Abstract
Aims
Pathological cardiac remodelling is characterized by cardiomyocyte (CM) hypertrophy and fibroblast activation, which can ultimately lead to maladaptive hypertrophy and heart failure (HF). Genome-wide expression analysis on heart tissue has been instrumental for the identification of molecular mechanisms at play. However, these data were based on signals derived from all cardiac cell types. Here, we aimed for a more detailed view on molecular changes driving maladaptive CM hypertrophy to aid in the development of therapies to reverse pathological remodelling.
Methods and results
Utilizing CM-specific reporter mice exposed to pressure overload by transverse aortic banding and CM isolation by flow cytometry, we obtained gene expression profiles of hypertrophic CMs in the more immediate phase after stress, and CMs showing pathological hypertrophy. We identified subsets of genes differentially regulated and specific for either stage. Among the genes specifically up-regulated in the CMs during the maladaptive phase we found known stress markers, such as Nppb and Myh7, but additionally identified a set of genes with unknown roles in pathological hypertrophy, including the platelet isoform of phosphofructokinase (PFKP). Norepinephrine-angiotensin II treatment of cultured human CMs induced the secretion of N-terminal-pro-B-type natriuretic peptide (NT-pro-BNP) and recapitulated the up-regulation of these genes, indicating conservation of the up-regulation in failing CMs. Moreover, several genes induced during pathological hypertrophy were also found to be increased in human HF, with their expression positively correlating to the known stress markers NPPB and MYH7. Mechanistically, suppression of Pfkp in primary CMs attenuated stress-induced gene expression and hypertrophy, indicating that Pfkp is an important novel player in pathological remodelling of CMs.
Conclusion
Using CM-specific transcriptomic analysis, we identified novel genes induced during pathological hypertrophy that are relevant for human HF, and we show that PFKP is a conserved failure-induced gene that can modulate the CM stress response.
Keywords: Hypertrophy, Pathological remodelling, RNA sequencing, Cardiomyocyte, Heart failure, PFKP
1. Introduction
Heart failure (HF) is a major cause of morbidity and mortality, that can originate from numerous diseases such as myocardial infarction and hypertension.1 Independent of the aetiology, cardiac hypertrophy is an immediate response comprising compensatory alterations in size, shape, and function of the myocardium to restore cardiac output. However, if left untreated, it can progress into maladaptive alterations, including a decline in cardiomyocyte (CM) contractility, fibrosis, ventricular dilatation, chronic inflammation, and increased cellular apoptosis, which can ultimately lead to HF.2 While the remodelling process involves all cardiac cell types, CMs function as key determinants of cardiac health and disease.3 Within the CMs, stress induces changes in gene expression, including reactivation of the fetal gene programme, that can trigger cell growth resulting in the initial CM hypertrophy. Next, there is a cellular metabolic shift towards carbohydrates as energy substrate and a change in mitochondria-associated pathways to adapt to the increase in ATP demand.2,4 As a result, CMs progress towards failure, presenting a significant reduction of mitochondria biogenesis.5 At the same time, cardiac function progressively deteriorates, ultimately leading to HF.
Current HF therapies target neurohormonal activation to improve cardiac relaxation and energetics or are antiarrhythmic approaches. While important advances have been made in the treatment of HF, so far these have been mainly directed at symptom management and prevention of sudden cardiac death. To date, there is no cure available to stop disease progression. RNA sequencing on heart tissue has been instrumental for understanding the molecular mechanisms important during hypertrophy and failure.6 These studies have shown us the relevance of genes encoding factors such as myocyte enhancer factor 2 (MEF2), periostin, angiotensin, and insulin-like growth factor during the transition into HF.7,8 While insightful, most RNA sequencing efforts to date have been limited by the fact that the obtained signal was derived from tissue. The complex cell type composition dilutes out any cell type-specific signals and fails to provide information on cellular origin of the detected gene expression changes. An improved strategy for the discovery of new targets could be to decipher the changes occurring during the transition from hypertrophic towards failing CMs specifically. Identification of novel genes and pathways that play a role during CM failure combined with functional studies addressing the mechanisms by which these contribute to disease progression could further aid in the identification of novel druggable targets and form the basis for development of enhanced therapies.
Here, we set out to obtain genome-wide gene expression profiles from CMs during the early and maladaptive phase of cardiac remodelling. To do so we used a recently developed CM isolation method9 combined with an animal model of pressure overload to comprehensively examine gene expression changes. In doing so, we identified specific molecular signatures for hypertrophic CM and CMs showing pathological hypertrophy and found multiple novel genes that are unknown for their role in pathological remodelling. We further showed that these pathological genes are also up-regulated in human end-stage HF and in stressed human-induced pluripotent stem cell (hiPSC)-derived CMs. Amongst the failure-induced genes, we identified phosphofructokinase-platelet (PFKP) as a stress-induced glycolytic enzyme in CMs, and its in vitro inhibition blocked expression of the gene encoding natriuretic peptide B (Nppb) expression and hypertrophy. Together, these findings may set the basis for accelerating the field of molecular cardiology towards the identification of new therapeutic targets for HF.
2. Methods
Additional detailed methods are reported in the Supplementary material online.
2.1 Animals
Animal studies were performed according to the guidelines from Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes. Animal experiments were approved by the institutional guidelines and regulations of the Animal Welfare Committee of the Royal Netherlands Academy of Arts and Sciences (HI 13.2304, AVD8011002015250 16.2305/IVD366) and following the guide for the care and use of laboratory animals. To characterize the effects of pressure overload in time, wild-type C57Bl/6J mice were used. To genetically label CMs in mice, Rosa26-tdTomato reporter mice (R26RTdT)10 were crossed with Myh6-Cre transgenic mice (Myh6cre; a generous gift from Jeffery Molkentin, Cincinnati Children’s Hospital Medical Center). All mice were maintained on a C57Bl/6J background.
2.1.1 Transverse aortic banding surgery
Pressure overload was induced in 8- to 9-week-old male mice by transaortic banding (TAB). In brief, mice were injected subcutaneously with Buprenorphine (0.05–0.1 mg/kg) as analgesic at least 30 min pre-surgery to alleviate pain or distress. Mice were anaesthetized with a mixture of Fentanyl (0.05 mg/kg), Midazolam (5 mg/kg), DEX-medetomidine (0.125 mg/kg) via an IP injection and if necessary 1–2% isoflurane was added as maintenance anaesthetic. A tracheal tube was placed, connecting the mouse to a ventilator. After disinfection with iodine and 70% ethanol, skin was incised left of the midline to allow access to the first intercostal space. Pectoral muscles were retracted and the intercostal muscles cut caudal to the first rib. Retractors were placed to separate the thymus from the heart. A 6.0 silk suture was placed around the aorta between the first and the second branch. The suture was closed with a 26 G needle in between to have a standard size aorta opening. The needle was removed immediately after closing the suture. Pectoral muscles were replaced and the skin was closed with a wound clip. After surgery, the tracheal tubing was removed and the mice were placed on a nose cone with 100% oxygen until waking up. During the whole procedure and recovery period, the mice were placed on a 38°C heating pad. Buprenorphine (0.05–0.1 mg/kg) was again administered postoperatively and the next morning to constantly alleviate pain. After the surgery, Revertidine was given to antagonize the sedative effects.
2.1.2 Blood flow velocity
Blood flow velocity was assessed using pulsed wave (PW)-mode, assisted by Colour Doppler mode in aortic arch view. For flow measurement near the location of the aortic band, descending aorta settings were used. The mean value of at least nine cardiac cycles was used to determine the peak velocity for each animal. For pressure gradient across the constriction, the modified Bernoulli’s equation was used: pressure gradient = 4 × Vmax2.
2.2 Digestion of the mouse heart and cardiomyocyte sorting
For cardiac tissue collection, mice were euthanized by cervical dislocation. The chest was opened to expose the heart. The heart was removed, washed in ice-cold PBS and weighed. For protein/RNA analysis, the heart was snap-frozen in liquid nitrogen. For CM sorting, mice were euthanized and the heart was immediately perfused by gradual injection of 10 mL cold perfusion buffer [135 mM NaCl, 4 mM KCl, 1 mM MgCl2, 10 mM HEPES, 0.33 mM NaH2PO4, 10 mM glucose, 10 mM 2,3-butanedione monoxime (Sigma, St Louis, MO, USA), 5 mM taurine (Sigma), pH 7.2] into the left ventricle. After perfusing the heart, the ventricles were collected and washed in cold perfusion buffer. Tissue was gently minced using a scalpel and transferred into a glass vial with 1.5 mL of cold digestion buffer [0.5 mg/mL Liberase TL, 20 µg/mL DNase I, Hepes 1 M in Dulbecco's modified Eagle's medium (DMEM)]. Cells were dissociated at 37°C with gentle shaking (100 rpm) for 15 min. The obtained cell suspension was gently pipetted up and down (10×), passed through a 100 µm cell strainer and collected by centrifugation (6 min at 4°C at 300 g). The cell pellet was suspended in 2–3 mL of DMEM with 4′,6-Diamidine-2′-phenylindole (DAPI) 1:1000 for cell sorting using a FACS Aria SORP (BD bioscience).9 Living (DAPI negative), single cells were sorted based on forward scatter and side scatter area (FSC-Area and SSC-Area). CMs were identified by high tdTomato expression and 488 nm auto-fluorescence. We enriched for elongated CMs using FSC-Width thresholding. CMs were collected in TRIzol and frozen down at −80°C.
2.3 RNA isolation and quality control
To isolate RNA, we used TRIzol® reagent (Life Technologies, Carlsbad, CA, USA) and RNA quality was assessed using Agilent RNA 6000 Pico Kit according to the manufacturer’s instructions.
2.4 RNA sequencing of sorted CMs
For RNA sequencing of sorted CM populations, CELseq111 preparation was done using the MessageAMP II aRNA amplification kit (ThermoFisher Scientific, Waltham, MA, USA) until IVT step. Final library preparation was continued according to the CELseq2 manual. RNA-Seq data has been made publicly available through the NCBI Gene Expression Omnibus (GEO), GEO accession number GSE138299.
2.5 Quantitative real-time PCR
Total RNA (1 μg) was reverse transcribed using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). Real-time PCR was performed using iQ SYBRgreen kit and CFX96 real-time PCR detection system (BioRAD).
2.6 Histology, immunohistochemistry, and in situ hybridization
Mouse hearts were excised from euthanized mice, washed in cold PBS and fixed with 4% formalin at room temperature (RT) for 48 h, embedded in paraffin and sectioned at 4 μm. Haematoxylin and eosin (H&E) and picrosirius red (SR) staining were used following standard procedures. For immunohistochemistry, sections were put to heat-induced antigen retrieval and blocking with 1% BSA. The sections were incubated with specific primary antibodies overnight at 4°C. After washing with PBS, the sections were incubated with secondary antibodies and/or fluorescein isothiocyanate (FITC) labelled wheat germ agglutinin lectin (100 μg/mL, Sigma-Aldrich, L4895) for 1 h at RT, washed and sealed with a mounting medium containing DAPI (Vector Laboratories, Burlingame, CA, USA). Cultured CMs on cover-slips were washed with PBS and fixed with 4% formaldehyde at RT for 20 min. After washing with PBS, cover-slips were incubated with Blocking buffer with 1% Fish gelatin and incubated with specific primary antibody 25 min at RT. After washing with blocking buffer, the cover-slips were incubated with secondary antibodies for 25 min at RT, washed with MQ water and sealed with a mounting medium containing DAPI. In situ hybridization was performed as described before12 on 8 µm mouse heart paraffin sections of mounted on Starfrost slides. Primers used for partial coding sequence amplification can be found in Supplementary material online, Table S4. Amplified sequences were ligated into pSPT18 vectors (Roche).
2.7 Human heart samples
Collection and use of human heart tissue were approved by the medical ethics committee and scientific advisory board of the biobank of University Medical Center Utrecht, Utrecht, the Netherlands (protocol no. 12/387). The study met the criteria of the code of conduct for responsible use of human tissue in the Netherlands and were conform to the principles outlined in the Declaration of Helsinki. Written informed consent was obtained or in certain cases waived by the ethics committee when not possible due to death of the patient. We included tissue from the left ventricular free wall or septum from explanted hearts of patients with HF and left ventricular free wall of non-failing donor hearts that were not used for transplantation.
Human heart RNA was isolated using ISOLATE II RNA Mini Kit (Bioline) according to the manufacturers’ instructions with minor adjustments.
2.8 Human-induced pluripotent stem cell-derived cardiomyocytes
LUMC0099iCTRL04 hiPSCs were used to generate CMs using defined medium with timed addition of growth factors and small molecules as described previously.13 hiPSCs were plated at a density of 50 000 cell/cm2 3 days prior to differentiation to allow for attachment to the Geltrex (Gibco) coated cell-culture plates. To enrich for CMs, purification was done for 4 days using selection medium [RPMI without d-glucose (Life Technologies, Carlsbad, CA, USA), supplemented with 0.5 mg/mL Albumin, 0.2 mg/mL L-Ascorbic Acid 2-phophate, 4 µM Sodium DL-lactate/HEPES buffer (Sigma)]. Differentiation efficiency was determined by flow cytometry for cardiac troponin T (cTnT) expression, using an antibody directed to cTnT (1:1000 ab45932, Abcam). CMs were dissociated after selection and reseeded at a density of 400 000 cells/cm2 on Geltrex coated cell-culture plates.
2.8.1 hiPSC-CMs NE/angiotensin II stimulation
CMs were dissociated and seeded at a cell density of 1 million cells per well in a Geltrex coated six-well plate for RNA or protein analysis or with a cell density of 50 000 cells per well on a Geltrex coated cover-slip in a 24-well plate for immunofluorescence or calcium transient analysis. CMs were cultured in control medium or medium with 10 µM L-Norepinephrine (NE, Sigma-Aldrich) and 1 µM Angiotensin II (AngII, Sigma-Aldrich) to induce stress for 7 days. The medium was refreshed every 48 h and collected to measure secreted NT-pro-BNP levels using the NT-pro-BNP kit (R&D systems, USA) according to manufacturer’s instructions.
2.8.2 Intracellular calcium transients
hiPSC-CMs plated on glass coverslips were loaded for 15 min with Fluo-4 AM 0.2 mM (Invitrogen). Fluorescent signals were acquired using a custom build upright microscope. Line-scan images were acquired at the sampling rate of 1 ms per line for 10 seconds using a high-speed camera (Andor Zyla 4.2 plus sCMOS). Cells were recorded at 37°C in Tyrode’s solution. Following background subtractions, data were analysed with a custom-written programme within MATLAB (Peaks, T.P. de Boer).
2.9 Primary cardiomyocyte culture
Neonatal rat ventricular cardiomyocytes (NRCMs) were isolated by enzymatic dissociation of neonatal rat hearts and cultured as described previously.14 In short, hearts from 1- to 2-day-old rat pups were collected, the atria were removed and the ventricular cells were enzymatically dissociated with trypsin (Life Technologies, Carlsbad, CA, USA) in a water (37˚C) jacketed spinner flask 15 min for 6–8 rounds. The single-cell suspension was filtered through a double gauze swab and pre-plated for one hour, divided over 2–3 15 cm plates to remove debris and non-myocytes, respectively. CMs were then plated on 24-well plates with cover-slips (125 000 cells per well) for immunostainings or on six-well plates (1 million cells per well) for RNA or Protein analysis.
2.9.1 β-Adrenergic stimulation of NRCM
NRCMs were exposed to culture medium with or without addition of 10 µM final concentration of Phenylephrine (PE, Sigma-Aldrich) for 48 h to induce control or stress conditions.
2.9.2 Transfection of CMs with siRNA oligo duplexes
27-nucleotide siRNA duplexes were obtained from OriGene. Transfection was performed with 10 µmol of siRNA and Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. The siRNA control used was the Universal-scrambled negative control from OriGene.
2.10 Statistical analysis
Values are presented as mean ± standard error of the mean. Outliers were identified and excluded using a ROUT test (GraphPad, using Q = 1%). Statistical significance was evaluated using an unpaired t-test for comparisons between two groups or one-way analysis of variance with Sidak post hoc test for comparison of multiple groups. Analyses were performed using GraphPad Prism Version 8.0 software. *P < 0.05 and was considered statistically significant compared to control.
3. Results
3.1 Identifying the early and pathological hypertrophic states during cardiac remodelling
To define the gene expression profiles that mark hypertrophic and failing CMs, we used TAB in mice to model pressure overload-induced pathological hypertrophy. To establish the suitable timepoints for analysis, we collected molecular data for up to 8 weeks after surgery on wild-type mice (Supplementary material online, Figure S1A). The aortic pressure gradient was significantly increased at 6 and 8 weeks post-banding (Supplementary material online, Figure S1B). Histological and morphological analysis showed an increase in heart size [heart weight to tibia length (HW/TL)] and CM cross-sectional area (CSA) already 1 week after banding that was maintained for the duration of the study (Supplementary material online, Figure S1C–E, Table S1). Additionally, collagen deposition was significantly increased after 4 weeks of banding (Supplementary material online, Figure S1F). At a molecular level, cardiac stress markers natriuretic peptide A (Nppa)15 and beta myosin heavy chain (Myh7) increased after TAB, while alpha myosin heavy chain (Myh6) decreased.16 Collagen type III alpha 1 (Col3a1)17 showed an initial increase after TAB, while there was a pronounced reduction in the expression of the metabolic regulators PPARG coactivator 1 alpha and beta (Pgc1α and Pgc1β)5 after banding (Supplementary material online, Figure S1G). Based on the increased heart size and signs of increased CM size at 1 week post-TAB, we concluded this timepoint to resemble early phase hypertrophy. The additional increase in fibrotic tissue and increased aortic pressure gradient at 8 weeks post-TAB indicated that disease state had advanced towards pathological remodelling and maladaptive hypertrophy at this timepoint.
3.2 Using flow cytometry to isolate CMs
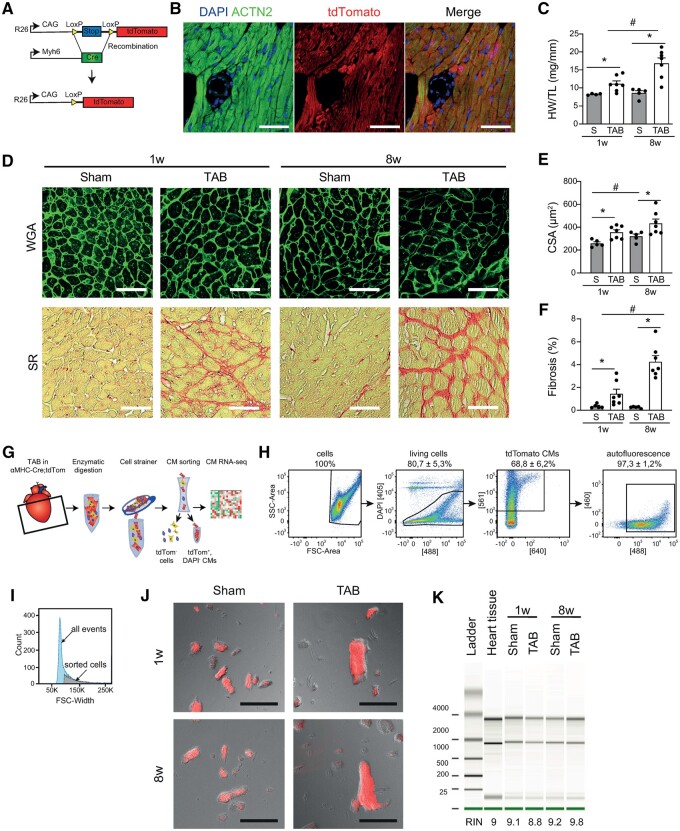
To genetically label CMs, we crossed Myh6-Cre transgenic mice18 with Rosa26-td-Tomato reporter mice (R26-lsl-tdTomato)10 (Figure1A andB). Mice were subjected to TAB or sham surgery and we collected heart tissue after either 1 or 8 weeks, after which we could confirm an increase in HW/TL ratio, CM size (CSA) and fibrosis (Figure 1C–F). We next used our optimized digestion protocol followed by FACS9 to collect CMs based on viability, tdTomato positivity, and sarcomere auto-fluorescence19 (Figure1G andH). To enrich for CMs, we selected cells with a higher forward scatter width (Figure 1I, Supplementary material online, Figure S2A and B). Microscopy confirmed the sorted cells were tdTomato positive, rod-shaped and striated cells, indicative for CMs (Figure 1J). RNA extracted from these cells appeared to be of good quality as indicated by RNA integrity number >8.0 (Figure 1K and Supplementary material online, Figure S2C and D).
Figure 1.
Sorting hypertrophic cardiomyocytes and cardiomyocytes showing signs of pathological remodelling. (A) Strategy for the generation of a CM-specific reporter mouse (Myh6-Cre-tdTomato). (B) Immunofluorescence indicating sarcomeric α-actinin (ACTN2; CMs) and tdTomato. Scale bar is 200 μm. (C) Heart weight to tibia length (HW/TL) ratios (n = 5–7). (D) Wheat germ agglutinin (WGA, upper row) and picrosirius red (SR, bottom row) staining of hearts 1 week or 8 weeks after transverse aortic banding (TAB) and corresponding sham controls. Scale bars are 200 µm. (E) Quantification of cross-sectional area (CSA) of CMs (≥50 cells per heart, n = 5–7). (F) Quantification of ventricular fibrosis (n = 5–7). (G) Schematic drawing of the CM isolation and sorting protocol. (H) Representative FACS plots showing the gating strategy to obtain tdTomato positive CMs. Selections are based on DAPI negativity, tdTomato positivity, and green auto-fluorescence. (I) Forward scatter (FSC)-width plot showing the fraction of sorted cells compared to all events. (J) Representative images of CMs after sort. (K) Bioanalyzer plot showing the RNA quality isolated from the sorted CMs 1 week or 8 weeks after TAB surgery and their respective sham controls. RIN, RNA Integrity Number. Data expressed as mean fold change ± SEM; *P < 0.05 compared to sham (S) in unpaired t-test; #P < 0.05 comparing sham 1 week to sham 8 weeks with unpaired t-test.
3.3 Hypertrophic and failing cardiomyocytes present a distinct gene expression profile
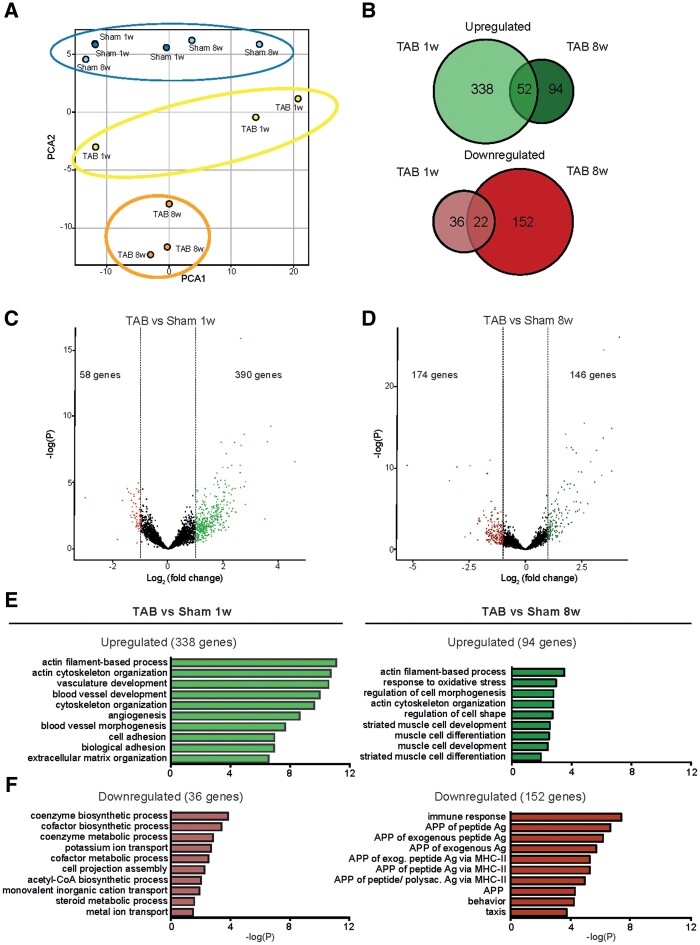
To comprehensively define the CM gene expression profiles in the compensatory hypertrophy state and in the pathological hypertrophy state, we performed RNA sequencing on sorted CM populations from hearts 1- and 8 weeks post-TAB and their respective sham controls. Principal component analysis (PCA) revealed high similarity between all sham transcriptomes, whereas CMs from TAB 1w animals, which will be termed ‘hypertrophic CMs’, and CMs from TAB 8w, which will be termed ‘pathological CMs’, had a clearly distinct gene expression pattern (Figure 2A). Moreover, component 2 of the PCA plot revealed higher similarity between control and hypertrophic CMs than with pathological CMs, indicating a progressive change in gene expression in response to stress. A total of 390 and 146 genes were up-regulated in hypertrophic and pathological CMs, respectively, with 52 overlapping genes between both conditions (Figure 2B–D). In contrast, 58 and 174 genes were down-regulated in hypertrophic and pathological CMs, respectively, with 22 genes down-regulated in both conditions (Figure 2B–D and Supplementary material online, Figure S3A–C). Functional annotation of differentially expressed genes revealed a strong enrichment of cytoskeleton organization and angiogenesis-related genes in the early hypertrophic state whereas failing CMs were enriched for genes involved in muscle cell morphology and development (Figure 2E). Down-regulated genes were involved in metabolic pathways and immune response, respectively (Figure 2F). Additionally, almost all mitochondrial genes were down-regulated in the failing CMs (Supplementary material online, Figure S4), indicating severe stress in this group of cells. Altogether, these results reinforce that hypertrophic and pathological CMs present a distinct transcriptomic profile, implicating different processes to be relevant during these different stages of disease.
Figure 2.
Bulk RNA sequencing analysis reveals differential gene regulation in hypertrophic and pathological CMs. (A) Principle component analysis (PCA) plot showing the distinct gene expression between groups. (B) Venn diagrams showing the intersection between significantly up-regulated genes (log2FC > 1), upper panel in green, and significantly down-regulated genes (log2FC < −1), lower panel in red, in CMs 1 week (TAB 1w) and 8 weeks (TAB 8w) after TAB when compared with corresponding control (sham 1w and sham 8w), respectively (n = 3 per group). (C) Volcano plot of all genes in TAB 1w samples compared to sham 1w. Significant up-regulated genes based on log2FC > 1; and significant down-regulated genes (red) based on log2FC < −1. (D) Volcano plot of all genes in TAB 8w samples compared to sham 8w. Significant up-regulated genes based on log2FC > 1; and significant down-regulated genes (red) based on log2FC < −1. (E) GO enrichment analysis for the up-regulated genes in CMs from TAB 1w and TAB 8w compared to corresponding control. (F) GO enrichment analysis for the down-regulated genes in CMs from TAB 1w and TAB 8w compared to corresponding control.
3.4 Identification of novel genes regulated in hypertrophic and failing CMs
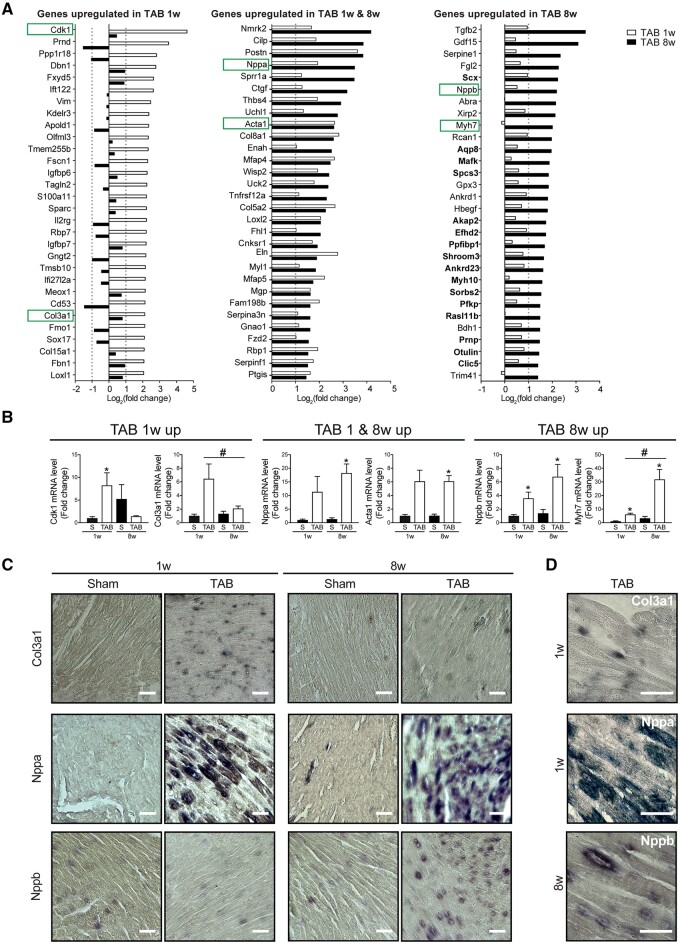
To explore genes that are key in driving maladaptive remodelling, we focused on the top 30 up-regulated genes in hypertrophic and/or pathological CMs (Figure 3A). Interestingly, genes specific to hypertrophic CMs (Figure 3A, left panel) presented a variable expression pattern during the pathological state. However, most of the genes specific to CM maladaptive remodelling (Figure 3A, right panel) were already showing some degree of up-regulation during hypertrophy, likely representing a gradual activation of this gene programme. Among the genes up-regulated in CM hypertrophy, we identified cyclin-dependent kinase 1 (Cdk1), a gene involved in cardiac hypertrophy.20 Interestingly, Col3a1, previously shown to be up-regulated in CMs,21,22 was also up-regulated in the hypertrophic cells. Since collagen genes are canonical fibroblast markers, we chose to confirm this finding in a single cell sequencing data set (Supplementary material online, Figure S5). Also in this dataset expression of Col3a1 increased five-fold after stress in CMs, indicating a possible relevant biological function during pathological remodelling (Supplementary material online, Figure S5C and D). Additionally, maladaptive genes included natriuretic peptide B (Nppb), the clinically used biomarker for HF,17 and Myh7, a known marker for diseased CMs.16
Figure 3.
Known cardiac stress markers are differentially regulated in hypertrophic and pathological CMs. (A) Top 30 differentially up-regulated genes in CMs from TAB 1w (left panel), TAB 1w and TAB 8w (middle panel), and TAB 8w (right panel) compared to corresponding control. (B) Real-time PCR on sorted CMs of differentially up-regulated cardiac stress markers. (C) In situ hybridization analysis for Col3a1 (upper panels), Nppa (middle panels), or Nppb (lower panels) mRNA. Scale bars represent 200 µm. (D) Larger inset of representative images in C showing CMs expressing Col3a1 (upper panel), Nppa (middle panel), or Nppb (lower panel), respectively. Scale bars represent 50 µm. Data are expressed as mean fold change ± SEM; *P < 0.05 compared to control (S) and #P < 0.05 compared to TAB1w in a one-way ANOVA or unpaired t-test (n = 3).
Real-time PCR on mRNA from sorted CMs confirmed the expression patterns indicated by RNA sequencing with up-regulation of Cdk1 and Col3a1 in hypertrophic CMs, and Nppb and Myh7 in failing CMs (Figure 3B). In addition, in situ hybridization confirmed Col3a1, Nppa, and Nppb mRNA expression in hypertrophic and/or pathological CMs (Figure3C andD).
3.5 Failure-induced genes are activated by stress in human CMs
In search for novel relevant players during maladaptive remodelling, we screened the top up-regulated genes during CM failure for genes that had not previously been functionally linked to hypertrophy and/or failure. In doing so we identified 16 candidate genes to further pursue (Figure 3A, right panel genes in bold).
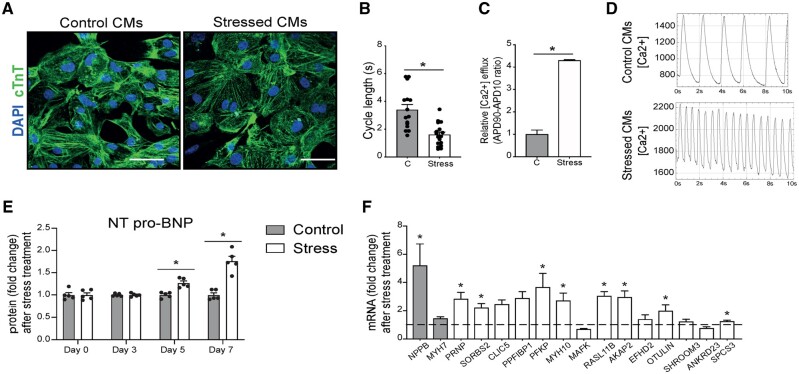
To investigate whether up-regulation of these genes in pathological CMs is conserved between species, we utilized human iPSC-CMs. To mimic pathological remodelling in a physiological manner, we subjected a highly pure CM population (Supplementary material online, Figure S6) to a 7-day-stress protocol with Norepinephrine (NE) and Angiotensin II (AngII).23,24 After stimulation, the morphology of the stressed CMs remained grossly normal compared to control (Figure 4A). However, NE/AngII treatment significantly reduced the calcium cycle length and induced faster calcium influx (Figure 4B–D), characteristics of a beta-adrenergic stimulation.17 Importantly, NE/AngII led to a strong induction of NT-pro-BNP protein secretion (Figure 4E), clinically used to detect and monitor HF in human patients.25NPPB mRNA levels were also highly induced, whereas MYH7 was not (Figure 4F), which is likely due to the relative immaturity of the hiPSC-CMs. Having established the stress response after stimulation, we next investigated the expression of the genes that were found to be up-regulated in failing mouse CMs and found eight of them to be significantly up-regulated in stressed hiPS-CMs and two were undetectable (Figure 4F). These data indicate that the failing gene expression programme is partly recapitulated in stressed hiPSC-CMs and suggest conservation of this response between mouse and human CMs.
Figure 4.
Genes induced in pathological CMs are increased in stressed human iPS-derived CMs. (A) Representative images of hiPS-CMs in control conditions or after NE and AngII treatment. Immunofluorescence for cardiac troponin T (cTnT) and nuclei DAPI. (B) Calcium transient analysis of the frequency of spontaneous calcium transients and (C) calcium transient analysis of the calcium release of control (C) or NE/Ang II treated (Stress) CMs. (D) Representative spontaneous calcium transients in control (upper panel) and stressed (lower panel) iPS-derived CMs. (E) Human NT-proBNP content in the supernatant of control and NE/AngII (stress) treated CMs determined by ELISA assay (n = 5). (F) Real-time PCR analysis of stress markers and the newly identified genes on NE-AngII treated CMs. Data are expressed as mean fold change ± SEM; *P < 0.05 compared to control (C) with unpaired t-test (n = 3–9) or in a two-way ANOVA.
3.6 Increased expression of the failure-induced genes in human end-stage HF
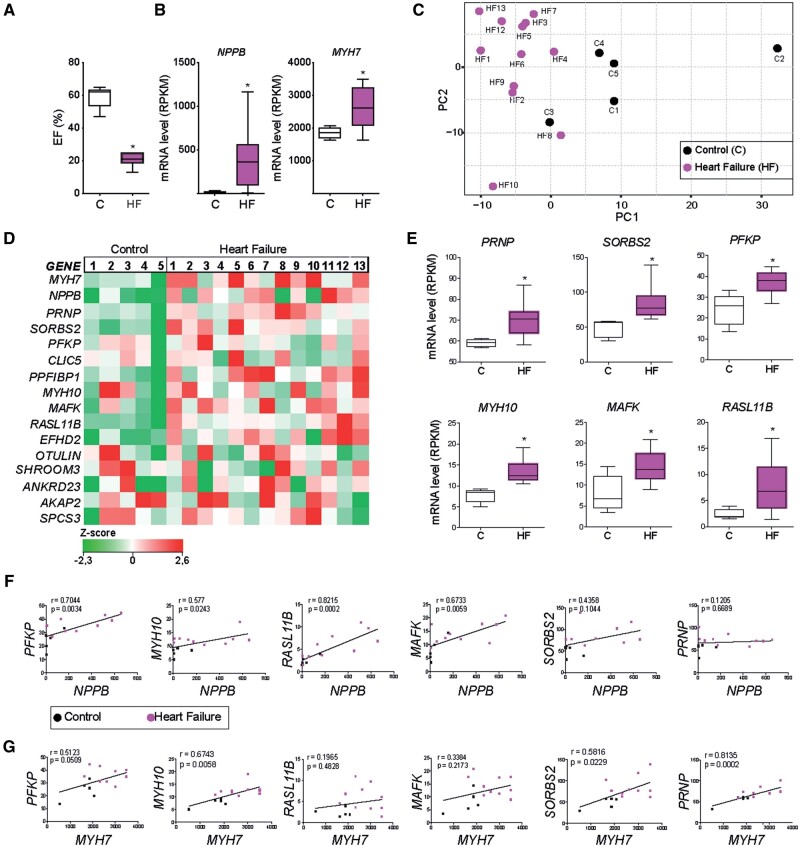
Next, we examined if the candidate genes induced during maladaptive remodelling were also differentially regulated in human failing hearts. To this end, we collected left ventricular biopsies from explanted hearts of patients presenting end-stage HF and non-failing donors. The selected patients presented a decline in cardiac function (Figure 5A), as well as an up-regulation of the failure markers NPPB and MYH7 (Figure 5B) in accordance with our findings in sorted failing mouse CMs (Figure 3A). RNA sequencing analysis of these hearts revealed transcriptomic clustering for most of the HF samples and a good separation between healthy control and patient samples (Figure 5C). The candidate genes induced during maladaptive remodelling presented heterogeneous expression levels between different patients, possibly due to different cell compositions of the tissue samples (Figure 5D). However, six candidate genes were significantly up-regulated in human HF (Figure 5E). These genes were the platelet isoform of phosphofructokinase (PFKP), prion protein (PRNP), sorbin and SH3 domain-containing protein 2 (SORBS2), myosin heavy chain 10 (MYH10), MAF BZIP transcription factor K (MAFK) and RAS like family 11 member B (RASL11b). Notably, except for MAFK, all these genes were also up-regulated in stressed hiPSC-CM (Figure 4F). To further examine the association of these genes with HF, we performed correlation analysis to NPPB and MYH7 expression. NPPB expression was positively correlated to PFKP, RASL11B, MAFK, and MYH10 (Figure 5F), while MYH7 was positively correlated to SORBS2, PRNP, and MYH10 (Figure 5G). Altogether, these results demonstrate that the induction of PFKP, PRNP, SORBS2, MYH10, MAFK, and RASL11B in CMs showing pathological hypertrophy is conserved between human and mouse and correlates to the expression of known markers of cardiac failure.
Figure 5.
Genes induced in pathological CMs are increased during human heart failure. (A) Average percentage ejection fraction (EF %) in non-failing controls (C) and heart failure patients (HF). (B) Expression analysis of cardiac failure markers NPPB and MYH7 from control and diseased hearts. RPKM, reads per kilobase million. (C) Unsupervised principal component analysis (PCA) plot. Each dot represents an expression profile of an individual sample plotted by PCA score showing diseased hearts cluster together and are distinct from controls. (D) Gene expression map representing the expression of MYH7 and NPPB and 14 novel genes identified in the failing CMs, in control and disease hearts. (E) RPKM levels of six novel genes showing a significant up-regulation in disease hearts (HF) when compared with control (C). (F) Correlation between the cardiac failure marker NPPB and the six significantly up-regulated genes identified in E. (G) Correlation between the cardiac failure marker MYH7 and the six significantly up-regulated genes identified in E. Spearman r values and P-values are shown in the graphs. Data are expressed as mean ± SEM or mean fold change ± SEM; *P < 0.05 compared to control in an unpaired t-test (n = 5–13).
3.7 PFKP protein is up-regulated in human failing hearts and involved in cardiomyocyte remodelling
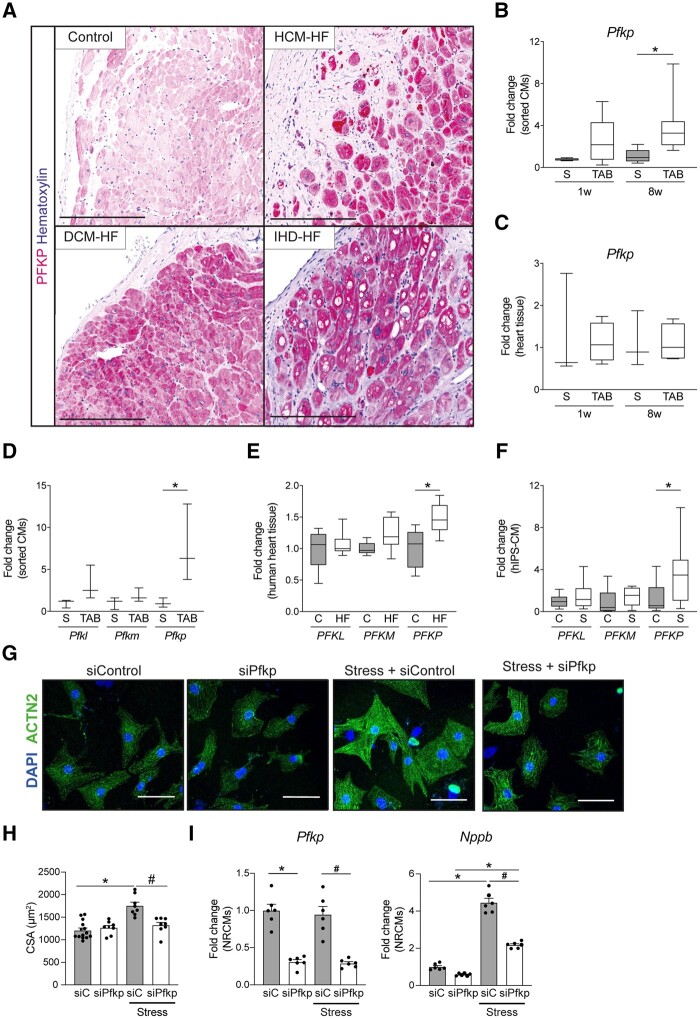
As PFKP is a rate-limiting enzyme of glycolysis,26 and there being a shift in metabolism during HF, we wanted to investigate the relevance of PFKP during CM failure in greater detail. To validate PFKP expression at protein level, we performed immunohistochemistry in human failing hearts. PFKP expression was induced in a subset of CMs in explanted hearts from patients with dilated cardiomyopathy, hypertrophic cardiomyopathy, and ischaemic heart disease (Figure 6A). Next, to further validate the RNA sequencing results of Pfkp expression on mouse isolated CMs, we performed real-time PCR analysis. We could confirm a higher Pfkp mRNA level in CMs showing pathological remodelling compared to sham, whereas this was not observed in hypertrophic CMs compared to sham (Figure 6B). Interestingly, no differential expression was found in the real-time results on full mouse heart tissue (Figure 6C), probably due to the high expression of this isoform in other cell types of the heart.
Figure 6.
PFKP is expressed in human failing CMs. (A) Immunohistochemistry of PFKP in healthy and HF dilated cardiomyopathy (DCM-HF), hypertrophic cardiomyopathy (HCM-HF), and ischaemic heart disease (IHD-HF). (B) Real-time PCR of PFKP on mouse sorted CMs (n = 3) on sham (S) or TAB conditions. (C) Real-time PCR of PFKP on mouse heart tissue (n = 4–7) on sham (S) or TAB conditions. (D) Expression analysis of PFK isoforms PFKM (phosphofructokinase-muscle) and PFKL (phosphofructokinase-liver) from bulk RNA sequencing of mouse sorted CMs 8w post TAB (n = 3). (E) Expression analysis of PFK isoforms PFKM (phosphofructokinase-muscle) and PFKL (phosphofructokinase-liver) from RNA sequencing of human failing hearts (n = 5–13). (F) Real-time PCR of PFK- isoforms on NE/AngII treated hiPS-derived CMs (n = 6–9). (G) Representative images of control or PE-treated NRCMs transfected with scramble siRNA control or Pfkp siRNA. Immunofluorescence for sarcomeric α actinin (ACTN2) and nuclei DAPI. (H) Cardiomyocyte cross-sectional area (CSA) quantification (n = >120), and (I) real-time PCR analysis of the cardiac stress markers Nppb and Pfkp of control or PE-treated NRCMs transfected with Pfkp siRNA (siPFKP) or scrambled siRNA control (si-C) (n = 6). (B–F) Data are expressed as average fold change with box (25–75 percentile) and whiskers (min–max); *P < 0.05 in one way ANOVA with Sidak post hoc test. (H and I) Data are expressed as mean fold change ± SEM; *P < 0.05 compared to control (C) or sham (S) and #P < 0.05 compared to siRNA control-stressed (si-C-Stress) in a one-way ANOVA or unpaired t-test.
In addition to the platelet isoform, there are two other known PFK isoforms in mammals: the muscle isoform PFKM, and the liver isoform PFKL.26 To determine if the differential regulation in failing CMs is specific for the platelet isoform, we analysed the expression of the liver and muscle isoforms in mouse and human samples. RNA-Seq analysis indicated that Pfkm and Pfkl were not differentially regulated in failing mouse CMs when compared to sham (Figure 6D). Moreover, PFKP was significantly up-regulated in human failing hearts compared to control, whereas the other isoforms PFKM and PFKL were not significantly changed (Figure 6E). Similarly, PFKP was also the only isoform up-regulated in hiPS-CMs after NE/AngII treatment (Figure 6F). Taken together, these results provide strong support for a conserved mechanism that leads to the up-regulation of the platelet isoform of PFK in response to pathological remodelling in CM.
To investigate whether PFKP is functionally involved in CM failure, we used NRCMs as they are a well-established model to induce hypertrophy upon pharmacological stimulation and also present efficient gene silencing results.27,28 To induce stress in NRCMs, we exposed the cells with 10 µm Phenylephrine (PE). This led to an increase in CM cell size (Figure6G andH) and induction of the failure marker Nppb (Figure 6I), indicating a strong stress response. Next, Pfkp gene silencing using small interfering RNAs (siRNA) efficiently reduced Pfkp mRNA expression with more than 70% in control and stress conditions (Figure 6I). Pfkp inhibition blocked the hypertrophic response and Nppb expression (Figure6H andI), indicating a functional link between Pfkp up-regulation and CM stress. Collectively, we identified PFKP as the stress-induced PFK isoform that may reinforce the metabolic switch towards glycolysis during pathological remodelling of CM and its suppression may be beneficial for blocking the progression towards cardiomyocyte failure.
4. Discussion
In this study, we identified gene programmes specific to hypertrophic CMs and CMs showing signs of pathological remodelling. Our data demonstrate that using flow sorting and deep sequencing of healthy and diseased CM populations allows for the identification of known and novel marker genes for disease, which would not be necessarily revealed by RNA sequencing on tissue. The relevance of the novel genes for pathological CMs is supported by the observations that (i) the majority of genes identified in pathological mouse CMs was also up-regulated in failing human CMs, (ii) multiple candidate genes are found to be up-regulated in human HF samples, and (iii) expression of these genes positively correlated to known markers of cardiac failure, like NPPB and MYH7, in human failing hearts. Using this approach, we were able to identify and confirm PFKP as the failure-induced isoform of PFK. Furthermore, inhibition of Pfkp blocked CM remodelling and Nppb expression, suggesting that PFKP is involved in the CM stress response.
We show that hypertrophic CMs and CMs showing signs of pathological remodelling differ widely in gene expression with more up-regulated genes in the hypertrophic cells and more down-regulated genes in the more diseased CMs. These results imply an active response of many pathways during the primary, compensatory response to overcome the initial stress and support cell growth. This is consistent with a previous study of single CM sequencing showing an increase of regulated genes during hypertrophy.29 Additionally, the genes that are significantly induced during the maladaptive phase, already show an early, less pronounced activation during the hypertrophic stage. This suggests that the pathological gene programme is already slightly activated during the compensatory phase, and becomes more pronounced during failure, but is initially still overridden by the early hypertrophic gene programme. Based on our results, we propose that these failure-induced genes are potential therapeutic targets, as inhibition strategies at earlier phases could block or reduce disease progression. In the failing CMs we also found almost all mitochondrial genes down-regulated, corroborating transcriptional regulation of the decline in mitochondrial biogenesis during pathological remodelling.5
Another observation that our CM-specific sequencing data allowed us to make, is that stress markers that have been linked to other cell types, like Col3a1, are also expressed in CMs, but at a significant lower level when compared with fibroblasts. The function of Col3a1 expression in CMs has not been studied, however, its up-regulation is in accordance with studies showing that multiple ECM genes expressed in fibroblasts are also expressed in CMs and regulated during stress.21,22 Furthermore, it was recently shown that fibrosis-like conditions can induce collagen gene expression in human iPS-CMs.30 Taken together, these observations further reinforce the relevance of cell-specific sequencing to identify meaningful factors in disease development.
Our study identified five new genes that could be linked to pathological remodelling in both mouse and human CMs. While they might have a known cardiac role, they have so far not been functionally linked to pathological hypertrophy. For instance, structural protein MYH10 is the only non-muscle myosin expressed in the heart.31 MYH10 is required for normal heart development, as loss of MYH10 in CMs during embryogenesis led to HF.31 However, the role of MYH10 in adult CMs has not been reported, and the relevance of its up-regulation in HF requires further functional studies. Another structural protein is SORBS2. It is considered an adapter protein involved in cytoskeleton organization, myofibril assembly and z-band signalling and it is highly expressed in CMs.32 Its up-regulation in pathological hypertrophy might be involved with the structural changes occurring in failing CMs, but this for now remains speculative. Moreover, RASL11B is a small GTPase protein with pro-apoptotic functions in cancer cells.33 No role has been reported in CMs but we could hypothesize a similar function considering it is overexpressed in myocytes that might be at risk of apoptosis. PRNP is a membrane glycoprotein associated to pathogenesis on muscle and nervous system when overexpressed.34 PRNP overexpression in mice led to muscle degradation with aging due to autophagy activation.34 These results suggest a detrimental effect of PRNP induction in failing CMs. Finally, PFKP is the platelet isoform of PFK, a glycolytic enzyme catalyzing one of the key regulatory and rate-limiting steps of glycolysis.
PFKP was selected for further study due to its function as a limiting enzyme in glycolysis. The failing heart is characterized by an increase in glucose uptake and glycolytic rates but not by an increase in glucose oxidation.4PFK activity is increased, enhancing the glucose flux into glycolysis and forming pyruvate and lactate which at elevated levels lead to contractile dysfunction.4 In a meta-analysis of human, mouse, rat, and dog microarray datasets, PFKP was found associated with HF.35 Those findings are in line with our finding that PFKP induction in failing CMs is conserved between mouse and humans. Recent studies in cancer have linked the up-regulation of PFKP with increased glycolysis and tumourigenesis of the cancer cells by the activation of AKT.36 This suggests that the up-regulation of PFKP in cancer cells, as in CMs, is stressed-induced and may have an important role in disease development. Accordingly, our data indicate that PFKP inhibition reduces stress responses in pathological CMs. For future targeted therapies, it is relevant to identify the molecular mechanisms behind PFKP overexpression in failing CMs along with the impact of PFKP inhibition on HF development.
While follow up analysis indicates the validity of our findings, using flow cytometry to isolate CMs may come with some limitations. There is a variation between sorts and the possibility that the sorted fraction is influenced by disease changing cell morphology and composition. For these reasons, it is key that findings are confirmed by additional studies, like the cell-based assays and immunohistochemistry used in our studies. In addition, to reduce age-related changes, we included proper sham controls at each timepoint.
To model aspects of CM failure in vitro we used NE/AngII to stress hiPS-CMs, which impaired calcium transients and induced expression of NPPB. However, the relatively immature phenotype of hiPSC-CMs makes it difficult to assess how well this resembles a truly failing CM. The same issue is partially true for neonatal CMs, although to a lesser extent. Advances in tissue engineering, maturation and disease modelling will help for improving these cell-based assays to better mimic the adult diseased conditions.
In summary, our study provides a resource for CM-specific genes related to hypertrophy pathological remodelling that might be relevant for the different stages of disease. Our data also show that PFKP is functionally involved in CM failure and remodelling. Using these data for follow-up functional studies might further reveal biological implications of the newly defined genes in the process of CM hypertrophy or maladaptive remodelling and may aid in the development novel therapeutic strategies aimed to reverse cardiac remodelling and reduce HF.
Data accessibility
RNA-Seq data has been made publicly available through the NCBI Gene Expression Omnibus (GEO), GEO accession number GSE138299.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
Conception and design of the research: M.V.G., C.D., M.G., T.V., C.B., E.V.R.; acquisition of data: M.V.G., C.D., D.V., H.R., I.P., L.K., F.A., A.V., M.H., A.B.; analysis and interpretation of the data: M.V.G., C.D., J.E., A.V., M.H., A.B., T.V., C.B., E.V.R.; statistical analysis: M.V.G., C.D., J.E., M.H.; supervising the experiments: M.G., T.V., C.B., E.V.R.; drafting the manuscript: M.V.G., C.D., C.B., E.V.R.; critical revision of the manuscript for important intellectual content: J.E., F.A., A.V., M.H., T.V.
Translational perspective
Maladaptive cardiac remodelling is a consequence of pathological hypertrophy which includes cardiomyocytes changes and a decline in contractility. Our cardiomyocyte-specific gene expression studies revealed a gene programme specific for pathological hypertrophy that is conserved in diseased mouse and human cardiomyocytes. We identified PFKP as a novel gene actively involved in cardiomyocyte remodelling, indicating PFKP as a potential therapeutic target to block the progression of heart failure.
Supplementary Material
Acknowledgements
We would like to thank Anko de Graaff and the Hubrecht Imaging Center for supporting the imaging. We gratefully acknowledge Jeroen Korving for help with histology. We thank Stefan van der Elst and the Hubrecht FACS facility for help with sorting cells and Judith Vivié for help with library preparation. We thank Petra van der Kraak, Joyce van Kuik, and Erica Sierra-de Koning for technical support with the immunostainings of the human tissue. We thank Michal Mokry (Epigenomic facility UMC Utrecht) and the Utrecht Sequencing Facility for support in processing human RNA-seq data.
Conflict of interest: none declared.
Time for primary review: 25 days
Funding
This work was supported by the Leducq Foundation (14CVD04) and the European Research Council under the European Union's Seventh Framework Program (ERC Grant Agreement CoG 615708 MICARUS), and the Dutch Cardiovascular Alliance (DCVA), an initiative with support of the Dutch Heart Foundation, DCVA2014-27 CVON-REMAIN and DCVA2017-18 CVON-PRIME to E.v.R.; UCL Hospitals National Institute of Health Research Biomedical Research Centre to F.A.; and the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 751988 to C.J.B.
References
- 1. Braunwald E. The war against heart failure: the Lancet lecture. Lancet 2015;385:812–824. [DOI] [PubMed] [Google Scholar]
- 2. Burchfield JS, Xie M, Hill JA.. Pathological ventricular remodeling: mechanisms: part 1 of 2. Circulation 2013;128:388–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fountoulaki K, Dagres N, Iliodromitis EK.. Cellular Communications in the Heart. Card Fail Rev 2015;1:64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bertero E, Maack C.. Metabolic remodelling in heart failure. Nat Rev Cardiol 2018;15:457–470. [DOI] [PubMed] [Google Scholar]
- 5. Rosca MG, Hoppel CL.. Mitochondria in heart failure. Cardiovasc Res 2010;88:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu Y, Morley M, Brandimarto J, Hannenhalli S, Hu Y, Ashley EA, Tang WH, Moravec CS, Margulies KB, Cappola TP, Li M; MAGNet consortium. RNA-Seq identifies novel myocardial gene expression signatures of heart failure. Genomics 2015;105:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. el Azzouzi H, van Oort RJ, van der Nagel R, Sluiter W, Bergmann MW, De Windt LJ.. MEF2 transcriptional activity maintains mitochondrial adaptation in cardiac pressure overload. Eur J Heart Fail 2010;12:4–12. [DOI] [PubMed] [Google Scholar]
- 8. Koentges C, Pepin ME, Musse C, Pfeil K, Alvarez SVV, Hoppe N, Hoffmann MM, Odening KE, Sossalla S, Zirlik A, Hein L, Bode C, Wende AR, Bugger H.. Gene expression analysis to identify mechanisms underlying heart failure susceptibility in mice and humans. Basic Res Cardiol 2018;113:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gladka MM, Molenaar B, de Ruiter H, van der Elst S, Tsui H, Versteeg D, Lacraz GPA, Huibers MMH, van Oudenaarden A, van Rooij E.. Single-cell sequencing of the healthy and diseased heart reveals cytoskeleton-associated protein 4 as a new modulator of fibroblasts activation. Circulation 2018;138:166–180. [DOI] [PubMed] [Google Scholar]
- 10. Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng HA.. robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 2010;13:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hashimshony T, Senderovich N, Avital G, Klochendler A, de Leeuw Y, Anavy L, Gennert D, Li S, Livak KJ, Rozenblatt-Rosen O, Dor Y, Regev A, Yanai I.. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol 2016;17:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lacraz GPA, Junker JP, Gladka MM, Molenaar B, Scholman KT, Vigil-Garcia M, Versteeg D, de Ruiter H, Vermunt MW, Creyghton MP, Huibers MMH, de Jonge N, van Oudenaarden A, van Rooij E.. Tomo-Seq identifies SOX9 as a key regulator of cardiac fibrosis during ischemic injury. Circulation 2017;136:1396–1409. [DOI] [PubMed] [Google Scholar]
- 13. Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, Plews JR, Abilez OJ, Cui B, Gold JD, Wu JC.. Chemically defined generation of human cardiomyocytes. Nat Methods 2014;11:855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Rooij E, Fielitz J, Sutherland LB, Thijssen VL, Crijns HJ, Dimaio MJ, Shelton J, De Windt LJ, Hill JA, Olson EN.. Myocyte enhancer factor 2 and class II histone deacetylases control a gender-specific pathway of cardioprotection mediated by the estrogen receptor. Circ Res 2010;106:155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gohar A, Rutten FH, Ruijter H, Kelder JC, Haehling S, Anker SD, Möckel M, Hoes AW.. Mid-regional pro-atrial natriuretic peptide for the early detection of non-acute heart failure. Eur J Heart Fail 2019;21:1219–1227. [DOI] [PubMed] [Google Scholar]
- 16. Morkin E. Regulation of myosin heavy chain genes in the heart. Circulation 1993;87:1451–1460. [DOI] [PubMed] [Google Scholar]
- 17. Harvey PA, Leinwand LA.. The cell biology of disease: cellular mechanisms of cardiomyopathy. J Cell Biol 2011;194:355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Agah R, Frenkel PA, French BA, Michael LH, Overbeek PA, Schneider MD.. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest 1997;100:169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chorvat D Jr, Kirchnerova J, Cagalinec M, Smolka J, Mateasik A, Chorvatova A.. Spectral unmixing of flavin autofluorescence components in cardiac myocytes. Biophys J 2005;89:L55–L57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tamamori M, Ito H, Hiroe M, Terada Y, Marumo F, Ikeda MA.. Essential roles for G1 cyclin-dependent kinase activity in development of cardiomyocyte hypertrophy. Am J Physiol 1998;275:H2036–H2040. [DOI] [PubMed] [Google Scholar]
- 21. DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, Hinson JT, Homsy J, Gray J, Pu W, Bruneau BG, Seidman JG, Seidman CE.. Single-cell resolution of temporal gene expression during heart development. Dev Cell 2016;39:480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cui Y, Zheng Y, Liu X, Yan L, Fan X, Yong J, Hu Y, Dong J, Li Q, Wu X, Gao S, Li J, Wen L, Qiao J, Tang F.. Single-cell transcriptome analysis maps the developmental track of the human heart. Cell Rep 2019;26:1934–1950. [DOI] [PubMed] [Google Scholar]
- 23. Eisenhofer G, Friberg P, Rundqvist B, Quyyumi AA, Lambert G, Kaye DM, Kopin IJ, Goldstein DS, Esler MD.. Cardiac sympathetic nerve function in congestive heart failure. Circulation 1996;93:1667–1676. [DOI] [PubMed] [Google Scholar]
- 24. Chinnaiyan KM, Alexander D, McCullough PA.. Role of angiotensin II in the evolution of diastolic heart failure. J Clin Hypertens (Greenwich) 2005;7:740–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Verdu JM, Comin-Colet J, Domingo M, Lupon J, Gomez M, Molina L, Casacuberta JM, Munoz MA, Mena A, Bruguera-Cortada J.. Rapid point-of-care NT-proBNP optimal cut-off point for heart failure diagnosis in primary care. Rev Esp Cardiol (Engl Ed) 2012;65:613–619. [DOI] [PubMed] [Google Scholar]
- 26. Wang J, Zhang P, Zhong J, Tan M, Ge J, Tao L, Li Y, Zhu Y, Wu L, Qiu J, Tong X.. The platelet isoform of phosphofructokinase contributes to metabolic reprogramming and maintains cell proliferation in clear cell renal cell carcinoma. Oncotarget 2016;7:27142–27157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bass GT, Ryall KA, Katikapalli A, Taylor BE, Dang ST, Acton ST, Saucerman JJ.. Automated image analysis identifies signaling pathways regulating distinct signatures of cardiac myocyte hypertrophy. J Mol Cell Cardiol 2012;52:923–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. LaMorte VJ, Thorburn J, Absher D, Spiegel A, Brown JH, Chien KR, Feramisco JR, Knowlton KU.. Gq- and ras-dependent pathways mediate hypertrophy of neonatal rat ventricular myocytes following alpha 1-adrenergic stimulation. J Biol Chem 1994;269:13490–13496. [PubMed] [Google Scholar]
- 29. Nomura S, Satoh M, Fujita T, Higo T, Sumida T, Ko T, Yamaguchi T, Tobita T, Naito AT, Ito M, Fujita K, Harada M, Toko H, Kobayashi Y, Ito K, Takimoto E, Akazawa H, Morita H, Aburatani H, Komuro I.. Cardiomyocyte gene programs encoding morphological and functional signatures in cardiac hypertrophy and failure. Nat Commun 2018;9:4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heras-Bautista CO, Mikhael N, Lam J, Shinde V, Katsen-Globa A, Dieluweit S, Molcanyi M, Uvarov V, Jutten P, Sahito RGA, Mederos-Henry F, Piechot A, Brockmeier K, Hescheler J, Sachinidis A, Pfannkuche K.. Cardiomyocytes facing fibrotic conditions re-express extracellular matrix transcripts. Acta Biomater 2019;89:180–192. [DOI] [PubMed] [Google Scholar]
- 31. Ma X, Takeda K, Singh A, Yu ZX, Zerfas P, Blount A, Liu C, Towbin JA, Schneider MD, Adelstein RS, Wei Q.. Conditional ablation of nonmuscle myosin II-B delineates heart defects in adult mice. Circ Res 2009;105:1102–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bang C, Batkai S, Dangwal S, Gupta SK, Foinquinos A, Holzmann A, Just A, Remke J, Zimmer K, Zeug A, Ponimaskin E, Schmiedl A, Yin X, Mayr M, Halder R, Fischer A, Engelhardt S, Wei Y, Schober A, Fiedler J, Thum T.. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest 2014;124:2136–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He H, Dai J, Zhuo R, Zhao J, Wang H, Sun F, Zhu Y, Xu D.. Study on the mechanism behind lncRNA MEG3 affecting clear cell renal cell carcinoma by regulating miR-7/RASL11B signaling. J Cell Physiol 2018;233:9503–9515. [DOI] [PubMed] [Google Scholar]
- 34. Joshi-Barr S, Bett C, Chiang WC, Trejo M, Goebel HH, Sikorska B, Liberski P, Raeber A, Lin JH, Masliah E, Sigurdson CJ.. De novo prion aggregates trigger autophagy in skeletal muscle. J Virol 2014;88:2071–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barth AS, Kumordzie A, Frangakis C, Margulies KB, Cappola TP, Tomaselli GF.. Reciprocal transcriptional regulation of metabolic and signaling pathways correlates with disease severity in heart failure. Circ Cardiovasc Genet 2011;4:475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee JH, Liu R, Li J, Zhang C, Wang Y, Cai Q, Qian X, Xia Y, Zheng Y, Piao Y, Chen Q, de Groot JF, Jiang T, Lu Z.. Stabilization of phosphofructokinase 1 platelet isoform by AKT promotes tumorigenesis. Nat Commun 2017;8:949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data has been made publicly available through the NCBI Gene Expression Omnibus (GEO), GEO accession number GSE138299.