Summary
Aberrant NOTCH3 signaling and overexpression is oncogenic, associated with cancer stem cells and drug resistance, yet therapeutic targeting remains elusive. Here, we develop NOTCH3-targeted antibody drug conjugates (NOTCH3-ADCs) by bioconjugation of an auristatin microtubule inhibitor through a protease cleavable linker to two antibodies with differential abilities to inhibit signaling. The signaling inhibitory antibody rapidly induces ligand-independent receptor clustering and internalization through both caveolin and clathrin-mediated pathways. The non-inhibitory antibody also efficiently endocytoses via clathrin without inducing receptor clustering but with slower lysosomal co-localization kinetics. In addition, DLL4 ligand binding to the NOTCH3 receptor mediates transendocytosis of NOTCH3-ADCs into ligand-expressing cells. NOTCH3-ADCs internalize into receptor and ligand cells independent of signaling and induce cell death in both cell types representing an atypical mechanism of ADC cytotoxicity. Treatment of xenografts with NOTCH3-ADCs leads to sustained tumor regressions, outperforms standard-of-care chemotherapy, and allows targeting of tumors that overexpress NOTCH3 independent of signaling inhibition.
Keywords: notch signaling, NOTCH3 receptor, DLL4, JAG1, antibody drug conjugates, cancer stem cells, tumor initiating cells, transendocytosis, oncology, biotherapeutic
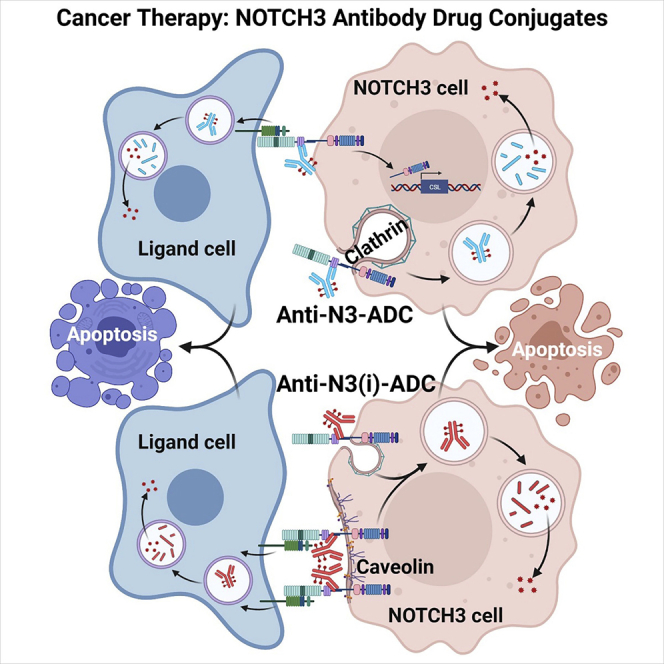
Graphical abstract
Highlights
NOTCH3 receptor is overexpressed in breast, lung, and ovarian tumors
Newly generated NOTCH3-targeted antibody drug conjugates are efficacious and safe
NOTCH3 antibodies internalize through different routes depending on signaling status
NOTCH3 antibody intercellular trafficking occurs by transendocytosis into ligand cells
Inhibition of Notch signaling in cancer is challenging due to dose-limiting, on-target toxicities associated with inhibition of its normal function. Geles et al. demonstrate that a NOTCH3-targeted antibody drug conjugate without signaling inhibition is efficacious by killing receptor and ligand expressing cells, and thus has a lower potential for side effects.
Introduction
Despite recent successes, a high unmet medical need still exists for cancer patients who are not responsive to targeted or immuno-oncology therapies and eventually succumb to metastatic disease. Cancer stem cells (CSCs) are considered a tumor progenitor cell population that drives tumor initiation, metastasis, chemotherapeutic resistance, and recurrence. Emerging studies have mainly focused on identifying subpopulations of cancer cells with tumor-initiating cell (TIC) properties and their corresponding targets for therapeutic intervention.1 Antibody drug conjugates (ADCs) are a therapeutic modality that harnesses the targeting specificity of a monoclonal antibody (mAb) in combination with potent cytotoxic payloads with the goal of improving their therapeutic indices.2 There have been several ADCs developed against TIC populations using targets such as DLL3,3,4 EFNA4,5 LGR5,6,7 PTK7,8 and TPBG (5T4).9
Oncogenic Notch signaling is associated with CSC features that are responsible for maintaining self-renewal, progenitor cell phenotypes, and TIC functions.10 Notch receptors are synthesized as precursor polypeptides that are cleaved at site 1 (S1) located between heterodimerization domain 1 (HD1) and HD2 while in the Golgi before being transported to the cell membrane. Bipartite Notch receptors are held together by non-covalent HD interactions and kept autoinhibited by the juxtamembrane negative regulatory region (NRR). Notch signaling is initiated by binding to Jagged (JAG) or Delta-like ligands (DLL) on adjacent cells. Ligand binding relieves the NRR autoinhibition thus permitting ADAM metalloproteinase cleavage at site S2 near the C-terminal region of HD2. The Notch extracellular domain (NECD) is shed and undergoes transendocytosis (TEC) into ligand-expressing cells. Gamma secretase mediates cleavage at site S3 releasing the Notch intracellular domain (NICD) from the cell membrane, which then translocates into the nucleus and activates the transcription of target genes.11,12 Upregulation and activation of Notch signaling is one mechanism by which cancer cells evade the growth inhibitory effects of hormone therapy,13, 14, 15 PI3K inhibitors,16,17 and chemotherapy.18, 19, 20 Tumors that develop with NOTCH3 overexpression or amplification are common in lung, breast, and ovarian cancer patients, and these alterations are associated with poor patient survival.21, 22, 23 In addition, NOTCH3 is highly expressed in ovarian cancer patients who have recurring disease and is correlated with worse clinical outcome.20 Triple-negative breast cancers (TNBC, ER–/PR–/HER2–) with Notch pathway alterations have particularly high levels of NOTCH3 expression.24
Anti-cancer therapies targeting the Notch pathway mainly focus on signaling inhibition.25, 26, 27, 28 However, certain Notch signaling inhibitors have undesirable side effects in patients that limit their therapeutic potential.29 Furthermore, clinical development of Notch inhibitors requires selection of patients likely to be responsive by identifying those with active Notch signaling or driver mutations.24 In contrast, the ADC therapeutic platform has the potential to enhance efficacy by delivering potent cytotoxics directly to tumors30 and may deplete them of NOTCH3-expressing tumor cells as well as adjacent ligand cells. In the current study, NOTCH3-targeted ADCs (NOTCH3-ADCs) were generated with mAbs that were classified as being signaling inhibitory or non-inhibitory. We demonstrate that efficacy can be achieved by utilizing a non-inhibitory mAb-based ADC to avoid potential on-target toxicities associated with the inhibition of Notch signaling. Unexpectedly, NOTCH3-ADCs were transendocytosed into DLL4 cells even in the absence of signaling and induced cell death. NOTCH3-ADCs exhibited robust anti-tumor activity in preclinical studies and induce tumor regressions independent of their ability to inhibit signaling.
Results
NOTCH3 is overexpressed in multiple human tumors
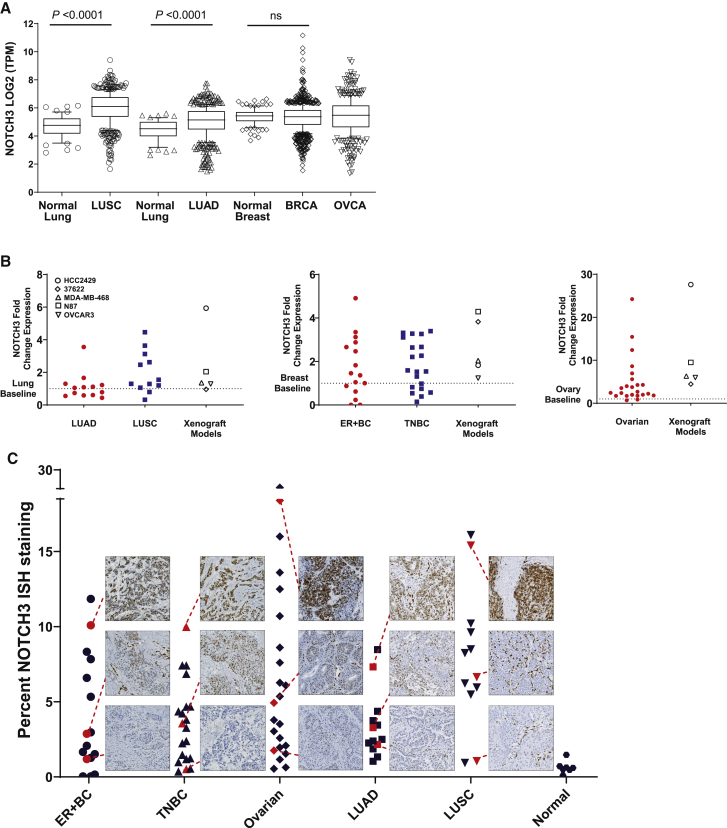
Gene-expression analysis of The Cancer Genome Atlas (TCGA) databases31, 32, 33 revealed that NOTCH3 expression is significantly upregulated in non-small cell lung cancer (NSCLC) for both lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) compared to normal lung tissue (p < 0.0001; Figure 1A). Additionally, a subset of breast and ovarian cancer (BRCA, OVCA, respectively) samples have high expression within the 90th percentile compared to normal or median levels (Figure 1A).
Figure 1.
NOTCH3 is overexpressed in multiple human tumors
(A) Expression of NOTCH3 mRNA in primary human tumors from TCGA compared to normal tissues. Box and whiskers plots are drawn with individual points below 10th and above 90th percentiles. Median values are drawn as a line in the middle of the box. TPM, transcripts per kilobase million, ns, non-significant.
(B) NOTCH3 mRNA fold change in primary human lung, breast, and ovarian tumors and xenografts compared to normal tissues (baseline, dashed line). Data represent mean (n = 2–4).
(C) Quantitation of NOTCH3 ISH staining as a measure of percentage of area stained on breast (ER+, TNBC), lung (LUAD, LUSC), ovarian, and normal (breast, lung, and ovary) tissue sections.
To confirm TCGA data in an independent set of tumor samples, NOTCH3 mRNA levels from lung, breast, and ovarian tumors were analyzed by qRT-PCR and compared to preclinical xenograft models as well as normal lung, breast, and ovarian tissues (the baseline). As demonstrated, there are tumor samples that have NOTCH3 mRNA levels at ∼2–5 times higher than baseline for lung cancer (LUAD and LUSC), ∼2 times higher than baseline for breast cancer (ER+ and ER–/PR–/HER2–), and from ∼2 to 25 times higher than baseline for ovarian cancer (Figure 1B). These data also demonstrate that the 37622 LUSC patient-derived xenograft (PDX)34 and cell line-derived xenograft (CLX) models (HCC2429 lung cancer cell line, MDA-MB-468 TNBC cell line, OVCAR3 ovarian cancer cell line, NCI-N87 gastric cancer cell line) selected for further study recapitulate a range in the levels of NOTCH3 mRNA seen in primary human tumor samples (Figure 1B). The HCC2429 CLX model had the highest levels of NOTCH3 mRNA overexpression, while the other xenograft models had low to moderate expression of NOTCH3 mRNA (Figure 1B).
An in situ hybridization (ISH) assay and image-based analysis was developed to quantitate NOTCH3 expression in the same set of tumor samples that were analyzed by qRT-PCR. Using the ISH technique, tumor tissues from 25 lung, 36 breast, and 23 ovarian cancers along with normal tissues from 3 lung, 3 breast, and 1 ovary were stained and quantified for the percentage of NOTCH3 positive ISH staining in the tissue sections (Figure 1C). Each tumor set was separated into tertiles defined as low, moderate, and high NOTCH3 expression based on ISH staining values and represented images are shown (Figures 1C and S1A). In addition, the ISH assay developed correlated with qRT-PCR from the same samples, demonstrating specificity and the ability to quantify a range of NOTCH3 expression (Figure S1B). For each tumor set, a range in the distribution of NOTCH3 levels was observed suggesting good biological representation of NOTCH3 expression in the three tumor cohorts evaluated.
Generation and characterization of therapeutic anti-NOTCH3 mAbs
The NOTCH3 NRR domain was chosen to generate therapeutic mAbs since they would not interfere with ligand binding but have the potential to inhibit signaling26,35 (Figure S2A). A screening strategy was designed to identify mAbs that would specifically bind to the NRR and either stabilize it in an auto-inhibited conformation, and thus termed signaling inhibitory, or permit S2 cleavage, and thus termed non-signaling inhibitory.
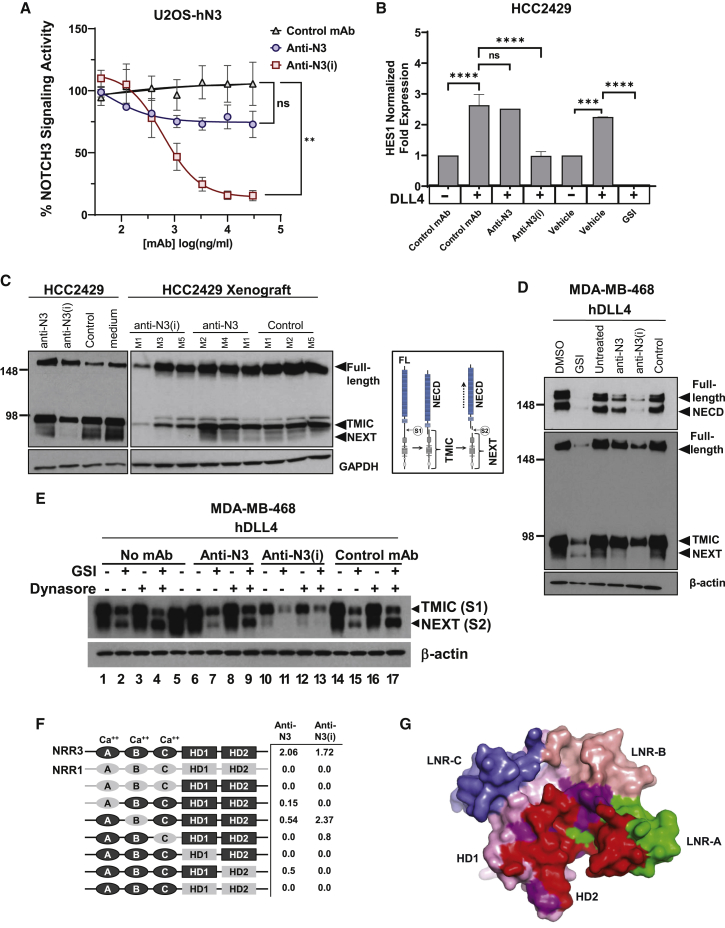
Ligand-dependent NOTCH3 signaling was measured by activation of a Notch-responsive promoter containing canonical CSL (CBF-1/Suppressor of Hairless/Lag-1) binding sites driving the expression of a luciferase (Luc) reporter gene in U2OS cells expressing human NOTCH3 (U2OS-hN3). U2OS-hN3 Luc-reporter cells were incubated with a titration of NOTCH3-targeted or control mAbs and then added to HEK293 cells expressing DLL4 (HEK-DLL4). Two lead mAbs were identified and termed anti-NOTCH3 inhibitory (anti-N3(i)) clone 75 and anti-NOTCH3 (anti-N3) clone 28. The anti-N3(i) mAb demonstrated potent signaling inhibition in a dose-dependent manner that was statistically significant, while the anti-N3 mAb did not block ligand-stimulated NOTCH3 signaling, similar to a non-targeted isotype control mAb (Figure 2A). Anti-N3(i) inhibited Notch-dependent reporter gene activity by ∼95%. A γ-secretase inhibitor (GSI), which inhibits signaling from any endogenous Notch receptor was used as a control and completely inhibited activation of the reporter gene (Figure S2B).
Figure 2.
Generation and characterization of therapeutic anti-NOTCH3 mAbs
(A) NOTCH3-dependent report gene assay with NOTCH3-targeted and control mAbs. Data represent mean ± SEM from 3 biological replicates (n = 4 per replicate), ns, non-significant.
(B) qRT-PCR of HES1 gene expression in treated cells. Data represent mean ± SD from 2 biological replicates (n = 3 per replicate), ns, non-significant.
(C) NOTCH3 immunoblot from mAb-treated cells and xenografts. GAPDH is shown as a loading control. Schematic diagram of NOTCH3 cleavage events and protein fragments. M, mouse number.
(D and E) Immunoblot using N- or C-terminal domain antibodies to detect NOTCH3 fragments after DLL4 activation in treated cells. β-actin is shown as a loading control.
(F) Epitope mapping of anti-NOTCH3 mAbs using NRR3-NRR1 domain swap chimeric constructs. NRR3 domains are shown in black and NRR1 domains are shown in gray. Representative data represent mean (n = 2).
(G) Binding interface of anti-N3(i) Fab on the NOTCH3-NRR domain as determined from the co-crystal structure is shown in red, LNR-A (green), LNR-B (beige), LNR-C (blue), HD1 (pink), and HD2 (magenta).
Additional characterization of NOTCH3-targeted mAbs demonstrated that only anti-N3 was cross-reactive to mouse NOTCH3 (mN3) expressed on U2OS cells (U2OS-mN3) (Figure S2C). Furthermore, anti-N3 had a greater saturation level of binding to the human NOTCH3 receptor than anti-N3(i) (Figure S2C) and ∼10-fold higher affinity to human NRR than anti-N3(i) (Figure S2D). Notch receptor specificity of lead mAbs was tested against U2OS cells expressing the human or mouse NOTCH1 receptor (U2OS-hN1 or USOS-mN1, respectively). No significant binding to NOTCH1-expressing U2OS cells was detected with either mAb (Figure S2E).
The effects of both anti-NOTCH3 mAbs on endogenous Notch-target gene expression was examined in HCC2429 lung cancer cells that overexpress NOTCH3.36, 37 Although HCC2429 cells have endogenous expression of JAG1, they were plated on recombinant DLL4 to further enhance activation of Notch signaling in the presence or absence of the anti-NOTCH3 mAbs and the GSI PF-0308401438 as a positive control for signaling inhibition. In the presence of DLL4, HES1 mRNA expression was increased by approximately 3-fold (Figure 2B). Anti-N3(i), but not anti-N3 or a control mAb, prevented DLL4-activated HES1 expression (Figure 2B). The GSI, a pan-Notch inhibitor, completely abolished HES1 expression (Figure 2B).
To further characterize mAb effects on proteolytic cleavage events during NOTCH3 signaling, MDA-MB-468 breast cancer and HCC2429 lung cancer cells were treated with anti-NOTCH3 mAbs and analyzed by immunoblot analysis. HCC2429 cells express endogenous JAG1 ligand37 and MDA-MB-468 cells also have low levels of JAG1 though both are reported to activate NOTCH3 signaling by ligand-independent mechanisms.39, 40, 41 A diagram of NOTCH3 cleavage events and resulting fragments is shown for reference (Figure 2C). We detected basal activation of NOTCH3 in HCC2429 and MDA-MB-468 cells as previously described39,40 (Figures 2C and S2F). Similar analyses were performed on HCC2429 xenografts generated in mice that were dosed with anti-NOTCH3 mAbs (Figure 2C). These data demonstrate that the anti-N3(i) mAb prevents S2 cleavage since the Notch extracellular truncation (NEXT) product was not detected (Figures 2C and S2F). As expected, S1 cleavage was not affected and the transmembrane intracellular (TMIC) domain along with full-length NOTCH3 was detected although at reduced levels. In control and anti-N3 mAb-treated cells and xenografts, the expected S1- and S2-proteolytic cleavage events were detected, indicating the absence of an inhibitory effect (Figures 2C and S2F).
When MDA-MB-468 cells are plated on DLL4-coated plates to enhance activated NOTCH3 signaling, the addition of the anti-N3(i) mAb resulted in a downregulation of NOTCH3 at the protein level similar to GSI-treated cells (Figure 2D). Again, the NEXT fragment was not detected in anti-N3(i) mAb-treated cells but present after GSI and anti-N3 treatments (Figure 2D). Cells that were treated with the anti-N3 mAb showed NOTCH3 protein levels more comparable to control-treated cells (Figure 2D).
In the presence of the anti-N3 mAb, we observed a decrease in reporter gene activity that was not statistically significant from control antibody and detected lower levels of the NEXT fragment by immunoblot analysis. Thus, we wanted to determine whether the anti-N3 mAb may be a partial antagonist or mediates internalization and downregulation of the receptor. We performed immunoblot analysis on anti-NOTCH3 mAb-treated MDA-MB-468 cells that were plated on DLL4 in the presence of GSI and Dynasore, a small-molecule dynamin inhibitor that blocks clathrin-mediated endocytosis (CME) (Figure 2E). When combined, these two treatments should prevent S3 cleavage and trafficking/degradation of the NEXT fragment, respectively. By comparing no mAb-treated (lane 4) and control mAb-treated cells (lane 17), we do not detect a change in the levels of the NEXT fragment in the presence of anti-N3 (lane 9) (Figure 2E). In contrast, the NEXT fragment is not detected in the anti-N3(i) mAb-treated cells (lane 13) as expected (Figure 2E). These results suggest that the anti-N3 mAb is not a partial antagonist but instead may mediate receptor internalization and degradation.
Epitope mapping was conducted to assess anti-NOTCH3 mAb binding to specific NRR sub-domains. As both anti-NOTCH3 mAbs lacked cross-reactivity with the NOTCH1 protein, domain swap chimeric constructs between the NOTCH1- and NOTCH3-NRR domains were generated. Relative binding of anti-N3 and anti-N3(i) to these chimeric fusion proteins was assessed by ELISA (Figure 2F). Anti-N3 interacted only with the C terminus of the N-terminal half of the NOTCH3-NRR heterodimer, more specifically, the Lin12-Notch repeat C (LNR-C) and HD1 domains, while anti-N3(i) interacts with both halves of the NRR heterodimer (LNR-A and both HD domains) (Figures 2F and S3A). Additionally, anti-N3(i) effectively competed against anti-N3 for binding to NOTCH3 expressed on U2OS-hN3 cells and thus the two antibodies bin together (Figure S3B). As the epitope mapping data demonstrated, the two antibodies both bind to the HD1 subdomain, which likely precluded their simultaneous binding to target NOTCH3 (Figures 2F and S3A). These findings are consistent with the two mAbs’ non-inhibitory or signaling inhibitory features.
To further understand anti-NOTCH3 mAb binding, we solved the co-crystal structure of the Fab domain of anti-N3(i) mAb in association with the human NOTCH3 NRR fragment (Table S1). The epitope for this mAb covers portions of both the HD domains and LNR-A with the anti-N3(i) light chain interacting mostly with HD1 and HD2 and the heavy chain primarily with LNR-A (shown in red in Figures 2G and S3C). The S2 cleavage site, near the C terminus of HD2, is directly adjacent to the epitope. Binding of anti-N3(i) would therefore be expected to stabilize the compact conformation of the NRR in which the interaction between the LNR domains and HD2 protects the S2 cleavage site from proteolysis. Using the X-ray structure of human NOTCH3-NRR in the co-crystal complex with anti-N3(i), we mapped the expected epitope for anti-N3 based on the epitope mapping described above. The binding epitope for anti-N3 encompasses the LNR-C and the neighboring HD-1 domain (Figure S3D). Consistent with the lack of signaling inhibition activity of anti-N3, no stabilization of the NRR heterodimer would be expected from this predicted structural view. Thus, specific binding epitopes within the NRR for both mAbs could be identified and showed differential domain requirements for binding to the NOTCH3-NRR, and consistent with their respective inhibitory and non-inhibitory functions.
Cell-membrane distribution of Anti-NOTCH3 mAbs depends on signaling status
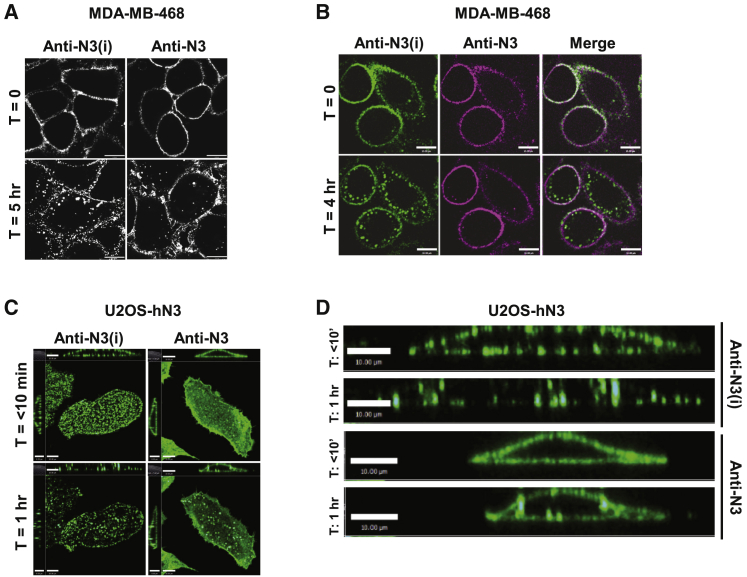
To determine whether binding of the anti-NOTCH3 mAbs to their respective epitopes on the NRR would alter the cell-membrane distribution or internalization of NOTCH3, we performed pulse-chase analysis and examined the subcellular localization of anti-N3 and anti-N3(i) in MDA-MB-468 cells by confocal microscopy. At time 0 h, staining of both mAbs appeared to be uniformly distributed across the cell membrane (Figure 3A). After 5 h at 37°C, anti-N3(i) engagement with the NOTCH3 resulted in clustering and more rapid internalization as visualized by the enhanced appearance of puncta at the cell membrane and within the cytoplasm. At the same 5 h time point, the anti-N3 mAb remained more uniformly distributed across the cell membrane (Figure 3A).
Figure 3.
Cell-membrane distribution of anti-NOTCH3 mAbs depends on signaling status
(A) Confocal images of anti-NOTCH3 mAbs on fixed cells. Scale bar, 10 μm.
(B) Single optical sections from live-cell confocal imaging of anti-NOTCH3 mAbs simultaneously bound to cells and imaged over time with anti-N3(i)-Alexa 488 (green) and anti-N3-DyLight647 (magenta). Scale bar, 10 μm.
(C) z stack of maximum intensity projections from live-cell confocal imaging of anti-NOTCH3 mAbs over time. Scale bar, left = 8 μm, right = 10 μm.
(D) Cross-section of cells bound by NOTCH3-targeted mAbs from images in Figure 2C. Scale bar, 10 μm.
The kinetics of clustering and internalization within the same cell was examined by live-cell imaging with anti-N3 and anti-N3(i) mAbs that were directly labeled with different fluorescent dyes and then both were bound to MDA-MB-468 cells (Figures 3B and S4A). At time 0 h, both anti-NOTCH3 mAbs appeared to have a uniform cell-membrane distribution. By 4 h, large membrane and intracellular punctate-like structures of anti-N3(i) (green) were observed, while few anti-N3-containing puncta (magenta) were detected. Antibody binding to the cell surface was diminished after NOTCH3-small interfering RNA (siRNA) knockdown in MDA-MB-468 cells confirming that both mAbs specifically bound to the NOTCH3 (Figures S4B and S4C).
To confirm the cell-membrane distribution and internalization of anti-NOTCH3 mAbs in another cell type, we used U2OS-hN3 cells because they have a large cytoplasmic volume. The kinetics of antibody internalization was conducted by live-cell imaging of anti-N3 and anti-N3(i) mAbs that were directly labeled with the same fluorescent dye and bound separately to USO2-hN3 cells. Within the first 10 min of binding, anti-N3(i) already appeared clustered as demonstrated by puncta formation, and by 1 h internalized as evident by reduced cell-membrane staining, while anti-N3 had a more uniform distribution across the cell surface similar to the results obtained with the MDA-MB-468 cells (Figure 3C). The differential cell-membrane distribution and clustering of the two anti-NOTCH3 mAbs are also shown in cross-section (Figure 3D). Taken together, these data indicate that at early time points the anti-N3(i) mAb internalized more rapidly than anti-N3 potentially as a consequence of receptor clustering.
Anti-NOTCH3 mAbs have differential rates of endocytosis and lysosomal trafficking
Inactive Notch receptors are constitutively internalized and degraded in the lysosome or recycled back to the membrane.42 We next wanted to determine the endocytic routes of internalization and lysosomal trafficking that would be necessary for efficient payload release from an ADC. Live-cell imaging was performed on MDA-MB-468 cells labeled with pHrodo Red dextran to identify low pH lysosomal compartments and anti-N3(i) or anti-N3 mAbs that were directly labeled (Figure S5A). The Pearson coefficient of correlation (PCC) and Manders colocalization coefficient (MCC) were calculated over time for both mAbs and pHrodo Red dextran compartments (Figure 4A). As seen with both quantitative measurements, over a 16 h time period there was more rapid and robust targeting of anti-N3(i) to lysosome compartments compared to anti-N3. When imaging is extended to 24 and 48 h, there is an increase in the amount of each mAb seen correlated with lysosomes, but there is no statistically significant difference between anti-N3(i) and anti-N3 (p > 0.05, Figure 4B). These data indicate that, although anti-N3(i) is more rapidly internalized and targeted to lysosomal compartments, over longer time periods (24–48 h) there is no distinguishable difference between the amount of anti-N3(i) and anti-N3 localized to lysosomes. This indicated that both mAbs are amenable to delivering cytotoxic payloads to the lysosome.
Figure 4.
Anti-NOTCH3 mAbs have differential rates of endocytosis and lysosomal trafficking
(A and B) Colocalization of mAbs with pHrodo Red dextran compartments calculated by PCC and MCC from live-cell confocal images. (A) Data represent mean ± SEM of 3 biological replicates (n = 3 fields imaged per replicate). (B) Data represent mean ± SEM of 2 biological replicates (n = 2 fields imaged per replicate), ns, non-significant.
(C) Colocalization of mAbs with CT-β were calculated by PCC and MCC from live-cell confocal images. Data represent mean ± SEM of 2 biological replicates (n = 5 fields imaged per replicate).
(D) Single optical sections of mAbs with CT-β from live-cell confocal imaging. Scale bar, 10 μm.
(E) Colocalization of mAbs with CAV-1 or clathrin were calculated by MCC in fixed cells. Data represent mean ± SEM (n = 3 fields imaged).
(F) Colocalization of mAbs with pHrodo Red dextran compartments were calculated by MCC from live-cell confocal images of MβCD-treated cells. Cross hairs were set at a value of 0.2 MCC to highlight more rapid colocalization of both mAbs with pHrodo Red dextran compartments. Data represent mean ± SEM of 2 biological replicates (n = 5 fields imaged per replicate).
Due to the differences observed in early internalization of the two anti-NOTCH3 mAbs, we hypothesized that this could be attributed to the mAb-receptor complexes utilizing different endocytic routes. MDA-MB-468 cells were incubated with labeled anti-N3(i) or anti-N3 mAb and labeled cholera toxin-beta subunit (CT-β), which selectively binds to lipid rafts on the cell membrane and internalizes via caveolae endocytosis (Figure S5B). Over time, the anti-N3(i) remains co-localized with CT-β, which was measured using the PCC and MCC, while the anti-N3 mAb was initially co-localized with CT-β at the membrane but this co-localization is lost over time (Figures 4C and 4D). This observation is due to anti-N3 remaining on the cell membrane over time, while CT-β is internalized along with anti-N3(i). The CT-β and anti-N3(i) then co-localize in vesicles inside the cell. These data demonstrate that anti-N3(i) is internalized, at least in part, at lipid rafts, while there is minimal internalization of anti-N3 at these membrane compartments.
Next, the co-localization of the anti-NOTCH3 mAbs with either clathrin or caveolin-1 (CAV-1) was directly measured to investigate internalization through clathrin-mediated endocytosis (CME) or CAV-1, respectively. MDA-MB-468 cells were incubated with anti-N3(i) or anti-N3 and then fixed at 0, 3, 8, and 24 h. Indirect immunofluorescence was performed with anti-CAV-1 or anti-clathrin antibodies and MCC values were calculated. Over time, anti-N3(i) co-localized with both CAV-1 and clathrin having similar MCC values for both colocalizations (Figure 4E). However, anti-N3 had a higher MCC colocalization value for clathrin than CAV-1 (Figure 4E). Additionally, anti-N3(i) had higher MCC colocalization values with CAV-1 than anti-N3 that were statistically significant at all time points after the 0 h, but not statistically significant from anti-N3 colocalization with clathrin (Figure S5C). These data indicate that anti-N3 is primarily internalized via CME, while anti-N3(i) has the ability to be internalized via caveolae and CME. Methyl-β-cyclodextrin (MβCD) was used for acute cholesterol depletion to inhibit lipid-raft dependent internalization of caveolae and clathrin-independent endocytosis (CIE). Upon MβCD treatment, the internalization of both anti-N3 and anti-N3(i) mAbs occurs at a faster rate and is more robust (Figure 4F; Figure S5D), but the mechanism for the increased rate and robustness of mAb internalization when caveolae and CIE are blocked remains unclear. However, these data suggest that when lipid-raft associated endocytic machinery is inhibited, both mAbs can still undergo internalization utilizing the efficient CME.
TEC of anti-NOTCH3 mAbs into DLL4 ligand cells
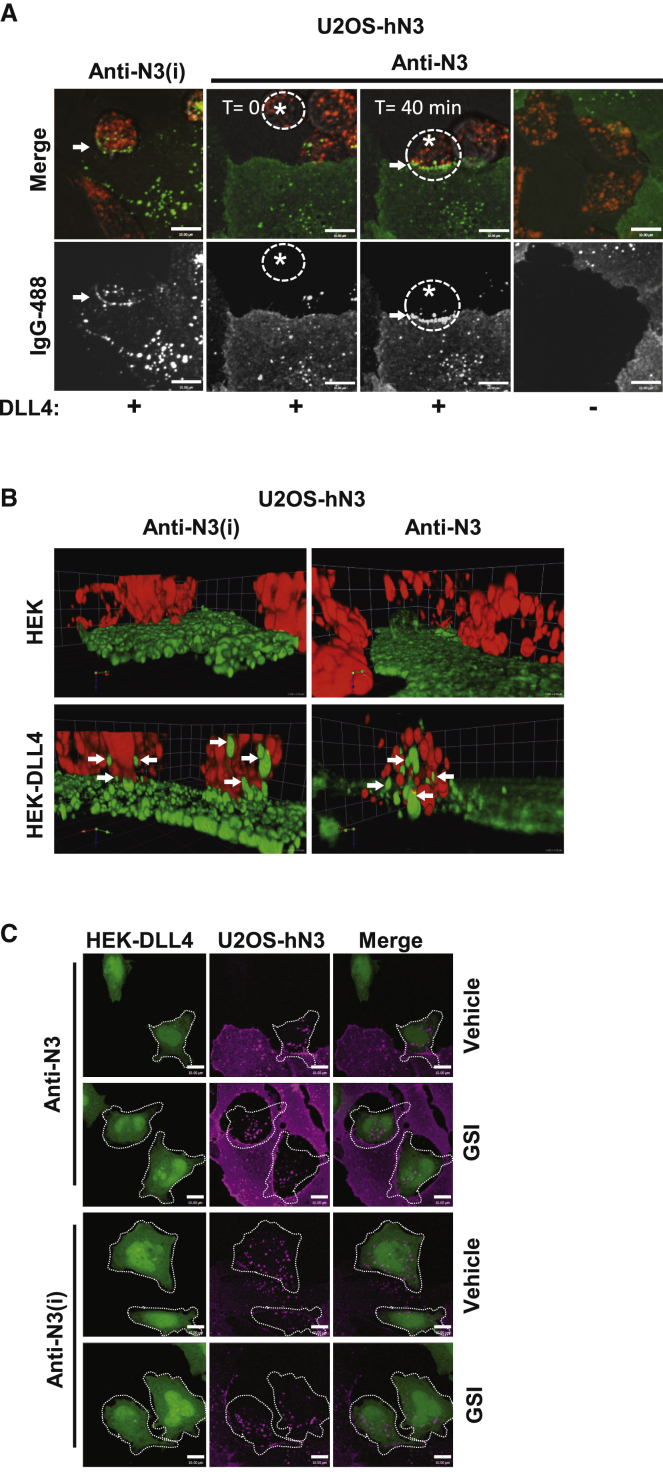
Previous studies indicate that TEC of the NECD was dependent on S1 cleavage and the formation of a heterodimeric Notch receptor, but not on ADAM proteolysis at S2.43,44 The anti-N3(i) mAb stabilizes the NRR in an autoinhibited conformation through interactions with both halves of the S1-cleaved HD domain and LNR-A. To investigate whether mAb binding to NOTCH3 affected receptor clustering and TEC in the presence of ligand, live-cell imaging was performed on co-cultures of U2OS-hN3 and HEK-DLL4 cells. Labeled anti-NOTCH3 mAbs were first bound to U2OS-hN3 cells and then excess mAb was removed by washing. HEK-DLL4 or HEK-control cells were labeled with pHrodo Red dextran and then added to the anti-NOTCH3 mAb-bound U2OS-hN3 cells, and live-cell imaging was performed (Figure 5A). Anti-N3(i) clusters were observed in the presence of DLL4 cells at sites of intercellular contact (arrow) and intracellular anti-N3(i) puncta (green in merged image) were also observed in both cell types. Prior to HEK-DLL4 cell contact with anti-N3-bound U2OS-hN3 cells (T = 0, asterisk), minimal membrane clustering was observed (Figure 5A; Figure S6A). When a migrating HEK-DLL4 cell contacted the anti-N3-bound U2OS-hN3 cell (T = 40 min, asterisk), anti-N3 now appeared to be clustered similar to anti-N3(i) (Figure 5A). Both anti-N3(i) and anti-N3 puncta (green in merged images) can be observed in adjacent HEK-DLL4 cells after binding mAb-bound U2OS-hN3 cells suggesting TEC into the ligand cells. The HEK-control cells without DLL4 ligand had no effect on NOTCH3 receptor clustering or internalization for either mAb (DLL4(–), right column, Figure 5A).
Figure 5.
Anti-NOTCH3 mAbs transendocytose into DLL4 ligand cells
(A and B) TEC of labeled mAbs (green) that were bound to U2OS-hN3 cells and then co-cultured with HEK-DLL4 or HEK-parental cells (red) from live-cell confocal imaging. (A) Single optical sections of a U2OS-hN3 cell and a migrating HEK-DLL4 cell (asterisk) before and after contact (arrows). Scale bar, 10 μm.
(B) z stack of maximum intensity projections. Arrows, anti-NOTCH3 mAbs inside HEK-DLL4 cells.
(C) Confocal images of maximum intensity projections acquired from indirect immunofluorescence of mAbs bound to U2OS-hN3 cells (magenta) and then co-cultured with HEK-DLL4 (green) from. Dashed white line demarcates anti-NOTCH3 mAbs inside HEK-DLL4 cells. Scale bar, 10 μm.
As shown in the 3D projections generated from live-cell imaging, the flat, green-labeled U2OS-hN3 cells co-cultured with round, red-labeled HEK-DLL4 cells on top demonstrates that the ligand cells contain the anti-NOTCH3 mAbs (arrows in bottom panels), again regardless of the signaling status of NOTCH3, while the HEK-control cells fail to transendocytose the anti-NOTCH3 mAbs due to their lack of ligand expression (Figure 5B, Videos S1, S2, S3, and S4). These results suggest that anti-N3(i) clusters the NOTCH3 receptor in the presence or absence of ligand cells resulting in rapid internalization and TEC but loss of NOTCH3 signaling. In contrast, anti-N3 did not efficiently cluster the NOTCH3 receptor in the absence of ligand but did allow clustering and TEC upon ligand binding and activation of NOTCH3 signaling.
Live cell imaging of TEC with anti-N3 mAb (green) bound to U20S-hN3 cells and co-cultured with HEK-DLL4 cells (red). Scale bar, 20 μm.
Live cell imaging of TEC with anti-N3(i) mAb (green) bound to U20S-hN3 cells and co-cultured with HEK-DLL4 cells (red). Scale bar = 20 μm.
Control live cell imaging of anti-N3 mAb (green) bound to U20S-hN3 cells and co-cultured with HEK-parental cells (red). Scale bar = 20 μm.
Control live cell imaging of anti-N3(i) mAb (green) bound to U20S-hN3 cells and co-cultured with HEK-parental cells (red). Scale bar = 20 μm.
To confirm the live-cell imaging results of anti-NOTCH3 mAb TEC, images were acquired from fixed co-cultures of U2OS-hN3 cells that were bound by the anti-N3 mAb and HEK-DLL4 or HEK-control cells that were labeled with MitoTracker Deep Red. Following incubation from 0.5 to 3 h, cells were fixed, and then indirect immunofluorescence was performed to detect the anti-N3 mAb. Again, TEC of the anti-N3 mAb can be observed only in HEK-DLL4 cells consistent with the live-cell imaging results (Figure S6B).
Since anti-N3(i) undergoes TEC into ligand cells despite stabilizing the NRR in an autoinhibited conformation and inhibiting signaling, we next examined the effects of simultaneously inhibiting both S2 and S3 cleavage events. To test simultaneous S2/S3 inhibition, anti-N3 or anti-N3(i) mAbs were bound to U2OS-hN3 cells and added to HEK-DLL4 cells in the presence or absence of GSI (Figure 5C). Surprisingly, regardless of NOTCH3 signaling status and ability of the anti-N3(i) mAb to stabilize the NRR in an inactive conformation, we still observed TEC of NOTCH3-targeted mAbs into the HEK-DLL4 cells (Figure 5C).
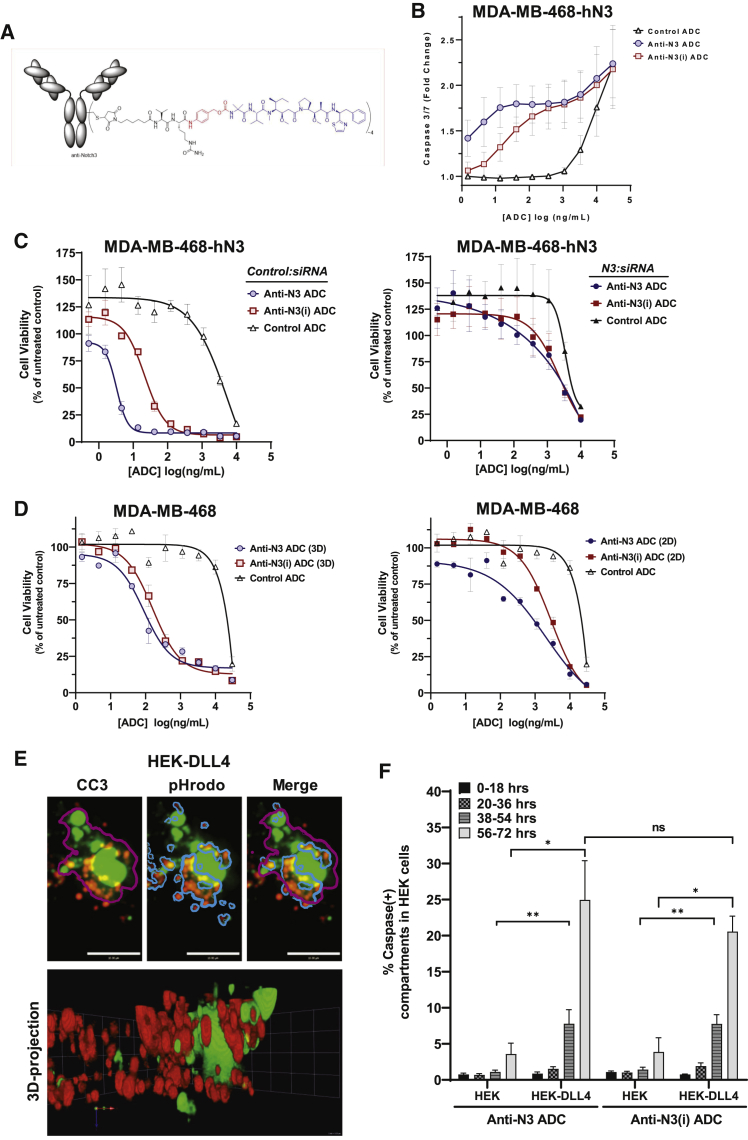
NOTCH3-ADCs induce cytotoxicity in both receptor and ligand cells
NOTCH3-ADCs were generated by bioconjugation to the Aur0101 payload, an auristatin microtubule inhibitor,45 via a protease cleavable valine-citrulline-based linker (mc-ValCitPABC or “vc”) to native cysteine residues (Figure 6A). Bioconjugation did not alter the cell-surface binding characteristics of either anti-NOTCH3 mAb (Figure S7A) and resulted in an average drug-to-antibody ratio (DAR) of 3.9 for anti-N3 ADC (anti-N3-vc0101) and DAR of 3.8 for anti-N3(i) ADC (anti-N3(i)-vc0101) (Figure S7B). Both the anti-N3 and anti-N3(i) ADCs induced an increase in caspase-3/7 activity in MDA-MB-468 cells overexpressing human NOTCH3 (MDA-MB-468-hN3) at 48 h, indicating cytotoxic activity of the ADCs (Figure 6B). To confirm the expected ADC mechanism of action, OVCAR3 ovarian cancer cells were treated with anti-N3 ADC and then subjected to immunofluorescence analysis by staining with an anti-α-tubulin antibody to mark microtubules and DAPI to stain DNA. Anti-N3 ADC treatment resulted in a disruption of microtubule structures, an abnormal chromosome configuration and cell-cycle arrest (Figure S7C). The result was similar for the free Aur0101 payload and thus implies that the anti-N3 ADC was conferring cytotoxicity by the same mechanism as the payload itself. In contrast, the control ADC and anti-N3 mAb did not result in altered microtubule or chromosome morphology (Figure S7C). This mechanism required cell binding through the NOTCH3 target since the control ADC did not exhibit the same effect and demonstrates that the anti-N3 ADC inhibits cell proliferation by disrupting microtubules.
Figure 6.
NOTCH3-targeted ADCs induce cytotoxicity in both receptor and ligand cells
(A) General structure of NOTCH3-targeted ADCs that were generated with mAbs, a cleavable dipeptide-based linker and the Aur0101 payload (blue).
(B) NOTCH3-ADC induction of caspase-3/7 activity. Data represent mean ± SEM of 3 biological replicates (n = 3 per replicate).
(C) In vitro cytotoxicity of NOTCH3-ADCs after control (Control:siRNA) or siRNA knockdown of NOTCH3 mRNA (N3:siRNA). Data represent mean ± SEM of 2 biological replicates (n = 3 per replicate).
(D) In vitro cytotoxicity of NOTCH3-ADCs using parental MDA-MB-468 cells under 2D and 3D culture conditions. Data represent mean ± SEM (n = 3).
(E and F) TEC of NOTCH3-ADCs induces caspase activity in HEK-DLL4 cells. (E) z stack of maximum intensity projections from live-cell confocal imaging of anti-N3(i) ADC bound to U2OS-hN3 cells and co-cultured with HEK-DLL4 cells labeled with pHrodo Red dextran. Caspase compartments (magenta line), pHrodo Red dextran compartments (blue line) and the merged image. (F) Percentage of the caspase-positive compartments that were calculated after treatment with NOTCH3-ADCs. Data represent mean ± SEM for 3 biological replicates (n = 15 fields imaged per replicate), ns, non-significant.
We tested the in vitro cytotoxic activity and specificity of these ADCs using MDA-MB-468-hN3 cells. Both the anti-N3 and anti-N3(i) ADCs efficiently killed NOTCH3 cells. When NOTCH3 was depleted in these cells using siRNA, the ADC activity was similar to that of a non-binding control ADC (Figure 6C). Finally, we tested ADC activity in parental MDA-MD-468 cells expressing endogenous levels of NOTCH3 in 2D and 3D cytotoxicity assays (Figure 6D). Both ADCs showed enhanced efficacy in the more physiologically relevant 3D model compared to monolayer cell culture.
Next, we wanted to determine whether the ADCs can kill ligand-expressing cells through TEC. To quantitate TEC of the ADCs using a significant number of cells, we co-cultured HEK-DLL4 or HEK-control and U2OS-hN3 cells in the presence of the anti-N3 or anti-N3(i) ADCs and then detected caspase activity in the ligand-expressing versus control HEK cells. HEK cells were labeled with pHrodo Red dextran, U2OS-hN3 cells were not labeled, and caspase activity was detected with CellEvent Caspase-3/7 detection reagent (green). Next, a series of 10 μm optical image sections were acquired over time, and a z stack of maximum intensity projections was generated to create 3D volumetric renderings (Figures 6E and S7D). Using imaging analysis software, the number of caspase positive compartments in the pHrodo Red dextran-labeled HEK-DLL4 cells were quantified to measure the cytotoxic effects of the ADCs in ligand-expressing cells. Compartmental analysis was used to identify all caspase positive compartments (magenta line, Figures 6E and S7D) and then pHrodo Red dextran-positive compartments in the HEK-DLL4 cells (gray line, Figures 6E and S7D). Next, we calculated the percentage of the caspase-positive compartments that were also pHrodo Red dextran positive (merged image, Figures 6E and S7D), which allowed us to exclude caspase activity in the unlabeled U2OS-hN3 cells (Figure 6F). There were minimal caspase-positive structures of HEK-control cells that were used as a negative control for non-specific ADC cytotoxicity (Figure 6F). However, there was a statistically significant increase in caspase-positive structures in HEK-DLL4 cells, particularly at 38–54 and 56–72 h with both NOTCH3-targeted ADCs (Figure 6F). These data are in agreement with our TEC data showing that ligand-expressing cells have the ability to transendocytose anti-NOTCH3 mAbs.
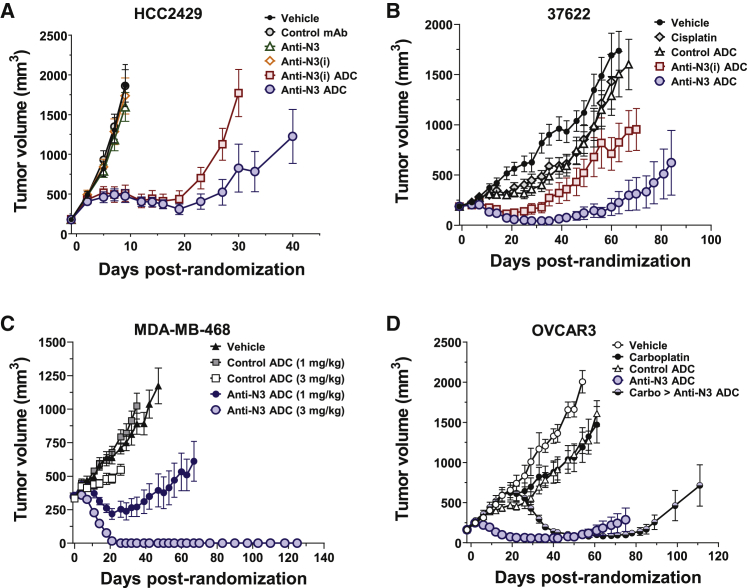
NOTCH3-ADCs induce prolonged tumor regressions
NOTCH3-ADCs were evaluated for in vivo efficacy in tumor xenograft models. Immunoblot analysis and immunohistochemistry (IHC) of NOTCH3 was conducted on a series of PDX and CLXs that were previously used to quantitate NOTCH3 mRNA levels. By immunoblot, NOTCH3-ECD (NECD) and C-terminal fragments were detected in all models (Figure S8A). JAG1 was detected in HCC2429 and OVCAR3 at higher levels than the 37622 NSCLC PDX, while MDA-MB-468 had the lowest levels (Figure S8A). Using a C-terminal specific anti-NOTCH3 antibody for IHC, NOTCH3 localization was detected at the cell membrane and within the nucleus of some tumor models (Figure S8B). Next, we examined the co-expression of NOTCH3 and JAG1 in the 37622 LUSC PDX model. As shown by IHC in the 37622 PDX, nuclear NOTCH3-ICD staining was detected in most tumor cells, while JAG1 staining was heterogeneous and mainly located on the membrane of tumor cells at the periphery of the tumor nest (Figure S8C).
In the HCC2429 lung CLX model with high levels of uniform NOTCH3 expression within the xenograft, NOTCH3-ADCs (3 mg/kg) suppress tumor growth, while the unconjugated mAbs (10 mg/kg) did not inhibit tumor growth (Figure 7A). The anti-N3 ADC demonstrated more tumor growth inhibition than the anti-N3(i) ADC. Thus, NOTCH3-ADCs inhibit tumor growth in a tumor model with high levels of NOTCH3 expression that was independent of signaling inhibition.
Figure 7.
NOTCH3-targeted ADCs induce prolonged tumor regressions
(A) Tumor growth inhibition of HCC2429 CLXs treated with control and NOTCH3-targeted mAbs and ADCs. Data represent mean ± SEM (n = 7–8 per group).
(B) Tumor growth inhibition of 37622 PDXs treated with NOTCH3-ADCs compared to cisplatin or control ADC. Data represent mean ± SEM (n = 9–10 per group).
(C) Tumor growth inhibition of MDA-MB-468 CLXs treated with control and anti-N3 ADCs. Data represent mean ± SEM (n = 7–8 per group).
(D) Tumor growth inhibition of OVCAR3 CLXs treated with control and anti-N3 ADCs compared with carboplatin alone, and carboplatin then anti-N3 ADC. Data represent mean ± SEM (n = 7–8 per group).
In the 37622 LUSC PDX model with heterogenous NOTCH3 expression and the JAG1 ligand, NOTCH3-ADCs have the potential for cytotoxic killing of both target and ligand cells through TEC. NOTCH3-targeted and control ADCs were dosed at 3 mg/kg. The anti-N3 ADC inhibited tumor growth more than the anti-N3(i) ADC (Figure 7B). Further, 37622A PDXs failed to respond to cisplatin administered at the maximum tolerated dose of 5 mg/kg; thus, both NOTCH3-ADCs outperformed cisplatin standard of care. There was no impact on athymic nu/nu mouse body weights for the NOTCH3-targeted ADCs indicating a lack of general payload toxicity at the doses tested (Figure S9A).
The MDA-MB-468 orthotopic (mammary fat pad) model also demonstrates heterogeneous NOTCH3 expression by IHC but has the lowest levels of the JAG1 compared to the other tumor models as determined by immunoblot (Figures S8A and S8B). NOTCH3-targeted and control ADCs were dosed at 1 and 3 mg/kg (Figure 7C; Figure S9B). The 3 mg/kg dose of anti-N3 ADC completely regressed tumors by day 26 through the end of the study on day 125 and all 8 animals remained tumor free. The 1 mg/kg dose of anti-N3 ADC completely suppressed tumor growth up to day 42 at which time the tumors began to slowly re-grow (Figure 7C). The control ADC did not inhibit tumor growth in any meaningful manner (Figure 7C). Similar results were observed with the anti-N3(i) ADC (Figure S9B). Since not all tumor cells express NOTCH3, we hypothesized that the protease-released, cell-permeable Aur0101 payload may exhibit a bystander effect similar to the recently reported anti-HER2-vc0101 ADC that contains the same linker payload,46 thereby inhibiting the growth of nearby tumor cells even if they lack NOTCH3 receptor and/or ligand expression. We conducted pharmacodynamic biomarker analysis with an anti-phospho-histone H3 (pHH3) antibody to examine cells arrested in mitosis, and anti-hIgG and anti-Aur0101 antibodies to identify cells bound by the anti-N3 ADC by IHC (Figure S9C). Both anti-hIgG and anti-Aur0101 antibody staining appeared heterogenous, while the pHH3 staining had a homogeneous distribution suggesting potential bystander activity of the Aur0101 payload as previously reported46 (Figure S9C). Again, there was no impact on SHO mouse body weights for the anti-N3 ADC indicating a lack of general payload toxicity at the doses tested (Figure S9D).
Due to the enhanced activity seen with the anti-N3 ADC over the anti-N3(i) ADC in some tumor models, the OVCAR3 ovarian cancer model was dosed with the anti-N3 ADC or control ADC at 3 mg/kg (Figure 7D). The 3 mg/kg dose of anti-N3 ADC regressed tumor growth, while the control ADC and carboplatin did not inhibit tumor growth in any meaningful manner. Next, OVCAR3 tumors were treated with carboplatin and then switched anti-N3 ADC, which again resulted in tumor regression until day 85. Thus, the anti-N3 ADC treatment outperformed carboplatin and inhibited the growth of carboplatin-resistant tumors.
Discussion
The use of inhibitory agents against the Notch family of receptors has revealed valuable information about the importance of Notch signaling during human development and disease.25 However, certain pan-Notch signaling inhibitors such as GSIs used to treat cancer patients produce dose-limiting on-target gastrointestinal (GI) toxicities in the clinic.29 Although receptor-specific inhibitory mAbs were developed in an attempt to mitigate safety issues,47 it was later determined that an anti-NOTCH1 mAb must contend with similar on-target toxicities in the clinic.48 Unlike NOTCH-1 and -2,49, 50, 51 NOTCH3 is not expressed or required for intestinal crypt stem cell functions in the GI tract, although there could be other unknown on-target issues from inhibition of NOTCH3 signaling. For instance, NOTCH3 has known roles in the maintenance of vascular smooth muscle cells (vSMCs),52 which is the underlying pathology for CADASIL syndrome.53 Therefore, inhibitory therapeutics for NOTCH3 could interfere with the important roles of NOTCH3 in the function and maintenance of adult blood vessels. The anti-N3 ADC was generated with the goal of achieving efficient internalization to induce cytotoxicity in proliferating cancer cells, while avoiding additional toxicities specifically associated with NOTCH3 signaling inhibition in normal cells. The majority of ADC toxicity is thought to be a consequence of the payload, but there have been instances of ADCs exhibiting toxicity at very low doses, suggesting an mAb-mediated effect rather than payload toxicity.54,55 NOTCH3-ADCs did not impact body weights of the mice in efficacy studies at the doses tested. However, we were unable to analyze potential on-target safety issues of the anti-N3(i) mAb or ADC since it was not cross-reactive with mouse NOTCH3.
We developed therapeutic mAbs against the NRR domain to allow the NOTCH3 receptor to retain binding to its ligands. Although the early rates of internalization and the endocytic machinery used appear to be different when cells are presented with the inhibitory versus non-inhibitory NOTCH3-targeted mAb, both ADCs had the ability to kill target- and ligand-expressing cells in vitro, and both were efficacious in vivo. However, the non-inhibitory anti-N3 mAb had several advantages over the anti-N3(i) mAb including lack of NOTCH3 downregulation, a greater saturation level of binding to cell-surface NOTCH3 and higher affinity to NOTCH3-NRR. In the 37622 LUSC PDX model, these attributes of the anti-N3 ADC appear to translate into enhanced efficacy compared to the anti-N3(i) ADC. Thus, in tumors with lower levels of NOTCH3-ECD, it was more beneficial to have higher levels of receptor binding by the anti-N3 ADC than inhibition of signaling with the anti-N3(i) ADC.
These data confirm that an ADC generated with a non-inhibitory mAb can show similar, and in some cases better efficacy than an ADC whose mAb portion has an inhibitory effect on receptor signaling. Although the non-inhibitory anti-N3 mAb demonstrated slower lysosomal co-localization kinetics compared with the inhibitory mAb, ligand binding stimulated receptor clustering allowing for efficient internalization and increased release of the cytotoxic payload. In fact, there have been biparatopic ADCs designed to target two different epitopes on the same receptor target to enhance clustering and internalization for increased efficacy; this has been demonstrated with the HER2 receptor.56 The NOTCH3 receptor-ligand interactions allows for clustering to occur without the need for a biparatopic mAb design.
Notch signaling is a cell-cell communication system, where a Notch receptor on one cell interacts in trans with a Notch ligand on an adjacent cell.57,58 Receptors that undergo TEC present an opportunity to target both receptor and ligand cells with an ADC. However, the ability of anti-N3(i) to stabilize the NRR in an autoinhibited conformation and still permit TEC was unexpected. We hypothesize that there may be two potential explanations for TEC of the mAb-receptor complex when bound to ligand, even in the absence of signaling. NOTCH3-mAb complexes may be transferred to ligand cells through extracellular vesicles or exosomes, which has been recently reported for NOTCH3.59 Alternatively, it is known that transmembrane proteins and lipids can be efficiently exchanged during direct cell-cell contact in a process termed intercellular transfer of cellular components (ICT).60 An ICT-type mechanism is possible since there are transient close contacts of neighboring Notch receptor-ligand cells that would permit local membrane fusion and exchange of the antibody-bound NOTCH3 receptor potentially including the TMIC fragment.
In the TME, Notch ligands such as JAG1 and DLL4 are known to be expressed by tumor cells and tumor-associated stromal, respectively.61 We hypothesize that in the context of the TME the non-inhibitory mAb would efficiently internalize and undergo TEC into ligand cells to allow for ADC cytotoxicity. Therefore, it is possible that NOTCH3-ADCs are able to mediate killing of NOTCH3-expressing tumor cells as well as ligand cells in the TME. Once released, the membrane permeable Aur0101 auristatin payload has the potential to kill neighboring target negative cells through a bystander effect. Bystander activity of Aur0101 has been described in detail using an anti-HER2-vc0101 ADC containing the same linker-payload as NOTCH3-ADCs.46 We hypothesize that NOTCH3-ADCs may have the potential to mediate the killing of non-Notch expressing cells in heterogenous NOTCH3-expressing tumors through a bystander effect. These findings support the design and development of additional ADCs targeting similar signaling-based receptor-ligand pairs. In summary, we targeted the CSC-associated Notch pathway without the need for signaling inhibition or reliance on the identification of tumors with signaling addiction using an ADC approach.
Limitations of study
Additional experiments will be needed to determine how the anti-N3(i) mAb mediates clustering of the NOTCH3 receptor, and whether it depends on mAb-mediated dimerization or impacts some other aspect of the global conformation of NOTCH3. Further studies will be needed to determine the exact mechanism of TEC as well as the impact on the NOTCH3 receptor: specifically, how TEC occurs if HD1/2 dissociation along with S2 cleavage is suppressed in the presence of anti-N3(i) mAb and which NOTCH3 domains (ECD, TMIC) are transendocytosed into the ligand cells.
The inhibitory anti-N3(i) mAb and ADC are not cross-reactive with mouse NOTCH3, and thus we were unable to further assess the potential impact of inhibiting NOTCH3 signaling along with delivering a cytotoxic payload in vivo. However, effects on mouse body weights were not observed for either ADC indicating a lack of general payload toxicity. Additional safety studies will need to be conducted in non-rodent species. Even so, the studies presented here demonstrate that targeting the NOTCH3 receptor with ADCs is safe and efficacious at the doses tested.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-N3-vc0101 ADC | Pfizer | PF-06650808 |
| Anti-N3(i)-vc0101 ADC | Pfizer | PF-06650810 |
| Rabbit anti-Notch3 Clone D11B8 | Cell Signaling | #5276; RRID: AB_10560515 |
| Mouse monoclonal anti-Notch3 Clone 1G5 | Abnova | H00004854-M01; RRID: AB_546554 |
| Rabbit monoclonal anti-Caveolin-1 | Cell Signaling | #3267; RRID: AB_2275453 |
| Rabbit monoclonal anti-Clathrin Heavy Chain | Cell Signaling | #4796; RRID: AB_10828486 |
| Rabbit monoclonal anti-GAPDH | Cell Signaling | #8884; RRID: AB_11129865 |
| Rabbit monoclonal anti-β−actin | Cell Signaling | #5125; RRID: AB_1903890 |
| Rabbit anti-phospho-Histone H3 Serine 10 | Cell Signaling | #9701; RRID: AB_331535 |
| Transferrin Receptor Monoclonal Antibody (MEM-75), Alexa Fluor 647 | Thermo Fisher | Cat# MA5-18151; RRID: AB_2539525 |
| Rabbit IgG Isotype Clone DA1E | Cell Signaling | #3900; RRID: AB_1550038 |
| Rabbit anti-Human IgG Pan | Epitomics | #3443-1; RRID: AB_10863040 |
| Rabbit anti-Jagged1 Clone EPR4290 | Abcam | ab109536; RRID: AB_10862281 |
| Mouse IgG Isotype Control | Life Technologies | #026502; RRID: AB_2532951 |
| Rabbit anti-Alexa Fluor 488 | Life Technologies | #A11094; RRID: AB_221544 |
| Mouse anti-Auristatin0101 | Pfizer | Lot 1 D8.C9 |
| SignalStain Boost Reagent HRP, Rabbit | Cell Signaling | #8114; RRID: AB_10544930 |
| MACH2 Rabbit AP Polymer | Biocare Medical | #ALP525 |
| Liquid DAB+ Substrate Chromogen System | DAKO | #K3468 |
| ImmPACT Vector Red AP Substrate | Vector Laboratories | #SK-5105 |
| Hematoxylin QS Counterstain | Vector Laboratories | #H-3404 |
| 10 mM Citrate Buffer pH 6.0 RTU | Invitrogen | #5001 |
| Borg Decloaker, pH 9.5 RTU | Biocare Medical | #BD1000 |
| Protein Block, Serum Free | DAKO | #X0909 |
| Permount Mounting Medium | Fisher | #SP15-500 |
| Retriever | Electron Microscopy Sciences | #62706 |
| RPMI | GIBCO | Cat#11875 |
| Sodium pyruvate | Cellgro | Cat#25-00-Cl |
| McCoy’s 5A medium | GIBCO | Cat#16600 |
| FBS | GIBCO | Cat#10082 |
| Penicillin-Streptomycin-Glutamine | GIBCO | Cat#10378 |
| Geneticin | GIBCO | Cat#10131 |
| Hygromycin | Invitrogen | Cat#10687-010 |
| Puromycin | Sigma | Cat#P9620 |
| DMEM | GIBCO | Cat#11995 |
| MEM | Mediatech | Cat#10-010-CV |
| TransIT-LT1 | Mirus Bio | Cat#MIR2300 |
| SDS-PAGE on a 4–15% polyacrylamide gel | Bio-Rad Criterion | Cat#345-0027 |
| Biological samples | ||
| Breast Tumor Tissue Samples | ILSBio, ProteoGenex, Tristar | N/A |
| Normal Breast Tissue Samples | Cleveland Clinic | N/A |
| Ovarian Tumor Tissue Samples | Cornell University, University of Michigan | N/A |
| Normal Ovary Tissue Samples | Cleveland Clinic | N/A |
| Lung Tumor Tissue Samples | Indivumed; Cornell University, University of Michigan, ProteoGenex, Inc. | N/A |
| Normal Lung Tissue Samples | Cleveland Clinic | N/A |
| Normal Breast RNA | Biochain | Cat # R1234086-50 |
| Normal Lung RNA | Life Technologies | Cat # AM7968 |
| Normal Lung RNA | Biochain | Cat # R1234152-50 |
| Chemicals, peptides, and recombinant proteins | ||
| Methyl-β−cyclodextrin | MilliporeSigma | M7439 |
| Dynasore | Sigma | D7693 |
| DBZ | Syncom | #18231 |
| γ-secretase inhibitor (GSI) | Pfizer | PF-03084014 |
| Freund’s complete adjuvant | ThermoFisher | 77140 |
| Recombinant human DLL4-Fc | R&D Systems | #10185-D4-050 |
| Recombinant human NOTCH3 NRR | Pfizer | N/A |
| Recombinant mouse NOTCH3 NRR | Pfizer | N/A |
| Critical commercial assays | ||
| QIAGEN RNeasy Mini Kit | QIAGEN | Cat#74106 |
| High Capacity RNA-to-cDNA kit | Life Technologies | Cat#4387406 |
| Alexa Fluor 647 Antibody Labeling Kit | Life Technologies | Cat#A20186 |
| Alexa Fluor 488 Antibody Labeling kit | Life Technologies | Cat#A30052 |
| DyLight647 Antibody Labeling Kit | Thermo Fisher | Cat#62295 |
| Notch3 probe targeting nucleotides 727-2210 of human NOTCH3 mRNA (NM_000435.2) | Advanced Cell Diagnostics | 558996 |
| VS ACD RNAscope FFPE Brown kit | Advanced Cell Diagnostics | 320600 |
| Ventana mRNA Detection kit | Roche | 760-225 |
| Cholera toxin subunit β (CT-β) Alexa Fluor® 647 | Thermo Fisher | Cat#C34778 |
| Deposited data | ||
| Atomic coordinates anti-N3(i) Fab-NOTCH3 NRR | Protein Data Bank | 6XSW |
| Experimental models: Cell lines | ||
| MDA-MB-468 | ATCC | HTB-132; RRID: CVCL_0419 |
| NCI-N87 | ATCC | CRL-5822; RRID: CVCL_1603 |
| OVCAR-3 | ATCC | HTB-161; RRID: CVCL_0465 |
| U-2 OS | ATCC | HTB-96; RRID: CVCL_0042 |
| HEK293 | ATCC | CRL-1573; RRID: CVCL_0045 |
| HCC2429 | UTSW | Dr. J. Minna |
| U2OS-hN3 Luc-reporter | Pfizer | N/A |
| MDA-MB-468-hN3 | Pfizer | N/A |
| HEK-DLL4 | Pfizer | N/A |
| 37622 PDX | Pfizer | N/A |
| U2OS-hN1 | Pfizer | N/A |
| U2OS-mN1 | Pfizer | N/A |
| U2OS-hN3 | Pfizer | N/A |
| U2OS-mN3 | Pfizer | N/A |
| CHO-PACE | Pfizer | N/A |
| Experimental models: Organisms/strains | ||
| Athymic (nu/nu) mice | Charles River Laboratories | |
| Sprague Dawley rats | Taconic | SD-F, SD-M |
| SHO mice | Charles River Laboratories | |
| Oligonucleotides | ||
| NOTCH3 (Hs01128541_m1) | ThermoFisher Scientific | Cat # 433182 |
| ACTB (Hs99999903_m1) | ThermoFisher Scientific | Cat # 433182 |
| HES1 (Hs00172878_m1) | ThermoFisher Scientific | Cat # 433182 |
| B2M (Hs00984230_m1) | ThermoFisher Scientific | Cat # 433182 |
| Recombinant DNA | ||
| Human NOTCH3 | Origene | Cat#RC224711 |
| Mouse NOTCH3 | Origene | Cat#MR212149 |
| Human NOTCH1 | Origene | Cat#RC211365 |
| Mouse NOTCH1 | Origene | Cat#MR212150 |
| Human DLL4 | Origene | Cat#RC212628 |
| pGL4.27 [Luc2P/MinP/Hygro] vector, containing 8 tandem copies of the CSL enhancer sequence (CGTGGGAAAAT) | Promega, Pfizer | Cat#E8451, N/A |
| Software and algorithms | ||
| Volocity | Quorum Technologies Inc. | Quorumtechnologies.com |
| Zen Black | Zeiss | Zeiss.com |
| GraphPad Prism | GraphPad Software | |
| Microsoft Excel | Microsoft | |
Resource availability
Lead Contact
Additional information and request for resources and reagents should be directed to and will be made available by the Lead Contact, Kenneth G. Geles (kgg11ggk@gmail.com).
Materials availability
All unique/stable reagents generated in this study are available through Pfizer, Inc. from the Lead Contact, subject to materials transfer agreement requirements.
Data and code availability
All data associated with this study are available in the main text or supplemental materials. The accession number for atomic coordinates of anti-N3(i) clone 75 Fab-NOTCH3 NRR structure reported in this paper is PDB: 6XSW.
Experimental models and subject details
Rodents
Female Sprague Dawley rats of around 8 weeks old for immunization, strain NTac:SD, were purchased from Taconic. Athymic (nu/nu) and SHO mice were purchased from Charles River Laboratories. All animals were housed in pathogen-free conditions in the vivarium at Pfizer Inc. in Pearl River, NY or Cambridge, MA in accordance with IACUC guidelines. All animals had ad libitum access to food and water for consumption and were maintained at 22°C on a 12-hour light/12-hour dark cycle with 2 rats per cage or 5 mice per cage. Rats were observed daily for health status during the immunization process which typically takes 1-2 months. At the end of immunization terms, rats were euthanized. For in vivo efficacy studies with human tumor xenografts, female mice were used and ranged in age from 6-16 weeks at the start of each study, though in a given study all animals were of similar age. Clinical conditions of mice were checked daily for health limitations associated with tumor progression such as loss of weight, mobility or food intake and were euthanized if they met IACUC criteria for euthanasia.
Human tumor tissues
Human lung, breast and ovary flash frozen primary tumors (≥500 mg) were acquired from Cornell Univ., Weill Medical College, New York City, NY; University of Michigan Health System, Ann Arbor, MI; Cleveland Clinic, Cleveland, OH; Integrated Laboratory Services-Biotech, LLC, Chestertown, MD; ProteoGenex, Inc., Culver City, CA; TriStar Technology Group, LLC, Washington, D.C.; and Indivumed, Inc., Baltimore, MD according to informed consent and ethics review board requirements in compliance with IRB/IEC-approved protocols.
Cell lines
Cell lines were purchased from American Type Culture Collection (ATCC): MDA-MB-468 cells (ATCC® HTB-132), NCI-N87 cells (ATCC® CRL-5822), OVCAR-3 cells (ATCC® HTB-161), U-2 OS cells (ATCC® HTB-96) and HEK293 cells (ATCC®, CRL-1573). ATCC cell lines were grown according to recommended conditions. The HCC2429 lung cancer cell line was obtained from Dr. J. Minna (University of Texas Southwestern Medical School). HCC2429 cells were cultured in RPMI (GIBCO®, cat. no. 11875) supplemented with 10% FBS (GIBCO®, cat. no. 10082), 1% penicillin-streptomycin-glutamine (GIBCO®, cat. no. 10378) and 1% sodium pyruvate (Cellgro®, cat. no. 25-000-Cl). Engineered U2OS-hN3 and U2OS-mN3 cells were cultured in McCoy’s 5A medium (GIBCO®, cat. no. 16600) supplemented with 10% FBS (GIBCO®, cat. no. 10082), 1% penicillin-streptomycin-glutamine (GIBCO®, cat. no. 10378) and 250 μg/ml Geneticin® (GIBCO®, cat. no. 10131). The engineered U2OS-hN3 Luc-reporter gene cell line was cultured in McCoy’s 5A medium (GIBCO®, cat. no. 16600) supplemented with 10% FBS (GIBCO®, cat. no. 10082), 1% penicillin-streptomycin-glutamine (GIBCO®, cat. no. 10378), 250 μg/ml Geneticin® (GIBCO®, cat. no. 10131), 300 μg/ml Hygromycin B (Invitrogen, cat. no. 10687-010) and 1 μg/ml Puromycin (Sigma®, cat. no. P9620). Engineered MDA-MB-468-hN3 cells were cultured in DMEM (GIBCO®, cat. no. 11995) supplemented with 10% FBS (GIBCO®, cat. no. 10082), 1% penicillin-streptomycin-glutamine (GIBCO®, cat. no. 10378) and 500 μg/ml Geneticin® (GIBCO®, cat. no. 10131). Engineered HEK293-DLL4 cells were cultured in MEM (Mediatech, cat. no. 10-010-CV) with 10 % FBS (GIBCO®, cat. no. 10082), 1% penicillin-streptomycin-glutamine (GIBCO®, cat. no. 10378) and 500 μg/mL Geneticin® (GIBCO®, cat. no. 10131). Cell line authentication of NCI-N87 and MDA-MB-468 was conducted at IDEXX using STR-based DNA profiling and multiplex PCR.
Method details
In situ hybridization and digital image analysis
Frozen human tumor and xenograft tissues were fragmented with a Cellcrusher (CellCrusher Limited) and random pieces of crushed tissue were chosen for fixation or RNA processing. Frozen fragmented human tumor tissue was transferred to 10% NBF at room temperature and allowed to fix for 24 hr with gentle agitation on a bench-top rocker. Tissues were processed on a Tissue-Tek VIP tissue processor (Sakura Finetek USA) according to standard methods.
Sections of FFPE tissues were cut at 5 μm with a microtome and placed on positively charged microscope slides (Fisherbrand SuperFrost Plus slides, Cat. No. 12-550-15). Slides were air-dried overnight and then baked for 1 hour at 60°C. Slides were de-paraffinized twice (6 minutes each) in fresh xylene, washed in fresh 100% ethanol twice (5 minutes each) and air-dried. Slides were then placed into boiling (100°-104°C) citrate buffer (Pretreatment 2 buffer) for 5 minutes (Advanced Cell Diagnostics (ACD), Newark, CA). Slides were immediately removed from the buffer, washed several times in water, then loaded onto a Ventana Discovery Ultra automated staining system (Ventana Medical Systems, VMS). Slides were covered with water prior to performing ISH. The synthetic ISH probe set (ACD; Cat. No. 558996) was designed to hybridize to nucleotides 727 to 2210 of human NOTCH3 mRNA (NM_000435.2). Positive control assays were performed using a PPIB probe (ACD; Cat. No. 313906), targeted against a human housekeeping gene to confirm mRNA preservation, and negative control assays were performed using an Escherichia coli DapB probe (ACD, Cat. No. 310048).
Automated ISH was performed using the VS ACD RNAscope FFPE Brown kit (ACD; Cat. No. 320600), and Ventana mRNA Detection Kit (VMS; Cat. No. 760-225), according to the manufacturer’s Ventana platform protocol (Doc# 323200-USM-ULT). Slides were counter-stained using hematoxylin for 16 minutes and bluing reagents for 4 minutes (ACD; Cat. No. 320630). Following the ISH processing, slides were washed in a solution of Dawn Original dish soap, rinsed several times in water and dehydrated through a graded ethanol series (95% ethanol for 1 minute, 3 times in 100% ethanol for 2 minutes each) and xylene (twice for 2 minutes each). Glass coverslips were applied with a Tissue-Tek Glas 6400 using Tissue-Tek Glas mounting medium (Sakura Finetek USA; Cat. No. 6419).
Stained tissue sections were digitally scanned using a Leica/Aperio AT2 whole slide digital scanner at 40X magnification. Definiens Tissue Studio within the Architect XD interface was used to quantify NOTCH3 ISH signal. For quantification of NOTCH3 ISH staining in tumor samples, a rule set was created in which tissue area is identified using optical density of the image. Following the identification of tissue area for each sample a threshold for DAB intensity was set. An algorithm was run to identify the NOTCH3 ISH positive tissue area based upon the defined DAB optical density threshold, and a value for NOTCH3 positive tissue area was reported for each sample. Using this value, as well as the total tissue area value, a % NOTCH3 tissue area was calculated.
Quantitative RT-PCR and data analysis
Total RNA was isolated with QIAGEN RNeasy Mini kit (QIAGEN, cat # 74106) according to manufacturer’s instructions from frozen tumor xenografts and human tumor tissue fragments that were crushed to powder and homogenized with a cooled (−80°C) 5 mm stainless steel bead (QIAGEN, cat # 69989) at 20 Hz using a TissueLyser II (QIAGEN, cat # 85300). Additionally, tumor tissue supernatants from this procedure were transferred to a QIAshredder spin column (QIAGEN, cat # 79654). DNA was removed from spin columns through the RNase-Free DNase Set (QIAGEN, cat # 79254). Normal lung samples were processed in the same manner described above, but were lysed in QIAzol Lysis Reagent (QIAGEN, cat # 79306) instead. Total RNA was eluted from the column in RNase free water (QIAGEN, cat #129112). RNA quantitation and quality assessment were performed using the HT RNA microfluidic LabChip assay and LabChip GX microfluidic capillary electrophoresis instrument (Perkin Elmer). Isolated RNA samples were reverse transcribed to cDNA using the Life Technologies, High Capacity RNA-to-cDNA Kit (cat # 4387406) following a protocol outlined by the manufacturer. The qRT-PCR reaction was performed using the TaqMan Probe-Based Gene Expression Analysis and ABI ViiA7 Real-Time PCR Systems (Life Technologies). Target gene and endogenous controls (see Key Resources Table) were run in quadruplicate for each probe set on prefabricated TaqMan low density array cards. TaqMan gene expression master mix was added (Life Technologies, cat # 4352042) and run under standard thermal cycling conditions.
Quantitation of NOTCH3 expression was assessed using the relative fold difference (RQ) or comparative Ct method, (2–ΔΔCt) method. Initially ΔCt values are generated for each sample by adjusting to the Ct value of a stably expressed endogenous control gene(s) (ΔCt = Ct, endogenous control – Ct, NOTCH3). Sample ΔCt values are then used to generate ΔΔCt values relative to a sample of comparison (ΔΔCt = ΔCt, sample - ΔCt, reference). RQ between sample and reference may then be calculated using the equation RQ = 2–ΔΔCt. All tumor sample ΔCt values were generated using ACTB as the endogenous control gene. The breast cancer RQ data reported represents fold difference NOTCH3 expression relative to the average of RQ values generated from 3 normal breast RNA samples purchased from BioChain (Newark, CA, cat # R1234086-50, lot# B610189, 75 year old female) and 2 samples provided by the Cleveland Clinic (Cleveland, OH). The ovarian cancer RQ data reported represents fold differences in NOTCH3 expression relative to RNA isolated from a normal ovary tissue (tissue ID #0204C011C) provided by the Cleveland Clinic (Cleveland, OH). The lung cancer RQ data reported represents fold differences in NOTCH3 expression relative to the average of RQ values generated from 5 normal lung RNAs purchased from ThermoFisher Scientific (cat #AM7968, lot # 1308017, 80 year old female), BioChain (cat # R1234152-50, lot#: B503032, 23 year old male) and 3 provided by the Cleveland Clinic (Cleveland, OH). All statistical and correlation analysis was performed using GraphPad Prism 6 (v6.03) (GraphPad Software, Inc. La Jolla, CA). Non-parametric Spearman’s (ρ) rank correlation values were generated to establish the directional correlation of each NOTCH3 quantitative endpoint (ISH, qRT-PCR) within a sample set.
HCC2429 cells were plated in wells with or without DLL4 ligand by first coating wells for 1 hour at room temperature with 2 μg/mL of recombinant human DLL4 Fc chimera protein (R&D Systems, # 10185-D4-050) or left uncoated and cultured overnight in complete growth medium according to ATCC instructions. NOTCH3-targeted and control mAbs were added at a concentration of 10 μg/ml. GSI (PF-03084014) was added at 1 μM or an equal volume of DMSO as a control. RNA was isolated using the RNeasy mini kit (QIAGEN, #74106), reverse transcribed with the iScript cDNA synthesis kit (BioRad, #1708891), and PCR amplified with Taqman Fast Advanced Master Mix (Applied Biosystems, #1705119) in MicroAmp fast optical 96-well reaction plates (Applied Biosystems, #4346906) using TaqMan gene expression assays B2M (Hs00984230_m1), HES1 (Hs00172878_m1), NOTCH3 (Hs01128541_m1) (ThermoFisher Scientific). Quantitation of HES1 and NOTCH3 expression was assessed using the relative fold difference (RQ) as described above but using β-microglobulin (B2M) as the endogenous control gene. The RQ values for each of the compound-treated cells is relative to control cells in the absence of DLL4 ligand.
Antibody generation and characterization
Sprague Dawley rats were immunized by subcutaneous injections of a mixture of 20 μg each of human and mouse NOTCH3 NRR-AviHis recombinant proteins in Freund’s complete adjuvant. Both immunogens were produced in their native heterodimeric conformations after the proteolytic cleavage at site 1 (S1), similar to that found on the cell surface. Immunizations were repeated at 2-week intervals for 12 weeks. Collected sera samples at day 0, 35, 49, and 63 were tested for circulating anti-NOTCH3 antibody titer activity by enzyme-linked immunosorbent assay (ELISA). When optimal titers were reached, a final dose of the protein mixture was injected intravenously (tail vein) into one rat that had the optimal antibody titer 4 days before it was euthanized and splenocyte collected for hybridoma fusion. Fusion were performed with standard procedure and hybridoma supernatants were tested for binding activity to human and mouse NOTCH3 NRR recombinant antigens, and full-length human and mouse NOTCH3 expressed on the surface of U2OS cells. Selected hybridoma clones that showed NOTCH3 binding with or without signaling inhibition activity were subjected to subcloning followed by antibody cDNA cloning for recombinant antibody production.
The heavy and light chain variable domains of rat anti-N3 and Anti-N3(i) antibodies generated from hybridoma technology were first fused to human IgG1 heavy and light chain constant domains respectively. This process generated the chimeric anti-N3 and Anti-N3(i) antibodies in which the variable domains are from rat origin whereas the constant domains are of human origin, namely the chimeric anti-N3 and anti-N3(i) antibodies. Subsequently, humanization of anti-N3 and anti-N3(i) antibodies was performed. CDR donor sequences from anti-N3 and anti-N3(i) were dropped in human acceptor framework DP-54 for the heavy chain and DPK9 for the light chain, and fused in frame with human IgG1 constant region for heavy chain and human kappa for light chain, respectively62. When necessary, backmutations to rat residues in framework regions were made to fully recover full parental rat antibodies activities.
Expression constructs encoding human NOTCH3-NOTCH1 NRR domain swap chimeras with C-terminal human IgG1 Fc-fusion were individually transfected into CHO-PACE cells63 and stable pools expressing each chimera were established. Conditioned media from each stable pool were applied to rmp protein A affinity chromatography (Cytiva, cat. # 17513801), followed by size exclusion chromatography (SEC) (Cytiva, cat no. 28989335). Purified preparations were dialyzed into TBS with 1 mM CaCl2 and analyzed on analytical SEC (Cytiva) to ensure quality of preparations. For assessing antibody binding to NOTCH3-NOTCH1 domain swap-Fc fusion proteins, 1 μg/ml recombinant proteins were individually coated on CoStar hi-bound 96-well ELISA plates (Corning Inc, Corning, NY; cat. no.3590) in 100 μL of PBS with 1 mM MgCl2 and CaCl2 (PBS-Mg/Ca) for overnight at 4°C. Plates were washed 3 times with PBS-Mg/Ca and then blocked for 1 hr with 1% bovine serum albumin (BSA) (Sigma, cat #10735086001) in PBS-Mg/Ca. Blocking solution was decanted from the plate and antibody solutions in blocking buffer were applied. Plates were incubated for 1 hr and washed before HRP-conjugated secondary goat anti-human IgG Fc antibody (Southern Biotech, Birmingham, AL, cat no. 2048-05) diluted at 1:5000 was applied for 1 hr. Plates were washed and TMB substrate solution (3,3′,5,5′-tetramethylbenzidine; BioFX Labs, Owing Mills, MD; cat no. TMBW0100-01) was added. Developing reaction was allowed for 10 minutes before stopping solution, 0.18M H2SO4, was added and absorbance at OD450 nM was measured.
NOTCH3-targeted mAbs and ADCs were screened for cell surface binding activity in a cell-based ELISA. NOTCH3 engineered cell lines (U2OS-hN3, U2OS-mN3) were plated at 50,000 cells/well in 96 well poly D-Lysine coated plates (white opaque, BD/VWR) the day before ELISA assay. On the day of the ELISA, culture media was removed from wells and serially diluted (1:3 in DPBS with calcium chloride and magnesium chloride and 1% BSA) antibody and ADC solutions were applied to the plate. Plates were incubated at room temperature for 1 hour before washed with DPBS with calcium chloride and magnesium chloride. HRP-conjugated secondary antibody was then applied and incubated with cells for 1 hour. Plates were washed with DPBS with calcium chloride and magnesium chloride before being developed with Pico Chemiluminescent substrate (Thermo Scientific), and chemiluminescence measurements were performed per manufacturer’s instructions. Data plotting and analyses were performed with Microsoft Excel and GraphPad-Prism software.
For the competition ELISA, U2OS-hN3 cells were plated at 5 × 104 cells/well in 96 well plates (white opaque, BD/VWR) the day before the ELISA assay. The following day, culture medium was removed from the wells, and 1:3 serially diluted test antibodies, specifically, Anti-N3 and Anti-N3(i), in blocking buffer (1% bovine serum albumin BSA in PBS with 0.9 mM Mg/Ca) were applied to the plate in the presence of 0.8 nM of biotinylated parental Anti-N3 antibody. Plates were incubated at room temperature for 2 hours and then washed 4 times with PBS-Mg/Ca. Afterward, HRP-conjugated streptavidin (Southern Biotech, Birmingham, AL) diluted 1:5000 in blocking buffer was applied to the plates. Incubation with HRP-streptavidin was allowed for 30 minutes before the plates were washed again with 4 times with PBS-Mg/Ca and developed with TMB solution (ThermoFisher N301) for 10 minutes. The developing reaction was then stopped by adding equal volume of 0.18 M H2SO4 and absorbance at 450 nm measured. Data plotting and analyses were performed with Microsoft Excel and Graphpad-Prism software.
Crystal structure of NOTCH3 NRR-anti-N3 complex
NOTCH3 NRR with an N-terminal His tag was expressed in HEK293 cells treated with kifunensine. The purified protein was mixed with anti-N3(i) Fab domain generated by treating the full-length antibody with papain. The complex was purified by size exclusion chromatography with Tris Buffered Saline containing 0.9 mM CaCl2 as the running buffer. The complex was then concentrated to 8.7 mg/ml and crystalized using the hanging drop method with 100 mM sodium Citrate pH 5.5, 12 % PEG 6000 as the well solution. X-ray data was collected at the Advanced Photon Source beamline 17ID. Diffraction data was processed using autoproc (Global Phasing). An initial electron density map was obtained by molecular replacement using the previously solved structure of the NOTCH1 NRR as a search model. The structure was refined through iterative rounds of manual building using COOT and reciprocal space refinement using autobuster (Global Phasing).
Affinity measurement of NOTCH3-targeted mAbs
The kinetic constants of anti-NOTCH3 and NOTCH3 NRR interactions were determined by surface plasmon resonance (Biacore T100, BIAcore Inc., NJ). Flow cells of a CM5 chip were immobilized with approximately 10,000 RU of anti-human IgG-Fc (BIAcore) in 10mM Glycine, pH 5.0 at 10 μl/min for 600 s. Next, 10 μg/ml of humanized anti-NOTCH3 antibodies, diluted in TBS with 1 mM CaCl2, were captured at 10 μl/min. Association of four concentrations of human NOTCH3 NRR_AviHis recombinant protein (from 3.7-100 nM) and a zero concentration (running buffer) at 100 μl/min were recorded for 3 minutes in TBS with 1 mM CaCl2. Dissociation of the complexes was measured for 10 minutes. The surface of the chip was regenerated by injecting 3 M MgCl2 with 3 mM EGTA for 60 s at 10 μl/min. Curves obtained after subtraction of the reference and buffer signals were fitted to a 1:1 Langmuir binding model with Biacore T100 Evaluation Software (BIAcore).
Engineered cell lines and reporter gene assay
To generate NOTCH3 expressing cell lines, stable transfections were performed in the U2OS human osteosarcoma cell line with full-length human NOTCH3 (Origene, catalog number RC224711) and mouse NOTCH3 (Origene, catalog number MR212149) with the TransIT-LT1 transfection reagent (Mirus Bio® LLC, Madison, WI; cat. no. MIR 2300). Cell were selected in Geneticin® and clonal lines were isolated. A stable transfection with full-length human NOTCH3 was performed in the MDA-MB-468 breast cancer cell lines in a manner similar.
To generate a NOTCH3-dependent reporter gene cells, a stable clonal line of U2OS-hN3 cells were retransfected with the pGL4.27 [Luc2P/MinP/Hygro] vector (Promega, Madison, WI) containing eight tandem copies of the CSL enhancer sequence (CGTGGGAAAAT), selected in Hygromycin B plus Geneticin® and clonal lines were isolated. Next, the human U2OS-hN3-8xCSL-Luc2P cells were transduced with Cignal Lenti Renilla Control lentiviral particles (QIAGEN, CA), selected in Puromycin, Hygromycin B and Geneticin®, and clonal lines were isolated. The Cignal Lenti Renilla control vector encoded the Renilla-luciferase gene that is constitutively expressed from a CMV promoter and served as an internal control. The triple stable transfected U2OS line hereinafter was termed U2OS-hN3 Luc-reporter cells. To generate the ligand-expressing cells, HEK293 cells were transfected with a vector for expression of human DLL4 in the pCMV6-AC-HA-His backbone (Origene, Rockville, MD) and termed HEK-DLL4. Following transfection, HEK293 cells were selected in 0.5 mg/ml Geneticin®, and clonal lines were isolated.
U2OS-hN3 Luc-reporter cells were added to 96 well culture plate (white opaque, BD/VWR®), in the presence of serially diluted (1:3 in complete assay media) NOTCH3-targeted or non-targeted control mAbs and incubated on the plates at room temperature for 1 hr before HEK-DLL4 or HEK parental cells were added to each well in a ratio of 3:1 to the reporter cells. Plates were incubated overnight and then the Dual-Glo Luciferase assay system (Promega) was used to measure the Firefly-luciferase and internal control Renilla-luciferase activity (per manufacturer’s instructions) on a Victor3R 1420 multi-label plate reader (PerkinElmer®). The luminescent readings from Firefly-luciferase were divided by the internal control Renilla-luciferase reading to normalize the signals (F/R ratio). To calculate the fold induction of NOTCH3 signaling, the F/R ratios generated from the reporter cells in the presence of DLL4 ligand were divided by the F/R ratios from reporter cells in the absence of ligand and termed relative luciferase unit (RLU). The percent NOTCH3 signaling activity was calculated by dividing treated/untreated RLUs x 100. Data was plotted and analyzed using Microsoft® Excel and Graphpad-Prism software.
Immunoblot analysis
HCC2429 and MDA-MB-468 cells were plated in microtiter plates in complete growth medium according to ATCC instructions or MDA-MB-468 cells were plated in wells that were first coated for 1 hr at room temperature with 2 μg/mL of recombinant human DLL4-Fc (R&D Systems, # 10185-D4-050) and cultured overnight. NOTCH3-targeted and control mAbs were added at a concentration of 5 or 10 μg/ml. GSI (PF-03084014) was added at 1 μM or an equal volume of DMSO as a control. Dynasore was added at 60 μM (Sigma). Cells were incubated at 37°C in a 5% carbon dioxide incubator for 24-48 hr and then directly lysed in extraction buffer containing 1% NP40, 0.5% sodium deoxycholate, 5 mM EDTA, 0.25 M NaCl, 0.025 M Tris-HCl, pH 7.5, containing Complete Mini Protease Inhibitor Cocktail (Roche, cat. no. 04-693-124-001) and 0.4 mM PMSF (Sigma®, cat. no. 93482). Extracts were resolved by denaturing SDS-PAGE on a 7.5% polyacrylamide gel (Bio-Rad criterion gel) and transferred to nitrocellulose paper using an iBlot Gel transfer system (Invitrogen). HCC2429 xenograft were harvested 24hr after the 3rd dose of NOTCH3-targeted or control mAbs that were dosed intravenously at 10 mg/kg every 4 days. Fragments were homogenized in extraction buffer as described above and resolved by denaturing SDS-PAGE on a 4%–15% polyacrylamide gel (Bio-Rad Criterion, No. 345-0027) and transferred to nitrocellulose paper using an iBlot Gel transfer system (Invitrogen, Cat. No. IB1001). The C terminus of NOTCH3 was detected with the D11B8 antibody (Cell Signaling Technology, #5276), the N-terminal extracellular domain of NOTCH3 was detected with a mouse monoclonal anti-NOTCH3 antibody clone 1G5 (Abnova, number H00004854-M01) and anti-GAPDH (Sigma) or anti-β-actin (Cell Signaling) as a loading control using standard western blot procedures.
Indirect immunofluorescence of anti-NOTCH3 mAbs
MDA-MB-468 cells were seeded in a Nunc Lab Tec II 8 chambered cover #1.5 μm borosilicate sterile slides (Thermo Fisher Scientific Inc., cat. #155409) at 1.2x103 cells per well and incubated overnight. Next day, cells were washed 1x with cold HBSS (GIBCO Life Technologies, cat. #14025-092) and incubated with 10 μg/ml anti-N3 or anti-N3(i) at 4°C for 30 minutes. Cells were washed 1x with cold HBSS and incubated with medium at 37°C up to 5 hr. After 5 hr incubation, cells were fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.5% Triton X-100 for 5 minutes, and blocked with 3% BSA for 30 minutes. Slides were washed 2x with PBS and incubated with labeled goat anti-human Alexa Fluor® 647 secondary antibody for 1 hr at room temperature, washed three times with PBS and imaged. Images were acquired through 63x oil immersion objective with Zeiss LSM710 confocal laser microscope.
U2OS-hN3 cells were plated in a 96 well plate at a concentration 0.2 x104 per well and 24 hr later treated with γ-secretase inhibitor DBZ (Syncom, #18231, Groningen, the Netherlands) at a concentration of 100 nM. DBZ remained present throughout the entire experiment. HEK parent and HEK-DLL4 cells were labeled overnight with 5 μM CellTracker Green CMFDA (Invitrogen, C7025). Next day, 10 μg/ml NOTCH3-targeted mAbs were added to GSI-treated U2OS-hN3 cells in 2%BSA/HBSS on ice for 30 minutes, washed twice with HBSS and then incubated in culture media. CellTracker Green CMFDA-labeled HEK-DLL4 cells were added at a 1:1 ratio to NOTCH3-targeted antibody bound and GSI-treated U2OS-hN3 cells. Plates were centrifuged for 1 minute at 100 g and incubated at 37°C for 2 hr. Cells were processed for indirect immunofluorescence as described above and imaged on the Zeiss LSM510 confocal microscope.
Simultaneous binding of NOTCH3-targeted mAbs to MDA-MB-468 cells
MDA-MB-468 cells were seeded at 1.2 × 103 cells per well in a Nunc Lab Tec II 8 chambered cover #1.5 μm borosilicate sterile slides (Thermo Fisher Scientific Inc. 155409) and incubated overnight. Anti-N3 was directly conjugated to DyLight647 (ThermoFisher Scientific, cat. #62295) and anti-N3(i) was directly conjugated to Alexa Fluor 488 (Life Technologies Protein Labeling kit A#30052) according to manufacturer’s instructions. Next day, cells were washed 1x with cold HBSS and incubated with 5 μg/ml anti-N3(i)-Alexa488 at 4°C for 10 minutes. At 10 minutes, while cells still on ice, we added 5 μg/ml anti-N3-DyLight647 and left to bind for 20 minutes on ice. After a total of 30 minutes binding, cells were then washed 2x with cold HBSS. Cold medium was added to the cells were taken to the Ultraview spinning disk confocal microscope (PerkinElmer) and imaged every 10 minutes for 20 hr.
Co-localization of NOTCH3-targeted mAbs with CAV-1 and Clathrin
MDA-MB-468 cells were seeded at 1.2 × 103 cells per well in a Nunc Lab Tec II 8 chambered cover #1.5 μm borosilicate sterile slides (Thermo Fisher Scientific Inc. 155409) and incubated overnight. Next day, cells were washed 1x with cold HBSS and incubated with 10 μg/ml anti-N3 or anti-N3(i) for 30 minutes. Cholera toxin subunit β (CT-β) Alexa Fluor® 647 conjugate (ThermoFisher Scientific, cat. #C34778) or anti-Transferrin Receptor (TfR) Alex Fluor 647 (ThermoFisher Scientific, cat. #MA5-18151) were also added to cells at 4°C for 30 minutes. CT-β was used as a positive imaging control for CAV-1 staining and anti-TfR antibody was used as a positive imaging control for clathrin, but neither co-localizations were quantitated. Slides then were washed 1x with cold HBSS and incubated with medium for 3 hr, 8 hr and 24 hr at 37°C for the indicated time points. At each time point slides were fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.5% Triton X-100 for 5 minutes, and blocked with 3% BSA for 30 minutes, all in PBS. Primary antibodies for clathrin rabbit monoclonal (Cell Signal #4796) at 1:50 and caveolin-1 rabbit monoclonal (Cell Signal #3267) at 1:400 incubated overnight at 4°C. The following day, slides were washed 2x with PBS and incubated with labeled goat anti-rabbit Alexa Fluor 555 and goat anti-human Alexa Fluor 488 secondary antibodies for 1 hr at room temperature, washed three times with PBS and imaged. Images were acquired through 63x oil immersion objective with Zeiss LSM710 confocal laser microscope. Quantitative analysis was done using Volocity 6.3 software, with calculation of thresholded Pearson’s correlation coefficients and Manders colocalization coefficients.
Live cell imaging
MDA-MB-468 or U2OS-hN3 cells were seeded in a Lab Tec II 8 chambered cover glass with cover #1.5 μm borosilicate sterile slides (Thermo Fisher Scientific Inc.) at 1.2-5 × 103 cells per well and incubated overnight. Following cell attachment overnight, cells were washed once with warm HBSS and incubated in serum-medium containing 10 μg/ml of pHrodo red dextran (Life Technologies cat. #P10361) for staining acidic vesicles overnight. On day 2, cells were washed twice with cold HBSS. Anti-N3 and anti-N3(i) mAbs were directly conjugated to Alexa Fluor 488 (Life Technologies Protein Labeling kit #A30052) according to manufacturer’s instructions. The anti-NOTCH3 fluorescently conjugated antibodies then were added to the cells at a concentration 10 μg/ml in 2% BSA/HBSS for 30 minutes on wet ice. After 30 minutes, cells were washed 2x with cold HBSS and cold medium was added. Cells were imaged with an Ultraview spinning disk confocal microscope (PerkinElmer) every 10 minutes for 20 hr. A series of 10 μm optical image sections were acquired over time and a z stack of maximum intensity projections were generated with the U2OS-hN3 cells. The above procedure was followed as well when cells were imaged live with anti-TfR-Alexa647 and CT-β-Alexa647 with the addition of anti-N3-Alexa488 or anti-N3(i)-Alexa488.
MDA-MB-468 cells were seeded at 1.2 × 103 cells per well and incubated overnight. Following day, cells were washed once with warm HBSS and incubated with 10 μg/ml of pHrodo Red dextran and 10 μg/ml anti-N3-Alexa488, anti-N3(i)-Alexa488. At 24 hr and 48 hr cells were washed once with HBSS and warm medium added before imaged live in a temperature and humidity regulated environment on Ultraview spinning disk confocal. Multiple fields were collected with z stacks 10 μm thickness and each optical slice was to 0.2 μm. Images were deconvolve using the iterative restoration software within Volocity 6.3. Colocalization analysis was performed using Volocity 6.3 software, with calculation of thresholded Pearson’s correlation coefficients and Manders colocalization coefficients.
Anti-NOTCH3 antibody TEC into ligand cells
For live imaging, U2OS-hN3 cells were plated in a Lab Tec II 2 chambered cover glass slides at 0.5 × 104 cells per well and incubated overnight. HEK-control and HEK-DLL4 cells were also plated in a Lab Tec II 2 chambered cover glass slides at 4 × 105 and incubated overnight at 37°C. Next day, HEK-control and HEK-hDLL4 were washed once with warm HBSS and incubated in serum-medium containing 10 μg/ml of pHrodo Red dextran overnight. The following day, U2OS-hN3 cells were washed once with cold HBSS and incubated with 1 μg/ml of anti-N3-Alexa488 or with 1 μg/ml of anti-N3(i)-Alexa488 for 30 minutes on wet ice. HEK control and HEK-DLL4 cells were also trypsinized and counted. After 30 minutes, U2OS-hN3 cells were washed once with cold HBSS and HEK control or HEK-hDLL4 cells were added in cold serum-containing medium at a concentration of 1.2 × 104 cells in each well. Slides were spun down for 1 minute at 200 g and then were imaged live every 10-20 minutes for 20-24 hr on the Ultraview spinning disk confocal microscope.
For indirect immunofluorescence, U2OS-hN3 cells were plated in a 96 well plate at a concentration 0.5 x104 per well and incubated overnight. Next day, HEK-control and HEK-DLL4 cells were stained with MitoTracker Deep Red (ThermoFisher cat. # M22426) at 100 nM for 30 minutes at 37°C. HEK-control and HEK-DLL4 cells were washed twice with cold HBSS (GIBCO Life Technologies, cat. #14025-092), trypsinized and counted. U2OS-hN3 cells were then washed once with cold HBSS and incubated with 1 μg/ml anti-N3 at 4°C for 30 minutes. After 30 minutes, U2OS-hN3 cells were washed once with cold HBSS and then HEK-control or HEK-DLL4 cells were added at a concentration of 1.2 × 104 cells. Plates were incubated at 37°C for 30 minutes, 1.5, 3 and 8 hr. At each time point, plates were fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.5% Triton X-100 for 5 minutes, and blocked with 3% BSA for 30 minutes. Then, plates were washed 2x with cold PBS and incubated with goat anti-human Alexa Fluor 488 secondary antibody for 1 hr at room temperature, washed three times with PBS and imaged. Images were acquired on the Zeiss LSM510 confocal microscope.
For GSI-treatments, U2OS-hNOTCH3 cells were plated in Lab Tec II 2 chambered cover glass slides at 0.5 × 104 cells per well and incubated overnight. After 24 hours the cells were treated with γ-secretase inhibitor (PF-03084014) and drug remained present throughout the entire experiment. Next day, HEK control and HEK-DLL4 cells were lablelled with CellTracker Green CMFDA (Invitrogen C7025) with 5 μM concentration. U2OS-hN3 cells were washed twice with HBSS (Life Technologies 14025-092) and then anti-NOTCH3 antibodies were added at 10 μg/ml in 2%BSA/HBSS on ice for 30 minutes. Cells were washed and then media was added. HEK control or HEK-DLL4 cells were added accordingly on top of U2OS-hN3 at approximately 1:1 ratio. Cells were spun down onto U2OS-hN3 cells for 1 minute at 100 g and incubated at 37°C for 2 hours, washed and then fixed with 4% paraformaldehyde (Electron Microscopy Sciences 15710) for 10 minutes. Cells were permeabilized with 0.5% Triton X-100 (Thermo Scientific 28314) 5 minutes, blocked with 3%BSA/DPBS for 1 hour and then washed. Secondary anti-hIgG Alexa Fluor 647 (Life technologies) antibody was added at 1:500 for 45 minutes at room temperature. Finally, 0.5 μg/ml of DAPI was added for 1 hour to the cells and then washed. Cells were imaged on the Zeiss LSM510 confocal microscope.
Mechanism of action for Anti-N3 ADC-treated cells
OVCAR-3 cells were seeded in a Lab Tec II 4 chambered cover glass with cover #1.5 μm borosilicate sterile slides (Thermo Fisher Scientific Inc.) and left to attached overnight. Next day, cells were treated with 1.0 μg/ml of the anti-N3 ADC and control ADC, anti-N3, free auristatin or left untreated. Forty-eight hours later, cells were washed twice with HBSS and fixed in 4% paraformaldehyde for 10 minutes. Cells were washed three times with PBS and permeabilized with 0.5 % Triton X-100 for 2 minutes. After washing cells three times with PBS, cells were blocked with 3%BSA/PBS for 30 minutes at room temperature. Cells were incubated with 2 μg/ml of anti-α-Tubulin (Millipore, cat#05-829) and anti-phospho-Histone-H3 (Cell Signaling, #9701) at 1:600 in 2%BSA/PBS for 2 hours at room temperature. After 2 hours, cells were washed twice with PBS and then incubated with a 1:500 dilution of goat anti-mouse Alexa Fluor 488, goat anti-rabbit Alexa Fluor 555 and 1 μg/ml of DAPI (Sigma) for 45 minutes at room temperature. Finally, cells were washed twice with PBS and then samples were imaged on Zeiss LSM710 confocal laser microscope.
Live imaging of Trans-endocytosis assay with ADCs
U2OS-hN3 cells were plated in a Lab Tec II 2 chambered cover glass slides at 0.5 × 104 cells per well and incubated overnight. HEK parent and HEK-DLL4 cells were also plated in a Lab Tec II 2 chambered cover glass slides at 4 × 105 and incubated overnight at 37°C. Next day, HEK parent and HEK-DLL4 were washed once with warm HBSS and incubated in serum-medium containing 10 μg/ml of pHrodo Red dextran overnight. The following day, U2OS-hN3 cells were washed once with cold HBSS and incubated with 1 μg/ml of anti-N3-Alexa488 and 4 μg/ml of Anti-N3 ADC or with 1 ug/ml of anti-N3(i)-Alexa488 and 4 ug/ml of anti-N3(i) ADC for 30 minutes on wet ice. HEK parent and HEK-DLL4 cells were also trypsinized and counted. After 30 minutes, U2OS-hN3 cells were washed once with cold HBSS and then added HEK parents or HEK-DLL4 cells in cold serum-containing medium and at a concentration of 1.2 × 104 cells in each well with the addition of 2 μg/ml of anti-N3 ADC or anti-N3(i) ADC and 5 μM CellEvent Caspase-3/7 detection reagent (LifeTechnologies #C10423). Slides were spun down for 1 minute at 200 g and then were imaged every 2 hr for 72 hr on Ultraview spinning disk confocal. A series of 10 μm optical image sections were acquired over time and a z stack of maximum intensity projections were generated with the U2OS-hN3 and HEK-DLL4 co-cultures to create 3D volumetric renderings. Using Volocity 6.3 software (Perkin Elmer), compartmentalization analysis was used to analyze caspase positive compartments containing pHrodo positive compartments in HEK cells when in co-culture with U2OS-hN3 and treated with N3-ADCs.
ADC bioconjugation
The N3-ADCs are composed of either the humanized anti-N3 clone 28 or anti-N3(i) clone 75 IgG1 antibodies conjugated to the cytotoxic agent auristatin Aur0101 (PF-06380101) via a valine-citrulline (mcValCitPABC) linker64. The control ADC generated from the human antibody 8.8 was previously described8. ADCs were prepared via partial reduction of the antibody with tris(2-carboxyethyl)phosphine (TCEP) followed by a reaction of reduced cysteine residues with the mcValCitPABC-Aur0101. Specifically, the antibody was partially reduced via addition of 2.7-2.8 fold molar excess of tris(2-carboxyethyl)phosphine (TCEP) in 100 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffer), pH 7.0 and 1 mM diethylenetriaminepentaacetic acid (DTPA) for 2 hours at 37°C. The mcValCitPABC-Aur0101 linker-payload was then added to the reaction mixture at a linker-payload/antibody molar ratio of about 7 and reacted for an additional 1 hour at 25°C in the presence of 15% v/v of dimethylacetamide (DMA). After the 1 hr incubation period, 3-fold excess of N-ethylmaleimide was added to cap the unreacted thiols and was allowed to react for 15 minutes, followed by addition of 6-fold excess L-Cysteine to quench any unreacted linker-payload. The reaction mixture was dialyzed overnight at 4°C in phosphate buffered saline (PBS), pH 7.4 for Anti-N3 ADC and 20 mM Histidine, 50 mM Sucrose, pH 6.0 for anti-N3(i) ADC, and purified via SEC (AKTA explorer, Superdex 200). The ADC was further characterized via SEC for purity, hydrophobic interaction chromatography (HIC), and liquid chromatography electrospray ionization tandem mass spectrometry (LC-ESI MS) to calculate drug:antibody ratio (loading). The protein concentration was determined via UV spectrophotometer.
In vitro pharmacology
MDA-MB-468-hN3 cells were seeded at a density of 5 × 104 cells per well 24 hr before treatment. For 3D assays, MDA-MB-468 parental cells were seeded at 5 × 104 cells per well in Growth Factor Reduced Matrigel (Corning, #356231) and allowed to grow for 4 days prior to treatment. Cells were treated with 3-fold serially diluted NOTCH3-targeted or control ADCs, in triplicates at 10 concentrations ranging from 0-30,000 ng/ml. For 3D assays, media was removed, and cells were re-treated with compounds after 3-4 days. Relative cell viability was determined as percentage of untreated control 96 hours for 2D and 168 hours for 3D cultures after treatment using the Cell Titer 96® Aqueous Non-radioactive Cell Proliferation Assay kit (MTS, Promega, cat. no. G5430). IC50 values were calculated by logistic non-linear regression, model no. 203 with XL fit v4.2 (IDBS, Guildford, Surry, UK) and presented as ng/mL. Caspase-3/7 activity was measured with CellEvent Caspase-3/7 Green Detection Reagent according to manufacturer’s instructions (ThermoFisher Scientific, #C10423).
NOTCH3 knockdown with siRNAs were generated using ON-TARGET plus SMART pool human NOTCH3 (L-011093-00), ON-TARGET plus Control Non-Targeting pool (D-001810-10) (Thermo Scientific Dharmacon) and Lipofectamine RNAiMAX Reagent (Invitrogen). MDA-MB-468-hN3 cells were plated in 10 cm dishes in growth medium without antibiotics the day before transfection. The next day fresh medium without antibiotics was added. The siRNAs and the Lipofectamine RNAiMAX were diluted in OPTI-MEM media and used as per the manufacturer’s specifications. The cells were incubated with the transfection mixture for 24 hr in a humidified, 37°C, 5% CO2 incubator. After 24 hr, cells were trypsinized and plated for assessment using MTS cellular viability indicator as described above. Immunoblot analysis on extracts prepared from control and NOTCH3 siRNA-treated cells was performed to confirm that NOTCH3 receptor knockdown occurred by 24 hr.
Immunohistochemistry
IHC was used to detect NOTCH3 in human tumor xenografts. A tissue fragment from each xenograft was formalin-fixed and paraffin embedded (FFPE) using standard histological procedures. Five-micron thick sections were cut, deparaffinized in xylene substitute and rehydrated with graded alcohols to distilled water. Antigens were retrieved in 10 mM Citrate buffer pH 6.0 (Invitrogen) in a pressure cooker (Retriever; Electron Microscopy Sciences) and cooled to room temperature. Endogenous peroxidase was blocked with 3.0% hydrogen peroxide for 10 minutes. Non-specific protein interactions were blocked with Protein block (DAKO) for 20 minutes. Tissue sections were incubated with 0.659 μg/mL rabbit anti-NOTCH3 (D11B8; Cell Signaling Technologies) for 1 hr at room temperature. Anti-NOTCH3 primary antibody was detected with Signalstain Boost reagent (Cell Signaling Technologies #8114) for 30 minutes at room temperature. DAB+ (3′,3′-Diaminobenzidine; DAKO) was used to develop color for 5 minutes. Sections were briefly counterstained in hematoxylin QS (Vector Laboratories), washed in water, dehydrated in graded alcohols, cleared in xylene substitute, mounted with Permount Mounting Medium (Fisher Chemicals, Fair Lawn, NJ) and coverslipped.
For pharmacodynamic biomarker analysis, anti-N3 ADC-treated MDA-MB-468 xenografts were analyzed by IHC. A tissue fragment from each xenograft was formalin-fixed and paraffin embedded (FFPE) using standard histological procedures. Five-micron thick sections were cut, deparaffinized in xylene substitute and rehydrated with graded alcohols to distilled water. Antigens were retrieved in 10 mM Citrate buffer pH 6.0 (Invitrogen) for NOTCH3 and phospho-Histone H3 detection; Borg Decloaker buffer pH 9.5 (Biocare Medical) for anti-human IgG and anti-Aur0101 detection in a pressure cooker (Retriever; Electron Microscopy Sciences) and cooled to room temperature. Endogenous peroxidase was blocked with 3.0% hydrogen peroxide for 10 minutes. Non-specific protein interactions were blocked with Protein block (DAKO) for 20 minutes. Tissue sections were incubated with primary antibody for 1 hour at room temperature. Primary antibodies were: 0.659 μg/mL rabbit anti-NOTCH3 (D11B8; Cell Signaling Technologies); 0.3 μg/mL anti-human Pan IgG antibody (Epitomics #3443-1); 10 μg/mL anti-Aur0101 (Pfizer Inc., Lot 1D8.C9); 0.13 μg/mL anti-phospho Histone H3 (Cell Signaling Technologies, #9701). Rabbit IgG Isotype mAb (Cell Signaling Technologies #3900) and mouse IgG (Life Technologies #026502) served as isotype controls. Rabbit primary antibodies were detected with Signalstain Boost reagent (Cell Signaling Technologies #8114) for 30 minutes at room temperature. Alexa Fluor 488 labeled primary antibodies were detected with 1 μg/ml rabbit anti-Alexa Fluor 488 (Life Technologies A11094) for 45 minutes at room temperature, followed by Signalstain Boost reagent (Cell Signaling Technologies #8114) for 30 minutes at room temperature. DAB+ (3′,3′-Diaminobenzidine; DAKO) was used to develop color for 5 minutes. Sections were briefly counterstained in hematoxylin QS (Vector Laboratories), washed in water, dehydrated in graded alcohols, cleared in xylene substitute, mounted with Permount Mounting Medium (Fisher Chemicals, Fair Lawn, NJ) and coverslipped. To avoid mouse on mouse detection, anti-Aur0101 and mouse IgG isotype antibodies were labeled with Alexa Fluor 488 using Life Technology Alexa Fluor 488 protein labeling kit (#A10235).
For sequential IHC chromogenic staining NOTCH3 with JAG1, a 37622 tumor xenograft was formalin-fixed for 48 hr, processed and paraffin embedded (FFPE) using standard histological procedures. Five-micron thick serial sections were cut, deparaffinized in xylene substitute and rehydrated with graded alcohols to distilled water. Antigens were retrieved in 10 mM Citrate buffer pH 6.0 (Invitrogen) in a pressure cooker (Retriever; Electron Microscopy Sciences) and cooled to room temperature. Endogenous peroxidase was blocked with 3.0% hydrogen peroxide for 10 minutes. Non-specific protein interactions were blocked with Protein block (DAKO) for 20 minutes. For sequential chromogenic staining, tissue sections were incubated with rabbit anti-NOTCH3 (1:1000, Clone D11B8, Cell Signaling Technologies, #5276) antibodies for 1 hour at room temperature. Primary antibodies were detected with Signalstain Boost HRP reagent (Cell Signaling Technologies, #8114) for 30 minutes at room temperature. DAB+ (3′,3′-Diaminobenzidine; DAKO) was used to develop brown color for 5 minutes. Sections were rinsed in distilled water and blocked for peroxidase activity and non-specific protein interactions as mentioned above. Tissue sections were incubated with rabbit anti-JAG1 (1:500, Clone EPR4290, Abcam, ab109536) antibody for 1 hr at room temperature. JAG1 primary antibody was detected with MACH2 Rabbit AP polymer (Biocare Medical, ALP525) for 30 minutes at room temperature. Magenta color was developed using ImmPACT Vector Red (Vector Laboratories, SK-5105) for 7 minutes at room temperature. Sections were rinsed in distilled water and briefly counterstained in hematoxylin QS (Vector Laboratories), washed in tap water, dehydrated in graded alcohols, cleared in xylene substitute, mounted with Permount Mounting Medium (Fisher Chemicals, Fair Lawn, NJ) and coverslipped.
In vivo efficacy studies
All procedures using mice were approved by the Pfizer Institutional Animal Care and Use Committee according to established guidelines and in compliance with the Guide for the Care and Use of Laboratory Animals. MDA-MB-468 (orthotopic mammary fat pad) and OVCAR3 (subcutaneous) were implanted into SHO mice and HCC2429 and 37622 PDX fragments were implantation subcutaneously into athymic (Nu/Nu) mice, xenografts were grown until they reached an average tumor volume of 130-350 mm3 depending on the model. After xenografts were staged according to tumor volumes, mice were randomized into groups of 8-10 animals and then treated with N3-ADCs and a control ADC at concentrations of 1 or 3 mg/kg based on antibody content. Unconjugated humanized anti-N3 and chimeric anti-N3(i) mAbs were dosed at 10 mg/mg in the HCC2429 model. The ADC or unconjugated antibody was diluted in phosphate buffered saline (PBS) and doses were administered via intravenous tail vein injection every 4 days for 4 cycles (q4Dx4) starting at day 0. Cytotoxic chemotherapy Carboplatin (Hospira, Lot Z061709AA) was diluted in PBS and dosed intraperitoneally at 20 mg/kg, q7Dx7, in the OVCAR3 model. Cytotoxic chemotherapy Cisplatin (TEVA, Lot 11K175SA) was diluted in PBS and dosed intraperitoneally at 5 mg/kg q4Dx4 in 37622 PDX model. Tumor measurements were recorded one to two times per week using digital calipers. Tumor volume was calculated as [length x width x width] x 0.5 = volume in mm3. Body weights were recorded once per week. Generally, animals bearing tumors with volumes greater than 10% of body weight (approximately 2,500 mm3) were euthanized according to IACUC guidelines. Although remaining animals in the group were kept on study, the average tumor volume for a given group was not plotted and no statistical analysis was performed after > 15% of the animals were lost from the group. Tumor volume is plotted as Mean ± SEM.
Quantification and statistical analysis
Statistics were determined using an unpaired two-tailed t tests, one-way ANOVA, and linear regression analysis in GraphPad Prism software unless otherwise stated. Data are calculated as the mean ± SEM for biological triplicates and the mean ± SD for technical replicates unless otherwise stated. Statistical significance is represented in figures by: ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001, ∗∗∗∗, p < 0.0001.
Acknowledgments
We would like to thank Chris Brown, Weijun Ma, Darren Ferguson, Kelvin Kerns, and other BioMedicine Design colleagues for the preparation of protein reagents; Marc Damelin and Michelle Baxter for HEK-DLL4 cells; Matthew Doroski, Alex Porte, Hud Risley, Zecheng Chen, and Edmund Graziani for synthesis of the linker-payload; Frank Loganzo, Nadira Prasad, and Elwira Muszynska for analytical characterization of the ADCs; Mike Cinque, Xin Han, Chris Hosselet, Kenny Kim, Ed Rosfjord, LuAnna Tardio, Eric Upeslacis, Sharon Yang, and Johnny Yao for in vivo studies; John Jakubczak for tumor samples from the Pfizer Tissue Bank; and Marc Damelin, Shuang Chen, and Jeanine Pignatelli for discussions and/or assistance with manuscript preparation. The graphical abstract was created with Biorender.com. The results included here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga.
Author contributions
Conceptualization, K.G.G., Y.G., A.G., A.M., J.L., S.P.S., and M.R.; writing – original draft, K.G.G., Y.G., and A.G.; visualization, K.G.G., Y.G., A.G., J.B., and J.G.; methodology, K.G.G., Y.G., A.G., J.B., J.G., A.M., J.L., S.P.S., and M.R.; investigation, Y.G., A.G., L.S., T.T.Y., J.Z., R.K., J.B., N.P.N., M.C., J.G., M.G., J.Q., and T.F.; formal analysis, K.G.G., Y.G., A.G., M.R., J.Q., T.F., and W.Z.; data curation, J.B. and W.Z.; supervision, C.J.O., L.T., H.P.G., and P.S.
Declaration of interests
All authors are/were employees and shareholders of Pfizer. US patents associated with the development of NOTCH3-ADCs include 8,828,401 and 9,433,687.
Published: May 18, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100279.
Contributor Information
Kenneth G. Geles, Email: kgg11ggk@gmail.com.
Puja Sapra, Email: psapra111@gmail.com.
Supplemental information
References
- 1.Zhou B.B., Zhang H., Damelin M., Geles K.G., Grindley J.C., Dirks P.B. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat. Rev. Drug Discov. 2009;8:806–823. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
- 2.Leal M., Sapra P., Hurvitz S.A., Senter P., Wahl A., Schutten M., Shah D.K., Haddish-Berhane N., Kabbarah O. Antibody-drug conjugates: an emerging modality for the treatment of cancer. Ann. N Y Acad. Sci. 2014;1321:41–54. doi: 10.1111/nyas.12499. [DOI] [PubMed] [Google Scholar]
- 3.Rudin C.M., Pietanza M.C., Bauer T.M., Ready N., Morgensztern D., Glisson B.S., Byers L.A., Johnson M.L., Burris H.A., 3rd, Robert F., SCRX16-001 investigators Rovalpituzumab tesirine, a DLL3-targeted antibody-drug conjugate, in recurrent small-cell lung cancer: a first-in-human, first-in-class, open-label, phase 1 study. Lancet Oncol. 2017;18:42–51. doi: 10.1016/S1470-2045(16)30565-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saunders L.R., Bankovich A.J., Anderson W.C., Aujay M.A., Bheddah S., Black K., Desai R., Escarpe P.A., Hampl J., Laysang A. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci. Transl. Med. 2015;7:302ra136. doi: 10.1126/scitranslmed.aac9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Damelin M., Bankovich A., Park A., Aguilar J., Anderson W., Santaguida M., Aujay M., Fong S., Khandke K., Pulito V. Anti-EFNA4 Calicheamicin Conjugates Effectively Target Triple-Negative Breast and Ovarian Tumor-Initiating Cells to Result in Sustained Tumor Regressions. Clin. Cancer Res. 2015;21:4165–4173. doi: 10.1158/1078-0432.CCR-15-0695. [DOI] [PubMed] [Google Scholar]
- 6.Gong X., Azhdarinia A., Ghosh S.C., Xiong W., An Z., Liu Q., Carmon K.S. LGR5-Targeted Antibody-Drug Conjugate Eradicates Gastrointestinal Tumors and Prevents Recurrence. Mol. Cancer Ther. 2016;15:1580–1590. doi: 10.1158/1535-7163.MCT-16-0114. [DOI] [PubMed] [Google Scholar]
- 7.Junttila M.R., Mao W., Wang X., Wang B.E., Pham T., Flygare J., Yu S.F., Yee S., Goldenberg D., Fields C. Targeting LGR5+ cells with an antibody-drug conjugate for the treatment of colon cancer. Sci. Transl. Med. 2015;7:314ra186. doi: 10.1126/scitranslmed.aac7433. [DOI] [PubMed] [Google Scholar]
- 8.Damelin M., Bankovich A., Bernstein J., Lucas J., Chen L., Williams S., Park A., Aguilar J., Ernstoff E., Charati M. A PTK7-targeted antibody-drug conjugate reduces tumor-initiating cells and induces sustained tumor regressions. Sci. Transl. Med. 2017;9:eaag2611. doi: 10.1126/scitranslmed.aag2611. [DOI] [PubMed] [Google Scholar]
- 9.Sapra P., Damelin M., Dijoseph J., Marquette K., Geles K.G., Golas J., Dougher M., Narayanan B., Giannakou A., Khandke K. Long-term tumor regression induced by an antibody-drug conjugate that targets 5T4, an oncofetal antigen expressed on tumor-initiating cells. Mol. Cancer Ther. 2013;12:38–47. doi: 10.1158/1535-7163.MCT-12-0603. [DOI] [PubMed] [Google Scholar]
- 10.Aster J.C., Pear W.S., Blacklow S.C. The Varied Roles of Notch in Cancer. Annu. Rev. Pathol. 2017;12:245–275. doi: 10.1146/annurev-pathol-052016-100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bray S.J. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 12.Kopan R., Ilagan M.X. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haughian J.M., Pinto M.P., Harrell J.C., Bliesner B.S., Joensuu K.M., Dye W.W., Sartorius C.A., Tan A.C., Heikkilä P., Perou C.M., Horwitz K.B. Maintenance of hormone responsiveness in luminal breast cancers by suppression of Notch. Proc. Natl. Acad. Sci. USA. 2012;109:2742–2747. doi: 10.1073/pnas.1106509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sansone P., Ceccarelli C., Berishaj M., Chang Q., Rajasekhar V.K., Perna F., Bowman R.L., Vidone M., Daly L., Nnoli J. Self-renewal of CD133(hi) cells by IL6/Notch3 signalling regulates endocrine resistance in metastatic breast cancer. Nat. Commun. 2016;7:10442. doi: 10.1038/ncomms10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simões B.M., O’Brien C.S., Eyre R., Silva A., Yu L., Sarmiento-Castro A., Alférez D.G., Spence K., Santiago-Gómez A., Chemi F. Anti-estrogen Resistance in Human Breast Tumors Is Driven by JAG1-NOTCH4-Dependent Cancer Stem Cell Activity. Cell Rep. 2015;12:1968–1977. doi: 10.1016/j.celrep.2015.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shepherd C., Banerjee L., Cheung C.W., Mansour M.R., Jenkinson S., Gale R.E., Khwaja A. PI3K/mTOR inhibition upregulates NOTCH-MYC signalling leading to an impaired cytotoxic response. Leukemia. 2013;27:650–660. doi: 10.1038/leu.2012.285. [DOI] [PubMed] [Google Scholar]
- 17.Muellner M.K., Uras I.Z., Gapp B.V., Kerzendorfer C., Smida M., Lechtermann H., Craig-Mueller N., Colinge J., Duernberger G., Nijman S.M. A chemical-genetic screen reveals a mechanism of resistance to PI3K inhibitors in cancer. Nat. Chem. Biol. 2011;7:787–793. doi: 10.1038/nchembio.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi C., Qian J., Ma M., Zhang Y., Han B. Notch 3 protein, not its gene polymorphism, is associated with the chemotherapy response and prognosis of advanced NSCLC patients. Cell Physiol Biochem. 2014;34:743–752. doi: 10.1159/000363039. [DOI] [PubMed] [Google Scholar]
- 19.McAuliffe S.M., Morgan S.L., Wyant G.A., Tran L.T., Muto K.W., Chen Y.S., Chin K.T., Partridge J.C., Poole B.B., Cheng K.H. Targeting Notch, a key pathway for ovarian cancer stem cells, sensitizes tumors to platinum therapy. Proc. Natl. Acad. Sci. USA. 2012;109:E2939–E2948. doi: 10.1073/pnas.1206400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J.T., Chen X., Tropè C.G., Davidson B., Shih IeM., Wang T.L. Notch3 overexpression is related to the recurrence of ovarian cancer and confers resistance to carboplatin. Am. J. Pathol. 2010;177:1087–1094. doi: 10.2353/ajpath.2010.100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye Y.Z., Zhang Z.H., Fan X.Y., Xu X.L., Chen M.L., Chang B.W., Zhang Y.B. Notch3 overexpression associates with poor prognosis in human non-small-cell lung cancer. Med. Oncol. 2013;30:595. doi: 10.1007/s12032-013-0595-7. [DOI] [PubMed] [Google Scholar]
- 22.Park J.T., Li M., Nakayama K., Mao T.L., Davidson B., Zhang Z., Kurman R.J., Eberhart C.G., Shih IeM., Wang T.L. Notch3 gene amplification in ovarian cancer. Cancer Res. 2006;66:6312–6318. doi: 10.1158/0008-5472.CAN-05-3610. [DOI] [PubMed] [Google Scholar]
- 23.Reedijk M., Odorcic S., Chang L., Zhang H., Miller N., McCready D.R., Lockwood G., Egan S.E. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 24.Wang K., Zhang Q., Li D., Ching K., Zhang C., Zheng X., Ozeck M., Shi S., Li X., Wang H. PEST domain mutations in Notch receptors comprise an oncogenic driver segment in triple-negative breast cancer sensitive to a γ-secretase inhibitor. Clin. Cancer Res. 2015;21:1487–1496. doi: 10.1158/1078-0432.CCR-14-1348. [DOI] [PubMed] [Google Scholar]
- 25.Andersson E.R., Lendahl U. Therapeutic modulation of Notch signalling--are we there yet? Nat. Rev. Drug Discov. 2014;13:357–378. doi: 10.1038/nrd4252. [DOI] [PubMed] [Google Scholar]
- 26.Li K., Li Y., Wu W., Gordon W.R., Chang D.W., Lu M., Scoggin S., Fu T., Vien L., Histen G. Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J. Biol. Chem. 2008;283:8046–8054. doi: 10.1074/jbc.M800170200. [DOI] [PubMed] [Google Scholar]
- 27.Lin L., Mernaugh R., Yi F., Blum D., Carbone D.P., Dang T.P. Targeting specific regions of the Notch3 ligand-binding domain induces apoptosis and inhibits tumor growth in lung cancer. Cancer Res. 2010;70:632–638. doi: 10.1158/0008-5472.CAN-09-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katoh M., Katoh M. Precision medicine for human cancers with Notch signaling dysregulation (Review) Int. J. Mol. Med. 2020;45:279–297. doi: 10.3892/ijmm.2019.4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takebe N., Nguyen D., Yang S.X. Targeting notch signaling pathway in cancer: clinical development advances and challenges. Pharmacol. Ther. 2014;141:140–149. doi: 10.1016/j.pharmthera.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Damelin M., Zhong W., Myers J., Sapra P. Evolving Strategies for Target Selection for Antibody-Drug Conjugates. Pharm. Res. 2015;32:3494–3507. doi: 10.1007/s11095-015-1624-3. [DOI] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas N., Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Damelin M., Geles K.G., Follettie M.T., Yuan P., Baxter M., Golas J., DiJoseph J.F., Karnoub M., Huang S., Diesl V. Delineation of a cellular hierarchy in lung cancer reveals an oncofetal antigen expressed on tumor-initiating cells. Cancer Res. 2011;71:4236–4246. doi: 10.1158/0008-5472.CAN-10-3919. [DOI] [PubMed] [Google Scholar]
- 35.Gordon W.R., Vardar-Ulu D., Histen G., Sanchez-Irizarry C., Aster J.C., Blacklow S.C. Structural basis for autoinhibition of Notch. Nat. Struct. Mol. Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- 36.Dang T.P., Gazdar A.F., Virmani A.K., Sepetavec T., Hande K.R., Minna J.D., Roberts J.R., Carbone D.P. Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. J. Natl. Cancer Inst. 2000;92:1355–1357. doi: 10.1093/jnci/92.16.1355. [DOI] [PubMed] [Google Scholar]
- 37.Konishi J., Kawaguchi K.S., Vo H., Haruki N., Gonzalez A., Carbone D.P., Dang T.P. Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res. 2007;67:8051–8057. doi: 10.1158/0008-5472.CAN-07-1022. [DOI] [PubMed] [Google Scholar]
- 38.Wei P., Walls M., Qiu M., Ding R., Denlinger R.H., Wong A., Tsaparikos K., Jani J.P., Hosea N., Sands M. Evaluation of selective gamma-secretase inhibitor PF-03084014 for its antitumor efficacy and gastrointestinal safety to guide optimal clinical trial design. Mol. Cancer Ther. 2010;9:1618–1628. doi: 10.1158/1535-7163.MCT-10-0034. [DOI] [PubMed] [Google Scholar]
- 39.Bernasconi-Elias P., Hu T., Jenkins D., Firestone B., Gans S., Kurth E., Capodieci P., Deplazes-Lauber J., Petropoulos K., Thiel P. Characterization of activating mutations of NOTCH3 in T-cell acute lymphoblastic leukemia and anti-leukemic activity of NOTCH3 inhibitory antibodies. Oncogene. 2016;35:6077–6086. doi: 10.1038/onc.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choy L., Hagenbeek T.J., Solon M., French D., Finkle D., Shelton A., Venook R., Brauer M.J., Siebel C.W. Constitutive NOTCH3 Signaling Promotes the Growth of Basal Breast Cancers. Cancer Res. 2017;77:1439–1452. doi: 10.1158/0008-5472.CAN-16-1022. [DOI] [PubMed] [Google Scholar]
- 41.Xu X., Choi S.H., Hu T., Tiyanont K., Habets R., Groot A.J., Vooijs M., Aster J.C., Chopra R., Fryer C., Blacklow S.C. Insights into Autoregulation of Notch3 from Structural and Functional Studies of Its Negative Regulatory Region. Structure. 2015;23:1227–1235. doi: 10.1016/j.str.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fortini M.E., Bilder D. Endocytic regulation of Notch signaling. Curr. Opin. Genet. Dev. 2009;19:323–328. doi: 10.1016/j.gde.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nichols J.T., Miyamoto A., Olsen S.L., D’Souza B., Yao C., Weinmaster G. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J. Cell Biol. 2007;176:445–458. doi: 10.1083/jcb.200609014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meloty-Kapella L., Shergill B., Kuon J., Botvinick E., Weinmaster G. Notch ligand endocytosis generates mechanical pulling force dependent on dynamin, epsins, and actin. Dev. Cell. 2012;22:1299–1312. doi: 10.1016/j.devcel.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maderna A., Doroski M., Subramanyam C., Porte A., Leverett C.A., Vetelino B.C., Chen Z., Risley H., Parris K., Pandit J. Discovery of cytotoxic dolastatin 10 analogues with N-terminal modifications. J. Med. Chem. 2014;57:10527–10543. doi: 10.1021/jm501649k. [DOI] [PubMed] [Google Scholar]
- 46.Graziani E.I., Sung M., Ma D., Narayanan B., Marquette K., Puthenveetil S., Tumey L.N., Bikker J., Casavant J., Bennett E.M. PF-06804103, A Site-specific Anti-HER2 Antibody-Drug Conjugate for the Treatment of HER2-expressing Breast, Gastric, and Lung Cancers. Mol. Cancer Ther. 2020;19:2068–2078. doi: 10.1158/1535-7163.MCT-20-0237. [DOI] [PubMed] [Google Scholar]
- 47.Wu Y., Cain-Hom C., Choy L., Hagenbeek T.J., de Leon G.P., Chen Y., Finkle D., Venook R., Wu X., Ridgway J. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 48.Ferrarotto R., Eckhardt G., Patnaik A., LoRusso P., Faoro L., Heymach J.V., Kapoun A.M., Xu L., Munster P. A phase I dose-escalation and dose-expansion study of brontictuzumab in subjects with selected solid tumors. Ann. Oncol. 2018;29:1561–1568. doi: 10.1093/annonc/mdy171. [DOI] [PubMed] [Google Scholar]
- 49.Riccio O., van Gijn M.E., Bezdek A.C., Pellegrinet L., van Es J.H., Zimber-Strobl U., Strobl L.J., Honjo T., Clevers H., Radtke F. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008;9:377–383. doi: 10.1038/embor.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carulli A.J., Keeley T.M., Demitrack E.S., Chung J., Maillard I., Samuelson L.C. Notch receptor regulation of intestinal stem cell homeostasis and crypt regeneration. Dev. Biol. 2015;402:98–108. doi: 10.1016/j.ydbio.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.VanDussen K.L., Carulli A.J., Keeley T.M., Patel S.R., Puthoff B.J., Magness S.T., Tran I.T., Maillard I., Siebel C., Kolterud Å. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Domenga V., Fardoux P., Lacombe P., Monet M., Maciazek J., Krebs L.T., Klonjkowski B., Berrou E., Mericskay M., Li Z. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 2004;18:2730–2735. doi: 10.1101/gad.308904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joutel A., Corpechot C., Ducros A., Vahedi K., Chabriat H., Mouton P., Alamowitch S., Domenga V., Cécillion M., Marechal E. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 54.Donaghy H. Effects of antibody, drug and linker on the preclinical and clinical toxicities of antibody-drug conjugates. MAbs. 2016;8:659–671. doi: 10.1080/19420862.2016.1156829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Annunziata C.M., Kohn E.C., LoRusso P., Houston N.D., Coleman R.L., Buzoianu M., Robbie G., Lechleider R. Phase 1, open-label study of MEDI-547 in patients with relapsed or refractory solid tumors. Invest. New Drugs. 2013;31:77–84. doi: 10.1007/s10637-012-9801-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J.Y., Perry S.R., Muniz-Medina V., Wang X., Wetzel L.K., Rebelatto M.C., Hinrichs M.J., Bezabeh B.Z., Fleming R.L., Dimasi N. A Biparatopic HER2-Targeting Antibody-Drug Conjugate Induces Tumor Regression in Primary Models Refractory to or Ineligible for HER2-Targeted Therapy. Cancer Cell. 2016;29:117–129. doi: 10.1016/j.ccell.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 57.Parks A.L., Klueg K.M., Stout J.R., Muskavitch M.A. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 2000;127:1373–1385. doi: 10.1242/dev.127.7.1373. [DOI] [PubMed] [Google Scholar]
- 58.Ahimou F., Mok L.P., Bardot B., Wesley C. The adhesion force of Notch with Delta and the rate of Notch signaling. J. Cell Biol. 2004;167:1217–1229. doi: 10.1083/jcb.200407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin X., Li S., Wang Y.J., Wang Y., Zhong J.Y., He J.Y., Cui X.J., Zhan J.K., Liu Y.S. Exosomal Notch3 from high glucose-stimulated endothelial cells regulates vascular smooth muscle cells calcification/aging. Life Sci. 2019;232:116582. doi: 10.1016/j.lfs.2019.116582. [DOI] [PubMed] [Google Scholar]
- 60.Niu X., Gupta K., Yang J.T., Shamblott M.J., Levchenko A. Physical transfer of membrane and cytoplasmic components as a general mechanism of cell-cell communication. J. Cell Sci. 2009;122:600–610. doi: 10.1242/jcs.031427. [DOI] [PubMed] [Google Scholar]
- 61.Dufraine J., Funahashi Y., Kitajewski J. Notch signaling regulates tumor angiogenesis by diverse mechanisms. Oncogene. 2008;27:5132–5137. doi: 10.1038/onc.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geles K.G., Gao Y., Sapra P., Tchistiakova L.G., Zhou B.-B.S. US patent; 2016. Anti-Notch3 antibodies and antibody-drug conjugates. 9,433,687. [Google Scholar]
- 63.Harrison S., Adamson S., Bonam D., Brodeur S., Charlebois T., Clancy B., Costigan R., Drapeau D., Hamilton M., Hanley K. The manufacturing process for recombinant factor IX. Semin. Hematol. 1998;35(2, Suppl 2):4–10. [PubMed] [Google Scholar]
- 64.Doroski M.D., Maderna A., O’Donnell C.J., Subramanyam C., Vetelino B., Dushin R.G., Strop P., Graziani E.I. US patent; 2014. Cytotoxic peptides and antibody drug conjugates thereof. 8,828,401. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Live cell imaging of TEC with anti-N3 mAb (green) bound to U20S-hN3 cells and co-cultured with HEK-DLL4 cells (red). Scale bar, 20 μm.
Live cell imaging of TEC with anti-N3(i) mAb (green) bound to U20S-hN3 cells and co-cultured with HEK-DLL4 cells (red). Scale bar = 20 μm.
Control live cell imaging of anti-N3 mAb (green) bound to U20S-hN3 cells and co-cultured with HEK-parental cells (red). Scale bar = 20 μm.
Control live cell imaging of anti-N3(i) mAb (green) bound to U20S-hN3 cells and co-cultured with HEK-parental cells (red). Scale bar = 20 μm.
Data Availability Statement
All data associated with this study are available in the main text or supplemental materials. The accession number for atomic coordinates of anti-N3(i) clone 75 Fab-NOTCH3 NRR structure reported in this paper is PDB: 6XSW.