Abstract
Coelomycetous fungi are ubiquitous in soil, sewage, and sea- and freshwater environments. However, freshwater coelomycetous fungi have been very rarely reported in the literature. Knowledge of coelomycetous fungi in freshwater habitats in Spain is poor. The incubation of plant debris, from freshwater in various places in Spain into wet chambers, allowed us to detect and isolate in pure culture several pycnidia-producing fungi. Fungal strains were phenotypically characterized, and a phylogenetic study was carried out based on the analysis of concatenated nucleotide sequences of the D1–D2 domains of the 28S nrRNA gene (LSU), the internal transcribed spacer region (ITS) of the nrDNA, and fragments of the RNA polymerase II subunit 2 (rpb2) and beta tubulin (tub2) genes. As a result of these, we report the finding of two novel species of Neocucurbitaria, three of Neopyrenochaeta, and one of Pyrenochaetopsis. Based on the phylogenetic study, we also transferred Neocucurbitaria prunicola to the genus Allocucurbitaria. This work makes an important contribution to the knowledge of the mycobiota of plant debris in freshwater habitats.
Keywords: Ascomycota, coelomycetous, freshwater fungi, phylogeny, plant debris, taxonomy
1. Introduction
Coelomycetous fungi are characterized by the production of conidia within a cavity lined by fungal or fungal-host tissue called conidiomata [1]. Conidiomata can be acervular (open, cup-shaped asexual fruiting bodies developing below the epidermis of the plant host tissue and bearing a series of adpressed conidiophores), pycnidial (globose, pyriform to flask-shaped asexual reproductive structures whose conidia are liberated through an usually apical opening [ostiolum]), or stromatic (consisting of undifferentiated sclerotic tissues, ostiolate or not, in which one or more lysigenic cavities develops, upholstered inside by conidiophores/conidiogenous cells forming conidia). Coelomycetous fungi are mostly parasites of terrestrial vascular plants but are also saprobic, growing at the expense of dead organic matter on the ground, especially on plant debris. These are ubiquitous on soil, sewage, and in salt- and freshwater environments. [2]. Freshwater coelomycetous fungi occur on stream-side plants or on submerged wood litter, and their conidia can also be recovered from foam and water samples [3]. Usually, they produce brown to blackish pycnidial fruiting bodies on submerged woody debris and stems of herbaceous plants, and produce several conidia from the conidiogenous cells [4]. Identification of coelomycetous fungi has gone through dramatic changes over the last decade, and currently involves DNA sequencing of several (four to six) genetic markers and the building of phylogenetic trees [5]. In Spain, there have been a few reports of coelomycetous fungi recovered from freshwater habitats. In 1990, Roldán and Honrubia reported Bartalinia robillardoides and Truncatella angustata [6], and Giralt described Diplolaviopsis ranula [7]. Up to 2014, only 16 coelomycetous fungi had been reported from freshwater habitats [4,8,9,10,11,12,13,14,15,16].
The main objective of this work was to characterize phenotypically and to identify molecularly those coelomycetous fungi found in different freshwater habitats in Spain.
2. Materials and Methods
2.1. Sampling and Fungal Isolation
A hundred samples of decomposing plant material submerged in freshwater habitats in Spain were collected: 3 from “Les Guilleries” (Barcelona province), 50 from “Cascadas del Huéznar” (Cazalla de la Sierra, Sevilla province, Spain), 17 from Riaza (Segovia province, Spain), and 30 from “Serra del Montsant” (Tarragona province, Spain). The samples were placed into self-sealing sterile plastic bags, which were closed and transported to the laboratory, and stored at room temperature (20−25 °C) until they were processed. The specimens were rinsed twice with 500 mL tap water, placed into Petri dishes or appropriate plastic containers lined inside with two sheets of filter paper, and moistened with sterile water with diehldrin® (1 mL of a solution of 20 mg diehldrin® in 20 mL of dimethyl-ketone/L of water), incubated at room temperature, and examined periodically under stereomicroscope for up to 2 months. Several propagules and/or fruiting bodies were taken and transferred using sterile disposable tuberculin-type needles to 55 mm diameter Petri dishes containing oatmeal agar (OA; 30 g of filtered oat flakes, 15 g agar-agar, 1 L tap water; [17]), and then incubated at room temperature. Once obtaining an axenic culture of each isolate, these were stored in the culture collection of the Faculty of Medicine of University Rovira i Virgili (FMR; Reus, Spain). Type specimens and ex-type cultures of the novel fungi were deposited in the Westerdijk Fungal Biodiversity Institute (CBS), Utrecht, The Netherlands (Table S1).
2.2. Phenotypic Study
Macroscopic characterization of the colonies was performed on OA and on malt extract agar (MEA; Difco, Detroit, MI, USA) incubated for 14 d in the dark at 25 ± 1 °C [17]. Colony colour was determined according to Kornerup and Wanscher [18]. The ability of the isolates to grow at cardinal temperatures was determined on potato dextrose agar (PDA; Pronadisa, Madrid, Spain) after 7 d in the dark, ranging from 5 to 35 ± 1 °C at 5 °C intervals, plus 37 ± 1 °C [19]. Morphological characterization of vegetative and reproductive structures was performed growing the fungal strains on OA in the same conditions as for colony characterization, and examining at least 30 individuals of each structure [20,21] on Shear’s mounting medium (3 g potassium acetate, 60 mL glycerol, 90 mL ethanol 95%, and 150 mL distilled water; [22]) using a Olympus BH-2 bright field microscope (Olympus Corporation, Tokyo, Japan). Photomicrographs were taken using a Zeiss Axio-Imager M1 microscope (Oberkochen, Germany) with a DeltaPix Infinity X digital camera using Nomarski differential interference contrast.
2.3. DNA Extraction, Amplification and Sequencing
Fungal strains were cultured on PDA for 7 days at 25 ± 1 °C in the dark. Total DNA was extracted using the FastDNA kit protocol (Bio101, Vista, CA, USA) with a FastPrep FP120 instrument (Thermo Savant, Holbrook, NY, USA) according to the manufacturer’s protocol. DNA was quantified by using Nanodrop 2000 (Thermo Scientific, Madrid, Spain). The following loci were amplified and sequenced: LSU, with the primer pair LR0R [23] and LR5 [24]; ITS, with the primer pair ITS5 and ITS4 [25]; a fragment of the beta-tubulin gene (tub2) with the primers TUB2Fw and TUB4Rd [26]; and a fragment of the RNA polymerase II subunit 2 gene (rpb2) with RPB2-5F2 [27] and fRPB2-7cR primers [28]. The PCR amplifications were performed in a total volume of 25 µL containing 5 µL 10× PCR Buffer (Invitrogen, CA, USA), 0.2 mM dNTPs, 0.5 µL of each primer, 1 U Taq DNA polymerase, and 1−10 ng genomic DNA. PCR conditions for LSU, ITS, and tub2 were set as follows: an initial denaturation at 95 °C for 5 min; followed by 35 cycles of denaturation, annealing, and extension; and a final extension step at 72 °C for 10 min. For the LSU and ITS amplification, the 35 cycles consisted of 45 s at 95 °C, 45 s at 53 °C, and 2 min at 72 °C; and for the tub2 region 30 s at 94 °C, 45 s at 56°C, and 1 min at 72 °C. The PCR program for rpb2 amplification consisted of 5 cycles of 45 s at 94 °C, 45 s at 60 °C, and 2 min at 72 °C; then 5 cycles with 58 °C annealing temperature; and 30 cycles with a 54 °C annealing temperature. PCR products were purified and stored at −20 °C until sequencing. The same pairs of primers were used to obtain the sequences at Macrogen Spain (Macrogen Inc., Madrid, Spain). The consensus sequences were obtained using the SeqMan software v. 7 (DNAStar Lasergene, Madison, WI, USA).
2.4. Phylogenetic Analysis
We made a preliminary molecular identification by comparing the LSU, ITS, tub2, and rpb2 sequences of our isolates with those of the National Center for Biotechnology Information (NCBI) using the Basic Local Alignment Search Tool (BLAST; https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 16 March 2021)). For tub2 sequences, a maximum level of identity (MLI) of <98% provides identification only at genus level, and a value >98% was considered to allow for species-level identification. Alignment for each locus was performed with the MEGA (Molecular Evolutionary Genetics Analysis) software v. 7.0. (Tamura et al. 2013), using the ClustalW algorithm [29] and refined with MUSCLE [30] or manually, if necessary, on the same platform. Individual and concatenated phylogenetic trees were built after a maximum likelihood (ML) analysis carried out using the RAxML v. 8.2.10 [31] software on the online Cipres Science gateway portal [32], and a Bayesian Inference (BI) analysis using MrBayes v. 3.2.6 [33]. For ML analyses, the best nucleotide substitution model was General Time Reversible with Gamma distribution. Support for internal branches was assessed by 1000 ML bootstrapped pseudoreplicates. For the BI phylogenetic analysis, the best nucleotide substitution model was determined using jModelTest [34]. For ITS we used the symmetrical model with gamma distribution (SYM + G), for LSU and tub2 the symmetrical model with proportion of invariable sites and gamma distribution (SYM + I + G), and for rpb2 the symmetrical model with gamma distribution (SYM + G). The parameter settings used were two simultaneous runs of 5 M generations and four Markov chain Monte Carlo (MCMC), sampled every 1000 generations. The 50% majority-rule consensus tree and posterior probability values (PP) were calculated after discarding the first 25% of the samples. Pleospora herbarum CBS 191.86 and P. typhicola CBS 132.69 served as outgroup taxa. Confident branch support is defined as Bayesian posterior probabilities (PP) >0.95 and maximum likelihood bootstrap support (BS) >70%. Sequences generated in this study were deposited in European Nucleotide Archive (ENA), the final matrix used for phylogenetic analyses in TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S28077 (accessed on 16 March 2021)) and the novel taxonomic descriptions and nomenclature in MycoBank (www.mycobank.org (accessed on 16 March 2021)).
3. Results
3.1. Blast Search
Blast search results are shown in Table S2 (Supplementary Material).
3.2. Phylogenetic Relationships among Freshvwater Fungi
The final concatenated dataset obtained with both ML and BI analyses contained 71 in-groups of strains with a total of 2252 characters including gaps (455 for ITS, 791 for LSU, 272 for tub2, and 734 for rpb2), of which 704 are parsimony informative (170 for ITS, 69 for LSU, 143 for tub2, and 322 for rpb2). The sequence datasets did not show conflict in the tree topologies for the 70% reciprocal bootstrap trees, which allowed us to combine the four genes for the multi-locus analysis. The ML analysis showed similar tree topology and was congruent with that obtained in the BI. For the BI multi-locus analysis, a total of 11,663 trees were sampled after removal of the burn-in and reaching a stop value of 0.01. The support values were slightly different with the two analysis methods. In the phylogenetic tree (Figure 1), our strains were spread into three well-supported main clades, representing the families Cucurbitariaceae (99% BS/1 PP), Neopyrenochaetaceae (98% BS/1 PP), and Pyrenochaetopsisaceae (100% BS/1 PP). The Cucurbitariaceae clade was divided into four well-supported clades corresponding to the accepted genera (Neocucurbitaria, 100% BS/1 PP; Paracucurbitaria, 100% BS/1 PP; Cucurbitaria, 100% BS/1 PP and Allocucurbitaria, 95% BS/1 PP). The Neocucurbitaria clade was represented by all previously described species and three of our strains, all placed in independent terminal branches. The clade corresponding to the genus Allocucurbitaria included the type species A. botulispora and the new combination A. prunicola (basionym Neocucurbitaria prunicola). The Neopyrenochaetaceae clade included 11 species of the genus Neopyrenochaeta. Five of our strains resulted as co-specific with N. annellidica and N. maesuayensis, whereas the other three strains were each placed into independent terminal branches. The family Pyrenochaetopsisaceae was divided in two clades, corresponding to the genera Neopyrenochaetopsis and Pyrenochaetopsis (100% BS/1PP). Pyrenochaetopsis encompassed 16 species and our strain FMR 17327, which is located in an independent branch. Single gene-based phylogenies are shown as Supplemental Material (Figures S1–S4) because they resulted in being less informative and resolutive than those based on the four-loci concatenated tree.
Figure 1.
ML phylogenetic tree of Cucurbitariaceae, Pseudopyrenochaetaceae, Neopyrenochaetaceae, and Pyrenochaetopsidaceae inferred from the combined sequences of ITS, LSU, tub2, and rpb2 loci. Support in nodes is indicated above branches and is represented by posterior probabilities (BI analysis) of 0.95 and higher, and/or bootstrap values (ML analysis) of 70% and higher. Full-supported branches (100% BS/1 PP) are indicated by thicker lines. T =ex-type strains. New species are indicated in blue. New strains isolated during this study are indicated in bold. Alignment length 2252 bp. The sequences not generated by us were retrieved from EMBL/GenBank and are indicated in Supplementary Table S1.
3.3. Taxonomy
Dothideomycetes.
Cucurbitariaceae G. Winter (as Cucurbitarieae), Rabenh. Krypt.-Fl., Edn 2 (Leipzig) 1.2: 308 (1885). MycoBank MB 80667.
Type genus: Cucurbitaria Gray, Nat. Arr. Brit. Pl. (London) 1: 519 (1821).
Neocucurbitaria Wanas., E.B.G. Jones & K.D. Hyde, in Wanasinghe, Phookamsak, Jeewon, Li, Hyde, Jones, Camporesi & Promputtha, Mycosphere 8(3): 408 (2017). MycoBank MB 552832.
Type species: Neocucurbitaria unguis-hominis (Punith. & M.P. English) Wanas., E.B.G. Jones & K.D. Hyde, in Wanasinghe, Phookamsak, Jeewon, Li, Hyde, Jones, Camporesi & Promputtha, Mycosphere 8(3): 412 (2017). MB 552835.
= Pyrenochaeta unguis-hominis Punith. & M.P. English, Transactions of the British Mycological Society 64 (3): 539 (1975). MB 322137.
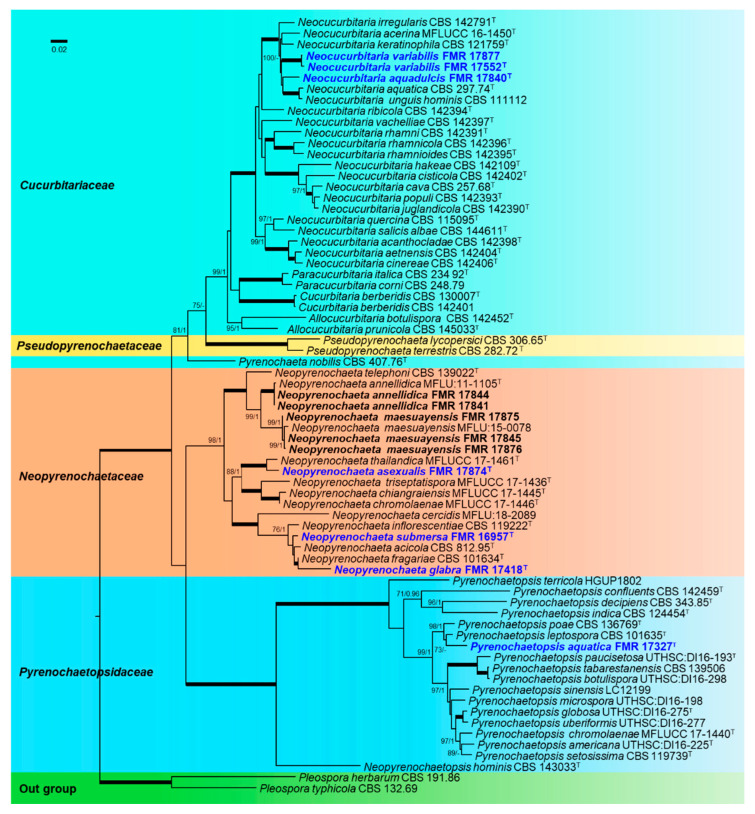
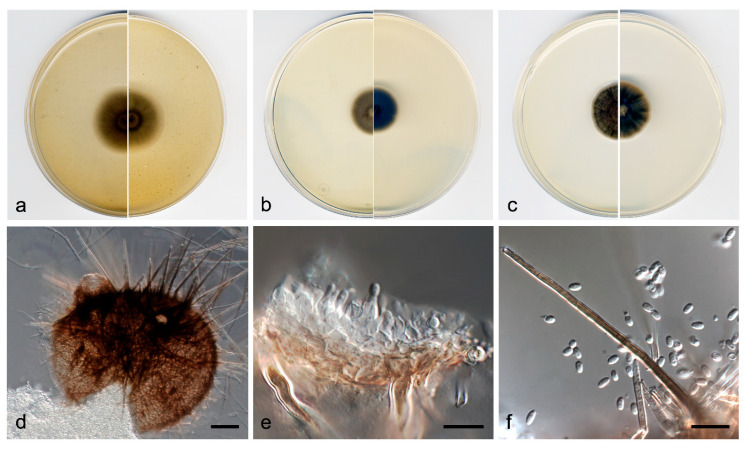
Neocucurbitaria variabilis V. Magaña-Dueñas, Stchigel & Cano, sp. nov. Figure 2. MycoBank MB 838833.
Figure 2.
Neocucurbitaria variabilis FMR 17552 T. (a) Colonies on OA, (b) MEA, and (c) PDA, after 2 weeks at 25 ± 1 °C (surface, left; reverse, right); (d) pycnidium; (e) conidiogenous cells (black arrow, elongated cylindrical; white arrow, flask-shaped); (f) conidia. Scale bars: d = 50 µm, e,f = 10 µm.
Etymology: From Latin variabilis, due to the variable shape of the conidiogenous cells.
Type: Spain, Segovia province, Riaza, from plant debris in freshwater, May 2018, Viridiana Magaña Dueñas, holotype CBS H-24739, culture ex-type FMR 17552.
Description: Hyphae hyaline to pale brown; septate; branched; smooth- and thin-walled; 2–5 µm wide; with short, finger-like lateral projections; anastomosing. Conidiomata pycnidial, brown to dark brown, immersed to semi-immersed, solitary, scattered, setose, ostiolate, subglobose to globose, 110−120 µm × 120–140 µm, ostiole 40−50 µm diameter. Setae pale brown, erect, septate, smooth- and thick-walled, rounded at the tip, 40−80 µm × 3−4 µm. Conidiomata wall composed of three to five layers of cells, 15–25 µm thick, covered by a mass of interwoven, pale brown to brown hyphae; outer layer of textura angularis, composed of brown to dark brown, flattened polygonal cells of 3.5–4.5 µm diameter. Conidiophores absent. Conidiogenous cells phialidic, determinate, hyaline and smooth-walled, flask-shaped, 5–6 µm × 2–3 µm, or elongate-cylindrical, straight, sinuous or slightly curved, 10−14 µm × 1.5−3 µm. Conidia one-celled, hyaline, smooth- and thin-walled, ellipsoidal, ovoid or kidney-shaped, 2.5–3.5 µm × 1.0–1.5 µm. Chlamydospores absent.
Culture characteristics: Colonies on PDA reaching 22 mm diameter after 7 days at 25 ± 1 °C, flattened, velvety, margin regular, surface and reverse yellowish grey (4B2). Colonies on OA reaching 20 mm diameter after 7 days at 25 ± 1 °C, flattened, floccose, margin regular, grey to brownish grey (4F1/4D2); reverse grey to yellowish grey (4F1/4B2). Colonies on MEA reaching 16 mm diameter after 7 days at 25 ± 1 °C, umbonate, velvety, margin regular, yellowish grey to olive brown (4B2/4D3); reverse brownish grey to yellowish grey (7F2/4B2). Exopigment absent. Cardinal temperatures of growth: minimum 5 °C, optimum 25 °C, and maximum 30 °C.
Other material examined: Spain, Sevilla province, Parque Natural Sierra Norte (37.994712, −5.668709), from plant debris in freshwater, May 2019, José F. Cano Lira, living cultures FMR 17877.
Diagnosis: In our phylogenetic tree, N. variabilis was placed in a terminal branch within the same subclade as N. acerina, N. aquadulcis, N. aquatica, N. irregularis, N. keratinophila, and N. unguis-hominis. Neocucurbitaria variabilis differs morphologically from all these species in having two kinds of enteroblastic conidiogenous cells, ampulliform and elongate-cylindrical, which are discrete; and doliiform in N. acerina, N. aquatica and N. irregularis, and integrated in acropleurogenous conidiophores (i.e., having terminal and lateral openings) in N. keratinophila and N. unguis-hominis [35].
Notes: Differences in nucleotide sequences (ITS-LSU-tub2-rpb2 concatenated dataset; 2252 bp) between N. variabilis and the species in the same terminal clade are: N. aquadulcis, 71 bp; N. keratinophila, 77 bp; N. irregularis, 78 bp; N. acerina, 80 bp; N. aquatica, 82 bp; and N. unguis-hominis, 83 bp.
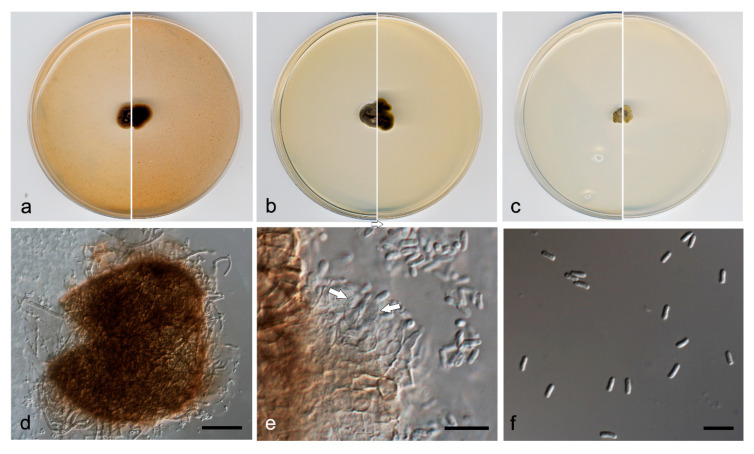
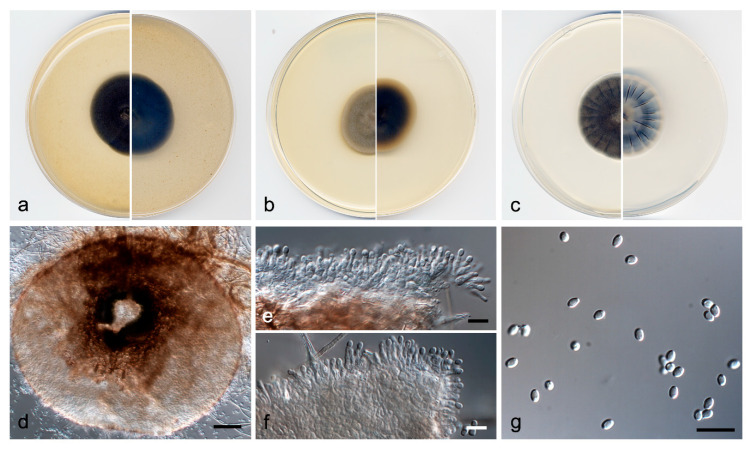
Neocucurbitaria aquadulcis V. Magaña-Dueñas, Cano & Stchigel, sp. nov. Figure 3. MycoBank MB 838834.
Figure 3.
Neocucurbitaria aquadulcis FMR 17840 T. (a) Colonies on OA, (b) MEA, and (c) PDA, after 2 weeks at 25 ± 1 °C (surface, left; reverse, right); (d) pycnidium; (e) conidiogenous cells (white arrows); (f) conidia. Scale bars: d = 50 µm, e,f = 10 µm.
Etymology: From Latin aqua-, water; and -dulcis, sweet, because of the origin of the fungus.
Type: Spain, Sevilla province, Parque Natural Sierra Norte (37.931670, −5.704493), from plant debris in freshwater, May 2019, José F. Cano Lira, holotype CBS H-24740, culture ex-type FMR 17840 = CBS 147605.
Description: Hyphae hyaline to light brown, septate, branched, smooth- and thin-walled, 2–3 µm wide, anastomosing. Conidiomata pycnidial, brown to dark brown, immersed to semi-immersed, solitary or confluent, scattered, ostiolate, covered by a mass of interwoven, pale brown to brown hyphae, ovoid to globose, 120–150 µm × 130–170 µm, ostiole of 15–20 µm diameter. Conidiomata wall composed of three to six layers of cells, 15–35 µm thick, outer layer of textura angularis, composed of brown to dark brown, flattened polygonal cells of 3–5.5 µm diameter. Conidiophores absent. Conidiogenous cells phialidic, determinate, hyaline, smooth-walled, ampulliform, 4–6 µm × 2–3 µm. Conidia one-celled, hyaline, smooth- and thin-walled, bacillary, slightly curved, 2.5–4.5 µm × 1.0–2.0 µm. Chlamydospores absent.
Culture characteristics: Colonies on PDA reaching 6–9 mm diameter after 7 days at 25 ± 1 °C, convex, granular, margins irregular, surface, and reverse olive brown (4D5). Colonies on OA reaching 10–11 mm diameter after 7 days at 25 ± 1 °C, flattened, velvety, margin regular, surface and reverse yellowish brown (5F5) to brownish grey (5B2). Colonies on MEA reaching 10–15 mm diameter after 7 days at 25 ± 1 °C, umbonate, velvety, margins regular, brownish grey to orange grey (5E2/5B2) reverse yellowish brown to orange grey (5E4/5B2). Exopigment not produced. Cardinal temperatures of growth: minimum 5 °C, optimum 25 °C, and maximum 30 °C.
Diagnosis: Neocucurbitaria aquadulcis, unlike N. variabilis, produces solely ampulliform phialides (see before) and bigger conidia (2.5–4.5 µm × 1.1–1.9 µm vs. 2.5–3.5 µm × 1.2–1.7 µm). Also, the conidiomata wall of N. aquadulcis is covered by hyphae, whereas it is setose in N. variabilis.
Notes: Differences in nucleotide sequences (ITS-LSU-tub2-rpb2 concatenated dataset; 2252 bp) between N. aquadulcis and the species in the same terminal clade are: N. keratinophila, 54 bp; N. irregularis, 57 bp; N. acerina, 61 bp; N. aquatica and N. unguis-hominis, 62 bp; and N. variabilis, 71 bp.
Allocucurbitaria Valenz.-Lopez, Stchigel, Guarro & Cano, in Valenzuela-Lopez, Cano-Lira, Guarro, Sutton, Wiederhold, Crous & Stchigel, Stud. Mycol. 90:51 (2017). MycoBank MB 821455.
Type species: Allocucurbitaria botulispora Valenz.-Lopez, Stchigel, Guarro & Cano, in Valenzuela-Lopez, Cano-Lira, Guarro, Sutton, Wiederhold, Crous & Stchigel, Stud. Mycol. 90:51 (2017). MycoBank MB 819770.
Allocucurbitaria prunicola (Crous & Akulov) V. Magaña-Dueñas, Stchigel & Cano, comb. nov. MycoBank MB 838843.
Basionym: Neocucurbitaria prunicola Crous & Akulov, in Crous, Schumacher, Akulov, Thangavel, Hernández-Restrepo, Carnegie, Cheewangkoon, R; Wingfield, Summerell, Quaedvlieg, Coutinho, Roux, Wood, Giraldo & Groenewald, Fungal Systematics and Evolution 3:91 (2019).
Description: Crous & Akulov 2019.
Notes: In 2019, Crous & Akulov introduced N. prunicola to the genus Neocucurbitaria, based on morphological and nucleotide sequence data analysis [36]. However, in our phylogenetic study, N. prunicola is clearly placed in the genus Allocucurbitaria. Therefore, we propose a new combination for that species.
Neopyrenochaetaceae Valenz.-Lopez, Crous, Stchigel, Guarro & Cano, in Valenzuela-Lopez, Cano-Lira, Guarro, Sutton, Wiederhold, Crous & Stchigel, Stud. Mycol. 90:54 (2017). MycoBank MB 820416.
Type genus: Neopyrenochaeta Valenz.-Lopez, Crous, Stchigel, Guarro & Cano, in Valenzuela-Lopez, Cano-Lira, Guarro, Sutton, Wiederhold, Crous & Stchigel, Stud. Mycol. 90:54 (2017). MycoBank MB 820313.
Type species: Neopyrenochaeta acicola (Moug. & Lév.) Valenz.-Lopez, Crous, Stchigel, Guarro & Cano, in Valenzuela-Lopez, Cano-Lira, Guarro, Sutton, Wiederhold, Crous & Stchigel, Stud. Mycol. 90:54 (2017). MycoBank MB 820314.
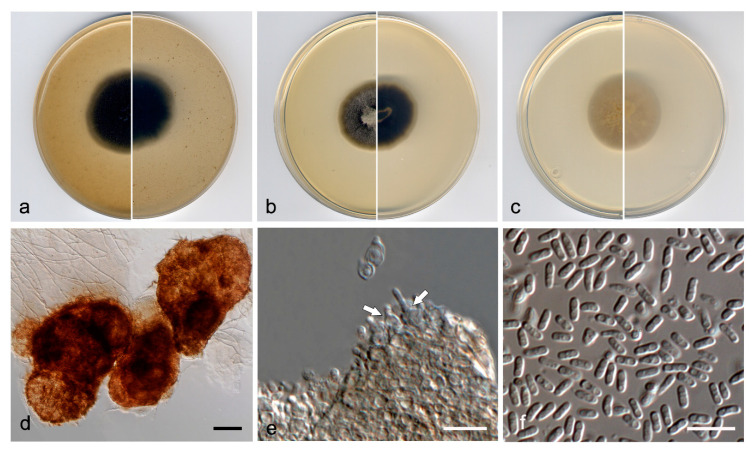
Neopyrenochaeta asexualis V. Magaña-Dueñas, Stchigel & Cano, sp. nov. Figure 4. MycoBank MB 838835.
Figure 4.
Neopyrenochaeta asexualis FMR 17874 T. (a) Colonies on OA, (b) MEA, and (c) PDA, after 2 weeks at 25 ± 1 °C (surface, left; reverse, right); (d) pycnidium; (e) conidiogenous cells (white arrow); (f) conidia. Scale bars: d = 50 µm, e,f = 10 µm.
Etymology: From Latin asexualis, without sex, because of lack of a known sexual morph.
Type: Spain, Sevilla province, Parque Natural Sierra Norte (37.994712, −5.668709), from plant debris in freshwater, May 2019, José F. Cano Lira, holotype CBS H-24741, culture ex-type FMR 17874 = CBS 147606.
Description: Hyphae hyaline to light brown, septate, branched, smooth-and thin-walled, 2–3 µm wide. Conidiomata pycnidial, brown to dark brown, immersed to semi-immersed, solitary or confluent, setose, ostiolate, globose to subglobose, 100–170 µm × 85–150 µm, ostiole 30–40 µm diameter. Setae pale brown to brown, septate, erect, nodose, narrowing towards the tip, thick-walled 40–80 µm × 2–4 µm, mainly disposed around the ostiole but also scattered, sometimes curved or recurved at the tip. Conidiomata wall composed of two to four layers of cells, 10–25 µm thick, outer layer of textura angularis, composed of brown to dark brown, flattened polygonal cells of 5–7 µm diameter. Conidiophores absent. Conidiogenous cells phialidic; determinate; hyaline; smooth-walled; doliiform; 5–6 µm × 4–5 µm; with mostly one, less frequently two conidiogenous loci. Conidia one-celled, hyaline, smooth- and thin-walled, ellipsoidal, 4–5 µm × 1.5–2.5 µm, sometimes slightly curved. Chlamydospores absent.
Culture characteristics: Colonies on PDA reaching 36 mm diameter after 7 days at 25 ± 1 °C, umbonate, velvety, margins regular, brownish grey (4D2), with patches of white; reverse olive brown (4D3). Colonies on OA reaching 40 mm diameter after 7 days at 25 ± 1 °C, flattened to slightly floccose, margin regular, with sparse aerial mycelium, grey (30C1); reverse greenish grey to yellowish grey (30E1/30F2). Colonies on MEA reaching 25 mm diameter after 7 days at 25 ± 1 °C, convex, velvety, margin regular, golden grey (4C2); reverse brownish grey to beige (4F2/4C3). Exopigment absent. Cardinal temperatures of growth: minimum 5 °C, optimum 25 °C, and maximum 30 °C.
Diagnosis: Neopyrenochaeta asexualis is grouped in the same terminal clade as N. thailandica, but as a distinct taxon. Morphological comparison between N. asexualis and N. thailandica is not possible because only the former produces the asexual morph and only the latter one forms ascomata [37]. However, it is noteworthy that N. asexualis produces conidiomata with doliiform phialides with up two conidiogenous loci.
Notes: The difference in nucleotide sequences (ITS-LSU-tub2-rpb2 concatenated dataset) between N. asexualis and N. thailandica is 38 bp.
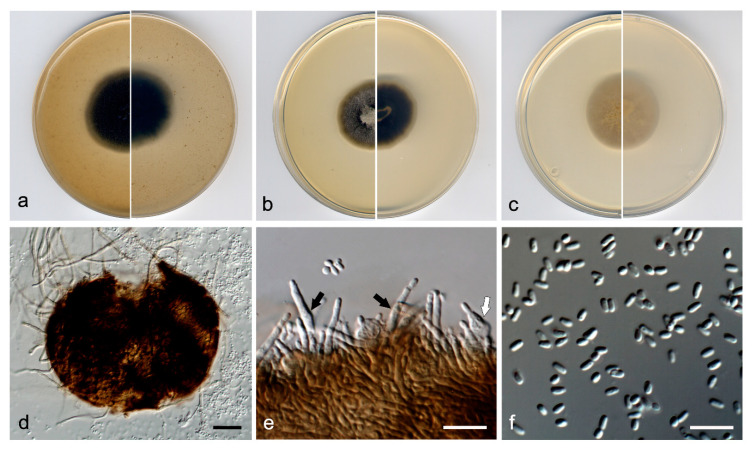
Neopyrenochaeta submersa V. Magaña-Dueñas, Cano & Stchigel, sp. nov. Figure 5. MycoBank MB 838840.
Figure 5.
Neopyrenochaeta submersa FMR 16957 T. (a) Colonies on OA, (b) MEA, and (c) PDA, after 2 weeks at 25 ± 1 °C (surface, left; reverse, right); (d) pycnidium; (e) conidiogenous cells; (f) conidia and setae. Scale bars: d = 50 µm, e,f = 10 µm.
Etymology: From Latin submersum, submerged, because the fungus was recovered from plant debris in freshwater.
Type: Spain, Barcelona province, Les Guilleries (41.9362028, 2.4122862), from plant debris in freshwater, Nov 2017, Eduardo Jose de Carvalho Reis, holotype CBS H-24742, culture ex-type FMR 16957 = CBS 147607.
Description: Hyphae pale to dark brown, septate, branched, smooth- and thin-walled, 2–3 µm wide. Conidiomata pycnidial, brown to dark brown, semi-immersed, solitary or confluent, scattered, ostiolate, setose, globose to subglobose, 140–200 µm × 180–240 µm, one to three ostioles per conidioma, 60–85 µm diameter. Setae brown to dark brown, septate, erect, rounded at the tip, thick-walled, 75−160 µm × 2−3 µm, narrowing towards the tip, and mostly disposed around the ostiole. Conidiomata wall composed of three to five layers of cells, 10–20 µm thick, with an outer layer of textura angularis, composed of brown to dark brown, flattened polygonal cells of 3–4 µm diameter. Conidiophores absent. Conidiogenous cells phialidic, determinate, hyaline, smooth-walled, ampulliform, 6–8 µm × 2.5–3.5 µm. Conidia one-celled, hyaline, smooth- and thin-walled, ellipsoidal, 3–4 µm × 2–3 µm. Chlamydospores absent.
Culture characteristics: Colonies on PDA reaching 23 mm diameter after 7 days at 25 ± 1 °C, umbonate, velvety, rugose, margin regular, surface greyish green to greenish grey (30E4/30C2), reverse greyish green to dull green (30B4/30E3), margin greenish grey (30C2). Colonies on OA reaching 28 mm diameter after 7 days at 25 ± 1 °C, convex, velvety, margin regular, surface and reverse grey (30C1). Colonies on MEA reaching 20 mm diameter after 7 days at 25 ± 1 °C, umbonate, velvety, margins regular, greyish green to dull green (30C3/30E3), margin white; reverse dark green (30F3), margins white. Exopigment absent. Cardinal temperatures of growth: minimum 5 °C, optimum 25 °C, and maximum 30 °C.
Diagnosis: In our phylogenetic analysis, N. submersa is located in the same terminal clade as N. acicola, N. fragariae, N. inflorescentiae, and N. glabra. With the exception of N. glabra, all these species are morphologically very similar. However, N. submersa grows faster than N. fragariae (reaching 14 mm and 11 mm diameter after 7 days at 25 °C on OA and MEA, respectively). Also, N. submersa does not produce exopigment on OA, which is lilac-rose in N. acicola [38] and orange in N. fragariae.
Notes: Differences in nucleotide sequences (ITS-LSU-tub2-rpb2 concatenated dataset) between N. submersa and the other species in the same terminal clade are: N. fragariae 28 bp; N. acicula, 33 pb; N. inflorescentiae, 51 pb; and N. glabra, 98 bp.
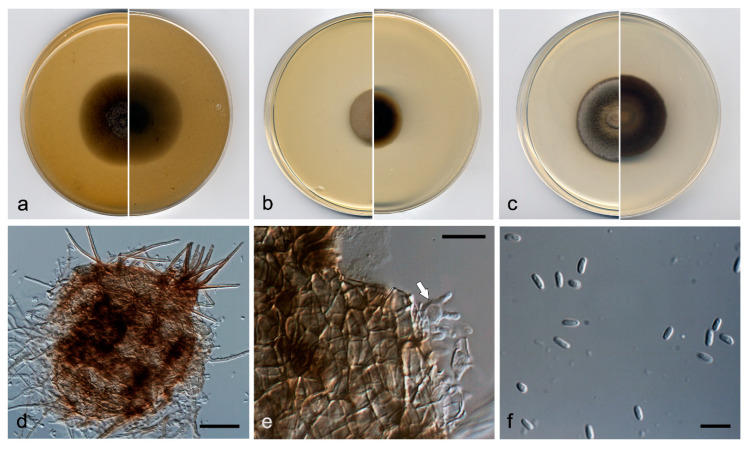
Neopyrenochaeta glabra V. Magaña-Dueñas, Stchigel & Cano, sp. nov. Figure 6. MycoBank MB 838841.
Figure 6.
Neopyrenochaeta glabra FMR 17418 T. (a) Colonies on OA, (b) MEA, and (c) PDA, after two weeks at 25 ± 1 °C (surface, left; reverse, right); (d) pycnidium; (e,f) conidiogenous cells; (g) conidia. Scale bars: d = 50 µm, e–g = 10 µm.
Etymology: From Latin glaber, hairless, relating to absence of setae.
Type: Spain, Segovia province, Riaza (41.238863, -3.435258), from freshwater submerged plant debris, May 2018, Viridiana Magaña Dueñas, holotype CBS H-24743, culture ex-type FMR 17418 = CBS 147608.
Description: Hyphae hyaline to pale brown, septate, branched, smooth- and thin-walled, 1.5–2.5 µm wide. Conidiomata pycnidial immersed to semi-immersed, solitary or confluent, scattered, ostiolate, glabrous, translucent, pale brown to brown, but carbonaceous around the ostiole, mostly subglobose, 140–200 µm × 180–240 µm, ostiole 60–85 µm diam. Conidiomata wall composed of four to six layers of cells, 10–25 µm thick, with an outer layer of textura angularis, composed of pale brown to brown, flattened polygonal cells of 3–4 µm diameter. Conidiophores absent. Conidiogenous cells phialidic, determinate, hyaline, smooth-walled, ampulliform, 7–9 µm × 3–4 µm. Conidia aseptate, hyaline, smooth- and thin-walled, ellipsoidal, 3–4 µm × 2–3 µm. Chlamydospores absent.
Culture characteristics: Colonies on PDA reaching 23 mm diameter after 7 days at 25 ± 1 °C, umbonate, velvety, rugose, margin regular, surface, and reverse grey to dark brown (8F1/8D1). Colonies on OA reaching 25 mm diameter after 7 days at 25 ± 1 °C, flattened, velvety, margin regular, surface and reverse grey (6F1). Colonies on MEA reaching 23 mm diam after 7 days at 25 ± 1 °C, convex, velvety, margins regular, grey to olive brown (4F1/4D3), margin yellowish grey (4B2); reverse brownish grey (5E2) margins orange grey (5B2). Exopigment absent. Cardinal temperatures of growth: optimum 25 °C, maximum 30 °C, minimum 5 °C.
Diagnosis: Morphologically, N. glabra differs from the phylogenetically closest species, N. acicula, N. fragariae, N. inflorescentiae, and N. submersa by lacking conidiomatous setae in the conidiomata walls.
Notes: Differences in nucleotide sequences (ITS-LSU-tub2-rpb2 concatenated dataset) between N. glabra and the other species of the same terminal clade are: N. fragariae, 89 bp; N. submersa, 98 bp; N. acicola, 108 bp; and N. inflorescentiae, 131 bp.
Pyrenochaetopsidaceae Valenz.-Lopez, Crous, Cano, Guarro & Stchigel, in Valenzuela-Lopez, Cano-Lira, Guarro, Sutton, Wiederhold, Crous & Stchigel, Stud. Mycol. 90:56 (2017). MycoBank MB 820308.
Type genus: Pyrenochaetopsis Gruyter, Aveskamp & Verkley, Mycologia 102 (5):1076 (2010). MycoBank MB 514653.
Type species: Pyrenochaetopsis lep.tospora (Sacc. & Briard) Gruyter, Aveskamp & Verkley, Mycologia 102 (5):1076 (2010). MycoBank MB 514654.
Pyrenochaetopsis aquatica V. Magaña-Dueñas, Cano & Stchigel, sp. nov. Figure 7. MycoBank MB 838842.
Figure 7.
Pyrenochaetopsis aquatica FMR 17327 T. (a) Colonies on OA, (b) MEA, and (c) PDA, after 2 weeks at 25 ± 1 °C (surface, left; reverse, right); (d) pycnidium; (e,f) conidiogenous cells (white arrows).
Etymology: From Latin aquaticus, referring to the habitat from which the fungus was recovered (freshwater).
Type: Spain, Tarragona province, Serra del Montsant (41.32871, 0.87105), from plant debris in freshwater, February 2018, Eduardo Jose de Carvalho Reis, holotype CBS H-24744, culture ex-type FMR 17327 = CBS 147609.
Description: Hyphae hyaline to pale brown, septate, branched, smooth- and thin-walled, 1.5–2 µm wide. Conidiomata pycnidial, brown, immersed to semi-immersed, solitary or confluent, scattered, ostiolate, mostly glabrous or covered with few short setae, pyriform, 200–300 µm × 130–180 µm, ostiole 60–80 µm diameter. Setae pale brown to brown, septate, erect, nodose, thick-walled, of 10–20 µm × 3–4 µm, tapering towards the apex, mainly disposed around the ostiole. Conidiomata wall composed of four to six layers of cells, 15–30 µm thick, with an outer layer of textura angularis, composed of brown to dark brown, flattened polygonal cells of 3.5–4.5 µm diameter. Conidiophores absent. Conidiogenous cells phialidic, determinate, hyaline, smooth-walled, ampulliform, 6–7 µm × 2.5–3 µm. Conidia aseptate, hyaline, smooth- and thin-walled, mostly long ellipsoidal, 3.5–5 µm × 1–1.8 µm, slightly constricted at the middle, sometimes slightly curved and irregularly shaped, biguttulate. Chlamydospores absent.
Culture characteristics: Colonies on PDA reaching 15 mm diameter after 7 days at 25 ± 1 °C, flattened, velvety, margin regular, grey to yellowish white (A1C/4A2); reverse greyish green to yellowish white (4B3/4A2). Colonies on OA reaching 20 mm diameter after 7 days at 25 ± 1 °C, flattened, velvety, margin regular, yellowish grey to yellowish white (4B2/4A2). Colonies on MEA reaching 20 mm diameter after 7 days at 25 ± 1 °C, umbonate, velvety, margins regular, yellowish grey to yellowish brown (4B2/5F4), margin orange grey (5B2); reverse orange grey (5B2). Exopigment absent. Cardinal temperatures of growth: optimum 25 °C, maximum 30 °C, minimum 5 °C.
Diagnosis: Pyrenochaetopsis aquatica differs morphologically from the phylogenetically nearest species P. leptospora and P. poae, because it is mostly glabrous or covered with few short setae, while the pycnidia of P. leptospora and P. poae are abundantly covered with long setae [39].
Notes: Differences in nucleotide sequences (ITS-LSU-tub2-rpb2 concatenated dataset) between P. aquatica and the other species of the same terminal clade are: P. leptospora, 57 bp; and P. poae, 67 bp.
4. Discussion
The genus Neocucurbitaria was introduced by Wanasinghe et al. [40] to accommodate N. acerina, N. quercina and N. unguis-hominis (the type species of the genus). Twenty-two species are currently accepted (Index of Fungi; http://www.indexfungorum.org/names/Names.asp (accessed on 16 March 2021)). Neocucurbitaria spp. has been isolated from human corneal and skin lesions, seawater, and trees and shrubs [5,40,41]. We described two new species for the genus, N. aquadulcis and N. variabilis, from submerged plant debris in freshwaters, the first report for this sort of habitat. It is remarkable that N. variabilis produces two sorts of conidiogenous cells (flask-shaped and long cylindrical) and that N. aquadulcis only produces ampulliform phialides, whereas the other species in the same subclade (N. acerina, N. aquatica, N. irregularis, N. keratinophila and N. unguis-hominis) produce doliiform phialides or well-developed conidiophores. In 2019, Crous & Akulov introduced N. prunicola to that genus [36]. However, in our phylogenic analysis, N. prunicola was located far from the type species of Neocucurbitaria (N. unguis-hominis), being located within the genus Allocucurbitaria. Consequently, we propose the new combination Allocucurbitaria prunicola.
A molecular study by Valenzuela-López et al. [5] allowed recognition of four new families of coelomycetous fungi included previously in the family Cucurbitariaceae: Neopyrenochaetaceae, Parapyrenochaetaceae, Pseudopyrenochaetaceae, and Pyrenochaetopsidaceae. In the latter family, the authors recognized four species belonging to the genus Neopyrenochaeta: N. acicola (basionym: Vermicularia acicola; originally described on decaying leaves of Pinus sylvestris, Vosges, France), N. fragariae (originally identified as Pyrenochaeta acicola; isolated from Fragaria (×) ananassa, The Netherlands), N. inflorescentiae (basionym: Pyrenochaeta inflorescentiae; from style of senescent flowerhead of Protea neriifolia, Western Cape Province, South Africa), and N. thelephonii (basionym: Pyrenochaeta telephonii; from surface of cell phone, Maharashtra, India) [40,42,43]. During 2019 and 2020, eight more species were described [37,44], three of them (Neopyrenochaeta annellidica, Neopyrenochaeta chiangraiensis and Neopyrenochaeta maesuayensis) from submerged decaying wood in Thailand. Interestingly, we also identified two of these latter three species in Spain (Figure 1). This implies that the geographical distribution of N. annellidica and N. maesuayensis is much broader than would be expected, since their original report was from tropical areas of Southeastern Asia. In the present study, we report the finding of three novel additional species from submerged plant debris in Spain: Neopyrenochaeta glabra, N. asexualis, and N. submersa. Neopyrenochaeta glabra is easily recognized by the absence of setae and the darker conidiomata wall around the ostiole. Neopyrenochaeta asexualis is distinguished from other species of the genus because it produces doliiform phialides with one or two conidiogenous loci. Otherwise, N. submersa is difficult to discriminate morphologically from N. acicola, N. fragariae, and N. inflorescentiae species phylogenetically related but differing molecularly.
The fungal genus Pyrenochaetopsis was introduced by De Gruyter et al. [45] to accommodate: P. decipens, P. indica, P. leptospora (type species of the genus), P. microspore, and P. pratorum. Currently 16 species are accepted (Index Fungorum 2020). The members of this genus have been found in terrestrial and marine environments, human dermatitis, sputum, and blood human samples, [5,37,46,47,48,49]. In our phylogenetic analysis, the strain FMR 17337, named here as P. aquatica, clustered within the Pyrenochaetopsis clade, is distant from other species of this genus, with the exception of P. leptospora and P. poae, which forms a sister clade. Both species differ phylogenetically and morphologically from P. aquatica in having more abundant and longer setae.
5. Conclusions
In the present study, we have isolated several coelomycetous fungi from submerged plant debris collected in different freshwater habitats in Spain by incubation of the samples in wet chambers. After a phenotypic characterization and a phylogenic study based on the analysis of nucleotide sequences of the ITS, LSU, tub2, and rpb2 loci, six new species have been described: Neocucurbitaria aquadulcis and N. variabilis; Neopyrenochaeta glabra, N. asexualis and N. submersa; and Pyrenochaetopsis aquatica. Also, thanks to the phylogenetic analysis, Neocucurbitaria prunicola was transferred to the genus Allocucurbitaria. In our opinion, the present study makes an important contribution to the knowledge of the coelomycetous fungi growing on decomposing plant material in aquatic habitats.
Acknowledgments
We thank Juan Ramón García Martínez (a non-professional mycologist from Granja de Torrehermosa, Badajoz, Spain) for collaborating in the Sierra Norte de Sevilla sampling and to the Sociedad Ibérica de Micología (SIM) for the organization of the sampling into Riaza. We also thank Phil Hoddy for editing and proofreading the final text.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jof7050368/s1, Table S1: coelomycetous fungi sequences used in this study; Table S2: Results of blast search of the new proposed species. Figure S1. ML phylogenetic tree of Cucurbitariaceae, Neopyrenochaetaceae, Pseudopyrenochaetaceae, and Pyrenochaetopsidaceae inferred from the ITS sequences (455 bp). Support in nodes is indicated above by bootstrap values of 70% or higher. T = ex-type strains. New species are indicated in blue. New strains isolated during this study are indicated in bold; Figure S2. ML phylogenetic tree of Cucurbitariaceae, Neopyrenochaetaceae, Pseudopyrenochaetaceae, and Pyrenochaetopsidaceae inferred from the LSU sequences (791 bp). Support in nodes is indicated above branches by bootstrap values of 70% or higher. T = ex-type strains. New species are indicated in blue. Strains isolated during this study are indicated in bold; Figure S3. ML phylogenetic tree of Cucurbitariaceae, Neopyrenochaetaceae, Pseudopyrenochaetaceae, and Pyrenochaetopsidaceae inferred from rpb2 sequences (734 bp). Support in nodes is indicated above branches by bootstrap values of 70% or higher. T = ex-type strains. New species are indicated in blue. New strains isolated during this study are indicated in bold; Figure S4. ML phylogenetic tree of Cucurbitariaceae, Neopyrenochaetaceae, Pseudopyrenochaetaceae, and Pyrenochaetopsidaceae inferred from tub2 sequences (272 bp). Support in nodes is indicated above branches by bootstrap values of 70% and higher. T = ex-type strains. New species are indicated in blue. Strains isolated during this study are indicated in bold. Alignment length.
Author Contributions
Conceptualization, A.M.S. and J.F.C.-L.; methodology, V.M.-D.; software, V.M.-D. and J.F.C.-L.; validation, A.M.S. and J.F.C.-L.; formal analysis, V.M.-D., A.M.S. and J.F.C.-L.; investigation, V.M.-D.; resources, J.F.C.-L.; data curation, A.M.S. and J.F.C.-L.; writing—original draft preparation, V.M.-D.; writing—review and editing, V.M.-D., A.M.S. and J.F.C.-L.; visualization, A.M.S. and J.F.C.-L.; supervision, A.M.S. and J.F.C.-L.; project administration, J.F.C.-L.; funding acquisition, J.F.C.-L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Spanish Ministerio de Economía y Competitividad, grant CGL2017-88094-P.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kirk P.M., Cannon P.F., Stalpers J.A., Minter D.W. Ainsworth & Bisby’s Dictionary of the Fungi. 10th ed. CAB International; Wallingford, UK: 2008. [Google Scholar]
- 2.Stchigel A.M., Sutton D.A. Coelomycete fungi in the clinical lab. Curr. Fungal Infect. Rep. 2013;7:171–191. doi: 10.1007/s12281-013-0139-9. [DOI] [Google Scholar]
- 3.Raja H.A., Shearer C.A., Tsui C.K.-M. eLS. John Wiley & Sons Ltd.; Hoboken, NJ, USA: 2021. Freshwater fungi. [Google Scholar]
- 4.Dian-Ming H., Cai L., Gareth Jones E.B., Zhang H., Boonyuen N., Hyde K.D. Taxonomy of filamentous asexual fungi from freshwater habitats, links to sexual morphs and their phylogeny. In: Garreth Jones E.B., Hyde K.D., Pang K.-L., editors. Freshwater Fungi: And Fungal-Like Organisms, 1st ed. De Gruyter; Berlín, Germany: 2014. pp. 109–131. [Google Scholar]
- 5.Valenzuela-Lopez N., Cano-Lira J.F., Guarro J., Sutton D.A., Wiederhold N., Crous P.W., Stchigel A.M. Coelomycetous Dothideomycetes with emphasis on the families Cucurbitariaceae and Didymellaceae. Stud. Mycol. 2018;90:1–69. doi: 10.1016/j.simyco.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roldán A., Honrubia M. Dos celomicetos, nuevos para la flora española, aislados en medio acuático. An. Jard. Bot. Madrid. 1990;47:3–9. [Google Scholar]
- 7.Giralt M., Hawksworth D.L. Diplolaeviopsis ranula, a new genus and species of lichenicolous coelomycetes growing on the Lecanora strobilina group in Spain. Mycol. Res. 1991;95:759–761. doi: 10.1016/S0953-7562(09)80828-9. [DOI] [Google Scholar]
- 8.Dyko B.J., Sutton B.C. Two new genera of water-borne coelomycetes from submerged leaf litter. Nova Hedwig. 1978;29:167–178. [Google Scholar]
- 9.Uecker F.A., Kulik M.M. Pseudorobillarda sojae, a New Pycnidial Coelomycete from Soybean Stems. Mycologia. 1986;78:449–453. doi: 10.1080/00275514.1986.12025268. [DOI] [Google Scholar]
- 10.Révay A., Gönczöl J. Longitudinal distribution and colonization patterns of wood inhabiting fungi in a mountain stream in Hungary. Nova Hedwig. 1990;51:505–520. [Google Scholar]
- 11.Hyde K.D. Tropical Australian freshwater fungi. VI. Tiarosporella paludosa and Clohesyomyces aquaticus gen. et sp. nov. Coelomycetes. Aust. Syst. Bot. 1993;6:169–173. [Google Scholar]
- 12.Czeczuga B., Orlowska M. Hyphomycetes in twenty springs of the Knyszyn-Bialystok Forest in various seasons. Int. Rev. Gesamten Hydrobiol. Hydrogr. 1996;81:417–433. doi: 10.1002/iroh.19960810311. [DOI] [Google Scholar]
- 13.Jeewon R., Cai L., Liew E.C.Y., Zhang K.Q., Hyde K.D. Dyrithiopsis lakefuxianensis gen. et sp. nov. from Fuxian Lake, Yunnan, China, and notes on the taxonomic confusion surrounding Dyrithium. Mycologia. 2003;95:911–920. doi: 10.1080/15572536.2004.11833050. [DOI] [PubMed] [Google Scholar]
- 14.Yonezawa H., Tanaka K. The second species of Neoheteroceras and additional characters of the genus. Mycoscience. 2008;49:152–154. doi: 10.1007/S10267-007-0398-9. [DOI] [Google Scholar]
- 15.Abdel-Aziz FA, Abdel-Wahab MA Lolia aquatica gen. et sp. nov. (Lindgomycetaceae, Pleosporales), a new coelomycete from freshwater habitats in Egypt. Mycotaxon. 2010;114:33–42. [Google Scholar]
- 16.Zhang H., Hyde K.D., Mckenzie E.H., Bahkali A.H., Zhou D. Sequence data reveals phylogenetic affinities of Acromalia aquatica sp. nov., Aquasubmersa mircensis gen. et sp. nov. and Clohesyomuces aquaticus. (freshwater Coelomycetes) Cryptogamie Mycol. 2012;33:333–346. doi: 10.7872/crym.v33.iss3.2012.333. [DOI] [Google Scholar]
- 17.Samson R.A., Houbraken J., Thrane U., Frisvad J.C., Andersen B. Food and Indoor Fungi. 2nd ed. CBS-KNAW Fungal Biodiversity Centre; Utrecht, The Netherlands: 2010. p. 390. (CBS Laboratory Manual Series). [Google Scholar]
- 18.Kornerup A., Wanscher J.H. Methuen Handbook of Colour. 3rd ed. Methuen; London, UK: 1978. [Google Scholar]
- 19.Hawksworth D.L., Kirk P.M., Sutton B.C., Pegler D.N. Ainsworth & Bisby’s Dictionary of the Fungi. 8th ed. CAB International; Oxon, UK: 1995. p. 616. [Google Scholar]
- 20.Aveskamp M.M., de Gruyter J., Woudenberg J.H.C., Verkley G.J.M., Crous P.W. Highlights of the Didymellaceae: A polyphasic approach to characterise Phoma and related pleosporalean genera. Stud. Mycol. 2010;65:1–60. doi: 10.3114/sim.2010.65.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Q., Jiang J.R., Zhang G.Z., Crous P.W. Resolving the Phoma enigma. Stud. Mycol. 2015;82:137–217. doi: 10.1016/j.simyco.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chupp C. Further notes on double cover-glass mounts. Mycologia. 1940;32:269–270. doi: 10.2307/3754504. [DOI] [Google Scholar]
- 23.Rehner S.A., Samuels G.J. Taxonomy and Phylogeny of Gliocladium Analysed from Nuclear Large Subunit Ribosomal DNA Sequences. Mycol. Res. 1994;98:625–634. doi: 10.1016/S0953-7562(09)80409-7. [DOI] [Google Scholar]
- 24.Vilgalys R., Hester M. Rapid Genetic Identification and Mapping of Enzymatically Amplified Ribosomal DNA from Several Cryptococcus Species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/JB.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White T.J., Bruns T., Lee S.J.W.T., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. 1st ed. Academic Press; San Diego, CA, USA: 1990. pp. 315–322. [Google Scholar]
- 26.Woudenberg J.H.C., Aveskamp M.M., de Gruyter J., Spiers A.G., Crous P.W. Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia. 2009;22:56–62. doi: 10.3767/003158509X427808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sung G.-H., Sung J.-M., Hywel-Jones N.L., Spatafora J.W. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenetics Evol. 2007;44:1204–1223. doi: 10.1016/j.ympev.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 29.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids. Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stamatakis A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller M.A., Pfeiffer W., Schwartz T. The CIPRES science gateway: Enabling High-Impact science for phylogenetics researchers with limited resources; Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the Extreme to the Campus and Beyond; Chicago, IL, USA. 16 July 2012; New York, NY, USA: Association for Computing Machinery; 2012. pp. 1–8. [Google Scholar]
- 33.Ronquist F., Teslenko M., Van Der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Posada D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 35.Punithalingam E., English M.P. Pyrenochaeta unguis-hominis sp. nov. on human toe-nails. Trans. Brit. Mycol. Soc. 1975;64:539–541. doi: 10.1016/S0007-1536(75)80163-X. [DOI] [Google Scholar]
- 36.Crous P.W., Schumacher R.K., Akulov A., Thangavel R., Hernández-Restrepo M., Carnegie A.J., Cheewangkoon R., Wingfield M.J., Summerell B.A., Quaedvlieg W., et al. New and Interesting Fungi 2. Fungal. Syst. Evol. 2019;3:57–134. doi: 10.3114/fuse.2019.03.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mapook A., Hyde K.D., McKenzie E.H.C., Gareth-Jones E.B., Jayarama Bhat D., Jeewon R., Stadler M., Samarakoon M.C., Malaithong M., Tanunchai B., et al. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed) Fungal Divers. 2020;101:1–175. doi: 10.1007/s13225-020-00444-8. [DOI] [Google Scholar]
- 38.Dorembosch M.M.J. Key to nine ubiquitous soil-borne phoma-like fungi. Persoonia. 1970;6:1–14. [Google Scholar]
- 39.Boerema G.H., de Gruyer J., Noordeloos M.E., Hamers M. Phoma Identification Manual. Differentiation of Specific and Infra-Specific Taxa in Culture. 1st ed. CABI Publishing; Wallingford, UK: 2004. 448p [Google Scholar]
- 40.Chen Q., Hou L.W., Duan W.J., Crous P.W., Cai L. Didymellaceae revisited. Stud. Mycol. 2017;87:105–159. doi: 10.1016/j.simyco.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wanasinghe D.N., Phookamsak R., Jeewon R., Li W.J., Hyde K.D., Jones E.B.G., Camporesi E., Promputtha I. A family level rDNA based phylogeny of Cucurbitariaceae and Fenestellaceae with descriptions of new Fenestella species and Neocucurbitaria gen. nov. Mycosphere. 2017;8:397–414. doi: 10.5943/mycosphere/8/4/2. [DOI] [Google Scholar]
- 42.Toh Y.F., Yew S.M., Chan C.L., Na S.L., Kok W.L., Chee-Choong H., Wain-Yan Y., Kee P.N., Chee S.K. Genome anatomy of Pyrenochaeta unguis-hominis UM 256, a multidrug resistant strain isolated from skin scraping. PLoS ONE. 2016;11:e0162095. doi: 10.1371/journal.pone.0162095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marincowitz S., Crous P.W., Groenewald J.Z., Wingfield M.J. Microfungi Occurring on Proteaceae in the Fynbos. Westerdijk Fungal Biodiversity Institute; Utrecht, The Netherlands: 2008. p. 166. (CBS Biodiversity Series No. 7). [Google Scholar]
- 44.Crous P.W., Wingfield M.J., Le Roux J.J., Richardson D.M., Strasberg D., Shivas R.G., Alvarado P., Edwards J., Moreno G., Sharma R., et al. Fungal Planet description sheets: 371–399. Persoonia. 2015;35:264–327. doi: 10.3767/003158515X690269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W., McKenzie E.H.C., Liu J.K., Bhat J., Dai D.Q., Camporesi E., Tian Q., Maharachchikumbura S.S.N., Luo Z.L., Shang Q.J., et al. Taxonomy and phylogeny of hyaline-spored coelomycetes. Fungal Divers. 2020;100:279–801. doi: 10.1007/s13225-020-00440-y. [DOI] [Google Scholar]
- 46.De Gruyter J., Woudenberg J.H., Aveskamp M.M., Gerard J.M.V., Groenewald J.Z., Crous P.W. Systematic reappraisal of species in Phoma section Paraphoma, Pyrenochaeta and Pleurophoma. Mycologia. 2010;102:1066–1081. doi: 10.3852/09-240. [DOI] [PubMed] [Google Scholar]
- 47.De Gruyter J., Woudenberg J.H., Aveskamp M.M., Verkley G.J.M., Groenewald J.Z., Crous P.W. Redisposition of phoma-like anamorphs in pleosporales. Stud. Mycol. 2013;75:1–36. doi: 10.3114/sim0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crous P.W., Shivas R.G., Quaedvlieg W., Van der Bank M., Zhang Y., Summerell B.A., Guarro J., Wingfield M.J., Wood A.R., Alfenas A.C., et al. Fungal Planet description sheets: 214–280. Peersonia. 2014;32:184–306. doi: 10.3767/003158514X682395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papizadeh M., Soudi M.R., Amini L., Wijayawardene N.N., Hyde K.D. Pyrenochaetopsis tabarestanensis (Cucurbitaceae, Pleosporales), a new species isolated from rice ferms in north Iran. Phytotaxa. 2017;297:15–28. doi: 10.11646/phytotaxa.297.1.2. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.