Abstract
The B cell scaffold protein with ankyrin repeats (BANK1) is expressed primarily in B cells and with multiple but discrete roles in B cell signaling, including B cell receptor signaling, CD40-related signaling, and Toll-like receptor (TLR) signaling. The gene for BANK1, located in chromosome 4, has been found to contain genetic variants that are associated with several autoimmune diseases and also other complex phenotypes, in particular, with systemic lupus erythematosus. Common genetic variants are associated with changes in BANK1 expression in B cells, while rare variants modify their capacity to bind efferent effectors during signaling. A BANK1-deficient model has shown the importance of BANK1 during TLR7 and TLR9 signaling and has confirmed its role in the disease. Still, much needs to be done to fully understand the function of BANK1, but the main conclusion is that it may be the link between different signaling functions within the B cells and they may act to synergize the various pathways within a cell. With this review, we hope to enhance the interest in this molecule.
Keywords: B cells, B cell receptor, CD40, autoimmunity, genetic association, B cell scaffold with ankyrin repeats, TLR7, BLK, LYN, TRAF6
1. Introduction
The importance of the gene B cell scaffold with ankyrin repeats (BANK1) was gained due to the genetic studies on systemic lupus erythematosus and other diseases that followed. The role of BANK1 in B cell signaling had been somewhat difficult to define; however, since the commencement of mouse studies, its role in the disease and TLR7 signaling has become evident.
BANK1 was first identified by Yokoyama et al. [1] when using a solid-phase phosphorylation screening in the search for Lyn substrates. A novel ankyrin repeat-containing protein was identified and sequenced. Based on the sequence information, the coding sequence of the protein was obtained by RT-PCR of RNA isolated from a Daudi cell line. In general, the major isoform of BANK1 of 755 amino acids was identified, with a molecular weight of 85,500 Da.
The initial gene expression analyses using northern blots and mouse tissues found that BANK1 was expressed in the spleen and lymph nodes of adult mice, but not in Rag-/- or SCID mice. When looking into the B cell development populations, BANK1 was observed in immature and recirculating B cells, but not in pro-B or pre-B cells. Looking into separated splenic B cells, BANK1 was expressed in IgMhiIgD−, IgMhiIgDhi and the most mature IgMloIgDhi populations. The authors suggested that the expression of BANK1 depended on the expression of the B cell receptor (BCR); however, there is evidence showing that BANK1 is also expressed in dendritic cells (BioGPS).
It was also found that BANK1 had some homology with other ankyrin-containing proteins, such as BCAP and the Drosophila protein Dof, although the overall homology with BCAP is 33%. The ankyrin repeats are located in amino acids 309-372 of BANK1, while a coiled-coil sequence of BANK1 is located in amino acids 647–675.
Thus, BANK1 was now entering the scientific publication sphere, but more was to come.
2. The Genetic Associations of Variants of BANK1 with Diseases
Table 1 shows the main genetic associations of BANK1 polymorphisms and the diseases where these associations have been found. A genome-wide association study identified BANK1 as a susceptibility gene for systemic lupus erythematosus (SLE) [2]. This study identified the D2 isoform of BANK1 by RT-PCR. It was observed that two closely linked single nucleotide polymorphisms (SNPs) were strongly associated in a European population, the non-synonymous variant rs10516487 located in the coding exon 2 and causing an R61H change in the protein, and rs17266594 located in the preceding intron 1 in a branch-point site. A third variant, rs3733197, creates an A383T change in the ankyrin domain and was weakly associated. The SNP association was also associated with a differential expression of the D2 isoform, whose expression was increased in homozygous for the protective allele of rs17266594. Initially, it was considered that the exonic variant could affect the binding of the inositol-3-phosphate receptor by BANK1; however, the role of BANK1 in TLR7 signaling appears to be more relevant. Another important feature of the genetic association of the gene BANK1 with SLE is that it was the major allele of rs10516487 that was showing the association, R61 being the major allele and, hence, the risk variant. This genetic association was fully replicated by Guo et al. [3], a study that also showed an association with hematological and immunological SLE classification criteria. Further studies also replicated the association in Finns [4], and other independent studies in European populations [5,6,7,8]. In one of these studies using a cohort of over 7000 SLE cases and nearly 16,000 controls, the major risk variant identified was rs10028805 [7], later confirmed in African Americans [9]. Further studies on the genetic association of BANK1 with SLE showed an association with the presence of anti-dsDNA autoantibodies [10]. The genetic association of BANK1 polymorphisms with SLE was also confirmed in Chinese, Hong Kong Chinese, and Thai populations [11,12,13]. However, BANK1 has been weakly associated in Mexican populations [14] or not at all in a population enriched for Amerindian ancestry [15].
Table 1.
List of genetic associations of polymorphisms of BANK1.
| Disease | SNPs 1 | Location in Protein | Study |
|---|---|---|---|
| SLE | rs10516487 | TIR domain | [2] |
| SLE | rs17266594 | -- | [2] |
| SLE | rs3733197 | Ankyrin repeats | [2] |
| SLE | rs17266594 | --- | [3] |
| SLE | rs10028805 | --- | [4,5,6,7,8,9,12,13,14,16,17,18,19] |
| SLE | W40C | TIR domain | [20] |
| Diffuse SSC | rs10516487 | TIR domain | [21] |
| Diffuse SSC | rs3733197 | Ankyrin repeats | [21] |
| RA | rs10516487 | TIR domain | [18,22] |
| Chronic Lymphocytic Leukemia | rs10028805 | --- | [23] |
| LDL Cholesterol | rs3733197 | Ankyrin repeats | [24] |
1 Single nucleotide polymorphisms associated in the study.
Of importance is the genetic association of BANK1 in African Americans. The genetic association is particularly strong [16,17], and two independent effects have been associated with BANK1 in SLE patients with AA ancestry [11].
Further association studies were performed with other diseases, for example, primary antiphospholipid syndrome, where the variants of BANK1 were not associated [25], or rheumatoid arthritis (RA), where an association was observed [22] but was not replicated in a second study [18]. Similarly, BANK1 genetic polymorphisms were associated with systemic sclerosis [26] and replicated in a second European population [21], particularly in diffuse SSC. A more recent study identified, using whole exome sequencing [18], potential causal variants for diffuse systemic sclerosis in patients who also had interstitial lung disease [27]. Regarding cancers, a genetic association of BANK1 SNP rs10028805 with chronic lymphocytic leukemia has been reported [23]. Finally, a study reported the well-replicated association of BANK1 with serum LDL cholesterol levels in Koreans, represented by the association with the ankyrin non-synonymous SNP rs3733197 [24]. Of interest, in this regard, a study tested up to 62 GWAS loci from a GWAS, investigating the association of body mass index [26] with functional knock-out screens in Drosophila melanogaster brain and fat tissue [28]. While the genetic association pointed to SLC39A8, the closest to the associated SNP rs13107325, BANK1 was one of the sets of genes functionally corroborated.
A genetic association has also been observed with primary Sjogren’s syndrome (pSS) [29]. This study analyzed salivary gland biopsies associating the allele frequencies with patients having ectopic germinal center structures and found a positive association with BANK1 polymorphisms as well as other genes, such as AICDA and BCL2. Association between variants of BANK1 and Sjogren’s syndrome was not replicated in a Chinese population [30] or any major GWAS of the disease [31], while BLK was indeed associated [30,31]; however, no study has analyzed the potential genetic role of BANK1 in the development of lymphoma in pSS or RA.
Several studies analyzed the genetic interaction between BANK1 and a second SLE-associated B cell gene, BLK [32,33], and one of the studies, as mentioned in the previous section, showed the physical interaction between the proteins of both genes [33]. Such interaction has been identified in several populations [19] but also in several diseases, apart from SLE, such as SSC [32] and RA [34]. One study that also replicated the interaction in a Chinese population showed that the relative expression level of mRNA for BLK was lower in presence of the risk allele rs2736340, and a significant linear association of relative mRNA expression of BLK and BANK1 was also detected as indicative of this interaction in SLE patients [35]. Finally, a study analyzing a non-synonymous mutation in BLK (A71T) was related with decreased levels of BLK protein and binding to BANK1 through the impairment of the function of the SH3 domain of BLK, confirming once again the existence of the interaction at the protein level [36].
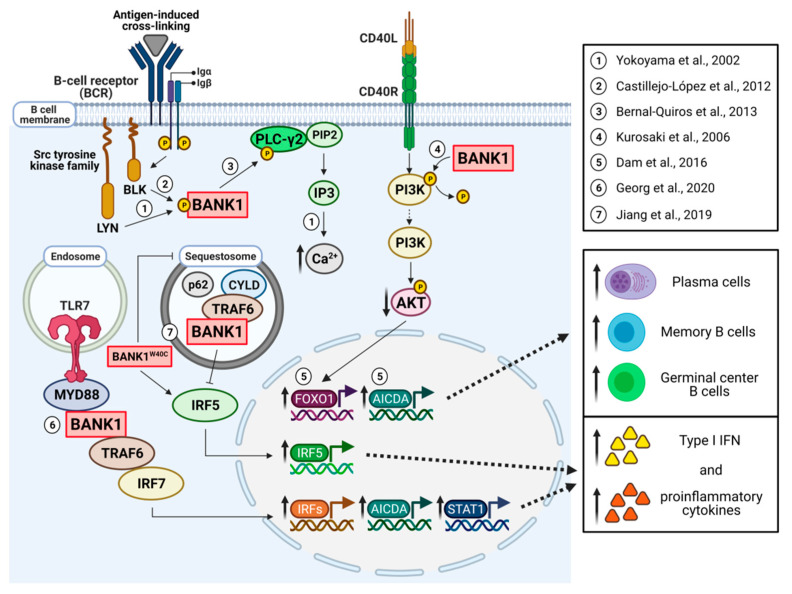
One study analyzed a family with multiple cases of SLE and found rare coding variants in both genes BANK1 and BLK that segregated in families [20]. The BANK1 variant was a W to C change in amino acid position 40 located in exon 2 (W40C). Importantly, this study found that the expression of BANK1 40C in HEK293T cells led to reduced formation of the so-called sequestosomes, and, therefore, when expressed with TRAF6 and IRF5, BANK1 40C could not repress TRAF6-mediated IRF5 nuclear localization, leading to increased IRF5 activation and induction of type I interferon, important in the pathogenesis of SLE (Figure 1). In a second study, using immunoprecipitation experiments, Georg et al. showed that the same change (40C) promoted the binding of BANK1 to MYD88 [37] and led to increased production of IL-8 in a transfection system (Figure 1). Both studies show the importance of BANK1 in mediating TRAF6 activity through several mechanisms and that the 40C variant is indeed a risk mutation, promoting the development of SLE in families.
Figure 1.
Model of the role of BANK1 in B cell signaling. BANK1 is primarily expressed in B cells and is involved in B cell signaling pathways. During BCR activation, BANK1 becomes tyrosine phosphorylated and binds the Src family kinases LYN and BLK (1-2) [1]. BANK1 also interacts with PLC-γ2 [44], regulating BCR-induced calcium mobilization. BANK1 attenuates CD40-mediated AKT activation (4) [45]. Following the PI3K/AKT pathway, the transcription factors FOXO1 and AICDA appear to increase in subjects with BANK1 risk variants, resulting in an increase in plasma cells, memory B cells, and germinal center B cells (5) [38]. BANK1 interacts with TRAF6 and MyD88 through endosomal TLR7 activation (6), triggering an IRF7-dependent production of IFNα and several proinflammatory cytokines. In the absence of exon 2, in BANK1–D2, there is a significant decrease in binding to MyD88 compared to BANK1–FL [37]. Finally, BANK1 binds to TRAF6, forming a complex with the sequestosome protein p62 and the deubiquitinating enzyme CYLD. In these structures, BANK1 promotes TRAF6 sequestration, diminishing IRF5 activation and type I IFN induction, and this is modified by a mutation in the amino acid position 40 in exon 2 (W40C) (7) [20].
Another study went deeper into the role of the BANK1 gene in alterations in peripheral B cell signaling [38]. This study found that B cells of BANK1 risk allele carriers of rs10516487 had increased the basal expression of the FOXO1 protein and increased the expression of FOXO1 target genes upon stimulation with anti-IgM and CD40. FOXO1 is an important transcriptional target of the PI3K/AKT pathway, and, as described previously, it is modulated in the absence of BANK1 (Figure 1). Activation of the PI3K/AKT pathway was severely decreased in individuals who were carriers of the risk alleles of BANK1. This was observed in reduced phosphorylation of AKT and PLCg2, showing a general reduction in BCR signaling. Of interest, upon BCR/CD40 stimulation, BANK1 full-length isoform expression was increased in risk allele carrier B cells and correlated strongly with mRNA levels of AICDA and SELL (CD62L) as well as FOXO1 (Figure 1). This is interesting, as AICDA is involved in class switching, while SELL or CD62L, on the other hand, is involved in B cell homing into the lymph nodes.
Of interest, BANK1 has also been found to be genetically linked to SLE in dogs, particularly with the presence of antinuclear antibodies and with similar gene expression patterns as found in the human disease [39].
3. The Role of BANK1 in B Cell Signaling
The BANK1 protein is constituted by three conserved domains: two double ankyrin repeat-like [12] motifs, a coiled-coil (CC) domain, and a Dof/BCAP/BANK (DBB) motif [40]. BANK1 also includes in its sequence several tyrosine- and proline-rich regions that could provide docking sites for SH2- and SH3-containing proteins. At the protein level, four isoforms have been reported, of which two have been the subject of several studies. The first isoform contains the canonical sequence (785 amino acids), is generated by alternative splicing, and is named the full-length isoform (FL). The other isoform of interest, named Delta2 (D2) (652 amino acids), lacks exon 2, which encodes for a putative conformational TIR domain [41]. Increased expression of this isoform and lower expression of the FL isoform have been linked, genetically, to protection against SLE [2]. It is unclear how splicing of the isoforms of BANK1 occurs and how the D2 is formed. One work suggests that the non-synonymous variant rs10516487 in exon 2 does influence splicing efficiency through the creation of an exonic splicing enhancer site that binds the SRp40 splicing factor [42]. Furthermore, the FL isoform containing the risk variant (R61) is capable of forming larger scaffold complexes in the cell cytoplasm, that is, has increased potential of multimerization. A functional TRAF6 binding motif has been reported as well as the confirmation of the presence of a functional TIR domain [37].
BANK1 is mainly expressed in B cells and, as a scaffold, it is involved in B cell signaling pathways. As mentioned previously, the first study demonstrated that BANK1 binds to the Scr tyrosine kinase LYN [1] (Figure 1). In this line, posterior studies showed that BANK1 also binds to BLK when tested in a B cell line and primary and naive B cells.
BANK1 also interacts with phospholipase 2C (PLCg2) (Figure 1). PLCg2 is part of a major group of signaling switch molecules involved in the formation of the second messenger inositol 1,4-5-triphosphate (IP3) and diacylglycerol. PLCg1 and PLCg2 have two Src homology domains (SH2) and an SH3 domain. PLCg2 is the one expressed in hematopoietic cells and is important in the regulation of immune activation [43]. This interaction of BANK1 and PLCg2 is promoted by engagement to the BCR and through binding with the proline-rich motifs and the phosphorylation of tyrosine residues on BANK1. The formation of the BANK1–PLCg2 complex is modulated by its sub-cellular location and the kinase activity of BLK, suggesting a role of BANK1 in modulating BCR signaling through a BANK1–BLK interaction [44].
The first study using the mouse deficient of BANK1 (BANK1−/−) [45] showed that that the IgM B cell responses to T-dependent antigens, numbers of mature B cells, spontaneously formed germinal centers, and levels of IgG2a were enhanced in these mice, a phenotype blocked by a double knock-out of BANK1 and CD40. In vitro, CD40-mediated proliferation and cell survival were increased in BANK1−/− mice accompanied by enhanced Akt activation [45].
BANK1 has a role in type I interferon signaling and cytokine production. BANK1 interacts with TRAF6 and MyD88 as demonstrated with immunoprecipitation studies [37] (Figure 1). TRAF6 forms a complex with MyD88 and IRF7 after TLR7, TLR8, or TLR9 stimulation, triggering the production of IFNalpha [46]. In in vitro studies using the HEK293 cell line, the absence of exon 2, the BANK1–D2 isoform lacking the TIR domain, exhibited a significant decrease in binding to MyD88 compared to BANK1–FL [37], demonstrating that the putative TIR domain of BANK1 is indeed functional. As mentioned above, this study identified TRAF6-binding motifs confirmed using point mutations and decoy peptide experiments. These experiments showed the requirement to form the complex through the C-terminal domain with TRAF6 and the TIR domain for MyD88. Polyubiquitination of BANK1 is mediated by lysing 63 (K63) through the TIR domain. This ubiquitination is important for the activation of the signaling pathway and cytokine production, as shown [37]. In the same cell line, others have demonstrated that BANK1 binds to TRAF6 forming a complex with the sequestosome protein p62 and with the deubiquitinating enzyme CYLD [20], which plays a critical role in regulating TLR signaling [47]. The authors proposed that BANK1 promotes TRAF6 sequestration diminishing IRF5 activation and IFN induction. The above results need to be further explored to better understand the BANK1 function in the various signaling pathways.
4. Mechanistic Aspects of the Role of BANK1 in Experimental Disease Model Systems
To date, very little is known about the role that BANK1 may play in different diseases. Following the report of the genetic association of BANK1 with SLE [2] and RA [22], a study discovered that BANK1 had a role in islet primary nonfunction (PNF), a serious problem in islet transplantation [48]. The transplantation of isolated islets from a donor pancreas is used to treat type 1 diabetes mellitus, an autoimmune disease characterized by selective destruction of pancreatic beta cells causing a gradual deficiency of insulin. This study investigated if DcR3, a TNF family receptor, could protect islets from apoptosis, as it may occur through the FasL, LIGHT, or TL1A pathways. By generating transgenic mice expressing human DcR3, two molecules, Adcyap1 and BANK1, were found to reduce β cell apoptosis by modulating their expression. With gene expression analysis, it was shown that overexpression of Adcyap1 or reduced expression of BANK1 in transgenic DcR3 mice prevented cells from undergoing cytokine-triggered apoptosis, suggesting the existence of a novel mechanism of islet protection and survival. There is evidence that IL-1β- and glucose-induced β cell apoptosis is calcium flux dependent [49]. Hence, a possible mechanism for DcR3 to protect β cells is to inhibit cytokine-induced BANK1 up-regulation, which in turn, prevents calcium mobilization, as previously described for BANK1 in B cells [1]. It must, however, be mentioned that the changes in calcium mobilization dependent on BANK1 have not been confirmed by any other study.
As mentioned above, BANK1 has a role in B cell signaling. During BCR activation, BANK1 becomes tyrosine phosphorylated and is able to bind the Src family kinases Lyn and Blk, acting as an adaptor or scaffold protein connecting these protein tyrosine kinases (PTKs) to IP3R receptors, which are in the same family as the B cell adapter BCAP [1,41]. In addition, it has been shown that exon 2 of human BANK1 encodes an N-terminal toll/IL-1 receptor [26] domain that is shared by BCAP. Toll-like receptors (TLRs) use adapters that contain the TIR domain, such as MyD88 and TRIF, to induce activation of transcription factors, including NF-κB, MAP kinases, and IFN regulatory factors [41]. In this regard, TLR9 is one of the most relevant endosomal TLRs in B cells, and TLR9 signaling is believed to have a critical role in autoimmunity [50]. Because of the role of BANK1 as a TIR-containing adaptor and the role of TLR9 in autoimmunity, the effect of BANK1 in TLR9 signaling was studied. Using BANK1 deficient mice and stimulation with the TLR9 agonist CpG, a reduction in MAPK p38 phosphorylation was observed in purified splenic B cells as compared with littermate control wild-type mice. BANK1 deficiency also led to the reduced production of the proinflammatory cytokine IL-6 in response to CpG alone or in combination with BCR ligation. However, there was no reduction in IL-6 mRNA, so an alternative mechanism had to be found. It was then found that BANK1 deficiency also reduced CpG-induced MNK1/2 and eIF4E phosphorylation kinases, which form the MNK1/2/eIF4E/eIF4G pathway of the translation initiation process, controlled by p38 [51], thus reducing translation of IL-6.
Subsequent studies were aimed at investigating the effects of BANK1 in the context of an autoimmune disease in a lupus mouse model. In order to do so, crosses of B6.Sle1.yaa with BANK1−/− mice were generated [52]. This model carries the Sle1 locus, which is analogous to a chromosomal region linked with SLE susceptibility in humans [53], and the yaa locus, known to be the result of the translocation of an X chromosome region to the Y chromosome resulting in a duplication of a large number of genes, including Tlr7 [54], another endosomal TLR receptor, which contains TIR domains and is associated with autoimmune pathogenesis [55]. Increased expression of TLR7 is enough to induce a lupus-like disease [56]. In this study [52], the effect of BANK1 deficiency on major lupus phenotypes was observed, among which were a reduction in mortality and in the production of total IgG and IgG anti-dsDNA antibodies, as well as the pathogenic isoform IgG2c, in B6.Sle1.yaa.BANK1−/− mice compared with B6.Sle1.yaa.BANK1+/+. The proinflammatory cytokine IL-6 was also reduced in the absence of BANK1 as previously seen [51]. By flow cytometry analysis, it was observed that BANK1 deficiency restores the cellular phenotypes of splenic lymphocytes and myeloid cells. The only effect observed in T cells was the normalization of the expression of CXCR4 on follicular T helper cells (Tfh), suggesting that BANK1 has effects on the formation of extrafollicular foci. In addition, in vitro experiments stimulating TLR7 and TLR8 agonists with imiquimod and resiquimod, respectively, showed that BANK1 regulates TLR7-induced signaling pathways in B cells, leading to the reduced expression of Aicda, as well as interferon response factor genes (Irf1, Irf7 and Irf9), Stat1, and type I IFN genes (Ifna4 and Ifnb) in B6.Sle1.yaa.BANK1−/− mice [52].
BANK1 has been implicated in B cell lymphomagenesis, a tumorigenesis of immature B cells [57]. The SL/KH mouse model, a model of spontaneous pre-B-lymphomas, was used. By qRT–PCR, it was observed that overexpression of Zfp521, a putative gene involved in the induction of B cell lymphomagenesis, caused a marked increase in the expression of pre-BCR-related genes, including BANK1. Knockdown of BANK1 reduced the proliferation of pre-B cells.
BANK1 has also been related with colitis, an inflammatory disease also known as Crohn’s disease. Employing a novel experimental procedure, based on the immuno-capture of MHCII complexes obtained from lymph nodes followed by a mass spectrometric analysis of eluted peptides, peptides derived from a subset of proteins, including BANK1, were identified in a dextran sodium sulfate (DSS)-induced colitis mouse model [58].
Finally, a study using collagen-induced arthritis (CIA) mice, the classic murine model of rheumatoid arthritis (RA), observed a reduction in BANK1 levels in the spleen, peripheral blood, and lymph nodes of CIA mice during the acute stage of arthritis, which had a negative correlation with disease severity and autoantibody production [59].
5. Conclusions
There is still much to be done to understand the role of BANK1 in B cell signaling. Moreover, some evidence suggests that BANK1 may also be expressed in myeloid cells and plasmacytoid dendritic cells, but no work has been performed on these aspects. Being a scaffold important in the signal transduction of various signaling pathways, how these pathways may converge or synergize through BANK1 is still completely unknown. BANK1 holds major risk alleles for several related antibody-mediated autoimmune conditions, and, thus, it warrants much greater attention.
Acknowledgments
Figure 1 was created with https://biorender.com/ on 29 April 2021.
Author Contributions
All authors wrote a section of the manuscript. M.E.A.-R. edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The preparation of this review was funded by the Ministerio de Economía y Competitividad (now Ministerio de Ciencia e Innovación) of the Government of Spain with number SAF2016-78631-P. G.G.H. is supported through this grant.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest. The authors are NOT employees of Pfizer.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yokoyama K., Su Ih I.H., Tezuka T., Yasuda T., Mikoshiba K., Tarakhovsky A., Yamamoto T. BANK regulates BCR-induced calcium mobilization by promoting tyrosine phosphorylation of IP(3) receptor. Embo J. 2002;21:83–92. doi: 10.1093/emboj/21.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozyrev S.V., Abelson A.K., Wojcik J., Zaghlool A., Linga Reddy M.V., Sanchez E., Gunnarsson I., Svenungsson E., Sturfelt G., Jonsen A., et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat. Genet. 2008;40:211–216. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 3.Guo L., Deshmukh H., Lu R., Vidal G.S., Kelly J.A., Kaufman K.M., Dominguez N., Klein W., Kim-Howard X., Bruner G.R., et al. Replication of the BANK1 genetic association with systemic lupus erythematosus in a European-derived population. Genes Immun. 2009;10:531–538. doi: 10.1038/gene.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarvinen T.M., Hellquist A., Zucchelli M., Koskenmies S., Panelius J., Hasan T., Julkunen H., D’Amato M., Kere J. Replication of GWAS-identified systemic lupus erythematosus susceptibility genes affirms B-cell receptor pathway signalling and strengthens the role of IRF5 in disease susceptibility in a Northern European population. Rheumatology. 2012;51:87–92. doi: 10.1093/rheumatology/ker263. [DOI] [PubMed] [Google Scholar]
- 5.Suarez-Gestal M., Calaza M., Endreffy E., Pullmann R., Ordi-Ros J., Sebastiani G.D., Ruzickova S., Jose Santos M., Papasteriades C., Marchini M., et al. Replication of recently identified systemic lupus erythematosus genetic associations: A case-control study. Arthritis Res. Ther. 2009;11:R69. doi: 10.1186/ar2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budarf M.L., Goyette P., Boucher G., Lian J., Graham R.R., Claudio J.O., Hudson T., Gladman D., Clarke A.E., Pope J.E., et al. A targeted association study in systemic lupus erythematosus identifies multiple susceptibility alleles. Genes Immun. 2011;12:51–58. doi: 10.1038/gene.2010.47. [DOI] [PubMed] [Google Scholar]
- 7.Bentham J., Morris D.L., Cunninghame Graham D.S., Pinder C.L., Tombleson P., Behrens T.W., Martin J., Fairfax B.P., Knight J.C., Chen L., et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langefeld C.D., Ainsworth H.C., Cunninghame Graham D.S., Kelly J.A., Comeau M.E., Marion M.C., Howard T.D., Ramos P.S., Croker J.A., Morris D.L., et al. Transancestral mapping and genetic load in systemic lupus erythematosus. Nat. Commun. 2017;8:16021. doi: 10.1038/ncomms16021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Bueno M., Oparina N., Dozmorov M.G., Marion M.C., Comeau M.E., Gilkeson G., Kamen D., Weisman M., Salmon J., McCune J.W., et al. Trans-Ethnic Mapping of BANK1 Identifies Two Independent SLE-Risk Linkage Groups Enriched for Co-Transcriptional Splicing Marks. Int. J. Mol. Sci. 2018;19:2331. doi: 10.3390/ijms19082331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung S.A., Tian C., Taylor K.E., Lee A.T., Ortmann W.A., Hom G., Graham R.R., Nititham J., Kelly J.A., Morrisey J., et al. European population substructure is associated with mucocutaneous manifestations and autoantibody production in systemic lupus erythematosus. Arthritis Rheum. 2009;60:2448–2456. doi: 10.1002/art.24707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang Y.K., Yang W., Zhao M., Mok C.C., Chan T.M., Wong R.W., Lee K.W., Mok M.Y., Wong S.N., Ng I.O., et al. Association of BANK1 and TNFSF4 with systemic lupus erythematosus in Hong Kong Chinese. Genes Immun. 2009;10:414–420. doi: 10.1038/gene.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W., Shen N., Ye D.Q., Liu Q., Zhang Y., Qian X.X., Hirankarn N., Ying D., Pan H.F., Mok C.C., et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6:e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan M., Yu B., Wan J., Zhang X., Wu Z., Zhong Q., Zhang W., Zou H. Identification of BANK1 polymorphisms by unlabelled probe high resolution melting: Association with systemic lupus erythematosus susceptibility and autoantibody production in Han Chinese. Rheumatology (Oxford) 2011;50:473–480. doi: 10.1093/rheumatology/keq353. [DOI] [PubMed] [Google Scholar]
- 14.Ramirez-Bello J., Jimenez-Morales S., Montufar-Robles I., Fragoso J.M., Barbosa-Cobos R.E., Saavedra M.A., Sanchez-Munoz F. BLK and BANK1 polymorphisms and interactions are associated in Mexican patients with systemic lupus erythematosus. Inflamm. Res. 2019;68:705–713. doi: 10.1007/s00011-019-01253-9. [DOI] [PubMed] [Google Scholar]
- 15.Alarcon-Riquelme M.E., Ziegler J.T., Molineros J., Howard T.D., Moreno-Estrada A., Sanchez-Rodriguez E., Ainsworth H.C., Ortiz-Tello P., Comeau M.E., Rasmussen A., et al. Genome-Wide Association Study in an Amerindian Ancestry Population Reveals Novel Systemic Lupus Erythematosus Risk Loci and the Role of European Admixture. Arthritis Rheumatol. 2016;68:932–943. doi: 10.1002/art.39504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez E., Comeau M.E., Freedman B.I., Kelly J.A., Kaufman K.M., Langefeld C.D., Brown E.E., Alarcon G.S., Kimberly R.P., Edberg J.C., et al. Identification of novel genetic susceptibility loci in African American lupus patients in a candidate gene association study. Arthritis Rheum. 2011;63:3493–3501. doi: 10.1002/art.30563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant S.F., Petri M., Bradfield J.P., Kim C.E., Santa E., Annaiah K., Frackelton E.C., Glessner J.T., Otieno F.G., Shaner J.L., et al. Association of the BANK 1 R61H variant with systemic lupus erythematosus in Americans of European and African ancestry. Appl. Clin. Genet. 2009;2:1–5. doi: 10.2147/TACG.S4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orozco G., Eyre S., Hinks A., Bowes J., Morgan A.W., Wilson A.G., Wordsworth P., Steer S., Hocking L., Thomson W., et al. Study of the common genetic background for rheumatoid arthritis and systemic lupus erythematosus. Ann> Rheum. Dis. 2011;70:463–468. doi: 10.1136/ard.2010.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou X.J., Qi Y.Y., Cheng F.J., Zhang H. Genetic interactions between BANK1 and BLK in Chinese patients with systemic lupus erythematosus. J. Rheumatol. 2013;40:1772–1773. doi: 10.3899/jrheum.130477. [DOI] [PubMed] [Google Scholar]
- 20.Jiang S.H., Athanasopoulos V., Ellyard J.I., Chuah A., Cappello J., Cook A., Prabhu S.B., Cardenas J., Gu J., Stanley M., et al. Functional rare and low frequency variants in BLK and BANK1 contribute to human lupus. Nat. Commun. 2019;10:2201. doi: 10.1038/s41467-019-10242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dieude P., Wipff J., Guedj M., Ruiz B., Melchers I., Hachulla E., Riemekasten G., Diot E., Hunzelmann N., Sibilia J., et al. BANK1 is a genetic risk factor for diffuse cutaneous systemic sclerosis and has additive effects with IRF5 and STAT4. Arthritis Rheum. 2009;60:3447–3454. doi: 10.1002/art.24885. [DOI] [PubMed] [Google Scholar]
- 22.Orozco G., Abelson A.K., Gonzalez-Gay M.A., Balsa A., Pascual-Salcedo D., Garcia A., Fernandez-Gutierrez B., Petersson I., Pons-Estel B., Eimon A., et al. Study of functional variants of the BANK1 gene in rheumatoid arthritis. Arthritis Rheum. 2009;60:372–379. doi: 10.1002/art.24244. [DOI] [PubMed] [Google Scholar]
- 23.Berndt S.I., Camp N.J., Skibola C.F., Vijai J., Wang Z., Gu J., Nieters A., Kelly R.S., Smedby K.E., Monnereau A., et al. Meta-analysis of genome-wide association studies discovers multiple loci for chronic lymphocytic leukemia. Nat. Commun. 2016;7:10933. doi: 10.1038/ncomms10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong K.W., Lyu J., Lee S.H., Choi B.Y., Kim S.S., Kim Y. A nonsynonymous SNP in BANK1 is associated with serum LDL cholesterol levels in three Korean populations. J. Hum. Genet. 2015;60:113–118. doi: 10.1038/jhg.2014.108. [DOI] [PubMed] [Google Scholar]
- 25.Yin H., Borghi M.O., Delgado-Vega A.M., Tincani A., Meroni P.L., Alarcon-Riquelme M.E. Association of STAT4 and BLK, but not BANK1 or IRF5, with primary antiphospholipid syndrome. Arthritis Rheum. 2009;60:2468–2471. doi: 10.1002/art.24701. [DOI] [PubMed] [Google Scholar]
- 26.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Lango Allen H., Lindgren C.M., Luan J., Magi R., et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mak A.C., Tang P.L., Cleveland C., Smith M.H., Kari Connolly M., Katsumoto T.R., Wolters P.J., Kwok P.Y., Criswell L.A. Brief Report: Whole-Exome Sequencing for Identification of Potential Causal Variants for Diffuse Cutaneous Systemic Sclerosis. Arthritis Rheumatol. 2016;68:2257–2262. doi: 10.1002/art.39721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baranski T.J., Kraja A.T., Fink J.L., Feitosa M., Lenzini P.A., Borecki I.B., Liu C.T., Cupples L.A., North K.E., Province M.A. A high throughput, functional screen of human Body Mass Index GWAS loci using tissue-specific RNAi Drosophila melanogaster crosses. PLoS Genet. 2018;14:e1007222. doi: 10.1371/journal.pgen.1007222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reksten T.R., Johnsen S.J., Jonsson M.V., Omdal R., Brun J.G., Theander E., Eriksson P., Wahren-Herlenius M., Jonsson R., Nordmark G. Genetic associations to germinal centre formation in primary Sjogren’s syndrome. Ann. Rheum. Dis. 2014;73:1253–1258. doi: 10.1136/annrheumdis-2012-202500. [DOI] [PubMed] [Google Scholar]
- 30.Sun F., Xu J., Wu Z., Li P., Chen H., Su J., You X., Li M., Zhao Y., Tian X., et al. Polymorphisms in the FAM167A-BLK, but not BANK1, are associated with primary Sjogren’s syndrome in a Han Chinese population. Clin. Exp. Rheumatol. 2013;31:704–710. [PubMed] [Google Scholar]
- 31.Lessard C.J., Li H., Adrianto I., Ice J.A., Rasmussen A., Grundahl K.M., Kelly J.A., Dozmorov M.G., Miceli-Richard C., Bowman S., et al. Variants at multiple loci implicated in both innate and adaptive immune responses are associated with Sjogren’s syndrome. Nat. Genet. 2013;45:1284–1292. doi: 10.1038/ng.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coustet B., Dieude P., Guedj M., Bouaziz M., Avouac J., Ruiz B., Hachulla E., Diot E., Kracowski J.L., Tiev K., et al. C8orf 13/BLK is a genetic risk locus for systemic sclerosis and has additive effect with BANK1: Results from a large French cohort and meta-analysis. Arthritis Rheum. 2011;63:2091–2096. doi: 10.1002/art.30379. [DOI] [PubMed] [Google Scholar]
- 33.Castillejo-Lopez C., Delgado-Vega A.M., Wojcik J., Kozyrev S.V., Thavathiru E., Wu Y.Y., Sanchez E., Pollmann D., Lopez-Egido J.R., Fineschi S., et al. Genetic and physical interaction of the B-cell systemic lupus erythematosus-associated genes BANK1 and BLK. Ann. Rheum. Dis. 2012;71:136–142. doi: 10.1136/annrheumdis-2011-200085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Genin E., Coustet B., Allanore Y., Ito I., Teruel M., Constantin A., Schaeverbeke T., Ruyssen-Witrand A., Tohma S., Cantagrel A., et al. Epistatic interaction between BANK1 and BLK in rheumatoid arthritis: Results from a large trans-ethnic meta-analysis. PLoS ONE. 2013;8:e61044. doi: 10.1371/journal.pone.0061044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dang J., Li J., Xin Q., Shan S., Bian X., Yuan Q., Liu N., Ma X., Li Y., Liu Q. Gene-gene interaction of ATG5, ATG7, BLK and BANK1 in systemic lupus erythematosus. Int. J. Rheum. Dis. 2016;19:1284–1293. doi: 10.1111/1756-185X.12768. [DOI] [PubMed] [Google Scholar]
- 36.Diaz-Barreiro A., Bernal-Quiros M., Georg I., Maranon C., Alarcon-Riquelme M.E., Castillejo-Lopez C. The SLE variant Ala71Thr of BLK severely decreases protein abundance and binding to BANK1 through impairment of the SH3 domain function. Genes Immun. 2016;17:128–138. doi: 10.1038/gene.2016.1. [DOI] [PubMed] [Google Scholar]
- 37.Georg I., Diaz-Barreiro A., Morell M., Pey A.L., Alarcon-Riquelme M.E. BANK1 interacts with TRAF6 and MyD88 in innate immune signaling in B cells. Cell Mol. Immunol. 2020;17:954–965. doi: 10.1038/s41423-019-0254-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dam E.M., Habib T., Chen J., Funk A., Glukhova V., Davis-Pickett M., Wei S., James R., Buckner J.H., Cerosaletti K. The BANK1 SLE-risk variants are associated with alterations in peripheral B cell signaling and development in humans. Clin. Immunol. 2016;173:171–180. doi: 10.1016/j.clim.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilbe M., Jokinen P., Truve K., Seppala E.H., Karlsson E.K., Biagi T., Hughes A., Bannasch D., Andersson G., Hansson-Hamlin H., et al. Genome-wide association mapping identifies multiple loci for a canine SLE-related disease complex. Nat. Genet. 2010;42:250–254. doi: 10.1038/ng.525. [DOI] [PubMed] [Google Scholar]
- 40.Battersby A., Csiszar A., Leptin M., Wilson R. Isolation of proteins that interact with the signal transduction molecule Dof and identification of a functional domain conserved between Dof and vertebrate BCAP. J. Mol. Biol. 2003;329:479–493. doi: 10.1016/S0022-2836(03)00489-3. [DOI] [PubMed] [Google Scholar]
- 41.Troutman T.D., Hu W., Fulenchek S., Yamazaki T., Kurosaki T., Bazan J.F., Pasare C. Role for B-cell adapter for PI3K (BCAP) as a signaling adapter linking Toll-like receptors (TLRs) to serine/threonine kinases PI3K/Akt. Proc. Natl. Acad. Sci. USA. 2012;109:273–278. doi: 10.1073/pnas.1118579109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kozyrev S.V., Bernal-Quiros M., Alarcon-Riquelme M.E., Castillejo-Lopez C. The dual effect of the lupus-associated polymorphism rs10516487 on BANK1 gene expression and protein localization. Genes Immun. 2012;13:129–138. doi: 10.1038/gene.2011.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang D., Feng J., Wen R., Marine J.C., Sangster M.Y., Parganas E., Hoffmeyer A., Jackson C.W., Cleveland J.L., Murray P.J., et al. Phospholipase Cgamma2 is essential in the functions of B cell and several Fc receptors. Immunity. 2000;13:25–35. doi: 10.1016/S1074-7613(00)00005-4. [DOI] [PubMed] [Google Scholar]
- 44.Bernal-Quiros M., Wu Y.Y., Alarcon-Riquelme M.E., Castillejo-Lopez C. BANK1 and BLK Act through Phospholipase C Gamma 2 in B-Cell Signaling. PLoS ONE. 2013;8:e59842. doi: 10.1371/journal.pone.0059842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aiba Y., Yamazaki T., Okada T., Gotoh K., Sanjo H., Ogata M., Kurosaki T. BANK negatively regulates Akt activation and subsequent B cell responses. Immunity. 2006;24:259–268. doi: 10.1016/j.immuni.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 46.Kawai T., Sato S., Ishii K.J., Coban C., Hemmi H., Yamamoto M., Terai K., Matsuda M., Inoue J., Uematsu S., et al. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 2004;5:1061–1068. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- 47.Kim J.Y., Ozato K. The sequestosome 1/p62 attenuates cytokine gene expression in activated macrophages by inhibiting IFN regulatory factor 8 and TNF receptor-associated factor 6/NF-kappaB activity. J. Immunol. 2009;182:2131–2140. doi: 10.4049/jimmunol.0802755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han B., Wu J. DcR3 protects islet beta cells from apoptosis through modulating Adcyap1 and Bank1 expression. J. Immunol. 2009;183:8157–8166. doi: 10.4049/jimmunol.0901165. [DOI] [PubMed] [Google Scholar]
- 49.Fei H., Zhao B., Zhao S., Wang Q. Requirements of calcium fluxes and ERK kinase activation for glucose- and interleukin-1beta-induced beta-cell apoptosis. Mol. Cell Biochem. 2008;315:75–84. doi: 10.1007/s11010-008-9791-8. [DOI] [PubMed] [Google Scholar]
- 50.Christensen S.R., Kashgarian M., Alexopoulou L., Flavell R.A., Akira S., Shlomchik M.J. Toll-like receptor 9 controls anti-DNA autoantibody production in murine lupus. J. Exp. Med. 2005;202:321–331. doi: 10.1084/jem.20050338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Y.Y., Kumar R., Haque M.S., Castillejo-Lopez C., Alarcon-Riquelme M.E. BANK1 Controls CpG-Induced IL-6 Secretion via a p38 and MNK1/2/eIF4E Translation Initiation Pathway. J. Immunol. 2013;191:6110–6116. doi: 10.4049/jimmunol.1301203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Y.Y., Kumar R., Iida R., Bagavant H., Alarcon-Riquelme M.E. BANK1 Regulates IgG Production in a Lupus Model by Controlling TLR7-Dependent STAT1 Activation. PLoS ONE. 2016;11:e0156302. doi: 10.1371/journal.pone.0156302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morel L., Blenman K.R., Croker B.P., Wakeland E.K. The major murine systemic lupus erythematosus susceptibility locus, Sle1, is a cluster of functionally related genes. Proc. Natl Acad. Sci. USA. 2001;98:1787–1792. doi: 10.1073/pnas.98.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Subramanian S., Tus K., Li Q.Z., Wang A., Tian X.H., Zhou J., Liang C., Bartov G., McDaniel L.D., Zhou X.J., et al. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc. Natl. Acad. Sci. USA. 2006;103:9970–9975. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santiago-Raber M.L., Dunand-Sauthier I., Wu T., Li Q.Z., Uematsu S., Akira S., Reith W., Mohan C., Kotzin B.L., Izui S. Critical role of TLR7 in the acceleration of systemic lupus erythematosus in TLR9-deficient mice. J. Autoimmun. 2010;34:339–348. doi: 10.1016/j.jaut.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 56.Fairhurst A.M., Hwang S.H., Wang A., Tian X.H., Boudreaux C., Zhou X.J., Casco J., Li Q.Z., Connolly J.E., Wakeland E.K. Yaa autoimmune phenotypes are conferred by overexpression of TLR7. Eur. J. Immunol. 2008;38:1971–1978. doi: 10.1002/eji.200838138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hiratsuka T., Takei Y., Ohmori R., Imai Y., Ozeki M., Tamaki K., Haga H., Nakamura T., Tsuruyama T. ZFP521 contributes to pre-B-cell lymphomagenesis through modulation of the pre-B-cell receptor signaling pathway. Oncogene. 2016;35:3227–3238. doi: 10.1038/onc.2015.385. [DOI] [PubMed] [Google Scholar]
- 58.Fugmann T., Sofron A., Ritz D., Bootz F., Neri D. The MHC Class II Immunopeptidome of Lymph Nodes in Health and in Chemically Induced Colitis. J. Immunol. 2017;198:1357–1364. doi: 10.4049/jimmunol.1601157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J., Ren J., Yang Y., Sun J., Zhou X., Zheng S., Xuan D., Xue Y., Fan H., Zhang J., et al. BANK1 alters B cell responses and influences the interactions between B cells and induced T regulatory cells in mice with collagen-induced arthritis. Arthritis Res. Ther. 2018;20:9. doi: 10.1186/s13075-017-1503-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.