Abstract
Rapid identification and characterization of multidrug-resistant Klebsiella pneumoniae strains is essential to diagnose severe infections in patients. In clinical routine practice, K. pneumoniae is frequently identified and characterized for outbreak investigation. Pulsed-field gel electrophoresis or multilocus sequence typing could be used, but, unfortunately, these methods are time-consuming, laborious, expensive, and do not provide any information about the presence of resistance and virulence genes. In recent years, the decreasing cost of next-generation sequencing and its easy use have led to it being considered a useful method, not only for outbreak surveillance but also for rapid identification and evaluation, in a single step, of virulence factors and resistance genes. Carbapenem-resistant strains of K. pneumoniae have become endemic in Italy, and in these strains the ability to form biofilms, communities of bacteria fixed in an extracellular matrix, can defend the pathogen from the host immune response as well as from antibiotics, improving its persistence in epithelial tissues and on medical device surfaces.
Keywords: Klebsiella pneumoniae, carbapenemase, ST405, biofilm
1. Introduction
Klebsiella pneumoniae, bacteria of the family Enterobacteriaceae, is a main human pathogen causing hospital- and community-acquired infections such as bacteremia, urinary tract infections, pneumonia, and pyogenic liver abscesses [1,2].
Some K. pneumoniae isolates have been shown to be resistant to first-line antibiotics. Due to the spread of carbapenem-resistant K. pneumoniae (CR-Kp), this pathogen represents one of the microorganisms constituting an urgent threat to human health. Antimicrobial resistance to carbapenem is commonly related to the spread of transmissible plasmids, and the acquisition of resistance genes normally occurs by horizontal gene transfer [3]. Antimicrobial resistance plasmids have been called “epidemic resistance plasmids” because of their tendency to acquire resistance genes and the rapid diffusion among members of the family Enterobacteriaceae. Antimicrobial resistance determinants of epidemic plasmids offer a benefit to high-risk clones and are probably essential to their spread [4].
Resistance to carbapenems involves multiple mechanisms, including the following: alterations in outer membrane permeability mediated by the loss of porins, upregulation of efflux systems along with hyperproduction of AmpC β-lactamases, extended-spectrum β-lactamases (ESBLs), or more commonly, the production of carbapenemases.
Twenty-two different KPC (Klebsiella pneumoniae carbapenemase) enzyme variants have been identified. KPC β-lactamases can hydrolyze all β-lactams, including carbapenems, cephalosporins, cephamycins, monobactams, and clavulanic acid.
KPCs have been found in many Gram-negative species, including both Enterobacteriaceae and non-fermenters. KPCs are frequently found in K. pneumoniae associated with nosocomial infections, such as urinary tract infections, septicemia, pneumonia, and intra-abdominal infections.
The global spread of CR-Kp has been largely associated with “high-risk” carbapenemase-producing clones, mostly sequence type (ST) 258 and its related variants (clonal complex 258 (CC258)). However, in healthcare facilities, the spread of CR-Kp isolates other than ST258 has already been described [5,6,7]. In the geographic area of Palermo, Italy, CC258 is still prevalent; however, several other STs (e.g., ST307, ST395, ST392, ST348, ST405, and ST101) are emerging and circulating. This indicates a more complex polyclonal spread of K. pneumoniae carbapenemase producers [8,9,10].
For pathogen survival, other than the acquisition of resistance, virulent traits are also important, and some reports suggest that they may have an essential role in the pathogenesis of K. pneumoniae infections [11,12]. However, the most important virulence factors responsible for K. pneumoniae pathogenesis are capsular polysaccharides, which can collaborate to form biofilm [13,14], supporting the bacterial attachment to living or abiotic surfaces and preventing the effects of antimicrobial agents [15,16].
Using a next-generation sequencing (NGS) approach, it is possible to obtain molecular characterization in terms of genotyping and analyses of the genetic repertoire, including antibiotic resistance, virulence-related factors, and plasmid content of strains [17,18].
On the basis of this indication, the determination of genetic diversity for dominant species identification and complete knowledge of biofilm formation is necessary for stopping the spread of infection in hospitals and for the control and management of associated infections. In addition, knowing the principal genotype of isolates can aid in the identification of the cause of an infection in order to realize protective actions and infection control.
The purpose of this study was to report the draft sequence of a carbapenemase- and biofilm-producing K. pneumoniae strain belonging to ST405 isolated from a clinical sample.
We hypothesize that despite the in vitro resistance to the antibiotic therapy used, the infection could be resolved by the early use of combined therapy at high doses of antibiotics and by the immediate removal of the peripherally inserted central catheter (PICC).
2. Results
2.1. Molecular Genotyping
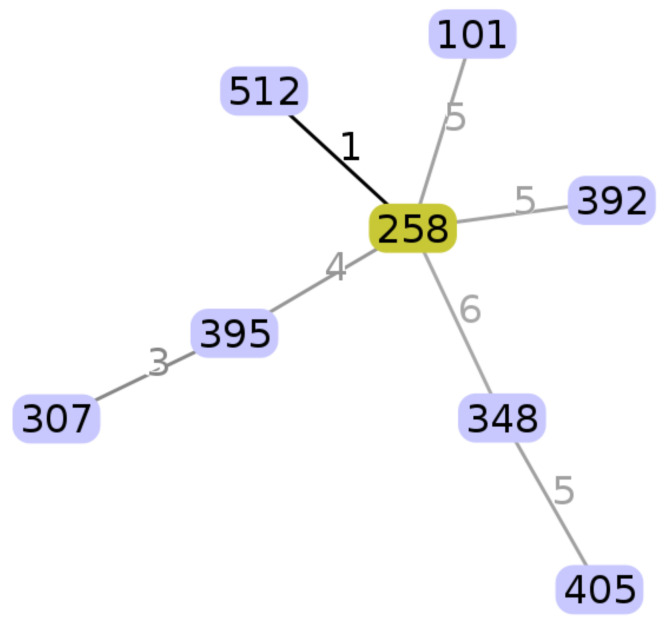
The in silico multilocus sequence typing (MLST) analysis revealed that the strain belonged to ST405. The image below shows the ST405 with respect to the carbapenem-resistant K. pneumoniae ST collected from March 2014 to March 2016 at the Azienda Ospedaliera Universitaria Policlinico “P. Giaccone” of Palermo [13].
In Figure 1, we report a radial diagram showing each sequence type (ST) represented by a circle with the numbers indicating the locus differences between the STs. The numbers in Table 1 show the corresponding housekeeping gene alleles for each ST.
Figure 1.
A radial diagram created using goeBURST.
Table 1.
Housekeeping gene alleles for each sequence type (ST).
| Locus | Putative Function of Gene | Lengh (bp) | ST 258 | ST 512 | ST 395 | ST 101 | ST 392 | ST 348 | ST 307 | ST 405 |
|---|---|---|---|---|---|---|---|---|---|---|
| rpoB | beta subunit of RNA polymerase B | 501 | 1 | 1 | 1 | 1 | 4 | 15 | 1 | 4 |
| gap A | Glyceraldehyde 3-phoshate dehydrogenase | 450 | 3 | 54 | 3 | 2 | 3 | 2 | 4 | 2 |
| mdh | Malate deydrogenase | 477 | 1 | 1 | 2 | 1 | 6 | 20 | 2 | 62 |
| Pgi | Phosphoglucose isomerase | 432 | 1 | 1 | 4 | 5 | 1 | 10 | 52 | 3 |
| pho E | Phosphoporine E | 420 | 1 | 1 | 1 | 4 | 7 | 12 | 1 | 10 |
| inf B | Translation initiation factor 2 | 318 | 3 | 3 | 1 | 6 | 4 | 1 | 1 | 1 |
| ton B | Periplasmic energy trasducer | 414 | 79 | 79 | 4 | 6 | 40 | 16 | 7 | 110 |
A resistome study confirmed the susceptibility test results, and genes encoding for factors associated with resistance to the main classes of antibiotic were identified in silico. In particular, we found the presence of genes associated with resistance to beta-lactamase (blaKPC-3, blaCTX-M-15, blaOXA-1, blaSHV-76, blaTEM-1), aminoglycoside (aac(3) lla-c-2, aac(6′)-Ib-cr, aph(3)pp Ib-2, aph(6) Id-1), heavy metal (pcoA-2, pcoB-3, pcoC-1, pcoD-2, pcoE-1, pcoR-1, pcoS-2, silC-3, silE-3, silR-2, silS-2), quinolone (gyrA-4, gyrB-1, parC-2, qnrB-1), and efflux systems and regulators (acrR-1, envR-15, fis-1, marA-2, marR-1, oqxR-4, rob-21, sdiA-6, soxR-2, soxS-4).
The screening for (putative) virulence profile revealed the presence of a variety of virulence-associated genes. In particular, genes encoding for type-3 fimbriae, biofilm formation and host cell adherence (mrkA-4, mrkB-1, mrkF-38, mrkH-15, mrkI-18, mrkJ-1, and cps cluster genes, wzi-143 and wzc-937, associated with the K type (K) and the K locus (KL)-151), the fyuA yersiniabactin receptor, irp1-148 and irp2-145 aerobactin, kvgA-2 regulator system component, yersiniabactin system (ybtA-1, ybtA-39, ybtE-69, ybtP-4, ybtQ-60, ybtS-6, ybtT-39, ybtU-2, ybtX-62), and microcin E492 system components (mceA-1, mceC-1, mceD-3, mceE-2, mceH-5) were detected.
The PlasmidFinder (http://www.genomicepidemiology.org) (accessed on 10 May 2021) database was used to describe the replicon plasmid content type of the K. pneumoniae isolate. Three plasmid Inc types were identified: IncFII(K) (CP000648), IncFIB(pQil) (JN233705), and IncFIB(K)-Kpn3 (JN233704). However, the intrinsic limits of short-read technologies (e.g., Illumina) in the WGS (Whole Genome Sequencing) method did not allow us to accurately reconstruct the genomic context surrounding the repeated sequences, typically related to antibiotic resistance and virulence determinants, in the plasmids.
The genes correlated with resistance and virulence are reported in Table 2.
Table 2.
Resistance genes and virulence factors detected by next-generation sequencing (NGS) analysis.
| Resistance Genes | ||||||
|---|---|---|---|---|---|---|
| Beta-Lactamase | Aminoglycoside | Heavy Metal | Quinolone | Efflux Systems and Regulator Systems | ||
| Gene alleles | blaKPC-3,blaCTX-M-15, blaOXA-1, blaSHV-76, blaTEM-1 | aac3 lla-c2, aac6p Ib-b-cr, aph3pp Ib-2, aph6 Id-1 | pcoA-2, pcoB-3, pcoC-1, pcoD-2, pcoE-1, pcoR-1, pcoS-2, silC-3, silE-3, silR-2, silS-2 | gyrA-4, gyrB-1, parC-2, qnrB-1 | acrR-1, envR-15, fis-1, marA-2, marR-1, oqxR-4, rob-21, sdiA-6, soxR-2, soxS-4 | |
| Virulence Factors | ||||||
| Gene alleles | type-3 fimbriae, biofilm formation, host cell adherence/capsule | yersiniabactin receptor | aerobactin/regulator system | yersiniabactin system | microcin E492 system components | |
| Gene alleles | mrkA-4, mrkB-1, mrkF-38, rkH-15, mrkI-18,mrkJ-1/wzi-143, wzc-937 | fyuA | irp1-148, irp2-145/kvgA-2 | ybtA-1, ybtA-39, ybtE-69, ybtP-4, ybtQ-60, ybtS-6, ybtT-39, ybtU-2, ybtX-62 | mceA-1, mceC-1, mceD-3, mceE-2, mceH-5 | |
2.2. Biofilm Formation Detection
In this study, the K. pneumoniae isolate tested was considered a moderate biofilm producer.
The obtained values of optical density (OD) of negative control, cut-off, ST 405, and two K. pneumoniae ATCC strains (ATCC 700603 and ATCC 13883 used as controls) are reported in Table 3.
Table 3.
Optical density (O.D.) values.
| OD 570 | |
|---|---|
| Negative control (median values) | 0.059 |
| Standard deviation | 0.005 |
| Cut-off (median values) | 0.071 |
| K. pneumoniae ST405 (median values) | 0.215 |
| ATCC 700603 (median values) | 0.765 |
| ATCC 13883 (median values) | 0.102 |
3. Discussion
In summary, the retrospective WGS analysis of the K. pneumoniae isolate allowed us to define a comprehensive overview of the genetic profile, which is useful for gaining insights into the molecular characterization of antibiotic resistance, virulence potential, and plasmid content [19].
The identified virulence determinants may have contributed to the bacterial attachment/penetration and consequently to the infection and/or colonization severity of K. pneumoniae, which are very well adapted to their host environment.
The manifestation of adhesins is principally significant in the colonization phase, when mechanical forces such as peristalsis and salivary secretions impede bacterial attack of the host. The manifestation of type-3 fimbriae is known to be involved in biofilm development on biotic and abiotic sides of medical devices in a hospital environment. Capsules can also play a central role in Klebsiella spp. persistence inside and outside human hosts by repelling complement-mediated lysis or phagocytosis and contributing some defense against environmental dehydration. They may also have a counteracting effect on antibodies as a result of the release of excessive capsular material. Iron scavenging is significant in infections since hosts have little free iron under physiological settings, and the existence of multiple iron acquisition systems may confirm optimal iron acquisition in diverse host environments. Microcin E492, a bacteriocin active against members of the Enterobacteriaceae family, has been exposed to induce apoptosis in human cell lines, which may offer a benefit during gastrointestinal colonization by helping selective environmental fitness or playing a direct part in virulence [20,21].
Our results confirm that despite the in vitro resistance to the antibiotic therapy used, the patient survived the infection thanks to the early use of combined therapy at high doses of tigecycline and the immediate removal of the peripherally inserted central catheter (PICC). Retrospective studies that also included patients with severe infections due to K. pneumoniae (KPC-Kp) that produce blaKPC have also described the importance of early onset of double and or triple combination therapy in reducing mortality at 14 days in pan-resistant isolates [22,23].
Finally, other studies have suggested that a double combination of CS (Colistin) plus TGC (Tigecycline) is synergic to reducing biofilms on in vitro catheter models, but only at high concentrations of both drugs is TGC effective in the reduction of biofilm cells [24,25].
4. Materials and Methods
4.1. Patient Information, Bacterial Isolation, and Identification
The strain was isolated from a peripherally inserted central catheter (PICC) applied in a 54-year-old female patient hospitalized in the University Hospital of Palermo, Italy, in 2014. The patient signed an informed consent form before the recovery.
The patient was admitted for emergency surgery on 16 September 2013, for colon surgery; after 18 days, she was transferred to the intensive care unit until 18 March 2014, when she was transferred to the internal medicine unit. After 5 days, a strain of K. pneumoniae was isolated from the PICC.
The same strain was isolated from all blood cultures performed.
At this time, K. pneumoniae sequence type (ST) 258 producing K. pneumoniae carbapenemase (KPC-Kp) bloodstream infection was reported in postoperative abdominal surgery patients [26]. Therefore, due to the patient’s critical condition and suspicion of surgical complication, an empiric double combination antibiotic treatment with high-dose tigecycline (100 mg every 12 h) and colistin at a dosage of 5 mg/kg/day divided in three equal doses was started, the PICC was removed, and a susceptibility test was requested. The patient was discharged on 13 May 2014.
A Becton-Dickinson Phoenix™ automated system (Becton Dickinson, Sparks, MD, USA) was used for species and antimicrobial susceptibility tests. Susceptibility profiles were understood according to the European Committee on Antimicrobial Susceptibility Testing breakpoints criteria, while susceptibility to colistin and tigecycline was confirmed by the MicroScan system (Beckman Coulter, Inc., Brea, CA, USA).
The strain showed resistance to multiple clinically used antibiotics: beta-lactams, aminoglycosides, antipseudomonal penicillin and beta-lactamase inhibitors, extended and non-extended spectrum cephalosporins, fluoroquinolones, sulfonamides, monobactams, penicillin and beta-lactamase inhibitors, antipseudomonal fluoroquinolones, and colistin. The isolate showed susceptibility for only tigecycline, as shown in Table 4.
Table 4.
MIC values obtained.
| Antibiotics | MIC Values µg/mL |
|---|---|
| IMP | >8 |
| MEP | >8 |
| ETP | >1 |
| CIP | >1 |
| AUG | >8/2 |
| CXM | >8 |
| CTX | >4 |
| CAZ | >8 |
| FEP | >8 |
| SXT | >4/76 |
| CN | >4 |
| ATM | >16 |
| TZP | >16/4 |
| FOS | 32 |
| TGC | 2 |
| CS | 4 |
MIC: minimal inhibitory concentration; IMI: imipenem; MEP: meropenem; ETP: ertapenem; CIP: ciprofloxacin; AUG: amoxicillin/clavulanic acid; CXM: cefuroxime; CTX: cefotaxime; CAZ: ceftazidime; FEP: cefepime; SXT: trimethoprim-sulfamethoxazole; CN: gentamycin; ATM: aztreonam; TZP: piperacillin/tazobactam; FOS: fosfomycin c/G6P; TGC: tigecycline; CS: colistin.
4.2. Molecular Genotyping
The bacteria grown on blood agar for 18 h were used for the DNA extraction and for sequencing using the QIAmp® DNA Mini kit Qiagen (QIAGEN GmbH, Hilden, Germany). A NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) was used to analyze the quantity and purity of the DNA.
A Nextera XT Library Prep Kit (Illumina, San Diego, CA, USA) was used for library preparation and a NextSeq Mid-Output kit v2 (300-cycles) for sequencing on a NextSeq 500 platform (Illumina). The sequencing generated 3,534,792 paired-end reads checked for quality using FASTQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/, accessed on 10 May 2021), trimmed with Sickle (https://github.com/najoshi/sickle, accessed on 10 May 2021), and assembled using SPAdes V3.7.0 [27]. The assembly yielded 207 contigs >500 bp with 137-fold coverage. The submitted sequences have a combined length of 5,571,183 bp with a G + C content of 58% and an N50 of 82,335 nucleotides. The contiguous sequences were annotated using the NCBI Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP; http://www.ncbi.nim.nih.gov/genome/annotation_prok/, accessed on 10 May 2021).
The BIGSdb-Kp database (http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html, accessed on 10 May 2021) was used to conduct multilocus sequence typing (MLST) analysis and to define antibiotic resistance and virulence-related mechanisms.
Phyloviz software based on the goeBURST algorithm was used to visualize the evolutionary relationships among the isolates [28,29].
The PlasmidFinder-1.3 web tool (Center for Genomic Epidemiology, Lyngby, Denmark; http://www.genomicepidemiology.org (accessed on 10 May 2021); ID 95%) was used to define the replicon plasmid content type.
4.3. Quantitative Biofilm Production Assay
Three wells of a 96-well flat-bottomed plastic tissue culture plate were filled with 180 μL of Luria–Bertani (LB) supplemented with 1% glucose and 20 μL of overnight culture diluted to a final optical density of 600 (OD 600) = 0·1 were used for quantification of biofilm production. As a negative control, sterile LB supplemented with 1% glucose was used, while the strain type K. pneumoniae ATCC 13883 was selected as positive control. After incubation at 37 °C for 18 h, each well was washed three times with PBS, dehydrated for 1 h at 60 °C, and marked for 15 min with 180 μL of 2% Hucker’s crystal violet (Sigma-Aldrich Corporation, St. Louis, MO, USA). The dye bound to the adherent cells was solubilized with 180 μL of 33% (v/v) glacial acetic acid, and the absorbance was measured at 570 nm (OD 570). The assay was done in triplicate and repeated four times [30,31].
The OD cut-off (ODc) was defined as three standard deviations above the mean OD of the negative control.
The ability of biofilm formation was evaluated on the basis of the adherence capabilities into the following categories: non-biofilm producers (OD ≤ ODc), weak biofilm producers (ODc < OD ≤ 2 × ODc), moderate biofilm producers (2 × ODc < OD ≤ 4 × ODc), and strong biofilm producers (4 × ODc < OD).
5. Conclusions
In conclusion, the routine clinical implementation of real-time WGS and evaluation of the ability to produce biofilm in hospital settings would help to surveil nosocomial pathogens and alert clinicians to their presence. This would provide well-founded and punctual data on the emergence and spread of antibiotic resistance with the consequent possibility of preventing outbreaks by applying infection control procedures and implementing a targeted antimicrobial stewardship program. The enhancement of the surveillance program is even more essential in the case of strains with antibiotic resistance and virulence factor co-existence that could lead to life-threatening, untreatable, and invasive infections.
Author Contributions
Conceptualization: T.F., P.D.C. and A.C.; methodology: M.R.T., P.D.C. and R.V.; software: A.C. and B.G.; formal analysis: G.L.P., A.C. and B.G.; investigation: M.R.T., B.G., D.M.P., G.L.P. and R.V.; data curation: T.F. and A.C.; writing—original draft preparation: T.F., D.M.P. and A.C.; writing—review and editing: T.F., A.C., B.G., A.G. and F.L.; visualization: A.G. and F.L.; supervision: A.G. and F.L.; project administration: A.G. and F.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
All data used in the study were anonymized, according to the requirements set by the Italian Data Protection Code (leg. Decree 196/2003) and by the general authorizations issued by the Data Protection Authority. Approval by the Ethics Committee was obtained by Azienda Ospedaliera Universitaria Policlinico “P. Giaccone” of Palermo (protocol n°07/2019).
Informed Consent Statement
K. pneumoniae strain was isolated during routine diagnostics and preserved according to the local epidemiological surveillance regulations. Written informed consent for routine diagnostic and medical procedures was collected from the patient.
Data Availability Statement
The whole-genome shotgun project of K. pneumoniae has been deposited at GenBank, accession number SUB5047324, SAMN10765330.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Podschun R., Ullmann U. Klebsiella spp. as Nosocomial Pathogens: Epidemiology, Taxonomy, Typing Methods, and Pathogenicity Factors. Clin. Microbiol. Rev. 1998;11:589–603. doi: 10.1128/CMR.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broberg C.A., Palacios M., Miller V.L. Klebsiella: A long way to go towards understanding this enigmatic jet-setter. F1000Prime Rep. 2014;6:64. doi: 10.12703/P6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derakhshan S., Peerayeh S.N., Bakhshi B. Association Between Presence of Virulence Genes and Antibiotic Resistance in ClinicalKlebsiella PneumoniaeIsolates. Lab. Med. 2016;47:306–311. doi: 10.1093/labmed/lmw030. [DOI] [PubMed] [Google Scholar]
- 4.Pitout J.D., Nordmann P., Poirel L. Carbapenemase-Producing Klebsiella pneumoniae, a Key Pathogen Set for Global Nos-ocomial Dominance. Antimicrob. Agents Chemother. 2015;29:5873–5874. doi: 10.1128/AAC.01019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muggeo A., Guillard T., Klein F., Reffuveille F., François C., Babosan A., Bajolet O., Bertrand X., de Champs C. Spread of Klebsiella pneumoniae ST395 non-susceptible to carbapenems and re-sistant to fluoroquinolones in North-Eastern France. J. Glob. Antimicrob. Resist. 2018;13:98–103. doi: 10.1016/j.jgar.2017.10.023. [DOI] [PubMed] [Google Scholar]
- 6.Andrade L.N., Curiao T., Ferreira J.C., Longo J.M., Clímaco E.C., Martinez R., Bellissimo-Rodrigues F., Basile-Filho A., Evaristo M.A., Del Peloso P.F., et al. Dissemination of blaKPC-2 by the Spread of Klebsiella pneumoniae Clonal Complex 258 Clones (ST258, ST11, ST437) and Plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae Species in Brazil. Antimicrob. Agents Chemother. 2011;55:3579–3583. doi: 10.1128/AAC.01783-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geraci D., Bonura C., Giuffrè M., Saporito L., Graziano G., Aleo A., Fasciana T., Di Bernardo F., Stampone T., Palma D., et al. Is the monoclonal spread of the ST258, KPC-3-producing clone being replaced in southern Italy by the dissemination of multiple clones of carbapenem-nonsusceptible, KPC-3-producing Klebsiella pneumoniae? Clin. Microbiol. Infect. 2015;21:e15–e17. doi: 10.1016/j.cmi.2014.08.022. [DOI] [PubMed] [Google Scholar]
- 8.Bonura C., Giuffrè M., Aleo A., Fasciana T., Di Bernardo F., Stampone T., Giammanco A., Palma D.M., Mammina C. The MDR-GN Working Group an Update of the Evolving Epidemic of blaKPC Carrying Klebsiella pneumoniae in Sicily, Italy, 2014: Emergence of Multiple Non-ST258 Clones. PLoS ONE. 2015;10:e0132936. doi: 10.1371/journal.pone.0132936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mammina C., Bonura C., Di Bernardo F., Aleo A., Fasciana T., Sodano C., Saporito M.A., Verde M.S., Tetamo R., Palma D.M. Ongoing spread of colistin-resistant Klebsiella pneumoniae in different wards of an acute general hospital, Italy, June to December 2011. Euro Surveill. 2012;17:20248. doi: 10.2807/ese.17.33.20248-en. [DOI] [PubMed] [Google Scholar]
- 10.Bengoechea J.A., Pessoa J.S. Klebsiella pneumoniae infection biology: Living to counteract host defences. FEMS Microbiol. Rev. 2019;43:123–144. doi: 10.1093/femsre/fuy043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mammina C., Bonura C., Aleo A., Fasciana T., Brunelli T., Pesavento G., Degl’Innocenti R., Nastasi A. Sequence type 101 (ST101) as the predominant carbapenem-non-susceptible Klebsiella pneumoniae clone in an acute general hospital in Italy. Int. J. Antimicrob. Agents. 2012;39:543–545. doi: 10.1016/j.ijantimicag.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 12.Gona F., Barbera F., Pasquariello A.C., Grossi P., Gridelli B., Mezzatesta M.L., Caio C., Stefani S., Conaldi P.G. In vivo multiclonal transfer of blaKPC-3 from Klebsiella pneumoniae to Escherichia coli in surgery patients. Clin. Microbiol. Infect. 2014;20:O633–O635. doi: 10.1111/1469-0691.12577. [DOI] [PubMed] [Google Scholar]
- 13.Fasciana T., Gentile B., Aquilina M., Ciammaruconi A., Mascarella C., Anselmo A., Fortunato A., Fillo S., Petralito G., Lista F., et al. Co-existence of virulence factors and antibiotic resistance in new Klebsiella pneumoniae clones emerging in south of Italy. BMC Infect. Dis. 2019;19:2019. doi: 10.1186/s12879-019-4565-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: New A Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cubero M., Marti S., Domínguez M.Á., González-Díaz A., Berbel D., Ardanuy C. Hy-pervirulent Klebsiella pneumoniae serotype K1 clinical isolates form robust biofilms at the air-liquid interface. PLoS ONE. 2019;14:e0222628. doi: 10.1371/journal.pone.0222628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huynh D.T.N., Kim A.Y., Kim Y.R. Identification of Pathogenic Factors in Klebsiella pneumoni-ae Using Impedimetric Sensor Equipped with Biomimetic Surfaces. Sensors. 2017;17:1406. doi: 10.3390/s17061406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nirwati H., Sinanjung K., Fahrunissa F., Wijaya F., Napitupulu S., Hati V.P., Hakim M.S., Meliala A., Aman A.T., Nuryastuti T. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC Proc. 2019;13(Suppl. 11):20. doi: 10.1186/s12919-019-0176-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khatoon Z., McTiernan C.D., Suuronen E.J., Mah T.F., Alarcon E.I. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon. 2018;4:e01067. doi: 10.1016/j.heliyon.2018.e01067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin J., Phan H.T., Findlay J., Stoesser N., Pankhurst L., Navickaite I., De Maio N., Eyre D.W., Toogood G., Orsi N.M., et al. Covert dissemination of car-bapenemase-producing Klebsiella pneumoniae (KPC) in a successfully controlled outbreak: Long- and short-read whole-genome sequencing demonstrate multiple genetic modes of transmission. J. Antimicrob. Chemother. 2017;72:3025–3034. doi: 10.1093/jac/dkx264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rimoldi S.G., Gentile B., Pagani C., Di Gregorio A., Anselmo A., Palozzi A.M., Fortunato A., Pittiglio V., Ridolfo A.L., Gismondo M.R., et al. Whole genome sequencing for the molecular characterization of carbapenem-resistant Klebsiella pneumoniae strains iso-lated at the Italian ASST Fatebenefratelli Sacco Hospital, 2012–2014. BMC Infect. Dis. 2017;17:666. doi: 10.1186/s12879-017-2760-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gentile B., Grottola A., Orlando G., Serpini G.F., Venturelli C., Meschiari M., Anselmo A., Fillo S., Fortunato A., Lista F., et al. A Retrospective Whole-Genome Sequencing Analysis of Carbapenem and Colistin-Resistant Klebsiella pneumoniae Nosocomial Strains Isolated during an MDR Surveillance Program. Antibiotics. 2020;9:246. doi: 10.3390/antibiotics9050246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tumbarello M., Trecarichi E.M., De Rosa F.G., Giannella M., Giacobbe D.R., Bassetti M., Losito A.R., Bartoletti M., Del Bono V., Corcione S., et al. Infections caused by KPC-producing Klebsiella pneumoniae: Differences in therapy and mortalityin a multicentre study. J. Antimicrob. Chemother. 2015;70:2133–2143. doi: 10.1093/jac/dkv086. [DOI] [PubMed] [Google Scholar]
- 23.Daikos G.L., Tsaousi S., Tzouvelekis L.S., Anyfantis I., Psichogiou M., Argyropoulou A., Stefanou I., Sypsa V., Miriagou V., Nepka M., et al. Carbapenemase-Producing Klebsiella pneumoniae Bloodstream Infections: Lowering Mortality by Antibiotic Combination Schemes and the Role of Carbapenems. Antimicrob. Agents Chemother. 2014;58:2322–2328. doi: 10.1128/AAC.02166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zusman O., Avni T., Leibovici L., Adler A., Friberg L., Stergiopoulou T., Carmeli Y., Paul M. Systematic review and me-ta-analysis of in vitro synergy of polymyxins and carbapenems. Antimicrob. Agents Chemother. 2013;57:5104–5111. doi: 10.1128/AAC.01230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mataraci Kara E., Ozbek Celik B. Investigation of the effects of various antibiotics against Klebsiella pneu-moniae biofilms on in vitro catheter model. J. Chemother. 2018;30:82–88. doi: 10.1080/1120009X.2017.1390633. [DOI] [PubMed] [Google Scholar]
- 26.Di Carlo P., Gulotta G., Casuccio A., Pantuso G., Raineri M., Farulla C.A., Bonventre S., Guadagnino G., Ingrassia D., Cocorullo G., et al. KPC—3 Klebsiella pneumoniaeST258 clone infection in postoperative abdominal surgery patients in an intensive care setting: Analysis of a case series of 30 patients. BMC Anesth. 2013;13:1–8. doi: 10.1186/1471-2253-13-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brhelova E., Antonova M., Pardy F., Kocmanova I., Mayer J., Racil Z., Lengerova M. Investigation of the next-generation sequencing data of Klebsiella pneumoniae using web-based tools. J. Med. Microbiol. 2017;66:1673–1683. doi: 10.1099/jmm.0.000624. [DOI] [PubMed] [Google Scholar]
- 28.Francisco A.P., Bugalho M., Ramirez M., Carrico J.A. Global Optimal eBURST analysis of Multilocus typing data using a raphic matroid approach. BMC Bioinform. 2009;10:1–15. doi: 10.1186/1471-2105-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Francisco A.P., Vaz C., Monteiro P.T., Melo-Cristino J., Ramirez M., Carriço J.A. PHYLOViZ: Phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinform. 2012;13:87. doi: 10.1186/1471-2105-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calà C., Amodio E., Di Carlo E., Virruso R., Fasciana T., Giammanco A. Biofilm production in Staphylococcus epidermidis strains, isolated from the skin of hospitalized patients: Genetic and phenotypic characteristics. New Microbiol. 2015;38:521–529. [PubMed] [Google Scholar]
- 31.Vuotto C., Longo F., Pascolini C., Donelli G., Balice M.P., Libori M.F., Tiracchia V., Salvia A., Varaldo P.E. Varaldo. Biofilm formation and antibiotic resistance in Klebsiella pneumoniae urinary strains. J. Appl. Microbiol. 2017;123:1003–1018. doi: 10.1111/jam.13533. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The whole-genome shotgun project of K. pneumoniae has been deposited at GenBank, accession number SUB5047324, SAMN10765330.