Abstract
In today’s world, we find ourselves struggling to fight one of the worst pandemics in the history of humanity known as COVID-2019 caused by a coronavirus. When the virus reaches the lungs, we observe ground-glass opacity in the chest X-ray due to fibrosis in the lungs. Due to the significant differences between X-ray images of an infected and non-infected person, artificial intelligence techniques can be used to identify the presence and severity of the infection. We propose a classification model that can analyze the chest X-rays and help in the accurate diagnosis of COVID-19. Our methodology classifies the chest X-rays into four classes viz. normal, pneumonia, tuberculosis (TB), and COVID-19. Further, the X-rays indicating COVID-19 are classified on a severity-basis into mild, medium, and severe. The deep learning model used for the classification of pneumonia, TB, and normal is VGG-16 with a test accuracy of 95.9 %. For the segregation of normal pneumonia and COVID-19, the DenseNet-161 was used with a test accuracy of 98.9 %, whereas the ResNet-18 worked best for severity classification achieving a test accuracy up to 76 %. Our approach allows mass screening of the people using X-rays as a primary validation for COVID-19.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42979-021-00695-5.
Keywords: Chest X-ray, COVID-19, Severity-based classification
Introduction
The arrival of novel coronavirus disease (COVID-19), which originated in Wuhan, China, in December 2019 marked the beginning of several unwanted circumstances, such as, economical crisis, loss of life, etc [1]. This disease then spread rapidly from one country to another in a few months. This led to the World Health Organization (WHO) declaring the COVID-19 outbreak as a global pandemic [2]. Animals are capable of transmitting the coronavirus. But, because of the zoonotic nature of the coronavirus [3], the coronavirus has the capability to enter into the human body. Such instances have been witnessed earlier also, i.e. in the case of the severe acute respiratory syndrome coronavirus (SARS-COV) and middle-east respiratory syndrome coronavirus (MERS-COV). They had a major impact on human life leading to many deaths. Some of the clinical symptoms of COVID-19 are muscle pain, shortness of breath, fever, cough, sore throat, headache, fatigue, etc. As soon as a patient experiences signs similar to those of COVID-19, they get tested via standard COVID-19 test. But the results usually arrive within one or two days period. In case of a serious infection, this delay can be harmful. Thus, chest X-rays can prove to be a cheaper, faster, and reliable alternative to the standard COVID-19 test. The diagnosis of COVID-19 currently involves various methods that use reverse-transcription polymerase chain reaction (RT-PCR). The sensitivity of RT-PCR is low and there are chances that few results go as a false negative. Hence there was a need for cross-verification examination, such as radiological images.
Clinical images play a vital role in the early detection of the virus. Chest computer tomography (CT) imaging using radiological techniques and X-ray proved to diagnose the initial stages of COVID-19 before the disease spreads further to the lungs causing greater damage. COVID-19 infection starts from the throat and spreads through the lungs at later stages. This makes it difficult for radiologists to detect COVID-19 at early stages. Further tests need to be carried out to confirm the results. Hence, automated diagnosis from X-Rays and CT scans can help detect COVID-19 at an earlier stage.
Machine learning (ML) [4] has wide applications in the medical field, as the machine can replace any professional if it is trained to do a particular task. Hence, machine learning models can be used to assist clinicians to perform tasks with higher speed and accuracy. Deep learning (DL) [5] models (sub-set of ML) are mathematically designed networks that are trained to take certain input and classify it into one of the specified categories. With this advancement in technology, we have deep learning algorithms that are applied to treat several diseases like arrhythmia, skin cancer, breast cancer, pneumonia, and many more. Deep learning models have also been deployed on chest X-ray images and fundus images to diagnose diseases. With the advancement in technology, the number of radiologists and hospitals can use deep learning models even when working remotely. In the current COVID-19 pandemic, fast DL models have to be trained and tested to provide timely assistance and accurately produce results. A major constraint so far is the insufficiency of the RT-PCR test and the test has a high cost and it takes relatively more time to produce the results. Deep learning on chest X-ray images can solve such issues and could help in eliminating the disadvantages of RT-PCR and CT scans.
Literature review
In recent years, medical imaging has proved to be an effective tool for the diagnosis of a patient [6] with diseases like pneumonia, TB, etc. In the Chest X-ray of a COVID-19 infected patient, ground-glass opacities (GGO) and airspace opacities are observed by Kong et al. [7]. Solitary pulmonary nodules (SPN) are discrete, rounded opacity structures that are usually observed during infection in the lungs, on a Chest X-ray. Yoon et al. [8] found that the presence of SPNs in a Chest X-ray may indicate the early stage of lung abnormalities. Zhao et al. [9] found ground-glass opacities in the patients and vascular dilation in lesions. GGO and consolidation interlobular septal thickening in X-ray images were found by Li et al. [10] based on the air bronchogram. Zu et al. [11] found that 33 % of the chest CTs had rounded lung opacities. Hemdan et al. [12] used deep learning to diagnose COVID-19 using X-ray images using COVIDX-Net. Wang et al. [13] used the deep model and the results showed 92.4 % accuracy for datasets of non-COVID, who had pneumonia in the COVID-19 phase. Ioannis et al. [14] used Chest X-ray images of 224 COVID-19 positive patients, 700 Pneumonia, and 504 healthy cases. They used VGG-19 (a 19 layer deep convolutional network) and achieved an accuracy of 93.48 %. Narin et al. [15] used ResNet-50 and deep convolutional neural networks (CNN) for processing the Chest X-ray for diagnosis and achieved an accuracy of 98 %. Sethy et al. [16] processed the chest X-ray images with the help of convolutional neural networks (CNN) using ResNet-50 and SVM.
Chest X-rays have been by far most effective in the detection of COVID-19 as said by Abbas et al. [17]. The authors have presented the Decompose-Transfer-Compose strategy to classify COVID-19 from Chest X-rays. With the help of Decision Tree classifiers, Yoo et al. [18] have shown three stages of binary classification to classify COVID-19 with the help of Chest X-ray Radiography. A deep learning approach was used by Ismael et al. [19] in detecting COVID-19 based on chest X-ray images. They have made the use of transfer learning for this purpose. Different pre-trained CNN models namely VGG-16, AlexNet, GoogleNet, MobileNet-V2, SqueezeNet, ResNet-34, ResNet-50 and Inception-V3 have been considered by Nayak et al. [20] for automated COVID-19 screening method using chest X-ray images. Novel deep learning based on CNN architecture, CVDNet, was used by Ouchicha et al. [21] to detect COVID-19 infection from normal and other pneumonia cases using chest X-ray images.
Methodology
Proposed system flow diagram has been shown in Fig. 1. The chest X-ray image after obtaining from the X-ray screening machine is passed through a VGG-16 model. The results are bracketed as normal, Pneumonia, or Tuberculosis. The Pneumonia images are then passed through a DenseNet-161 model and classified into normal Pneumonia or COVID-19. Then the COVID-19 X-Ray images are passed through a ResNet-18 model and categorized as severe, medium, or mild. The details of the models and the data-set used for the proposed work have been described further below.
Fig. 1.
Proposed system flow diagram. When a person passes through the X-ray screening machine, his X-ray image is passed as an input for further processing. The input chest X-ray image is passed through a VGG-16 model and labeled as either normal, tuberculosis, or pneumonia. Further, the pneumonia affected X-ray images are passed through a DenseNet-161 model and categorized as normal pneumonia or COVID-19. The COVID-19 images are passed through a ResNet-18 model and classified as severe, medium, or mild
A total of 2271 chest X-ray images (895 1024 3 pixels) were obtained from Clinico Diagnostic Lab, Mumbai India. This data was verified and labeled into the different classes by three experts, and this data was later used to train the models. The experts were qualified with postgraduate degree of Doctor of Medicine (MD) in Radiology. These experts independently assessed the data and in case of any disagreement, the class label received from two out of the three experts was considered. No case arose where all the three experts gave a different opinion. The image was converted from RGB to gray. The resolution of images was reduced to (64 64 1). The images in the dataset varied in height and width. Resizing the images to 64 64 is beneficial as it increases response time and at the same time preserves features in the original image. Gray scaling images from RGB images reduces the number of layers from 3 to 1 and the values of the pixels of the grayscale image range from 0 to 255. This improves feature capture as well as standardized the input layer size before passing to further layers. A total of 1071 images of all three types (i.e. normal, TB, Pneumonia) were split into 80 % training images and the remaining 20 % test images. We tested the accuracy of each model for different ratios of train and test as shown in Table 1. The best testing accuracy was obtained for the 80:20 ratio, so we chose this ratio to train the models. A graph of varying values of learning rate starting from 0 was plotted against the loss function value of the model after training for each learning rate. The learning rate values are slowly ramped up in a log-linear way. We then pick the point at which the curve is dropping fastest per step. This is the point where the model can learn quickly, and hence that point is chosen as the learning rate. Input images were fed to Visual Geometry Group (VGG) VGG-16. The training dataset consisted of 388 normal images, 500 pneumonia images, and 303 TB images. The pneumonia images consisted of both normal pneumonia and COVID-19 images. The resolution of pneumonia images was kept the same i.e. (64 64 1). The resultant images were sent to the next model which labelled them as normal pneumonia or COVID-19. The dataset used for the training consisted of 1000 images, 500 of which represented pneumonia and 500 represented COVID-19. It used DenseNet-161 and predicted 233 COVID-19 and 152 pneumonia images. The images that indicated COVID-19 were passed to the model for classifying the X-rays based on the severity, which used ResNet-18. The size of the input images was kept at (64 64 1) and a total of 80 images were used for training the ResNet-18. The resultant classes determined on the basis of the severity of COVID-19 were mild, severe, and medium.
Table 1.
Accuracy for different train and test ratio
| Ratio | DenseNet161 | VGG16 | ResNet18 |
|---|---|---|---|
| 70 : 30 | 97.1 | 93.33 | 53.2 |
| 75 : 25 | 96.68 | 92.57 | 48 |
| 80 : 20 | 98.9 | 95.9 | 76 |
As shown in Fig. 2 We have used a pre-trained network called visual geometry group (VGG) VGG-16 [22] for the classification of chest X-rays into normal, TB, and Pneumonia. VGG-16 is a 19 layer model. The model makes use of imagenet weights. The input size to the model is (64, 64, 1) which is fed to a convolutional layer. This convolutional layer increases the channels of the image and gives an output with 64 channels, keeping the dimensions of the image the same as before. The next layer is also a Convolutional layer which gives an output of (64, 64, 1). After this, we apply max-pooling, which changes the image dimensions to (112, 112). The next two layers are convolutional layers, which keep the image channels as (112, 112, 28). Next, we apply a max-pooling layer, which reduces the image dimensions to (56, 56, 128). Then, we have a block of three convolutional layers and a max-pool layer, whose output is a (28, 28, 256) image. Next, we have a set of three convolutional layers with output dimensions of (28, 28, 512) and a max-pooling layer with an output of (14, 14, 512). The final three convolutional layers have an output size of (14, 14, 512) and the max-pooling layer has an output size of (7, 7, 512). Since we are doing a 3-way classification, we flatten the output of the Max pooling layer and then have a dense layer with three neurons. The learning rate was kept at 0.001 with Adam as the optimizer. We trained the model for 12 epochs.
Fig. 2.
Configuration and performance of VGG-16 a Model of VGG-16. The input size of the image is 64 64 1. After this, the image passes through a 2D convolution layer with dimensions 224x224. Then it passes through the next layer with dimensions 112 112. After this, it passes to consecutive convolution layers along with MaxPooling with dimension changes 56 56, 28 28, and 14 14. Then it goes to a Max Pooling layer with dimension change 7 7. After a series of 2500, 4096, and 4096 fully connected layers, the X-rays are classified into normal X-ray, Tuberculosis affected X-ray, and Pneumonia affected X-Ray. b The graph of the learning rate versus a loss in the range of 0.8 and 1.7 (red dot shows selected learning rate). c A total of 322 images were tested. 127 were correctly labelled as COVID-Pneumonia, 104 were correctly labelled as normal and 78 were correctly labelled as Tuberculosis
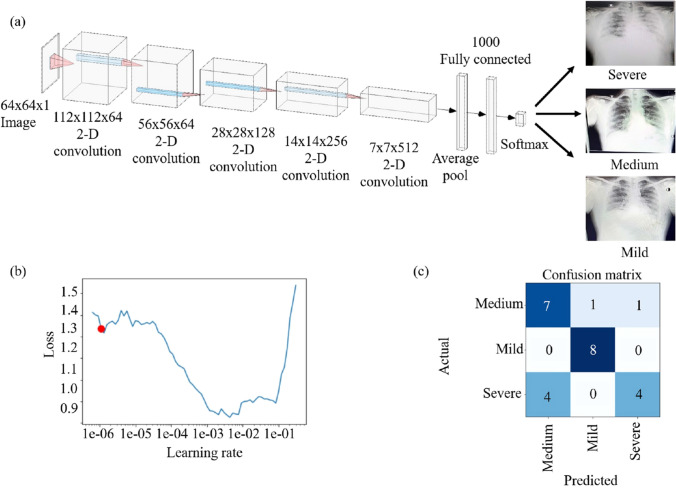
For the next step in binary classification, we have used the DenseNet-161 [23] pre-trained model (Fig. 3). An image of (64, 64, 1) is fed to the model. We have a total of 4 Dense blocks with 3 transitional layers in between. Each transitional layer consists of a 1 1 convolutional layer and an average pool layer of size (2, 2) with a stride of 2. Each dense block consists of two convolutional layers. The first block has a 1 1 6 layer and a 3 3 6 layer. The second block has a 1 1 12 and a 3 3 12 layer. The third block consists of a 1 1 48 and a 3 3 48 layer. The final dense block consists of a 1 1 31 layer and a 3 3 31 layer. The final classification layer is a 7 7 global average pool layer and finally a softmax layer of 2 neurons. The learning rate was chosen to be 0.002 and the model was trained for 15 epochs. We have performed a 3-way classification between severe, mild, and medium severity of the COVID-19 X-ray images. As shown in Fig. 4, pre-trained ResNet-18 [24] was used for this purpose. The input image for this model is a 64 64 3 image. This model consists of 5 convolutional blocks. The first convolutional block consists of a 7 7 64 kernel. In the next convolutional block, we have a 3 3 max pool layer with a stride of 2. For the second block, we have two (3 3 64) blocks. The third, fourth, and fifth convolutional blocks have the filter dimensions of 128, 256, 512 respectively with each of the kernel sizes as 3 3. There is an average pooling layer after these and a fully connected layer with three neurons for a 3-way classification. The learning rate used was 0.002 and the model was trained for 15 epochs.
Fig. 3.
Configuration and performance of DenseNet-161 a Model of DenseNet-161. The input size of the image is 64 64 1 after which the image passes through a 2-D convolutional layer with dimensions 112 112. Then it passes through the next layer with dimensions 56 56 After passing through the consecutive layers the dimensions change from 56 56 to 28 28, 14 14, and finally 7 7. After 1000 fully connected layers, the X-rays are classified into normal pneumonia and COVID-19. b The graph of the learning rate versus a loss in the range of 0.5–0.9 (red dot shows selected learning rate). c Confusion matrix of the output for DenseNet-161. A total of 235 images were tested, out of which 232 were correctly classified as COVID, and 3 were found to be wrongly classified. From 150 images, 149 were correctly categorized as pneumonia whereas 1 was wrongly classified
Fig. 4.
Configuration and performance of ResNet-18 a Model of ResNet-18. The input size of the image is 64 64 1. After this, the image passes through a 2D convolution layer with dimensions 112 112 64. Then it passes through the next layer with dimensions 56 56 64. After this, it passes to consecutive 2D convolution layers with dimension changes 28 28 128, 14 14 256, and 7 7 512. After 1000 fully connected layers, the X-rays are classified into severe, medium, and mild. b The graph of the learning rate versus a loss in the range of 0.9 and 1.6 (red dot shows selected learning rate). c Confusion matrix of the output for ResNet-18. A total of 25 images were tested, out of which 7 were correctly labelled as a medium, 8 were correctly labelled as mild and 4 were correctly labelled as severe
Results and Discussion
A total of 1000 images were chosen out of which 500 were regular pneumonia X-ray images and the remaining 500 were COVID infected X-ray images. These images were used to train the DenseNet-161 Network and the ratio of training and testing was 80:20 and we found that with 99 % accuracy, we can segregate pneumonia from COVID infected chest X-rays. It has been quite difficult for expert radiologists to differentiate pneumonia from COVID-infected X-rays and making it possible through deep learning was a very big achievement. At the request of radiologists, we also tried to segregate COVID X-rays based on the severity of the infection into three subclasses viz. mild, medium, and severe. For this, we only had 80 labelled images of each subclass. These images were used to train and test on multiple networks and we found that out of all the ones that were trained and tested, ResNet-18 performed well with an accuracy of up to 76 % to classify between the three subclasses. Overall, there is still a scope of improvement in accuracy for classifying the COVID-19 infected X-rays into three different classes.
For Table 2, NPT stands for Normal, Pneumonia, and Tuberculosis. RC stands for Regular Pneumonia and COVID Pneumonia. SMm stands for Severe, Medium and mild. To validate the results, we first trained the VGG-16 network using 970 labelled images via supervised learning. 270 images were used to test the pre-trained VGG-16 network which resulted in an accuracy of 98 % between pneumonia infected, TB infected, and non-infected X-rays.
Table 2.
Comparison of output accuracy (in percentage) with respect to different deep learning networks
| Deep Learning Networks | NPT | RC | SMm |
|---|---|---|---|
| Alexnet | 50 | 70 | 50.4 |
| Resnet-18 | 72.3 | 78.2 | 76 |
| Resnet-50 | 60.4 | 89.5 | 60 |
| VGG-13 | 70.2 | 87.8 | 40.9 |
| VGG-16 | 95.9 | 78.9 | 40 |
| DenseNet-161 | 89.8 | 98.9 | 54.2 |
| Densenet-169 | 78.6 | 94.7 | 65.8 |
| Densenet-201 | 85.9 | 93.2 | 60 |
Figure 2b depicts the graph of the learning rate versus a loss in the range of 0.8 and 1.7. The red dot shows the selected learning rate. Figure 2c shows that a total of 322 images were tested. Out of these images, 127 were correctly labelled as COVID-Pneumonia, 104 were correctly labelled as normal and 78 were labelled as Tuberculosis. The graph of the learning rate versus a loss in the range of 0.5 to 0.9 can be seen in Fig. 3b. The red dot shows the selected learning rate. Figure 3c demonstrates the Confusion matrix of the output for DenseNet-161. A total of 235 images were tested, out of which 232 were correctly classified as COVID, and 3 were found to be wrongly classified. From 150 images, 149 were correctly categorized as pneumonia whereas 1 was wrongly classified. Figure 4b illustrates the graph of the learning rate versus a loss in the range of 0.9 and 1.4. The red dot shows the selected learning rate. The confusion matrix of the output for ResNet-18 is observed in Fig. 4c. A total of 25 images were tested, out of which 7 were correctly labelled as a medium, 8 were correctly labelled as mild and 4 were correctly labelled as severe.
For the classification of X-ray into three types i.e. TB, Pneumonia, and normal, we used AlexNet first. The disadvantage of AlexNet was that it had a lesser number of layers and thus, the accuracy obtained was 50 %. Secondly, we trained and tested the ResNet-50 model which comparatively showed a better accuracy of 60.4 % but the time it took to train was too long and the model was too large, so we tried VGG-13 and obtained an accuracy of 70.2 % and then VGG-16 was also trained and tested which gave by far the best results. VGG-16 gave an accuracy of by far the highest which is 95.9 %. Other models like ResNet-18, DenseNet-161, DenseNet-169, DenseNet-201 were trained and tested but did not show any improvement in accuracy. For further classification of images into Pneumonia or COVID-19, we started first with AlexNet since the model is smaller in size. We got 70 % accuracy. We then trained a ResNet-50 model which gave us an accuracy of 89.5%. We looked for much bigger models to train and hence used DenseNet-201 which gave us an accuracy of 93.2 % but the model size was quite large and the training took long. Other models like ResNet-18, DenseNet-161, DenseNet69 were trained and tested but did not show any improvement in accuracy. Hence, we tried a smaller version of the DenseNet, i.e. DenseNet-161 which gave us an accuracy of 98.9 %. For further classification of COVID-19 positive X-ray into three types i.e. mild, severe, and medium, we used AlexNet first. The disadvantage of AlexNet was that it had a lesser number of layers and thus the accuracy obtained was 50.4 %. Secondly, we trained and tested the DenseNet-161 model which gave an accuracy of 54.2%. Further, we trained DenseNet-169 which comparatively showed a better accuracy of 65.8 %. ResNet-18 gave an accuracy of by far the highest of 76 %. Other models like ResNet-50, DenseNet-161, DenseNet-201 were trained and tested but did not show any improvement in accuracy. Table 3 gives a brief summary of the different parameters for assessing the models. For each of the model, the total images used, the training and testing size, the sensitivity, specificity and the F1-score are shown.
Table 3.
The training and testing size, sensitivity, specificity, and the F1-Score for each of the models, i.e. VGG16, DenseNet161, and ResNet18
| VGG16 | Normal | Tuberculosis | Pneumonia affected |
|---|---|---|---|
| Total Images | 492 | 382 | 638 |
| Training size | 388 | 303 | 500 |
| Testing size | 104 | 79 | 138 |
| Sensitivity | 100% | 98% | 92% |
| Specificity | 97% | 97% | 99% |
| F1 Score | 0.97 | 0.96 | 0.95 |
| Densenet161 | Pneumonia | COVID-19 | |
| Total Images | 750 | 1175 | |
| Training size | 600 | 1025 | |
| Testing size | 150 | 235 | |
| Sensitivity | 99% | 98% | |
| Specificity | 98% | 99% | |
| F1 Score | 0.98 | 0.99 | |
| ResNet-18 | Medium | Mild | Severe |
| Total Images | 30 | 26 | 26 |
| Training size | 21 | 18 | 18 |
| Testing size | 9 | 8 | 8 |
| Sensitivity | 77% | 100% | 50% |
| Specificity | 75% | 94% | 94% |
| F1 Score | 0.7 | 0.94 | 0.61 |
Comparison with Existing Literature
Table 4 shows that some of the reported research groups have used X-ray reports whereas some have used CT reports to attain COVID-19 results. We have sequentially used three methods to attain the required results. The accuracy of the VGG-16 model, DenseNet-161 model, and ResNet-18 gave us an accuracy of 95.9 %, 98.9 % and 76 % respectively. We achieved the highest accuracy which can further be improved.
Table 4.
Comparison with other methods. Most of the methods classify X-rays only into pneumonia, COVID, and normal, our method further classifies COVID-19 into 3 categories
| Method | Type of images | Number of cases | Method used | Accuracy (%) |
|---|---|---|---|---|
| Ioannis et al. [14] | Chest X-ray | 1428 | VGG-19 | 93.48 |
| Wang et al. [13] | Chest X-ray | 13645 | COVID-Net | 92.4 |
| Sethy et al. [16] | Chest X-ray | 50 | ResNet50+SVM | 95.38 |
| Hemdan et al. [12] | Chest X-ray | 50 | COVIDX-Net | 90 |
| Narin et al. [15] | Chest X-ray | 100 | Deep CNN | 98 |
| Ying et al. [25] | Chest CT | 1485 | DRE-Net | 86 |
| Zheng et al. [9] | Chest CT | 542 | Unet+3D Deep Network | 90.8 |
| Xu et al. [11] | Chest CT | 618 | ResNet+ Location Attention | 86.7 |
| Proposed method | Chest X-ray | 22 | VGG-16 DenseNet-161 ResNet-18 | 95.9 98.9 76 |
Discussions
Due to the initial stage of the disease, the number of labeled data points that could be obtained was limited. Hence, the size of the dataset and the number of data points used were less. Because of this, there could be chances of overfitting. Better results can be obtained if the dataset size is increased. Currently, our proposed work consists of three different models used to carry out the mentioned classification tasks. In the future, an architecture could also be prepared to carry out a successful grouping of the chest X-rays at once in the mentioned classes. We considered the VGG-16, Densenet-161, Resnet-18 models for the classification tasks. We also tried various models and obtained different levels of accuracy as shown in Table 2. The different pre-implemented architectures such as GoogleNet, Inception could be used to get better results. Our proposed work does not take into consideration the same patient data for classifying the COVID-19 positive X-ray images into severe, medium and mild. The severity classification would become more useful if the same patient data could be obtained that would show the progression of the disease and thereby classify the images into the three classes.
The chest X-ray machines can act as a source of detecting COVID-19 and further diagnosis can be carried out using RT-PCR, saving the hard-earned money of patients. Furthermore, X-ray machines are well available in bulk, and in most regular hospitals, the X-ray machine is an integral part of the setup and it will be easy to make this available in local hospitals as well. Adding to that, X-ray machines require less maintenance compared to RT-PCR in terms of their reagents, and hence the cost of operation is comparatively low. There are some side effects of X-ray scanning, such as people carrying metals and pregnant women, for such cases, we recommend that X-rays should not be used, otherwise mass-screening through X-ray is possible.
Conclusion
Deep learning can be a very useful tool in the medical industry for the detection of diseases just by processing the images of the chest X-rays and feeding them as an input to the model. The chest X-rays were initially classified into different classes such as normal, pneumonia, tuberculosis, and COVID-19. The deep learning model used for the classification of pneumonia, TB, and normal was VGG-16 and an accuracy of 95.9 % was achieved. After that, we distinguished normal pneumonia from the COVID-19 positive patient X-ray using the DenseNet-161 and we achieved an accuracy of 98.9 %. As a proof of concept, we have shown that COVID-19 X-rays can be classified based on the severity of the COVID-19 into different sub-units such as severe, mild, and medium stage, where the ResNet-18 worked best for severity classification achieving an accuracy up to 76 %.
Mass screening of people for detection of COVID-19 can be done effectively with our proposed model. It will help yield faster and accurate results and will be cost-effective as compared to the conventional RT-PCR method. Being an integral part of medical equipment setup, X-ray machines are generally present in hospitals. Thus, this diagnostic treatment can be made available in local areas as well.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Authors would like to thank Clinico Diagnostic Lab for providing the images and Dr. Madhura Mehendale for their support in labeling the supervisory dataset.
Author Contributions
Conceptualization was done by Ninad Mehendale (NM) and Ankita Shelke (AS). All the experiments were performed by AS and Talha Chafekar (TC). Manuscript writing original draft preperation Aafiya Hussain (AH), Vruddhi Shah (VS) and Amanshu Tiwari (AT). Manuscript review and editing was done by VS, AH, NM. Visualization work carried out by Madhura Inamdar (MI) and NM.
Funding
No funding was involved in the present work.
Conflict of Interest
Authors A. Shelke, M. Inamdar, V. Shah, A. Tiwari, A. Hussain, T. Chafekar and N. Mehendale declare that they have no conflict of interest.
Involvement of Human Participant and Animals
This article does not contain any studies with animals or Humans performed by any of the authors. All the necessary permissions were obtained from the Institute Ethical Committee and concerned authorities.
Informed Consent
Informed consent was not required as there were no participant
Footnotes
This article is part of the topical collection “Artificial Intelligence for HealthCare” guest edited by Lydia Bouzar-Benlabiod, Stuart H. Rubin and Edwige Pissaloux.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Acter T, Uddin N, Das J, Akhter A, Choudhury TR, Kim S. Evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as coronavirus disease 2019 (COVID-19) pandemic: A global health emergency. Sci Total Environ. 2020:138996. [DOI] [PMC free article] [PubMed]
- 2.Zhang D, Hu M, Ji Q. Financial markets under the global pandemic of COVID-19. Finance Res Lett. 2020 Oct 1;36:101528. [DOI] [PMC free article] [PubMed]
- 3.Ye ZW, Yuan S, Yuen KS, Fung SY, Chan CP, Jin DY. Zoonotic origins of human coronaviruses. Int J Biol Sci. 2020;16(10):1686. [DOI] [PMC free article] [PubMed]
- 4.King DE. Dlib-ml: A machine learning toolkit. J Mach Learn Res. 2009 Dec 1;10:1755–8.
- 5.Goodfellow I, Bengio Y, Courville A, Bengio Y. Deep learning, vol. 1. Cambridge: MIT Press; 2016.
- 6.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020 Jun 1;121:103792. [DOI] [PMC free article] [PubMed]
- 7.Kong W, Agarwal PP. Chest imaging appearance of COVID-19 infection. Radiol: Cardiothorac Imaging. 2020 Feb 13;2(1):e200028. [DOI] [PMC free article] [PubMed]
- 8.Yoon SH, Lee KH, Kim JY, Lee YK, Ko H, Kim KH, Park CM, Kim YH. Chest radiographic and CT findings of the 2019 novel coronavirus disease (COVID-19): analysis of nine patients treated in Korea. Korean J Radiol. 2020; 21(4):494. [DOI] [PMC free article] [PubMed]
- 9.Zhao W, Zhong Z, Xie X, Yu Q, Liu J. Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: a multicenter study. American J Roentgenol. 2020;214(5):1072–7. [DOI] [PubMed]
- 10.Li Y, Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. American J Roentgenol. 2020 214(6):1280–6. [DOI] [PubMed]
- 11.Zu ZY, Jiang MD, Xu PP, Chen W, Ni QQ, Lu GM, Zhang LJ. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiol. 2020 Aug;296(2):E15–25. [DOI] [PMC free article] [PubMed]
- 12.Hemdan EE, Shouman MA, Karar ME. Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv:2003.11055. Accessed 24 Mar 2020.
- 13.Wang L, Lin ZQ, Wong A. Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci Rep. 2020;10(1):1–2. [DOI] [PMC free article] [PubMed]
- 14.Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020 Jun;43(2):635–40. [DOI] [PMC free article] [PubMed]
- 15.Narin A, Kaya C, Pamuk Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. Pattern Anal Appl. 2021 May 9:1–4. [DOI] [PMC free article] [PubMed]
- 16.Sethy PK, Behera SK, Ratha PK, Biswas P. Detection of coronavirus disease (COVID-19) based on deep features and support vector machine. 2020.
- 17.Abbas A, Abdelsamea MM, Gaber MM. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl Intell. 2021 Feb;51(2):854–64. [DOI] [PMC free article] [PubMed]
- 18.Yoo SH, Geng H, Chiu TL, Yu SK, Cho DC, Heo J, Choi MS, Choi IH, Cung Van C, Nhung NV, Min BJ. Deep learning-based decision-tree classifier for COVID-19 diagnosis from chest X-ray imaging. Front Med. 2020 Jul 14;7:427. [DOI] [PMC free article] [PubMed]
- 19.Ismael AM, Şengür A. Expert Syst Appl. 2021;164:114054. doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nayak SR, Nayak DR, Sinha U, Arora V, Pachori RB. Application of deep learning techniques for detection of COVID-19 cases using chest X-ray images: A comprehensive study. Biomed Signal Process Control. 2021;64:102365. [DOI] [PMC free article] [PubMed]
- 21.Ouchicha C, Ammor O, Meknassi M. Chaos Solitons Fractals. 2020;140:110245. doi: 10.1016/j.chaos.2020.110245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tammina S. Transfer learning using VGG-16 with deep convolutional neural network for classifying images. Int J Sci Res Publ. 2019 Oct;9(10):143–50.
- 23.Minaee S, Kafieh R, Sonka M, Yazdani S, Soufi GJ. Deep-covid: Predicting covid-19 from chest x-ray images using deep transfer learning. Med image Anal. 2020;65:101794. [DOI] [PMC free article] [PubMed]
- 24.Chen PH, Bak PR. Imaging Informatics for Healthcare, Research, and Applications. In Proc. of SPIE Vol 2019 (Vol. 10954, pp. 1095401–1).
- 25.Song Y, Zheng S, Li L, Zhang X, Zhang X, Huang Z, Chen J, Wang R, Zhao H, Zha Y, Shen J. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE/ACM Trans Comput Biol Bioinf. 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.