Abstract
Tendon plays a critical role in the joint movement by transmitting force from muscle to bone. This transmission of force is facilitated by its specialized structure, which consists of highly aligned extracellular matrix consisting predominantly of type I collagen. Tenocytes, fibroblast-like tendon cells residing between the parallel collagen fibers, regulate this specialized tendon matrix. Despite the importance of collagen structure and tenocyte function, the biological mechanisms regulating fibrillogenesis and tenocyte maturation are not well understood. Here we examine the function of Reticulocalbin 3 (Rcn3) in collagen fibrillogenesis and tenocyte maturation during postnatal tendon development using a genetic mouse model. Loss of Rcn3 in tendon caused decreased tendon thickness, abnormal tendon cell maturation, and decreased mechanical properties. Interestingly, Rcn3 deficient mice exhibited a smaller collagen fibril distribution and over-hydroxylation in C-telopeptide cross-linking lysine from α1(1) chain. Additionally, the proline 3-hydroxylation sites in type I collagen were also over-hydroxylated in Rcn3 deficient mice. Our data collectively suggest that Rcn3 is a pivotal regulator of collagen fibrillogenesis and tenocyte maturation during postnatal tendon development.
Subject terms: Developmental biology, Genetics
Introduction
Tendons are critical for native function of the musculoskeletal system. They are primarily composed of type I collagen and individual collagen fibrils arrange themselves into the highly organized bundles that can resist high tensile forces1. Tendons undergo significant changes in tissue and cell morphology throughout postnatal development. Tendon structure becomes denser and more organized via matrix production and collagen fibrillogenesis. Cell density becomes lower as matrix production increases and cell proliferation decreases2. The shape of tendon cell nuclei transform from round to a short spindles, and eventually to a long, flat spindles3. Previous studies have shown that several signaling pathways and transcription factors are critical regulators of embryonic and postnatal tendon development, but the precise regulatory mechanisms for cellular maturation and collagen fibrillogenesis during postnatal tendon development are still not clear4–11.
Developmental stage of tendons is marked by an increase in the number of collagen fibrils deposited in the matrix12. However, postnatal stages are shown to increase the diameter of existing collagen fibrils rather than an increase in collagen fibril number12. This transition occurs in the rough endoplasmic reticulum and involves a complex process wherein small collagen fibrils undergo lateral fusion to form thicker fibrils13. Mutations in several genes that regulate this process, including Decorin, Fibromodulin, and Lumican can cause defects in tendon maturation14–19.
Collagens consist of repeating Gly-X-Y triplets. Procollagen biosynthesis occurs in the ER and has characteristic post-translational modifications, including hydroxylation and glycosylation. Two modifications of proline residues are known, prolyl 4-hydroxylation and prolyl 3-hydroxylation13. Almost all proline residues found in the Y position undergo prolyl 4-hydroxylation. Prolyl 4-hydroxylation residues are believed to provide stability to the collagen triple helix through hydrogen bonds and water bridges20,21. Several tissue-specific prolyl 3-hydroxylation modification sites have been identified in tendon type I collagen, including α1(I) Pro986, α1(I) Pro707, α2(I) Pro707, and a C-terminal (GPP)n motif that are mainly modified by P3H2 and α1(I) Pro986 that is modified by P3H122. The total prolyl 3-hydroxylation content in the adult Achilles tendon collagen was higher than that in fetal Achilles tendon collagen23. Several studies suggested that prolyl 3-hydroxylation is involved in collagen fibril assembly22,24,25.
Reticulocalbin 3 (Rcn3), a 45 kDa ER lumen protein, is a member of the CREC (Cab45/reticulocalbin/ERC45/calumenin) family of multiple EF-hand Ca2+-binding proteins, including Rcn1, ERC-55 (Rcn2), Rcn3, Cab45, and calumenin26. Proteomic studies have suggested that Rcn3 functions as a chaperone protein in the secretory pathway and has emerged as a new potential regulator of collagen production27,28. Recent mouse genetic studies have demonstrated that Rcn3 plays a critical role in perinatal lung maturation by regulating synthesis and secretion of surfactant and is involved in pulmonary injury remodeling via regulation of alveolar epithelial cell (AEC) apoptosis and ER stress29,30. Despite this emerging role of Rcn3 in the secretory pathway, the physiological function of Rcn3 in various tissues producing a large amount of extracellular matrix is not yet fully established.
In this study, we investigated the physiological function of Rcn3 in tendon by conditionally deleting Rcn3 in mouse tendon cells. Loss of Rcn3 caused impaired postnatal tendon maturation with markedly decreased tendon thickness. Consistently, structural properties were reduced in Rcn3 deficient mice, but material properties were mostly unchanged. We further found that Rcn3 deficiency resulted in impaired collagen fibrillogenesis with over-modification of type 1 collagen and reduced fibril diameter distribution. Collectively, these findings, for the first time, establish the biological function of Rcn3 in postnatal tendon development and provide an exciting mouse genetic model for studying collagen fibrillogenesis and modification in tendon.
Results
Loss of Rcn3 caused decreased tendon thickness
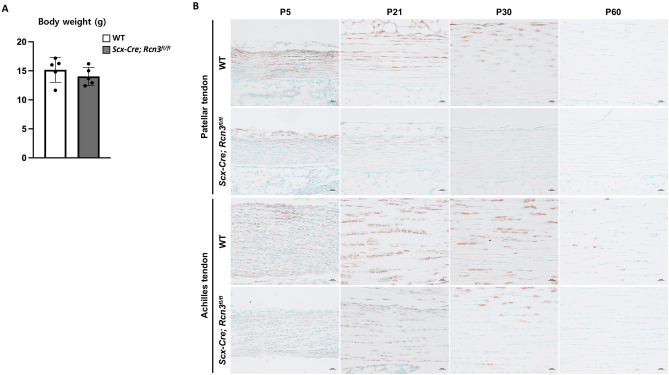
The endogenous Scleraxis (Scx) begins to be expressed in the lateral plate mesoderm and the body wall from E9.5, and its expression is restricted to the developing tendons at E14.531. Besides, Scx is expressed in tenocytes and directly regulates the expression of type I collagen (Col1a1) that is highly expressed in differentiated tenocytes32–34. Therefore, to understand the function of Rcn3 in tendons, we generated Scx-Cre; Rcn3fl/fl mice in which Rcn3 was specifically deleted in Scx-expressing cells that are abundant in tendons35,36. The growth of Scx-Cre; Rcn3fl/fl mice was normal compared with wild-type mice, which was evident with normal body weight (Fig. 1A). To access the knockout efficiency of Rcn3 in tendons, we performed immunohistochemistry analysis against Rcn3 (Fig. 1B). Rcn3 was highly expressed in patellar and Achilles tendons in wild-type mice at the early postnatal stage, including P5, P21, and P30, but its expression was decreased at a later stage, P60 (Fig. 1B). Immunohistochemistry analysis demonstrated that the deletion of Rcn3 in Scx-Cre; Rcn3fl/fl tendons were efficient in both patellar and Achilles tendons throughout the postnatal maturation (Fig. 1B). In the patellar tendon, we observed residual Rcn3 expression at P5 and P21, but the expression was not detected at P30 and P60. In the Achilles tendon, we found residual expression till P30, but the expression was not detected at P60. These data suggest that the deletion of Rcn3 is efficient in the patellar and Achilles tendons.
Figure 1.
The weight of wild-type mice and Scx-Cre; Rcn3fl/fl mice at P30 (A). Immunohistochemical analysis of Rcn3 on patellar and Achilles tendons from wild-type mice and Scx-Cre; Rcn3fl/fl littermates during postnatal tendon maturation (B). (Brown color indicates Rcn3, Scale bar indicates 20 µm (B) and n = 5 (A)).
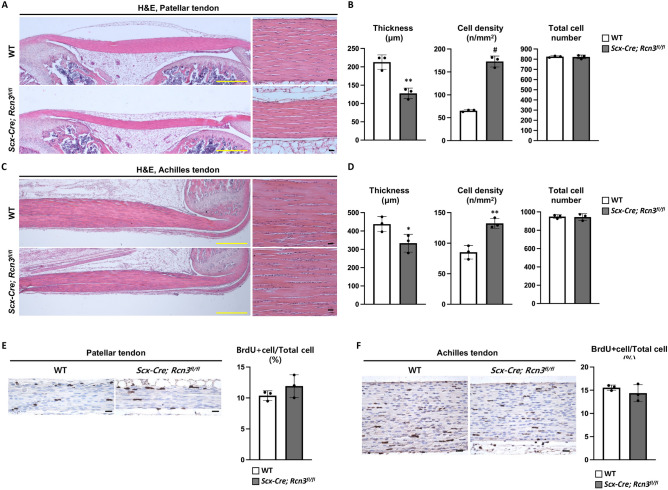
We performed histological analyses on patellar and Achilles tendons to assess the tendon phenotypes in Scx-Cre; Rcn3fl/fl mice. Although Scx-Cre; Rcn3fl/fl mice had no difference in body weight compared to wild-type mice (Fig. 1A), Scx-Cre; Rcn3fl/f mice clearly displayed decreased tendon thickness at P30 compared to wild-type littermate in both patellar (Fig. 2A) and Achilles tendon (Fig. 2C). Our quantification results confirmed the 40% reduced thickness in the patellar tendon and 20% reduced thickness in the Achilles tendon (Fig. 2B,D). We also found that the cellularity of tendons in Scx-Cre; Rcn3fl/fl mice was higher than wild-type mice, with threefold and 1.5-fold increases in the patellar and Achilles tendons, respectively (Fig. 2B,D). The increased cell density without the change in total cell number (Fig. 2B,D) prompted us to investigate cellular proliferation in Scx-Cre; Rcn3fl/fl mice. We found no differences in cell proliferation of Scx-Cre; Rcn3fl/fl mice (Fig. 2E,F), suggesting that reduced matrix formation likely caused increased cell density. These results collectively suggest that Rcn3 is critical for postnatal tendon development for both patellar and Achilles tendons.
Figure 2.
Histology of patellar and Achilles tendons from wild-type mice and Scx-Cre; Rcn3fl/fl littermates during postnatal tendon development (A,C). The thickness, cell density and total cell number of patellar tendon and Achilles tendon at P30 (B,D). The BrdU staining and quantification of BrdU positive cell ratio in patellar and Achilles tendon at P10 (E,F) (Yellow scale bar indicates 500 µm (A) and black scale bar 20 µm (A,C,E,F), * indicates P < 0.05, ** indicates P < 0.01 and # indicates P < 0.001 between genotypes, n = 3).
Loss of Rcn3 reduced collagen fibril diameter
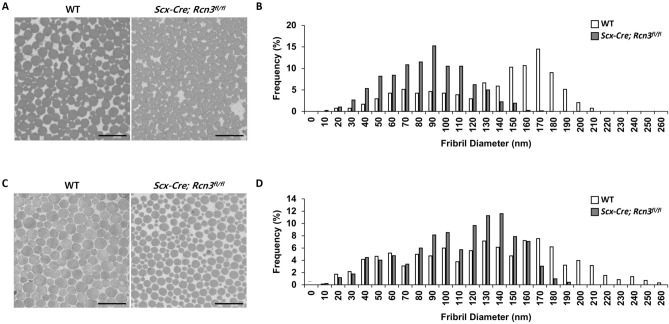
The decreased tendon thickness prompted us to examine collagen fibril ultrastructure because collagen fibrillogenesis is critical for tendon growth. We performed transmission electron microscopy (TEM) using patellar and Achilles tendons at 2 months of age to measure collagen fibril diameter. Scx-Cre; Rcn3fl/fl mice exhibited decreased fibril diameter in both patellar (Fig. 3A) and Achilles tendons (Fig. 3C) compared with wild-type mice. Specifically, quantification results showed that the distribution of collagen fibril diameter in patellar tendon ranged from 20 to 210 nm in wild-type mice, whereas Scx-Cre; Rcn3fl/fl mice showed a narrower collagen fibril distribution (10–170 nm) in the patellar tendon lacking the larger fibrils found in WT mice (Fig. 3B). In the patella tendon, control mice had the peak number of collagen fibril diameter at 170 nm, and Scx-Cre; Rcn3fl/fl mice had the peak number at 90 nm. Consistent with the patellar tendon, the Achilles tendon also showed the same trend with a shifted distribution in the mutants lacking the larger fibrils (i.e., 190–260 nm) found in WT mice (Fig. 3D). In the Achilles tendon, control mice had the peak number of collagen fibril diameter at 170 nm, and Scx-Cre; Rcn3fl/fl mice had the peak number at 140 nm. These results suggest that Rcn3 is required for proper collagen fibrillogenesis, and decreased tendon thickness could be due to the smaller diameter of collagen fibril in Scx-Cre; Rcn3fl/fl mice.
Figure 3.
EM image of patellar (A) and Achilles (C) tendons and frequency graph of patellar (B) and Achilles (D) tendons of wild-type mice and Scx-Cre; Rcn3fl/fl littermates at P60. (Scale bar indicates 500 nm (A, C), n = 3 animals).
Loss of Rcn3 increased collagen modification
Collagen post-translational modifications and cross-linking analysis were investigated in Scx-Cre; Rcn3fl/fl mice at 1 month of age. The modification state of several key residues was examined using peptide mass spectrometry. Notable findings include the C-telopeptide cross-linking lysine from α1(I) chain, which was slightly over-hydroxylated in Scx-Cre; Rcn3fl/fl patellar (20% more hydroxylated) and Achilles (9% more hydroxylated) tendons compared to wild-type mice (Table 1A). These data support that partial telopeptide lysine hydroxylation in tendon is being shifted to fuller hydroxylation in Scx-Cre; Rcn3fl/fl mice. This is a useful mark of altered collagen cross-linking and is consistent with an increase in pyridinoline cross-links. Indeed, the cross-linking analysis revealed an increase in hydroxylysylpyridinoline in both tendons of the Scx-Cre; Rcn3fl/fl mice. It should be noted that while hydroxylysylpyridinoline provides a useful measure of stable (irreversible) cross-links, it is not a measure of the total cross-links (Table 1B). The pattern of collagen over-modification was also observed across the sites of proline 3-hydroxylation in type I collagen in Scx-Cre; Rcn3fl/fl Achilles and patellar tendons (Table 1). A function has yet to be discovered for this rare modification, but it has been suggested that 3-hydrxyproline can regulate the postnatal lateral fusion of smaller fibrils in developing tendons23.
Table 1.
Collagen cross-link and mass spectral analysis to investigate the hydroxylation levels (A) of lysine and proline in type 1 collagen (n = 1 for patellar and n = 2 for Achilles tendon) and the collagen cross-linking levels (B) (* indicates P < 0.05 between genotypes, n = 1 for patellar and n = 3 for Achilles tendon) from wild-type mice and Scx-Cre; Rcn3fl/fl (tendon-specific Rcn3 loss-of-function model) littermates at P30.
| A | ||||
|---|---|---|---|---|
| Hydroxylation analysis | Patellar tendon (n = 1) (%) | Achilles tendon (n = 2) (%) | ||
| Site | WT | Scx-Cre; Rcn3fl/fl | WT | Scx-Cre; Rcn3fl/fl |
| Hydroxylysine | ||||
| α1(I)K87 | 100 | 100 | 100 | 100 |
| 100 | 100 | |||
| α2(I)K87 | 100 | 100 | 100 | 100 |
| 100 | 100 | |||
| α1(I) C-telo Hyl | 45 | 65 | 33 | 53 |
| 44 | 42 | |||
| 3-Hydroxyproline | ||||
| α1(I)P986 | 92 | 96 | 93 | 93 |
| 91 | 95 | |||
| α1(I)P707 | 60 | 80 | 50 | 76 |
| 50 | 65 | |||
| α2(I)P707 | 45 | 65 | 79 | 85 |
| 74 | 88 | |||
| B | ||||
|---|---|---|---|---|
| Cross-linking analysis | Patellar tendon (n = 1) | Achilles tendon (n = 3) | ||
| Site | WT | Scx-Cre; Rcn3fl/fl | WT | Scx-Cre; Rcn3fl/fl |
| Hydroxylysylpyridinoline (mol/mol) | 0.25 | 0.56 | 0.07 | 0.11 * |
| 0.07 | 0.21 | |||
| 0.075 | 0.19 | |||
Loss of Rcn3 caused abnormal tendon cell morphology
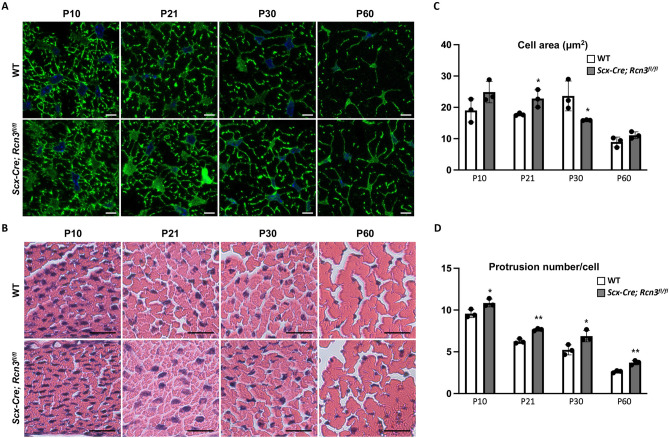
To investigate the cellular phenotype caused by the deletion of Rcn3, we analyzed tendon cell morphology using confocal microscopy after Zo1 and phalloidin double staining (Fig. 4A). Tendon cell areas became largest at P30 and dramatically decreased at P60 in wild-type mice (Fig. 4C, white bar). Interestingly, tendon cells in Scx-Cre; Rcn3fl/fl mice exhibited increased cell area at P10 and P21 but significantly decreased cell areas at P30 compared to wild-type mice (Fig. 4C). We also found changes in protrusion number in tendon cells of Scx-Cre; Rcn3fl/fl mice. These protrusions from the cell body are critical for tendon cell interaction with adjacent cells and the ECM. The protrusion number, in these transverse sections, decreased during postnatal tendon maturation in wild-type mice (Fig. 4D, white bar). The Scx-Cre; Rcn3fl/fl mice showed a significantly increased protrusion number compared with wild-type mice at all stages of postnatal tendon development (Fig. 4D). Collectively, these results suggest that the function of Rcn3 is indispensable for the morphological maturation of tendon cells.
Figure 4.
Immunofluorescence staining of ZO1 and phalloidin double staining on cross-sectional patellar tendon from wild-type mice and Scx-Cre; Rcn3fl/fl (tendon-specific Rcn3 loss-of-function model) littermates during postnatal tendon maturation (A). H&E staining of the cross-sectional patellar tendon (B). Cell area (C) and protrusion number (D) of the patellar tendon. (Scale bar indicates 5 µm (A) and 20 µm (B), * indicates P < 0.05, and ** indicates P < 0.01 between genotypes, n = 3).
Loss of Rcn3 increased the gene expression related to tenocyte differentiation
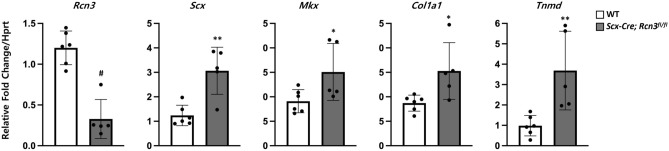
To further understand the cellular phenotype at the molecular level, we performed qRT-PCR using Achilles tendon from wild-type mice and Scx-Cre; Rcn3fl/fl mice. First, we confirmed the efficient deletion of Rcn3 in the Achilles tendon of Scx-Cre; Rcn3fl/fl mice (Fig. 5). The expression of tenogenic markers, such as Scx, Mkx, Col1a1, and Tnmd, were significantly increased in Scx-Cre; Rcn3fl/fl mice compared with wild-type mice (Fig. 5). These results suggest that the loss of Rcn3 enhanced tendon cell differentiation or delayed full tenocyte maturation.
Figure 5.
Quantitative real-time PCR of Achilles tendons from wild-type mice and Scx-Cre; Rcn3fl/fl (tendon-specific Rcn3 loss-of-function model) littermates at P28 (C). (* indicates P < 0.05, ** indicates P < 0.01 and # indicates P < 0.001 between genotypes, wild-type mice n = 6, Scx-Cre; Rcn3fl/fl mice n = 5).
Loss of Rcn3 decreased mechanical properties
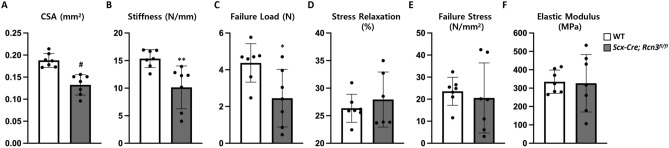
To measure the functional outcome of Rcn3 loss in tendons, we performed uniaxial biomechanical testing of the Achilles tendon at 2 months of age. Consistent with our histological analysis, cross-sectional area (CSA) was significantly reduced in Scx-Cre; Rcn3fl/fl tendons (Fig. 6A). Concomitantly, stiffness and failure load were also significantly lower, suggesting a loss of structural capability in the Scx-Cre; Rcn3fl/fl mice (Fig. 6B,C). No difference was observed in percent stress relaxation between the two groups, suggesting that Rcn3 does not perturb the viscoelastic response of the Achilles tendon (Fig. 6D). In addition, no differences were observed in failure stress or tissue elastic modulus, which indicates that loss of Rcn3 did not cause an inherent material change in the tendon tissue (Fig. 6E,F). These data demonstrate that reduced mechanical properties caused by loss of Rcn3 are due to decreased structural properties but not material properties.
Figure 6.
Mechanical property, cross-sectional area (A), stiffness (B), failure load (C), stress relaxation (D), failure stress (E), and elastic modulus (F) of Achilles tendons from wild-type mice and Scx-Cre; Rcn3fl/fl (tendon-specific Rcn3 loss-of-function model) littermates at P60. (* indicates P < 0.05, ** indicates P < 0.01, and # indicates P < 0.001 between genotypes, n = 7).
Discussion
We investigated the function of Rcn3 in postnatal tendon development by characterizing a tendon-specific Rcn3 knockout mouse (Scx-Cre; Rcn3fl/fl) model. The Scx-Cre; Rcn3fl/fl mice exhibited impaired postnatal tendon development, including decreased tendon thickness, increased cellularity, impaired tenocyte maturation, and reduced collagen fibril diameter with altered collagen modification. Our study identified the novel function of Rcn3 in tendon and provides genetic approaches that can be used to investigate the underlying mechanisms regulating cellular and matrix maturation in tendon.
Rcn3 is known as an ER protein regulating the secretion of proteins, but its critical molecular function is not clear. Our study showed that Rcn3 deficiency caused impaired collagen fibrillogenesis with over-modification of collagen. These results suggest that Rcn3 is a critical regulator of collagen fibrillogenesis via regulation of collagen modification. However, the precise molecular mechanism by which Rcn3 regulates collagen modification is still not clear. One possibility is that Rcn3, as a chaperon protein, stabilizes collagen for proper modification. Previous studies also suggested that Rcn3 functions as a chaperon protein27,28. The other possibility is that Rcn3 mediates the binding of collagen modification enzymes to collagen molecules. Further molecular analyses will be required to elucidate the precise function of Rcn3 in collagen modification in tendon.
To measure morphological changes of tendon cells at a single-cell level, we developed a novel confocal imaging method (Fig. 4). We found that the protrusion number was decreased throughout the postnatal tendon development in wild-type mice. Interestingly, this result is different from the current finding using serial block face-scanning electron microscopy (SBF-SEM)12. Kalson et al. showed that a similar number of protrusions contacting adjacent cells between the tendons of newborn and 6-week-old mice. There are several variations between our and Kalson’s study. First, we have analyzed the patellar tendon, but they analyzed the tail tendon. Second, the age is different. We have analyzed age from P10 to P60. They analyzed newborn and 6-week-old mice. These age and tendon types could cause different results of tendon phenotype in each study. Further studies analyzing various tendon types and ages using our confocal imaging technique and SBF-SEM will be necessary to fully understand the morphological maturation of tenocytes.
During normal tendon development, the average cross-sectional cell area became largest at P30 in wildtype (Fig. 4). However, Scx-Cre; Rcn3fl/fl mice showed a prematurely increased cell area with increased cell area at P10 and P21, while significantly decreased cell area at P30 compared to wildtype. Interestingly, the protrusion numbers in mutant mice are significantly increased at all stages, suggesting the delay of tenocyte maturation. These cellular changes correspond with the molecular analysis (Fig. 5) that shows increased expression of early tenogenic markers in Rcn3 deficient mice. It is also consistent with the immature growth of collagen fibril. Based on these data, we carefully suggest that Rcn3 positively regulates the maturation of differentiated tenocytes. Therefore, deletion of Rcn3 caused the delay of full maturation of tenocytes which cause increased protrusion number, increase expression of differentiated tenocyte genes, and immature growth of collagen fibril diameter. Investigating the mechanism by which Rcn3 regulates tenocyte maturation will be an interesting future direction of this study.
Scx-Cre; Rcn3fl/fl mice displayed an increase in post-translational modifications and cross-linking in type I collagen. Specifically, Scx-Cre; Rcn3fl/fl mice showed an increase in telopeptide lysine hydroxylation as well as hydroxylysylpyridinoline in both patellar and Achilles tendons. Hydroxylysylpyridinoline is a stable collagen cross-link, typically found in adult bone and tendon, that is known to increase in concentration as these tissues mature. Besides, Scx-Cre; Rcn3fl/fl mice exhibited over-hydroxylated proline 3-hydroxylation in type I collagen. The function of this rare modification is not clear, but it has been suggested that 3-hydrxyproline can regulate the postnatal lateral fusion of smaller fibrils in developing tendons23. Based on these collagen modification data, we carefully suggest that the reduced collagen fibril diameter in Scx-Cre; Rcn3fl/fl mice is due to the inhibition of lateral fusion caused by over-modification on type I collagen.
In conclusion, our results uncover the novel function of Rcn3 in postnatal tendon development. This study will be the basis of future mechanistic studies for tenocyte maturation, collagen modification, and fibrillogenesis by providing useful mouse genetic models.
Methods
Animals
All studies were approved by the Institutional Animal Care and Use Committee (IACUC) and University Laboratory Animal Resource (ULAR) at the University of Pennsylvania (Philadelphia, Pennsylvania, USA) and we confirm that all the methods in current studies were performed in accordance with the relevant guidelines and regulations following the IACUC approved protocol. The study was carried out in compliance with the ARRIVE guidelines37. The Scx-Cre mouse line was previously described38. Mouse sperms carrying Knockout-first Rcn3 allele (Rcn3 tm1a(EUCOMM)Hmgu) were obtained from the Knockout Mouse Programme (KOMP), and mouse carrying Rcn3 tm1a(EUCOMM)Hmgu was generated by in vitro fertilization at Baylor College of Medicine. Rcn3 tm1a(EUCOMM)Hmgu mice were crossed with Rosa26-Flippase (Flp) mice to delete β-gal and the neo cassette in order to generate the Rcn3 conditional knockout allele (Rcn3fl/fl).
Immunohistochemistry
For immunohistochemisty, mouse hindlimbs were collected during postnatal tendon maturation from P10 to P60 and fixed in 10% neutral buffered formalin overnight at 4 °C. Samples were paraffin-embedded and sectioned at 6 μm, following decalcification in 10% (w/v) EDTA (pH 7.4) for 2 weeks with daily solution changes. The paraffin sections of the patellar and Achilles tendon incubated with antibodies against Rcn3 (Abcam, ab204178) at 4 °C overnight. The sections were then incubated in anti-rabbit-biotin antibody (Jackson Immuno, 711-065-152) for 30 min at RT, rinsed with PBS, and incubated with streptavidin-HRP (Jackson Immuno, 016-030-084) for 30 min at RT. Bounded antibodies were detected using NovaRed substrate (Vector Laboratories, SK-4800).
Histological analyses
Mouse hindlimbs were collected from specific stages and fixed in 10% neutral buffered formalin overnight at 4 °C. Samples were paraffin-embedded and sectioned at 6 μm, following decalcification in 10% (w/v) EDTA (pH 7.4) for 2 weeks with daily solution changes. The paraffin sections of the patellar and Achilles tendons were used for Hematoxylin and Eosin (H&E). Histological evaluation was blindly performed by 2 independent individuals.
BrdU assay
For BrdU labeling in tendon, a single stock solution of 10 mg/mL BrdU (Sigma, B5002) and 1.2 mg/mL FdU (Sigma, F0503) was prepared in 1X PBS. BrdU/FdU stock (0.01 ml/g of body weight) was injected into the intraperitoneal space of mice at P8 and P9. And then, mouse hindlimbs were collected at P10 and fixed in 10% neutral buffered formalin overnight at 4 °C. Samples were paraffin-embedded and sectioned at 6 μm, following decalcification in 10% (w/v) EDTA (pH 7.4) for 1 week with daily solution changes. BrdU Immunohistochemistry Kit (Abcam, ab125306) was used to detect BrdU incorporation into the cells according to the manufacturer's instructions.
Collagen EM analysis
Mouse hindlimbs were fixed in freshly prepared 1.5% glutaraldehyde/1.5% paraformaldehyde (Tousimis) with 0.05% tannic acid (Sigma) in DPBS at 4 °C with agitation overnight. The following day, dissect the relevant tendons out of the limb in PBS as follows: for the Achilles, take the mid-tendons that lies below the myotendinous junction and above the enthesis (~ 1–1.5 mm in length); and for the patellar tendon, take the complete mid-tendon between the patella and tibial enthesis. Following dissection, samples were post-fixed in 1% OsO4, rinsed in DMEM and dehydrated in a graded series of ethanol to 100%. Samples were then rinsed in propylene oxide, infiltrated in Spurrs epoxy, and polymerized at 70 °C overnight. TEM images were acquired using a FEI G20 TEM at multiple magnifications to visualize transverse sections of collagen fibrils. Collagen fibril diameter was measured using ImageJ. TEM images of tendons from 3 mice per group were analyzed. Total 545 control fibrils and 905 Rcn3 mutant fibrils from patellar tendons were counted. Total 1568 control fibrils and 1871 Rcn3 mutant fibrils from Achilles tendons were counted.
Collagen extraction
Intact type I collagen was solubilized from mouse patellar and Achilles tendons by acid extraction in 3% acetic acid for 24 h at 4 °C. Acid-extracted collagen α-chains were resolved by 6% SDS-PAGE under non-reducing conditions and stained with Coomassie Blue R-250.
Collagen cross-linking analysis
Collagen pyridinoline cross-link content was determined by fluorescence monitoring with reverse-phase HPLC. Pyridinoline cross-links were analyzed in mouse tendons by HPLC after acid hydrolysis in 6 M HCl for 24 h at 108 °C. Dried samples were dissolved in 1% (v/v) n-heptafluorobutyric acid for quantitation of hydroxylysyl pyridinoline (HP) by reverse-phase HPLC and fluorescence monitoring as previously described39.
Mass spectrometry
Collagen α-chains were cut from SDS-PAGE gels and subjected to in-gel trypsin digestion as previously described24. Electrospray mass spectrometry was carried out on the trypsin-digested peptides using an LTQ XL linear quadrupole ion-trap mass spectrometer equipped with in-line Accela 1250 liquid chromatography and automated sample injection (ThermoFisher Scientific). Proteome Discoverer software (ThermoFisher Scientific) was used for peptide identification. Tryptic peptides were also identified manually by calculating the possible MS/MS ions and matching these to the actual MS/MS spectrum using Thermo Xcalibur software. Differences in post-translational modifications were determined manually by averaging the full scan MS over several LCMS minutes to include all the post-translational variations of a given peptide. Protein sequences used for MS analysis were obtained from the Ensembl genome database.
Immunofluorescent staining
For immunofluorescent staining, mouse hindlimbs were collected during postnatal tendon maturation from P10 to P60 and fixed in 4% PFA overnight at 4 °C. Samples were OCT compound embedded and sectioned at 10 μm, following decalcification in 10% (w/v) EDTA (pH 7.4) for 10 days with daily solution changes. The frozen sections of the patellar tendon blocked with blocking solution (Thermo, 8120) for 1 h RT, followed by incubation at 4 °C overnight with antibodies against ZO1 (Fisher, 61-7300) and then incubated for 1 h at RT with Phalloidin Alexa Fluor 488 conjugate. Finally, sections were mounted in antifade solution with DAPI (Invitrogen, P36935).
qRT-PCR
RNA was extracted with Trizol and Direct-zol kit (Zymo Research, R2060) from primary tendon cells. And cDNA was synthesized from 1 μg RNA using iScript Reverse Transcription (Bio-rad, 1708841). qRT-PCR was performed by QuantStudio 6 Flex real-time PCR system (ThermoFisher Scientific) using Fast SYBR Green Master Mix (Pec, 4385612). The sequences of primers are listed in Table 2.
Table 2.
Primer sequences for the quantitative real-time PCR.
| Gene | Primer sequences (5′–3′) | |
|---|---|---|
| mRcn3 | F: | CTA CTC GGG AAG AGC TGA CG |
| R: | ACC TGC ACG TAG CCA TCT TT | |
| mScx | F: | AAG ACG GCG ATT CGA AGT TAG AAG |
| R: | TCT CTC TGT TCA TAG GCC CTG CTC ATA G | |
| mMkx | F: | GAT GGC GAC TCC TGC TCT GA |
| R: | CGG TCT GCC GCC AGC TTT TA | |
| mCol1a1 | F: | TTG GGG CAA GAC AGT CAT CGA AT |
| R: | TTG GGG TGG AGG GAG TTT ACA CGA A | |
| mTnmd | F: | CTT TAC TAG GCT ACT ACC CAT ACC CCT ACT |
| R: | ATA TAT TGG CTA ACA GAA GGT TAA GCG TTT | |
Uniaxial biomechanical test
All mice assigned for mechanical testing were frozen at − 20 °C untill the day of testing. Mice were thawed at room temperature, and calcaneal bone—Achilles tendon—muscle complexes were grossly dissected. Under microscope guidance, all extraneous soft tissues and muscles were finely dissected with care to ensure no damage to the tendinous tissue. Following dissection, a custom laser device was used to measure the cross-sectional area (CSA) of the Achilles tendon. The myotendinous junction was sandwiched between two sandpaper tabs with cyanoacrylate glue to prevent slippage. The calcaneal bone was gripped with a custom fixture, and the construct was mounted onto a material testing machine (Instron 5542, Instron Inc., Norwood, MA). All testing was conducted in phosphate buffered saline bath at room temperature. Each sample was preloaded to 0.02 N followed by 10 cycles of preconditioning between 0.02 and 0.04 N. After a resting period of 300 s at 0 N, the sample underwent stress relaxation after a ramp to 5% strain (assuming a gauge length of 5 mm) for 600 s. This was followed by a rest of 60 s at zero load. Finally, the sample was quasi-statically ramped to failure at a strain rate of 0.03%/s. All data were collected at 100 Hz. Ensuing force–displacement curves were analyzed to obtain failure load (N) and tissue stiffness (N/mm, defined as the slope of the linear region). Cross-sectional areas (mm2) and gauge length (mm) values were used to obtain stress–strain curves for each sample. Elastic modulus (N/mm2) was calculated as the slope of the linear region of the stress–strain curve and failure stress (N/mm2) as the maximum stress value observed. Stress relaxation (%) was defined as the percent change in stress between the peak and equilibrium stress during the stress relaxation period of 600 s.
Statistical analysis
Results are expressed as mean ± SD. At least three mice per group were analyzed. Differences between values were analyzed by Student’s t test. P < 0.05, 0.01 and 0.001 are considered significant.
Acknowledgements
We would like to thank Penn Center for Musculoskeletal Disorders (PCMD) Histology and Biomechanics Core for technical assistance with the histology and biomechanical testing, respectively. We also thank Cell and Developmental Biology (CDB) Microscopy Core for technical assistance with confocal microscopy. We thank Dr. Ronen Schweitzer for providing Scx-Cre mouse line and Dr. Brendan Lee for providing Rcn3fl/fl mouse line.
Author contributions
Study conception and design: K.S.J., N.A.D., N.R.P., N.Q.; Acquisition of data: N.R.P., S.S., I.B., D.R.K., S.T., D.M.H., M.A.; Analysis and interpretation of data: N.R.P., D.M.H., D.R.K., L.J.S., N.A.D., K.S.J.; Drafting of the manuscript: K.S.J., N.R.P., D.M.H., D.R.K.
Funding
This work was supported in part by NIH/ NIAMS K01 AR069002 (K. S. J.). The PCMD Cores were supported by NIH/ NIAMS P30AR069619.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wang JH-C, Guo Q, Li B. Tendon biomechanics and mechanobiology—A minireview of basic concepts and recent advancements. J. Hand Ther. Off. J. Am. Soc. Hand Ther. 2012;25:133–140. doi: 10.1016/j.jht.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grinstein M, et al. A distinct transition from cell growth to physiological homeostasis in the tendon. Elife. 2019;8:e48689. doi: 10.7554/eLife.48689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J, et al. Characterization and comparison of post-natal rat Achilles tendon-derived stem cells at different development stages. Sci. Rep. 2016;6:22946. doi: 10.1038/srep22946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu W, et al. The atypical homeodomain transcription factor Mohawk controls tendon morphogenesis. Mol. Cell Biol. 2010;30:4797–4807. doi: 10.1128/MCB.00207-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang AH, et al. Requirement for scleraxis in the recruitment of mesenchymal progenitors during embryonic tendon elongation. Development. 2019;146:dev182782. doi: 10.1242/dev.182782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan G-K, et al. Tgfβ signaling is critical for maintenance of the tendon cell fate. Elife. 2020;9:e52695. doi: 10.7554/eLife.52695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim J, et al. mTORC1 signaling is a critical regulator of postnatal tendon development. Sci. Rep. 2017;7:17175. doi: 10.1038/s41598-017-17384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pryce BA, et al. Recruitment and maintenance of tendon progenitors by TGFbeta signaling are essential for tendon formation. Development. 2009;136:1351–1361. doi: 10.1242/dev.027342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito Y, et al. The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10538–10542. doi: 10.1073/pnas.1000525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murchison ND, et al. Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development. 2007;134:2697–2708. doi: 10.1242/dev.001933. [DOI] [PubMed] [Google Scholar]
- 11.Cong XX, et al. Activation of AKT-mTOR signaling directs tenogenesis of mesenchymal stem cells. Stem Cells. 2018;36:527–539. doi: 10.1002/stem.2765. [DOI] [PubMed] [Google Scholar]
- 12.Kalson NS, et al. A structure-based extracellular matrix expansion mechanism of fibrous tissue growth. Elife. 2015;4:e05958. doi: 10.7554/eLife.05958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004;20:33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Yoon JH, Halper J. Tendon proteoglycans: Biochemistry and function. J. Musculoskelet. Neuronal Interact. 2005;5:22–34. [PubMed] [Google Scholar]
- 15.Mao JR, Bristow J. The Ehlers-Danlos syndrome: On beyond collagens. J. Clin. Investig. 2001;107:1063–1069. doi: 10.1172/JCI12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majava M, et al. Novel mutations in the small leucine-rich repeat protein/proteoglycan (SLRP) genes in high myopia. Hum. Mutat. 2007;28:336–344. doi: 10.1002/humu.20444. [DOI] [PubMed] [Google Scholar]
- 17.Docheva D, Hunziker EB, Fässler R, Brandau O. Tenomodulin is necessary for tenocyte proliferation and tendon maturation. Mol. Cell Biol. 2005;25:699–705. doi: 10.1128/MCB.25.2.699-705.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dex S, et al. Tenomodulin is required for tendon endurance running and collagen I fibril adaptation to mechanical load. EBioMedicine. 2017;20:240–254. doi: 10.1016/j.ebiom.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ezura Y, Chakravarti S, Oldberg A, Chervoneva I, Birk DE. Differential expression of lumican and fibromodulin regulate collagen fibrillogenesis in developing mouse tendons. J. Cell Biol. 2000;151:779–788. doi: 10.1083/jcb.151.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenbloom J, Harsch M, Jimenez S. Hydroxyproline content determines the denaturation temperature of chick tendon collagen. Arch. Biochem. Biophys. 1973;158:478–484. doi: 10.1016/0003-9861(73)90539-0. [DOI] [PubMed] [Google Scholar]
- 21.Myers JC, Kivirikko S, Gordon MK, Dion AS, Pihlajaniemi T. Identification of a previously unknown human collagen chain, alpha 1(XV), characterized by extensive interruptions in the triple-helical region. Proc. Natl. Acad. Sci. U. S. A. 1992;89:10144–10148. doi: 10.1073/pnas.89.21.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eyre DR, Weis M, Hudson DM, Wu J-J, Kim L. A novel 3-hydroxyproline (3Hyp)-rich motif marks the triple-helical C terminus of tendon type I collagen. J. Biol. Chem. 2011;286:7732–7736. doi: 10.1074/jbc.C110.195768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taga Y, Kusubata M, Ogawa-Goto K, Hattori S. Developmental stage-dependent regulation of prolyl 3-hydroxylation in tendon type I collagen. J. Biol. Chem. 2016;291:837–847. doi: 10.1074/jbc.M115.686105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weis MA, et al. Location of 3-hydroxyproline residues in collagen types I, II, III, and V/XI implies a role in fibril supramolecular assembly. J. Biol. Chem. 2010;285:2580–2590. doi: 10.1074/jbc.M109.068726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hudson DM, et al. Post-translationally abnormal collagens of prolyl 3-hydroxylase-2 null mice offer a pathobiological mechanism for the high myopia linked to human LEPREL1 mutations. J. Biol. Chem. 2015;290:8613–8622. doi: 10.1074/jbc.M114.634915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honoré B. The rapidly expanding CREC protein family: Members, localization, function, and role in disease. BioEssays. 2009;31:262–277. doi: 10.1002/bies.200800186. [DOI] [PubMed] [Google Scholar]
- 27.Tsuji A, et al. A proteomic approach reveals transient association of reticulocalbin-3, a novel member of the CREC family, with the precursor of subtilisin-like proprotein convertase, PACE4. Biochem. J. 2006;396:51–59. doi: 10.1042/BJ20051524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martínez-Martínez E, et al. Differential proteomics identifies reticulocalbin-3 as a novel negative mediator of collagen production in human cardiac fibroblasts. Sci. Rep. 2017;7:12192. doi: 10.1038/s41598-017-12305-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin J, et al. Neonatal respiratory failure with retarded perinatal lung maturation in mice caused by reticulocalbin 3 disruption. Am. J. Respir. Cell Mol. Biol. 2016;54:410–423. doi: 10.1165/rcmb.2015-0036OC. [DOI] [PubMed] [Google Scholar]
- 30.Jin J, et al. Reticulocalbin 3 deficiency in alveolar epithelium exacerbated bleomycin-induced pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2018;59:320–333. doi: 10.1165/rcmb.2017-0347OC. [DOI] [PubMed] [Google Scholar]
- 31.Schweitzer R, et al. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development. 2001;128:3855–3866. doi: 10.1242/dev.128.19.3855. [DOI] [PubMed] [Google Scholar]
- 32.Léjard V, et al. Scleraxis and NFATc regulate the expression of the pro-alpha1(I) collagen gene in tendon fibroblasts. J. Biol. Chem. 2007;282:17665–17675. doi: 10.1074/jbc.M610113200. [DOI] [PubMed] [Google Scholar]
- 33.Huang AH, Lu HH, Schweitzer R. Molecular regulation of tendon cell fate during development. J. Orthop. Res. Off. Publ. Orthop. Res. Soc. 2015;33:800–812. doi: 10.1002/jor.22834. [DOI] [PubMed] [Google Scholar]
- 34.Carlberg AL, Tuan RS, Hall DJ. Regulation of scleraxis function by interaction with the bHLH protein E47. Mol. Cell Biol. Res. Commun. 2000;3:82–86. doi: 10.1006/mcbr.2000.0195. [DOI] [PubMed] [Google Scholar]
- 35.Sugimoto Y, Takimoto A, Hiraki Y, Shukunami C. Generation and characterization of ScxCre transgenic mice. Genesis. 2013;51:275–283. doi: 10.1002/dvg.22372. [DOI] [PubMed] [Google Scholar]
- 36.Pryce BA, Brent AE, Murchison ND, Tabin CJ, Schweitzer R. Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2007;236:1677–1682. doi: 10.1002/dvdy.21179. [DOI] [PubMed] [Google Scholar]
- 37.du Sert NP, et al. Reporting animal research: Explanation and elaboration for the ARRIVE guidelines 2.0. PLoS Biol. 2020;18:e3000411. doi: 10.1371/journal.pbio.3000411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blitz E, et al. Bone ridge patterning during musculoskeletal assembly is mediated through SCX regulation of Bmp4 at the tendon-skeleton junction. Dev. Cell. 2009;17:861–873. doi: 10.1016/j.devcel.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eyre D. Collagen cross-linking amino acids. Methods Enzymol. 1987;144:115–139. doi: 10.1016/0076-6879(87)44176-1. [DOI] [PubMed] [Google Scholar]