Abstract
Osteosarcoma is one of the most aggressive bone tumors in children and adolescents. Development of effective therapeutic options is still lacking due to the complexity of the genomic background. In previous work, we applied a proteomics-guided drug repurposing to explore potential treatments for osteosarcoma. Our follow-up study revealed an FDA-approved immunosuppressant drug, mycophenolate mofetil (MMF) targeting inosine-5′-phosphate dehydrogenase (IMPDH) enzymes, has an anti-tumor effect that appeared promising for further investigation and clinical trials. Profiling of IMPDH2 and hypoxanthine–guanine phosphoribosyltransferase (HPRT), key purine-metabolizing enzymes, could deepen understanding of the importance of purine metabolism in osteosarcoma and provide evidence for expanded use of MMF in the clinic. In the present study, we investigated levels of IMPDH2, and HPRT in biopsy of 127 cases and post-chemotherapy tissues in 20 cases of high-grade osteosarcoma patients using immunohistochemical (IHC) analysis. Cox regression analyses were performed to determine prognostic significance of all enzymes. The results indicated that low levels of HPRT were significantly associated with a high Enneking stage (P = 0.023) and metastatic status (P = 0.024). Univariate and multivariate analyses revealed that patients with low HPRT expression have shorter overall survival times [HR 1.70 (1.01–2.84), P = 0.044]. Furthermore, high IMPDH2/HPRT ratios were similarly associated with shorter overall survival times [HR 1.67 (1.02–2.72), P = 0.039]. Levels of the enzymes were also examined in post-chemotherapy tissues. The results showed that high IMPDH2 expression was associated with shorter metastasis-free survival [HR 7.42 (1.22–45.06), P = 0.030]. These results suggest a prognostic value of expression patterns of purine-metabolizing enzymes for the pre- and post-chemotherapy period of osteosarcoma treatment.
Subject terms: Bone cancer, Prognostic markers, Cancer metabolism, Sarcoma, Cancer therapy
Introduction
Osteosarcoma is a malignant bone cancer that is more common in children and adolescents. Currently standard treatment includes a combination of chemotherapy and surgery. Since the introduction of chemotherapy in combination with surgery in 1970, the survival rate of osteosarcoma cancer patients with localized disease has increased to 70%1,2. However, the five-year survival rate of patients not responding to chemotherapy is remains 30%3,4. Our previous study reported 37.9% of the 5-year overall survival rates in Northern Thailand5. This finding is comparable with studies conducted within Asia–Pacific region reporting 27–48% of the 5-year overall survival rates that were much lower in comparison to western regions with rates between 60 and 70%6. Major factors of the poor clinical outcomes were presence of metastasis and not completing treatment regarding patients’ non-compliance to treatment regime.
The main reason treatment for osteosarcoma has not improved the survival rate over the past 30 years is due to an inability to specify the key mechanisms involve in the occurrence and spread of osteosarcoma. Information obtained from whole genome and exome sequencing has indicated that osteosarcoma has a highly chaotic genetic background including chromosome aberrations, chromothripsis, kataegis, and genome instability with a high rate of copy number alterations (CNAs)7,8. There is also evidence of epigenetic changes resulting from histone modifications and DNA methylation9. These abnormalities result in changes in protein levels in both oncogenic proteins and tumor suppressors.
Cancer cells divide rapidly with a significantly higher rate of DNA and RNA replication. Cell division is controlled by many types of transcription factors and involves changes in volume, energy, and anabolic metabolism to pass through the S-phase in the cell cycle10. Cancer cells require both energy metabolism and increased nucleotide biosynthetic pathways. Purine nucleotides are the most important source of cellular energy and are involved in diverse signaling pathways11. Biosynthesis of purine nucleotides occur via two mechanisms, the “salvage pathway” that reuses biomolecules derived from DNA and RNA degradation and the “de novo pathway”, synthesis of purines from small molecules derived from cellular biochemical processes12,13. The de novo pathway produces greater amounts of purine nucleotides than the salvage pathway, but it uses more energy and involves more complex reactions14. It has been found that purine nucleotides are produced under many conditions, mainly through the de novo pathway. The most obvious example is the activation of the de novo pathway in active B and T lymphocytes15, as well as in various types of cancer16. Nucleotide metabolism in cancer cells has been studied for several decades. In 1975, a study by Jackson et al. first demonstrated the importance of the de novo pathway in nucleotide synthesis in liver cancer cells17. They reported higher activity of glutamine PRPP amidotransferase enzyme (GPAT; EC 2.4.2.14), an enzyme regulating synthesis of inosine monophosphate (IMP) in liver cancer cells, with significantly lower IMP degradation. They also found increasing activity of inosine-5′-phosphate (IMP) dehydrogenase (IMPDH), the enzyme with the highest activity level in the fastest dividing liver cancer cells tested in the experiment. IMPDH is an enzyme that is important in the process of guanine nucleotide synthesis through the de novo pathway. It accelerates the conversion of IMP to xanthosine 5′-phosphate (XMP), the rate-limiting step in the guanine nucleotide synthesis process. Proliferation of cancer cells depends largely on the amount of nucleotides within the cell as well as the activity of key enzymes regulating the rate-limiting step of the nucleotide synthesis process. Furthermore, cancer cells strongly rely on the glycolysis and the TCA cycle to obtain enough ribose-5-phosphate, a primary precursor of de novo nucleotide synthesis, to boost their intracellular nucleotide content. Ribose-5-phosphate is synthesized from the pentose phosphate pathway (PPP) through the nonoxidative branch, in which cancer cells vigorously stimulate the PPP through glycolytic flux by oncogenic signaling of the PI3K-Akt pathway18. Taken together, this makes the de novo purine nucleotide synthesis pathway an attractive target for cancer treatment.
Inosine-5′-phosphate (IMP) dehydrogenase (IMPDH) is a well-known target for antileukemic and immunosuppressive therapy19. Inhibiting IMPDH results in decreased cellular guanine nucleotide pools and subsequently modulates the synthesis of DNA and RNA which is lethal for cells. Human IMPDH consists of 2 isoforms, IMPDH1 and IMPDH2, that are encoded from different genes. Both IMPDH isoforms share a similar amino acid sequence (84%), structure (95%), kinetic properties, and substrate binding site20. Expression of IMPDH2 is enhanced significantly in rapidly proliferating cells including various types of cancers including colorectal, bladder, prostate, kidney, nasopharyngeal carcinoma and leukemia21–24. Decades ago, researchers explored a link between Myc oncogene and production of nucleotides to support a high growth rate of cancer cells. Oncogenic signaling through the Myc pathway directly controls glutamine uptake and increases expression of various rate-limiting enzymes in de novo nucleotide synthesis including, but not limited to, IMPDH1 and IMPDH2 genes14,25.
Hypoxanthine guanine phosphoribosyltransferase (HPRT) is a key enzyme in salvage pathway responsible for the formation of GMP and IMP by recycling guanine and inosine from degraded DNA and RNA, respectively26. To maintain cellular GTP level in somatic cells, HPRT is reliably expressed in low level27. Unlike in normal cells, recent study showed that HPRT expression is highly variable in various cancer tissues including lung, breast, colon, prostate and pancreas27. HPRT activity was also detected in human osteosarcoma xenografts, in which the activity ranging from 0.97 to 4.06 nmol/min/mg of protein at pH 7.428. Further study of human osteosarcoma tissues showed that all samples contained substantial activities of HPRT enzyme within the range of xenografts of human osteosarcoma29.
Mycophenolate mofetil (MMF) is an immunosuppressive agent approved for use in patients receiving allogeneic renal, cardiac or hepatic transplants to prevent organ rejection30. MMF is a prodrug of mycophenolic acid (MPA), an inhibitor of IMPDH31. The main reason MPA is a potent immunosuppressive agent is that activated T- and B-lymphocytes are highly divided based on de novo purine nucleotide synthesis30. A previous study by the authors unveiled the potency of MPA, a pan-IMPDH inhibitor, as an anti-tumor agent for osteosarcoma32. That study showed MPA effectively inhibits osteosarcoma cell viability and also reduces tumor mass in animal models at clinically achievable doses32. The study also demonstrated an effect of MPA in the modulation of pulmonary metastasis of osteosarcoma.
To enhance understanding of guanine nucleotide synthesis at population levels, this longitudinal study investigated expression patterns of individual key enzymes regulating de novo and salvage guanine nucleotide pathways in osteosarcoma samples. Furthermore, accumulating studies have reported that cancer cells have an increase in the activation of nucleotide anabolism and a lower rate of nucleotide catabolism14. Integrative analysis of IMPDH2 and HPRT will provide valuable data on an association of rewired GTP synthesis system in osteosarcoma and clinical outcomes of the patients.
Methods
Patient characteristics
The present study recruited 127 osteosarcoma patients who had been diagnosed and treated at Maharaj Nakorn Chiang Mai Hospital, Thailand, between 1999 and 2018. Patients were followed-up for survival and metastasis-free survival until 11 January 2020. Clinicopathological parameters, including date of diagnosis, date of metastasis, Enneking staging, location of tumor, and metastatic status were retrieved from hospital records and pathology reports (Table 1). All primary biopsy and post-operative chemotherapy tissue slides were evaluated histologically by a bone and soft tissue pathologist (JS). Inclusion criteria for post-operative chemotherapy studies included post-operative necrosis report available and percentage of tumor necrosis ≤ 80%.
Table 1.
Characteristics of osteosarcoma patients in study cohort.
| Factor | All patients | IMPDH2 expression | HPRT expression | ||||
|---|---|---|---|---|---|---|---|
| Low (IRS ≤ 8) | High (IRS > 8) | P value | Low (IRS ≤ 2) | High (IRS > 2) | P value | ||
| Age at diagnosis (years) | |||||||
| ≤ 15 | 65 | 37 (57%) | 28 (43%) | > 0.999 | 19 (29%) | 46 (71%) | 0.220 |
| > 15 | 62 | 36 (58%) | 26 (42%) | 12 (19%) | 50 (81%) | ||
| Gender | |||||||
| Male | 68 | 35 (52%) | 33 (48%) | 0.154 | 18 (26%) | 50 (74%) | 0.680 |
| Female | 59 | 38 (64%) | 21 (36%) | 13 (22%) | 46 (78%) | ||
| Enneking stage | |||||||
| IIB | 72 | 41 (57%) | 31 (43%) | 0.545 | 13 (18%) | 59 (82%) | 0.023 |
| III | 39 | 25 (64%) | 14 (36%) | 15 (38%) | 24 (62%) | ||
| Site | |||||||
| Extremities | 104 | 61 (59%) | 43 (41%) | > 0.999 | 14 (33%) | 80 (77%) | 0.750 |
| Axial | 15 | 9 (60%) | 6 (40%) | 4 (27%) | 11 (73%) | ||
| Metastasis | |||||||
| No | 36 | 21 (58%) | 15 (42%) | 0.742* | 4 (11%) | 32 (89%) | 0.024* |
| At follow-up | 36 | 20 (56%) | 16 (44%) | 9 (25%) | 27 (75%) | ||
| At diagnosis | 39 | 25 (64%) | 14 (36%) | 15 (38%) | 24 (62%) | ||
P values were calculated with Fisher’s exact test.
*P values were calculated with Chi-square test.
This study protocol has been approved by the Research Ethics Committee of the Faculty of Medicine, Chiang Mai University. All patients and/or parent provided informed consent for patient information to be stored in the hospital database. All methods were carried out in accordance with good clinical practices (GCPs) and relevant guidelines.
Immunohistochemistry and scoring
Immunohistochemistry was performed on formalin-fixed paraffin-embedded (FFPE) tissues from archival biopsy and post-chemotherapy paraffin blocks at the Department of Pathology, Faculty of Medicine, Chiang Mai University. We applied the Ventana automated staining system and an Ultraview Universal DAB Detection Kit (both from Ventana Medical Systems, Tucson, AZ, USA) to detect primary antibodies including anti-IMPDH2 (dilution 1:500, Abcam Cat# ab131158, RRID:AB_11156264) and anti-HPRT (dilution 1:300, Abcam Cat# ab10479, RRID:AB_297217) (all from Abcam, Cambridge, UK). Cytoplasmic staining of IMPDH and HPRT was independently evaluated by two observers (by PC and JS), blind to patient outcome data, and scored by using a semi-quantitative immunoreactive scoring system9. The percentage of immunoreactive cells was estimated and scored as follows: negative = 0, positive staining < 10% = 1, positive staining ≥ 10 and < 33% = 2, positive staining ≥ 33% and < 66% = 3, positive staining ≥ 66% = 4. Intensity of staining was scored on a scale of 0–3: no color reaction = 0, weak staining = 1, moderate staining = 2, and strong staining = 3. Immunoreactive score (IRS) was derived by multiplying immunoreactive cell scores and intensity of staining scores to compute an immunoreactive score ranging from 0 to 12 defined as negative; IRS = 0, weak; IRS > 0–4, moderate; IRS > 4–8, and strong; IRS > 8–12.
Statistical analysis
Statistical analysis was performed using STATA version 11 (Stata, RRID:SCR_012763) and GraphPad Prism version 8.4.2 (GraphPad Prism, RRID:SCR_002798) (GraphPad Software, Inc., La Jolla, CA, USA). The statistical significance of correlations between expressions of IMPDH and HPRT and clinicopathologic data were determined by Fisher’s exact and Chi-square test. Receiver operating characteristic (ROC) curve analysis was applied to define the optimal cutoff points for IMPDH2 and HPRT expression levels as well as IMPDH2/HPRT ratio. Associations of IMPDH, HPRT and IMPDH2/HPRT ratio with overall survival and metastasis-free survival of osteosarcoma patients were evaluated by the Kaplan–Meier method with the log-rank test. Multivariate survival analysis was determined using Cox regression of proportional hazards to probe for significance at the 95% confidence interval (CI). P values < 0.05 were considered to be statistically significant.
Results
Expression patterns of IMPDH2 and HPRT in the osteosarcoma cohort
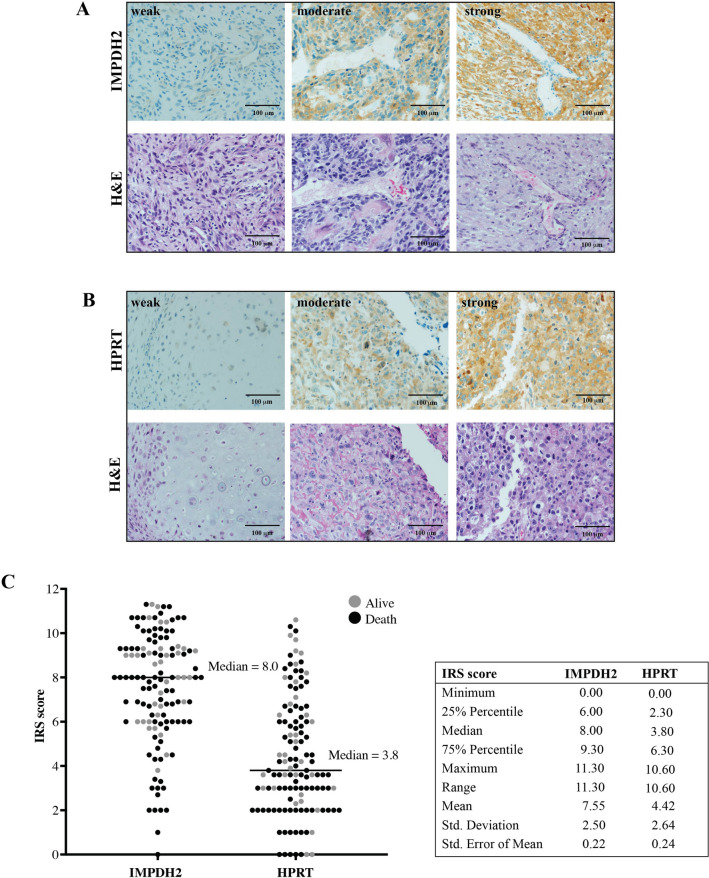
To examine expression profiles and staining patterns of IMPDH2 and HPRT enzymes, we conducted immunohistochemical analysis of formalin-fixed paraffin-embedded tissues from biopsy samples of 127 osteosarcoma cases and 20 post-chemotherapy samples. We found IMPDH2 and HPRT expressed mainly in the cytosol of the osteosarcoma cells (Fig. 1A, B). The majority were positive for IMPDH2 (99%) and HPRT (94%) staining. Interestingly, we observed a reverse overall distribution between IMPDH isoforms and HPRT staining patterns. We found that the 42% of patients had strong IMPDH2 staining, whereas only 11% had strong HPRT staining (IRS > 8). Expression patterns of IMPDH2 and HPRT stanning are shown in Fig. 1C.
Figure 1.
Immunohistochemical staining of (A) IMPDH2 and (B) HPRT in osteosarcoma tissues (Χ400). (C) Distribution of IMPDH2 and HPRT levels in osteosarcoma cohort (N = 127). H&E; hematoxylin and eosin.
Correlations of IMPDH2 and HPRT staining patterns with clinicopathological data
Characteristics of 127 osteosarcoma patients in the cohort are shown in Table 1. By comparing between expression levels of IMPDH2 and HPRT in biopsy tissues and individual primary characteristics of patients, we found a negative correlation between HPRT levels with Enneking stage (P = 0.023) as well as metastatic status (P = 0.024).
Survival analysis of IMPDH2 and HPRT in osteosarcoma
In the survival analysis, we excluded data from 8 osteosarcoma patients who were lost to follow-up. Overall, survival time of 119 patients ranged from 1 to 209 months after initial diagnosis (median = 18 months). Seventy percent (84 cases) of the patients died after 1 to 102 months (median = 13 months). The 1-year and 5-year survival rates were 70% and 32%, respectively.
Immunoreactive score (IRS) cutoff points for each of the purine metabolizing enzymes were defined by receiver operating characteristic (ROC) curves: IMPDH2, IRS = 8; HPRT, IRS = 2. Then, univariate survival analysis of individual enzymes (IMPDH2 and HPRT) was conducted using Kaplan–Meier survival analysis and log rank tests. We found that low expression levels of HPRT were associated with shorter overall survival times (P = 0.0196), but not for IMPDH2 results (Fig. 2A; Tables 2, 3). The median survival time of patients with low HPRT was 13.6 months, whereas patients with high HPRT expression had a median survival of 21.9 months. Multivariate survival analysis showed that low expression of HPRT [HR 1.70 (1.01–2.84), P = 0.044] was an independent prognostic factor for low survival time (Table 4). Other significant prognostic factors for low survival time in this cohort included advanced Enneking stage (P = 0.001), not completing treatment (P < 0.0001) and the presence of metastasis (P < 0.0001).
Figure 2.
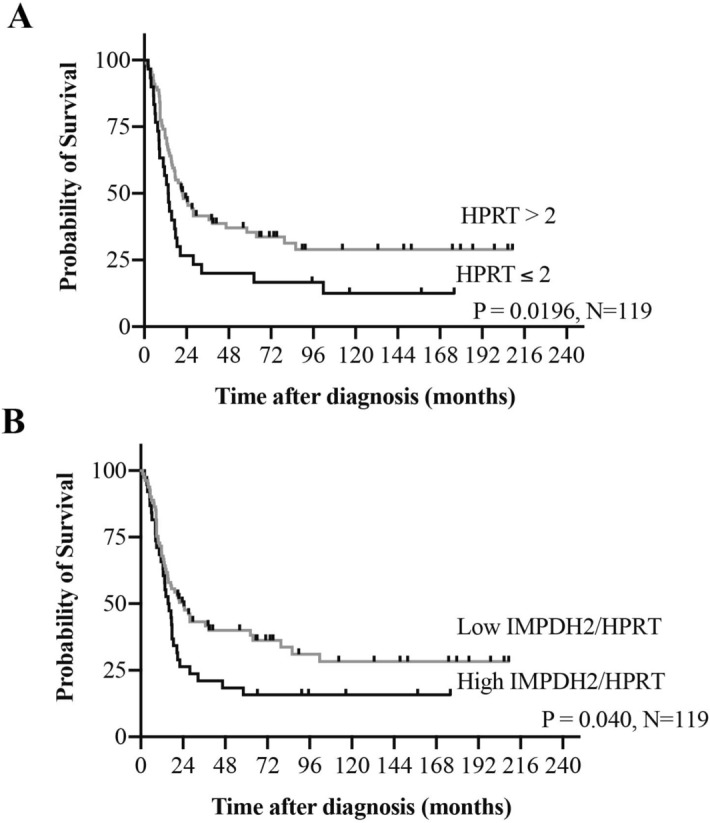
Kaplan–Meier curves showing overall survival by (A) HPRT and (B) IMPDH2/HPRT ratio in osteosarcoma cases.
Table 2.
Patient survival analysis (N = 119).
| Factor | Patients | Events (death) | Median survival (months) | P value (Log rank test) |
|---|---|---|---|---|
| Age at diagnosis (years) | ||||
| ≤ 15 | 58 | 40 | 20.5 | 0.384 |
| > 15 | 61 | 44 | 17.0 | |
| Gender | ||||
| Male | 64 | 50 | 17.7 | 0.258 |
| Female | 55 | 34 | 20.5 | |
| Enneking stage | ||||
| IIB | 71 | 42 | 27.1 | 0.001 |
| III | 38 | 32 | 12.5 | |
| Site | ||||
| Extremities | 102 | 70 | 20.6 | 0.183 |
| Axial | 13 | 10 | 12.7 | |
| Metastasis | ||||
| No | 36 | 12 | Undefined | < 0.0001 |
| Yes | 73 | 62 | 15.4 | |
| Treatment | ||||
| Completed treatment | 74 | 44 | 27.8 | < 0.0001 |
| Not completed treatment | 45 | 40 | 9.0 | |
| IMPDH2 expression | ||||
| Low (IRS ≤ 8) | 70 | 53 | 15.5 | 0.119 |
| High (IRS > 8) | 49 | 31 | 21.4 | |
| HPRT expression | ||||
| Low (IRS ≤ 2) | 30 | 26 | 13.6 | 0.0196 |
| High (IRS > 2) | 89 | 58 | 21.9 | |
| IMPDH2/HPRT ratio | ||||
| Low IMPDH2/HPRT | 81 | 52 | 24.4 | 0.040 |
| High IMPDH2/HPRT | 38 | 32 | 15.8 | |
P values were calculated using the log-rank test. P values < 0.05 shown in bold.
Table 3.
Multivariate analysis of factors associated with overall survival in the osteosarcoma cohort with inclusion of IMPDH2 expression.
| Factor | Overall survival | |
|---|---|---|
| HR (95% CI) | P value | |
| Age at diagnosis (years) | ||
| ≤ 15 | 1.00 | – |
| > 15 | 1.30 (0.80–2.11) | 0.289 |
| Site | ||
| Extremities | 1.00 | – |
| Axial | 1.04 (0.49–2.20) | 0.915 |
| Stage | ||
| IIB | 1.00 | – |
| III | 2.43 (1.48–3.99) | < 0.001 |
| Treatment completion | ||
| Completed | 1.00 | – |
| Not completed | 3.10 (1.89–5.08) | < 0.001 |
| IMPDH2 expression | ||
| Low (IRS ≤ 8) | 1.00 | – |
| High (IRS > 8) | 1.40 (0.87–2.27) | 0.169 |
Table 4.
Multivariate analysis of factors associated with overall survival in the osteosarcoma cohort with inclusion of HPRT expression.
| Factor | Overall survival | |
|---|---|---|
| HR (95% CI) | P value | |
| Age at diagnosis (years) | ||
| ≤ 15 | 1.00 | – |
| > 15 | 1.30 (0.80–2.12) | 0.286 |
| Site | ||
| Extremities | 1.00 | – |
| Axial | 1.14 (0.53–2.48) | 0.736 |
| Stage | ||
| IIB | 1.00 | – |
| III | 2.38 (1.44–3.91) | 0.001 |
| Treatment completion | ||
| Completed | 1.00 | – |
| Not completed | 3.20 (1.94–5.27) | < 0.001 |
| HPRT expression | ||
| High (IRS > 2) | 1.00 | – |
| Low (IRS ≤ 2) | 1.70 (1.01–2.84) | 0.044 |
Following up on the significant association between low HPRT and poor prognosis, we further analyzed our cohort by considering the combinational effect of IMPDH2 and HPRT enzymes. The results indicated a remarkable strong correlation between high IMPDH2/HPRT ratios and shorter survival time (Fig. 2B). Median survival of patients with a high IMPDH2/HPRT ratio was 15.8 months, whereas patients with a low IMPDH2/HPRT had a median survival of 24.4 months. The multivariate survival analysis results showed that a high IMPDH2/HPRT ratio [HR 1.67 (1.02–2.72), P = 0.039] was an independent poor prognosis factor (Table 5).
Table 5.
Multivariate analysis of factors associated with overall survival in osteosarcoma with inclusion of IMPDH2/HPRT ratio.
| Factor | Overall survival | |
|---|---|---|
| HR (95% CI) | P value | |
| Age at diagnosis (years) | ||
| ≤ 15 | 1.00 | – |
| > 15 | 1.36 (0.83–2.23) | 0.216 |
| Site | ||
| Extremities | 1.00 | – |
| Axial | 0.92 (0.43–1.97) | 0.841 |
| Stage | ||
| IIB | 1.00 | – |
| III | 2.38 (1.45–3.91) | 0.001 |
| Treatment completion | ||
| Completed | 1.00 | – |
| Not completed | 3.03 (1.85–4.97) | < 0.001 |
| IMPDH2/HPRT ratio | ||
| Low | 1.00 | – |
| High | 1.67 (1.02–2.72) | 0.039 |
Metastasis-free survival analysis of IMPDH2 and HPRT expression in post-chemotherapy tissues
In this study, we assessed expression of IMPDH2 and HPRT enzymes in biopsies as well as in post-operative chemotherapy tissues of 20 osteosarcoma patients. The results demonstrated that expression of IMPDH2 and HPRT was maintained at the same level in post-operative chemotherapy tissues as in biopsy samples (Fig. 3A). We further analyzed the association between the levels of each of the three enzymes and metastasis-free survival. Univariate and multivariate analysis results showed that high levels of IMPDH2 (IRS > 6) in post-chemotherapy tissues was an indicator of a poor prognosis [HR 7.42 (1.22–45.06), P = 0.030] (Fig. 3B; Tables 6, 7), whereas no association between HPRT level or IMPDH2/HPRT ratio and metastasis-free survival was observed in post-chemotherapy tissues (Tables 6, 7, 8, 9). Other significant prognostic factors for short metastasis-free period in this cohort included female gender (P = 0.027) and advanced Enneking stage (P < 0.001).
Figure 3.
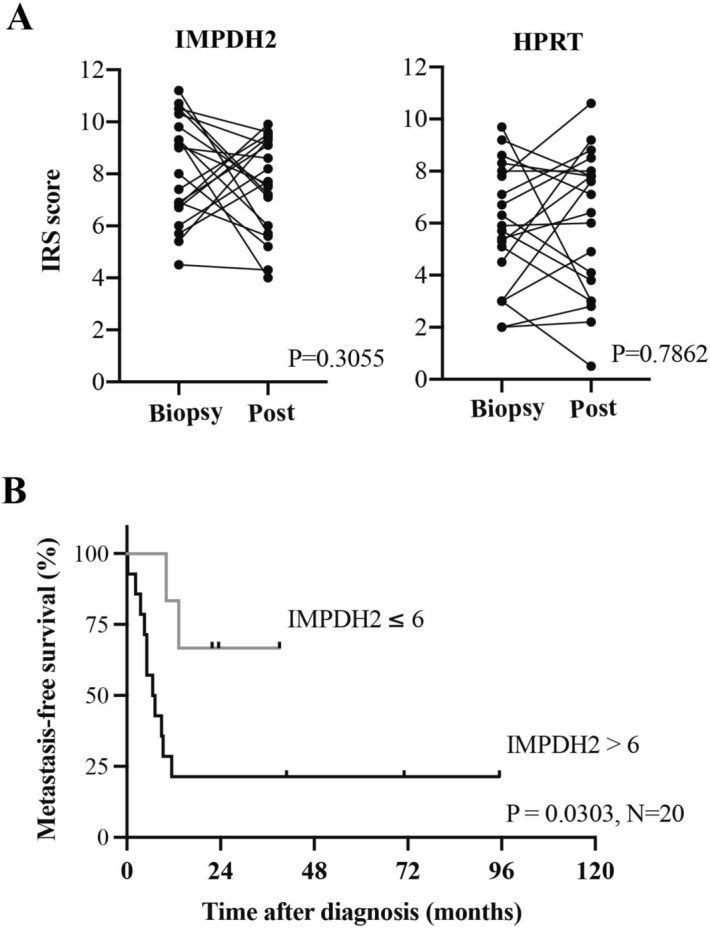
Expression levels of IMPDH2 and HPRT enzymes in longitudinal samples. (A) Expression of IMPDH2 and HPRT in biopsy and post-chemotherapy tissues. (B) Metastasis-free survival analysis of IMPDH2 in post-chemotherapy tissues of osteosarcoma.
Table 6.
Metastasis-free survival analysis (N = 20).
| Factor | Patients | Events (metastasis) | Median (months) | P-value (log rank test) |
|---|---|---|---|---|
| Age at diagnosis (years) | ||||
| ≤ 15 | 6 | 4 | 11.1 | 0.984 |
| > 15 | 14 | 9 | 9.7 | |
| Gender | ||||
| Male | 11 | 10 | 8.9 | 0.027 |
| Female | 9 | 3 | Undefined | |
| Enneking stage | ||||
| IIB | 18 | 11 | 10.8 | < 0.001 |
| III | 2 | 2 | 1.2 | |
| Site | ||||
| Extremities | 16 | 11 | 9.5 | 0.478 |
| Axial | 4 | 2 | 24.2 | |
| IMPDH2 expression | ||||
| Low (IRS ≤ 6) | 6 | 2 | Undefined | 0.030 |
| High (IRS > 6) | 14 | 11 | 6.9 | |
| HPRT expression | ||||
| Low (IRS ≤ 6) | 9 | 5 | 11.5 | 0.640 |
| High (IRS > 6) | 11 | 8 | 9.3 | |
| IMPDH2/HPRT ratio | ||||
| Low IMPDH2/HPRT | 16 | 11 | 9.1 | 0.472 |
| High IMPDH2/HPRT | 4 | 2 | 53.4 | |
P values were calculated using the log-rank test. P values < 0.05 shown in bold.
Table 7.
Multivariate analysis of metastasis-free survival in osteosarcoma patients and IMPDH2 expression in post-chemotherapy tissues (n = 20).
| Factor | Overall survival | |
|---|---|---|
| HR (95% CI) | P value | |
| Age at diagnosis (years) | ||
| ≤ 15 | 1.00 | – |
| > 15 | 0.66 (0.15–2.84) | 0.576 |
| Gender | ||
| Male | 1.00 | – |
| Female | 0.21 (0.05–0.88) | 0.033 |
| Site | ||
| Extremities | 1.00 | – |
| Axial | 0.71 (0.14–3.73) | 0.687 |
| IMPDH2 expression | ||
| Low (IRS ≤ 6) | 1.00 | – |
| High (IRS > 6) | 7.42 (1.22–45.06) | 0.030 |
Table 8.
Multivariate analysis of metastasis-free survival in osteosarcoma patients and HPRT expression in post-chemotherapy tissues (n = 20).
| Factor | Overall survival | |
|---|---|---|
| HR (95% CI) | P value | |
| Age at diagnosis (years) | ||
| ≤ 15 | 1.00 | – |
| > 15 | 1.69 (0.47–6.11) | 0.423 |
| Gender | ||
| Male | 1.00 | – |
| Female | 0.24 (0.06–0.95) | 0.042 |
| Site | ||
| Extremities | 1.00 | – |
| Axial | 0.58 (0.12–2.96) | 0.517 |
| HPRT expression | ||
| Low (IRS ≤ 6) | 1.00 | – |
| High (IRS > 6) | 1.07 (0.32–3.55) | 0.909 |
Table 9.
Multivariate analysis of metastasis-free survival in osteosarcoma patients and IMPDH2/HPRT ratio in post-chemotherapy tissues (n = 20).
| Factor | Overall survival | |
|---|---|---|
| HR (95% CI) | P value | |
| Age at diagnosis (years) | ||
| ≤ 15 | 1.00 | – |
| > 15 | 2.22 (0.56–8.84) | 0.257 |
| Gender | ||
| Male | 1.00 | – |
| Female | 0.25 (0.06–1.00) | 0.051 |
| Site | ||
| Extremities | 1.00 | – |
| Axial | 0.45 (0.08–2.38) | 0.348 |
| IMPDH2/HPRT ratio | ||
| Low IMPDH2/HPRT | 1.00 | – |
| High IMPDH2/HPRT | 0.47 (0.08–2.55) | 0.380 |
Discussion
Unrestricted proliferation of cancer cells is limited by the abundance of total nucleotide pools as well as the level and the activity of rate-limiting enzymes of nucleotide synthesis33. Nucleotide biosynthesis is formed in two major ways (1) by a de novo biosynthetic pathway through biochemical reactions of simple metabolites including glucose, amino acids, glycine, CO2, and one-carbon units derived from the serine-glycine pathway, and (2) nucleotide salvage pathway that utilizes free bases recycled from the degradation of DNA and RNA or from exogenous dietary intake34.
To our knowledge, this is the first report on an association of HPRT expression with clinical outcomes and survival rate of osteosarcoma patients. Present study showed that HPRT was independent indicators of a poor prognosis, where a low HPRT level in biopsy tissues indicated shorter survival of the patient. Recent study showed that to meet high metabolic demand for cell growth, cancer cells tune the GTP biosynthesis by stimulating de novo pathway with reduced salvage pathway34,35. Sasaki et al. demonstrated that IMPDH2 was upregulated in glioblastoma tissues, while HPRT1 were reciprocally downregulated showing significant correlation with IMPDH2 levels35. Our results also revealed that IMPDH2/HPRT ratio is an independent prognostic marker, where a high IMPDH2/HPRT ratio associated with shorter survival of the patient. However, the result from multivariate analysis revealed a significant association between HPRT levels and survival rate of osteosarcoma patients, but not for IMPDH2 expression results (Table 3). Therefore, a prognostic power of IMPDH2/HPRT ratio was, at least in part, influenced by an alteration of HPRT levels.
Until now, there have not been many studies of the nucleotide metabolic switch in osteosarcoma, but there are some indications of the importance of nucleotide metabolism, in particular, de novo guanine nucleotide synthesis. Fellenberg et al. showed that IMPDH2 was one of eight drug-regulated genes highly expressed in chemo-resistant osteosarcoma cells36. A later study also demonstrated an association between high IMPDH2 level and poor response to chemotherapy and low metastasis-free survival in osteosarcoma patients37. IMPDH2 expression is also an independent prognostic biomarker for other cancer types. It has been reported that IMPDH2 expression is related to advanced stage and distant metastasis in nasopharyngeal carcinoma38. High IMPDH2 levels also significantly indicate shorter overall survival and disease-free survival. Enhanced IMPDH2 expression is significantly associated with advanced stage, metastasis, and higher Gleason scores in cases of prostate cancer39.
Our findings also suggest that high levels of IMPDH2 in post-chemotherapy tissues are significantly associated with shorter metastasis-free survival. Multivariate analysis showed the independent prognostic power of IMPDH2 expression. Several reports have previously demonstrated the role of IMPDH and de novo guanine nucleotide synthesis in cancer metastasis and progression. A study by Zhou et al. showed that the level of IMPDH2 was enhanced in prostate cancer with metastasis39. Interestingly, one study demonstrated GTP-biosynthetic enzymes, including IMPDH1 and IMPDH2, were highly accumulated at the leading edge of renal carcinoma cells40. This plasma membrane localization of IMPDH enzymes might promote localized nucleotide metabolism which augments cancer cell migration and metastasis. Bianchi-Smiraglia et al. reported an increased level of guanosine monophosphate synthase (GMPS), an enzyme that catalyzes the amination of xylitol monophosphate (XMP) to guanosine monophosphate (GMP) in de novo guanine nucleotide synthesis pathways in metastatic human melanoma samples41. GMPS inhibitor could also potentially modulate melanoma cell invasion in vitro.
The main reason MPA, an IMPDH inhibitor, is a potent immunosuppressant agent is that lymphocytes are lacking salvage capability to restore level of GTP pool in the cells30. The present findings on an association of low HPRT level and unfavorable clinical outcomes suggest a potential metabolic vulnerability of poor prognostic patients. Currently, we are running phase II multi-center clinical trial to evaluate the efficacy and safety of mycophenolate mofetil (MMF) in patients with high-grade locally advanced or metastatic osteosarcoma (ESMMO) (Thai Clinical Trials Registry number: TCTR20190701 001)42. A total of 27 high-grade locally advanced or metastatic osteosarcoma patients have been recruited into the study from collaborating hospitals in Thailand.
In summary, our findings strengthen the concept that nucleotide metabolic switches and de novo guanine synthesis in tumorigenesis and osteosarcoma progression could be used as prognostic markers and to indicate actionable targets for osteosarcoma treatment.
Acknowledgements
This work was supported by the Faculty of medicine, Chiang Mai University, the National Science and Technology Development Agency (NSTDA) code P-18-51991, the National Research University (NRU) fund, and the Musculoskeletal Science and Translational Research Center. The authors would also like to express their sincere thanks to Dr. G. Lamar Robert, Ph.D., and Assoc. Prof. Dr. Chongchit Sripun Robert, Ph.D., for editing the English manuscript.
Author contributions
P.C.: Conceptualization, Methodology, Investigation, Writing—Original Draft, Writing—Review and Editing. A.P.: Statistical analysis. N.S.: Investigation, Validation. J.K.: Investigation, Validation. V.T.: Investigation, Validation. P.T.: Investigation, Validation. P.P.: Statistical analysis. D.P.: Supervision, Conceptualization, Writing—Review and Editing. J.S.: Supervision: Investigation, Writing—Review and Editing. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dumnoensun Pruksakorn, Email: dumnoensun.p@cmu.ac.th.
Jongkolnee Settakorn, Email: jsettakorn@gmail.com.
References
- 1.Pruksakorn D, et al. Age standardized incidence rates and survival of osteosarcoma in Northern Thailand. Asian Pac. J. Cancer Prev. 2016;17:3455–3458. [PubMed] [Google Scholar]
- 2.Wittig JC, et al. Osteosarcoma: a multidisciplinary approach to diagnosis and treatment. Am. Fam. Phys. 2002;65:1123–1132. [PubMed] [Google Scholar]
- 3.Meyers PA, et al. Osteosarcoma: a randomized, prospective trial of the addition of ifosfamide and/or muramyl tripeptide to cisplatin, doxorubicin, and high-dose methotrexate. J. Clin. Oncol. 2005;23:2004–2011. doi: 10.1200/JCO.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 4.Bishop MW, Janeway KA, Gorlick R. Future directions in the treatment of osteosarcoma. Curr. Opin. Pediatr. 2016;28:26–33. doi: 10.1097/MOP.0000000000000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pruksakorn D, et al. Survival rate and prognostic factors of conventional osteosarcoma in Northern Thailand: A series from Chiang Mai University Hospital. Cancer Epidemiol. 2015;39:956–963. doi: 10.1016/j.canep.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 6.Yasin NF, Abdul Rashid ML, Ajit Singh V. Survival analysis of osteosarcoma patients: a 15-year experience. J. Orthop. Surg. (Hong Kong) 2020 doi: 10.1177/2309499019896662. [DOI] [PubMed] [Google Scholar]
- 7.Cortes-Ciriano I, et al. Comprehensive analysis of chromothripsis in 2658 human cancers using whole-genome sequencing. Nat. Genet. 2020;52:331–341. doi: 10.1038/s41588-019-0576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kovac M, et al. Exome sequencing of osteosarcoma reveals mutation signatures reminiscent of BRCA deficiency. Nat. Commun. 2015;6:8940. doi: 10.1038/ncomms9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaiyawat P, et al. Expression patterns of class I histone deacetylases in osteosarcoma: a novel prognostic marker with potential therapeutic implications. Mod. Pathol. 2018;31:264–274. doi: 10.1038/modpathol.2017.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lane AN, Fan TW. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015;43:2466–2485. doi: 10.1093/nar/gkv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedley AM, Benkovic SJ. A new view into the regulation of purine metabolism: the purinosome. Trends Biochem. Sci. 2017;42:141–154. doi: 10.1016/j.tibs.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray AW. The biological significance of purine salvage. Annu. Rev. Biochem. 1971;40:811–826. doi: 10.1146/annurev.bi.40.070171.004115. [DOI] [PubMed] [Google Scholar]
- 13.Yamaoka T, et al. Amidophosphoribosyltransferase limits the rate of cell growth-linked de novo purine biosynthesis in the presence of constant capacity of salvage purine biosynthesis. J. Biol. Chem. 1997;272:17719–17725. doi: 10.1074/jbc.272.28.17719. [DOI] [PubMed] [Google Scholar]
- 14.Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr. Opin. Genet. Dev. 2009;19:32–37. doi: 10.1016/j.gde.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allison AC, Hovi T, Watts RW, Webster AD. The role of de novo purine synthesis in lymphocyte transformation. Ciba Found Symp. 1977 doi: 10.1002/9780470720301.ch13. [DOI] [PubMed] [Google Scholar]
- 16.Collart FR, Chubb CB, Mirkin BL, Huberman E. Increased inosine-5′-phosphate dehydrogenase gene expression in solid tumor tissues and tumor cell lines. Cancer Res. 1992;52:5826–5828. doi: 10.2172/10148922. [DOI] [PubMed] [Google Scholar]
- 17.Jackson RC, Weber G, Morris HP. IMP dehydrogenase, an enzyme linked with proliferation and malignancy. Nature. 1975;256:331–333. doi: 10.1038/256331a0. [DOI] [PubMed] [Google Scholar]
- 18.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allison AC, Eugui EM. Mycophenolate mofetil and its mechanisms of action. Immunopharmacology. 2000;47:85–118. doi: 10.1016/s0162-3109(00)00188-0. [DOI] [PubMed] [Google Scholar]
- 20.Wu TY, et al. Pharmacogenetics of the mycophenolic acid targets inosine monophosphate dehydrogenases IMPDH1 and IMPDH2: gene sequence variation and functional genomics. Br. J. Pharmacol. 2010;161:1584–1598. doi: 10.1111/j.1476-5381.2010.00987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen K, et al. Differential sensitivities of fast- and slow-cycling cancer cells to inosine monophosphate dehydrogenase 2 inhibition by mycophenolic acid. Mol Med. 2015 doi: 10.2119/molmed.2015.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagai M, et al. Selective up-regulation of type II inosine 5′-monophosphate dehydrogenase messenger RNA expression in human leukemias. Cancer Res. 1991;51:3886–3890. [PubMed] [Google Scholar]
- 23.Zou J, et al. Elevated expression of IMPDH2 is associated with progression of kidney and bladder cancer. Med. Oncol. 2015;32:373. doi: 10.1007/s12032-014-0373-1. [DOI] [PubMed] [Google Scholar]
- 24.Natsumeda Y, et al. Two distinct cDNAs for human IMP dehydrogenase. J. Biol. Chem. 1990;265:5292–5295. doi: 10.1016/S0021-9258(19)34120-1. [DOI] [PubMed] [Google Scholar]
- 25.Liu YC, et al. Global regulation of nucleotide biosynthetic genes by c-Myc. PLoS ONE. 2008;3:e2722. doi: 10.1371/journal.pone.0002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Townsend MH, Robison RA, O'Neill KL. A review of HPRT and its emerging role in cancer. Med. Oncol. 2018;35:89. doi: 10.1007/s12032-018-1144-1. [DOI] [PubMed] [Google Scholar]
- 27.Townsend MH, et al. Falling from grace: HPRT is not suitable as an endogenous control for cancer-related studies. Mol. Cell Oncol. 2019;6:1575691. doi: 10.1080/23723556.2019.1575691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer WH, Houghton JA, Lutz PJ, Houghton PJ. Hypoxanthine:guanine phosphoribosyltransferase activity in xenografts of human osteosarcoma. Cancer Res. 1986;46:4896–4899. [PubMed] [Google Scholar]
- 29.Meyer WH, Houghton JA, Houghton PJ. Hypoxanthine:guanine phosphoribosyltransferase activity in primary human osteosarcomas. A rationale for therapy with methotrexate-thymidine rescue? J. Clin. Oncol. 1987;5:657–661. doi: 10.1200/JCO.1987.5.4.657. [DOI] [PubMed] [Google Scholar]
- 30.Bentley R. Mycophenolic Acid: a one hundred year odyssey from antibiotic to immunosuppressant. Chem. Rev. 2000;100:3801–3826. doi: 10.1021/cr990097b. [DOI] [PubMed] [Google Scholar]
- 31.Carr SF, Papp E, Wu JC, Natsumeda Y. Characterization of human type I and type II IMP dehydrogenases. J. Biol. Chem. 1993;268:27286–27290. doi: 10.1016/S0021-9258(19)74247-1. [DOI] [PubMed] [Google Scholar]
- 32.Klangjorhor J, et al. Mycophenolic acid is a drug with the potential to be repurposed for suppressing tumor growth and metastasis in osteosarcoma treatment. Int. J. Cancer. 2020;146:3397–3409. doi: 10.1002/ijc.32735. [DOI] [PubMed] [Google Scholar]
- 33.Kohnken R, Kodigepalli KM, Wu L. Regulation of deoxynucleotide metabolism in cancer: novel mechanisms and therapeutic implications. Mol. Cancer. 2015;14:176. doi: 10.1186/s12943-015-0446-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villa E, Ali ES, Sahu U, Ben-Sahra I. Cancer cells tune the signaling pathways to empower de novo synthesis of nucleotides. Cancers (Basel). 2019 doi: 10.3390/cancers11050688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kofuji S, et al. IMP dehydrogenase-2 drives aberrant nucleolar activity and promotes tumorigenesis in glioblastoma. Nat. Cell Biol. 2019;21:1003–1014. doi: 10.1038/s41556-019-0363-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fellenberg J, Dechant MJ, Ewerbeck V, Mau H. Identification of drug-regulated genes in osteosarcoma cells. Int. J. Cancer. 2003;105:636–643. doi: 10.1002/ijc.11135. [DOI] [PubMed] [Google Scholar]
- 37.Fellenberg J, Bernd L, Delling G, Witte D, Zahlten-Hinguranage A. Prognostic significance of drug-regulated genes in high-grade osteosarcoma. Mod. Pathol. 2007;20:1085–1094. doi: 10.1038/modpathol.3800937. [DOI] [PubMed] [Google Scholar]
- 38.Xu Y, et al. High expression of IMPDH2 is associated with aggressive features and poor prognosis of primary nasopharyngeal carcinoma. Sci. Rep. 2017;7:745. doi: 10.1038/s41598-017-00887-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou L, et al. Enhanced expression of IMPDH2 promotes metastasis and advanced tumor progression in patients with prostate cancer. Clin. Transl. Oncol. 2014;16:906–913. doi: 10.1007/s12094-014-1167-9. [DOI] [PubMed] [Google Scholar]
- 40.Wolfe K, et al. Dynamic compartmentalization of purine nucleotide metabolic enzymes at leading edge in highly motile renal cell carcinoma. Biochem. Biophys. Res. Commun. 2019;516:50–56. doi: 10.1016/j.bbrc.2019.05.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bianchi-Smiraglia A, et al. Pharmacological targeting of guanosine monophosphate synthase suppresses melanoma cell invasion and tumorigenicity. Cell Death Differ. 2015;22:1858–1864. doi: 10.1038/cdd.2015.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koonrungsesomboon N, et al. Phase II, multi-center, open-label, single-arm clinical trial evaluating the efficacy and safety of Mycophenolate mofetil in patients with high-grade locally advanced or metastatic osteosarcoma (ESMMO): rationale and design of the ESMMO trial. BMC Cancer. 2020;20:268. doi: 10.1186/s12885-020-06751-2. [DOI] [PMC free article] [PubMed] [Google Scholar]