation in regularisation parameter
See Table C.1.
Appendix D. Information Architecture for the Ai-CovScan Smartphone App
See Fig. D.1.
Fig. D.1.
Information Architecture for the Ai-CovScan App.
References
- 1.World Health Organization . World Heal. Organ.; 2020. Q & A on Coronaviruses (COVID-19) https://www.who.int/emergencies/diseases/novel-coronavirus-2019/question-and-answers-hub/q-a-detail/q-a-coronaviruses. (Accessed 12 June 2020) [Google Scholar]
- 2.Khan A.I., Shah J.L., Bhat M.M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020;196 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakrapani A., Kumar T., Shivakumar S., Bhaumik R., Bhalla K., Prajapati S. 2020 IEEE Int. Smart Cities Conf. IEEE; 2020. A Pandemic-specific ‘Emergency Essentials Kit’ for Children in the Migrant BoP communities; pp. 1–8. [DOI] [Google Scholar]
- 4.United Nations Development Programme A. United Nations Dev. Program.; 2020. COVID-19 Pandemic. https://www.undp.org/content/undp/en/home/coronavirus.html. (Accessed 5 August 2020) [Google Scholar]
- 5.Miller L.E., Bhattacharyya R., Miller A.L. Data regarding country-specific variability in Covid-19 prevalence, incidence, and case fatality rate. Data Br. 2020;32 doi: 10.1016/j.dib.2020.106276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmad S. Potential of age distribution profiles for the prediction of COVID-19 infection origin in a patient group. Inform. Med. Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Worldometer S. 2020. COVID-19 coronavirus pandemic, worldometer. https://www.worldometers.info/coronavirus/. (Accessed 31 December 2020) [Google Scholar]
- 8.Detoc M., Bruel S., Frappe P., Tardy B., Botelho-Nevers E., Gagneux-Brunon A. Intention to participate in a COVID-19 vaccine clinical trial and to get vaccinated against COVID-19 in France during the pandemic. Vaccine. 2020;38:7002–7006. doi: 10.1016/j.vaccine.2020.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazinani M., Rude B.J. The novel zoonotic coronavirus disease 2019 (COVID-19) pandemic: Health perspective on the outbreak. J. Healthc. Qual. Res. 2020 doi: 10.1016/j.jhqr.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization M. WHO - Interim Guid.; 2020. Laboratory Testing for 2019 Novel Coronavirus (2019-NCoV) in Suspected Human Cases; pp. 1–7. https://www.who.int/publications/i/item/10665-331501. [Google Scholar]
- 11.McFee R.B. COVID-19 - a brief review of radiology testing. Dis. Mon. 2020;66 doi: 10.1016/j.disamonth.2020.101059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silva Júnior J.V.J., Merchioratto I., de Oliveira P.S.B., Rocha Lopes T.R., Brites P.C., de Oliveira E.M., Weiblen R., Flores E.F. End-point RT-PCR: a potential alternative for diagnosing coronavirus disease 2019 (COVID-19) J. Virol. Methods. 2020 doi: 10.1016/j.jviromet.2020.114007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kilic T., Weissleder R., Lee H. Molecular and immunological diagnostic tests of COVID-19: Current status and challenges. IScience. 2020;23 doi: 10.1016/j.isci.2020.101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaye A.D., Okeagu C.N., Pham A.D., Silva R.A., Hurley J.J., Arron B.L., Sarfraz N., Lee H.N., Ghali G.E., Gamble J.W., Liu H., Urman R.D., Cornett E.M. Economic impact of COVID-19 pandemic on healthcare facilities and systems: International perspectives. Best Pract. Res. Clin. Anaesthesiol. 2020 doi: 10.1016/j.bpa.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adebisi Y.A., Lucero Prisno III D.E. Towards an inclusive health agenda: People who inject drugs and the COVID-19 response in Africa. Public Health. 2020 doi: 10.1016/j.puhe.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan H.P., Samala R.K., Hadjiiski L.M., Zhou C. Deep learning in medical image analysis. Adv. Exp. Med. Biol. 2020;1213:3–21. doi: 10.1007/978-3-030-33128-3_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bala K., Sharma S., Kaur G. A study on smartphone based operating system. Int. J. Comput. Appl. 2015;121 [Google Scholar]
- 18.Moschovis P.P., Sampayo E.M., Cook A., Doros G., Parry B.A., Lombay J., Kinane T.B., Taylor K., Keating T., Abeyratne U., Porter P., Carl J. The diagnosis of respiratory disease in children using a phone-based cough and symptom analysis algorithm: The smartphone recordings of cough sounds 2 (SMARTCOUGH-C 2) trial design. Contemp. Clin. Trials. 2021;101 doi: 10.1016/j.cct.2021.106278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sait U., Shivakumar S., Gokul Lal K.V., Kumar T., Ravishankar V.D., Bhalla K. 2019 IEEE Glob. Humanit. Technol. Conf. GHTC 2019. 2019. A mobile application for early diagnosis of pneumonia in the rural context. [DOI] [Google Scholar]
- 20.2020. Digital around the world, data reportal. https://datareportal.com/global-digital-overview. (Accessed 1 January 2021) [Google Scholar]
- 21.Statista U. Share of mobile phone users that use a smartphone in India from 2014 to 2019. Statista. 2019 https://www.statista.com/statistics/257048/smartphone-user-penetration-in-india/. (Accessed 30 December 2020) [Google Scholar]
- 22.Wanga H., Joseph T., Chuma M.B. Social distancing: Role of smartphone during coronavirus (COVID–19) pandemic era. Int. J. Comput. Sci. Mob. Comput. 2020;9:181–188. [Google Scholar]
- 23.Yasaka T.M., Lehrich B.M., Sahyouni R. Peer-to-Peer contact tracing: development of a privacy-preserving smartphone app. JMIR MHealth UHealth. 2020;8 doi: 10.2196/18936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLachlan S., Lucas P., Dube K., Hitman G.A., Osman M., Kyrimi E., Neil M., Fenton N.E. 2020. Bluetooth Smartphone Apps: Are they the most private and effective solution for COVID-19 contact tracing? ArXiv Prepr. arXiv:2005.06621. [Google Scholar]
- 25.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 26.Budd J., Miller B.S., Manning E.M., Lampos V., Zhuang M., Edelstein M., Rees G., Emery V.C., Stevens M.M., Keegan N. Digital technologies in the public-health response to COVID-19. Nat. Med. 2020;26:1183–1192. doi: 10.1038/s41591-020-1011-4. [DOI] [PubMed] [Google Scholar]
- 27.Currie D.J., Peng C.Q., Lyle D.M., Jameson B.A., Frommer M.S. Stemming the flow: how much can the Australian smartphone app help to control COVID-19. Public Heal. Res Pr. 2020;30 doi: 10.17061/phrp3022009. [DOI] [PubMed] [Google Scholar]
- 28.Collado-Borrell R., Escudero-Vilaplana V., Villanueva-Bueno C., Herranz-Alonso A., Sanjurjo-Saez M. Features and functionalities of smartphone apps related to COVID-19: systematic search in app stores and content analysis. J. Med. Internet Res. 2020;22 doi: 10.2196/20334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong N., Zhao L., Wu C.H., Chang J.F. Inception v3 based cervical cell classification combined with artificially extracted features. Appl. Soft Comput. 2020;93 doi: 10.1016/j.asoc.2020.106311. [DOI] [Google Scholar]
- 30.Milani P.M. 2017. The power of inception: Tackling the tiny ImageNet challenge; p. 7678. http://cs231n.stanford.edu/reports/2017/pdfs/928.pdf. [Google Scholar]
- 31.Chhikara P., Singh P., Gupta P., Bhatia T. Deep convolutional neural network with transfer learning for detecting pneumonia on chest x-rays. Adv. Intell. Syst. Comput. 2020;1064:155–168. doi: 10.1007/978-981-15-0339-9_13. [DOI] [Google Scholar]
- 32.Horry M.J., Chakraborty S., Paul M., Ulhaq A., Pradhan B., Saha M., Shukla N. COVID-19 detection through transfer learning using multimodal imaging data. IEEE Access. 2020;8:149808–149824. doi: 10.1109/ACCESS.2020.3016780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pal A., Sankarasubbu M. 2020. Pay attention to the cough: Early diagnosis of COVID-19 using interpretable symptoms embeddings with cough sound signal processing; pp. 1–15. http://arxiv.org/abs/2010.02417. [Google Scholar]
- 34.Padilla-Ortiz A.L., Ibarra D. Lung and heart sounds analysis: state-of-the-art and future trends. Crit. Rev. Biomed. Eng. 2018;46 doi: 10.1615/CritRevBiomedEng.2018025112. [DOI] [PubMed] [Google Scholar]
- 35.Kandaswamy A., Kumar C.S., Ramanathan R.P., Jayaraman S., Malmurugan N. Neural classification of lung sounds using wavelet coefficients. Comput. Biol. Med. 2004;34:523–537. doi: 10.1016/S0010-4825(03)00092-1. [DOI] [PubMed] [Google Scholar]
- 36.Wilkins R.L., Dexter J.R., Murphy R.L.H., DelBono E.A. Lung sound nomenclature survey. Chest. 1990;98:886–889. doi: 10.1378/chest.98.4.886. [DOI] [PubMed] [Google Scholar]
- 37.Aygün H., Apolskis A. The quality and reliability of the mechanical stethoscopes and Laser Doppler Vibrometer (LDV) to record tracheal sounds. Appl. Acoust. 2020;161 doi: 10.1016/j.apacoust.2019.107159. [DOI] [Google Scholar]
- 38.hui Huang Y., jun Meng S., Zhang Y., sheng Wu S., Zhang Y., wei Zhang Y., xiang Ye Y., feng Wei Q., gui Zhao N., ping Jiang J., ying Ji X., xia Zhou C., Zheng C., Zhang W., zhong Xie L., chao Hu Y., quan He J., Chen J., yue Wang W., hua Zhang C., Cao L., Xu W., Lei Y., hua Jian Z., ping Hu W., juan Qin W., yu Wang W., long He Y., Xiao H., fang Zheng X., Hu Y.Q., Pan W.S., feng Cai J. 2020. The respiratory sound features of COVID-19 patients fill gaps between clinical data and screening methods. [DOI] [Google Scholar]
- 39.Brown C., Chauhan J., Grammenos A., Han J., Hasthanasombat A., Spathis D., Xia T., Cicuta P., Mascolo C. Proc. ACM SIGKDD Int. Conf. Knowl. Discov. Data Min. 2020. Exploring automatic diagnosis of COVID-19 from crowdsourced respiratory sound data; pp. 3474–3484. [DOI] [Google Scholar]
- 40.Imran A., Posokhova I., Qureshi H.N., Masood U., Riaz M.S., Ali K., John C.N., Hussain M.D.I., Nabeel M. AI4COVID-19: AI enabled preliminary diagnosis for COVID-19 from cough samples via an app. Inf. Med. Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li F., Valero M., Shahriar H., Khan R.A., Ahamed S.I. Wi-COVID: A COVID-19 symptom detection and patient monitoring framework using WiFi. Smart Health. 2021;19 doi: 10.1016/j.smhl.2020.100147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vinod D.N., Prabaharan S.R.S. Data science and the role of Artificial intelligence in achieving the fast diagnosis of Covid-19. Chaos Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmad Alhiyari M., Ata F., Islam Alghizzawi M., Bint I Bilal A., Salih Abdulhadi A., Yousaf Z. Post COVID-19 fibrosis, an emerging complicationof SARS-CoV-2 infection. IDCases. 2021;23 doi: 10.1016/j.idcr.2020.e01041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quirino M.G., Colli C.M., Macedo L.C., Sell A.M., Visentainer J.E.L. Methods for blood group antigens detection: cost-effectiveness analysis of phenotyping and genotyping. Hematol. Transfus. Cell Ther. 2019;41:44–49. doi: 10.1016/j.htct.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belkacem A.N., Ouhbi S., Lakas A., Benkhelifa E., Chen C. 2020. End-to-end AI-based point-of-care diagnosis system for classifying respiratory illnesses and early detection of COVID-19; pp. 1–11. ArXiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alsabek M.B., Shahin I., Hassan A. 2020. Studying the similarity of COVID-19 sounds based on correlation analysis of MFCC; pp. 1–5. [DOI] [Google Scholar]
- 47.Hu Y., Deng H., Huang L., Xia L., Zhou X. Analysis of characteristics in death patients with COVID-19 pneumonia without underlying diseases. Acad. Radiol. 2020;27:752. doi: 10.1016/j.acra.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daniel Kermany M.G., Zhang Kang. Mendeley; 2018. Labeled Optical Coherence Tomography (OCT) and Chest X-Ray Images for Classification. https://data.mendeley.com/datasets/rscbjbr9sj/2. (Accessed 14 May 2019) [Google Scholar]
- 49.Hassantabar S., Ahmadi M., Sharifi A. Diagnosis and detection of infected tissue of COVID-19 patients based on lung x-ray image using convolutional neural network approaches. Chaos Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Panwar H., Gupta P.K., Siddiqui M.K., Morales-Menendez R., Singh V. Application of deep learning for fast detection of COVID-19 in X-rays using nCOVnet. Chaos Solitons Fractals. 2020;138 doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abbas A., Abdelsamea M.M., Gaber M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl. Intell. 2020 doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gunraj H., Wang L., Wong A. 2020. COVIDNet-CT: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest CT images; pp. 1–12. http://arxiv.org/abs/2009.05383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rahman T. Kaggle; 2020. COVID-19 Radiography Database. https://www.kaggle.com/tawsifurrahman/covid19-radiography-database. (Accessed 5 September 2020) [Google Scholar]
- 54.Khosravi B., Aghaghazvini L., Sorouri M., Naybandi Atashi S., Abdollahi M., Mojtabavi H., Khodabakhshi M., Motamedi F., Azizi F., Rajabi Z., Kasaeian A., Sima A.R., Davarpanah A.H., Radmard A.R. Predictive value of initial CT scan for various adverse outcomes in patients with COVID-19 pneumonia. Hear. Lung. 2020 doi: 10.1016/j.hrtlng.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alafif T., Tehame A.M., Bajaba S., Barnawi A., Zia S. 2020. Machine and deep learning towards COVID-19 diagnosis and treatment: Survey, challenges, and future directions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pathan S., Siddalingaswamy P.C., Ali T. Automated Detection of Covid-19 from Chest X-ray scans using an optimized CNN architecture. Appl. Soft Comput. 2021;104 doi: 10.1016/j.asoc.2021.107238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Demir F. DeepCoroNet: A deep LSTM approach for automated detection of COVID-19 cases from chest X-ray images. Appl. Soft Comput. 2021;103 doi: 10.1016/j.asoc.2021.107160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feki I., Ammar S., Kessentini Y., Muhammad K. Federated learning for COVID-19 screening from Chest X-ray images. Appl. Soft Comput. 2021;106 doi: 10.1016/j.asoc.2021.107330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rahaman M.M., Li C., Yao Y., Kulwa F., Rahman M.A., Wang Q., Qi S., Kong F., Zhu X., Zhao X. Identification of COVID-19 samples from chest X-Ray images using deep learning: A comparison of transfer learning approaches. J. X-Ray Sci. Technol. 2020;28:821–839. doi: 10.3233/XST-200715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pulm. Assoc. Brand.; 2017. Pulmonary Associates of Brandon, What’s the Difference between a Chest X-Ray and a Chest CT Scan?https://floridachest.com/pulmonary-blog/chest-x-ray-chestct-scan-differences (Accessed April 2021) [Google Scholar]
- 61.GitHub, Inc.; 2019. GitHub, Google Images Download. https://github.com/hardikvasa/google-images-download. (Accessed 13 May 2019) [Google Scholar]
- 62.SAIT U., GL k.V., Prajapati S., Bhaumik R., Kumar T., S S., Bhalla K. Mendeley; 2020. Curated Dataset for COVID-19 Posterior-Anterior Chest Radiography Images (X-Rays) https://data.mendeley.com/datasets/9xkhgts2s6/1. (Accessed 15 October 2020) [Google Scholar]
- 63.MD U., Sarkar Malay, Madabhavi Irappa, Niranjan Narasimhalu. Auscultation of the respiratory system. Ann. Thorac. Med. 2015;10(3):158–168. doi: 10.4103/1817-1737.160831. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4518345/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prajapati S.P., Bhaumik R., Kumar T., Sait U. In: An AI-Based Pedagogical Tool for Creating Sketched Representation of Emotive Product Forms in the Conceptual Design Stages BT - ICT Systems and Sustainability. Tuba M., Akashe S., Joshi A., editors. Springer Singapore; Singapore: 2021. pp. 649–659. [Google Scholar]
- 65.Sait U., Kv G., Shivakumar S., Kumar T., Bhaumik R., Prakash Prajapati S., Bhalla K. 2020. Spectrogram images of breathing sounds for COVID-19 and other pulmonary abnormalities. [DOI] [Google Scholar]
- 66.Grepl J., Penhaker M., Kubíček J., Liberda A., Selamat A., Majerník J., Hudák R. Real time breathing signal measurement: Current methods. IFAC-PapersOnLine. 2015;48:153–158. doi: 10.1016/j.ifacol.2015.07.024. [DOI] [Google Scholar]
- 67.Gunatilaka C.C., Schuh A., Higano N.S., Woods J.C., Bates A.J. The effect of airway motion and breathing phase during imaging on CFD simulations of respiratory airflow. Comput. Biol. Med. 2020;127 doi: 10.1016/j.compbiomed.2020.104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.2017. Last second medicine, complete breath sounds types & causes. https://www.youtube.com/watch?v=TlgP8MzlMaw. (Accessed 30 November 2020) [Google Scholar]
- 69.Sait U., Kv G., Shivakumar Sanjana, Kumar T., Bhaumik R., Prakash Prajapati S., Bhalla K. 2021. Spectrogram images of breathing sounds for COVID-19 and other pulmonary abnormalities, Mendeley Data. [DOI] [Google Scholar]
- 70.Shin H.-C., Roth H.R., Gao M., Lu L., Xu Z., Nogues I., Yao J., Mollura D., Summers R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging. 2016;35:1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chang J., Yu J., Han T., Chang H., Park E. 2017 IEEE 19th Int. Conf. E-Health Networking, Appl. Serv. IEEE; 2017. A method for classifying medical images using transfer learning: A pilot study on histopathology of breast cancer; pp. 1–4. [Google Scholar]
- 72.Talo M., Baloglu U.B., Yıldırım Ö., Acharya U.R. Application of deep transfer learning for automated brain abnormality classification using MR images. Cogn. Syst. Res. 2019;54:176–188. [Google Scholar]
- 73.Yu Y., Lin H., Meng J., Wei X., Guo H., Zhao Z. Deep transfer learning for modality classification of medical images. Information. 2017;8:91. [Google Scholar]
- 74.Wang J., Zhu H., Wang S.-H., Zhang Y.-D. A review of deep learning on medical image analysis. Mob. Netw. Appl. 2020:1–30. [Google Scholar]
- 75.2020. Google cloud, advanced guide to inception v3 on cloud TPU. https://cloud.google.com/tpu/docs/inception-v3-advanced. (Accessed 6 January 2021) [Google Scholar]
- 76.Treder M., Lauermann J.L., Eter N. Automated detection of exudative age-related macular degeneration in spectral domain optical coherence tomography using deep learning. Graefe’s Arch. Clin. Exp. Ophthalmol. 2018;256:259–265. doi: 10.1007/s00417-017-3850-3. [DOI] [PubMed] [Google Scholar]
- 77.Rampasek L., Goldenberg A. Learning from everyday images enables expert-like diagnosis of retinal diseases. Cell. 2018;172:893–895. doi: 10.1016/j.cell.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 78.Kingma D.P., Ba J. 2014. Adam: A method for stochastic optimization. ArXiv Prepr. arXiv:1412.6980. [Google Scholar]
- 79.Eckle K., Schmidt-Hieber J. A comparison of deep networks with relu activation function and linear spline-type methods. Neural Netw. 2019;110:232–242. doi: 10.1016/j.neunet.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 80.Brownlee J. Use early stopping to halt the training of neural networks at the right time. Mach. Learn. Mastery. 2018;9:2018. [Google Scholar]
- 81.Wang M., Li C., Zhang W., Wang Y., Feng Y., Liang Y., Wei J., Zhang X., Li X., Chen R. Support vector machine for analyzing contributions of brain regions during task-state fMRI. Front. Neuroinformatics. 2019;13:10. doi: 10.3389/fninf.2019.00010. https://www.frontiersin.org/article/10.3389/fninf.2019.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brownlee J. How to use weight decay to reduce overfitting of neural network in keras. Mach. Learn. Mastery. 2018 https://machinelearningmastery.com/how-to-reduce-overfitting-in-deep-learning-with-weight-regularization/. (Accessed 19 April 2021) [Google Scholar]
- 83.Fangoh A.M., Selim S. 2020 15th Int. Conf. Comput. Eng. Syst. 2020. Using CNN-XGBoost deep networks for COVID-19 detection in chest X-ray images; pp. 1–7. [DOI] [Google Scholar]
- 84.2021. Wikipedia, precision and recall. https://en.wikipedia.org/wiki?curid=14343887. (Accessed 20 April 2021) [Google Scholar]
- 85.2021. Qualcomm technologies, snapdragon 430 mobile platform. https://www.qualcomm.com/products/snapdragon-430-mobile-platform. (Accessed 20 April 2021) [Google Scholar]
- 86.2021. Qualcomm technologies, snapdragon 835 mobile platform. https://www.qualcomm.com/products/snapdragon-835-mobile-platform. (Accessed 20 April 2002) [Google Scholar]
- 87.2021. Qualcomm technologies, snapdragon 855+/860 mobile platform. https://www.qualcomm.com/products/snapdragon-855-plus-and-860-mobile-platform. (Accessed 20 April 2021) [Google Scholar]