Abstract
Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) has been responsible for the cause of global pandemic Covid-19 and to date, there is no effective treatment available. The spike ‘S’ protein of SARS-CoV-2 and ACE2 of the host cell are being targeted to design new drugs to control Covid-19. Similarly, a transmembrane serine protease, TMPRSS2 of the host cell plays a significant role in the proteolytic cleavage of viral ‘S’ protein helpful for the priming of ACE2 receptors and viral entry into human cells. However, three-dimensional structural information and the inhibition mechanism of TMPRSS2 is yet to be explored experimentally. Hence, we have used a molecular dynamics (MD) simulated homology model of TMPRSS2 to study the inhibition mechanism of experimentally known inhibitors Camostat mesylate, Nafamostat and Bromhexine hydrochloride (BHH) using molecular modeling techniques. Prior to docking, all three inhibitors were geometry optimized by semi-empirical quantum chemical RM1 method. Molecular docking analysis revealed that Camostat mesylate and its structural analogue Nafamostat interact strongly with residues His296 and Ser441 present in the catalytic triad of TMPRSS2, whereas BHH binds with Ala386 along with other residues. Comparative molecular dynamics simulations revealed the stable behavior of all the docked complexes. MM-PBSA calculations also revealed the stronger binding of Camostat mesylate to TMPRSS2 active site residues as compared to Nafamostat and BHH. Thus, this structural information could be useful to understand the mechanistic approach of TMPRSS2 inhibition, which may be helpful to design new lead compounds to prevent the entry of SARS-Coronavirus 2 in human cells.
Keywords: Covid-19, SARS-CoV-2, TMPRSS2, Molecular docking, Molecular dynamics simulation
1. Introduction
The rapid outbreak of Severe Acute Respiratory Syndrome coronavirus-2 (SARS-CoV-2) caused a serious global public health threat [1]. Earlier, other beta coronaviruses such as Severe Acute Respiratory Syndrome (SARS-CoV) and Middle East Respiratory Syndrome (MERS-CoV) caused respiratory diseases in humans [2,3]. The disease caused by SARS-CoV-2 was named as COVID-19 and has been declared as a global pandemic by WHO [1,4]. As of today, more than 195 countries have been affected by SARS-CoV-2. After America, Brazil, India, and a few European countries are severely affected with more mortality rates in Italy. The incubation period of COVID-19 is approximately 5.2 days [5], but it is shorter in elderly patients (age >70) [5]. The most common symptoms after the onset of COVID-19 infection are cough, fever, and fatigue, while other symptoms include headache, sputum production, diarrhea, hemoptysis, lymphopenia, and dyspnea [[6], [7], [8], [9]]. Previously, it has been suggested that SARS-CoV-2 was originated from a bat, but transmitted primarily by person-to-person contacts and through droplet nuclei formed after coughing or sneezing of infected person [10].
SARS-CoV-2 is an enveloped virus with a positive-sense RNA genome, belongs to the family Coronaviridae of the order Nodovirales and genera Betacoronavirus [11]. Recently, it has been shown that SARS-CoV-2 enters the host cell by interacting its spike glycoprotein with receptor present on epithelial cells i.e. Angiotensin-Converting Enzyme-2 (ACE-2) [12]. A high level of expression of ACE-2 has been observed in lungs, kidneys and heart cells [13,14]. However, most of the fatalities are observed due to damage to the lungs. The development and further use of vaccines against SARS-CoV-2 could face some kind of difficulties due to mutations in spike glycoprotein. In-vitro study has been reported on Nafamostat, an inhibitor of TMPRSS2 to block the MERS-CoV infection [15]. Similarly, an ingredient of mucolytic cough suppressant bromhexine hydrochloride could also be used for the treatment of influenza virus and coronavirus infections as an inhibitor of TMPRSS2 [16]. However, several potential targets for the treatment of influenza virus and coronavirus infections have been reported earlier [17]. Steardo et al., 2020 reported that the coronavirus can infect brain cells resulting in a more complex clinical scenario [18].
Currently, various broad-spectrum antiviral drugs are being used to treat the COVID-19 patients. Antimalarial drugs such as Chloroquine and its derivative hydroxychloroquine have been shown positive effects to control the infection [19]. The glycosylated spike ‘S’ protein of SARS-CoV-2 and ACE2 of the host cell have been studied thoroughly [20,21] and would be a useful target to design and discover new lead molecules to control the SARS-CoV-2. Another drug target, transmembrane serine protease (TMPRSS2) of the host cell known to cleave the viral spike ‘S’ protein, the priming event of S protein is essential for the fusion of host and viral membrane for viral entry [22,23]. The excellent work by Hoffman and coworkers suggested that influenza virus and coronavirus entry can be blocked by targeting the host cell protease such as TMPRSS2, without toxicity towards the Calu-3 cell lines [24]. Hence, the inhibition of TMPRSS2 could be a promising therapy to block the viral entry into the human cell to control SARS-CoV-2 infection.
However, there is no clear literature available about the three-dimensional structure showing involvement of different domains and the specific residues in the inhibition mechanism of TMPRSS2 in detail at the molecular level. Hence, in the present study, we have generated a three-dimensional model of TMPRSS2 using homology modeling technique and performed explicit molecular dynamics simulation (MD) to get a stable structure. Further, this MD simulated model was then used to investigate the molecular interactions between TMPRSS2 and its experimentally known inhibitors such as Camostat mesylate, Nafamostat, and Bromhexine hydrochloride using molecular docking technique. Molecular docking analysis revealed that the Camostat mesylate, Nafamostat, and Bromhexine hydrochloride interacts with the amino acids present at the active site pocket of TMPRSS2. Thus, this inhibition mechanism of TMPRSS2 could be useful to design new approaches to control the SARS-CoV-2 entry into the human cells.
2. Material and methods
2.1. Sequence retrieval and homology modeling of TMPRSS2
The amino acid sequence of transmembrane serine protease TMPRSS2 (Accession No C9JKZ3) was retrieved from UniProt [25]. Further, BLASTp program was used to search for a suitable template to build a homology model of TMPRSS2 [26]. Homology modeling of TMPRSS2 was done using online server PRIMO [27]. Three-dimensional structure of TMPRSS2 was predicted by using templates such as human plasma kallikrein (5TJX.pdb) and Hepsin (5CE1.pdb) [28]. Then, the predicted model was refined by using ModRefiner [29]. The refined model of TMPRSS2 was validated using different online servers such as PROSA [30], PROCHECK [31], and PDBsum [32].
The verified homology model of TMPRSS2 having good quality was further subjected to energy minimization using Steepest-Descent method, equilibration MD and finally, 500 ns molecular dynamics (MD) simulation in order to get a stable structure of TMPRSS2 for molecular docking studies. The detailed procedure has been discussed in the subsequent section.
2.2. Preparation, parameterization of inhibitors
Three-dimensional coordinates of Camostat mesylate (CID5284360), Nafamostat (CID4413), and Bromhexine hydrochloride (CID5702220) were extracted from PubChem Database (https://pubchem.ncbi.nlm.nih.gov/) in SDF format and then converted into PDB format with the help of Open Babel [33]. Dock Prep tool of chimera was used to calculate the charges and prepare these molecules for docking procedure [34].
2.3. Geometry optimization of inhibitors by semi-empirical quantum chemical method
Dock Prep prepared inhibitors Camostat mesylate, Nafamostat, and Bromhexine hydrochloride were then subjected to full geometry optimization by semi-empirical quantum chemical RM1 method, using commercially available molecular modeling software SPARTAN version 18 [35].
2.4. Prediction of binding pocket of TMPRSS2
Computed Atlas of Surface Topography of proteins CASTp [36], online server was used to predict the binding pocket of TMPRSS2. A potential binding pocket was selected based on consensus residues present in the related serine proteases.
2.5. Molecular dynamic simulation of a homology model of TMPRSS2
In order to get a stable model for the molecular docking procedure, explicit molecular dynamic (MD) simulation was performed on predicted TMPRSS2 homology model using GROMACS 2018.2 on Linux environment [37]. Optimized potentials for liquid simulations all atoms force field (OPLS-AA) [38] was used to generate topology TMPRRS2 receptor to study its stability in an aqueous environment using single point charge (SPC) solvent model. The protonation state has been assigned according to the physiological pH for both C-terminus and N-terminus for TMPRSS2 prior to simulation by using a tool in Gromacs. The TMPRSS2 protein was centered into a 6.9 nm from the edge of the cubic box and the system was then solvated by using Single Point Charge (SPC216) water model. The NaCl concentration was maintained at 100 mM after the addition of six numbers of chlorine ions for neutralization of the system. Periodic boundary condition (PBC) was applied in all directions, followed by 50000 steps of steepest descent energy minimization. The short-range nonbonded Lennard–Jones potential interactions were truncated to 1.0 nm. Long-range electrostatic interactions were calculated by Particle Mesh Ewald (PME) [39]. The Linear Constraint Solver (LINCS) algorithm was used to constrain all bonds [40].
Two steps equilibration (NVT and NPT) was performed, first under NVT ensemble (constant number of particles, volume, and temperature) for 100 ps at 310K temperature using Nose-Hoover thermostat [41] and NPT ensemble for 100 ps (constant number of particles, pressure, and temperature) using Nose-Hoover thermostat and Parrinello-Rahman barostat at 310K by applying position restrained [42]. Production MD run was performed for 500 ns at 2 fs time step without position restrained and trajectories were saved at every 20 ps interval for further analysis. To perform MD simulation of TMPRSS2 complexed with Camostat mesylate, Nafamostat, and Bromhexine hydrochloride, the topology of protein (TMPRSS2) and ligand molecules were generated separately from respective complexes. Topology for protein was generated by GROMACS 2018.2 using OPLS-AA force field and for ligand by using an online PRODRG server [43]. This prepared system further treated similarly as mentioned above for the TMPRSS2 model as a control.
2.6. Analysis of MD trajectories
Global stability of TMPRSS2 protein and the docked complexes with all inhibitors were evaluated by inbuilt tools of GROMACS such as g_rms, g_rmsf, g_hbond, g_gyrate, g_energy, g_sas, and do_dssp. Solvent accessibility surface area was calculated by g_sasa of Gromacs module [37], while van der Waals and hydrophobic contacts were assayed by using Discovery studio.
2.7. Molecular docking of TMPRSS2 with its inhibitors
MD simulated stable model of TMPRSS2 was used for molecular docking process with three inhibitors viz. Camostat mesylate, Nafamostat, and Bromhexine hydrochloride. Molecular docking studies were carried out using the MD simulated TMPRSS2 model with least potential energy obtained from the average structure of top cluster from MD trajectories (average structure). MD simulated model of TMPRSS2 docked with Camostat mesylate by using online docking server “Achilles”, a blind docking server (uses Autodock vina) available at http://bio-hpc.eu/software/blind-docking-server/. TMPRSS2 model as a receptor and Camostat mesylate in PDB format separately sent to the server to perform docking calculations. Series of docking calculations were performed across the whole protein to find out the binding sites, and results were clustered by using a pose clustering algorithm.
2.8. Molecular docking by AutoDock
Homology modeling and molecular docking techniques have been found useful to investigate the folding patterns and molecular interactions between several enzymes and ligands [[44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54]]. Binding affinities of Camostat mesylate, Nafamostat, and Bromhexine hydrochloride to the active site of modeled MD simulated protein i.e. TMPRSS2 were confirmed by using Autodock 4.2 with Lamarckian Genetic Algorithm (LGA) [55]. Here, blind docking was performed by taking the protease domain in a grid box. All the residues of TMPRSS2 were kept rigid. Grid dimensions were set to 70 Å × 70 Å × 70 Å to accommodate the ligand with 0.375 Å grid spacing. Grid centre was selected at X = 59.832 Y = 50.088 Z = 42.894 coordinates with 0.02 rate of mutation & 0.8 crossing over rate. Population size was fixed to 150 to generate 50 conformations for 27000 generations and for 25000 evaluations. The best docked complex was clustered based on the default RMSD tolerance range of 2.0 Å. The inhibition constant (Ki) of best docked pose was evaluated using an in-built program of AutoDock. Ligand-receptor interactions were visualized with AutoDock and UCSF Chimera [33].
2.9. Binding free energy calculations of docked complex by MM-PBSA
Binding affinities of all the three inhibitors with TMPRSS2 were evaluated by using Molecular Mechanics Poisson−Boltzmann Surface Area (MM-PBSA) [56]. MM-PBSA approach was used to estimate binding free energies of TMPRSS2 complexed with all three inhibitors respectively. The binding free energy was calculated by using the g_mmpbsa tool of GROMACS, for this total 20 snaps were collected from 300 to 500 ns MD trajectories. The program g_mmpbsa tool decomposes the total binding energy of the system into ΔGbinding = ΔGMM + ΔGSolv. The contribution of each residue in binding free energy was calculated by the MmPbSaDecomp.py python script. This has helped to determine the residues involved in interactions of the protein-ligand complex.
The binding free energy was calculated as:
| ΔGbinding = ΔGTMPRSS2 complex − (ΔGTMPRSS2 + ΔGInhibitors) |
| ΔGbinding = ΔEMM + ΔGSolv |
| ΔEMM = ΔEvdw + ΔEelec |
| ΔGSolv = ΔGnps + ΔGps |
3. Results and discussions
3.1. Structural analysis of homology model of TMPRSS2
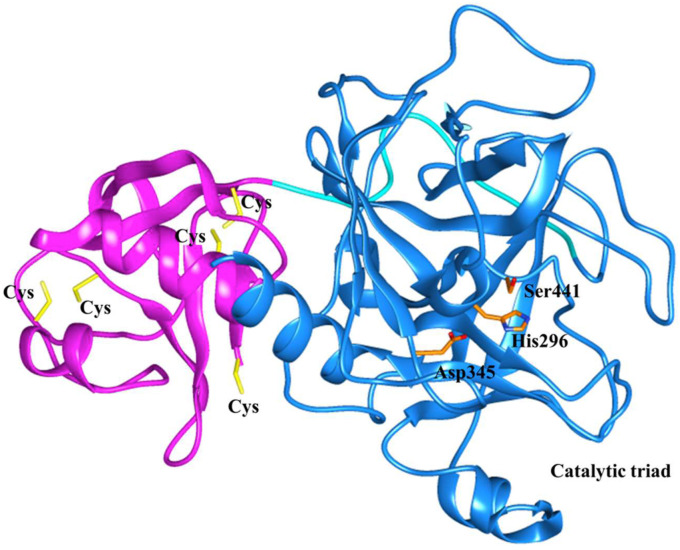
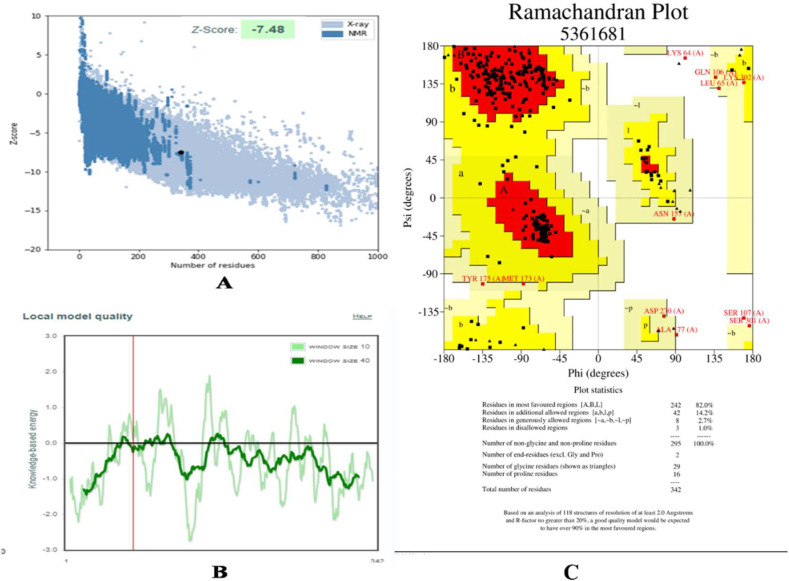
Three-dimensional structure of TMPRSS2 (Accession Number C9JKZ3) with 489 amino acids was predicted by using multiple templates, human plasma kallikrein (5TJX.pdb), and Hepsin (5CE1.pdb). The plasma kallikrein template showed the highest 42.56% identity with the TMPRSS2 sequence. TMPRSS2 consists of an intracellular domain (residues 1 to 84), transmembrane spanning domain (residues 84–106), and low-density lipoprotein receptor domain (LDLRA: residues 133–147). The present homology model of TMPRSS2 possesses two extracellular domains; cysteine-rich domain (residues 148–242) and serine protease domain (residues 255–489) (Fig. 1 ). The residues of catalytic triad such as His296, Asp345, and Ser441 have been found in the terminal serine domain of TMPRSS2 model (Fig. 1). The CATSp analysis showed His296, Asp345, and Ser441 amino acid residues in the binding pocket along with several other residues (Fig. 2 , Fig. S1 Supplementary material). TMPRSS2 model was subjected to model refinement and energy minimization by online server ModRefiner [29]. The predicted model was assessed by various online servers, the PROSA [29] analysis showed that the predicted model of TMPRSS2 has a Z score of −7.48 (Fig. 3A) as compared to the template having a Z score of −6.64, (Supplementary Fig. S1), which is within the range of X-ray and NMR native structures. Most of the amino acid residues of the TMPRSS2 model showed negative interaction energy suggesting the good quality of the predicted 3-D structure (Fig. 3B). Further, PROCHECK analysis was carried out in order to check the quality of the predicted TMPRSS2 model [31]. This analysis shows that 99% of residues are present in allowed regions and only 1% residues in the disallowed region (Fig. 3 C ), suggesting the good quality of the TMPRSS2 model.
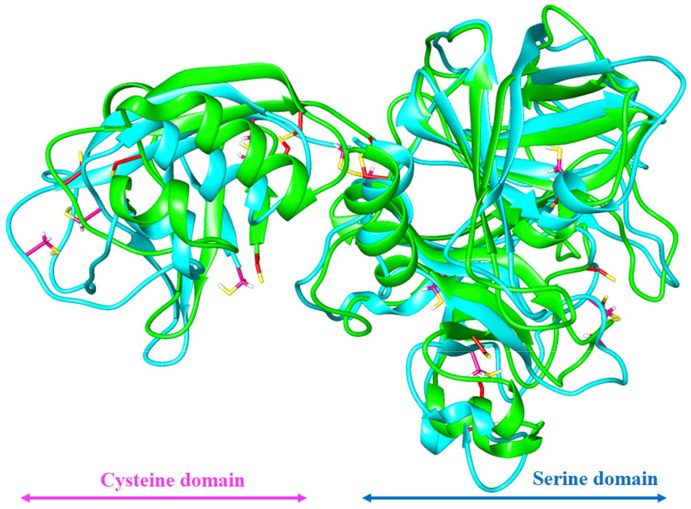
Fig. 1.
Predicted model of TMPRSS2 showing SRCR: Scavenger receptor cysteine rich domain (magenta) and catalytic triad His296, Asp345, and Ssr441 (orange) in serine protease domain (blue). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
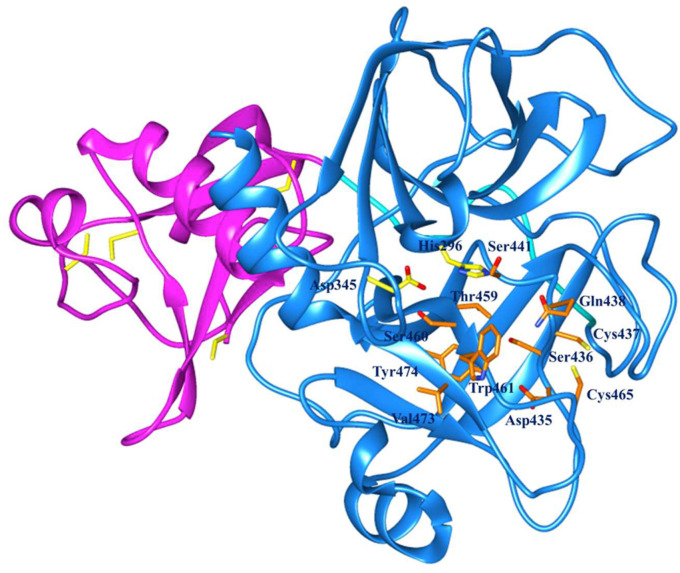
Fig. 2.
Active site residues in orange in binding pocket of TMPRSS2 predicted by CATSp. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 3.
PROSA analysis of TMPRSS2 model A) Z Score, B) Local model quality. C) Ramchandran plot of TMPRSS2 model.
3.2. Active site prediction
Active site of serine proteases generally consists Ser, His, and Asp residues in the catalytic triad [57]. TMPRSS2 active site residues were predicted using CASTp online server [36]. The CASTp server showed several pockets, out of which, a pocket was selected in such a way that at least one residue from the catalytic triad remain present in the selected pocket, these resulted in the identification of three binding pockets in TMPRSS2. As TMPRSS2 is a serine protease, hence pockets having Ser, Thr, His, Asp residues, were selected for further study. However, from the selected pockets, we observed that the residues His296, Glu299, Asp435, Gln438, Ser441, Asp345, Ser346, Thr459, Ser460, and Thr461 could have involvement in the TMPRSS2 activity (Supplementary Fig. 2).
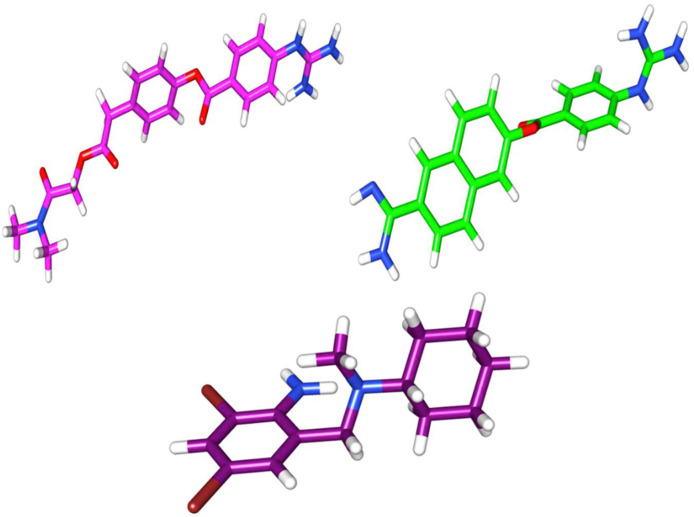
3.3. Full geometry optimization of inhibitors using semi-empirical quantum chemical method
The semi-empirical quantum chemical RM1 method was applied to optimize the geometry of Camostat mesylate, Nafamostat, and Bromhexine hydrochloride in this study. The full geometry optimization was done to get proper 3-D structures of inhibitors in terms of bond distances, bond angles, and torsion angles. These geometry-optimized inhibitors using semi-empirical quantum chemical RM1 method (Fig. 4 ) were then further used for molecular docking studies with the MD simulated stable model of TMPRSS2 (Fig. 5 ).
Fig. 4.
Three dimensional structure of TMPRSS2 inhibitors Camostat mesylate (Magenta), Nafamostat (green) and Bromhexine hydrochloride (purple). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 5.
Superimposition of TMPRSS2 model before (green) and after (cyan) MD simulation with disulfide bonds in stick before (red) and after (magenta) MD simulation with cysteine domain (magenta) and Serine domain (blue). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. Molecular interactions of TMPRSS2 with inhibitors in the docked complexes
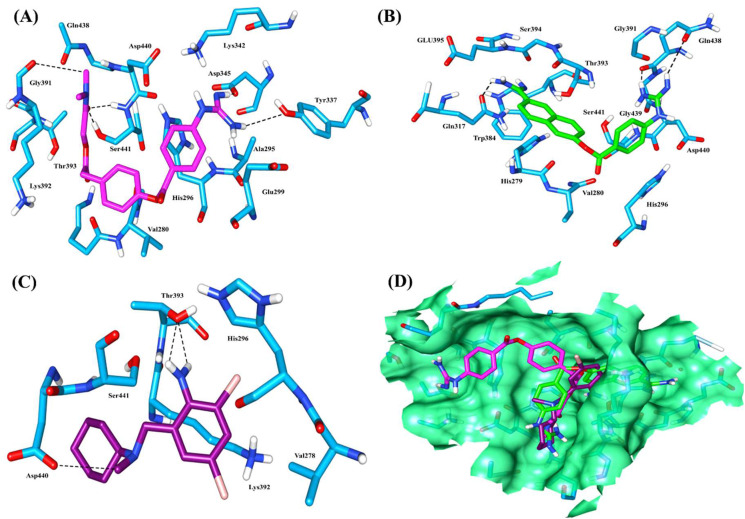
Semi-empirical quantum chemically optimized structures of Camostat mesylate, Nafamostat, and Bromhexine hydrochloride inhibitors (Fig. 4) were used to perform molecular docking with MD simulated stable model of TMPRSS2 (Fig. 5). Initially, although the active site was predicted by CATSp server, further to get binding probabilities of inhibitors of TMPRSS2, blind docking was performed by using an online blind docking server. For Camostat mesylate, we obtained a total of 29 possible clusters using 2.0 Å RMSD tolerances of binding poses. Out of which, the one having −6.23 kcal/mol lowest binding energy pose was observed with the active residues. The analysis of docked complex by AutoDock showed that residues Tyr337, Asp345, Gly391, and Ser441 of TMPRSS2 form hydrogen bonds; whereas Val280, Ala295, His296, Glu299, Lys342, Lys392, Thr393, Gln438, and Asp440 are involved in Van der Waals interactions. A molecular docking study revealed that Camostat mesylate can fit into the pocket of the serine protease domain of TMPRSS2 as shown in different confirmations (Fig. 6 A ).
Fig. 6.
Docking interaction of active site residues in stick of TMPRSS (cyan) with A) Camostat mesylate (Magenta); B) Nafamostat (green); C) Bromhexine hydrochloride (purple); D) Super imposition of docked complex of all three inhibitor showing Camostat mesylate (Magenta), Nafamostat (green) and Bromhexine hydrochloride (purple) within active site of TMPRSS2 active site residues (cyan). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The docked complex of TMPRSS2 with Nafamostat, a structural analogue of Camostat mesylate also showed similar type of hydrogen bonding interactions with the catalytic residues (Fig. 6 B, and Table 1, Table 2 ). However, bromhexine hydrochloride interacts with Asp440 and Thr393 with single and double hydrogen bonds respectively, whereas Nafamostat shows hydrogen bonding with Gln317 and double hydrogen bonds with Gln438 of TMPRSS2 (Fig. 6 C, Table 1, Table 2). Docked complex analysis revealed that Camostat mesylate and Nafamostat binds in the same pocket of TMPRSS2.
Table 1.
Molecular docking results of TMPRSS2 with its inhibitors.
| Sr | Name | CID | TMPRSS2 Residues involved in interactions | Binding Energy (Kcal/mol) | Ki | No. Hydrogen bonds |
|---|---|---|---|---|---|---|
| 1 | Camostat mesylate | 5284360 | Val280, Ala295, His296, Glu299, Tyr337, Lys342, Asp345,Gly391, Lys392, Thr393, Gln438, Asp440, Ser441, Ala295 GLU299 Tyr337 |
−6.23 | 26.98 uM | 5 |
| 2 | Nafamostat | 4413 | His279, Val280, His296, Gln317, Trp384, Gly391, Thr393, Ser394, Glu395, Gly439, Asp440, Ser441, Gln438, | −7.20 | 5.25 uM | 3 |
| 3 | Bromhexine hydrochloride | 5702220 | Val278, His296, Lys392, Thr393, Asp440, Ser441. | −5.51 | 91.26 uM | 3 |
Table 2.
Hydrogen bonding interactions between TMPRSS2 and Camostat mesylate, Nafamostat and Bromhexine hydrochloride after molecular docking.
| Sr. No. | Interactions between active site residues of TMPRSS2 with Camostat mesylate. | Distance in Å |
|---|---|---|
| 1 | Asp 345 OD2 ------ Lig. 1.A H: | 1.92 |
| Ser 441 HG ------ Lig. 1.A O: | 1.94 | |
| 2 | Ser 441 HN ------ Lig. 1.A O: | 2.21 |
| 3 | ||
| 4 | Tyr 337 OH ------ Lig. 1.A H: | 3.07 |
| 5 | Gly391 O ------ Lig. 1.A C: | 3.73 |
| Sr. No. | Interactions between active site residues of TMPRSS2 with Nafamostat. | Distance in Å |
| 1 | Gln 438 O ------ Lig. 1.A H: | 1.79 |
| 2 | Gln 317 O ------ Lig. 1.A H: | 2.17 |
| 3 | Gln 438 OE1 ------ Lig. 1.A H: | 3.12 |
| Sr. No. | Interactions between active site residues of TMPRSS2 with Bromhexine hydrochloride. | Distance in Å |
| 1 | Thr 393 OG ------ Lig. 1.A H: | 2.26 |
| 2 | Thr 393 OG ------ Lig. 1.A H: | 2.51 |
| 3 | Asp 440 OD2 ------ Lig. 1.A C: | 3.35 |
The docked complex analysis of Bromhexine hydrochloride (BHH) with TMPRSS2 shows fewer hydrogen bonding interactions as compared to Camostat mesylate and Nafamostat. A nitrogen atom of BHH interacts with Thr393 of TMPRSS2 (Fig. 6C; Table 1, Table 2). However, the residues such as Val278, His279, Val280, His296, Gly391, Lys392, Gln438, Asp440, and Ser441 of TMPRSS2 are providing additional hydrophobic interactions (Fig. 6, Table 1). The docked complex of TMPRSS2 with Camostat mesylate showed strong hydrogen bonding interactions between the guanidino group of Camostat mesylate with active site residues present in the catalytic triad such as Ser441 and Asp345 (Fig. 6 A and Table 1). Residue Ser441 of TMPRSS2 interacts with Camostat mesylate oxygen and hydrogen atoms with strong interatomic distances of 2.21 and 1.94 Å respectively. Similarly, other interacting residues of TMPRSS2 like Glu299, Thr393, Gln438, and Asp440 also showed hydrogen bonding ability. Hence, these interactions can stabilize the Camostat mesylate into the binding pocket of the serine protease domain present in TMPRSS2 (Fig. 6 A and Table 1). The binding energies of MD simulated TMPRSS2 model with Camostat mesylate, Nafamostat and Bromhexine hydrochloride complexes found as −6.23 kcal/mol, −7.20 kcal/mol, and −5.51 kcal/mol respectively (Table 1). The docked complexes of TMPRSS2 with Camostat mesylate and Nafamostat show lower binding energy with a minor difference as compared to Bromhexine hydrochloride. The inhibitor constant (Ki) of Camostat mesylate, Nafamostat, and Bromhexine hydrochloride is 26.98 μM, 5.25 μM, and 91.26 μM respectively as shown in Table 1.
3.5. Molecular dynamic (MD) simulation of a homology model of TMPRSS2
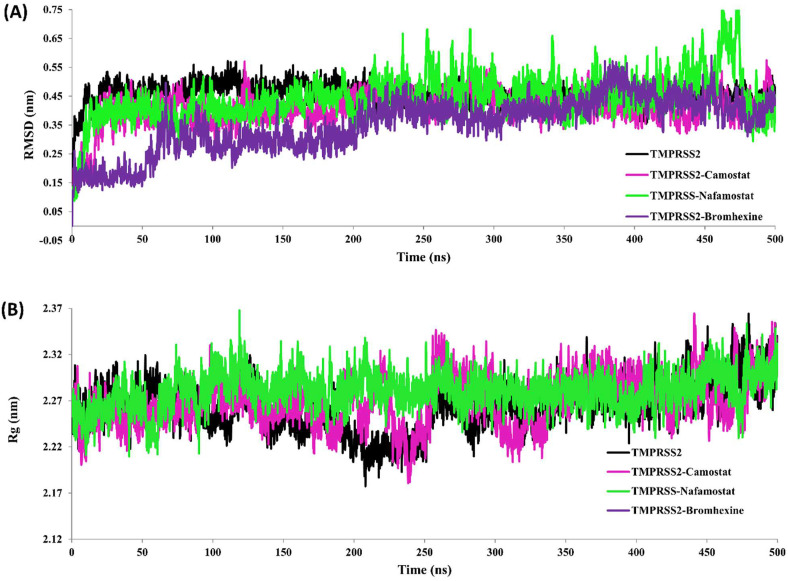
In order to get a stable 3-D structure, a 500 ns MD simulation was performed on a generated homology model of TMPRSS2 using GROMACS 2018.2 OPLS-AA all-atom force field. Dynamic stability of the TMPRSS2 model was evaluated mainly based on the root mean square deviation (RMSD), RMSF, and radius of gyration (Rg). The superimposed image of homology model and MD simulated model has been shown in Fig. 5. Backbone RMSD of TMPRSS2 without inhibitor shows stable behaviour over 500 ns with RMSD value 0.45 ± 0.03 nm (Fig. 7 A). Structural stability of the protein has also been defined based on a radius of gyration (Rg). Rg measures compactness of the protein due to spatial arrangement of secondary structures. Rg value of the whole TMPRSS2 was found in the range of 2.22–2.36 nm with an average 2.248 ± 0.002 nm representing the compactness due to proper folding of TMPRSS2 (Fig. 7B).
Fig. 7.
MD simulation of TMPRSS2 and TMPRSS2 docked complex. (A) Root mean square deviation (RMSD) during simulation. (B) Radius of gyration (Rg): TMPRSS2 without inhibitor (black) TMPRSS2 in complex TMPRSS2-Camostat mesylate (magenta), TMPRSS2-Nafamostat (green) and TMPRSS2-Bromhexine complex during simulation. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
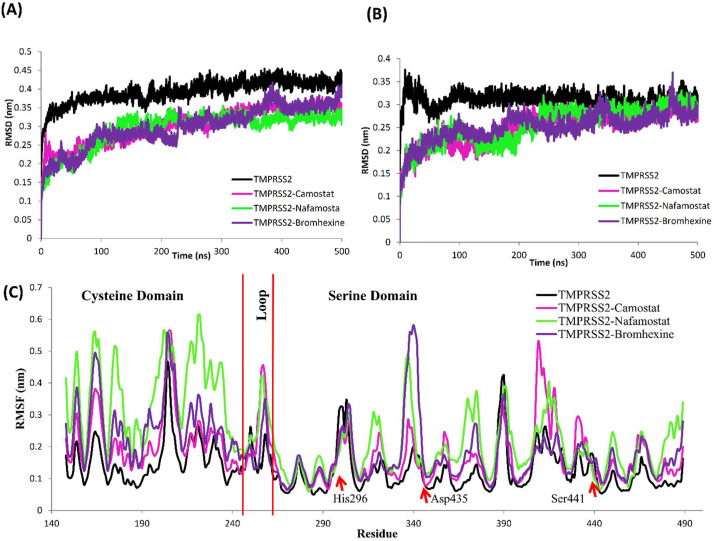
TMPRSS2 is a membrane protein present on human cells which consists of multiple domains [17]. TMPRSS2 model with two domains that is cysteine domain and catalytic serine domain shows 0.31 ± 0.01 nm and 0.39 ± 0.01 nm average RMSD values respectively (Fig. 8 A and B). The difference in RMSD values between the whole TMPRSS2 model (Fig. 7A) and specific domains (Fig. 8A and B) could be due to the presence of a loop between two domains viz. cysteine domain and catalytic serine domain (Fig. 5). RMSF analysis of TMPRSS2 shows more fluctuations in amino acids of cysteine domain and the loop region with an average RMSF 0.158 nm as compared to 0.120 nm of serine domain in TMPRSS2 (Fig. 8C). However, large peaks with 0.46 nm in the RMSF plot represents fluctuations in the loop region, whereas catalytic triad residues His296, Asp435, and Ser441, including other secondary structures of TMPRSS2 show less fluctuations (Fig. 8C). Secondary structure analysis of TMPRSS2 using DSSP tool of GROMACS shows more number of beta-sheets as compared to helices. Overall, this 3D structure of TMPRSS2 is stabilized by secondary structural elements along with five disulphide bonds. This MD simulated stable 3-D model of TMPRSS2 was then used further for molecular docking procedure.
Fig. 8.
(A) Root mean square deviation (RMSD) during simulation of serine domain and (B) Cysteine domain (C) Root mean square fluctuation (RMSF) during simulation of TMPRSS2 with both serine domain, loop region and cysteine domain along with catalytic residues RMSF indicated red .TMPRSS2 in absence of inhibitor (black), TMPRSS2 in complex TMPRSS2-Camostat mesylate (magenta), TMPRSS2-Nafamostat (green) and TMPRSS2-Bromhexine complex (purple). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. MD simulation of TMPRSS2 complexed with known inhibitors
MD simulations of low energy docked complex of TMPRSS2 with three inhibitors e.g. TMPRSS2-Camostat mesylate, TMPRSS2-Nafamostat, and TMPRSS2-Bromhexine hydrochloride were performed up to 500 ns each by using GROMACS 2018.2. The stability of TMPRSS2 complexed with these inhibitors has been evaluated by considering backbone RMSD, RMSF, and Rg values (Fig. 7, Fig. 8, Table 3 ). All three complexes viz. TMPRSS2-Camostat mesylate, TMPRSS2-Nafamostat, and TMPRSS2-Bromhexine hydrochloride show stable RMSD for the entire simulation time. The average backbone RMSD value of TMPRSS2 in complex with Camostat mesylate has been found 0.41 nm. The RMSD value of TMPRSS2-Nafamostat was obtained as 0.44 nm (Fig. 7A Table 3) with a slight deviation in last 50 ns that is 0.74 nm whereas, RMSD value for TMPRSS2-Bromhexine hydrochloride complex is 0.35 nm. The complex TMPRSS2-Nafamostat also shows stable RMSD after 50 ns and for TMPRSS2-Bromhexine hydrochloride, it was 70 ns (Fig. 7A). Overall, RMSD values of TMPRSS2 complexed with three inhibitors showed the more stability of TMPRSS2 as compared to TMPRSS2 without inhibitors (Fig. 7A). All three complexes depicted stable behaviour during the simulation run (Fig. 7A–B).
Table 3.
Analysis of MD trajectories for RMSD, RMSF, Rg and hydrogen bonding interactions of TMPRSS2 with inhibitors of 500 ns.
| Sr. No. |
MD Properties of TMPRSS2 | TMPRSS2 | TMPRSS2- Camostat mesylate | TMPRSS2- Nafamostat | TMPRSS2- Bromhexine hydrochloride |
|---|---|---|---|---|---|
| 1 | Root Mean Square Deviation (RMSD) (nm) | 0.455 ± 0.03 | 0.410 ± 0.05 | 0.447 ± 0.07 | 0.352 ± 0.09 |
| 2 | Root Mean Square Fluctuation (RMSF) (nm) | 0.132 ± 0.06 | 0.180 ± 0.09 | 0.243 ± 0.12 | 0.195 ± 0.09 |
| 3 | Radius of gyration (Rg) | 2.264 ± 0.02 | 2.268 ± 0.02 | 2.280 ± 0.02 | 2.280 ± 0.02 |
| 4 | Hydrogen Bond with water | 710 | 694 | 732 | 708 |
3.7. RMSD analysis of domains of TMPRSS2 in presence of inhibitors
RMSD analysis of two domains separately presented in Fig. 8 and Table 3, in which serine domain (255–489) shows stable behaviour over the entire simulation period for all three complexes because of interactions of inhibitors to TMPRSS2 as compared to TMPRSS2 without inhibitors (Fig. 8A). The difference in RMSD values for serine domain of TMPRSS2 with and without inhibitor is 0.09 nm for TMPRSS2-Camostat mesylate complexes and 0.10 nm for TMPRSS2-Nafamostat and 0.04 nm for TMPRSS2-Bromhexine hydrochloride complex (Table .3).
Analysis of Cysteine domain RMSD shows a similar type of scenario. RMSD of TMPRSS2 model without inhibitor has been found more as compared to the docked complexes with three inhibitors (Fig. 8B). RMSF analyses of TMPRSS2 without inhibitor were also calculated and compared with all three complexes (Fig. 8C). The results of RMSF analysis show that residues from a cysteine domain of TMPRSS2 fluctuate more as compared to the serine domain of TMPRSS2 alone as well as in complex with all three inhibitors (Fig. 8C). RMSF values of serine domain of TMPRS2 model without inhibitor and in presence of Camostat mesylate, Nafamostat, BHH are 0.12 nm, 0.16 nm, 0.19 nm and 0.16 nm respectively (Fig. 8C, Table 3). Similarly, residues in a cysteine domain of TMPRSS2-Nafamostat complex shows more fluctuations with RMSF 0.37 nm as compared to all complexes with RMSF 0.22 nm and 0.26 nm for TMPRSS2-Camostat mesylate and TMPRSS2-Bromhexine respectively, four peaks in RMSF graph with ~0.46 nm RMFS value corresponds to loops of TMPRSS2, fewer fluctuations were observed in catalytic residues His296, Asp345 and Ser441 (Fig. 8C).
Compactness and structural changes of all three complexes were evaluated on the basis of a radius of gyration (Rg). The average values of Rg for TMPRSS2 in complex with Camostat mesylate, Nafamostat, and Bromhexine hydrochloride are 2.26, 2.28, and 2.28 respectively (Fig. 7B). Binding of all three inhibitors to TMPRSS2 shows similar type of structural changes as compared to the apo form of TMPRSS2. Hence, to get further insights, we calculated the RMSD and Rg values of TMPRSS2 along with inhibitors. Secondary structure analysis of TMPRSS2 with and without inhibitors showed increase in helix and β-sheet contents with respect to decrease in coils and turns, while other secondary structures i.e beta-bridge, bend elements slightly affected as compared to TMPRSS2 alone (Table 4 ).
Table 4.
Percentage of secondary structural elements of TMPRSS2 and all three inhibitors in complex.
| Secondary structure element | TMPRSS2 | TMPRSS2- Camostat mesylate | TMPRSS2- Nafamostat | TMPRSS2- Bromhexine hydrochloride |
|---|---|---|---|---|
| Coil | 34 | 29 | 28 | 28 |
| β-Sheet | 27 | 31 | 31 | 30 |
| β-Bridge | 3 | 2 | 2 | 2 |
| Bend | 15 | 16 | 17 | 17 |
| Turn | 15 | 13 | 13 | 11 |
| Alpha Helix | 2 | 5 | 6 | 6 |
| 3-Helix | 4 | 4 | 3 | 5 |
3.8. Molecular interactions of camostat mesylate, nafamostat and bromhexine to inhibit TMPRSS2
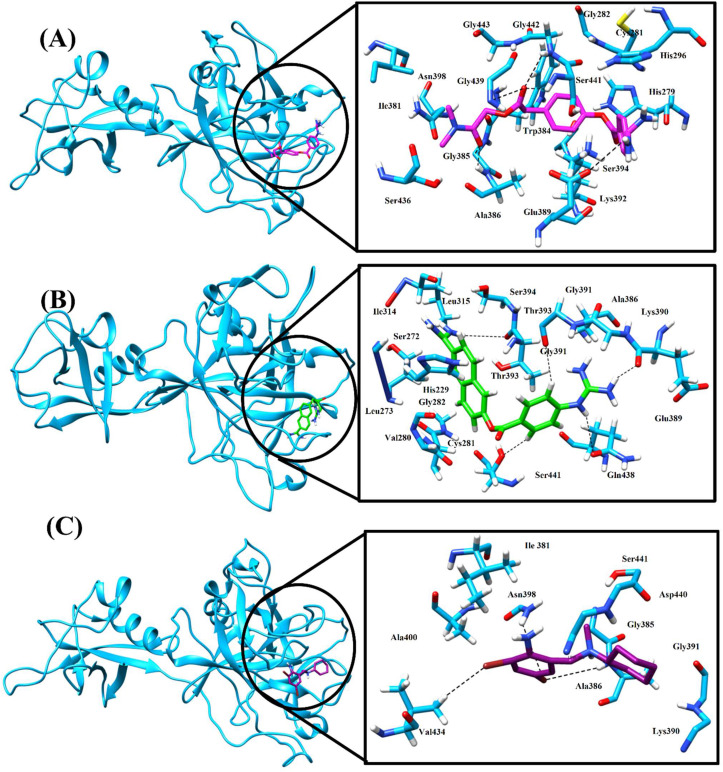
MD trajectories of TMPRSS2 complexes were clustered and the representative structure of topmost clusters extracted and then used for the analysis of molecular interactions between TMPRSS2 and inhibitors. The analysis of MD trajectories revealed that camostat mesylate moves slightly within the pocket of TMPRSS2 up to 50 ns, and further stably interacts with TMPRSS2 (Fig. 7, Fig. 9 A). After the analysis of trajectories, the representative structure from the top cluster showed that the Camostat mesylate interacts by hydrophobic as well as hydrogen bonding interactions with the TMPRSS2 catalytic site. The amino acid residues Ile381, His296, and His279 are involved in hydrophobic interactions, whereas Asn398, Gly282, His296, and Cys281 provide Van der Waals interactions, and His279, Lys392, Trp384 interact with benzene ring of camostat mesylate with Pi-alkyl interaction (Fig. 9A). The side-chain oxygen of Glu389 of TMPRSS2 interacts with guanidine nitrogen of Camostat mesylate through hydrogen bonds and backbone amide nitrogen of Ala386, Ser441, and Gly442 form hydrogen bond with the carbonyl oxygen of camostat mesylate. Similarly, week hydrogen bonding was also observed between His279, Gly385, and Gly439 residues of TMPRSS2 and the carbonyl oxygen of camostat mesylate which further strengthen the interactions (Fig. 9A and Table 5 ).
Fig. 9.
Molecular interactions of Average structure of TMPRSS2 from top cluster after MD simulation, right side whole TMPRSS2 with Camostat, Nafamostat and Bromhexine (A), (B) and (C) respectively. At left side molecular interaction of TMPRSS2 which contributes for binding of (A) Camostat mesylate (magenta) (B) Nafamostat (green) and (C) Bromhexine(purple). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 5.
Molecular interactions between TMPRSS2 with its inhibitors after MD simulation of 500 ns.
| Sr. No. |
Name of Inhibitors | TMPRSS2 Residues involve in various interactions | No. Hydrogen bonds | Interacting residues in hydrogen bond |
|---|---|---|---|---|
| 1 | Camostat mesylate | His279, Cys281, Gly282, His296, Ile381, Gly385, Ala386, Glu389, Ser394, Asn398, Ser436, Gly439, Ser441, Gly442, Gly443, Ala295 GLU299 Tyr337 |
4 | Ala386, Glu389, Ser441, Gly442, |
| 2 | Nafamostat | His229, Ser272, Leu273, Val280, Cys281, Gly282, Ile314, Leu315, Ala386, Glu389, Lys390, Gly391, Thr393, Ser394, Gln438, Ser441, | 5 | Glu389, Gly391, Thr393, Gln438, Ser441, |
| 3 | Bromhexine hydrochloride | Ile381, Gly385, Ala386, Lys390, Gly391, Asn398, Ala400, Val434, Asp440, Ser441, | 3 | Asn398, Ala386, Val434, |
Nafamostat bind at the catalytic site of TMPRSS2 with the involvement of a total five hydrogen bonding interactions. The hydrogen bond was observed between the amide nitrogen of Gln438 and the guanidine group of nafamostat observed within the first 100 ns simulation, another hydrogen bond was observed between the hydroxyl group of Ser441 with benzene ring of nafamostat. A similar type of hydrogen bond was also observed in between the backbone carbonyl oxygen of Gly391 and the benzene ring of Nafamostat. The involvement of the guanidine group of nafamostat in foming hydrogen bonding interactions with oxygen atoms of backbone carbonyl groups of Glu389 and Thr393 provides an additional stability to the complex (Fig. 9B).
Residues Leu273, Val280, Cys281, Ile314, Leu315, and Ala386 are mainly involved in Van der Waals and hydrophobic interactions, which further confirmed by binding energy calculation using MM-PBSA (Fig. 9B; Table 5). Analysis of TMPRS2-Bromhexine complex (Fig. 9C) shows fewer interactions as compared to Camostat mesylate and Nafamostat. The backbone nitrogen atom of residue Ala386 forms hydrogen bonding interactions with bromine of BHH. Similarly, another bromine of BHH involved in pi-alkyl interactions with Ile381, Val434, and residues Asn398, Ala386, Ala400, Val434, Ser441 dipected Van der Waals and hydrophobic interactions (Fig. 9C, Table 5). Analysis of all three complexes shows that along with other active site residues, at least one residue from the catalytic triad has been found interacting with Camostat mesylate, Nafamostat, and bromhexine hydrochloride within the catalytic pocket of TMPRSS2 which may impede the binding of its substrate in the active site.
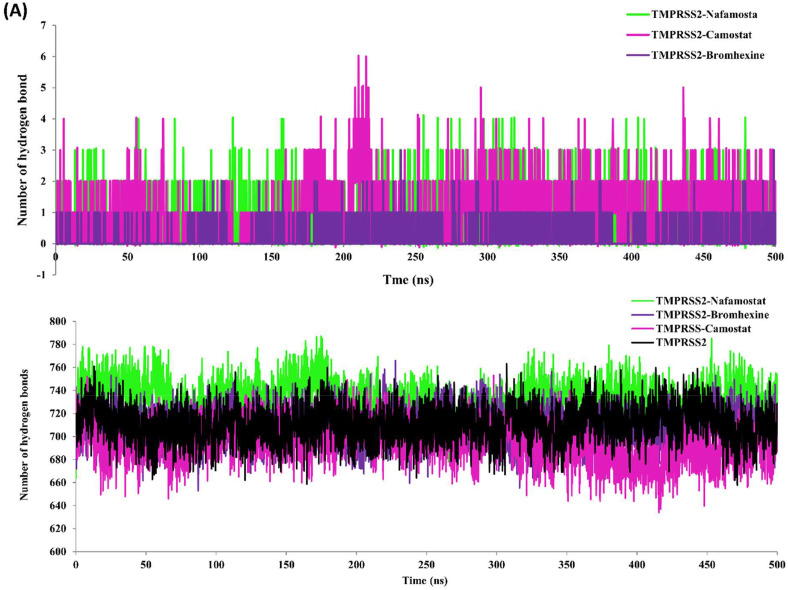
3.9. Hydrogen bonding interactions during MD simulation
Hydrogen bonding analysis of all three complexes showed that camostat mesylate forms three hydrogen bonding interactions during MD simulation (Fig. 9, Fig. 10 A), similar to our docking results (Fig. 6A). Two most stable hydrogen bonding patterns were observed in between Gln438 and Ser441 residues in TMPRSS2-Nafamostat complex, while one stable hydrogen bond involved in binding of bromhexine hydrochloride with TMPRSS2 for its inhibition (Fig. 9B–C and Fig. 10A). Earlier study has reported that hydrogen bonding is an important contribution for molecular interactions in order to bind inhibitors to TMPRSS2, which replaces the native hydrogen bonds of TMPRSS2 with water upon binding at the active site [58].
Fig. 10.
Hydrogen bond analysis: (A) Time dependent total hydrogen bond between TMPRSS2 with Camostat mesylate (magenta), Nafamostat (green) and Bromhexine (purple) in respective complex. (B) Time dependent total hydrogen bond between TMPRSS2 and water (black) TMPRSS2 with water in complex with Camostat mesylate (magenta), Nafamostat (green) and Bromhexine (purple) in respective complex. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
This hydrogen bond penalty can be evaluated for the estimation of binding efficiency of inhibitors with a protein receptor. TMPRSS2 without inhibitor can from an average of 717 hydrogen bonds with water, upon binding of Camostat mesylate, and bromhexine hydrochloride decreases in number of hydrogen bonds to 694 and 708 respectively whereas, a slight increase in hydrogen bonds 732 with water of TMPRSS in complex with Nafamostat was observed. This replacement of hydrogen bonding pattern was more intense upon camostat binding followed with bromhexine hydrochloride (Fig. 10B, Table 3).
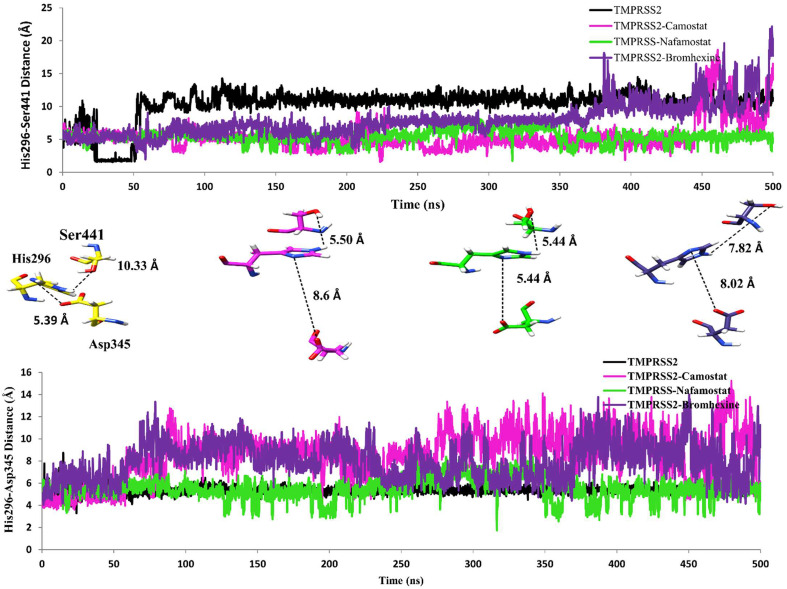
3.10. Impact of inhibitors on geometry of catalytic triad and inhibition of TMPRSS2
Catalytic triad (His296, Asp345, and Ser441), an active site of TMPRSS2 has been involved in the catalysis of spike ‘S’ protein of SARS-CoV-2 as a substrate, which is an important event of the viral entry. Thus, the geometry of these residues is crucial for the catalytic reaction. Hence, in order to understand the impact of inhibitor binding on the geometry of the catalytic triad, we investigated distances in between His296…Asp345 and His296…Ser441 residues present in the catalytic triad with respect to time. The hydrogen bond distance between the nitrogen atom of His296 and the carbonyl oxygen of Asp345, whereas the hydrogen atom of imidazole ring in His296 and hydroxyl oxygen atom of Ser441 has been shown in (Fig. 11 ). The binding of Camostat mesylate in the active site of TMPRSS2 increases the intermolecular distances between His296 to Asp345 with 8.6 Å as compared to the apo form of TMPRSS2 i.e. 5.39 Å (Fig. 11). These interactions show more fluctuations especially after 60 ns (Fig. 11). The bond distance between His296 to Asp345 has been found increased upon binding of Nafamostat at the active site of TMPRSS2 up to 5.44 Å, which fluctuates after 80 ns, and also found increased in TMPRSS2 in complex with bromhexine hydrochloride with 8.02 Å (Fig. 11). His296 to Ser441 distance was found decreased to 5.50 Å, 5.44 Å, and 7.82 Å upon binding of Camostat mesylate, Nafamostat, and bromhexine hydrochloride respectively as compared to the earlier distance 10.33 Å in case of TMPRSS2 without inhibitor (Fig. 11). An increase in intermolecular distance between His296 and Asp345 has been observed during simulations of all three inhibitors, which are actually involved in the catalysis. Thus, an increase in the distance between His296----Asp345 of catalytic triad highlights disturbances in the geometry of catalytic triad upon binding to inhibitors (Fig. 11). These results clearly depict that binding of these inhibitors at the active site or in the vicinity of the active site could destabilize the geometry of the catalytic triad of TMPRSS2 and subsequently to the substrate binding such as spike ‘S’ protein of the SARS CoV-2.
Fig. 11.
Intermolecular distance between (Nitrogen) of His296 and hydroxyl group (-OH) of Ser441 top panel and (-NH) of His296 and carbonyl (=OC) of Asp345 shown bottom panel, at middle residues in stick showing geometry of catalytic triad at active site, TMPRSS2 alone (yellow), in TMPRSS2 complex with Camostat mesylate (magenta), in TMPRSS2 complex with Nafamostat (green) and in TMPRSS2 complex with Bromhexine (purple). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.11. Binding energy calculations and residue contribution of TMPRSS2 with inhibitors
In order to investigate the binding mechanism, binding free energy between all three inhibitors and TMPRSS2 residues was calculated using Molecular Mechanics-Poisson Boltzmann Surface Area (MM-PBSA). MM-PBSA results show that the total binding energy for binding of camostat mesylate to TMPRSS2 was found to be ΔGBinding −151.36 kJ/mol, which is highest as compare to Nafamostat ΔGBinding −134.121 kJ/mol and for bromhexine is ΔGBinding −107.674 kJ/mol (Table .6 ). Analysis of components of binding energy shows that non-bonded Van der Waals interactions are predominant driving force for binding of all three inhibitors with TMPRSS2 as shown in Table 6, while the minor contribution of electrostatic (ΔEle) and non-polar (Δnp) energy was also favored for binding of respective inhibitors. These results indicate that Camostat mesylate strongly interacts with TMPRSS2 followed by Nafamostat and bromhexine hydrochloride (Table 6).
Table 6.
The binding free energy (KJ/mol) between TMPRSS2 with all three inhibitors calculated by MM-PBSA method.
| Complex | ΔEvdw | ΔEelec | ΔGpolar | ΔGnon-polar | ΔGbinding |
|---|---|---|---|---|---|
| TMPRSS2-Camostat mesylate | −219.02 ± 33.63 | −17.08 ± 8.31 | 104.399 ± 15.38 | −19.656 ± 1.17 | −151.36 ± 33.87 |
| TMPRSS2-Nafamostat | −188.60 ± 11.88 | −3.02 ± 5.74 | 77.16 ± 10.05 | −19.6 ± 0.72 | −134.12 ± 17.01 |
| TMPRSS2-Bromhexine hydrochloride | −128.13 ± 13.98 | −9.26 ± 7.377 | 42.77 ± 9.9 | −13.03 ± 1.30 | −107.67 ± 14.46 |
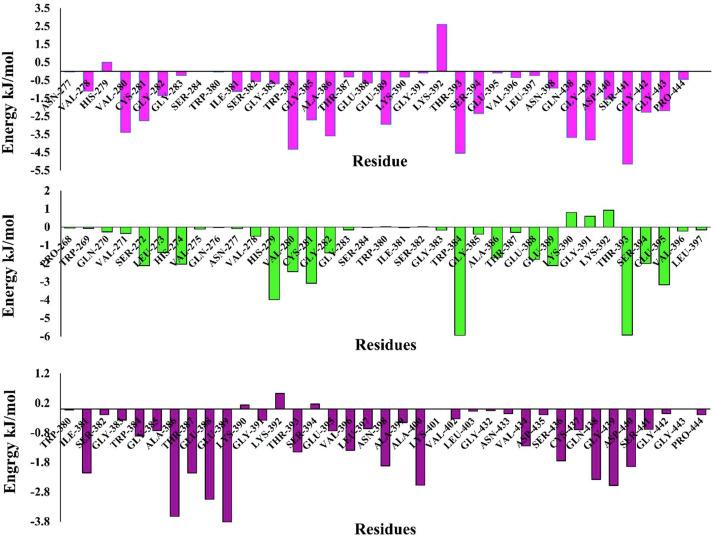
Analysis of residue wise decomposition of binding energy showed that residues Val278, His279, Val280, Cys281, Gly282, Ile381, Ser382, Gly383, Trp384, Gly385, Ala386, Ser394, Gln438, Gly439, Asp440, Ser441, Gly442, and Gly443 are involved in binding of Camostat mesylate with TMPRSS2 by various interactions (Fig. 12 A). However, some part of this study has been published as a preprint version previously [59] and other studies have screened several compounds which would be useful inhibitors of TMPRSS2 [60,61]. Residue wise decomposition energy results of TMPRSS2-Nafamostat complex shows that significant residues such as Ser272, Leu273, His274, His279, Val280, Cys281, Gly282 Trp384, Gly385, Ala386, Thr393, Ser394, and Glu395 negatively contributes to binding energy, while Lys390 and Lys392, positively contributes to the total binding energy (Fig. 12B). Analysis of TMPRSS2-BHH complex shows that residues Ile381, Trp384, Gly385, Ala386, Thr387, Glu388, Glu389, Thr393, Val396, Leu397, Asn398, Ala400, Val434, Ser436, Gln438, Gly439, and Asp440 significantly contributes negatively in total binding energy, while residues Lys392 positively contributes in total binding energy suggesting non-favorable interactions (Fig. 12C). Thus, these results show that similar types of residues are involved in the binding of Camostat mesylate and bromhexine hydrochloride to TMPRSS2, while slight variations in binding residues were observed in case of nafamostat. Binding energy analysis of all three inhibitors by the MM-PBSA method shows that camostat mesylate strongly interacts with TMPRSS2 as compare to nafamostat and bromhexine hydrochloride. Van der Wall energy is the main driving force responsible for binding free energy of all three inhibitors at the active site.
Fig. 12.
Energetic contribution of individual residues in (A) TMPRSS2-Camostat, (magenta) (blue) to binding energy in KJ/mol, (B) all residue contribution of TMPRSS2-Nafamostat complex (green) in binding energy (KJ/mol). (C) Residue contribution of TMPRSS2-Bromhexine complex (purple) in binding energy (KJ/mol). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Overall, Camostat mesylate, Nafamostat, and Bromhexine hydrochloride could be good inhibitors of TMPRSS2. The interactions of Camostat mesylate, Nafamostat, and Bromhexine hydrochloride may prevent the priming ability of transmembrane serine protease TMPRSS2 to activate the viral ‘S’ protein to the receptor ACE2 to facilitate the entry of SARS-CoV-2 in a human cell. The docked complex of TMPRSS2 with Camostat mesylate and Nafamostat shows lower binding energy with minor differences and strong hydrogen bonding interactions suggesting stable complexes. Hence, from this bioinformatics studies, we suggest that Camostat mesylate, Nafamostat, and BHH could be used as strong inhibitors of TMPRSS2 to control the SARC CoV-2 entry into the host cell.
4. Conclusion
Understanding the mechanism of effective drug targets in detail at the molecular level becomes pivotal to combat SARS-CoV-2 infection. Hence, in the present study, we used MD simulated three-dimensional structure of TMPRSS2 and studied its inhibition mechanism by Camostat mesylate, Nafamostat, and Bromhexine hydrochloride inhibitors using various molecular modeling techniques. MD simulation of a homology model of TMPRSS2 shows overall good quality structure with cysteine and serine domains. The docking and MD results revealed that Camostat mesylate and its structural analogue Nafamostat strongly interact with His296 and Ser441 residues present in the catalytic triad of TMPRSS2. Guanidine group of Camostat and Nafamostat are crucial for binding to TMPRSS2 catalytic pocket. Bromhexine hydrochloride interacts weakly with the active site through hydrophobic contacts.
Additionally, MM-PBSA analysis revealed that the Camostat mesylate and Nafamostat bind strongly in the catalytic pocket of TMPRSS2 as compared to Bromhexine hydrochloride. The binding of these inhibitors at the active site might disturb the geometry of the catalytic triad of TMPRSS2, which could impede the binding of spike ‘S’ protein, which is a crucial event of SARS Coronavirus-2 entry. Thus, this structural information obtained from the present study would be useful to design new inhibitors through ligand based drug designing approach to control the outbreak caused by SARS coronavirus-2.
Author contributions
Conceptualization: KDS.
Formal analysis: KDS, SSB, MJD, ASD.
Investigation: KDS, SSB, MJD.
Methodology: KDS, SSB, MJD, PMF, SAK, AAM, AMM, NMN, VBM.
Project administration: KDS.
Supervision: KDS.
Validation: KDS, SSB, MJD.
Writing ± original draft: KDS, SSB, MJD, SRW, NHN.
Writing ± review & editing: KDS, SSB, MJD, SRW.
Financial & competing interests’ disclosure
NA.
Ethical conduct of research
NA.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
KDS gratefully acknowledges DST-SERB, New Delhi for financial support (Ref. No. EMR/2017/002688/BBM dated 25th October 2018). KDS is also thankful to University Grants Commission, New Delhi for financial support under the UGC-SAP-DRS Phase-II scheme sanctioned to Department of Biochemistry, Shivaji University, Kolhapur. Authors thankfully acknowledge Department of Science and Technology, Government of India, New Delhi for financial support through DST-PURSE-Phase II (Ref no. SR/PURSE-PHASE-II/24(C) dated 8/3/2018). Authors are very grateful to the Structural Bioinformatics unit, Department of Biochemistry, Shivaji University, Kolhapur and Department of Microbiology, Shivaji University, Kolhapur and for extending the laboratory facilities to complete the investigation. SSB and VBM are thankful to SARTHI for providing fellowship. All authors are thankful to Computer center, Shivaji University, Kolhapur for providing necessary computational facilities.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2021.100597.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Zhao S., Lin Q., Ran J. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: a data-driven analysis in the early phase of the outbreak. Int J Infect Dis. 2020;92:214–217. doi: 10.1016/j.ijid.2020.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De W.E., Van D.N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song Z., Xu Y., Bao L. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019 Jan 14;11(1):59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO WHO Director-General opening remarks at the media briefing on COVID-19-11 March 2020. 2020. https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 [Online] Availabble.
- 5.Li Q., Guan X., Wu P. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang W., Tang J., Wei F. Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China. J Med Virol. 2020;92(4):441–447. doi: 10.1002/jmv.25689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren L.L., Wang Y.M., Wu Z.Q. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J. 2020 May 5;133(9):1015–1024. doi: 10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham C.W., Dela Cruz C.S., Cao B. Novel Wuhan (2019-nCoV) coronavirus. Am J Respir Crit Care Med. 2020 Feb 15;201(4) doi: 10.1164/rccm.2014P7. P7–P8. [DOI] [PubMed] [Google Scholar]
- 10.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020 May;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seah I., Su X., Lingam G. Revisiting the dangers of the coronavirus in the ophthalmology practice. Eye. 2020;34:1155–1157. doi: 10.1038/s41433-020-0790-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu C., Liu Y., Yang Y. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020 May;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donoghue M., Hsieh F., Baronas E. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ Res. 2000 Sep 1;87(5):E1–9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 14.Tipnis S.R., Hooper N.M., Hyde R. A human homolog of angiotensin-converting enzyme: cloning and functional expression as a captopril-insensitive carboxypeptidase. J Biol Chem. 2000 Oct 27;275(43):33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto M., Matsuyama S., Li X. Identification of nafamostat as a potent inhibitor of middle east respiratory syndrome Coronavirus s protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob Agents Chemother. 2016 Oct 21;60(11):6532–6539. doi: 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucas J.M., Heinlein C., Kim T., Hernandez S.A., Malik M.S., True L.D., Morrissey C., Corey E., Montgomery B., Mostaghel E., Clegg N., Coleman I., Brown C.M., Schneider E.L., Craik C., Simon J.A., Bedalov A., Nelson P.S. The androgen-regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis. Canc Discov. 2014 Nov;4(11):1310–1325. doi: 10.1158/2159-8290.CD-13-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen L.W., Mao H.J., Wu Y.L., Tanaka Y., Zhang W. TMPRSS2: a potential target for treatment of influenza virus and coronavirus infections. Biochimie. 2017 Nov;142:1–10. doi: 10.1016/j.biochi.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steardo L., Steardo L., Jr., Zorec R., Verkhratsky A. Neuroinfection may contribute to pathophysiology and clinical manifestations of COVID-19. Acta Physiol. 2020 Jul;229(3) doi: 10.1111/apha.13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Touret F., de Lamballerie X. Of chloroquine and COVID-19. Antivir Res. 2020 May;177:104762. doi: 10.1016/j.antiviral.2020.104762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Senathilake K., Samarakoon S., Tennekoon K. 2020030042. 2020. Virtual Screening of Inhibitors against Spike Glycoprotein of 2019 Novel Corona Virus: A Drug Repurposing Approach. Preprints. [DOI] [Google Scholar]
- 21.Xia S., Liu M., Wang C. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:30343–30355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glowacka I., Bertram S., Muller M.A. Evidence that TMPRSS2 activates the severe Acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011 May;85(9):4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shulla A., Heald-Sargent T., Subramanya G. A transmembrane serine protease is linked to the severe Acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol. 2011 Dec;85(2):873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffmann M., Kleine-Weber H., Schroeder S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 Apr 16;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bateman A. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019 Jan 8;47(D1):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul S.F., Gish W., Miller W. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Hatherley R., Brown D.K., Glenister M., Bishop Ö.T. PRIMO: an interactive homology modeling pipeline. PLoS One. 2016 Nov 17;11(11) doi: 10.1371/journal.pone.0166698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z., Partridge J., Silva-Garcia A. Structure-guided design of novel, potent, and selective macrocyclic plasma kallikrein inhibitors. ACS Med Chem Lett. 2016 Dec 6;8(2):185–190. doi: 10.1021/acsmedchemlett.6b00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu D., Zhang Y. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J. 2011 Nov 16;101(10):2525–2534. doi: 10.1016/j.bpj.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007 Jul;35 doi: 10.1093/nar/gkm290. (Web Server issue):W407-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 32.Laskowski R.A., Jabłońska J., Pravda L. PDBsum: structural summaries of PDB entries. Protein Sci. 2018 Jan;27(1):129–134. doi: 10.1002/pro.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Boyle N.M., Banck M., James C.A. Open Babel: an open chemical toolbox. J Cheminf. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettersen E.F., Goddard T.D., Huang C.C. UCSF Chimera - a visualization system for exploratory research and analysis. J Comput Chem. 2004 Oct;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 35.Hehre W., Radom L., Schleyer P., Pople J. Ab initio molecular orbital theory. J Comput Chem. 1986;7(3):379–383. doi: 10.1002/jcc.540070314. [DOI] [Google Scholar]
- 36.Tian W., Chen C., Lei X. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018 Jul 2;46(W1):W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pronk S., Páll S., Schulz R. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013 Apr 1;29(7):845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaminski G., Friesner R., Tirado-Rives J., Jorgensen W. Evaluation and Reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B. 2001 April 19;105(28):6474–6487. doi: 10.1021/jp003919d. [DOI] [Google Scholar]
- 39.Essmann U., Perera L., Berkowitz M.L. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 40.Hess B. P-LINCS: a parallel linear constraint solver for molecular simulation. J Chem Theor Comput. 2008;4(1):116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 41.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys Rev A. 1985;31(3):1695–1697. doi: 10.1103/PhysRevA.31.1695. [DOI] [PubMed] [Google Scholar]
- 42.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 43.Schüttelkopf A.W., Van Aalten D.M.F. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004 Aug;60(Pt 8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 44.Jalkute C.B., Barage S.H., Dhanavade M.J., Sonawane K.D. Identification of angiotensin converting enzyme inhibitor: an in silico perspective. Int J Pept Res Therapeut. 2015;21:107–115. doi: 10.1007/s10989-014-9434-8. [DOI] [Google Scholar]
- 45.Jalkute C.B., Barage S.H., Dhanavade M.J., Sonawane K.D. Molecular dynamics simulation and molecular docking studies of angiotensin converting enzyme with inhibitor lisinopril and amyloid beta peptide. Protein J. 2013 Jun 01;32(5):356–364. doi: 10.1007/s10930-013-9492-3. [DOI] [PubMed] [Google Scholar]
- 46.Barage S.H., Jalkute C.B., Dhanavade M.J., Sonawane K.D. Simulated interactions between endothelin converting enzyme and aβ peptide: insights into subsite recognition and cleavage mechanism. Int J Pept Res Therapeut. 2014;20:409–420. doi: 10.1007/s10989-014-9403-2. [DOI] [Google Scholar]
- 47.Dhanavade M.J., Parulekar R.S., Kamble S.A., Sonawane K.D. Molecular modeling approach to explore the role of cathepsin B from Hordeum vulgare in the degradation of Aβ peptides. Mol Biosyst. 2016;12:162–168. doi: 10.1039/c5mb00718f. [DOI] [PubMed] [Google Scholar]
- 48.Dhanavade M.J., Sonawane K.D. Insights into the molecular interactions between aminopeptidase and amyloid beta peptide using molecular modeling techniques. Amino Acids. 2014;46:1853–1866. doi: 10.1007/s00726-014-1740-0. [DOI] [PubMed] [Google Scholar]
- 49.Dhanavade M.J., Jalkute C.B., Barage S.H., Sonawane K.D. Homology modeling, molecular docking and MD simulation studies to investigate role of cysteine protease from Xanthoras campestris in degradation of Aβ peptide. Comput Biol Med. 2013;43(12):2063–2070. doi: 10.1016/j.compbiomed.2013.09.021. [DOI] [PubMed] [Google Scholar]
- 50.Parulekar R.S., Barage S.H., Jalkute C.B. Homology modeling, molecular docking and DNA binding studies of nucleotide excision repair uvrc protein from M. tuberculosis. Protein J. 2013 Aug;32(6):467–476. doi: 10.1007/s10930-013-9506-1. [DOI] [PubMed] [Google Scholar]
- 51.Sonawane K.D., Dhanavade M.J. Molecular docking technique to understand enzyme-ligand interactions. Published in book “methods and algorithms for molecular docking-based drug design and discovery. Pharmaceutical sciences: breakthroughs in research and practice, edited by information resources management association. IGI Global. 2017:727–746. doi: 10.4018/978-1-5225-1762-7.ch028. [DOI] [Google Scholar]
- 52.Sonawane K.D., Dhanavade M.J. Computational approaches to understand cleavage mechanism of amyloid beta (Aβ) peptide. Neuromethods. 2018;132:263–282. doi: 10.1007/978-1-4939-7404-7. [DOI] [Google Scholar]
- 53.Barale S.S., Parulekar R.S., Fandilolu P.M. Molecular insights into destabilization of Alzheimer's aβ protofibril by arginine containing short peptides: a molecular modeling approach. ACS Omega. 2019 Jan 10;4(1):892–903. doi: 10.1021/acsomega.8b02672. [DOI] [Google Scholar]
- 54.Kavathekar V.K., Dhanavade M.J., Sonawane K.D., Balakrishnan A. Role of cell surface vimentin in Chandipura virus replication in Neuro-2a cells. Virus Res. 2020 Aug;285:198014. doi: 10.1016/j.virusres.2020.198014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009 Dec;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumari R., Kumar R. Open source drug discovery consortium, lynn A. g_mmpbsa--a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014 May 28;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 57.Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002 Nov 23;102(12):4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 58.Zhao H., Huang D. Hydrogen bonding penalty upon ligand binding. PLoS One. 2011 June 17;6(6) doi: 10.1371/journal.pone.0019923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sonawane K.D., Barale S.S., Dhanavade M.J., Waghmare S.R., Nadaf N.H., Kamble S.A. 2020. Homology Modeling and Docking Studies of TMPRSS2 with Experimentally Known Inhibitors Camostat mesylate, Nafamostat and Bromhexine hydrochloride to Control SARS-Coronavirus-2. ChemRxiv. [Preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rahman N., Basharat Z., Yousuf M., Castaldo G., Rastrelli L., Khan H. Virtual screening of natural products against type II transmembrane serine protease (TMPRSS2), the priming agent of coronavirus 2 (SARS-CoV-2) Molecules. 2020 May 12;25(10):2271. doi: 10.3390/molecules25102271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar V., Dhanjal J.K., Bhargava P. Withanone and Withaferin-A are predicted to interact with transmembrane protease serine 2 (TMPRSS2) and block entry of SARS-CoV-2 into cells. J Biomol Struct Dyn. 2020 Jun 16:1–13. doi: 10.1080/07391102.2020.1775704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.