Abstract
Background:
Phosphatidylethanol (PEth) homologs are ethanol metabolites used to identify and monitor alcohol drinking in humans. In this study, we measured levels of the two most abundant homologs, PEth 16:0/18:1 and 16:0/18:2, in whole blood samples from rhesus macaque monkeys that drank ethanol daily ad libitum to assess the relationship between PEth levels and recent ethanol exposure in this animal model.
Methods:
Blood samples were obtained from The Monkey Alcohol Tissue Research Resource. The monkeys were first induced to consume 4% (w/v) ethanol in water from a panel attached to their home cage. Then, monkeys were allowed to drink ethanol and water ad libitum 22 hours daily for 12 months and the daily amount of ethanol each monkey consumed was measured. Whole, uncoagulated blood was collected from each animal at the end of the entire experimental procedure. PEth 16:0/18:1 and 16:0/18:2 levels were analyzed by HPLC with tandem mass spectrometry and the ethanol consumed during the preceding 14 days was measured. Combined PEth was the sum of concentrations of both homologs.
Results:
Our results show that 1) PEth accumulates in the blood of rhesus monkeys after ethanol consumption; 2) PEth homolog levels were correlated with the daily average ethanol intake during the 14-day period immediately preceding blood collection; 3) application of established human PEth 16:0/18:1 cutoff concentrations indicative of light social and no ethanol consumption (< 20 ng/ml), moderate ethanol consumption (≥ 20 and < 200 ng/ml) and heavy consumption (≥ 200 ng/ml) predicted significantly different ethanol intake in these animals. PEth homologs were not detected in ethanol-naïve controls.
Conclusions:
This study confirms that PEth is a sensitive biomarker for ethanol consumption in rhesus macaque monkeys. This non-human primate model may prove useful to test for sources of variability previously shown between ethanol consumption and PEth homolog levels amongst humans.
Keywords: Phosphatidylethanol (PEth), Blood, Ethanol, Rhesus Monkey, Phospholipase D (PLD)
INTRODUCTION
Alcohol use disorder presents a serious public health and economic burden, and biomarkers that allow monitoring of drinking over time are important in clinical and forensic applications. Phosphatidylethanol (PEth), a metabolite of ethanol, is a direct biomarker of alcohol consumption in common use today (Gnann et al., 2010; Helander and Zheng, 2009; Varga et al., 1998). PEth accumulates in human red blood cells (Aradottir et al 2004) and is conveniently quantified in whole blood samples. As an ethanol biomarker, PEth is specific because it is formed exclusively after ethanol has been consumed (Alling et al., 1983; Gustavsson and Alling, 1987; Kobayashi and Kanfer, 1987). It is sensitive because it is detectable after ingesting one or two standard alcohol drinks (Javors et al., 2016). Because of this sensitivity and specificity, PEth has been used to detect recent alcohol use (Bajunirwe et al., 2014, Magidson et al., 2019; Muyindike et al., 2017) and to follow outcomes in clinical trials of alcohol interventions (Eyawo et al., 2018; Hahn et al., 2018).
PEth is a cell membrane phospholipid that is synthesized by the enzyme phospholipase D (PLD) in many human and animal tissues (Aradottir et al., 2002). While PEth is rapidly eliminated in all species and tissues studied to date, PEth accumulates in human red blood cells because these cells lack a degrading mechanism (Aradottir et al 2004). To date, 48 homologs of PEth have been identified (Gnann et al., 2010). These homologs are differentiated by the number of carbons and double bonds present in their two lipid tails. The two most abundant of the 48 PEth homologs are PEth 16:0/18:1 and PEth 16:0/18:2 (Helander and Zhang, 2009) and their elimination half-lives are 7.8 days and 6.4 days, respectively (Javors et al., 2016; Lopez-Cruzan et al., 2018).
Although PEth levels observed in blood are functionally related to recent alcohol consumption in humans, it is becoming clear that PEth accumulation differs across individuals, even after they ingest the same amount of ethanol over the same period of time (Gnann et al., 2012; Hahn et al., 2016; Helander et al., 2012; Hill-Kapturczak et al., 2018; Javors et al., 2016; Lopez-Cruzan et al., 2018; Schrock et al., 2017). Identifying the sources of this variation and refining the utility of PEth remains a critical limitation in the application of PEth to reveal recent patterns of drinking accurately in clinical and forensic situations (Hahn et al., 2016). In spite of this demonstrated variability of PEth synthesis versus ethanol consumption among humans, specific concentration cutoffs of PEth 16:0/18:1 have been established to estimate 3 categories of consumption, 1) light social and no drinking (<20 ng/ml), 2) moderate drinking (≥20 and <200 ng/ml) and 3) heavy, harmful drinking (≥200 ng/ml) (Helander et al, 2012; Ulwelling and Smith, 2018).
Preclinical models of ethanol consumption provide greater experimental control over ethanol dose and access compared with human clinical studies. Thus, a preclinical model of PEth, as a biomarker of recent drinking, could help us understand the sources of variability and improve our interpretation of PEth levels found in blood samples of humans. Rodents are commonly used as a preclinical model of ethanol consumption, but in rodent blood, PEth is rapidly eliminated by the action of degrading enzymes (Aradottir et al., 2004), which, as previously mentioned, are absent in human blood (Aradottir et al., 2004; Selle et al., 1992). This prevents using rodents to examine long-term changes in PEth as a function of ethanol exposure.
Nonhuman primates, specifically rhesus macaque (Macaca mulatta) monkeys, are a less common preclinical model of ethanol consumption; however, they chronically consume intoxicating amounts of ethanol, have similar ethanol absorption and metabolism (Grant and Bennett, 2003) and they exhibit remarkably high genome homology to humans (Gibbs et al., 2007). Whether PEth accumulation in rhesus monkeys is comparable to that observed in humans remains unclear. Thus, in this study we sought to assess 1) whether PEth accumulates in rhesus red blood cells after long-term, stable daily ethanol consumption and 2) whether PEth levels measured in blood obtained from these monkeys are related to ethanol consumption during the previous two weeks. Demonstrating such a relationship would strengthen the argument for using nonhuman primates as a preclinical model of PEth accumulation, allowing more controlled studies of this relationship than are possible in human studies.
MATERIALS AND METHODS
Blood Samples and Rhesus Macaques
Subjects in these experiments were 39 male rhesus macaques (Macaca mulatta), with an average age of 7.80 years (SD ± 1.8) and a mean weight of 9.12 kg (SD ± 1.2). Of the 39 subjects, 31 were ethanol drinkers and 8 were ethanol-naïve controls. Animals were born and raised at the Oregon National Primate Research Center. All animals were handled according to the Guide for the Care and Use of Laboratory Animals and the protocols approved by the Oregon Health & Science University Institutional Animal Care and Use Committee.
The monkeys were trained to comply with venipuncture without the use of anesthesia, allowing acquisition of the blood sample (3 ml) used to measure PEth homologs after approximately 12 months of open-access drinking. Blood samples were collected from the femoral vein into chilled ethylenediaminetetraacetic acid (EDTA) tubes (Bectin Dickinson, Franklin Lakes, NJ). Samples were stored on ice until centrifuged (3,200xg, 20 minutes, 4°C; Model Allegra 21R, Beckman Coulter, Fullerton, CA, USA). Plasma and packed blood cells were aliquoted and frozen at −80°C in 2-ml microtubes. All blood samples were obtained from the Monkey Alcohol Tissue Research Resource (MATRR, Baylor University, Waco, Texas). MATRR is a biobank that provides longitudinal and postmortem tissue, including blood samples, from monkeys under an ethanol self-administration protocol. The subjects used in these experiments were enrolled in cohorts 4, 5, 7a and 7b (see www.MATRR.com for details).
Ethanol Self-Administration
Self-administration training and experimental procedures have been described (Baker et al., 2014; Grant et al., 2008). Briefly, food and fluid delivery were provided via an operant panel present in each cage. Monkeys obtained access to water and ethanol by pulling a dowel that opened a solenoid valve for each fluid. Food was delivered when the dowel pull was paired with a finger poke, registered by disrupting an infrared beam. After animals learned to obtain all food and fluids from the operant panel, drinking was induced by timed, regular delivery of food pellets. Over approximately 30 days, monkeys were induced to drink water by delivering a 1 g banana-flavored pellet (Granville Milling Co., Creedmoor, NC) every 5 minutes until monkeys drank 47.5 ml/kg (equivalent to the volume of 4% w/v ethanol), resulting in a dose of 1.5 g/kg. Subsequently, monkeys were induced to drink increasing doses of ethanol (4% w/v in water) by increasing the amount of fluid each animal consumed before timed pellet delivery ceased, resulting in monkeys consuming 0.5 g/kg/day, 1.0 g/kg/day and 1.5 g/kg/day for approximately 30 days at each dose. Following induction, food was available in three meals, each separated by two hours, and monkeys had open access to ethanol and water for 22 hours each day (Grant et al., 2008).
PEth 16:0/18:1 Cutoffs to Reflect Ethanol Drinking
Recommended and published concentration cutoffs of PEth 16:0/18:1 have been established by consensus to estimate 3 categories of alcohol consumption in humans (Helander et al, 2012; Ulwelling and Smith, 2018): 1) light social and no drinking (<20 ng/ml), 2) moderate drinking (≥20 and <200 ng/ml) and 3) heavy, harmful drinking (≥200 ng/ml).
Sample Preparation for HPLC with Mass Spectrometry Detection
PEth 16:0/18:1 and 16:0/18:2 homologs were quantified in EDTA–treated whole blood samples using high-performance liquid chromatography (HPLC) with tandem mass spectrometry detection (MS/MS) as previously described (Javors et al., 2016). All solvents and reagents were HPLC analytical grade and purchased from Millipore Sigma (Sigma-Aldrich Corp., St. Louis, MO). Analytical solutions were prepared with Milli-Q Plus water (Millipore Sigma, EMD Millipore, Billerica, MA). PEth 16:0/18:1 (1-palmitoyl-2-oleoyl-phosphatidylethanol), PEth 16:0/18:2 (1-palmitoyl-2-linoleoyl-phosphatidylethanol) analytes and the deuterated dPEth 16:0/18:1 internal standard were purchased from Avanti Polar Lipids (Alabaster, AL).
Samples were thawed at room temperature on the analysis day, and 300 μl of each sample were spiked with 5 μl of each internal standard. Then, 600 μl of isopropanol were added to each sample. Specimens were vortexed for 1 minute, combined with 1 ml of hexane, shaken for 15 minutes and centrifuged for 20 minutes at 3,200xg at 4°C. The resulting supernatants were transferred to new tubes and evaporated with a gentle stream of nitrogen at 30°C to obtain the lipid residues. These residues were then dissolved in 100 μl of mobile phase A (20% 10 mM ammonium acetate with 80% acetonitrile), transferred to microfilter tubes and centrifuged at 1,000xg for 10 minutes. The eluted samples were transferred to 300 μl polypropylene autosampler vials and 10 μl were injected into the HPLC/MS/MS system.
HPLC/MS/MS Detection Method
HPLC method—
The HPLC/MS/MS method was performed as described in Javors et al., 2016. The HPLC system consisted of a Shimadzu SCL controller, 2 LC-20AD pumps with a DGU-20A degassing unit and mixing chamber, a SIL-20ACHT autosampler, a CTO-20AC column oven (all from Shimadzu Scientific Instruments, Inc., Houston, TX) and an AB Sciex API 4000 Q-TRAP mass spectrometer with turbo ion spray (AB Sciex LLC., Framingham, MA). The analytical column was a High Purity C4 (Thermo Fisher Scientific, Waltham, MA). Mobile phase A was 20% 10 mM ammonium acetate with 80% acetonitrile. For the separation and detection of the PEth analytes, a gradient elution was used in which mobile phase B (100% isopropanol) was 25% for 2 minutes, linear gradient 50% from 2 to 3 minutes, linear gradient to 100% from 3 to 6 minutes, 100% B from 6 to 7 minutes, linear gradient to 25% from 7 to 8 minutes and then, equilibration of the column at 25% for 10 minutes before the next injection.
Tandem Mass Spectrometry—
The electrospray ionization interface operated in negative ion mode using selected reaction monitoring to detect the major ion products from the deprotonated molecules of PEth 16:0/18:1 (m/z 701.7→281.5), PEth 16:0/18:2 (m/z 699.7→279.0) and deuterated dPEth 16:0/18:1 (m/z 706.7→286.5). The injection volume was 10 μl and the flow rate was 200 μl/min. The MS conditions were curtain gas 15 psi, collision gas 8 psi, ion spray voltage −4,500 V, temperature 550°C, nebulizer gas 15 psi, auxiliary gas 40 psi, declustering potential −61 V, focusing potential −350 V, entrance potential −10 V and collision energy −40 V. The lower limit of detection was estimated to be 5 ng/ml as determined using product ions m/z 281.5 for PEth 16:0/18:1 and m/z 279.0 for PEth 16:0/18:2.
Rhesus Macaque Monkey and Human PLD2 Comparison
PLD2 amino acid sequences from rhesus macaque (Macaca mulatta) and human (Homo sapiens) were obtained from the National Center for Biotechnology (NCBI) Protein Database [Macaca mulatta accession EHH24401 (Yan et al., 2011); Homo sapiens accession 014939 (Lopez et al., 1998)]. The protein sequences were uploaded to MultAlin web-based software (Corpet, 1988).
Statistical Analysis
Experimental results were statistically analyzed using paired t-test, correlation analysis and one-way ANOVA followed by Tukey’s multiple test comparisons corrected for multiple comparisons, where applicable. Statistical analysis was performed with GraphPad Prism software, version 8 (GraphPad Software Inc., La Jolla, California, USA) and R software, version 3.6.3 (Foundation for Statistical Computing, Institute for Statistics and Mathematics, Vienna, Austria). Results were considered statistically significant at P < 0.05.
RESULTS
Ethanol Consumption
Daily ethanol consumption for the 14 days prior to blood collection ranged from 0.46 to 4.8 g/kg, with a mean (SD) daily intake of 2.5 (1.0) g/kg/day and a median of 2.6 g/kg/day. The mean (SD) daily ethanol consumption for the 12-month period (consecutive daily sessions: 491 – 499 for cohort 4, 393 – 396 for cohort 5, 420 – 427 for cohort 7a and 377 – 380 for cohort 7b) ranged from 0.47 to 4.1 g/kg/day, with a mean of 2.4 (0.8) g/kg/day and a median of 2.4 g/kg/day.
Mean daily ethanol consumption during the 14-day period immediately preceding the collection of the PEth blood samples closely matched that observed during the approximately 12-months of open-access (Fig. 1) [paired t-test: t (30) = 1.4; P = 0.17; 95% CI = −0.054 to 0.29].
Fig 1: Ad-libitum Alcohol Consumption.
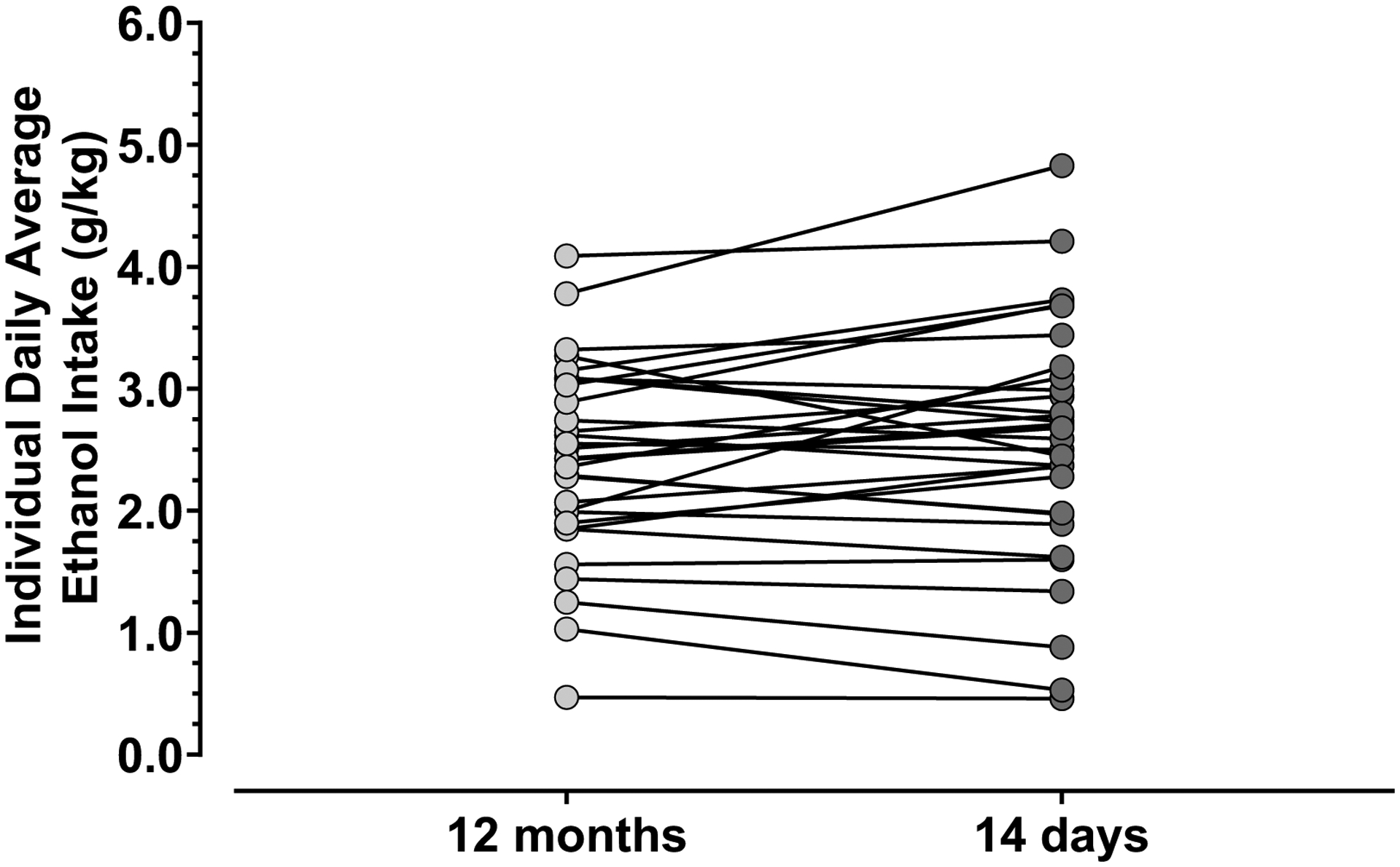
Average daily ethanol intake of each monkey (N = 31) is shown for the 12 months (light circles) of open-access drinking and the 14 days (dark circles) prior to PEth assessment. Each line between light and dark circles corresponds to the same monkey’s average ethanol consumed each day in both time periods. The average daily ethanol intake was similar between the 12-month and 14-day periods of evaluation. Statistical analysis performed with paired t-test. Ethanol-naïve controls were excluded in the graph.
Ethanol Intake and PEth Homolog Levels
PEth homologs and combined PEth were compared against daily average ethanol consumption during the 14 days prior to blood sample collection (Fig. 2). The 14-day period of ethanol consumption was chosen based on the published range of mean PEth half-lives varying from 6.4 to 7.8 days (Hill-Kapturczak et al., 2018; Javors et al., 2016; Lopez-Cruzan et al., 2018).
Fig. 2: Correlation Between Daily Average Ethanol Drinking and PEth Levels.
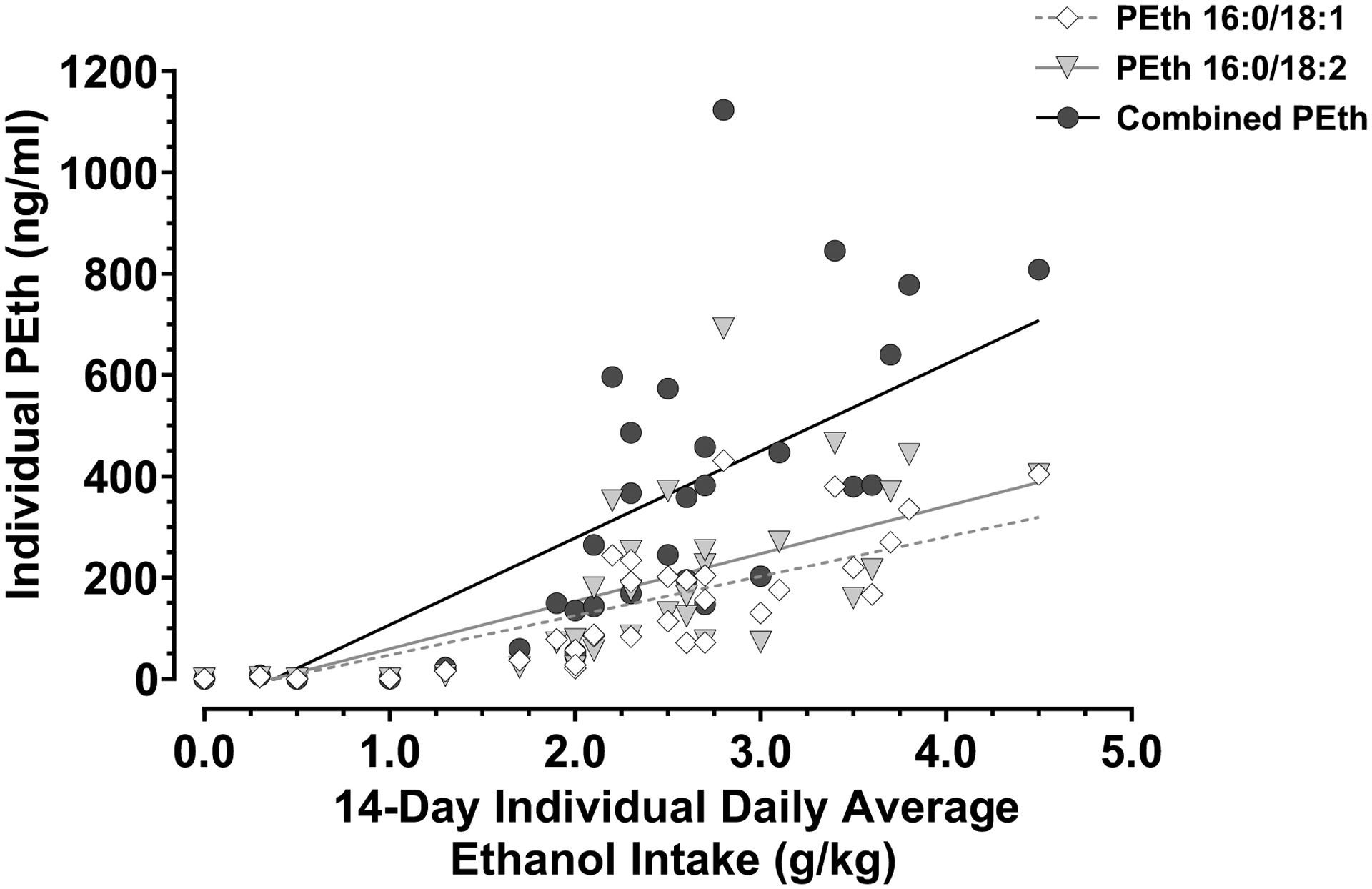
Individual PEth 16:0/18:1 (diamonds), PEth 16:0/18:2 (triangles) and combined PEth (circles) measures were analyzed against the individual daily average amount of ethanol consumed in the 14-day period before the blood draw. Pearson’s correlation analysis was performed. Ethanol-naïve controls (N = 8) were excluded from the statistical analysis, but they are shown in the graph at zero in the X and Y axes without being part of the best fit line. N = 31.
The concentrations of PEth 16:0/18:1, PEth 16:0/18:2 and the sum of PEth 16:0/18:1 and PEth 16:0/18:2 (Combined PEth) in blood were significantly correlated with the daily average ethanol consumption during the 14-day period before blood collection (Fig. 2), as shown by Pearson’s correlation: for PEth 16:0/18:1, R = 0.81, P = < 0.0001; for PEth 16:0/18:2, R = 0.72, P = < 0.0001; and for Combined PEth, R = 0.77; P = <0.0001; α = 0.05. For this correlation analysis, only samples from ethanol-exposed monkeys were considered, but ethanol-naïve controls were included in the graph to show that they lacked any PEth levels in their blood.
Ethanol Intake of Rhesus Monkeys Among Human PEth 16:0/18:1 Concentration Cutoffs
The individual mean (SD) daily ethanol consumption during the 14-day period preceding blood collection significantly differed (Fig. 3) when their blood PEth 16:0/18:1 levels were stratified into values < 20 ng/ml [0.26 (0.45) g/kg/day, range 0 to 1.3 g/kg/day, median 0.0 g/kg/day]; values ≥ 20 ng/ml and < 200 ng/ml [2.4 (0.50) g/kg/day, range 1.7 to 3.6 g/kg/day, median 2.3 g/kg/day]; and values ≥ 200 ng/ml [3.1 (0.75) g/kg/day, range 2.2 to 4.5 g/kg/day, median 3.1 g/kg/day]; [F (2, 36) = 83; P < 0.0001]. The daily average ethanol intake of monkeys with blood PEth 16:0/18:1 levels < 20 ng/ml was significantly lower than the daily average ethanol intake of monkeys with blood PEth 16:0/18:1 levels ≥ 20 ng/ml and < 200 ng/ml [0.26 (0.45) g/kg/day vs. 2.4 (0.50) g/kg/day; mean difference = 2.2; 95% CI of difference = 1.6 to 2.7; adjusted P < 0.0001]; and lower than the daily average ethanol intake of monkeys with blood PEth 16:0/18:1 levels ≥ 200 ng/ml [0.26 (0.45) g/kg/day vs. 3.1 (0.75) g/kg/day; mean difference = 2.9; 95% CI of difference = 2.3 to 3.5; adjusted P < 0.0001]. Also, the daily average ethanol intake of monkeys with blood PEth 16:0/18:1 levels ≥ 20 ng/ml and < 200 ng/ml was significantly lower than the daily average ethanol intake of monkeys with blood PEth 16:0/18:1 levels ≥ 200 ng/ml [2.4 (0.50) g/kg/day vs. 3.1 (0.75) g/kg/day; mean difference = 0.72; 95% CI of difference = 0.17 to 1.3; adjusted P = 0.0077]. Although there was a statistically significant difference between the means of these two groups, there was considerable overlap of the individual intake.
Fig. 3: PEth 16:0/18:1 discrimination of drinking amounts.
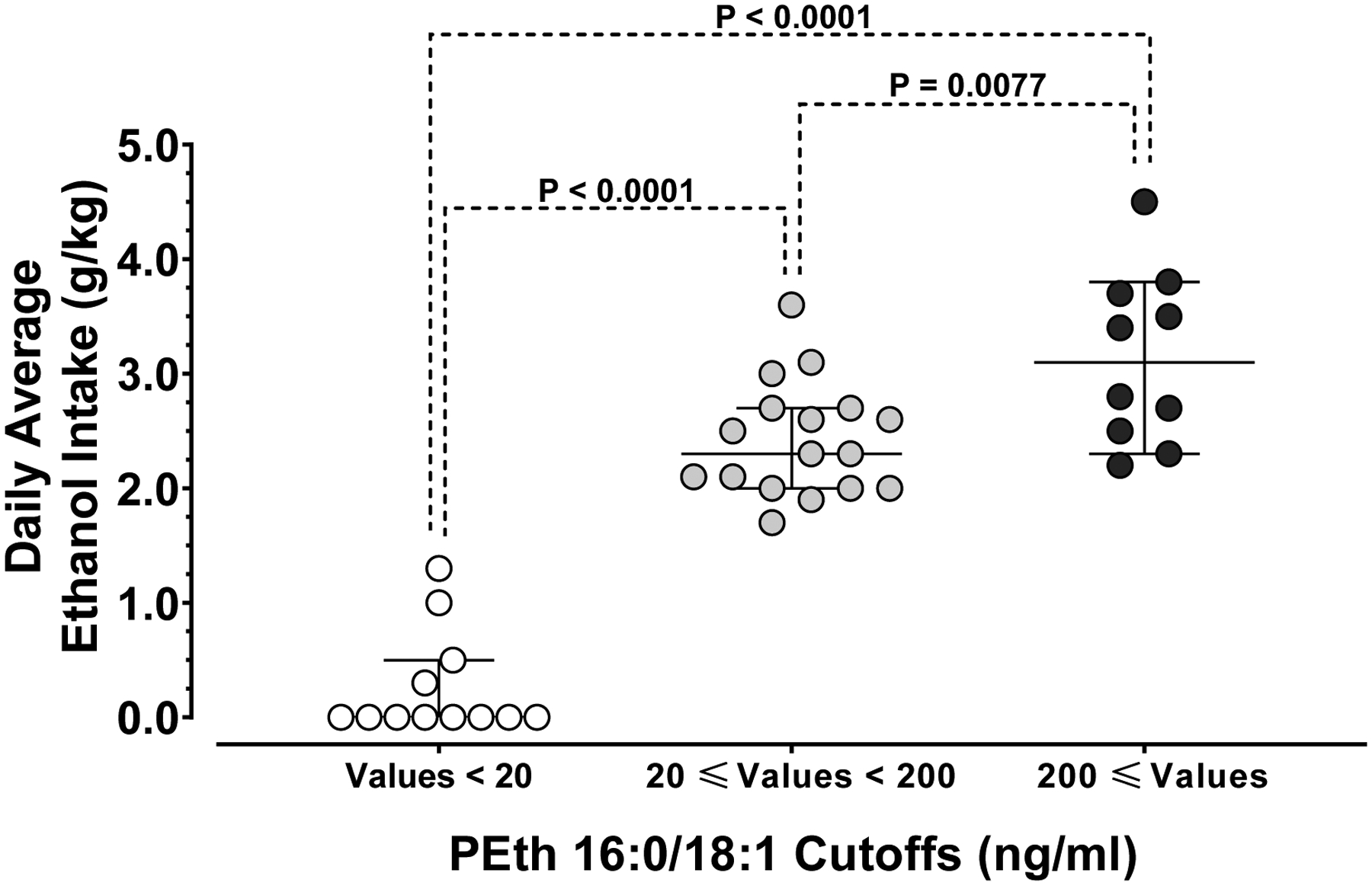
PEth 16:0/18:1 cutoffs used in human clinical or forensic evaluations (see text) were used to analyze 14-day individual daily ethanol drinking amounts in rhesus monkeys. Individual daily average ethanol intake of rhesus monkeys PEth 16:0/18:1 levels below 20 ng/ml (white circles), levels equal or above 20 ng/ml and below 200 ng/ml (grey circles) and levels equal or above 200 ng/ml. N = 12, 17 and 10, respectively. Medians are shown as a horizontal line for each homolog and error bars represent 95% CI.
Comparison of Rhesus and Human PLD2 Protein Amino Acid Sequence
The alignment and comparison of the rhesus PLD2 amino acid sequence against human PLD2 resulted in high conserved homology (Fig. 4). Out of the 933 amino acids present in both enzymes, only 22 amino acids were substituted (2.4%) and 911 residues were conserved (98%). There were no substitutions in the first active site of the enzyme and the second active site had only one substitution, V759M. In the Phox homology (PX) phospholipid-binding domain, there were three amino acid residues substituted: S97T, R137Q, G140A and four substitutions were shown in the Pleckstrin homology (PH) domain: R202C, M208T, H288Y, Q311R.
Fig. 4: Comparison of rhesus monkey and human PLD2 amino acid sequences.
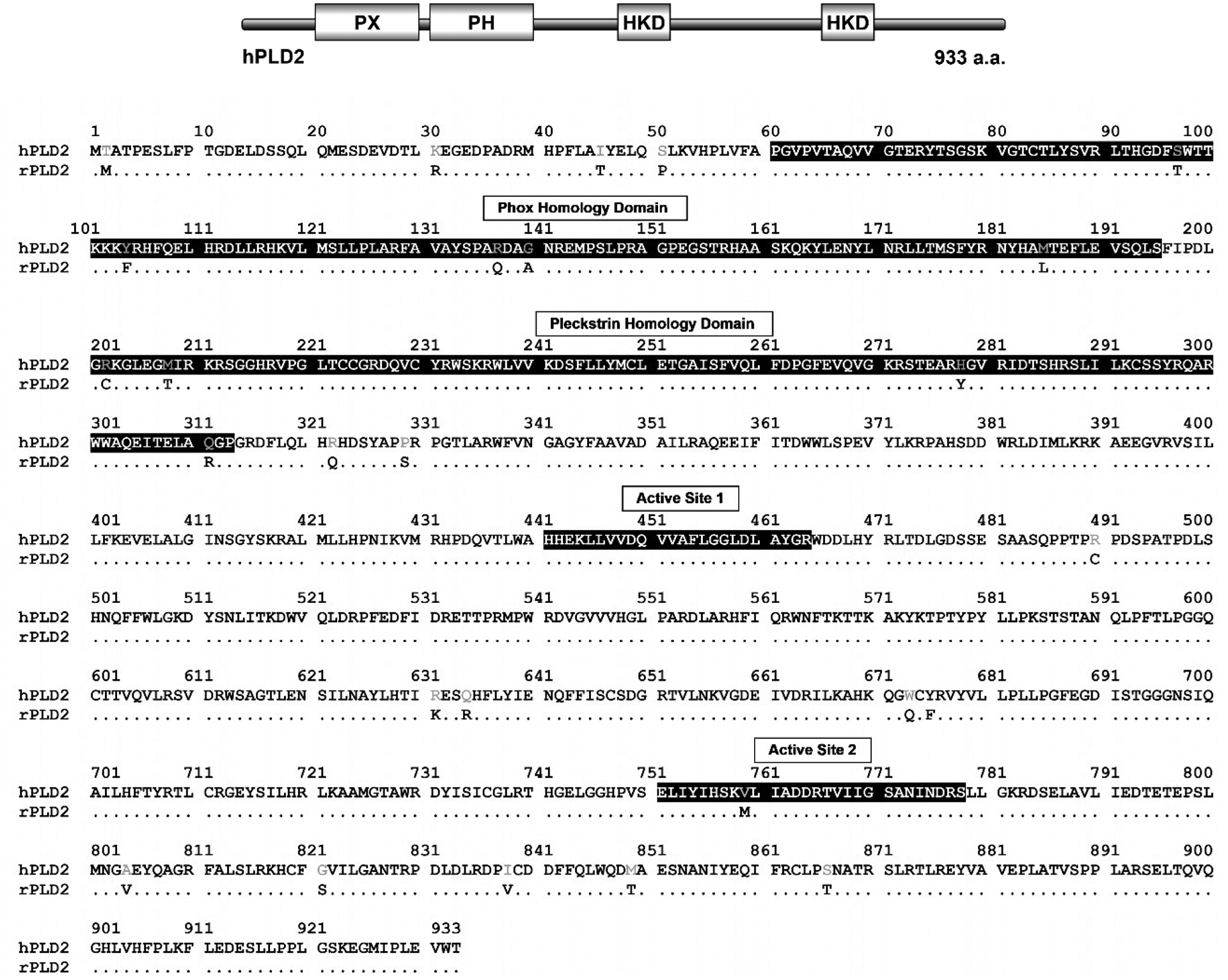
On the top is the schematic drawing of human PLD2 with its domains. PLD2 amino acid sequence from rhesus macaque was aligned with the human PLD2 protein. The human PLD2 amino acid residues are all shown. Only those residues that are not conserved in the rhesus enzyme are shown, thus, conserved residues are depicted as dots. Important protein domains are highlighted in black.
DISCUSSION
For the first time in a non-human primate model, our present study reports the functional relationship between the PEth homologs 16:0/18:1 and 16:0/18:2 and recent ethanol consumption. Specifically, our results show that 1) blood PEth concentrations in rhesus monkeys are detectable and proportional to the daily average ethanol consumption in the previous 14 days, while PEth homolog levels were undetected in ethanol-naïve monkeys and 2) when individual daily average ethanol drinking was categorized into light or no drinking, moderate and heavy consumption, the means of the individual PEth 16:0/18:1 homolog levels were able to distinguish monkeys consuming ethanol in those categories.
In addition, when we analyzed PEth 16:0/18:1 concentration cutoff levels used to predict patterns of drinking in humans, these cutoffs also detected a difference between no or light and moderate and heavy ethanol intake in these monkeys. Although statistical significance was found between the means of alcohol doses in the ≥20 ng/ml to <200 ng/ml and ≥200 ng/ml PEth 16:0/18:1 cutoff groups, there was considerable overlap. We have observed similar overlap in PEth levels among human volunteers after they consumed a single 0.4 or 0.8 g/kg ethanol dose (Hill-Kapturczak et al., 2018). Indeed, better understanding the underlying cause(s) of this discrepancy is a major motivation for our interest in developing this non-human primate model of PEth formation. Future research will include more controlled experimental designs to reveal the sources of variance that lead to this overlap we have observed in both monkeys and in humans.
The mechanism of PEth formation in rhesus macaques is unknown. In humans, PEth is formed through the catalysis of phosphatidylcholine by the enzyme PLD in the presence of ethanol (Kobayashi and Kanfer, 1987). Because PLD, specifically PLD2, catalyzes the formation of PEth in human blood, we hypothesized that a counterpart of PLD2 could be also present in rhesus blood. We could find neither studies published with rhesus PLD2 nor the crystal structure of the enzyme. Therefore, we searched for the rhesus PLD2 protein in the NIH NCBI protein database and performed an alignment of its amino acid sequence with the human PLD2 enzyme and found a difference in 2.4% of their amino acids. None of the substituted residues would alter in theory the phospholipid-binding PX or catalytic sites of the enzyme. Thus, we suspect the mechanism of PEth formation in rhesus monkeys is similar to that in humans.
The mechanism for the elimination of PEth is also unknown. In rodents, PEth is quickly degraded in all organs studied including red blood cells (Aradottir et al., 2004; Bruhl et al., 2003). In humans, PEth is rapidly degraded in all tissues studied to date except in red blood cells. The life of a human red blood cell is approximately 115 days (Franco, 2012), hence, red blood cell turnover cannot account for the average 7-day half-life of PEth. It appears that one or more of the enzymes that metabolize PEth are absent or inactive in human red blood cells (Bruhl et al., 2003; Viel, 2012), but it is unclear which enzyme or enzymes are responsible for PEth degradation. The exact elimination mechanism of PEth in monkey red blood cells will be the subject of future research.
As it has been observed in humans (Gnann et al., 2012; Hahn et al., 2016; Helander et al, 2012; Hill-Kapturczak et al., 2018; Javors et al., 2016; Lopez-Cruzan et al., 2018; and Schrock et al., 2017), there are potential sources of interindividual variability in PEth synthesis and elimination in monkeys reflected by the PEth variability seen in the present results. The specific mechanisms responsible for this variability remain unclear and should be the focus of future research. Those sources of variability might include differences in individual ethanol absorption, metabolism, and differences in the components responsible for the formation of PEth, such as concentration of the PEth substrate phosphatidylcholine, protein expression of the PLD enzyme or its activity (Hahn et al., 2016). From our results, this variability between ethanol consumption and PEth levels observed in monkeys supports our goal of establishing a monkey model for PEth biomarker discovery.
This is a preliminary study with some limitations. The monkey blood samples used to measure PEth levels were obtained from blood tissues kept in a biobank rather than from animals assigned to a hypothesis-based experiment designed for the assessment of PEth in a controlled format. Another limitation is that the mechanisms of PEth formation and degradation in monkeys are yet unknown and that there are no reports showing the pharmacokinetics of PEth in this animal model. Finally, only males were included in this study and it will be important to examine any potential differences between sexes. Despite these limitations, this model has numerous strengths that will be critical for understanding the relationship between ethanol consumption and PEth formation and degradation. For example, the contribution of patterns of ethanol consumption on variability of PEth levels, whether within-subject variation is impacted by the duration of daily ethanol intake on PEth concentration and the genetic contribution of PEth variation. Furthermore, because of the similarities in ethanol consumption, metabolism and degradation as well as genetics, we can evaluate proposed sources of PEth variability observed in humans.
Future research should assess PEth after assigning monkeys to drinking categories with fixed doses of ethanol to reduce variability originating from the current self-administration protocol. Additionally, future studies using this model should 1) test for sources of PEth variability observed in humans, such as quantitative differences in the key players of the formation and elimination of PEth and the effect that the individual ethanol absorption has on PEth levels; 2) refine the use of PEth homologs 16:0/18:1 and 16:0/18:2, as well as others such as 16:0/20:4 (Lopez-Cruzan et al., 2018) to evaluate ethanol drinking patterns and 3) examine novel treatments for alcohol disorders while consumption is monitored by PEth.
In conclusion, these data validate the macaque model of ethanol self-administration as a tool to assess PEth as a biomarker of ethanol consumption.
ACKNOWLEDGEMENTS
This publication was supported in part by a grant from NIH National Institute on Alcohol Abuse and Alcoholism (R01AA022361) to Drs. Donald Dougherty and Marty Javors, a grant from the NIH National Institute on Alcohol Abuse and Alcoholism to Dr. Brett Ginsburg (R01AA025664) and by a grant from the NIH National Institute on Alcohol Abuse and Alcoholism to Dr. Kathleen Grant (R24AA019431). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
CONFLICT OF INTEREST
None of the authors have conflicts of interest concerning this manuscript.
REFERENCES
- Alling C, Gustavsson L, Anggard E (1983) An abnormal phospholipid in rat organs after ethanol treatment. FEBS Lett 152:24–28. [DOI] [PubMed] [Google Scholar]
- Aradottir S, Lundqvist C, Alling C (2002) Phosphatidylethanol in rat organs after ethanol exposure. Alcohol Clin Exp Res 26:514–518. [PubMed] [Google Scholar]
- Aradottir S, Moller K, Alling C (2004) Phosphatidylethanol formation and degradation in human and rat blood. Alcohol Alcohol 39:8–13. [DOI] [PubMed] [Google Scholar]
- Bajunirwe F, Haberer JE, Boum Y, Hunt P, Mocello R, Martin JN, Bangsberg DR, Hahn JA (2014) Comparison of self-reported alcohol consumption to phosphatidylethanol measurement among HIV-infected patients initiating antiretroviral treatment in southwestern Uganda. PLoS One 9:e113152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EJ, Farro J, Gonzales S, Helms C, Grant KA (2014) Chronic Alcohol Self-Administration in Monkeys Shows Long-Term Quantity/Frequency Categorical Stability. Alcohol Clin Exp Res 38:2835–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brühl A, Faldum A, Löffelholz K (2003) Degradation of phosphatidylethanol counteracts the apparent phospholipase D-mediated formation in heart and other organs. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 1633:84–89. [DOI] [PubMed] [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyawo O, MacGinnis Kathleen A, Justice AC, Fiellin DA, Hahn JA, Williams EC, Gordon AJ, Marshall BD, Kraemer KL, Crystal S, Gaither JR, Edelman JE, Bryant KJ, Tate JP (2018) Alcohol and Mortality: Combining Self-Reported (AUDIT-C) and Biomarker Detected (PEth) Alcohol Measures Among HIV Infected and Uninfected. J Acquir Immune Defic Syndr. 77:135–14e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco RS (2012) Measurement of red cell lifespan and aging. Transfus Med Hemother 39:302–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnann H, Engelmann C, Skopp G, Winkler M, Auwärter V, Dresen S, Ferreirós N, Wurst FM, Weinmann W (2010) Identification of 48 homologues of phosphatidylethanol in blood by LC-ESI-MS/MS. Analytical and Bioanalytical Chemistry 396:2415–2423. [DOI] [PubMed] [Google Scholar]
- Gnann H, Weinmann W, Thierauf A (2012) Formation of Phosphatidylethanol and Its Subsequent Elimination During an Extensive Drinking Experiment Over 5 Days. Alcoholism: Clinical and Experimental Research 36:1507–1511. [DOI] [PubMed] [Google Scholar]
- Grant KA, Bennett AJ (2003) Advances in nonhuman primate alcohol abuse and alcoholism research. Pharmacol Ther 100:235–255. [DOI] [PubMed] [Google Scholar]
- Grant KA, Leng X, Green HL, Szeliga KT, Rogers LSM, Gonzales SW (2008) Drinking Typography Established by Scheduled Induction Predicts Chronic Heavy Drinking in a Monkey Model of Ethanol Self-Administration. Alcoholism: Clinical and Experimental Research 32:1824–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson L, Alling C (1987) Formation of phosphatidylethanol in rat brain by phospholipase D. Biochemical and Biophysical Research Communications 142:958–963. [DOI] [PubMed] [Google Scholar]
- Hahn JA, Anton RF, Javors MA (2016) The Formation, Elimination, Interpretation, and Future Research Needs of Phosphatidylethanol for Research Studies and Clinical Practice. Alcohol Clin Exp Res . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn JA, Cheng DM, Emenyonu NI, Lloyd-Travaglini C, Fatch R, Shade SB, Ngabirano C, Adong J, Bryant K, Muyindike WR, Samet JH (2018) Alcohol Use and HIV Disease Progression in an Antiretroviral Naive Cohort. J Acquir Immune Defic Syndr 77:492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helander A, Peter O, Zheng Y (2012) Monitoring of the alcohol biomarkers PEth, CDT and EtG/EtS in an outpatient treatment setting. Alcohol Alcohol 47:552–557. [DOI] [PubMed] [Google Scholar]
- Helander A, Zheng Y (2009) Molecular species of the alcohol biomarker phosphatidylethanol in human blood measured by LC-MS. Clin Chem 55:1395–1405. [DOI] [PubMed] [Google Scholar]
- Hill‐Kapturczak Dougherty DM, Roache JD Karns‐Wright TE, Javors MA (2018) Differences in the Synthesis and Elimination of Phosphatidylethanol 16:0/18:1 and 16:0/18:2 after Acute Doses of Alcohol. Alcohol Clin Exp Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javors MA, Hill-Kapturczak N, Roache JD, Karns-Wright T, Dougherty DM (2016) Characterization of the Pharmacokinetics of Phosphatidylethanol 16:0/18:1 and 16:0/18:2 in Human Whole Blood After Alcohol Consumption in a Clinical Laboratory Study. Alcohol Clin Exp Res 40:1228–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Kanfer JN (1987) Phosphatidylethanol formation via transphosphatidylation by rat brain synaptosomal phospholipase D. J Neurochem 48:1597–1603. [DOI] [PubMed] [Google Scholar]
- Lopez I, Arnold RS, Lambeth JD (1998) Cloning and initial characterization of a human phospholipase D2 (hPLD2). ADP-ribosylation factor regulates hPLD2. J Biol Chem 273:12846–12852. [DOI] [PubMed] [Google Scholar]
- Lopez-Cruzan M, Roache JD, Hill-Kapturczak N, Karns-Wright TE, Dougherty DM, Sanchez JJ, Koek W, Javors MA (2018) Pharmacokinetics of Phosphatidylethanol 16:0/20:4 in Human Blood After Alcohol Intake. Alcohol Clin Exp Re 42:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson JF, Fatch R, Orrell C, Amanyire G, Haberer JE, Hahn JA, META team (2019) Biomarker-Measured Unhealthy Alcohol Use in Relation to CD4 Count Among Individuals Starting ART in Sub-Saharan Africa. AIDS Behav 23:1656–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyindike WR, Lloyd-Travaglini C, Fatch R, Emenyonu NI, Adong J, Ngabirano C, Cheng DM, Winter MR, Samet JH, Hahn JA (2017) Phosphatidylethanol confirmed alcohol use among ART-naive HIV-infected persons who denied consumption in rural Uganda. AIDS Care 29:1442–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhesus Macaque Genome Sequencing and Analysis Consortium, Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, Batzer MA, Bustamante CD, Eichler EE, Hahn MW, Hardison RC, Makova KD, Miller W, Milosavljevic A, Palermo RE, Siepel A, Sikela JM, Attaway T, Bell S, Bernard KE, Buhay CJ, Chandrabose MN, Dao M, Davis C, Delehaunty KD, Ding Y, Dinh HH, Dugan-Rocha S, Fulton LA, Gabisi RA, Garner TT, Godfrey J, Hawes AC, Hernandez J, Hines S, Holder M, Hume J, Jhangiani SN, Joshi V, Khan ZM, Kirkness EF, Cree A, Fowler RG, Lee S, Lewis LR, Li Z, Liu YS, Moore SM, Muzny D, Nazareth LV, Ngo DN, Okwuonu GO, Pai G, Parker D, Paul HA, Pfannkoch C, Pohl CS, Rogers YH, Ruiz SJ, Sabo A, Santibanez J, Schneider BW, Smith SM, Sodergren E, Svatek AF, Utterback TR, Vattathil S, Warren W, White CS, Chinwalla AT, Feng Y, Halpern AL, Hillier LW, Huang X, Minx P, Nelson JO, Pepin KH, Qin X, Sutton GG, Venter E, Walenz BP, Wallis JW, Worley KC, Yang SP, Jones SM, Marra MA, Rocchi M, Schein JE, Baertsch R, Clarke L, Csuros M, Glasscock J, Harris RA, Havlak P, Jackson AR, Jiang H, Liu Y, Messina DN, Shen Y, Song HX, Wylie T, Zhang L, Birney E, Han K, Konkel MK, Lee J, Smit AF, Ullmer B, Wang H, Xing J, Burhans R, Cheng Z, Karro JE, Ma J, Raney B, She X, Cox MJ, Demuth JP, Dumas LJ, Han SG, Hopkins J, Karimpour-Fard A, Kim YH, Pollack JR, Vinar T, Addo-Quaye C, Degenhardt J, Denby A, Hubisz MJ, Indap A, Kosiol C, Lahn BT, Lawson HA, Marklein A, Nielsen R, Vallender EJ, Clark AG, Ferguson B, Hernandez RD, Hirani K, Kehrer-Sawatzki H, Kolb J, Patil S, Pu LL, Ren Y, Smith DG, Wheeler DA, Schenck I, Ball EV, Chen R, Cooper DN, Giardine B, Hsu F, Kent WJ, Lesk A, Nelson DL, O’brien WE, Prufer K, Stenson PD, Wallace JC, Ke H, Liu XM, Wang P, Xiang AP, Yang F, Barber GP, Haussler D, Karolchik D, Kern AD, Kuhn RM, Smith KE, Zwieg AS (2007) Evolutionary and biomedical insights from the rhesus macaque genome. Science 316:222–234. [DOI] [PubMed] [Google Scholar]
- Schröck A, Thierauf-Emberger A, Schürch S, Weinmann W (2017) Phosphatidylethanol (PEth) detected in blood for 3 to 12 days after single consumption of alcohol—a drinking study with 16 volunteers. Int J Legal Med 131:153–160. [DOI] [PubMed] [Google Scholar]
- Selle H, Chapman BE, Kuchel PW (1992) Release of choline by phospholipase D and a related phosphoric diester hydrolase in human erythrocytes. 1H spin-echo n.m.r. studies. Biochem J 284:61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulwelling W, Smith K (2018) The PEth Blood Test in the Security Environment: What it is; Why it is Important; and Interpretative Guidelines. J Forensic Sci 63:1634–1640. [DOI] [PubMed] [Google Scholar]
- Varga A, Hansson P, Lundqvist C, Alling C (1998) Phosphatidylethanol in Blood as a Marker of Ethanol Consumption in Healthy Volunteers: Comparison with Other Markers. Alcoholism: Clinical and Experimental Research 22:1832–1837. [PubMed] [Google Scholar]
- Viel G, Boscolo-Berto R, Cecchetto G, Fais P, Nalesso A, Ferrara SD (2012) Phosphatidylethanol in Blood as a Marker of Chronic Alcohol Use: A Systematic Review and Meta-Analysis. International Journal of Molecular Sciences 13:14788–14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G, Zhang G, Fang X, Zhang Y, Li C, Ling F, Cooper DN, Li Q, Li Y, van Gool AJ, Du H, Chen J, Chen R, Zhang P, Huang Z, Thompson JR, Meng Y, Bai Y, Wang J, Zhuo M, Wang T, Huang Y, Wei L, Li J, Wang Z, Hu H, Yang P, Le L, Stenson PD, Li B, Liu X, Ball EV, An N, Huang Q, Zhang Y, Fan W, Zhang X, Li Y, Wang W, Katze MG, Su B, Nielsen R, Yang H, Wang J, Wang X, Wang J (2011) Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nat Biotechnol 29:1019–1023. [DOI] [PubMed] [Google Scholar]


