Summary
Prior immunological exposure to dengue virus can be both protective and disease-enhancing during subsequent infections with different dengue virus serotypes. We provide here a systematic, longitudinal analysis of B cell, T cell, and antibody responses in the same patients. Antibody responses as well as T and B cell activation differentiate primary from secondary responses. Hospitalization is associated with lower frequencies of activated, terminally differentiated T cells and higher percentages of effector memory CD4 T cells. Patients with more severe disease tend to have higher percentages of plasmablasts. This does not translate into long-term antibody titers, since neutralizing titers after 6 months correlate with percentages of specific memory B cells, but not with acute plasmablast activation. Overall, our unbiased analysis reveals associations between cellular profiles and disease severity, opening opportunities to study immunopathology in dengue disease and the potential predictive value of these parameters.
Keywords: dengue, longitudinal study, disease severity, immune cell phenotype, CyTOF, neutralizing antibodies, T cells, memory B cells, plasmablasts
Graphical abstract

Highlights
T cell, B cell phenotypes, and antibodies are associated with dengue disease severity
CXCR3+ CD8 T cell responses are associated with memory B cell formation
Treg responses are associated with plasmablast responses
Memory B cell numbers correlate with long-lasting neutralizing antibody titers
Rouers et al. examine the phenotype of dengue immune responses in a longitudinal patient cohort and find associations between cellular profiles and disease severity. Immune cells that are associated with long-lasting neutralizing antibodies up to 1 year after disease onset are described.
Introduction
Dengue virus (DENV) is an arbovirus transmitted by Aedes mosquitoes. There are four serotypes of DENV that are endemic in all tropical countries globally. An estimated 390 million persons are infected per year, out of which 100 million show clinical symptoms.1 Although the four serotypes can co-circulate, outbreaks are usually dominated by one serotype.2,3 In Singapore, alternating DENV-1 and -2 outbreaks have occurred since 2004, with a small contribution of DENV-3 and only a sporadic appearance and low infection force of DENV-4.3
Temporally limited herd immunity is thought to be the crucial factor behind the cyclical occurrence of dengue outbreaks. Similarly, populations in low endemic countries are more susceptible to outbreaks due to low herd immunity.4 Furthermore, immunity against dengue can be both protective and disease enhancing during repeated infection with a different serotype. The risk of enhanced disease is highest within a narrow range of cross-reactive, sub-neutralizing antibody (Ab) concentrations.5 This and other studies6,7 have defined a time window of susceptibility to enhanced disease, but they have not provided information about immune cells and cell phenotypes involved in disease severity.
Asymptomatic infection is associated with a higher percentage of activated T cells and an increased adaptive response compared to symptomatic infection.8 It has also been proposed that the plasmablast response correlates with more severe disease.8,9 However, other studies found no association.10 During secondary infection, a large proportion of plasmablasts produce cross-reactive Abs that bind to conserved epitopes of the DENV family.11, 12, 13 While cross-reactive Abs can be protective at high concentrations, they have the potential to worsen disease at sub-neutralizing concentrations.14
While the plasmablast response has been studied extensively during the acute phase after infection, we still know little about the protective value and longevity of memory B cells (MBCs) in primary and secondary infection. MBCs are specific for both cross-reactive and serotype-specific epitopes, and only a small percentage of MBCs encodes highly neutralizing and potentially serotype cross-neutralizing Abs, mostly to quaternary epitopes present on the whole virus particle but not on monomers of the surface glycoprotein.15, 16, 17
T cell responses during repeat infections were associated with disease severity.18,19 However, from human studies and mouse models, T cells have been recognized to play an important protective role.20, 21, 22 A subset of CD4 T cells with a cytotoxic effector phenotype after in vitro peptide stimulation was enriched in donors with more than one previous dengue infection. More than 50% of interferon-γ+ (IFN-γ+) cells showed a cytotoxic phenotype associated with CX3CR1, CD57, perforin, granzyme, TBET, and eomes expression, and correlated with a protective histocompatibility leukocyte antigen-DR isotype (HLA-DR) allele.23 Besides associations with HLA types with susceptibility to infection,24 a more vigorous T cell response and cytokine-mediated polyfunctionality was associated preferentially with HLA-B types.21 In addition, the T cell receptor repertoire determined by germline alleles may affect the ability to respond to certain DENV serotypes.25
Overall, there is accumulating evidence that multiple components of the adaptive immune response are critical for long-term immunity to dengue and protection from re-infection. However, host-response studies have focused on either the T or the B cell response. We were therefore particularly interested in correlations between T and B cell phenotypes and/or Ab profiles, the long-term effect of acute disease phenotype, and differences between primary and secondary responses. To study these parameters, we collected blood samples from primary and secondary dengue patients at day <6 after fever onset, days 6–10 (acute disease), days 14–28 (post-febrile), 6 months, and 1 year after fever onset. MBC response remained elevated until 6 months after disease, and DENV-specific T cells analyzed 6 months or 1 year after disease onset showed multiple distinct fates, including cells with terminally differentiated effector-memory phenotypes, as recently reported.26 A multiparameter analysis revealed several T and B cell phenotypes associated with disease severity and long-term immunity.
Results
Longitudinal patient cohort and parameters assessed
We followed a cohort of 68 adult patients in Singapore, starting with the collection of a blood sample <6 days after fever onset, and 4 additional samples at days 6–10 (acute disease, visit 2), days 14–28 (post-febrile, visit 3), 6 months (visit 4), and 1 year after fever onset (visit 5) (Figure 1A; Table 1; Table S1).
Figure 1.
Design of the longitudinal study and correlation of T and B cell readouts
(A) Study overview in the context of a typical course of dengue infection and the immune response generated. Viremia (red curve), IgG level (primary infection: light blue curve, secondary infection: dark blue curve), and B and T cell memory (green curve) are represented as arbitrary units from the fever onset until 12 months after fever. Each time point studied is indicated with an arrow: <6 days (visit 1; V1), 6–10 days (visit 2, acute disease), 14–28 days (visit 3, post-febrile), 6 months (visit 4), 1 year (visit 5). Readouts obtained at each time point are listed.
(B) Viremia measured by qRT-PCR at the first visit (>6 days after fever onset) in inpatients (n = 8), outpatients (n = 27), and outpatient/inpatients (n = 7); in primary (n = 20) versus secondary (n = 22) infection cases; in patients with (n = 24) and without (n = 18) warning signs (WSs). Blue symbols: primary infection, red symbols: secondary infection. p = ns (Kruskal-Wallis or unpaired Student’s t test). Means ± SD are shown.
(C) Network representing the correlations between all readouts. One node represents 1 readout. Readouts are classified in colored groups. The prefix CyTOF/flow cytometry denotes the technique used to identify the cell type. Dengue CD8+ T cells: dengue-specific T cells identified using tetramers, MBC: memory B cells, MBC ELISA: IgG detected in re-stimulated MBC culture supernatant, ELISA plasma: readouts from plasma antibodies. Betweenness centrality was used to prepare the network: the node that is commonly shared among the shortest paths is given a bigger node size than the other nodes in the paths.
Table 1.
Summary of the patient cohort
| Age, y (mean ± SD) | Male/female | Time after fever onset |
|||||
|---|---|---|---|---|---|---|---|
| <6 days | 6–10 days | 14–28 days | 6 months | 1 y | |||
| Entire cohort (N = 68) | 37.3 ± 11.7 | 51/17 | 68 | 55 | 46 | 41 | 37 |
| Serotype of infectiona | |||||||
| DENV-1 (n = 21) | 34.6 ± 12.1 | 17/4 | 21 | 18 | 15 | 13 | 11 |
| DENV-2 (n = 23) | 40.2 ± 11.2 | 18/5 | 23 | 17 | 14 | 13 | 13 |
| DENV-3 (n = 4) | 37.8 ± 13.5 | 2/2 | 4 | 3 | 3 | 3 | 3 |
| DENV-4 (n = 2) | 32 ± 11.3 | 0/2 | 2 | 2 | 2 | 1 | 0 |
| Details according to the status of infection | |||||||
| Serotype of infectiona | |||||||
| Primary (n = 30) | 34.4 ± 10.3 | 23/7 | 30 | 24 | 19 | 17 | 15 |
| DENV-1 (n = 14) | 31.8 ± 11.1 | 11/3 | 14 | 12 | 9 | 9 | 8 |
| DENV-2 (n = 10) | 38.0 ± 9.9 | 8/2 | 10 | 7 | 7 | 6 | 5 |
| DENV-3 (n = 1) | 28 ± 0 | 1/0 | 1 | 0 | 0 | 0 | 0 |
| DENV-4 (n = 0) | – | 0/0 | 0 | 0 | 0 | 0 | 0 |
| Serotype of infectiona | |||||||
| Secondary (n = 38) | 39.6 ± 12.4 | 28/10 | 38 | 31 | 27 | 24 | 22 |
| DENV-1 (n = 7) | 40.4 ± 12.9 | 6/1 | 7 | 6 | 6 | 4 | 3 |
| DENV-2 (n = 13) | 41.9 ± 13.2 | 10/3 | 13 | 10 | 7 | 7 | 8 |
| DENV-3 (n = 3) | 41 ± 14.4 | 1/2 | 3 | 3 | 3 | 3 | 3 |
| DENV-4 (n = 2) | 32 ± 11.3 | 0/2 | 2 | 2 | 2 | 1 | 0 |
DENV, dengue virus.
In addition to the confirmed serotypes, there were suspected primary DENV-2 (n = 2), suspected secondary DENV-2 (n = 6), suspected secondary DENV-3 (n = 2) based on very weak RT-PCR signals. There were unclear serotype primary (n = 2), and unclear serotype secondary (n = 5) based on unclear serology. For a detailed list, refer to Table S1.
The relatively low dengue endemicity in Singapore provided a unique opportunity to study both acute primary and secondary infections in adults from the same cohort. To distinguish the severity status of the patients enrolled in this cohort, we applied two independent criteria: (1) the 2009 World Health Organization (WHO) classification that includes a set of clinical parameters to diagnose patients as having dengue fever (DF) without warning signs (WSs), DF with WSs, and severe dengue (SD) (see Method details), and (2) treatment as outpatient versus hospitalization. There was a third group of patients who were treated as outpatients initially, but eventually had to be hospitalized due to worsening symptoms (outpatient/inpatient). The two methods of severity classification agreed significantly when tested in a Fisher test (Table S2). Viremia at the first visit could be measured by qRT-PCR for most patients and was not different between the different patient groups (Figure 1B).
Blood samples were collected for plasma Ab analysis (ELISA and neutralization assays) and for peripheral blood mononuclear cell (PBMC) isolation. With the exception of flow cytometry analysis of plasmablasts, which was performed with fresh cells, all other cellular readouts were conducted with frozen-thawed PBMCs (CyTOF, B cell ELISpot, T cell ELISpot, flow cytometry). An overview of the different readouts is illustrated in Figure 1A.
Uniform manifold approximation and projection (UMAP) dimensionality reduction27 and Louvain clustering analyses using PhenoGraph28 were performed on the CyTOF datasets. Clusters identified by PhenoGraph were named according to the median expression of surface markers and transcription factors (for Ab panels, see Table S3), and were subsequently quantified for all samples using manual gating (Figures S1–S4). From CD45+CD14− immune cells, we could identify the following subsets: CD4 T cells, CD8 T cells, mucosal-associated invariant T (MAIT) cells, γδT cells, B cells, plasmablasts, natural killer (NK) cells, and Lineage− cells (Figure S1). Twelve phenotypically distinct subsets were defined for total CD4 T cells (Figure S2), 10 subsets for total CD8 T cells (Figure S3), and 7 subsets for CD19+ B cells (Figures S4A–S4C). In addition, 12 B cell subsets were defined and analyzed by flow cytometry (Figure S4D). To identify dengue-specific CD8 T cells (called “Dengue CD8 T cells” in figures), multiplexed HLA tetramers presenting peptides derived from DENV-1 and -2 structural and non-structural proteins, as described previously26 and in the Method details section, were used.
Combining the results from all readouts and all time points, we conducted a bivariate correlation analysis. Using a false discovery rate (FDR) of 0.05 and a correlation coefficient cutoff of 0.6 for graphic illustration, the biggest cluster with 318 nodes included both T and B cells parameters (Figure 1C). A stratified analysis according to primary versus secondary infection and severity or disease progression allowed us to associate immune parameters with long-term immunity and disease outcomes, as described in the following paragraphs.
Immune cell phenotypes associated with disease severity
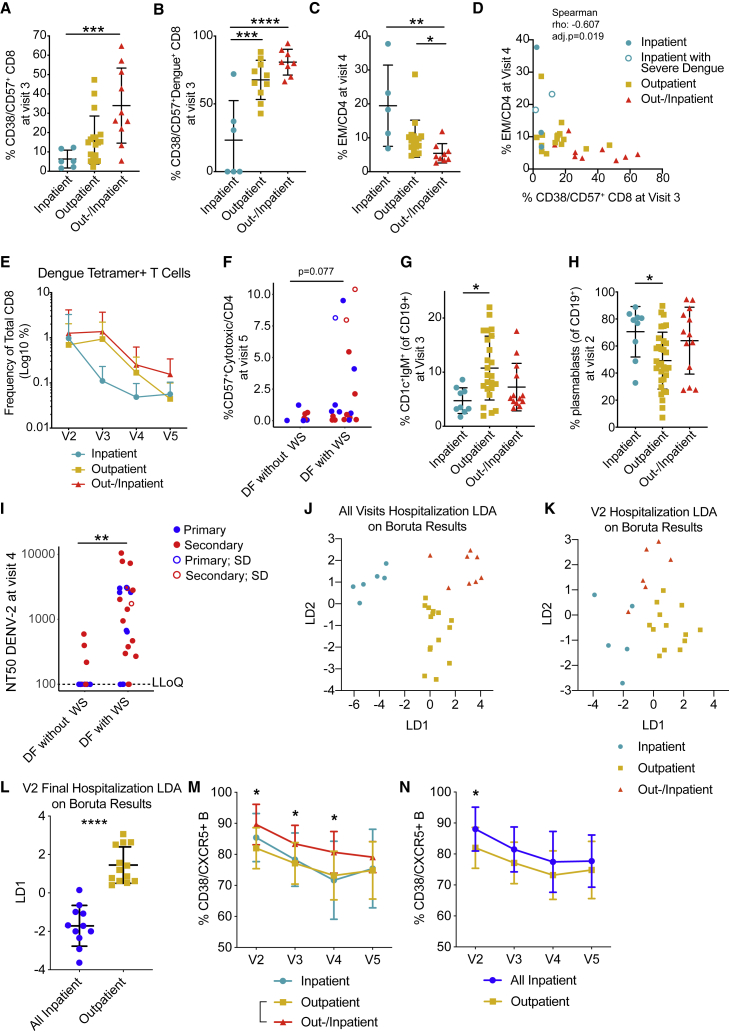
We analyzed the dataset for significant correlations between disease severity and T cell phenotypes, B cell phenotypes, or Ab responses. When considering hospitalization status, the most significant immune cell associations (non-adjusted p values) were related to T cell phenotypes. Patients who were hospitalized showed lower frequencies of CD38 expressing CD57+ CD8 T cells at the post-febrile (visit 3) time point compared to outpatients (Figure 2A). These CD57+ CD8 T cells also showed high expression of GPR56, CD45RA, killer cell lectin-like receptor subfamily G member 1 (KLRG1), TBET, and granzyme B (Figure S3). For dengue-specific CD57+ CD8 T cells, we observed a similar decrease in CD38+ cells for inpatient samples at the same stage (Figure 2B). However, the frequencies of CD8 T cells with this activated phenotype were highest for the patients in the outpatient/inpatient group, suggesting that either too-low or too-high frequencies of these cells are detrimental to disease outcomes.
Figure 2.
Adaptive immune response readouts associated with disease severity
(A–C) Frequencies of T cell populations by hospitalization: inpatient (n = 6), outpatient (n = 24), and outpatient/inpatient (n = 10).
(A) Frequency of CD38+/CD57+ CD8 T cells at V3; ∗∗∗p = 0.0007.
(B) Frequency of CD38+/CD57+ dengue+ CD8 cells at V3; ∗∗∗p = 0.0003, ∗∗∗∗p < 0.0001.
(C) Frequency of effector memory (EM) CD4+ T cells at V4; ∗p = 0.0408, ∗∗p = 0.0028.
(D) Correlation of frequency of EM CD4+ T cells at V4 with frequency of CD38+ CD57+ CD8+ T cells at V3.
(E) Frequencies of total dengue tetramer+ T cells by hospitalization groups. Mixed-effects analysis with Geisser-Greenhouse correction.
(F) Frequency of CD57+ cytotoxic T /CD4 cells at V5 for patients with dengue fever (DF) but no WSs (DF without WS) and patients with DF and WSs (DF with WS). Each symbol represents 1 patient: primary (blue), secondary (red), empty symbols represent severe dengue (SD) cases.
(G) Frequency of CD1c+IgM+ cells among CD19+ B cells at V3 by hospitalization: inpatient (n = 9), outpatient (n = 24), and outpatient/inpatient (n = 13); ∗p = 0.0108.
(H) Frequency of plasmablasts (CD38+CD27+) among CD19+ B cells at V2 by hospitalization: inpatient (n = 9), outpatient (n = 24), and outpatient/inpatient (n = 13); ∗p = 0.0335.
(I) Neutralization antibody titer (NT50) against DENV-2 at V4 for patients with DF without WS and patients with DF with WS. Empty symbols represent SD cases. ∗∗p = 0.0012, Mann-Whitney test. LLoQ: lowest limit of quantification.
(J) LDA plot of features selected by Boruta feature selection that was performed on all-visit data and classified by hospitalization status (n = 29).
(K) LDA plot of data selected by Boruta feature selection that was performed on V2 data and classified by hospitalization (n = 24).
(L) LDA plot of data selected by Boruta feature selection that was performed on V2 data and classified by final hospitalization status (n = 24). Mann-Whitney test.
(M) Frequencies of CD38+/CXCR5+ B cells over time by hospitalization status.
(N) Frequencies of CD38+/CXCR5+ B cells over time by final hospitalization status.
For all graphs, except where specified otherwise: error bars indicate means ± SD. Kruskal-Wallis with Dunn’s multiple comparison test.
Six months after disease, inpatients had significantly higher frequencies of effector memory (EM) CD4 T cells compared to subjects in the outpatient/inpatient group (Figure 2C). There was no significant difference (p = 0.07, Kruskal-Wallis test) in the percentages of CD38+CD57+ CD8 cells during the acute phase when comparing inpatients versus outpatients (Figure S5A). It is possible that non-specific T cells were activated only during the post-febrile stage or massive cell activation and potential cell death during acute disease masked correlations at this time point.
CD38+CD57+ CD8 and EM CD4 T cells were correlated negatively (Figure 2D), with more severe cases falling into the extreme ends of the correlation, suggesting that a balanced response of these activated cells was beneficial for disease outcomes. CD57+ CD8 T cells have been described to proliferate less and produce more inflammatory cytokines compared to CD57− CD8 T cells,29 possibly contributing to excessive inflammation.
Severity and secondary infection status are potentially linked and could affect whether immune features are associated with infection status or severity. In our cohort, 15 primary infections and 22 secondary infections fell into the category of DF with WSs. The separation according to hospitalization status was clearly affected by previous infection: 6 primary and 19 secondary patients were hospitalized as inpatients or outpatients/inpatients (Table S1). Both primary and secondary patients showed large numbers of CD38+CD57+ CD8 T cells during the acute phase that was sustained into the post-febrile stage (Figure S5B). Percentages of EM CD4 T cells per patient were largely consistent across all time points (Figure S5C), which may indicate that high percentages of these cells in severe patients were already present before infection and could influence the susceptibility of these individuals toward a detrimental outcome.
While total frequencies of tetramer+ cells are not necessarily reflective of the magnitude of the response because of the variation in epitope usage and HLA alleles, the average responses per patient group are still informative. While the frequency of dengue-specific CD8 T cells was similar for all of the patients during acute disease, these cells decreased faster in inpatients at visits 3 and 4 than in the other 2 hospitalization groups (not statistically significant, 2-way ANOVA) (Figure 2E).
One year after fever onset, the frequency of CD57+ cytotoxic CD4 T cells was higher in patients with WSs compared to those without WSs (Figure 2F). The patients classified as SD (empty symbols) were among those with the highest proportions of CD57+ cytotoxic CD4 T cells. Two secondary patients with WSs showed consistently high percentages of CD57+ cytotoxic CD4 T cells over 1 year (Figures S5D–S5G).
B cell phenotypes were also associated with disease outcomes. Frequencies of CD1c+IgM+ B cells were highest in the outpatient group at the post-febrile stage (Figure 2G). CD1c+IgM+ B cells have been proposed to represent marginal zone-like B cells30 and could play a protective role due to their capacity to rapidly produce IgM (immunoglobulin M). Plasmablast numbers at acute phase were higher in hospitalized patients compared to outpatients (Figure 2H). Patients with WSs had higher DENV-2 NT50s 6 months after fever (Figure 2I). Since this group consisted of both primary and secondary patients, high convalescent NT50 titers and WSs appeared to be independent of infection status. A similar correlation was not observed for DENV-1-specific NT50 6 months after fever, although equal numbers of patients with WSs had a DENV-1 or DENV-2 infection (Table S1). It is possible that the DENV-2 neutralization assay is biased toward cross-reactive Abs with higher enhancement potential (see Discussion).
Because the number of variables is far larger than the number of unique patients, we performed a first pass of feature selection using the Boruta algorithm, followed by linear discriminant analysis (LDA), to separate the patients according to whether they were hospitalized. Boruta is an all-relevant feature selection algorithm based on the random forest classification algorithm.31 LDA is a supervised linear transformation that reduces the dimensionality of the dataset by finding axes of projection that maximize the separation between classes.32 Boruta, performed on a curated all-visit dataset containing 29 patients and 377 variables, selected 14 features that may contribute to hospitalization status (Table S4A). CD38+/CD57+CD8 T cells, EM CD4 T cells, and plasmablasts were also identified by the bivariate analysis (Figures 2A–2C). We performed LDA on these 14 features using hospitalization status as a classifier and found a clean separation between the different groups, confirming that this cluster of features distinguishes different hospitalization statuses (Figure 2J).
While the post-febrile time point appeared to have the strongest association with hospitalization status, it is relatively late in the dengue infection time course (14–28 days after fever onset), when most patients are recovering. To assess earlier biomarkers, we used Boruta and LDA to analyze only the data from the acute time point (visit 2), classified by recorded hospitalization status (Figure 2K; Table S4B). We created a category called ”final hospitalization,” in which all of the patients with an outpatient/inpatient status were given an inpatient status (Figure 2L; Table S4C). The frequency of CD38+ CXCR5+ B cells was significantly distinct between outpatient and outpatient/inpatient samples (Figure 2M), as well as between outpatients and patients in the final hospitalization group (Figure 2N).
To validate immune phenotypes associated with severity with a simple flow cytometry panel (Figure S6A), we leveraged samples from an independent cohort of hospitalized dengue patients showing different degrees of disease severity (validation cohort, Table S5). A principal-components analysis (PCA) on immune cell phenotypes selected based on Figure 2 and the LDA analysis in Table S4B (V2 hospitalization was considered because the validation cohort samples were collected in the same time window) clearly separated patients with more than two or less than two WSs recorded in the study visits before and including the time point of analysis (Figures S6B and S6C). Although this validation cohort is not perfect since a separation based on hospitalization was not possible (all patients were hospitalized), the PCA confirmed a combined contribution of CD38+CD57+ CD8 T cells (negative association with inpatient status; Figure 2A), CD38+CXCR5+ B cells (positive association with inpatient status; Figure 2N) and EM cells re-expressing CD45RA (EMRA) T cells to the separation of the 2 severity groups.
Overall, 14 T and B cell features of the adaptive response separated patients according to their hospitalization status. Correlations of immune cell phenotypes with severity measured at the late time point and consistency in phenotypes over 1 year suggested that an individual’s general predisposition and capacity to produce particular cell phenotypes could affect disease outcome.
Potential predictive value of dengue-specific T cell responses for long-term immunity
A panel of 165 distinct tetramers was used to identify dengue-specific cells.26 For controls, Flu, Epstein-Barr virus (EBV)-specific, and cytomegalovirus (CMV)-specific CD8 T cells were analyzed as well, as described previously.33,34 DENV-specific cells showed a highly activated phenotype during acute disease and post-febrile stage that was very distinct from the phenotype observed at 6 months and 1 year (Figures 3A and 3B).26 In contrast, EBV- and Flu-specific CD8 T cell phenotypes were largely consistent in dengue patients over 1 year. However, based on CD38 expression and granzyme B expression, we noted bystander activation for CMV-specific cells during acute disease (Figure 3B).
Figure 3.
Predictive value of dengue-specific T cell responses for long term immunity
(A) UMAP was performed on a combined dataset of 201 tetramer+ samples. A total of 27 patients are represented for 1–4 time points (and 3 healthy controls: V0) with a maximum of 300 cells per sample. UMAP plots display tetramer+ cells coded by disease (CMV, dengue, EBV, Flu) and time point of dengue infection (V2–V5) (left) and expression intensities of important markers GPR56, Ki67, CD57, CXCR5, and CD127 among dengue tetramer+ CD8 T cells (right).
(B) Mean frequencies of marker-expressing cells within dengue, EBV, flu, and CMV tetramer+ CD8 T cells, summarized as a heatmap.
(C) Highly significant correlations between percentages of phenotypically distinct dengue-specific T cell populations and other dengue-specific or total T cell populations.
Among dengue tetramer+ CD8 T cells, many significant correlations were for a given phenotype between early and late time points, suggesting consistency over time and potential predictive value of early phenotypes for long-term immunity (Table S6). PD1+ dengue-specific CD8 T cells were highly significantly correlated between acute disease and 6 months (Figure 3C). Other notable correlations for dengue-specific cells were between total central memory CD4 T cells during acute disease and CXCR5+ tetramer+ CD8 cells at 1 year (Figure 3D), total T helper 17 (Th17) cells during acute disease and CXCR5+ tetramer+ CD8 cells at 1 year (Figure 3E), GPR56+ tetramer+ CD8 cells during acute disease, and total EM CD8 cells at 1 year (negative correlation) (Figure 3F). GPR56 expression has been previously associated with dengue-specific CD4 T cells with cytotoxic features.35 GPR56+tetramer+ CD8 cells at V4 correlated significantly with viremia (adjusted p close to 0, ρ 0.88; Data S1), suggesting that a higher viral load led to a higher expansion and/or longer persistence of these specific cells. In addition, viremia correlated with CyTOF granzyme B+/dengue-specific CD8 cells at V4 (adjusted p = 0.013).
Overall, we found that the percentages of many T cell phenotypes in individual patients were consistent over 1 year. Correlations between the specific phenotype of total and dengue-specific cells between acute disease and 1 year supported the hypothesis that the individual capacity of patients to generate given T cell types may directly or indirectly affect the outcome of dengue-specific T cell immunity.
Connections between B and T cell readouts and implications for immunity
B cell ELISpots involved a 6-day pre-culture of B cells with R848 (TLR 7/8 ligand) and interleukin-2 (IL-2) to differentiate MBCs into Ab -secreting cells. IgG specific for E protein and UV-inactivated DENV from both DENV-1 and DENV-2 were analyzed by ELISA (Figures S7A–S7D). The area under the curve (AUC) and the optical density (OD) detected for a low quantity of IgG (0.25 μg) were included in the bivariate correlation analysis. These readouts reflect the quantity and/or affinity of DENV-specific IgG secreted by MBCs. Ab readouts from in vitro differentiated MBCs that connected T and B cell readouts in the network shown in Figure 1B: the AUC for Abs secreted by late time point MBCs specific for whole virus particles correlated negatively with EM CD8 T cells at the post-febrile stage (Figure 4A) (adjusted p = 0.025), indicating a potential negative impact of CD8 T cells on MBC formation specific for virus particles. In turn, Ki67/CD57+ cytotoxic CD4 T cells at the post-febrile stage correlated positively with DENV-2 E protein-specific Abs secreted by MBCs at 6 months (Figure 4B) (adjusted p = 0.024). Both the OD and AUC of post-febrile DENV-2 E protein-specific Abs correlated significantly with total Ki67+-naive CD8 T cells at 1 year (Figures S7E and S7F). Hospitalized patients tended to have higher proportions of Ki67+-naive CD8 T cells at 1 year and also a higher proportion of granzyme B+ CD4 T cells at 6 months (Figures S7G and S7H).
Figure 4.
Connections between T and B cell readouts
(A) Correlation between MBC-secreted antibodies binding to UV-inactivated DENV-2 (AUC) at visit 5 and the percentage of EM (EM: CD8+ CD45RA− CD45RO+ CCR7− CD27−) CD8 T cells at visit 3.
(B) Correlation between MBC-secreted antibodies binding to E protein DENV-2 (AUC) at visit 4 and the percentage of Ki67+ CD57+ cytotoxic CD4 T cells at visit 3.
(C) Correlation between the percentages of PD-1+ of dengue-specific CD8 T cells and IgG+ MBCs of CD19+ B cells at visit 2.
(D) Correlation between the percentages of Treg CD4+ T cells and Ki67+ plasmablasts (CD27hi CD38hi) at visit 2.
(E) Correlation between the percentages of CD38+CCR7− Tregs and PD-1+ plasmablasts (CD27hi CD38hi) at visit 2. Spearman test with adjusted p value. Each dot represents 1 patient.
The most significant correlation of B cell readouts with a dengue-specific CD8 T cell readout was a negative correlation between IgG+ MBCs and PD-1+ dengue-specific CD8 T cells during acute disease (Figure 4C) (ρ = −0.7856; p = 5.40E−6; adjusted p = 0.0008) (Table S7). PD-1+ CD8 T cells are known to be dysfunctional and inefficient at clearing viral infection.36 The presence of this population early during dengue infection may have a negative impact on MBC establishment. B cells express the ligands of programmed cell death protein-1 (PD-L1 and PD-L2). PD-L1hi B cells have been described for their key role in the regulation of humoral response by limiting the differentiation and function of T follicular helper cells (Tfhs),37 but nothing is known about their potential interactions with PD-1+ CD8 T cells.
Ki67+ plasmablasts during acute disease correlated positively with total CD4 T regulatory cells (Tregs) (Figure 4D) (ρ = 0.7666; p = 8.0797E−9; adjusted p = 4.28E−6), whereas PD-1-expressing plasmablasts correlated negatively with CD38+ CCR7− Tregs (Figure 4E) (ρ = −0.5266; p = 0.0006; adjusted p = 0.025). CCR7− Tregs are the EM arm of the Treg population,38 and expression of CD38 on Tregs confers a more suppressive function.39 CCR7− Treg cells may therefore have an inhibitory impact on at least a subset of the plasmablast response during acute disease, whereas total CD4 Tregs directly or indirectly support the plasmablast response.
In summary, although few correlations between B and T cell readouts were significant based on adjusted p values, a clear connection between the early T cell response and B cell memory was observed, with a potential regulatory role for Tregs on plasmablast formation and a negative correlation between total IgG+ MBCs and dengue-specific PD-1+ CD8+ T formation. Alternatively, as the total IgG+ MBC population decreased with increasing plasmablast formation during acute disease, the correlation with PD-1+ CD8+ T cells could also be driven by plasmablast formation.
Cellular phenotypes associated with B cell effector functions
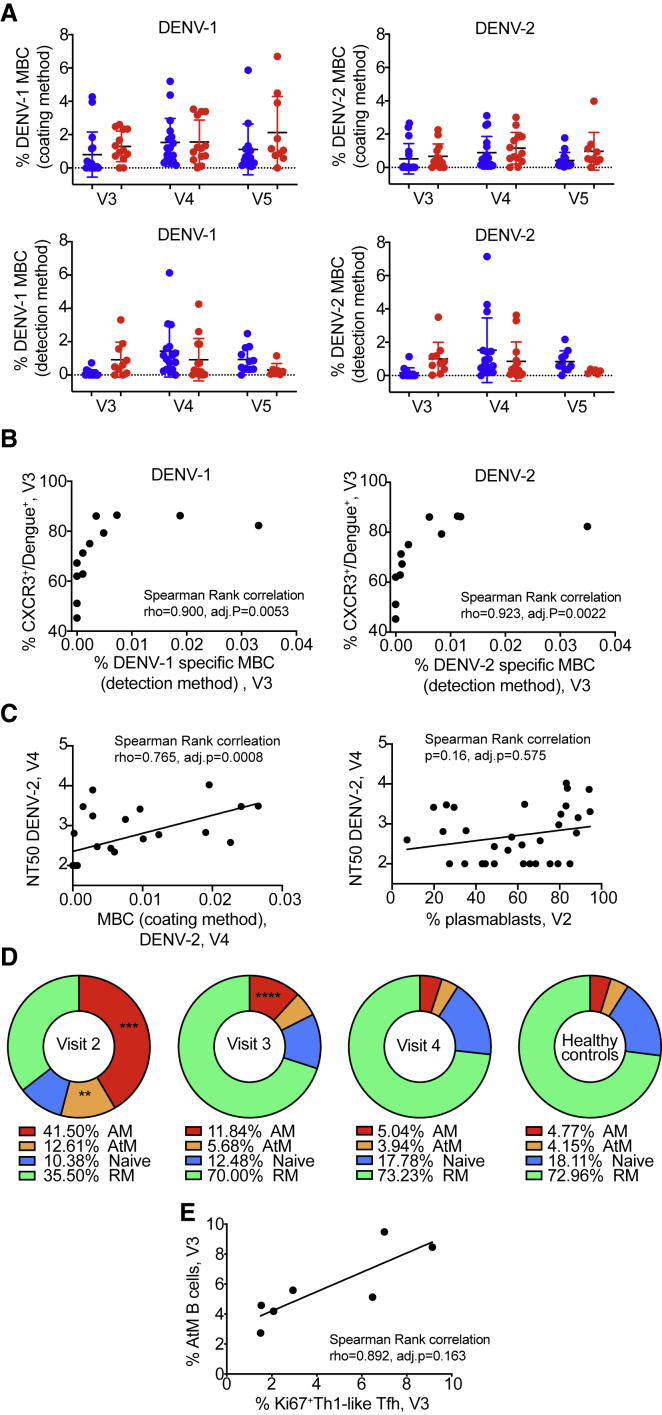
Up to 4% of total MBCs are dengue specific between 8 days and 3 weeks after fever onset.40,41 For the current cohort, we used two methods with different sensitivity: (1) E protein was coated on ELISpot plates and binding IgG was detected using fluorescent anti-human IgG (coating method) or (2) all Igs secreted by MBCs were captured on the plate, followed by probing with E protein and a tagged anti-E Ab for quantification by fluorospot (detection method). Below 1% of total IgG MBCs were dengue specific for the coating and <0.5% for the detection method at the post-febrile stage in primary patients, whereas secondary patients showed slightly higher responses at this time point (Figure 5A). At the 1-year time point, patients with primary infection tended to have higher numbers of specific MBCs for the possibly more specific detection method, whereas the opposite was observed for the coating method (Figure 5A).
Figure 5.
Cellular phenotypes associated with B cell effector functions
(A) Frequencies of dengue-specific MBCs. MBCs were quantified using 2 types of B cell ELISpot assays: (top row) coating of the E protein (coating method) or (bottom row) probing with E protein and a tagged anti-E antibody (detection method). (Left column) DENV-1- and (right column) DENV-2-specific MBCs. The data are normalized for total IgG secreted by MBCs for each patient. Each symbol represents 1 patient (primary: blue, secondary: red). Error bars indicate means ± SD.
(B) Correlation between CXCR3+/Tet+ CD8 at V3 and frequency of MBCs specific for DENV-1 (left) or DENV-2 (right).
(C) Correlation between DENV-2 NT50 at V4 and frequency of DENV-2 MBCs measured by the coating method at V3 (left) or frequency of plasmablasts at V2 (right).
(D) Pie charts representing the proportions of activated memory (AM: CD27+CD21−), atypical memory (AtM: CD27−CD21−), naive (CD27−CD21+), and resting memory (RM: CD27+CD21+) among CD19+ B cells at V2 (n = 9), V3 (n = 10), V4 (n = 10), and for healthy controls (n = 8). The frequency of each population is specified below each chart. Comparison between dengue patients and healthy control: t test.
(E) Correlation between frequency of AtM B cells and frequency of Ki67+/Th1-like Tfh cells at V3.
(B), (C), and (E) Each symbol represents 1 patient. Statistics: Spearman rank correlation test.
In addition to the observed impact of T cell phenotypes on B cell memory formation, a significant correlation between percentages of specific MBCs at the post-febrile stage and the frequency of CXCR3+ DENV-specific CD8 T cells at the same time point existed (Figure 5B). Dengue patients have high plasma concentrations of IP10, the ligand of CXCR3,42,43 which could explain the recruitment of this specific subset of T cells. The presence of protective, DENV-specific CD8 T cells may support a micro-environment that is favorable to the establishment of MBC responses.
One limitation of the ELISpot is that it was only done for E protein-binding B cells and cannot provide information on potentially more protective virus particle-specific MBCs. MBCs specific for the E protein of DENV-2 at the post-febrile stage (coating method) correlated with plasma DENV-2 neutralization 6 months after fever (Figure 5C). Too few data points were available for correlations with DENV-1 NT50 at 6 months. Nevertheless, the correlation suggested a link between the generation of an MBC response and a lasting ability to neutralize the virus. In turn, there was no correlation between the neutralizing titers at 6 months and the frequency of plasmablasts during acute disease (Figure 5C).
We next assessed the formation of atypical MBCs (AtM) (CD19+CD21−CD27−), which have been described in the context of chronic infection with HIV, malaria, mycobacterium tuberculosis, and hepatitis C virus (HCV).44 A phenotypically similar cell type is also observed during acute infection, as described for influenza and Ebola virus infection.45 This population of B cells does not respond well to stimuli and contributes to a defective immune response.
In the dengue infection context, we found an expansion of AtMs, in parallel with an expansion of activated MBCs (AM, CD19+CD27+CD21−) during the acute phase (Figure 5D). The CD19+CD27+CD21+ activated memory population also included plasmablasts. Both AtM and AM populations collapsed over time. Six months after fever, the percentages were comparable to those observed in healthy individuals (Figure 5D). AtM B cells expressed mostly IgM (Figure S8A). The proportions of AtM B cells were positively correlated with the frequency of plasmablasts at the post-febrile stage, but not during the acute phase (Figures S8B and S8C). The plasmablast frequency decreased faster compared to the AtM frequency, providing a potential explanation for the opposite trend of the correlation at acute and post-febrile stages. Steady-state plasmablasts and DENV-2-specific MBCs correlated negatively with AtMs (Figures S8D and S8E). The positive correlation at the post-febrile stage, when both plasmablasts and AtMs are more likely to be dengue specific, supports the hypothesis of a defective B cell response contributing to severity, in addition to a potential role of a high frequency of plasmablasts early in the course of dengue infection being associated with severity.
To assess the impact of T cell help on B cell phenotypes, we assessed circulating c)Tfhs: Th1-like CXCR3+ and Th2-like CXCR3− (in Data S1, the latter are called “TFH”). It has been proposed that Th2-like CXCR3− cTfh are more efficient in supporting protective B cell responses compared to Th1-like CXCR3+ cTfhs.46, 47, 48 Ki67+Th1-like cTfhs correlated positively with AtM B cells at visit 3 (Figure 5E). This may suggest that Th1-like cTfh cells, by providing suboptimal B cell help, could lead to the formation of non-functional AtM B cells, affecting the overall outcome of the response. There was no significant correlation for any cTfh subset with neutralizing titers. Due to the limitation of samples available for the analysis of B cell phenotypes, correlations with cTfh phenotypes need to be validated further.
Overall, these data confirmed a role for CD8 T cells in the formation of B cell memory. In addition, Th1-like CXCR3+ Tfh cells seemed to directly or indirectly affect the formation of AtMs.
Primary and secondary status of infection is linked to B cell and Ab readouts
The status of infection was highly significantly correlated with most of the ELISA parameters measuring the quantity of DENV-1 or DENV-2 Abs in the plasma (Figure 1B; Data S1). At least for the first time point this is expected as preexisting Abs were used to categorize primary versus secondary patients. Secondary patients had a higher titer of DENV-specific Abs compared to primary-infected patients until the post-febrile stage (Figures 6A and 6B). DENV-2 half-maximal neutralization titer (NT50) values at the acute stage correlated positively with the IgG (IgA−IgM−) plasmablast response measured by flow cytometry (p = 0.001, adjusted p = 0.039). The IgA plasmablast response correlated negatively, but highly significantly with DENV-2 NT50 titers (p = 7.45 × 10−5, adjusted p = 0.006) (Data S1). Similar to ELISA readouts, DENV-2 neutralization titers at <6 days correlated with acute phase and post-febrile titers. The neutralization readouts correlated with the status of infection, due to the higher (cross-) neutralizing ability of Abs in the serum from secondary patients compared to primary patients.
Figure 6.
Primary and secondary status of infection is linked to B cell/antibody readouts
(A) NT50 values of plasma collected at V1–V5 against DENV-1 (left column) or DENV-2 (right column) and for primary patients (top row, blue) or secondary patients (bottom row, red). Each set of connected dots represents 1 patient.
(B) Binding of plasma antibodies from V1–V5. ELISA plates were coated with E protein (mostly monomeric, top row) or with UV-inactivated polyethylene glycol (PEG)-precipitated DENV (UV-DENV) (bottom row). E protein ELISA: line indicates the lowest dilution tested (1:500). Endpoint titers for the negative control plasma were ≪500. UV-DENV ELISA: line indicates the endpoint titer of the negative control plasma (2,500). Lowest dilution tested was 1:100. Primary: blue, secondary: red. Means ± SD are indicated.
(C) Frequency of plasmablasts (CD27+CD38+) among CD19+ B cells at V2 for primary and secondary patients. ∗∗∗p = 0.0007, unpaired t test. Bars indicate the mean.
(D) Frequencies of IgA+, IgM+, and IgG+ (IgA−IgM−) plasmablasts during acute disease, stratified according to hospitalization status. Bars indicate the median.
(E) IgA expressed at the surface by the plasmablasts in (C); ∗∗∗∗p < 0.0001, unpaired t test. Bars indicate the mean
(F) Frequency of HLA-DR+ cells among CD8 T cells at V2 in primary and secondary patients. ∗∗p = 0.002, unpaired t test. Bars indicate the mean.
(G) Frequency of CXCR3+ Tfh (circle) or CXCR3− Tfh (triangle) among CD4 T cells from V2–V5 for primary (red) and secondary (blue) patients. Bars indicate the mean.
At the cellular level, secondary-infected patients displayed higher proportions of plasmablasts during acute disease (Figure 6C). The plasmablasts of primary-infected subjects expressed mostly IgM or IgA, whereas in subjects with secondary dengue infection, the plasmablasts were predominantly IgM− and IgA− (Figure 6D). Plasmablasts without any surface expression also fall in this IgM−IgA− gate, leading to a possible overestimation of the IgG+ fraction.
Status of infection was highly correlated with the frequencies of acute phase CD19+ B cells (p = 6 × 10−4), IgA+ plasmablasts (p = 0.0001), IgA−IgM− plasmablasts (p = 0.001), and IgM+ plasmablasts (p = 0.02) (Kruskal-Wallis test, adjusted p values). In particular, IgA+ plasmablasts significantly distinguished primary from secondary patients, providing a potential biomarker to assess immune status during early infection (Figure 6E).
When considering adjusted p values, no T cell phenotypes were significantly different between primary and secondary infection. The best correlation was seen between status of infection and acute phase HLA-DR+ CD8 T cells (Figure 6F).
Percentages of total Th1-like cTfhs at acute disease tended to be higher in secondary compared to primary cases, suggesting that these cTfhs contained reactivated, dengue-specific cells during secondary infection (Figure 6G).
In summary, B cell- and Ab-related readouts were more distinct between primary and secondary patients compared to T cell phenotype readouts.
Discussion
This study provides a comprehensive, longitudinal analysis of B and T cell responses in acutely diagnosed dengue patients. Our cohort comprised a balanced mix between primary and secondary patients and between patients with and without WSs.
We made the following observations:
-
(1)
Hospitalized patients showed the lowest frequency of CD57+ T cells with an activated (CD38+) phenotype at the post-febrile stage and the highest frequency of EM CD4 T cells at 6 months, representing steady state. In contrast, patients with outpatient/inpatient status had the highest frequencies of these CD38+CD57+ CD8 T cells at the post-febrile stage and the lowest frequency of EM CD4 T cells at 6 months. It is possible that the treatment and care received at the hospital contributed to decreasing the inflammation in the inpatient group. Alternatively, the profile in the outpatient/inpatient group could reflect an exaggerated or delayed response. Considering the overall longitudinal consistency of cell phenotypes (Table S6), the severity-dependent percentages of activated and pro-inflammatory CD8 and EM CD4 T cells may be due to a combination of a genetically and/or environmentally influenced propensity to produce particular T cell phenotypes in individual patients. This is in line with the suggestion that insufficient regulation of the adaptive response contributes to symptomatic dengue instead of asymptomatic infection.8
-
(2)
Cytotoxic CD4 T cells were associated with greater disease severity. Cytotoxic/activated CD4 T cells were maintained over the 1-year observation period and, if they preexisted dengue infection, could have predisposed the patient toward severe disease. We also found that granzyme B+ CD4 T cells were linked to MBCs via the Abs they produced and to Ki67+ plasmablasts (Table S7). Cytotoxic CD4 T cells appeared to have a negative impact on the establishment of a protective, virus particle-specific MBC response, while supporting plasmablast formation.
Tfhs are the main CD4 T cells supporting B cell maturation, including Ab affinity maturation. We hypothesize that the environment conferred by cytotoxic CD4 T cells affected the ability of Tfhs to support protective B cell responses, possibly by contributing to the formation of Th1-like cTfhs via unknown mechanisms.
-
(3)
Severe clinical outcome was associated with higher neutralizing titers at 6 months. This is counter-intuitive, as neutralizing Abs are associated with protection.5,49,50 There are two elements that could have contributed to this observation: (1) more cases with WSs were secondary cases, which are expected to have higher titers due to the reactivation of MBCs. (2) The time point of the NT50 measurement is crucial. When detected 6 months after fever, the plasma can still contain a large amount of mostly cross-reactive, plasmablast-derived Abs (the average t½ of plasma IgG is 21 days), which may still be protective. However, percentages of plasmablasts during acute disease did not correlate with neutralizing titers at 6 months, possibly due to non-specifically activated B cells that can contribute to the plasmablast pool.11
-
(4)
Higher acute or post-febrile dengue-specific T cell activation correlated with a higher abundance of T cells with the effector phenotype 6 months and 1 year after fever onset. In particular, there was a significant correlation between acute phase percentages of GPR56+ tetramer+ CD8 T cells with total EM CD57+ CD8 T cells at the 1-year time point (Figure 3F). Separately, the percentage of CXCR5+tetramer+ CD8 T cells at 1 year were highly significantly correlated with total central memory (CM) CD4 T cells and with total Th17 CD4 T cells during acute disease (Figures 3D and 3E). GPR56 is an adhesion molecule expressed on a subset of CD4+ terminally differentiated effector memory (TEMRA) cells with higher clonality and some enrichment of virus-specific, IFN-γ+ cells, when compared to GPR56− TEMRA cells.35 In the context of both CD4 and CD8 T cells, GPR56 is considered a cytotoxicity marker.51 KLRG1 expression has been associated with short-lived effector CD8 T cells during human CMV infection.52 Since it is unlikely that dengue-specific cells affect the phenotype of total T cells, it seems more likely that the phenotypes of total T cells observed during acute phase or at 1 year had an impact on the phenotype and function of dengue-specific CD8 T cells.
-
(5)
AtM were expanded early after dengue infection and could be associated with a biased Tfh profile, based on the correlation between these two cell types (Figure 5E). Due to the important role of Tfh in shaping the B cell response, we analyzed CXCR3+ and CXCR3− circulating Tfh. In the context of HIV, PD1+CXCR3− Tfh cells were correlated with broadly neutralizing Ab (bnAbs) formation in HIV-infected individuals Tfh cells.48 In our cohort, activated, Ki67+ CXCR3+ Th1-like Tfh correlated positively with AtM B cells (Spearman ρ = 0.892, adjusted p = 0.163; Figure 5E), while Ki67+ CXCR3− Tfh did not (Spearman ρ = 0.643, adjusted p = 0.53). There was no correlation with neutralizing Ab titers.
-
(6)
We observed a strong correlation of CXCR3+ dengue-specific CD8 T cells with the MBC response at the post-febrile stage (Figure 5B). It is possible that memory CD8 T cell activation plays a supportive role during MBC generation since the total number of MBCs tended to be higher for secondary cases at the post-febrile stage (Figure 5A). The correlation of MBCs with lasting neutralizing titers (Figure 5C) suggested that MBCs may also be recruited to become Ab-secreting cells at convalescence, contributing to the plasma Ab titer.
Beyond the insights to the impact of CD4 and CD8 T cells on B cell memory, our study suggests that certain T cell phenotypes in individuals may preexist and could explain patient-specific outcomes independent of viral load and serotype of infection. The importance of baseline immune phenotype affecting immune responses has been demonstrated in the context of influenza vaccination (transcriptional profile).53 This phenomenon appears to apply similarly to dengue infection, whereby preexisting phenotypes, together with the status of infection, could affect disease outcomes. Individual bias to generate certain immune phenotypes, such as CD38+CD57+ CD8 T cells, EM CD4 T cells, and CD38+CXCR5+ B cells, which correlated with hospitalization status, could provide prognostic disease biomarkers that will, however, need to be assessed and validated in independent cohorts.
Limitations of study
In deeply profiling the characteristics of both the T and B cell-mediated immune responses at up to several time points across this cohort of 68 acute dengue infection patients, we believe that the large dataset provided here will be valuable as a resource for the generation of new hypotheses related to the immunological basis for dengue. We have highlighted a number of associations that we found to be most striking and potentially useful as biomarkers of disease severity. Future prospective studies will be needed to validate these findings since the sample size of this study does not permit us to fully dissociate each of the clinical parameters. For instance, we compared profiles of hospitalized versus outpatients to look for correlations with disease severity. However, it is also possible that this difference in setting and early hospital care received would have an effect on the immunological parameters we have measured. Furthermore, the overrepresentation of patients with secondary infection among those hospitalized did not allow us to independently assess the effect of infection status on immune cell phenotypes. Based on cellular profiles from late time points, we have also hypothesized that baseline cellular profiles could be predictive of disease severity. This would need to be tested by analyzing blood samples taken before infection, which were not available for this cohort. Lastly, we imagine that future parallel analyses of various acute viral infections will allow for the identification of immunological parameters that are specifically associated with severe dengue or with systemic viral infections in general.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human-IgD PECy7 (IA6-2) | Biolegend | 348210 |
| Anti-human-IgM APCCy7 (MHM-88) | Biolegend | 314520 |
| Anti-human-CD20 BV711 (2H7) | BD Biosciences | 563126 |
| Anti-human-CD27 PE (M-T271) | BD Biosciences | 555441 |
| Anti-human-CD38 APC (HIT-2) | BD Biosciences | 555462 |
| Anti-human-CD3 V450 (UCHT1) | BD Biosciences | 560365 |
| Anti-human-CD16 V500 (3G8) | BD Biosciences | 561394 |
| Anti-human-CD14 ECD (RMO52) | Beckman Coulter | IM2707U |
| Anti-human-IgA Viobright (IS11-8E10) | Miltenyi Biotec | 130-104-726 |
| Anti-HLA-DR eFluor605NC (LN3) | ThermoFisher Scientific | 47-9956-42 |
| Anti-human-CD19 BV650 (HIB19) | Biolegend | 302237 |
| Anti-human-IgG purified (G18-145) | BD PharMingen | 555784 |
| Anti-human-Ig purified (polyclonal) | Thermofisher Scientific | H1700 |
| Anti-human-IgG AF488 (polyclonal) | Invitrogen | A11013 |
| Anti-human-IgA purified (polyclonal) | Jackson Immunoresearch | 109-005-011 |
| Anti-human-IgM Pacific Blue (MHM-88) | Biolegend | 314513 |
| Anti-human-CD1c PerCPCy5.5 (L161) | Biolegend | 331514 |
| Monoclonal Anti-Flavivirus Group Antigen (D1-4G2-4-15) | ATCC | VR-1852 |
| Anti-human-CD45 89Y (HI30) | DVS Sciences | 3089003B |
| Anti-human-CD14 Qdot800 (TuK4) | Molecular Probes (Invitrogen) | Q10064 |
| Anti-human-CD57 (HCD57) | Biolegend | 322302 |
| Anti-human-CD19 purified (HIB19) | Biolegend | 302202 |
| Anti-human-HLA-DR purified (L243) | Biolegend | 307602 |
| Anti-human-CLA purified (HECA-452) | Biolegend | 321302 |
| Anti-human-CD27 purified (LG.7F9) | eBioscience (ThermoFisher) | 14-0271-85 |
| Anti-human-Granzyme B purified (CLB-GB11) | Abcam | ab103159 |
| Anti-human-CD45RA purified (HI100) | BD Biosciences | 555486 |
| Anti-human-CD8α purified (SK1) | Biolegend | 344702 |
| Anti-human-CD45RO purified (UCHL1) | Biolegend | 304202 |
| Anti-human-CD4 purified (SK3) | Biolegend | 344602 |
| Anti-human-CD161 purified (HP-3G10) | Biolegend | 339902 |
| Anti-human-CD3 purified (UCHT1) | Biolegend | 300402 |
| Anti-human-Ki67 purified (B56) | BD PharMingen | 550609 |
| Anti-human-CXCR3 purified (1C6/CXCR3) | BD PharMingen | 557183 |
| Anti-human-CD38 purified (HIT2) | Biolegend | 303502 |
| Anti-human-ICOS purified (C398.4A) | Biolegend | 313502 |
| Anti-human-CD69 purified (FN50) | Biolegend | 310902 |
| Anti-human-PD-1 purified (J105) | eBioscience (ThermoFisher) | 14-2799-80 |
| Anti-human-CX3CR1 purified (K0124E1) | Biolegend | 355702 |
| Anti-human-GPR56 purified (CG4) | Biolgend | 358202 |
| Anti-human-CD71 purified (OKT-9) | eBioscience (ThermoFisher) | 14-0719-82 |
| Anti-human-KLRG1 purified (13F12F2) | eBioscience (ThermoFisher) | 16-9488-85 |
| Anti-human-CD39 purified (A1) | Biolegend | 328202 |
| Anti-human-CD25 purified (M-A251) | BD PharMingen | 555430 |
| Anti-human-CXCR5 purified (RF8B2) | BD PharMingen | 552032 |
| Anti-human-CD127 purified (A019D5) | Biolegend | 351302 |
| Anti-human-CD16 209Bi (3G8) | DVS Sciences | 3209002B |
| Anti-human-CD56 purified (REA196) | Miltenyi Biotec | 130-108-016 |
| Anti-human-CCR4 purified (205410) | R&D Systems | MAB1567 |
| Anti-human-TCRγδ PE (5A6.E9) | Life Technologies (ThermoFisher) | MHGD04 |
| Anti-PE purifed (PE001) | Biolegend | 408102 |
| Anti-human-T-BET purified (4B10) | eBioscience (ThermoFisher) | 14-5825-82 |
| Anti-human-CRTH2 APC (BM16) | Biolegend | 350110 |
| Anti-APC (APC003) | Biolegend | 408002 |
| Anti-human-FOXP3 162Dy (259D/C7) | Fluidigm | 3162024A |
| Anti-human-EOMES (WD1928) | eBioscience (ThermoFisher) | 14-4877-82 |
| Anti-human-RORγt 168Er (600214) | Fluidigm | 3168018B |
| Anti-human-Helios purified (22F6) | Biolegend | 137202 |
| Anti-human-BCL-2 purified (100) | Biolegend | 658702 |
| Anti-human-TCR Vδ2 purified (B6) | Biolegend | 331402 |
| Anti-human-TCR Vδ1 FITC (REA173) | Miltenyi Biotec | 130-122-285 |
| Anti-FITC purified (FIT-22) | Biolegend | 408302 |
| Anti-human-TCR Vα7.2 (3C10) | Biolegend | 351702 |
| Bacterial and virus strains | ||
| DENV-1 strain D1/SG/05K2916DK1/2005 | Environmental Health Institute EHI, Singapore | Genbank: EU081234.1 |
| DENV-2 strain DENV-2-TSV01 | Novartis Institute of Tropical Diseases, Singapore | Genbank: AY037116.1 |
| DENV-1 strain DENV-1-WestPac74 | Novartis Institute of Tropical Diseases, Singapore | Genbank: U88535.1 |
| Chemicals, peptides, and recombinant proteins | ||
| LIVE/DEAD stain | ThermoFisher Scientific | L34957 |
| TetraMethylBenzidine (TMB) | Sigma Aldrich (Merck) | 54827-17-7 |
| Resiquimod (R848) | InVivogen | tltl-r848 |
| IL-2 | Prospecbio | CYT-209 |
| KLH | Sigma Aldrich (Merck) | 9013-72-3 |
| Critical commercial assays | ||
| SD. BIOLINE Dengue Duo kit for NS1 detection | Abbott | 11FK46 |
| PanBio Dengue IgG Indirect ELISA | Abbott | 01PE30 |
| Experimental models: Cell lines | ||
| Vero cells | ATCC | ATCC® CCL-81 |
| Oligonucleotides | ||
| GAACATGGRACAAYTGCAACYAT | Integrated DNA Technologies | DENV-1 D1MGBEn469s-forward |
| CCGTAGTCDGTCAGCTGTATTTCA | Integrated DNA Technologies | DENV-1 D1MGBEn536r-reverse |
| ACACCACAGAGTTCCATCACAGA | Integrated DNA Technologies | DENV-2 Den 2.2F |
| CATCTCATTGAAGTCNAGGCC | Integrated DNA Technologies | DENV-2 Den 2.2R |
| ATGAGATGYGTGGGAGTRGGAAAC | Integrated DNA Technologies | DENV-3 D3MGBEn1sFWD |
| CAC CAC DTC AAC CCA CGT AGC T | Integrated DNA Technologies | DENV-3 D3MGBEn71rREV |
| GGTGACRTTYAARGTHCCTCAT | Integrated DNA Technologies | DENV-4 D4TEn711sFWD |
| WGARTGCATRGCTCCYTCCTG | Integrated DNA Technologies | DENV-4 D4TEn786cREV |
| GARAGACCAGAGATCCTGCTGTCT | Integrated DNA Technologies | DENV-1 to 4 DV1-4_realt_fwd |
| ACCATTCCATTTTCTGGCGTT | Integrated DNA Technologies | DENV-1 to 4 DV1-4_realt_rev |
| Software and algorithms | ||
| Boruta package | CRAN | N/A |
| Gephi package | N/A | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data generated from the study are included in the manuscript or in the supplemental information in the form of figures and tables. Data analysis methods and packages used are described in the section Quantification and statistical analysis.
Experimental model and subject details
Patient cohort details and classification of severity
All patients were recruited at Tan Tock Seng Hospital (TTSH) in Singapore following IRB approval from the National Healthcare Group Domain Specific Review Board (NHG DSRB Ref: 2015/0528 and NHG DSRB Ref: 2016/00982). Patients presenting symptoms of dengue and with fever of less than 6 days in duration were tested for dengue infection using the SD. BIOLINE Dengue Duo kit (Abbott, previously Alere). NS1-positive patients were then enrolled into the study after obtaining their consent. In total, 68 subjects were enrolled into the study and collection of blood was planned at 5 time points: less than 6 days after fever onset (Visit 1), 6-10 days (acute, Visit 2), 14-28 days (post-febrile, Visit 3), 6 months (Visit 4), and 1 year (Visit 5). Table 1 summarizes the number of subjects that were initially enrolled and those that returned for the subsequent time-points according to their status and serotype of infection. Severity of disease was judged based on hospitalization status. In parallel, patients were assessed according to the 2009 WHO guidelines,54 which introduced the use of warning signs to facilitate identification of patients with diagnosis “dengue fever” at risk of developing severe dengue: Abdominal pain or tenderness, persistent vomiting, clinical fluid accumulation, mucosal bleed, lethargy, restlessness, liver enlargement > 2cm, increase in HTC concurrent with rapid decresase in platelet count. Patients with HIV, HBV and HCV infection were excluded from the study. Patient LNA047 had a Zika-coinfection based on PCR, whereas no Zika-specific IgM and IgG titers could be detected at visit 1. None of the patients were positiv for Zika IgM at visit 1 (Anti-Zika Virus ELSIA, EUROIMMUNE).
Sample processing
Blood samples were collected in EDTA vacutainer tubes. PBMCs were isolated from peripheral blood by Ficoll gradient purification and cryopreserved in 90% fetal calf serum (FCS) + 10% DMSO. Plasma obtained from the blood drawn at the first visit was used to extract viral RNA to confirm dengue virus infection status and serotype by PCR. Additionally, PanBio Dengue IgG kit was used to assess whether the dengue virus infection is a primary or secondary infection.
Method details
Confirmation of DENV infection and serotype by PCR
RNA was isolated from 70 μL plasma using High Pure Viral Nucleic Acid Kit (Roche) following the manufacturer’s protocol, except that additional 100 μL binding buffer was not added after the first 10min incubation and only one washing step was performed. RT-PCR was performed using the TaqMan RNA-to-CTTM 1-step kit, Taqman FAM probes (Applied Biosystems), and primers at 1uM. Sequences of the primers (1st BASE, Axil Scientific) are listed in the Key resources table. Programme was run with Applied BioSystems 7500 machine as follow: 30mins at 48°C followed by 10mins at 95°C and 40 cycles 15secs at 95°C + 1min at 57°C.
Determination of status of infection with PanBio Dengue IgG kit
PanBio Dengue IgG kit was purchased from Abbot (previously Alere) and used according to the manufacturer’s protocol. This kit classifies patients as primary or secondary-infected by dengue based on the capture and quantification of dengue-specific IgG by ELISA. Patients that were too close to the detection limit were classified “unclear” first and classified later according to the longitudinal profile (sharp increase after visit 2 titer indicates secondary infection) based on in house dengue-specific ELISA and microneutralization assays.
Mass cytometry
All antibody and tetramer cocktails were prepared on the day of cell staining. Antibodies used for pre-staining (Table S3) were combined in a 30 kDa filter (Amicon Ultra, Millipore), washed and centrifuged with PBS twice to remove azide content. Before staining, all tetramer and antibody cocktails were filtered using a 0.1 μM filter (Ultrafree, Millipore) to remove aggregates. Cryopreserved cells were thawed and washed twice with complete RPMI media (10% FCS, 1% penicillin/streptomycin/L-glutamine) (GIBCO, Invitrogen). 80% of the cells were split evenly into two wells for Panel 1 staining with two different tetramer configurations and the remaining 20% of cells were used for staining with Panel 2. Cells were incubated with 50 nM dasatinib and pre-stain antibodies in complete RPMI for 30 min at 37°C. After pre-staining, cells were washed in cold CyFACS buffer (PBS with 4% FCS, 2 mM EDTA, and 0.05% Azide) and incubated on ice with 200 nM Cisplatin (Sigma) for 5 min. Cells were then washed twice with CyFACs and incubated for 1 hour at room temperature with tetramer cocktail. Subsequently, cells were washed and stained with primary surface antibodies and/or EasySep Human T cell Isolation Cocktail (STEMCELL) for 15 min at 4°C. Cells were washed and stained with secondary surface antibodies and/or EasySep Dextran RapidSpheres (STEMCELL) for 15 min at 4°C.
For intracellular and intranuclear antibody staining, cells were washed and fixed with Foxp3/Transcription Factor Fix/Perm buffer (eBiosciences) for 30 min on ice. Cells were then washed with 1X Permeabilization buffer (eBiosciences) and stained with primary intracellular antibodies for 30 min on ice, washed twice, followed by incubation with secondary intracellular antibodies for 30 min on ice. Finally, cells were washed with 1X Permeabilization buffer and fixed in 2% PFA (paraformaldehyde, Electron Microscopy Science) at 4°C overnight.
On the next day, PFA was removed and cells were incubated with combinations of two metal barcodes for 30 min on ice as previously described.55 Cells were then washed first with perm buffer, followed by CyFACS. Cellular DNA was labeled at room temperature with 250 nM iridium interchelator (Fluidigm) diluted in 2% PFA for 20 min. Subsequently, cells were washed with CyFACS, combined and enriched for CD3+ T cells using a magnet (EasySep). Immediately before mass cytometry acquisition, cells were washed twice with MilliQ water and passed into a filter top FACS tube. 1.5% of Four EQ beads (Fluidigm DVS) were mixed with the cell suspension. Samples were then acquired on a Helios instrument.
Flow cytometry
Analysis of plasmablasts at visit 2 and 3 was with freshly isolated PBMCs. Cell viability was evaluated using LIVE/DEAD® (ThermoFisher Scientific) and the following antibodies were used: CD19-BV650 (HIB19), CD1c-PerCPCy5.5 (L161), IgD-PECy7 (IA6-2) and IgM-APCCy7 (MHM-88) from Biolegend, CD20-BV711 (2H7), CD27-PE (M-T271), CD38-APC (HIT-2), CD3-V450 (UCHT1), CD14-ECD (RMO52), CD16-V500 (3G8) from BD Biosciences, IgA-VioBright-FITC (IS11-8E10) from Miltenyi Biotec, and HLA-DR- eFluor605NC (LN3) from eBiosciences (ThermoFisher Scientific). Samples were processed on a LSRII cytometer using FACSDiva software (BD Biosciences) and further analyzed using FlowJo_v10 software (Tree Star).
DENV-specific IgG antibodies detection by ELISA
Recombinant monomeric E protein from DENV-1 or DENV-2 (E80 provided by Merck) and UV-inactivated virus from DENV-1 (strain DENV-1-D1/SG/05K2916DK1/2005 (EU081234.1)) or DENV-2 (strain DENV-2-TSV01 (AY037116.1)) were used as antigens to coat 96-well half-area plates (675061, Greiner bio-one) overnight at 4°C. Virus was concentrated by PEG precipitation and UV-inactivated for 10min using a germicidal UV light source (UV lamp VL-206.G 2 × 6 X – 254 nm tube, Vilber Lourmat) . Plates were washed and saturated with PBS 1X + 0.05% Tween-20 + 3% skim milk and incubated either with serially diluted plasma (1/500, 1/2500, 1/7500, 1/22500, 1/67500, 1/337500) or supernatant from B cell culture (volume added normalized to total IgG: 4ug/ml, 2ug/ml, 1ug/ml, 0.5ug/ml or 0.25ug/ml) for 1.5hrs at room temperature. Plates were then washed and an anti-human-IgG-HRP was added for 1h at room temperature. Finally, 3,3′,5,5′ TetraMethylBenzidine (TMB) substrate was added into the plates for 5 min and the reaction was stopped by adding hydrochloric acid (HCl). Optical density (OD) was read within 30 min at 450nm (reference 570nm) using Infinite M200 Elisa reader (TECAN).
DENV-specific IgG antibodies microneutralization assay
Plasma containing antibodies were serially diluted 10-fold over 8 wells of a 96 well plate, starting from 1/10 dilution in serum free RPMI medium. An equal volume of dengue virus DENV-1 (DENV-1-WestPac74(U88535.1) or DENV-2 (TSV01) was added in each well and incubated for 1hr at 37°C - 5% CO2. 100 μL of the antibody-virus mixture was then added to Vero cells, seeded at a density of 20,000 cells per well the day before, and incubated for 1hr at 37°C - 5% CO2. 100 μL of 10% FCS RPMI medium with 1% Penicillin/Streptomycin was then added in each well and the plate was incubated for 4 days at 37°C - 5% CO2. On day 4, cells were fixed with 3.7% formalin, permeabilized with 0.1% Triton X-100, and blocked by adding 100 μL of 10% FCS RPMI medium. Mouse anti-DENV antibody (4G2) was incubated at 1 μg/ml in each well for 2hrs at 37°C. Cells were washed before adding secondary antibody goat-anti-mouse-HRP antibody for 1hr at 37°C. Finally, TMB substrate was added into the plates for 5 min and the reaction was stopped by adding HCl. OD was read at 450nm using Infinite M200 Elisa reader (TECAN).
B cell fluorospot assay
PBMCs were thawed and cultured in RPMI medium supplemented with 10% FCS, 0.5 μg/mL of TLR7/8 ligand (R848, InVivoGen) and 6ng/ml recombinant human IL-2 (Prospecbio). Cells were cultured at 1 × 106 cells/mL in 6-wells plate. After 6 days, cells were harvested, and the proportion and phenotype of B cells were analyzed using flow cytometry before being used for the B cell Fluorospot assay.
ELISPOT plates (Millipore MSIPN4550) were pre-wet with 35% ethanol (1 min), washed with PBS and coated overnight at 4°C with 10 μg/mL polyclonal anti-Ig antibodies (H1700, ThermoFisher) or E protein from DENV-1 or DENV-2 (E80 provided by Merck) diluted in PBS. Keyhole limpet hemocyanin coating (KLH, 10 μg/mL, Sigma-Aldrich) was used as negative control. Plates were washed with PBS and saturated with RPMI containing 10% FCS. Six days post activation, 1,500 to 3,000 cells/well were plated for total IgG, IgA and IgM detection. 1.5 × 105 to 3 × 105 cells/well were plated for DENV-E-protein-specific detection. Cells were incubated overnight at 37°C in RPMI+10% FCS. Plates were then washed with PBS+0.05% Tween-20, followed by incubation with polyclonal AF488-anti-IgG (1ug/ml, Invitrogen #A11013), polyclonal AF488-anti-IgA (3ug/ml, Goat anti-human IgA from Jackson Immunoresearch #109-005-011 labeled with Alexa Fluor 488 Antibody Labeling Kit from ThermoFisher #A20181) or PB-anti-IgM (2ug/ml, Biolegend, clone MHM-88, #314513) for 1h at 37°C.
An additional method was used in parallel to detect DENV-E-protein-specific IgG-secreting B cells. Plates were coated with purified anti-IgG (10ug/ml, BD PharMingen, clone G18-145 #555784), incubate the cells (1.5 × 105 to 3 × 105 cells/well), washed and 10ug/ml of E-protein was added. Secondary antibody mouse-anti-flavivirus antibody (4G2) coupled to AF488 was then added at 2ug/ml for the detection. Plates were finally washed with PBS+0.05% Tween, followed by water and dried before reading with ImmunoSpot Elispot reader (Cellular Technology Limited, CTL).
Quantification and statistical analysis
Data analysis
Bivariate correlation analysis and network was built on the entire dataset (Data S1). We used the Gephi package to plot the network.56 Betweenness centrality was used as a feature to represent the nodes in the network. A betweenness centrality of a node defines the degree to which a given node is connected to other nodes. It is calculated by counting the fraction of shortest paths going through the node. Nodes with a higher betweenness centrality (larger in size) interact with many other nodes and represent key nodes in the network, having high control over other nodes communicating via them.
For linear discriminant analysis (LDA) or Boruta feature selection, to limit the impact of missing values and time points, only samples with dengue tetramer+ T cell data at visits 2 or 3 were included for “all visit” analysis (n = 29). For just analysis on visit 2, only samples with dengue tetramer+ T cell data at visit 2 were used (n = 24). For the final hospitalization status, all “outpatient/inpatient” statuses were changed to Inpatient. Columns with more than 30% of values missing were removed. The R package missMDA was used to impute any remaining missing values using the regularized iterative PCA algorithm.57
Using the curated dataset(s), all-relevant feature selection was performed using the Boruta package.31 Linear Discriminant Analysis was performed using the MASS package32 on a dataset consisting of only the features selected by Boruta. The results of LDA and feature selection were compared with the unadjusted Kruskal-Wallis test performed on the overall dataset. ∗p < 0.05.
Figures showing cell frequencies use the original dataset (Data S1). p values are adjusted for multiple hypothesis testing using the Benjamini-Hochberg method. Time course data was analyzed by fitting a mixed model with Geisser-Greenhouse correction, as implemented in GraphPad Prism 8.0. This mixed model uses a compound symmetry covariance matrix, and is fit using Restricted Maximum Likelihood (REML), and takes into account non-independent patient-specific variability over time.
Acknowledgments
The work was supported by an MRL/Singapore project grant (062015). K.F. and E.W.N. were supported by SIgN core funding. B.L. is part of the SIgN immunomonitoring platform (supported by a BMRC IAF 311006 grant, BMRC transition funds #H16/99/b0/011, a BMRC IAF-PP H19/01/a0/024 SIGNAL grant, and an NRF SIS NRF2017_SISFP09 grant).
Author contributions
A.R., M.H.Y.C., K.K., Y.X.T., D.S., and T.L. conducted the experiments and analyzed the data. B.L., M.P.R., and L.T.-K. analyzed the data. Y.-S.L. provided the patient samples and clinical data and determined the patient classification. A.S., T.W.Y., V.W.X.L., T.-L.T., Y.-S.L., K.A.V., D.C., and B.L. provided material and intellectual input and facilitated the study. L.R., E.W.N., and K.F. conceived the study and designed the experiments. A.R., M.H.Y.C., E.W.N., and K.F. wrote the manuscript.
Declaration of interests
E.N. is a founder and scientific advisor for Immunoscape. K.A.V., D.C., and B.L. are or were employees of Merck at the time of the study and may have received stocks as a part of their annual compensation.
Published: May 18, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100278.
Supplemental information
References
- 1.Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., Drake J.M., Brownstein J.S., Hoen A.G., Sankoh O. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett S.N., Drummond A.J., Kapan D.D., Suchard M.A., Muñoz-Jordán J.L., Pybus O.G., Holmes E.C., Gubler D.J. Epidemic dynamics revealed in dengue evolution. Mol. Biol. Evol. 2010;27:811–818. doi: 10.1093/molbev/msp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajarethinam J., Ang L.W., Ong J., Ycasas J., Hapuarachchi H.C., Yap G., Chong C.S., Lai Y.L., Cutter J., Ho D. Dengue in Singapore from 2004 to 2016: cyclical epidemic patterns dominated by serotypes 1 and 2. Am. J. Trop. Med. Hyg. 2018;99:204–210. doi: 10.4269/ajtmh.17-0819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ooi E.E., Goh K.T., Gubler D.J. Dengue prevention and 35 years of vector control in Singapore. Emerg. Infect. Dis. 2006;12:887–893. doi: 10.3201/10.3201/eid1206.051210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katzelnick L.C., Montoya M., Gresh L., Balmaseda A., Harris E. Neutralizing antibody titers against dengue virus correlate with protection from symptomatic infection in a longitudinal cohort. Proc. Natl. Acad. Sci. USA. 2016;113:728–733. doi: 10.1073/pnas.1522136113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson K.B., Gibbons R.V., Cummings D.A., Nisalak A., Green S., Libraty D.H., Jarman R.G., Srikiatkhachorn A., Mammen M.P., Darunee B. A shorter time interval between first and second dengue infections is associated with protection from clinical illness in a school-based cohort in Thailand. J. Infect. Dis. 2014;209:360–368. doi: 10.1093/infdis/jit436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montoya M., Gresh L., Mercado J.C., Williams K.L., Vargas M.J., Gutierrez G., Kuan G., Gordon A., Balmaseda A., Harris E. Symptomatic versus inapparent outcome in repeat dengue virus infections is influenced by the time interval between infections and study year. PLoS Negl. Trop. Dis. 2013;7:e2357. doi: 10.1371/journal.pntd.0002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simon-Lorière E., Duong V., Tawfik A., Ung S., Ly S., Casadémont I., Prot M., Courtejoie N., Bleakley K., Buchy P. Increased adaptive immune responses and proper feedback regulation protect against clinical dengue. Sci. Transl. Med. 2017;9:eaal5088. doi: 10.1126/scitranslmed.aal5088. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Bates T.M., Cordeiro M.T., Nascimento E.J.M., Smith A.P., Soares de Melo K.M., McBurney S.P., Evans J.D., Marques E.T., Jr., Barratt-Boyes S.M. Association between magnitude of the virus-specific plasmablast response and disease severity in dengue patients. J. Immunol. 2013;190:80–87. doi: 10.4049/jimmunol.1103350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wrammert J., Onlamoon N., Akondy R.S., Perng G.C., Polsrila K., Chandele A., Kwissa M., Pulendran B., Wilson P.C., Wittawatmongkol O. Rapid and massive virus-specific plasmablast responses during acute dengue virus infection in humans. J. Virol. 2012;86:2911–2918. doi: 10.1128/JVI.06075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Appanna R., Kg S., Xu M.H., Toh Y.X., Velumani S., Carbajo D., Lee C.Y., Zuest R., Balakrishnan T., Xu W. Plasmablasts During Acute Dengue Infection Represent a Small Subset of a Broader Virus-specific Memory B Cell Pool. EBioMedicine. 2016;12:178–188. doi: 10.1016/j.ebiom.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Priyamvada L., Cho A., Onlamoon N., Zheng N.-Y., Huang M., Kovalenkov Y., Chokephaibulkit K., Angkasekwinai N., Pattanapanyasat K., Ahmed R. B Cell Responses during Secondary Dengue Virus Infection Are Dominated by Highly Cross-Reactive, Memory-Derived Plasmablasts. J. Virol. 2016;90:5574–5585. doi: 10.1128/JVI.03203-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toh Y.X., Gan V., Balakrishnan T., Zuest R., Poidinger M., Wilson S., Appanna R., Thein T.L., Ong A.K., Ng L.C. Dengue serotype cross-reactive, anti-e protein antibodies confound specific immune memory for 1 year after infection. Front. Immunol. 2014;5:388. doi: 10.3389/fimmu.2014.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.St John A.L., Rathore A.P.S. Adaptive immune responses to primary and secondary dengue virus infections. Nat. Rev. Immunol. 2019;19:218–230. doi: 10.1038/s41577-019-0123-x. [DOI] [PubMed] [Google Scholar]
- 15.Dejnirattisai W., Supasa P., Wongwiwat W., Rouvinski A., Barba-Spaeth G., Duangchinda T., Sakuntabhai A., Cao-Lormeau V.M., Malasit P., Rey F.A. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat. Immunol. 2016;17:1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith S.A., Zhou Y., Olivarez N.P., Broadwater A.H., de Silva A.M., Crowe J.E.J., Jr. Persistence of circulating memory B cell clones with potential for dengue virus disease enhancement for decades following infection. J. Virol. 2012;86:2665–2675. doi: 10.1128/JVI.06335-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fibriansah G., Lok S.M. The development of therapeutic antibodies against dengue virus. Antiviral Res. 2016;128:7–19. doi: 10.1016/j.antiviral.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Mongkolsapaya J., Dejnirattisai W., Xu X.N., Vasanawathana S., Tangthawornchaikul N., Chairunsri A., Sawasdivorn S., Duangchinda T., Dong T., Rowland-Jones S. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat. Med. 2003;9:921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- 19.Duangchinda T., Dejnirattisai W., Vasanawathana S., Limpitikul W., Tangthawornchaikul N., Malasit P., Mongkolsapaya J., Screaton G. Immunodominant T-cell responses to dengue virus NS3 are associated with DHF. Proc. Natl. Acad. Sci. USA. 2010;107:16922–16927. doi: 10.1073/pnas.1010867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yauch L.E., Zellweger R.M., Kotturi M.F., Qutubuddin A., Sidney J., Peters B., Prestwood T.R., Sette A., Shresta S. A protective role for dengue virus-specific CD8+ T cells. J. Immunol. 2009;182:4865–4873. doi: 10.4049/jimmunol.0801974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiskopf D., Angelo M.A., de Azeredo E.L., Sidney J., Greenbaum J.A., Fernando A.N., Broadwater A., Kolla R.V., De Silva A.D., de Silva A.M. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. USA. 2013;110:E2046–E2053. doi: 10.1073/pnas.1305227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elong Ngono A., Chen H.W., Tang W.W., Joo Y., King K., Weiskopf D., Sidney J., Sette A., Shresta S. Protective Role of Cross-Reactive CD8 T Cells Against Dengue Virus Infection. EBioMedicine. 2016;13:284–293. doi: 10.1016/j.ebiom.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiskopf D., Bangs D.J., Sidney J., Kolla R.V., De Silva A.D., de Silva A.M., Crotty S., Peters B., Sette A. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. Proc. Natl. Acad. Sci. USA. 2015;112:E4256–E4263. doi: 10.1073/pnas.1505956112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen T.P., Kikuchi M., Vu T.Q., Do Q.H., Tran T.T., Vo D.T., Ha M.T., Vo V.T., Cao T.P., Tran V.D. Protective and enhancing HLA alleles, HLA-DRB1∗0901 and HLA-A∗24, for severe forms of dengue virus infection, dengue hemorrhagic fever and dengue shock syndrome. PLoS Negl. Trop. Dis. 2008;2:e304. doi: 10.1371/journal.pntd.0000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Culshaw A., Ladell K., Gras S., McLaren J.E., Miners K.L., Farenc C., van den Heuvel H., Gostick E., Dejnirattisai W., Wangteeraprasert A. Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8+ T cell response. Nat. Immunol. 2017;18:1228–1237. doi: 10.1038/ni.3850. [DOI] [PubMed] [Google Scholar]
- 26.Chng M.H.Y., Lim M.Q., Rouers A., Becht E., Lee B., Macary P.A., Lye D.C., Leo Y.S., Chen J., Fink K. Large-Scale HLA Tetramer Tracking of T Cells during Dengue Infection Reveals Broad Acute Activation and Differentiation into Two Memory Cell Fates. Immunity. 2019;51:1119–1135.e5. doi: 10.1016/j.immuni.2019.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Becht E., McInnes L., Healy J., Dutertre C.A., Kwok I.W.H., Ng L.G., Ginhoux F., Newell E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018;37:38–47. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 28.Levine J.H., Simonds E.F., Bendall S.C., Davis K.L., Amir A.D., Tadmor M.D., Litvin O., Fienberg H.G., Jager A., Zunder E.R. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brenchley J.M., Karandikar N.J., Betts M.R., Ambrozak D.R., Hill B.J., Crotty L.E., Casazza J.P., Kuruppu J., Migueles S.A., Connors M. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood. 2003;101:2711–2720. doi: 10.1182/blood-2002-07-2103. [DOI] [PubMed] [Google Scholar]
- 30.Weller S., Braun M.C., Tan B.K., Rosenwald A., Cordier C., Conley M.E., Plebani A., Kumararatne D.S., Bonnet D., Tournilhac O. Human blood IgM “memory” B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire. Blood. 2004;104:3647–3654. doi: 10.1182/blood-2004-01-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kursa M.B., Rudnicki W.R. Feature selection with the boruta package. J. Stat. Softw. 2010;36:1–13. [Google Scholar]
- 32.Venables W., Ripley B. Fourth Edition. Springer; 2002. Modern Applied Statistics with S. [Google Scholar]
- 33.Simoni Y., Chng M.H.Y., Li S., Fehlings M., Newell E.W. Mass cytometry: a powerful tool for dissecting the immune landscape. Curr. Opin. Immunol. 2018;51:187–196. doi: 10.1016/j.coi.2018.03.023. [DOI] [PubMed] [Google Scholar]
- 34.Newell E.W., Sigal N., Nair N., Kidd B.A., Greenberg H.B., Davis M.M. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nat. Biotechnol. 2013;31:623–629. doi: 10.1038/nbt.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian Y., Babor M., Lane J., Schulten V., Patil V.S., Seumois G., Rosales S.L., Fu Z., Picarda G., Burel J. Unique phenotypes and clonal expansions of human CD4 effector memory T cells re-expressing CD45RA. Nat. Commun. 2017;8:1473. doi: 10.1038/s41467-017-01728-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahn E., Araki K., Hashimoto M., Li W., Riley J.L., Cheung J., Sharpe A.H., Freeman G.J., Irving B.A., Ahmed R. Role of PD-1 during effector CD8 T cell differentiation. Proc. Natl. Acad. Sci. USA. 2018;115:4749–4754. doi: 10.1073/pnas.1718217115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan A.R., Hams E., Floudas A., Sparwasser T., Weaver C.T., Fallon P.G. PD-L1hi B cells are critical regulators of humoral immunity. Nat. Commun. 2015;6:5997. doi: 10.1038/ncomms6997. [DOI] [PubMed] [Google Scholar]
- 38.Tosello V., Odunsi K., Souleimanian N.E., Lele S., Shrikant P., Old L.J., Valmori D., Ayyoub M. Differential expression of CCR7 defines two distinct subsets of human memory CD4+CD25+ Tregs. Clin. Immunol. 2008;126:291–302. doi: 10.1016/j.clim.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 39.Chen J., Chen Y.-G., Reifsnyder P.C., Schott W.H., Lee C.-H., Osborne M., Scheuplein F., Haag F., Koch-Nolte F., Serreze D.V., Leiter E.H. Targeted disruption of CD38 accelerates autoimmune diabetes in NOD/Lt mice by enhancing autoimmunity in an ADP-ribosyltransferase 2-dependent fashion. J. Immunol. 2006;176:4590–4599. doi: 10.4049/jimmunol.176.8.4590. [DOI] [PubMed] [Google Scholar]
- 40.Balakrishnan T., Bela-Ong D.B., Toh Y.X., Flamand M., Devi S., Koh M.B., Hibberd M.L., Ooi E.E., Low J.G., Leo Y.S. Dengue virus activates polyreactive, natural IgG B cells after primary and secondary infection. PLoS ONE. 2011;6:e29430. doi: 10.1371/journal.pone.0029430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woda M., Friberg H., Currier J.R., Srikiatkhachorn A., Macareo L.R., Green S., Jarman R.G., Rothman A.L., Mathew A. Dynamics of Dengue Virus (DENV)-Specific B Cells in the Response to DENV Serotype 1 Infections, Using Flow Cytometry With Labeled Virions. J. Infect. Dis. 2016;214:1001–1009. doi: 10.1093/infdis/jiw308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malavige G.N., Huang L.C., Salimi M., Gomes L., Jayaratne S.D., Ogg G.S. Cellular and cytokine correlates of severe dengue infection. PLoS ONE. 2012;7:e50387. doi: 10.1371/journal.pone.0050387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Her Z., Kam Y.W., Gan V.C., Lee B., Thein T.L., Tan J.J.L., Lee L.K., Fink K., Lye D.C., Rénia L., SIgN Immunomonitoring Platform Severity of Plasma Leakage Is Associated With High Levels of Interferon γ-Inducible Protein 10, Hepatocyte Growth Factor, Matrix Metalloproteinase 2 (MMP-2), and MMP-9 During Dengue Virus Infection. J. Infect. Dis. 2017;215:42–51. doi: 10.1093/infdis/jiw494. [DOI] [PubMed] [Google Scholar]
- 44.Portugal S., Obeng-Adjei N., Moir S., Crompton P.D., Pierce S.K. Atypical memory B cells in human chronic infectious diseases: an interim report. Cell. Immunol. 2017;321:18–25. doi: 10.1016/j.cellimm.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ellebedy A.H., Jackson K.J., Kissick H.T., Nakaya H.I., Davis C.W., Roskin K.M., McElroy A.K., Oshansky C.M., Elbein R., Thomas S. Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat. Immunol. 2016;17:1226–1234. doi: 10.1038/ni.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morita R., Schmitt N., Bentebibel S.-E., Ranganathan R., Bourdery L., Zurawski G., Foucat E., Dullaers M., Oh S., Sabzghabaei N. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmitt N., Bentebibel S.E., Ueno H. Phenotype and functions of memory Tfh cells in human blood. Trends Immunol. 2014;35:436–442. doi: 10.1016/j.it.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Locci M., Havenar-Daughton C., Landais E., Wu J., Kroenke M.A., Arlehamn C.L., Su L.F., Cubas R., Davis M.M., Sette A., International AIDS Vaccine Initiative Protocol C Principal Investigators Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39:758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buddhari D., Aldstadt J., Endy T.P., Srikiatkhachorn A., Thaisomboonsuk B., Klungthong C., Nisalak A., Khuntirat B., Jarman R.G., Fernandez S. Dengue virus neutralizing antibody levels associated with protection from infection in thai cluster studies. PLoS Negl. Trop. Dis. 2014;8:e3230. doi: 10.1371/journal.pntd.0003230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corbett K.S., Katzelnick L., Tissera H., Amerasinghe A., de Silva A.D., de Silva A.M. Preexisting neutralizing antibody responses distinguish clinically inapparent and apparent dengue virus infections in a Sri Lankan pediatric cohort. J. Infect. Dis. 2015;211:590–599. doi: 10.1093/infdis/jiu481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng Y.-M., van de Garde M.D.B., Cheng K.-F., Baars P.A., Remmerswaal E.B.M., van Lier R.A.W., Mackay C.R., Lin H.H., Hamann J. Specific expression of GPR56 by human cytotoxic lymphocytes. J. Leukoc. Biol. 2011;90:735–740. doi: 10.1189/jlb.0211092. [DOI] [PubMed] [Google Scholar]
- 52.Remmerswaal E.B.M., Hombrink P., Nota B., Pircher H., Ten Berge I.J.M., van Lier R.A.W., van Aalderen M.C. Expression of IL-7Rα and KLRG1 defines functionally distinct CD8+ T-cell populations in humans. Eur. J. Immunol. 2019;49:694–708. doi: 10.1002/eji.201847897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Avey S., Cheung F., Fermin D., Frelinger J., Gaujoux R., Gottardo R., HIPC-CHI Signatures Project Team. HIPC-I Consortium Multicohort analysis reveals baseline transcriptional predictors of influenza vaccination responses. Sci. Immunol. 2017;2:eaal4656. doi: 10.1126/sciimmunol.aal4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.World Health Organization . 2009. Dengue: guidelines for diagnosis, treatment, prevention and control.https://www.who.int/tdr/publications/documents/dengue-diagnosis.pdf [PubMed] [Google Scholar]
- 55.Wong M.T., Chen J., Narayanan S., Lin W., Anicete R., Kiaang H.T.K., De Lafaille M.A., Poidinger M., Newell E.W. Mapping the Diversity of Follicular Helper T Cells in Human Blood and Tonsils Using High-Dimensional Mass Cytometry Analysis. Cell Rep. 2015;11:1822–1833. doi: 10.1016/j.celrep.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 56.Bastian M., Heymann S., Jacomy M. Proceedings of the 3rd AAAI International Conference on Weblogs and Social Media (ICWSM 2009) Association for the Advancement of Artificial Intelligence; 2009. Gephi: An Open Source Software for Exploring and Manipulating Networks; pp. 361–362. [Google Scholar]
- 57.Josse J., Husson F. missMDA: A package for handling missing values in multivariate data analysis. J. Stat. Softw. 2016;70 doi: 10.18637/jss.v070.i01. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated from the study are included in the manuscript or in the supplemental information in the form of figures and tables. Data analysis methods and packages used are described in the section Quantification and statistical analysis.