Abstract
Diffuse large B-cell lymphoma (DLBCL) represents a group of tumors characterized by substantial heterogeneity in terms of their pathological and biological features, a causal factor of their varied clinical outcome. This variation has persisted despite the implementation of rituximab in treatment regimens over the last 20 years. In this context, prognostic biomarkers are of great importance in order to identify high-risk patients that might benefit from treatment intensification or the introduction of novel therapeutic agents. Herein, we review current knowledge on specific immunohistochemical or genetic biomarkers that might be useful in clinical practice. Gene-expression profiling is a tool of special consideration in this effort, as it has enriched our understanding of DLBCL biology and has allowed for the classification of DLBCL by cell-of-origin as well as by more elaborate molecular signatures based on distinct gene-expression profiles. These subgroups might outperform individual biomarkers in terms of prognostication; however, their use in clinical practice is still limited. Moreover, the underappreciated role of the tumor microenvironment in DLBCL prognosis is discussed in terms of prognostic gene-expression signatures, as well as in terms of individual biomarkers of prognostic significance. Finally, the efficacy of novel therapeutic agents for the treatment of DLBCL patients are discussed and an evidence-based therapeutic approach by specific genetic subgroup is suggested.
Keywords: ABC, biomarkers, COO, DLBCL, double-expressor, double-hit, GCB, GEP, prognosis, tumor microenvironment
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common lymphoid neoplasm, accounting for ~30% of all non-Hodgkin lymphomas (NHLs). DLBCL is not a single entity; rather, it represents a heterogeneous group of disorders with distinct clinical, pathological, and biological features. The broadest category is termed DLBCL-not otherwise specified (DLBCL, NOS). By definition, these patients do not have specific clinical or pathological characteristics, but they can be further divided into several morphological, molecular, and immunohistochemical subgroups.1 The addition of rituximab to standard chemotherapy, namely cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP), has undoubtedly improved the outcomes of all DLBCL patients and has been widely accepted as the standard of care. Despite this, a considerable proportion of patients either relapse or experience primary refractory disease and eventually succumb to the disease.2,3
The International Prognostic Index (IPI) is currently the most robust prognostic tool for patients with DLBCL. The IPI was introduced and validated in the pre-rituximab era.4 Although the IPI’s prognostic value has been re-assessed in the rituximab era and has been deemed trustworthy, it fails to identify patients with less than 50% of 3-year event-free survival (EFS) who could potentially benefit from other treatment modalities instead of R-CHOP.5,6
Even among patients within the same IPI risk group there is a high variability of outcome. This may reflect the marked genetic and molecular heterogeneity that underlies disease aggressiveness. Therefore, many studies have focused on the identification of biomarkers that may contribute to this phenomenon. Several individual prognostic biomarkers had already been described before the introduction of rituximab; albeit, these are incapable of capturing the great complexity of the underlying biological processes. In the early 2000s, gene expression profiling (GEP) represented an important step towards the elucidation of DLBCL biology and heterogeneity, further optimizing its prognostic stratification.7–9 GEP studies have identified different molecular DLBCL subtypes related to the cell of origin (COO) as well as several gene expression signatures related to the tumor microenvironment (TME). Both are of prognostic significance. In addition, GEP studies have highlighted the prognostic value of many genes and have led to the discovery of several molecular pathways that may serve as therapeutic targets.
The addition of rituximab to the CHOP regimen has altered the significance of certain established prognostic factors, either as a result of statistical reasons (the marked improvement in outcome of patients with DLBCL leads to fewer events) or directly through its mechanism of action. Therefore, previously well-described prognostic biomarkers have been re-evaluated in the rituximab era. The emergence of novel agents for the treatment of DLBCL patients highlights the need for the establishment of their prognostic relevance for patients treated with these therapeutic modalities.
The present review summarizes the current knowledge regarding biological prognostic factors in DLBCL-NOS in the rituximab era. In addition, it provides insights into the efficacy of novel agents in the frontline therapy of high-risk DLBCL patients.
Genetic subgroups of prognostic significance
COO
GEP assessed by DNA microarray allows for the simultaneous profiling of the expression of thousands of genes in cells while obtaining a detailed record of their expression. Alizadeh et al. identified two distinct molecular subgroups of DLBCL with gene expression patterns indicative of different stages of B-cell differentiation, as well as highly distinct overall survival (OS). The first subgroup was composed of DLBCL with a gene expression signature resembling that of germinal center B-cells (Germinal Center B-like, GCB), whereas the second contained DLBCL with expression of genes which are induced during in vitro activation of peripheral blood B-cells (Activated B-cell, ABC).7 Rosenwald et al. demonstrated the presence of a third molecular subgroup, called type 3 or unclassified DLBLC, that included cases not expressing either set of genes characteristic of GCB or ABC subgroups. The distribution of cases among these different subgroups was 47.9%, 30.4%, and 21%, for GCB, ABC, and type 3 DLBCL, respectively. 5-year OS rates were significantly higher for the GCB DLBCL patients compared with the other subgroups independent of their IPI. Furthermore, four distinct gene-expression signatures [GCB, proliferation, major histocompatibility complex (MHC) class II, and lymph node] with prognostic significance were identified.8 The prognostic value of DLBCL subtyping by GEP analysis has been re-evaluated in the rituximab era. Lenz et al.9 reported that GCB DLBCL patients had significantly higher OS and progression-free survival (PFS) than ABC DLBCL patients, a finding highly consistent in several studies.10,11
Based on the above studies, the molecular classification of DLBCL by COO has been recognized as the gold-standard approach for the molecular classification of DLBCL and provides valuable prognostic information independently of the IPI. However, these techniques are not available for routine use and require fresh or frozen tissue samples with adequate amounts of RNA. To overcome these limitations, many researchers have tried to determine COO by applying GEP techniques to formalin-fixed paraffin-embedded tissues (FFPET) with high accuracy.12–19 Among the suggested approaches, Lymph2Cx, a digital GEP assay based on a panel of 20 genes, has been validated and demonstrated its non-inferiority to GEP determination of COO.16
The unquestionable prognostic significance of COO in DLBCL led many researchers to develop prediction models based on simpler techniques such as immunohistochemistry (IHC) in FFPET. The IHC algorithms uses antibodies specific to GCB and ABC-markers. It assesses protein expression in order to classify DLBCL cases as either GCB or non-GCB. The most widely utilized of them, Hans algorithm, uses CD10, BCL6 and MUM1.20 Although in accordance with GEP, its prognostic significance in the rituximab era has been disputed. A recent meta-analysis showed that COO determined by the Hans algorithm was predictive of PFS but not OS in patients treated with rituximab.21 A recent study by Adulla et al. in 359 DLBCL patients confirmed the inferiority of the Hans algorithm to predict OS.22 Most strikingly, a recent study by Cho et al.23 failed to demonstrate any prognostic effect of the Hans algorithm in PFS or OS; in contrast, COO determination by Lymph2Cx was strongly predictive of both outcomes. The Choi algorithm, developed in the rituximab era, utilizes CD10, MUM1, GCET, FOXP1 and BCL6, allowing for a more accurate COO designation;24 however, similar to the Hans algorithm, its prognostic significance might be limited.21 Several other algorithms (Colomo, modified Hans, modified Choi, Visco–Young, Tally, Muris, Natkuman, and Nyman) have been proposed;25–30 albeit, results regarding the prognostic significance of these algorithms remain equivocal.31
The poor prognostic performance of the IHC algorithms could be attributed to their inherently binary nature, as they classify cases as GCB or non-GCB. Therefore, cases unclassified by GEP (type 3) are inevitably misclassified by the algorithms, hindering their prognostic value. Moreover, as shown by the Lunenburg Lymphoma Biomarker Consortium Study (LLBC), these results could be explained by sampling techniques and technical issues, as well as by inter-observer variation.32 The optimization of staining techniques and scoring criteria has failed to improve the prognostic value of IHC algorithms.33 To summarize, the IHC algorithms remain suboptimal for a prognostically relevant classification of DLBCL, and GEP represents the gold-standard for COO classification. Of note, novel assays such as Lymph2Cx are applicable in FFPET, overcoming limitations of earlier GEP assays. Other novel FFPET-based approaches utilize multiplex quantitative real-time polymerase chain reaction (qRT-PCR) and next-generation sequencing (NGS) in order to target a specific panel of genes. These approaches are highly accordant to GEP and highly predictive of PFS and OS.34,35
Gene-expression models
Findings from GEP analysis drove researchers to pursue prognostic models that incorporate the expression of several genes. Lossos et al.36 evaluated a qRT-PCR model based on the expression of six genes (LMO2, BCL6, FN1, CCND2, SCYA3, and BCL2) that is also applicable in FFPET. In the rituximab era, the model has been shown to predict OS but not EFS.37,38 Another model incorporating four genes of the COO signature (LMO2, MME, LPP, and FOXP1) and two immune-related genes (APOBEC3G and RAB33A) has been proposed; however, as it has been based in a small cohort of elderly patients and has not been externally validated, no conclusions regarding its prognostic significance can be drawn.11 In a more simplified approach, Alizadeh et al. created a two-gene model based on the expression of LMO2 and a TME-related gene (TNFRSF9) in FFPET. The two-gene model was an independent predictor of OS, independent of COO and IPI. A composite score integrating these gene-expression with IPI could stratify patients in low-, intermediate- and high-risk groups with distinct PFS and OS.39 More recently, Green et al. proposed a model incorporating the expression of LMO2 and HLADQA1 as well as three gene interactions for GCSAMxMIB1, GCSAMxCTGF, and FOXP1xPDE4B that predicted PFS and OS independently of IPI. As the complexity of this model might hinder its applicability, a simplified version has been proposed, comprising LMO2, BCL2 expression, and IPI. This showed comparable performance to the more complex model and was validated in an independent cohort.40
Apart from qRT-PCR, other gene-expression assays have been evaluated for prognostication in DLBCL. Among them, quantitative S1 nuclease protection assay (qNPA) in FFPET has been used to assess the expression of several genes. In this context, Rimsza et al.12 demonstrated that a model comprising HLA-DRB and MYC expression assessed by qNPA could predict OS and PFS.
In conclusion, gene-expression assays applicable in FFPET have allowed for the development of prognostic models incorporating gene-expression information as well as clinical factors. However, lacking external validation, the results of these studies should be interpreted cautiously. Moreover, no consensus on the optimal combination of genes for the prediction of the clinical outcome as well as the methodology for gene-expression assessment have been reached. This has largely hindered the reproducibility of results. Of interest among the investigated genes, LMO2 has been consistently associated with favorable outcome; however, further studies are needed to elucidate the appropriate gene combination that would comprise a widely accepted prognostic model.
Novel molecular subgroups
Recent reports have highlighted the presence of residual heterogeneity in DLBCL prognosis, even among the well-characterized COO subgroups. Several studies have tried to refine the molecular classification of DLBCL in this context. Reddy et al. integrated whole exome sequencing and transcriptome sequencing to identify 150 driver genes in 1001 DLBCL patients. Their mutational and gene-expression profiles were used to construct a prognostic model that outperformed other established prognostic approaches such as COO determination and IPI. According to this model, 39 subgroups emerged, with significant discrepancies in OS. The subgroup with the most dismal prognosis comprised cases with MYC genetic and/or gene-expression aberrations irrespective of COO. In contrast, GCB-DLBCL with CD70 alterations represented the subgroup with the most favorable outcome.41
In another approach, Schmitz et al. utilized whole-exome and transcriptome sequencing, array-based DNA copy-number analysis, and targeted resequencing of 372 genes in 574 DLBCL cases. They managed to classify 44.8% of cases into four distinct subgroups: MCD (combined MYD88L265P and CD79B mutations), BN2 (BCL6 fusions and NOTCH2 mutations), N1 (NOTCH1 mutations), and EZB (EZH2 and BCL2 rearrangements). The MCD and N1 subtypes where mostly composed of ABC-DLBCL, EZB composed mostly of GCB-DLBCL, whereas BN2 was equally prevalent in all COO groups. The four subtypes had statistically significant differences in PFS and OS; the 5-year OS for the MCD, N1, BN2, and EZB subtypes were 26%, 36%, 65%, and 68%, respectively. Within the ABC subgroup, BN2 represented the subtype with the most favorable OS and PFS, whereas N1 and MCD had dismal prognosis compared with ABC-NOS and BN2; within the GCB subtype, EZB subtype demonstrated inferior survival compared with GCB-NOS. Notably, MCD and BN2 demonstrated recurrent B-cell receptor (BCR)-dependent NF-κB activation, and N1 revealed a T-cell gene-expression signature with potential therapeutic implications.42
Most recently, Chapuy et al. analyzed 304 DLBCL samples for recurrent low-frequency alterations, mutations, somatic copy number alterations (SCNAs), and structural variants (SVs). They identified five DLBCL subsets (C1-C5) with distinct clinical behavior. Within the ABC group, C1 (BCL6 SVs and NOTCH2 mutations) represents a subgroup with favorable prognosis, whereas C5 (gains in BCL2 and/or mutations in MYD88L265P, CD79B, ETV6, PIM1, GRHPR, TBL1XR1, and BTG1) showed inferior outcome. On the other hand, two subgroups were identified within the GCB group, those being C3 (BCL2 mutations and SVs along with mutations in epigenetic modifiers, KMT2D, CREBBP, and EZH2) which was characterized by inferior outcome, and C4 with favorable prognosis characterized by aberrations in BCR/PI3K signaling, NF-κB and RAS/JAK/STAT pathway (mutations in CD83, CD58, and CD70, RHOA, GNA13, and SGK1, CARD11, NFKBIE, and NFKBIA, and BRAF, STAT3).
The remaining cluster, named C2, is composed of COO-independent DLBCL with biallelic inactivation of TP53 as well as copy loss of CDKN2A, and RB1. It demonstrated an intermediate OS between C1, C4 and C3, C5.43 It should be noted that the C1, C3, and C5 groups partially overlap with the BN2, EZB, and MCD groups outlined by Schmitz et al.42
Lacy et al. proposed a similar classification scheme based on the targeted sequencing of 928 DLBCL FFPET samples. Five distinct subsets were identified (MYD88, BCL2, SOCS1/SGK1, TET2/SGK1, and NOTCH2), which significantly overlap with those described by Schmitz et al.42 and Chapuy et al.43 Indeed, MYD88 overlaps with MCD and C5, BCL2 with EZB and C3, and NOTCH2 with BN2 and C1. Regarding SOCS1/SGK1 and TET2/SGK1, overlap is seen with the C4 subgroup; however, they might represent distinct subgroups, based on the augmented expression of TET2 and BRAF in the latter. Moreover, although both subgroups have relatively good prognosis, the former group is associated with a more favorable outcome.44
Almost concurrently, Wright et al. proposed a refinement of the classification scheme by Schmitz et al.,42 aiming to eliminate the previously unclassifiable cases. In this context, they proposed two additional subgroups named A53 and ST2. The former aligns with the C2 subgroup by Chapuy et al.,43 as it is composed of cases enriched for TP53 mutations, whereas the ST2 aligns with the TET2/SGK1 subgroup which was described previously. Similarly, to the previous classification schemes, significant differences in clinical outcome were noted among different subgroups.45
A considerable portion of DLBCL remains unclassified, even by the implementation of the novel approaches discussed so far; as a result, prognostic ability is hampered. Alkodsi et al. proposed a classification scheme which is based on the somatic hypermutation (SHM) patterns of 36 target genes. They managed to identify four distinct subgroups named SHM1-4 that allowed prognostic stratification of patients within the ABC and GCB subtypes, but also within unclassified DLBCL. In this scheme, ABC is subdivided into SHM2 with aberrant activation of the BCR signaling pathway and the worse outcome among all SHM subgroups, while SHM4 is characterized by BCL6 fusions as well as CD70 and BCL10 mutations. On the other hand, GCB group is subdivided into SHM1 with high frequency of BCL2 and MYC aberrations in addition to mutation of chromatin modifying genes, showing poor outcome with conventional immunochemotherapy, and SHM3 exhibits aberrant JAK/STAT signaling and the most favorable outcome compared with the other subgroups.46
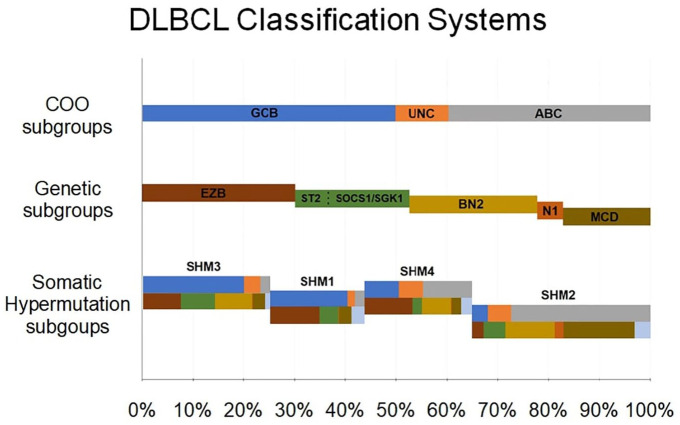
The inter-correlation of the novel genetic subgroups is depicted in Figure 1. Key genetic features of each subgroup are summarized in Tables 1 and 2. To summarize, genomic studies have disentangled the complex genomic infrastructure of DLBCL, allowing for the subclassification of cases in prognostically relevant subgroups with shared genetic aberrations. Although most of the techniques used in the described studies might be time-consuming and excessively expensive to be applied in clinical practice, targeted NGS, which in addition, is applicable in FFPET, might represent an appealing approach for genetic classification in general practice. For this to occur, validation in prospective studies is needed. Nonetheless, classification in well-characterized genetic subgroups might provide the basis for designing of meaningful preclinical and clinical studies.
Figure 1.
Schematic representation of the relationship between cell-of-origin (COO), genetic (Wright et al.45), and somatic hypermutation (Alkodsi et al.46) subgroups in diffuse large B-cell lymphoma (DLBCL). The upper panel depicts the relative proportion of germinal center-like B cells (GCBs), activated B cell (ABC), and unclassified, among all DLBCL cases. The intermediate panel depicts the molecular subgroups, identified by gene-expression profiling; their relative position to the upper panel correlates to the association of these subgroups with COO (the A53 subgroup is not depicted on this Figure as there is no correlation with COO). The lower panel depicts the somatic hypermutation subgroups; the chromatic code of the upper part of each bar depicts the correlation of each subgroup to COO, whereas the chromatic code of the lower part depicts correlation to molecular subgroups. The relative width of each bar corresponds to the relative proportion of each subgroup among all DLBCL cases.
Table 1.
Key genetic features and 5-year overall survival by molecular diffuse large B-cell lymphoma (DLBCL) subgroups, identified by gene-expression profiling, by Schmitz et al.,42 Chapuy et al.,43 Lacy et al.,44 and Wright et al.45.
| Molecular subgroups | Key genetic aberrations | 5-year overall survival | |||
|---|---|---|---|---|---|
| Schmitz et al.42 | Chapuy et al.43 | Lacy et al.44 | Wright et al.45 | ||
| BN2 | C1 | NOTCH2 | BN2 | BCL6, NOTCH2, TNFAIP3, SPEN, BCL10, TMEM30A | 67% |
| C2 | A53 | TP53, TP53BP1 | 63% | ||
| EZB | C3 | BCL2 | EZB-MYC+ | BCL2, EZH2, KMT2D, TNFRSF14, CREBBP, GNA13, MEF2B, IRF8 | 48% (DHITsig-positive) |
| EZB-MYC- | 82% (DHITsig-negative) | ||||
| C4 | TET2/SGK1 | ST2 | SGK1, TET2, ZFP36L1 | 84% | |
| SOCS1/SGK1 | SOCS1, SGK1, CD38 | 80% | |||
| MCD | C5 | MYD88 | MCD | MYD88, CD79A/B, PIM1, TBL1XR1, PRDM1, SPIB, BTG1/2, CDKN2A | 40% |
| N1 | N1 | NOTCH1, ID3, KLHL6 | 27% | ||
Table 2.
Key genetic aberrations of somatic hypermutation subgroups in diffuse large B-cell lymphoma (DLBCL), identified by Alkodsi et al.46.
| Subgroup | Key genetic features |
|---|---|
| SHM1 | EZH2, KMT2D, CREBBP, MYC, BCL2, GNA13, GNA12, P2RY8, Chr7, Chr8 gains |
| SHM2 | MYD88, CD79B, CDKN2A, PIM1, MPEG1, ETV6, IRF4, Chr3, Chr18 gains |
| SHM3 | SOCS1, STAT3, STAT6, TNFAIP3, SGK1, IRF8, Chr3, Chr7 Chr18 gains |
| SHM4 | BCL6, CD70, BCL10, SPEN, MYD88 (not L265P), HLA-A,B,C |
Chr, Chromosome.
The tumor microenvironment
Although the role of the TME has been widely established in other lymphoid malignancies such as Hodgkin lymphoma, its role in DLBCL remains controversial. In 2008 Lenz et al. identified two gene-expression signatures, stromal-1 and stromal-2, which reflected discrepant composition of TME in DLBCL. The favorable stromal-1 signature, associated with a phenotype characterized by abundant extracellular matrix and infiltration by histocytes, was enriched for genes encoding for the major components of the extracellular matrix and the anti-angiogenic factor thrombospondin, along with modifiers of collagen synthesis and proteins implicated in the remodeling of extracellular matrix. In contrast, the less favorable signature stromal-2 was mainly enriched for genes encoding for markers of endothelial cells and regulators of angiogenesis, and it was characterized by high blood-vessel density.9 However, the lack of reproducible methodology in FFPET hampered its applicability, despite the clear prognostic implications of TME. Nonetheless, vascular endothelial growth factor receptor 2 (VEGFR2) expression and high microvessel density, assessed by IHC, correlate with poor outcome,47,48 as opposed to expression of VEGFR1, which has been associated with a more favorable prognosis.48 Notably, IHC expression of HIF1a might confer improved prognosis in DLBCL, despite promoting angiogenesis, through upregulation of several genes within the favorable stromal-1 signature.49 As expected, IHC expression of SPARC, overexpressed within the favorable stromal-1 signature, has been associated with improved OS and PFS independently of IPI; however, its prognostic effect is restricted within the ABC subgroup.50 An IHC-based predictive model incorporating the non-GCB subtype, low expression of SPARC (<5%), and high microvessel density has been suggested.51
Several studies have assessed the prognostic role of immune composition of TME in DLBCL. Ciavarella et al. demonstrated that higher proportions of myofibroblasts, dendritic cells, and CD4+ T cells correlated with superior OS, whereas activated natural killer (NK) and plasma cells (PCs) correlated with inferior outcome. TME gene-expression profiling identified three clusters (low, intermediate, high-expression) that predicted OS independently of COO. A classification scheme integrating COO and TME subtypes has also been proposed.52 A high number of FOXP3+ regulatory T-cells have been associated with inferior outcomes in most of these studies.53,54 Moreover, lymphoma-associated macrophages (LAMs) play a crucial part within the TME; however, distinct subsets of LAMs may occur in opposing modes. M2 macrophages are immunosuppressive and promote tumor evasion, whereas M1 macrophages induce immune response and exert anti-lymphomatic action. Therefore, studies of individual macrophage markers have yielded conflicting results.55 To overcome this inherent limitation, Stagger et al.56 constructed a LAM interaction signature (LAMIS) that was applied to 466 FFPET samples, demonstrating that high expression of this signature was predictive of inferior PFS and OS, irrespective of IPI and COO.
Most recently, the role of the programmed cell death protein 1 (PD-1)/PD-L(ligand)1 axis has been highlighted as a key mechanism of immune evasion, both in solid tumors and in DLBCL. When PD-L1 is expressed by tumor cells, it interacts with PD-1 in T-cells leading to T-cell anergy and immune evasion.57 PD-L1 overexpression is observed in ~20% of DLBCL cases due to gains, amplification, or rearrangements affecting the PD-L1 locus.58 Several studies have shown that the overexpression of PD-L1 by tumor cells correlates with poor OS and PFS, independent of IPI and COO.59–61 Notably, PD-L1 expression strongly correlates with Epstein–Barr virus infection and the ABC subtype;61 on the other hand, PD-1 expression by T-cells within the TME might predict a more favorable outcome.62,63 Other mechanisms of immune evasion include the downregulation of several genes comprising the MHC class II and inactivating mutations of the B2M gene, encoding for β2-microglobulin as well as downregulation of CD58, which is involved in NK cell responses.64 An association between these immune evasion mechanisms and OS has been noted.65–68
To evaluate the role of different subsets of immune cells in the TME, Keane et al. assessed the expression of immune effector and checkpoint genes in 252 FFPET DLBCL. They demonstrated that the expression of immune effectors (T/NK) correlates with the expression of markers associated with macrophages and the PD/PD-L1 axis. Thus, the anti-lymphomatic action exerted by the former cells is truncated. Therefore, the CD4*CD8:M2*PD-L1 ratio, assessed by digital hybridization, was used to stratify patients in two prognostic groups irrespective of IPI and COO. Patients with a high ratio experienced more favorable PFS and OS, as well as better response rates to R-CHOP compared with patients with low ratio.69
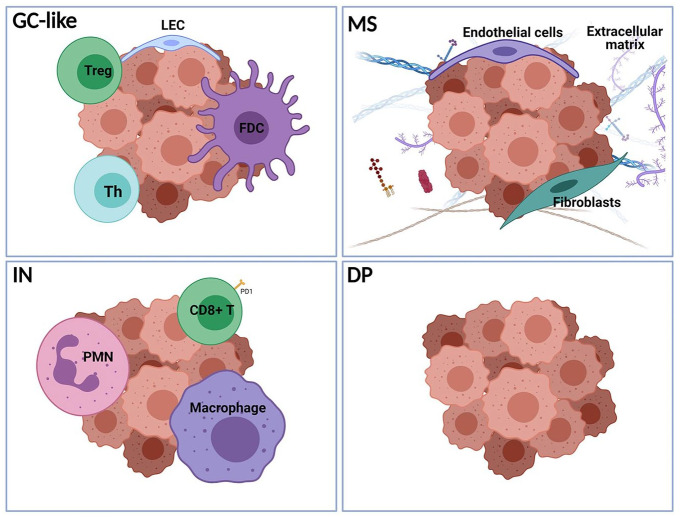
In a pivotal transcriptomic study of more than 4000 DLBCL samples, Kotlov et al. characterized four clusters termed germinal center-like (GC-like), mesenchymal (MS), inflammatory (IN), and depleted (DP). GC-like cluster resembles the cellular composition of normal germinal center, whereas MS is characterized by increased endothelial cells and fibroblasts as well as abundant extracellular matrix. Both clusters, enriched within the GCB subgroup, are associated with favorable PFS and OS. On the other hand, IN cluster, characterized by a highly inflammatory TME rich in neutrophils and macrophages and the DP cluster, showing a deserted TME, are associated by inferior prognosis, irrespective of COO designation. Notably, the TME clusters are distributed across all genetic subgroups, suggesting that a classification system based on the composition of TME may serve an auxiliary role to the genetic classification for the prognostic characterization, and therapeutic management of DLBCL patients.70 The validation of recent findings in large prospective studies and the development of simplified techniques for application in FFPET is needed for the adoption of TME clustering in clinical practice. Until then, the considerable prognostic role of TME can be assessed by IHC of specific markers implicated in angiogenesis or immune response as well as targeted sequencing. A graphical representation of the TME clusters based on their cellular composition of TME is depicted in Figure 2.
Figure 2.
Schematic representation of the predominant cells in the microenvironment of the four DLBCL subgroups with prognostic significance, described by Kotlov et al.70 (GC-like, germinal center-like; MS, mesenchymal; IN, inflammatory; and DP, depleted).
LEC, lymphatic endothelial cells; PMN, polymorphonuclear cells; Th, T-helper cells; Treg, T-regulatory cells. (Created with BioRender.com).
Double-hit, triple-hit, and double-expressor lymphomas
BCL2 is overexpressed in 47-58% of DLBCL patients.71 In the GCB subgroup and particularly within the EZB genetic subgroup, BCL2 upregulation is mainly attributed to the rearrangement t(14;18)(q32;q21).72 In contrast, the ABC subgroup is characterized by the 18q21 chromosome locus gain/amplification.73 In the rituximab era, BCL2 rearrangement remain predictive of significantly inferior OS among patients with the GCB subtype, irrespective of MYC status. On the other hand, BCL2 amplification or gains are predictive of inferior OS and PFS within the ABC subgroup.74,75 BCL2 overexpression has retained its prognostic ability solely within the GCB subgroup.76,77 MYC rearrangements can be identified in 5-14% of DBCL patients, and more commonly with the GCB subgroup.78 The rearrangement t(8;14)(q24;q32) represents the most typical, bridging MYC to the immunoglobulin heavy chain gene locus; however, in 53% of these cases, the partner is not an immunoglobulin (IG) gene.79 Although numerous studies have demonstrated the negative prognostic effect of MYC rearrangements on PFS and OS, as well as on central nervous system (CNS) relapse risk, the prognostic significance of isolated MYC rearrangements has been disputed.80–83 It has been shown that cases with isolated MYC rearrangements demonstrate an OS and PFS approximating that of non-rearranged cases. This highlights that the detrimental effect of MYC rearrangements is highly dependent on a second genetic hit, particularly in BCL2 or BCL6 and TP53, which are found in up to 80% of cases.84,85 Similarly, overexpression of the MYC protein, which is demonstrated in ~30% of DLBCL cases, has been considered an independent prognostic factor for OS and PFS irrespective of the underlying mechanism; however, its prognostic effect is modified by concurrent BCL2 or BCL6 genetic aberrations or overexpression of the respective proteins.85 BCL6, located in the 3q27 chromosome, represents a major marker of GCB origin;20,86 albeit, BCL6 rearrangements are twice as common within the ABC subgroup87 and confer a negative effect to OS and PFS as is evident in a recent meta-analysis.88 On the other hand, BCL6 overexpression, mainly attributed to gene mutations, represents a prominent feature of the GCB subgroup.87 High BCL6 mRNA and protein expression have been, and still are, strong predictors of favorable outcome in DLBCL patients.89,90 It should be noted, however, that the prognostic significance of BCL6 rearrangements and overexpression might reflect its higher prevalence within the prognostically significant GCB and ABC subgroups, respectively.
In 58–63% of the cases, the MYC rearrangement is accompanied by at least one additional rearrangement, most commonly of BCL2 or BCL6. Cases harboring MYC and BCL2 or BCL6 rearrangements are termed double-hit (DH) lymphomas, whereas concurrent rearrangement of all three genes characterizes the subset of triple-hit (TH) lymphomas.91 In the 2016 revision of WHO classifications, DH and TH lymphomas with DLBCL morphological features were excluded from the DLBCL-NOS category, and have been assigned to a new diagnostic entity termed high-grade B-cell lymphomas (HGBL) with MYC and BCL2 and/or BCL6 (HGBL-DH/TH).1 HGBL-DH/TH accounts for 7.9% of tumors with DLBCL morphology; among them, DH-BCL2 and TH lymphomas represent more than 80% of cases, whereas DH-BCL6 lymphomas are relatively rare, accounting for 18.6% of cases. Most strikingly, DH-BCL2 and TH lymphomas are almost invariably associated with the GCB subgroup, whereas DH-BCL6 is distributed equally among COO subgroups.92
DH and TH have been associated with an inferior outcome, predicting an aggressive clinical course and poor response to R-CHOP.82,85,93,94 As 5-year OS and PFS has been reported to be rather poor (27% and 18%, respectively) in R-CHOP treated patients,85 more aggressive therapeutic approaches have been suggested; however, several ongoing controversies should be highlighted. First, DH and TH are not invariably associated with overexpression of the respective proteins. Several studies have demonstrated that these cases, which represent a non-negligible proportion of ~20% of DHs, have a more favorable prognostic profile.85,95,96 Moreover, the prognostic significance of DH-BCL6 cases remains equivocal. Older studies demonstrated that DH-BCL6 is associated with dismal outcomes;97,98 in contrast, more recent studies have showed that the co-occurrence of MYC and BCL6 re-arrangements is not associated with an inferior outcome in DLBCL.99,100 Recent findings have also underscored the differential role of the partner gene in MYC rearrangement in prognosis among DH and TH DLBCL patients. A recent large study by Rosenwald et al.101 showed that DH and TH cases harboring MYC rearrangements to non-immunoglobulin genes showed no significant differences in terms of OS and PFS, compared with non-DH/TH cases. Moreover, cases with gene amplifications rather than rearrangements have been identified, however the prognostic significance of these abnormalities remains controvertial.102
Previous limitations have led researchers to utilize GEP to identify DH and TH cases with genuine prognostic significance. Ennishi et al. identified a 104-gene DH signature (DHITsig) which characterizes most DH/TH cases. This signature was identified in 27% of cases within the GCB subgroup; among them, only one half were DH/TH by fluorescence in situ hybridization (FISH). Most strikingly, it was shown that DHITsig-positive (DHITsig +ve) cases had dismal outcome, accompanied by poor response rates to R-CHOP, irrespective of their MYC, BCL2, and BCL6 rearrangement status.103 Further analysis using whole-genome sequencing identified genetic alterations to MYC and BCL2 which are undetectable by conventional FISH in most of the non-DH/TH DHITsig +ve cases. Notably, six out of 20 analyzed cases harbored rearrangements cryptic to conventional FISH, whereas genetic events affecting both MYC and BCL2 were identified in seven additional cases.104 Almost concurrently, Sha et al. identified a molecular high-grade (MHG) gene expression signature characteristic of DH/TH cases which extends beyond them, within the GCB subgroup. This signature was predictive of inferior outcome irrespective of DH/TH status.105 The two genetic signatures are highly correlated and characterize tumors originating from the intermediate germinal center zone, particularly enriched within the EZB subgroup. Tumors within the EZB subgroup can be further classified by the presence of DHIT signature into EZB-MYC+ and EZB-MYC-. EZB-MYC+ might represent highly aggressive tumors arising from a dark zone with a 5-year OS of 48%. In contrast, EZB-MYC- tumors, arising from the light zone, have a more favorable prognosis (5-year OS: 82%).45 DHITsig +ve DLBCL shows high proliferation and immune evasion, owing to the frequent loss of MHC antigens and their lymphocyte-depleted microenvironment.103,105 Indeed, the DHITsig +ve subgroup was significantly enriched within the DP TME subgroup by Kotlov et al.70 Notably, stratification by TME composition retains its prognostic significance even in this subgroup.
Lymphomas with a high co-expression of MYC and BCL2 proteins are called double expressor lymphomas (DE) and should not be confused with DH/TH lymphomas as they do not represent a distinct biological subgroup but a prognostically relevant subcategory. MYC and BCL2 protein co-expression by IHC is present in 21%-29% of DLBCL patients and undoubtedly confers poor prognosis (5-year OS: ~40%).85,106 Furthermore, MYC/BCL2 co-expression was also associated with poor prognosis in another large study of 893 DLBCL patients treated with R-CHOP (5-year OS: 30% versus 75%).107 DEs are more common within the ABC subgroup, particularly when HGBL-DH/TH are excluded, contributing to the inferior prognosis of cases fitting in this subgroup.80 The underlying mechanism of DE differs among COO subgroups; within GCB, overexpression is attributed to gene re-arrangements, whereas in the ABC subgroup, overexpression represents the sequela of a complex genetic interplay involving gene amplifications and aberrations in B-cell receptor and NF-kB signaling.77 Recently Horn et al. proposed a prognostic model based on MYC protein expression and MYC rearrangement status in combination with BCL2 and BCL6 expression status. MYC rearrangements, MYChigh, BCL2high, and BCL6low protein expression were predictive of inferior survival independently of IPI.90 Recently, the incorporation of CD37, MYC, and BCL2 to the R-IPI has shown to augment its prognostic power.108 The distribution of genuine and cryptic DH, DE, and DHITsig +ve cases among GCB and ABC subgroups along with their overlap is depicted in Figure 3.
Figure 3.
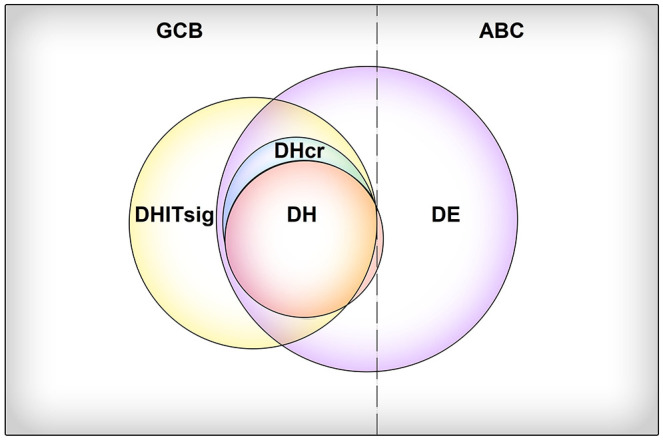
Schematic representation of the distribution of double-hit (DH), double-expressor (DE), and double-hit gene expression signature (DHITsig) diffuse large B-cell lymphoma (DLBCL) cases among the germinal center B-cell (GCB) and activated B cell (ABC) subgroups. DHITsig+ cases, harboring BCL2 and MYC rearrangements cryptic to conventional fluorescence in situ hybridization (FISH) are termed cryptic DH (DHcr). The area of each circle corresponds to the relative prevalence of each group among DLBCL cases.
In conclusion, DE and DH lymphomas seem to predict a more aggressive clinical course, underlying the need for early identification, and potentially treatment intensification as well as the introduction of novel agents; however, limitations of the current IHC and FISH methods hamper the classification in biologically distinct subgroups. In this context, gene-expression signatures might serve for the accurate distinction between these subgroups. Notably, a DHITsig module has been incorporated in the Lymph3Cx assay for COO characterization, allowing for the application in FFPET in clinical practice.
Other biomarkers
TP53 mutations, found in ~20% of DLBCL patients among both COO subgroups, tend to be more common among cases with MYC rearrangements. TP53 mutations correlate with unfavorable disease characteristics and predict inferior OS and PFS independent of IPI and COO.109–111 In contrast, the prognostic significance of TP53 deletions and or del(17p) in the absence of a mutated allele remains controversial.109,112 In regards to IHC, strong TP53 expression (in at least 50% of the malignant cells) might be an independent predictor of shorter OS;113 however, the absence of a concurrent TP53 mutation negates the prognostic significance of the respective protein overexpression.109
High proliferation rate, reflected by high expression of Ki-67, has been predictive of inferior outcomes in DLBCL, as demonstrated by a recent meta-analysis.114 In addition, recent research has shown that the prognostic value of Ki-67 might be more pronounced within the non-GCB subgroup.115,116
De novo CD5+ DLBCL, accounting for 5–22% of DLBCL cases, represents a distinct immunohistochemical subgroup within DLBCL-NOS.117 Most commonly of ABC origin (82%), this subgroup highly correlates with double MYC/BCL2 overexpression.118 CD5+ cases tend to present with more advanced disease, whereas CNS recurrence is particularly high (13% versus 5% for CD5- DLBCL).119 Despite the introduction of rituximab, the prognosis for CD5+ DLBCL remains dismal, with 5-year OS and PFS rates of 35.5% and 29.6% respectively, and high CNS relapse rates.120–122 The aggressiveness of CD5+ DLBCL has been attributed to several mechanisms, including the inhibition of BCR signaling as well as the overexpression of IL-10, BCL2, cyclin D2, and CXCR4.117
Patients with reduced CD20 expression and high CD19 expression (discordant CD20), identified through flow cytometry (FCM), have been shown to have inferior OS independently of their IPI.123,124 Notably, IHC assessment might not be a reliable method for estimation of CD20 expression level compared with FCM; albeit, the latter requires fresh tissue samples. To overcome the inherent limitation of FCM, a semi-quantitative IHC method has been developed for the assessment of CD20 expression in FFPET, verifying the prognostic significance of low CD20 expression.125
CD30 was overexpressed in 14% of patients and was correlated with superior 5-year OS and PFS independent of COO and IPI. CD30+ DLBCL demonstrated a distinct GEP signature, characterized by the downregulation of NF-κB and BCR pathways, potentially explaining the favorable profile of this DLBCL subset. Interestingly, a strong correlation between CD30 expression and EBV infection has been observed. As a side note, EBER seems to negate the favorable effect of CD30, as cases co-expressing EBER and CD30 had a dismal outcome.126
With regards to molecules implicated in apoptosis, the role of BCL2 has been thoroughly assessed previously, in contrast to other genes that have not been evaluated as much. Recently, it has been shown that high BCL2L12 expression, assessed both at the mRNA level and via IHC, confers a more favorable outcome in patients with DLBCL, irrespective of COO and IPI.127 Expression of other anti-apoptotic genes such as BIRC5 (survivin) and XIAP has been reported to confer an adverse effect on prognosis,128,129 while results regarding CFLAR (c-FLIP) are contradictory.130,131 On the other hand, the expression of CASP3 and CDKN1A, a downstream effector of TP53, may correlate with favorable outcome.132,133 Markovic et al.134 have created an apoptotic score based on the IHC expression of CASP3, CD95, CFLAR, BIRC5, XIAP, and BCL2 that predicts OS, whereas Pasanen et al.135 have designed a prognostic score among GCB DLBCL patients which is based on the cell cycle-regulating proteins PDKN1A, PDKN1B, PDKN2A, and TP53.
The expression of PKCβ and p-AKT, two components of the PIK3/AKT signaling pathway, correlates with adverse outcome.136,137 Moreover, expression of phosphotyrosine STAT3, enriched in ABC cases, has been associated with inferior outcomes in DLBCL patients. Notably, an 11-gene STAT3 activation signature has been shown to predict decreased OS, both in the entire DLBCL cohort as well as in the ABC subgroup as described by Huang et al.138 Adverse prognostic significance has also been attributed to the expression of indoleamine 2,3-dioxygenase (IDO)139 and SKP2.140,141
Circulating cell-free DNA
Circulating cell-free DNA (cfDNA) represents DNA fragments released from apoptotic or necrotic cells into the circulation. As DLBCL is characterized by high cell turnover, several studies have evaluated the role of cfDNA in DLBCL prognosis. High levels of cfDNA at diagnosis have been shown to correlate with high tumor burden, advanced stage, high LDH levels, and high IPI score, as well as inferior OS and PFS in DLBCL.142 In the largest prospective study of 217 DLBCL patients, Kurtz et al. demonstrated that cfDNA levels at diagnosis assessed through deep sequencing (CAPP-seq) were predictive of EFS independently of IPI. Applying the same technique, Scherer et al.143 achieved stratification of DLBCL cases among the COO subgroups, demonstrating high accordance with COO designated by IHC in FFPET. Notably, early decreases in cfDNA 21 days into treatment were highly predictive of the response to R-CHOP and EFS.144 Global methylation patterns in cfDNA have also been found to predict OS and response to treatment.145,146 Most importantly, several studies have shown that targeted NGS might be applicable in cfDNA. These studies, apart from validating the prognostic and predictive role of overall cfDNA burden, provide evidence that cfDNA could be used for the genetic characterization of DLBCL cases.147 Intriguingly, it was recently shown that cfDNA could be used for the stratification of patients in the prognostic genetic subgroups proposed by Wright et al.,45 allowing for an in-depth, minimally-invasive prognostic evaluation of patients.148
Therapeutic implications
The addition of rituximab to the standard CHOP regimen has improved survival of DLBCL patients irrespective of COO; however, the ABC subtype still confers adverse prognosis compared with the GCB-subtype, retaining its significant prognostic effect even in the relapsed/refractory (R/R) setting. Therefore, current research focuses on the design of novel therapies that target specific oncogenic pathways which are activated and play a crucial role in the pathogenesis of the disease.
A hallmark of ABC DLBCL is constitutive activation of the NF-kB pathway through aberrant BCR signaling and MYD88 activation.149 Although thought to represent independent pathways converging to NF-κB activation, co-occurrence of CD79B and MYD88L265P mutations in a significant subset of ABC DLBCL (namely the MCD subgroup) suggest at a potential interplay between the two pathways. Most recently, the My-T-BCR supercomplex was identified, comprising BCR, MYD88, and TLR9, leading to NF-κB and mTOR pathway activation.150
The significance of the NF-kB pathway in the pathogenesis of DLBCL led to the investigation of the proteasome inhibitor bortezomib, which inhibits NF-κB by preventing proteasomic degradation of IκBα.151 Although initial results had been promising, a large randomized phase III trial showed that the addition of bortezomib in the standard R-CHOP did not confer any benefit to the PFS or OS of newly diagnosed DLBCL patients, irrespective of the COO.152 The disappointing performance of this agent in DLBCL could reflect its unspecific mode of action, highlighting the need for more targeted treatment modalities.
Lenalidomide is an immunomodulatory drug with multiple effects, including inhibition of the NF-kB activity through the downregulation of IRF4 and SPIB.153 Results of the ECOG-ACRIN1412 phase II trial demonstrated that the addition of lenalidomide to R-CHOP could reduce the risk of progression or death by 33%, irrespective of COO. It should be noted that the effect of lenalidomide was more robust within the ABC subgroup.154 Surprisingly, the ROBUST phase III trial which was based on 570 newly diagnosed ABC DLBCL patients did not show any difference between the lenalidomide-R-CHOP (R2-CHOP) arm and the arm of standard R-CHOP treatment in terms of PFS.155 There may be many reasons that explain this difference in the two trials apart from their inherent differences in the study design, such as the higher dosage of lenalidomide in the ACRIN trial or the significantly longer time lag between the diagnosis and initiation of treatment in the ROBUST trial.156 Nonetheless, lenalidomide may represent a promising agent for tumors within the MCD and BN2 subgroups which consistently overexpress IRF4. More studies focusing on these subgroups are needed.
Several components of the BCR pathway have been proposed as potential therapeutic targets in DLBCL. Among them, the inhibition of Bruton’s tyrosine kinase (BTK) by ibrutinib is the most well studied. The recently published results of the phase III Phoenix trial, which compares ibrutinib-R-CHOP with R-CHOP for newly diagnosed patients with ABC DLCBL demonstrated that the addition of ibrutinib prolongs PFS and OS in younger (<60 years) patients with ABC DLBCL. The differential effect of ibrutinib by age could be explained by the increased number of serious adverse events in older patients, leading to deviation from treatment schedule or treatment discontinuation.157 In terms of genetic subgroups, MCD, BN2, and A53 might represent the most BCR-dependent tumors among the ABC subgroup; therefore, ibrutinib might be particularly beneficial for tumors falling within these subgroups. Notably, co-occurrence of CD79B and MYD88L265P mutations, a hallmark of the MCD subgroup, predicts high sensitivity to ibrutinib.158 A recent phase II study of ibrutinib and lenalidomide, in combination with R-CHOP, showed promising results.159 Other inhibitors of the proximal components of the BCR pathway, such as fostamatinib (syk inhibitor) and enzastaurin (PCKβ inhibitor) have shown limited effect in DLBCL;160,161 on the other hand, JNJ-67856633, a MALT-1 inhibitor, showed efficacy in preclinical studies and is currently investigated in a phase I trial in DLBCL patients (NCT03900598).162
Activation of the PI3K pathway represents an important oncogenic event in most DLBCL cases. Within the ABC subgroup, PI3K activation occurs mainly as a sequela of BCR activation and leads to NF-κB activation; in contrast, in GCB DLBCL it represents the result of PTEN inactivating mutations and leads to activation of the AKT/mTOR pathway.163 Idelalisib, a selective PI3Kδ inhibitor, showed disappointing results in DLBCL; however, preclinical data have demonstrated that simultaneous inhibition of PI3Kα and δ is needed to exert cytotoxicity in ABC DLBCL.164 Consistently, copanlisib, which is a PI3Kα/δ inhibitor, has shown encouraging results as a monotherapy in the R/R setting, particularly for the ABC subgroup.165 Buparlisib, a pan-PI3K inhibitor, has also been evaluated in a phase II trial; albeit, the effect in DLBCL has been limited.166 Preclinical data on the synergetic effect of PI3Bα/δ and BTK inhibitors triggered researchers to investigate the efficacy of the combination of PI3K inhibitors and ibrutinib.167 For MK-2206, an AKT inhibitor which had shown promising results in preclinical models, the results in the clinical setting have been rather disappointing.168 Regarding mTOR inhibitors, everolimus and temsirolimus have demonstrated activity in the R/R setting; however, in the frontline setting, adjuvant therapy with everolimus after R-CHOP did not improve the disease-free survival (DFS) of high-risk patients.169 Recently, a phase I trial evaluated the safety of everolimus in combination with R-CHOP for newly diagnosed DLBCL; although the combination has been deemed safe, its superiority to standard R-CHOP treatment has not yet been evaluated.170 In conclusion, more studies are needed to evaluate the effect of PI3K/mTOR inhibitors in DLBCL. It should be noted that, based on GEP studies, the MCD, BN2, ST2 and EZB subgroups might benefit more from this therapeutic approach.45
BCL2 plays an essential role in DLBCL pathogenesis, particularly within the MCD, BN2, and EZB genetic subgroups. In this context, venetoclax, a selective BCL2 inhibitor, has been evaluated in DLBCL. A recent phase II study of 208 newly diagnosed patients demonstrated that the addition of venetoclax to R-CHOP provided improved OS and PFS compared with standard treatment. Notably, venetoclax was effective even in cases not expressing BCL2, although the effect was more robust in BCL2+ patients.171 Based on this finding, venetoclax might be beneficial in the treatment of DH/TH lymphomas, although this should be confirmed by randomized trials.
The JAK/STAT pathway is also implicated in the pathogenesis of a subset of DLBCL, corresponding to the MCD and ST2 genetic subgroups. JAK inhibition might represent a promising treatment approach in this subset.45 A preliminary phase I trial has shown modest efficacy of pacritinib, a JAK1/2 inhibitor, in R/R DLBCL patients.172
The finding that EZH2 is mutated in up to 22% of GCB DLBCLs, comprising the EZB subgroup, has drawn attention to the role of hypomethylating agents in DLBCL treatment.173 An EZH2 inhibitor called tazemetostat has shown promising results. Interim results of a phase II trial in R/R DLBCL showed an ORR of 40% in patients with DLBCL harboring EZH2 mutations, compared with 18% in patients with wild-type EZH2.174 A phase I trial has also shown the feasibility and safety of tazemetostat in combination with R-CHOP in the frontline setting.175
In contrast to Hodgkin lymphoma and solid tumors, checkpoint inhibitors have yielded disappointing results in NHL, potentially because of the low prevalence of PD-L1 overexpression in DLBCL.176 However, checkpoint inhibitors combined with other agents might be effective in a subset of DLBCL patients with high PD-L1 expression. Durvalumab has recently been evaluated in combination with R-CHOP or R2-CHOP for the frontline treatment of high-risk patients, including a considerable number of DH/TH. The combination demonstrated its efficacy and safety, but randomized phase III trials are needed to establish its efficacy.177 A potential evidence-based approach for treatment selection, which takes into account the molecular subgroups of DLBCL, is presented in Table 3.
Table 3.
Summary of major prognostic biomarkers in DLBCL.
| Prognostic factor | Effect on prognosis | Comments |
|---|---|---|
| Cell of origin (COO) | ||
| ABC by GEP, Lymph2Cx7–19 | UF | |
| Non-GCB by IHC20–31 | UF# | Inferior to GEP in prognostication |
| LMO236–40 | F | |
| Molecular subgroups 45 | ||
| MCD, N1, A53 | UF | |
| BN2, ST2 | F | |
| EZB | F, if DHITsig-negative | |
| UF, if DHITsig-positive | ||
| Somatic hypermutation subgroups 46 | ||
| SHM1, SHM2 | UF | |
| SHM3, SHM4 | F | |
| BCL2 | ||
| Overexpression76,77 | UF# | In absence of MYC overexpression: no effect on OS |
| Rearrangement74,75 | UF# | In absence of MYC rearrangement: no effect on OS |
| MYC | ||
| Overexpression85 | UF# | In absence of BCL2 overexpression: no effect on OS |
| Rearrangement80–85 | UF# | In absence of BCL2 rearrangement: no effect on OS |
| BCL6 | ||
| Overexpression89,90 | F# | Strong correlation with ABC subgroup: potential confounder |
| Rearrangement88 | UF# | Strong correlation with GCB subgroup: potential confounder |
| DH/TH 82,85,93,94 | UF | The role of DH-BCL6 is equivocal.97–110 |
| Non-IG partner gene in MYC rearrangement: no effect in OS101 | ||
| Double-expressor 85,86,107 | UF | |
| DHITsig/MHG 103–105 | UF | |
| TP53 | ||
| Mutations109–111 | UF | |
| Overexpression109,113 | UF# | Overexpression in the absence of TP53 mutation: No association with OS |
| CD5 120–122 | UF | |
| Low CD20 123–125 | UF | |
| CD30 126 | F# | Potential role of brentuximab vedotin, UF in EBER-positive cases |
| Ki-67 114 | UF | |
| TME composition | ||
| GB-like, MS subgroups70 | F | |
| IN, DP subroups70 | UF | |
| Stromal-2 expression9 | UF | |
| Stromal-1 expression9 | F | |
| High CD4*CD8:M2*PD-L1 ratio69 | F | |
| High LAMIS expression56 | UF | |
| VEGFR2/VEGFR147,48 | UF | |
| HIF-1a49 | F | |
| SPARC50,51 | F | |
| MHC-II loss65–68 | UF | |
| PD-L1 (expressed by tumor cells)59–61 | UF | Potential role of immune checkpoint inhibitors |
| PD-1 (expressed in TME)62,63 | F | |
| FOXP353,54 | UF# | |
| Cell-cycle regulation and apoptosis | ||
| BCL2L12127 | UF | |
| BIRC5129 | UF | |
| XIAP128 | UF | |
| Other | ||
| PKCβ137 | UF | |
| p-AKT136 | UF | |
| STAT3138 | UF | |
| Circulating cell-free DNA 142–148 | UF | |
Studies show conflicting results regarding the prognostic effect of this biomarker.
ABC, activated B-cell; COO, cell of origin; DH/TH, double/triple-hit lymphomas; DHITsig, double-hit signature; DLBCL, diffuse large B-cell lymphoma; DP, depleted; F, favorable; GB-like, germinal center-like; GCB, germinal center B-cell; GEP, gene-expression profiling; IG, immunoglobulin; IHC, immunohistochemistry; IN, inflammatory; LAMIS, lymphoma-associated macrophage interaction signature; MHG, molecular high-grade; MS, mesenchymal; OS, overall survival; TME, tumor microenvironment; UF, unfavorable.
Therapeutic approach for DH/TH lymphomas
Based on their aggressive nature, DH/TH lymphomas require a more intensified therapeutic approach. A meta-analysis of retrospective studies compared with OS and PFS of DH lymphoma patients treated with the standard R-CHOP on the one hand, to more intensified treatment protocols such as dose-adjusted R-EPOCH (rituximab, etoposide, doxorubicin, cyclophosphamide, vincristine, prednisone), Hyper-CVAD, and R-CODOX-M/IVAC on the other. Treatment with dose-adjusted R-EPOCH yielded a median PFS of 22.2 months, compared with 12.1 months with R-CHOP, as well as a 34% reduction in progression-risk; however, no effect on OS was noted.178 Most recently, the phase III ALLIANCE trial did not demonstrate a survival benefit for patients treated with dose-adjusted R-EPOCH compared with treatment with R-CHOP; however, MYC-rearranged cases, and DH/TH cases were significantly underrepresented within the study population. Therefore, extrapolation of the results in this subgroup would not be advised.179 Nonetheless, the results of a phase II trial on MYC-rearranged DLBCL cases showed promising results, with 2-year PFS and OS of 71% and 76.7% respectively.180 Given the lack of randomized trials focusing on DH/TH HGBL, dose-adjusted R-EPOCH represents an encouraging frontline treatment approach for these patients.
Other agents have been tried in order to mitigate the inferior prognosis of DH/TH. Venetoclax, in combination with dose-adjusted R-EPOCH, has been evaluated in a phase I trial which demonstrated acceptable safety and efficacy, leading to its current evaluation in a phase II/III trial.181 Tazemetostat and other epigenetic modulators might also prove effective in the treatment of DH/TH. Other novel agents, such as bromodomain, and external domain (BET) inhibitors and Aurora kinase inhibitors might also be effective, as they work by disrupting downstream MYC signaling. These agents are still in preclinical or early clinical trials.182,183 The therapeutic agents that might be effective in the subset of DH/TH are summarized in Table 4.
Table 4.
Potential therapeutic agents by molecular subgroup of DLBCL45. Potential therapeutic approaches for double/triple-hit lymphomas (DH/TH) are also noted.
| Subgroup | Potential therapeutic agents |
|---|---|
| BN2 | BTK inhibitors (ibrutinib, acalabrutinib, zanibrutinib) |
| Lenalidomide | |
| PI3K/mTOR inhibitors (copanlisib, buparlisib, everolimus) | |
| A53 | BTK inhibitors |
| ST2 | JAK/STAT inhibitors (ruxolitinib, pacritinib) |
| PI3K inhibitors | |
| MCD | BTK inhibitors |
| Lenalidomide, | |
| JAK/STAT inhibitors | |
| N1 | Immune checkpoint inhibitors (nivolumab, pembrolizumab, durvalumab) |
| EZB | EZH2 inhibitors (tazemetostat) |
| PI3K inhibitors | |
| BCL2 inhibitors (venetoclax) | |
| DH/TH | R-da-EPOCH (rituximab, dose-adjusted etoposide, vincristine, cyclophosphamide, doxorubicin, prednisone) |
| BCL2 inhibitors | |
| EZH2 inhibitors | |
| PI3K inhibitors |
BCL2, B-cell lymphoma 2; BTK, Bruton’s tyrosine kinase; DH/TH, double/triple-hit; EZH2, enhancer of zest homolog 2; JAK, Janus kinase; mTOR, mechanistic target of rapamycin; PI3K, Phosphoinositide-3 kinase; STAT, signal transducer and activator of transcription.
Conclusions
Several lines of evidence have been published with regards to the prognostic biomarkers of DLBCL in the rituximab era. Past established prognostic factors have been disputed in the rituximab era, whilst there are still conflicting data on the prognostic value of innovative biomarkers. The retrospective nature of most studies, the lack of validation within large prospective trials, the lack of reproducible techniques, and the use of different cut-offs (especially regarding certain IHC markers) are some of the reasons that studies have failed to reflect the underlying complexity of the disease pathophysiology. Moreover, significant inter-correlation of individual biomarkers as well as correlation between biomarkers and IPI categories confound the results of the studies. In the effort to evaluate these prognostic biomarkers, a great variety of methods, including IHC, GEP, NGS, and genomic hybridization have been trailed, but very few are applicable in clinical practice due to cost-related factors and lack of reproducibility.
Among the evaluated prognostic biomarkers, COO, concurrent rearrangements of MYC/BCL2/BCL6, the characterization of DH/TH HGBL, and the overexpression of MYC/BCL2, characterizing DE lymphomas, remain the more robust tools to identify high-risk patients that might need treatment intensification and incorporation of novel target treatment modalities. However, it should be acknowledged that most studies have failed to demonstrate a survival benefit by differentiating the therapeutic approach in these patients. The wide genetic heterogeneity of tumors, even within the same COO subgroup, might explain why individualized treatment simply based on COO classification has providing disappointing results. Most recently, GEP studies have managed to partially elucidate the complex genetic and transcriptomic landscape of DLBCL identifying gene-expression signatures, allowing for the classification of DLBCL cases in prognostically relevant genetic subgroups. The same method has been employed for disentangling the complex composition of the TME and elucidating its prognostic significance. Efforts to translate the results of these studies into techniques applicable in clinical practice are being made. In this context, Lymph2Cx, the gold standard for COO determination, can be expanded to allow for identification of DHITsig+ cases in true need of a more intensified treatment approach. Similarly, targeted NGS can be used to stratify patients among the novel genetic subgroups that might benefit from specific novel agents. Notably, these techniques have been validated for application in FFPET; therefore, their use can be expanded in clinical practice. More intriguingly, liquid biopsies and targeted NGS in cfDNA might revolutionize prognostication in DLBCL. Genetic characterization of cases, and classification in COO and genetic subgroups through studies in cfDNA might overcome limitations pertaining to the quantity and quality of biotic samples and allow for the evaluation of dynamic changes in the genetic landscape of DLBCL during treatment and follow-up. Considering the TME, the translation of the recent findings into clinically applicable methods for stratifying patients in prognostic subgroups is eagerly anticipated.
As knowledge regarding the complex genetic landscape of DLBCL accumulates, the ultimate goal is a comprehensive evaluation of the gene-expression and mutational profile, both in the tumor cells and TME of each DLBCL case, which might allow for more precise prognostication and provide the basis for individually-tailored treatment of DLBCL patients.
Footnotes
Conflict of interest statement: The authors declare that there is no conflict of interest.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Sotirios G. Papageorgiou  https://orcid.org/0000-0002-3376-837X
https://orcid.org/0000-0002-3376-837X
Contributor Information
Sotirios G. Papageorgiou, Second Department of Internal Medicine and Research Unit, University General Hospital ‘Attikon’, 1 Rimini Street, Haidari, Athens 12462, Greece.
Thomas P. Thomopoulos, Second Department of Internal Medicine and Research Unit, Hematology Unit, University General Hospital, ‘Attikon’, Haidari, Athens, Greece
Ioannis Katagas, Second Department of Internal Medicine and Research Unit, Hematology Unit, University General Hospital, ‘Attikon’, Haidari, Athens, Greece.
Anthi Bouchla, Second Department of Internal Medicine and Research Unit, Hematology Unit, University General Hospital, ‘Attikon’, Haidari, Athens, Greece.
Vassiliki Pappa, Second Department of Internal Medicine and Research Unit, Hematology Unit, University General Hospital, ‘Attikon’, Haidari, Athens, Greece.
References
- 1. Swerdlow SH, Campo E, Harris NL, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. Rev 4th ed. Lyon: IARC, 2017. [Google Scholar]
- 2. Morrison VA, Shou Y, Bell JA, et al. Evaluation of treatment patterns and survival among patients with diffuse large B-cell lymphoma in the USA. Future Oncol 2019; 15: 1021–1034. [DOI] [PubMed] [Google Scholar]
- 3. Friedberg JW. Relapsed/refractory diffuse large B-cell lymphoma. Hematology 2011; 2011: 498–505. [DOI] [PubMed] [Google Scholar]
- 4. International Non-Hodgkin’s Lymphoma Prognostic Factors Project. A predictive model for aggressive non-Hodgkin’s lymphoma. New Engl J Med 1993; 329: 987–994. [DOI] [PubMed] [Google Scholar]
- 5. Sehn LH, Berry B, Chhanabhai M, et al. The revised International Prognostic Index (R-IPI) is a better predictor of outcome than the standard IPI for patients with diffuse large B-cell lymphoma treated with R-CHOP. Blood 2007; 109: 1857–1861. [DOI] [PubMed] [Google Scholar]
- 6. Ziepert M, Hasenclever D, Kuhnt E, et al. Standard International prognostic index remains a valid predictor of outcome for patients with aggressive CD20+B-cell lymphoma in the rituximab era. J Clin Oncol 2010; 28: 2373–2380. [DOI] [PubMed] [Google Scholar]
- 7. Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000; 403: 503–511. [DOI] [PubMed] [Google Scholar]
- 8. Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. New Engl J Med 2002; 346: 1937–1947. [DOI] [PubMed] [Google Scholar]
- 9. Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. New Engl J Med 2008; 359: 2313–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gutiérrez-García G, Cardesa-Salzmann T, Climent F, et al. Gene-expression profiling and not immunophenotypic algorithms predicts prognosis in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Blood 2011; 117: 4836–4843. [DOI] [PubMed] [Google Scholar]
- 11. Jais JP, Haioun C, Molina TJ, et al. The expression of 16 genes related to the cell of origin and immune response predicts survival in elderly patients with diffuse large B-cell lymphoma treated with CHOP and rituximab. Leukemia 2008; 22: 1917–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rimsza LM, LeBlanc ML, Unger JM, et al. Gene expression predicts overall survival in paraffin-embedded tissues of diffuse large B-cell lymphoma treated with R-CHOP. Blood 2008; 112: 3425–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rimsza LM, Wright G, Schwartz M, et al. Accurate classification of diffuse large B-cell lymphoma into germinal center and activated B-cell subtypes using a nuclease protection assay on formalin-fixed, paraffin-embedded tissues. Clin Cancer Res 2011; 17: 3727–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barrans SL, Crouch S, Care MA, et al. Whole genome expression profiling based on paraffin embedded tissue can be used to classify diffuse large B-cell lymphoma and predict clinical outcome. Br J Haematol 2012; 159: 441–453. [DOI] [PubMed] [Google Scholar]
- 15. Masqué-Soler N, Szczepanowski M, Kohler CW, et al. Molecular classification of mature aggressive B-cell lymphoma using digital multiplexed gene expression on formalin-fixed paraffin-embedded biopsy specimens. Blood 2013; 122: 1985–1986. [DOI] [PubMed] [Google Scholar]
- 16. Scott DW, Wright GW, Williams PM, et al. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood 2014; 123: 1214–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jais JP, Molina TJ, Ruminy P, et al. Reliable subtype classification of diffuse large B-cell lymphoma samples from GELA LNH2003 trials using the Lymph2Cx gene expression assay. Haematologica 2017; 102: e404–e406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vitolo U, Trneˇný M, Belada D, et al. Obinutuzumab or rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone in previously untreated diffuse large B-cell lymphoma. J Clin Oncol 2017; 35: 3529–3537. [DOI] [PubMed] [Google Scholar]
- 19. Scott DW, Mottok A, Ennishi D, et al. Prognostic significance of diffuse large B-cell lymphoma cell of origin determined by digital gene expression in formalin-fixed paraffin-embedded tissue biopsies. J Clin Oncol 2015; 33: 2848–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004; 103: 275–282. [DOI] [PubMed] [Google Scholar]
- 21. Read JA, Koff JL, Nastoupil LJ, et al. Evaluating cell-of-origin subtype methods for predicting diffuse large B-cell lymphoma survival: a meta-analysis of gene expression profiling and immunohistochemistry algorithms. Clin Lymphoma Myeloma Leuk 2014; 14: 460–467.e462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abdulla M, Hollander P, Pandzic T, et al. Cell-of-origin determined by both gene expression profiling and immunohistochemistry is the strongest predictor of survival in patients with diffuse large B-cell lymphoma. Am J Hematol 2020; 95: 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cho I, Yoon N, Hyeon J, et al. Comparison of the lymph2Cx assay and hans algorithm in determining the cell-of-origin of diffuse large B-cell lymphomas, not otherwise specified. Appl Immunohistochem Mol Morphol 2020; 28: 731–740. [DOI] [PubMed] [Google Scholar]
- 24. Choi WWL, Weisenburger DD, Greiner TC, et al. A new immunostain algorithm classifies diffuse large B-Cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res 2009; 15: 5494–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Muris J, Meijer C, Vos W, et al. Immunohistochemical profiling based on Bcl-2, CD10 and MUM1 expression improves risk stratification in patients with primary nodal diffuse large B cell lymphoma. J Pathol 2006; 208: 714–723. [DOI] [PubMed] [Google Scholar]
- 26. Nyman H, Jerkeman M, Karjalainen-Lindsberg ML, et al. Prognostic impact of activated B-cell focused classification in diffuse large B-cell lymphoma patients treated with R-CHOP. Mod Pathol 2009; 22: 1094–1101. [DOI] [PubMed] [Google Scholar]
- 27. Natkunam Y, Farinha P, Hsi ED, et al. LMO2 protein expression predicts survival in patients with diffuse large B-cell lymphoma treated with anthracycline-based chemotherapy with and without rituximab. J Clin Oncol 2008; 26: 447–454. [DOI] [PubMed] [Google Scholar]
- 28. Visco C, Li Y, Xu-Monette ZY, et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia 2012; 26: 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meyer PN, Fu K, Greiner TC, et al. Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. J Clin Oncol 2011; 29: 200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Colomo L, López-Guillermo A, Perales M, et al. Clinical impact of the differentiation profile assessed by immunophenotyping in patients with diffuse large B-cell lymphoma. Blood 2003; 101: 78–84. [DOI] [PubMed] [Google Scholar]
- 31. Perfecto-Avalos Y, Garcia-Gonzalez A, Hernandez-Reynoso A, et al. Discriminant analysis and machine learning approach for evaluating and improving the performance of immunohistochemical algorithms for COO classification of DLBCL. J Transl Med 2019; 17: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Jong D, Xie W, Rosenwald A, et al. Retracted: immunohistochemical prognostic markers in diffuse large B-cell lymphoma: validation of tissue microarray as a prerequisite for broad clinical applications (a study from the Lunenburg Lymphoma Biomarker Consortium). J Clin Oncol 2007; 25: 805–812. [DOI] [PubMed] [Google Scholar]
- 33. Salles G, de Jong D, Xie W, et al. Prognostic significance of immunohistochemical biomarkers in diffuse large B-cell lymphoma: a study from the Lunenburg Lymphoma Biomarker Consortium. Blood 2011; 117: 7070–7078. [DOI] [PubMed] [Google Scholar]
- 34. Yan W-H, Jiang X-N, Wang W-G, et al. Cell-of-origin subtyping of diffuse large B-cell lymphoma by using a qPCR-based gene expression assay on formalin-fixed paraffin-embedded tissues. Front Oncol 2020; 10: 803–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu-Monette ZY, Zhang H, Zhu F, et al. A refined cell-of-origin classifier with targeted NGS and artificial intelligence shows robust predictive value in DLBCL. Blood Adv 2020; 4: 3391–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lossos IS, Czerwinski DK, Alizadeh AA, et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. New Engl J Med 2004; 350: 1828–1837. [DOI] [PubMed] [Google Scholar]
- 37. Malumbres R, Chen J, Tibshirani R, et al. Paraffin-based 6-gene model predicts outcome in diffuse large B-cell lymphoma patients treated with R-CHOP. Blood 2008; 111: 5509–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tekin N, Omidvar N, Morris TP, et al. Protocol for qRT-PCR analysis from formalin fixed paraffin embedded tissue sections from diffuse large b-cell lymphoma: validation of the six-gene predictor score. Oncotarget 2016; 7: 83319–83329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alizadeh AA, Gentles AJ, Alencar AJ, et al. Prediction of survival in diffuse large B-cell lymphoma based on the expression of 2 genes reflecting tumor and microenvironment. Blood 2011; 118: 1350–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Green TM, Jensen AK, Holst R, et al. Multiplex polymerase chain reaction-based prognostic models in diffuse large B-cell lymphoma patients treated with R-CHOP. Br J Haematol 2016; 174: 876–886. [DOI] [PubMed] [Google Scholar]
- 41. Reddy A, Zhang J, Davis NS, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell 2017; 171: 481–494.e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schmitz R, Wright GW, Huang DW, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. New Engl J Med 2018; 378: 1396–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chapuy B, Stewart C, Dunford AJ, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med 2018; 24: 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lacy SE, Barrans SL, Beer PA, et al. Targeted sequencing in DLBCL, molecular subtypes, and outcomes: a Haematological Malignancy Research Network report. Blood 2020; 135: 1759–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wright GW, Huang DW, Phelan JD, et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell 2020; 37: 551–568.e514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alkodsi A, Cervera A, Zhang K, et al. Distinct subtypes of diffuse large B-cell lymphoma defined by hypermutated genes. Leukemia 2019; 33: 2662–2672. [DOI] [PubMed] [Google Scholar]
- 47. Cardesa-Salzmann TM, Colomo L, Gutierrez G, et al. High microvessel density determines a poor outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus chemotherapy. Haematologica 2011; 96: 996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gratzinger D, Advani R, Zhao S, et al. Lymphoma cell VEGFR2 expression detected by immunohistochemistry predicts poor overall survival in diffuse large B cell lymphoma treated with immunochemotherapy (R-CHOP). Br J Haematol 2010; 148: 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Evens AM, Sehn LH, Farinha P, et al. Hypoxia-inducible factor-1 α Expression Predicts Superior Survival in patients with diffuse large B-cell lymphoma treated with R-CHOP. J Clin Oncol 2010; 28: 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Meyer PN, Fu K, Greiner T, et al. The stromal cell marker SPARC predicts for survival in patients with diffuse large B-cell lymphoma treated with rituximab. Am J Clin Pathol 2011; 135: 54–61. [DOI] [PubMed] [Google Scholar]
- 51. Perry AM, Cardesa-Salzmann TM, Meyer PN, et al. A new biologic prognostic model based on immunohistochemistry predicts survival in patients with diffuse large B-cell lymphoma. Blood 2012; 120: 2290–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ciavarella S, Vegliante MC, Fabbri M, et al. Dissection of DLBCL microenvironment provides a gene expression-based predictor of survival applicable to formalin-fixed paraffin-embedded tissue. Ann Oncol 2018; 29: 2363–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tzankov A, Meier C, Hirschmann P, et al. Correlation of high numbers of intratumoral FOXP3+ regulatory T cells with improved survival in germinal center-like diffuse large B-cell lymphoma, follicular lymphoma and classical Hodgkin’s lymphoma. Haematologica 2008; 93: 193–200. [DOI] [PubMed] [Google Scholar]
- 54. Lee NR, Song EK, Jang KY, et al. Prognostic impact of tumor infiltrating FOXP3 positive regulatory T cells in diffuse large B-cell lymphoma at diagnosis. Leuk Lymphoma 2008; 49: 247–256. [DOI] [PubMed] [Google Scholar]
- 55. Kridel R, Steidl C, Gascoyne RD. Tumor-associated macrophages in diffuse large B-cell lymphoma. Haematologica 2015; 100: 143–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Staiger AM, Altenbuchinger M, Ziepert M, et al. A novel lymphoma-associated macrophage interaction signature (LAMIS) provides robust risk prognostication in diffuse large B-cell lymphoma clinical trial cohorts of the DSHNHL. Leukemia 2020; 34: 543–552. [DOI] [PubMed] [Google Scholar]
- 57. Keir ME, Butte MJ, Freeman GJ, et al. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol 2008; 26: 677–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Georgiou K, Chen L, Berglund M, et al. Genetic basis of PD-L1 overexpression in diffuse large B-cell lymphomas. Blood 2016; 127: 3026–3034. [DOI] [PubMed] [Google Scholar]
- 59. Hu LY, Xu XL, Rao HL, et al. Expression and clinical value of programmed cell death-ligand 1 (PD-L1) in diffuse large B cell lymphoma: a retrospective study. Chin J Cancer 2017; 36: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dong L, Lv H, Li W, et al. Co-expression of PD-L1 and p-AKT is associated with poor prognosis in diffuse large B-cell lymphoma via PD-1/PD-L1 axis activating intracellular AKT/mTOR pathway in tumor cells. Oncotarget 2016; 7: 33350–33362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kiyasu J, Miyoshi H, Hirata A, et al. Expression of programmed cell death ligand 1 is associated with poor overall survival in patients with diffuse large B-cell lymphoma. Blood 2015; 126: 2193–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kwon D, Kim S, Kim PJ, et al. Clinicopathological analysis of programmed cell death 1 and programmed cell death ligand 1 expression in the tumour microenvironments of diffuse large B cell lymphomas. Histopathology 2016; 68: 1079–1089. [DOI] [PubMed] [Google Scholar]
- 63. Fang X, Xiu B, Yang Z, et al. The expression and clinical relevance of PD-1, PD-L1, and TP63 in patients with diffuse large B-cell lymphoma. Medicine 2017; 96: e6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Challa-Malladi M, Lieu Y, Califano O, et al. Combined genetic inactivation of β2-Microglobulin and CD58 reveals frequent escape from immune recognition in diffuse large B cell lymphoma. Cancer Cell 2011; 20: 728–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bernd H-W, Ziepert M, Thorns C, et al. Loss of HLA-DR expression and immunoblastic morphology predict adverse outcome in diffuse large B-cell lymphoma - analyses of cases from two prospective randomized clinical trials. Haematologica 2009; 94: 1569–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rimsza LM, Roberts RA, Miller TP, et al. Loss of MHC class II gene and protein expression in diffuse large B-cell lymphoma is related to decreased tumor immunosurveillance and poor patient survival regardless of other prognostic factors: a follow-up study from the Leukemia and Lymphoma Molecular Profiling Project. Blood 2004; 103: 4251–4258. [DOI] [PubMed] [Google Scholar]
- 67. Yamamoto W, Nakamura N, Tomita N, et al. Human leukocyte antigen-DR expression on flow cytometry and tumor-associated macrophages in diffuse large B-cell lymphoma treated by rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone therapy: retrospective cohort study. Leuk Lymphoma 2014; 55: 2721–2727. [DOI] [PubMed] [Google Scholar]
- 68. Higashi M, Tokuhira M, Fujino S, et al. Loss of HLA-DR expression is related to tumor microenvironment and predicts adverse outcome in diffuse large B-cell lymphoma. Leuk Lymphoma 2016; 57: 161–166. [DOI] [PubMed] [Google Scholar]
- 69. Keane C, Vari F, Hertzberg M, et al. Ratios of T-cell immune effectors and checkpoint molecules as prognostic biomarkers in diffuse large B-cell lymphoma: a population-based study. Lancet Haematol 2015; 2: e445–e455. [DOI] [PubMed] [Google Scholar]
- 70. Kotlov N, Bagaev A, Revuelta MV, et al. Clinical and biological subtypes of B-cell lymphoma revealed by microenvironmental signatures. Cancer Discov 2021; 0839.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vaidya R, Witzig TE. Prognostic factors for diffuse large B-cell lymphoma in the R(X)CHOP era. Ann Oncol 2014; 25: 2124–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Iqbal J, Sanger WG, Horsman DE, et al. BCL2 translocation defines a unique tumor subset within the germinal center B-cell-like diffuse large B-cell lymphoma. Am J Pathol 2004; 165: 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Iqbal J, Neppalli VT, Wright G, et al. BCL2 expression is a prognostic marker for the activated B-cell–like type of diffuse large B-cell lymphoma. J Clin Oncol 2006; 24: 961–968. [DOI] [PubMed] [Google Scholar]
- 74. Ennishi D, Mottok A, Ben-Neriah S, et al. Genetic profiling of MYC and BCL2 in diffuse large B-cell lymphoma determines cell-of-origin–specific clinical impact. Blood 2017; 129: 2760–2770. [DOI] [PubMed] [Google Scholar]
- 75. Visco C, Tzankov A, Xu-Monette ZY, et al. Patients with diffuse large B-cell lymphoma of germinal center origin with BCL2 translocations have poor outcome, irrespective of MYC status: a report from an International DLBCL rituximab-CHOP Consortium Program Study. Haematologica 2013; 98: 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Iqbal J, Meyer PN, Smith LM, et al. BCL2 predicts survival in germinal center B-cell–like diffuse large B-cell lymphoma treated with CHOP-like therapy and rituximab. Clin Cancer Res 2011; 17: 7785–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Meriranta L, Pasanen A, Alkodsi A, et al. Molecular background delineates outcome of double protein expressor diffuse large B-cell lymphoma. Blood Adv 2020; 4: 3742–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Karube K, Campo E. MYC alterations in diffuse large B-cell lymphomas. Semin Hematol 2015; 52: 97–106. [DOI] [PubMed] [Google Scholar]
- 79. Dang CV, O’Donnell KA, Zeller KI, et al. The c-Myc target gene network. Semin Cancer Biol 2006; 16: 253–264. [DOI] [PubMed] [Google Scholar]
- 80. Savage KJ, Johnson NA, Ben-Neriah S, et al. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood 2009; 114: 3533–3537. [DOI] [PubMed] [Google Scholar]
- 81. Tzankov A, Xu-Monette ZY, Gerhard M, et al. Rearrangements of MYC gene facilitate risk stratification in diffuse large B-cell lymphoma patients treated with rituximab-CHOP. Mod Pathol 2014; 27: 958–971. [DOI] [PubMed] [Google Scholar]
- 82. Barrans S, Crouch S, Smith A, et al. Rearrangement of MYC is associated with poor prognosis in patients with diffuse large B-cell lymphoma treated in the era of rituximab. J Clin Oncol 2010; 28: 3360–3365. [DOI] [PubMed] [Google Scholar]
- 83. Akyurek N, Uner A, Benekli M, et al. Prognostic significance of MYC, BCL2, and BCL6 rearrangements in patients with diffuse large B-cell lymphoma treated with cyclophosphamide, doxorubicin, vincristine, and prednisone plus rituximab. Cancer 2012; 118: 4173–4183. [DOI] [PubMed] [Google Scholar]
- 84. Clipson A, Barrans S, Zeng N, et al. The prognosis of MYC translocation positive diffuse large B-cell lymphoma depends on the second hit. J Pathol Clin Res 2015; 1: 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Johnson NA, Slack GW, Savage KJ, et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol 2012; 30: 3452–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yang H, Green MR. Epigenetic programing of B-Cell lymphoma by BCL6 and its genetic deregulation. Front Cell Dev Biol 2019; 7: 272–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Iqbal J, Greiner TC, Patel K, et al. Distinctive patterns of BCL6 molecular alterations and their functional consequences in different subgroups of diffuse large B-cell lymphoma. Leukemia 2007; 21: 2332–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Li S, Wang Z, Lin L, et al. BCL6 rearrangement indicates poor prognosis in diffuse large B-cell lymphoma patients: a meta-analysis of cohort studies. J Cancer 2019; 10: 530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Winter JN, Weller EA, Horning SJ, et al. Prognostic significance of Bcl-6 protein expression in DLBCL treated with CHOP or R-CHOP: a prospective correlative study. Blood 2006; 107: 4207–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Horn H, Ziepert M, Becher C, et al. MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B-cell lymphoma. Blood 2013; 121: 2253–2263. [DOI] [PubMed] [Google Scholar]
- 91. Aukema SM, Siebert R, Schuuring E, et al. Double-hit B-cell lymphomas. Blood 2011; 117: 2319–2331. [DOI] [PubMed] [Google Scholar]
- 92. Scott DW, King RL, Staiger AM, et al. High-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements with diffuse large B-cell lymphoma morphology. Blood 2018; 131: 2060–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Oki Y, Noorani M, Lin P, et al. Double hit lymphoma: the MD Anderson Cancer Center clinical experience. Br J Haematol 2014; 166: 891–901. [DOI] [PubMed] [Google Scholar]
- 94. Petrich AM, Gandhi M, Jovanovic B, et al. Impact of induction regimen and stem cell transplantation on outcomes in double-hit lymphoma: a multicenter retrospective analysis. Blood 2014; 124: 2354–2361. [DOI] [PubMed] [Google Scholar]
- 95. Johnson NA, Savage KJ, Ludkovski O, et al. Lymphomas with concurrent BCL2 and MYC translocations: the critical factors associated with survival. Blood 2009; 114: 2273–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Herrera AF, Mei M, Low L, et al. Relapsed or refractory double-expressor and double-hit lymphomas have inferior progression-free survival after autologous stem-cell transplantation. J Clin Oncol 2017; 35: 24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Li S, Desai P, Lin P, et al. MYC/BCL6 double-hit lymphoma (DHL): a tumour associated with an aggressive clinical course and poor prognosis. Histopathology 2016; 68: 1090–1098. [DOI] [PubMed] [Google Scholar]
- 98. Pillai RK, Sathanoori M, Van Oss SB, et al. Double-hit B-cell lymphomas with BCL6 and MYC translocations are aggressive, frequently extranodal lymphomas distinct from BCL2 double-hit B-cell lymphomas. Am J Surg Pathol 2013; 37: 323–332. [DOI] [PubMed] [Google Scholar]
- 99. Copie-Bergman C, Cuillière-Dartigues P, Baia M, et al. MYC-IG rearrangements are negative predictors of survival in DLBCL patients treated with immunochemotherapy: a GELA/LYSA study. Blood 2015; 126: 2466–2474. [DOI] [PubMed] [Google Scholar]
- 100. Ye Q, Xu-Monette ZY, Tzankov A, et al. Prognostic impact of concurrent MYC and BCL6 rearrangements and expression in de novo diffuse large B-cell lymphoma. Oncotarget 2016; 7: 2401–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Rosenwald A, Bens S, Advani R, et al. Prognostic significance of MYC rearrangement and translocation partner in diffuse large B-cell lymphoma: a study by the lunenburg lymphoma biomarker consortium. J Clin Oncol 2019; 37: 3359–3368. [DOI] [PubMed] [Google Scholar]
- 102. Gebauer N, Bernard V, Gebauer W, et al. TP53 mutations are frequent events in double-hit B-cell lymphomas with MYC and BCL2 but not MYC and BCL6 translocations. Leuk Lymphoma 2015; 56: 179–185. [DOI] [PubMed] [Google Scholar]
- 103. Ennishi D, Jiang A, Boyle M, et al. Double-hit gene expression signature defines a distinct subgroup of germinal center B-cell-like diffuse large B-cell lymphoma. J Clin Oncol 2019; 37: 190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Hilton LK, Tang J, Ben-Neriah S, et al. The double-hit signature identifies double-hit diffuse large B-cell lymphoma with genetic events cryptic to FISH. Blood 2019; 134: 1528–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sha C, Barrans S, Cucco F, et al. Molecular high-grade B-cell lymphoma: defining a poor-risk group that requires different approaches to therapy. J Clin Oncol 2019; 37: 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Green TM, Young KH, Visco C, et al. Immunohistochemical double-hit score is a strong predictor of outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol 2012; 30: 3460–3467. [DOI] [PubMed] [Google Scholar]
- 107. Hu S, Xu-Monette ZY, Tzankov A, et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program. Blood 2013; 121: 4021–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Xu-Monette ZY, Li L, Byrd JC, et al. Assessment of CD37 B-cell antigen and cell of origin significantly improves risk prediction in diffuse large B-cell lymphoma. Blood 2016; 128: 3083–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Xu-Monette ZY, Wu L, Visco C, et al. Mutational profile and prognostic significance of TP53 in diffuse large B-cell lymphoma patients treated with R-CHOP: report from an International DLBCL Rituximab-CHOP Consortium Program Study. Blood 2012; 120: 3986–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Young KH, Leroy K, Møller MB, et al. Structural profiles of TP53 gene mutations predict clinical outcome in diffuse large B-cell lymphoma: an international collaborative study. Blood 2008; 112: 3088–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Zenz T, Kreuz M, Fuge M, et al. TP53 mutation and survival in aggressive B cell lymphoma. Int J Cancer 2017; 141: 1381–1388. [DOI] [PubMed] [Google Scholar]
- 112. Cao Y, Zhu T, Zhang P, et al. Mutations or copy number losses of CD58 and TP53 genes in diffuse large B cell lymphoma are independent unfavorable prognostic factors. Oncotarget 2016; 7: 83294–83307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Farinha P, Sehn L, Skinnider B, et al. Strong p53 expression is an independent predictor of outcome in de novo diffuse large B cell lymphoma (DLBCL) treated with either CHOP or CHOP-R. Blood 2006; 108: 812–812.16537807 [Google Scholar]
- 114. He X, Chen Z, Fu T, et al. Ki-67 is a valuable prognostic predictor of lymphoma but its utility varies in lymphoma subtypes: evidence from a systematic meta-analysis. BMC Cancer 2014; 14: 153–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Li ZM, Huang JJ, Xia Y, et al. High Ki-67 expression in diffuse large B-cell lymphoma patients with non-germinal center subtype indicates limited survival benefit from R-CHOP therapy. Eur J Haematol 2012; 88: 510–517. [DOI] [PubMed] [Google Scholar]
- 116. Koh YW, Hwang HS, Park CS, et al. Prognostic effect of Ki-67 expression in rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone-treated diffuse large B-cell lymphoma is limited to non-germinal center B-cell-like subtype in late-elderly patients. Leuk Lymphoma 2015; 56: 2630–2636. [DOI] [PubMed] [Google Scholar]
- 117. Xu Y, Sun W, Li F. De Novo CD5(+) Diffuse large B-cell lymphoma: biology, mechanism, and treatment advances. Clin Lymphoma Myeloma Leuk 2020; 20: e782–e790. [DOI] [PubMed] [Google Scholar]
- 118. Na HY, Choe J-Y, Shin SA, et al. Characteristics of CD5-positive diffuse large B-cell lymphoma among Koreans: high incidence of BCL2 and MYC double-expressors. PLoS One 2019; 14: e0224247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Yamaguchi M, Nakamura N, Suzuki R, et al. De novo CD5+ diffuse large B-cell lymphoma: results of a detailed clinicopathological review in 120 patients. Haematologica 2008; 93: 1195–1202. [DOI] [PubMed] [Google Scholar]
- 120. Hyo R, Tomita N, Takeuchi K, et al. The therapeutic effect of rituximab on CD5-positive and CD5-negative diffuse large B-cell lymphoma. Hematol Oncol 2010; 28: 27–32. [DOI] [PubMed] [Google Scholar]
- 121. Miyazaki K, Yamaguchi M, Suzuki R, et al. CD5-positive diffuse large B-cell lymphoma: a retrospective study in 337 patients treated by chemotherapy with or without rituximab. Ann Oncol 2011; 22: 1601–1607. [DOI] [PubMed] [Google Scholar]
- 122. Xu-Monette ZY, Tu M, Jabbar KJ, et al. Clinical and biological significance of de novo CD5+ diffuse large B-cell lymphoma in Western countries. Oncotarget 2015; 6: 5615–5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Johnson NA, Boyle M, Bashashati A, et al. Diffuse large B-cell lymphoma: reduced CD20 expression is associated with an inferior survival. Blood 2009; 113: 3773–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Suzuki Y, Yoshida T, Wang G, et al. Association of CD20 levels with clinicopathological parameters and its prognostic significance for patients with DLBCL. Ann Hematol 2012; 91: 997–1005. [DOI] [PubMed] [Google Scholar]
- 125. Choi CH, Park YH, Lim JH, et al. Prognostic Implication of Semi-quantitative Immunohistochemical Assessment of CD20 Expression in Diffuse Large B-Cell Lymphoma. J Pathol Transl Med 2016; 50: 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Hu S, Xu-Monette ZY, Balasubramanyam A, et al. CD30 expression defines a novel subgroup of diffuse large B-cell lymphoma with favorable prognosis and distinct gene expression signature: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Blood 2013; 121: 2715–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Papageorgiou SG, Kontos CK, Foukas PG, et al. BCL2L12 protein overexpression is associated with favorable outcome in diffuse large B-cell lymphoma patients in the rituximab era. Leuk Lymphoma 2016; 57: 2199–2203. [DOI] [PubMed] [Google Scholar]
- 128. Hussain AR, Uddin S, Ahmed M, et al. Prognostic significance of XIAP expression in DLBCL and effect of its inhibition on AKT signalling. J Pathol 2010; 222: 180–190. [DOI] [PubMed] [Google Scholar]
- 129. Zhang Y, Wang J, Sui X, et al. Prognostic and clinicopathological value of survivin in diffuse large B-cell lymphoma: a meta-analysis. Medicine 2015; 94: e1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Harris J, Ibrahim H, Amen F, et al. Cellular (FLICE) like inhibitory protein (cFLIP) expression in diffuse large B-cell lymphoma identifies a poor prognostic subset, but fails to predict the molecular subtype. Hematol Oncol 2012; 30: 8–12. [DOI] [PubMed] [Google Scholar]
- 131. Markovic O, Marisavljevic D, Cemerikic-Martinovic V, et al. c-FLIP does not correlate with response to immunochemotherapy treatment and outcome of patients with nodal diffuse large B-cell lymphoma. Biomed Pharmacother 2013; 67: 445–449. [DOI] [PubMed] [Google Scholar]
- 132. Winter JN, Li S, Aurora V, et al. Expression of p21 protein predicts clinical outcome in DLBCL patients older than 60 years treated with R-CHOP but not CHOP: a prospective ECOG and Southwest Oncology Group correlative study on E4494. Clin Cancer Res 2010; 16: 2435–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Provencio M, Martín P, García V, et al. Caspase 3a: new prognostic marker for diffuse large B-cell lymphoma in the rituximab era. Leuk Lymphoma 2010; 51: 2021–2030. [DOI] [PubMed] [Google Scholar]
- 134. Markovic O, Marisavljevic D, Cemerikic V, et al. Clinical and prognostic significance of apoptotic profile in patients with newly diagnosed nodal diffuse large B-cell lymphoma (DLBCL). Eur J Haematol 2011; 86: 246–255. [DOI] [PubMed] [Google Scholar]
- 135. Pasanen AK, Haapasaari K-M, Peltonen J, et al. Cell cycle regulation score predicts relapse-free survival in non-germinal centre diffuse large B-cell lymphoma patients treated by means of immunochemotherapy. Eur J Haematol 2013; 91: 29–36. [DOI] [PubMed] [Google Scholar]
- 136. Hong JY, Hong ME, Choi MK, et al. The impact of activated p-AKT expression on clinical outcomes in diffuse large B-cell lymphoma: a clinicopathological study of 262 cases. Ann Oncol 2014; 25: 182–188. [DOI] [PubMed] [Google Scholar]
- 137. Nandurkar H, Chaiwatanatorn K, Stamaratis G, et al. Protein Kinase C Beta II Expression in Diffuse Large B-Cell Lymphoma Predicts for Inferior Outcome of Anthracycline-Based Chemotherapy With And Without Rituximab. Clin Lymphoma Myeloma 2009; 9: E31. [DOI] [PubMed] [Google Scholar]
- 138. Huang X, Meng B, Iqbal J, et al. Activation of the STAT3 signaling pathway is associated with poor survival in diffuse large B-cell lymphoma treated with R-CHOP. J Clin Oncol 2013; 31: 4520–4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Ninomiya S, Hara T, Tsurumi H, et al. Indoleamine 2,3-dioxygenase in tumor tissue indicates prognosis in patients with diffuse large B-cell lymphoma treated with R-CHOP. Ann Hematol 2011; 90: 409–416. [DOI] [PubMed] [Google Scholar]
- 140. Okamura T, Seki R, Nagafuji K, et al. Prognostic significance of S-phase kinase-associated protein 2 (Skp2) in DLBCL patients treated with upfront autologous peripheral blood stem cell transplantation. Blood 2013; 122: 3022–3022. [Google Scholar]
- 141. Seki R, Ohshima K, Fujisaki T, et al. Prognostic significance of S-phase kinase-associated protein 2 and p27kip1 in patients with diffuse large B-cell lymphoma: effects of rituximab. Ann Oncol 2010; 21: 833–841. [DOI] [PubMed] [Google Scholar]
- 142. Eskandari M, Manoochehrabadi S, Pashaiefar H, et al. Clinical significance of cell-free DNA as a prognostic biomarker in patients with diffuse large B-cell lymphoma. Blood Res 2019; 54: 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Scherer F, Kurtz DM, Newman AM, et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med 2016; 8: 364ra155–364ra155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Kurtz DM, Scherer F, Jin MC, et al. Circulating tumor DNA measurements as early outcome predictors in diffuse large B-cell lymphoma. J Clin Oncol 2018; 36: 2845–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Chen HY, Zhang WL, Zhang L, et al. 5-Hydroxymethylcytosine profiles of cfDNA are highly predictive of R-CHOP treatment response in diffuse large B cell lymphoma patients. Clin Epigenetics 2021; 13: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Chiu BCH, Zhang Z, You Q, et al. Prognostic implications of 5-hydroxymethylcytosines from circulating cell-free DNA in diffuse large B-cell lymphoma. Blood Adv 2019; 3: 2790–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Rossi D, Diop F, Spaccarotella E, et al. Diffuse large B-cell lymphoma genotyping on the liquid biopsy. Blood 2017; 129: 1947–1957. [DOI] [PubMed] [Google Scholar]
- 148. Rivas-Delgado A, Nadeu F, Enjuanes A, et al. Mutational landscape and tumor burden assessed by cell-free DNA in diffuse large B-cell lymphoma in a population-based study. Clin Cancer Res 2021; 27: 513–521. [DOI] [PubMed] [Google Scholar]
- 149. Pasqualucci L, Zhang B., et al. Genetic drivers of NF-κB deregulation in diffuse large B-cell lymphoma. Semin Cancer Biol 2016; 39: 26–31. [DOI] [PubMed] [Google Scholar]
- 150. Phelan JD, Young RM, Webster DE, et al. A multiprotein supercomplex controlling oncogenic signalling in lymphoma. Nature 2018; 560: 387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Dunleavy K, Pittaluga S, Czuczman MS, et al. Differential efficacy of bortezomib plus chemotherapy within molecular subtypes of diffuse large B-cell lymphoma. Blood 2009; 113: 6069–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Davies A, Cummin TE, Barrans S, et al. Gene-expression profiling of bortezomib added to standard chemoimmunotherapy for diffuse large B-cell lymphoma (REMoDL-B): an open-label, randomised, phase 3 trial. Lancet Oncol 2019; 20: 649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Yang Y, Shaffer A, Emre NC, et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell 2012; 21: 723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Nowakowski GS, Hong F, Scott DW, et al. Addition of lenalidomide to R-CHOP (R2-CHOP) improves outcomes in newly diagnosed diffuse large B-cell lymphoma (DLBCL): first report of ECOG-ACRIN1412 a randomized phase 2 US intergroup study of R2-CHOP vs R-CHOP. Hematol Oncol 2019; 37: 37–38. [Google Scholar]
- 155. Vitolo U, Witzig TE, Gascoyne RD, et al. ROBUST: first report of phase III randomized study of lenalidomide/R-CHOP (R2 -CHOP) vs placebo/R-CHOP in previously untreated ABC-type diffuse large B-cell lymphoma. Hematol Oncol 2019; 37: 36–37. [Google Scholar]
- 156. Nowakowski GS, Laplant BR, Pederson L, et al. Lenalidomide combined with R-CHOP (R2CHOP) overcomes negative prognostic impact of ABC molecular subtype in newly diagnosed diffuse large B-cell lymphoma. Blood 2016; 128: 3035–3035. [Google Scholar]
- 157. Younes A, Sehn LH, Johnson P, et al. Randomized phase III trial of ibrutinib and rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone in non-germinal center B-cell diffuse large B-cell lymphoma. J Clin Oncol 2019; 37: 1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Wilson WH, Young RM, Schmitz R, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med 2015; 21: 922–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Westin JR, Nastoupil LJ, Fayad L, et al. Smart start: rituximab, lenalidomide, and ibrutinib alone and in combination with standard chemotherapy for patients with newly diagnosed diffuse large B-cell lymphoma: final phase II results. Blood 2019; 134: 1581–1581. [Google Scholar]
- 160. Flinn IW, Bartlett NL, Blum KA, et al. A phase II trial to evaluate the efficacy of fostamatinib in patients with relapsed or refractory diffuse large B-cell lymphoma (DLBCL). Eur J Cancer 2016; 54: 11–17. [DOI] [PubMed] [Google Scholar]
- 161. Crump M, Leppä S, Fayad L, et al. Randomized, double-blind, phase III trial of enzastaurin versus placebo in patients achieving remission after first-line therapy for high-risk diffuse large B-cell lymphoma. J Clin Oncol 2016; 34: 2484–2492. [DOI] [PubMed] [Google Scholar]
- 162. Philippar U, Lu T, Vloemans N, et al. Abstract 5690: discovery of JNJ-67856633: a novel, first-in-class MALT1 protease inhibitor for the treatment of B cell lymphomas. Cancer Res 2020; 80: 5690–5690. [Google Scholar]
- 163. Erdmann T, Klener P, Lynch JT, et al. Sensitivity to PI3K and AKT inhibitors is mediated by divergent molecular mechanisms in subtypes of DLBCL. Blood 2017; 130: 310–322. [DOI] [PubMed] [Google Scholar]
- 164. Paul J, Soujon M, Wengner AM, et al. Simultaneous Inhibition of PI3Kδ and PI3Kα Induces ABC-DLBCL Regression by Blocking BCR-Dependent and -Independent Activation of NF-κB and AKT. Cancer Cell 2017; 31: 64–78. [DOI] [PubMed] [Google Scholar]
- 165. Lenz G, Hawkes E, Verhoef G, et al. Single-agent activity of phosphatidylinositol 3-kinase inhibition with copanlisib in patients with molecularly defined relapsed or refractory diffuse large B-cell lymphoma. Leukemia 2020; 34: 2184–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Younes A, Salles G, Martinelli G, et al. Pan-phosphatidylinositol 3-kinase inhibition with buparlisib in patients with relapsed or refractory non-Hodgkin lymphoma. Haematologica 2017; 102: 2104–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Jain N, Singh S, Laliotis G, et al. Targeting phosphatidylinositol 3 kinase-β and -δ for Bruton tyrosine kinase resistance in diffuse large B-cell lymphoma. Blood Adv 2020; 4: 4382–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Oki Y, Fanale M, Romaguera J, et al. Phase II study of an AKT inhibitor MK2206 in patients with relapsed or refractory lymphoma. Br J Haematol 2015; 171: 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Witzig TE, Tobinai K, Rigacci L, et al. Adjuvant everolimus in high-risk diffuse large B-cell lymphoma: final results from the PILLAR-2 randomized phase III trial. Ann Oncol 2018; 29: 707–714. [DOI] [PubMed] [Google Scholar]
- 170. Johnston PB, LaPlant B, McPhail E, et al. Everolimus combined with R-CHOP-21 for new, untreated, diffuse large B-cell lymphoma (NCCTG 1085 [Alliance]): safety and efficacy results of a phase 1 and feasibility trial. Lancet Haematol 2016; 3: e309–e316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Morschhauser F, Feugier P, Flinn IW, et al. A phase II study of venetoclax plus R-CHOP as first-line treatment for patients with diffuse large B-cell lymphoma. Blood 2020; 137: 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Younes A, Romaguera J, Fanale M, et al. Phase I study of a novel oral Janus kinase 2 inhibitor, SB1518, in patients with relapsed lymphoma: evidence of clinical and biologic activity in multiple lymphoma subtypes. J Clin Oncol 2012; 30: 4161–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Cogan JC, Liu Y, Amengual JE. Hypomethylating agents in lymphoma. Curr Treat Options Oncol 2020; 21: 61. [DOI] [PubMed] [Google Scholar]
- 174. Morschhauser F, Salles G, McKay P, et al. Interim report from a phase 2 mutlicenter study of tazemetostat, an EZH2 inhibitor, in patients with relapsed or refractory B-cell non-Hodgkin lymphoma. Hematol Oncol 2017; 35: 24–25. [Google Scholar]
- 175. Sarkozy C, Morschhauser F, Dubois S, et al. A LYSA phase Ib study of tazemetostat (EPZ-6438) plus R-CHOP in patients with newly diagnosed diffuse large B-cell lymphoma (DLBCL) with poor prognosis features. Clin Cancer Res 2020; 26: 3145–3153. [DOI] [PubMed] [Google Scholar]
- 176. Ansell SM, Minnema MC, Johnson P, et al. Nivolumab for relapsed/refractory diffuse large B-cell lymphoma in patients ineligible for or having failed autologous transplantation: a single-arm, phase II study. J Clin Oncol 2019; 37: 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Nowakowski GS, Willenbacher W, Greil R, et al. Safety and efficacy of PD-L1 inhibitor durvalumab with R-CHOP or R2-CHOP in subjects with previously untreated, high-risk DLBCL. J Clin Oncol 2019; 37: 7520–7520. [Google Scholar]
- 178. Howlett C, Snedecor SJ, Landsburg DJ, et al. Front-line, dose-escalated immunochemotherapy is associated with a significant progression-free survival advantage in patients with double-hit lymphomas: a systematic review and meta-analysis. Br J Haematol 2015; 170: 504–514. [DOI] [PubMed] [Google Scholar]
- 179. Bartlett NL, Wilson WH, Jung SH, et al. Dose-Adjusted EPOCH-R compared with R-CHOP as frontline therapy for diffuse large B-cell lymphoma: clinical outcomes of the phase III intergroup trial alliance/CALGB 50303. J Clin Oncol 2019; 37: 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Dunleavy K, Fanale MA, Abramson JS, et al. Dose-adjusted EPOCH-R (etoposide, prednisone, vincristine, cyclophosphamide, doxorubicin, and rituximab) in untreated aggressive diffuse large B-cell lymphoma with MYC rearrangement: a prospective, multicentre, single-arm phase 2 study. Lancet Haematol 2018; 5: e609–e617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Rutherford SC, Abramson JS, Bartlett NL, et al. Phase I study of the Bcl-2 inhibitor venetoclax with DA-EPOCH-R as initial therapy for aggressive B-cell lymphomas. J Clin Oncol 2020; 38: 8003–8003. [Google Scholar]
- 182. Kelly KR, Friedberg JW, Park SI, et al. Phase I study of the investigational aurora a kinase inhibitor alisertib plus rituximab or rituximab/vincristine in relapsed/refractory aggressive B-cell lymphoma. Clin Cancer Res 2018; 24: 6150–6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Li W, Gupta SK, Knudson R, et al. Targeting bromodomain and external domain epigenetic reader protein as effective strategy for double-hit and triple-hit B-cell lymphoma. Blood 2017; 130: 1472–1472. [Google Scholar]