Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes coronavirus disease 2019 (COVID-19), has the characteristic accessory protein ORF8. Although clinical reports indicate that ORF8 variant strains (Δ382 and L84S variants) are less likely to cause severe illness, functional differences between wild-type and variant ORF8 are unknown. Furthermore, the physicochemical properties of the ORF8 protein have not been analyzed. In this study, the physicochemical properties of the wild-type ORF8 and its L84S variant were analyzed and compared. Using the tobacco BY-2 cell production system, which has been successfully used to produce the wild-type ORF8 protein with a single conformation, was used to successfully produce the ORF8 L84S variant protein at the same level as wild-type ORF8. The produced proteins were purified, and their temperature and pH dependencies were examined using nuclear magnetic resonance spectra. Our data suggested that the wild-type and L84S variant ORF8 structures are highly stable over a wide temperature range. Both proteins displayed an aggregated conformation at higher temperature that reverted when the temperature was decreased to room temperature. Moreover, ORF8 precipitated at acidic pH and this precipitation was reversed when the solution pH was shifted to neutral. Interestingly, the L84S variant exhibited greater solubility than wild-type ORF8 under acidic conditions. Thus, the finding indicated that conformational stability and reversibility of ORF8 are key properties related to function in oppressive environments.
Keywords: L84S variant, ORF8, Protein conformation, Protein stability, Reversibility, SARS-CoV-2
Abbreviations: COVID-19, coronavirus disease 2019; IFN, interferon; IL-1RA, interleukin-1 receptor antagonist; MHC, myosin heavy chain; MCP, monocyte chemoattractant protein; NMR, nuclear magnetic resonance; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; ToMV, tobamovirus
1. Introduction
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), emerged in 2019. As of May 2021, approximately 160,000,000 cases of COVID-19, including nearly 3.3 million fatalities, have been reported globally (Johns Hopkins Coronavirus Resource Center, https://coronavirus.jhu.edu/map.html). It has been reported that patients with underlying diseases such as diabetes and adiposity are more susceptible to severe COVID-19 [[1], [2], [3]]. To facilitate the development of therapeutic agents against COVID-19, it is necessary to determine the mode of action of SARS-CoV-2. The genome of SARS-CoV-2 has been sequenced [[4], [5], [6], [7]], and the proteins produced by SARS-CoV-2 have also been predicted. Interestingly, among the proteins produced by SARS-CoV-2, the accessory protein ORF8 displays lower homology than those of closely related SARS viruses [4,8]. Subsequently, Flower et al. revealed the three-dimensional structure of ORF8 via X-ray crystallography [9]. ORF8 forms a dimer via disulfide bonds [9,10], and it is transported out of cells [11]. ORF8 is highly immunogenic because anti-ORF8 antibodies are formed in the early stage of SARS-CoV-2 infection [12], and a significant T-cell response to ORF8 is observed in recovered patients [13]. Furthermore, SARS-CoV-2 mutants carrying ORF8 variants (Δ382 variant and L84S variant) were reported to be less likely to cause severe disease [14,15]. Patients infected with the Δ382 variant displayed higher levels of interferon (IFN)-γ and lower levels of chemokines (IFN-γ–inducible protein 10, monocyte chemoattractant protein [MCP]-1, and MCP-1β) and anti-inflammatory proteins (interleukin 1 receptor antagonist [IL-1RA]) than those infected with the wild-type virus [14]. Thus, ORF8 is considered to be closely related to COVID-19 symptoms caused by SARS-CoV-2.
Recent studies have begun to reveal the function of ORF8. ORF8 might prevent the induction of immunity following SARS-CoV-2 infection by strongly suppressing myosin heavy chain (MHC)-I [16], and it might be involved in the induction of the cytokine storm [17]. However, no studies have examined the physicochemical properties of ORF8 protein. To analyze the physicochemical properties of ORF8 protein, a large amount of homogeneous protein folded into its native conformation must be produced. A plant-based production system can be used to mass-produce physiologically active eukaryotic proteins. We previously developed a highly efficient chemically inducible protein production system using tobacco BY-2 cells by utilizing the extremely high protein production capacity of tobamovirus (ToMV) [18]. We successfully produced large amounts of plant- and animal-derived proteins in a bioactive form using this system [[19], [20], [21]]. Recently, we succeeded in the mass production of ORF8 protein folded into a single conformation [10].
Studying the physicochemical properties of ORF8 is important for completely understanding its structure–function relationship. In this study, we succeeded in the mass production of wild-type ORF8 and its L84S variant with a single conformation in tobacco BY-2 cells. Using these proteins, we employed nuclear magnetic resonance (NMR) to examine the conformational stability of ORF8. NMR data suggested that ORF8 is stable against temperature and pH changes. Although ORF8 aggregated and precipitated at higher temperature and lower pH, respectively, it regained its original conformation when the temperature was lowered to room temperature and pH was increased to neutral.
2. Material and methods
2.1. Plasmid construction and transformation of BY-2 cells
Tobacco BY-2 cells were cultured in Linsmaier and Skoog medium supplemented with 3% sucrose and 0.2 mg/L 2,4-dichlorophenoxyacetic acid at 26 °C [22].
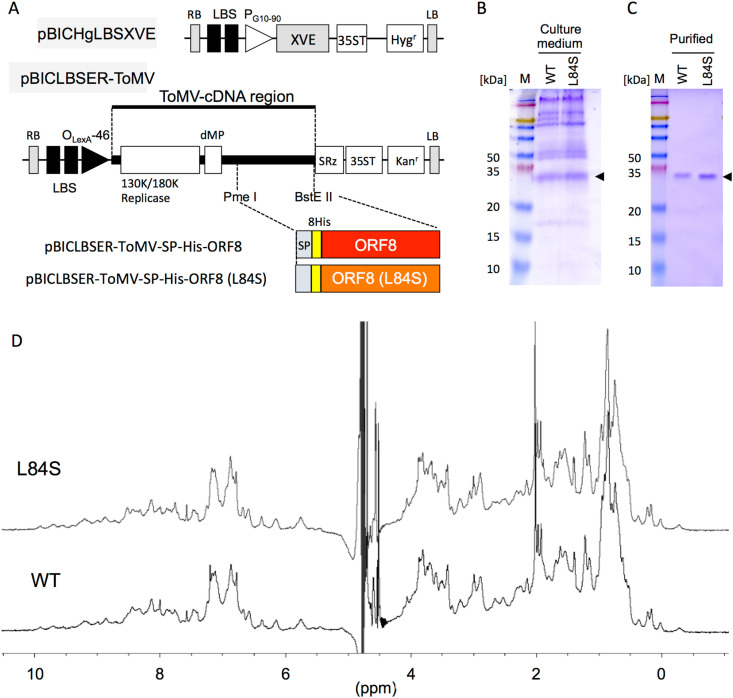
To construct a plasmid for the producing His-tagged ORF8 L84S variant protein, His-tagged ORF8 L84S, which was fused with the Arabidopsis chitinase signal peptide at its N-terminus, was generated by PCR. PrimeSTAR GXL DNA polymerase (TaKaRa, Kusatsu, Japan) and oligonucleotides containing a restriction enzyme cleavage site were used for PCR amplification (Supplementary Table 1). The template for PCR was pBICLBSER-ToMV-SP-His-ORF8, which produced 8×His-taged-DDDDK-ORF8 protein [10]. The procedure for amplification comprised initial denaturation at 94 °C for 2 min, followed by 35 cycles at 98 °C for 10 s, 55 °C for 15 s, and 68 °C for 30 s. The amplified fragment was introduced into a chemically inducible ToMV vector (pBICLBSER-ToMV) to generate pBICLBSER-ToMV-SP-His-ORF8(L84S) (Fig. 1 A). BigDye terminator chemistry and an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Foster City, CA, USA) were used to sequence the resulting plasmids.
Fig. 1.
Production of ORF8 protein in tobacco BY-2 cells. (A) Schematic representation of tobamovirus (ToMV)-mediated chemically induced expression plasmids. XVE, transcriptional activator that responds to estrogen; SP, signal peptide of Arabidopsis chitinase; 8His, 8 × His-tag; ORF8; mature ORF8 coding sequence (CDS); ORF8(L84S), ORF8 L84S variant CDS; LBS, LexA binding site; PG10-90, synthetic constitutive promoter; PLexA-46, fusion promoter controlled by XVE; dMP, partial movement protein; SRz, ribozyme sequence from tobacco ringspot virus satellite RNA; 35ST, 35S terminator; RB, right border; and LB, left border. Hygr and Kanr denote the expression cassette of the hygromycin and kanamycin resistance genes, respectively. (B) Detection of wild-type ORF8 and its L84S variant via Coomassie brilliant blue staining without dithiothreitol. Culture medium represents samples from cells incucubated in culture medium supplemented with 17β-estradiol for 7 days. (C) Purified indicates ORF8s purified using a Ni2+ affinity column and size exclusion column. Arrowheads indicate ORF8. M, protein size marker. Numbers indicate molecular weights (kDa). (D) 1H nuclear magnetic resonance (NMR) spectra of wild-type ORF8 (WT) and its L84S variant (L84S). The spectra were recorded on a 500 MHz-NMR spectrometer. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Next, pBICHgLBSXVE (Fig. 1A) expressing the artificial transcription factor XVE, which activates transcription by binding with 17β-estradiol [18], and pBICLBSER-ToMV-SP-His-ORF8(L84S) were introduced into tobacco BY-2 cells via Agrobacterium-mediated transformation [23]. The His-tagged ORF8 variant-producing cell line was selected on agar medium containing 50 mg/L hygromycin, 100 mg/L kanamycin, and 500 mg/L carbenicillin. His-tagged ORF8-producing cell line was generated in a previous study [10]. Suspension cells developed from calli were cultured in 3 mL of liquid medium in six-well culture plates during primary screening, after which they were transferred to 150 mL of liquid medium in 500-mL flasks under constant shaking conditions at 135 rpm. After initial culture for 2–3 weeks, the suspension cells were maintained without selective agents. These cell lines were suspension-cultured in normal MS medium.
2.2. Protein production and purification
Protein production was induced by adding 10 μM 17β-estradiol [18]. After 7 days, desired proteins accumulated in the culture medium and the culture medium was collected via centrifugation. In the first purification step using a nickel column (His GraviTrap, GE Healthcare, Chicago, Illinois, USA), proteins in the culture medium were eluted with phosphate buffer containing 500 mM imidazole (pH 6.8). Next, the eluate was purified by gel filtration chromatography using AKTA prime plus (GE Healthcare) to obtain a single protein. Gel filtration chromatography was performed using Superdex 75 10/300 GL (Amersham Biosciences, Uppsala, Sweden) as the column and 500 mM imidazole in phosphate buffer (pH 6.8) as the buffer. The collected fraction was concentrated using a centrifugal concentrator (CC-105, Tomy Seiko Inc., Tokyo, Japan) and replaced with 0.02 × PBS (pH 7.4) by dialysis. After concentration, protein samples were stored at 4 °C until further use.
2.3. NMR study
For both wild-type ORF8 and its L84S variant, 2 mg/mL of protein sample was dissolved in the NMR buffer containing 2.74 mM NaCl, 162 μM Na2HPO4, 53.6 μM KCl, 29.4 μM KH2PO4, and 10% D2O. The pH of the sample was adjusted to 7.8 for the temperature experiments. Regarding pH titration experiments, the pH of the sample was changed by adding DCl or NaOD. The pH value of the sample was directly measured using a Toko TPX-90i pH meter (Toko Co., Japan) without considering the isotope effect. Each sample was packed in a Shigemi NMR tube (Shigemi, Japan). NMR measurements were performed using a Bruker AVANCE III 500 equipped with a BBO probe or a Bruker AVANCE III 800 equipped with a TCI cryogenic probe. In both machines, the sample temperature was controlled using a BVT3000 temperature controller (Bruker Biospin, Germany). Data processing was performed using Bruker TopSpin ver. 3.6.2.
2.4. Modeling the three-dimensional structure of the L84S variant
The three-dimensional structure of the L84S variant was modeled on the Phyre2 web server [24]. SARS-CoV-2 ORF8 (PDB code; 7JTL) was selected as a template for homology modeling and 88.4% of amino acid residues were involved in the modeling.
3. Results
3.1. ORF8 protein production in BY-2 cells
To compare the physicochemical properties of wild-type ORF8 and its L84S variant, we attempted to mass-produce L84S. After 7 days, the production of wild-type ORF8 and its L84S variant was confirmed in the culture medium (Fig. 1B). Both proteins were present at concentrations of approximately 10 mg/L culture medium. Following purification, wild-type ORF8 and its L84S variant in the culture medium were obtained as single proteins and both existed as dimeric forms (Fig. 1C).
3.2. Conformation of the L84S variant
Fig. 1D presents 1H NMR spectra for wild-type ORF8 and its L84S variant at 30 °C. The existence of higher field-shifted methyl signals and lower field-shifted amide proton signals suggested that these samples had folded conformations. These two spectra were almost identical, indicating that their three-dimensional structures are extremely similar. The result indicates that the mutation introduced at residue 84 does not affect the overall conformation of ORF8. Because residue 84 is located in the solvent exposed loop, the result seems acceptable. To obtain supporting information, we also attempted to model the three-dimensional structure of the L84S variant (Supplementary Fig. 1). The L84S variant had an identical structure to that of ORF8 excluding the orientation of the ORF-specific flexible loop. This result supports our interpretation of the NMR spectra.
3.3. Thermal stability
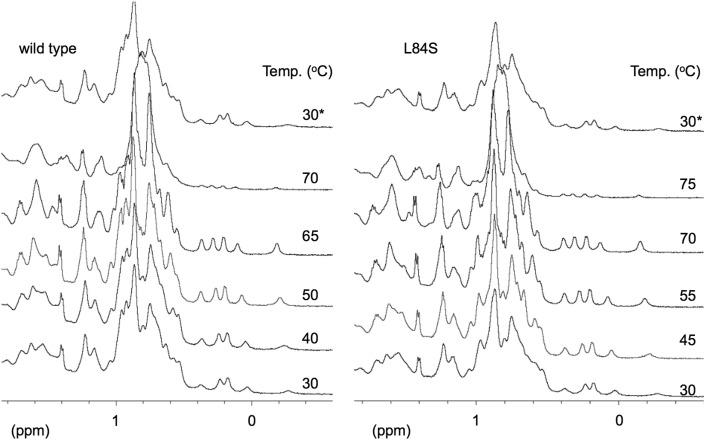
Fig. 2 presents the methyl region of the 1H NMR spectra at various temperatures. The left panel in Fig. 2 presents the NMR spectral changes of wild-type ORF8. Extremely small spectral changes were observed at <65 °C. This indicated that ORF8 retains its conformation between room temperature and 65 °C. However, the spectrum at 70 °C was completely different from those at lower temperatures. The signals around 0 ppm were nearly absent, and the overlapping large peak appearing from 0.5 to 1 ppm broadened. This spectrum was also different from that of random coils, which display sharp NMR peaks. At 70 °C, the NMR sample obtained from the NMR magnet appeared to be clear. Thus, the sample at 70 °C was presumably in equilibrium between the aggregated state and the original dimeric form. After cooling the sample to room temperature, ORF8 displayed an identical NMR spectrum to the spectrum observed at 30 °C. This finding clearly indicated that the conformation of ORF8 was reversible against heating.
Fig. 2.
Methyl region of the 1H nuclear magnetic resonance (NMR) spectra at various temperatures. Wild-type ORF8 (left) and the L84S variant (right) were analyzed. The spectra labeled with 30∗ were those recorded at 30 °C after heating.
As presented in the right panel of Fig. 2, the L84S variant was also heat-stable. Unlike wild-type ORF8, the L84S variant had a clear 1H NMR spectrum at 70 °C, but not at 75 °C. It was found that the denaturing temperature of L84S variant is slightly higher than that of wild type. The mutation from Leu to Ser at residue 84 is expected to reduce the hydrophobicity of the molecular surface, because L84 is located at a loop exposed to solvent [9]. Thus, this mutation is likely a factor that prevents heat-induced aggregation. When the sample was cooled to room temperature, the L84S variant displayed a spectrum identical to that observed for the initial sample at 30 °C. Thus, the L84S variant exhibited the same ability to regain its original conformation following temperature fluctuation as wild-type ORF8.
3.4. pH dependency
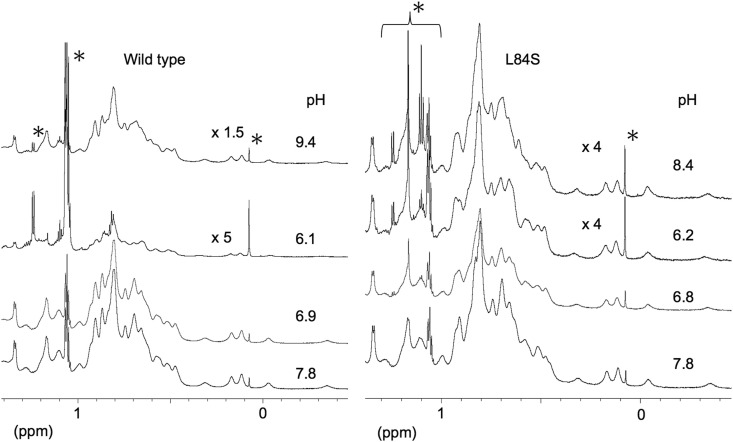
Fig. 3 presents the methyl region of the 1H NMR spectra under various pH conditions. The left panel in Fig. 3 presents the spectral changes of ORF8. Starting from an initial pH of 7.8, the NMR spectrum was unchanged until the pH reached approximately 6.3. When we further reduced the pH of the sample, flocculation was observed in the sample solution. Because of this flocculation, the concentration of the dissolved ORF8 decreased, resulting in diminished NMR signal intensity at pH 6.1. The spectrum itself was similar to that observed at pH 7.8; thus, the dissolved ORF8 retained its conformation. After the NMR measurement, we attempted to alter the pH of the sample from acidic to alkaline. Surprisingly, the NMR sample became clear. Moreover, the sample at pH 9.4 had an NMR spectrum that was nearly identical as the initial sample at pH 7.8. These results strongly suggested that precipitated ORF8 returned to its soluble state with proper conformation when the pH of the sample is controlled.
Fig. 3.
Methyl region of 1H nuclear magnetic resonance (NMR) spectra at various pH values. Wild-type ORF8 (left) and the L84S variant (right) were analyzed. The peaks labeled with ∗ were resonated from impurity. The label “xN” means that the vertical axis was magnified N times as indicated by the height of an impurity peak at 0.08 ppm. The spectra were recorded at 25 °C on an 800 MHz-NMR spectrometer.
The solution of the L84S variant also became cloudy at lower pH. However, the precipitate was wellsuspended unlike that of wild-type ORF8. As presented in the right panel of Fig. 3, the peak intensity of the L84S variant at pH 6.2 was much greater than that of wild-type ORF8 at pH 6.1. Therefore, the L84S variant had greater solubility at reduced pH (6.1–6.2) than wild-type ORF8. When the pH of the sample was altered from acidic to alkaline, the sample precipitate dissolved, and the solution became clear. The NMR spectrum of the redissolved sample at pH 8.4 was identical to that of the L84S variant at pH 7.8. Thus, the L84S variant exhibited conformational reversibility following pH fluctuation similar to wild-type ORF8.
After the pH change experiments, both wild-type and variant NMR samples were subjected to SDS-PAGE analysis, as presented in Fig. 4 . The results clearly indicated that both wild-type ORF8 and its L84S variant maintained their dimeric nature, indicating that stability of conformation and retention were achieved even in the redissolved state after precipitation.
Fig. 4.
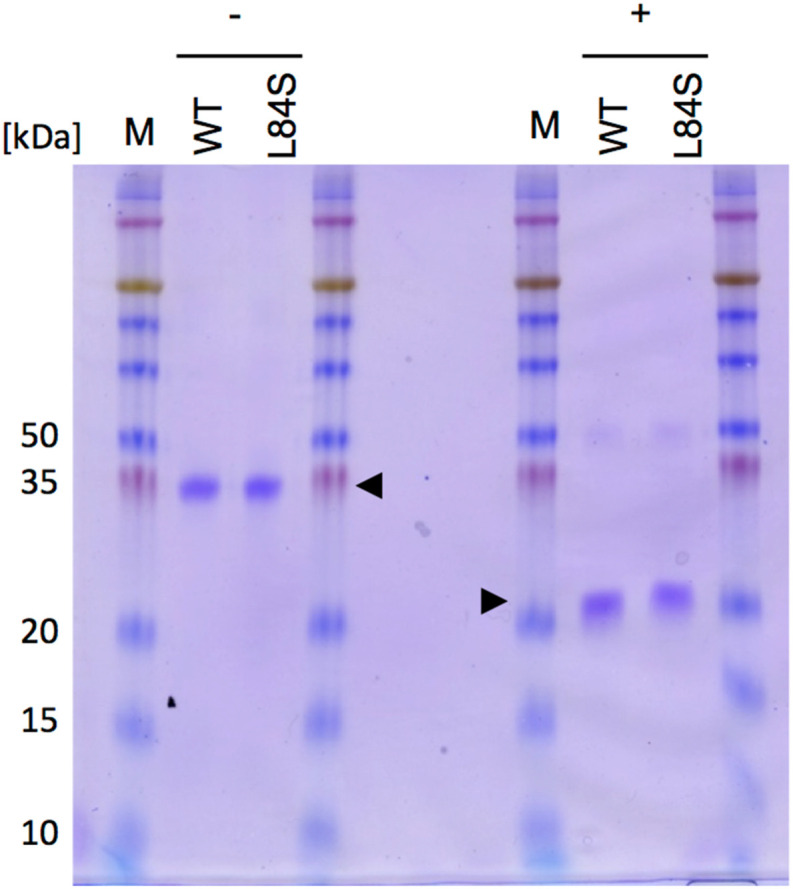
Resolubilization of ORF8 proteins. Detection of wild-type ORF8 and its L84S variant via Coomassie brilliant blue staining. WT, wild-type ORF8; L84S, L84S variant. + and − indicate the presence and absence of 10 mM dithiothreitol, respectively. Arrowheads indicate ORF8. M, protein size marker. Numbers indicate molecular weights (kDa). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
In this study, we analyzed the physicochemical properties of ORF8 to determine its involvement in COVID-19. Using a BY-2 cell-based protein production system, we mass-produced wild-type ORF8 and its L84S variant with a single conformation. The wild-type and L84S variant proteins were purified and enriched, and their thermal and pH stability were evaluated using NMR. The thermal stability of both proteins suggested that they would retain their activity at higher temperatures. With respect to pH stability, the proteins precipitated and aggregated under weakly acidic conditions. The precipitated and aggregated states of ORF8 could be reverted to the soluble state with proper conformation when the surrounding environment became neutral. Intriguingly, the L84S variant displayed slightly greater conformational stability against temperature and pH variation than wild-type ORF8.
In prior research, no patients infected with SARS-CoV-2 strain lacking the ORF8 gene developed severe disease [14]. Patients infected with strain carrying the L84S variant exhibited significantly less severe illness than those infected with wild-type ORF8-expressing strain [15]. These reports suggest that ORF8 influences the severity of COVID-19. Recent functional analyses indicated that ORF8 evades immunity by inhibiting the extracellular presentation of MHC-I [16], and it possibly participates in cytokine storm induction by binding to the proinflammatory cytokine receptor IL-17RA [17]. However, no reports have described differences in the function of wild-type ORF8 and its L84S variant that would explain differences in disease severity. This study revealed that ORF8 precipitates and aggregates under weakly acidic conditions (pH 6.1). Furthermore, the L84S variant was more stable and soluble than the wild-type protein under weakly acidic conditions. In eukaryotic cells, the intracellular pH is different in each organelle [25]. The organelles with more acidic environments include the trans-Golgi network (pH of approximately 6), lysosome (pH 4.7), endosome (pH 5.5–6.5), and secretory granules (pH 5.2) [25]. The presence of ORF8 in these acidic organelles may result in precipitation resulting in impaired functioning of organelles. In fact, it has been reported that the transfer of MHC-I to the cell surface, which involves passages through the Golgi network, is impaired in cells overexpressing ORF8 [16]. In addition, the L84S variant, which is more soluble than the wild-type protein under weakly acidic conditions, is expected to cause less damage to acidic organelles. This might explain the low incidence of severe disease in patients infected with SARS-CoV-2 strains carrying the L84S variant.
The L84S variant exhibited greater solubility than the wild-type protein under weakly acidic conditions. Modeling of the three-dimensional structure revealed no significant differences in the conformation of the wild-type ORF8 and its L84S variant (Supplementary Fig. 1). Therefore, the difference in the solubility of the two proteins under weakly acidic conditions can be attributed to the difference in the side chain of the amino acid residue 84. Because this residue is located on the surface of the protein [9], the Lue to Ser mutation certainly changes the molecular surface properties such as hydrophobicity. However, it is unclear why protein stability is enhanced by this single mutation currently. Additionally, it is noteworthy that in BY-2 cells, the production of an ORF8 variant lacking the intermolecular disulfide bond (ORF8 C20S variant) was significantly lower than that of wild-type ORF8 and its L84S variant (Supplementary Table 2), probably because of reduced dimerizing ability. These facts suggested that some residues contribute to the conformational stability and reversibility of ORF8. Further studies based on the new findings reported in this study will provide clues to understand the structure–function relationship of ORF8, which may be related to the severe disease.
We believe that the information in this study will aid the identification and/or design of chemical compounds that can modulate the stability and reversibility of ORF8, which could prompt the development of novel therapeutic agents.
Author contributions
MM conducted this study. MM and SO designed the experiments. All authors performed the experiments. SO, TI, and MM wrote the manuscript. All authors have read and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was partly supported by the Nanotechnology Platform of MEXT, Japan. The authors deeply thank all technicians at Center for Nano Materials and Technology (CNMT), JAIST for maintenance of the NMR machines used in this study. The authors also thank Akiko Mizuno, Hiroko Hayashi, and Mami Awatani for providing technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2021.05.074.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Huang I., Lim M.A., Pranata R. Diabetes mellitus is associated with increased mortality and severity of disease in COVID-19 pneumonia - a systematic review, meta-analysis, and meta-regression. Diabetes Metab. Syndr. 2020;14:395–403. doi: 10.1016/j.dsx.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williamson E.J., Walker A.J., Bhaskaran K., et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGurnaghan S.J., Weir A., Bishop J., et al. Risks of and risk factors for COVID-19 disease in people with diabetes: a cohort study of the total population of Scotland. Lancet Diabetes Endocrinol. 2021;9:82–93. doi: 10.1016/s2213-8587(20)30405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu R., Zhao X., Li J., et al. Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/s0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu F., Zhao S., Yu B., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou P., Yang X.L., Wang X.G., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan Y., Schneider T., Leong M., et al. Novel immunoglobulin domain proteins provide insights into evolution and pathogenesis of SARS-CoV-2-related viruses. mBio. 2020;11 doi: 10.1128/mBio.00760-20. e00760-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flower T.G., Buffalo C.Z., Hooy R.M., et al. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proc. Natl. Acad. Sci. U.S.A. 2021;118 doi: 10.1073/pnas.2021785118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imamura T., Isozumi N., Higashimura Y., et al. Production of ORF8 protein from SARS-CoV-2 using an inducible virus-mediated expression system in suspension-cultured tobacco BY-2 cells. Plant Cell Rep. 2021;40:433–436. doi: 10.1007/s00299-020-02654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., Lam J.Y., Wong W.M., et al. Accurate diagnosis of COVID-19 by a novel immunogenic secreted SARS-CoV-2 orf8 protein. mBio. 2020;11 doi: 10.1128/mbio.02431-20. e02431-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hachim A., Kavian N., Cohen C.A., et al. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 2020;21:1293–1301. doi: 10.1038/s41590-020-0773-7. [DOI] [PubMed] [Google Scholar]
- 13.Grifoni A., Weiskopf D., Ramirez S.I., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. e1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young B.E., Fong S.W., Chan Y.H., et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. 2020;396:603–611. doi: 10.1016/s0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagy Á., Pongor S., Győrffy B. Different mutations in SARS-CoV-2 associate with severe and mild outcome. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.106272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Zhang J., Chen Y., et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through potently downregulating MHC-I. bioRxiv [Preprint] 2020 doi: 10.1101/2020.05.24.111823. [DOI] [Google Scholar]
- 17.Lin X., Fu B., Yin S., et al. ORF8 contributes to cytokine storm during SARS-CoV-2 infection by activating IL-17 pathway. iScience. 2021;24 doi: 10.1016/j.isci.2021.102293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dohi K., Nishikiori M., Tamai A., et al. Inducible virus-mediated expression of a foreign protein in suspension-cultured plant cells. Arch. Virol. 2006;151:1075–1084. doi: 10.1007/s00705-005-0705-8. [DOI] [PubMed] [Google Scholar]
- 19.Costa L.M., Marshall E., Tesfaye M., et al. Central cell-derived peptides regulate early embryo patterning in flowering plants. Science. 2014;344:168–172. doi: 10.1126/science.1243005. [DOI] [PubMed] [Google Scholar]
- 20.Ohki S., Dohi K., Tamai A., et al. Stable-isotope labeling using an inducible viral infection system in suspension-cultured plant cells. J. Biomol. NMR. 2008;42:271–277. doi: 10.1007/s10858-008-9283-x. [DOI] [PubMed] [Google Scholar]
- 21.Ohki S., Takeuchi M., Mori M. The NMR structure of stomagen reveals the basis of stomatal density regulation by plant peptide hormones. Nat. Commun. 2011;2:512. doi: 10.1038/ncomms1520. [DOI] [PubMed] [Google Scholar]
- 22.Nagata T., Nemoto Y., Hasezawa S. In: International Review of Cytology. Jeon K.W., Friedlander M., editors. Academic Press; Cambridge: 1992. Tobacco BY-2 cell line as the “HeLa” Cell in the cell biology of higher plants; pp. 1–30. [Google Scholar]
- 23.Hagiwara Y., Komoda K., Yamanaka T., et al. Subcellular localization of host and viral proteins associated with tobamovirus RNA replication. EMBO J. 2003;22:344–353. doi: 10.1093/emboj/cdg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelley L.A., Mezulis S., Yates C.M., et al. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casey J.R., Grinstein S., Orlowski J. Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 2010;11:5–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.