Abstract
We aimed to examine the circulating microRNA (miRNA) profile of hospitalized COVID-19 patients and evaluate its potential as a source of biomarkers for the management of the disease. This was an observational and multicenter study that included 84 patients with a positive nasopharyngeal swab Polymerase chain reaction (PCR) test for SARS-CoV-2 recruited during the first pandemic wave in Spain (March-June 2020). Patients were stratified according to disease severity: hospitalized patients admitted to the clinical wards without requiring critical care and patients admitted to the intensive care unit (ICU). An additional study was completed including ICU nonsurvivors and survivors. Plasma miRNA profiling was performed using reverse transcription polymerase quantitative chain reaction (RT-qPCR). Predictive models were constructed using least absolute shrinkage and selection operator (LASSO) regression. Ten circulating miRNAs were dysregulated in ICU patients compared to ward patients. LASSO analysis identified a signature of three miRNAs (miR-148a-3p, miR-451a and miR-486-5p) that distinguishes between ICU and ward patients [AUC (95% CI) = 0.89 (0.81-0.97)]. Among critically ill patients, six miRNAs were downregulated between nonsurvivors and survivors. A signature based on two miRNAs (miR-192-5p and miR-323a-3p) differentiated ICU nonsurvivors from survivors [AUC (95% CI) = 0.80 (0.64–0.96)]. The discriminatory potential of the signature was higher than that observed for laboratory parameters such as leukocyte counts, C-reactive protein (CRP) or D-dimer [maximum AUC (95% CI) for these variables = 0.73 (0.55–0.92)]. miRNA levels were correlated with the duration of ICU stay. Specific circulating miRNA profiles are associated with the severity of COVID-19. Plasma miRNA signatures emerge as a novel tool to assist in the early prediction of vital status deterioration among ICU patients.
Abbreviations: AUC, area under the ROC curve; CRP, C-reactive protein; Cq, quantification cycle; ICU, intensive care unit; LASSO, least absolute shrinkage and selection operator; LDH, lactate dehydrogenase; miRNA, microRNA; MSE, mean square error; ncRNA, noncoding RNA; PCA, principal component analysis; ROC, Receiver operating characteristic; RT, Reverse transcription
At A Glance Commentary.
de Gonzalo-Calvo D, et al.
Background
The identification of biomarkers to assist physicians in the management of COVID-19 constitutes a priority in research. MiRNAs have recently emerged as clinical indicators to assist in medical decision-making. We aimed to evaluate the peripheral blood miRNA signature of hospitalized COVID-19 patients, and to explore its potential role as a source of biomarkers for the management of the disease.
Translational Significance
This study identifies plasma miRNA expression profiles that are associated with COVID-19 severity and mortality at ICU. Plasma miRNA signatures emerge as a novel tool to assist in the early prediction of vital status deterioration among ICU patients.
Alt-text: Unlabelled box
Introduction
Approximately 20 to 30% of hospitalized COVID-19 patients develop a severe phenotype of the disease that requires intensive care unit (ICU) admission with a varying ICU death rate ranging from 25 to 50%.1 , 2 In this scenario, there is an urgent demand for reliable tools to define patient status and predict clinical outcomes. Although risk modeling based on clinical characteristics and/or biomarkers has made remarkable progress, early prediction of vital status deterioration remains a challenge for clinicians.3 Furthermore, most COVID-19 studies have focused on proteomic, metabolomic and cellular biomarkers.4 , 5 The investigation of alternative clinical indicators, including those provided by emerging omics technologies, may shed light on the development of novel tools for the management of COVID-19.
In the past decade, noncoding RNAs (ncRNAs), and the class of microRNAs (miRNAs) in particular, have risen to prominence as a novel tool to assist in medical decision-making.6 miRNAs are short (19-25 nt), single-stranded noncoding RNAs that regulate gene expression at the posttranscriptional level by binding to target mRNA and leading to its degradation or translational repression. These small ncRNAs are present in body fluids, including blood, are highly resistant to degradation and can be easily measured through standard techniques already employed in clinical laboratories, such as real-time quantitative polymerase chain reaction (RT-qPCR). The use of circulating miRNAs as clinical biomarkers in liquid biopsies has been explored under a variety of conditions,7, 8, 9 including viral infections.10 The results suggest that miRNAs are sensitive, robust and cost-effective biomarkers that offer additional information to clinical variables and already established clinical indicators.11 , 12 Indeed, several miRNA-based diagnostic products are already available for clinical practice.13
Here, we aimed to examine the circulating miRNA profile of hospitalized COVID-19 patients and explore the potential role and clinical significance as biomarkers of disease severity. To the best of our knowledge, the current study is the largest to date that has profiled the circulating miRNA profile in the context of COVID-19.
Methods
Ethics statement
The study was performed in full compliance with the Declaration of Helsinki. The study protocol was approved by the respective ethics committee. Participants, or their legal representatives, provided oral consent when possible. In the remaining cases, an informed consent waiver was authorized by the ethics committee.
Patients
This is a preliminary report on the epigenetic substudy of the ongoing multicenter study CIBERESUCICOVID registered at www.clinicaltrials.gov with the identification NCT04457505.
This was an observational and multicenter study that included 84 participants. Patients aged 18 years or older with a positive nasopharyngeal swab PCR test for SARS-CoV-2 were recruited during the first pandemic wave in Spain (March-May 2020). The centers included Hospital Universitario Arnau de Vilanova y Santa María (Lleida), Hospital Clínico Universitario (Valladolid), Hospital del Río Hortega (Valladolid), Hospital General Universitario Gregorio Marañón (Madrid) and Hospital Universitario Infanta Leonor (Madrid). The patients were divided according to the severity of the disease into the following groups: (1) hospitalized patients admitted to the pneumology, infectious diseases or internal medicine wards without requiring critical care, and ; (2) hospitalized patients admitted to the ICU. An additional study including ICU nonsurvivors and survivors was performed.
Comprehensive demographic, clinical, pharmacological and laboratory data were abstracted manually from the electronic medical records. Data collected at the time of hospital or ICU admission were recorded by dedicated clinical research assistants and entered into a REDCap database. Incoherent or missing data were identified using automatic checks. Abnormal data were reviewed by researchers/clinicians.
Blood samples were collected in ethylenediaminetetraacetic acid (EDTA) tubes before or following admission to the clinical ward or the ICU (Supplemental Figure S1). Blood samples were centrifuged to separate plasma (1500 × g for 10 minutes). All specimens were immediately aliquoted, frozen and stored at -80°C. Samples collected at the Hospital Universitario Arnau de Vilanova y Santa María (Lleida, Spain) were obtained with support by IRBLleida Biobank (B.0000682) and “Plataforma Biobancos PT17/0015/0027". Except for those samples collected at Hospital Universitario Arnau de Vilanova y Santa María (Lleida, Spain), frozen plasma aliquots were shipped on dry ice to the Biomedical Research Institute of Lleida (Lleida, Spain). No freeze-thaw cycles were performed during the experiments.
RNA isolation and microRNA quantification
miRNA quantification was performed in the same laboratory and under standardized conditions by experienced staff blinded to the clinical data. A panel of 41 circulating miRNAs was selected after an extensive review of the literature by experienced researchers (FB and DdGC). The panel included miRNAs previously associated with molecular pathways potentially altered in COVID-19 (immune/inflammatory response, viral infections, lung damage or fibrosis, myocardial damage and coagulation) in in vitro, in vivo and patient-based approaches and investigated as biomarkers of mechanisms linked to COVID-19 pathophysiology by independent groups, including ours14 (Supplemental Figure S2).
Total RNA was isolated from 180 μL of plasma using a miRNeasy Serum/Plasma Advanced Kit (Qiagen, Hilden, Germany). For normalization, synthetic Caenorhabditis elegans cel-miR-39-3p, lacking sequence homology to human miRNAs, was added as an external reference miRNA (1.6 × 108 copies/μL). The mixture was supplemented with 1 µg of MS2 carrier RNA (Roche, Merck, Darmstadt, Germany) to improve extracellular RNA yield and the RNA Spike-In Kit (UniSp2, UniSp4 and UniSp5) (Qiagen) to monitor RNA isolation. All reagents were spiked into samples during RNA isolation after incubation with the denaturing solution. RNA was eluted into 20 μL of nuclease-free water and stored at -80°C.
Since the overall amount of RNA that is present in plasma is low and the RNA concentration cannot be accurately determined in plasma samples, the input RNA amount for subsequent analysis was based on the starting volume rather than RNA quantity. A consistent input amount was used for all samples. miRNA quantification was performed according to the protocol for the miRCURY LNA Universal RT microRNA PCR System (Qiagen), which offers optimal accuracy and reproducibility.15 Reverse transcription (RT) cDNA synthesis was performed using a miRCURY LNA RT Kit (Qiagen) in a total reaction volume of 10 μL. An additional spike-in UniSp6 (Qiagen) was added to monitor the RT reaction. Heparin, an anticoagulant usually administered to COVID-19 patients during hospital stays,16 inhibits miRNA quantification.17 , 18 To avoid this potential inhibitory effect, heparinase (New England BioLabs, Massachusetts, USA) was added to RT reactions, as previously described.19 The RT reactions were performed in a total volume of 10 μL under the following conditions: incubation for 60 minutes at 42°C, inactivation for 5 minutes at 95°C, and immediate cooling to 4°C. Then, cDNA was stored at 20°C.
Plasma miRNA signatures were analyzed using miRCURY LNA miRNA Custom Panels (384-well plates) (Qiagen). qPCR was carried out using the Applied Biosystems QuantStudio 7 Flex Real-Time PCR System in a total volume of 10 μL. RT-qPCR conditions were 95°C for 2 minutes, followed by 40 cycles of 95°C for 10 seconds and 56°C for 1 minute, followed by melting curve analysis. Synthetic UniSp3 was analyzed as an interplate calibrator and qPCR control. Amplification curves were evaluated using QuantStudio Software v1.3 (Thermo Fisher Scientific, Massachusetts, USA). The quantification cycle (Cq) was defined as the fractional cycle number at which the fluorescence exceeded a given threshold. The specificity of the qPCR was corroborated by melting curve analysis. To ensure the optimal quality of the data, we first analyzed spike-in RNA templates to monitor the uniformity of the RNA extraction procedure and the efficiency of the RT and PCRs. The ΔCq (miR-23a-3p - miR-451a) method was used to exclude hemolysis contamination, as previously described.20 Cqs above 35 cycles were considered undetectable and were censored at the minimum level observed for each miRNA. Relative quantification was performed using the 2−ΔCq method (ΔCq = CqmiRNA-Cqcel-miR-39-3p). Expression levels were log-transformed for statistical purposes.
Statistical analysis
Descriptive statistics were used to summarize the characteristics of the study population. The normality of the distribution was analyzed using the Shapiro–Wilk test. Data are presented as the median [Quartile 1;Quartile 3] for continuous variables and as frequencies (percentage) for categorical variables. Continuous variables were compared between groups using the Mann–Whitney U test. Categorical variables were compared between groups using Fisher's exact test. The Spearman rank correlation test was used to assess the correlation between continuous variables. Differences in miRNA expression between groups were evaluated using linear models for arrays.21 Differential miRNA expression between study groups is displayed in volcano plots. Principal component analysis (PCA) and hierarchical clustering included the differentially expressed miRNAs. miRNAs with statistical differences according to severity groups were also evaluated after adjustment for confounding factors. Age, sex and medication use were included in the lineal models. This analysis was not performed for the comparison survivor versus nonsurvivor due to the sample size.
A predictive model with miRNAs was constructed using a relaxed least absolute shrinkage and selection operator (LASSO) model. Fivefold cross-validation was carried out to determine the lambda parameter of the LASSO model. Lambda was selected as the value that minimized the mean square error (MSE). miRNA levels were standardized prior to fitting the LASSO model. Receiver operating characteristic (ROC) curves were constructed for circulating miRNAs and laboratory parameters using the area under the ROC curve (AUC) as the global discrimination value measure. The P-value threshold defining statistical differential expression was set at <0.05. All statistical analyses were performed using R software, version 4.0.2.
Results
Impact of COVID-19 severity on the circulating microRNA profile
The study flowchart is presented in Fig 1 . We first analyzed the impact of COVID-19 severity on the circulating miRNA profile. To that end, forty-one miRNAs known to be implicated in molecular pathways linked to COVID-19, or proposed as biomarkers of mechanisms associated with the disease, were measured in plasma samples from hospitalized patients admitted to the clinical wards without requiring critical care and from critically ill patients admitted to the ICU. Blood samples were available from 84 patients. Patients with hemolyzed samples [ΔCq(miR-23a-3p - miR-451a) ≥ 6] or in which miRNA quantification did not pass the quality control (high variability in spike-ins) were excluded (n=5) (Supplementary Figure S3A & S3B). Seven miRNAs, miR-9-5p, miR-34b-5p, miR-34c-5p, miR-124-3p, miR-208a-3p, miR-208b-3p and miR-499a-5p, were below the limit of detection (Cq ≥ 35) in more than 80% of samples and therefore were not considered in further analyses.
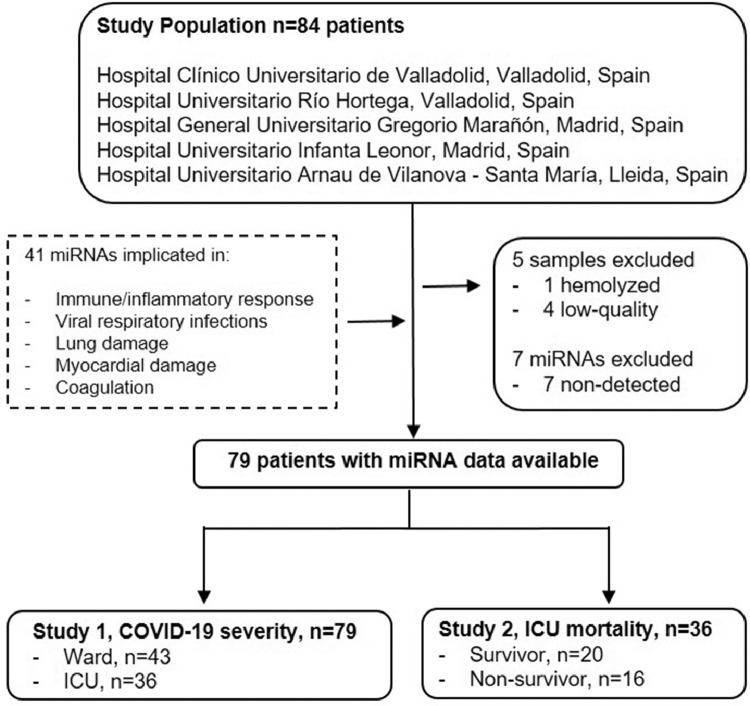
Fig 1.
Study flowchart. The study included 84 hospitalized patients with a positive nasopharyngeal swab PCR test for SARS-CoV-2 recruited during the first pandemic wave in Spain (March-June 2020). The centers included were Hospital Clínico Universitario (Valladolid), Hospital del Río Hortega (Valladolid), Hospital General Universitario Gregorio Marañón (Madrid), Hospital Universitario Infanta Leonor (Madrid) and Hospital Universitario Arnau de Vilanova y Santa María (Lleida). A panel of 41 circulating microRNAs was selected after an extensive review of the literature. The panel included microRNAs previously associated with molecular pathways potentially altered in COVID-19 (immune/inflammatory response, viral infections, lung damage or fibrosis, myocardial damage and coagulation) in in vitro, in vivo and patient-based approaches and investigated as biomarkers of mechanisms linked to COVID-19 pathophysiology. Patients with hemolyzed or low-quality samples were excluded (n=5). Seven microRNAs, miR-9-5p, miR-34b-5p, miR-34c-5p, miR-124-3p, miR-208a-3p, miR-208b-3p and miR-499a-5p, were below the limit of detection (Cq ≥ 35) in more than 80% of samples and therefore were not considered in further analysis. Patients were stratified according to disease severity: hospitalized patients admitted to the clinical wards without requiring critical care (n=43) and patients admitted to the ICU (n=36). An additional study was completed including ICU nonsurvivors (n=16) and survivors (n=20).
The main demographic, clinical, pharmacological and biochemical data of the study groups are summarized in Table I . Patients admitted to the ICU were typically men. As expected, the use of pharmacological therapies, including hydroxychloroquine, tocilizumab, antibiotics and corticoids, the requirement for invasive ventilation and the duration of the hospital stay were higher in critically ill patients than in ward patients. ICU patients also had higher glucose, D-dimer, lactate dehydrogenase (LDH), ferritin, and C-reactive protein (CRP) concentrations along with elevated levels of leukocytes and neutrophils and reduced lymphocyte counts.
Table 1.
Characteristics of the study population (ward vs ICU patients)
| All | Ward | ICU | P-value | n | |
|---|---|---|---|---|---|
| n = 79 | n = 43 | n = 36 | |||
| Demographic characteristics | |||||
| Age, years | 68.0 (56.5;77.0) | 68.0 (56.5;84.0) | 68.0 (56.8;72.2) | 0.116 | 79 |
| Sex | 0.013 | 79 | |||
| Male | 44 (55.7) | 18 (41.9) | 26 (72.2) | ||
| Female | 35 (44.3) | 25 (58.1) | 10 (27.8) | ||
| Clinical characteristics | |||||
| Cardiovascular disease | 10 (12.8) | 5 (11.9) | 5 (13.9) | 1.000 | 78 |
| Obesity | 18 (24.7) | 11 (29.7) | 7 (19.4) | 0.455 | 73 |
| Hypertension | 40 (50.6) | 26 (60.5) | 14 (38.9) | 0.092 | 79 |
| Type II diabetes mellitus | 15 (19.0) | 10 (23.3) | 5 (13.9) | 0.442 | 79 |
| COPD | 4 (5.06) | 4 (9.30) | 0 (0.00) | 0.121 | 79 |
| Asthma | 3 (3.85) | 3 (7.14) | 0 (0.00) | 0.245 | 78 |
| Chronic kidney disease | 11 (14.1) | 6 (14.3) | 5 (13.9) | 1.000 | 78 |
| Chronic liver disease | 2 (2.53) | 1 (2.33) | 1 (2.78) | 1.000 | 79 |
| Autoimmune disease | 2 (2.53) | 1 (2.33) | 1 (2.78) | 1.000 | 79 |
| Smoking | 6 (8.57) | 4 (11.4) | 2 (5.71) | 0.673 | 70 |
| Alcoholism | 1 (1.79) | 0 (0.00) | 1 (2.86) | 1.000 | 56 |
| Measurements at admission | |||||
| Oxygen saturation, % | 94.0 (91.8;97.0) | 94.0 (92.5;97.0) | 93.0 (91.0;96.0) | 0.262 | 68 |
| Glucose, mg/dL | 126 (110;165) | 118 (108;137) | 164 (126;192) | <0.001 | 75 |
| Creatinine, mg/dL | 0.94 (0.70;1.32) | 0.94 (0.71;1.27) | 0.93 (0.63;1.37) | 0.496 | 76 |
| Leukocyte count, x103/µL | 7.45 (5.47;10.2) | 6.38 (4.48;8.50) | 9.84 (7.40;13.6) | <0.001 | 76 |
| Lymphocyte count, x103/µL | 0.74 (0.50;1.14) | 0.96 (0.72;1.23) | 0.50 (0.30;0.70) | <0.001 | 75 |
| Neutrophil count, x103/µL | 5.79 (3.87;8.68) | 5.00 (3.20;6.90) | 8.36 (6.09;12.6) | <0.001 | 75 |
| Platelet count, x103/µL | 201 (149;268) | 207 (146;258) | 198 (161;290) | 0.603 | 75 |
| D-dimer, ng/mL | 1.94 (0.73;8.99) | 0.82 (0.27;1.74) | 6.66 (2.71;24.8) | <0.001 | 71 |
| LDH, U/L | 415 (307;582) | 363 (270;492) | 488 (359;666) | 0.005 | 67 |
| Ferritin, µg/L | 527 (297;1124) | 397 (209;565) | 1378 (863;2998) | <0.001 | 41 |
| CRP, mg/dL | 108 (48.0;180) | 80 (19.8;114) | 155 (84.0;265) | 0.001 | 75 |
| Troponin I, ng/L | 11.3 (6.3;20.4) | 8.0 (7.5;8.5) | 11.9 (6.0;34.3) | 0.390 | 12 |
| Creatine kinase, U/L | 78.0 (42.8;139) | 82.5 (55.8;141) | 62.0 (29.5;114) | 0.418 | 20 |
| NT-proBNP, pg/mL | 214 (120;548) | 534 (309;3414) | 203 (142;388) | 0.569 | 10 |
| Complications during hospitalization | |||||
| Acute respiratory distress syndrome | 44 (55.7) | 9 (20.9) | 35 (97.2) | <0.001 | 79 |
| Hospital/ICU mortality | 18 (22.8) | 2 (4.65) | 16 (44.4) | <0.001 | 79 |
| Hospital stay, days | 12.5 (6.00;21.2) | 8.00 (4.00;14.5) | 21.0 (17.0;42.0) | <0.001 | 64 |
| Treatment during hospitalization | |||||
| Hydroxychloroquine | 54 (68.4) | 19 (44.2) | 35 (97.2) | <0.001 | 79 |
| Tocilizumab | 8 (10.1) | 1 (2.33) | 7 (19.4) | 0.020 | 79 |
| Antibiotic | 67 (87.0) | 33 (78.6) | 34 (97.1) | 0.018 | 77 |
| Corticoids | 36 (45.6) | 13 (30.2) | 23 (63.9) | 0.006 | 79 |
| Remdesivir | 1 (1.27) | 0 (0.00) | 1 (2.78) | 0.456 | 79 |
| Invasive mechanical ventilation | 32 (41.0) | 1 (2.38) | 31 (86.1) | <0.001 | 78 |
| Noninvasive mechanical ventilation | 15 (20.3) | 8 (21.1) | 7 (19.4) | 1.000 | 74 |
Abbreviations: COPD, chronic obstructive pulmonary disease; CRP, C-reactive protein; ICU, Intensive care unit; LDH, lactic acid dehydrogenase; NT-proBNP, N-terminal prohormone of brain natriuretic peptide.
Continuous variables are expressed as median (Q1;Q3) and categorical as n (%).
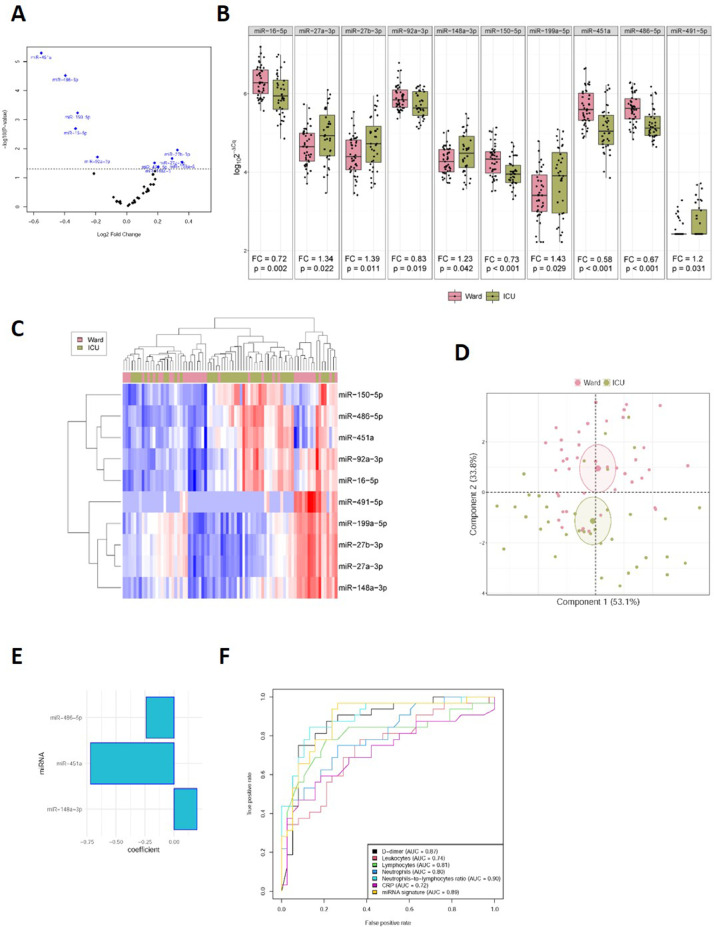
The expression levels of circulating miRNAs were compared between ward and ICU patients. Ten of 41 miRNAs were differentially detected. miR-27a-3p [fold change (FC)=1.34], miR-27b-3p (FC=1.39), miR-148a-3p (FC=1.23), miR-199a-5p (FC=1.43) and miR-491-5p (FC=1.20) were upregulated in ICU patients compared to ward patients. Decreased levels of miR-16-5p (FC=0.72), miR-92a-3p (FC=0.83), miR-150-5p (FC=0.73), miR-451a (FC=0.58) and miR-486-5p (FC=0.67) were also observed in critically ill patients (Fig 2 A and B). As shown in the heat map and PCA (Fig 2 C and D), the levels of the ten miRNAs were able to segregate patients based on disease severity, that is, ward versus ICU patients. No differences were observed for other candidates (Supplemental Table S1).
Fig 2.
Impact of COVID-19 severity on the circulating microRNA profile. A, Volcano plot of fold change and corresponding P-values for each microRNA after comparison of ward patients and ICU patients (unadjusted). Each point represents one microRNA. Blue dots represent the microRNA candidates that showed significant differences; B, Boxplot including plasma levels of microRNA candidates that showed differences between ward patients and ICU patients. Between-group differences were analyzed using linear models for arrays. P-values describe the significance level for each comparison; C, Heat map showing the unsupervised hierarchical clustering. Each column represents a patient (ward or ICU patient). Each row represents a microRNA. The color scale illustrates the relative expression level of microRNAs. The expression intensity of each microRNA in each sample varies from red to blue, which indicates relatively high or low expression, respectively. D, Principal component analysis. Each point represents a patient. E, Predictive model constructed using a variable selection process based on LASSO regression. miRNA levels were standardized prior to fitting the LASSO regression model. Estimated regression coefficients are shown. F, ROC curves for laboratory parameters and the microRNA signature. Expression levels were quantified by RT-qPCR. Relative quantification was performed using cel-miR-39-3p as the external standard. Relative quantification was performed using the 2−ΔCq method (ΔCq = CqmicroRNA-Cqcel-miR-39-3p). Expression levels were log-transformed for statistical purposes. microRNA levels are expressed as arbitrary units. “For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.
The ideal scenario for miRNA testing seems to be based on the concept of “several miRNAs-one disease”, contrary to the traditional “one miRNA-one disease” concept.12 Therefore, we explored whether COVID-19 severity could be associated with a specific miRNA signature. Multivariate predictive models were constructed using a variable selection process based on LASSO regression. This approach identified a signature of three miRNAs, miR-148a-3p, miR-486-5p and miR-451a, associated with ICU stay (Fig 2E). ROC curves and AUCs were used to assess the discriminative accuracy of the plasma miRNA signature. The AUC (95% CI) for discriminating ward vs. ICU patients was 0.89 (0.81-0.97) (Fig 2F). The AUC was comparable to, or even higher than, that observed for the contemporaneous test proposed as biomarkers of COVID-19 severity, such as leukocyte counts, D-dimer or CRP [AUC (95% CI) from 0.72 (0.59 – 0.84) to 0.90 (0.82 – 0.97)] (Fig 2F).
The impact of different pharmacological treatments on the levels of circulating miRNAs, or on the quantification method, has been previously described, for example, antiplatelet therapy, statins or heparin.22 , 23 Furthermore, in the context of such an emerging disease, the effect of the therapeutic drugs used to treat COVID-19 patients on the circulating levels of miRNAs remains unknown. Therefore, we further checked the influence of medications widely used to treat hospitalized COVID-19 patients. A significant effect was observed for miR-16-5p, miR-92a-3p and miR-150-5p (Supplemental Table S2). The greatest impact was caused by antibiotics (for the three miRNAs), corticoid use (for miR-92-3p) and hydroxychloroquine (for miR-150-5p) (Supplemental Figure S4). We did not observe any impact of these confounding factors on other miRNAs. No effect of age or sex was reported.
Next, we evaluated the association between laboratory parameters and circulating miRNAs (Supplemental Figure S5). A correlation was observed between the dysregulated miRNAs and the counts of leukocytes (miR-27a-3p, miR-27b-3p and miR-148a-3p), neutrophils (miR-27a-3p, miR-27b-3p, miR-148a-3p, miR-451a and miR-486-5p), lymphocytes (miR-16-5p, miR-92-3p, miR-150-5p, miR-451a and miR-486-5p), platelets (miR-16-5p, miR-27a-3p, miR-27b-3p, miR-92a-3p, miR-148-3p, miR-199a-5p and miR-491-5p) and the concentrations of D-dimer (miR-16-5p, miR-92a-3p, miR-150-5p, miR-451a and miR-486-5p), ferritin (miR-16-5p, miR-92a-3p, miR-150-5p, miR-451a and miR-486-5p) and CRP (miR-27a-3p, miR-27b-3p, miR-148a-3p, miR-199-5p, miR-451a and miR-491-5p).
Circulating microRNAs as biomarkers for mortality in critically ill COVID-19 patients
The comparison between hospitalized patients admitted to the clinical wards without requiring critical care and hospitalized patients admitted to the ICU, although useful for exploring the impact of COVID-19 severity on the miRNA profile, does not provide real clues about the clinical significance of these small ncRNAs as biomarkers of COVID-19. Consequently, we performed an additional study to test whether the circulating miRNA signature constitutes a predictor of mortality in critically ill patients. The characteristics of the study population according to ICU survival are detailed in Table II .
Table 2.
Characteristics of the study population (ICU survivors vs ICU nonsurvivors)
| Survivor | Nonsurvivor | P-value | n | |
|---|---|---|---|---|
| n = 20 | n = 16 | |||
| Demographic characteristics | ||||
| Age, years | 60.0 (48.0;68.2) | 70.5 (68.0;73.2) | 0.002 | 36 |
| Sex | 0.722 | 36 | ||
| Male | 15 (75.0) | 11 (68.8) | ||
| Female | 5 (25.0) | 5 (31.2) | ||
| Clinical characteristics | ||||
| Cardiovascular disease | 3 (15.0) | 2 (12.5) | 1.000 | 36 |
| Obesity | 5 (25.0) | 2 (12.5) | 0.426 | 36 |
| Hypertension | 7 (35.0) | 7 (43.8) | 0.848 | 36 |
| Type II diabetes mellitus | 3 (15.0) | 2 (12.5) | 1000 | 36 |
| COPD | 0 (0%) | 0 (0%) | · | 36 |
| Asthma | 0 (0%) | 0 (0%) | · | 36 |
| Chronic kidney disease | 3 (15.0) | 2 (12.5) | 1.000 | 36 |
| Chronic liver disease | 0 (0.00) | 1 (6.25) | 0.444 | 36 |
| Autoimmune disease | 1 (5.00) | 0 (0.00) | 1.000 | 36 |
| Smoking | 1 (5.26) | 1 (6.25) | 1.000 | 35 |
| Alcoholism | 0 (0.00) | 1 (6.25) | 0.457 | 35 |
| Measurements at admission | ||||
| Systolic blood pressure | 136 (129;152) | 139 (118;150) | 0.755 | 34 |
| Diastolic blood pressure | 77.0 (60.0;89.5) | 64.0 (53.0;67.5) | 0.041 | 34 |
| Oxygen saturation, % | 93.5 (92.0;95.8) | 93.0 (88.0;97.0) | 0.679 | 36 |
| Glucose, mg/dL | 163 (116;173) | 164 (126;234) | 0.317 | 32 |
| Creatinine, mg/dL | 0.88 (0.60;1.27) | 0.97 (0.70;1.39) | 0.539 | 33 |
| Leukocyte count, x103/µL | 9.78 (7.53;14.3) | 9.84 (6.05;13.2) | 0.448 | 33 |
| Lymphocyte count, x103/µL | 0.60 (0.50;0.80) | 0.40 (0.22;0.52) | 0.031 | 32 |
| Neutrophil count, x103/µL | 8.39 (6.40;12.7) | 8.32 (5.10;12.6) | 0.806 | 32 |
| Platelet count, x103/µL | 243 (169;309) | 179 (160;205) | 0.062 | 32 |
| D-dimer, ng/mL | 4.31 (2.02;22.0) | 9.42 (5.07;23.3) | 0.286 | 33 |
| LDH, U/L | 464 (320;531) | 590 (434;716) | 0.116 | 33 |
| Ferritin, µg/L | 1124 (521;3087) | 1633 (888;1967) | 0.947 | 14 |
| CRP, mg/dL | 170 (91.5;251) | 143 (84;282) | 0.928 | 33 |
| Troponin I, ng/L | 11.6 (6.9;55.2) | 13.4 (8.9;27.3) | 0.732 | 10 |
| Creatine kinase, U/L | 74.0 (50.0;106) | 30.0 (25.5;84.5) | 0.456 | 8 |
| NT-proBNP, pg/mL | 203 (99.0;224) | 368 (276;460) | 0.699 | 7 |
| SOFA score | 5.00 (4.00;8.00) | 6.00 (4.50;8.00) | 0.698 | 20 |
| APACHE-II score | 14.0 (11.0;17.5) | 17.0 (15.0;20.5) | 0.201 | 35 |
| Complications during hospitalization | ||||
| Acute respiratory distress syndrome | 19 (95.0) | 16 (100) | 1.000 | 36 |
| Treatment during hospitalization | ||||
| Hydroxychloroquine | 19 (95.0) | 16 (100) | 1.000 | 36 |
| Tocilizumab | 5 (25.0) | 2 (12.5) | 0.426 | 36 |
| Antibiotic | 18 (90.0) | 16 (100) | 0.492 | 36 |
| Corticoids | 0 (0.00%) | 1 (6.25%) | 0.444 | 36 |
| Remdesivir | 0 (0.00%) | 1 (6.25%) | 0.444 | 36 |
| Catecholamines | 8 (61.5%) | 5 (38.5%) | 0.867 | 34 |
| Invasive mechanical ventilation | 16 (80.0) | 15 (93.8) | 0.355 | 36 |
| Invasive mechanical ventilation, days | 24.0 (10.5;30.0) | 21.0 (14.5;24.0) | 0.930 | 29 |
| Noninvasive mechanical ventilation | 5 (25.0) | 2 (12.5) | 0.426 | 36 |
| Positive end-expiratory pressure, cm H2O | 12.0 (9.00;12.0) | 12.0 (10.0;12.0) | 0.231 | 28 |
| Peak pressure, cm H2O | 35.2 (33.0;37.8) | 34.4 (33.5;36.5) | 0.830 | 10 |
| Plateau pressure, cm H2O | 23.4 (21.3;27.0) | 24.0 (23.1;27.4) | 1.000 | 10 |
Abbreviations: APACHE-II, Acute Physiology and Chronic Health disease Classification System II; COPD, chronic obstructive pulmonary disease; CRP, C-reactive protein; ICU, Intensive care unit; LDH, lactic acid dehydrogenase; NT-proBNP, N-terminal prohormone of brain natriuretic peptide; SOFA, Sepsis related Organ Failure Assessment.
Continuous variables are expressed as median (Q1;Q3) and categorical as n (%).
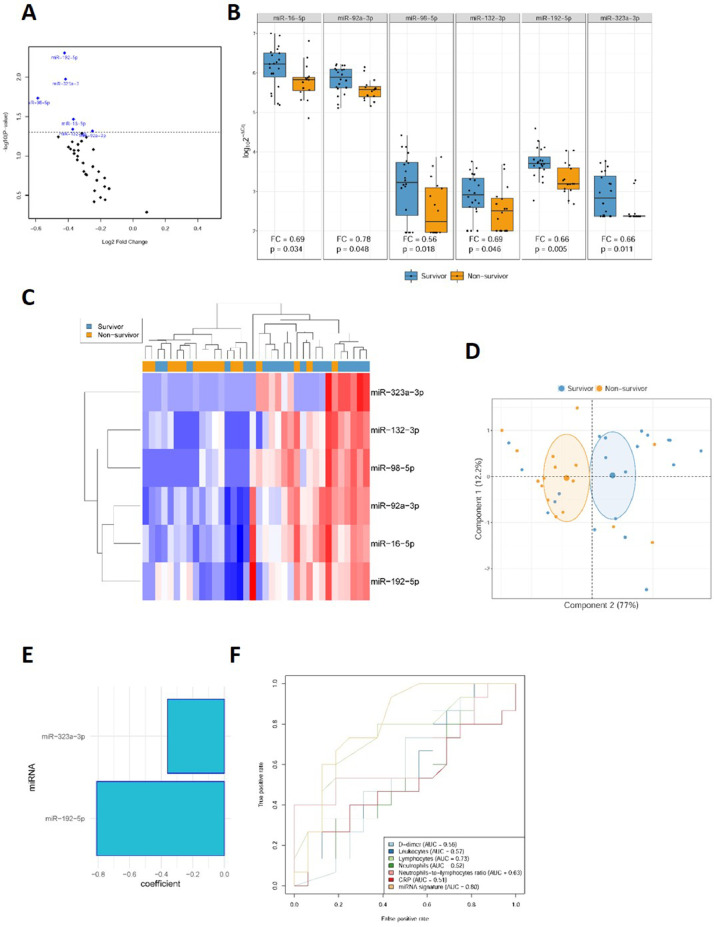
As shown in Fig 3 A and B, miR-16-5p (FC=0.69), miR-92a-3p (FC=0.78), miR-98-5p (FC=0.56), miR-132-3p (FC=0.69), miR-192-5p (FC=0.66) and miR-323a-3p (FC=0.66) showed significant suppression in patients who did not survive the ICU stay. Unsupervised hierarchical clustering and PCA based on the expression profile of these 6 miRNAs separated survivors from nonsurvivors (Fig 3 C and D). No other significant differences were found (Supplemental Table S3).
Fig 3.
Circulating microRNAs as biomarkers for ICU mortality in COVID-19 patients. A, Volcano plot of fold change and corresponding P-values for each microRNA after comparison of nonsurvivors and survivors (unadjusted). Each point represents one microRNA. Blue dots represent the microRNA candidates that showed significant differences; D, Box plot including plasma levels of microRNA candidates that showed differences between nonsurvivor and survivor patients. Between-group differences were analyzed using linear models for arrays. P-values describe the significance level for each comparison; C, Heat map showing the unsupervised hierarchical clustering. Each column represents a patient (nonsurvivor or survivor). Each row represents a microRNA. The color scale illustrates the relative expression level of microRNAs. The expression intensity of each microRNA in each sample varies from red to blue, which indicates relatively high or low expression, respectively. D, Principal component analysis. Each point represents a patient. E, Predictive model constructed using a variable selection process based on LASSO regression. miRNA levels were standardized prior to fitting the LASSO regression model. Estimated regression coefficients are shown. F, ROC curves for laboratory parameters and the microRNA signature. Expression levels were quantified by RT-qPCR. Relative quantification was performed using cel-miR-39-3p as the external standard. Relative quantification was performed using the 2−ΔCq method (ΔCq = CqmicroRNA-Cqcel-miR-39-3p). Expression levels were log-transformed for statistical purposes. microRNA levels are expressed as arbitrary units.
A predictor selection procedure was performed using the panel of miRNAs. The multivariable analysis selected a signature based on two miRNAs, miR-192-5p and miR-323a-3p, that was found to be a relevant predictor of mortality during the ICU stay [AUC = 0.80 (0.64 – 0.96)] (Fig 3 E and F). The derived miRNA signature was compared to commonly used parameters. The discriminatory potential of the miRNA signature was higher than that observed for clinical laboratory parameters such as CRP, D-dimer or leukocyte counts, including neutrophil count, lymphocyte count and the neutrophil-to-lymphocyte ratio [maximum AUC for these variables = 0.73 (0.55 – 0.92)] (Fig 3F).
An additional study was performed to explore the association between circulating miRNAs and the duration of ICU stay in critically ill survivors. Plasma levels of miR-16-5p (rho= -0.38), miR-92a-3p (rho= -0.36), miR-93-5p (rho= -0.35), miR-150-5p (rho= -0.31) and miR-214-3p (rho= -0.36) were inversely correlated with the total number of days of ICU stay. No significant correlation between miRNA levels and other relevant clinical variables was found (Supplemental Table S4).
Discussion
COVID-19 has had a considerable impact on public health and the global economy, and it is expected to continue in the short and medium terms. Vaccines should dramatically reduce the number of COVID-19 cases in the long term. Nevertheless, COVID-19 will continue to be treated in the ICU in the near future. As such, there is an urgent need to find new tools to manage the disease. In this context, limited information is currently available regarding the host circulating ncRNA signature of COVID-19. Given the potential of miRNAs as clinical indicators and their hypothesized role in cell-to-cell communication, we analyzed the impact of COVID-19 severity on a panel of miRNAs previously associated with or proposed as biomarkers of molecular mechanisms linked to the disease. To explore their role as clinical indicators in more detail, we also evaluated the potential of the miRNA panel to predict ICU mortality. We report two major findings: (1) severe COVID-19 induces characteristic molecular changes in the circulating miRNA profile; and (2) miRNAs, particularly a signature composed of miR-192-5p and miR-323a-3p, are relevant predictors of patient outcome in the clinically severe phase.
Characterization of the circulating miRNA pattern in plasma samples from hospitalized patients admitted to the clinical wards without requiring critical care and patients admitted to the ICU showed that COVID-19 severity was associated with a specific circulating miRNA profile. In addition, our findings underscore the potential for using miRNA profiling to guide patient care. In ROC analysis, a 2-miRNA panel was able to discriminate between survivors and nonsurvivors with high accuracy. Interestingly, the performance of our 2-miRNA panel as a biomarker seems to be superior to contemporary laboratory tests, such as leukocyte counts, CRP and D-dimer. We also report a correlation between miRNA levels and duration of ICU stay. These results are especially relevant in the current scenario in which COVID-19 mortality is mainly concentrated in ICU patients (ranging between 25 and 50% depending on age group, sex and medical history) and the lack of robust information on the prognosis of critically ill patients.24 The use of molecular methods based on miRNAs could refine risk assessment and provide a straightforward approach to guide clinical decisions in terms of patient care, monitoring and treatment.
The miRNA profiles could inform molecular pathways implicated in disease severity, suggesting targets for therapeutic guidance. Previous data on the mechanisms regulated by the miRNA panel, together with correlation analysis, suggest a possible role in the disease. miRNAs for which significant alterations were observed are implicated in the regulation of the immune and/or inflammatory pathways at different levels: cytokine and chemokine synthesis (miR-16-5p, miR-192-5p, miR-451a), T cell development, differentiation and activation (miR-17-92 cluster, miR-27), or B-cell development, differentiation and activation (miR-17-92 cluster, miR-150-5p), among others.25, 26, 27 In the context of viral acute respiratory infections, viruses have been reported to induce alterations in the miRNA expression profile in cells of the respiratory tract, including the downregulation of miR-192-5p in human metapneumovirus infection.28 Pen et al. 29 demonstrated that expressed miRNAs in respiratory epithelial cells, such as miR-486-5p, participate in antiviral mechanisms against influenza A viruses. These results are in agreement with those from Song et al., 30 who suggested that miR-323a-3p inhibits replication of the H1N1 influenza A virus targeting the PB1 gene. Interestingly, some of the dysregulated candidates have been proposed as host miRNA targets in the viral genome of SARS-CoV. This is the case for miR-148a-3p, which targets the ORF1a, E, S and M genes.31 The same authors reported a downregulation of miR-98 induced by viral components, which facilitates the onset of infection.31 Both miRNAs have also been predicted to bind SARS-CoV-2-encoded transcripts.32 A direct association between our miRNA candidates and lung damage has been reported. miR-27a-3p has been described as a negative regulator of lung fibrosis by inhibiting the differentiation of lung fibroblasts into myofibroblasts.33 Low plasma levels of miR-92a-3p have been found in patients with pulmonary fibrosis.34 Depletion of miR-16-5p levels upon admission due to community-acquired pneumonia is a predictor of 30-day mortality.35 Furthermore, miR-486-5p promotes acute lung injury by inducing inflammation by targeting the gene OTUD7B.36 Other pathological mechanisms may also be implicated. Plasma levels of miR-192-5p have been recently associated with venous thrombosis.37 Nevertheless, we are aware that this discussion should be carried out with caution. Due to the experimental design used, causal relations could not be drawn from our data. Mechanistic in vitro and in vivo and/or Mendelian randomization studies are necessary to elucidate the extent to which the changes in the circulating miRNA profile are translated into changes in the corresponding biological pathways. It also remains unclear whether changes in circulating miRNA levels are mediators or consequences of the pathological mechanisms linked to COVID-19. The secretion and transport of circulating miRNAs is not well understood.38 Moreover, the identification of the cellular origin is challenging: (1) miRNAs appear in the bloodstream by active secretion or passive release during cell damage; (2) dysregulated miRNAs have been described in a wide variety of tissues;39 and (3) the extracellular miRNA pattern does not necessarily correlate with the intracellular miRNA expression pattern.40 Additionally, the identified miRNA signatures were constructed using automated methods that limit causal inferences. However, from the clinical perspective, these points are not critical for clinical application. A biomarker does not need to provide mechanistic information if it is useful for decision-making.
Comparison of our results with those of other authors is hampered by the paucity of studies that have evaluated the potential clinical translation of the circulating miRNA signature for the management of COVID-19 patients. Our data are consistent with those from Li et al.,41 who reported differential miRNA expression (35 upregulated/38 downregulated) in peripheral blood samples from COVID-19 patients compared to healthy controls. Using multi‐transcriptome sequencing of red blood cell‐depleted whole blood, Tang et al. 42 demonstrated a reduction in the levels of miR‐21‐5p, miR‐142‐3p and miR‐146a‐5p in severe COVID-19 patients compared to moderate COVID-19 patients. Sabbatinelli et al. 43 showed that COVID-19 patients who did not respond to tocilizumab had lower serum concentrations of miR-146a-5p after treatment and that in this patient subgroup, those with lower levels of the miRNA were at high risk of adverse outcomes. The results are also in agreement with those from Garg et al.,44 who demonstrated an alteration in the serum concentration of cardiovascular disease/inflammation-relevant miRNAs in mechanically ventilated COVID-19 patients compared to healthy subjects and influenza-ARDS patients. Strikingly, although cardiac injury has been well described in COVID-19 patients,45 all cardiomyocyte-specific miRNAs (miR-208a-3p, miR-208b-3p and miR-499a-5p) included in the current study were below the limit of detection of our quantification method, even in critically ill patients with high troponin I concentrations. The differences in the study populations, sample matrix, quantification platforms and data analysis may explain these results.46
From the perspective of miRNA-based biomarker studies, our data provide interesting findings. The results support the need for multivariable approaches when evaluating miRNAs as clinical indicators. The use of miRNA signatures/panels as biomarkers makes biological sense. miRNAs orchestrate physiological and pathological responses in a coordinated mechanism. The altered expression of individual miRNAs has a limited impact on the gene expression program. Furthermore, diseases are the consequence of abnormalities in entire gene expression networks. Concerning medication use, our results address a relevant point in the field. It has been clearly demonstrated that pharmacological therapies affect the circulating miRNA profile. For instance, antiplatelet therapy administration has a direct effect on circulating levels of platelet-enriched miRNAs.23 , 47 In the context of COVID-19, tocilizumab therapy has been associated with an increase in miR-146 levels.48 Consequently, we analyzed in detail the interaction of commonly used therapeutic drugs in COVID-19 patients with miRNA candidates. The results showed an impact of the treatments on the levels of three miRNAs with possible value as biomarkers: miR-16-5p, miR-92a-3p and miR-150-5p. Future investigations should take into account the effect of pharmacological therapies on the circulating miRNA pattern in this emerging disease.
The strengths of the study include the multicentric design, the inclusion of hospitalized patients and the comparison of biomarker performance with laboratory parameters currently available to clinicians. In contrast to the comparison of extreme cases and healthy controls, which could overestimate the performance of new biomarkers due to the inadequate consideration of medications, comorbidities and other confounding factors, we included a cohort of hospitalized patients. Nevertheless, the inclusion of asymptomatic patients or patients with less severe clinical phenotypes would have been desirable to provide a comprehensive view of COVID-19 pathophysiology. Comparisons with commonly used clinical measurements are also fundamental for obtaining robust evidence on biomarker performance.
Our results should be interpreted in the context of certain limitations. First, the findings need to be confirmed in larger populations. Second, the patients were not consecutively recruited, which limits the generalizability of our findings. Investigations using a “real clinical setting” including unbiased series of patients are fundamental. Indeed, our candidates will be validated in the ongoing CIBERESUCICOVID study (www.clinicaltrials.gov, NCT04457505). Third, the saturation of critical care capacity during the first pandemic wave in Spain may have an impact on the composition of the ward and ICU groups. Nevertheless, the ward group included hospitalized patients admitted to the pneumology, infectious diseases or internal medicine wards without requiring critical care. Forth, we used a targeted rather than untargeted approach. Our intention was to evaluate the biomarker potential of circulating miRNAs rather than to perform miRNA screening. Other miRNAs may have clinical significance. Consequently, we may have underestimated the role of miRNAs as clinical indicators. Fifth, potential confounding factors cannot be discarded despite adjustment. Sixth, we cannot exclude the impact on our findings of conditions/treatments that were not recorded. Seventh, given the exploratory nature of the study, the impact of type I error should not been discarded. Furthermore, optimism corrected AUCs were not considered in the statistical analysis.
In conclusion, the severity of COVID-19 impacts the circulating miRNA profile. Plasma miRNA profiling emerges as a useful tool for risk-based patient stratification in critically ill COVID-19 patients. Additional studies in larger samples and functional approaches will be required to validate these findings and provide further insight into the role of circulating miRNAs as biomarkers and functional mediators of COVID-19.
Acknowledgments
Conflict of interest: All authors have read the journal's authorship agreement and policy on disclosure of potential conflicts of interest. The manuscript has been reviewed by and approved by all authors. FB and DdGC have filed patents on microRNAs as biomarkers. Other authors declare that they have no competing interests.
Supported by ISCIII (CIBERESUCICOVID, COV20/00110), co-funded by ERDF, “Una manera de hacer Europa”. DdGC acknowledges receiving financial support from Instituto de Salud Carlos III (ISCIII), Miguel Servet 2020 (CP20/00041), co-funded by the European Social Fund (ESF), “Investing in your future”. LP is the recipient of a predoctoral fellowship from the Ministry of Universities of Spain (FPU19/03526). This work partially supported by IRBLleida Biobank (B.0000682) and “Plataforma Biobancos PT17/0015/0027/”.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.trsl.2021.05.004.
Appendix. Supplementary materials
References
- 1.Grasselli G, Greco M, Zanella A, et al. Risk Factors Associated with Mortality among Patients with COVID-19 in Intensive Care Units in Lombardy, Italy. JAMA Intern Med. 2020;180(10):1345–1355. doi: 10.1001/jamainternmed.2020.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grasselli G, Zangrillo A, Zanella A, et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 Admitted to ICUs of the lombardy region, Italy. JAMA - J Am Med Assoc. 2020;323(16):1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta RK, Marks M, Samuels THA, et al. Systematic evaluation and external validation of 22 prognostic models among hospitalized adults with COVID-19: an observational cohort study. Eur Respir J. 2020;56(6) doi: 10.1183/13993003.03498-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen B, Yi X, Sun Y, et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182(1):59–72. doi: 10.1016/j.cell.2020.05.032. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin B, Liu J, Liu Y, Qin X. Progress in understanding COVID-19: insights from the omics approach. Crit Rev Clin Lab Sci. 2020;58(4):1–18. doi: 10.1080/10408363.2020.1851167. [DOI] [PubMed] [Google Scholar]
- 6.De Gonzalo-Calvo D, Vea A, Bär C, et al. Circulating non-coding RNAs in biomarker-guided cardiovascular therapy: a novel tool for personalized medicine? Eur Heart J. 2019;40(20):1643–1650. doi: 10.1093/eurheartj/ehy234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prattichizzo F, Matacchione G, Giuliani A, et al. Extracellular vesicle-shuttled miRNAs: a critical appraisal of their potential as nano-diagnostics and nano-therapeutics in type 2 diabetes mellitus and its cardiovascular complications. Theranostics. 2020;11(3):1031–1045. doi: 10.7150/thno.51605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalani MYS, Alsop E, Meechoovet B, et al. Extracellular microRNAs in blood differentiate between ischaemic and haemorrhagic stroke subtypes. J Extracell Vesicles. 2020;9(1) doi: 10.1080/20013078.2020.1713540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lampignano R, Kloten V, Krahn T, Schlange T. Integrating circulating miRNA analysis in the clinical management of lung cancer: present or future? Mol Aspects Med. 2020;72 doi: 10.1016/j.mam.2020.100844. [DOI] [PubMed] [Google Scholar]
- 10.Tribolet L, Kerr E, Cowled C, et al. MicroRNA biomarkers for infectious diseases: from basic research to biosensing. Front Microbiol. 2020;11:1197. doi: 10.3389/fmicb.2020.01197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walter E, Dellago H, Grillari J, Dimai HP, Hackl M. Cost-utility analysis of fracture risk assessment using microRNAs compared with standard tools and no monitoring in the Austrian female population. Bone. 2018;108:44–54. doi: 10.1016/j.bone.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 12.de Gonzalo-Calvo D, Martínez-Camblor P, Bär C, et al. Improved cardiovascular risk prediction in patients with end-stage renal disease on hemodialysis using machine learning modeling and circulating microribonucleic acids. Theranostics. 2020;10(19):8665–8676. doi: 10.7150/thno.46123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonneau E, Neveu B, Kostantin E, Tsongalis GJ, De Guire V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC. 2019;30(2):114–127. /pmc/articles/PMC6599191/?report=abstract. Accessed January 12, 2021. [PMC free article] [PubMed] [Google Scholar]
- 14.De Gonzalo-Calvo D, Dávalos A, Montero A, et al. Circulating inflammatory miRNA signature in response to different doses of aerobic exercise. J Appl Physiol. 2015;119(2):124–134. doi: 10.1152/japplphysiol.00077.2015. [DOI] [PubMed] [Google Scholar]
- 15.Mestdagh P, Hartmann N, Baeriswyl L, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (mirQC) study. Nat Methods. 2014;11(8):809–815. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- 16.Levi M, Thachil J, Iba T, Levy JH. Coagulation abnormalities and thrombosis in patients with COVID-19. Lancet Haematol. 2020;7(6):e438–e440. doi: 10.1016/S2352-3026(20)30145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.WA Al-Soud, Rådström P. Purification and characterization of PCR-inhibitory components in blood cells. J Clin Microbiol. 2001;39(2):485–493. doi: 10.1128/JCM.39.2.485-493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willems M, Moshage H, Nevens F, Fevery J, Yap SH. Plasma collected from heparinized blood is not suitable for HCV-RNA detection by conventional RT-PCR assay. J Virol Methods. 1993;42(1):127–130. doi: 10.1016/0166-0934(93)90184-s. [DOI] [PubMed] [Google Scholar]
- 19.Plieskatt JL, Feng Y, Rinaldi G, Mulvenna JP, Bethony JM, Brindley PJ. Circumventing qPCR inhibition to amplify miRNAs in plasma. Biomark Res. 2014;2(1):13. doi: 10.1186/2050-7771-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blondal T, Jensby Nielsen S, Baker A, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods. 2013;59(1):S1–S6. doi: 10.1016/j.ymeth.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 21.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willeit P, Zampetaki A, Dudek K, et al. Circulating MicroRNAs as novel biomarkers for platelet activation. Circ Res. 2013;112(4):595–600. doi: 10.1161/CIRCRESAHA.111.300539. [DOI] [PubMed] [Google Scholar]
- 23.De Boer HC, Van Solingen C, Prins J, et al. Aspirin treatment hampers the use of plasma microRNA-126 as a biomarker for the progression of vascular disease. Eur Heart J. 2013;34(44):3451–3457. doi: 10.1093/eurheartj/eht007. [DOI] [PubMed] [Google Scholar]
- 24.Torres A, Arguimbau M, Bermejo-Martín J, et al. CIBERESUCICOVID: un proyecto estratégico para una mejor comprensión y manejo clínico de la COVID-19 en pacientes críticos. Arch Bronconeumol. 2021;57:1–2. doi: 10.1016/j.arbres.2020.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29(7):343–351. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 26.O'Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30(1):295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 27.Guo YE, Riley KJ, Iwasaki A, Steitz JA. Alternative capture of noncoding RNAs or protein-coding genes by herpesviruses to alter host T cell function. Mol Cell. 2014;54(1):67–79. doi: 10.1016/j.molcel.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng J, Ptashkin RN, Wang Q, et al. Human metapneumovirus infection induces significant changes in small noncoding rna expression in airway epithelial cells. Mol Ther - Nucleic Acids. 2014;3(5):e163. doi: 10.1038/mtna.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng S, Wang J, Wei S, et al. Endogenous cellular microRNAs mediate antiviral defense against influenza A virus. Mol Ther - Nucleic Acids. 2018;10:361–375. doi: 10.1016/j.omtn.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song L, Liu H, Gao S, Jiang W, Huang W. Cellular microRNAs inhibit replication of the H1N1 influenza A Virus in infected cells. J Virol. 2010;84(17):8849–8860. doi: 10.1128/JVI.00456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallick B, Ghosh Z, Chakrabarti J. MicroRNome analysis unravels the molecular basis of SARS infection in bronchoalveolar stem cells. PLoS One. 2009;4(11):e7837. doi: 10.1371/journal.pone.0007837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ali Hosseini Rad SM, McLellan AD. Implications of SARS-COV-2 mutations for genomic RNA structure and host microRNA targeting. Int J Mol Sci. 2020;21(13):1–18. doi: 10.3390/ijms21134807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui H, Banerjee S, Xie N, et al. MicroRNA-27a-3p is a negative regulator of lung fibrosis by targeting myofibroblast differentiation. Am J Respir Cell Mol Biol. 2016;54(6):843–852. doi: 10.1165/rcmb.2015-0205OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wuttge DM, Carlsen AL, Teku G, et al. Specific autoantibody profiles and disease subgroups correlate with circulating micro-RNA in systemic sclerosis. Rheumatol (United Kingdom) 2015;54(11):2100–2107. doi: 10.1093/rheumatology/kev234. [DOI] [PubMed] [Google Scholar]
- 35.Galván-Román JM, Lancho-Sánchez Á, Luquero-Bueno S, et al. Usefulness of circulating microRNAs miR-146a and miR-16-5p as prognostic biomarkers in community-acquired pneumonia. PLoS One. 2020;15(10) doi: 10.1371/journal.pone.0240926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo Q, Zhu J, Zhang Q, Xie J, Yi C, Li T. MicroRNA-486-5p promotes acute lung injury via inducing inflammation and apoptosis by targeting OTUD7B. Inflammation. 2020;43(3):975–984. doi: 10.1007/s10753-020-01183-3. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Rius A, Lopez S, Martinez-Perez A, Souto JC, Soria JM. Identification of a Plasma MicroRNA Profile Associated with Venous Thrombosis. Arterioscler Thromb Vasc Biol. 2020;40(5):1392–1399. doi: 10.1161/ATVBAHA.120.314092. [DOI] [PubMed] [Google Scholar]
- 38.Bär C, Thum T, de Gonzalo-Calvo D. Circulating miRNAs as mediators in cell-to-cell communication. Epigenomics. 2019;11(2):111–113. doi: 10.2217/epi-2018-0183. [DOI] [PubMed] [Google Scholar]
- 39.Ludwig N, Leidinger P, Becker K, et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44(8):3865–3877. doi: 10.1093/nar/gkw116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Gonzalo-Calvo D, Cenarro A, Garlaschelli K, et al. Translating the microRNA signature of microvesicles derived from human coronary artery smooth muscle cells in patients with familial hypercholesterolemia and coronary artery disease. J Mol Cell Cardiol. 2017;106:55–67. doi: 10.1016/j.yjmcc.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 41.Li C, Hu X, Li L, hui Li J. Differential microRNA expression in the peripheral blood from human patients with COVID-19. J Clin Lab Anal. 2020;34(10):e23590. doi: 10.1002/jcla.23590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang H, Gao Y, Li Z, et al. The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19. Clin Transl Med. 2020;10(6):e200. doi: 10.1002/ctm2.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sabbatinelli J, Giuliani A, Matacchione G, et al. Decreased serum levels of the inflammaging marker miR-146a are associated with clinical non-response to tocilizumab in COVID-19 patients. Mech Ageing Dev. 2021;193 doi: 10.1016/j.mad.2020.111413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garg A, Seeliger B, Derda AA, et al. Circulating cardiovascular microRNAs in critically ill COVID-19 patients Short title: microRNA signatures in COVID-19. Eur J Heart Fail. 2021;23(3):468–475. doi: 10.1002/ejhf.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doyen D, Dupland P, Morand L, et al. Characteristics of cardiac injury in critically ill patients with coronavirus disease 2019. Chest. 2021;159(5):1974–1985. doi: 10.1016/j.chest.2020.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pinilla L, Benitez ID, González J, Torres G, Barbé F, de Gonzalo-Calvo D. Peripheral blood microRNAs and the COVID-19 patient: methodological considerations, technical challenges and practice points. RNA Biol. 2021;18(5):688–695. doi: 10.1080/15476286.2021.1885188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willeit P, Zampetaki A, Dudek K, et al. Circulating microRNAs as novel biomarkers for platelet activation. Circ Res. 2013;112(4):595–600. doi: 10.1161/CIRCRESAHA.111.300539. [DOI] [PubMed] [Google Scholar]
- 48.Sabbatinelli J, Giuliani A, Matacchione G, et al. Decreased serum levels of the inflammaging marker miR-146a are associated with non-clinical response to tocilizumab in COVID-19 patients. Mech Ageing Dev. 2021;193:111413. doi: 10.1016/j.mad.2020.111413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.