Abstract
Background:
The genetic variant rs9923231 (VKORC1) is associated with differences in the coagulation of blood and consequentially with sensitivity to the drug warfarin. Variation in VKORC1 has been linked in a gene-based test to dementia/Alzheimer’s disease in the parents of participants, with suggestive evidence for an association for rs9923231 (p = 1.8×10–7), which was included in the genome-wide significant KAT8 locus.
Objective:
Our study aimed to investigate whether the relationship between rs9923231 and dementia persists only for certain dementia sub-types, and if those taking warfarin are at greater risk.
Methods:
We used logistic regression and data from 238,195 participants from UK Biobank to examine the relationship between VKORC1, risk of dementia, and the interplay with warfarin use.
Results:
Parental history of dementia, APOE variant, atrial fibrillation, diabetes, hypertension, and hypercholesterolemia all had strong associations with vascular dementia (p < 4.6×10–6). The T-allele in rs9923231 was linked to a lower warfarin dose (βperT - allele = –0.29, p < 2×10–16) and risk of vascular dementia (OR = 1.17, p = 0.010), but not other dementia sub-types. However, the risk of vascular dementia was not affected by warfarin use in carriers of the T-allele.
Conclusion:
Our study reports for the first time an association between rs9923231 and vascular dementia, but further research is warranted to explore potential mechanisms and specify the relationship between rs9923231 and features of vascular dementia.
Keywords: Alzheimer disease, genetics, vascular dementia, warfarin
INTRODUCTION
Warfarin is the most prescribed anticoagulant worldwide [1] and is commonly used as a treatment for atrial fibrillation (AF) [2]. The drug functions by inhibiting the enzyme vitamin K epoxide reductase (VKOR), effectively interfering with the vitamin K cycle required for coagulation of blood [3]. As a result of variations in age, height, weight, genotype, and other factors [4–6], patients vary up to 20-fold in their sensitivity to warfarin [7]. Clinically, the optimum dose is estimated using tests of blood coagulation, commonly the International Normalized Ratio (INR). The strongest genetic predictor of warfarin sensitivity is the gene VKORC1, which encodes for the vitamin K epoxide reductase subunit 1 (VKORC1) and acco-unts for approximately a third of the variance in warfarin sensitivity [3]. Three VKORC1 SNPs, rs9923231, rs9934438, and rs2359612—which are in very high linkage disequilibrium—are the best gen-etic predictors of warfarin sensitivity [3, 7, 8].
In a recent genome-wide association study (GWAS) meta-analysis of parental dementia and case-control Alzheimer’s dementia (ADem) [9], VKORC1 was associated (after Bonferroni correction) with ADem in a gene-based test (p = 5.1×10–8); the T-allele in rs9923231, which is related to the need for a lower dose of warfarin, was not a genome-wide significant finding, but was both located within a genome-wide significant locus and nominally associated with an increased risk of ADem (p = 1.8×10–7). Pure Alzheimer’s disease pathology, characterized by amyloid plaques and neurofibrillary tangles in the grey matter, is uncommon, and most patients exhibit a mixed pathology in which vascular factors often play a prominent role [10]. In fact, there is extensive evidence directly linking vascular dysfunction to ADem [11]. Thus, a possible explanation for the findings [9] is that vascular factors played a crucial role in a proportion of the ADem cases/family history cases observed. If that is the case, then there should be an even stronger relationship between VKORC1 and vascular dementia (VaD) that is mostly due to cardiovascular factors.
Most strokes in western countries are due to occlusions in blood vessels (ischemic), and some are due to ruptures in blood vessels (hemorrhagic) [12]. If carriers of the T-allele in rs9923231 experience a reduction of blood coagulation and subsequent seq-uential minor hemorrhagic strokes, the resulting pa-thology could manifest in dementia and explain the observed link. Furthermore, compared to non-carriers of the T-allele, patients with AF that carry the T-allele could be at an increased risk of intracerebral hemorrhage and consequentially VaD when prescribed warfarin. Here, we study the same UK Biobank cohort as previously [9], but consider both individual and parental dementia status. We test whether T-allele status is associated with an increased risk of VaD and explore whether carriers of the T-allele are at a greater risk of VaD than non-carriers when prescribed warfarin.
METHODS
Sample
We used data from UK Biobank, a large and detailed prospective study of over 500,000 participants aged 37–73 that were recruited between the years 2006 and 2010. UK Biobank has been described in detail before [13]. The Research Ethics Committee (REC) granted ethical approval for the study (reference 11/NW/0382) and the current analysis was conducted under data application 10279.
Genotyping
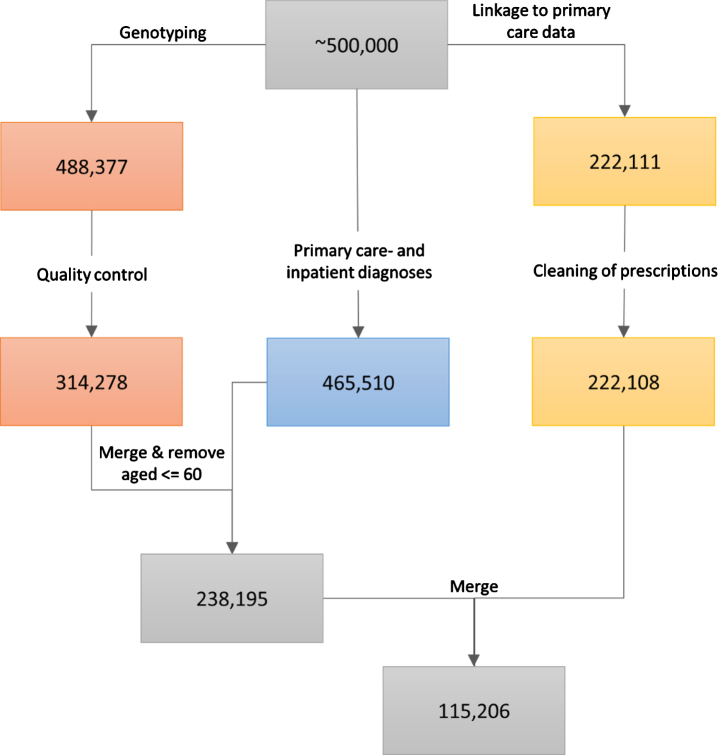
Details on genotyping in the UK Biobank have been reported before [14, 15]. Briefly, for 49,950 participants, genotyping was performed using the UK BiLEVE Axiom Array, and for 438,427 participants, genotyping was performed using the UK Biobank Axiom Array. The released data contained 805,426 markers for 488,377 participants. Further quality control steps were performed as previously reported [9]. They included the removal of outliers, of incongruent data points, and of related participants using a relationship cut-off of 0.025 (GCTA GREML) [16]. This left an unrelated cohort of 314,278 individuals of white British ancestries (Fig. 1).
Fig. 1.
The data cleaning procedure. The left path (orange boxes) represents the genotyping and associated quality control, the middle path (blue box) represents the ascertainment of primary care- and inpatient diagnoses, and the right path (yellow boxes) represents the linkage to primary care prescriptions and the cleaning of the latter. The last two steps (grey boxes) involve the inclusion of only those participants that were older than 60 at the end of sampling and who passed through the left and middle paths (first grey box, 238,195 participants), or through all three data-cleaning paths (second grey box, 115,206 participants). All analyses that did not include prescribing data in the models were performed using the 238,195 participants, while the analyses that utilized warfarin prescription history used the 115,206 participants.
Warfarin prescription data
The UK Biobank obtained data on prescriptions for 222,111 participants via primary care computer system suppliers (EMIS Health and Vision for Scotland, and Wales, Vision and The Phoenix Partnership for England) and has engaged other intermediaries (Albasoft, a third-party data processor, for Scotland and the SAIL databank for Wales). All participants provided written consent for linkage to their health records upon recruitment to UK Biobank. The data were extracted in May 2017 for Scotland, in September 2017 for Wales, and in June, in July, and in August 2017 for England. The data include the exact dates of prescriptions, drug codes (BNF, Read v2, CTV3, and dm + d), names of drugs as written on the prescription, and, where available, the dosages of prescribed drugs. Empty prescriptions, prescriptions without a date, and duplicate prescriptions (defined as identical prescriptions issued to the same person on the same day) were removed from the sample. This resulted in the removal of 1,467,547 prescriptions. Three participants were completely removed from the dataset (Fig. 1). Warfarin prescriptions were extracted by searching for the word “warfarin” under the name/content of each prescription. For each participant, we calculated warfarin use by summing the number of days on which warfarin was prescribed, and warfarin dose by averaging the prescribed dose over all prescriptions of warfarin.
Disease status
Data on diagnoses for 465,510 participants were obtained by the UK Biobank from two sources: 1) from primary care similarly to the prescriptions described above, and 2) from hospital inpatient admissions data. Inpatients are defined as people who are admitted to hospital and occupy a hospital bed. These data included Hospital Episode Statistics for England, Scottish Morbidity Records for Scotland, and the Patient Episode Database for Wales. People with record of any dementia were included in a broad dementia category of “general dementia” that included ADem and VaD, as well as other types of dementia. Furthermore, narrower, more specific categories (ADem, VaD) were also identified. Information on the codes used in the extraction of each diagnosis is provided in Supplementary Table 1. We excluded from our analyses all participants that were 60 years old or younger on the last date of sampling (June 30, 2020) since dementia risk increases steeply with age. Parental diagnoses were ascertained during the initial assessment by asking participants about the presence of “Alzheimer’s disease/dementia” for both mother and father. In our analyses, the parental diagnosis of dementia was considered positive if at least one parent was reported to have suffered from the disorder.
Models
All analyses where the outcome variable was continuous were performed using linear regression; all models where the outcome variable was binary were performed using logistic regression. All models were controlled for the assessment center in which the par-ticipant was tested, the genotyping- batch and array, 40 genetic principal components, the age, sex, edu-cation, socioeconomic deprivation, alcohol consumption, smoking, physical activity, and body mass index (BMI) of the participants. The models predicting VaD were subsequently additionally controlled for APOE variant, concentration of triglycerides (mmol/L), and the diagnoses of hypertension, hypercholesterolemia, and diabetes. All covariates were ascertained immediately prior to or during the participants’ recruitment to the UK Biobank. For education, a binary classification was used that indicated whether a graduate degree had been attained. For socioeconomic deprivation, the Townsend index [17] was used, where higher values indicate greater socioeconomic deprivation (range in the sample: –6.3–10.8). For alcohol consumption, a 6-level scale of frequency of alcohol consumption was used, where 1: “daily or almost daily”, 2: “three or four times a week”, 3:“one or two times a week”, 4: “one to three times a month”, 5: “special occasions only”, 6: “never”. For smoking, the participants were classified as non-smokers, past smokers, or current smokers. For physical activity, the scale provided by the UK Biobank was reduced to a 3-level scale, indicating light, moderate, or strenuous physical activity, as has been used before [18]. For APOE genotype based on the nucleotides at SNP positions rs429358 and rs7412, participants with the ɛ3/ɛ3 haplotype were denoted as carrying variant ɛ3, participants with the ɛ2/ɛ2 or ɛ2/ɛ3 haplotypes were denoted as carrying variant ɛ2, and participants with the ɛ3/ɛ4 or ɛ4/ɛ4 haplotypes were denoted as carrying variant ɛ4. Brain imaging data, including the volume of white matter hyperintensities (WMH), were available for 18,251 participants in the sample. For analysis where WMH was modelled as an outcome, WMH was log-transformed and corrected for intracranial volume. For analyses where parental diagnoses were modelled as outcomes, the ages of each parent (current age or age at death) were included in the models. In all cases where we tested for associations between rs9923231 (VKORC1) and any form of dementia, we assumed an additive genetic effect for rs99232331. All covariates were simultaneously added to the model and the models were not corrected for multiple comparisons. The effects are reported in odds ratios (OR’s) or unstandardized beta-coefficients. All analyses were performed in R version 3.6.3. The code for preparing and analyzing the data is available at https://github.com/JuM24/VKORC1-and-VaD.
RESULTS
Sample characteristics
Among the 238,195 participants, 129,034 (54.2%) were female and 109,161 (45.8%) were male (Table 1). The age range at recruitment was 46–74 years and the median age was 60.9 years (IQR = 9.1). The demographic characteristics of the sub-sample used for analyses utilizing prescription history (Fig. 1) were very similar to the entire sample (Supplementary Table 2). A total of 13,361 (5.6%) participants had been diagnosed with AF and among the 115,206 participants with data on prescriptions, 5,513 (4.8%) had a history of being prescribed warfarin (Supplementary Table 3).
Table 1.
Demographic characteristics of the sample
| Variable | Level | Median (IQR) or n (%) | |||
| All (n = 238,195) | General dementia (n = 4259) | ADem (1531) | VaD (669) | ||
| Age | 60.9 (9.1) | 65.5 (5.8) | 65.9 (5.3) | 66.3 (4.7) | |
| Sex | Female | 129,034 (54.2) | 1,939 (45.5) | 795 (51.9) | 267 (39.9) |
| Male | 109,161 (45.8) | 2,320 (54.5) | 736 (48.1) | 402 (60.1) | |
| Education | Graduate degree | 72,385 (30.7) | 947 (22.6) | 313 (20.9) | 115 (17.6) |
| No graduate degree | 163,563 (69.3) | 3,235 (77.4) | 1,199 (79.1) | 539 (82.4) | |
| Deprivation | –2.5 (3.6) | –2.2 (4.2) | –2.3 (4.1) | –2.2 (3.8) | |
| Alcohol consumption | Daily or almost daily | 54,261 (22.8) | 990 (23.3) | 311 (20.3) | 152 (22.8) |
| 3 or 4 times a week | 57,255 (24.1) | 814 (19.1) | 313 (20.5) | 117 (17.5) | |
| 1 or 2 times a week | 60,106 (25.2) | 952 (22.4) | 357 (23.3) | 147 (22.0) | |
| 1–3 times a month | 24,778 (10.4) | 396 (9.3) | 156 (10.2) | 59 (8.8) | |
| Special occasions only | 25,409 (10.7) | 576 (13.5) | 215 (14.1) | 91 (13.6) | |
| Never | 16,239 (6.8) | 524 (12.3) | 177 (11.6) | 101 (15.1) | |
| Smoking | Current smoker | 21,470 (9.0) | 413 (9.8) | 115 (7.6) | 74 (11.2) |
| Previous smoker | 89,608 (37.8) | 1,839 (43.4) | 647 (42.6) | 310 (46.8) | |
| Non-smoker | 126,288 (53.2) | 1,983 (46.8) | 757 (49.8) | 278 (42.0) | |
| Physical activity | Strenuous | 19,441 (8.7) | 186 (4.9) | 74 (5.2) | 27 (4.7) |
| Moderate | 148,812 (66.6) | 2,350 (62.1) | 908 (64.2) | 359 (62.2) | |
| Light | 55,137 (24.7) | 1,247 (33.0) | 432 (30.6) | 191 (28.6) | |
| BMI | 26.8 (5.7) | 27.1 (6.0) | 26.8 (5.6) | 27.8 (7.1) | |
| APOE variant | ɛ2 | 30,818 (13.3) | 306 (8.2) | 79 (5.3) | 51 (7.8) |
| ɛ3 | 138,634 (59.7) | 1,559 (42.0) | 505 (33.6) | 275 (42.1) | |
| ɛ4 | 62,665 (27.0) | 1,846 (49.7) | 917 (61.1) | 327 (50.1) | |
There were 145,186 (61.0%) carriers of the T-allele in the sample: 111,756 (46.9%) were heterozygous for the T-allele, and 33,430 (14.0%) were homozygous for the T-allele; the allele frequencies were in Hardy-Weinberg equilibrium (χ2 = 0.23, df = 1, p = 0.63). Among the participants, 4,259 (1.8%) had suspected general dementia, 1,531 (0.64%) had suspected ADem (Supplementary Table 4, Supplementary Figure 1), and 669 (0.28%) had suspected VaD (Supplementary Table 4, Supplementary Figure 1); 152 participants (0.03%) had been diagnosed with both ADem and VaD. People with at least one parent with dementia were more likely develop ADem (OR = 3.0, 95% CI = 2.6–3.4, p < 2.0×10–16) and more likely to develop VaD (OR = 2.1, 95% CI = 1.7–2.7, p < 1.9×10–9).
rs9923231 polymorphism and warfarin dose
Carrying the T-allele was negatively associated with the average dose of warfarin (βperT - allele = –0.29, SE = 0.015, p < 2.0×10–16). Individuals heterozygous for the T-allele were prescribed a dose of war-farin that was on average 0.23 mg smaller than the dose prescribed to non-carriers (SE = 0.022, p < 2.0×10–16), while individuals homozygous for the T-allele were prescribed a dose of warfarin that was on average 0.62 mg smaller than the dose presc-ribed to non-carriers (SE = 0.032, p < 2.0×10–16). The average dose of warfarin was also negatively associated with age (β= –0.010, SE = 2.0×10–3, p = 4.1×10–7), and was higher in males (β= 0.062, SE = 0.022, p = 5.7×10–3).
rs9923231 polymorphism and dementia risk
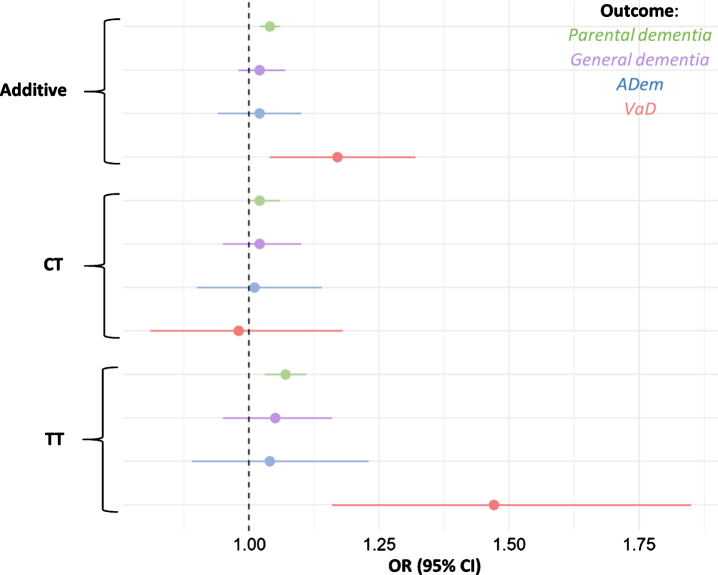
Parents of carriers of the T-allele were more li-kely to have developed dementia (additive effect per T-allele: OR = 1.04, 95% CI = 1.02–1.06, p = 3.7×10–5). When the presence of the T-allele was used to predict general dementia in participants, the effect was not significant, nor was the effect significant when the presence of the T-allele was used to predict ADem in participants (Table 2, Fig. 2).
Table 2.
Results of the additive models with T as the effect allele, using rs9923231 to predict parental dementia, general dementia, ADem, and VaD
| Effect per T allele | ||||
| rs9923231 | OR | 95% CI | p | n cases |
| Parental dementia | 1.04 | 1.02–1.06 | 3.7×10–5 | 34,737 |
| General dementia | 1.02 | 0.98–1.07 | 0.33 | 4,259 |
| ADem | 1.02 | 0.94–1.10 | 0.60 | 1,531 |
| VaD | 1.17 | 1.04–1.32 | 0.010 | 669 |
Fig. 2.
Odds ratios for parental dementia, ADem, and VaD per rs9923231 genotype status. Depicted are the additive effect and the effects of each allele group. The tails represent 95% confidence intervals for the ORs.
When limited to the specific outcome of VaD, the additive effect of the T-allele was much larger (OR = 1.17, 95% CI = 1.04–1.32, p = 0.010, Table 2, Fig. 2). The full breakdown of all allele groups is shown in Supplementary Table 5. We repeated the models for VaD, with rs9923231 as a predictor and with the simultaneous addition of concentration of triglycerides, APOE variant, diagnoses of hypertension (n = 84,694), hypercholesterolemia (n = 40,363), and diabetes (n = 20,990) as additional covariates. While triglycerides, APOE variant, hypertension, hypercholesterolemia, and diabetes were significant predictors, this did not affect the relationship between rs9923231 and VaD (Supplementary Table 6). Beca-use of the importance of cardiovascular events in the etiology of VaD, the T-allele was also used to predict stroke, with the full set of covariates as above. The models were not significant for ischemic (n = 8,087, OR = 0.98, 95% CI = 0.94–1.01, p = 0.21), nor for hemorrhagic (n = 2,146, OR = 0.94, 95% CI = 0.88–1.01, p = 0.073) stroke. Due to the likely causal link between WMH and dementia [19], rs9923231 was related to WMH in the sample. When all the above covariates were included in the model, the association was significant, with the T-allele negatively associated with WMH (β = –2.3×10–8, SE = 7.5×10–9, p = 2.8×10–3).
Warfarin use and VaD in carriers of the T-allele
In our sample, participants diagnosed with AF were at greater risk for ADem (OR = 1.55, 95% CI = 1.31–1.81, p = 1.7×10–7) and for VaD (OR = 2.92, 95% CI = 2.38–3.57, p < 2.0×10–16). The effect remained significant for both ADem (OR = 1.29, 95% CI = 1.08–1.53, p = 4.5×10–3) and VaD (OR = 2.17, 95% CI = 1.74–2.69, p = 1.9×10–12) when APOE status, triglycerides, and diagnoses of hypercholesterolemia, hypertension, and diabetes were included in the model as covariates. To test whether warfarin use in T-allele carriers diagnosed with AF increases the risk of VaD, we performed a logistic model with AF, warfarin use, and rs9923231 predicting VaD, with the inclusion of a 3-way interaction term between AF, warfarin, and rs9923231. The interaction between AF, warfarin use, and rs9923231 was not significant (OR = 0.99, 95 % CI = 0.98–1.00, p = 0.063). The two-way interactions between the above variables were also not significant and effect sizes (main effects) were not substantially attenuated by the addition of the other variables into the models (Supplementary Tables 7and 8). Due to the small number of people with VaD and very limited statistical power for these analyses (Supplementary Text 1), we repeated the analysis by modelling parental dementia as an outcome and including the 3-way interaction term as above; parental dementia was thus treated as a proxy for VaD in the participa-nts. The interaction between AF, warfarin use, and carrier-status was not significant (OR = 0.999, 95% CI = 0.996–1.00, p = 0.77).
DISCUSSION
In this study, we explored the relationship between suspected dementia, atrial fibrillation, warfarin use, and rs9923231, whose T-allele is associated with a reduction in the dose of warfarin [3, 7, 8]. We found a significant association between rs9923231 and suspected VaD, but not between rs9923231 and either suspected general dementia or suspected ADem. While AF was linked to VaD, the use of warfarin in patients that have AF and carry the T-allele did not increase the risk for VaD.
While there have been reports of variants for monogenic forms of VaD [20], data on the genetics of sporadic VaD are sparse. To our knowledge, only two GWAS have been conducted to investigate this: One (n = 5,700) [21] found only rs12007229 on the X-chromosome to be linked to incident VaD, while the other (n = 284) [22] did not find any significant associations for VaD. A systematic review of all genetic association studies for the broader term of vascular cognitive impairment found an association for 6 SNPs in 6 genes: APOE, ACT, ACE, MTHFR, PON1, and PSEN-1.
Previous research has associated variation in rs9923231 with warfarin dose [3, 7, 8, 24], and with various adiposity-related traits, such as hip circumference, arm- and leg fat mass, and BMI (Gene Atlas [25]). To our knowledge the present study for the first time describes an association between rs9923231 and VaD, although it is important to note that this is not at a genome-wide significant threshold. The lack of a relationship between rs9923231 and either ADem or general dementia in the present study suggests that the association between the T-allele and ADem, as reported previously [9], might have been partly due to the classification of parental dementia. The UK Biobank questionnaire administered to participants did not distinguish between different types of dementia and it is not known how many of the 42,034 parents that were reportedly diagnosed with “Alzheimer’s/dementia” [9] may have suffered from VaD. This hypothesis is further supported by the estimated effect sizes for the association between rs9923231 and ADem, which were not num-erically larger than those for the association between rs9923231 genotype and parental dementia. Since parental dementia was used as a proxy for ADem in participants, the effect for ADem in participants sho-uld have been substantially greater than for parental dementia (even in the absence of statistical significance) if there truly was an association between rs9923231 and ADem (as opposed to an association between rs9923231 and VaD). Furthermore, in a rec-ent GWAS of clinically diagnosed ADem (n = 94,437) [26] there was no association between rs9923231 and ADem.
Based on our results and considering the impo-rtance of cardiovascular abnormalities in the pathology of dementia [10, 11], any future studies exploring the association between rs9923231 and dementia must strongly consider the role of cardiovascular factors; The relationship between genotype and dem-entia might hold only for cases of pure VaD or for those in which vascular pathology represents the main cause of the disorder.
There is an established association between dem-entia and both stroke [27] and WMH [28]; thus, stroke or WMH could act as mediators between rs9923231 genotype and VaD. However, we found no evidence for a positive association between either rs9923231 and stroke or rs9923231and WMH. Moreover, the latter association was statistically significant and negative in direction, suggesting participants carrying the T-allele were less likely to exhibit WMH. While we did not directly test for the effects of other relevant processes, including microbleeds and covert stroke, in the relationship between rs9923231 and VaD, given the lack of evidence for an association between rs9923231 and stroke, they are unlikely to act as prominent mediators. These results further complicate the potential relationship between rs9923231 and VaD and reinforce the need for additional studies to confirm this association and to test alternative mechanism distinct from stroke or WMH.
Interplay between AF, VaD and warfarin use
AF has been previously associated with cognitive decline and dementia. In our study, AF was associated with VaD and with ADem, even after controlling for hypertension and hypercholesterolemia. The association between AF and VaD is unsurprising, considering the inclusion of either vascular disease or history of stroke in almost all definitions of VaD [29]. Despite a substantial overlap of risk factors for AF and ADem, there is some evidence for an independent relationship between the two disorders [30, 31]
Due to the positive association between AF and VaD, the relationship between rs9923231 and VaD, and between rs9923231 and required warfarin dose, T-allele carriers that take warfarin to treat their AF might be at an increased risk of VaD than non-carriers due to warfarin-related brain hemorrhages. To test this, we studied an interaction between warfarin use, AF, and VKORC1 genotype with VaD. We observed no variation in dementia risk by different combinations of these predictors. Due to reduced coagulation of blood in carriers of the T-allele, these individuals could be at greater risk of internal bleeding when taking warfarin. However, the required dose of warfarin is regularly estimated and adjusted using tests of blood coagulation and based on the results of the present paper, this approach is just as efficient in patients carrying the T-allele.
Limitations and future directions
The present study has the advantages of having used a well-characterized sample with access to both inpatient- and primary-care diagnoses. However, we acknowledge several limitations. First, despite the large number of people recruited to UK Biobank, the age range at the end of the sampling period for the cohort is 60–83 years, resulting in a low incidence and prevalence of dementia. This heavily reduced the size of our sample, especially when testing for interactions, and led to wide confidence intervals for the estimated odds ratios. Second, despite it not being the only vitamin K antagonist anticoagulant on the UK market, only warfarin was included in the analysis. Third, the dose of warfarin ingested by participants was assumed to correspond to the average of their prescribed dose, despite some individuals possibly taking more or less of the medicine depending on their individual drug regimes. Fourth, clinical diagnoses of dementia subtypes are difficult and are prone to errors due to the presence of comorbidities and cardiovascular factors [32]. In the present paper, imaging data to confirm the diagnoses was unavailable and all diagnoses were based solely on records from primary care and hospitals. Finally, while most definitions of VaD include both dementia and a history of stroke or cardiovascular disease, VaD is very heterogeneous [33]; in the present study, we did not explore potential mechanisms and mediators of the association between rs9923231 and VaD, nor did we test the relationship for different subtypes of VaD.
The knowledge of genetic risk factors for diseases enables the generation of more accurate hypotheses about underlying biological mechanisms and illuminates potential targets for pharmacological intervention. Moreover, it allows for more informed stratification of participants in clinical trials. Studies that build on our research should aim to replicate the findings in a bigger sample and with greater precision determine the effect size for the association between rs9923231 and VaD. Additionally, further work is required to identify possible associations between rs9923231 and features of VaD, such as lacunar infarction, intracerebral hemorrhage, and white matter hyperintensities.
Supplementary Material
ACKNOWLEDGMENTS
DLM and REM are supported by Alzheimer’s Research UK major project grant ARUK-PG2017B-10. JM is supported by funding from the Wellcome Trust 4-year PhD in Translational Neuroscience—training the next generation of basic neuroscientists to embrace clinical research [108890/Z/15/Z]. PMV acknowledges funding from the Australian National Health and Medical Research Council (1113400) and the Australian Research Council (FL180100072). JM and TCR are members of the Alzheimer Scotland Dementia Research Centre funded by Alzheimer Scotland. TCR is employed by NHS Lothian and the Scottish Government. SRC is supported by Age UK (Disconnected Mind project), the UK Medical Research Council [MR/R024065/1] and a National Institutes of Health (NIH) research grant R01AG054628. The authors thank all participants of the UK Biobank for providing data for the study and Dr Michelle Luciano (Department of Psychology, University of Edinburgh) for managing UK Biobank data application 10279.
Authors’ disclosures available online (https://www.j-alz.com/manuscript-disclosures/20-1256r1).
SUPPLEMENTARY MATERIAL
The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/JAD-201256.
REFERENCES
- [1]. Ross KA, Bigham AW, Edwards M, Gozdzik A, Suarez-Kurtz G, Parra EJ (2010) Worldwide allele frequency distribution of four polymorphisms associated with warfarin dose requirements. J Hum Genet 55, 582–589. [DOI] [PubMed] [Google Scholar]
- [2]. Zimetbaum P (2017) Atrial fibrillation. Ann Intern Med 166, ITC33–ITC48. [DOI] [PubMed] [Google Scholar]
- [3]. Wadelius M, Chen LY, Downes K, Ghori J, Hunt S, Eriksson N, Wallerman O, Melhus H, Wadelius C, Bentley D, Deloukas P (2005) Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J 5, 262–270. [DOI] [PubMed] [Google Scholar]
- [4]. Saleh MI (2016) Clinical predictors associated with warfarin sensitivity. Am J Ther 23, e1690–e1694. [DOI] [PubMed] [Google Scholar]
- [5]. Sconce EA, Khan TI, Wynne HA, Avery P, Monkhouse L, King BP, Wood P, Kesteven P, Daly AK, Kamali F (2005) The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: Proposal for a new dosing regimen. Hemostasis, Thromb Vasc Biol 106, 2329–2333. [DOI] [PubMed] [Google Scholar]
- [6]. Wells PS, Holbrook AM, Crowther NR, Hirsh J (1994) Interactions of warfarin with drugs and food. Ann Intern Med 121, 676–683. [DOI] [PubMed] [Google Scholar]
- [7]. Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, Whittaker P, Ranganath V, Kumanduri V, McLaren W, Holm L, Lindh J, Rane A, Wadelius M, Deloukas P (2009) A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet 5, e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Cha PC, Mushiroda T, Takahashi A, Kubo M, Minami S, Kamatani N, Nakamura Y (2010) Genome-wide association study identifies genetic determinants of warfarin responsiveness for Japanese. Hum Mol Genet 19, 4735–4744. [DOI] [PubMed] [Google Scholar]
- [9]. Marioni RE, Harris SE, Zhang Q, McRae AF, Hagenaars SP, Hill WD, Davies G, Ritchie CW, Gale CR, Starr JM, Goate AM, Porteous DJ, Yang J, Evans KL, Deary IJ, Wray NR, Visscher PM (2018) GWAS on family history of Alzheimer’s disease. Transl Psychiatry 8, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10]. Van Der Flier WM, Skoog I, Schneider JA, Pantoni L, Mok V, Chen CLH, Scheltens P (2018) Vascular cognitive impairment. Nat Rev Dis Prim 4, 1–16. [DOI] [PubMed] [Google Scholar]
- [11]. Sweeney MD, Montagne A, Sagare AP, Nation DA, Schneider LS, Chui HC, Harrington MG, Pa J, Law M, Wang DJJ, Jacobs RE, Doubal FN, Ramirez J, Black SE, Nedergaard M, Benveniste H, Dichgans M, Iadecola C, Love S, Bath PM, Markus HS, Salman RA, Allan SM, Quinn TJ, Kalaria RN, Werring DJ, Carare RO, Touyz RM, Williams SCR, Moskowitz MA, Katusic ZS, Lutz SE, Lazarov O, Minshall RD, Rehman J, Davis TP, Wellington CL, González HM, Yuan C, Lockhart SN, Hughes TM, Chen CLH, Sachdev P, O’Brien JT, Skoog I, Pantoni L, Gustafson DR, Biessels GJ, Wallin A, Smith EE, Mok V, Wong A, Passmore P, Barkof F, Muller M, Breteler MMB, Román GC, Hamel E, Seshadri S, Gottesman RF, van Buchem MA, Arvanitakis Z, Schneider JA, Drewes LR, Hachinski V, Finch CE, Toga AW, Wardlaw JM, Zlokovic B V. (2019) Vascular dysfunction—The disregarded partner of Alzheimer’s disease. Alzheimers Dement 15, 158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Kalaria RN, Akinyemi R, Ihara M (2016) Stroke injury, cognitive impairment and vascular dementia. Biochim Biophys Acta 1862, 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13]. Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, Pell J, Silman A, Young A, Sprosen T, Peakman T, Collins R (2015) UK Biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 12, e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, Cortes A, Welsh S, Young A, Effingham M, McVean G, Leslie S, Allen N, Donnelly P, Marchini J (2018) The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]. Wain L V., Shrine N, Miller S, Jackson VE, Ntalla I, Artigas MS, Billington CK, Kheirallah AK, Allen R, Cook JP, Probert K, Obeidat M, Bossé Y, Hao K, Postma DS, Paré PD, Ramasamy A, Mägi R, Mihailov E, Reinmaa E, Melén E, O’Connell J, Frangou E, Delaneau O, Freeman C, Petkova D, McCarthy M, Sayers I, Deloukas P, Hubbard R, Pavord I, Hansell AL, Thomson NC, Zeggini E, Morris AP, Marchini J, Strachan DP, Tobin MD, Hall IP, Farrall M, Barroso I, Anderson CA, Botía J, Vandrocova J, Guelfi S, D’Sa K, Ryten M, Trabzuni D, Matarin M, Hardy JA, Weale ME, Varghese V, Forabosco P, Farmer A, McGuffin P, Zgaga L, Wilson JF, Wild SH, Campbell H, Rudan I, Smith C, Walker R, Liu JZ, Tozzi F, Muglia P, Waterworth DM, Pillai SG, Yuan X, Mooser V, Middleton L, Kooner J, Chambers JC, Berrettini W, Knouff CW, Waeber G, Vollenweider P, Preisig M, Wareham NJ, Zhao JH, Loos RJF, Khaw KT, Grundy S, Barter P, Mahley R, Kesaniemi A, McPherson R, Vincent JB, Strauss J, Kennedy JL, Day R, Matthews K, Bakke P, Gulsvik A, Lucae S, Ising M, Brueckl T, Horstmann S, Wichmann HE, Rawal R, Wichmann HE, Lamina C, Dahmen N, Polasek O, Kolcic I, Huffman J, Campbell S, Vitart V, Hayward C, Wright AF, Burnett MS, Devaney JM, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Epstein S, Reilly MP, Li M, Qu L, Wilensky R, Matthai W, Hakonarson HH, Rader DJ, Ellinghaus D, Lieb W, Franke A, Uda M, Busonero F, Terracciano A, Schlessinger D, Xiao X, Scheet P, St Clair D, Rujescu D, Abecasis GR, Grabe HJ, Teumer A, Völzke H, Petersmann A, John U, Wright BJ, Thompson JR, Balmforth AJ, Hall AS, Samani NJ, Ahmad T, Mathew CG, Parkes M, Satsangi J, Caulfield M, Munroe PB, Dominiczak A, Worthington J, Thomson W, Eyre S, Barton A, Francks C (2015) Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): A genetic association study in UK Biobank. Lancet Respir Med 3, 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet 88, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17]. Townsend P (1987) Deprivation. J Soc Policy 16, 125–146. [Google Scholar]
- [18]. Hanlon P, Quinn TJ, Gallacher KI, Myint PK, Jani BD, Nicholl BI, Lowrie R, Soiza RL, Neal SR, Lee D, Mair FS (2020) Assessing risks of polypharmacy involving medications with anticholinergic properties. Ann Fam Med 18, 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. Prins ND, Scheltens P (2015) White matter hyperintensities, cognitive impairment and dementia: An update. Nat Rev Neurol 11, 157–165. [DOI] [PubMed] [Google Scholar]
- [20]. Ikram MA, Bersano A, Manso-Calderón R, Jia JP, Schmidt H, Middleton L, Nacmias B, Siddiqi S, Adams HHH (2017) Genetics of vascular dementia - review from the ICVD working group. BMC Med 15, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Schrijvers EMC, Schürmann B, Koudstaal PJ, Van Den Bussche H, Van Duijn CM, Hentschel F, Heun R, Hofman A, Jessen F, Kölsch H, Kornhuber J, Peters O, Rivadeneira F, Rüther E, Uitterlinden AG, Riedel-Heller S, Dichgans M, Wiltfang J, Maier W, Breteler MMB, Ikram MA (2012) Genome-wide association study of vascular dementia. Stroke 43, 315–319. [DOI] [PubMed] [Google Scholar]
- [22]. Kim Y, Kong M, Lee C (2013) Association of intronic sequence variant in the gene encoding spleen tyronase kinase with susceptability to vascular dementia. World J Biol Psychiatry 14, 220–226. [DOI] [PubMed] [Google Scholar]
- [23]. Dwyer R, Skrobot OA, Dwyer J, Munafo M, Kehoe PG (2013) Using Alzgene-like approaches to investigate susceptibility genes for vascular cognitive impairment. J Alzheimers Dis 34, 145–154. [DOI] [PubMed] [Google Scholar]
- [24]. Buniello A, Macarthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, Suveges D, Vrousgou O, Whetzel PL, Amode R, Guillen JA, Riat HS, Trevanion SJ, Hall P, Junkins H, Flicek P, Burdett T, Hindorff LA, Cunningham F, Parkinson H (2019) The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47, D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25]. Canela-Xandri O, Rawlik K, Tenesa A (2018) An atlas of genetic associations in UK Biobank. Nat Genet 50, 1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, Boland A, Vronskaya M, van der Lee SJ, Amlie-Wolf A, et al. (2019) Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet 51, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Kuźma E, Lourida I, Moore SF, Levine DA, Ukoumunne OC, Llewellyn DJ (2018) Stroke and dementia risk: A systematic review and meta-analysis. Alzheimers Dement 14, 1416–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]. Bos D, Wolters FJ, Darweesh SKL, Vernooij MW, de Wolf F, Ikram MA, Hofman A (2018) Cerebral small vessel disease and the risk of dementia: A systematic review and meta-analysis of population-based evidence. Alzheimers Dement 14, 1482–1492. [DOI] [PubMed] [Google Scholar]
- [29]. Gorelick PB, Scuteri A, Black SE, Decarli C, Greenberg SM, Iadecola C, Launer LJ, Laurent S, Lopez OL, Nyenhuis D, Petersen RC, Schneider JA, Tzourio C, Arnett DK, Bennett DA, Chui HC, Higashida RT, Lindquist R, Nilsson PM, Roman GC, Sellke FW, Seshadri S (2011) Vascular contributions to cognitive impairment and dementia: A statement for healthcare professionals from the American Heart Association/American Stroke Association. Stroke 42, 2672–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Aldrugh S, Sardana M, Henninger N, Saczynski JS, McManus DD (2017) Atrial fibrillation, cognition and dementia: A review. J Cardiovasc Electrophysiol 28, 958–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Dietzel J, Haeusler KG, Endres M (2018) Does atrial fibrillation cause cognitive decline and dementia? Europace 20, 408–419. [DOI] [PubMed] [Google Scholar]
- [32]. Barragán Martínez D, García Soldevilla MA, Parra Santiago A, Tejeiro Martínez J (2019) Alzheimer’s disease. Medicine 12, 4338–4346. [Google Scholar]
- [33]. Perneczky R, Tene O, Attems J, Giannakopoulos P, Ikram MA, Federico A, Sarazin M, Middleton LT (2016) Is the time ripe for new diagnostic criteria of cognitive impairment due to cerebrovascular disease? Consensus report of the International Congress on Vascular Dementia working group. BMC Med 14, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.