Abstract
To combat the ongoing public health threat of antibiotic-resistant infections, a technology that can quickly identify infecting bacterial pathogens and concurrently perform antimicrobial susceptibility testing (AST) in point-of-care settings is needed. Here we develop a technology for point-of-care AST with a low-magnification solution scattering imaging system and a real time video-based object scattering intensity detection method. The low magnification (1–2X) optics provide sufficient volume for direct imaging of bacteria in urine samples, avoiding the time-consuming process of culture-based bacterial isolation and enrichment. Scattering intensity from moving bacteria and particles in the sample is obtained by subtracting both spatial and temporal background from a short video. The time profile of scattering intensity is correlated with the bacterial growth rate and bacterial response to antibiotic exposure. Compared to the image-based bacterial tracking and counting method we previously developed, this simple image processing algorithm accommodates a wider range of bacterial concentrations, simplifies sample preparation, and greatly reduces the computational cost of signal processing. Furthermore, development of this simplified processing algorithm eases implementation of multiplexed detection and allows real-time signal readout, which are essential for point-of-care AST applications. To establish the method, 130 clinical urine samples were tested, and the results demonstrated an accuracy of ~92% within 60–90 min for UTI diagnosis. Rapid AST of 55 positive clinical samples revealed 98% categorical agreement with both the clinical culture results and the on-site parallel AST validation results. This technology provides opportunities for prompt infection diagnosis and accurate antibiotic prescriptions in point-of-care settings.
Keywords: antimicrobial susceptibility testing, AST, optical imaging, intrinsic features, object intensit, background removal, antibiotic resistance, E. coli, urinary tract infections, UTI
Graphical Abstract
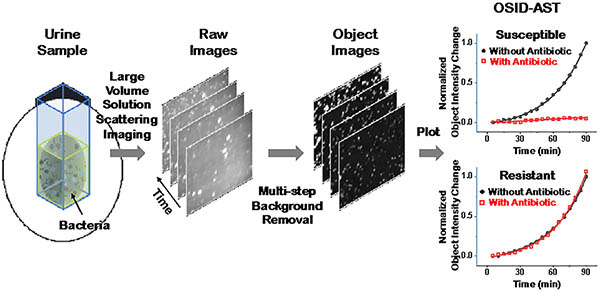
A large volume solution scattering imaging (LVSi) system and a real time background removal algorithm for object scattering intensity detection are developed for direct antimicrobial susceptibility testing on raw clinical urine samples. The rapid determination of antibiotic resistance helps with the precise antibiotic prescriptions and proper treatment of the patient within a single clinic visit.
The misuse and overuse of the broad-spectrum antibiotics has led to widespread development of antimicrobial resistance, posing long-term threats to public health.1–3 Multidrug-resistant bacteria persist in many healthcare settings, leading to a wide range of acute and nosocomial infections with high mortality rates. Each year, resistant infections cause at least 2.8 million hospitalizations and 35,000 deaths in the US alone.1 Urinary tract infections (UTIs) are the most frequent bacterial infection in the outpatient setting, affecting 50% of women during their lifetime.4, 5 Although many UTIs are uncomplicated, progression into life-threatening infections, such as sepsis, can occur. The problem of antimicrobial resistance is aggravated by the empirical prescription of the antibiotics for UTI treatment.6, 7 Current standard methods for both identification and antimicrobial susceptibility testing (AST) are slow, typically taking 2–4 days for results to be reported to the patient.8–10 The standard method for UTI screening and AST are culture based, which takes 48 h or more to produce results, while dipstick tests and manual microscopy are faster but less reliable.11, 12 Therefore, a rapid, affordable, and highly sensitive detection platform, capable of significantly reducing the time needed for antibiotic susceptibility determination and optimizing targeted therapy in point-of-care (POC) settings, is urgently needed.
Various emerging rapid AST technologies have been developed using either genotypic or phenotypic approaches.13–17 The former detects genes responsible for conferring drug resistance, which is powerful, but requires prior knowledge of the genes.14, 18–22 Genotypic approaches are nucleic acid amplification testing (NAT)-based detection methods that require a series of sample preparation steps and use of primers and enzymes.23 Phenotypic AST technologies usually detect phenotypic features (e.g., bacterial physiology, size, length, number, and morphology) for direct bacterial cell growth measurements11, 24–32, among which, optical detection, including real time microscopy, live cell imaging, flow cytometry and scattering microscopy, have been leading rapid AST assay development. However, most of these technologies require pure cultures of isolated bacteria. Without sample isolation and enrichment, very few bacteria can be imaged with high magnification optics. Furthermore, single cell imaging with microfluidics is confronted with critical issues such as clogging, bubble formation, and precise fluid management when used to directly analyze clinical samples. Thus, a simple, faster, and affordable method is needed for direct bulk sample analysis in POC settings to promptly avoid antibiotic usage in case of nonbacterial infections and promote accurate antibiotic prescription for resistant pathogens.
Here, we introduce rapid AST with free-solution, forward-scattering imaging that directly assesses clinical urine samples in a cuvette without microfluidics. The effect of antibiotics on bacterial growth rate is quantified by a background-free, video-based Object Scattering Intensity Detection method (referred to as OSID-AST). Similar to the light scattering-based bacterial growth detection methods (e.g., BacterioScan33), our method simply detects the total light intensity scattered from the sample. However, with real-time imaging capability, we removed background noises, tracking only the scattered light from the moving bacterial cells and particles from the clinical urine sample, thereby providing accurate information on the responsiveness of cells to antibiotic exposure. We describe the optical setup and principle of the OSID-AST, validate it with cultured, stationary-phase Escherichia coli and Staphylococcus saprophyticus, and apply it to 130 clinical urine samples from patients with suspected UTIs.
RESULTS AND DISCUSSION
Detection Principle
Measurement of optical density (OD) at 600 nm (OD600), commonly used to monitor relative bacterial growth in liquid suspension, is based on light scattered in random directions by the bacteria suspended in solution. This indirect assessment of bacterial growth and cell concentration requires pure sample inoculation and time for the cells in suspension to reach appropriate density levels.34 With the improvement of the light source and optical configurations, other light scattering methods were developed to increase the detection limit.35–37 However, the methods collect total scattered light from the sample, including background noises from vial defects, wall contamination, and surface reflection, and still require enrichment and substantial cell growth, and thus, longer sample-to-answer time. To minimize sample pre-processing time, we introduced a low magnification solution that incorporates forward-scattering imaging with dark field illumination, direct real sample detection, and an efficient background removal algorithm, thereby employing rapid AST based on object scattering intensity (Fig. 1).
Figure 1. Principle of rapid AST with integrated object scattering intensity quantification.
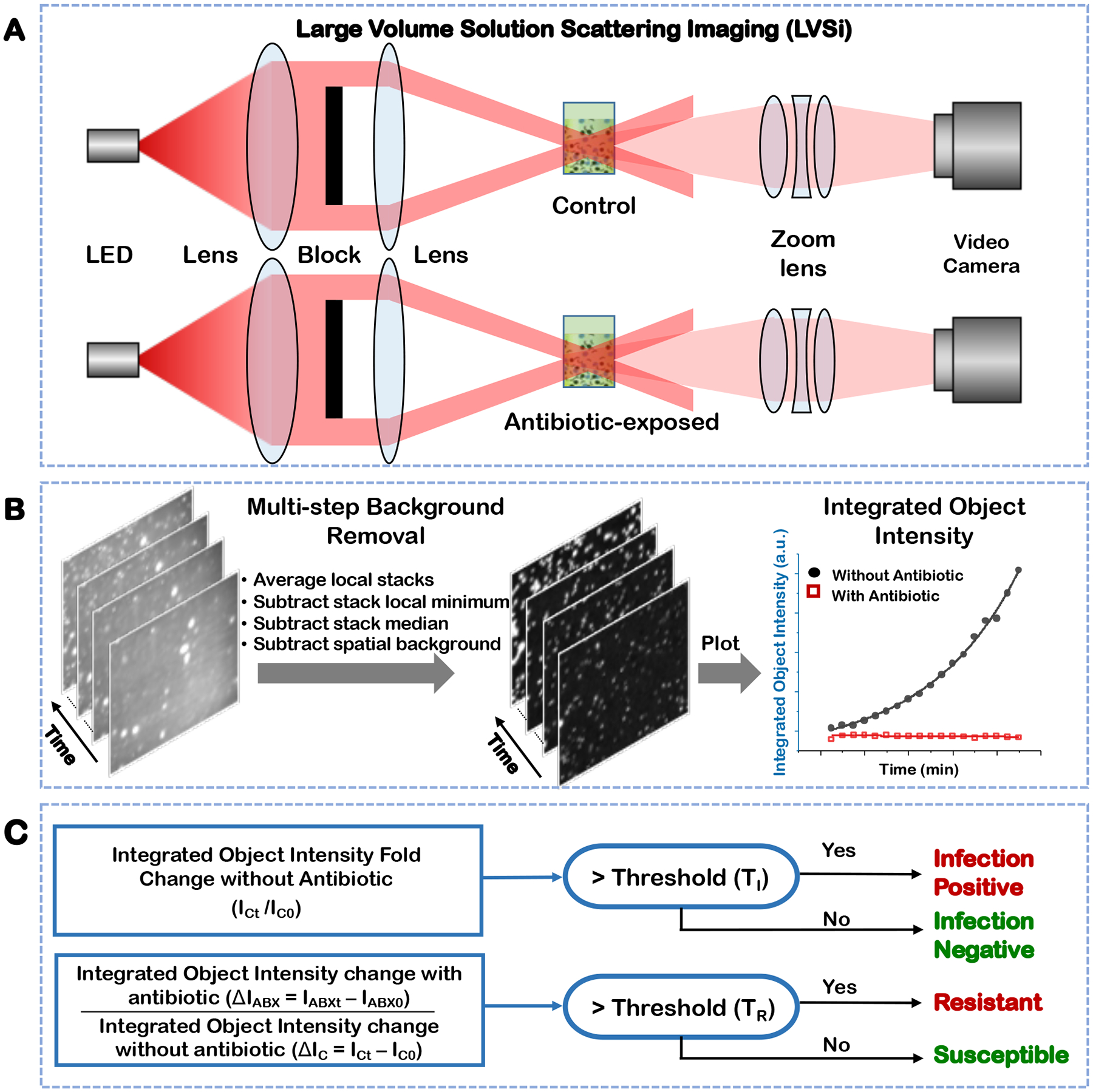
Schematic illustration of the dual-channel experimental setup for video-based solution scattering imaging of a clinical sample (A). Multi-step image processing for background removal and integrated object intensity plotting (B). The detailed image processing flow chart is in Supporting Information S1. Clinical decision determination based on integrated object scattering intensity. ICt is the integrated object scattering intensity of samples without antibiotic at time t. IC0 is the initial intensity of the same sample. ΔIC = ICt -IC0 and ΔIABX = IABXt -IABX0, represent the integrated object scattering intensity change at time t for antibiotic-unexposed (control) and -exposed samples, respectively (C).
The dual channel large volume solution scattering imaging (LVSi) system can simultaneously measure antibiotic-exposed and control samples. The system uses a forward scattering geometry to minimize image-intensity blinking associated with the rotation of cells in the solution. Furthermore, a beam block prevents the incident light from directly entering the camera, enabling high contrast and low background dark field images of the bacterial cells (Fig. 1A). Bacterial cells and similar-sized particles from a urine sample are imaged as individual bright spots. Due to thermal drift, the bright spots move slowly within the image field as the basal platform heats the sample to 37 °C. We developed an automated image processing protocol that removes all background noises in the video in four simple steps (Fig. 1B, Supporting Information Figure S1). In step one, the raw video stacks (1 min duration, 10 fps) are averaged for every four frames to simultaneously increase signal-to-noise ratios and reduce data file sizes. In step two, background noises from cuvette-based defects and interferences are removed every 10 frames via pixel-level, temporal, local minimium subtraction. In step three, dynamic background noise, caused by thermal and mechanical drift that induces moving reflections and scattering, is primarily removed by subtracting the whole stack temporal median image. In step four, the remaining background is removed by subtracting the local spatial background, which is calculated by rolling ball average with radius of 10 pixels for all pixels in the image. The typical computational time for processing a 1 min video is ~ 45 seconds with a desktop personal computer. After background removal, the video intensity is dominated by the object intensity of all particles, including both bacterial cells and particles. Since particle intensities are stable over time, the change of the object scattering intensity is correlated with bacterial growth.
The integrated object scattering intensity quantification enables detection of bacterial infection and AST for infection-positive samples with the algorithm graphically presented in Fig. 1C. Two test solutions are prepared with a patient’s urine sample, one solution with medium and antibiotics for AST and the other one with medium only as a control. To detect bacterial infection, the integrated object scattering intensity in the control group is quantified over time (e.g., 1 min video measure of object scattering intensity every 5 min for a total of 90 min). As shown in Fig. 1C, if the fold increase of integrated object intensity over the initial value (ICt/IC0) is above an infection threshold (TI) (Material and Methods and Supporting Information S2), or ICt/IC0 > TI, the sample is identified as infection positive. Otherwise, the sample is determined as infection negative. For antibiotic resistance detection, the integrated object intensity changes over time in the control urine sample (ΔIC = ICt -IC0) and in the antibiotic-exposed sample (ΔIABX = IABXt -IABX0) are calculated. If the ratio between the two (ΔIABX/ΔIC) is above a resistance threshold (TR), ΔIABX/ΔIC > TR, indicating insufficient bacterial inhibition by the antibiotic, the sample is identified as resistant. If the ratio is at or below TR, bacterial growth is inhibited by the antibiotic, and the sample is defined as susceptible to that antibiotic.
Testing OSID-AST with pure E. coli and S. saprophyticus cultures
To establish the method, E. coli and S. saprophyticus cultures (see Materials and Methods) with and without antibiotics were imaged for bacterial growth measurements. E. coli is the most predominant pathogen causing 60–80% of community-acquired UTIs, while S. saprophyticus is the second-most common cause of community-acquired UTIs. In urine samples, bacterial cells are likely in stationary phase due to recent release of intracellular bacterial communities from infected epithelial cells and/or nutrient depletion.38 We worked directly with E. coli and S. saprophyticus stationary phase cultures diluted in fresh culture medium without additional subculturing. The individual bacterial cells were imaged as bright spots moving dynamically in the video. The integrated object intensity of all cells was quantified, after real-time background removal processing, with a 1 min duration video at 5 min intervals over a total of 90 min. The bacterial growth, as integrated object intensity, of both control and antibiotic-exposed samples were plotted for antibiotic susceptibility determination (Fig. 2 and Fig. 3).
Figure 2. Integrated scattering intensity detection of antibiotic-susceptible E. coli.
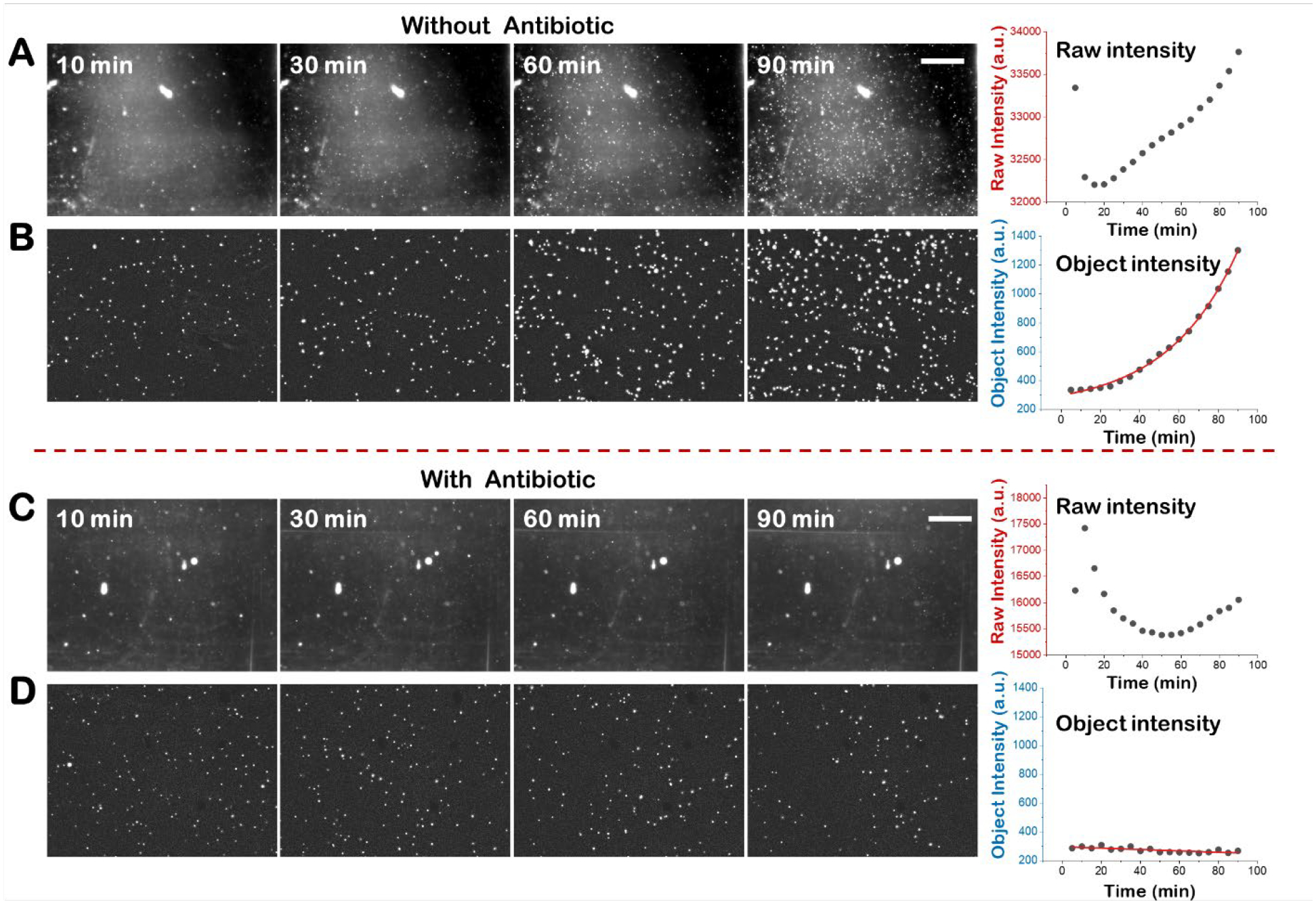
Snapshot images of pure E. coli culture without antibiotic at different time points and the corresponding image intensity plot vs time before (A) and after multi-step background removal (B). Snapshot images of pure E. coli culture with antibiotic (32 μg/mL nitrofurantoin) at different time points and the corresponding image intensity plot vs time before (C) and after multi-step background removal (D). Scale bar, 400 μm.
Figure 3. Integrated scattering intensity detection of antibiotic susceptible S. saprophyticus.
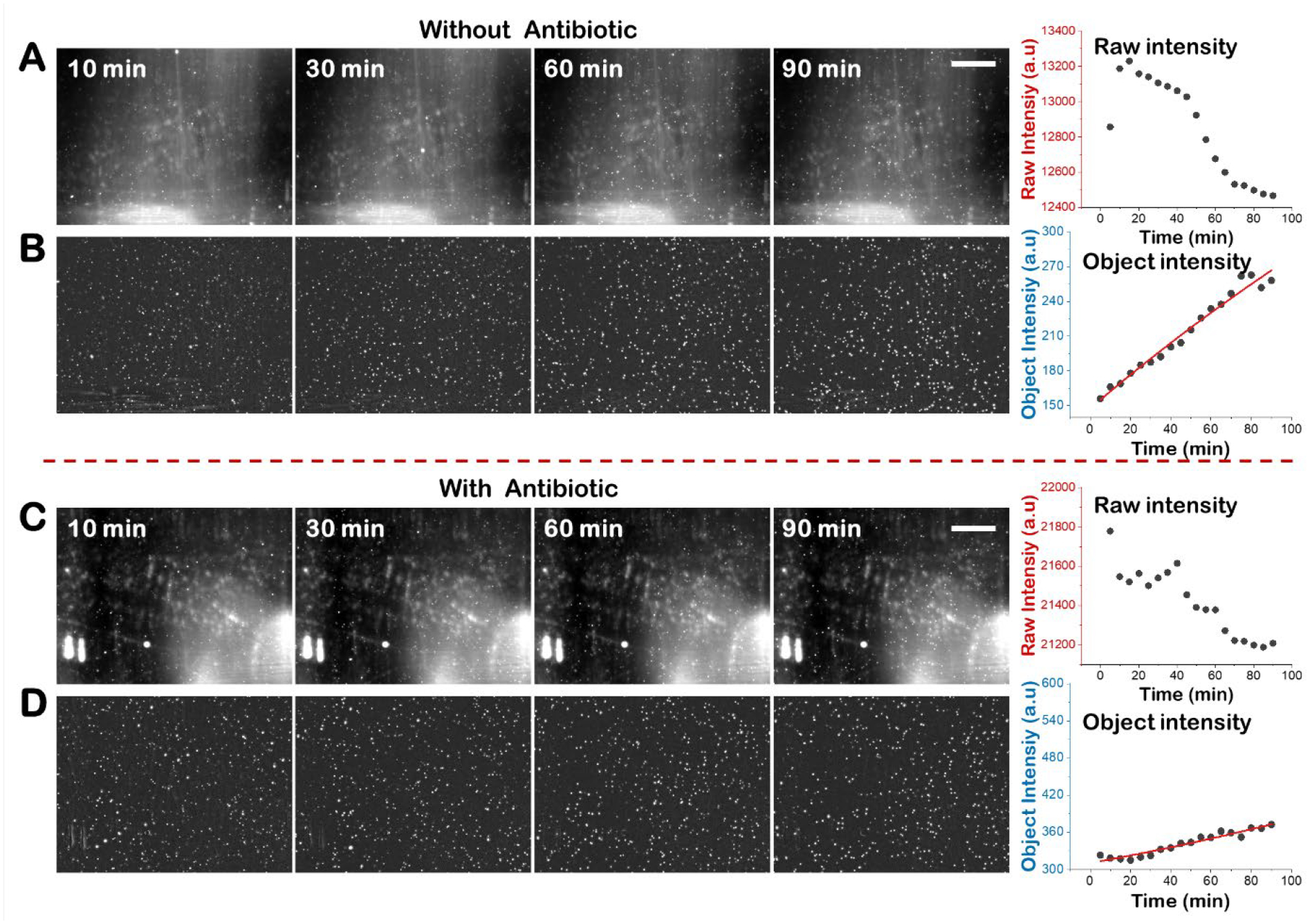
Snapshot images of pure S. saprophyticus culture without antibiotic at different time points and the corresponding image intensity plot vs time before (A) and after multi-step background removal (B). Snapshot images of pure S. saprophyticus culture with antibiotic (2 μg/mL ciprofloxacin) at different time points and the corresponding image intensity plot vs time before (C) and after multi-step background removal (D). Scale bar, 400 μm.
The object intensity detection and the raw intensity detection for antibiotic susceptibility testing with pure E. coli cultures, are compared in Fig. 2. The raw intensity detection is equivalent to the traditional optical density light scattering measurement with spectrometry. In the absence of antibiotics, E. coli multiplies over time as indicated by the increase in both the raw intensity and integrated object intensity (Fig. 2A and 2B). However, the raw video presents obvious background noises including both static and dynamic optical background from residual background illumination, scattering from defects and dust on the cuvette wall, and ghost light (Fig. 2A), which interfere with the intensity increase induced by E. coli growth. In contrast, after background removal, the intensity changes are more evident as the bacterial cell scattered light dominates the images (Fig. 2B). To accurately track E. coli growth, the averaged intensity of a 1 min video was quantified every 5 min. With the raw video intensity, E. coli growth was detected in 90 min, but the growth curve does not follow an obvious exponential increase and the intensity increase is small compared with the initial background intensity. The growth curve plotted with the integrated object intensity shows more evident exponential growth (Fig. 2B). Similarly, for the antibiotic-exposed sample (32 μg/mL nitrofurantoin), the raw video shows obvious background noises, while the raw intensity shows a fluctuate curve (Fig. 2C) due to drifting background noise. Conversely, the background-free integrated object intensity curve (Fig. 2D) shows a flat line over time, indicating no bacterial growth in the presence of the antibiotic and revealing a clear antibiotic susceptible result.
Similarly, the object intensity detection and the raw intensity detection for pure S. saprophyticus cultures were performed and compared (Fig. 3). S. saprophyticus is a Gram-positive bacterium that forms grape-like clusters of coccus-shaped cells during growth. S. saprophyticus grows slower than E. coli and lacks flagella, so increases in scattered light intensity often occurs without accompanying detection of increases in individual cells. During the 90 min detection, the raw intensity of both control and antibiotic- (2 μg/mL ciprofloxacin) exposed samples decreases with time as the background noise intensity dominates, thereby negating raw intensity usage for bacterial growth quantification. In contrast, obvious growth was observed with object intensity detection in the control sample with intensity nearly doubling (~1.7X), while the antibiotic-exposed sample intensity increased minimally (~1.1X). As ΔIABX/ΔIC is approximately 0.14, well below the resistance threshold of 0.5, the S. saprophyticus culture is deemed antibiotic (ciprofloxacin) susceptible with our OSID-AST.
To further validate the robustness of the object intensity detection method, we statistically analyzed independent culture replicates. To compare the results from independent experiments, both raw intensity and object intensity at each time point were normalized to the initial video intensity to generate the fold increase of cell growth. Five independent experimental results with and without antibiotics are plotted in Fig. 4A–C (32 μg/mL nitrofurantoin) and Fig. 4D–F (2 μg/mL ciprofloxacin) for E. coli cultures and Fig. 4G–I (2 μg/mL ciprofloxacin) for S. saprophyticus cultures. Prior to background removal, sample-to-sample intensity detection variations between control and antibiotic-exposed samples made it difficult to determine antibiotic susceptibility (Fig. 4A, D, G). In contrast, object intensity detection after background removal revealed obvious and consistent growth differences between control and antibiotic-exposed samples for both antibiotics and strains tested. OSID-AST of E. coli cells was determined within 40 min for nitrofurantoin (Fig. 4B) and 60 min for ciprofloxacin (Fig. 4E). Due to a slower growth rate and object intensity increases in both control and antibiotic-exposed samples, S. saprophyticus cultures require longer incubation times for antibiotic susceptibility determination (Fig. 4H). Statistical analysis of all the samples at different time points (0, 60, 90 min) indicates reliable AST with integrated object intensity detection (Fig. 4C, F, I). While raw image intensities show no significant difference between samples with and without antibiotics, the integrated object intensities show significant differences between the two groups after 60 min for both E. coli and S. saprophyticus (Fig. 4C, F, I).
Figure 4. Statistical analysis of the integrated object intensity detection for rapid AST with different antibiotics and different bacterial strains.
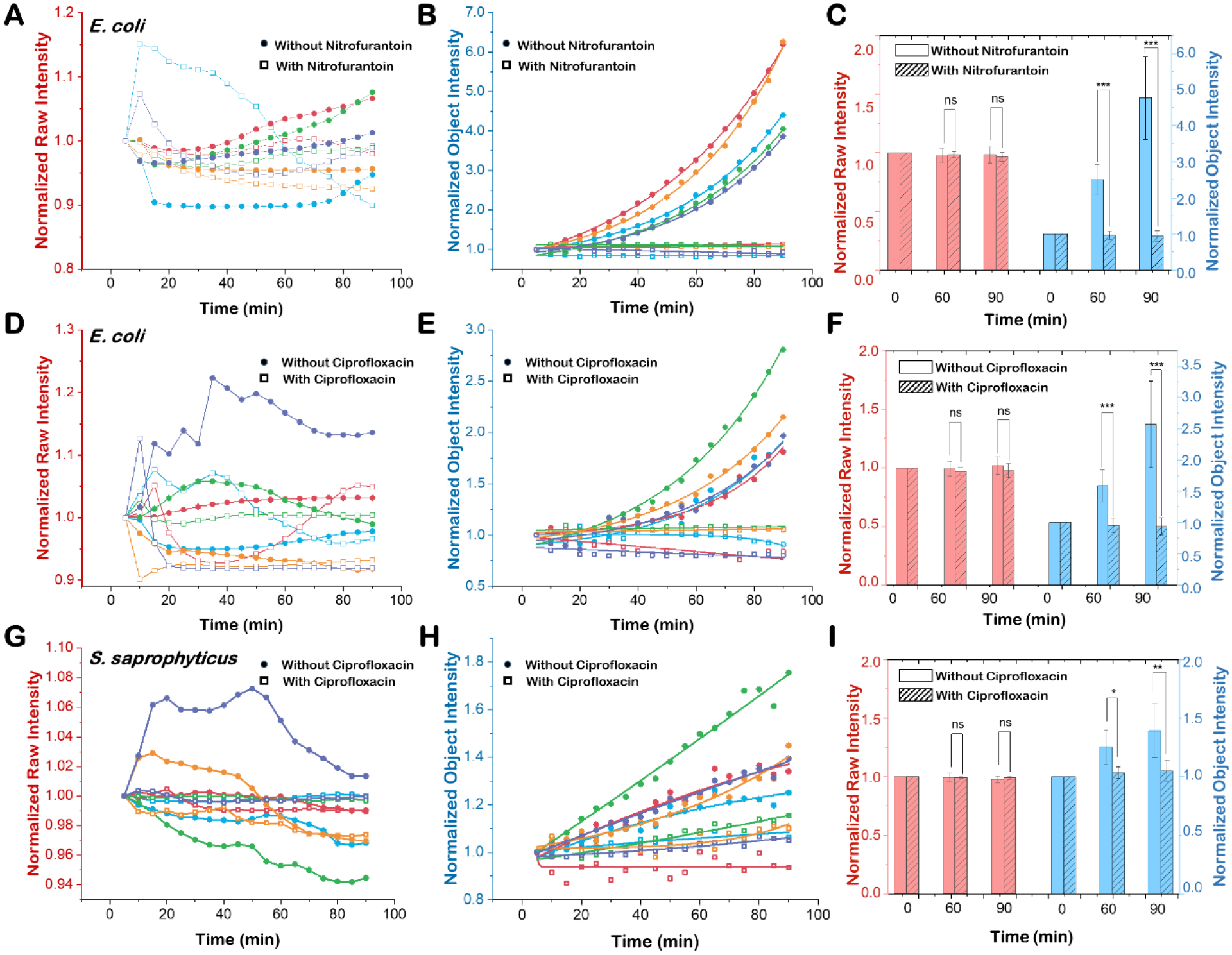
Original image intensity results of five independent replicates of E. coli cultures exposed to (A) nitrofurantoin (32 μg/mL) or (D) ciprofloxacin (2 μg/mL) and S. saprophyticus cultures exposed to (G) ciprofloxacin (2 μg/mL). Image intensity results after multi-step background removal of five independent replicates of E. coli cultures exposed to (B) nitrofurantoin (32 μg/mL) or (E) ciprofloxacin (2 μg/mL) and S. saprophyticus cultures exposed to (H) ciprofloxacin (2 μg/mL). Comparisons of bacterial intensity results for E. coli (C, F) and S. saprophyticus (I) from original images (red bars) and background-corrected images (blue bars) at 0, 30, and 60 min with (hatched bars) and without (open bars) antibiotics. ns: not significant, *: p<0.01, **: p<0.005, ***: p<0.001.
Compared to our previous single cell counting method,39, 40 video-based object intensity detection does not need to identify and track individual scattering objects. Therefore, video-based OSID can function with a higher density of particles and thus, a wider range of bacterial concentrations (e.g., 104 – 107 cells/mL), as long as the original intensities do not saturate during detection (Supporting Information Figure S3). The wider dynamic range of the object intensity detection platform enables detecting bacteria in clinical urine samples with a single unified dilution step, thereby simplifying the sample preparation process by eliminating particle concentration premeasurement and additional dilution steps. In addition, the total AST time decreases with increasing bacterial concentration in the working sample (Supporting Information Figure S4). Furthermore, the background removal process, which eliminates artifacts and dramatically reduces computational costs, does not require manual threshold detections or complicated tracking processes. Also, object intensity detection quantitates relative growth for both E. coli and S. saprophyticus, while single cell counting cannot quantify S. saprophyticus, which aggregates and forms clusters during growth (Supporting Information S5). The object intensity detection platform allows fully automated, real time image processing and results reporting and quantifies both rod- and coccus-shaped bacteria that are motile (E. coli) and nonmotile (S. saprophyticus).
The integrated object intensity directly correlates with changes in cell number and cell size for bacterial growth determination. For bactericidal antibiotics, such as nitrofurantoin and ciprofloxacin, that kill bacteria, the object intensity measurements provide rapid and evident results for AST determination. However, the bactericidal mechanisms of some antibiotics, such as ampicillin, induce cell elongation and accompanying increases in object scattering intensity prior to killing, and thus might necessitate longer detection times for AST determination. For antibiotics that induce elongation or morphological changes, quantifying cell number changes or tracking division events can effectively determine antibiotic susceptibility, but with increased computational costs.
UTI infection detection and AST with clinical urine samples
After validation of OSID-AST with pure cultures, we applied it to clinical urine samples for both UTI infection detection and AST of the UTI-causing bacteria. Infection detection measures the integrated object intensity increase over time in the antibiotic free (control) sample. A positive infection is identified when the intensity increase is higher than the infection threshold, which indicates active bacterial growth. AST compares the intensity changes in samples incubated with and without antibiotics. One hundred and thirty de-identified clinical urine samples collected from hospitalized patients at Mayo Clinic were measured by OSID-AST. The results for all samples are validated with on-site initial plating and parallel AST plating for CFU quantitation and compared with clinical lab results measured by BD Phoenix. The workflow for the clinical sample preparation, testing and validation is illustrated in Support Information S6, Figure S6.
UTI detection:
We tested 130 clinical urine samples and identified samples with UTIs (Fig. 5). Fifty-five clinical samples exhibited significant object intensity increases, despite sample-to-sample variability, and were identified as infection positive (Fig. 5A, Supporting Information S7, Table S1), while the remaining 75 samples showed minimal object intensity changes and were classified as infection negative (Fig. 5B, Supporting Information S7, Table S1). For cross validation, the normalized integrated object intensity of each sample at 60 min and 90 min (Figs. 5C and 5D) were compared with the BD Phoenix gold standard method results obtained in the Microbiology Lab at Mayo Clinic, where the samples were collected. At 60 min, the integrated object intensity falls into two separated clusters: one cluster of samples with normalized integrated object intensities greater than 1 and one cluster of samples with normalized object intensities less than 1 (Fig. 5D). The two clusters were further separated with increased incubation times, indicating increasing UTI detection accuracy with time. By setting the infection threshold at 1.1 (Materials and Methods), 75 samples were determined to be infection negative and 55 samples were determined to be infection positive with accuracies that increased from ~90% at 60 min to ~91.5% at 90 min. At the 90 min time point, 11 false negative samples were determined from the 130 samples tested. The on-site parallel plating results shows that these false negative samples have initial bacterial concentrations between 103–104 CFU/mL (2 samples), 104–105 CFU/mL (7 samples), and >105 CFU/mL (2 samples) (Supporting Information S8).
Figure 5. Rapid infection detection with 130 clinical urine samples.
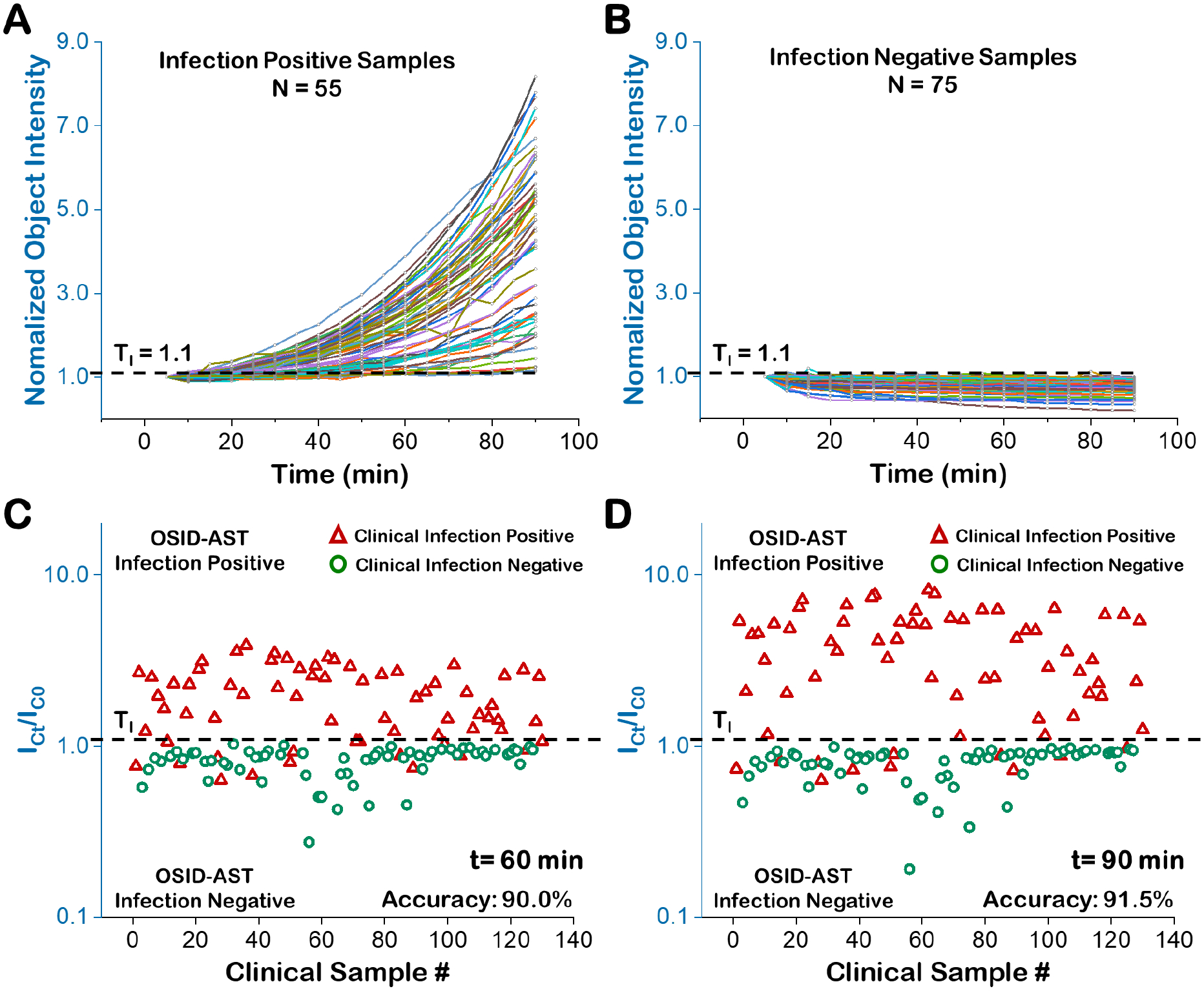
The normalized integrated object intensity processing results (90 min) of 55 infection positive clinical samples (A) and 75 infection negative clinical samples (B). The comparison of reference method (BD Phoenix, red triangles are infection positive and green circles are infection negative) and OSID-AST determinations of infection at 60 min (C) and 90 min (D) with infection threshold TI = 1.1 (black hatched line). ICt is the integrated object scattering intensity of samples without antibiotic at time t. IC0 is the initial intensity of the same sample.
The present LVSi system images a volume of 5 μL, allowing for sufficient numbers of bacterial cells from urine samples to be recorded at clinically relevant concentrations (104 – 107 CFU/mL). However, when the bacterial concentration is below 1000 cells/mL, less than 5 cells would be visible, and a longer detection time is required for AST (Supporting Information Figure S3). Among the 11 false negative samples, two samples yielded bacterial concentrations below the clinical threshold of 104 CFU/mL when validated with on-site plating. Samples are typically collected and stored at 4° C for 2 days before being received and processed on-site; we expect such reductions in viable bacteria to be negligible when OSID-AST is performed in POC settings with fresh urine. The remaining nine false negative samples exhibited initial bacterial concentrations above the clinical threshold, however 7 of the 9 had concentrations between 104–105 CFU/mL. Parallel plating validation, performed alongside OSID-AST after sample handling, including prewarm, filtration and dilution, revealed low concentrations of bacterial cells (below 1000 cells/mL, Supporting Information S9). Therefore, most of these false negative results (9/11) are due to low bacterial concentrations from sample handling. While these samples were diluted 10 to 1000 times for both single cell counting and object scattering intensity analysis, the OSID-AST method functions with a higher particle concentration range, thus these false negative results can likely be avoided with an optimized dilution scheme and improved sample handling process. In this work, we compared two analytical methods to rapidly determine infection, a single cell counting method and the OSID method. We found that measuring bacterial cell growth with integrated object scattering intensity to be more accurate with simpler processing requirements. While the single cell counting method resulted in 17 false negative samples of the 130 tested samples, the OSID method resulted in only 11 false negatives (Supporting Information S10). While single cell counting tracks the numerical increases only, OSID method measures both size and numerical increases during cell growth. It can be noted that cell enlargement, and therefore intensity, typically increases before a cell divides. This could account for some of the improved accuracy of the OSID method. Aside from accuracy, this method also allows automated data processing in real time with lower computational cost compared to that of the single cell counting method. Extra manual threshold selection for cell detection and tracking processing, prevent fully automated data processing for the single cell counting method and highlight the simplicity of sample processing for the OSID method. Additionally, for samples with high particle densities, cell counting is not accurate and requires an extra imaging and dilution determination steps for each sample. Increased dilution could result in a false negative infection determination when insufficient bacteria are present in the viewing volume. Furthermore, the counting method does not accurately enumerate bacterial cells that aggregated after division, such as with S. saprophyticus in UTIs, while the OSID method accounts for this type of cell enlargement and growth morphology (Supporting information S5).
Rapid AST:
We performed OSID-AST on 55 UTI positive clinical samples by comparing integrated object intensity change in antibiotic-exposed samples (ΔIABX) with that of the control samples (ΔIC) following the algorithm defined in Fig. 1C. When ΔIABX is consistently lower than ΔIC, the sample is determined to be susceptible to the antibiotic tested (Fig. 6A). In contrast, when ΔI ABX and ΔI C values are similar, the sample is identified as resistant to the antibiotic tested (Fig. 6B). To explore AST accuracy over time, we set the resistance threshold (TR) at 0.5, corresponding to 50% reduction in bacterial growth rate, and compared the results with those obtained from reference method (BD Phoenix) (Fig. 6C–D). With the 60-min detection, six clinically determined susceptible samples localized within the resistance zone or at the threshold, demonstrating a category accuracy of ~89% (Fig. 6C), while with the 90-min detection increased the category accuracy to 98% with 17 samples identified as resistant to ciprofloxacin and the remaining 38 identified as susceptible (Fig. 6D). These results were in 98% agreement with both BD Phoenix results from clinical microbiology testing and the on-site parallel AST plating validation, with one susceptible sample mis-categorized as resistant (Sample # 80, Supporting Information S11, Table S2). From clinical report, Sample #80 contains >100,000 CFU/mL Citrobacter freundii Complex, and the high magnification imaging (Insets of Figure S10 C & D) confirmed the elongation of this bacteria under 2 μg/mL ciprofloxacin. The elongation induced object intensity increase in OSID-AST system was similar to the control sample within the 90 min measurement time and longer detection time is needed for accurate AST in this case (Supporting Information S12).
Figure 6. Direct AST with infection positive clinical samples.
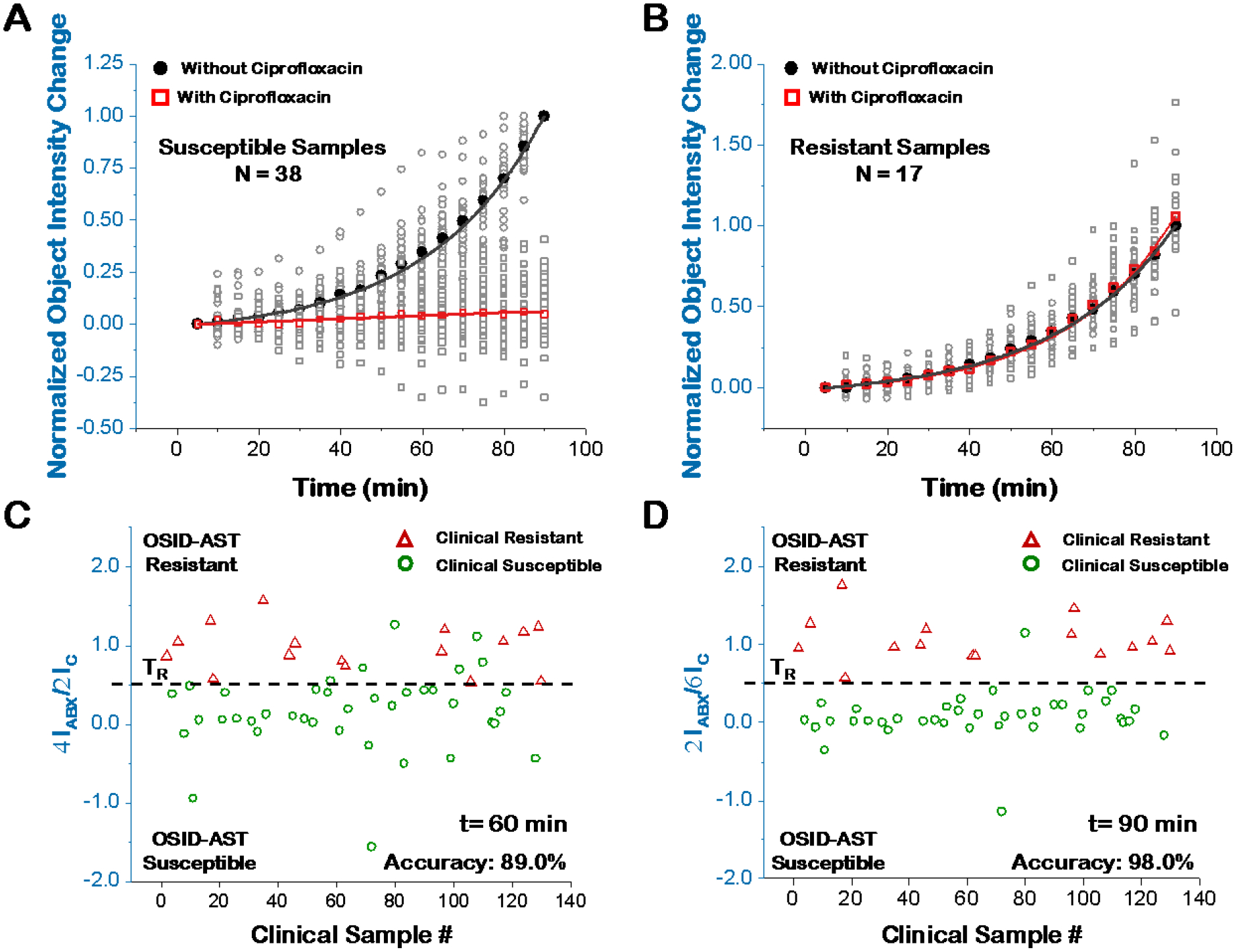
The normalized integrated object intensity change (normalized to the integrated object intensity of control sample at 90 min) over 90 min of 38 susceptible samples (A) and 17 resistant samples (B). Open grey circles/squares are individual sample values, while filled black dots and open red squares represent mean values. Comparison of reference method (BD Phoenix) and direct AST for susceptibility determination at 60 min (C) and 90 min (D). ΔIC = ICt -IC0 and ΔIABX = IABXt -IABX0, represent the integrated object scattering intensity change at time t for antibiotic-unexposed (control) and -exposed samples, respectively.
The current protocol (Figure S6), from sample collection to result, occurs over several days due to study and location constraints. Clinical samples are collected, refrigerated, and transported on ice to the lab. To mimic a fresh and warm urine sample, each sample was prewarmed for 30 min before testing. Next, to remove large particles, the sample was filtered then diluted with microbiological media to supply nutrients. To avoid oversaturation, a quick check of particle concentration is performed in LSVi after the initial dilution of the sample. Additional dilutions of the sample will be carried out if the particle concentration is above the detection limit. These simple steps averaged approximately 2–5 min for each sample. Therefore, the current total assay time for OSID-AST of a clinical sample includes 30 min sample prewarming, 2–5 min sample pre-treatment (filtration/dilution), and 60–90 min video-based object scattering detection. Ultimately, in a POC setting, the pre-warm step would not be necessary for fresh urine samples and the sample pre-treatment time could be reduced with an improved sample collection device that integrates a filter. Thus, the total time for OSID method of infection determination and AST time can be as short as ~1 hour with real-time growth curve determination.
Due to the throughput limit of current imaging setup, we focused on E. coli infection detection and AST with ciprofloxacin at the clinical breakpoint concentration (CLSI regulation) for proof-of-concept demonstration. The breakpoint concentration was above the minimum inhibitory concentration (MIC) of each antibiotic determined by OSID-AST (Supporting Information S13) for validated growth inhibition. Our future development plan includes improvement of detection throughput with multiplexed sample detections, and study of additional strains of bacteria and different types of antibiotics for complete coverage of UTI/AST diagnosis. Furthermore, the present work focuses on direct AST without specific bacterial strain identification, which differs from the current clinical practice. Our ongoing studies include bacterial identification using LVSi, but these investigations are beyond the scope of this paper. Nevertheless, by improving system throughput and integrating multiple phenotypic features (e.g. counts, intensity, and division), we anticipate this technique can be used for bacterial cell identification and multiplexed AST directly on the raw urine samples.
Conclusions
OSID-AST is a low-magnification, video-based object scattering imaging detection technique for rapid detection of bacterial infection and determination of antimicrobial susceptibility for clinical urine samples. To quantify the growth of the bacterial cells with high sensitivity within 60–90 min, accurate object scattering intensities of particles within the sample were obtained by removing the video background with simple spatial and temporal filters. The scattering intensities correlate with changes in both bacterial size and quantity and are more sensitive than bacterial cell counting alone with an increased dynamic range by two orders of magnitude. We first tested the method with pure cultures and achieved direct AST in 60 min. Then, we applied the technique to 130 clinical urine samples and accurately identified 91.5% of the clinically confirmed infection-positive samples in 90 min. We also performed AST on these patient samples with ciprofloxacin and achieved 98% categorical agreements with the clinical lab results within 90 min. Our technique can test clinical samples directly without overnight culturing and can detect object scattering intensities in real time for point-of-care AST. In summary, OSID-AST simplifies sample preparation and testing procedures, improves precision, and shortens the sample-to-result measurement time from days to under two hours, thereby promoting more judicious use of antibiotics, reducing the emergence of antibiotic resistance, and ultimately saving lives.
Materials and Methods
Materials.
E. coli ATCC 25922 and S. saprophyticus ATCC 15305 were purchased from American Type Culture Collection (ATCC) and stored at −80°C in 5% glycerol. Ciprofloxacin and nitrofurantoin were purchased from Sigma-Aldrich. The antibiotic powders were stored in the dark at 2 to 8°C.
Bacterial preparation.
E. coli and S. saprophyticus were grown overnight (~15 h) in Luria−Bertani (LB) broth (per liter: 10 g peptone 140, 5 g yeast extract, and 5 g sodium chloride) and Mueller Hinton Broth (MHB, per liter: 2.0 g beef infusion solids, 1.5 g starch, and 17.5 g casein hydrolysate) at 37°C and 150 rpm. E. coli and S. saprophyticus cultures were diluted in fresh LB broth or MHB, respectively, to a concentration ranging from 104 to 107 cells/mL. To mimic the real sample conditions and speed up AST, we worked directly with E. coli and S. saprophyticus stationary phase cultures in fresh culture medium. An antibiotic at the standard breakpoint concentration (32 μg/mL and 2 μg/mL for nitrofurantoin and ciprofloxacin, respectively) was added to one of two preparations. Each bacterial suspension (70 μL), one with and one without antibiotic, was transferred into a cuvette at 37 °C for imaging.
Clinical urine samples.
De-identified excess and residual clinical urine samples were obtained from the clinical microbiology laboratory at Mayo Clinic Hospital, Phoenix, Arizona (Approved by Mayo Clinic Biospecimen Subcommittee BIO00015462). Clinical urine samples were stored at 4°C and transported in an insulated box with ice packs. Prior to processing, urine samples were pre-warmed for 30 min at 37°C to mimic the temperature of fresh obtained urine samples in POC setting. The sample processing and testing workflow for these samples is illustrated in figure S6 with the following steps: The warmed samples are passed through a 5 μm syringe filter (MilliporeSigma, Burlington, MA) to remove large substances. Then, a 10-fold dilution with LB broth was performed for nutrient supplementary. To avoid the saturation of the initial scattering intensity, each sample was quick imaged and counted with LSVi for further dilution if needed. The final concentration of the diluted clinical samples ranges from 104 to 107 particles/mL with and without ciprofloxacin (2 μg/mL, final concentration). After mixing, diluted samples (70 μL) were transferred to cuvettes (Uvette, Eppendorf, Germany), and subjected to OSID-AST. A total of 130 urine samples were tested using both object intensity tracking and on-site parallel validating plating. Urine samples were prepared and transferred to researchers in a blinded fashion. Upon completion of weekly batches of samples, the OSID-AST and parallel plating results were compared with clinical microbiology culture results from the Mayo Clinic Hospital Microbiology Lab.
On-site initial plating and parallel plating validation.
Once clinical urine specimens were received on-site, aliquots were taken for serial dilution and plating on LB agar plates for enumeration of initial CFU/mL post-storage and transport.
The parallel validation plating was performed after prewarming, preprocessing, and supplementation with LB. Similarly, post-prepared samples were subjected to serial dilution for estimation of CFU/mL at the start of OSID-AST and after 90 min.
LVSi.
The dual channel large volume scattering imaging system (Fig. 1A) consists of two 800 mW, 780 nm infrared (IR) LEDs (M780LP1, Thorlabs, Inc., USA), each with collimating and focusing lens and a central blocking aperture to focus a ring-shaped illumination through the sample or the reference cuvettes. Wide-view and deep field depth scattering images were recorded by two CMOS camera (BFS-U3–16S2M-CS, Point Grey Research Inc., Canada) at 10 fps through two variable zoom lenses (NAVITAR 12X, Navitar, USA) with zoom factors set at 2.0X for the sample and reference cuvettes. The image volume was determined by the viewing size and focal depth of the optics. For the experiments described in this study, the viewing volume of 2.5 mm × 1.9 mm × 1.0 mm was equivalent to 4.8 μL at 2.0X magnifying power. The imaging system was enclosed in a thermally-isolated housing unit with a controlled temperature (37°C).
Biosafety.
All sample preparations and measurements were performed in biosafety level 2 (BSL2) laboratories following an IBC-approved BSL2 protocol.
Video Processing.
The automated image processing protocol to remove all background noises in the video has 4 steps (Fig. 1B, Supporting Information Figure S1): Step one, the raw video stacks (1 min duration, 10 fps) are averaged for every 4 frames to increase signal to noise ratio and to reduce data size. The size of local stack average is set to avoid cell motion induced blur. Step two, static and slow drifting background noises from cuvette defects scattering and cuvette wall reflection are removed with pixel level temporal local minimum subtraction for every 10 frames. Temporal local minimum is calculated by project the minimum intensity over time for a small stack for each pixel. The stack size is set with all bacterial cells moved to avoid signal lost. Step three, dynamic background noise caused by thermal and mechanical drift induced moving reflection and scattering is mostly removed by subtracting the whole stack temporal median image. Stack median is calculated by project the median intensity of each pixel for the entire video stacks. Step four, the remaining background is removed by subtracting the local spatial background calculated by rolling ball average with radius of 10 pixels for all pixels in the image. The radius should be set to at least the size of the largest object that is not part of the background. The typical computational time for processing a 1 min video is only ~ 45 seconds. After background removal, the video intensity is dominated by the object intensity of all particles including both bacterial cells and particles. The algorithms were implemented using ImageJ software.
Setting thresholds for data interpretation.
To determine the infection threshold, the results were evaluated using the receiver operating characteristic (ROC) curve constructed using It/I0 as a predictor. From the ROC curve for first 20 clinical samples (Supporting Information S2), of which 10 were positive and 10 were negative, we determined the best infection threshold at 1.1 with a sensitivity of 100% and a specificity of 100% at a 90-minute testing time. Therefore, the final threshold for infection identification (TI) was set as 1.1 for all samples. The resistance threshold (TR) was set to 0.5, corresponding to 50% growth inhibition in the antibiotic-exposed samples compared to the unexposed control preparation at 90 minutes.
Dynamic range of OSID-AST.
To determine the dynamic range of OSID-AST method, pure E. coli cultures with concentrations ranging from 103 to 107 CFU/mL were prepared and tested. E. coli was grown overnight (~15 h) in LB broth at 37°C and 150 rpm, and the bacterial concentration after serial dilutions (1–105) were calibrated and validated with overnight agar plating. The initial bacterial concentration was determined to be about 109 CFU/mL, and the diluted samples were calculated to be range from 104 to 109 CFU/mL with relevant dilution factors (Supporting Information Figure S4A). When the bacterial concentration exceeds 107 CFU/mL, the initial object scattering intensity saturates with current LVSi system. Therefore, the OSID-AST detection of samples ranging from 103 to 107 CFU/mL were performed and positive correlation was observed between AST time and E. coli concentration (Supporting Information S3 and S4B–C). At the low concentration of 103 CFU/mL, no obvious increase was detected within 90 min. At concentrations between 104 and 107 CFU/mL, OSID method works well with the total AST time decreasing with increasing cell concentrations. Thus, the detection range of object intensity method is between 104 and 107 CFU/mL, while the raw intensity detection only accurately detects growth with concentrations above 106 CFU/mL. In contrast, the single cell counting method needs lower concentrations, between 104 and 105 CFU/mL, to accurately enumerate increases in bacterial cells. Therefore, OSID-AST can accept a wider dynamic range of bacterial loads, which simplifies the sample preparation process while providing robust results.
Statistical analysis.
An unpaired two-sided student t-test was used to compare the group differences. A p value of <0.05 was considered as statistically significant.
Supplementary Material
ACKNOWLEDGMENT
The authors acknowledge the financial support from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01AI138993). The authors thank Noel De Lucia and Kathrine McAulay at Mayo Clinic Arizona for clinical sample coordination.
Footnotes
Supporting Information
Supporting Information Available: Background removal processing flow chart; ROC curve for infection threshold determination; OSID-AST with E. coli cultures at different concentrations; calibration curve between bacterial CFU concentrations and AST time; comparison of object intensity detection and single cell counting for E. coli and S. saprophyticus cultures; flow chart of the clinical sample preparation, testing and validation process; clinical urine sample ID results; initial sample validation results; initial and parallel plating validation result of 11 false negative samples; examples of false negative samples by single cell counting, positive by OSID; clinical urine sample AST results; clinical sample # 80; MIC determination for OSID-AST.
The authors declare no competing financial interest.
REFERENCES
- 1.CDC. Antibiotic Resistance Threats in the United States; Atlanta, GA: U.S., 2019. [Google Scholar]
- 2.Laxminarayan R; Duse A; Wattal C; Zaidi AK; Wertheim HF; Sumpradit N; Vlieghe E; Hara GL; Gould IM; Goossens H; Greko C; So AD; Bigdeli M; Tomson G; Woodhouse W; Ombaka E; Peralta AQ; Qamar FN; Mir F; Kariuki S; Bhutta ZA; Coates A; Bergstrom R; Wright GD; Brown ED; Cars O, Antibiotic resistance-the need for global solutions. Lancet Infect Dis 2013, 13 (12), 1057–98. [DOI] [PubMed] [Google Scholar]
- 3.Tacconelli E; Carrara E; Savoldi A; Harbarth S; Mendelson M; Monnet DL; Pulcini C; Kahlmeter G; Kluytmans J; Carmeli Y; Ouellette M; Outterson K; Patel J; Cavaleri M; Cox EM; Houchens CR; Grayson ML; Hansen P; Singh N; Theuretzbacher U; Magrini N; Group, W. H. O. P. P. L. W., Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 2018, 18 (3), 318–327. [DOI] [PubMed] [Google Scholar]
- 4.Dielubanza EJ; Schaeffer AJ, Urinary tract infections in women. Med Clin North Am 2011, 95 (1), 27–41. [DOI] [PubMed] [Google Scholar]
- 5.Foxman B, Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am 2014, 28 (1), 1–13. [DOI] [PubMed] [Google Scholar]
- 6.Barber AE; Norton JP; Spivak AM; Mulvey MA, Urinary tract infections: current and emerging management strategies. Clin Infect Dis 2013, 57 (5), 719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flores-Mireles AL; Walker JN; Caparon M; Hultgren SJ, Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 2015, 13 (5), 269–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu C; Bayer A; Cosgrove SE; Daum RS; Fridkin SK; Gorwitz RJ; Kaplan SL; Karchmer AW; Levine DP; Murray BE; M JR; Talan DA; Chambers HF, Clinical practice guidelines by the infectious diseases society of america for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children: executive summary. Clin Infect Dis 2011, 52 (3), 285–92. [DOI] [PubMed] [Google Scholar]
- 9.Muscedere J; Dodek P; Keenan S; Fowler R; Cook D; Heyland D; Committee VAPG; the Canadian Critical Care Trials, G., Comprehensive evidence-based clinical practice guidelines for ventilator-associated pneumonia: diagnosis and treatment. J Crit Care 2008, 23 (1), 138–47. [DOI] [PubMed] [Google Scholar]
- 10.Bhargava D; Deshpande A; Sreekumar K; Koneru G; Rastogi S, Guidelines of the Infectious Diseases Society of America for the Treatment of Methicillin-Resistant Staphylococcus aureus Infections: As Applied to Oral and Maxillofacial Clinical Practice. J Maxillofac Oral Surg 2013, 12 (3), 354–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van den Broek D; Keularts IM; Wielders JP; Kraaijenhagen RJ, Benefits of the iQ200 automated urine microscopy analyser in routine urinalysis. Clin Chem Lab Med 2008, 46 (11), 1635–40. [DOI] [PubMed] [Google Scholar]
- 12.Mori R; Yonemoto N; Fitzgerald A; Tullus K; Verrier-Jones K; Lakhanpaul M, Diagnostic performance of urine dipstick testing in children with suspected UTI: a systematic review of relationship with age and comparison with microscopy. Acta Paediatr 2010, 99 (4), 581–4. [DOI] [PubMed] [Google Scholar]
- 13.Bauer KA; Perez KK; Forrest GN; Goff DA, Review of rapid diagnostic tests used by antimicrobial stewardship programs. Clin Infect Dis 2014, 59 Suppl 3, S134–45. [DOI] [PubMed] [Google Scholar]
- 14.Khan ZA; Siddiqui MF; Park S, Current and Emerging Methods of Antibiotic Susceptibility Testing. Diagnostics 2019, 9 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leonard H; Colodner R; Halachmi S; Segal E, Recent Advances in the Race to Design a Rapid Diagnostic Test for Antimicrobial Resistance. ACS Sens 2018, 3 (11), 2202–2217. [DOI] [PubMed] [Google Scholar]
- 16.van Belkum A; Burnham CD; Rossen JWA; Mallard F; Rochas O; Dunne WM Jr., Innovative and rapid antimicrobial susceptibility testing systems. Nat Rev Microbiol 2020, 18 (5), 299–311. [DOI] [PubMed] [Google Scholar]
- 17.Vasala A; Hytonen VP; Laitinen OH, Modern Tools for Rapid Diagnostics of Antimicrobial Resistance. Front Cell Infect Microbiol 2020, 10, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fluit AC; Visser MR; Schmitz FJ, Molecular detection of antimicrobial resistance. Clin Microbiol Rev 2001, 14 (4), 836–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frye JG; Lindsey RL; Rondeau G; Porwollik S; Long F; McClelland M; Jackson CR; Englen MD; Meinersmann RJ; Berrang ME; Davis JA; Barrett JB; Turpin JB; Thitaram SN; Fedorka-Cray PJ, Development of a DNA microarray to detect antimicrobial resistance genes identified in the National Center for Biotechnology Information database. Microb Drug Resist 2010, 16 (1), 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park S; Zhang Y; Lin S; Wang TH; Yang S, Advances in microfluidic PCR for point-of-care infectious disease diagnostics. Biotechnol Adv 2011, 29 (6), 830–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Athamanolap P; Hsieh K; Chen LB; Yang S; Wang TH, Integrated Bacterial Identification and Antimicrobial Susceptibility Testing Using PCR and High-Resolution Melt. Anal Chem 2017, 89 (21), 11529–11536. [DOI] [PubMed] [Google Scholar]
- 22.Machowski EE; Kana BD, Genetic Mimetics of Mycobacterium tuberculosis and Methicillin-Resistant Staphylococcus aureus as Verification Standards for Molecular Diagnostics. Journal of Clinical Microbiology 2017, 55 (12), 3384–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shifman O; Steinberger-Levy I; Aloni-Grinstein R; Gur D; Aftalion M; Ron I; Mamroud E; Ber R; Rotem S, A Rapid Antimicrobial Susceptibility Test for Determining Yersinia pestis Susceptibility to Doxycycline by RT-PCR Quantification of RNA Markers. Front Microbiol 2019, 10, 754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Syal K; Iriya R; Yang Y; Yu H; Wang S; Haydel SE; Chen HY; Tao N, Antimicrobial Susceptibility Test with Plasmonic Imaging and Tracking of Single Bacterial Motions on Nanometer Scale. ACS Nano 2016, 10 (1), 845–52. [DOI] [PubMed] [Google Scholar]
- 25.Cermak N; Olcum S; Delgado FF; Wasserman SC; Payer KR; A Murakami M; Knudsen SM; Kimmerling RJ; Stevens MM; Kikuchi Y; Sandikci A; Ogawa M; Agache V; Baléras F; Weinstock DM; Manalis SR, High-throughput measurement of single-cell growth rates using serial microfluidic mass sensor arrays. Nat Biotechnol 2016, 34 (10), 1052–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pancholi P; Carroll KC; Buchan BW; Chan RC; Dhiman N; Ford B; Granato PA; Harrington AT; Hernandez DR; Humphries RM; Jindra MR; Ledeboer NA; Miller SA; Mochon AB; Morgan MA; Patel R; Schreckenberger PC; Stamper PD; Simner PJ; Tucci NE; Zimmerman C; Wolk DM, Multicenter Evaluation of the Accelerate PhenoTest BC Kit for Rapid Identification and Phenotypic Antimicrobial Susceptibility Testing Using Morphokinetic Cellular Analysis. Journal of Clinical Microbiology 2018, 56 (4), e01329–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lissandrello C; Inci F; Francom M; Paul MR; Demirci U; Ekinci KL, Nanomechanical motion of Escherichia coli adhered to a surface. Appl Phys Lett 2014, 105 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pantel A; Monier J; Lavigne J-P, Performance of the Accelerate Pheno™ system for identification and antimicrobial susceptibility testing of a panel of multidrug-resistant Gram-negative bacilli directly from positive blood cultures. Journal of Antimicrobial Chemotherapy 2018, 73 (6), 1546–1552. [DOI] [PubMed] [Google Scholar]
- 29.Choi J; Yoo J; Lee M; Kim EG; Lee JS; Lee S; Joo S; Song SH; Kim EC; Lee JC; Kim HC; Jung YG; Kwon S, A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci Transl Med 2014, 6 (267), 267ra174. [DOI] [PubMed] [Google Scholar]
- 30.Longo G; Alonso-Sarduy L; Rio LM; Bizzini A; Trampuz A; Notz J; Dietler G; Kasas S, Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nat Nanotechnol 2013, 8 (7), 522–526. [DOI] [PubMed] [Google Scholar]
- 31.Besant JD; Sargent EH; Kelley SO, Rapid electrochemical phenotypic profiling of antibiotic-resistant bacteria. Lab Chip 2015, 15 (13), 2799–807. [DOI] [PubMed] [Google Scholar]
- 32.Schoepp NG; Schlappi TS; Curtis MS; Butkovich SS; Miller S; Humphries RM; Ismagilov RF, Rapid pathogen-specific phenotypic antibiotic susceptibility testing using digital LAMP quantification in clinical samples. Science Translational Medicine 2017, 9 (410), eaal3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hassan F; Bushnell H; Taggart C; Gibbs C; Hiraki S; Formanek A; Gripka M; Selvarangan R, Evaluation of BacterioScan 216Dx in Comparison to Urinalysis as a Screening Tool for Diagnosis of Urinary Tract Infections in Children. J Clin Microbiol 2019, 57 (9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutton S, Measurement of microbial cells by optical density. Journal of Validation Technology 2011. Winter, 2011, p 46+. [Google Scholar]
- 35.Roberts AL; Joneja U; Villatoro T; Andris E; Boyle JA; Bondi J, Evaluation of the BacterioScan 216Dx for Standalone Preculture Screen of Preserved Urine Specimens in a Clinical Setting. Lab Med 2017, 49 (1), 35–40. [DOI] [PubMed] [Google Scholar]
- 36.Hayden RT; Clinton LK; Hewitt C; Koyamatsu T; Sun Y; Jamison G; Perkins R; Tang L; Pounds S; Bankowski MJ, Rapid Antimicrobial Susceptibility Testing Using Forward Laser Light Scatter Technology. Journal of Clinical Microbiology 2016, 54 (11), 2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boland L; Streel C; De Wolf H; Rodriguez H; Verroken A, Rapid antimicrobial susceptibility testing on positive blood cultures through an innovative light scattering technology: performances and turnaround time evaluation. BMC Infectious Diseases 2019, 19 (1), 989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosen DA; Hooton TM; Stamm WE; Humphrey PA; Hultgren SJ, Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med 2007, 4 (12), e329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mo MN; Yang YZ; Zhang FN; Jing WW; Iriya R; Popovich J; Wang SP; Grys T; Haydel SE; Tao NJ, Rapid Antimicrobial Susceptibility Testing of Patient Urine Samples Using Large Volume Free-Solution Light Scattering Microscopy. Anal Chem 2019, 91 (15), 10164–10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang F; Jiang J; McBride M; Yang Y; Mo M; Iriya R; Peterman J; Jing W; Grys T; Haydel SE; Tao N; Wang S, Direct Antimicrobial Susceptibility Testing on Clinical Urine Samples by Optical Tracking of Single Cell Division Events. Small (Weinheim an der Bergstrasse, Germany) 2020, e2004148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


