Abstract
Marine bird populations have been declining globally with the factors driving this decline not fully understood. Viral diseases, including those caused by poxviruses, are a concern for endangered seabird species. In this study we have characterised a novel avipoxvirus, tentatively designated albatrosspox virus (ALPV), isolated from a skin lesion of an endangered New Zealand northern royal albatross (Diomedea sanfordi). The ALPV genome was 351.9 kbp in length and contained 336 predicted genes, seven of which were determined to be unique. The highest number of genes (313) in the ALPV genome were homologs of those in shearwaterpox virus 2 (SWPV2), while a further 10 were homologs to canarypox virus (CNPV) and an additional six to shearwaterpox virus 1 (SWPV1). Phylogenetic analyses positioned the ALPV genome within a distinct subclade comprising recently isolated avipoxvirus genome sequences from shearwater, penguin and passerine bird species. This is the first reported genome sequence of ALPV from a northern royal albatross and will help to track the evolution of avipoxvirus infections in this endangered species.
Keywords: avipoxvirus, complete genome, endangered, evolution, northern royal albatross
1. Introduction
Marine bird populations have been declining globally [1] with the sustainability of the albatrosses (family Diomedeidae) and large petrels (Macronectes and Procellaria spp.) being of particular concern [2,3,4]. This group includes some of the world’s most endangered bird species, with rapidly decreasing populations and their conservation status markedly deteriorating in recent years [5,6]. The northern royal albatross (Diomedea sanfordi), which is one of the largest seabirds in the world, is categorised as an “endangered” species under the International Union for Conservation of Nature (IUCN) Red List and is ranked as Category B for conservation priority [7]. The northern royal albatrosses range widely throughout the Southern Ocean, though rarely into Antarctic waters. The breeding range is restricted to the Chatham Islands and Taiaroa Head on the Otago Peninsula, Dunedin, New Zealand. The total breeding population in the Chatham Islands colonies (99% of the total) is estimated at approximately 6500–7000 pairs, which equates to a total population of 17,000 mature individuals [8]. Northern royal albatrosses are normally solitary foragers, but they may congregate at food sources at sea. Most of their food is thought to be obtained by seizing dead or dying prey from the surface and also by scavenging discards and offal from fishing boats. Breeding birds forage over the continental shelves to shelf edges in New Zealand waters. Non-breeding and young birds can be found anywhere in the Southern Ocean throughout the year, with the main wintering areas off the coasts of southern South America [8].
Human activities such as fisheries and pollution have been documented as threats for incidental mortality of these species [7,9,10,11,12,13]. Invasive alien species, degradation or loss of nesting habitats, storms and flooding, and marine pollution or plastic ingestion are also significant factors in population declines [6,7]. Infectious diseases, including those caused by avipoxviruses, have been identified as an important risk factor in the conservation of small and endangered bird populations, including albatrosses [14,15,16,17,18,19]. The impact of the introduction of avipoxviruses has been severe for the avifauna of various archipelagos [20]. For example, the emergence of an avipoxvirus with a high prevalence (88%) in Hawaiian Laysan albatrosses (Phoebastria immutabilis) enabled one of the first detailed studies of the epidemiology and population-level impact of the disease in these seabirds [21].
Avipoxviruses are large, double-stranded DNA (dsDNA) viruses comprising the genus Avipoxvirus. They occur worldwide and are known to infect a large number of wild and domestic avian species across 76 families and 20 orders [22,23,24]. The behaviour of wild birds allows avian poxviruses to reach new hosts through bird migration, species introductions, and habitat change. Avipoxviruses have been identified as an important risk factor in the conservation of endangered bird populations [19,25]. In affected birds, avipoxvirus infection can cause two different forms of disease, defined as cutaneous or diphtheritic. The cutaneous form is characterised by proliferative ‘wart-like’ lesions that commonly develop on unfeathered body areas, including the eyes, feet, legs, face and around the beak. The less common diphtheritic form is characterised by soft and yellowish cankers and proliferative lesions on the mucous membranes of the upper alimentary and respiratory tracts [23,26,27].
Little is known about the effects of poxviruses on some bird taxa, particularly for seabird species including the northern royal albatross (D. sanfordi). The aim of the present study was to characterise the genome sequence of a novel poxvirus, which was isolated from a skin lesion that was collected in 1997 from an endangered northern royal albatross on the Otago Peninsula, near Dunedin, on the South Island of New Zealand.
2. Results
2.1. Genome of ALPV
The complete genome of ALPV was assembled into a contiguous sequence of linear double-stranded DNA 351,909 bp in length (the second-largest avipoxvirus genome so far characterised) and submitted to GenBank under accession number MW365933. Like many other avipoxviruses [25,28,29], the ALPV genome contained a well-conserved central coding region surrounded by two identical inverted terminal repeat (ITR) regions, comprising 4069 bp each (coordinates 1–4069 sense and 347,841–351,909 antisense orientation). The nucleotide composition of the ALPV genome was A + T rich (69.9%), which was in agreement with other avipoxviruses isolated from yellow-eyed penguin [19], shearwater [25] and passerine bird species [30,31] (Table 1). The ALPV genome showed the highest nucleotide identities with penguinpox virus 2 (PEPV2, GenBank accession no. MW296038) (98.92%), followed by shearwaterpox virus 2 (SWPV2, GenBank accession no. KX857216) (95.75%), canarypox virus (CNPV, GenBank accession no. AY318871) (92.71%) and mudlarkpox virus (MLPV, GenBank accession no. MK903864) (88.47%) (Table 1).
Table 1.
Comparative analysis of representative avipoxviruses and ALPV based on complete genome nucleotide sequences.
| Avipoxviruses (Abbreviation) | GenBank Accession Numbers | Genome Identity (%) | Genome Length (kbp) | A + T Content (%) | Number of ORFs | References |
|---|---|---|---|---|---|---|
| Albatrosspox virus (ALPV) | MW365933 | 352 | 69.9 | 336 | This study | |
| Penguinpox virus 2 (PEPV2) | MW296038 | 98.92 | 350 | 69.9 | 327 | [19] |
| Shearwaterpox virus 2 (SWPV2) | KX857215 | 95.75 | 351 | 69.8 | 312 | [25] |
| Canarypox virus (CNPV) | AY318871 | 92.71 | 360 | 69.6 | 328 | [32] |
| Mudlarkpox virus (MLPV) | MT978051 | 88.47 | 343 | 70.2 | 352 | [30] |
| Magpiepox virus (MPPV) | MK903864 | 78.75 | 293 | 70.4 | 301 | [31] |
| Shearwaterpox virus 1 (SWPV1) | KX857216 | 61.44 | 327 | 72.4 | 310 | [25] |
| Penguinpox virus (PEPV) | KJ859677 | 49.83 | 307 | 70.5 | 285 | [33] |
| Fowlpox virus (FWPV) | AF198100 | 48.89 | 289 | 69.1 | 260 | [29] |
| Pigeonpox virus (FeP2) | KJ801920 | 47.54 | 282 | 70.5 | 271 | [33] |
| Flamingopox virus (FGPV) | MF678796 | 46.51 | 293 | 70.5 | 285 | [24] |
| Turkeypox virus (TKPV) | KP728110 | 33.39 | 189 | 70.2 | 171 | [34] |
2.2. Genome Annotation and Comparative Analyses of ALPV
The ALPV genome encoded 336 putative genes, 45 to 1936 amino acids in length, that have been numbered from left to right (Figure 1 and Table 2). Among them, four ORFs were located within the inverted terminal repeats (ITRs) and were therefore present as diploid copies. Comparative analysis of the predicted ORF sequences was performed, and a significant number of ORFs (329) were found to be homologs with other chordopoxvirus (ChPV) gene products (Table 2). Among these conserved ChPV gene products, the highest number of protein-coding genes (313) in ALPV were homologs to the recently isolated SWPV2 [25]. The remaining ten gene products (ALPV-079, -155, -163, -165, -166, -167, -168, -175, -233 and -236) were homologous to ORFs of CNPV, and a further six gene products (ALPV-003, -009, -090, -127, -229 and -334) were homologs to SWPV1 (Table 2). All conserved genes of ALPV showed the highest sequence similarity to homologs of avipoxviruses isolated from Pacific shearwater, canary and yellow-eyed penguin bird species, implying a common evolutionary history [19,25,32]. In comparison to SWPV2, two gene products (SWPV2-121 and -213) were absent from the ALPV genome, and a further nineteen genes were predicted to be truncated/fragmented (Figure 1 and Table 2). In comparison to vaccinia virus strain Copenhagen (VACV-Cop), 138 ORFs of ALPV showed homology to VACV-Cop and the sequence identities ranging from 20.9–76.7% (Table 2).
Figure 1.
Comparative genomic illustration of the novel ALPV. A sequence alignment using MAFFT in Geneious (version 10.2.2) was performed to compare ORFs between albatrosspox virus (ALPV, GenBank accession no. MW365933) and shearwaterpox virus 2 (SWPV2, GenBank accession no. KX857215). The arrows symbolise genes and open reading frames (ORFs), with orientation indicating their direction of transcription. Each gene or ORF is colour coded, as indicated by the key in the legend. The top graph represents the mean pairwise sequence identity over all pairs in the column between ALPV and SWPV2 (green: 100% identity; mustard: ≥30% and <100% identity; red: <30% identity).
Table 2.
Albatrosspox virus (ALPV) genome annotations and comparative analysis of ORFs.
| ALPV Synteny | ALPV Genome Coordinates | SWPV2 Synteny | ALPV AA Size | SWPV2 AA Size | SWPV2 BLAST Hits | ALPV AA Identity (%) Compared to Avipoxviruses | ALPV AA Identity (%) Compared to VACV-Cop | VACV BLAST Hits | Notes |
|---|---|---|---|---|---|---|---|---|---|
| ALPV-001 | 827-1342 | SWPV2-001 | 171 | 171 | SWPV2-001 hypothetical protein | 100 | identical to ALPV-336 | ||
| ALPV-002 | 2271-1645 | SWPV2-002 | 208 | 208 | SWPV2-002 C-type lectin-like protein | 100 | identical to ALPV-335 | ||
| ALPV-003 | 2553-2326 | 75 | 56.1 | SWPV1-002 C-type lectin-like protein, identical to ALPV-334 | |||||
| ALPV-004 | 2679-3347 | SWPV2-003 | 222 | 222 | SWPV2-003 conserved hypothetical protein | 100 | identical to ALPV-333 | ||
| ALPV-005 | 3464-3925 | SWPV2-004 | 153 | 134 | SWPV2-004 conserved hypothetical protein | 86.9 | identical to ALPV-332 | ||
| ALPV-006 | 4899-4042 | SWPV2-005 | 285 | 169 | SWPV2-005 C-type lectin-like protein | 46.9 | |||
| ALPV-007 | 5455-4946 | SWPV2-005 | 169 | 169 | SWPV2-005 C-type lectin-like protein | 100 | |||
| ALPV-008 | 7809-5743 | SWPV2-006 | 688 | 688 | SWPV2-006 ankyrin repeat protein | 100 | 23.9 | B4R | |
| ALPV-009 | 8828-8184 | 214 | 43.1 | SWPV1-006 ankyrin repeat protein | |||||
| ALPV-010 | 11218-9458 | SWPV2-007 | 586 | 586 | SWPV2-007 ankyrin repeat protein | 100 | 28.8 | M1L | |
| ALPV-011 | 11483-12052 | SWPV2-008 | 189 | 189 | SWPV2-008 conserved hypothetical protein | 100 | |||
| ALPV-012 | 12765-12259 | SWPV2-009 | 168 | 168 | SWPV2-009 conserved hypothetical protein | 100 | |||
| ALPV-013 | 14555-13083 | SWPV2-010 | 490 | 490 | SWPV2-010 Ig-like domain protein | 100 | |||
| ALPV-014 | 14719-16368 | SWPV2-011 | 549 | 528 | SWPV2-011 ankyrin repeat protein | 96.2 | 28.7 | M1L | |
| ALPV-015 | 16429-16935 | SWPV2-012 | 168 | 168 | SWPV2-012 C-type lectin-like protein | 100 | 24.0 | A40R | |
| ALPV-016 | 17039-18478 | SWPV2-013 | 479 | 479 | SWPV2-013 ankyrin repeat protein | 100 | 26.7 | B4R | |
| ALPV-017 | 19149-18577 | SWPV2-014 | 190 | 190 | SWPV2-014 IL-10-like protein | 100 | |||
| ALPV-018 | 20576-19266 | SWPV2-015 | 436 | 436 | SWPV2-015 ankyrin repeat protein | 100 | 27.5 | M1L | |
| ALPV-019 | 20767-22026 | SWPV2-016 | 419 | 419 | SWPV2-016 ankyrin repeat protein | 100 | 38.2 | M1L | |
| ALPV-020 | 23769-22162 | SWPV2-017 | 535 | 535 | SWPV2-017 ankyrin repeat protein | 100 | 21.5 | B4R | |
| ALPV-021 | 24886-23810 | SWPV2-018 | 358 | 358 | SWPV2-018 putative serpin | 100 | 27.9 | C12L | |
| ALPV-022 | 26242-24968 | SWPV2-019 | 424 | 424 | SWPV2-019 vaccinia C4L/C10L-like protein | 100- | |||
| ALPV-023 | 26520-27056 | SWPV2-020 | 178 | 178 | SWPV2-020 hypothetical protein | 100 | |||
| ALPV-024 | 28172-27270 | SWPV2-021 | 300 | 300 | SWPV2-021 alpha-SNAP-like protein | 100 | |||
| ALPV-025 | 29443-28295 | SWPV2-022 | 382 | 382 | SWPV2-022 ankyrin repeat protein | 100 | 22.0 | C9L | |
| ALPV-026 | 31392-29512 | SWPV2-023 | 626 | 626 | SWPV2-023 ankyrin repeat protein | 100 | |||
| ALPV-027 | 32608-31511 | SWPV2-024 | 365 | 365 | SWPV2-024 ankyrin repeat protein | 100 | |||
| ALPV-028 | 33146-32718 | SWPV2-025 | 142 | 142 | SWPV2-025 C-type lectin-like protein | 100 | |||
| ALPV-029 | 34224-33202 | SWPV2-026 | 340 | 340 | SWPV2-026 ankyrin repeat protein | 100 | 26.8 | B4R | |
| ALPV-030 | 34487-34287 | 66 | hypothetical protein, unique to ALPV, containing a transmembrane helix | ||||||
| ALPV-031 | 34467-34826 | SWPV2-027 | 119 | 119 | SWPV2-027 hypothetical protein | 100 | |||
| ALPV-032 | 35782-35054 | SWPV2-028 | 242 | 242 | SWPV2-028 Ig-like domain putative IFN-gamma binding protein | 100 | |||
| ALPV-033 | 36598-35858 | SWPV2-029 | 246 | 246 | SWPV2-029 Ig-like domain protein | 100 | |||
| ALPV-034 | 38673-36694 | SWPV2-030 | 659 | 659 | SWPV2-030 ankyrin repeat protein | 100 | 23.5 | K1L | |
| ALPV-035 | 39392-38991 | SWPV2-031 | 133 | 133 | SWPV2-031 C-type lectin-like protein | 100 | |||
| ALPV-036 | 39776-39489 | SWPV2-032 | 95 | 95 | SWPV2-032 conserved hypothetical protein | 100 | |||
| ALPV-037 | 39840-40379 | SWPV2-033 | 179 | 179 | SWPV2-033 conserved hypothetical protein | 100 | |||
| ALPV-038 | 41625-40384 | SWPV2-034 | 413 | 413 | SWPV2-034 vaccinia C4L/C10L-like protein | 99.8 | 24.3 | C10L | |
| ALPV-039 | 41743-42726 | SWPV2-035 | 327 | 327 | SWPV2-035 G protein-coupled receptor-like protein | 100 | |||
| ALPV-040 | 44520-42745 | SWPV2-036 | 591 | 591 | SWPV2-036 ankyrin repeat protein | 99.7 | 23.2 | B4R | |
| ALPV-041 | 45885-44593 | SWPV2-037 | 430 | 430 | SWPV2-037 ankyrin repeat protein | 100 | 27.7 | B18R | |
| ALPV-042 | 47751-45934 | SWPV2-038 | 605 | 605 | SWPV2-038 ankyrin repeat protein | 100 | 21.6 | B4R | |
| ALPV-043 | 48462-47857 | SWPV2-039 | 201 | 201 | SWPV2-039 conserved hypothetical protein | 100 | |||
| ALPV-044 | 49946-48504 | SWPV2-040 | 480 | 480 | SWPV2-040 ankyrin repeat protein | 100 | 25.2 | B18R | |
| ALPV-045 | 50212-51210 | SWPV2-041 | 332 | 332 | SWPV2-041 G protein-coupled receptor-like protein | 100- | |||
| ALPV-046 | 52589-51237 | SWPV2-042 | 450 | 450 | SWPV2-042 ankyrin repeat protein | 100 | 29.7 | VACV-Cop-B4R | |
| ALPV-047 | 53030-52656 | SWPV2-043 | 124 | 124 | SWPV2-043 conserved hypothetical protein | 100 | |||
| ALPV-048 | 55603-53198 | SWPV2-044 | 801 | 801 | SWPV2-044 alkaline phosphodiesterase-like protein | 100 | |||
| ALPV-049 | 56143-55691 | SWPV2-045 | 150 | 150 | SWPV2-045 hypothetical protein | 100 | |||
| ALPV-050 | 57255-56197 | SWPV2-046 | 352 | 352 | SWPV2-046 ankyrin repeat protein | 100 | |||
| ALPV-051 | 58528-57302 | SWPV2-047 | 408 | 408 | SWPV2-047 DNase II-like protein | 100 | |||
| ALPV-052 | 59069-58554 | SWPV2-048 | 171 | 171 | SWPV2-048 C-type lectin-like protein | 100 | |||
| ALPV-053 | 59689-59249 | SWPV2-049 | 146 | 146 | SWPV2-049 conserved hypothetical protein | 100 | |||
| ALPV-054 | 60104-59682 | SWPV2-050 | 140 | 140 | SWPV2-050 conserved hypothetical protein | 100 | |||
| ALPV-055 | 60647-60156 | SWPV2-051 | 163 | 163 | SWPV2-051 conserved hypothetical protein | 100 | |||
| ALPV-056 | 61081-60644 | SWPV2-052 | 145 | 145 | SWPV2-052 CNLV056 dUTPase | 100 | 52.8 | F2L | |
| ALPV-057 | 62028-61108 | SWPV2-053 | 306 | 306 | SWPV2-053 putative serpin | 100 | |||
| ALPV-058 | 62601-62059 | SWPV2-054 | 180 | 180 | SWPV2-054 bcl-2 like protein | 100 | |||
| ALPV-059 | 63674-62658 | SWPV2-055 | 338 | 338 | SWPV2-055 putative serpin | 100 | |||
| ALPV-060 | 64555-63743 | SWPV2-056 | 270 | 206 | SWPV2-056 conserved hypothetical protein | 99.5 | |||
| ALPV-061 | 66342-64645 | SWPV2-057 | 565 | 565 | SWPV2-057 DNA ligase | 100 | 46.9 | A50R | |
| ALPV-062 | 67433-66381 | SWPV2-058 | 350 | 350 | SWPV2-058 putative serpin | 100 | 25.0 | K2L | |
| ALPV-063 | 68580-67504 | SWPV2-059 | 358 | 358 | SWPV2-059 hydroxysteroid dehydrogenase-like protein | 100 | 38.2 | A44L | |
| ALPV-064 | 69493-68642 | SWPV2-060 | 283 | 283 | SWPV2-060 TGF-beta-like protein | 100 | |||
| ALPV-065 | 71324-69573 | SWPV2-061 | 583 | 583 | SWPV2-061 semaphorin-like protein | 100 | 31.7 | A39R | |
| ALPV-066 | 71606-71424 | SWPV2-062 | 60 | 399 | SWPV2-062 hypothetical protein | 100 | |||
| ALPV-067 | 71608-71784 | 59 | hypothetical protein, unique to ALPV | ||||||
| ALPV-068 | 72000-71728 | SWPV2-062 | 90 | 399 | SWPV2-062 hypothetical protein | 90.8 | |||
| ALPV-069 | 72255-72082 | SWPV2-063 | 57 | 57 | SWPV2-063 hypothetical protein | 100 | |||
| ALPV-070 | 72415-73188 | SWPV2-064 | 257 | 257 | SWPV2-064 GNS1/SUR4-like protein | 100 | |||
| ALPV-071 | 73281-73748 | SWPV2-065 | 155 | 155 | SWPV2-065 late transcription factor VLTF-2 | 100 | 46.6 | A1L | |
| ALPV-072 | 73765-75420 | SWPV2-066 | 551 | 551 | SWPV2-066 putative rifampicin resistance protein, IMV assembly | 100 | 56.2 | D13L | |
| ALPV-073 | 75452-76321 | SWPV2-067 | 289 | 289 | SWPV2-067 mRNA capping enzyme small subunit | 100 | 57.5 | D12L | |
| ALPV-074 | 76342-77283 | SWPV2-068 | 313 | 132 | SWPV2-068 CC chemokine-like protein | 100 | |||
| ALPV-075 | 77690-77361 | SWPV2-069 | 109 | 109 | SWPV2-069 hypothetical protein | 100 | |||
| ALPV-076 | 77761-79668 | SWPV2-070 | 635 | 635 | SWPV2-070 NPH-I, transcription termination factor | 100 | 60.8 | D11L | |
| ALPV-077 | 80351-79665 | SWPV2-071 | 228 | 228 | SWPV2-071 mutT motif putative gene expression regulator | 100 | 38.0 | D10R | |
| ALPV-078 | 81048-80335 | SWPV2-072 | 237 | 232 | SWPV2-072 mutT motif | 97.9 | 46.3 | D9R | |
| ALPV-079 | 81601-81287 | 104 | 100 | CNPV-077 hypothetical protein | |||||
| ALPV-080 | 81796-82065 | 89 | hypothetical protein, unique to ALPV | ||||||
| ALPV-081 | 82693-82448 | 81 | hypothetical protein, unique to ALPV | ||||||
| ALPV-082 | 83172-82690 | SWPV2-073 | 160 | 160 | SWPV2-073 RNA polymerase subunit RPO18 | 100 | 57.5 | D7R | |
| ALPV-083 | 84332-83508 | SWPV2-074 | 274 | 274 | SWPV2-074 Ig-like domain protein | 100 | |||
| ALPV-084 | 86356-84455 | SWPV2-075 | 633 | 633 | SWPV2-075 early transcription factor small subunit VETFS | 100 | 72.2 | D6R | |
| ALPV-085 | 87561-86560 | SWPV2-076 | 333 | 334 | SWPV2-076 Ig-like domain protein | 98.8 | |||
| ALPV-086 | 90271-87887 | SWPV2-077 | 794 | 794 | SWPV2-077 NTPase, DNA replication | 100 | 54.3 | D5R | |
| ALPV-087 | 91091-90426 | SWPV2-078 | 221 | 221 | SWPV2-078 CC chemokine-like protein | 100 | |||
| ALPV-088 | 91830-91174 | SWPV2-079 | 218 | 218 | SWPV2-079 uracil DNA glycosylase | 99.5 | 54.2 | D4R | |
| ALPV-089 | 92626-91871 | SWPV2-080 | 251 | 303 | SWPV2-080 putative RNA phosphatase | 94.7 | 34.1 | H1L | |
| ALPV-090 | 93861-92674 | 395 | 71.9 | SWPV1-075 conserved hypothetical protein | |||||
| ALPV-091 | 93946-94311 | SWPV2-081 | 121 | 112 | SWPV2-081 TNFR-like protein | 80.8 | 33.9 | B28R | |
| ALPV-092 | 96471-96866 | SWPV2-082 | 131 | 131 | SWPV2-082 putative glutathione peroxidase | 100 | |||
| ALPV-093 | 96891-97193 | SWPV2-083 | 100 | 100 | SWPV2-083 conserved hypothetical protein | 100 | |||
| ALPV-094 | 97677-97198 | SWPV2-084 | 159 | 159 | SWPV2-084 conserved hypothetical protein | 100 | |||
| ALPV-095 | 98047-97664 | SWPV2-085 | 127 | 127 | SWPV2-085 conserved hypothetical protein | 100 | |||
| ALPV-096 | 98384-98133 | SWPV2-086 | 83 | 83 | SWPV2-086 HT motif protein | 100 | |||
| ALPV-097 | 99122-98736 | SWPV2-087 | 128 | 146 | SWPV2-087 conserved hypothetical protein | 87.7 | |||
| ALPV-098 | 100027-99224 | SWPV2-088 | 267 | 267 | SWPV2-088 virion protein | 100 | |||
| ALPV-099 | 100102-100929 | SWPV2-089 | 275 | 275 | SWPV2-089 T10-like protein | 100 | |||
| ALPV-100 | 101074-100937 | SWPV2-090 | 45 | 45 | SWPV2-090 conserved hypothetical protein | 100 | |||
| ALPV-101 | 101313-101056 | SWPV2-091 | 85 | 85 | SWPV2-091 ubiquitin | 100 | |||
| ALPV-102 | 102422-101445 | SWPV2-092 | 325 | 339 | SWPV2-092 conserved hypothetical protein | 95.9 | |||
| ALPV-103 | 102692-102450 | SWPV2-093 | 80 | 80 | SWPV2-093 hypothetical protein | 100 | |||
| ALPV-104 | 103285-102698 | SWPV2-094 | 195 | 195 | SWPV2-094 beta-NGF-like protein | 100 | |||
| ALPV-105 | 103815-103309 | SWPV2-095 | 168 | 168 | SWPV2-095 putative interleukin binding protein | 100 | |||
| ALPV-106 | 104127-103870 | SWPV2-096 | 85 | 85 | SWPV2-096 hypothetical protein | 100 | |||
| ALPV-107 | 104455-104138 | SWPV2-097 | 105 | 105 | SWPV2-097 conserved hypothetical protein | 100 | |||
| ALPV-108 | 105044-104472 | SWPV2-098 | 190 | 190 | SWPV2-098 N1R/p28-like protein | 100 | |||
| ALPV-109 | 105246-105623 | SWPV2-099 | 125 | 125 | SWPV2-099 putative glutaredoxin 2, virion morphogenesis | 100 | 32.2 | G4L | |
| ALPV-110 | 106270-105566 | SWPV2-100 | 234 | 234 | SWPV2-100 putative elongation factor | 100 | |||
| ALPV-111 | 106264-106572 | SWPV2-101 | 102 | 102 | SWPV2-101 conserved hypothetical protein | 100 | |||
| ALPV-112 | 106708-106941 | SWPV2-102 | 77 | 77 | SWPV2-102 hypothetical protein | 100 | |||
| ALPV-113 | 107186-109084 | SWPV2-103 | 632 | 632 | SWPV2-103 putative metalloprotease, virion morphogenesis | 100 | 43.9 | G1L | |
| ALPV-114 | 111113-109068 | SWPV2-104 | 681 | 681 | SWPV2-104 NPH-II, RNA helicase | 100 | 43.8 | I8R | |
| ALPV-115 | 111148-112416 | SWPV2-105 | 422 | 422 | SWPV2-105 virion core proteinase | 100 | 54.4 | I7L | |
| ALPV-116 | 112421-113596 | SWPV2-106 | 391 | 391 | SWPV2-106 DNA-binding protein | 100 | 34.8 | I6L | |
| ALPV-117 | 113597-113842 | SWPV2-107 | 81 | 81 | SWPV2-107 putative IMV membrane protein | 100 | |||
| ALPV-118 | 113864-114403 | SWPV2-108 | 179 | 179 | SWPV2-108 thymidine kinase | 100 | 52.0 | J2R | |
| ALPV-119 | 114524-114772 | SWPV2-109 | 82 | 82 | SWPV2-109 HT motif protein | 100 | |||
| ALPV-120 | 114842-115711 | SWPV2-110 | 289 | 289 | SWPV2-110 DNA-binding phosphoprotein | 99.7 | 33.5 | I3L | |
| ALPV-121 | 115712-115921 | SWPV2-111 | 69 | 69 | SWPV2-111 conserved hypothetical protein | 100 | |||
| ALPV-122 | 115928-116860 | SWPV2-112 | 310 | 310 | SWPV2-112 DNA-binding virion protein | 100 | 58.0 | I1L | |
| ALPV-123 | 117040-118998 | SWPV2-113 | 652 | 652 | SWPV2-113 conserved hypothetical protein | 100 | 20.9 | O1L | |
| ALPV-124 | 118928-119323 | SWPV2-114 | 131 | 131 | SWPV2-114 virion core protein | 100 | 34.98 | E11L | |
| ALPV-125 | 119601-119320 | SWPV2-115 | 93 | 93 | SWPV2-115 putative IMV redox protein, virus assembly | 100 | 51.6 | E10R | |
| ALPV-126 | 119628-122594 | SWPV2-116 | 988 | 988 | SWPV2-116 DNA polymerase | 100 | 50.3 | E9L | |
| ALPV-127 | 123413-122586 | 275 | 80.4 | 48.2 | E8R | SWPV1-111 putative membrane protein | |||
| ALPV-128 | 125130-123415 | SWPV2-117 | 571 | 502 | SWPV2-117 conserved hypothetical protein | 87.2 | 49.9 | E6R | |
| ALPV-129 | 130930-125192 | SWPV2-118 | 1912 | 1916 | SWPV2-118 variola B22R-like protein | 99.8 | |||
| ALPV-130 | 136264-130997 | SWPV2-119 | 1755 | 1767 | SWPV2-119 variola B22R-like protein | 99.3 | |||
| ALPV-131 | 142243-136544 | SWPV2-120 | 1899 | 1839 | SWPV2-120 variola B22R-like protein | 95.5 | |||
| ALPV-132 | 142444-142992 | SWPV2-122 | 182 | 182 | SWPV2-122 RNA polymerase subunit RPO30 | 100 | 57.0 | E4L | |
| ALPV-133 | 143024-145189 | SWPV2-123 | 721 | 721 | SWPV2-123 conserved hypothetical protein | 100 | 28.0 | E2L | |
| ALPV-134 | 145182-146600 | SWPV2-124 | 472 | 472 | SWPV2-124 poly(A) polymerase large subunit PAPL | 100 | 50.5 | E1L | |
| ALPV-135 | 146953-146594 | SWPV2-125 | 119 | 119 | SWPV2-125 DNA-binding virion core protein | 100 | 38.8 | F17R | |
| ALPV-136 | 147029-147652 | SWPV2-126 | 207 | 207 | SWPV2-126 conserved hypothetical protein | 100 | |||
| ALPV-137 | 147746-148192 | SWPV2-127 | 148 | 148 | SWPV2-127 conserved hypothetical protein | 100 | 40.4 | F15L | |
| ALPV-138 | 148426-148725 | SWPV2-128 | 99 | 99 | SWPV2-128 conserved hypothetical protein | 100 | |||
| ALPV-139 | 154234-148796 | SWPV2-129 | 1812 | 1801 | SWPV2-129 variola B22R-like protein | 99.4 | |||
| ALPV-140 | 154384-155520 | SWPV2-130 | 378 | 378 | SWPV2-130 putative palmitoylated EEV envelope lipase | 100 | 38.0 | F13L | |
| ALPV-141 | 155598-157475 | SWPV2-131 | 625 | 625 | SWPV2-131 putative EEV maturation protein | 100 | 26.3 | F12L | |
| ALPV-142 | 157518-158906 | SWPV2-132 | 462 | 462 | SWPV2-132 conserved hypothetical protein | 100 | 27.0 | F11L | |
| ALPV-143 | 158997-160331 | SWPV2-133 | 444 | 444 | SWPV2-133 putative serine/threonine protein kinase, virus assembly | 100 | 53.6 | F10L | |
| ALPV-144 | 160306-160947 | SWPV2-134 | 213 | 213 | SWPV2-134 conserved hypothetical protein | 100 | 31.5 | F9L | |
| ALPV-145 | 161030-161230 | SWPV2-135 | 66 | 66 | SWPV2-135 conserved hypothetical protein | 100 | |||
| ALPV-146 | 161556-162110 | SWPV2-136 | 184 | 184 | SWPV2-136 HAL3-like domain protein | 100 | |||
| ALPV-147 | 162371-163336 | SWPV2-137 | 321 | 321 | SWPV2-137 N1R/p28-like protein | 100 | |||
| ALPV-148 | 163448-165463 | SWPV2-138 | 671 | 671 | SWPV2-138 ankyrin repeat protein | 100 | 26.0 | M1L | |
| ALPV-149 | 165489-167159 | SWPV2-139 | 556 | 556 | SWPV2-139 ankyrin repeat protein | 100 | 26.1 | B4R | |
| ALPV-150 | 167380-168702 | SWPV2-140 | 440 | 440 | SWPV2-140 conserved hypothetical protein | 100 | 33.3 | G5R | |
| ALPV-151 | 168710-168898 | SWPV2-141 | 62 | 62 | SWPV2-141 RNA polymerase subunit RPO7 | 98.4 | 55.2 | G5.5R | |
| ALPV-152 | 168891-169457 | SWPV2-142 | 188 | 188 | SWPV2-142 conserved hypothetical protein | 100 | 31.8 | G6R | |
| ALPV-153 | 170468-169422 | SWPV2-143 | 348 | 348 | SWPV2-143 virion core protein | 100 | 34.6 | G7L | |
| ALPV-154 | 171554-170634 | SWPV2-144 | 306 | 306 | SWPV2-144 putative thioredoxin binding protein | 100 | |||
| ALPV-155 | 171682-171915 | 77 | 97.1 | CNPV-150 ankyrin repeat protein | |||||
| ALPV-156 | 173269-172031 | SWPV2-145 | 412 | 412 | SWPV2-145 ankyrin repeat protein | 100 | 42.9 | M1L | |
| ALPV-157 | 173945-173496 | SWPV2-146 | 149 | 149 | SWPV2-146 hypothetical protein | 100 | |||
| ALPV-158 | 175111-174173 | SWPV2-147 | 312 | 312 | SWPV2-147 Rep-like protein | 100 | |||
| ALPV-159 | 181355-175545 | SWPV2-148 | 1936 | 875 | SWPV2-148 variola B22R-like protein | 98.2 | |||
| ALPV-160 | 186885-181408 | SWPV2-149 | 1825 | 1831 | SWPV2-149 variola B22R-like protein | 99.7 | |||
| ALPV-161 | 187202-189685 | SWPV2-150 | 827 | 834 | SWPV2-150 hypothetical protein | 90.5 | |||
| ALPV-162 | 190813-189782 | SWPV2-151 | 343 | 343 | SWPV2-151 TGF-beta-like protein | 100 | |||
| ALPV-163 | 190815-191294 | 159 | 64.7 | CNPV-157 TGF-beta-like protein | |||||
| ALPV-164 | 191763-192263 | SWPV2-150 | 166 | 834 | SWPV2-150 hypothetical protein | 47.8 | |||
| ALPV-165 | 192733-193446 | 237 | 97.2 | CNPV-159 N1R/p28-like protein | |||||
| ALPV-166 | 193490-194494 | 334 | 91.3 | CNPV-169 N1R/p28-like protein | |||||
| ALPV-167 | 194549-195412 | 287 | 78.7 | CNPV-169 N1R/p28-like protein | |||||
| ALPV-168 | 195417-195821 | 134 | 98.5 | CNPV-160 N1R/p28-like protein | |||||
| ALPV-169 | 197008-195920 | SWPV2-152 | 362 | 358 | SWPV2-152 TGF-beta-like protein | 98.9 | |||
| ALPV-170 | 197058-197507 | SWPV2-153 | 149 | 149 | SWPV2-153 TGF-beta-like protein | 100 | |||
| ALPV-171 | 197843-198883 | SWPV2-154 | 346 | 320 | SWPV2-154 N1R/p28-like protein | 95.3 | |||
| ALPV-172 | 199118-200155 | SWPV2-155 | 345 | 345 | SWPV2-155 Ig-like domain protein | 99.7 | |||
| ALPV-173 | 200427-200933 | SWPV2-156 | 168 | 168 | SWPV2-156 Ig-like domain protein | 97 | |||
| ALPV-174 | 201028-202059 | SWPV2-157 | 343 | 350 | SWPV2-157 N1R/p28-like protein | 93.6 | |||
| ALPV-175 | 202011-202415 | 134 | 92.5 | CNPV-226 N1R/p28-like protein | |||||
| ALPV-176 | 202488-203126 | SWPV2-158 | 212 | 212 | SWPV2-158 thymidylate kinase | 100 | 45.2 | A48R | |
| ALPV-177 | 203179-203961 | SWPV2-159 | 260 | 260 | SWPV2-159 late transcription factor VLTF-1 | 100 | 66.2 | G8R | |
| ALPV-178 | 203975-204982 | SWPV2-160 | 335 | 335 | SWPV2-160 putative myristylated protein | 100 | 38.0 | G9R | |
| ALPV-179 | 204983-205714 | SWPV2-161 | 243 | 243 | SWPV2-161 putative myristylated IMV envelope protein | 100 | 54.7 | L1R | |
| ALPV-180 | 205774-206064 | SWPV2-162 | 96 | 96 | SWPV2-162 conserved hypothetical protein | 100 | |||
| ALPV-181 | 206965-206054 | SWPV2-163 | 303 | 303 | SWPV2-163 conserved hypothetical protein | 100 | 42.0 | L3L | |
| ALPV-182 | 206991-207749 | SWPV2-164 | 252 | 252 | SWPV2-164 DNA-binding virion core protein | 100 | 36.3 | L4R | |
| ALPV-183 | 207750-208142 | SWPV2-165 | 130 | 130 | SWPV2-165 conserved hypothetical protein | 100 | 41.6 | L5R | |
| ALPV-184 | 208096-208542 | SWPV2-166 | 148 | 148 | SWPV2-166 putative IMV membrane protein | 100 | 42.5 | J1R | |
| ALPV-185 | 208576-209484 | SWPV2-167 | 302 | 302 | SWPV2-167 poly(A) polymerase small subunit PAPS | 100 | 56.0 | J3R | |
| ALPV-186 | 209481-210041 | SWPV2-168 | 186 | 186 | SWPV2-168 RNA polymerase subunit RPO22 | 100 | 55.0 | J4R | |
| ALPV-187 | 210444-210034 | SWPV2-169 | 136 | 136 | SWPV2-169 conserved hypothetical protein | 100 | 47.9 | J5L | |
| ALPV-188 | 210487-214353 | SWPV2-170 | 1288 | 1288 | SWPV2-170 RNA polymerase subunit RPO147 | 100 | 70.6 | J6R | |
| ALPV-189 | 214856-214356 | SWPV2-171 | 166 | 166 | SWPV2-171 putative protein-tyrosine phosphatase, virus assembly | 100 | 47.6 | H1L | |
| ALPV-190 | 214872-215441 | SWPV2-172 | 189 | 189 | SWPV2-172 conserved hypothetical protein | 100 | 48.9 | H2R | |
| ALPV-191 | 216503-215517 | SWPV2-173 | 328 | 328 | SWPV2-173 ankyrin repeat protein | 100 | |||
| ALPV-192 | 217538-216546 | SWPV2-174 | 330 | 330 | SWPV2-174 putative IMV envelope protein | 100 | 32.2 | H3L | |
| ALPV-193 | 220029-217630 | SWPV2-175 | 799 | 799 | SWPV2-175 RNA polymerase associated protein RAP94 | 100 | 55.2 | H4L | |
| ALPV-194 | 220198-220710 | SWPV2-176 | 170 | 170 | SWPV2-176 late transcription factor VLTF-4 | 100 | |||
| ALPV-195 | 220711-221661 | SWPV2-177 | 316 | 316 | SWPV2-177 DNA topoisomerase | 100 | 58.0 | H6R | |
| ALPV-196 | 221666-222127 | SWPV2-178 | 153 | 153 | SWPV2-178 conserved hypothetical protein | 100 | 33.8 | H7R | |
| ALPV-197 | 222401-222090 | SWPV2-179 | 103 | 103 | SWPV2-179 conserved hypothetical protein | 100 | |||
| ALPV-198 | 222409-224949 | SWPV2-180 | 846 | 846 | SWPV2-180 mRNA capping enzyme large subunit | 100 | 53.9 | D1R | |
| ALPV-199 | 225020-225340 | SWPV2-181 | 106 | 106 | SWPV2-181 HT motif protein | 100 | |||
| ALPV-200 | 225759-225337 | SWPV2-182 | 140 | 140 | SWPV2-182 virion protein | 100 | |||
| ALPV-201 | 225813-226247 | SWPV2-183 | 144 | 144 | SWPV2-183 hypothetical protein | 100 | |||
| ALPV-202 | 226312-226884 | SWPV2-184 | 190 | 190 | SWPV2-184 conserved hypothetical protein | 100 | |||
| ALPV-203 | 226950-227777 | SWPV2-185 | 275 | 275 | SWPV2-185 N1R/p28-like protein | 100 | |||
| ALPV-204 | 228314-227844 | SWPV2-186 | 156 | 156 | SWPV2-186 C-type lectin-like protein | 100 | 30.8 | A34R | |
| ALPV-205 | 228622-229299 | SWPV2-187 | 225 | 225 | SWPV2-187 deoxycytidine kinase-like protein | 100 | |||
| ALPV-206 | 229305-229805 | SWPV2-188 | 166 | 166 | SWPV2-188 Rep-like protein | 99.4 | |||
| ALPV-207 | 229864-230367 | SWPV2-189 | 167 | 167 | SWPV2-189 conserved hypothetical protein | 100 | |||
| ALPV-208 | 230421-231251 | SWPV2-190 | 276 | 276 | SWPV2-190 N1R/p28-like protein | 100 | |||
| ALPV-209 | 231324-232472 | SWPV2-191 | 382 | 382 | SWPV2-191 N1R/p28-like protein | 100 | |||
| ALPV-210 | 232528-232713 | SWPV2-192 | 61 | 61 | SWPV2-192 conserved hypothetical protein | 100 | |||
| ALPV-211 | 232932-233888 | SWPV2-193 | 318 | 318 | SWPV2-193 N1R/p28-like protein | 100 | |||
| ALPV-212 | 233949-235367 | SWPV2-194 | 472 | 472 | SWPV2-194 putative photolyase | 100 | |||
| ALPV-213 | 235424-235579 | 51 | hypothetical protein, unique to ALPV, containing a transmembrane helix | ||||||
| ALPV-214 | 235557-236078 | SWPV2-195 | 173 | 173 | SWPV2-195 N1R/p28-like protein | 100 | |||
| ALPV-215 | 236184-236723 | SWPV2-196 | 179 | 200 | SWPV2-196 conserved hypothetical protein | 89 | |||
| ALPV-216 | 236767-237699 | SWPV2-197 | 310 | 310 | SWPV2-197 N1R/p28-like protein | 100 | |||
| ALPV-217 | 237747-238142 | SWPV2-198 | 131 | 131 | SWPV2-198 N1R/p28-like protein | 100 | |||
| ALPV-218 | 238197-238361 | SWPV2-199 | 54 | 54 | SWPV2-199 conserved hypothetical protein | 100 | |||
| ALPV-219 | 238421-238951 | SWPV2-200 | 176 | 176 | SWPV2-200 N1R/p28-like protein | 100 | |||
| ALPV-220 | 239662-239012 | SWPV2-201 | 216 | 216 | SWPV2-201 deoxycytidine kinase-like protein | 100 | |||
| ALPV-221 | 239836-240906 | SWPV2-202 | 356 | 356 | SWPV2-202 vaccinia C4L/C10L-like protein | 100 | 28.1 | C10L | |
| ALPV-222 | 241181-241795 | SWPV2-203 | 204 | 204 | SWPV2-203 CC chemokine-like protein | 100 | |||
| ALPV-223 | 241885-243090 | SWPV2-204 | 401 | 401 | SWPV2-204 conserved hypothetical protein | 100 | |||
| ALPV-224 | 243204-244196 | SWPV2-205 | 330 | 330 | SWPV2-205 N1R/p28-like protein | 100 | |||
| ALPV-225 | 244284-245555 | SWPV2-206 | 423 | 223 | SWPV2-206 N1R/p28-like protein | 99.5 | |||
| ALPV-226 | 245795-245619 | 58 | hypothetical protein, unique to ALPV, containing a transmembrane helix | ||||||
| ALPV-227 | 246013-245858 | 51 | hypothetical protein, unique to ALPV, containing a transmembrane helix | ||||||
| ALPV-228 | 246363-247412 | SWPV2-207 | 349 | 349 | SWPV2-207 N1R/p28-like protein | 100 | |||
| ALPV-229 | 247466-248308 | 280 | 80.9 | SWPV1-198 N1R/p28-like protein | |||||
| ALPV-230 | 248847-248644 | SWPV2-208 | 67 | 85 | SWPV2-208 N1R/p28-like protein | 62.7 | |||
| ALPV-231 | 249295-249483 | SWPV2-209 | 62 | 213 | SWPV2-209 N1R/p28-like protein | 100 | |||
| ALPV-232 | 250029-250292 | SWPV2-210 | 87 | 285 | SWPV2-210 N1R/p28-like protein | 100 | |||
| ALPV-233 | 250283-250615 | 110 | 98.2 | CNPV-227 N1R/p28-like protein | |||||
| ALPV-234 | 253677-251134 | SWPV2-211 | 847 | 847 | SWPV2-211 ankyrin repeat protein | 99.9 | 31.6 | B4R | |
| ALPV-235 | 253931-254650 | SWPV2-212 | 239 | 239 | SWPV2-212 hypothetical protein | 100 | |||
| ALPV-236 | 254650-255027 | 125 | 73.3 | CNPV-227 N1R/p28-like protein | |||||
| ALPV-237 | 255062-255346 | SWPV2-214 | 94 | 126 | SWPV2-214 N1R/p28-like protein | 96.4 | |||
| ALPV-238 | 257103-255799 | SWPV2-215 | 434 | 434 | SWPV2-215 ankyrin repeat protein | 100 | 27.3 | B4R | |
| ALPV-239 | 257301-257498 | SWPV2-216 | 65 | 65 | SWPV2-216 hypothetical protein | 100 | |||
| ALPV-240 | 257446-257922 | SWPV2-217 | 158 | 158 | SWPV2-217 MyD116-like domain protein | 100 | |||
| ALPV-241 | 257952-258566 | SWPV2-218 | 204 | 204 | SWPV2-218 CC chemokine-like protein | 98 | |||
| ALPV-242 | 258748-260163 | SWPV2-219 | 471 | 471 | SWPV2-219 ankyrin repeat protein | 100 | 31.4 | M1L | |
| ALPV-243 | 260183-261709 | SWPV2-220 | 508 | 508 | SWPV2-220 ankyrin repeat protein | 100 | 37.0 | M1L | |
| ALPV-244 | 261780-263084 | SWPV2-221 | 434 | 432 | SWPV2-221 conserved hypothetical protein | 99.5 | |||
| ALPV-245 | 263129-264100 | SWPV2-222 | 323 | 323 | SWPV2-222 ribonucleotide reductase small subunit | 100 | 70.9 | F4L | |
| ALPV-246 | 264281-265606 | SWPV2-223 | 441 | 441 | SWPV2-223 ankyrin repeat protein | 99.8 | 33.3 | B4R | |
| ALPV-247 | 266342-265665 | SWPV2-224 | 225 | 225 | SWPV2-224 late transcription factor VLTF-3 | 100 | 76.7 | A2L | |
| ALPV-248 | 266557-266330 | SWPV2-225 | 75 | 75 | SWPV2-225 virion redox protein | 100 | |||
| ALPV-249 | 268550-266571 | SWPV2-226 | 659 | 659 | SWPV2-226 virion core protein P4b | 100 | 54.1 | A3L | |
| ALPV-250 | 269284-268637 | SWPV2-227 | 215 | 215 | SWPV2-227 immunodominant virion protein | 100 | |||
| ALPV-251 | 269323-269832 | SWPV2-228 | 169 | 169 | SWPV2-228 RNA polymerase subunit RPO19 | 100 | 55.2 | A5R | |
| ALPV-252 | 270948-269827 | SWPV2-229 | 373 | 373 | SWPV2-229 conserved hypothetical protein | 100 | 39.3 | A6L | |
| ALPV-253 | 273084-270955 | SWPV2-230 | 709 | 709 | SWPV2-230 early transcription factor large subunit VETFL | 100 | 59.6 | A7L | |
| ALPV-254 | 273148-274050 | SWPV2-231 | 300 | 300 | SWPV2-231 intermediate transcription factor VITF-3 | 100 | 38.7 | A8R | |
| ALPV-255 | 274242-274015 | SWPV2-232 | 75 | 75 | SWPV2-232 putative IMV membrane protein | 100 | |||
| ALPV-256 | 276924-274243 | SWPV2-233 | 893 | 893 | SWPV2-233 virion core protein P4a | 100 | 39.6 | A10L | |
| ALPV-257 | 276942-277781 | SWPV2-234 | 279 | 279 | SWPV2-234 conserved hypothetical protein | 100 | 39.5 | A11R | |
| ALPV-258 | 278284-277778 | SWPV2-235 | 168 | 168 | SWPV2-235 virion protein | 99.4 | 32.9 | A12L | |
| ALPV-259 | 278299-278556 | SWPV2-236 | 85 | 56 | SWPV2-236 conserved hypothetical protein | 87.0 | |||
| ALPV-260 | 278754-278545 | SWPV2-237 | 69 | 69 | SWPV2-237 putative IMV membrane protein | 100 | |||
| ALPV-261 | 279080-278802 | SWPV2-238 | 92 | 92 | SWPV2-238 putative IMV membrane protein | 100 | 27.5 | A14L | |
| ALPV-262 | 279258-279097 | SWPV2-239 | 53 | 53 | SWPV2-239 putative IMV membrane virulence factor | 100 | 35.3 | A 14.5L | |
| ALPV-263 | 279564-279274 | SWPV2-240 | 96 | 96 | SWPV2-240 conserved hypothetical protein | 100 | |||
| ALPV-264 | 280654-279548 | SWPV2-241 | 368 | 368 | SWPV2-241 predicted myristylated protein | 100 | 43.0 | A16L | |
| ALPV-265 | 281248-280670 | SWPV2-242 | 192 | 192 | SWPV2-242 putative phosphorylated IMV membrane protein | 100 | 33.2 | A17L | |
| ALPV-266 | 281266-282654 | SWPV2-243 | 462 | 462 | SWPV2-243 DNA helicase, transcriptional elongation | 100 | 50.2 | A18R | |
| ALPV-267 | 282891-282622 | SWPV2-244 | 89 | 89 | SWPV2-244 conserved hypothetical protein | 100 | 42.9 | A19L | |
| ALPV-268 | 283237-282899 | SWPV2-245 | 112 | 112 | SWPV2-245 conserved hypothetical protein | 100 | 45.3 | A21L | |
| ALPV-269 | 283236-284540 | SWPV2-246 | 434 | 434 | SWPV2-246 DNA polymerase processivity factor | 100 | 25.9 | A20R | |
| ALPV-270 | 284537-284995 | SWPV2-247 | 152 | 152 | SWPV2-247 Holliday junction resolvase protein | 100 | 45.0 | A22R | |
| ALPV-271 | 285012-286163 | SWPV2-248 | 383 | 383 | SWPV2-248 intermediate transcription factor VITF-3 | 100 | 51.3 | A23R | |
| ALPV-272 | 286189-289662 | SWPV2-249 | 1157 | 1157 | SWPV2-249 RNA polymerase subunit RPO132 | 100 | 74.6 | A24R | |
| ALPV-273 | 291456-289651 | SWPV2-250 | 601 | 601 | SWPV2-250 A type inclusion-like protein | 100 | |||
| ALPV-274 | 292918-291491 | SWPV2-251 | 475 | 475 | SWPV2-251 A type inclusion-like/fusion protein | 100 | 56.4 | A27L | |
| ALPV-275 | 293341-292919 | SWPV2-252 | 140 | 140 | SWPV2-252 conserved hypothetical protein | 100 | 41.8 | A28L | |
| ALPV-276 | 294263-293346 | SWPV2-253 | 305 | 305 | SWPV2-253 RNA polymerase subunit RPO35 | 100 | 42.6 | A29L | |
| ALPV-277 | 294465-294238 | SWPV2-254 | 75 | 75 | SWPV2-254 conserved hypothetical protein | 100 | |||
| ALPV-278 | 294590-294931 | SWPV2-255 | 113 | 113 | SWPV2-255 conserved hypothetical protein | 100 | 31.6 | A31R | |
| ALPV-279 | 294940-295302 | SWPV2-256 | 120 | 120 | SWPV2-256 conserved hypothetical protein | 100 | |||
| ALPV-280 | 296145-295291 | SWPV2-257 | 284 | 284 | SWPV2-257 DNA packaging protein | 100 | 47.1 | A32L | |
| ALPV-281 | 296260-296805 | SWPV2-258 | 181 | 181 | SWPV2-258 C-type lectin-like EEV protein | 100 | |||
| ALPV-282 | 297030-297854 | SWPV2-259 | 274 | 274 | SWPV2-259 conserved hypothetical protein | 100 | |||
| ALPV-283 | 297914-298723 | SWPV2-260 | 269 | 269 | SWPV2-260 putative tyrosine protein kinase | 100 | |||
| ALPV-284 | 298766-299782 | SWPV2-261 | 338 | 338 | SWPV2-261 putative serpin | 100 | |||
| ALPV-285 | 300562-299804 | SWPV2-262 | 252 | 252 | SWPV2-262 conserved hypothetical protein | 100 | |||
| ALPV-286 | 300672-301604 | SWPV2-263 | 310 | 310 | SWPV2-263 G protein-coupled receptor-like protein | 100 | |||
| ALPV-287 | 301615-301905 | SWPV2-264 | 96 | 96 | SWPV2-264 conserved hypothetical protein | 100 | |||
| ALPV-288 | 301971-302501 | SWPV2-265 | 176 | 169 | SWPV2-265 beta-NGF-like protein | 96 | |||
| ALPV-289 | 302911-302519 | SWPV2-266 | 130 | 130 | SWPV2-266 HT motif protein | 100 | |||
| ALPV-290 | 303015-303659 | SWPV2-267 | 214 | 214 | SWPV2-267 conserved hypothetical protein | 100 | |||
| ALPV-291 | 304032-303670 | SWPV2-268 | 120 | 120 | SWPV2-268 HT motif protein | 100 | |||
| ALPV-292 | 304198-304533 | SWPV2-269 | 111 | 111 | SWPV2-269 CC chemokine-like protein | 100 | |||
| ALPV-293 | 304612-305193 | SWPV2-270 | 193 | 193 | SWPV2-270 putative interleukin binding protein | 100 | |||
| ALPV-294 | 305303-305683 | SWPV2-271 | 126 | 126 | SWPV2-271 EGF-like protein | 100 | 34.8 | C11R | |
| ALPV-295 | 305685-306602 | SWPV2-272 | 305 | 305 | SWPV2-272 putative serine/threonine protein kinase | 100 | 41.1 | B1R | |
| ALPV-296 | 306645-307127 | SWPV2-273 | 160 | 160 | SWPV2-273 conserved hypothetical protein | 100 | |||
| ALPV-297 | 307210-307677 | SWPV2-274 | 155 | 147 | SWPV2-274 C-type lectin-like protein | 99.3 | 22.6 | A34R | |
| ALPV-298 | 307720-308139 | SWPV2-275 | 139 | 139 | SWPV2-275 putative interleukin binding protein | 100 | |||
| ALPV-299 | 308208-308435 | SWPV2-276 | 75 | 75 | SWPV2-276 conserved hypothetical protein | 100 | |||
| ALPV-300 | 308637-310421 | SWPV2-277 | 594 | 594 | SWPV2-277 ankyrin repeat protein | 99.8 | 22.3 | B4R | |
| ALPV-301 | 310445-310669 | SWPV2-278 | 74 | 74 | SWPV2-278 hypothetical protein | 100 | |||
| ALPV-302 | 310712-311566 | SWPV2-279 | 284 | 284 | SWPV2-279 ankyrin repeat protein | 99.6 | 33.9 | B24R | |
| ALPV-303 | 311621-312913 | SWPV2-280 | 430 | 430 | SWPV2-280 ankyrin repeat protein | 99.8 | C18L | ||
| ALPV-304 | 313106-314296 | SWPV2-281 | 396 | 396 | SWPV2-281 ankyrin repeat protein | 100 | 28.6 | M1L | |
| ALPV-305 | 314299-315675 | SWPV2-282 | 458 | 458 | SWPV2-282 ankyrin repeat protein | 100 | 32.5 | B4R | |
| ALPV-306 | 315784-317997 | SWPV2-283 | 737 | 737 | SWPV2-283 ankyrin repeat protein | 100 | 25.8 | M1L | |
| ALPV-307 | 318053-319768 | SWPV2-284 | 571 | 571 | SWPV2-284 ankyrin repeat protein | 100 | 28.8 | B4R | |
| ALPV-308 | 319772-320674 | SWPV2-285 | 300 | 300 | SWPV2-285 putative serine/threonine protein kinase | 100 | 24.6 | M1L | |
| ALPV-309 | 320747-321481 | SWPV2-286 | 244 | 244 | SWPV2-286 ankyrin repeat protein | 100 | 32.8 | B1R | |
| ALPV-310 | 322082-323665 | SWPV2-287 | 527 | 527 | SWPV2-287 ankyrin repeat protein | 100 | 23.7 | M1L | |
| ALPV-311 | 324261-323680 | SWPV2-288 | 193 | 193 | SWPV2-288 conserved hypothetical protein | 100 | 26.1 | B18R | |
| ALPV-312 | 324329-325831 | SWPV2-289 | 500 | 500 | SWPV2-289 ankyrin repeat protein | 100 | |||
| ALPV-313 | 326047-327447 | SWPV2-290 | 466 | 466 | SWPV2-290 ankyrin repeat protein | 100 | 28.3 | M1L | |
| ALPV-314 | 327518-328306 | SWPV2-291 | 262 | 262 | SWPV2-291 N1R/p28-like protein | 100 | 28.2 | B4R | |
| ALPV-315 | 328368-328586 | SWPV2-292 | 72 | 72 | SWPV2-292 hypothetical protein | 100 | |||
| ALPV-316 | 329054-328590 | SWPV2-293 | 154 | 154 | SWPV2-293 C-type lectin-like protein | 100 | |||
| ALPV-317 | 329231-330304 | SWPV2-294 | 357 | 357 | SWPV2-294 ankyrin repeat protein | 100 | 34.4 | A40R | |
| ALPV-318 | 330452-331042 | SWPV2-295 | 196 | 196 | SWPV2-295 ankyrin repeat protein | 100 | |||
| ALPV-319 | 331147-332760 | SWPV2-296 | 537 | 537 | SWPV2-296 ankyrin repeat protein | 100 | 37.4 | M1L | |
| ALPV-320 | 332794-333168 | SWPV2-297 | 124 | 124 | SWPV2-297 EFc-like protein | 100 | |||
| ALPV-321 | 333178-333678 | SWPV2-298 | 166 | 166 | SWPV2-298 conserved hypothetical protein | 100 | |||
| ALPV-322 | 333750-334406 | SWPV2-299 | 218 | 218 | SWPV2-299 Ig-like domain protein | 100 | |||
| ALPV-323 | 334433-336322 | SWPV2-300 | 629 | 629 | SWPV2-300 ankyrin repeat protein | 99.8 | |||
| ALPV-324 | 336421-337368 | SWPV2-301 | 315 | 315 | SWPV2-301 G protein-coupled receptor-like protein | 100 | 40.8 | B4R | |
| ALPV-325 | 337435-339069 | SWPV2-302 | 544 | 544 | SWPV2-302 ankyrin repeat protein | 99.8 | |||
| ALPV-326 | 339250-339417 | SWPV2-303 | 55 | 55 | SWPV2-303 hypothetical protein | 100 | 33.9 | B4R | |
| ALPV-327 | 339592-341124 | SWPV2-304 | 510 | 514 | SWPV2-304 ankyrin repeat protein | 99.2 | |||
| ALPV-328 | 341457-343424 | SWPV2-305 | 655 | 637 | SWPV2-305 ankyrin repeat protein | 96.6 | 36.8 | M1L | |
| ALPV-329 | 343616-345025 | SWPV2-306 | 469 | 469 | SWPV2-306 Ig-like domain protein | 100 | 25.8 | M1L | |
| ALPV-330 | 345157-345531 | SWPV2-307 | 124 | 124 | SWPV2-307 EFc-like protein | 100 | |||
| ALPV-331 | 345865-347868 | SWPV2-308 | 667 | 689 | SWPV2-308 ankyrin repeat protein | 96.5 | |||
| ALPV-332 | 348446-347985 | SWPV2-309 | 153 | 186 | SWPV2-309 conserved hypothetical protein | 86.9 | identical to ALPV-005 | ||
| ALPV-333 | 349231-348563 | SWPV2-310 | 222 | 222 | SWPV2-310 conserved hypothetical protein | 100 | identical to ALPV-004 | ||
| ALPV-334 | 349357-349584 | 75 | 56.1 | SWPV1-002 C-type lectin-like protein, identical to ALPV-003 | |||||
| ALPV-335 | 349639-350265 | SWPV2-311 | 208 | 208 | SWPV2-311 C-type lectin-like protein | 100 | identical to ALPV-002 | ||
| ALPV-336 | 351083-350568 | SWPV2-312 | 171 | 171 | SWPV2-312 hypothetical protein | 100 | identical to ALPV-001 |
Note: ALPV, albatrosspox virus (GenBank accession no. MW365933); SWPV1, shearwaterpox virus 1 (GenBank accession no. KX857216); SWPV2, shearwaterpox virus 2 (GenBank accession no. KX857215); CNPV, canarypox virus (GenBank accession no. AY318871). Avipoxviruses as being identity to SWPV2 unless indicated in the note column. Truncated or fragmented ORFs of ALPV compared to SWPV2 are highlighted in italic font.
Interestingly, ALPV contained seven predicted protein-coding genes (ORF030, -067, -080, -081, -213, -226 and -227) that were not present in any other characterised poxvirus genomes, nor did they match any sequences in the NR protein database using BLASTX and BLASTP; these unique ORFs encoded proteins of 51 to 89 amino acids in length (Table 2). Furthermore, four of these unique protein-coding genes (ALPV-ORF030, -213, -226 and -227) were predicted to contain a single transmembrane helix (TMH) using the software packages employed in this study (Table 2). However, we did not find any known motif, nor significant homology with known proteins, for the unique ORFs encoded in the ALPV genome when using the Phyre2, HHpred and SWISS-MODEL, which might be due to the lack of closely related structures in the database.
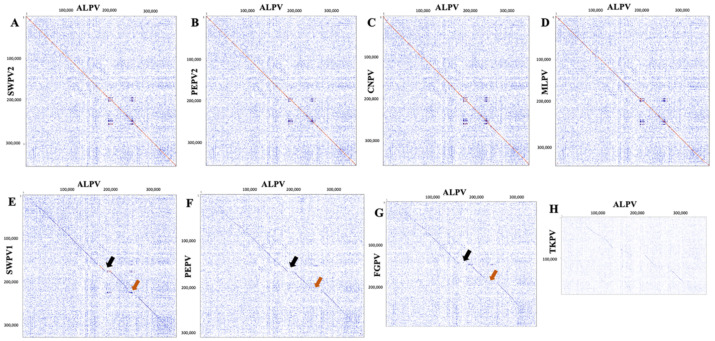
Comparison of the ALPV genome to that of other avipoxviruses was performed using dot plot analyses. The ALPV genome was shown to be highly syntenic with SWPV2, PEPV2, CNPV and MLPV (Figure 2A–D) and demonstrated significant differences compared to SWPV1, PEPV, FGPV and TKPV (black and orange arrows, Figure 2E–H).
Figure 2.
Dot plots of the ALPV genome (x-axis) vs. other poxvirus genomes (y-axis). (A) ALPV vs SWPV2, (B) ALPV vs PEPV2, (C) ALPV vs CNPV, (D) ALPV vs MLPV, (E) ALPV vs SWPV1, (F) ALPV vs PEPV, (G) ALPV vs FGPV and (H) ALPV vs TKPV (refer to Table 2 for virus details and GenBank accession numbers). The Classic colour scheme was chosen in Geneious (version 10.2.2) for the dot plot lines according to the length of the match, from blue for short matches to red for matches over 100 bp long. Window size = 12.
2.3. Evolutionary Relationships of ALPV
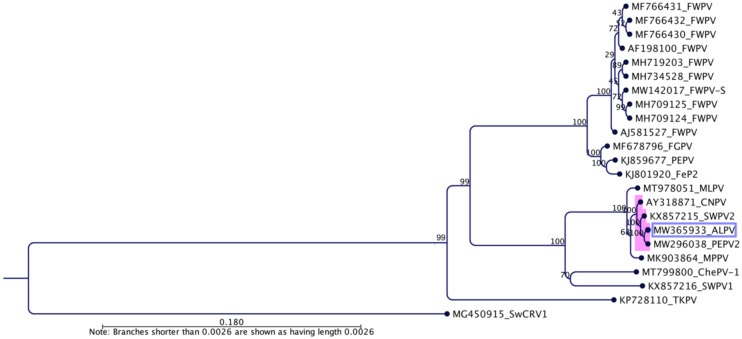
Phylogenetic analysis using concatenated amino acid sequences of the selected nine core poxvirus proteins supported the inclusion of the newly assembled ALPV in the genus Avipoxvirus. In the maximum likelihood (ML) tree, ALPV was located within a sub-clade comprising SWPV2, PEPV2 and CNPV with strong bootstrap support (100%) (Figure 3), suggesting that it may represent an ancient evolutionary lineage within the genus. Using the same set of concatenated protein sequences, we found that the maximum inter-lineage sequence identity values were 100% among ALPV, SWPV2 and PEPV2, which mirrored the phylogenetic position of this novel avipoxvirus sequenced from an endangered northern royal albatross. A large number of poxviruses were positioned in the phylogenetic tree when we used partial nucleotide sequences of the DNA polymerase gene (Supplementary Figure S1) and p4b gene (Supplementary Figure S2). We discovered that several other avipoxviruses were represented within the ALPV, PEPV2, CNPV, SWPV2, MPPV and MLPV clade. This included a poxvirus isolated from a common bullfinch (Pyrrhula pyrrhula) in Belgium [35] and a northern harrier (Circus cyaneus) in Spain [35], which is almost identical to ALPV within this relatively small fragment of the genome.
Figure 3.
Phylogenetic relationships between ALPV and other chordopoxviruses. A maximum likelihood (ML) tree was constructed from multiple alignments of the concatenated amino acid sequences of the selected nine poxvirus core proteins using CLC Genomic Workbench (version 9.5.4, CLC bio, a QIAGEN Company, Prismet, Aarhus C, Denmark). The numbers on the left show bootstrap values as percentages. The ML tree is displayed as a phylogram. The labels at branch tips refer to original ChPV GenBank accession numbers followed by abbreviated species names. Saltwater crocodilepox virus (SwCRV1) [36] was used as an outgroup. The position of the novel ALPV is highlighted using a purple box and the subclade relevant to ALPV is shown with pink shading.
3. Discussion
This study presents the characterisation of the complete genome sequence of a novel avipoxvirus, ALPV, isolated from cutaneous pox lesions in a juvenile northern royal albatross. In addition to the highest number of genes being homologous to SWPV2, a further ten were homologs to CNPV and six to SWPV1. An additional seven genes were not present in any other known poxvirus, nor did they match any sequences in the NR protein database. Given this genome structure, gene content, genome nucleotide similarities and phylogenetic relationships, the authors postulate that the ALPV genome is most closely related to avipoxviruses isolated from shearwater, penguin and canary bird species.
It has been reported that an avipoxvirus may have caused the death of some northern royal albatrosses in 1997 [37]. However, as far as we are aware, there have been no further scientific studies investigating the epidemiology and characteristics of avipoxviruses circulating in this population. Moreover, avipoxviruses have been reported to be a significant cause of chick mortality in several other albatross species. For instance, an avipoxvirus spread by bird fleas has caused high chick mortalities in some seasons within colonies of shy albatrosses (Thalassarche cauta) in Tasmania [38] and may act as a potential threat to adults and chicks of Buller’s albatross (Thalassarche bulleri) [37]. An avipoxvirus has also been described as a key threat for the rapid population decline of a critically endangered seabird, the waved Albatross (Phoebastria irrorata), in the Galápagos Islands, Ecuador [39]. Given the conservation status of the northern royal albatross, it would be important to improve understanding of the epidemiology, transmission and genetic diversity of circulating avipoxviruses, including ALPV, and the threat that they pose to this and other albatross species.
Identifying the transmission mode is essential to characterise incidence, ecology, and effective control of disease in wild populations. However, it is not yet known how this avipoxvirus is transmitted. Mechanical transmission by biting arthropods is thought to play a role in the transmission of avipoxviruses within wild bird populations. Ticks, fleas [40], hippoboscid flies [41], and mosquitos [20,42] are all potential mechanical vectors. It would therefore seem likely that, as for other avipoxviruses, transmission of ALPV in the northern royal albatross is also mediated by insect vectors. Moreover, poxvirus infection can also occur through ingestion, parenteral inoculation, or droplet or aerosol exposure to mucous membranes or broken skin. Some poxviruses can be transmitted by fomites (inanimate objects) [43]. For example, studies have revealed that sheeppox and goatpox viruses are predominantly transmitted via aerosols [44]; whereas poxviruses from the genus Parapoxvirus can pass from one animal to another through direct or indirect contact. However, unfortunately, there are no available studies addressing whether closely related avipoxviruses employ similar or different routes of spread.
There are a number of factors that threaten the populations of large sea birds, particularly albatrosses. Amongst these are longline fishing, climate change and diseases such as those caused by avipoxviruses. It is well-established in the literature that avipoxviruses are mechanically transmitted by biting insects. Although yet to be confirmed, it is therefore expected that this will also be the case for ALPV.
4. Materials and Methods
4.1. Sampling and Virus Isolation
Cutaneous pox lesions were collected from an endangered juvenile northern royal albatross (Diomedea sanfordi), located on the Otago Peninsula, near Dunedin, on the South Island of New Zealand. Sampling was conducted in March 1997 by Wallaceville Animal Research Centre, New Zealand, and lesions sent to the Australian Animal Health Laboratory, Geelong, Victoria, Australia (sample ID: SL 08/05/1997). Virus isolation was undertaken by homogenisation of the tissue samples (~10% w/v) in the presence of antibiotics. This material was then inoculated onto the chorioallantoic membranes (CAMs) of 10 to 12 day old embryonated chicken eggs. The CAMs were harvested 3 to 5 days later and examined for the presence of pock lesions. The infected CAMs were similarly homogenised and passaged onto fresh CAMs. Similarly, the homogenised tissue samples were inoculated onto monolayers of chicken embryo skin cells and examined for 7 to 10 days for the development of cytopathic effect. Additional passages were undertaken with frozen and thawed tissue culture cells inoculated onto fresh chicken embryo skin cells. All passages were stored frozen at −80 °C.
4.2. DNA Extraction and Sequencing
Infected cell culture pellets were digested with DNAase and RNAase, and then with trypsin. Released virus was pelleted through a 36% sucrose cushion for 80 min at 20,000 rpm. Poxvirus cores were released from the pelleted virus with 1% Triton X100 and mercaptoethanol by incubation for 10 min on ice. The released cores were pelleted through a 36% sucrose cushion and the viral DNA released by Proteinase K/RNAase digestion followed by phenol/chloroform extraction and ethanol precipitation, as reported previously [45,46]. Sequencing was undertaken using TruSeq (Illumina) protocols and standard multiplex adaptors available in March 2011. A paired-end 100-base-read protocol was used for sequencing on an Illumina GAIIx instrument using a previously established protocol [47].
4.3. Genome Assembly and Annotation
The resulting 3,343,202 paired-end raw sequence reads were used to assemble the complete genome of ALPV as described previously [25,31,48,49] using CLC Genomics Workbench (version 9.5.4, CLC bio, a QIAGEN Company, Prismet, Aarhus C, Denmark) and Geneious (version 10.2.2, Biomatters, New Zealand). Briefly, the sequences were processed to remove Illumina adapters, low quality reads and ambiguous bases. Trimmed sequence reads were mapped against the chicken genome (Gallus gallus, GenBank accession number NC_006088) to remove likely host DNA contamination. In addition, reads were further mapped to Escherichia coli bacterial genomic sequence (GenBank accession no. U00096) to remove possible bacterial contamination. Unmapped reads were used as input data for de novo assembly using CLC Genomics Workbench (version 9.5.4). This resulted in the generation of a 351,909 bp genome. Clean raw reads (1.15 million) were mapped back to the assembled ALPV genome and resulted in an average coverage of 136.33x. The genome was annotated according to the previously published protocol [19] using Geneious software (version 10.2.2, Biomatters, Auckland, New Zealand). Open reading frames (ORFs) longer than 50 amino acids, with a methionine start codon (ATG) and minimal overlap with other ORFs (not exceeding 50% of one of the genes), were selected and annotated. ORFs shorter than 50 amino acids that had been previously annotated in other poxvirus genomes were also included. Similarity BLAST searches were performed on the predicted ORFs and were annotated as potential genes if predicted ORFs showed significant sequence similarity to known viral or cellular genes (BLAST E value ≤ e−5) [50]. Additional BLAST searches were performed on the predicted ORFs of ALPV against VACV-Cop [51].
To predict the function of unique ORFs tentatively identified in this study, the derived protein sequence of each ORF was searched by multiple applications to identify conserved domains or motifs. Transmembrane helices were searched using the TMHMM package (version 2.0) [52] and TMpred [53]. Additionally, searches for conserved secondary structure (HHpred) [54] and protein homologs using Phyre2 [55] were used to predict the function of unique ORFs identified in this study. To identify the likely promoter sequences of predicted unique ORFs of ALPV, a promoter motif search analysis was conducted using CLC Genomic Workbench (version 9.5.4), where vaccinia virus unique promoter sequences were used [51,56,57,58].
4.4. Comparative Genomics
Genomic features of the newly sequenced ALPV were visualised using Geneious (version 10.2.2). Sequence similarity percentages between representative ChPV and ALPV complete genome sequences were determined using tools available in Geneious (version 10.2.2). Dot plots were created based on the EMBOSS dottup program in Geneious software, with word size = 12 [59].
4.5. Phylogenetic Analyses
Phylogenetic analyses were performed using the novel ALPV genome sequence determined in this study, together with other selected ChPV genome sequences available in GenBank (Table 3). Nucleotide sequences of the partial DNA polymerase and partial p4b genes, as well as concatenated amino acid sequences of the selected nine poxvirus core proteins, were aligned as described previously [30] using the MAFTT L-INS-I algorithm implemented in Geneious (version 7.388) [60]. To determine the best-fit model to construct phylogenetic analyses, a model test was performed using CLC Genomics Workbench (version 9.5.4), which favoured a general-time-reversible model with gamma distribution rate variation and a proportion of invariable sites (GTR+G+I). Phylogenetic analyses for nucleotide sequences were performed using the GTR substitution model with 1000 bootstrap support in CLC Genomics Workbench (version 9.5.4), but the LG substitution model was chosen for concatenated amino acid sequences in Geneious (version 10.2.2).
Table 3.
Related poxvirus genome sequences used in further analysis of ALPV.
| Virus | Abbreviation | GenBank Accession Number | Reference |
|---|---|---|---|
| Albatrosspox virus | ALPV | MW365933 | This study |
| Canarypox virus | CNPV | AY318871 | [32] |
| Cheloniidpox virus 1 | ChePV-1 | MT799800 | [61] |
| Fowlpox virus | FWPV |
AF198100, MF766430-32, MH709124-25, MH719203, MH734528, AJ581527, MW142017 |
[28,29,62,63] |
| Flamingopox virus | FGPV | MF678796 | [24] |
| Magpiepox virus | MPPV | MK903864 | [31] |
| Mudlarkpox virus | MLPV | MT978051 | [30] |
| Nile crocodilepox virus | CRV | DQ356948 | [64] |
| Penguinpox virus | PEPV | KJ859677 | [33] |
| Penguinpox virus 2 | PEPV2 | MW296038 | [19] |
| Pigeonpox virus | FeP2 | KJ801920 | [33] |
| Saltwater crocodilepox virus 1 | SwCRV1 | MG450915 | [36,65] |
| Shearwaterpox virus 1 | SWPV1 | KX857216 | [25] |
| Shearwaterpox virus 2 | SWPV2 | KX857215 | [25] |
| Turkeypox virus | TKPV | NC_028238 | [34] |
5. Conclusions
This study reports the genomic characterisation of a novel avipoxvirus, ALPV, isolated from an endangered northern royal albatross. The ALPV genome sequence was sufficiently divergent from other known avipoxviruses to be considered a novel species within the genus Avipoxvirus, family Poxviridae. This discovery has enhanced our understanding of the pathogen landscape relevant to northern royal albatrosses in New Zealand. Obtaining and sequencing additional poxvirus isolates will also be important to further investigate the epidemiology, transmission, pathogenesis and host specificity of ALPV infections in this endangered bird species.
Acknowledgments
Subir Sarker is the recipient of an Australian Research Council Discovery Early Career Researcher Award (grant number DE200100367) funded by the Australian Government. We also gratefully acknowledge the funding contributed by the Australian Biosecurity CRC for Emerging Infectious Disease in support of this work. The authors are thankful to the New Zealand Ministry for Primary Industries, Animal Health Laboratory, for providing the skin lesion from which ALPV was isolated.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/pathogens10050575/s1, Figure S1: Maximum likelihood (ML) phylogenetic tree from partial nucleotide sequences of the DNA polymerase gene of selected avipoxviruses, Figure S2: Maximum likelihood phylogenetic tree from partial nucleotide sequences of the p4b gene of selected avipoxviruses.
Author Contributions
Conceptualization, S.S., T.R.B. and D.B.B.; Formal analysis, S.S., A.A., T.N. and D.B.B.; Funding acquisition, S.S. and D.B.B.; Investigation, S.S. and D.B.B.; Methodology, S.S. and D.B.B.; Writing—original draft, S.S.; Writing—review and editing, S.S., A.A., T.N., T.R.B. and D.B.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The complete genome sequence and associated datasets generated during this study were deposited in GenBank under the accession number MW365933.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Croxall J.P., Butchart S.H.M., Lascelles B.E.N., Stattersfield A.J., Sullivan B.E.N., Symes A., Taylor P. Seabird conservation status, threats and priority actions: A global assessment. Bird Conserv. Int. 2012;22:1–34. doi: 10.1017/S0959270912000020. [DOI] [Google Scholar]
- 2.Croxall J.P., Gales R. An assessment of the conservation status of albatrosses. In: Robertson G., Gales R., editors. Albatross Biology and Conservation. Surrey Beatty & Sons; Chipping Norton, UK: 1998. pp. 46–65. [Google Scholar]
- 3.Cooper J., Baker G.B., Double M.C., Gales R., Papworth W., Tasker M.L., Waugh S.M. The agreement on the conservation of albatrosses and petrels: Rationale, history, progress and the way forward. Mar. Ornithol. 2006;34:1–5. [Google Scholar]
- 4.Marcela M., Uhart L.G., Flavio Q. Review of diseases (pathogen isolation, direct recovery and antibodies) in albatrosses and large petrels worldwide. Bird Conserv. Int. 2017;28:1–28. doi: 10.1017/S0959270916000629. [DOI] [Google Scholar]
- 5.Paleczny M., Hammill E., Karpouzi V., Pauly D. Population Trend of the World’s Monitored Seabirds, 1950–2010. PLoS ONE. 2015;10:e0129342. doi: 10.1371/journal.pone.0129342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips R.A., Gales R., Baker G.B., Double M.C., Favero M., Quintana F., Tasker M.L., Weimerskirch H., Uhart M., Wolfaardt A. The conservation status and priorities for albatrosses and large petrels. Biol. Conserv. 2016;201:169–183. doi: 10.1016/j.biocon.2016.06.017. [DOI] [Google Scholar]
- 7.BirdLife International. Diomedea Sanfordi. The IUCN Red List of Threatened Species 2018: E.T22728323A132656392. [(accessed on 30 January 2021)];2018 doi: 10.2305/IUCN.UK.2018-2.RLTS.T22728323A132656392.en. Available online: [DOI]
- 8.Sugishita J. Northern royal albatross. In: Miskelly C.M., editor. New Zealand Birds Online. 2013. [(accessed on 20 January 2021)]. Available online: http://www.nzbirdsonline.org.nz/species/northern-royal-albatross. [Google Scholar]
- 9.Halpern B.S., Walbridge S., Selkoe K.A., Kappel C.V., Micheli F., D’Agrosa C., Bruno J.F., Casey K.S., Ebert C., Fox H.E., et al. A global map of human impact on marine ecosystems. Science. 2008;319:948–952. doi: 10.1126/science.1149345. [DOI] [PubMed] [Google Scholar]
- 10.Tuck G.N., Polacheck T., Croxall J.P., Weimerskirch H. Modelling the impact of fishery by-catches on albatross populations. J. Appl. Ecol. 2001;38:1182–1196. doi: 10.1046/j.0021-8901.2001.00661.x. [DOI] [Google Scholar]
- 11.Baker G.B., Gales R., Hamilton S., Wilkinson V. Albatrosses and petrels in Australia: A review of their conservation and management. Emu Austral Ornithol. 2002;102:71–97. doi: 10.1071/MU01036. [DOI] [Google Scholar]
- 12.Lewison R.L., Crowder L.B., Read A.J., Freeman S.A. Understanding impacts of fisheries bycatch on marine megafauna. Trends Ecol. Evol. 2004;19:598–604. doi: 10.1016/j.tree.2004.09.004. [DOI] [Google Scholar]
- 13.Rolland V., Barbraud C., Weimerskirch H. Assessing the impact of fisheries, climate and disease on the dynamics of the Indian yellow-nosed Albatross. Biol. Conserv. 2009;142:1084–1095. doi: 10.1016/j.biocon.2008.12.030. [DOI] [Google Scholar]
- 14.Department of Sustainability, Environment, Water, Population and Communities (2011) Background Paper, Population Status and Threats to Albatrosses and Giant Petrels Listed as Threatened under the Environment Protection and Biodiversity Conservation Act 1999. Commonwealth of Australia, Hobart. [(accessed on 29 January 2021)];2011 Available online: https://www.environment.gov.au/resource/background-paper-population-status-and-threats-albatrosses-and-giant-petrels-listed.
- 15.Illera J.C., Emerson B.C., Richardson D.S. Genetic characterization, distribution and prevalence of avian pox and avian malaria in the Berthelot’s pipit (Anthus berthelotii) in Macaronesia. Parasitol. Res. 2008;103:1435–1443. doi: 10.1007/s00436-008-1153-7. [DOI] [PubMed] [Google Scholar]
- 16.Lecis R., Secci F., Antuofermo E., Nuvoli S., Scagliarini A., Pittau M., Alberti A. Multiple gene typing and phylogeny of avipoxvirus associated with cutaneous lesions in a stone curlew. Vet. Res. Commun. 2017:1–7. doi: 10.1007/s11259-016-9674-5. [DOI] [PubMed] [Google Scholar]
- 17.Woolaver L.G., Nichols R.K., Morton E.S., Stutchbury B.J.M. Population genetics and relatedness in a critically endangered island raptor, Ridgway’s Hawk Buteo ridgwayi. Conserv. Genet. 2013;14:559–571. doi: 10.1007/s10592-013-0444-4. [DOI] [Google Scholar]
- 18.Thiel T., Whiteman N.K., Tirape A., Baquero M.I., Cedeno V., Walsh T., Uzcategui G.J., Parker P.G. Characterization of canarypox-like viruses infecting endemic birds in the Galapagos Islands. J. Wildl. Dis. 2005;41:342–353. doi: 10.7589/0090-3558-41.2.342. [DOI] [PubMed] [Google Scholar]
- 19.Sarker S., Athukorala A., Bowden T.R., Boyle D.B. Genomic Characterisation of a Novel Avipoxvirus Isolated from an Endangered Yellow-Eyed Penguin (Megadyptes antipodes) Viruses. 2021;13:194. doi: 10.3390/v13020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Riper C., III, van Riper S.G., Hansen W.R. Epizootiology and Effect of Avian Pox on Hawaiian Forest Birds. Auk. 2002;119:929–942. doi: 10.1093/auk/119.4.929. [DOI] [Google Scholar]
- 21.Young L.C., VanderWerf E.A. Prevalence of avian pox virus and effect on the fledging success of Laysan Albatross. J. Field Ornithol. 2008;79:93–98. doi: 10.1111/j.1557-9263.2008.00149.x. [DOI] [Google Scholar]
- 22.Bolte A.L., Meurer J., Kaleta E.F. Avian host spectrum of avipoxviruses. Avian Pathol. 1999;28:415–432. doi: 10.1080/03079459994434. [DOI] [PubMed] [Google Scholar]
- 23.van Riper C., Forrester D.J. Avian Pox. In: Thomas N.J., Hunter D.B., Atkinson C.T., editors. Infectious Diseases of Wild Birds. Wiley Blackwell Publishing; Oxford, UK: 2007. pp. 131–176. [Google Scholar]
- 24.Carulei O., Douglass N., Williamson A.-L. Comparative analysis of avian poxvirus genomes, including a novel poxvirus from lesser flamingos (Phoenicopterus minor), highlights the lack of conservation of the central region. BMC Genom. 2017;18:947. doi: 10.1186/s12864-017-4315-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarker S., Das S., Lavers J.L., Hutton I., Helbig K., Imbery J., Upton C., Raidal S.R. Genomic characterization of two novel pathogenic avipoxviruses isolated from pacific shearwaters (Ardenna spp.) BMC Genom. 2017;18:298. doi: 10.1186/s12864-017-3680-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tripathy D.N., Schnitzlein W.M., Morris P.J., Janssen D.L., Zuba J.K., Massey G., Atkinson C.T. Characterization of poxviruses from forest birds in Hawaii. J. Wildl. Dis. 2000;36:225–230. doi: 10.7589/0090-3558-36.2.225. [DOI] [PubMed] [Google Scholar]
- 27.Niemeyer C., Favero C.M., Kolesnikovas C.K.M., Bhering R.C.C., Brandão P., Catão-Dias J.L. Two different avipoxviruses associated with pox disease in Magellanic penguins (Spheniscus magellanicus) along the Brazilian coast. Avian Pathol. 2013;42:546–551. doi: 10.1080/03079457.2013.849794. [DOI] [PubMed] [Google Scholar]
- 28.Joshi L.R., Bauermann F.V., Hain K.S., Kutish G.F., Armién A.G., Lehman C.P., Neiger R., Afonso C.L., Tripathy D.N., Diel D.G. Detection of Fowlpox virus carrying distinct genome segments of Reticuloendotheliosis virus. Virus Res. 2019;260:53–59. doi: 10.1016/j.virusres.2018.10.017. [DOI] [PubMed] [Google Scholar]
- 29.Afonso C.L., Tulman E.R., Lu Z., Zsak L., Kutish G.F., Rock D.L. The genome of fowlpox virus. J. Virol. 2000;74:3815–3831. doi: 10.1128/JVI.74.8.3815-3831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarker S., Athukorala A., Raidal S.R. Molecular characterisation of a novel pathogenic avipoxvirus from an Australian passerine bird, mudlark (Grallina cyanoleuca) Virology. 2021;554:66–74. doi: 10.1016/j.virol.2020.12.011. [DOI] [PubMed] [Google Scholar]
- 31.Sarker S., Batinovic S., Talukder S., Das S., Park F., Petrovski S., Forwood J.K., Helbig K.J., Raidal S.R. Molecular characterisation of a novel pathogenic avipoxvirus from the Australian magpie (Gymnorhina tibicen) Virology. 2020;540:1–16. doi: 10.1016/j.virol.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 32.Tulman E.R., Afonso C.L., Lu Z., Zsak L., Kutish G.F., Rock D.L. The Genome of Canarypox Virus. J. Virol. 2004;78:353–366. doi: 10.1128/JVI.78.1.353-366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Offerman K., Carulei O., van der Walt A.P., Douglass N., Williamson A.-L. The complete genome sequences of poxviruses isolated from a penguin and a pigeon in South Africa and comparison to other sequenced avipoxviruses. BMC Genom. 2014;15:1–17. doi: 10.1186/1471-2164-15-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banyai K., Palya V., Denes B., Glavits R., Ivanics E., Horvath B., Farkas S.L., Marton S., Balint A., Gyuranecz M., et al. Unique genomic organization of a novel Avipoxvirus detected in turkey (Meleagris gallopavo) Infect Genet. Evol. 2015;35:221–229. doi: 10.1016/j.meegid.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 35.Gyuranecz M., Foster J.T., Dán Á., Ip H.S., Egstad K.F., Parker P.G., Higashiguchi J.M., Skinner M.A., Höfle U., Kreizinger Z., et al. Worldwide Phylogenetic Relationship of Avian Poxviruses. J. Virol. 2013;87:4938–4951. doi: 10.1128/JVI.03183-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarker S., Isberg S.R., Milic N.L., Lock P., Helbig K.J. Molecular characterization of the first saltwater crocodilepox virus genome sequences from the world’s largest living member of the Crocodylia. Sci. Rep. 2018;8:5623. doi: 10.1038/s41598-018-23955-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graeme A.T. Action Plan for Seabird Conservation in New Zealand. Part A: Threatened Seabirds. Threatened Species Occasional Publication, 1170-3709; 16, Biodiversity Recovery Unit, Department of Conservation, Wellington, New Zealand. [(accessed on 30 January 2021)];2000 Available online: https://www.doc.govt.nz/documents/science-and-technical/tsop16.pdf.
- 38.Woods R. In: Results of a Preliminary Disease Survey in Shy Albatross (Thalassarche Cauta Gould 1841) Chicks at Albatross Island, Bass Strait, Tasmania. Woods R., editor. Australian Association of Veterinary Conservation Biologists; Canberra, Australia: 2004. pp. 98–104. [Google Scholar]
- 39.Tompkins E.M., Anderson D.J., Pabilonia K.L., Huyvaert K.P. Avian Pox Discovered in the Critically Endangered Waved Albatross (Phoebastria irrorata) from the Galápagos Islands, Ecuador. J. Wildl. Dis. 2017;53:891–895. doi: 10.7589/2016-12-264. [DOI] [PubMed] [Google Scholar]
- 40.Kane O.J., Uhart M.M., Rago V., Pereda A.J., Smith J.R., Van Buren A., Clark J.A., Boersma P.D. Avian pox in Magellanic Penguins (Spheniscus magellanicus) J. Wildl. Dis. 2012;48:790–794. doi: 10.7589/0090-3558-48.3.790. [DOI] [PubMed] [Google Scholar]
- 41.Warner R.E. The Role of Introduced Diseases in the Extinction of the Endemic Hawaiian Avifauna. Condor. 1968;70:101–120. doi: 10.2307/1365954. [DOI] [Google Scholar]
- 42.Annuar B.O., Mackenzie J.S., Lalor P.A. Isolation and characterization of avipoxviruses from wild birds in Western Australia. Arch. Virol. 1983;76:217–229. doi: 10.1007/BF01311106. [DOI] [PubMed] [Google Scholar]
- 43.Aleksandr K., Olga B., David W.B., Pavel P., Yana P., Svetlana K., Alexander N., Vladimir R., Dmitriy L., Alexander S. Non-vector-borne transmission of lumpy skin disease virus. Sci. Rep. 2020;10:7436. doi: 10.1038/s41598-020-64029-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kitching R.P., Taylor W.P. Transmission of capripoxvirus. Res. Vet. Sci. 1985;39:196–199. doi: 10.1016/S0034-5288(18)31744-2. [DOI] [PubMed] [Google Scholar]
- 45.Nakano E., Panicali D., Paoletti E. Molecular genetics of vaccinia virus: Demonstration of marker rescue. Proc. Natl. Acad. Sci. USA. 1982;79:1593–1596. doi: 10.1073/pnas.79.5.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prideaux C.T., Boyle D.B. Fowlpox virus polypeptides: Sequential appearance and virion associated polypeptides. Arch. Virol. 1987;96:185–199. doi: 10.1007/BF01320959. [DOI] [PubMed] [Google Scholar]
- 47.Sarker S., Roberts H.K., Tidd N., Ault S., Ladmore G., Peters A., Forwood J.K., Helbig K., Raidal S.R. Molecular and microscopic characterization of a novel Eastern grey kangaroopox virus genome directly from a clinical sample. Sci. Rep. 2017;7:16472. doi: 10.1038/s41598-017-16775-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Boyle D.B., Bulach D.M., Amos-Ritchie R., Adams M.M., Walker P.J., Weir R. Genomic sequences of Australian blue-tongue virus prototype serotypes reveal global relationships and possible routes of entry into Australia. J. Virol. 2012;86:6724–6731. doi: 10.1128/JVI.00182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarker S., Isberg S.R., Athukorala A., Mathew R., Capati N., Haque M.H., Helbig K.J. Characterization of a Complete Genome Sequence of Molluscum Contagiosum Virus from an Adult Woman in Australia. Microbiol. Resour. Announc. 2021;10:e00939-20. doi: 10.1128/MRA.00939-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2013;41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Z., Bruno D.P., Martens C.A., Porcella S.F., Moss B. Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing. Proc. Natl. Acad. Sci. USA. 2010;107:11513. doi: 10.1073/pnas.1006594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 53.Hofmann K., Stoffel W. TMBASE—A database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler. 1993;374:166. [Google Scholar]
- 54.Zimmermann L., Stephens A., Nam S.Z., Rau D., Kubler J., Lozajic M., Gabler F., Soding J., Lupas A.N., Alva V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018;430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 55.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Z., Reynolds S.E., Martens C.A., Bruno D.P., Porcella S.F., Moss B. Expression profiling of the intermediate and late stages of poxvirus replication. J. Virol. 2011;85:9899–9908. doi: 10.1128/JVI.05446-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Z., Maruri-Avidal L., Sisler J., Stuart C.A., Moss B. Cascade regulation of vaccinia virus gene expression is modulated by multistage promoters. Virology. 2013;447:213–220. doi: 10.1016/j.virol.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goebel S.J., Johnson G.P., Perkus M.E., Davis S.W., Winslow J.P., Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 59.Maizel J.V., Jr., Lenk R.P. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc. Natl. Acad. Sci. USA. 1981;78:7665–7669. doi: 10.1073/pnas.78.12.7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katoh K., Standley D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarker S., Hannon C., Athukorala A., Bielefeldt-Ohmann H. Emergence of a Novel Pathogenic Poxvirus Infection in the Endangered Green Sea Turtle (Chelonia mydas) Highlights a Key Threatening Process. Viruses. 2021;13:219. doi: 10.3390/v13020219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Croville G., Le Loc’h G., Zanchetta C., Manno M., Camus-Bouclainville C., Klopp C., Delverdier M., Lucas M.-N., Donnadieu C., Delpont M., et al. Rapid whole-genome based typing and surveillance of avipoxviruses using nanopore sequencing. J. Virol. Methods. 2018;261:34–39. doi: 10.1016/j.jviromet.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 63.Sarker S., Athukorala A., Bowden T.R., Boyle D.B. Characterisation of an Australian fowlpox virus carrying a near-full-length provirus of reticuloendotheliosis virus. Arch. Virol. 2021;166:1485–1488. doi: 10.1007/s00705-021-05009-x. [DOI] [PubMed] [Google Scholar]
- 64.Afonso C.L., Tulman E.R., Delhon G., Lu Z., Viljoen G.J., Wallace D.B., Kutish G.F., Rock D.L. Genome of Crocodilepox Virus. J. Virol. 2006;80:4978–4991. doi: 10.1128/JVI.80.10.4978-4991.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sarker S., Isberg R.S., Moran L.J., Araujo D.R., Elliott N., Melville L., Beddoe T., Helbig J.K. Crocodilepox Virus Evolutionary Genomics Supports Observed Poxvirus Infection Dynamics on Saltwater Crocodile (Crocodylus porosus) Viruses. 2019;11:1116. doi: 10.3390/v11121116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete genome sequence and associated datasets generated during this study were deposited in GenBank under the accession number MW365933.