Abstract
The fecundity of female mammals is resolved by the limited size of the primordial follicle (PF) pool formed perinatally. The establishment of PF pool is accompanied by a significant programmed oocyte death. Long non-coding RNAs (lncRNA) are central modulators in regulating cell apoptosis or autophagy in multiple diseases, however, the significance of lncRNAs governing perinatal oocyte loss remains unknown. Here we find that Yin-Yang 1 (YY1) directly binds to the lncRNA X-inactive-specific transcript (Xist) promoter and facilitates Xist expression in the perinatal mouse ovaries. Xist is highly expressed in fetal ovaries and sharply downregulated along with the establishment of PF pool after birth. Gain or loss of function analysis reveals that Xist accelerates oocyte autophagy, mainly through binding to pre-miR-23b or pre-miR-29a in the nucleus and preventing the export of pre-miR-23b/pre-miR-29a to the cytoplasm, thus resulting in decreased mature of miR-23b-3p/miR-29a-3p expression and upregulation miR-23b-3p/miR-29a-3p co-target, STX17, which is essential for timely control of the degree of oocyte death in prenatal mouse ovaries. Overall, these findings identify Xist as a key non-protein factor that can control the biogenesis of miR-23b-3p/miR-29a-3p, and this YY1-Xist-miR-23b-3p/miR-29a-3p-STX17 regulatory axis is responsible for perinatal oocyte loss through autophagy.
Subject terms: Autophagy, Oogenesis
Introduction
The establishment of the primordial follicle (PF) pool represents the first stage of folliculogenesis and this non-renewable PF resource is crucial for maintaining female reproductive life1. As the principal functional reproductive unit that provides mature oocytes, the population of PFs is perinatally established, in a process termed cyst breakdown2. Ensuring proper primordial follicle generation and early follicle development is under precise control1. However, multiple factors can induce excessive loss or overactivation of PFs in the PF pool, which is closely related to primary premature ovarian insufficiency (POI) and even to female infertility3,4. A better understanding of the regulatory mechanisms underlying PF pool formation will provide us with deep insights into the potential pathogenic mechanism of POI5–7.
Till now, the main studies on PF pool formation or even the pathogenesis of POI associated with PF formation are restricted to protein-coding genes8,9. However, only a small proportion of POI cases can be interpreted by causative genetic defects, such as chromosomal abnormalities and gene mutations4,8,10,11. Eukaryotic genomes have a wide range of transcriptional properties, producing a large number of RNA transcripts. Among them, only <2% of these RNAs will serve as mRNA templates, and the remaining part of transcripts are non-coding RNAs (ncRNAs). Obviously, there is a lack of study on the roles of ncRNAs, especially long non-coding RNAs (lncRNAs) at the early stage of folliculogenesis, since these ncRNAs have recently been proved to play essential roles in both physiological and pathological conditions in female reproduction9.
LncRNAs are a category of ncRNAs with a length of over 200 nucleotides without protein-coding potential12. They can function as a signal, guide, decoy, microRNA sponging, or scaffold to modulate gene expression at transcription and post-transcription levels13,14. Several recent studies have linked lncRNAs to the process of folliculogenesis15–18, and dysregulation of their expression can cause ovarian diseases, including POI16,17,19,20. Besides, transcriptional profiling of lncRNAs in ovarian cortical tissues from women with POI has recently been performed, and 20 differently expressed lncRNAs were identified to be involved in ovarian follicular development21. However, whether lncRNAs are participating in the PF pool formation is nearly completely unknown. A recent transcriptome analysis of lncRNA expression in human primordial, primary and small antral follicles was performed and lncRNA X-inactive-specific transcript (Xist) is revealed to be highly transcribed throughout the stages tested, suggesting that Xist has emerged from the dormant primordial stage of human follicle development15. But its biological function and the molecular mechanism underlying the formation of the PF pool have not been clarified.
MicroRNAs (miRNAs) are another class of short ncRNAs with a length of 20–24 nucleotides, which are generated through the initial nuclear cleavage and subsequent cytoplasmic cleavage events22,23. The miRNA gene is initially transcribed from the genome into primary transcript (pri-miRNA), followed by cleavage into ~70 nucleotide precursors miRNA (pre-miRNAs). The pre-miRNA is then exported into the cytoplasm where it is further cleaved into an RNA duplex analogous. The mature miRNA is finally generated once one strand is discarded. It is well acknowledged that miRNAs can negatively control the expression of target genes in a wide range of cell signaling pathways related to different physiological and pathological processes24,25, including PF formation and even POI26–29. However, these studies focus more on how these miRNAs regulate their downstream targets and participate in the regulation of PF formation, and reports of miRNA biogenesis and regulation are missing.
In this study, we showed a time-specific decrease in Xist levels from the fetal to neonatal period in the ovaries, which correlates with the upregulated expression of miR-23b-3p/miR-29a-3p, and a stepwise decrease of STX17. We then demonstrated that Xist can block miR-23b-3p/miR-29a-3p processing, resulting in decreased mature of miR-23b-3p/miR-29a-3p expression, leading to upregulation of STX17, which is essential for timely control of the degree of oocyte death induced by autophagy in prenatal mouse ovaries.
Results
Xist is essential for PF pool formation in the perinatal mouse ovaries
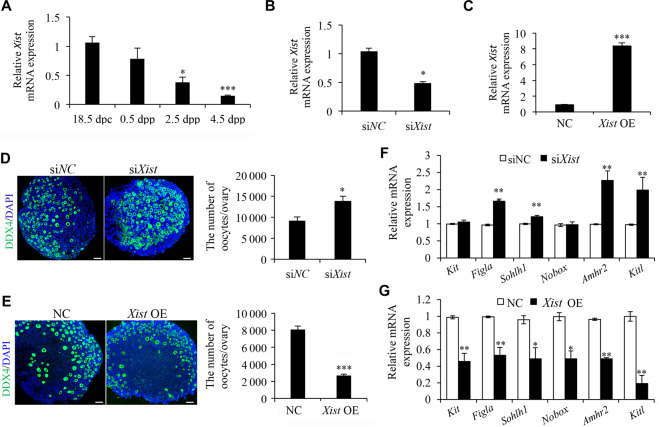
To address the physiological significance of Xist during the establishment of PF pool in the ovaries of newborn mice, we first showed by qRT-PCR analyses that Xist transcripts started to decrease in a time-specific manner from 18.5 d postcoitus (dpc) to 4.5 d postpartum (dpp) (Fig. 1A). This dynamic change of Xist just coincides with the timing of massive oocyte loss, indicating that Xist may be involved in regulating oocyte loss. To assess this possibility, gain or loss of function experiments were carried out into 0.5 dpp ovaries by separately transfecting with Xist siRNA (siXist) and the control siRNA (siNC), or Xist-pcDNA3.1 (Xist OE) and the empty control (NC). The qRT-PCR analysis confirmed that siXist transfection decreased, and Xist overexpression increased Xist expression in neonatal mouse ovaries after 96 h of in vitro culture (Fig. 1B, C). Subsequent histological analysis showed a significant increase of oocyte number upon Xist knockdown compared with the control (Fig. 1D), while Xist OE resulted in significantly less available oocytes than did NC (Fig. 1E). In agreement with the changes of oocyte quantification, expression of the genes essential for PF formation also showed an increase in siXist-treated ovaries (Fig. 1F). Alternatively, expression of these genes was inhibited upon Xist OE (Fig. 1G). These results indicate that Xist may be required for oocyte loss in the newborn ovaries and a decrease of Xist is indispensable for PF pool formation.
Fig. 1. Xist expression in perinatal ovaries is essential for PF pool formation.
a qRT-PCR analysis of Xist expression at the indicated time points. Gapdh was used to normalize samples. b, c qRT-PCR analysis of Xist expression in newborn ovaries transfected with siXist (b), and Xist OE (c), relative to respective controls. d, e Representative images of DDX4 immunofluorescence staining (left) and quantification of follicles (right) in newborn ovaries transfected with siXist (d), and Xist OE (e). Scale bars: 50 μm. f, g qRT-PCR analysis of genes involved in PF survival and development in siXist (f), and Xist OE (g) transfected ovaries, relative to respective controls. Student’s t-test: *P < 0.05, **P < 0.01, ***P < 0.001.
Xist promotes perinatal oocyte loss mainly through oocyte autophagy
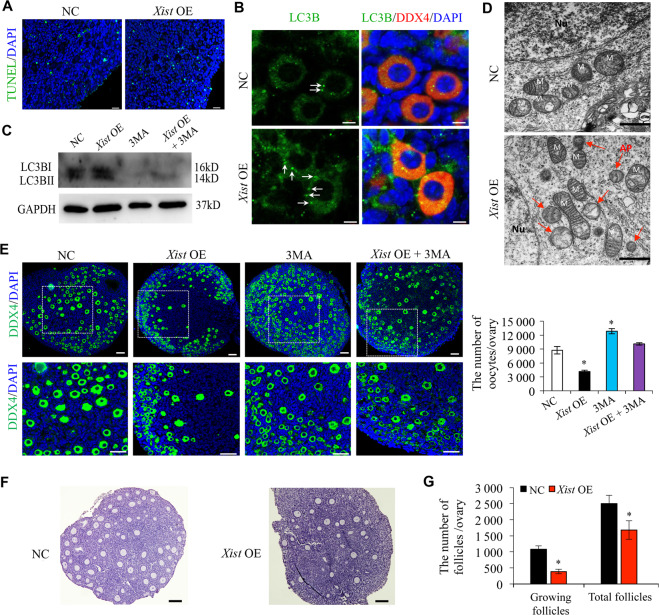
It is well acknowledged that apoptosis and autophagy are involved in the process of oocyte death during PF pool establishment30,31. To clarify whether Xist is involved in the programmed oocyte death through apoptosis and autophagy, we first performed TUNEL analysis to evaluate oocyte apoptosis. Overexpression of Xist did not increase the number of TUNEL-positive cells, suggesting that oocyte apoptosis may not be the major cause of oocyte loss upon Xist OE (Fig. 2A). As autophagy is also reported to be active during follicular assembly and contributes to perinatal oocyte loss5, we, therefore, tested whether Xist could induce autophagy in perinatal mouse ovaries. As the microtubule-associated protein light chain 3B (LC3B) is a marker of autophagosomes, we observed more LC3B puncta presented in the cytoplasm of Xist-overexpressed oocytes (Fig. 2B). We then incubated the 0.5 dpp ovaries with 3MA, an autophagy inhibitor, and 3MA treatment nearly completely inhibited LC3B protein expression, while Xist overexpression could slightly eliminate the negative effect of 3MA on LC3B expression (Fig. 2C). Furthermore, TEM analysis identified the large double-membrane vacuoles filled with degraded organelles in Xist-overexpressed oocytes (Fig. 2D), which is the typical appearance of autophagosomes32. To further confirm that autophagy is essential for oocyte death under Xist OE overexpression, we calculated the number of oocytes in 0.5 dpp ovaries treated with 3MA. The number of oocytes that survived in the 3MA group was higher than that in the control group, while Xist overexpression partly abrogated this stimulatory effect of 3MA (Fig. 2E). Finally, the 0.5 dpp ovaries transfected with pcDNA-Xist were cultured for 96 h and then transplanted under the kidney capsule of ovariectomized adult recipient mice (8-wk old) and monitored for 14 d. Ovarian histology analysis revealed a significant loss of activated follicles in the Xist-overexpressed group compared to the controls (Fig. 2F, G). Together, these results suggested that Xist promotes perinatal oocyte loss through oocyte autophagy.
Fig. 2. Xist promotes oocyte autophagy during PF formation.
a TUNEL staining in newborn ovaries transfected with Xist-pcDNA3.1 (Xist OE) or the empty vector control (NC). The nucleus was stained by DAPI (blue). Scale bars: 20 μm. b Immunofluorescence staining of LC3B (green), and DDX4 (red) in newborn ovaries transfected with Xist-OE compared to NC control. Scale bars: 5 μm. Arrows indicating LC3B puncta. c WB analysis of active LC3B expression in newborn ovaries treated under indicated condition. d TEM analysis of autophagosomes in newborn ovaries transfected with Xist-OE compared to NC control. Red arrow indicating autophagosomes. Scale bars: 1 μm. e Representative images of DDX4 immunofluorescence staining (left) and quantification of follicles (right) in newborn ovaries treated under indicated condition. The nucleus was stained by DAPI (blue). Scale bars: 50 μm. f, g Representative images (f) and quantification of follicles (g) in the ovary transfected under indicated condition. Scale bar, 20 μm. Student’s t-test: *P < 0.05.
Xist modulates miR-23b-3p and miR-29a-3p maturationin perinatal mouse ovaries
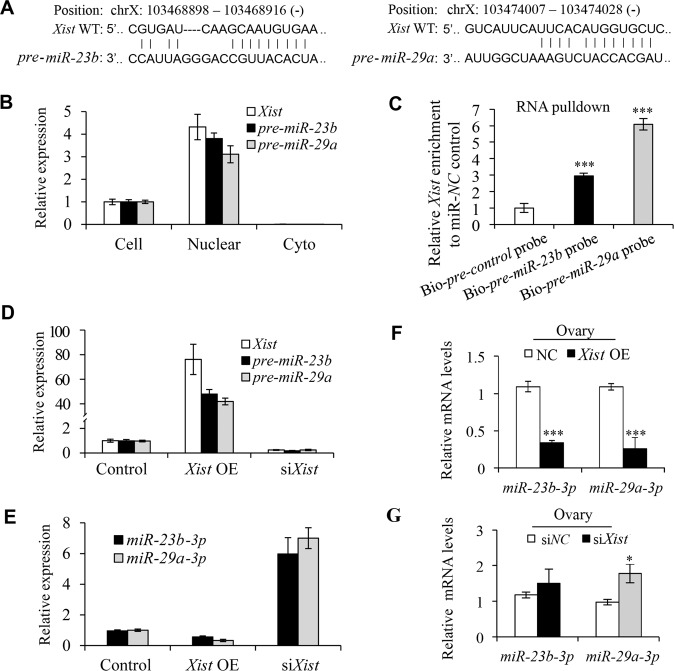
We next followed upon the underlying mechanism of Xist in regulating oocyte autophagy during perinatal PF formation. Recent studies have demonstrated that nuclear lncRNA regulates miRNA expression by suppressing its maturation process in the nucleus33,34. Given that Xist is also a nuclear lncRNA and that miRNAs are essential to promote oocyte survival and decrease apoptosis26–29, we wondered whether Xist may also play such a role in regulating oocyte death during PF formation. Previous studies have profiled differentially expressed miRNAs for association with premature ovarian failure development in both animal models and patients, including miR-23b-3p and miR-29a-3p35,36. We particularly selected these two miRNAs for further validation since the RNA fragments of the pre-miR-23b and pre-miR-29a are part of the Xist sequences (Fig. 3A), implying that Xist may regulate miR-23b-3p and miR-29a-3p expression. Additionally, accumulating studies have demonstrated that both miR-23b-3p and miR-29a-3p can regulate autophagy in a wide range of cells37–39, which may help raise the possibility that expression of miR-23b-3p and miR-29a-3p modulated by Xist in perinatal mouse ovaries is also associated with perinatal oocyte autophagy. To test this, we first confirmed that pre-miR-23b and pre-miR-29a were enriched in the nuclear fraction of cells from 0.5 dpp ovaries (Fig. 3B and Supplementary Fig. S1). Then, we applied a biotin-avidin pulldown assay to further delineate the direct interaction between Xist and pre-miR-23b and pre-miR-29a in the nucleus. Briefly, a biotin-labeled-specific anti-pre-miR-23b or anti-pre-miR-29a probe was synthesized and incubated with nuclear lysate from 3T3 cells. Pre-miR-23b or pre-miR-29a was co-precipitated via avidin-conjugated agarose beads, and the levels of Xist in the pulldown complex were analyzed by qRT-PCR. As shown in Fig. 3C, Xist was significantly enriched in the biotin-labeled pre-miR-23b or pre-miR-29a pulldown product compared to Bio-pre-NC control probe, suggesting that Xist can directly bind to pre-miR-23b or pre-miR-29a in the nucleus, thus preventing the export of pre-miR-23b/pre-miR-29a to the cytoplasm. Therefore, when we modulated Xist expression in 3T3 cells, we observed that both pre-miR-23b and pre-miR-29a levels were elevated upon Xist overexpression, and decreased upon Xist knockdown, compared to their respective controls (Fig. 3D). Correspondingly, overexpression of Xist decreased while depletion of Xist increased the expression of mature miR-23b-3p and miR-29a-3p (Fig. 3E). The expression of Xist positively correlated with the expression of pre-miR-23b and pre-miR-29a, but negatively with mature miR-23b-3p and miR-29a-3p, suggesting that Xist may regulate miR-23b and miR-29a maturation process.
Fig. 3. Xist regulates miR-23b and miR-29a maturation process.
a The predicted pre-miR-23b and pre-miR-29a binding sites in the Xist transcript. b qRT-PCR analysis of Xist, pre-miR-23b, and pre-miR-29a expression in nuclear and cytoplasmic fraction relative to the whole cells from 0.5 dpp ovaries. c Enrichment of Xist pulled down by biotin-pre-miR-23b, biotin-pre-miR-29a, or biotin-pre-miR-NC control in the nuclear fraction from 3T3 cells. d qRT-PCR analysis of Xist, pre-miR-23b, and pre-miR-29a expression in Xist overexpression or Xist knockdown 3T3 cells. e qRT-PCR analysis of miR-23b-3p, and miR-29a-3p expression in Xist overexpression or Xist knockdown 3T3 cells. f, g Relative levels of miR-23b-3p and miR-29a-3p in cultured newborn ovaries transfected with Xist or empty control (f), and in ovaries transfected with siXist or scrambled control (g). Student’s t-test: *P < 0.05, ***P < 0.001.
To assess this hypothesis in vivo, we transfected 0.5 dpp mouse ovaries with pcDNA3.1-Xist or siXist, and found that overexpression of Xist significantly increased the expression of pre-miR-23b and pre-miR-29a, while knockdown of Xist obviously decreased the expression of pre-miR-23b and pre-miR-29a in the nuclei (Supplementary Fig. S2). Correspondingly, both miR-23b-3p and miR-29a-3p levels were significantly decreased upon Xist overexpression (Fig. 3F), and upregulated upon Xist knockdown, compared to their respective controls (Fig. 3G). These results indicated that Xist may negatively regulate miR-23b-3p/miR-29a-3p expression in the neonatal mouse ovaries. Together, these data demonstrate that the maturation of miR-23b-3p and miR-29a-3p is associated with the expression of Xist in perinatal ovaries.
MiR-23b-3p/miR-29a-3p are involved in the regulation of PF formation in the perinatal mouse ovaries
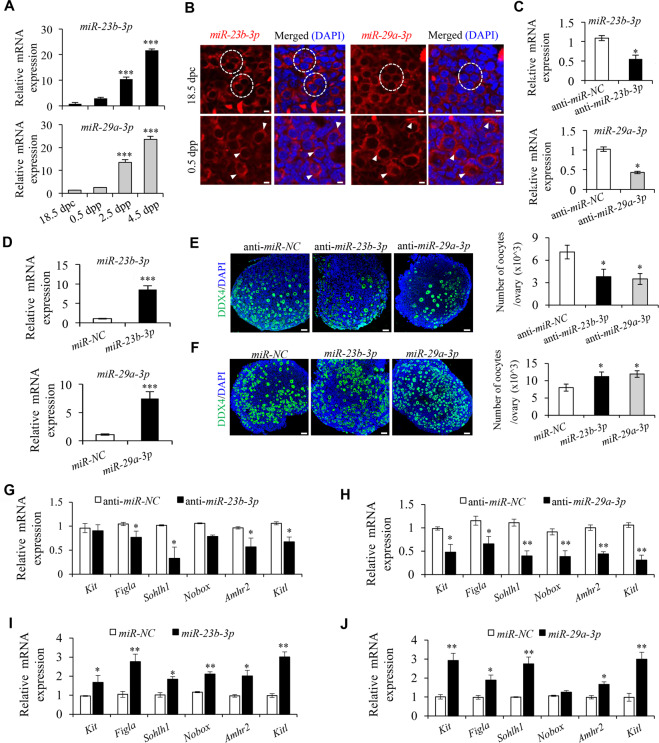
Given Xist negatively regulates mature miR-23b-3p/miR-29a-3p, we speculated to observe an inversed expression pattern of miR-23b-3p/miR-29a-3p in perinatal mouse ovaries to that of Xist. Actually, qRT-PCR analysis just confirmed that miR-23b-3p/miR-29a-3p expression increased significantly along with the establishment of the PF pool from 18.5 dpc to 4.5 dpp (Fig. 4A). In addition, the cytoplasmic localization of the miR-23b-3p/miR-29a-3p by RNA-FISH analysis in oocytes from both cysts and PFs can be readily detected (Fig. 4B). These data raised a possibility that the promoting effect of Xist on oocyte loss during PF formation is likely mediated by its negative regulation on miR-23b-3p/miR-29a-3p expression. To further assess the effect of miR-23b-3p/miR-29a-3p in PF formation, we silenced or overexpressed them in 0.5 dpp ovaries, respectively. The silencing or overexpression efficiency was then confirmed 96 h post transfection (Fig. 4C, D). Then, the cultured ovaries were subjected to histological examination. Immunohistochemistry analysis with anti-DDX4 antibody showed that anti-miR-23b-3p or anti-miR-29a-3p significantly decreased oocyte number compared to the controls (Fig. 4E), and overexpression of miR-23b-3p or miR-29a-3p exhibited more oocytes than the miRNA controls 96 h post transfection (Fig. 4F). Analysis of LC3B expression confirmed that inhibiting miR-23b-3p or miR-29a-3p in the newborn ovaries obviously elevated, while overexpression of miR-23b-3p or miR-29a-3p decreased oocyte autophagy compared to their respective controls (Supplementary Fig. S3). Furthermore, expression of the genes related to PF formation decreased in anti-miR-23b-3p or anti-miR-29a-3p treated ovaries (Fig. 4G, H), while increased in miR-23b-3p or miR-29a-3p overexpressed group (Fig. 4I, J). These results indicated that miR-23b-3p/miR-29a-3p have the opposite effect to that of Xist, which may promote PF formation in perinatal mouse ovaries, possibly by preventing oocyte loss.
Fig. 4. Expression of miR-23b-3p and miR-29a-3p are essential for PF formation in the perinatal mouse ovaries.
a qRT-PCR analysis of miR-23b-3p (top) and miR-29a-3p (bottom) expression at the indicated time points. Gapdh was used to normalize samples. b RNA-FISH analysis of miR-23b-3p and miR-29a-3p expression in perinatal mouse ovaries from 18.5 dpc, and 0.5 dpp. The nucleus was stained by DAPI (blue). Dashed circle indicating representative cysts. Arrows heads indicating oocytes. Scale bars: 5 μm. c, d qRT-PCR analysis of miR-23b-3p (top) and miR-29a-3p (bottom) expression in newborn ovaries transfected with anti-miR-23b-3p, anti-miR-29a-3p, or anti-miR-NC (c), and mimics for miR-23b-3p, miR-29a-3p, or miR-NC (d). e, f Representative DDX4 immunofluorescence images (left) and quantification of follicles (right) in newborn ovaries transfected with anti-miR-23b-3p, anti-miR-29a-3p, or anti-miR-NC (e), and mimics for miR-23b-3p, miR-29a-3p, or miR-NC (f). Scale bars: 50 μm. g–j qRT-PCR analysis of genes involved in PF survival and development in newborn ovaries transfected with anti-miR-23b-3p (g), anti-miR-29a-3p (h), or anti-miR-NC, and mimics for miR-23b-3p (i), miR-29a-3p (j), or miR-NC. Student’s t-test: *P < 0.05, **P < 0.01, ***P < 0.001.
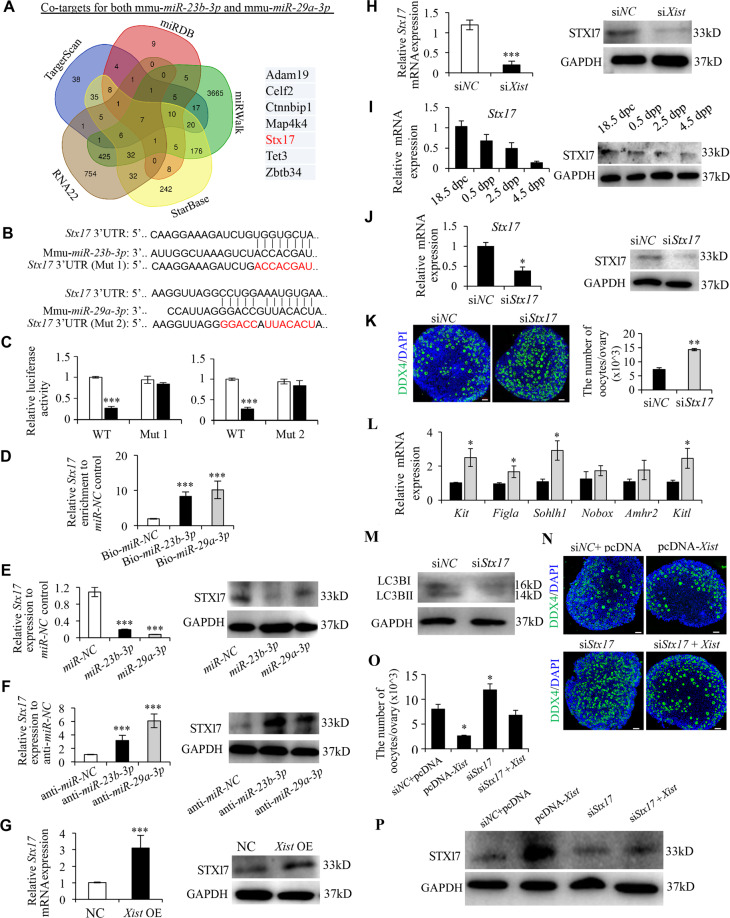
Xist relieves the inhibition of miR-23b-3p/miR-29a-3p on their common target STX17
To find out downstream genes sharing the regulatory role of miR-23b-3p/miR-29a-3p with Xist, we used five online bioinformatic tools (TargetScan, miRDB, miRWalk, Starbase, and RNA22) to predict the potential co-target genes of miR-23b-3p/miR-29a-3p (Fig. 5A). Among the seven co-target genes with sequence complementarity to both miR-23b-3p/miR-29a-3p, STX17 protein has attracted our particular attention, since it is directly implicated in the autophagy process40. Syntaxin 17 (STX17) is a soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) protein that drives the maturation of autophagosomes acquiring degradative enzymes by fusing with the lysosome41,42. STX17 localizes to the outer membrane of autophagosomes, and binds directly to the autophagosomal protein, LC343. Failure of autophagosome-lysosome binding leads to the blockade of the autophagic flux and abnormal degradation of autophagosomes44. Therefore, STX17 may function as the downstream target for miR-23b-3p/miR-29a-3p in regulating oocyte autophagy in the perinatal ovaries, and thus was selected for further analysis. We first synthesized wild-type (WT) and mutant Stx17 3′UTR (Mut1 corresponding to miR-23b-3p, and Mut2 to miR-29a-3p), and constructed them into luciferase reporters, separately (Fig. 5B). Luciferase reporter assays demonstrated that overexpression of miR-23b-3p or miR-29a-3p repressed luciferase activity in HEK293 cells transfected with WT Stx17 3′UTR reporter plasmid, whereas such effect was not observed in the mutant reporters (Fig. 5C). Next, biotin-labeled miRNA pulldown assays verified that Stx17 is the target gene for miR-23b-3p/miR-29a-3p (Fig. 5D). Further analyses revealed that STX17 expression could be downregulated by overexpression of miR-23b-3p or miR-29a-3p, while upregulated by inhibitors for miR-23b-3p or miR-29a-3p (Fig. 5E, F). Since Xist inhibits the maturation of miR-23b-3p/miR-29a-3p, upregulation of Xist positively affected STX17 expression through modulating miR-23b-3p/miR-29a-3p (Fig. 5G), while an opposite effect was observed upon Xist inhibition (Fig. 5H). Together, these findings confirmed the existence of a Xist-miR-23b-3p/miR-29a-3p-STX17 regulatory axis in the perinatal ovaries.
Fig. 5. STX17 is a downstream target of Xist-miR-23b-3p/miR-29a-3p axis and regulates PF formation.
a The Venn diagram shows overlapping of the co-target genes of miR-23b-3p/miR-29a-3p from five representative databases. b The predicted binding sites of miR-23b-3p and miR-29a-3p binding sites in the 3′-UTR of Stx17. The red nucleotides represent mutant sequences of target sites. c Luciferase activities in HEK293 cells transfected with WT or mutant Stx17 plasmid together with miR-23b-3p or miR-29a-3p, or miR-NC. d Enrichment of Stx17 pulled down by biotin-miR-23b-3p, biotin-miR-29a-3, or biotin-miR-NC control in 3T3 cells. e–h Relative Stx17 mRNA (left), and STX17 protein (right) expression in cultured newborn ovaries transfected with mimics for miR-23b-3p, miR-29a-3p, or miR-NC (e), anti-miR-23b-3p, anti-miR-29a-3p, or anti-miR-NC (f), Xist or empty control (g), and siXist or scrambled control (h). i Relative Stx17 mRNA (left), and STX17 protein (right) expression at the indicated time points. j Relative Stx17 mRNA (left), and STX17 protein (right) expression in cultured newborn ovaries transfected with siStx17, or siNC as a control. k Representative images (left) and quantification of follicles (right) in newborn ovaries transfected with siStx17, or siNC. Scale bars: 50 μm. l qRT-PCR analysis of genes involved in PF survival and development in newborn ovaries transfected with siStx17, and siNC as a control. m WB analysis of LC3B expression in newborn ovaries treated under indicated condition. n Representative images of DDX4 in newborn ovaries co-transfected with pcDNA-Xist or empty vector together with siStx17 or scrambled control. Scale bars: 50 μm. o Quantification of follicles in n. p WB analysis of STX17 expression in newborn ovaries treated under indicated condition. GAPDH was used as a loading control. Student’s t-test: *P < 0.05, **P < 0.01, ***P < 0.001.
STX17 is responsible for Xist-mediated perinatal oocyte loss
To further verify STX17 is the downstream effector in Xist-miR-23b-3p/miR-29a-3p-STX17 regulatory axis in regulating perinatal oocyte loss, we first showed a similar expression pattern of STX17 to that of Xist in the perinatal mouse ovaries (Fig. 5I). Next, silencing STX17 in 0.5 dpp ovaries significantly increased the oocyte number compared to controls, after 96 h of in vitro culture (Fig. 5J, K). Additionally, expression of the genes related to PF formation also increased in siStx17-treated ovaries (Fig. 5L). Furthermore, STX17-silenced neonatal ovaries showed a suppressed expression pattern of LC3B (Fig. 5M and Supplementary Fig. S4A). Conversely, we also overexpressed STX17 in the neonatal ovaries (Supplementary Fig. S4B), and observed that STX17 overexpression significantly decreased the oocyte number compared to controls after 96 h of in vitro culture (Supplementary Fig. S4C). Meanwhile, STX17-overexpressed neonatal ovaries showed an increased expression pattern of LC3B (Supplementary Fig. S4D). These data suggested that STX17 is involved in regulating PF pool formation through accelerating oocyte autophagy. Finally, to verify the ability of Xist to promote perinatal oocyte loss in an STX17-dependent manner, we treated 0.5 dpp ovaries with pcDNA3.1-Xist, siStx17, or both in combination. The results demonstrated that knockdown of STX17 partially rescued the suppressive effects of Xist on PF pool formation (Fig. 5N–P).
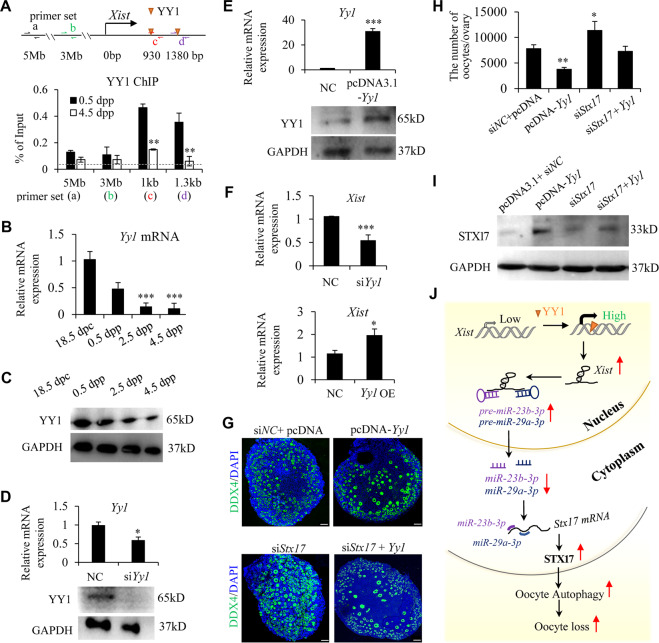
YY1 is essential for Xist transcription during PF pool formation
It is essential for transcription factor Yin-Yang 1 (YY1) to bind directly to Xist promoter and activate Xist transcription in multiple cells45,46. To test whether YY1 facilitates Xist expression in the perinatal ovaries, we first performed ChIP assays using YY1 antibody and detected YY1 binding at Xist promoter with qPCR primers flanking the YY1 binding sites (Fig. 6A). We observed a strong enrichment of YY1 to this region in 0.5 dpp ovaries, while YY1 binding dramatically decreased in 4.5 dpp ovaries (Fig. 6A). This binding pattern was closely related to a rapid decrease of Xist expression from 18.5 dpc to 4.5 dpp ovaries, suggesting that YY1 binding may positively regulate Xist expression in the perinatal ovaries. Next, we examined the expression levels of YY1 during PF formation, and found a gradual decrease of YY1 expression in a time-specific manner from 18.5 dpc to 4.5 dpp (Fig. 6B, C). Given previous studies have shown that depletion of YY1 specifically inhibits Xist expression in other female cells45,46, our data raised a possibility that the decreased YY1 in perinatal ovaries may contribute to the downregulated Xist. To confirm this causal relationship between YY1 and Xist expression, gain or loss of function experiments were performed in 0.5 dpp ovaries by silencing or overexpressing YY1. The qRT-PCR and WB analyses showed that siYy1 transfection decreased, and pcDNA-Yy1 transfection increased YY1 expression in 4.5 dpp ovaries (Fig. 6D, E). YY1 suppression significantly inhibited Xist expression, while overexpression of YY1 increased Xist expression (Fig. 6F). As a transactivator of Xist, YY1 was shown to play a similar role during PF pool formation to that of Xist, in an STX17-dependent manner (Fig. 6G–I). In agreement with the previous studies showing that YY1 is implicated in autophagy in multiple cells47,48, we then confirmed that YY1 positively regulated the LC3B protein expression in the newborn ovaries (Supplementary Fig. S5A). As we showed that Xist promotes perinatal oocyte loss through autophagy in an STX17-dependent manner (Figs. 2 and 5), YY1 overexpression also rescued the decreased LC3B expression mediated by STX17 depletion (Supplementary Fig. S5B). These data supported the existence of a YY1-Xist-miR-23b-3p/miR-29a-3p-STX17 regulatory axis during PF formation.
Fig. 6. YY1 is essential for Xist expression and functions as an upstream regulator for Xist-miR-23b-3p/miR-29a-3p-Stx17 axis during PF formation.
a Schematic representation for YY1 binding motif within the proximal promoter region of Xist, with respective primer sets to amplify YY1 binding region. The upstream control region is also represented (top). Bottom: ChIP analysis of YY1 binding in newborn mouse ovaries at 0.5 dpp, and 4.5 dpp. The dashed line represents the basal IgG binding. b, c Relative Yy1 mRNA (b), and YY1 protein (c) expression in perinatal ovaries at indicted time. d, e Relative Yy1 mRNA (top), and YY1 protein (bottom) expression in cultured newborn ovaries transfected with siYy1 or control (d), pcDNA3.1-Yy1, and NC control (e). f Relative Xist mRNA expression in cultured newborn ovaries transfected with siYy1 or siRNA control (top), pcDNA3.1-Yy1 or NC control (bottom). g, h Representative DDX4 immunofluorescence images (g) and quantification of follicles (h) in newborn ovaries transfected under indicated condition. Scale bars: 50 μm. i WB analysis of STX17 expression in newborn ovaries treated under indicated condition. j Proposed model for Xist-miR-23b-3p/miR-29a-3p-STX17 axis in regulating oocyte loss in the perinatal mouse ovaries. Student’s t-test: *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
It is well accepted that cellular apoptosis and autophagy processes are responsible for programmed oocyte loss occurring briefly around the time of birth5,31,40. Meanwhile, lncRNAs are emerging as central regulators in controlling cell autophagy or apoptosis in various diseases49,50. However, there is no such lncRNA report in the process of PF formation. Here, we demonstrated that Xist promotes perinatal oocyte loss mainly through oocyte autophagy. Mechanically, we showed explicitly that, during the early period of PF pool formation, YY1 activates Xist transcription; Xist blocks miR-23b-3a/miR-29a-3a biogenesis process, resulting in decreased mature miR-23b-3a/miR-29a-3a expression; Given STX17 is a co-target for miR-23b-3a/miR-29a-3a and that STX17 is a critical factor in the process of autophagy, Xist relieving the inhibition of miR-23b-3p/miR-29a-3p on STX17 finally leads to an increased STX17 expression, which promotes massive oocyte loss through oocyte autophagy (Fig. 6J). These findings indicate that Xist is an indispensable primordial folliculogenesis factor that controls PF pool formation.
Xist, whose gene product is a lncRNA, is exclusively expressed on the inactivated X-chromosome and its abnormal expression has been linked strongly to the process of X-chromosome inactivation51,52. It is speculated that variation in expression of Xist gene that leads to a preferential silencing of genes on the X-chromosome related to the maintenance of ovarian function may consider being a susceptibility factor for POI52. Clinically, the inadequate number of follicles in the perinatally generated PF pool is one of the causative factors for POI, leading to the shortened reproductive life span. But whether Xist is involved in PF pool formation is unclear. The only available clue is coming from the transcriptome analysis of lncRNA expression in human primordial, primary and small antral follicles, with Xist being identified as one of highly expressed lncRNAs15. Our finding that interference of Xist expression affects the oocyte loss during the formation of PF pool may lend support to this hypothesis and help better understand the novel functional relationship between Xist and pathogenesis of POI.
Accumulating studies have demonstrated that, in addition to protein-mediated post-transcriptional control in miRNA biogenesis, nuclear ncRNAs can also function as negative regulators for miRNA biogenesis as miRNA maturation process begins in the nucleus. For example, colon cancer-associated transcript-2 (Ccat2), a lncRNA mainly located in the nucleus, has been reported to selectively block miR-145 maturation by inhibiting pre-miR-145 export to the cytoplasm in colon cancer cells33. Tang et al.53 reported that, in the cell nucleus, miR-709 directly binds to pri-miR-15a/16-1 and prevents its processing into pre-miR-15a/16-1, leading to a suppression of miR-15a/16-1 biogenesis. In line with these reports, our findings that expression of Xist in the perinatal mouse ovaries positively correlated with expression of pre-miR-23b/pre-miR-29a, but inversely correlated with mature miR-23b-3a/miR-29a-3a, and that Xist binds to the pre-miR-23b/pre-miR-29a in the nucleus, imply that Xist may block the export of pre-miR-23b/pre-miR-29a to the cytoplasm, resulting in decreased mature miR-23b-3a/miR-29a-3. In the process of miRNA biogenesis, exportin-554, and exportin-155 are identified to be nuclear export factors to transport pre-miRNAs from the nucleus to the cytoplasm. It is not clear whether Xist affects the function of either exportin-5 or exportin-1 and this requires further investigation. Additionally, recent studies found that Xist functions as a competing endogenous RNA (ceRNA) to sponge the common miRNAs, and therefore insulates the miRNAs and facilitates the corresponding miRNA-targeted transcripts in tumorigenesis56,57, or in the regulation of inflammation and apoptosis58. Together, these studies significantly expand the biological function of Xist/miRNA/mRNA axis in regulating a broad spectrum of biological processes.
Transcriptional regulation of Xist has been widely investigated and several transcription factors are identified to be required for its transcription51. A recent study has carried out in silico analysis of Xist promoter regions in seven species of eutherian mammals and revealed clustered YY1 consensus binding sites evolutionarily well conserved in all species tested, including human and mouse. Furthermore, YY1 can directly bind to the Xist promoter region and promote Xist expression during initiation and maintenance of X-chromosome inactivation, suggesting that YY1 binding triggers the activity of the Xist promoter46. In consistence with this activity of YY1, our study in the perinatal ovaries also verified the binding of YY1 at Xist proximal promoter region, which is necessary for Xist transactivation during PF pool formation. Additionally, our findings that the same expression pattern of YY1 as that of Xist was observed in the perinatal ovaries, and that modulation of YY1 expression induced a similar phenotype during PF formation as that of Xist, strongly suggest that YY1 may function as an important upstream regulator in Xist-miR-23b-3p/miR-29a-3p axis.
Collectively, we elucidate a novel mechanism by which lncRNA Xist, activated by transcription factor YY1, blocks miR-23b-3p/miR-29a-3p maturation process by inhibiting pre-miR-23b/pre-miR-29a export to the cytoplasm, and relieves the inhibition of miR-23b-3p/miR-29a-3p on STX17, resulting in massive oocyte loss through oocyte autophagy in the perinatal ovaries. Our work may offer new insights into the mysteries of early folliculogenesis in mammalian ovaries, and highlight the potential clinical value of Xist-miR-23b-3p/miR-29a-3p-STX17 axis in the diagnosis and treatment of POI.
Materials and methods
Mice and ovary culture
All wild-type mice were purchased from the Animal Core Facility of Nanjing Medical University. The Animal Care and Use Committee of Nanjing Medical University approved all the animal experiments. The mice were housed under a 12/12-h dark/light cycle with free access to food and water at 20–22 °C. Mice were mated using timed mating, and the presence of a vaginal plug was defined as 0.5 dpp. Embryonic ovaries were collected at 18.5 dpc, and neonatal ovaries were collected at 0.5, 2.5, and 4.5 dpp. For ovary culture, 4 ovaries were randomly selected and placed in a 24-well dish and cultured in DMEM/F-12 medium (Gibco, Life Technologies, NY, USA) supplemented with penicillin-streptomycin and ITS at 37 °C under 5% CO2. The miR-23b-3p/miR-29a-3p mimics, inhibitors, and their corresponding scramble controls were ordered from GenePharma (Shanghai, China). The Xist, and Yy1 overexpression plasmids, siRNAs against Xist, and Yy1 were from General Biosystems (Anhui, China). The oligonucleotide sequences were listed in Supplementary Table 1. For ovaried treated with 2.5 mM of 3MA (S2767, Selleck, USA), dimethylsulfoxide (DMSO) was used as a control.
Cell culture
Human embryonic kidney (HEK) 293 cells and mouse NIH/3T3 cells were purchased from the Cell Resource Center of the Shanghai Institute for Biological Sciences (Shanghai, China). Both cell types cultured in Dulbecco’s Modified Eagle’s medium (DMEM; HyClone, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, USA) and 1% penicillin/streptomycin (P/S; Thermo Fisher Scientific, Rockford, IL, USA) and were incubated in a humidified incubator at 37 °C with 5% CO2.
Quantitative real-time PCR
RNA was extracted from mouse ovaries using TRIzol (#15596026, Invitrogen, MA, USA), followed by reverse transcription with First Strand cDNA Synthesis Kit (K1621, Thermo Scientific, MA, USA), and qRT-PCR analysis with AceQ qPCR SYBR Green Master Mix (Q141-02, Vazyme, Jiangsu, China). The primers are summarized in Supplementary Table 2. The relative fold change of gene expression was calculated using the relative standard curve method (2−ΔΔCt).
Isolation of nuclear and cytoplasmic fractions
Cells were washed three times with cold PBS, followed by centrifugation at 500 rpm for 5 min at 4 °C. Cell pellets were then resuspended in 1× Hypotonic Buffer (20 mM Tris, pH 8.0; 10 mM NaCl, 3 mM MgCl2, and 10% NP-40), and incubated on ice for 15 min. After centrifugation at 3000 rpm for 10 min at 4 °C. the supernatant portion (cytoplasmic extract) was transferred into the precooled tube. The pellet is the crude nuclear fraction. Nuclear pellets were then washed twice with cold PBS.
Western blotting
Ovaries were lysed with RIPA buffer (CW2333, CWBIO, Beijing, China) with 1× protease inhibitor (CW2200, CWBIO). The supernatant was collected and about 30 μg of denatured protein was separated on a 10% SDS-PAGE gel, and transferred to a nitrocellulose membrane. The blots were rinsed in TBS containing 0.5% Tween-20 and blocked with 5% nonfat dry milk. After incubation with the primary antibodies overnight at 4 °C, including anti-LC3B (#2775, Cell Signaling Technology, 1:1000), anti-STX17 (#31261, Cell Signaling Technology, 1:1000), anti-YY1 (#22156-1-AP, Proteintech, 1:1000), anti-Lamin B1 (#12586, Cell Signaling Technology, 1:1000), anti-GAPDH (ab8245, Abcam, 1:2000), and anti-FLAG (#14793, Cell Signaling Technology, 1:1000), the blots were then washed and incubated with corresponding peroxidase-conjugated secondary antibody for 1 h at room temperature. The signals were visualized using an Enhanced Chemiluminescence Detection Kit (#32106, Thermo Scientific) on a Bio-Rad gel imaging system.
Luciferase reporter assay
The wild-type or mutant Xist fragment containing the predicted binding sites of pre-miR-23b or pre-miR-29a was cloned into the modified pGL3 luciferase reporter vector (gift from Dr. Chun Lu from Nanjing Medical University). Luciferase reporter plasmid and 40 nM of pre-miR-23b, pre-miR-29a-3p, or pre-control were co-transfected into HEK293 cells. The luciferase reporter assay system (E2920, Promega) was used to measure luciferase activity.
RNA fluorescent in situ hybridization (RNA-FISH)
5′-FAM-labeled miR-23b-3p or miR-29a-3p probes were designed and synthesized by General Biosystems (Anhui, China). Hybridizations were carried out using FISH Kit from GenePharma (Shanghai, China). Briefly, the ovaries were fixed in 4% paraformaldehyde and processed for serial paraffin sectioning at 5 μm thickness. After permeabilization, sections were hybridized with specific probes, and 6-diamidino-2-phenylindole (DAPI) was used to stain nuclei. All fluorescence images were captured using Point Scanning Laser Confocal Microscope (ZEISS, Germany).
Biotin-labeled miRNA pulldown assay
Briefly, the 3′-end biotinylated pre-miR-23b-3p, pre-miR-29a-3p, or pre-miRNA control probe (50 nM, General Biosystems, Anhui, China) was transfected into 3T3 cell, and then cell nuclear fraction was extracted 48 h post transfection. Dynabeads MyOne Streptavidin C1 kit (Invitrogen) was used to enrich the biotin-coupled RNA complex. The beads were then pelleted and washed to remove unbound materials. Beads-bound RNA was extracted with TRIzol reagent. The abundance of Xist in the isolated fractions was tested by qRT-PCR.
Immunofluorescence
Sections were deparaffinized, rehydrated, followed by antigen retrieval by boiling the sections in 0.01 M citrate buffer, pH 6.0 for 15 min. Then the sections were blocked in 10% normal goat serum, and incubated with primary antibodies overnight at 4 °C, including anti-DDX4 (Ab13840, Abcam, 1:200), anti-DDX4(Ab27591, Abcam, 1:200), and anti-LC3B (#2775, Cell Signaling Technology, 1:100). After 5 washes with PBS, the sections were incubated with secondary antibodies for 1 h at room temperature. DAPI was used to stain nuclei.
Chromatin immunoprecipitation (ChIP)
MAGNA ChIP kit (#17371RF, Millipore, USA) was used to perform the ChIP assay. Cross-linked chromatin from mouse ovaries was prepared and immunoprecipitations were followed with primary antibodies, including anti-Ago2 (#2897, Cell Signaling Technology, 4 μg/sample), anti-YY1 (#22156-1-AP, Proteintech, 4 μg/sample) or rabbit IgG (#12–370, Millipore, 4 μg/sample). The enriched chromatin DNA was quantified by qPCR. Primers used are listed in Supplementary Table 2.
TUNEL assay
Oocyte apoptosis was measured by TUNEL staining with a TUNEL Apoptosis Detection Kit (FITC) (Yeasen, Shanghai, China) on ovary sections. Nuclei DNA was stained with DAPI. The images were photographed using a Carl Zeiss (Oberkochen, Germany) lens.
Transmission electron microscopy (TEM)
Ovaries were fixed in 2.5% glutaraldehyde in 0.2 M PBS (pH = 7.2) overnight at 4 °C, and processed and wrapped in epoxypropane resin following standard TEM procedures.
Histological evaluation of follicle numbers
Theo varies were fixed, embedded in paraffin, and serially sectioned at a thickness of 5 μm. After staining, follicles were counted in every fifth section. To avoid duplicate counts, the only oocyte with a visible nucleus was counted. Germ cells not surrounded by GCs were scored as unassembled cysts. Germ cells surrounded by GCs or a mixture of squamous and cuboidal somatic cells were scored as primordial follicles. The total number of oocytes in each ovary was calculated by multiplication by 5.
Statistical analysis
All experiments were repeated at least three independent biological replicates and presented as the mean ± the standard error of the mean. Statistical analysis was performed using Student’s t-tests or one-way ANOVA to compare the difference. A value of P < 0.05 was considered statistically significant.
Supplementary information
Author contributions
M.Z., X.L., and X.Z. designed the research. M.Z., Q.E., Y.S., S.L., and X.Z. performed the research. M.Z., X.L., and X.Z. analyzed the data and wrote the manuscript. All authors have seen and approved the final version.
Funding
This work was supported by the National Key Research and Development Program of China [2018YFC1003703, 2018YFC1004203], and NMU Science and Technology Innovation Project [2017NJMUCX007].
Ethics statement
All animal experiments were approved by the Animal Care and Use Committee of Nanjing Medical University.
Conflict of interest
The authors declare no competing interests.
Footnotes
Edited by V. Dötsch
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Meng Zhou, Xiaoqiu Liu
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-021-03831-4.
References
- 1.Wang C, Zhou B, Xia G. Mechanisms controlling germline cyst breakdown and primordial follicle formation. Cell Mol. Life Sci. 2017;74:2547–2566. doi: 10.1007/s00018-017-2480-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pepling ME. From primordial germ cell to primordial follicle: mammalian female germ cell development. Genesis. 2006;44:622–632. doi: 10.1002/dvg.20258. [DOI] [PubMed] [Google Scholar]
- 3.Hsueh AJ. Fertility: the role of mTOR signaling and KIT ligand. Curr. Biol. 2014;24:R1040–R1042. doi: 10.1016/j.cub.2014.09.033. [DOI] [PubMed] [Google Scholar]
- 4.Qin Y, Jiao X, Simpson JL, Chen ZJ. Genetics of primary ovarian insufficiency: new developments and opportunities. Hum. Reprod. Update. 2015;21:787–808. doi: 10.1093/humupd/dmv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He M, et al. LSD1 contributes to programmed oocyte death by regulating the transcription of autophagy adaptor SQSTM1/p62. Aging Cell. 2020;19:e13102. doi: 10.1111/acel.13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy P, Zheng W, Liu K. Mechanisms maintaining the dormancy and survival of mammalian primordial follicles. Trends Endocrinol. Metab. 2010;21:96–103. doi: 10.1016/j.tem.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Sun YC, Sun XF, Dyce PW, Shen W, Chen H. The role of germ cell loss during primordial follicle assembly: a review of current advances. Int. J. Biol. Sci. 2017;13:449–457. doi: 10.7150/ijbs.18836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins G, Patel B, Thakore S, Liu J. Primary ovarian Insufficiency: current concepts. South Med. J. 2017;110:147–153. doi: 10.14423/SMJ.0000000000000611. [DOI] [PubMed] [Google Scholar]
- 9.Tu J, et al. Long non-coding RNAs in ovarian granulosa cells. J. Ovarian Res. 2020;13:63. doi: 10.1186/s13048-020-00663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart J. Premature ovarian insufficiency is a lifelong condition. Clin. Endocrinol. 2017;86:168–169. doi: 10.1111/cen.13260. [DOI] [PubMed] [Google Scholar]
- 11.Tucker EJ, Grover SR, Bachelot A, Touraine P, Sinclair AH. Premature ovarian insufficiency: new perspectives on genetic cause and phenotypic spectrum. Endocr. Rev. 2016;37:609–635. doi: 10.1210/er.2016-1047. [DOI] [PubMed] [Google Scholar]
- 12.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 13.Nachtergaele S, He C. The emerging biology of RNA post-transcriptional modifications. RNA Biol. 2017;14:156–163. doi: 10.1080/15476286.2016.1267096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst EH, Nielsen J, Ipsen MB, Villesen P, Lykke-Hartmann K. Transcriptome analysis of long non-coding RNAs and genes encoding paraspeckle proteins during human ovarian follicle development. Front. Cell Dev. Biol. 2018;6:78. doi: 10.3389/fcell.2018.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, et al. Increased new lncRNA-mRNA gene pair levels in human cumulus cells correlate with oocyte maturation and embryo development. Reprod. Sci. 2015;22:1008–1014. doi: 10.1177/1933719115570911. [DOI] [PubMed] [Google Scholar]
- 17.Xiong Y, et al. Cyclophosphamide promotes the proliferation inhibition of mouse ovarian granulosa cells and premature ovarian failure by activating the lncRNA-Meg3-p53-p66Shc pathway. Gene. 2017;596:1–8. doi: 10.1016/j.gene.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 18.Zheng L, et al. Differentially expressed lncRNAs after the activation of primordial follicles in mouse. Reprod. Sci. 2019;26:1094–1104. doi: 10.1177/1933719118805869. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, et al. Long noncoding RNA HCP5 participates in premature ovarian insufficiency by transcriptionally regulating MSH5 and DNA damage repair via YB1. Nucleic Acids Res. 2020;48:4480–4491. doi: 10.1093/nar/gkaa127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J, et al. Polycystic ovary syndrome: novel and Hub lncRNAs in the insulin resistance-associated lncRNA-mRNA network. Front. Genet. 2019;10:772. doi: 10.3389/fgene.2019.00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao G, et al. Transcriptional profiling of long noncoding RNAs and their target transcripts in ovarian cortical tissues from women with normal menstrual cycles and primary ovarian insufficiency. Mol. Reprod. Dev. 2019;86:847–861. doi: 10.1002/mrd.23158. [DOI] [PubMed] [Google Scholar]
- 22.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 23.Kim VN. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol. 2004;14:156–159. doi: 10.1016/j.tcb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Ratti M, et al. MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) as new tools for cancer therapy: first steps from bench to bedside. Target Oncol. 2020;15:261–278. doi: 10.1007/s11523-020-00717-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 26.Guo Y, Sun J, Lai D. Role of microRNAs in premature ovarian insufficiency. Reprod. Biol. Endocrinol. 2017;15:38. doi: 10.1186/s12958-017-0256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, et al. microRNA 376a regulates follicle assembly by targeting Pcna in fetal and neonatal mouse ovaries. Reproduction. 2014;148:43–54. doi: 10.1530/REP-13-0508. [DOI] [PubMed] [Google Scholar]
- 28.Wang S, et al. MiR-125b regulates primordial follicle assembly by targeting activin receptor type 2a in neonatal mouse ovary. Biol. Reprod. 2016;94:83. doi: 10.1095/biolreprod.115.131128. [DOI] [PubMed] [Google Scholar]
- 29.Li T, Liu X, Gong X, E Q, Zhang X, Zhang X. microRNA 92b-3p regulates primordial follicle assembly by targeting TSC1 in neonatal mouse ovaries. Cell Cycle. 2019;18:824–833. doi: 10.1080/15384101.2019.1593648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alton M, Taketo T. Switch from BAX-dependent to BAX-independent germ cell loss during the development of fetal mouse ovaries. J. Cell Sci. 2007;120:417–424. doi: 10.1242/jcs.03332. [DOI] [PubMed] [Google Scholar]
- 31.Gawriluk TR, et al. Autophagy is a cell survival program for female germ cells in the murine ovary. Reproduction. 2011;141:759–765. doi: 10.1530/REP-10-0489. [DOI] [PubMed] [Google Scholar]
- 32.Eskelinen EL, Reggiori F, Baba M, Kovács AL, Seglen PO. Seeing is believing: the impact of electron microscopy on autophagy research. Autophagy. 2011;7:935–956. doi: 10.4161/auto.7.9.15760. [DOI] [PubMed] [Google Scholar]
- 33.Yu Y, Nangia-Makker P, Farhana L, Majumdar APN. A novel mechanism of lncRNA and miRNA interaction: CCAT2 regulates miR-145 expression by suppressing its maturation process in colon cancer cells. Mol. Cancer. 2017;16:155. doi: 10.1186/s12943-017-0725-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang X, et al. PTBP3 splicing factor promotes hepatocellular carcinoma by destroying the splicing balance of NEAT1 and pre-miR-612. Oncogene. 2018;37:6399–6413. doi: 10.1038/s41388-018-0416-8. [DOI] [PubMed] [Google Scholar]
- 35.Dang Y, et al. MicroRNA-22-3p is down-regulated in the plasma of Han Chinese patients with premature ovarian failure. Fertil. Steril. 2015;103:802–807. doi: 10.1016/j.fertnstert.2014.12.106. [DOI] [PubMed] [Google Scholar]
- 36.Kuang H, et al. Profiling of differentially expressed microRNAs in premature ovarian failure in an animal model. Gynecol. Endocrinol. 2014;30:57–61. doi: 10.3109/09513590.2013.850659. [DOI] [PubMed] [Google Scholar]
- 37.Zhou W, Xu J, Wang C, Shi D, Yan Q. miR-23b-3p regulates apoptosis and autophagy via suppressing SIRT1 in lens epithelial cells. J. Cell Biochem. 2019;120:19635–19646. doi: 10.1002/jcb.29270. [DOI] [PubMed] [Google Scholar]
- 38.YiRen H, et al. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol. Cancer. 2017;16:174. doi: 10.1186/s12943-017-0743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi JY, Chen C, Xu X, Lu Q. miR-29a promotes pathological cardiac hypertrophy by targeting the PTEN/AKT/mTOR signalling pathway and suppressing autophagy. Acta Physiol. 2019;227:e13323. doi: 10.1111/apha.13323. [DOI] [PubMed] [Google Scholar]
- 40.Vats S, Manjithaya R. A reversible autophagy inhibitor blocks autophagosome-lysosome fusion by preventing Stx17 loading onto autophagosomes. Mol. Biol. Cell. 2019;30:2283–2295. doi: 10.1091/mbc.E18-08-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Tsuboyama K, et al. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 2016;354:1036–1041. doi: 10.1126/science.aaf6136. [DOI] [PubMed] [Google Scholar]
- 43.Kumar S, et al. Mechanism of Stx17 recruitment to autophagosomes via IRGM and mammalian Atg8 proteins. J. Cell Biol. 2018;217:997–1013. doi: 10.1083/jcb.201708039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yim WW, Mizushima N. Lysosome biology in autophagy. Cell Discov. 2020;6:6. doi: 10.1038/s41421-020-0141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chapman AG, Cotton AM, Kelsey AD, Brown CJ. Differentially methylated CpG island within human XIST mediates alternative P2 transcription and YY1 binding. BMC Genet. 2014;15:89. doi: 10.1186/s12863-014-0089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Makhlouf M, et al. A prominent and conserved role for YY1 in Xist transcriptional activation. Nat. Commun. 2014;5:4878. doi: 10.1038/ncomms5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng L, et al. YY1-MIR372-SQSTM1 regulatory axis in autophagy. Autophagy. 2014;10:1442–1453. doi: 10.4161/auto.29486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang D, et al. YY1-PVT1 affects trophoblast invasion and adhesion by regulating mTOR pathway-mediated autophagy. J. Cell Physiol. 2020;235:6637–6646. doi: 10.1002/jcp.29560. [DOI] [PubMed] [Google Scholar]
- 49.Zhao X, Su L, He X, Zhao B, Miao J. Long noncoding RNA CA7-4 promotes autophagy and apoptosis via sponging MIR877-3P and MIR5680 in high glucose-induced vascular endothelial cells. Autophagy. 2020;16:70–85. doi: 10.1080/15548627.2019.1598750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang K, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat. Commun. 2015;6:6779. doi: 10.1038/ncomms7779. [DOI] [PubMed] [Google Scholar]
- 51.Hendrich BD, Plenge RM, Willard HF. Identification and characterization of the human XIST gene promoter: implications for models of X chromosome inactivation. Nucleic Acids Res. 1997;25:2661–2671. doi: 10.1093/nar/25.13.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon SH, Choi YM. Analysis of C43G mutation in the promoter region of the XIST gene in patients with idiopathic primary ovarian insufficiency. Clin. Exp. Reprod. Med. 2015;42:58–61. doi: 10.5653/cerm.2015.42.2.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang R, et al. Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis at the posttranscriptional level in the nucleus: evidence for a microRNA hierarchy system. Cell Res. 2012;22:504–515. doi: 10.1038/cr.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez I, et al. An Exportin-1-dependent microRNA biogenesis pathway during human cell quiescence. Proc. Natl Acad. Sci. USA. 2017;114:E4961–E4970. doi: 10.1073/pnas.1618732114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Q, Chen B, Liu P, Yang J. XIST promotes gastric cancer (GC) progression through TGF-β1 via targeting miR-185. J. Cell Biochem. 2018;119:2787–2796. doi: 10.1002/jcb.26447. [DOI] [PubMed] [Google Scholar]
- 57.Chen DL, et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J. Exp. Clin. Cancer Res. 2016;35:142. doi: 10.1186/s13046-016-0420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng Q, Wang L. LncRNA XIST serves as a ceRNA to regulate the expression of ASF1A, BRWD1M, and PFKFB2 in kidney transplant acute kidney injury via sponging hsa-miR-212-3p and hsa-miR-122-5p. Cell Cycle. 2020;19:290–299. doi: 10.1080/15384101.2019.1707454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.