Abstract
A computer-aided diagnosis (CAD) system that employs a super learner to diagnose the presence or absence of a disease has been developed. Each clinical dataset is preprocessed and split into training set (60%) and testing set (40%). A wrapper approach that uses three bioinspired algorithms, namely, cat swarm optimization (CSO), krill herd (KH) ,and bacterial foraging optimization (BFO) with the classification accuracy of support vector machine (SVM) as the fitness function has been used for feature selection. The selected features of each bioinspired algorithm are stored in three separate databases. The features selected by each bioinspired algorithm are used to train three back propagation neural networks (BPNN) independently using the conjugate gradient algorithm (CGA). Classifier testing is performed by using the testing set on each trained classifier, and the diagnostic results obtained are used to evaluate the performance of each classifier. The classification results obtained for each instance of the testing set of the three classifiers and the class label associated with each instance of the testing set will be the candidate instances for training and testing the super learner. The training set comprises of 80% of the instances, and the testing set comprises of 20% of the instances. Experimentation has been carried out using seven clinical datasets from the University of California Irvine (UCI) machine learning repository. The super learner has achieved a classification accuracy of 96.83% for Wisconsin diagnostic breast cancer dataset (WDBC), 86.36% for Statlog heart disease dataset (SHD), 94.74% for hepatocellular carcinoma dataset (HCC), 90.48% for hepatitis dataset (HD), 81.82% for vertebral column dataset (VCD), 84% for Cleveland heart disease dataset (CHD), and 70% for Indian liver patient dataset (ILP).
1. Introduction
Data related to symptoms observed on a patient at a point of time are stored in electronic health records (EHRs). Interesting patterns can be extracted from the data that are stored in EHRs, and the extracted patterns can be represented as knowledge, and this knowledge can assist the physicians to diagnose the presence or absence of a disease. Data mining tasks, namely, association rule mining, classification, and clustering are used to mine valuable patterns from the data stored in EHRs. Clinical decision support systems (CDSSs) that assist the physicians to diagnose the presence or absence of a disease can be developed from data stored in EHRs using bioinspired algorithms and data mining techniques. Although several algorithms have been proposed by researchers for association rule mining, classification, and clustering, no algorithm can be deliberated to be the “universal best.” Quality of data and data distribution are the two key factors that determine the effectiveness of a data mining task. The performance of a data mining task depends on how effective data preprocessing has been done. Classification plays a major role in the development of CDSSs. Classification is a two-step process, first, building the classifier and second, model usage. Building the classifier is the process of training the classifier with a supervised learning algorithm. Model usage is the process of estimating the accuracy of the classifier using testing instances commonly referred to as testing set. Overfitting and underfitting are two major problems associated with building the classifier.
Clinical dataset (s) (Cs) used for classifier construction is split into a training set (Ts) and a testing set (Tt). Researchers have proposed different methods to identify the Ts and Tt. One common method is to split 80% of the dataset into Ts and 20% of the dataset into Tt. For clinical decision-making, a balanced dataset is essential for building a prediction model. Clinical datasets are normally not balanced, and classification methods perform poorly on minority class samples when the dataset is tremendously imbalanced. For example, consider a Cs with n instances, each instance associated with a class label c1 or c2. Among the n instances that 75% of the instances in Cs are associated with class label c1, and 25% of the instances in Cs are associated with class label c2, it is evident that the class labels in Cs are not equally represented and therefore, the Cs is imbalanced. In this context, c1 is the majority class, and c2 is the minority class, and hence, constructing a classifier with class-imbalanced data will lead to bias in favor of the majority class. One method to handle class imbalance in a Cs is to generate additional instances from the minority class. The Synthetic Minority Oversampling Technique (SMOTE) [1] is one of the prevailing methods used to generate additional training and testing instances.
A training instance can be defined as a tuple ti(f1, f2, ⋯fm), where ti represents a training instance, and (f1, f2, ⋯fj) represents the features corresponding to a training instance. The subscript i in ti can range from 1 to n, where n is the number of instances. The subscript j in fj can range from 1 to m, where m is the number of features. Using irrelevant features to train a classifier will affect its performance. Selecting the optimal features from the Cs and then training the classifier will enhance the accuracy of the classifier. Feature selection methods can be supervised, unsupervised, and semisupervised depending upon whether the training set is labeled or not. Commonly used supervised feature selection methods are filter and wrapper methods. The filter method considers the dependency of each feature to the class label and is independent of any classification algorithm. Measures, namely, information gain [2], gain ratio [3], Gini index [4], Laplacian score [5], and cosine similarity [6] can be used to rank the features. Other measures to rank the features can also be used in filter method. The wrapper method considers the classification accuracy of a learning algorithm to select the relevant features. Researchers are using a confluence of disciplines to develop computer-aided diagnostic (CAD) systems to assist physicians.
Knowledge mining using rough sets for feature selection and backpropagation neural network (BPNN) for classifying clinical datasets has been proposed in [7]. A CDSS to diagnose Urticaria using Bayes classification is proposed in [8]. CDSSs to diagnose lung disorders are proposed in [9–14]. A CDSS to diagnose the severity of gait disturbances using a Q-backpropogated time delay neural network on patients affected by Parkinson's disease is proposed in [15]. A statistical tolerance rough set induced decision tree classifier to classify multivariate time series clinical data is proposed in [16]. A CDSS to diagnose gestational diabetes mellitus using the fuzzy logic and radial basis function neural network is proposed in [17]. Use of fuzzy sets and extreme learning machine to classify clinical datasets is proposed in [18]. Wind-driven swarm optimization, a metaheuristic method to classify clinical datasets, is proposed in [19]. A computer-aided diagnostic system that uses a neural network classifier trained using differential evolution, particle swarm optimization, and gradient descent backpropagation algorithms is proposed in [20]. A radial basis function neural network to classify clinical datasets using k-means clustering algorithm and quantum-behaved particle swarm optimization is proposed in [21]. Classifying clinical unevenly spaced time series data by imputing missing values has been proposed in [22]. A framework to classify unevenly spaced time series clinical data using improved double exponential smoothing, rough sets, neural network, and fuzzy logic is proposed in [23].
An outline of nature-inspired algorithms for optimization is presented in [24]. The cooperative intellectual actions of insects or animal groups in nature, for example, colonies of ants, schools of fish, flock of birds, swarms of bees, and termites, have fascinated the thoughtfulness of researchers. Entomologists have studied the collective actions of insects or animals to model biological swarms, and engineers have applied these models as a framework to solve complex real-world problems.
In this work, a CAD system that employs a super learner to diagnose the presence or absence of a disease has been proposed. The bioinspired algorithms used in this work are cat swarm optimization (CSO), krill herd (KH), and bacterial foraging optimization (BFO). The classifiers used in this work are support vector machine (SVM) and BPNN trained using the conjugate gradient algorithm.
The rest of the paper is organized as follows: the abbreviation used in the manuscript is presented in Section 2. An outline of the related work is presented in Section 3. An outline of the datasets used is presented in Section 4. The framework of the proposed classifier is presented in Section 5. The results and discussions are presented in Section 6. Finally, conclusion and scope for future work are presented in section 7.
2. Abbreviations Used
Table 1 presents the abbreviation used in the rest of the manuscript in alphabetic order.
Table 1.
Abbreviations used.
| Abbreviation | Phrase |
|---|---|
| ABCO | Artificial bee colony optimization |
| ACO | Ant colony optimization |
| ANN | Artificial neural networks |
| BCS | Binary cuckoo search |
| BFA | Binary firefly algorithm |
| BFO | Bacterial foraging optimization |
| BP | Back propagation |
| BPNN | Back propagation neural network |
| CAD | Computer-aided diagnosis |
| CDC | Counts of dimension to change |
| CDSS | Clinical decision support system |
| CFCSA | Hybrid crow search optimization algorithm |
| CGA | Conjugate gradient algorithm |
| CHD | Cleveland heart disease |
| CMVO | Chaotic multiverse optimization |
| CSM | Cosine similarity measure |
| CSO | Cat swarm optimization |
| CT | Computed tomography |
| DE | Differential evolution |
| DGA | Distance-based genetic algorithm |
| DISON | Diverse intensified strawberry optimized neural network |
| DNN | Deep neural network |
| E.coli | Escherichia Coli Bacteria |
| ECSA | Enhanced crow search algorithm |
| ELM | Extreme learning machine |
| FBFO | Feature selected by bacterial foraging optimization |
| FCM | Fuzzy C-means |
| FCSO | Feature selected by cat swarm optimization |
| FFO | Firefly optimization |
| FKH | Feature selected by krill herd |
| GA | Genetic algorithm |
| GSO | Glowworm swarm optimization |
| HCC | Hepatocellular carcinoma |
| HD | Hepatitis |
| IBPSO | Improved binary particle swarm optimization |
| ILP | Indian liver patient |
| ISSA | Improved Salp swarm algorithm |
| KH | Krill herd |
| k-NN | k-nearest neighbors |
| LO | Lion optimization |
| LR | Logistic regression |
| MCC | Mathew's correlation coefficient |
| MFO | Moth-flame optimization |
| ML | Machine learning |
| MPNN | Multilayer perceptron neural network |
| MR | Mixed ratio |
| NB | Naive Bayes |
| PCC | Pearson correlation coefficient |
| PID | Pima Indian diabetes |
| PSO | Particle swarm optimization |
| RD | Random diffusion |
| RDM | Rough dependency measure |
| RF | Random forest |
| RoIs | Regions of interest |
| SHD | Statlog heart disease |
| SMOTE | Synthetic minority oversampling technique |
| SMP | Seeking memory pool |
| SPC | Self-position consideration |
| SRD | Seeking range of the selected dimension |
| SVC | Support vector classification |
| SVM | Support vector machine |
| TS | Thoracic surgery |
| UCI | University of California Irvine |
| VCD | Vertebral column dataset |
| WBC | Wisconsin breast cancer |
| WDBC | Wisconsin diagnostic breast cancer |
| WOA | Whale optimization algorithm |
3. Literature Survey
Leema et al. [25] in their work have experimented the significance of fixing the appropriate values of parameters to train artificial neural networks using the backpropagation algorithm. The parameters are initial weight selection, bias, activation function used, number of hidden layers, number of neurons per hidden layer, number of training epochs, minimum error, and momentum term. Twelve backpropagation learning algorithms have been used in this study. Experimentation has been carried out using three clinical datasets from the UCI ML repository, namely, PID, hepatitis, and WBC datasets.
Elgin et al. [26] in their work have proposed a clinical-decision making system to diagnose allergic rhinitis. A wrapper approach that uses GA and the accuracy of ELM classifier as the fitness function has been used for feature selection. The selected features have been trained using ELM classifier. Intradermal skin test dataset of 872 patients collected from Good Samaritan Lab Services and Allergy Testing Centre, Chennai, has been used in this work, and an accuracy of 97.7% has been achieved.
Sreejith et al. [27] in their work have proposed a framework for classifying clinical datasets which uses an embedded approach for feature selection and a DISON for classification. The feature selection is performed by computing the feature importance of every attribute using an extremely randomized tree classifier. Classification is performed using DISON which is a feed forward neural network whose weights and bias are optimized in two stages first, by using a strawberry optimization algorithm and then by using a gradient descent BP algorithm. Vertebral column, PID, CHD, and SHD datasets from the UCI ML repository have been used for experimentation. The framework has achieved an accuracy of 87.17% for vertebral column, 90.92% for PID, 93.67% for CHD, and 94.5% for SHD.
Sreejith et al. [28] in their work have proposed a framework for CDSS which addresses the data imbalance problems associated with clinical dataset. The datasets are rebalanced using SMOTE enhanced using Orchard's algorithm. The feature selection is performed using a wrapper approach where CMVO is used to select the feature subsets, and RF classifier is used to evaluate the goodness of the features. The arithmetic mean of MCC and F-score computed using the RF classifier is used as the fitness function. Finally, an RF classifier, comprising of 100 decision trees which uses information gain ratio as the split criteria, is used for classifying the clinical data. Three clinical datasets from the UCI ML repository, namely, ILP, TS, and PID datasets, have been used for experimentation. The proposed framework achieved 0.65 MCC, 0.84 F-score, and 82.46% accuracy for ILP; 0.74 MCC, 0.87 F-score, and 86.88% accuracy for TS; and 0.78 MCC, 0.89 F-score, and 89.04% accuracy for PID datasets.
Isaac et al. [29] in their work have proposed a CAD system to diagnose pulmonary emphysema from chest CT slices. Spatial intuitionistic fuzzy C-means clustering algorithm has been used to segment the lung parenchyma and extracting the RoIs. From the RoIs, shape, texture, and run-length features have been extracted, and feature selection has been performed using a wrapper approach using four bioinspired algorithms with the classification accuracy of SVM as the fitness function. The bioinspired algorithms used are MFO, FFO, ABCO, and ACO. Tenfold crossvalidation technique has been used, and each feature set has been trained using an ELM classifier. Two independent datasets, one dataset consisting of CT slices collected from hospitals and the second dataset consisting of CT slices from a benchmark repository, have been used for classification. A maximum classification accuracy of 89.19% for MFO, 91.89% for FFO, 83.78% for ABCO, 86.49% for ACO, and 75.68% without feature selection have been achieved.
Elgin et al. [30] in their work have performed feature selection and instance selection using a wrapper approach that employs cooperative coevolution with the classification accuracy of the random forest classifier as the fitness function. The optimal feature set is used to train a random forest classifier. Seven datasets, namely, WDBC, HD, PID, CHD, SHD, VCD, and HCC from the UCI ML repository have been used for experimentation. An accuracy of 97.1%, 82.3%, 81.01%, 93.4%, 96.8%, 91.4%, and 72.2% for datasets WDBC, HD, PID, CHD, SHD, VCD, and HCC datasets have been achieved, respectively.
Anter et al. [31] in their work have developed CFCSA by integrating chaos theory and the FCM method to find the optimal feature subset. Ten clinical datasets from the UCI ML repository have been used for experimentation. The features of each clinical dataset have been normalized, and then random chaotic motion has been incorporated into CFCSA in the form of chaotic maps. The objective function of the FCM has been used as the fitness function, in which the crow with the best fitness has been considered the best solution. Comparison has been done with chaotic ant lion optimization, binary ant lion optimization, and the binary crow search algorithm, and it has been inferred that CFCSA outperforms these algorithms in all the datasets used for experimentation.
Elgin et al. [32] in their work have proposed a correlation-based ensemble feature selection using a wrapper approach that employs three bioinspired algorithms using differential evolution, lion optimization, and glowworm swarm optimization with the accuracy of the AdaboostSVM classifier as the fitness function. Tenfold crossvalidation technique has been used, and the optimal features selected have been used to train a gradient descent BP neural network with variable learning rates. Two clinical datasets from the UCI ML repository, namely, hepatitis and WDBC have been used for experimentation. An accuracy of 93.902% for hepatitis and 98.734% for WDBC datasets have been achieved.
Sweetlin et al. [33] in their work have proposed a CAD system to diagnose pulmonary tuberculosis from chest CT slices. The region growing algorithm has been used for segmenting the lung fields followed by edge reconstruction. The manifestations of pulmonary tuberculosis, namely, cavities, consolidations, and nodules have been considered to be RoIs. After extracting the RoIs, and from the RoI, texture features, run-length features and shape features have been extracted, and feature selection has been performed using a wrapper approach that employs the BCS algorithm with the accuracy of one-against-all multiclass SVM classifier as the fitness function. The Cuckoo search algorithm has been implemented in two ways, first, by using entropy measure and second, without using entropy measure. Using the selected feature training is performed using one-against-all multiclass SVM classifier. An accuracy of 85.54% for BCS algorithm with entropy measure and 84.65% accuracy for BCS algorithm without entropy measure have been achieved.
Sweetlin et al. [34] in their work have proposed a CAD system to diagnose pulmonary hamartoma nodules from chest CT slices. Otsu's thresholding method has been used to segment lung parenchyma from the CT slices. Nodules are considered to be the RoIs and from the RoIs, texture features, shape features and run-length features have been extracted. Feature selection has been performed using filter evaluation measures, namely, CSM and RDM with the ACO algorithm. The features selected by ACO-CSM and ACO-RDM have been used to train three classifiers, namely, SVM, NB, and J48 decision tree classifiers. Maximum classification accuracy of 94.36% for SVM classifier trained with 38 features selected using ACO-RDM has been achieved.
Sweetlin et al. [35] in their work have proposed a CAD system to diagnose pulmonary bronchitis from CT slices of the lung. Optimal thresholding has been used to segment the left and right lung fields from the lung CT slices. The RoIs are identified, and from the RoIs, texture and shape features have been extracted. Feature selection has been performed using a hybrid ACO algorithm combined with tandem run recruitment based on cosine similarity, and the accuracy of the SVM classifier has been used as the fitness function. The selected features have been used to train a SVM classifier. An accuracy of 81.66% for ACO with tandem run strategy, 78.10% for ACO without tandem run strategy, and 75.14% without feature selection has been achieved.
Raj et al. [36] in their work have proposed DGA for feature selection to develop a CAD system to diagnose lung disorders from chest CT slices. The entire dataset has been split into two sets one set containing 90% of the entire dataset and the other set containing 10% of the entire dataset. Out of the 90%, 50% has been used as training set and the other 50% as validation set for evaluating the objective function. The set containing 10% of the entire dataset has been used as testing set. The objective function has been defined as the sum of the squared deviation of each data in the training set of each class from each data in the validation set of the corresponding class. GA has been used for feature selection by minimizing the proposed objective function, resulting in the proposed DGA. The GA has been iterated over several generations to obtain individuals that are best fit with respect to the objective function. Classification has been performed using k-NN classifier to classify the RoIs into one of four classes, namely, bronchiectasis, tuberculosis, pneumonia, and normal. An average accuracy of 88.16% with feature selection and an average accuracy of 86.46% without feature selection have been achieved.
Zawbaa et al. [37] in their work have performed feature selection using a wrapper approach that uses the MFO algorithm with the accuracy of k-NN classifier as the fitness function. Eighteen datasets from the UCI ML repository have been used for experimentation among which four are clinical datasets. Comparison has been done with PSO and GA, and it has been inferred that MFO outperforms in fourteen datasets among which three are clinical datasets.
Shu-Chuan et al. [38] in their work have presented an algorithm called CSO by modeling the natural behavior of cats. The CSO algorithm considered two biological characteristics of cats, namely, seeking mode and tracking mode. Cats spend utmost of the time when they are awake on resting. Nevertheless, during their rests, their perception is really high, and they are well aware of what is happening around them. Cats continuously observe their environment wisely and consciously and when they perceive a prey, they advance towards it rapidly. Although resting, they move their position cautiously and slowly, occasionally even stay in the original position. Seeking mode has been used to represent this behavior into the CSO, and the tracing mode has been used to represent the behavior of cats advancing towards a prey into the CSO. The performance of CSO has been evaluated by applying CSO, standard PSO, and PSO with weighting factor into six benchmark functions. The results obtained reveal that the proposed CSO performs better compared to PSO and PSO with weighting factor.
Gandomi et al. [39] in their work have proposed a swarm intelligence algorithm named KH algorithm to solve optimization tasks and is centered on the imitation of the herding behavior of krill swarms with respect to precise biological and environmental processes. The fitness function of each krill individual has been defined as the least distance of each individual krill from food and from the highest density of the herd. Three vital actions considered to define the time-dependent position of an individual krill are, one, movement induced by other krill individuals, two, foraging activity, and three, random diffusion. The KH algorithm is tested using twenty benchmark functions and compared with eight algorithms. Experimentation results indicate that the KH algorithm can outperform these familiar algorithms.
Chen et al. [40] have proposed a cooperative bacterial foraging optimization algorithm (CBFO). Two cooperative methods are used to solve complex optimization problems in the original BFO [41] and achieved significant improvement. The serial heterogeneous cooperation on the implicit space decomposition level and the hybrid space decomposition level are the two methods used to improve the original BFO. The authors have compared the performance of two CBFO variants with the original BFO, PSO, and GA on four commonly used benchmark functions. The experimental results indicated that the CBFO achieved a better performance over the original BFO, PSO, and GA.
Chen et al. [42] have proposed an adaptive bacterial foraging optimization (ABFO) for optimizing functions. The adaptive foraging approaches are used to increase the performance of the original BFO. It is achieved by enabling the original BFO to adjust the run-length unit parameter dynamically during the time of algorithm implementation. The experimental results are compared with the original BFO, PSO, and GA using 4 benchmark functions. The proposed ABFO indicates the better performance over the original BFO and competitive with the PSO and GA.
From the literature, it is evident that classifier training using relevant features enhances the accuracy of the classifier. It can also be inferred that wrapper-based feature selection that employs bioinspired algorithms performs better in numerous cases compared to traditional feature selection methods.
4. Outline of the Datasets Used
Seven clinical datasets from the UCI ML repository, namely, WDBC, SHD, HCC, HD, VCD, CHD, and ILP have been used for binary classification. An outline of each dataset used is presented in Table 2.
Table 2.
Outline of the datasets used.
| Dataset name | No. of instances | No. of features∗ | No. of missing values | Class labels with no. of instances associated with each class label | Interpretation of class labels |
|---|---|---|---|---|---|
| WDBC | 569 | 31 | Nil | M (212)/B (357) | M-malignant, B-benign |
| SHD | 270 | 13 | Nil | 2 (120)/1 (150) | 2-present, 1-absent |
| HCC | 165 | 49 | 826 | 0 (63)/1 (102) | 0-dies, 1-lives |
| HD | 155 | 18 | 167 | 1 (32)/2 (123) | 1-die, 2-live |
| VCD | 310 | 6 | Nil | 0 (210)/1 (100) | 0-abnormal, 1-normal |
| CHD | 303 | 13 | Nil | 1 (139)/2 (164) | 1-presence, 2-absence |
| ILP | 583 | 10 | Nil | 1 (416)/2 (167) | 1-diseased, 2-nondiseased |
∗ without class label.
5. System Framework
The framework for feature selection and classification of clinical datasets using bioinspired algorithms and super learner is presented in Figure 1. The major building blocks of the framework are data preprocessing, feature selection, classifier training, classifier testing, and dataset construction for super learner, super learner training, and testing. Each building block is outlined below.
Figure 1.
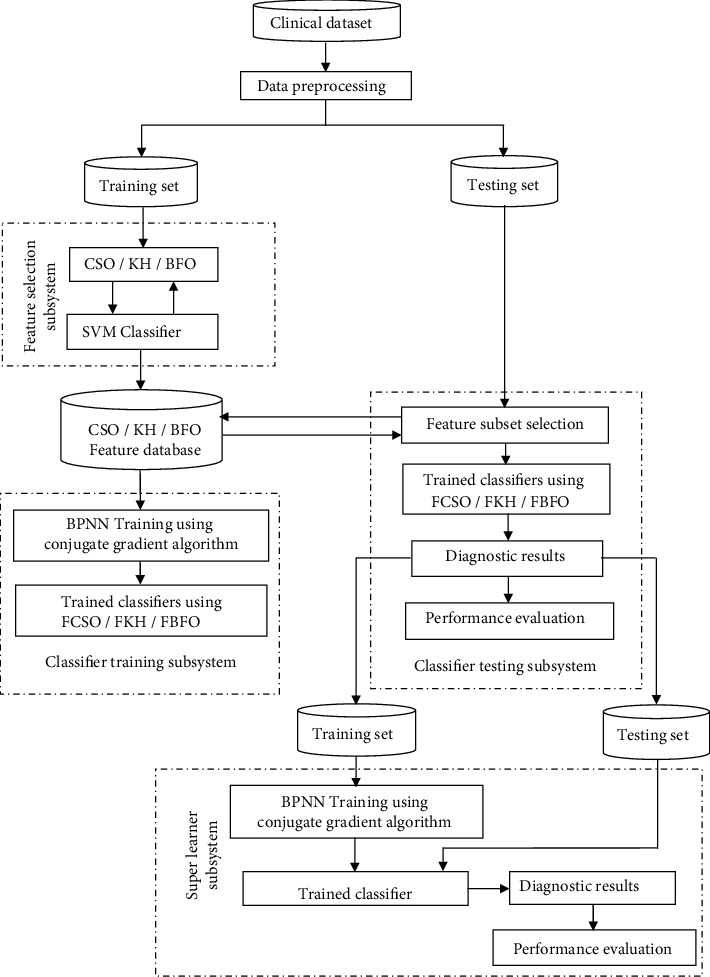
System framework.
5.1. Preprocessing
Each (Cs) has been subjected to preprocessing prior to feature selection to enhance the quality of data. Mean imputation has been used to handle missing values, and SMOTE is used to handle the class imbalance problem in each Cs by generating additional instances from the minority class.
Normalization has been used to scale the value of a feature so that the value will fall in a specified range and is predominantly useful for constructing a classifier involving a neural network. Training a classifier using normalized data will speedup learning. In this work, the range is 0 to 1, and min-max normalization is being used. When an attribute “A” in a clinical dataset Cs is subject to min-max normalization, the minimum value (minA) and maximum value (maxA) in the value set of “A” are first identified, and normalization is performed using the formula presented in equation (1).
| (1) |
If the formula “a′” is the normalized value of an attribute “a,” when a is drawn from the value set of “A.” Since min-max normalization is being used to normalize the values in the range 0 to 1, the value of newmaxA is 1 and newminA is 0.
The number of instances in each Cs used for constructing and testing the classifier prior to generating additional samples using SMOTE, the number of instances in each Cs after generating additional samples using SMOTE, the number of instances in the training set (Ts), and the number of instances in the testing set (Tt) is presented in Table 3. After preprocessing, each Cs is split into training set (60%) and testing set (40%).
Table 3.
Outline of training and testing instances of each Cs.
| Instances | WDBC dataset | SHD dataset | HCC dataset | HD dataset | VCD dataset | CHD dataset | ILP dataset |
|---|---|---|---|---|---|---|---|
| Total number of instances before SMOTE | 569 | 270 | 165 | 155 | 310 | 303 | 583 |
| Total number of instances after SMOTE | 780 | 270 | 228 | 251 | 410 | 303 | 750 |
| Number of training instances for FCSO/FKH/FBFO classifiers | 468 | 162 | 137 | 151 | 246 | 182 | 450 |
| 60% of the total number of instances after SMOTE | |||||||
| Number of testing instances for FCSO/FKH/FBFO classifiers | 312 | 108 | 91 | 100 | 164 | 121 | 300 |
| 40% of the total number of instances after SMOTE | |||||||
| Number of training instances for super learner | 250 | 86 | 73 | 80 | 131 | 97 | 240 |
| 80% of the total testing instances∗ for FCSO/FKH/FBFO classifiers | |||||||
| Number of testing instances for | 62 | 22 | 18 | 20 | 33 | 24 | 60 |
| Super learner | 20% of the total testing instances∗ for FCSO/FKH/FBFO classifiers | ||||||
∗Each instance refers to the classification result pertaining to each instance of the testing set for FCSO, FKH, and FBFO classifiers and the class label corresponding to each instance of the testing set.
5.2. Feature Selection
Feature selection is performed on each Ts used for experimentation to select the optimal features for training the classifier. Selecting the optimal features from the Ts will improve the classification accuracy. A wrapper approach that uses three bioinspired algorithms, namely, CSO, KH, and BFO with the accuracy of the SVM classifier is used to perform feature selection. An outline of CSO, KH, and BFO used for feature selection is presented below.
5.2.1. Outline of the CSO Algorithm for Feature Selection
CSO is inspired and modeled based on two main postures of cats, namely, resting and tracing. Mimicking the resting behavior of a cat is named as seeking mode, and mimicking the tracing behavior of a cat is named as tracing mode. The seeking mode relates to a local search process, whereas the tracing mode relates to a global search process. The vital parameters that play an important role in CSO are outlined in Table 4. Tracing mode relates to cat's movement while chasing a prey, for example, chasing a rat.
Table 4.
Outline of the parameters used in CSO.
| Parameter | Description |
|---|---|
| SMP | SMP is used to define the size of the seeking memory of each cat. Each cat selects possible neighborhood position from a set of solutions. |
| SRD | SRD is used to define the seeking range of the selected dimension. |
| CDC | CDC is a count of dimensions to be changed in seeking mode. |
| SPC | SPC indicates whether the cat is in the current position or not. |
| N | Number of cats |
| MR | Mixed ratio of cats |
| C | Constant value |
| D | Size of dimension |
| R | Random number in the range of [0,1] |
The steps to select the optimal feature subset using CSO is outlined below (Algorithm 1):
Algorithm 1.

5.2.2. Outline of the KH Algorithm for Feature Selection
The KH algorithm is centered on the imitation of the herding behavior of krill swarms with respect to precise biological and environmental processes. Krill density is reduced by predators, namely, seals, penguins, or seabirds. The herding of the krill individuals includes, one, increasing the krill density and two, reaching the food. The fitness function of each krill individual has been defined as the least distance of each individual krill from food and from the highest density of the herd.
Three vital actions considered to define the time-dependent position of an individual krill are one, movement induced by other krill individuals, two, foraging activity, and three, random diffusion.
Krill individuals attempt to maintain a high density and hence move due to their mutual effect. Local swarm density, target swarm density, and repulsive swarm density are used to estimate the direction of motion. Food location and prior experience about the food location are the two parameters used to estimate the foraging motion. Random diffusion is used for the exploration of the search space. In the KH algorithm, the population diversity is improved by means of the diffusion function, which is integrated into the krill individuals. Random diffusion is the net movement of each krill individual from high-density to low-density regions.
The motion velocity of krill particle applies the Lagrangian model [43] as shown in Equation (5).
| (5) |
In the above formula, dxi/dt is the motion velocity of krill particle i, Ni is the induced motion, Fi is the foraging motion, and RDi is the random diffusion of the ith krill individual. The vital parameters that play an important role in the KH algorithm are outlined in Table 5.
Table 5.
Outline of the parameters used in the KH algorithm.
| Parameter | Definition | Value |
|---|---|---|
| V f | Maximum foraging speed | V f = 0.02 m/s−1 |
| RDmax | Maximum random diffusion speed | RDmax ∈ (0.002 − 0.01 ) m/s−1 |
| N max | Maximum induction speed | N max = 0.01 m/s−1 |
| w n | Inertia weight of the motion induced | w n ∈ (0, 1) |
| w f | Inertia weight of the foraging motion | w f ∈ (0, 1) |
| C t | Step-length scaling factor | Constant no.between [0, 2] |
| δ | Random directional vector | Random numbers [−1, 1] |
The steps to select the optimal feature subset using KH is outlined below (Algorithm 2):
Algorithm 2.

5.2.3. Outline of the BFO Algorithm for Feature Selection
The bacterial foraging optimization (BFO) algorithm imitates the pattern exhibited during the foraging process of Escherichia coli bacteria, that includes chemotaxis, swarming, reproduction, and elimination-dispersal operations [41]. The basic idea behind the foraging strategy of E. coli bacteria is to obtain the maximum nutrition in a unit time. The chemotaxis strategy involves the searching of nutrition by taking small movements such as tumbling, moving, and swimming, using its locomotory organ called flagella. The swarming strategy deals with the communication between bacteria. When the bacteria discover high amount of nutrients, they will release chemical substances to attract other bacteria. If they are in danger, they will tend to prevent other bacteria. The reproduction process involves splitting of healthier bacterium into two bacteria, and the low healthy bacteria are set to die. Finally, the elimination-dispersal strategy involves replacing the low health bacterium by randomly generated new ones. The vital parameters that play an important role in the BFO algorithm are outlined in Table 6.
Table 6.
Outline of the parameters used in the BFO algorithm.
| Parameter | Description |
|---|---|
| p | Number of features |
| S | Number of bacteria |
| S r | Number of bacteria in the reproduction steps |
| N re | No. of reproductive steps |
| N ed | No. of elimination-dispersal steps |
| N c | No. of chemotactic steps |
| N s | No. of swimming steps |
| L (i) | Bacteria step size length |
| P ed | Elimination probability |
| ∅(i) | Direction of ith bacteria |
| x | Index of the chemotactic process |
| y | Index of the reproduction process. |
| z | Index of the elimination-dispersal process |
| θ i | The ith bacterium position |
| θ | A bacterium on the optimization domain |
| J last | The highest objective function value |
| ∆(i) | A random vector and its value lie between -1 and 1 |
| Jcc(θ, θi(x, y, z)) | Cell-to-cell attractant effect to nutrient concentration |
The steps involved in finding the optimal feature subset using the BFO algorithm is outlined below:
5.3. Classifier Training
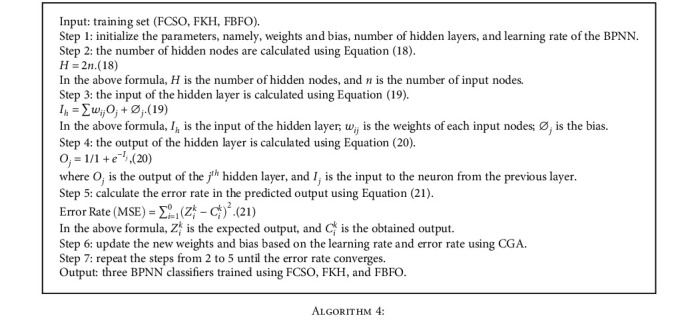
Each Cs is preprocessed and split into training set (Ts − 60%) and testing set (Tt− 40%). A wrapper approach that uses three bioinspired algorithms CSO, KH, and BFO with the classification accuracy of SVM as the fitness function has been used for feature selection. The features selected by each bioinspired algorithm are used to train three BPNNs independently using CGA. The number of hidden layers for each BPNN is 1, and the activation function used in the hidden layer is sigmoid. The learning rate is 1e–07, and the maximum number of iterations is 100. Since the classification is binary, each BPNN has only one output node, and the activation function used in the output layer is sigmoid. Figure 2 elaborates the process of training BPNN classifiers.
Figure 2.
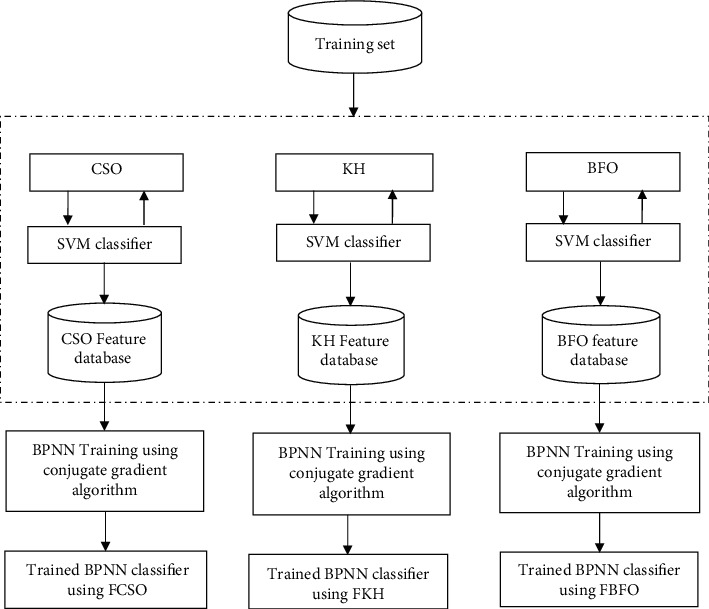
Classification using BPNN.
The number of training instances for FCSO, FKH, and FBFO classifiers is presented in Table 3. Though majority of the features selected by each bioinspired algorithm overlap, it has been inferred that the number of features selected by each algorithm is not the same. The parameter settings for each classifier is presented in Table 7.
Table 7.
Parameter settings for BPNN.
| BPNN parameter | Bioinspired algorithm | WDBC dataset | SHD dataset | HCC dataset | HD dataset | VCD dataset | CHD dataset | ILP dataset |
|---|---|---|---|---|---|---|---|---|
| Number of input nodes | CSO | 15 | 9 | 20 | 16 | 3 | 6 | 5 |
| KH | 17 | 10 | 39 | 10 | 3 | 10 | 8 | |
| BFO | 18 | 9 | 35 | 19 | 2 | 11 | 5 | |
| Number of hidden nodes | CSO | 30 | 18 | 40 | 32 | 6 | 12 | 10 |
| KH | 34 | 20 | 78 | 20 | 6 | 20 | 16 | |
| BFO | 36 | 18 | 70 | 38 | 4 | 22 | 10 |
The steps to train the BPNN classifier using three BPNN classifier and trained using CSO, KH, and BFO algorithms are outlined below:
5.4. Classifier Testing and Dataset Construction for Super Learner
After training the classifier with 60% of the preprocessed Cs (Ts), classifier testing is performed using the remaining 40% of the of the preprocessed Cs (Tt). Figure 3 elaborates the process of testing the three classifiers and also throws light on the process of training the super learner.
Figure 3.
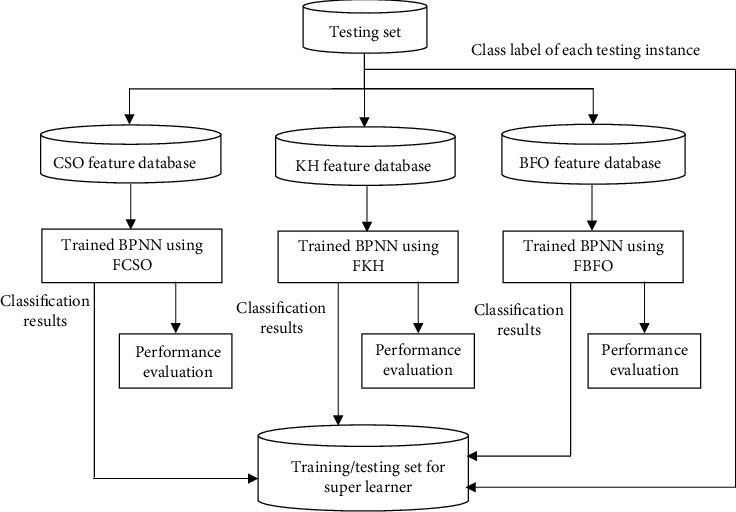
Classifier testing.
Feature selection is performed on the testing set by querying the FCSO, FKH, and FBFO databases. The instances of the testing set containing the features selected by the CSO are used to test the FCSO classifier; similarly, the instances of the testing set containing the features selected by the KH and BFO are used to test the FKH and FBFO classifier. The performance of the FCSO, FKH, and FBFO classifiers are evaluated using the results obtained from the testing set.
The classification result of each instance of the testing set for FCSO, FKH, and FBFO classifiers and the class label corresponding to each instance of the testing set will be the candidate instances for training and testing the super learner.
5.5. Super Learner Training and Testing
As outlined in Section 5.4, the classification result pertaining to each instance of the testing set for FCSO, FKH, and FBFO classifiers and the class label corresponding to each instance of the testing set will be the candidate instances for training and testing the super learner. Figure 4 elaborates the process of training and testing of the super learner. The training set comprises of 80% of the instances, and the testing set comprises of 20% of the instances. The number of training and testing instances for the super learner is presented in Table 3.
Figure 4.
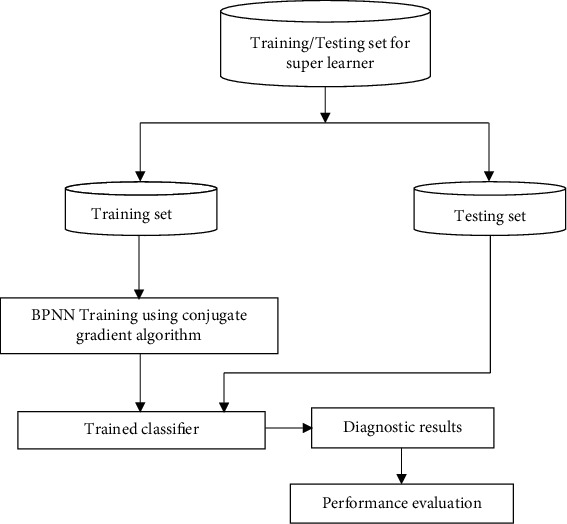
Super learner training and testing.
Super learner is a type of ensemble classifier [44]. In this work, a BPNN classifier trained using CGA is used as the super learner. The parameter settings for the super learner are presented in Table 8.
Table 8.
Parameter settings for super learner.
| Name of the parameter | WDBC dataset | SHD dataset | HCC dataset | HD dataset | VCD dataset | CHD dataset | ILP dataset |
|---|---|---|---|---|---|---|---|
| Initial population size | 250 | 86 | 73 | 80 | 131 | 97 | 240 |
| Number of input nodes | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| Number of hidden nodes | 6 | 6 | 6 | 6 | 6 | 6 | 6 |
The super learner is trained using the steps presented in Section 5.3 for training the BPNN classifier using CGA, and the performance of the super learner is evaluated using the testing set.
6. Results and Discussions
Seven clinical datasets from the UCI ML repository, namely, WDBC, SHD, HCC, HD, VCD, CHD, and ILP have been used for experimentation. The performance of the FCSO, FKH, and FBFO classifiers and super learner is evaluated in terms of accuracy, sensitivity, specificity, precision, and F-score, which are calculated based on true positive (TP), true negative (TN), false positive (FP), and false negative (FN) using Equations (22), (23), (24), (25), and (26).
| (22) |
In the above formula, TP is the number of positive instances predicted as positive by the classifier, TN is the number of negative instances predicted as negative by the classifier, FP is the number of negative instances predicted as positive by the classifier, and FN is the number of positive instances predicted as negative by the classifier.
| (23) |
| (24) |
| (25) |
| (26) |
Accuracy, sensitivity, specificity, precision, and F−score obtained using FCSO, FKH, and FBFO classifiers and super learner for the datasets WDBC, SHD, HCC, HD, VCD, CHD, and ILP are presented in Tables 9–15.
Table 9.
Performance of FCSO, FKH, and FBFO classifiers and super learner on WDBC dataset.
| Feature selection algorithm | Size of feature subset | TN | FP | FN | TP | Accuracy | Sensitivity | Specificity | Precision | F-score |
|---|---|---|---|---|---|---|---|---|---|---|
| CSO | 15 | 137 | 4 | 6 | 165 | 96.79 | 96.49 | 97.16 | 97.63 | 0.97 |
| KH | 17 | 139 | 2 | 5 | 166 | 97.76 | 97.08 | 98.58 | 98.81 | 0.98 |
| BFO | 18 | 139 | 2 | 8 | 163 | 96.79 | 95.32 | 98.58 | 98.79 | 0.97 |
| Super learner | — | 22 | 0 | 2 | 39 | 96.83 | 95.12 | 100.00 | 100.00 | 0.98 |
Table 10.
Performance of FCSO, FKH, and FBFO classifiers and super learner on Statlog dataset.
| Feature selection algorithm | Size of feature subset | TN | FP | FN | TP | Accuracy | Sensitivity | Specificity | Precision | F-score |
|---|---|---|---|---|---|---|---|---|---|---|
| CSO | 9 | 53 | 8 | 9 | 38 | 84.26 | 80.85 | 86.89 | 82.61 | 0.82 |
| KH | 10 | 53 | 8 | 11 | 36 | 82.41 | 76.60 | 86.89 | 81.82 | 0.79 |
| BFO | 9 | 51 | 10 | 10 | 37 | 81.48 | 78.72 | 83.61 | 78.72 | 0.79 |
| Super learner | — | 10 | 1 | 2 | 9 | 86.36 | 81.82 | 90.91 | 90.00 | 0.86 |
Table 11.
Performance of FCSO, FKH, and FBFO classifiers and super learner on HCC dataset.
| Feature selection algorithm | Size of feature subset | TN | FP | FN | TP | Accuracy | Sensitivity | Specificity | Precision | F-score |
|---|---|---|---|---|---|---|---|---|---|---|
| CSO | 20 | 43 | 9 | 9 | 31 | 80.43 | 77.50 | 82.69 | 77.50 | 0.78 |
| KH | 39 | 48 | 4 | 13 | 27 | 81.52 | 67.50 | 92.31 | 87.10 | 0.76 |
| BFO | 35 | 47 | 5 | 20 | 20 | 72.83 | 50.00 | 90.38 | 80.00 | 0.62 |
| Super learner | — | 10 | 1 | 0 | 8 | 94.74 | 100.00 | 90.91 | 88.89 | 0.94 |
Table 12.
Performance of FCSO, FKH, and FBFO classifiers and super learner on hepatitis dataset.
| Feature selection algorithm | Size of feature subset | TN | FP | FN | TP | Accuracy | Sensitivity | Specificity | Precision | F-score |
|---|---|---|---|---|---|---|---|---|---|---|
| CSO | 16 | 47 | 2 | 10 | 42 | 88.12 | 80.77 | 95.92 | 95.45 | 0.88 |
| KH | 10 | 45 | 4 | 6 | 46 | 90.10 | 88.46 | 91.84 | 92.00 | 0.90 |
| BFO | 19 | 47 | 2 | 12 | 40 | 86.14 | 76.92 | 95.92 | 95.24 | 0.85 |
| Super learner | — | 8 | 1 | 1 | 11 | 90.48 | 91.67 | 88.89 | 91.67 | 0.92 |
Table 13.
Performance of FCSO, FKH, and FBFO classifiers and super learner on vertebral column dataset.
| Feature selection algorithm | Size of feature subset | TN | FP | FN | TP | Accuracy | Sensitivity | Specificity | Precision | F-score |
|---|---|---|---|---|---|---|---|---|---|---|
| CSO | 3 | 74 | 10 | 17 | 63 | 83.54 | 78.75 | 88.10 | 86.30 | 0.82 |
| KH | 3 | 81 | 3 | 13 | 67 | 90.24 | 83.75 | 96.43 | 95.71 | 0.89 |
| BFO | 2 | 80 | 4 | 17 | 63 | 87.20 | 78.75 | 95.24 | 94.03 | 0.86 |
| Super learner | — | 19 | 2 | 4 | 8 | 81.82 | 66.67 | 90.48 | 80.00 | 0.73 |
Table 14.
Performance of FCSO, FKH, and FBFO classifiers and super learner on Cleveland heart disease dataset.
| Feature selection algorithm | Size of feature subset | TN | FP | FN | TP | Accuracy | Sensitivity | Specificity | Precision | F-score |
|---|---|---|---|---|---|---|---|---|---|---|
| CSO | 6 | 60 | 8 | 12 | 42 | 83.61 | 77.78 | 88.24 | 84.00 | 0.81 |
| KH | 10 | 56 | 12 | 11 | 43 | 81.15 | 79.63 | 82.35 | 78.18 | 0.79 |
| BFO | 11 | 53 | 15 | 13 | 41 | 77.05 | 75.93 | 77.94 | 73.21 | 0.75 |
| Super learner | — | 13 | 2 | 2 | 8 | 84.00 | 80.00 | 86.67 | 80.00 | 0.80 |
Table 15.
Performance of FCSO, FKH, and FBFO classifiers and super learner on Indian liver patient dataset.
| Feature selection algorithm | Size of feature subset | TN | FP | FN | TP | Accuracy | Sensitivity | Specificity | Precision | F-score |
|---|---|---|---|---|---|---|---|---|---|---|
| CSO | 5 | 103 | 62 | 33 | 102 | 68.33 | 75.56 | 62.42 | 62.20 | 0.68 |
| KH | 8 | 104 | 61 | 40 | 95 | 66.33 | 70.37 | 63.03 | 60.90 | 0.65 |
| BFO | 5 | 101 | 64 | 34 | 101 | 67.33 | 74.81 | 61.21 | 61.21 | 0.67 |
| Super learner | — | 26 | 15 | 3 | 16 | 70.00 | 84.21 | 63.41 | 51.61 | 0.64 |
The super learner has achieved a classification accuracy of 96.83% for WDBC, 86.36% for SHD, 94.74% for HCC, 90.48% for HD, 81.82% for VCD, 84.0% for CHD, and 70.0% for ILP. The classification accuracy of the proposed work has been compared with the performance of the existing work on clinical datasets and the comparison results summarized in Table 16.
Table 16.
Comparison of the proposed work and existing work using clinical dataset.
| Author/year | Method/reference | Accuracy % | ||||||
|---|---|---|---|---|---|---|---|---|
| WDBC | SHD | HCC | HD | VCD | CHD | ILP | ||
| Ayon et al. (2020) | DNN [45] | — | 98.15 | — | — | — | 94.39 | — |
| SVM [45] | — | 97.41 | — | — | — | 97.36 | — | |
| Bai Ji et al. (2020) | IBPSO with k-NN [46] | 96.14 | — | — | — | — | — | — |
| Elgin et al. (2020) | Cooperative coevolution and RF [30] | 97.1 | 96.8 | 72.2 | 82.3 | 91.4 | 93.4 | — |
| Magesh et al. (2020) | Cluster-based decision tree [47] | — | — | — | — | — | 89.30 | — |
| Rabbi et al. (2020) | PCC and AdaBoost [48] | — | — | — | — | — | — | 92.19 |
| Rajesh et al. (2020) | RF classifier [49] | — | — | 80.64 | — | — | — | — |
| Salima et al. (2020) | ECSA with k-NN [50] | 95.76 | 82.96 | — | — | — | — | — |
| Singh J et al. (2020) | Logistic regression [51] | — | — | — | — | — | — | 74.36 |
| Sreejith et al. (2020) | CMVO and RF [28] | — | — | — | — | — | — | 82.46 |
| Sreejith et al. (2020) | DISON and ERT[27] | — | 94.5 | — | — | 87.17 | 93.67 | — |
| Tougui et al. (2020) | ANN with Matlab [52] | — | — | — | — | — | 85.86 | — |
| Tubishat et al. (2020) | ISSA with k-NN [53] | — | 88.1 | — | — | 89.0 | — | — |
| Abdar et al. (2019) | Novel nested ensemble nu-SVC [54] | — | — | — | — | — | 98.60 | — |
| Anter et al. (2019) | CFCSA with chaotic maps [31] | 98.6 | — | — | 68.0 | — | 88.0 | 68.4 |
| Aouabed et al. (2019) | Nested ensemble nu-SVC, GA and multilevel balancing [55] | — | — | — | — | — | 98.34 | — |
| Elgin et al. (2019) | DE, LO and GSO with Adaboost SVM [32] | 98.73 | — | — | 93.9 | — | — | — |
| Książek et al. (2019) | SVM [56] | — | 97.41 | — | — | — | 97.36 | — |
| Sayed et al. (2019) | Novel chaotic crow search algorithm with k-NN [57] | 90.28 | 78.84 | — | 83.7 | — | — | 71.68 |
| Abdar et al. (2018) | MPNN and C5.0 [58] | — | — | — | — | — | — | 94.12 |
| Abdullah et al. (2018) | k-NN [59] | — | — | — | — | 85.32 | — | — |
| RF [59] | — | — | — | — | 79.57 | — | — | |
| Sawhney et al. (2018) | BFA and RF [60] | — | — | 83.50 | — | — | — | — |
| Abdar et al. (2017) | Boosted C5.0 [61] | — | — | — | — | — | — | 93.75 |
| CHAID [61] | — | — | — | — | — | — | 65.0 | |
| Zamani et al. (2016) | WOA with k-NN [62] | — | 77.05 | — | 87.10 | — | — | — |
| Abdar (2015) | SVM with rapid miner [63] | — | — | — | — | — | — | 72.54 |
| C5.0 with IBM SPSS modeller [63] | — | — | — | — | — | — | 87.91 | |
| Santos et al. (2015) | Neural networks and augmented set approach [64] | — | — | 75.2 | — | — | — | — |
| Chiu et al. (2013) | ANN and LR [65] | — | — | 85.10 | — | — | — | — |
| Mauricio et al. (2013) | ABCO with SVM [66] | — | 84.81 | — | 87.10 | — | 83.17 | — |
| Proposed | CSO, KH, BFO, and super learner | 96.83 | 86.36 | 94.74 | 90.48 | 81.82 | 84.00 | 70.00 |
7. Conclusion and Scope for Future Work
A CAD system that employs a super learner to diagnose the presence or absence of a disease has been implemented in this work. Seven Cs from the UCI ML repository, namely, WDBC, SHD, HCC, HD, VCD, CHD, and ILP have been used for experimentation. Each Cs is preprocessed, and the preprocessed Cs is split into training and testing sets. A wrapper-based feature selection approach using three bioinspired algorithms, namely, CSO, KH, and BFO, with the accuracy of SVM classifier has been used to select the optimal feature subsets. The selected feature subsets are used to train three BPNN classifiers using CGA, and the performance of the trained classifiers is evaluated. The classification results obtained for each instance of the testing set of the three classifiers and the class label associated with each instance of the testing set will be the candidate instances for training and testing the super learner. The super learner achieved a classification accuracy of 96.83% for WDBC, 86.36% for SHD, 94.74% for HCC, 90.48% for HD, 81.82% for VCD, 84.0% for CHD, and 70.0% for ILP.
CAD systems to diagnose disorders in the human body from different imaging modalities such as X-ray, computed tomography, magnetic resonance imaging, and positron emission tomography are gaining importance. This work can be extended by developing CAD systems to diagnose disorders from the medical images acquired through different imaging modalities. Features based on shape, texture, and run length can be extracted from the images, and the feature selection algorithms used in this work can be used to select the relevant features. The relevant features can be used to build classifier models to predict the presence or absence of disorders from the images.
Algorithm 3.

Algorithm 4.
Data Availability
The data supporting this study are from previously reported studies and datasets, which have been cited. The datasets used in this research work are available at UCI Machine Learning Repository.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
References
- 1.Chawla N. V., Bowyer K. W., Hall L. O., Kegelmeyer W. P. SMOTE: synthetic minority over-sampling technique. Journal of Artificial Intelligence Research. 2002;16:321–357. doi: 10.1613/jair.953. [DOI] [Google Scholar]
- 2.Prati R. C. Combining feature ranking algorithms through rank aggregation. The 2012 International Joint Conference on Neural Networks (IJCNN); 2012; Brisbane, QLD, Australia. pp. 1–8. [DOI] [Google Scholar]
- 3.Karegowda A. G., Manjunath A. S., Jayaram M. A. Comparative study of attribute selection using gain ratio and correlation based feature selection. International Journal of Information Technology and Knowledge Management. 2010;2:271–277. [Google Scholar]
- 4.Shang W., Huang H., Zhu H., Lin Y., Qu Y., Wang Z. A novel feature selection algorithm for text categorization. Expert Systems with Applications. 2007;33(1):1–5. doi: 10.1016/j.eswa.2006.04.001. [DOI] [Google Scholar]
- 5.He X., Cai D., Niyogi P. Laplacian score for feature selection. Advances in Neural Information Processing Systems. 2005;18:507–514. [Google Scholar]
- 6.Suebsing A., Nualsawat H. A novel technique for feature subset selection based on cosine similarity. Applied Mathematical Sciences. 2012;6:6627–6655. [Google Scholar]
- 7.Nahato K. B., Harichandran K. N., Arputharaj K. Knowledge mining from clinical datasets using rough sets and backpropagation neural network. Computational and Mathematical Methods in Medicine. 2015;2015:13. doi: 10.1155/2015/460189.460189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christopher J. J., Nehemiah H. K., Arputharaj K., Moses G. L. Computer-assisted medical decision-making system for diagnosis of Urticaria. MDM Policy & Practice. 2016;1(1) doi: 10.1177/2381468316677752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elizabeth D. S., Retmin Raj C. S., Nehemiah H. K., Kannan A. Computer-aided diagnosis of lung cancer based on analysis of the significant slice of chest computed tomography image. IET Image Processing. 2012;6(6):697–705. doi: 10.1049/iet-ipr.2010.0521. [DOI] [Google Scholar]
- 10.Elizabeth D. S., Nehemiah H. K., Raj C. S. R., Kannan A. A novel segmentation approach for improving diagnostic accuracy of CAD systems for detecting lung cancer from chest computed tomography images. Journal of Data and Information Quality (JDIQ) 2012;3:1–16. doi: 10.1145/2184442.2184444. [DOI] [Google Scholar]
- 11.Elizabeth D. S., Kannan A., Nehemiah H. K. Computer aided diagnosis system for the detection of bronchiectasis in chest computed tomography images. International Journal of Imaging Systems and Technology. 2009;19(4):290–298. doi: 10.1002/ima.20205. [DOI] [Google Scholar]
- 12.Darmanayagam S. E., Harichandran K. N., Cyril S. R. R., Arputharaj K. A novel supervised approach for segmentation of lung parenchyma from chest CT for computer-aided diagnosis. Journal of Digital Imaging. 2013;26(3):496–509. doi: 10.1007/s10278-012-9539-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Retmin Raj C. S., Nehemiah H. K., Elizabeth D. S., Kannan A. A novel feature-significance based k-nearest neighbour classification approach for computer aided diagnosis of lung disorders. Current Medical Imaging Reviews. 2018;14(2):289–300. doi: 10.2174/1573405613666170504152628. [DOI] [Google Scholar]
- 14.Titus A., Nehemiah H. K., Kannan A. Classification of interstitial lung diseases using particle swarm optimized support vector machine. International Journal of Soft Computing. 2015;10:25–36. [Google Scholar]
- 15.Nancy Jane Y., Khanna Nehemiah H., Arputharaj K. A Q-backpropagated time delay neural network for diagnosing severity of gait disturbances in Parkinson's disease. Journal of Biomedical Informatics. 2016;60:169–176. doi: 10.1016/j.jbi.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 16.Nancy J. Y., Khanna N. H., Kannan A. A bio-statistical mining approach for classifying multivariate clinical time series data observed at irregular intervals. Expert Systems with Applications. 2017;78:283–300. doi: 10.1016/j.eswa.2017.01.056. [DOI] [Google Scholar]
- 17.Leema N., Khanna Nehemiah H., Kannan A., Jabez Christopher J. Computer aided diagnosis system for clinical decision making: experimentation using Pima Indian diabetes dataset. Asian Journal of Information Technology. 2016;15:3217–3231. [Google Scholar]
- 18.Nahato K. B., Nehemiah K. H., Kannan A. Hybrid approach using fuzzy sets and extreme learning machine for classifying clinical datasets. Informatics in Medicine Unlocked. 2016;2:1–11. doi: 10.1016/j.imu.2016.01.001. [DOI] [Google Scholar]
- 19.Christopher J. J., Nehemiah H. K., Kannan A. A swarm optimization approach for clinical knowledge mining. Computer Methods and Programs in Biomedicine. 2015;121(3):137–148. doi: 10.1016/j.cmpb.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Leema N., Nehemiah H. K., Kannan A. Neural network classifier optimization using differential evolution with global information and back propagation algorithm for clinical datasets. Applied Soft Computing. 2016;49:834–844. doi: 10.1016/j.asoc.2016.08.001. [DOI] [Google Scholar]
- 21.Leema N., Nehemiah H. K., Kannan A. Quantum-behaved particle swarm optimization based radial basis function network for classification of clinical datasets. International Journal of Operations Research and Information Systems (IJORIS) 2020;9:32–52. doi: 10.4018/978-1-7998-2460-2.ch065. [DOI] [Google Scholar]
- 22.Nancy J. Y., Khanna N. H., Arputharaj K. Imputing missing values in unevenly spaced clinical time series data to build an effective temporal classification framework. Computational Statistics & Data Analysis. 2017;112:63–79. doi: 10.1016/j.csda.2017.02.012. [DOI] [Google Scholar]
- 23.Jane N. Y., Nehemiah K., Arputharaj K. A temporal mining framework for classifying un-evenly spaced clinical data: an approach for building effective clinical decision-making system. Applied Clinical Informatics. 2016;7(1):1–21. doi: 10.4338/ACI-2015-08-RA-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fister I., Jr., Yang X. S., Fister I., Brest J., Fister A Brief Review of Nature-Inspired Algorithms for Optimization. Elektrotehniski Vestnik. 2013;80(3):1–7. [Google Scholar]
- 25.Leema N., Nehemiah K. H., Elgin Christo V. R., Kannan A. Evaluation of parameter settings for training neural networks using bBackpropagation algorithms. International Journal of Operations Research and Information Systems. 2020;11(4):62–85. doi: 10.4018/IJORIS.2020100104. [DOI] [Google Scholar]
- 26.Christo V. R. E., Nehemiah H. K., Nahato K. B., Brighty J., Kannan A. Computer assisted medical decision-making system using genetic algorithm and extreme learning machine for diagnosing allergic rhinitis. International Journal of Bio-Inspired Computation. 2020;16(3):148–157. doi: 10.1504/IJBIC.2020.111279. [DOI] [Google Scholar]
- 27.Sreejith S., Khanna Nehemiah H., Kannan A. A classification framework using a diverse intensified strawberry optimized neural network (DISON) for clinical decision-making. Cognitive Systems Research. 2020;64:98–116. doi: 10.1016/j.cogsys.2020.08.003. [DOI] [Google Scholar]
- 28.Sreejith S., Khanna Nehemiah H., Kannan A. Clinical data classification using an enhanced SMOTE and chaotic evolutionary feature selection. Computers in Biology and Medicine. 2020;126:p. 103991. doi: 10.1016/j.compbiomed.2020.103991. [DOI] [PubMed] [Google Scholar]
- 29.Isaac A., Nehemiah H. K., Isaac A., Kannan A. Computer-aided diagnosis system for dDiagnosis of pulmonary emphysema using bio-inspired aAlgorithms. Computers in Biology and Medicine. 2020;124:p. 103940. doi: 10.1016/j.compbiomed.2020.103940. [DOI] [PubMed] [Google Scholar]
- 30.Christo V. E., Nehemiah H. K., Brighty J., Kannan A. Feature selection and instance selection from clinical datasets using co-operative co-evolution and classification using random Forest. IETE Journal of Research. 2020:1–14. doi: 10.1080/03772063.2020.1713917. [DOI] [Google Scholar]
- 31.Anter A. M., Ali M. Feature selection strategy based on hybrid crow search optimization algorithm integrated with chaos theory and fuzzy C-means algorithm for medical diagnosis problems. Soft Computing. 2020;24(3):1565–1584. doi: 10.1007/s00500-019-03988-3. [DOI] [Google Scholar]
- 32.Elgin Christo V. R., Khanna Nehemiah H., Minu B., Kannan A. Correlation-based ensemble feature selection using bioinspired algorithms and classification Using backpropagation neural network. Computational and mathematical methods in medicine. 2019;2019:17. doi: 10.1155/2019/7398307.7398307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sweetlin J. D., Nehemiah H. K., Kannan A. Computer aided diagnosis of drug sensitive pulmonary tuberculosis with cavities, consolidations and nodular manifestations on lung CT images. International Journal of Bio-Inspired Computation. 2019;13(2):71–85. doi: 10.1504/IJBIC.2019.098405. [DOI] [Google Scholar]
- 34.Dhalia Sweetlin J., Nehemiah H. K., Kannan A. Computer aided diagnosis of pulmonary hamartoma from CT scan images using ant colony optimization based feature selection. Alexandria Engineering Journal. 2018;57(3):1557–1567. doi: 10.1016/j.aej.2017.04.014. [DOI] [Google Scholar]
- 35.Sweetlin J. D., Nehemiah H. K., Kannan A. Feature selection using ant colony optimization with tandem-run recruitment to diagnose bronchitis from CT scan images. Computer Methods and Programs in Biomedicine. 2017;145:115–125. doi: 10.1016/j.cmpb.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 36.Sunil Retmin Raj C., Khanna Nehemiah H., Shiloah Elizabeth D., Kannan A. Distance based genetic algorithm for feature selection in computer aided diagnosis systems. Current Medical Imaging Reviews. 2017;13(3):284–298. doi: 10.2174/1573405612666160503164115. [DOI] [Google Scholar]
- 37.Zawbaa H. M., Emary E., Parv B., Sharawi M. Feature selection approach based on moth-flame optimization algorithm. 2016 IEEE Congress on Evolutionary Computation (CEC); July 2016; Vancouver, BC, Canada. pp. 4612–4617. [DOI] [Google Scholar]
- 38.Chu S.-C., Tsai P.-W., Pan J.-S. PRICAI 2006: Trends in Artificial Intelligence. Springer; 2006. Cat swarm optimization; pp. 854–858. [DOI] [Google Scholar]
- 39.Gandomi A. H., Alavi A. H. Krill herd: a new bio-inspired optimization algorithm. Communications in Nonlinear Science and Numerical Simulation. 2012;17(12):4831–4845. doi: 10.1016/j.cnsns.2012.05.010. [DOI] [Google Scholar]
- 40.Chen H., Zhu Y., Hu K. Cooperative bacterial foraging optimization. Discrete Dynamics in Nature and Society. 2009;2009:17. doi: 10.1155/2009/815247.815247 [DOI] [Google Scholar]
- 41.Passino K. M. Biomimicry of bacterial foraging for distributed optimization and control. IEEE Control Systems Magazine. 2002;22(3):52–67. doi: 10.1109/MCS.2002.1004010. [DOI] [Google Scholar]
- 42.Chen H., Zhu Y., Hu K. Adaptive bacterial foraging optimization. Abstract and Applied Analysis. 2011;2011:27. doi: 10.1155/2011/108269.108269 [DOI] [Google Scholar]
- 43.Hofmann E. E., Haskell A. G. E., Klinck J. M., Lascara C. M. Lagrangian modelling studies of antarctic krill (Euphausia superba) swarm formation. ICES Journal of Marine Science. 2004;61(4):617–631. doi: 10.1016/j.icesjms.2004.03.028. [DOI] [Google Scholar]
- 44.van der Laan M. J., Polley E. C., Hubbard A. E. Super learner. Statistical Applications in Genetics and Molecular Biology. 2007;6(1):p. 25. doi: 10.2202/1544-6115.1309. [DOI] [PubMed] [Google Scholar]
- 45.Ayon S. I., Islam M. M., Hossain M. R. Coronary artery heart disease prediction: a comparative study of computational intelligence techniques. IETE Journal of Research. 2020:1–20. doi: 10.1080/03772063.2020.1713916. [DOI] [Google Scholar]
- 46.Ji B., Lu X., Sun G., Zhang W., Li J., Xiao Y. Bio-inspired feature selection: an improved binary particle swarm optimization approach. IEEE Access. 2020;8:85989–86002. doi: 10.1109/ACCESS.2020.2992752. [DOI] [Google Scholar]
- 47.Magesh G., Swarnalatha P. Optimal feature selection through a cluster-based DT learning (CDTL) in heart disease prediction. Evolutionary Intelligence. 2020:1–11. doi: 10.1007/s12065-019-00336-0. [DOI] [Google Scholar]
- 48.Rabbi M. F., Hasan S. M., Champa A. I., Asif Zaman M., Hasan M. K. Prediction of liver disorders using machine learning algorithms: a comparative study. 2020 2nd International Conference on Advanced Information and Communication Technology (ICAICT); November 2020; Dhaka, Bangladesh. pp. 111–116. [DOI] [Google Scholar]
- 49.Rajesh S., Choudhury N. A., Moulik S. Hepatocellular carcinoma (HCC) liver cancer prediction using machine learning algorithms. 2020 IEEE 17th India Council International Conference (INDICON); December 2020; New Delhi, India. pp. 1–5. [DOI] [Google Scholar]
- 50.Ouadfel S., Abd Elaziz M. Enhanced crow search algorithm for feature selection. Expert Systems with Applications. 2020;159:p. 113572. doi: 10.1016/j.eswa.2020.113572. [DOI] [Google Scholar]
- 51.Singh J., Bagga S., Kaur R. Software-based prediction of liver disease with feature selection and classification techniques. Procedia Computer Science. 2020;167:1970–1980. doi: 10.1016/j.procs.2020.03.226. [DOI] [Google Scholar]
- 52.Tougui I., Jilbab A., el Mhamdi J. Heart disease classification using data mining tools and machine learning techniques. Health and Technology. 2020;10(5):1137–1144. doi: 10.1007/s12553-020-00438-1. [DOI] [Google Scholar]
- 53.Tubishat M., Idris N., Shuib L., Abushariah M. A. M., Mirjalili S. Improved Salp swarm algorithm based on opposition based learning and novel local search algorithm for feature selection. Expert Systems with Applications. 2020;145:p. 113122. doi: 10.1016/j.eswa.2019.113122. [DOI] [Google Scholar]
- 54.Abdar M., Acharya U. R., Sarrafzadegan N., Makarenkov V. NE-nu-SVC: a new nested ensemble clinical decision support system for effective diagnosis of coronary artery disease. IEEE Access. 2019;7:167605–167620. doi: 10.1109/ACCESS.2019.2953920. [DOI] [Google Scholar]
- 55.Aouabed Z., Abdar M., Tahiri N., Gareau J. C., Makarenkov V. International Conference Europe Middle East & North Africa Information Systems and Technologies to Support Learning. Springer; 2019. A novel effective ensemble model for early detection of coronary artery disease; pp. 480–489. [DOI] [Google Scholar]
- 56.Książek W., Abdar M., Acharya U. R., Pławiak P. A novel machine learning approach for early detection of hepatocellular carcinoma patients. Cognitive Systems Research. 2019;54:116–127. doi: 10.1016/j.cogsys.2018.12.001. [DOI] [Google Scholar]
- 57.Sayed G. I., Hassanien A. E., Azar A. T. Feature selection via a novel chaotic crow search algorithm. Neural Computing and Applications. 2019;31(1):171–188. doi: 10.1007/s00521-017-2988-6. [DOI] [Google Scholar]
- 58.Abdar M., Yen N. Y., Hung J. C. S. Improving the diagnosis of liver disease using multilayer perceptron neural network and boosted decision trees. Journal of Medical and Biological Engineering. 2018;38(6):953–965. doi: 10.1007/s40846-017-0360-z. [DOI] [Google Scholar]
- 59.Abdullah A. A., Yaakob A., Ibrahim Z. Prediction of spinal abnormalities using machine learning techniques. 2018 International Conference on Computational Approach in Smart Systems Design and Applications (ICASSDA); August 2018; Kuching, Malaysia. pp. 1–6. [DOI] [Google Scholar]
- 60.Sawhney R., Mathur P., Shankar R. Computational Science and Its Applications – ICCSA 2018. Vol. 10960. Springer; 2018. A firefly algorithm based wrapper-penalty feature selection method for cancer diagnosis; pp. 438–449. [DOI] [Google Scholar]
- 61.Abdar M., Zomorodi-Moghadam M., Das R., Ting I. H. Performance analysis of classification algorithms on early detection of liver disease. Expert Systems with Applications. 2017;67:239–251. doi: 10.1016/j.eswa.2016.08.065. [DOI] [Google Scholar]
- 62.Zamani H., Nadimi-Shahraki M. H. Feature selection based on whale optimization algorithm for diseases diagnosis. International Journal of Computer Science and Information Security. 2016;14:1243–1247. [Google Scholar]
- 63.ABDAR M. A survey and compare the performance of IBM SPSS modeler and rapid miner software for predicting liver disease by using various data mining algorithms. Cumhuriyet Üniversitesi Fen-Edebiyat Fakültesi Fen Bilimleri Dergisi. 2015;36:3230–3241. [Google Scholar]
- 64.Santos M. S., Abreu P. H., García-Laencina P. J., Simão A., Carvalho A. A new cluster-based oversampling method for improving survival prediction of hepatocellular carcinoma patients. Journal of Biomedical Informatics. 2015;58:49–59. doi: 10.1016/j.jbi.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 65.Chiu H.-C., Ho T.-W., Lee K.-T., Chen H.-Y., Ho W.-H. Mortality predicted accuracy for hepatocellular carcinoma patients with hepatic resection using artificial neural network. The Scientific World Journal. 2013;2013:10. doi: 10.1155/2013/201976.201976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schiezaro M., Pedrini H. Data feature selection based on artificial bee colony algorithm. EURASIP Journal on Image and Video Processing. 2013;2013(1) doi: 10.1186/1687-5281-2013-47.47 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting this study are from previously reported studies and datasets, which have been cited. The datasets used in this research work are available at UCI Machine Learning Repository.


