Abstract
In eukaryotes, N‐ethylmaleimide‐sensitive factor (NSF) is a conserved AAA+ATPase and a key component of the membrane trafficking machinery that promotes the fusion of secretory vesicles with target membranes. Here, we demonstrate that the Arabidopsis thaliana genome contains a single copy of NSF, AtNSF, which plays an essential role in the regulation of leaf serration. The AtNSF knock‐down mutant, atnsf‐1, exhibited more serrations in the leaf margin. Moreover, polar localization of the PIN‐FORMED1 (PIN1) auxin efflux transporter was diffuse around the margins of atnsf‐1 leaves and root growth was inhibited in the atnsf‐1 mutant. More PIN1‐GFP accumulated in the intracellular compartments of atnsf‐1 plants, suggesting that AtNSF is required for intracellular trafficking of PIN between the endosome and plasma membrane. Furthermore, the serration phenotype was suppressed in the atnsf‐1 pin1‐8 double mutant, suggesting that AtNSF is required for PIN1‐mediated polar auxin transport to regulate leaf serration. The CUP‐SHAPED COTYLEDON2 (CUC2) transcription factor gene is up‐regulated in atnsf‐1 plants and the cuc2‐3 single mutant exhibits smooth leaf margins, demonstrating that AtNSF also functions in the CUC2 pathway. Our results reveal that AtNSF regulates the PIN1‐generated auxin maxima with a CUC2‐mediated feedback loop to control leaf serration.
Keywords: AtNSF, Auxin, CUC2, leaf serration, PIN1, vesicle trafficking
N‐ethylmaleimide‐sensitive factor (NSF) promotes the fusion of secretory vesicles in eukaryotes. In Arabidopsis, AtNSF negatively regulates leaf serration by affecting trafficking of the PIN‐FORMED1 auxin transporter and interfering with the PIN‐FORMED1 and CUP‐SHAPED COTYLEDON2‐mediated feedback loop for leaf serration.
INTRODUCTION
In eukaryotic cells, vesicle trafficking mediates the transport of cargoes (i.e., biological molecules such as proteins) to their destinations. This process usually includes vesicle budding, movement, tethering, docking, and fusion. A number of proteins are involved in membrane trafficking. Soluble N‐ethylmaleimide sensitive factor (NSF) attachment protein receptors (SNAREs) mostly participate in vesicle docking and fusion (Kim and Brandizzi, 2012; Yoon and Munson, 2018). In eukaryotes, SNAREs function as key components in protein complexes together with other co‐factors to drive membrane fusion and cargo release (Jahn and Scheller, 2006; Wickner and Schekman, 2008). Based on their localization, SNAREs can be classified into two types, vesicle‐SNAREs (v‐SNAREs), which are present on vesicle membranes, and target‐SNAREs (t‐SNAREs), which are present on target membranes. When transport vesicles move into the vicinity of the target organelles, v‐SNAREs interact with their corresponding t‐SNAREs to form a trans‐SNARE complex that draws the two membranes toward each other, and together with related factors, executes membrane fusion (Hu et al., 2007). After membrane fusion, SNARE complexes are disassembled into individual SNARE proteins for recycling, which requires NSF proteins to hydrolyze ATP and thus drive the disassembly reaction (Baker and Hughson, 2016).
NSF belongs to the ATPases associated with diverse cellular activities (the AAA+ATPase protein family), which are present in all organisms and are involved in cellular processes such as protein degradation, membrane fusion, microtubule severing, and signal transduction (Hanson and Whiteheart, 2005; Baker and Hughson, 2016). The NSF protein contains three domains: the N‐terminal domain, the first ATP‐binding domain D1, and the second ATP‐binding domain D2 (Zhao et al., 2012). The N‐terminal domain is essential for interactions with the α‐SNAP–SNARE complex, whereas domains D1 and D2 possess ATPase activity and mediate the formation of a functional NSF homohexamer (Nagiec et al., 1995).
The role of NSF in vesicle trafficking has been elucidated in many eukaryotes. For example, the yeast (Saccharomyces cerevisiae) NSF, which is encoded by SEC18, plays a role in membrane trafficking. The temperature‐sensitive sec18‐1 mutant displays defective endoplasmic reticulum (ER) morphology and accumulates small vesicles at the restrictive temperature, suggesting that the membrane fusion step is blocked (Mittenbühler and Holzer, 1991). In mammalian cells, the dominant‐negative NSF mutant NSF‐E329Q results in cell death due to disruption of the Golgi apparatus (Dalal et al., 2004). In zebrafish (Danio rerio), mutations in NSF cause ER‐like membrane aggregation (Kurrasch et al., 2009). These studies have demonstrated that NSF is crucial for vesicle trafficking and cell survival. Recently, a forward‐genetic screen led to the identification of an NSF missense mutant in Arabidopsis thaliana that showed a pleiotropic Golgi defect, which suggests that NSF has an important role in maintaining the morphology of the Golgi (Tanabashi et al., 2018). In addition, an atypical NSF and SNAP in soybean (Glycine max) was shown to confer resistance to a highly damaging nematode pathogen (Bayless et al., 2018). However, the mechanism of NSF in regulating membrane fusion and vesicle transport in plant cells still needs to be elucidated.
Plant leaf shape is defined by serration and indentation of the leaf margin. Leaf shape is regulated by different developmental and genetic cues, which have been well characterized in Arabidopsis (Tsukaya, 2006). For example, microRNAs play an essential role in leaf serration. The JAW gene encodes a microRNA that controls leaf development by targeting the mRNAs of several genes encoding TEOSINTE BRANCHED 1/CYCLOIDEA/PCF (TCP) transcription factors for cleavage. Compared with the wild‐type, dominant jaw‐D mutants show more leaf serration and lower expression levels of certain TCP genes (Palatnik et al., 2003). The Arabidopsis SERRATE (SE) gene regulates leaf polarity and leaf margin development by regulating microRNA165/166, and se‐1 mutant leaves also have more serrations than wild‐type leaves (Yang et al., 2006). Another microRNA, miR164A, controls leaf margin serration by regulating the expression level of the transcription factor gene CUP‐SHAPED COTYLEDON2 (CUC2). The leaves of the mir164a‐4 mutant have more and deeper serrations than wild‐type leaves, whereas plants that overexpress MIR164A have leaves with smooth margins, a phenotype similar to that of the cuc2‐3 mutant (Nikovics et al., 2006). Recently, a computational model of leaf serration was developed based on the findings that CUC2 and auxin flux form a feedback loop that regulates leaf serration in Arabidopsis (Bilsborough et al., 2011). Furthermore, interaction between the secreted peptide EPIDERMAL PATTERNING FACTOR‐LIKE2 (EPFL2) and ERECTA was shown to promote leaf tooth growth by regulating auxin levels during leaf margin morphogenesis (Tameshige et al., 2016).
Auxin affects the growth and development of organs and its effect depends on the PIN auxin efflux transporters (Vieten et al., 2007; Vanneste and Friml, 2009). In the leaf margin, PIN‐FORMED1 (PIN1) polarity directs auxin flux, and teeth initiate from sites of auxin maxima in the leaf margin (Bilsborough et al., 2011). Consistent with this, plants treated with the auxin polar transport inhibitor N‐1‐naphthylphthalamic acid (NPA) and mutant plants that lack PIN1 have smooth leaf margins, whereas serration can be restored by expressing PIN1 in pin1 mutant leaf epidermal cells (Hay et al., 2006; Bilsborough et al., 2011). Therefore, PIN1‐mediated auxin polar transport is crucial for leaf serration outgrowth. However, PIN1 polarity is dynamic and changes in response to different cues. PIN1 is actively shuttled between the endosome and the plasma membrane (PM) through ARF‐GEF (GNOM) and clathrin‐dependent endocytosis (Geldner et al., 2001; Ischebeck et al., 2013). Conserved small GTPases of the Rab and ROP families play important roles in PIN1 trafficking (Feraru et al., 2012; Nagawa et al., 2012). For example, BREFELDIN A (BFA)‐VISUALIZED ENDOCYTIC TRAFFICKING DEFECTIVE 1 (BEN1), and BFA‐VISUALIZED EXOCYTIC TRAFFICKING DEFECTIVE 1 (BEX1) and 5 (BEX5) regulate PIN1 trafficking (Tanaka et al., 2009; Feraru et al., 2012; Tanaka et al., 2014), indicating that PIN1 is recycled between the endosome and PM via a conserved membrane fusion and secretion pathway. In this study, we address whether another essential player in protein trafficking regulates PIN1 intracellular transport.
The Arabidopsis thaliana genome contains a single copy of NSF (AtNSF). We isolated an AtNSF knock‐down mutant, atnsf‐1, which exhibits increased serration in the leaf margin. Root growth is inhibited in atnsf‐1 plants, especially upon treatment with the vesicle inhibitor BFA. We observed that endocytosis and exocytosis, as well as auxin distribution and PIN1 trafficking were defective in the atnsf‐1 mutant. The serration phenotype of atnsf‐1 leaves was completely suppressed by the auxin polar transport inhibitor NPA and by PIN1 depletion. Furthermore, the atnsf‐1 cuc2‐3 double mutant exhibited smooth leaf margins. Collectively, these results show that AtNSF regulates leaf serration by modulating PIN1 polarity to interfere with the PIN1‐ and CUC2‐mediated feedback loop in Arabidopsis.
RESULTS
Characterization of Arabidopsis NSF
To elucidate the function of NSF in plants, we identified an NSF homolog in Arabidopsis, AtNSF, which encodes a protein of 742 amino acids and shares 41.99% and 40.57% identities with human and yeast orthologs, respectively (Figure 1A). Similar to the human and yeast NSFs, AtNSF contains three conserved domains: the N‐terminal, D1, and D2 domains (Figure 1A). To determine whether AtNSF has ATPase activity, recombinant AtNSF protein was expressed and purified from Escherichia coli and subjected to an enzyme activity assay. When AtNSF was added, the concentration of inorganic phosphate (Pi), as an indicator of ATP hydrolysis activity, was much higher than that when AtNSF was absent, and the level of Pi increased along with increased AtNSF concentration (Figure 1B), indicating that AtNSF possesses ATPase activity in vitro. The ATPase activity was enhanced by the addition of the SNARE protein, SNAP (Figure 1B), which is similar to AAA+ATPase orthologs in yeast and mammals (Barnard et al., 1997).
Figure 1.
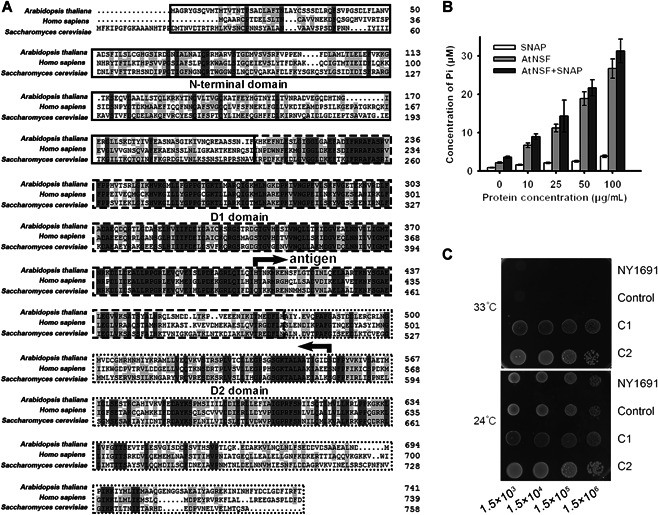
Identification and characterization of AtNSF
(A) Protein sequence alignment of three N‐ethylmaleimide‐sensitive factors (NSFs) from Arabidopsis thaliana, Homo sapiens, and Saccharomyces cerevisiae. Identical amino acid residues (dark gray) and amino acid residues with over 50% similarity (light gray) are highlighted. The N‐terminal domain (solid‐line box), D1 domain (dashed‐line box), and D2 domain (dotted‐line box) are marked. The oppositely facing arrows indicate the polypeptide region used for antibody production. (B) Enzyme activity assay. Showing that AtNSF has ATPase activity in vitro, and this activity was enhanced by SNAP. Error bars represent the SD for four biological replicates. (C) Growth phenotype of yeast mutant NY1691 (sec18‐1) transformed with AtNSF. Serial dilution of yeast cells containing the construct with the AtNSF‐coding sequence (C1 and C2 represent two different clones) or transformed with the empty vector (Control) in normal conditions (24°C) and high temperature (33°C).
To determine whether AtNSF has a typical NSF function, we conducted a functional complementation test by expressing AtNSF in the yeast nsf mutant, a SEC18 gene deletion strain NY1691 (also called sec18‐1). The NY1691 mutant is temperature sensitive and cannot survive at a high temperature (33°C) (Carr et al., 1999). Transfer of the AtNSF coding sequence into the NY1691 yeast mutant led to normal growth at this temperature (Figure 1C). Taken together, these results suggest that AtNSF is homologous to human and yeast NSF and possesses AAA+ATPase activity in Arabidopsis.
AtNSF inhibits secondary serration in the leaf margin
To further investigate the physiological function of AtNSF, we isolated an AtNSF‐defective mutant in Arabidopsis, atnsf‐1 (Salk_038536), which contains a T‐DNA insertion in the 5′‐untranslated region 305 bp upstream from the ATG start codon (Figure 2A). The atnsf‐1 mutant exhibited more leaf serration than the wild‐type, and more serration in older leaves than in younger leaves (Figures 2B, C, S1A). Furthermore, secondary teeth emerged between two outgrown teeth (Figure 2E, F), suggesting that serration formation was disordered.
Figure 2.

AtNSF regulates serration formation in the leaf margin
(A) Schematic diagram of the AtNSF gene (At4g04910) and the T‐DNA insertion sites in the nsf mutants. Twenty‐one exons and 20 introns are represented by gray boxes and solid lines, respectively. Two white boxes represent the 5′ and 3′‐UTRs. The location of the T‐DNA insertions in nsf mutants are indicated with triangles. (B–D) Leaf phenotype of wild‐type (WT) (B), atnsf‐1 (C), and a complemented plant (expressing 35S pro ::AtNSF) (D). Seedlings were grown in long‐day conditions (16 h light/8 h dark) for 4 weeks. Bars = 1 cm. (E, F) Leaf serration of the seventh leaf of WT (E) and atnsf‐1 (F) plants. The arrowhead in (F) indicates a secondary tooth formed between two outgrown teeth. Bars = 1 mm. (G) Protein abundance of AtNSF in WT, atnsf‐1, and complemented seedlings. The protein was detected using anti‐AtNSF antibody. Coomassie brilliant blue staining for Rubisco was used as a loading control.
To confirm whether the phenotype of atnsf‐1 was caused by AtNSF deficiency, we quantified the AtNSF transcript level, but this did not differ between the wild‐type and the mutant (Figure S1B). Considering the severe serration phenotype, we hypothesized that the AtNSF protein may be absent or present at low levels in the atnsf‐1 mutant. To address this, we designed a specific antibody against the AtNSF peptide from amino acids 407–560 (Figure 1A, antigen) and validated its efficacy in E. coli cells (Figure S1C). Then, we used this antibody to examine the level of AtNSF in the mutant. As we expected, the AtNSF level was significantly decreased in the atnsf‐1 mutant compared with that in the wild‐type (Figure 2G), indicating that the atnsf‐1 mutant is a knock‐down allele and AtNSF expression is attenuated at the post‐transcription level.
To further verify that the atnsf‐1 serration phenotype results from the knock‐down of AtNSF, we complemented the atnsf‐1 mutant by overexpressing AtNSF (35S pro ::AtNSF), and the defective leaf serration phenotype as well as the decreased AtNSF level was fully complemented (Figure 2D, G). We also expressed AtNSF via its native promoter (AtNSF pro ::AtNSF) in atnsf‐1 for complementation and obtained the same result (Figure S2). Taken together, these data confirmed the indispensable role of AtNSF in regulating the leaf margin.
We isolated two additional T‐DNA insertion lines. The atnsf‐2 (Salk_138721), in which the T‐DNA is inserted 74 bp upstream of the ATG start codon, did not show any mutant phenotype (Figures 2A, S3). According to reverse transcription polymerase chain reaction (RT‐PCR), we found that the expression level of AtNSF in the atnsf‐2 line was similar to that in wild‐type (Figure S3D), which together with the absence of any phenotypic differences, indicated that atnsf‐2 is a nonsense mutant. For atnsf‐3 (Sail_1155_C06), in which the T‐DNA was inserted 59 bp upstream of the ATG start codon (Figure 2A), we failed to isolate a homozygous line, suggesting that the homozygous mutant is lethal. Although the mechanism by which T‐DNA insertions affect the expression of AtNSF remains unclear, the decreased AtNSF protein level in the atnsf‐1 mutant and the restoration of the wild‐type phenotype in the complementation line confirmed that the serration phenotype results from the knock‐down of AtNSF. Therefore, we used the atnsf‐1 mutant for further analysis.
The role of AtNSF in root development
In addition to demonstrating an indispensable role for AtNSF in the serration of the leaf margin, we next investigated the role of AtNSF in root development. When seedlings were grown vertically on half‐strength Murashige and Skoog (1/2 MS) medium for 7 d, the roots of atnsf‐1 were slightly shorter than those of the wild‐type (Figure 3A). In 7 d‐old seedlings, the lengths of the mutant roots were 72.08% ± 9.24% (n ≥ 25) that of wild‐type roots, suggesting that AtNSF functions in root development, although this phenotype was not as obvious as in the leaf.
Figure 3.
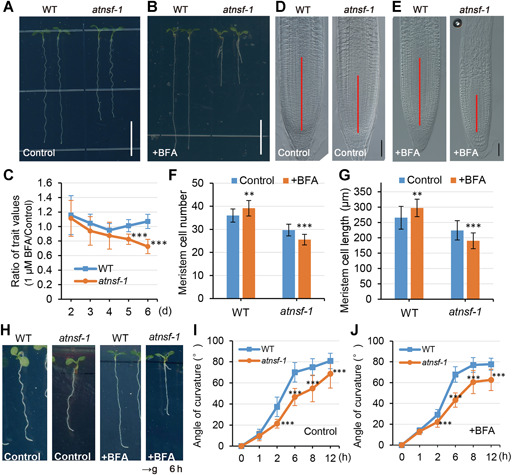
Functional characterization of AtNSF in root growth and the gravitropic response
(A, B) Root phenotype of 7‐d‐old wild‐type (WT) and atnsf‐1 seedlings in half‐strength Murashige‐Skoog (1/2 MS) medium with (+BFA) (B) or without (Control) (A) 1 μM BFA. Bars = 1 cm. (C) The rate of inhibition of WT and atnsf‐1 root growth for different numbers of days. Seedlings were grown on 1/2 MS medium with or without 1 μM BFA for varying lengths of time and the ratio of trait values (root length of 1 μM BFA/Control) was measured. Error bars represent the SD of biological replicates (n ≥ 20). Asterisks indicate significant differences (***P < 0.001) compared with the WT (t‐test). (D, E) Nomarski images of root tips in WT and atnsf‐1. Seedlings were grown in 1/2 MS medium with (+BFA) (E) or without (Control) (D) 1 μM BFA for 7 d. Red solid lines indicate the meristem. Bars = 40 μm. (F, G) Meristem cell number (F) and length (G) of 7‐d‐old WT and atnsf‐1 seedlings in 1/2 MS medium (Control) or the medium with 1 μM BFA (+BFA). Error bars represent means ± SD (n ≥ 20). Asterisks represent significant differences (**P < 0.01; ***P < 0.001; t‐test). (H) Images of root curvature of WT and atnsf‐1 seedlings in 1/2 MS medium (Control) or medium with 1 μM BFA (+BFA) after gravity stimulation for 6 h. Plates containing 6 d‐old seedlings were rotated by 90° to assess their gravitropic response. The black arrow at the bottom indicates the direction of the gravity vector after reorientation. (I, J) Quantification of the root gravitropic response in WT and atnsf‐1 seedlings in the medium with (+BFA) (J) or without 1 μM BFA (Control) (I) after reorientation for different times. Data are means ± SD (n ≥ 30). Asterisks indicate significant differences relative to the WT (***P < 0.001, t‐test).
Because NSF is important for disassembly of the SNARE complex to mediate vesicle trafficking in many eukaryotes (Mittenbühler and Holzer, 1991; Dalal et al., 2004; Kurrasch et al., 2009), we investigated whether the atnsf‐1 mutant is hypersensitive to BFA. BFA is a vesicle trafficking inhibitor that inhibits the function of ADP‐ribosylation factor GTPases (ARF GTPases) by interacting with their associated guanine nucleotide exchange factors (GEFs) and thereby results in membranous aggregates known as BFA compartments (Geldner et al., 2001; Huang et al., 2014; Ditengou et al., 2018). In a study by Tanaka et al. (2014), wild‐type roots were resistant to lower concentrations of BFA (1 μM), but higher BFA concentrations inhibited primary root growth (Tanaka et al., 2014). Thus, we treated wild‐type and atnsf‐1 plants with a low BFA concentration (1 μM BFA in 1/2 MS medium). In the wild‐type, the primary root length was not inhibited and was even slightly enhanced by BFA treatment (Figure 3A–C), which is in line with the previous report (Tanaka et al., 2014). However, this concentration of BFA significantly inhibited root length in atnsf‐1, especially at longer treatments (5 and 6 d Figure 3A–C). For 6 d‐old atnsf‐1 seedlings, root length was only 48.77% ± 8.14% (n ≥ 25) (Figure 3B) that of the wild‐type roots.
The remarkable root growth inhibition phenotype of atnsf‐1 prompted us to examine its root apical meristem architecture. As expected, meristem cell number and cell length were increased in the wild‐type following 1 μM BFA treatment (Figure 3D–G), suggesting that wild‐type seedlings are resistant to low concentrations of BFA. In contrast, both measured indices were significant decreased in atnsf‐1 (Figure 3D–G), demonstrating sensitivity to even low concentrations of BFA. Collectively, the results suggest that AtNSF also affects root growth, especially in the presence of the vesicle inhibitor BFA.
Next, we examined the effect of AtNSF in the auxin‐mediated root gravitropism response, because this process involves PIN2 trafficking. Quantitative evaluation of root growth following gravistimulation revealed an attenuated gravitropic response in atnsf‐1, both in the medium with or without low BFA concentration (1 μM) (Figure 3H–J). Notably, following gravity stimulation for 6 h, wild‐type roots showed a clear hook, whereas the atnsf‐1 roots showed a less‐pronounced response, especially in the presence of BFA (Figure 3H). This observation suggests that AtNSF also functions in the gravitropism response.
Auxin‐induced lateral root formation is another well‐studied process in root development (Olatunji et al., 2017; Du and Scheres, 2018). To analyze whether AtNSF is involved in lateral root development, mutant and wild‐type seedlings were grown in 1/2 MS medium with or without 1 μM BFA for 14 d and the number of emerged lateral roots was counted. The lateral root number increased in both plants with BFA treatment, but atnsf‐1 showed a hypersensitive response compared to the wild‐type (Figure S4). Taken together, our data indicate that AtNSF is involved not only in the primary root growth and gravitropic response, but also in lateral root formation.
AtNSF expression pattern
We examined the expression profile of AtNSF via quantitative real‐time polymerase chain reaction (qRT‐PCR). AtNSF was expressed in almost all tissues, at higher levels in the shoot, leaf, and flower (Figure S5A). The expression pattern was further analyzed in transgenic plants expressing β‐glucuronidase (GUS) driven by the AtNSF promoter (AtNSF pro ::GUS) in the wild‐type background for GUS staining. Strong GUS signal was observed in the vascular tissue in the cotyledons (C1), and in the vascular tissue and tip of the first true leaf (L3), which are appendages that lack margin serration (Figure 4A). From the second true leaf onwards (L5–7), GUS expression was restricted to the leaf tip and tips of the teeth, and weak signal was also present in the vascular tissue (Figure 4A). We further evaluated AtNSF expression during development of the seventh leaf and observed a restricted domain of GUS signal that accumulated in the tips of teeth during leaf serration formation (Figure S5B). This expression pattern further implicates an important role for AtNSF in the regulation of leaf serration.
Figure 4.
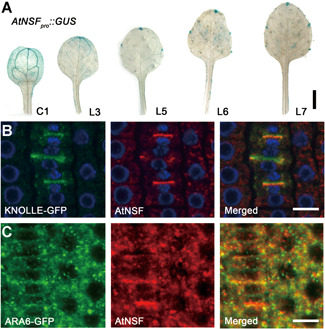
AtNSF expression pattern and AtNSF localization
(A) AtNSF tissue‐specific expression. Histochemical staining of GUS activity in transgenic plant leaves expressing AtNSF pro ::GUS. GUS activity was detected in the vascular tissue in the cotyledons (C1) and young leaves (L3) and was gradually restricted to the leaf tip in mature leaves (L5–L7). L3–L7, third to seventh leaf. Bar = 0.5 cm. (B, C) Immunolocalization of AtNSF (red) using transgenic plants expressing KNOLLE‐GFP (B) and ARA6‐GFP (C). The blue color in (B) represents the DAPI staining of DNA. Bars = 10 µm.
To determine the subcellular localization of AtNSF, we performed immunolocalization using transgenic plants expressing KNOLLE‐GFP, which is a cytokinesis marker that is specifically expressed in the M‐phase of the cell cycle, and ARA6‐GFP, a late endosome marker (Furutani et al., 2007; Touihri et al., 2011). AtNSF co‐localized or partially co‐localized with KNOLLE‐GFP and ARA6‐GFP (Figure 4B, C). This suggests that AtNSF is localized to the endosome and cell plate and may mediate membrane fusion between different organelles.
A defective Golgi phenotype was previously observed in an AtNSF mutant (Tanabashi et al., 2018). To determine the Golgi structure in atnsf‐1, we observed the cellular ultrastructure by transmission electron microscopy (TEM). Wild‐type cells contained the typical six stacked cisternae, but the Golgi shape was malformed and smaller in the atnsf‐1 mutant (Figure S6). This is consistent with the previous report (Tanabashi et al., 2018), indicating that AtNSF indeed plays a role in the structure of the Golgi apparatus, and is involved in vesicle trafficking from the Golgi to other endosomes and/or the PM.
AtNSF regulates multiple vesicle trafficking pathways
To investigate the function of AtNSF in membrane trafficking in plants, we evaluated the expression of the secreted green fluorescent protein (secGFP; a GFP fusion that is transported from the ER to the extracellular space) (Leucci et al., 2007) in the atnsf‐1 background. We also monitored intracellular accumulation of the endocytic tracer Fei Mao 4‐64 (FM4‐64) to investigate the internalization of vesicles in the secGFP genetic background of the wild‐type and atnsf‐1. We did not observe co‐localization of the FM dye with the secGFP. This is not a complete surprise since FM4‐64 is an endocytosis marker and secGFP in trafficked along the secretory pathway. One could suspect that the signal of those two markers could overlap but analysis did not visualize that. This might be explained by the fact that secGFP is secreted into the apoplast and it might be to some extent quenched by the acidic pH of the extracellular space. Notably however, the secGFP agglomerations were more numerous and larger in atnsf‐1 than those in the wild‐type (Figure 5A–C), suggesting that the secretion pathway was defective in the mutant. Moreover, more secGFP agglomerations were present at the cell periphery in atnsf‐1 (Figure 5A, B), indicating less‐efficient secretion. To investigate vesicle trafficking further, we performed FM4‐64 staining in the presence of the vesicle trafficking inhibitor BFA. Following treatment with 4 μM FM4‐64 for 5 min, intracellular accumulation was clearly observed in atnsf‐1 but was hardly present in the wild‐type >(Figure 5D, G, J). When seedlings were pre‐treated with FM4‐64 for 3 min, then immersed in water for 5 min followed by treatment with 50 μM BFA for 3 h, considerable FM4‐64 internalization was observed in BFA bodies in the wild‐type but this was even more severe in the atnsf‐1 mutant (Figure 5E, H, J). This observation established that AtNSF is required for endocytosis. In addition, when seedlings were washed out for 2 h, the internalized FM4‐64 in wild‐type was almost fully relocated to the PM, while it still remained in BFA bodies in the mutant (Figure 5F, I, J). These results indicate that vesicle recycling between the endosome and the PM is affected in the atnsf‐1 mutant.
Figure 5.
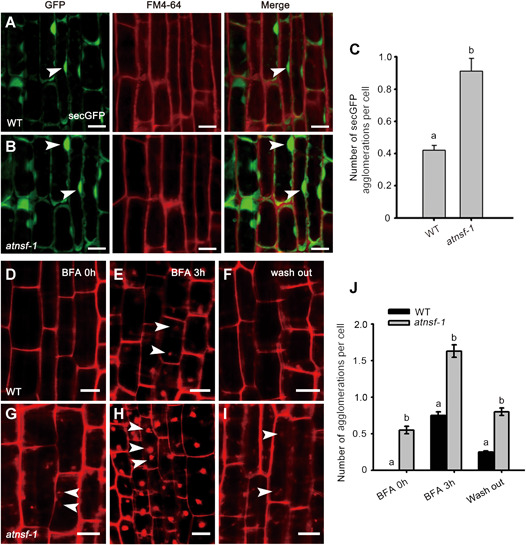
AtNSF regulates membrane trafficking
(A, B) Expression of the secretion marker secreted green fluorescent protein (secGFP) in wild‐type (WT) and atnsf‐1 seedlings. Seedlings were grown on half‐strength Murashige‐Skoog (1/2 MS) medium for 7 d and incubated in 4 µM FM4‐64 for 3 min before observation with confocal microscopy. Arrowheads indicate secGFP internalized in root cells. Bars = 10 µm. (C) Statistics for the number of secGFP agglomerations in WT and atnsf‐1 seedlings. Error bars represent SD (n ≥ 62). Different letters indicate statistically significant differences (P < 0.05, t‐test). (D–I) Recycling of FM4‐64 in root stele cells of WT (D–F) and atnsf‐1 (G–I) seedlings. Seven‐d‐old seedlings were treated with 4 μM FM4‐64 and immersed in water for 5 min (D, G) followed by treatment with 50 μM BFA for 3 h (E, H), and washing in water for 2 h (F, I). White arrowheads represent agglomerated BFA bodies. Bars = 10 µm. (J) Statistical analysis of the number of agglomerations in root stele cells in WT and atnsf‐1 seedlings with or without BFA treatment and 2 h of washing. The data represent means ± SD. Different letters indicate statistically significant differences (P < 0.05, t‐test, n ≥ 47).
Furthermore, we stained wild‐type and atnsf‐1 seedlings with FM4‐64 to observe short‐term endocytosis (30 min). In the first 5 min, hardly internalized FM4‐64 agglomeration was observed in wild‐type seedlings, whereas some aggregates were observed in the mutant (Figure S7A, D), indicating that endocytosis was accelerated in atnsf‐1 mutant. In the next 15 min, internalization gradually appeared in wild‐type roots, but was much more severe in the mutant (Figure S7B, E, G). This accelerated internalization became even more pronounced after 30 min (Figure S7C, F, G). These data show that endocytosis was strongly accelerated in atnsf‐1 and that AtNSF might regulate multiple vesicle trafficking pathways in Arabidopsis.
The atnsf‐1 mutant is defective in PIN1 protein cycling
Dynamic PIN1 cycling occurs between the endosome and the PM (Geldner et al., 2001; Tanaka et al., 2013), and PM‐localized PIN1 is rapidly internalized in response to BFA treatment (Geldner et al., 2001). To evaluate PIN1 distribution in atnsf‐1, a PIN1‐GFP fusion construct driven by the PIN1 promoter (PIN1 pro ::PIN1‐GFP) was introduced into wild‐type and the mutant by crossing, and roots expressing PIN1‐GFP were treated with the protein synthesis inhibitor cycloheximide (CHX) for 1 h followed by co‐treatment with CHX and BFA for 3 h. Before BFA treatment, hardly any intracellular accumulation of PIN1‐GFP was present in stele cells in the wild‐type but was more pronounced in atnsf‐1, indicating that PIN1 trafficking was defective in the mutant (Figure 6A, D, G). Upon treatment with 50 μM BFA, more PIN1‐GFP BFA agglomerations were observed in the mutant than in the wild‐type (Figure 6B, E, G). Even after washout for 2 h, PIN1 internalization was still higher in atnsf‐1 than in the wild‐type (Figure 6C, F, G), which was similar to the results for FM4‐64 staining after BFA treatment (Figure 5D–J). These results demonstrate that knock‐down of AtNSF disrupts PIN1 trafficking, which may in turn, disrupt auxin‐related development.
Figure 6.
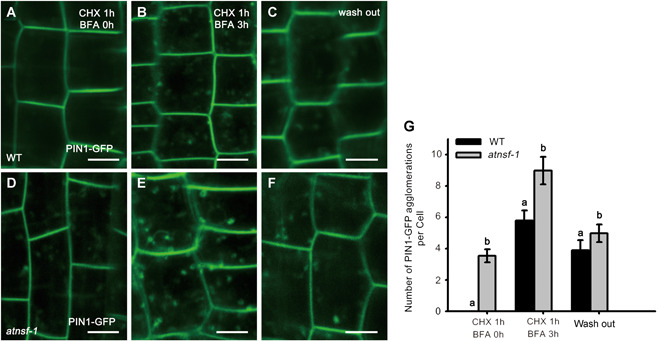
AtNSF regulates protein recycling of the PM‐localized auxin efflux carrier PIN1
(A–F) Recycling of PIN1‐GFP in wild‐type (WT) (A–C) and atnsf‐1 (D–F) root cells after CHX and BFA treatment. Seedlings were pre‐treated with 50 μM CHX for 1 h (A, D) and then treated with 50 μM CHX and 50 μM BFA for 3 h (B, E), and were then washed in water for 2 h (C, F). Bars = 10 µm. (G) Statistics for the number of PIN1‐GFP agglomerations in WT and atnsf‐1 roots before and after BFA treatment. The data represent means ± SD. Different letters indicate statistically significant differences (P < 0.05, t‐test, n ≥ 36).
In addition to analyzing PIN1 localization, we used the anti‐PIN2 antibody conjugated with the fluorescent dye Alexa Fluor 555 to observe the polar distribution of PIN2 in wild‐type and atnsf‐1 plants. PIN2 was localized to the apical membrane but was barely detected in the cytosol in wild‐type roots, whereas in atnsf‐1, PIN2 was internalized in the cytoplasm (Figure S8). However, the polar localization of PIN2 in the apical membrane of epidermal cells was not affected in atnsf‐1, indicating that AtNSF is involved in PIN2 trafficking. Together, these observations show that AtNSF is not PIN1 specific and also facilitates PIN1 and PIN2 cycling, which may affect auxin‐related developmental processes.
PIN1‐mediated polar auxin transport is required for the AtNSF regulation of leaf serration
Auxin is essential for organogenesis and auxin maxima are usually correlated with organ formation. In the leaf margin, auxin response maxima are usually restricted to the tips of initiating and growing teeth and are crucial for serration formation (Bilsborough et al., 2011). To determine the distribution of auxin response maxima in atnsf‐1 leaves, we monitored expression of the auxin‐responsive effector DR5 fused with GFP (DR5 rev ::GFP) in the atnsf‐1 and wild‐type background. In developing leaves, GFP signal was located in the leaf tips and tips of the teeth in both genotypes (Figure 7A–D), but in atnsf‐1, additional DR5 rev ::GFP maxima were observed between two outgrown teeth (Figure 7B, D, arrowheads). Therefore, more serrations were marked by DR5 rev ::GFP signal in leaf tips in atnsf‐1 than in wild‐type (Figure 7E), which is consistent with the enhanced leaf serration in the mutant.
Figure 7.
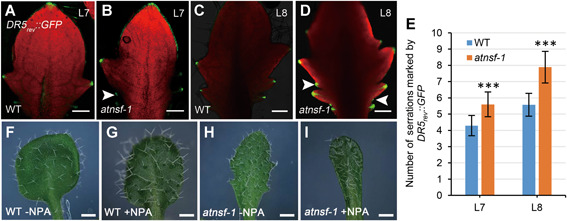
Auxin response is altered in atnsf‐1
(A, B) DR5 rev ::GFP expression in the seventh leaf (L7) in WT and atnsf‐1 plants. Bars = 200 µm. (C, D) DR5 rev ::GFP expression in the eighth leaf (L8) of WT and atnsf‐1 plants. Arrowheads in (B) and (D) indicate the restriction of green fluorescent protein (GFP) signal to the tip of secondary teeth formed between two outgrown teeth. Bars = 200 µm. (E) The number of serrations marked by DR5 rev ::GFP expression in the seventh leaf (L7) and the eight leaf (L8). Error bars represent SD (n ≥ 25). Asterisks indicate significant differences compared with the WT (P < 0.001, t‐test). (F–I) The serration phenotype of the seventh leaf of WT (F, G) and atnsf‐1 (H, I) plants with 1 μM NPA (G, I) or without NPA (F, H) treatment. Bars = 0.3 cm.
Polar auxin transport (PAT) mediated by PIN and P‐glycoprotein (PGP) is important for the formation of auxin concentration gradients (Cho et al., 2007). To test whether the atnsf‐1 leaf phenotype is linked to PAT, we treated wild‐type and atnsf‐1 seedlings with NPA, a PAT inhibitor. This treatment led to smooth leaf margins in the wild‐type, as was also reported previously (Mattsson et al., 2003), and serration was inhibited in the mutant in a manner similar to that in the treated wild‐type leaves (Figure 7F–I), suggesting that AtNSF regulates leaf serration in a PAT‐dependent manner.
Auxin influx proteins and efflux carrier PIN proteins facilitate PAT, and PIN1‐mediated auxin maxima is instructive in leaf serration (Bilsborough et al., 2011). Considering the auxin response in the superfluous teeth margin regions (Figure 7A–E) and the disturbed cycling of PIN1 (Figure 6) in atnsf‐1, we analyzed PIN1‐GFP distribution in the seventh leaf of wild‐type and mutant plants. Confocal imaging indicated a polar PIN1‐GFP distribution in the PM and a convergence point in the regions for outgrowth initiation in the wild‐type leaf (Figure 8A, B), whereas in the mutant, the polar localization of PIN1 was disturbed and showed clumped agglomerations in margin cells (Figure 8C–E), which resembled those in root stele cells. Therefore, AtNSF is required for PIN1 cycling, which generates PIN1 convergence points that are necessary for localized auxin maxima and for serration outgrowth.
Figure 8.
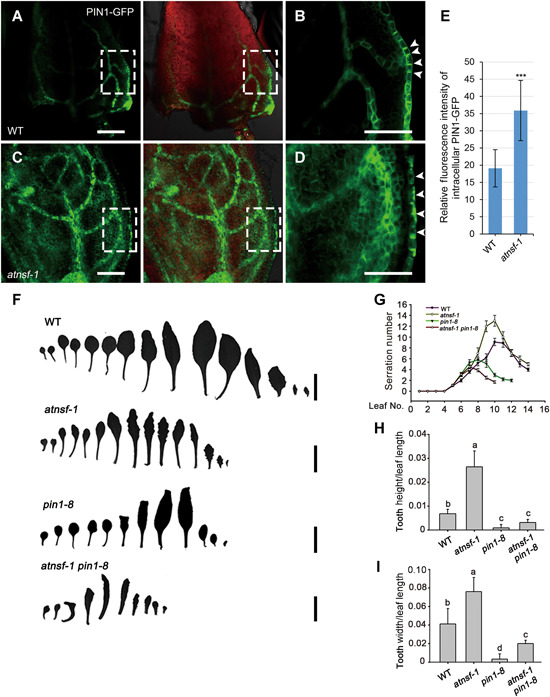
PIN1‐mediated auxin polar transport is required for the AtNSF‐mediated regulation of leaf serration
(A–D) Confocal micrographs of PIN1‐GFP expression in the margins of the seventh leaf in WT (A, B) and atnsf‐1 (C, D) plants. (B) and (D) represent the magnification of the areas framed in (A) and (C), respectively. The arrowheads in (B) and (D) indicate the impaired PIN1‐GFP localization in the leaf margin. Bars = 50 µm. (E) Relative fluorescence intensity of intracellular PIN1‐GFP in leaf margin cells. Error bars represent the SD (n ≥ 20). Asterisks indicate significant differences compared with the WT (P < 0.001, t‐test). (F) of a series of rosette leaves in WT, atnsf‐1, pin1‐8, and atnsf‐1 pin1‐8 plants. Bars = 1 cm. (G) Statistics for the number of serrations at various stages of leaf development in WT, atnsf‐1, pin1‐8, and atnsf‐1 pin1‐8 plants. Error bars represent means ± SD (n ≥ 20). (H, I) Relative tooth height (tooth height/leaf length) (H) and width (tooth width/leaf length) (I) of secondary teeth in the seventh leaves. Error bars represent SD. Different letters indicate statistically significant differences (P < 0.05, t‐test, n = 30).
To further determine the genetic relationship between AtNSF and PIN1, we first identified a new pin1 mutant (GABI_051A10) from GABI‐Kat with a T‐DNA insertion in PIN1. This mutant, named pin1‐8, produces an inflorescence meristem that does not initiate flowers, similar to that of other pin1 mutants. We identified pin1‐8 homozygous plants by PCR using three different primers and generated the atnsf‐1 pin1‐8 double mutant by crossing, and evaluated its phenotype. First, we counted the number of teeth in different leaves. In contrast to atnsf‐1, which has more teeth than the wild‐type, the atnsf‐1 pin1‐8 double mutant had fewer teeth than the wild‐type and a similar number to pin1‐8 (Figure 8F, G). We then measured the height and width of the teeth. In contrast to atnsf‐1, which had larger teeth than the wild‐type, the atnsf‐1 pin1‐8 mutant had smaller teeth, similar to those in pin1‐8 (Figure 8H, I). These results suggest that PIN1‐mediated polar auxin transport functions downstream of AtNSF and is required for AtNSF regulation of leaf serration.
CUC2 is required for the regulation of leaf serration by AtNSF
The growth repressor CUC2 also plays an essential role in regulating the extent of leaf serration and is regulated by the expression of MIR164A (Nikovics et al., 2006). CUC2 integrates into PIN1‐mediated auxin flux to operate a feedback loop that defines regions of growth promotion and retardation (Bilsborough et al., 2011). To determine the regulatory relationship between AtNSF and CUC2 during serration development, we first evaluated CUC2 expression in atnsf‐1 by qRT‐PCR. In atnsf‐1 leaves, CUC2 expression was significantly higher than that in wild‐type leaves, whereas it was lower than that in wild‐type in the atnsf‐1 complementation line that overexpressed AtNSF (Figure 9A). Consistent with this result, histochemical staining of GUS in atnsf‐1 that expressed GUS driven by the CUC2 promoter (CUC2 pro ::GUS) indicated areas of increased CUC2 expression in leaves (Figure 9B, C). Elevated CUC2 levels were also observed with the CUC2‐VENUS reporter marker in atnsf‐1 (Figure S9). These results indicate that AtNSF inhibits the expression of CUC2 in leaves.
Figure 9.
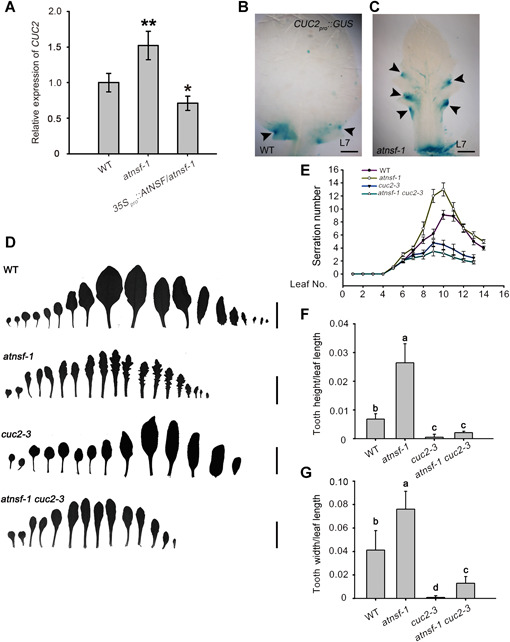
The regulation of leaf serration by AtNSF requires CUC2
(A) Relative transcript level of CUC2 determined by quantitative real‐time polymerase chain reaction (qRT‐PCR). Wild‐type, atnsf‐1, and AtNSF‐overexpressing atnsf‐1 plants were grown on half‐strength Murashige‐Skoog (1/2 MS) medium for 14 d and true leaves were collected for gene expression analysis. Error bars represent means ± SD. Asterisks indicate significant differences compared with the wild‐type (WT) (*P < 0.05; **P < 0.01, t‐test). (B, C) GUS expression in the margins of the seventh rosette leaf of WT (B) and atnsf‐1 (C) plants expressing CUC2 pro ::GUS. Arrowheads indicate restriction of GUS expression in the sinus. Bars = 200 µm. (D) Silhouettes of a series of rosette leaves showing the serration phenotype of WT, atnsf‐1, cuc2‐3, and atnsf‐1 cuc2‐3 plants. Bars = 1 cm. (E) Summary of leaf tooth number at various leaf developmental stages of WT, atnsf‐1, cuc2‐3, and atnsf‐1 cuc2‐3 plants. The data represent means ± SD (n ≥ 30). (F, G) Relative tooth height (tooth height/leaf length) (F) and width (tooth width/leaf length) (G) of secondary teeth in the seventh leaf in WT, atnsf‐1, cuc2‐3, and atnsf‐1 cuc2‐3 plants. Error bars represent SD. Different letters indicate statistically significant differences (P < 0.05, t‐test, n ≥ 20).
We further investigated the genetic relationship between AtNSF and CUC2 by crossing the single mutants atnsf‐1 and cuc2‐3. In contrast to atnsf‐1, which had more teeth than wild‐type, the atnsf‐1 cuc2‐3 double mutant had fewer teeth, similar to the number in the cuc2‐3 single mutant (Figure 9D, E). Moreover, in contrast to atnsf‐1, which had larger teeth than the wild‐type, the atnsf‐1 cuc2‐3 mutant had smaller teeth than wild‐type, similar to those in cuc2‐3 (Figure 9F, G) (Hasson et al., 2011). These results indicate that the leaf serration phenotype of atnsf‐1 was suppressed when CUC2 function is abolished. Taken together, these results show that CUC2 is required for AtNSF‐mediated regulation of leaf serration and they both regulate leaf serration in the same pathway, in which AtNSF functions upstream of CUC2 and inhibits its expression.
DISCUSSION
The NSF protein is a member of the AAA+ATPase family and plays a key function in intracellular membrane fusion events (Hanson and Whiteheart, 2005; Baker and Hughson, 2016). Although NSF has been well studied in mammals and yeast, the physiological function of NSF in plants remains unclear. In this study, we characterized the role of AtNSF in leaf serration formation and root development in Arabidopsis using a reverse genetic approach.
The function of AtNSF is conserved among eukaryotes
In Arabidopsis, we identified a single AtNSF gene that encodes a highly conserved NSF protein, and analyzed the characteristics and physiological function of AtNSF. AtNSF contains conserved AAA+ATPase domains and ATPase activity (Figure 1A, B) and rescued the yeast NSF‐deletion mutant (Figure 1C). In yeast, NSF plays a role in membrane trafficking. The sec18‐1 mutant displays defects in ER morphology and vesicle fusion at restrictive temperature (Mittenbühler and Holzer, 1991). The mammalian NSF regulates vesicle transport and the E329Q mutant has defects in Golgi function and causes cell death (Dalal et al., 2004). Furthermore, the zebrafish NSF‐defective mutants show intracellular membrane aggregation (Kurrasch et al., 2009). Consistent with the functions of these NSF orthologs, AtNSF also plays an essential role in membrane trafficking. The AtNSF protein is localized to the Golgi apparatus, endosome, and cell plate, and its mutation caused pleiotropic Golgi defects (Figures 4B, C, S6; Tanabashi et al., 2018), suggesting it is critically involved in vesicle trafficking between endomembranes. Defects in Golgi morphology in atnsf‐1 were also revealed by TEM images (Figure S6). Consistent with this, the knock‐down mutant atnsf‐1 showed impaired endocytosis and exocytosis (Figures 5A–C, S7) and disrupted PIN1 cycling and polar localization (Figures 6, 8A–E). Another PIN protein, PIN2, was also internalized into the intracellular compartment in normal conditions (1/2 MS medium) when AtNSF was knocked down (Figure S8). This demonstrates that the trafficking defect in atnsf‐1 is not PIN1 specific and may result from the general defects in the Golgi apparatus observed in atnsf‐1, which may affect other endocytic cargoes, such as PM ATPase. Collectively, these data confirm the conserved role of NSF in the Golgi apparatus and vesicle trafficking. We attempted to identify additional AtNSF knock‐down or knock‐out alleles; however, we failed to isolate a homozygous atnsf‐3 T‐DNA line and, similar to in mammals, the abolition of AtNSF is potentially embryo lethal in Arabidopsis. Therefore, the effect of T‐DNA insertions upstream of the AtNSF ATG start codon on AtNSF expression remains inconclusive. Taken together, our results show that AtNSF is a highly conserved plant NSF protein that functions in membrane trafficking and is critical for plant development.
AtNSF regulates polar auxin transport and PIN1 protein trafficking
The PIN1 auxin efflux carrier exhibits polar PM localization and cycles between the endosome and the PM. PIN proteins are internalized via clathrin‐mediated endocytosis, and the establishment of PIN1 basal polarity by the GNL1/GNOM‐mediated early secretory pathway is essential for plant development (Geldner et al., 2003; Doyle et al., 2015). However, the molecular mechanism that underlies PIN1 trafficking remains unknown. Arabidopsis BEX1/ARF1A1C, a small GTP‐binding protein of the ARF family, is important for PIN1 recruitment to the PM. Treatment of the bex1 mutant with a low concentration of BFA resulted in PIN1‐GFP aggregation in intracellular compartments and reduced its PM localization (Tanaka et al., 2014). By contrast, in the wild‐type, PIN1‐GFP mainly localized to the PM. The BEX5/RabA1b protein, a member of the large RabA GTPase class, regulates PIN1 trafficking from a trans‐Golgi network/early endosome (TGN/EE) compartment to the PM (Feraru et al., 2012). The bex5 mutant exhibits altered PIN1‐GFP endocytic recycling, due to impaired exocytosis and transcytosis (Feraru et al., 2012). By contrast, BEN1 encodes an ARF GEF vesicle trafficking regulator from the functionally uncharacterized BIG class. The ben1 mutant showed diminished internalization and agglomeration of PIN1‐GFP after BFA treatment (Tanaka et al., 2009). These data suggest that PIN1 trafficking is mediated via the conserved membrane secretion system in which SNAREs and related proteins play an essential role.
The function of SNAREs depends on the NSF‐mediated disassembly of SNARE complex subunits (Kim and Brandizzi, 2012; Baker and Hughson, 2016). Therefore, we queried whether PIN1 trafficking is mediated by an AtNSF‐dependent pathway. The phenotype of atnsf‐1 resembled an auxin‐defective phenotype and included more sites of DR5‐GFP maxima and increased leaf serration (Figures 2B–F, 7A–E), indicating that AtNSF regulates the distribution of auxin response maxima. Furthermore, atnsf‐1 exhibited defective PIN1 trafficking from endosomes to the PM, which further altered polar PIN localization in the leaf margin (Figures 6, 8A–E), suggesting that AtNSF affects auxin polar transport by regulating intracellular PIN1 trafficking. Treatment with the PAT inhibitor NPA disrupted the polar localization of PIN1 and led to wild‐type leaves with a smooth margin (Figure 7F, G). However, trafficking of PIN1 in atnsf‐1 was defective, but the expression of other regulators such as CUC2 was elevated (Figure 9A). The mechanism that regulates leaf serration is complex and the importance of the role of AtNSF is for PIN1 localization and polarity in leaf development remains to be elucidated. This could be performed by expressing AtNSF from the PIN1 promoter. Besides, PIN2 also accumulated in the intracellular compartment in normal conditions in atnsf‐1 (Figure S8), demonstrating that AtNSF may also be involved in the trafficking of other PIN proteins and further affect auxin‐related phenotypes. An appropriate root gravitropic response involves correct PIN2 polarity. Although the mechanism of this response is complex, it has been reported to involve endocytosis and exocytosis and the ubiquitination‐dependent degradation of PIN2 in the PM (Abas et al., 2006; Kleine‐Vehn et al., 2011). The defects in PIN2 trafficking may contribute to delayed bending in atnsf‐1 (Figure 3H, I). However, this phenotype might also be influenced by other processes; therefore, it remains to be determined which trafficking processes or which pathways contribute most to the bending defect in response to gravity in atnsf‐1. In primary root development, the distal auxin response maximum regulates pattern formation and the initiation of lateral root primordia is also associated with changes in auxin transport (Blilou et al., 2005; Olatunji et al., 2017; Du and Scheres, 2018). Here, we observed a decreased meristem length and supernumerary lateral roots in atnsf‐1 (Figures 3A–G, S4). Further studies to demonstrate that AtNSF regulates PIN1‐mediated auxin transport for meristem maintenance and lateral root development need to be considered.
The hypersensitivity of atnsf‐1 to BFA observed here resembles the previously reported responses of bex1 and bex5 (Feraru et al., 2012; Tanaka et al., 2014). BEX1/ARF1A1C and BEX5/RabA1b localize to the trans‐Golgi network/early endosome and function in BFA‐sensitive PIN trafficking. Further investigation into the relationships between AtNSF, BEX1, and BEX5 might reveal the mechanism of trafficking from the Golgi network to the PM or endosomes. Therefore, whether BEX1 and BEX5 are involved in leaf serration or function together with AtNSF in this process needs to be elucidated further. Collectively, the data here reveal that PIN1 trafficking is regulated by the conserved membrane secretion machinery via an AtNSF‐dependent pathway.
AtNSF interferes with the balance of the PIN1 and CUC2‐mediated feedback loop for leaf serration
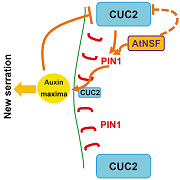
Leaf serration involves the CUC2 transcription factor, whose activity is negatively regulated by microRNA164A (Nikovics et al., 2006), and the cuc2‐3 mutant has smooth leaf margins (Nikovics et al., 2006). Our findings indicate that CUC2 expression was up‐regulated in the atnsf‐1 mutant, and mutation of CUC2 completely suppressed the atnsf‐1 phenotype (Figure 9D–G). These results demonstrate that AtNSF regulates leaf serration through the CUC2 pathway. Leaf margin development is regulated by CUC2 and PIN1, which form a feedback loop (Bilsborough et al., 2011). The first loop regulates auxin transport by the PIN1 efflux transporter. In a second loop, CUC2 promotes the generation of PIN1‐mediated auxin maxima while auxin represses CUC2 expression. The balanced activity of the feedback loop establishes stable serration patterns in the leaf margin (Figure 10C; Bilsborough et al., 2011). The data here improve our understanding of this regulatory loop by elucidating the role of AtNSF. First, AtNSF directly influences local auxin concentrations by controlling PIN1 vesicle trafficking. Knock‐down of AtNSF disrupts the polar localization of PIN1 in leaf margins and disperses or internalizes PIN1 in more leaf margin cells (Figure 10A–C). Ultimately, more auxin maxima are generated in the leaf margin, which lead to increased serrations. Second, AtNSF knock‐down increases CUC2 expression, which alters the balance of the PIN1/CUC2 feedback loop (Figure 10C). We hypothesize that AtNSF regulates CUC2 expression in two potential ways: AtNSF either indirectly suppresses CUC2 expression by PIN1‐mediated auxin distribution, or AtNSF represses CUC2 expression in a PIN1‐independent manner, and these hypotheses need to be addressed in future studies (Figure 10C). In conclusion, AtNSF is required for the PIN1/CUC2 feedback loop in the regulation of leaf serration. The ligand–receptor pair, EPFL2 and ERECTA, promote leaf tooth growth by forming a feedback loop between the peptide‐receptor system and auxin response (Tameshige et al., 2016). Thus, a potential relationship between the AtNSF pathway and the EPFL2‐ERECTA machinery is intriguing. Whether the secreted peptide EPFL2 is regulated by the AtNSF membrane secretion pathway is a topic that deserves further study.
Figure 10.
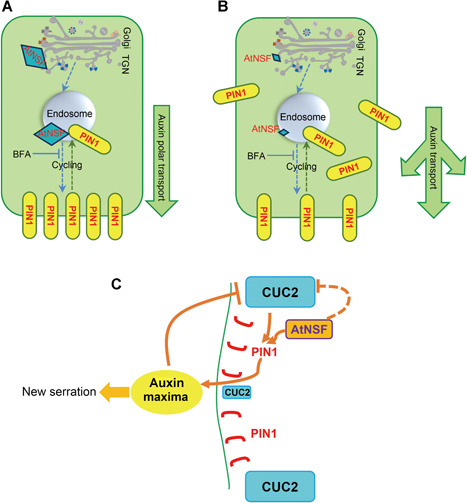
Hypothesis for the regulation of AtNSF‐mediated serration
(A) AtNSF‐mediated cycling of the auxin efflux carrier PIN1 in wild‐type (WT) cells. PIN1 is transported from the trans‐Golgi network (TGN) to endosomes and from endosomes to the PM in an AtNSF‐dependent manner, and ultimately localize to the basal membrane and establish the polar direction of auxin transport. Yellow boxes represent PIN1 protein and large blue diamonds represent AtNSF expression. The arrow to the right shows the downward flux of auxin. (B) AtNSF‐mediated PIN1 transport and cycling in atnsf‐1 mutant cells. When AtNSF is knocked‐down, the cycling of PIN1 between endosomes and the PM is disrupted, which leads to more PIN1‐dense vesicles in the intracellular compartment. Thus, the basal polar localization of PIN1 and further auxin transport is disturbed. Yellow boxes represent PIN1 protein and the small blue diamonds represent knocked‐down levels of AtNSF. The direction of the arrows shows the disturbed auxin flux. (C) An hypothesis for AtNSF‐, PIN1‐, and CUC2‐mediated leaf serration regulation based on Bilsborough et al. (2011). We have added the function of AtNSF in the regulation of the PIN1 and CUC2‐mediated feedback loop to this model. Serration development is based on the previously published computational model (Bilsborough et al., 2011). Auxin transport by PIN1 (polarly localized in red) leads to the formation of an auxin maximum (the yellow oval), which generates a new serration. CUC2 (blue boxes) drives this mechanism and promotes the establishment of PIN1 convergence. Auxin, in turn, inhibits CUC2 to stabilize the position of the auxin maximum (large and small blue boxes represent high and low expression of CUC2 in different zones, respectively). These interactions generate a pattern of auxin maxima interspersed with CUC2 expression along the leaf margin and a new serration is formed in the area of the auxin maxima. In this feedback loop, AtNSF (orange box) contributes to PIN1 cycling to establish PIN1 polar localization. AtNSF inhibits CUC2 expression to further regulate the balance between the loops. The solid lines in this schematic diagram represent direct regulation that has been reported or demonstrated in this study, the dotted line from AtNSF to CUC2 represents potential regulation that may involve other proteins and/or an indirect pathway that needs to be elucidated.
MATERIALS AND METHODS
Plant materials and growth conditions
Arabidopsis thaliana wild‐type ecotype Columbia‐0 (Col‐0) was used in this study. The T‐DNA insertion lines Salk_038536 (atnsf‐1), Salk_138721 (atnsf‐2), Sail_1155_C06 (atnsf‐3), and GABI_051A10 (pin1‐8) were obtained from the Nottingham Arabidopsis Stock Centre. Some plant materials used in this study have been previously described: DR5 rev ::GFP (Benková et al., 2003), PIN1 pro ::PIN1‐GFP (PIN1‐GFP) (Benková et al., 2003), CUC2 pro ::GUS (Nikovics et al., 2006), CUC2‐VENUS (Bilsborough et al., 2011), cuc2‐3 (Nikovics et al., 2006), ARA6‐GFP (Furutani et al., 2007), KNOLLE‐GFP (Touihri et al., 2011) and secGFP (Leucci et al., 2007).
Surface‐sterilized seeds were sown on plates containing 1/2 MS medium supplemented with 1% sucrose and 1.2% agar, and were stratified at 4°C in the dark for 2 d, and grown at 22°C in a 16 h:8 h (light:dark) cycle and a light intensity of approximately 100 µmol m−2 s−1. Seedlings were transferred to soil between 8 and 12 d after germination and were grown under the same conditions in a phytochamber.
AtNSF alignment analysis
The Arabidopsis thaliana (At4g04910), human (Homo sapiens, BAH12933.1), and yeast (Saccharomyces cerevisiae, AJP85518.1) NSF protein sequences were aligned using ClustalW as implemented in BioEdit. Sequence identity of 100% was highlighted in dark gray. Identity or similarity between 50% and 100% was highlighted in light gray. The conserved domains were marked with boxes as explained in Figure 1A.
Plasmid construction and plant transformation
The AtNSF coding sequence was amplified from clone RAFL09‐22‐D11 (R22030; RIKEN) with AtNSF‐CDS‐F and AtNSF‐CDS‐R primers (Table S1). The corresponding PCR fragment was recombined into the pDONR207 entry vector using attB × attP (BP) in a recombination reaction according to the manufacturer's instructions (Invitrogen). After sequence confirmation, the AtNSF coding sequence was transferred into the GATEWAY recombination sites of pMDC32 by attL × attR (LR) recombination reactions using LR clonase (Invitrogen), resulting in the 35S pro ::AtNSF construct. To construct the native promoter‐driven gene (the AtNSF pro ::AtNSF construct), the AtNSF promoter was amplified from the Arabidopsis genome with primers AtNSF‐P3 and AtNSF‐P4 (Table S1), and the 35S pro of the 35S pro ::AtNSF construct was replaced with AtNSF pro using PmeI and KpnI sites. To create the AtNSF pro ::GUS construct, the native promoter was amplified from Arabidopsis seedling genomic DNA with primers AtNSF‐P1 and AtNSF‐P2 (Table S1). The PCR product was recombined into the pDONR207 vector and transferred into gateway vector pMDC162 to generate the AtNSF pro ::GUS construct. The resulting binary vectors were introduced into Agrobacterium tumefaciens strain GV3101 and plants were transformed via the floral‐dip method.
Phenotypic analysis
Leaf clearing was performed by incubating leaves in excess 7:1 ethanol:acetic acid solution overnight at room temperature. Leaves were then transferred to chloral hydrate solution (chloral hydrate:water:glycerol = 8:3:1) and mounted on slides with clean solution before imaging with dark‐field microscopy.
For root length analysis, seedlings were scanned and measured using Image J software (https://fiji.sc). For Nomarski imaging of roots, seedlings were fixed and mounted in chloral hydrate solution (chloral hydrate:water:glycerol = 8:3:1) and images were captured by an AxioVision microscope (Carl Zeiss). The root apical meristem size was quantified by Image J, based on the region between the quiescent center and the first elongating cell in the cortex.
To measure root gravity responses, vertical plates on which seedlings had grown for 6 d were rotated by 90° and photographs were taken at 1, 2, 6, 8, 12 h after reorientation. The degree of root curvature was measured using Image J.
For TEM, wild‐type and atnsf‐1 seedlings were fixed with 2.5% glutaraldehyde and 1% osmium tetroxide for 12 h at 4°C, then washed five times by phosphate‐buffered saline (PBS; pH 7.2) and subjected to an ethanol gradient for dehydration. Seedlings were then transferred to propylene epoxide and gradually infiltrated with acrylic resin. Ultrathin sections of the samples were cut with a diamond knife and images were record by a transmission electron microscope (JEM‐1400Plus).
The T‐DNA insertions within AtNSF in Salk_038536, Salk_138721, and Sail_1155_C06 and in PIN1 of GABI_051A10 were validated by PCR‐based genotyping (http://signal.salk.edu/). The primers used are listed in Table S1.
Yeast complementation
The yeast mutant strain sec18‐1, NY1691 (MATα ura3‐52 leu2‐3, 112 trp1 his3Δ200 SEC1::SEC1MYC 3 URA3 sec18‐1) was obtained from Dr. Peter J. Novick. The open reading frame of AtNSF was cloned into the yeast expression vector pDR196. The resulting construct and empty vector were then transformed into the yeast NY1691 mutant strain. Two positive clones from each transformation procedure were selected for culture on yeast peptone dextrose medium containing 1% yeast extract, 2% Bacto peptone, and 2% glucose at 24°C for 72 h. The cultures were diluted in water and cultured on a plate at 33°C to assess the ability of the clone to grow.
Measurement of ATPase enzyme activity
The full‐length coding sequences of AtNSF and SNAPα were obtained by PCR amplification from the cDNA clones RAFL09‐22‐D11 and RALF19‐63‐F08 (RIKEN), using the primer pairs AtNSF‐F/AtNSF‐R and SNAP‐F/SNAP‐R, respectively. The primer sequences are shown in Table S1. The PCR product was digested with SacI and NotI and ligated into the protein expression vector pET28a (Novagen). AtNSF and SNAPα were expressed in E. coli BL21 and purified with Ni‐NTA resin (Thermo Fisher Scientific). ATPase activity was analyzed with the ATPase assay kit (Innova Biosciences) following the manufacturer's instructions.
Histochemical analysis
GUS staining was performed as previously described (Yu et al., 2013). An AxioVision microscope (Carl Zeiss imager A1) was used for high‐magnification imaging of stained samples and an AxioVision stereomicroscope (SteREO Discovery.V20) coupled to an AxioCam MRc digital camera (Carl Zeiss Microimaging) was used for low‐magnification imaging.
RNA isolation, RT‐PCR, and qRT‐PCR
Total RNA was extracted from different organs, including roots, leaves, shoots, flowers, and siliques, and from 10 d‐old wild‐type or atnsf‐1 true leaves using the RNeasy plant mini kit (Qiagen) and was digested with TURBO DNase (Ambion). The extracted RNA was reverse transcribed into cDNA using the High Capacity cDNA reverse‐transcription kit (Applied Biosystems). Reverse transcription‐PCR was used to analyze AtNSF expression in T‐DNA insertion mutants. TUBULIN (At5g62690) served as a control to confirm equal amounts of cDNA in each reaction. Quantitative RT‐PCR was performed with a SYBR green kit (Applied Biosystems) to detect AtNSF and CUC2 transcripts. Amplified samples were normalized against TUBULIN (At5g62690) transcript levels. All primer pairs used are listed in Table S1 online.
AtNSF antibody generation
To generate AtNSF‐specific polyclonal antibodies, the two primers Anti‐P1 and Anti‐P2 were used to clone the AtNSF cDNA region of 1,219–1,680 bp, which corresponds to amino acids 407–560, into the EcoRI/NotI sites of pET28a (Novagen). After expression in E. coli strain BL21 golden star, the recombinant protein containing the 6×His‐tag was affinity purified according to the manufacturers’ instructions (Qiagen) and evaluated by sodium dodecylsulfate‐polyacrylamide gel electrophoresis. The antigen peptide was used to immunize rabbits (Eurogentec). The polyclonal antiserum was affinity purified against the recombinant AtNSF peptide.
Immunolocalization
Plants were fixed with 4% paraformaldehyde in PBS (pH 7.3) and used for whole‐mount in situ immunolocalization in roots as previously described (Friml et al., 2002). AtNSF was detected using a rabbit anti‐AtNSF polyclonal antibody (1:100), and KNOLLE‐GFP and ARA6‐GFP were detected with a mouse anti‐GFP monoclonal antibody (Roche) (1:500). After washing out the primary antibodies, the samples were incubated with the secondary antibodies (Alexa Fluor 555 goat anti‐rabbit, and Alexa Fluor 488 goat anti‐mouse, respectively, from Invitrogen; all at a 1:500 dilution).
For PIN2 immunolocalization, PIN2 was detected using a guinea pig anti‐PIN2 antibody (1:500) (a gift from J. Friml), and the Alexa Fluor 555 goat anti‐guinea pig was used as the secondary antibody (1:500, Invitrogen).
Pharmacological treatment
The FM4‐64 treatment was performed as described previously (Feng et al., 2017). A 4 mM FM4‐64 stock solution was firstly prepared in dimethyl sulfoxide (DMSO). For the FM4‐64 internalization assay, 7‐d‐old seedlings were incubated with 4 μM FM4‐64 for 3 min and then rinsed twice with water for 5 min, 15 min, and 30 min for short‐term analysis.
For the BFA treatment to observe the trafficking in root cells, 7‐d‐old seedlings were immersed in 50 μM BFA and 50 μM CHX for 3 h (pretreated with 50 μM CHX for 1 h to block new protein synthesis) and BFA bodies were observed by confocal laser scanning microscopy (Zeiss LSM 880). To observe the FM4‐64‐stained cells, seedlings were first incubated with 4 μM FM4‐64 for 3 min and washed in water for 5 min, followed by 50 μM BFA for 3 h. In the washout experiments, the BFA‐treated seedlings were incubated in water for 2 h and the BFA agglomeration was imaged by confocal microscopy (Zeiss LSM 880) and the number of BFA bodies per cell was analyzed by statistically.
For the long‐term BFA treatment to analyze root growth, gravitropic bending, and the lateral root phenotype, seedlings were grown on 1/2 MS medium with or without 1 μM BFA for different numbers of days as indicated.
For the NPA treatment, wild‐type and atnsf‐1 seedlings were grown in 1/2 MS medium with or without 1 μM NPA for 10 d, and leaves were then imaged to observe the degree of margin serration.
Confocal microscopy
DAPI was excited with a 405 nm laser and collected using a 420–480 nm band‐pass filter. For GFP, VENUS, and Alexa Fluor 488, the 488 nm laser line was used for excitation, and emission was detected at 505–550 nm. For Alexa Fluor 555, we used a 543 nm laser excitation and 590–635 nm emission. Images were captured by a Zeiss LSM 880 confocal microscope. ZEN 2009 (Carl Zeiss Microimaging) was used to process and extract the images.
AUTHOR CONTRIBUTIONS
X.L. and K.P. designed the research and supervised the experiments; X.L. and Lixin L. wrote the manuscript; L.P.T., Y.Y., and Y.H.S. performed most of the experiments; H.W. and L. L. isolated the atnsf‐1 mutant. Y.L. and J. Y. performed immunolocalization and root‐phenotype analysis. L. L. and X.Y.Z. contributed reagents, materials, and analytical tools. All authors read and approved of the article.
Supporting information
Additional Supporting Information may be found online in the supporting information tab for this article: http://onlinelibrary.wiley.com/doi/10.1111/jipb.13043/suppinfo
Figure S1. Leaf serration phenotype of atnsf‐1 and anti‐NSF (N‐ethylmaleimide‐sensitive factor) antibody characterization
(A) Number of teeth (serrations) in growing leaves of wild‐type (WT) and atnsf‐1 plants. n ≥ 20. (B) Quantification of AtNSF expression in the WT and atnsf‐1. Total RNA was isolated from 10‐d‐old seedlings grown on half‐strength Murashige‐Skoog (1/2 MS) medium and analyzed by reverse transcription polymerase chain reaction (RT‐PCR) based on 25 cycles of amplification. TUBULIN was used as a control to show the equal amount of cDNA in each reaction. (C) The induction of prokaryotic expression of AtNSF in E. coli. AtNSF protein (81.62 kDa) was induced by 1 mM isopropylthio‐β‐galactoside (IPTG) for 6 h. Total protein from IPTG‐induced E. coli was subjected to sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) followed by Coomassie blue staining (left panel). Purified AtNSF protein from E. coli was subjected to immunoblotting with anti‐AtNSF antibody (right panel).
Figure S2. Leaf phenotype of the complementation line using the native AtNSF promoter
Four‐week‐old seedlings of the wild‐type, atnsf‐1, and the complementation line. Bar = 1 cm
Figure S3. Identification of the atnsf‐2 mutant
(A, B) Leaf phenotype of the wild‐type (WT) (A) and atnsf‐2 (B). Bars = 1 cm. (C) Genotyping of the atnsf‐2 mutant. (D) Expression level of AtNSF in WT and atnsf‐2 plants determined by reverse transcription polymerase chain reaction (RT‐PCR) using 5‐week‐old plants. TUBULIN was used as a control.
Figure S4. Lateral root phenotype of the atnsf‐1 mutant
(A) Root phenotype of wild‐type (WT) and atnsf‐1 plants grown in half‐strength Murashige‐Skoog (1/2 MS) medium with or without 1 μM BFA. (B) Statistical analysis of the lateral root number as in (A). Error bars represent the SD of biological replicates (n ≥ 20). Asterisks indicate significant differences compared with the WT (*P < 0.05; **P < 0.01; t‐test). Bar = 1 cm.
Figure S5. AtNSF is strongly expressed in leaf serrations
(A) Relative transcript levels of AtNSF in the root, rosette leaf, cauline leaf, shoot, flower, and silique determined by quantitative real‐time polymerase chain reaction (qRT‐PCR). The data represent means for three independent experiments per sample and three replicates per experiment. (B) The dynamic expression pattern of AtNSF pro::GUS in the seventh leaf throughout development. 1, the youngest leaf; 7, the oldest leaf.
Figure S6. The Golgi apparatus is defective in the atnsf‐1 mutant
Transmission electron microscopy images of the wild‐type (A) and the atnsf‐1 mutant (B, C). Images show the Golgi apparatus phenotype. Bars = 200 nm.
Figure S7. Short‐term internalization of FM4‐64 in wild‐type (WT) and atnsf‐1 plants
(A–F) Internalization of FM4‐64 in WT (A–C) and atnsf‐1 (D–F) plants. Images were taken at 5 min (A, D), 15 min (B, E), and 30 min (C, F) after FM4‐64 staining. Bars = 10 μm. (G) Statistical analysis of the number of agglomerations per cell in WT and atnsf‐1 plants for 5, 15, and 30 min. The data represent the means ± SD (n ≥ 10 cells). Asterisks indicate significant differences compared with the WT (***P < 0.001; t‐test).
Figure S8. Immunolocalization of PIN2 in wild‐type (WT) and atnsf‐1 epidermal cells
Seedlings were grown on half‐strength Murashige‐Skoog (1/2 MS) medium for 7 d and PIN2 localization in root epidermal cells was observed by an immunoassay. Arrows represent internalized PIN2 protein in the atnsf‐1 mutant. Bars = 5 μm
Figure S9. CUC2‐VENUS expression in atnsf‐1. CUC2‐VENUS expression in the seventh rosette leaf margins of wild‐type (WT) (A) and atnsf‐1 (B) plants. Arrowheads indicate the restriction of VENUS expression to the sinus. Bars = 200 µm.
Table S1. Primers used for quantitative real‐time polymerase chain reaction (qRT‐PCR) and expression constructs
ACKNOWLEDGEMENTS
We thank Dr. Peter J. Novick for providing the yeast sec18‐1 mutant strain NY1691 and Dr. Patrick Laufs for seeds. We also thank Dr. Renxiao Wang for his early work during the project. We thank Edwin Groot for helpful discussion about the work and critical reading of the manuscript. This work was supported by the National Natural Science Foundation of China (31570291, 31570246, 31872669); Funds of Shandong “Double Tops” Program (SYL2017YSTD03); Shandong “Foreign experts double hundred” Program (WST2017008); Taishan Scholar Project; DFG (SFB 746); Excellence Initiative of the German Federal and State Governments (EXC 294); Bundesministerium für Forschung und Technik (BMBF, MICROSYSTEMS); the Deutsches Zentrum für Luft und Raumfahrt (DLR 50WB1022), and Natural Science Foundation of Heilongjiang Province (C2016002).
Biographies


Tang, L.P., Yang, Y., Wang, H., Li, L., Liu, L., Liu, Y., Yuan, J., Zhao, X.Y., Palme, K., Su, Y.H., and Li, X. (2021). AtNSF regulates leaf serration by modulating intracellular trafficking of PIN1 in Arabidopsis thaliana . J. Integr. Plant Biol. 63: 737–754.
Edited by: Zhizhong Gong, China Agricultural University, China
Contributor Information
Ying Hua Su, Email: suyh@sdau.edu.cn.
Xugang Li, Email: xgli@sdau.edu.cn.
REFERENCES
- Abas, L. , Benjamins, R. , Malenica, N. , Paciorek, T. , Wiśniewska, J. , Moulinier‐Anzola, J.C. , Sieberer, T. , Friml, J. , and Luschnig, C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin‐efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8: 249–256. [DOI] [PubMed] [Google Scholar]
- Baker, R.W. , and Hughson, F.M. (2016). Chaperoning SNARE assembly and disassembly. Nat. Rev. Mol. Cell Biol. 17: 465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard, R.J. , Morgan, A. , and Burgoyne, R.D. (1997). Stimulation of NSF ATPase activity by alpha‐SNAP is required for SNARE complex disassembly and exocytosis. J. Cell Biol. 139: 875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless, A.M. , Zapotocny, R.W. , Grunwald, D.J. , Amundson, K.K. , Diers, B.W. , and Bent, A.F. (2018). An atypical N‐ethylmaleimide sensitive factor enables the viability of nematode‐resistant Rhg1 soybeans. Proc. Natl. Acad. Sci. U.S.A 115: E4512–E4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková, E. , Michniewicz, M. , Sauer, M. , Teichmann, T. , Seifertová, D. , Jürgens, G. , and Friml, J. (2003). Local, efflux‐dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Bilsborough, G.D. , Runions, A. , Barkoulas, M. , Jenkins, H.W. , Hasson, A. , Galinha, C. , Laufs, P. , Hay, A. , Prusinkiewicz, P. , and Tsiantis, M. (2011). Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. USA 108: 3424–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou, I. , Xu, J. , Wildwater, M. , Willemsen, V. , Paponov, I. , Friml, J. , Heidstra, R. , Aida, M. , Palme, K. , and Scheres, B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. [DOI] [PubMed] [Google Scholar]
- Carr, C.M. , Grote, E. , Munson, M. , Hughson, F.M. , and Novick, P.J. (1999). Sec1p binds to SNARE complexes and concentrates at sites of secretion. J. Cell Biol. 146: 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, M. , Lee, S.H. , and Cho, H.T. (2007). P‐glycoprotein4 displays auxin efflux transporter‐like action in Arabidopsis root hair cells and tobacco cells. Plant Cell 19: 3930–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal, S. , Rosser, M.F. , Cyr, D.M. , and Hanson, P.I. (2004). Distinct roles for the AAA ATPases NSF and p97 in the secretory pathway. Mol. Biol. Cell 15: 637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditengou, F.A. , Gomes, D. , Nziengui, H. , Kochersperger, P. , Lasok, H. , Medeiros, V. , Paponov, I.A. , Nagy, S.K. , Nádai, T.V. , Mészáros, T. , Barnabas, B. , Ditengou, B.I. , Rapp, K. , Qi, L. , Li, X. , Becker, C. , Li, C. , Doczi, R. , and Palme, K. (2018). Characterization of auxin transporter PIN6 plasma membrane targeting reveals a function for PIN6 in plant bolting. New Phytol. 217: 1610–1624. [DOI] [PubMed] [Google Scholar]
- Doyle, S.M. , Haeger, A. , Vain, T. , Rigal, A. , Viotti, C. , Łangowska, M. , Ma, Q. , Friml, J. , Raikhel, N.V. , Hicks, G.R. , et al. (2015). An early secretory pathway mediated by GNOM‐LIKE 1 and GNOM is essential for basal polarity establishment in Arabidopsis thaliana . Proc. Natl. Acad. Sci. USA 112: E806–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Y. , and Scheres, B. (2018). Lateral root formation and the multiple roles of auxin. J. Exp. Bot. 69: 155–167. [DOI] [PubMed] [Google Scholar]
- Feng, Q.N. , Song, S.J. , Yu, S.X. , Wang, J.G. , Li, S. , and Zhang, Y. (2017). Adaptor protein‐3‐dependent vacuolar trafficking involves a subpopulation of COPII and HOPS tethering proteins. Plant Physiol. 174: 1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraru, E. , Feraru, M.I. , Asaoka, R. , Paciorek, T. , De Rycke, R. , Tanaka, H. , Nakano, A. , and Friml, J. (2012). BEX5/RabA1b regulates trans‐Golgi network‐to‐plasma membrane protein trafficking in Arabidopsis . Plant Cell 24: 3074–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml, J. , Benková, E. , Blilou, I. , Wisniewska, J. , Hamann, T. , Ljung, K. , Woody, S. , Sandberg, G. , Scheres, B. , Jürgens, G. , et al. (2002). AtPIN4 mediates sink‐driven auxin gradients and root patterning in Arabidopsis . Cell 108: 661–673. [DOI] [PubMed] [Google Scholar]
- Furutani, M. , Kajiwara, T. , Kato, T. , Treml, B.S. , Stockum, C. , Torres‐Ruiz, R.A. , and Tasaka, M. (2007). The gene MACCHI‐BOU 4/ENHANCER OF PINOID encodes a NPH3‐like protein and reveals similarities between organogenesis and phototropism at the molecular level. Development 134: 3849–3859. [DOI] [PubMed] [Google Scholar]
- Geldner, N. , Anders, N. , Wolters, H. , Keicher, J. , Kornberger, W. , Muller, P. , Delbarre, A. , Ueda, T. , Nakano, A. , and Jürgens, G. (2003). The Arabidopsis GNOM ARF‐GEF mediates endosomal recycling, auxin transport, and auxin‐dependent plant growth. Cell 112: 219–230. [DOI] [PubMed] [Google Scholar]
- Geldner, N. , Friml, J. , Stierhof, Y.D. , Jürgens, G. , and Palme, K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428. [DOI] [PubMed] [Google Scholar]
- Hanson, P.I. , and Whiteheart, S.W. (2005). AAA+ proteins: Have engine, will work. Nat. Rev. Mol. Cell Biol. 6: 519–529. [DOI] [PubMed] [Google Scholar]
- Hasson, A. , Plessis, A. , Blein, T. , Adroher, B. , Grigg, S. , Tsiantis, M. , Boudaoud, A. , Damerval, C. , and Laufs, P. (2011). Evolution and diverse roles of the CUP‐SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell 23: 54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, A. , Barkoulas, M. , and Tsiantis, M. (2006). ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis . Development 133: 3955–3961. [DOI] [PubMed] [Google Scholar]
- Hu, S.H. , Latham, C.F. , Gee, C.L. , James, D.E. , and Martin, J.L. (2007). Structure of the Munc18c/Syntaxin4 N‐peptide complex defines universal features of the N‐peptide binding mode of Sec1/Munc18 proteins. Proc. Natl. Acad. Sci. USA 104: 8773–8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J.B. , Liu, H. , Chen, M. , Li, X. , Wang, M. , Yang, Y. , Wang, C. , Huang, J. , Liu, G. , Liu, Y. , Xu, J. , Cheung, A.Y. , and Tao, L.Z. (2014). ROP3 GTPase contributes to polar auxin transport and auxin responses and is important for embryogenesis and seedling growth in Arabidopsis . Plant Cell 26: 3501–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck, T. , Werner, S. , Krishnamoorthy, P. , Lerche, J. , Meijón, M. , Stenzel, I. , Löfke, C. , Wiessner, T. , Im, Y.J. , Perera, I.Y. , Iven, T. , Feussner, I. , Busch, W. , Boss, W.F. , Teichmann, T. , Hause, B. , Persson, S. , and Heilmann, I. (2013). Phosphatidylinositol 4,5‐bisphosphate influences PIN polarization by controlling clathrin‐mediated membrane trafficking in Arabidopsis . Plant Cell 25: 4894–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn, R. , and Scheller, R.H. (2006). SNAREs‐‐engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7: 631–643. [DOI] [PubMed] [Google Scholar]
- Kim, S.J. , and Brandizzi, F. (2012). News and views into the SNARE complexity in Arabidopsis . Front. Plant Sci. 3: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine‐Vehn, J. , Wabnik, K. , Martinière, A. , Łangowski, Ł. , Willig, K. , Naramoto, S. , Leitner, J. , Tanaka, H. , Jakobs, S. , Robert, S. , Luschnig, C. , Govaerts, W. , Hell, S.W. , Runions, J. , and Friml, J. (2011). Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol. Syst. Biol. 7: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurrasch, D.M. , Nevin, L.M. , Wong, J.S. , Baier, H. , and Ingraham, H.A. (2009). Neuroendocrine transcriptional programs adapt dynamically to the supply and demand for neuropeptides as revealed in NSF mutant zebrafish. Neural Dev. 4: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucci, M.R. , Di Sansebastiano, G.P. , Gigante, M. , Dalessandro, G. , and Piro, G. (2007). Secretion marker proteins and cell‐wall polysaccharides move through different secretory pathways. Planta 225: 1001–1017. [DOI] [PubMed] [Google Scholar]
- Mattsson, J. , Ckurshumova, W. , and Berleth, T. (2003). Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 131: 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittenbühler, K. , and Holzer, H. (1991). Characterization of different forms of yeast acid trehalase in the secretory pathway. Arch. Microbiol. 155: 217–220. [DOI] [PubMed] [Google Scholar]
- Nagawa, S. , Xu, T. , Lin, D. , Dhonukshe, P. , Zhang, X. , Friml, J. , Scheres, B. , Fu, Y. , and Yang, Z. (2012). ROP GTPase‐dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin‐dependent endocytosis. PLoS Biol. 10: e1001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagiec, E.E. , Bernstein, A. , and Whiteheart, S.W. (1995). Each domain of the N‐ethylmaleimide‐sensitive fusion protein contributes to its transport activity. J. Biol. Chem. 270: 29182–29188. [DOI] [PubMed] [Google Scholar]
- Nikovics, K. , Blein, T. , Peaucelle, A. , Ishida, T. , Morin, H. , Aida, M. , and Laufs, P. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis . Plant Cell 18: 2929–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olatunji, D. , Geelen, D. , and Verstraeten, I. (2017). Control of endogenous auxin levels in plant root development. Int. J. Mol. Sci. 18: 2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik, J.F. , Allen, E. , Wu, X. , Schommer, C. , Schwab, R. , Carrington, J.C. , and Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425: 257–263. [DOI] [PubMed] [Google Scholar]
- Tameshige, T. , Okamoto, S. , Lee, J.S. , Aida, M. , Tasaka, M. , Torii, K.U. , and Uchida, N. (2016). A secreted peptide and its receptors shape the auxin response pattern and leaf margin morphogenesis. Curr. Biol. 26: 2478–2485. [DOI] [PubMed] [Google Scholar]
- Tanabashi, S. , Shoda, K. , Saito, C. , Sakamoto, T. , Kurata, T. , Uemura, T. , and Nakano, A. (2018). A missense mutation in the NSF gene causes abnormal golgi morphology in Arabidopsis thaliana . Cell Struct. Funct. 43: 41–51. [DOI] [PubMed] [Google Scholar]
- Tanaka, H. , Kitakura, S. , De Rycke, R. , De Groodt, R. , and Friml, J. (2009). Fluorescence imaging‐based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 19: 391–397. [DOI] [PubMed] [Google Scholar]
- Tanaka, H. , Kitakura, S. , Rakusová, H. , Uemura, T. , Feraru, M.I. , De Rycke, R. , Robert, S. , Kakimoto, T. , and Friml, J. (2013). Cell polarity and patterning by PIN trafficking through early endosomal compartments in Arabidopsis thaliana . PLoS Genet. 9: e1003540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, H. , Nodzyłski, T. , Kitakura, S. , Feraru, M.I. , Sasabe, M. , Ishikawa, T. , Kleine‐Vehn, J. , Kakimoto, T. , and Friml, J. (2014). BEX1/ARF1A1C is required for BFA‐sensitive recycling of PIN auxin transporters and auxin‐mediated development in Arabidopsis . Plant Cell Physiol. 55: 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touihri, S. , Knöll, C. , Stierhof, Y.D. , Müller, I. , Mayer, U. , and Jürgens, G. (2011). Functional anatomy of the Arabidopsis cytokinesis‐specific syntaxin KNOLLE. Plant J. 68: 755–764. [DOI] [PubMed] [Google Scholar]
- Tsukaya, H. (2006). Mechanism of leaf‐shape determination. Annu. Rev. Plant Biol. 57: 477–496. [DOI] [PubMed] [Google Scholar]
- Vanneste, S. , and Friml, J. (2009). Auxin: A trigger for change in plant development. Cell 136: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Vieten, A. , Sauer, M. , Brewer, P.B. , and Friml, J. (2007). Molecular and cellular aspects of auxin‐transport‐mediated development. Trends Plant Sci. 12: 160–168. [DOI] [PubMed] [Google Scholar]
- Wickner, W. , and Schekman, R. (2008). Membrane fusion. Nat. Struct. Mol. Biol. 15: 658–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L. , Liu, Z. , Lu, F. , Dong, A. , and Huang, H. (2006). SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis . Plant J. 47: 841–850. [DOI] [PubMed] [Google Scholar]
- Yoon, T.Y. , and Munson, M. (2018). SNARE complex assembly and disassembly. Curr. Biol. 28: R397–R401. [DOI] [PubMed] [Google Scholar]
- Yu, X. , Pasternak, T. , Eiblmeier, M. , Ditengou, F. , Kochersperger, P. , Sun, J. , Wang, H. , Rennenberg, H. , Teale, W. , Paponov, I. , Zhou, W. , Li, C. , Li, X. , and Palme, K. (2013). Plastid‐localized glutathione reductase2‐regulated glutathione redox status is essential for Arabidopsis root apical meristem maintenance. Plant Cell 25: 4451–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C. , Smith, E.C. , and Whiteheart, S.W. (2012). Requirements for the catalytic cycle of the N‐ethylmaleimide‐Sensitive Factor (NSF). Biochim. Biophys. Acta 1823: 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found online in the supporting information tab for this article: http://onlinelibrary.wiley.com/doi/10.1111/jipb.13043/suppinfo
Figure S1. Leaf serration phenotype of atnsf‐1 and anti‐NSF (N‐ethylmaleimide‐sensitive factor) antibody characterization
(A) Number of teeth (serrations) in growing leaves of wild‐type (WT) and atnsf‐1 plants. n ≥ 20. (B) Quantification of AtNSF expression in the WT and atnsf‐1. Total RNA was isolated from 10‐d‐old seedlings grown on half‐strength Murashige‐Skoog (1/2 MS) medium and analyzed by reverse transcription polymerase chain reaction (RT‐PCR) based on 25 cycles of amplification. TUBULIN was used as a control to show the equal amount of cDNA in each reaction. (C) The induction of prokaryotic expression of AtNSF in E. coli. AtNSF protein (81.62 kDa) was induced by 1 mM isopropylthio‐β‐galactoside (IPTG) for 6 h. Total protein from IPTG‐induced E. coli was subjected to sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) followed by Coomassie blue staining (left panel). Purified AtNSF protein from E. coli was subjected to immunoblotting with anti‐AtNSF antibody (right panel).
Figure S2. Leaf phenotype of the complementation line using the native AtNSF promoter
Four‐week‐old seedlings of the wild‐type, atnsf‐1, and the complementation line. Bar = 1 cm
Figure S3. Identification of the atnsf‐2 mutant
(A, B) Leaf phenotype of the wild‐type (WT) (A) and atnsf‐2 (B). Bars = 1 cm. (C) Genotyping of the atnsf‐2 mutant. (D) Expression level of AtNSF in WT and atnsf‐2 plants determined by reverse transcription polymerase chain reaction (RT‐PCR) using 5‐week‐old plants. TUBULIN was used as a control.
Figure S4. Lateral root phenotype of the atnsf‐1 mutant
(A) Root phenotype of wild‐type (WT) and atnsf‐1 plants grown in half‐strength Murashige‐Skoog (1/2 MS) medium with or without 1 μM BFA. (B) Statistical analysis of the lateral root number as in (A). Error bars represent the SD of biological replicates (n ≥ 20). Asterisks indicate significant differences compared with the WT (*P < 0.05; **P < 0.01; t‐test). Bar = 1 cm.
Figure S5. AtNSF is strongly expressed in leaf serrations
(A) Relative transcript levels of AtNSF in the root, rosette leaf, cauline leaf, shoot, flower, and silique determined by quantitative real‐time polymerase chain reaction (qRT‐PCR). The data represent means for three independent experiments per sample and three replicates per experiment. (B) The dynamic expression pattern of AtNSF pro::GUS in the seventh leaf throughout development. 1, the youngest leaf; 7, the oldest leaf.
Figure S6. The Golgi apparatus is defective in the atnsf‐1 mutant
Transmission electron microscopy images of the wild‐type (A) and the atnsf‐1 mutant (B, C). Images show the Golgi apparatus phenotype. Bars = 200 nm.
Figure S7. Short‐term internalization of FM4‐64 in wild‐type (WT) and atnsf‐1 plants
(A–F) Internalization of FM4‐64 in WT (A–C) and atnsf‐1 (D–F) plants. Images were taken at 5 min (A, D), 15 min (B, E), and 30 min (C, F) after FM4‐64 staining. Bars = 10 μm. (G) Statistical analysis of the number of agglomerations per cell in WT and atnsf‐1 plants for 5, 15, and 30 min. The data represent the means ± SD (n ≥ 10 cells). Asterisks indicate significant differences compared with the WT (***P < 0.001; t‐test).
Figure S8. Immunolocalization of PIN2 in wild‐type (WT) and atnsf‐1 epidermal cells
Seedlings were grown on half‐strength Murashige‐Skoog (1/2 MS) medium for 7 d and PIN2 localization in root epidermal cells was observed by an immunoassay. Arrows represent internalized PIN2 protein in the atnsf‐1 mutant. Bars = 5 μm
Figure S9. CUC2‐VENUS expression in atnsf‐1. CUC2‐VENUS expression in the seventh rosette leaf margins of wild‐type (WT) (A) and atnsf‐1 (B) plants. Arrowheads indicate the restriction of VENUS expression to the sinus. Bars = 200 µm.
Table S1. Primers used for quantitative real‐time polymerase chain reaction (qRT‐PCR) and expression constructs


