Abstract
Neonatal jaundice is a common disease that affects up to 60% of newborns. Herein, we performed a comparative analysis of the gut microbiome in neonatal jaundice and non-neonatal jaundice infants (NJIs) and identified gut microbial alterations in neonatal jaundice pre- and post-treatment. We prospectively collected 232 fecal samples from 51 infants at five time points (0, 1, 3, 6, and 12 months). Finally, 114 samples from 6 NJIs and 19 non-NJI completed MiSeq sequencing and analysis. We characterized the gut microbiome and identified microbial differences and gene functions. Meconium microbial diversity from NJI was decreased compared with that from non-NJI. The genus Gemella was decreased in NJI versus non-NJI. Eleven predicted microbial functions, including fructose 1,6-bisphosphatase III and pyruvate carboxylase subunit B, decreased, while three functions, including acetyl-CoA acyltransferase, increased in NJI. After treatments, the microbial community presented significant alteration-based β diversity. The phyla Firmicutes and Actinobacteria were increased, while Proteobacteria and Fusobacteria were decreased. Microbial alterations were also analyzed between 6 recovered NJI and 19 non-NJI. The gut microbiota was unique in the meconium microbiome from NJI, implying that early gut microbiome intervention could be promising for the management of neonatal jaundice. Alterations of gut microbiota from NJI can be of great value to bolster evidence-based prevention against ‘bacterial dysbiosis’.
Keywords: Gut microbiota, MiSeq sequencing, Neonatal jaundice, Treatment
Introduction
Neonatal jaundice is the most common condition after birth and often occurs during the first week of life. Approximately 60% of term infants and 80% of preterm infants develop jaundice in the first week of life [1]. There are studies on the association between neonatal jaundice and adverse long-term health outcomes, such as childhood asthma, type 1 diabetes, and impaired visual function [2–4]. Neonatal jaundice is characterized by the presence of high total serum bilirubin levels [5]. Recent progress in understanding the microbiota reveals the role of bacteria in bilirubin metabolism. A study in mouse models of germ-free multidrug resistance 2 knockouts showed increased serum bilirubin levels [6]. There are studies on the possible association between the increase in direct bilirubin and bacteria such as Bifidobacterium [7,8]. Thus, it is hypothesized that the gut microbiota is associated with neonatal jaundice, but the gut microbial characteristics in neonatal jaundice remain limited. It is essential to understand the relationship between the gut microbiome and neonatal jaundice and to search for gut microbial alterations in neonatal jaundice pre- and post-treatment.
In the present study, a total of 51 infants were enrolled, and fecal samples were separately collected at 0 month (meconium) and 1, 3, 6, and 12 months postpartum. After confirmation, two premature infants, five infants with incomplete information, and eight infants receiving antibiotic therapy were excluded. After DNA extraction, 16S rRNA gene sequencing and data quality control, 11 neonatal jaundice infant (NJI) meconium samples with insufficient quantities were further discarded. Finally, 6 NJI with treatment and 19 non-NJI were included in the final analysis.
We characterized the meconium microbiome between NJI and non-NJI and identified microbial differences. Furthermore, we assessed gut microbial alterations in neonatal jaundice pre- and post-treatment and reported differences in the gut microbiota of recovered NJI versus non-NJI. The study of NJI gut microbiota development can be of great value to bolster evidence-based prevention against ‘bacterial dysbiosis’, especially in such a populous territory as China.
Materials and methods
Participants’ information
The study complied with the ethical guidelines of the Helsinki Declaration and Rules of Good Clinical Practice. The Institutional Review Board of the First Affiliated Hospital of Zhengzhou University approved the studies (2017-KY-12). Informed consent from all participants was obtained before data and stool samples were collected. Mode of delivery and infant medication use were obtained from hospital electronic medical records. Mothers were asked to complete a questionnaire at each time point, including infant diet (exclusive, partial or no breastfeeding), height and weight of the infant, antibiotic use, and Yinzhihuang oral liquid use. Yinzhihuang oral liquid, a well-known Chinese herbal formula, is officially listed in the Chinese Pharmacopoeia, and is a clinical drug for the treatment of neonatal jaundice [9].
The diagnostic criteria for neonatal jaundice followed international guidelines [10,11]. In our study, all NJI received the same brand and manufacturer of Yinzhihuang oral liquid treatment. All NJI received Yinzhihuang oral liquid combined with phototherapy (blue light) treatment (wavelength range: 425–475 nm); and did not receive other medical interventions. Throughout the study, the manufacturer of the light was consistent. All infants included in the study were born by vaginal delivery. The inclusion criteria were as follows: exclusively breastfed, birth weight 2500–4000 g, gestational age 37–42 weeks, and fifth-minute Apgar score 8–10. The exclusion criteria were as follows: hemolysis of newborns, hereditary metabolic diseases, and infectious diseases such as neonatal pneumonia.
Sample collection and DNA extraction
The study population was recruited before mothers’ parturition. Fecal samples were collected for the first time before infants exhibited jaundice in the hospital. The other fresh fecal samples were collected at their homes, and samples were immediately delivered to the laboratory on dry ice using foam containers. In the laboratory, the sample was stored at −80°C until DNA was extracted. Any samples that stayed at room temperature for more than 2 h were discarded. DNA was extracted from fecal samples according to the manufacturer’s instructions in the E.Z.N.A.® Stool DNA Kit (Omega Bio-tek, Inc., GA). The DNA concentration was determined by a NanoDrop 2000 spectrophotometer (Thermo Scientific), and its molecular size was estimated using agarose gel electrophoresis (0.8%). To assess DNA purity, absorbance ratios were determined at wavelengths of 260 nm relative to 280 nm and 260 nm relative to 230 nm.
PCR amplification and MiSeq sequencing
The extracted DNA was used as the template to amplify the V3 to V4 regions of the 16S rRNA gene. The forward primer (341F) was 5′-CCTACGGGNGGCWGCAG-3′, and the reverse primer (805R) was 5′-GACTACHVGGGTATCTAATCC-3′. PCR amplification was performed in an EasyCycler 96 PCR system (Analytik Jena Corp., AG) using the following program: 1 cycle at 95°C for 3 min; 21 cycles at 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s; and 1 cycle at 72°C for 5 min. Finally, PCR products were held at 4°C to prevent degradation following the PCR cycle. The products from different samples were mixed at equal ratios for sequencing according to the manufacturer’s instructions, and sequencing was performed on the Illumina MiSeq platform at Shanghai Mobio Biomedical Technology Co. Ltd. Raw Illumina read data for all samples were deposited in the European Bioinformatics Institute European Nucleotide Archive database under accession numbers PRJNA680178 and PRJNA665920.
Operational taxonomic units and taxonomy annotation
Equal numbers of reads were randomly chosen from all samples, and then operational taxonomic units (OTUs) were binned using the UPARSE pipeline [12]. Sequences with 97% similarity level were clustered into OTUs. RDP classifier version 2.6 software [13] was used to assign sequences to the new bacterial taxonomy.
Bacterial diversity and taxonomic analysis
Bacterial diversity was assessed by sampling-based analysis of OTUs and presented by the ACE index, which was calculated using the R program package ‘vegan’. Principal coordinates analysis (PCoA) and non-metric multidimensional scaling (NMDS) based on OTU abundance and distribution were conducted by the vegan R package (http://www.R-project.org/) to analyze microbial communities [14]. The weighted and unweighted UniFrac distances were calculated with the phyloseq package [15]. A heatmap of the identified key variables was generated by Heatmap Builder.
Bacterial differences were compared at the phylum and genus levels. Fecal microbial characteristics were analysed by the linear discriminant analysis (LDA) effect size (LEfSe) method (http://huttenhower.sph.harvard.edu/lefse/) [16]. Using a normalized relative abundance matrix, LEfSe performs the Kruskal–Wallis rank sum test to determine characteristics with significantly different abundances between assigned bacteria and uses LDA to assess the effect size of each characteristic.
Functional annotation of the gut microbial 16S rRNA gene
To predict the functional profiles of microbial communities based on 16S rRNA gene sequences, we utilized the phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) version 1.0.0 pipeline [17] and human version 0.99 [18] to establish KEGG orthology (KO) and KEGG pathway/module profiles.
Statistical analysis
The Wilcoxon rank sum test was used to compare continuous variables between both groups. Fisher’s exact test was used to compare categorical variables. The infant weight-for-length z-score was calculated according to World Health Organization standards [19]. The analyses were performed using SPSS version 21.0. The statistical significance was set at P<0.05.
Results
Characteristics of the participants
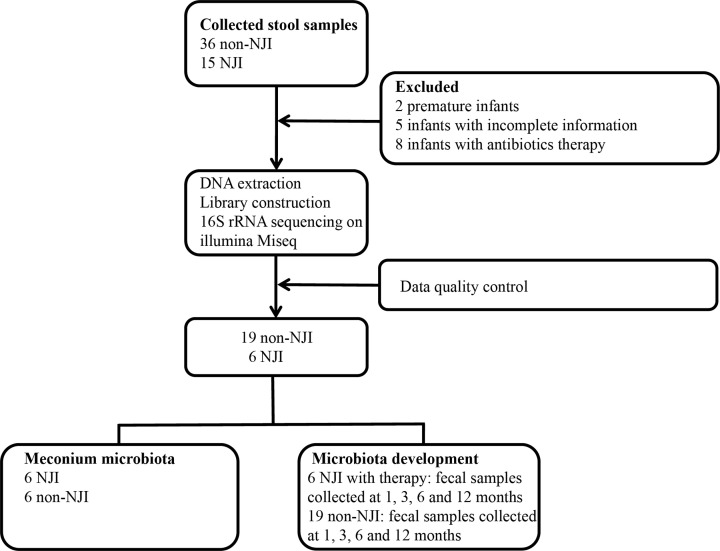
A total of 51 infants (232 fecal samples) were collected from Zhengzhou, Central China. After a strict pathological diagnosis and exclusion process, a total of 114 fecal samples from 6 NJI with treatment and 19 non-NJI were included (Figure 1). In the meconium cohort, all individuals included were required to be infants resulting from full-term pregnancies and vaginal deliveries and to be exclusively breastfed.
Figure 1. Study design and flow diagram.
A total of 232 fecal samples from 15 NJI and 36 non-NJI were collected. After a strict pathologic diagnosis and exclusion process, the remaining samples were used for DNA extraction, 16S rRNA sequencing and data quality control. Finally, 6 NJI with treatment and 19 non-NJI were utilized for bioinformatics analysis.
The demographic, clinical, and anthropometric characteristics of the 25 neonates are presented in Table 1. Among all participants, there were no significant differences in age, sex, birth weight, or birth length between NJI and non-NJI. The agarose gel electrophoresis results are shown in Supplementary Figure S1. DNA was extracted from 438 fecal samples. And exclusively breastfed infants, combined feeding infants, infants born by cesarean section, and infants born by vaginal delivery were included in Supplementary Figure S1.
Table 1. Characteristics of the study population.
| Variables | Cases (n=6) | Controls (n=19) | P-value |
|---|---|---|---|
| Maternal age (years) | 29.67 ± 3.01 | 32.26 ± 4.32 | 0.19 |
| Prenatal BMI (kg/m2) | 25.72 ± 3.73 | 28.68 ± 3.55 | 0.09 |
| Pregnancy weight gain (kg) | 13.78 ± 4.52 | 15.88 ± 3.17 | 0.21 |
| Newborn sex | |||
| Male | 4 (0.67) | 12 (0.63) | 0.64 |
| Female | 2 (0.33) | 7 (0.37) | |
| Birth weight (g) | 3241.67 ± 363.89 | 3489.47 ± 233.68 | 0.06 |
| Birth length (cm) | 51.67 ± 1.03 | 51.53 ± 1.22 | 0.80 |
| Delivery mode | |||
| Vaginal delivery | 5 (0.83) | 12 (0.63) | 0.35 |
| Cesarean section delivery | 1 (0.17) | 7 (0.37) | |
| Feeding patterns | |||
| Exclusive breastfeeding | 4 (0.67) | 15 (0.79) | 1.00 |
| Non-exclusive breastfeeding | 2 (0.33) | 4 (0.21) | |
| Apgar score | |||
| 1 min | 9.33 ± 0.82 | 9.37 ± 0.76 | 0.92 |
| 5 min | 9.50 ± 0.55 | 9.63 ± 0.50 | 0.59 |
| Gestational age (days) | 282.17 ± 6.65 | 280.11 ± 8.03 | 0.58 |
Continuous variables were compared using the Wilcoxon rank sum test. Fisher’s exact test was used to compare categorical variables.
Gut microbiomes are unique in NJI at 0 month
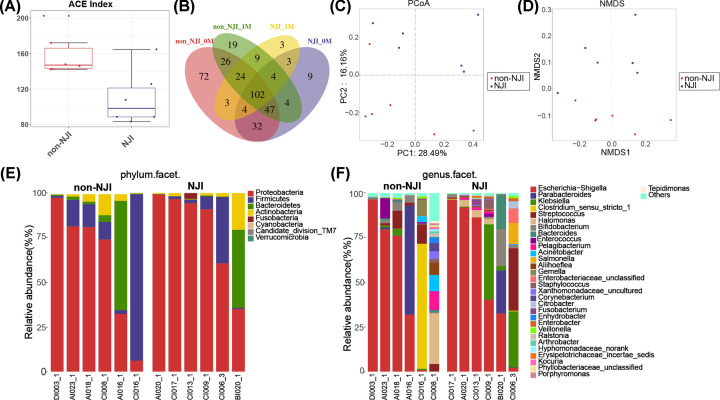
We collected meconium from NJI and matched non-NJI at 0 month. Compared with the non-NJI group, fecal microbial diversity, as estimated by the ACE estimator, was markedly decreased in NJI (P<0.05, Figure 2A, Supplementary Table S1). The Venn analysis showed that 185 of 330 OTUs were shared between NJI-0M and non-NJI-0M (Figure 2B). To assess similarity among microbial communities, we performed PCoA and NMDS analysis based on unweighted UniFrac distance (Figure 2C,D, Supplementary Tables S2 and S3).
Figure 2. Altered meconium microbiota composition in NJI.
(A) Microbial α diversity decreased in NJI, as shown by the ACE estimator. (B) Shared and unique genera among NJI-0M, non-NJI-0M, NJI-1M, and non-NJI-1M. Overall diversity was calculated using unweighted UniFrac by PCoA (C) and NMDS (D), indicating a separation of samples between NJI and non-NJI. Fecal microbiota composition at the phylum level (E) and genus level (F) between NJI and non-NJI.
A heatmap of the identified key variables demonstrated that a total of 27 key OTUs were significantly different between NJI and non-NJI (Supplementary Figure S2 and Table S4). We further analyzed the infant gut microbiota composition and alterations at the phylum and genus levels between the two groups. The fecal bacterial composition in each sample at the phylum and genus levels is shown in Figure 2E,F (Supplementary Tables S5 and S6). The average composition of the microbial community at the phylum and genus levels between the two groups is shown in Supplementary Figure S3A,B and Tables S7 and S8. The bacterial phyla Proteobacteria, Firmicutes, and Bacteroidetes, together accounting for up to 90% of sequences on average, were three dominant populations in the two groups (Figure 2E). Bacterial genera of Escherichia-Shigella, Parabacteroides, Klebsiella, and Clostridium sensu stricto 1, together accounting for up to 70% of sequences on average, were the four dominant populations in the two groups (Figure 2F). The average amount of Gemella was markedly decreased in NJI versus non-NJI (P<0.05, Supplementary Figure S3C and Table S9). Although there were no significant differences in the average relative abundances of Klebsiella and Clostridium between NJI and non-NJI (both P>0.05, Supplementary Table S9), changes in average relative abundances of Klebsiella and Clostridium were still noted in NJI (Supplementary Figure S3B).
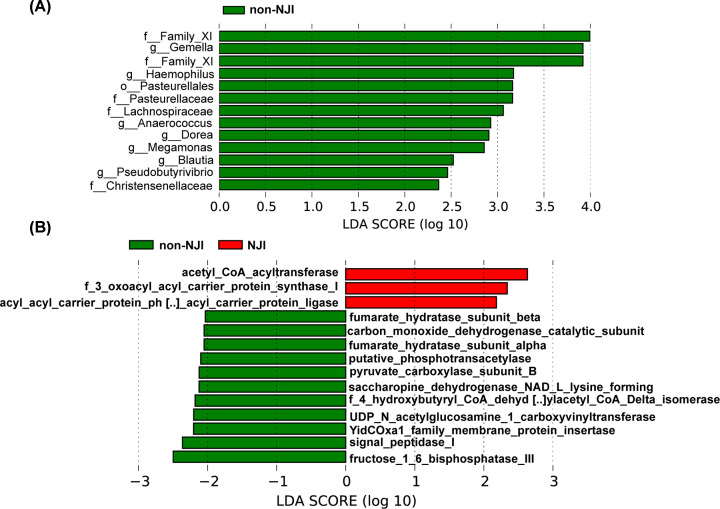
We used LDA effective size (LEfSe) to determine the specific bacterial taxa related to neonatal jaundice. A cladogram representative of fecal microbial structure and their predominant bacteria displayed the greatest differences in taxa between NJI and non-NJI (all P<0.05, Supplementary Figure S4 and Table S10). Meanwhile, the cladogram of fecal microbial structure between NJI and non-NJI also showed the greatest differences in taxa (all P<0.05, LDA > 2, Figure 3A, Supplementary Table S10), which suggested gut microbial alterations in NJI.
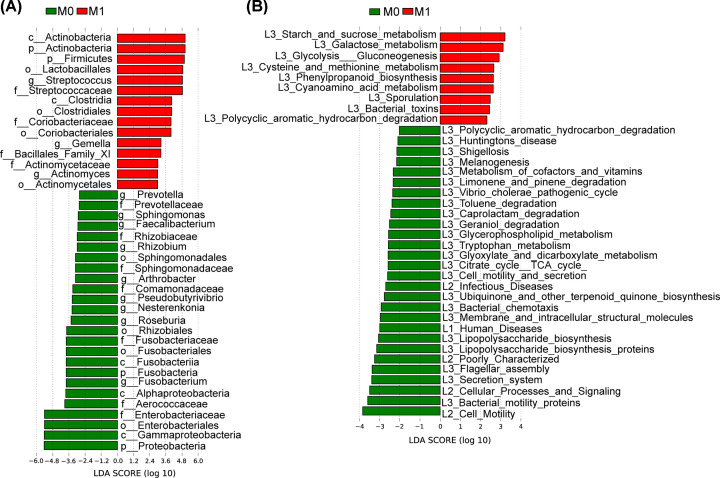
Figure 3. Identification of specific bacterial taxa and microbial functions associated with neonatal jaundice.
(A) The greatest differences in taxa between NJI and non-NJI are presented according to the LDA scores (log10). (B) Differences in gut microbial functions between NJI and non-NJI based on the LDA scores (log10).
Microbial metabolic function predictions using the PICRUSt pipeline [17] assessed the potential microbial functions associated with neonatal jaundice and again showed significant differences between the two groups. Based on LDA selection, 3 predicted microbial functions, mainly acetyl-CoA acyltransferase and 3 oxoacyl acyl carrier protein synthase I, were enriched, while 11 functions, mainly fructose 1,6-bisphosphatase III, signal peptidase I and YidCOxa1 family membrane protein insertase, were reduced in NJI versus non-NJI (all P<0.05, LDA > 2, Figure 3B, Supplementary Table S11).
Gut microbial alterations before and after treatment
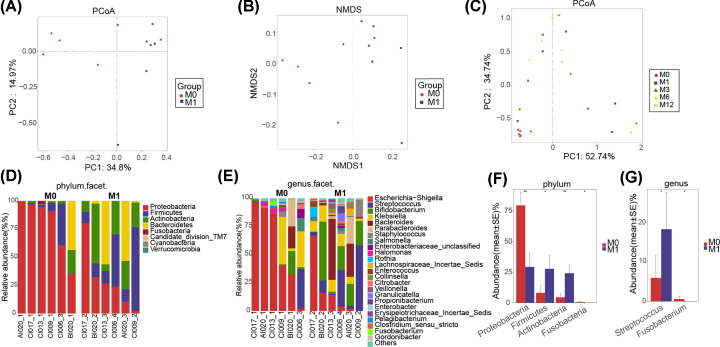
The Venn analysis showed that 113 of 244 OTUs were shared between pre-treatment (NJI-0M) and post-treatment (NJI-1M) (Figure 2B). To display microbiome space between pre-treatment and post-treatment, we performed PCoA and NMDS analysis based on unweighted UniFrac distance (P<0.05, Figure 4A,B, Supplementary Tables S12 and S13). Moreover, PCoA was conducted based on weighted UniFrac distances to assess the microbial distribution at 0, 1, 3, 6, and 12 months (Figure 4C, Supplementary Table S14). PCoA indicated that the samples tended to be uniform at 0 and 12 months, and no obvious separation was observed in NJI with treatment. Notably, samples were most heterogeneous at the age of 1–6 months.
Figure 4. Gut microbial differences of infants between pre-treatment (NJI-0M) and post-treatment (NJI-1M).
Overall diversity was calculated using unweighted UniFrac by PCoA (A) and NMDS (B), indicating a separation of samples between pre-treatment and post-treatment. (C) PCoA based on the weighted UniFrac distance in NJI with treatment to assess the microbial distribution among 0, 1, 3, 6, and 12 months, indicating that samples tended to be uniform at 0 and 12 months, and samples are most heterogeneous at 1–6 months. Fecal microbiota composition at the phylum level (D) and genus level (E) between pre-treatment and post-treatment. (F) Compared with pre-treatment, two phyla were significantly increased, while two phyla were significantly decreased post-treatment (all P<0.05). (G) One genus was increased whereas one genus was decreased in pre-treatment versus post-treatment (all P<0.05). Abbreviations: M0, 0 month; M1, 1 month; M3, 3 months; M6, 6 months; M12, 12 months.
A heatmap of the identified key variables demonstrated that a total of 26 key OTUs were significantly different between the two groups (Supplementary Figure S5 and Table S15). Fecal bacterial composition and differences at the phylum and genus levels between the two groups are shown in Figure 4D–G, respectively (all P<0.05, Supplementary Tables S16–S19).
We detected the greatest differences in taxa between pre-treatment and post-treatment using the LEfSe method and LDA scores, as shown in Figure 5A and Supplementary Figure S6A (all P<0.05, LDA > 2.4, Supplementary Table S20).
Figure 5. Identification of specific bacterial taxa and microbial functions between pre-treatment (0 month) and post-treatment (1 month).
(A) The greatest differences in taxa between pre-treatment and post-treatment are presented according to the LDA scores (log10). (B) Differences in gut microbial functions between pre-treatment and post-treatment based on the LDA scores (log10). Abbreviations: M0, 0 month; M1, 1 month.
The predominant fecal microbial functions between pre-treatment and post-treatment are shown by a cladogram and LDA (all P<0.05, LDA > 2, Figure 5B and Supplementary Figure S6B and Table S21). These data revealed significant differences between the two groups.
Gut microbiota alterations in recovered NJI and non-NJI
The Venn analysis showed that 139 of 248 OTUs were shared between recovered NJI (NJI-1M) and non-NJI (non-NJI-1M) at 1 month (Figure 2B). To display the microbiome space between recovered NJI and non-NJI at 0, 1, 3, 6, and 12 months, we performed PCoA based on weighted UniFrac distances (Figure 6A, Supplementary Table S22). These data revealed that distinctly separate bacterial communities were present between recovered NJI and non-NJI at an early age, while the microbial communities became more uniform over time.
Figure 6. Gut microbial alterations of infants between recovered NJI and non-NJI.
(A) PCoA based on the weighted UniFrac distance between recovered NJI and non-NJI at 0, 1, 3, 6, and 12 months, indicating that the microbial communities became more uniform over time. Fecal microbiota composition at the phylum level (B) and genus level (C) between recovered NJI and non-NJI. (D) Compared with recovered NJI, one phylum was significantly decreased in non-NJI (P<0.05). (E) Three genera were increased in non-NJI versus recovered NJI (all P<0.05). Abbreviations: M0, 0 month; M1, 1 month; M3, 3 months; M6, 6 months; M12, 12 months.
A heatmap of the identified key variables revealed that a total of 13 key OTUs were significantly different between recovered NJI and non-NJI at 1 month (Supplementary Figure S7 and Table S23). Fecal bacterial composition and differences at the phylum and genus levels between the two groups at 1 month are shown in Figure 6B–E (all P<0.05, Supplementary Table S24–S27).
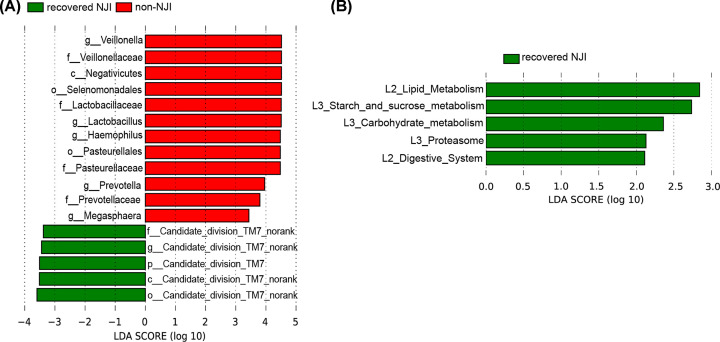
We detected the greatest differences in taxa between recovered NJI and non-NJI at 1 month using the LEfSe method and LDA scores, as shown in Figure 7A and Supplementary Figure S8A (all P<0.05, LDA > 3, Supplementary Table S28).
Figure 7. Identification of specific bacterial taxa and microbial functions between recovered NJI and non-NJI.
(A) The greatest differences in taxa between recovered NJI and non-NJI are presented according to the LDA scores (log10). (B) Differences in gut microbial functions between recovered NJI and non-NJI based on the LDA scores (log10).
The predominant fecal microbial functions between recovered NJI and non-NJI patients at 1 month are shown by a cladogram and LDA (all P<0.05, LDA > 2, Figure 7B and Supplementary Figure S8B and Table S29).
Effect of medication on infant growth
These observations prompted us to explore the potential relationship between NJI treatment and infant growth. Thus, we examined whether the weight-for-length z-score at 12 months differed between recovered NJI and non-NJI. A t test for independent samples was used to compare the development and growth of recovered NJI and non-NJI at 12 months of age. The infant weight-for-length z-score was calculated according to World Health Organization standards [19]. The infant weight-for-length z-score at 12 months did not differ significantly between recovered NJI and non-NJI (P<0.05). Our study demonstrated that growth at 12 months of age is independent of NJI treatment in early life.
Discussion
We illustrated that neonatal jaundice was associated with the altered composition and function of gut microbiota, as well as a decrease in α-diversity. Dong et al. [20] characterized meconium microbiome of NJIs and found that a-diversity were lower in the NJIs compared with controls, which is consistent with our study. Recent studies have reported that a high level of α-diversity was associated with a low risk of necrotizing enterocolitis, atopic eczema, and neonatal sepsis [21–23]. Our study suggested that neonatal jaundice can cause gut microbiota dysbiosis. And we observed a higher α-diversity of gut microbes at birth. Moreover, the bacterial community of neonates at risk of jaundice was separated from that of non-NJIs.
The gut microbiota is indispensable to the health of the host. Healthy infants may share some key microbiota structural features, whereas neonates at risk of jaundice may have aberrant patterns and lack some key bacteria, leading to a ‘dysbiosis’ state. We found that although the relative abundance of Bifidobacterium was lower in the non-NJI compared with NJI at 0 and 1 month, but P-values were not significant (P>0.05). Bifidobacterium was identified to exert antimicrobial activity against pathogens in infant gut, which could benefit the colonization of healthy bacteria [24,25]. Tuzun et al. [8] indicated that down-regulation of members of the Bifidobacterium genus, such as B. longum, B. bifidum, and B. adolescentis, was associated with an increase in serum bilirubin level. Our study showed no significant difference in the relative abundance of Bifidobacterium between two groups. We speculated that this is because all infants included were exclusively breastfed, and breast milk promoted the colonization of Bifidobacterium [26].
We observed that there was only one feature more abundant in the non-NJI (Gemella), and this bacterial genera can produce short-chain fatty acids (SCFAs) [27]. SCFAs (particularly propionate and butyrate) initiate several complementary mechanisms within the intestinal mucosa that activate intestinal gluconeogenesis [28]. Notably, butyrate plays an important role in bacterial energy metabolism and intestinal mucosa health in humans, is the major energy source of the intestinal mucosa, and is an important regulator of gene expression, inflammation, differentiation, and apoptosis in host cells [29–32]. LEfSe showed that some butyrate-producing bacteria, such as Blautia and Pseudobutyrivibrio, were increased in non-NJI versus NJI. A study indicated that Pseudobutyrivibrio are producers of butyrate, lactic acid, and formic acid [33]. In addition, members of the genus Blautia produce acetate, ethanol, hydrogen, lactate, or succinate, which can provide energy for the host [34]. Moreover, non-NJI harbor more beneficial populations, such as Lachnospiraceae, one of the major taxonomic groups of the gut microbiota, which degrade complex polysaccharides to SCFAs, including acetate, butyrate, and propionate, which can be used by the host as sources of energy [35]. These results indicated that gut microbial community alteration might play a key role during neonatal jaundice initiation.
Different microbial functions and metabolites are determined by different microbial communities, thereby contributing to the pathogenesis and development of different diseases [7,36,37]. Gut microbial functions involving fructose 1,6-bisphosphatase III and pyruvate carboxylase subunit B were enriched in non-NJI according to the LDA scores (log10). Fructose 1,6-bisphosphatase III, a key enzyme in intestinal gluconeogenesis, exerts important physiological functions in the regulation of energy metabolism and glucose homeostasis [38–40]. Previous studies have shown that an increase in lactate was found in cholestasis and are consistent with our results [41,42]. Lactate is a substrate of intestinal gluconeogenesis. Low levels of expression of the fructose 1,6-bisphosphatase III and pyruvate carboxylase subunit B enzymes involved in gluconeogenesis decreased the clearance of lactate. In addition, in infancy, the body has a great demand for energy, and the body may produce more ATP through glycolysis to maintain energy metabolism, resulting in increased lactate. Thus, neonates with limited capacity to metabolize lactate via intestinal gluconeogenesis may be associated with neonatal jaundice.
Our study analyzed the development of NJI with treatment at 0, 1, 3, 6, and 12 months. We found that gut microbiota differences within the NJI and treatment groups were completely decreased over time, suggesting that treatment can only temporarily perturb the NJI gut microbiota. Moreover, we conducted a longitudinal study of recovered NJI and non-NJI, and PCoA revealed that the microbial community of recovered NJI was clustered with that of non-NJI over time, suggesting that the microbiota of recovered NJI tends to recover to similar levels as non-NJI gradually and that the effect of treatment on gut microbiota is temporary. Importantly, infants’ growth and development at 12 months did not differ significantly between recovered NJI and non-NJI. Taken together, these results demonstrated that the gut microbiota composition was influenced by treatment at early stages, that these differences were absent over time and that the gut microbiota gradually recovered. Thus, we propose that treatment may have little long-term effect on infant health.
In summary, the present study comprehensively characterized the meconium microbiome between NJI and non-NJI and identified microbial differences. The combination of early gut microbiome intervention and currently used treatment methods may further benefit NJI. Moreover, we illustrated the gut microbial alterations and development in NJI with treatment, which may provide a solid foundation for future health outcomes through microbiota intervention.
Supplementary Material
Acknowledgements
We thank clinical doctors from the First Affiliated Hospital of Zhengzhou University. We also thank the generous volunteer subjects who enrolled in the study.
Abbreviations
- ACE
abundance-based coverage estimator
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LDA
linear discriminant analysis
- LEfSe
LDA effect size
- NJI
neonatal jaundice infant
- NMDS
non-metric multidimensional scaling
- OTU
operational taxonomic unit
- PCoA
principal coordinates analysis
- PICRUSt
phylogenetic investigation of communities by reconstruction of unobserved states
- SCFA
short-chain fatty acid
Data Availability
The raw Illumina read data for all samples were deposited in the European Bioinformatics Institute European Nucleotide Archive database under the accession numbers PRJNA680178 and PRJNA6659201.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the China Postdoctoral Science Foundation [grant numbers 2020T130609, 2020T130109ZX]; the National Key Research and Development Program of China [grant number 2018YFC2000500]; the Henan Provincial Medical Science and Technology Project [grant number SBGJ2018004]; the Henan Province Science and Technology Project [grant numbers 182102310404, 202102310055]; and the Key Scientific Research Projects of Higher Education Institutions in Henan Province [grant numbers 21A320055, 20A320056]. The funding sources had no role in the design of the present study nor any role during its execution, analyses, data interpretation or decision to submit results.
Author Contribution
J.D. and X.M. conceptualized and designed the study, drafted the initial manuscript, and reviewed and revised the manuscript. W.L., Q.X., L.H., X.Z., A.L., Z.L., and H.R. designed the data collection instruments, collected data, carried out the initial analyses, and reviewed and revised the manuscript. Z.R. conceptualized and designed the study, coordinated and supervised data collection, and critically reviewed the manuscript for important intellectual content. All authors approved the final manuscript as submitted and agreed to be accountable for all aspects of the work.
Ethics Approval and Consent to Participate
The present study was approved by the Institutional Review Board of the First Affiliated Hospital of Zhengzhou University (2017-XY-012). The study complied with the ethical guidelines of Helsinki Declaration and Rules of Good Clinical Practice. All participants signed written informed consents after the study protocol was fully explained.
Consent for Publication
Informed consent from all participants was obtained before data and fecal samples were collected.
References
- 1.Rennie J., Burman-Roy S. and Murphy M.S. (2010) Neonatal jaundice: summary of NICE guidance. BMJ 340, c2409, 10.1136/bmj.c2409 [DOI] [PubMed] [Google Scholar]
- 2.Huang L., Bao Y., Xu Z., Lei X., Chen Y., Zhang Y.et al. (2013) Neonatal bilirubin levels and childhood asthma in the US Collaborative Perinatal Project, 1959-1965. Am. J. Epidemiol. 178, 1691–1697, 10.1093/aje/kwt248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McNamee M.B., Cardwell C.R. and Patterson C.C. (2012) Neonatal jaundice is associated with a small increase in the risk of childhood type 1 diabetes: a meta-analysis of observational studies. Acta Diabetol. 49, 83–87, 10.1007/s00592-011-0326-5 [DOI] [PubMed] [Google Scholar]
- 4.Hou C., Norcia A.M., Madan A. and Good W.V. (2014) Visuocortical function in infants with a history of neonatal jaundice. Investig. Ophthalmol. Vis. Sci. 55, 6443–6449, 10.1167/iovs.14-14261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dennery P.A., Seidman D.S. and Stevenson D.K. (2001) Neonatal hyperbilirubinemia. N. Engl. J. Med. 344, 581–590, 10.1056/NEJM200102223440807 [DOI] [PubMed] [Google Scholar]
- 6.Tabibian J.H., O'Hara S.P., Trussoni C.E., Tietz P.S., Splinter P.L., Mounajjed T.et al. (2016) Absence of the intestinal microbiota exacerbates hepatobiliary disease in a murine model of primary sclerosing cholangitis. Hepatology 63, 185–196, 10.1002/hep.27927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou S., Wang Z., He F., Qiu H., Wang Y., Wang H.et al. (2019) Association of serum bilirubin in newborns affected by jaundice with gut microbiota dysbiosis. J. Nutr. Biochem. 63, 54–61, 10.1016/j.jnutbio.2018.09.016 [DOI] [PubMed] [Google Scholar]
- 8.Tuzun F., Kumral A., Duman N. and Ozkan H. (2013) Breast milk jaundice: effect of bacteria present in breast milk and infant feces. J. Pediatr. Gastroenterol. Nutr. 56, 328–332, 10.1097/MPG.0b013e31827a964b [DOI] [PubMed] [Google Scholar]
- 9.Qian Z.Z., Yang D., Liu Y.Z. and Peng Y. (2010) Pharmacopoeia of the People’s Republic of China (2010 Edition): a milestone in development of China’s healthcare. Chinese Herb. Med. 02, 157–160 [Google Scholar]
- 10.American Academy of Pediatrics Subcommittee on Hyperbilirubinemia . (2004) Management of hyperbilirubinemia in the newborn infant 35 or more weeks of gestation. Pediatrics 114, 297–316 10.1542/peds.114.1.297 [DOI] [PubMed] [Google Scholar]
- 11.Amos R.C., Jacob H. and Leith W. (2017) Jaundice in newborn babies under 28 days: NICE guideline 2016 (CG98). Arch. Dis. Childhood Educ. Pract. Ed. 102, 207–209, 10.1136/archdischild-2016-311556 [DOI] [PubMed] [Google Scholar]
- 12.Edgar R.C. (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998, 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- 13.Wang Q., Garrity G.M., Tiedje J.M. and Cole J.R. (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267, 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paradis E., Claude J. and Strimmer K. (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290, 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- 15.McMurdie P.J. and Holmes S. (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217, 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S.et al. (2011) Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60, 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langille M.G., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A.et al. (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821, 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abubucker S., Segata N., Goll J., Schubert A.M., Izard J., Cantarel B.L.et al. (2012) Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 8, e1002358, 10.1371/journal.pcbi.1002358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WHO Multicentre Growth Reference Study Group . (2006) WHO Child Growth Standards based on length/height, weight and age. Acta Paediatr. 450, 76–85 [DOI] [PubMed] [Google Scholar]
- 20.Dong T., Chen T., White R.A. III, Wang X., Hu W., Liang Y.et al. (2018) Meconium microbiome associates with the development of neonatal jaundice. Clin. Transl. Gastroenterol. 9, 182, 10.1038/s41424-018-0048-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warner B.B., Deych E., Zhou Y., Hall-Moore C., Weinstock G.M., Sodergren E.et al. (2016) Gut bacteria dysbiosis and necrotising enterocolitis in very low birthweight infants: a prospective case-control study. Lancet 387, 1928–1936, 10.1016/S0140-6736(16)00081-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abrahamsson T.R., Jakobsson H.E., Andersson A.F., Bjorksten B., Engstrand L. and Jenmalm M.C. (2012) Low diversity of the gut microbiota in infants with atopic eczema. J. Allergy Clin. Immunol. 129, 434–440, 10.1016/j.jaci.2011.10.025 [DOI] [PubMed] [Google Scholar]
- 23.Madan J.C., Salari R.C., Saxena D., Davidson L., O’Toole G.A., Moore J.H.et al. (2012) Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch. Dis. Child. Fetal Neonatal Ed. 97, F456–F462, 10.1136/fetalneonatal-2011-301373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liévin V., Peiffer I., Hudault S., Rochat F., Brassart D., Neeser J.R.et al. (2000) Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut 47, 646–652, PubMed PMID: 11034580. Pubmed Central PMCID: PMC1728100. 10.1136/gut.47.5.646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Čitar M., Hacin B., Tompa G., Štempelj M., Rogelj I., Dolinšek J.et al. (2015) Human intestinal mucosa-associated Lactobacillus and Bifidobacterium strains with probiotic properties modulate IL-10, IL-6 and IL-12 gene expression in THP-1 cells. Benef. Microb. 6, 325–336, 10.3920/BM2014.0081 [DOI] [PubMed] [Google Scholar]
- 26.Andreas N.J., Kampmann B. and Mehring Le-Doare K. (2015) Human breast milk: a review on its composition and bioactivity. Early Hum. Dev. 91, 629–635 10.1016/j.earlhumdev.2015.08.013 [DOI] [PubMed] [Google Scholar]
- 27.Wood H., Acharjee A., Pearce H., Quraishi M.N., Powell R., Rossiter A.et al. (2021) Breastfeeding promotes early neonatal regulatory T-cell expansion and immune tolerance of non-inherited maternal antigens. Allergy 10.1111/all.14736 [DOI] [PubMed] [Google Scholar]
- 28.De Vadder F., Kovatcheva-Datchary P., Goncalves D., Vinera J., Zitoun C., Duchampt A.et al. (2014) Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 156, 84–96 10.1016/j.cell.2013.12.016 [DOI] [PubMed] [Google Scholar]
- 29.Louis P. and Flint H.J. (2009) Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294, 1–8 10.1111/j.1574-6968.2009.01514.x [DOI] [PubMed] [Google Scholar]
- 30.Li F., Hinderberger J., Seedorf H., Zhang J., Buckel W. and Thauer R.K. (2008) Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J. Bacteriol. 190, 843–850 10.1128/JB.01417-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamer H.M., Jonkers D., Venema K., Vanhoutvin S., Troost F.J. and Brummer R.J. (2008) Review article: the role of butyrate on colonic function. Aliment. Pharmacol. Ther. 27, 104–119 10.1111/j.1365-2036.2007.03562.x [DOI] [PubMed] [Google Scholar]
- 32.Seedorf H., Fricke W.F., Veith B., Bruggemann H., Liesegang H., Strittmatter A.et al. (2008) The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc. Natl. Acad. Sci. U.S.A. 105, 2128–2133, 10.1073/pnas.0711093105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paillard D., McKain N., Chaudhary L.C., Walker N.D., Pizette F., Koppova I.et al. (2007) Relation between phylogenetic position, lipid metabolism and butyrate production by different Butyrivibrio-like bacteria from the rumen. Antonie Van Leeuwenhoek 91, 417–422, 10.1007/s10482-006-9121-7 [DOI] [PubMed] [Google Scholar]
- 34.Liu C., Finegold S.M., Song Y. and Lawson P.A. (2008) Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 58, 1896–1902 10.1099/ijs.0.65208-0 [DOI] [PubMed] [Google Scholar]
- 35.Biddle A., Stewart L., Blanchard J. and Leschine S. (2013) Untangling the genetic basis of fibrolytic specialization by lachnospiraceae and ruminococcaceae in diverse gut communities. Diversity 5, 627–640, 10.3390/d5030627 [DOI] [Google Scholar]
- 36.Sartor R.B. (2008) Microbial influences in inflammatory bowel diseases. Gastroenterology 134, 577–594 10.1053/j.gastro.2007.11.059 [DOI] [PubMed] [Google Scholar]
- 37.Wang Z., Klipfell E., Bennett B.J., Koeth R., Levison B.S., Dugar B.et al. (2011) Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63, 10.1038/nature09922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benkovic S.J. and deMaine M.M. (1982) Mechanism of action of fructose 1,6-bisphosphatase. Adv. Enzymol. Relat. Areas Mol. Biol. 53, 45–82, PubMed PMID: 6277165. 10.1002/9780470122983.ch2 [DOI] [PubMed] [Google Scholar]
- 39.Penhoat A., Fayard L., Stefanutti A., Mithieux G. and Rajas F. (2014) Intestinal gluconeogenesis is crucial to maintain a physiological fasting glycemia in the absence of hepatic glucose production in mice. Metabolism 63, 104–111, Epub 2013/10/19. eng. 10.1016/j.metabol.2013.09.005 [DOI] [PubMed] [Google Scholar]
- 40.Soty M., Gautier-Stein A., Rajas F. and Mithieux G. (2017) Gut-brain glucose signaling in energy homeostasis. Cell Metab. 25, 1231–1242, 10.1016/j.cmet.2017.04.032 [DOI] [PubMed] [Google Scholar]
- 41.Long Y., Dong X., Yuan Y., Huang J., Song J., Sun Y.et al. (2015) Metabolomics changes in a rat model of obstructive jaundice: mapping to metabolism of amino acids, carbohydrates and lipids as well as oxidative stress. J. Clin. Biochem. Nutr. 57, 50–59, 10.3164/jcbn.14-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fabbri C., de Cassia Mascarenhas-Netto R., Lalwani P., Melo G.C., Magalhaes B.M., Alexandre M.A.et al. (2013) Lipid peroxidation and antioxidant enzymes activity in Plasmodium vivax malaria patients evolving with cholestatic jaundice. Malar. J. 12, 315, 10.1186/1475-2875-12-315 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw Illumina read data for all samples were deposited in the European Bioinformatics Institute European Nucleotide Archive database under the accession numbers PRJNA680178 and PRJNA6659201.