Abstract
Objective
Viral nucleic acid detection by real-time reverse transcription polymerase chain reaction (qPCR) is the current standard method for diagnosis of SARS-CoV-2 infection. However, due to low viral load in some COVID-19 patients, false negative results from this method have been repeatedly reported.
Method
In this study, we compared the sensitivity and specificity of digital PCR (dPCR) in simulated samples and clinical samples with qPCR assay through a series of vigorous tests.
Results
The results showed that dPCR was more sensitive than qPCR especially for samples with low viral load (≤3 copies). In addition, dPCR had similar specificity as qPCR and could effectively distinguish other human coronaviruses and influenza virus from SARS-CoV-2. More importantly, dPCR was more sensitive than qPCR in detecting the virus in the “negative” samples from recurrent COVID-19 patients.
Conclusions
In summary, dPCR could serve as a powerful complement to the current qPCR method for SARS-CoV-2 detection, especially for the samples with extremely low viral load, such as recurrent COVID-19 patients.
Keywords: SARS-CoV-2, Digital PCR (dPCR), Sensitivity, Specificity, Recurrent COVID-19 patient
1. Introduction
The ongoing outbreak of 2019 novel coronavirus disease (COVID-19) caused by a new coronavirus, severe acute respiratory syndrome (SARS) coronavirus 2 (SARS-CoV-2), has spread rapidly across the world and became a public health emergency of international concern (Huang et al., 2020; Di Gennaro et al., 2020). Both human-to-human and asymptomatic transmission of SARS-CoV-2 infection through close contacts have been reported (Lai et al., 2020; Rothe et al., 2020). Early diagnosis of COVID-19 is critical for prevention and control of virus infection and can help patients receive early treatment and prevent disease progression (Tahamtan and Ardebili, 2020). To date, viral nucleic acid detection by real-time reverse transcription polymerase chain reaction (qPCR) is regarded as the gold standard method for diagnosis of SARS-CoV-2 infection (Davila et al., 2014; Zhu et al., 2020; Liu et al., 2020). However, the sensitivity and reliability of qPCR has been questioned due to its false negative results in some clinically diagnosed patients who have exposure history, typical clinical symptoms, and typical image of CT scanning (Xiao et al., 2020; Pan et al., 2020). Furthermore, recently, the recurrence of SARS-CoV-2 in discharged COVID-19 patients has also been reported (Xiao et al., 2020; Yuan et al., 2020a). Even though its mechanism is still unclear, the possible explanations might be false negative of detection, and reactivation or re-infection of the virus. Taking into account the fact that recurrence and asymptomatic COVID-19 patients usually have a very low viral load, a more sensitive detection method is urgently needed to improve the accuracy in identifying SARS-CoV-2 infected patients, in order to effectively prevent the virus transmission.
Digital PCR (dPCR) is an absolute quantitative method that partitions the PCR reaction into a large number of smaller reactions and collect the intensity of fluorescence signals at the end point of each reaction (Wang et al., 2016; Basu, 2017). Unlike qPCR, dPCR does not depend on standard curves or relative threshold (CT) values for quantitation (Pavsic et al., 2016) and is more tolerant to PCR inhibitors (Dingle et al., 2013). The absolute quantification results come from Poisson statistics by counting the negative reactions followed by calculating the positive wells or the nucleic acid copies in the sample (Strain et al., 2013). The high precision and the low limit of detection of dPCR could be used for detection of small variation of DNA or RNA copy number (Whale et al., 2012), rare allele mutation (Zhou et al., 2018a) and gene fusion (Zhou et al., 2018b). Recently, several studies reported that dPCR showed higher sensitivity and specificity in detecting low viral load of SARS-CoV-2 compared with qPCR (Vasudevan et al., 2021; Alteri et al., 2020; Falzone et al., 2020). However, the detectability for the samples with extremely low viral load, such as obtained from recurrent COVID-19 patients, remains unclear.
In this study, we compared the sensitivity and specificity of dPCR with qPCR on simulated and clinical sputum samples. To analyze the detection efficacy of the low viral load samples, dPCR quantitative assay of SARS-CoV-2 were explored in clinically recurrent COVID-19 patients. Our findings demonstrated that in comparison with qPCR, dPCR was more sensitive with low viral load of both simulated and clinical SARS-CoV-2 samples and could accurately reflect the variation in copy number of viral RNA. The dPCR also showed high specificity in distinguishing the SARS-CoV-2 from other respiratory pathogens such as common influenza virus and other human coronaviruses. In addition, dPCR shows superiority over qPCR in detection of “recurrent” COVID-19 patients, which could be used as an important complement method for difficult or confusing samples in the clinic.
2. Materials and methods
2.1. Patients and preparation of simulated and clinical samples
We enrolled 21 COVID-19 patients with confirmed SARS-CoV-2 infection including 17 recurrent patients and 4 non-recurrent patients from Jan 21 to Feb 29, 2020 at the First Affiliated Hospital of University of Science and Technology of China (USTC), Huainan First Hospital, Zongyang Hospital, Lixin Hospital, Bozhou Hospital, Fuyang Second Hospital, and Guoyang Hospital in Anhui Province, China. The throat swab, sputum, and/or anal swab were collected. In the meantime, six negative lower respiratory tract samples from 6 healthy donors were also collected and mixed together to serve as a negative matrix for preparation of different simulated virus samples with various LOD. In addition, 13 sputum samples containing influenza virus and other human coronavirus were collected from the corresponding patients to serve as negative controls. Following throat and anal swabs and sputum samples being collected and the simulated samples being prepared, the viral RNAs were extracted using the Nucleic acid purification kit (TIANLONG Technology CO. LTD., Cat. No: T014) according to the manufacturer’s instructions and were subsequently subjected to the detection by qPCR and dPCR. The reference SARS-CoV-2 RNA [GBW(E)091112] was obtained from Shanghai Institute of Measurement and Testing Technology, in which the target gene covers the full length of N gene (GenBank No.MT027064.1) with the RNA concentration of 3.0 107 copies/μL. The normalized RNA concentration was confirmed by dPCR method for authentic SARS-CoV-2.
The study protocol was approved by the medical ethics board of Anhui Center for Disease Control and Prevention (CDC). Please note that the study was performed on existing samples collected during standard diagnostic tests, posing no extra burden to patients.
2.2. Digital PCR (dPCR)
The dPCR assay was performed via the digital PCR chips (Thermofisher, cat#A26317) following the manufacturer’s protocols, and QuantStudio 3D Digital PCR System (Thermo Fisher, Waltham, MA) using the Gnomegen COVID-19 RT-PCR Detection Kit (Gnomegen, San Diego, CA). The kit allowed the detection of the N gene and a positive RNase P reference gene, which was approved for Emergency Use Authorization (EUA) by FDA, pursuant to Section 564 of the Federal Food, Drug, and Cosmetic Act (the Act) (21 U.S.C. §360bbb-3). Briefly, 14.5 μl of reaction mix was prepared for each reaction, comprised of 7.25 μl of digital PCR master mix (Thermofisher, A26358), 0.20 μl Superscript II (Thermofisher, cat# 18064014), 0.20 μl RNaseOUT (Thermofisher, cat# 10777019), 0.725 μl COVID-19 Assay, 2.125 μl Molecular Grade Nuclease Free Water and 4 μl RNA samples. The reaction mixture was then loaded on the chips using the QuantStudio™ 3D Digital PCR Chip Loader (Thermofisher, cat# 4482592), and cycled in ProFlex™ 2×Flat Block Thermal Cycler (Thermofisher, cat#4484078) under the following protocol: step 1, 42 °C for 20 min (reverse transcription); step 2, 96 °C for 10 min (DNA polymerase activation, denaturation) and 60 °C for 2 min (annealing); step 3, 39 cycles of 98 °C for 30 s (denaturation) and 60 °C for 2 min (annealing); step 4, hold at 20 °C. Finally, the chips were imaged by QuantStudio™ 3D Digital PCR Instrument with Power Cord (Thermofisher, cat# 4489084) and analyzed using the QuantStudio 3D Analysis Suite Cloud Software (Current version: 3.1.4-PRC-build1) to measures the concentration of N gene and RNase P gene, respectively.
The result was considered positive when the FAM signals of the SARS-CoV-2 target ≥ 3 with the VIC signals of the RNase P reference target ≥ 0, and negative when the FAM signals ≤ 2 with the VIC signals ≥ 60. If the FAM signals ≤ 2 with the VIC signals <60, the results were considered inconclusive and excluded. Concentrations of the target RNA sequences, along with their Poisson-based 95 % confidence intervals were calculated by using the QuantStudio 3D Analysis Suite Cloud.
2.3. Fluorescent quantitative PCR (qPCR)
qPCR was conducted with primers and probes targeting the ORF1ab (FAM) and N gene (VIC). Reaction system and amplification conditions were carried out according to the manufacturer's specifications (BioGerm, China). A 25μl of reaction mixture contained 12 μl of reaction buffer, 4 μl of mixed enzymes, 4 μl of ORF1ab/N reaction mixture and 5 μl of nucleic acid from each sample. Thermal cycling was performed at 50 °C for 10 min for reverse transcription, followed by 95 °C for 5 min (pre-denaturation) and then 40 cycles of 95 °C for 10 s (denaturation) and 55 °C for 40 s (extension) in Applied Biosystems ABI 7500 real time PCR system (Thermo Fisher, Waltham, MA). The baseline setting was 3–10 or 6–15 cycles of fluorescence signal, and the threshold setting principle was that the threshold line just exceeded the highest point of the amplification curve of the negative control substance, which could also be adjusted according to the noise of the instrument. The result was considered positive when the CT value of ORF1ab or N gene <35, and negative when there was no CT value or the CT value of ORF1ab or N gene ≥ 40. If the CT value of ORF1ab or N gene ≥ 35 and < 40, the result was inconclusive and the sample was retested.
3. Results
3.1. Both dPCR and qPCR could detect low and moderate viral load of simulated SARS-CoV-2 samples
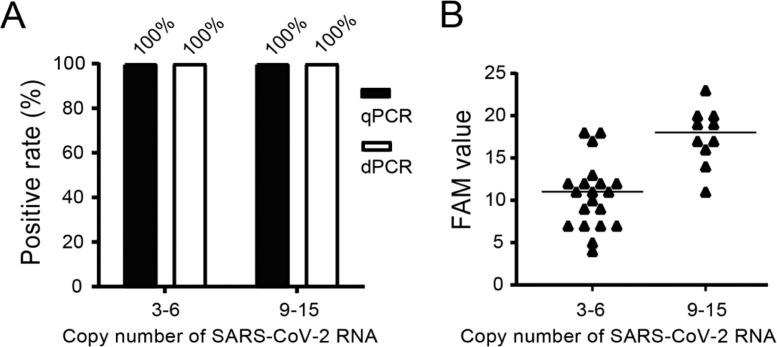
In the present study, we first investigated whether dPCR and qPCR assays could equally effectively detect SARS-CoV-2 virus in low and moderate concentrations. To do so, low and moderate positive simulated samples were prepared by diluting the authenticSARS-CoV-2 into the negative matrix to 3–6 copies per reaction as low viral load samples (20 samples) and 9–15 copies per reaction as moderate viral load samples (10 samples) which were subjected to both dPCR and qPCR assays. As shown in Fig. 1 A, the positive rates of the low and moderate positive samples were 100 % for either dPCR or qPCR assays. When analyzing the values of the low and moderate positive samples detected by dPCR assay, it showed they were largely in the range of the copy number of the virus in the respective samples (Fig. 1B).
Fig. 1.
Comparison of dPCR assay with qPCR assay on low and moderate viral load of simulated SARS-CoV-2 samples. (A) The positive rate of low (20 samples) and moderate (10 samples) viral load samples detected by dPCR and qPCR assays. (B) The FAM values of dPCR assay targeting N gene of SARS-CoV-2 of low and moderate viral load samples.
3.2. dPCR was more sensitive than qPCR in detecting very low viral load of simulated SARS-CoV-2 samples
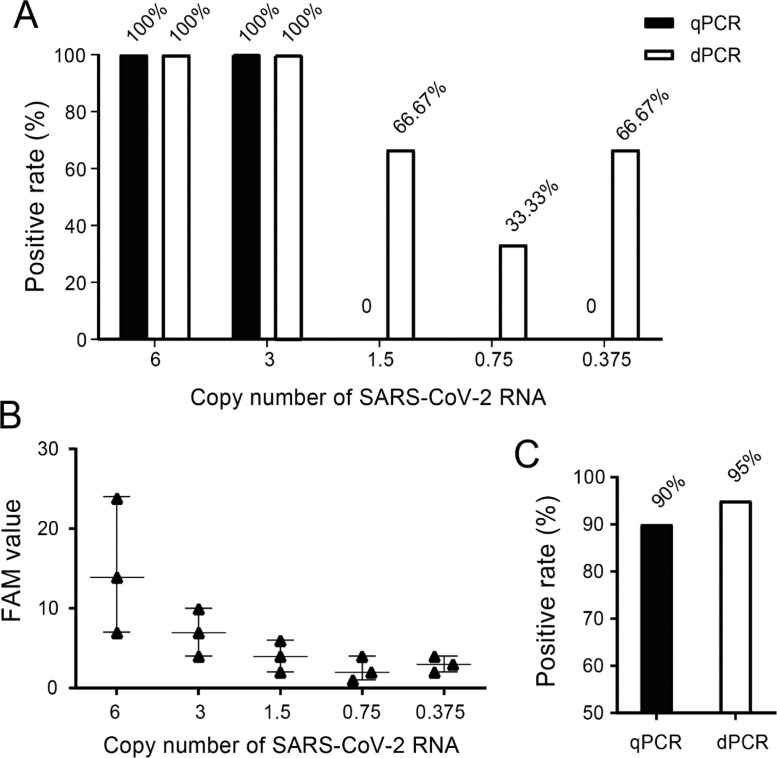
We next compared the dPCR and qPCR for their limit of sensitivity of detection of SARS-CoV-2 by using serial dilution of authentic SARS-CoV-2 spiked into the negative matrix. A two-phase approach was used to determine the limit of detection (LOD). In phase I, the preliminary LOD was established through diluting the authentic SARS-CoV-2 into different concentrations. To do so, a total of 5 dilutions were created at 0.375, 0.75, 1.5, 3 and 6 copies of viral RNA with three replicates per concentration. As shown in Fig. 2 A and B, when the virus was diluted between 3 and 6 copies per reaction, the positive rate was 100 % for both dPCR and qPCR. At concentration of 1.5, 0.75, and0.375 copies per reaction, dPCR was still able to obtain the positive results (66.67 % for 1.5 copies, 33.33 % for 0.75 copies, and 66.7 % for 0.375 copies) while qPCR failed to detect any positive signals at any concentration of the samples, indicating that dPCR is more sensitive in detecting SARS-CoV-2 when the viral copy number is below 3. To further elaborate the sensitivity of the assays, both dPCR and qPCR were used to test 20 replicates on simulated samples of 3 copies of SARS-CoV-2. The dPCR assay showed positive rate at 95 % (19/20) while qPCR at 90 % (18/20) (Fig. 2C), indicating that the dPCR was more sensitive than qPCR for samples with viral load around 3 copies.
Fig. 2.
Comparison of sensitivity between qPCR and dPCR assays in detecting simulated SARS-CoV-2 samples. (A) Positive rate at each concentration of 6.0, 3.0, 1.5, 0.75, 0.375 copies per reaction. (B) dPCR detection of serial dilution of authentic SARS-CoV-2 spiked into negative matrix. Expected copies of viral RNA were plotted on the X axis versus measured FAM values on the Y axis from dPCR assay targeting N gene of SRAS-CoV-2. Data are representative of 5 independent experiments with 3 replicates for each concentration of 6, 3, 1.5, 0.75, and 0.375 copies of viral RNA. (C) Further confirmation of positive rates detected by qPCR and dPCR assays on 20 replicates of low viral load (3 copies) simulated samples.
3.3. dPCR was specific in detecting simulated SARS-CoV-2 samples
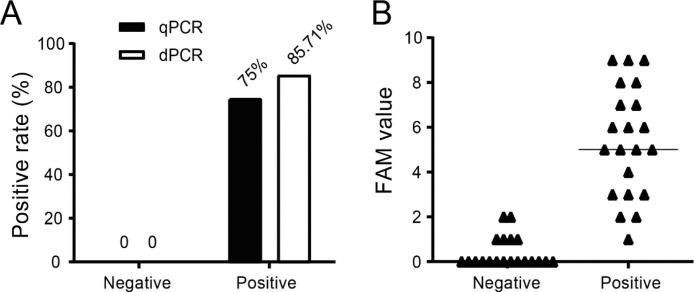
To test the specificity of dPCR and qPCR assays, six negative lower respiratory tract clinical samples were mixed together to act as a matrix of negative samples in which the quantified reference N gene RNA was added to prepare the low positive simulated samples. The matrix negative samples (20 replicates for dPCR and 21 replicates for qPCR) and low positive simulated samples (3–6 copies per reaction, 20 replicates for both dPCR and qPCR) were used to compare the specificity between qPCR and dPCR. The negative samples tested by qPCR and dPCR were all “negative”. Among the positive samples, 85.71 % (18/21) were tested positive (≥3 copies) by dPCR while only 75 % (15/20) were tested positive (CT < 35) by qPCR (Fig. 3 A). As shown in Fig. 3B, the FAM signals of dPCR were all below the threshold of 3. Thus, the dPCR assay showed a consistent specificity while performing higher sensitivity than qPCR.
Fig. 3.
Comparison of specificity between qPCR and dPCR assays. (A) The positive rate of the negative and positive samples detected by dPCR and qPCR. (B) The FAM values of the negative and low positive samples detected by dPCR.
3.4. dPCR was more sensitive than qPCR in detecting clinical SARS-CoV-2 samples
To investigate the possible difference of dPCR and qPCR assays in clinical use, we performed a parallel analysis using positive clinical sputum specimens with CT value at 20 from previous qPCR assay of four confirmed COVID-19 patients. The samples were serially diluted at 1:10 dilution, and a total of 5 dilutions for each specimen were prepared and subjected to dPCR and qPCR assays. The results showed that the dilutions ranged from 10−1 to 10-5 were all detected positive by both dPCR and qPCR. However, for the dilutions of 10-6, there were 4/4 (100 %) samples were detected positive by dPCR, while 2/4 (50 %) positive by qPCR (Table 1 ).
Table 1.
Results on gradient dilution samples of confirmed COVID-19 patients by qPCR and dPCR.
| Sample | CT | qPCR | Chip | # of Neg (FAM) | # of Neg (VIC) | FAM | VIC | dPCR | |
|---|---|---|---|---|---|---|---|---|---|
| 262 | 10−1 | 19.9841 | +* | 200418_074027_D02NNC.eds | 973 | 4133 | 15,427 | 12,267 | + |
| 10−2 | 23.3934 | + | 200418_074139_D040ID.eds | 5596 | 11,344 | 9970 | 4222 | + | |
| 10−3 | 27.3696 | + | 200418_074241_D0415Z.eds | 11,801 | 12,200 | 661 | 262 | + | |
| 10−4 | 30.4212 | + | 200418_074431_D03ZXH.eds | 16,648 | 16,691 | 91 | 48 | + | |
| 10−5 | 34.5454 | + | 200418_074532_D03ZXE.eds | 15,094 | 15,086 | 6 | 14 | + | |
| 10−6 | 35.6356 | + | 200418_074634_D040HO.eds | 15,684 | 15,664 | 3 | 23 | + | |
| 265 | 10−1 | 19.3003 | + | 200418_074814_D02NM5.eds | 114 | 14,126 | 15,629 | 1617 | + |
| 10−2 | 23.0511 | + | 200418_074945_D0415 G.eds | 4770 | 14,438 | 9855 | 187 | + | |
| 10−3 | 27.0236 | + | 200418_075046_D03ZYD.eds | 13,801 | 15,025 | 1250 | 26 | + | |
| 10−4 | 30.2578 | + | 200418_155345_D03JQP.eds(补) | 16,044 | 16,043 | 15 | 16 | + | |
| 10−5 | 33.4663 | + | 200418_075522_D040C0.eds | 16,190 | 16,180 | 6 | 16 | + | |
| 10−6 | 35.6604 | + | 200418_075638_D040A9.eds | 15,870 | 15,869 | 7 | 8 | + | |
| 341 | 10−1 | 22.7873 | + | 200418_075734_D03NQJ.eds | 4843 | 13,045 | 10,261 | 2059 | + |
| 10−2 | 26.5013 | + | 200418_075845_D02PET.eds | 14,131 | 15,675 | 1793 | 249 | + | |
| 10−3 | 30.0149 | + | 200418_080012_D03Z9U.eds | 15,468 | 15,562 | 133 | 39 | + | |
| 10−4 | 33.4111 | + | 200418_080133_D04169.eds | 15,366 | 15,218 | 30 | 178 | + | |
| 10−5 | 35.3413 | + | 200418_080234_D02NNZ.eds | 15,381 | 15,378 | 7 | 10 | + | |
| 10−6 | undetected | _* | 200418_080402_D02PCR.eds | 15,944 | 15,905 | 1 | 40 | _ | |
| 344 | 10−1 | 18.5843 | + | 200418_080526_D02NON.eds | 53 | 165 | 15,186 | 15,074 | + |
| 10−2 | 22.1338 | + | 200418_080626_D02NP7.eds | 4817 | 11,980 | 11,670 | 4507 | + | |
| 10−3 | 26.1415 | + | 200418_080741_D02PCD.eds | 13,589 | 14,388 | 1121 | 322 | + | |
| 10−4 | 29.7896 | + | 200418_080915_D02PDF.eds | 15,088 | 15,175 | 150 | 63 | + | |
| 10−5 | 32.833 | + | 200418_081047_D02LBX.eds | 14,567 | 14,562 | 14 | 19 | + | |
| 10−6 | undetected | _ | 200418_081157_D02NPA.eds | 16,320 | 16,305 | 3 | 18 | + |
Note: * The “+” is positive, the “-” is negative.
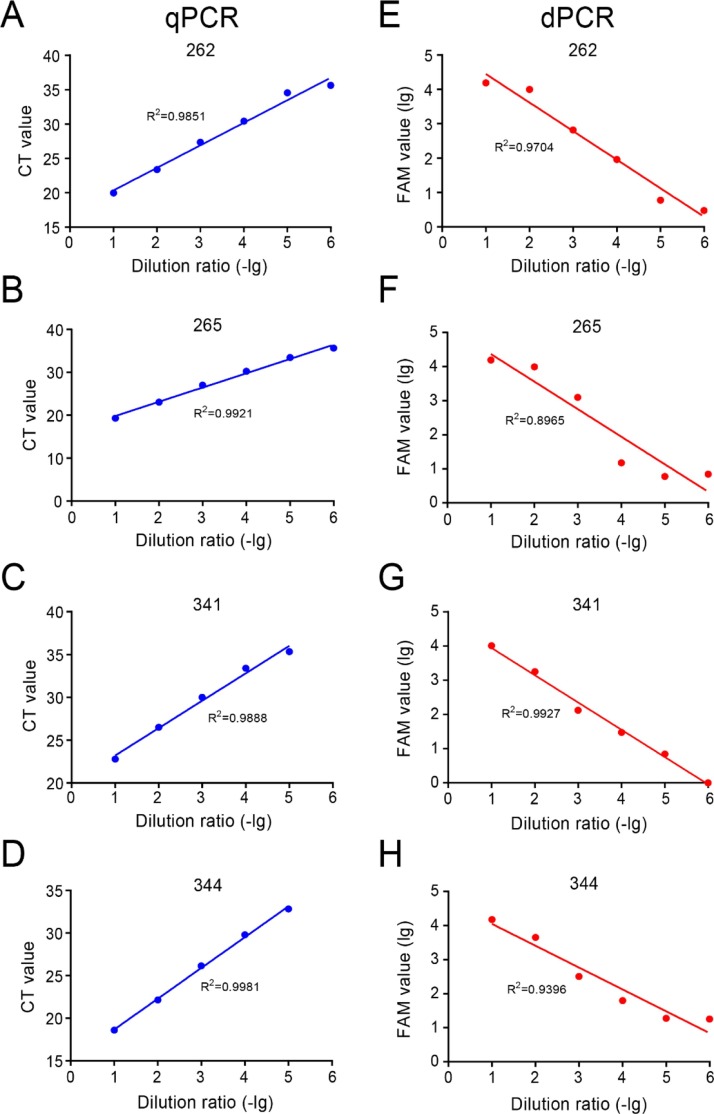
The CT value of qPCR was strongly correlated with the serial dilutions from 10−1 to 10-5 for the 4 samples (R2 = 0.99102 ± 0.00552). In the meantime, the FAM value of dPCR was also strongly correlated, but to a less degree than qPCR, with the serial dilutions (R2 = 0.9498 ± 0.04167) (Fig. 4 ). These results indicated that both dPCR and qPCR were able to accurately reflect the variation in concentration of SARS-CoV-2 samples.
Fig. 4.
Evaluation of linearity using clinical samples by dPCR and qPCR assay. The linearity of CT value of qPCR, FAM positive of dPCR with the serial dilution point ranges from 10−1 to 10−6 for the #262, #265, #341 and #344 COVID-19 patient samples.
3.5. The specificity of dPCR assay in detecting non-SARS-CoV-2 specimens
To further validate the specificity of dPCR assay, the samples of influenza virus and other human coronaviruses of respiratory pathogen were collected and detected by dPCR. As shown in Table 2 , none of 13 tested samples containing influenza virus or other human coronavirus demonstrated positive results, indicating that dPCR possessed high specificity to distinguish the SARS-CoV-2 from other respiratory pathogens such as common influenza virus and other human coronaviruses.
Table 2.
The test results of clinical samples with respiratory pathogen by dPCR.
| Name of sample | Pathogens | Chip | FAM | Result |
|---|---|---|---|---|
| SVIRUS1 | H1pdm2009 | 200327_015214_D02NP5.eds | 0 | -* |
| SVIRUS2 | Influenza-B | 200327_015325_D02LDH.eds | 2 | – |
| SVIRUS3 | Influenza-H9N2 | 200327_015425_D02PDB.eds | 1 | – |
| S19105 | Rhinovirus | 200418_155601_D0409Q.eds | 2 | – |
| SHAD1 | Parainfluenza-Ш | 200418_160706_D040W9.eds | 0 | – |
| S19144 | Adenoviridae | 200418_160157_D0409O.eds | 1 | – |
| SVIRUS4 | Measles virus | 200327_015525_D02PF0.eds | 0 | – |
| SSAM1 | Rubella virus | 200327_015620_D02LCC.eds | 1 | – |
| S19060 | hCoV-HKU1 | 200327_015718_D02NNQ.eds | 0 | – |
| S19111 | hCoV-NL63 | 200418_155850_D0408 J.eds | 0 | – |
| S19116 | hCoV-OC43 | 200418_160059_D03ZT4.eds | 1 | – |
| S19158 | HMPV | 200418_160608_D0409B.eds | 0 | – |
| S19201 | SFTSV | 200418_160816_D0407 N.eds | 2 | – |
Note: *The “-” is negative.
3.6. dPCR and qPCR assays on the “negative” samples from recurrent COVID-19 patients
Finally, to test whether dPCR can be applied to effectively detect the samples from suspected recurrent COVID-19 patients, a total of 18 “negative” samples from 17 such patients including 9 throat swabs, 6 sputum samples, and 3 anal swabs, were tested by dPCR and re-tested by qPCR. Two positive samples were used as controls (Number 1 and 2). The results of dPCR assay were contrastively analyzed with the qPCR results. As shown in Table 3 , 7 samples, including 1 (1/6, 16.7 %) sputum samples, 4 (4/9, 44.4 %) throat swab samples and 2 (2/3, 66.7 %) anal swab samples, were identified positive by dPCR assay (FAM ≥ 3) but not by qPCR assay while 2 throat swab samples (2/9, 22.2 %) were tested positive by qPCR assay (CT value < 35) but not by dPCR assay. These results demonstrated that the dPCR assay has an advantage in identifying the clinical samples with extremely low viral load of SARS-CoV-2 virus especially the samples from the recurrent patients and will be better ruling out the false negative results when used together with the qPCR assay in the clinic.
Table 3.
The test results of samples from suspicious recurrent COVID-19 patients by qPCR and dPCR.
| Number | Sample type | qPCR (ORF1ab) | qPCR (N) | dPCR (FAM) |
|---|---|---|---|---|
| 1# | Sputum | 27.75 | 25.83 | 20 |
| 2# | Sputum | 23.19 | 23.21 | 871 |
| 3 | Sputum | -* | – | 0 |
| 4 | Sputum | – | – | 2 |
| 5 | Sputum | – | – | 1 |
| 6 | Sputum | – | – | 13 |
| 7 | Sputum | 37.10 | 35.64 | 0 |
| 8 | Sputum | – | – | 0 |
| 9 | Throat swab | – | – | 1 |
| 10 | Throat swab | 34.88 | 34.91 | 2 |
| 11 | Throat swab | 37.43 | – | 3 |
| 12 | Throat swab | – | – | 3 |
| 13 | Throat swab | 34.99 | 33.04 | 2 |
| 14 | Throat swab | 35.27 | 36.54 | 6 |
| 15 | Throat swab | – | – | 0 |
| 16 | Throat swab | – | 37.27 | 6 |
| 17 | Throat swab | 38.106 | – | 2 |
| 18 | Anal swab | – | – | 6 |
| 19 | Anal swab | 35.67 | 35.96 | 5 |
| 20 | Anal swab | 36.85 | 37.41 | 1 |
Note: # Positive control samples; * The “-” is negative.
4. Discussion
It was reported that around 60 % of SARS-CoV-2 infections were asymptomatic (Qiu, 2020). Moreover, due to the limitations of RT-PCR technology, only 30 %−60 % positive results could be obtained in COVID-19 patients that further confirmed by chest CT and other diagnostic aid (Wu and McGoogan, 2020). In addition, it is very important to acquire the true negative results from the convalescent COVID-19 patients who are about to discharge and out of quarantine, in order to avoid virus transmission and recurrence. Therefore, it is essential to find more sensitive method(s) to ensure the accuracy of detection of SARS-CoV-2 virus. Compared with qPCR, digital PCR quantification is less affected by poor amplification efficiency and inhibitors of amplification that may present in samples (Vasudevan et al., 2021; Falzone et al., 2020; Jiang et al., 2020). The process of sample partitioning also effectively concentrates template molecules within the micro reactions, improving analytical sensitivity for rare species by reducing competition between different targets for amplification reagents in the reaction mixture (Sanders et al., 2013; White et al., 2009; Pohl and Shih, 2004).
In this study, we first tested the detection efficacy of dPCR and qPCR assays on low (3–6 copies) and middle (9–15 copies) simulated SARS-CoV-2 samples, and found that the two assays gave consistent results for both samples and dPCR was able to accurately reflect the copy number in the respective samples. We then compared the sensitivity of dPCR with qPCR on simulated and clinical sputum samples and the results showed that dPCR was more sensitive when the viral copy number is below 3 (lower threshold than qPCR) in the simulated or clinical samples. In the meantime, the serial dilutions with CT value and FAM value are both strongly correlated with the former slightly higher, indicating that both dPCR and qPCR could accurately reflect the variation in the concentration of SARS-CoV-2 virus in the samples.
In regard to the testing specificity, dPCR obtained absolute negative signals (FAM value = 0) for 14/20 (70 %) of negative samples. However, it did give certain positive signals (1–2 copies per reaction) in 6/20 (30 %) negative samples (Fig. 3B). Nevertheless, when using 2 copies as the cutoff value, dPCR showed 100 % specificity, same as qPCR, in testing negative clinical samples (Fig. 3A). By using this threshold, dPCR could effectively distinguish the SARS-CoV-2 from other respiratory pathogens such as common influenza virus and other human coronaviruses (Table 3). In addition, three of 21 positive samples were not detected by dPCR while 5 of them were not detected by qPCR, which indicated that the sensitivity of dPCR is higher than qPCR in detecting SARS-CoV-2 (Fig. 3B). Notably, the FAM value of 3 “negative samples” was not absolute negative signals. It is suggested that we should attaches more importance to “negative samples” with FAM value signals.
Recently, the recurrence of positive SARS-CoV-2 viral RNA in recovered COVID-19 patients is receiving more attention. Yuan B., et al. reported that 20 (10.99 %) out of 182 recovered COVID-19 patients were detected to be re-positive of SARS-CoV-2 RNA, although none showed any recurrent clinical symptoms (Yuan et al., 2020b). In the meantime, there is an argument on the possible reason for recurrence of COVID-19 patients. At present, it is believed that RNA negative conversion generally takes 2–3 weeks. A recent study indicated that SARS-CoV-2 nucleic acid existed in feces for nearly 50 days (Wu et al., 2020). Another recent study found that SARS-CoV-2 viral particles remained in the lungs of patients whose nasopharyngeal swab test results were negative at three consecutive times (Yao et al., 2020). These studies suggest that during the convalescent stage of the infection, the virus might remain in the patient at a very low level.
In this study, we compared the difference of the two assays in detecting the “negative” samples from clinically recurrent COVID-19 patients. Seven “negative” samples, including 1 sputum samples, 4 throat swab samples and 2 anal swab samples, were identified positive by dPCR assay but not by qPCR assay. On the contrary, only 2 throat swab samples were tested positive by qPCR assay but not by dPCR assay. More importantly, most of the dPCR identified positive samples could not be detected even subjected to the second qPCR test. Therefore, our study clearly demonstrated that dPCR was superior over qPCR in detecting difficult or confusing samples with extremely low viral load from “recurrent” or convalescent COVID-19 patients, suggesting that the more sensitive assay, e.g. dPCR, is a powerful tool to help rule out the false negative results to avoid discharge of COVID-19 patients who might still have the virus becoming active source of the disease.
5. Conclusion
Our study demonstrated that in comparison with qPCR, dPCR is more sensitive in detecting both simulated and clinical SARS-CoV-2 samples with low viral load and can accurately reflect the variation in the copy number of viral RNA. The dPCR is also highly specific in distinguishing SARS-CoV-2 from other respiratory pathogens such as common influenza virus and other human coronaviruses. More importantly, dPCR shows superiority over qPCR in detecting samples from “recurrent” COVID-19 patients, which can be used as an important complement method for difficult or confusing samples in the clinical settings.
Funding
This work is funded by Special Project for Emergency Scientific and Technological Research on New Coronavirus Infection (YG, No. YD9110002001) supported by “The Fundamental Research Funds for the Central Universities” and Emergency Scientific and Technological Research Project for Novel Coronavirus Infection supported by Anhui Science and Technology Bureau (202004a07020002; 202004a0702000)
Authors' contributions
YS, CD, QC, JX, JY and CJ collected the epidemiological, and performed the experiments. YS, ZZ, HH, YG and WL contributed to clinical and laboratory data acquisition and analysis. JH, CD and YG drafted the manuscript. YS, JH and YG had the idea for the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Ethics approval and consent to participate
The study was approved by the Institutional Review Board Ethics Committee of University of Science and Technology of China. Written informed consent was obtained from each patient.
Declaration of Competing Interest
The authors have no conflicts of interests to declare.
Acknowledgements
We acknowledge all health-care workers involved in the diagnosis and treatment of patients in Anhui province. We thank the Chinese National Health Commission for coordinating data collection for patients with COVID-19
References
- Alteri C., Cento V., Antonello M., Colagrossi L., Merli M., Ughi N., Renica S., Matarazzo E., Di Ruscio F., Tartaglione L., et al. Detection and quantification of SARS-CoV-2 by droplet digital PCR in real-time PCR negative nasopharyngeal swabs from suspected COVID-19 patients. PLoS One. 2020;15(9):10. doi: 10.1371/journal.pone.0236311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A.S. Digital assays part I: partitioning statistics and digital PCR. SLAS Technol. 2017;22(4):369–386. doi: 10.1177/2472630317705680. [DOI] [PubMed] [Google Scholar]
- Davila M.L., Riviere I., Wang X., Bartido S., Park J., Curran K., Chung S.S., Stefanski J., Borquez-Ojeda O., Olszewska M., et al. Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci. Transl. Med. 2014;6(224) doi: 10.1126/scitranslmed.3008226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gennaro F., Pizzol D., Marotta C., Antunes M., Racalbuto V., Veronese N., Smith L. Coronavirus diseases (COVID-19) current status and future perspectives: a narrative review. Int. J. Environ. Res. Public Health. 2020;17(8) doi: 10.3390/ijerph17082690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle T.C., Sedlak R.H., Cook L., Jerome K.R. Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin. Chem. 2013;59(11):1670–1672. doi: 10.1373/clinchem.2013.211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzone L., Musso N., Gattuso G., Bongiorno D., Palermo C.I., Scalia G., Libra M., Stefani S. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int. J. Mol. Med. 2020;46(3):957–964. doi: 10.3892/ijmm.2020.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Wang H., Hao S., Chen Y., He J., Liu Y., Chen L., Yu Y., Hua S. Digital PCR is a sensitive new technique for SARS-CoV-2 detection in clinical applications. Clin. Chim. Acta. 2020;511:346–351. doi: 10.1016/j.cca.2020.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55(3) doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Jiang Z.C., Shao C.X., Zhang H.G., Yue H.M., Chen Z.H., Ma B.Y., Liu W.Y., Huang H.H., Yang J., et al. Preliminary study of the relationship between novel coronavirus pneumonia and liver function damage: a multicenter study. Zhonghua Gan Zang Bing Za Zhi. 2020;28(2):107–111. doi: 10.3760/cma.j.issn.1007-3418.2020.02.003. [DOI] [PubMed] [Google Scholar]
- Pan Y., Long L., Zhang D., Yan T., Cui S., Yang P., Wang Q., Ren S. Potential false-negative nucleic acid testing results for Severe Acute Respiratory Syndrome Coronavirus 2 from thermal inactivation of samples with low viral loads. Clin. Chem. 2020 doi: 10.1093/clinchem/hvaa091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavsic J., Zel J., Milavec M. Assessment of the real-time PCR and different digital PCR platforms for DNA quantification. Anal. Bioanal. Chem. 2016;408(1):107–121. doi: 10.1007/s00216-015-9107-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl G., Shih L.M. Principle and applications of digital PCR. Expert Rev. Mol. Diagn. 2004;4(1):41–47. doi: 10.1586/14737159.4.1.41. [DOI] [PubMed] [Google Scholar]
- Qiu J. Covert coronavirus infections could be seeding new outbreaks. Nature. 2020 doi: 10.1038/d41586-020-00822-x. [DOI] [PubMed] [Google Scholar]
- Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N. Engl. J. Med. 2020;382(10):970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders R., Mason D.J., Foy C.A., Huggett J.F. Evaluation of digital PCR for absolute RNA quantification. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0075296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strain M.C., Lada S.M., Luong T., Rought S.E., Gianella S., Terry V.H., Spina C.A., Woelk C.H., Richman D.D. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0055943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev. Mol. Diagn. 2020:1–2. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan H.N., Xu P., Servellita V., Miller S., Liu L., Gopez A., Chiu C.Y., Abate A.R. Digital droplet PCR accurately quantifies SARS-CoV-2 viral load from crude lysate without nucleic acid purification. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-020-80715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Yamahara K.M., Cao Y., Boehm A.B. Absolute quantification of enterococcal 23S rRNA gene using digital PCR. Environ. Sci. Technol. 2016;50(7):3399–3408. doi: 10.1021/acs.est.5b05747. [DOI] [PubMed] [Google Scholar]
- Whale A.S., Huggett J.F., Cowen S., Speirs V., Shaw J., Ellison S., Foy C.A., Scott D.J. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Res. 2012;40(11):e82. doi: 10.1093/nar/gks203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R.A., III, Blainey P.C., Fan H.C., Quake S.R. Digital PCR provides sensitive and absolute calibration for high throughput sequencing. BMC Genomics. 2009:10. doi: 10.1186/1471-2164-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Wu Y.J., Guo C., Tang L.T., Hong Z.S., Zhou J.H., Dong X., Yin H., Xiao Q., Tang Y.P., Qu X.J., et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5(5):434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A.T., Tong Y.X., Zhang S. False-negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: rather than recurrence. J. Med. Virol. 2020 doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X.-H., He Z.-C., Li T.-Y., Zhang H.-R., Wang Y., Mou H., Guo Q., Yu S.-C., Ding Y., Liu X., et al. Pathological evidence for residual SARS-CoV-2 in pulmonary tissues of a ready-for-discharge patient. Cell Res. 2020;30(6):541–543. doi: 10.1038/s41422-020-0318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Kou S., Liang Y., Zeng J., Pan Y., Liu L. PCR assays turned positive in 25 discharged COVID-19 patients. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B., Liu H.-Q., Yang Z.-R., Chen Y.-X., Liu Z.-Y., Zhang K., Wang C., Li W.-X., An Y.-W., Wang J.-C., et al. Recurrence of positive SARS-CoV-2 viral RNA in recovered COVID-19 patients during medical isolation observation. Sci. Rep. 2020;10(1):11887. doi: 10.1038/s41598-020-68782-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Cai Y., Li Z., Shen S., Sha M., Head S.R., Wang Y. A digital PCR assay development to detect EGFR T790M mutation in NSCLC patients. Front. Lab. Med. 2018;2(3):89–96. [Google Scholar]
- Zhou R., Cai Y., Shen S., Sha M., Li Z., Head S.R., Wang Y. A digital PCR based assay to detect all ALK fusion species. Front. Lab. Med. 2018;2(2):49–54. [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]